Abstract
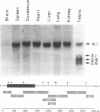
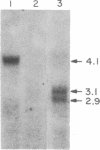
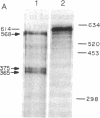
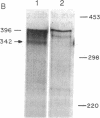
Previously we have shown that the gene encoding murine beta 1,4-galactosyltransferase (beta 1,4-GT; UDPgalactose:N-acetyl-D-glucosaminyl-glycopeptide 4-beta-D-galactosyltransferase, EC 2.4.1.38) is unusual in that it specifies two sets of mRNAs of about 3.9 and 4.1 kilobases (kb). Translation of the 3.9- and 4.1-kb mRNAs results in the predicted synthesis of two related membrane-bound forms of the protein of 386 amino acids (short form) and 399 amino acids (long form), respectively. In this study we have examined the expression of beta 1,4-GT during murine spermatogenesis. Spermatogonia contain a 4.1-kb transcript that is comparable in size to the beta 1,4-GT mRNA identified in somatic cells. During differentiation from spermatogonia (2n) to pachytene spermatocytes (4n), the amount of beta 1,4-GT mRNA is reduced to barely detectable levels. Continued differentiation to round spermatids (n) is coincident with a renewed production of beta 1,4-GT mRNA to levels comparable with those detected in spermatogonia. However, the characteristic 4.1-kb mRNA detected in spermatogonia is replaced by two truncated transcripts of 2.9 and 3.1 kb. By S1 nuclease analysis, the 2.9- and 3.1-kb transcripts were shown to encode the same open reading frame as the 4.1-kb transcript found in somatic cells. The shorter round spermatid transcripts arise as a consequence of the use of alternative poly(A) signals. Lastly, we show that, in direct contrast to all somatic tissues and cell lines examined to date, male germ cells synthesize only the long form of the beta 1,4-GT polypeptide.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bayna E. M., Shaper J. H., Shur B. D. Temporally specific involvement of cell surface beta-1,4 galactosyltransferase during mouse embryo morula compaction. Cell. 1988 Apr 8;53(1):145–157. doi: 10.1016/0092-8674(88)90496-5. [DOI] [PubMed] [Google Scholar]
- Birnstiel M. L., Busslinger M., Strub K. Transcription termination and 3' processing: the end is in site! Cell. 1985 Jun;41(2):349–359. doi: 10.1016/s0092-8674(85)80007-6. [DOI] [PubMed] [Google Scholar]
- Bleil J. D., Wassarman P. M. Galactose at the nonreducing terminus of O-linked oligosaccharides of mouse egg zona pellucida glycoprotein ZP3 is essential for the glycoprotein's sperm receptor activity. Proc Natl Acad Sci U S A. 1988 Sep;85(18):6778–6782. doi: 10.1073/pnas.85.18.6778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobner P. R., Kislauskis E., Wentworth B. M., Villa-Komaroff L. Alternative 5' exons either provide or deny an initiator methionine codon to the same alpha-tubulin coding region. Nucleic Acids Res. 1987 Jan 12;15(1):199–218. doi: 10.1093/nar/15.1.199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dugaiczyk A., Haron J. A., Stone E. M., Dennison O. E., Rothblum K. N., Schwartz R. J. Cloning and sequencing of a deoxyribonucleic acid copy of glyceraldehyde-3-phosphate dehydrogenase messenger ribonucleic acid isolated from chicken muscle. Biochemistry. 1983 Mar 29;22(7):1605–1613. doi: 10.1021/bi00276a013. [DOI] [PubMed] [Google Scholar]
- Farquhar M. G., Palade G. E. The Golgi apparatus (complex)-(1954-1981)-from artifact to center stage. J Cell Biol. 1981 Dec;91(3 Pt 2):77s–103s. doi: 10.1083/jcb.91.3.77s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geisow M. J. Cell-surface receptors: puzzles and paradigms. Bioessays. 1986 Apr;4(4):149–151. doi: 10.1002/bies.950040403. [DOI] [PubMed] [Google Scholar]
- Heidaran M. A., Showman R. M., Kistler W. S. A cytochemical study of the transcriptional and translational regulation of nuclear transition protein 1 (TP1), a major chromosomal protein of mammalian spermatids. J Cell Biol. 1988 May;106(5):1427–1433. doi: 10.1083/jcb.106.5.1427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hennig B. Change of cytochrome c structure during development of the mouse. Eur J Biochem. 1975 Jun 16;55(1):167–183. doi: 10.1111/j.1432-1033.1975.tb02149.x. [DOI] [PubMed] [Google Scholar]
- Hill R. L., Brew K., Vanaman T. C., Trayer I. P., Mattock P. The structure, function, and evolution of alpha-lactalbumin. Brookhaven Symp Biol. 1968 Jun;21(1):139–154. [PubMed] [Google Scholar]
- Hollis G. F., Douglas J. G., Shaper N. L., Shaper J. H., Stafford-Hollis J. M., Evans R. J., Kirsch I. R. Genomic structure of murine beta-1,4-galactosyltransferase. Biochem Biophys Res Commun. 1989 Aug 15;162(3):1069–1075. doi: 10.1016/0006-291x(89)90782-1. [DOI] [PubMed] [Google Scholar]
- Kleene K. C., Distel R. J., Hecht N. B. Translational regulation and deadenylation of a protamine mRNA during spermiogenesis in the mouse. Dev Biol. 1984 Sep;105(1):71–79. doi: 10.1016/0012-1606(84)90262-8. [DOI] [PubMed] [Google Scholar]
- Kozak M. Comparison of initiation of protein synthesis in procaryotes, eucaryotes, and organelles. Microbiol Rev. 1983 Mar;47(1):1–45. doi: 10.1128/mr.47.1.1-45.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letts P. J., Meistrich M. L., Bruce W. R., Schachter H. Glycoprotein glycosyltransferase levels during spermatogenesis in mice. Biochim Biophys Acta. 1974 Mar 20;343(1):192–207. doi: 10.1016/0304-4165(74)90251-7. [DOI] [PubMed] [Google Scholar]
- Lopez L. C., Bayna E. M., Litoff D., Shaper N. L., Shaper J. H., Shur B. D. Receptor function of mouse sperm surface galactosyltransferase during fertilization. J Cell Biol. 1985 Oct;101(4):1501–1510. doi: 10.1083/jcb.101.4.1501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLauchlan J., Gaffney D., Whitton J. L., Clements J. B. The consensus sequence YGTGTTYY located downstream from the AATAAA signal is required for efficient formation of mRNA 3' termini. Nucleic Acids Res. 1985 Feb 25;13(4):1347–1368. doi: 10.1093/nar/13.4.1347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meijer D., Hermans A., von Lindern M., van Agthoven T., de Klein A., Mackenbach P., Grootegoed A., Talarico D., Della Valle G., Grosveld G. Molecular characterization of the testis specific c-abl mRNA in mouse. EMBO J. 1987 Dec 20;6(13):4041–4048. doi: 10.1002/j.1460-2075.1987.tb02749.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meistrich M. L., Longtin J., Brock W. A., Grimes S. R., Jr, Mace M. L. Purification of rat spermatogenic cells and preliminary biochemical analysis of these cells. Biol Reprod. 1981 Dec;25(5):1065–1077. doi: 10.1095/biolreprod25.5.1065. [DOI] [PubMed] [Google Scholar]
- Nakazawa K., Ando T., Kimura T., Narimatsu H. Cloning and sequencing of a full-length cDNA of mouse N-acetylglucosamine (beta 1-4)galactosyltransferase. J Biochem. 1988 Aug;104(2):165–168. doi: 10.1093/oxfordjournals.jbchem.a122434. [DOI] [PubMed] [Google Scholar]
- Oppi C., Shore S. K., Reddy E. P. Nucleotide sequence of testis-derived c-abl cDNAs: implications for testis-specific transcription and abl oncogene activation. Proc Natl Acad Sci U S A. 1987 Dec;84(23):8200–8204. doi: 10.1073/pnas.84.23.8200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfarr D. S., Rieser L. A., Woychik R. P., Rottman F. M., Rosenberg M., Reff M. E. Differential effects of polyadenylation regions on gene expression in mammalian cells. DNA. 1986 Apr;5(2):115–122. doi: 10.1089/dna.1986.5.115. [DOI] [PubMed] [Google Scholar]
- Ponzetto C., Wolgemuth D. J. Haploid expression of a unique c-abl transcript in the mouse male germ line. Mol Cell Biol. 1985 Jul;5(7):1791–1794. doi: 10.1128/mcb.5.7.1791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Propst F., Rosenberg M. P., Iyer A., Kaul K., Vande Woude G. F. c-mos proto-oncogene RNA transcripts in mouse tissues: structural features, developmental regulation, and localization in specific cell types. Mol Cell Biol. 1987 May;7(5):1629–1637. doi: 10.1128/mcb.7.5.1629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Propst F., Rosenberg M. P., Vande Woude G. F. Proto-oncogene expression in germ cell development. Trends Genet. 1988 Jul;4(7):183–187. doi: 10.1016/0168-9525(88)90073-x. [DOI] [PubMed] [Google Scholar]
- Reeves R., Elton T. S., Nissen M. S., Lehn D., Johnson K. R. Posttranscriptional gene regulation and specific binding of the nonhistone protein HMG-I by the 3' untranslated region of bovine interleukin 2 cDNA. Proc Natl Acad Sci U S A. 1987 Sep;84(18):6531–6535. doi: 10.1073/pnas.84.18.6531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roth J., Berger E. G. Immunocytochemical localization of galactosyltransferase in HeLa cells: codistribution with thiamine pyrophosphatase in trans-Golgi cisternae. J Cell Biol. 1982 Apr;93(1):223–229. doi: 10.1083/jcb.93.1.223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roth J., Lentze M. J., Berger E. G. Immunocytochemical demonstration of ecto-galactosyltransferase in absorptive intestinal cells. J Cell Biol. 1985 Jan;100(1):118–125. doi: 10.1083/jcb.100.1.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scully N. F., Shaper J. H., Shur B. D. Spatial and temporal expression of cell surface galactosyltransferase during mouse spermatogenesis and epididymal maturation. Dev Biol. 1987 Nov;124(1):111–124. doi: 10.1016/0012-1606(87)90464-7. [DOI] [PubMed] [Google Scholar]
- Shaper N. L., Hollis G. F., Douglas J. G., Kirsch I. R., Shaper J. H. Characterization of the full length cDNA for murine beta-1,4-galactosyltransferase. Novel features at the 5'-end predict two translational start sites at two in-frame AUGs. J Biol Chem. 1988 Jul 25;263(21):10420–10428. [PubMed] [Google Scholar]
- Shaper N. L., Mann P. L., Shaper J. H. Cell surface galactosyltransferase: immunochemical localization. J Cell Biochem. 1985;28(3):229–239. doi: 10.1002/jcb.240280305. [DOI] [PubMed] [Google Scholar]
- Shaw G., Kamen R. A conserved AU sequence from the 3' untranslated region of GM-CSF mRNA mediates selective mRNA degradation. Cell. 1986 Aug 29;46(5):659–667. doi: 10.1016/0092-8674(86)90341-7. [DOI] [PubMed] [Google Scholar]
- Shur B. D. Evidence that galactosyltransferase is a surface receptor for poly(N)-acetyllactosamine glycoconjugates on embryonal carcinoma cells. J Biol Chem. 1982 Jun 25;257(12):6871–6878. [PubMed] [Google Scholar]
- Sorrentino V., McKinney M. D., Giorgi M., Geremia R., Fleissner E. Expression of cellular protooncogenes in the mouse male germ line: a distinctive 2.4-kilobase pim-1 transcript is expressed in haploid postmeiotic cells. Proc Natl Acad Sci U S A. 1988 Apr;85(7):2191–2195. doi: 10.1073/pnas.85.7.2191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka S., Fujimoto H. A postmeiotically expressed clone encodes lactate dehydrogenase isozyme X. Biochem Biophys Res Commun. 1986 Apr 29;136(2):760–766. doi: 10.1016/0006-291x(86)90504-8. [DOI] [PubMed] [Google Scholar]
- Waters S. H., Distel R. J., Hecht N. B. Mouse testes contain two size classes of actin mRNA that are differentially expressed during spermatogenesis. Mol Cell Biol. 1985 Jul;5(7):1649–1654. doi: 10.1128/mcb.5.7.1649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickens M., Stephenson P. Role of the conserved AAUAAA sequence: four AAUAAA point mutants prevent messenger RNA 3' end formation. Science. 1984 Nov 30;226(4678):1045–1051. doi: 10.1126/science.6208611. [DOI] [PubMed] [Google Scholar]