ABSTRACT
RAD6, an E2 ubiquitin-conjugating enzyme, is a key node for determining different DNA damage repair pathways, controlling both the error-prone and the error-free DNA damage repair pathways through differential regulation of the ubiquitination of the proliferating cell nuclear antigen (PCNA) protein. However, whether other pathways are involved in the RAD6-mediated regulation of DNA damage repair is still unclear. To deeply understand the molecular mechanisms of RAD6 in DNA damage repair, we performed a proteomic analysis and identified the changes of the protein-protein interaction (PPI) networks of RAD6 before and after X-ray irradiation. Furthermore, our study indicated that a proteasome-related event is likely involved in the DNA damage repair process. Moreover, we found that RAD6 promotes proteasome activity and nuclear translocation by enhancing the degradation of PSMF1 and the lamin B receptor (LBR). Therefore, we provide a novel pathway that is employed by RAD6 in response to DNA damage.
KEYWORDS: interactome analysis, RAD6, proteasome, PSMF1, PSMD3, LBR, DNA damage
INTRODUCTION
If DNA is not repaired before replication, damaged DNA is deleterious to normal cell cycle progression and a variety of life processes (1). The accumulation of unrepaired DNA usually triggers the activation of multiple cell death pathways or oncogenic pathways, eventually resulting in cell death, aging, tumorigenesis, or other diseases (2). Therefore, efficient repair of damaged DNA is essential for the normal development of organisms. Two types of DNA damage tolerance (DDT) pathways, the so-called error-prone and error-free DNA damage repair pathways, have evolved in mammals and are highly conserved from yeast (Saccharomyces cerevisiae) to humans (3, 4). The error-prone DNA damage repair pathway employs the translesion DNA synthesis (TLS) mechanism to insert correct or incorrect nucleotides to repair the damaged DNA, which often leads to insertions, deletions, or other types of chromosomal rearrangements. However, the error-free DNA damage repair pathway uses the undamaged sister duplex as the repair template, resulting in a precise repair and the correct inheritance of genetic information (3, 4).
Ubiquitination is a critical posttranslational protein modification that is catalyzed by a series of enzymes, including the E1 ubiquitin activation enzyme, E2 ubiquitin-conjugating enzyme, and E3 ubiquitin-protein ligase (5). RAD6 is an E2 ubiquitin-conjugating enzyme that functions as a key regulator in the control of DNA damage repair by regulating the ubiquitination of the proliferating cell nuclear antigen (PCNA) protein (3). RAD6 cooperates with the E3 ligase RAD18, which recruits RAD6 to chromatin (6) to regulate the monoubiquitination of PCNA at the site of lysine 164. The monoubiquitinated PCNA is essential for error-prone DNA damage repair (3). In addition, RAD18 associates with RAD5, and both contain a RING finger domain. The RAD18-RAD5 complex further recruits the heterodimeric ubiquitin-conjugating enzymes UBC13 and MMS2 to form a large complex containing RAD6, RAD18, RAD5, UBC13, and MMS2 that catalyzes the polyubiquitination of PCNA at the same site, which is required for error-free DNA damage repair (3, 7). Therefore, RAD6 functions as a node to direct different DNA damage repair pathways to repair damaged DNA in an error-prone or an error-free manner.
Histone H2B is another substrate of RAD6. By collaborating with the heterodimeric E3 ligase RNF20/RNF40, RAD6 catalyzes the monoubiquitination of H2B (8, 9). As a direct downstream target of RAD6, H2B monoubiquitination also participates in the regulation of DNA damage repair (10, 11). RNF20 is localized to double-strand DNA breaks (DSBs) and is required for DSB-induced H2B monoubiquitination. The loss of H2B monoubiquitination compromises the recruitment of RAD51 and BRCA1 to the DSBs, further resulting in significant defects in homologous recombination (HR) repair (10). In addition, another study indicated that DSB-induced H2B monoubiquitination is essential for both HR repair and nonhomologous end-joining (NHEJ) repair by affecting the recruitment of the components of both pathways (11).
In addition to the pathways described above, our recent work also showed that RAD6 participates in the regulation of DNA damage repair through regulating the autophagy-mediated protein degradation of heterochromatin protein 1 alpha (HP1α). We found that RAD6 regulates the ubiquitination and degradation of HP1α in human cells and the degradation of HP1α by RAD6 results in relatively open chromatin conditions, further facilitating the DNA damage repair efficiency, especially the HR repair efficiency (12). Multiple pathways are likely involved in the RAD6-mediated DNA damage repair process. It is essential to further understand the roles of RAD6 in the control of DNA damage repair.
Because RAD6 plays such a significant role in the regulation of DNA damage repair, the loss of RAD6 results in hypersensitivity to DNA damage reagents (3, 4). Although both the PCNA pathway and the H2B monoubiquitination pathway likely contribute to RAD6-mediated DNA damage repair, a comprehensive understanding of the functions of RAD6 in DNA damage repair is essential. To know whether other pathways are involved in RAD6-mediated DNA damage repair, we performed a proteomic analysis and identified the protein-protein interaction (PPI) dynamics of RAD6 in response to X-ray irradiation. Our results indicated that a proteasome event is likely involved in the RAD6-mediated DNA damage response. Further study suggested that RAD6 enhances proteasome activity and nuclear translocation by promoting the degradation of the proteasome inhibitor PSMF1 and the nuclear lamin B receptor (LBR).
RESULTS
Proteomic analysis of the RAD6 interactome in the DNA damage response.
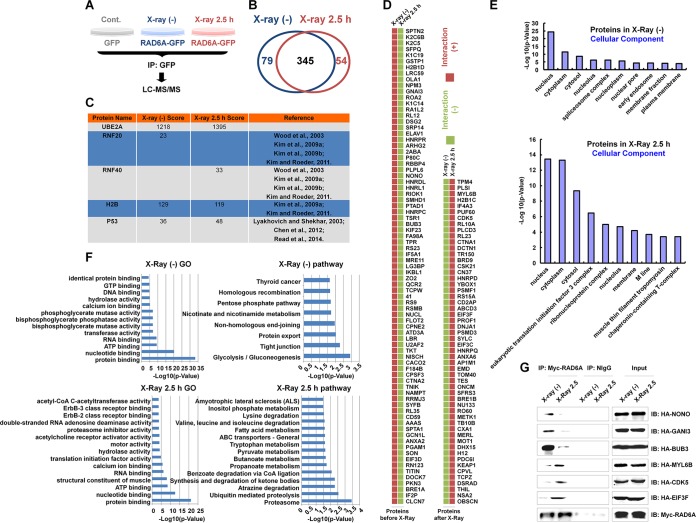
To further understand the roles of RAD6 in DNA damage repair, we performed a proteomic analysis to dissect the protein-protein interaction (PPI) dynamics of RAD6 in the DNA damage response. HEK293T cells transfected with green fluorescent protein (GFP)-tagged RAD6A (RAD6A-GFP) were either not irradiated or irradiated with X-rays (80 kV for 5 min), and then the cells were allowed to recover for 2.5 h. Total proteins were prepared and subjected to coimmunoprecipitation (co-IP) assays with anti-GFP antibodies. The precipitated proteins were eventually used for liquid chromatography (LC)-tandem mass spectrometry (MS/MS) analysis. Cells transfected with an empty vector expressing GFP were used as a negative control to determine the GFP-specific binding background (Fig. 1A). The original data from the mass spectrometry analysis are not shown here. Upon subtraction of the level of background binding, we identified 424 RAD6A binding proteins under normal conditions and 399 RAD6A binding proteins under X-ray irradiation conditions. There were 345 common binding proteins observed under both the nonirradiated control conditions and the X-ray irradiation conditions. By comparing these two groups of binding proteins, we identified 79 RAD6A-specific binding proteins under the control conditions and 54 RAD6A-specific binding proteins under the X-ray irradiation conditions (Fig. 1B). Interestingly, among our indentified RAD6A binding proteins, several well-established RAD6 binding proteins were found, including RNF20 and RNF40 (9, 13–15), H2B (9, 15), and p53 (16–18), strongly supporting the reliability of our proteomic data (Fig. 1C). All RAD6A-specific binding proteins identified under both normal conditions and irradiation conditions are listed in Fig. 1D.
FIG 1.
Proteomic analysis of RAD6 interaction networks in the DNA damage response. (A) Schematic of the experimental procedures used to identify RAD6-interacting partners before and after DNA damage. HEK293T cells transfected with the GFP control or GFP-tagged RAD6A were treated with X-ray irradiation (X-ray 2.5 h) or not treated with X-ray irradiation [X-ray (−)], as indicated, at a dosage of 80 kV for 5 min, and the cells were recovered after 2.5 h. Cell extracts were prepared and subjected to co-IP assays with anti-GFP antibodies. The precipitated proteins were then subjected to mass spectrometry analysis. (B) After depletion of the nonspecific binding background by comparison with the GFP control, a number of RAD6A-interacting proteins were identified in cells without or with X-ray irradiation. The data are summarized in the Venn diagram. (C) The well-established RAD6-interacting proteins that were identified in our proteomics analysis are listed. The score refers to the obtained value analyzed by Mascot software on the basis of the original mass spectrum data. The following references mentioned in the figure appear at the indicated reference numbers in the References section: Wood et al., 2003, reference 13; Kim et al., 2009a, reference 9; Kim et al., 2009b, reference 14; Kim and Roeder, 2011, reference 15; Lyakhovich and Shekhar, 2003, reference 17; Chen et al., 2012, reference 16; and Read et al., 2014, reference 18. (D) All RAD6A-interacting proteins that appeared specifically under the control or X-ray irradiation conditions are listed. (E) The results of cellular component analysis of the RAD6A-interacting proteins that appeared under both control and X-ray irradiation conditions are shown. (F) The results of GO and pathway analyses of the RAD6A-interacting partners that appeared under both control and X-ray irradiation conditions are shown. (G) Some of the interacting proteins were selected and subjected to experimental validation. HEK293T cells were transfected with a Myc-tagged RAD6A construct combined with different randomly selected HA-tagged RAD6-interacting partners identified in our immunoprecipitation-mass spectrometry analysis for 48 h. Cells were then lysed and subjected to co-IP assays with anti-Myc antibodies. The normal mouse IgG (NIgG) antibodies were used as a negative control. The precipitated proteins were then subjected to Western blot analysis with the indicated anti-HA antibodies. CoA, coenzyme A; IP, immunoprecipitation; IB, immunoblotting.
To learn the basic biological properties of the identified binding proteins, we performed a series of bioinformatics analyses. Most RAD6A binding proteins identified under both control conditions and irradiation conditions were localized in the nucleus, cytoplasm, and cytosol (Fig. 1E). This finding was expected, as RAD6 is distributed throughout the cell (data not shown). In addition, the results of gene ontology (GO) and pathway analyses are shown in Fig. 1F. The pathway analysis suggested that the RAD6A binding proteins were tightly related to the NHEJ and HR DNA damage repair pathways under normal control conditions, while proteolysis and degradation events were significantly correlated to the RAD6A binding proteins observed under the irradiation conditions (Fig. 1F), suggesting that the proteolysis- and degradation-related pathways are likely involved in DNA damage repair.
We next randomly selected a series of RAD6A binding proteins and constructed the corresponding hemagglutinin (HA)-tagged expression plasmids. The Myc-tagged RAD6A plasmid was cotransfected with the HA-tagged interacting candidates, and the cell extracts were prepared and subjected to co-IP assays with anti-Myc antibodies under both control conditions and X-ray irradiation conditions to verify the protein-protein interactions observed in our mass spectrometry analyses. The results are shown in Fig. 1G.
Proteasome-related events are likely involved in DNA damage repair.
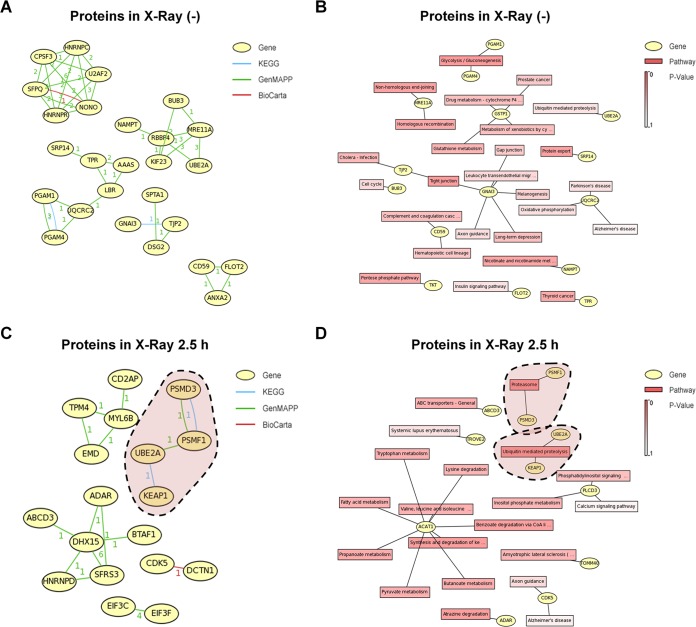
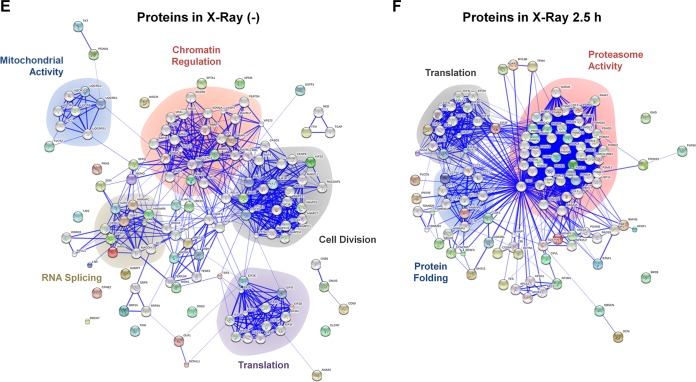
To analyze the relationship of the identified binding proteins, gene correlation and KEGG pathway analyses were performed (Fig. 2A to D). Interestingly, consistent with the findings of our GO and pathway analyses (Fig. 1F), proteasome-related events were tightly correlated to the RAD6A binding proteins observed under the X-ray irradiation conditions (Fig. 2C and D; highlighted by areas with dashed outlines), further suggesting that proteasome-related events likely play significant roles in the regulation of DNA damage repair. In addition, we performed an interaction network analysis with the STRING online service (http://string-db.org/). The RAD6A-associated proteins observed under control conditions were mainly enriched in the pathways of chromatin regulation, control of mitochondrial activity, cell division, RNA splicing, and protein translation (Fig. 2E). Intriguingly, most of these biological functions have been reported by others (9, 12, 15, 19–22), further validating the data from our mass spectrometry analysis and the related assays. However, when cells were irradiated with X rays, there was an obvious functional shift of the interaction networks of the RAD6A interaction partners. Most RAD6A binding proteins were strikingly enriched in the biological process of protein degradation (Fig. 2F), further indicating that protein degradation-related events likely play significant roles in the DNA damage response.
FIG 2.
Proteasome-related events potentially participate in the RAD6-mediated DNA damage response. (A) Network analysis of the RAD6-interacting proteins before X-ray irradiation. The numbers near the connecting lines between proteins refer to the number of times that the two specific proteins appeared in one pathway, which was analyzed by use of the indicated database. (B) KEGG pathway analysis of the RAD6-interacting proteins before X-ray irradiation. Drug metabolism-cytochrome P4 …, drug metabolism-cytochrome P450; Metabolism of xenobiotics by cy …, metabolism of xenobiotics by cytochrome P450; Leukocyte transendothelial migr, leukocyte transendothelial migration; Complement and coagulation casc, complement and coagulation cascade; Nicotinate and nicotinamide met, nicotinate and nicotinamide metabolism. (C) Network analysis of the RAD6-interacting proteins after X-ray irradiation. The numbers near the connecting lines between proteins refer to the number of times that the two specific proteins appeared in one pathway, which was analyzed by use of the indicated database. (D) KEGG pathway analysis of the RAD6-interacting proteins after X-ray irradiation. Phosphatidylinositol signaling …, phosphatidylinositol signaling system; Valine, leucine and isoleucine …, valine, leucine, and isoleucine degradation; Benzoate degradation via CoA li …, benzoate degradation via coenzyme A ligase; Synthesis and degradation of ke …, synthesis and degradation of ketone bodies; Amyotrophic lateral sclerosis …, amyotrophic lateral sclerosis (ALS). (E) STRING protein network analysis of the RAD6-interacting proteins before X-ray irradiation. (F) STRING protein network analysis of the RAD6-interacting proteins after X-ray irradiation.
Proteasome activity is essential for the efficiency of DNA damage repair.
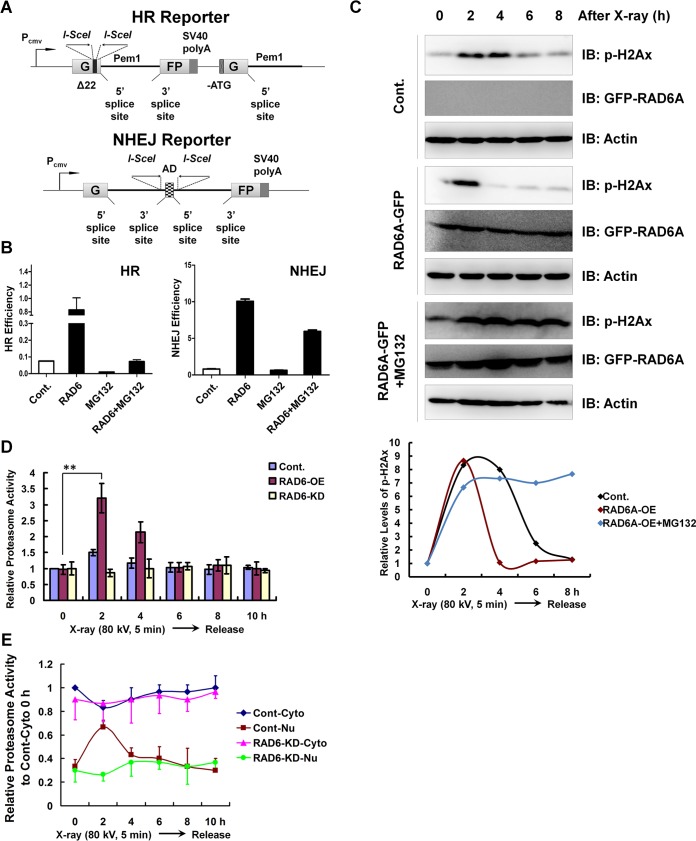
To experimentally verify whether proteasome activity is involved in the regulation of DNA damage repair, two well-established HR and NHEJ DNA damage repair reporter systems were employed (Fig. 3A). These two reporter systems were described in our previous report (12). RAD6A overexpression promoted both HR and NHEJ repair efficiency, consistent with the findings of our previous work (12). Moreover, RAD6A-promoted HR and NHEJ repair, especially HR repair, was abolished by proteasome inhibition (treatment with the proteasome-specific inhibitor MG132), indicating a novel role of proteasomes in the regulation of DNA damage repair (Fig. 3B). Phosphorylated H2Ax (p-H2Ax) is a well-established molecular marker indicating the progression of DNA damage repair. We therefore examined the effects of proteasome inhibition on p-H2Ax levels in response to DNA damage. p-H2Ax levels increased at 2 to 4 h after X-ray irradiation and recovered to basal levels at 8 h after X-ray irradiation, suggesting the completion of DNA damage repair. Consistent with the positive effect of RAD6A on HR and NHEJ repair (Fig. 3B), RAD6A overexpression promoted DNA damage repair, as p-H2Ax levels recovered more quickly than they did in the control cells. However, proteasome inhibition abolishes DNA damage repair progression, as evaluated by determination of p-H2Ax levels, even under conditions in which RAD6A is overexpressed, further supporting the essential role of proteasome activity in the regulation of DNA damage repair (Fig. 3C). In addition, we also observed that at 0 h p-H2Ax levels in MG132-treated cells were much higher than those in the other two types of cells not treated with MG132 (control [Cont.] and RAD6A-GFP-overexpressing cells) (Fig. 3C). We suspected that it is probably because MG132 treatment likely impaired the degradation of H2Ax, therefore resulting in the accumulation of H2Ax and its phosphorylated form, p-H2Ax.
FIG 3.
Proteasome activities are essential for RAD6-mediated HR and NHEJ repair. (A) Diagrams of the HR and NHEJ reporter systems. Detailed descriptions are provided in Materials and Methods. Pcmv, cytomegalovirus promoter; SV40, simian virus 40. G and FP indicate the separated parts of the GFP gene. (B) Elevated levels of HR and NHEJ repair regulated by RAD6 depend on the presence of proteasome activities. Cells carrying the HR or NHEJ reporters were transfected with or without Myc-tagged RAD6A, and cells were treated with 25 μM MG132 for another 8 h or not treated, as indicated. The HR and NHEJ repair efficiencies were calculated using the specific reporters as previously described (12). The error bars indicate the standard deviations from three biological replicates. (C) RAD6 overexpression promotes DNA damage repair, while this accelerating effect was abolished by the inhibition of proteasome activity. HEK293T cells transfected with an empty vector expressing GFP as a control or GFP-tagged RAD6A were treated with 25 μM MG132 for 8 h or not treated, as indicated. Cells were then subjected to X-ray irradiation at a dosage of 80 kV for 5 min and recovered at the indicated times. Lastly, cells were harvested and lysed for Western blot analyses with the indicated antibodies. (D) RAD6 promotes the upregulation of proteasome activities during DNA damage repair, while knockdown of RAD6 expression abolishes the proteasome activity increase during DNA damage repair. Control HEK293T cells and cells transfected with Myc-tagged RAD6A or RAD6A/B-specific siRNAs were subjected to X-ray irradiation as described above. Cells were harvested at specific recovery times, and total cell extracts were prepared. The proteasome activities were examined using the Amplit fluorimetric proteasome 20S activity assay kit (AAT Bioquest, CA; catalog number 13456) according to the manufacturer’s protocol. The error bars indicate the standard deviations from three biological replicates. **, P < 0.05. (E) X-ray irradiation induces an increase in nuclear proteasome activities, depending on the presence of RAD6. HEK293T cells were treated with X-ray irradiation as described above. Nuclear (Nu) and cytoplasmic (Cyto) fractions were prepared using a nuclear and cytoplasmic protein extraction kit (product number P0028; Beyotime, Jiangsu Province, People's Republic of China) according to the manufacturer's instruction. Then, the proteasome activities of each fraction were examined. The error bars indicate the standard deviations from three biological replicates. Cont, control.
In addition, proteasome activities were slightly elevated in the early stages after DNA damage (2 h to 4 h) and also recovered to basal levels in the late stages (6 h to 10 h) (Fig. 3D). Moreover, the elevation of proteasome activities was enhanced by RAD6A overexpression (RAD6-OE) and blocked by RAD6 depletion, i.e., RAD6 knockdown (RAD6-KD), suggesting that RAD6 is involved in the regulation of DNA damage-induced proteasome activities (Fig. 3D). As DNA damage repair is a nuclear event, we examined the proteasome activities in the cytoplasm and nucleus separately. Proteasome activities increased in the nuclear fraction after DNA damage (2 h) and decreased in the cytoplasmic fraction (2 h), suggesting that the nuclear translocation of the proteasomes might occur during the DNA damage response (Fig. 3E). This result is consistent with a previous report indicating that nuclear translocation is essential for the DNA damage response in yeast (23).
RAD6 regulates the degradation of the proteasome inhibitor PSMF1 induced by X-ray irradiation.
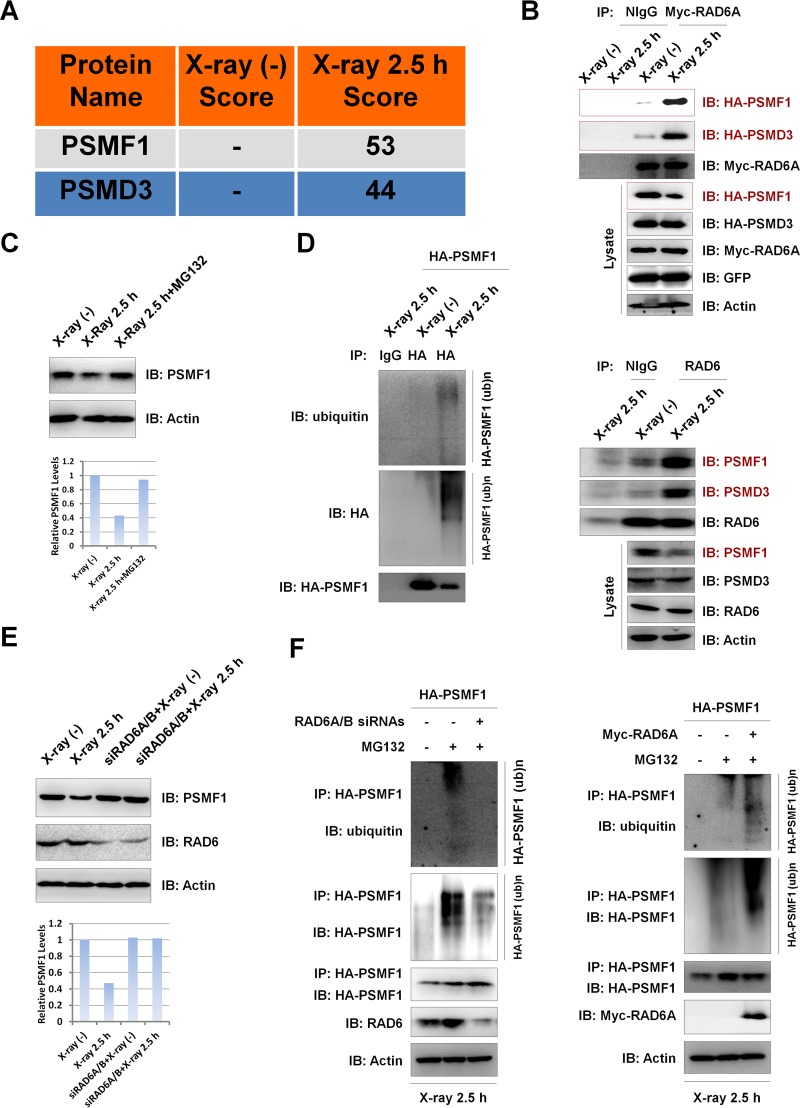
Intriguingly, from the data from our mass spectrometry analysis, we identified the proteasome-inhibitory protein PSMF1 (24, 25) and the proteasome 26S subunit PSMD3 to be two potential RAD6A-interacting partners under DNA damage conditions (Fig. 4A). Together with the reported roles of RAD6 in the control of protein degradation (12, 16, 18, 20, 26–29), we propose that the observed increase in the proteasome activities in RAD6-overexpressing cells under X-ray irradiation conditions (Fig. 3D) was achieved through the enhanced degradation of PSMF1 by RAD6. To test this hypothesis, we first verified the interaction between RAD6A and PSMF1. HEK293T cells were transfected with Myc-tagged RAD6A and HA-tagged PSMF1, and the cells were treated with (2.5 h) or without (0 h) X-ray irradiation, as in the assay whose results are presented in Fig. 1. Cells were harvested and subjected to co-IP assays with anti-Myc antibodies. RAD6A interacted with PSMF1, especially under X-ray irradiation conditions (Fig. 4B, top). Meanwhile, consistent with the data from our mass spectrometry analysis, RAD6A also interacted with PSMD3 in HEK293T cells under X-ray irradiation conditions (Fig. 4B, top). In addition, the results of the endogenous co-IP assays with anti-RAD6 antibody also support these results (Fig. 4B, bottom). Interestingly, we also detected a significant decrease in the PSMF1 protein levels but not in the PSMD3 protein levels after X-ray irradiation, both exogenous and endogenous (Fig. 4B, Lysate). This observed downregulation of PSMF1 protein levels after X-ray irradiation correlates well with the upregulated proteasome activity induced by X-ray irradiation (Fig. 3D), suggesting that the X-ray irradiation-induced increase in proteasome activity likely resulted from the decreased PSMF1 protein levels.
FIG 4.
X-ray irradiation promotes the degradation of PSMF1 through the ubiquitin-proteasome pathway regulated by RAD6. (A) PSMF1 and PSMD3 are potential RAD6-interacting partners according to the data from our mass spectrometry analysis in Fig. 1. Score refers to the obtained value analyzed by Mascot software on the basis of the original mass spectrum data. (B) DNA damage stimulates the interactions of RAD6A with PSMF1 or PSMD3 and results in a decrease in PSMF1 protein levels. (Top) HEK293T cells were transfected with an empty vector expressing GFP, Myc-tagged RAD6A, and HA-tagged PSMF1 and PSMD3, as indicated, and cells were subjected to X-ray irradiation (X-ray 2.5 h) or not irradiated (as a control), as indicated. Cell extracts were prepared and subjected to co-IP assays with anti-Myc antibodies, followed by Western blot analyses with the indicated antibodies. The empty vector expressing GFP was used as an external control to indicate that the observed changes were not due to the different transfection efficiencies. (Bottom) For endogenous co-IP assays, HEK293T cells were harvested and subjected to co-IP assays with anti-RAD6 antibodies, followed by Western blot analyses with the indicated antibodies. (C) The X-ray-induced decrease in PSMF1 depends on proteasome activity, suggesting that the observed PSMF1 downregulation occurs through the proteasome-mediated protein degradation pathway. HEK293T cells treated with 25 μM MG132 for 8 h or not treated were or were not subjected to X-ray irradiation, as indicated. Cell extracts were prepared and subjected to Western blot assays with the indicated antibodies (top), and the bands were semiquantified (bottom). (D) X-ray irradiation promotes the ubiquitination of PSMF1. HEK293T cells transfected with HA-tagged PSMF1 were subjected to X-ray irradiation or not irradiated, as indicated. HA-PSMF1 was precipitated with anti-HA antibodies under denaturing conditions, and the precipitates were subjected to Western blot analyses with the indicated antibodies. (E) X-ray irradiation-induced PSMF1 degradation depends on the presence of RAD6. HEK293T cells transfected with a control siRNA or RAD6A/B-specific siRNAs were treated with X-ray irradiation or not irradiated as previously described. Cell extracts were prepared and subjected to Western blot analyses with the indicated antibodies (top), and the bands were semiquantified (bottom). (F) RAD6 regulates PSMF1 ubiquitination under X-ray irradiation conditions. HEK293T cells expressing HA-tagged PSMF1 were transfected with a control siRNA (−) or RAD6A/B-specific siRNAs (+) for 48 h, and cells were treated with MG132 for 8 h (+) or not treated (−), as indicated. Cells were then subjected to X-ray irradiation for 5 min and recovered after another 2.5 h. Immunoprecipitation assays were performed under denaturing conditions with anti-HA antibodies followed by Western blotting analyses with the indicated antibodies (left). HEK293T cells were transfected with HA-tagged PSMF1 with or without a Myc-tagged RAD6A plasmid for 48 h, and cells were treated with MG132 for 8 h or not treated, as indicated. Cells were then subjected to X-ray irradiation for 5 min and recovered after another 2.5 h. Immunoprecipitation assays were performed under denaturing conditions with anti-HA antibodies, followed by Western blot analyses with the indicated antibodies (right). (ub)n, ubiquitination.
To further explore the potential mechanisms of X-ray irradiation-induced downregulation of PSMF1 protein levels, we first examined whether this occurred through a posttranslational mechanism. Blocking proteasome activity with MG132 significantly abolished the X-ray irradiation-induced decrease in PSMF1 protein levels (Fig. 4C), and X-ray irradiation also promoted the ubiquitination of PSMF1 (Fig. 4D). Moreover, the X-ray irradiation-induced downregulation of PSMF1 protein levels depended on the presence of RAD6, as RAD6 knockdown blocked the X-ray irradiation-induced decrease in PSMF1 protein levels. There was no obvious effect on the PSMF1 protein levels under normal conditions without X-ray irradiation upon RAD6 depletion (Fig. 4E). This result is consistent with the observation that the interaction between RAD6A and PSMF1 was obvious only with X-ray irradiation (Fig. 4B). As RAD6 is an E2 ubiquitin-conjugating enzyme that is tightly involved in the regulation of protein degradation (12, 16, 18, 20, 26–29), we next examined the effect of RAD6 on PSMF1 ubiquitination. Under X-ray irradiation conditions, RAD6 depletion significantly blocked the ubiquitination of PSMF1 (Fig. 4F, left), while overexpression of RAD6A strikingly increased the ubiquitination of PSMF1 (Fig. 4F, right). However, there was only a very slight effect of RAD6A on PSMF1 ubiquitination under control conditions without irradiation (data not shown), which is consistent with their irradiation-specific interaction.
Together, our results suggest that RAD6 regulates the degradation of the proteasome-inhibitory protein PSMF1 via the ubiquitin-proteasome pathway in human cells upon X-ray irradiation, which likely contributes to the observed increase in proteasome activity induced by X-ray irradiation.
RAD6 regulates the degradation of LBR and contributes to the nuclear translocation of proteasomes.
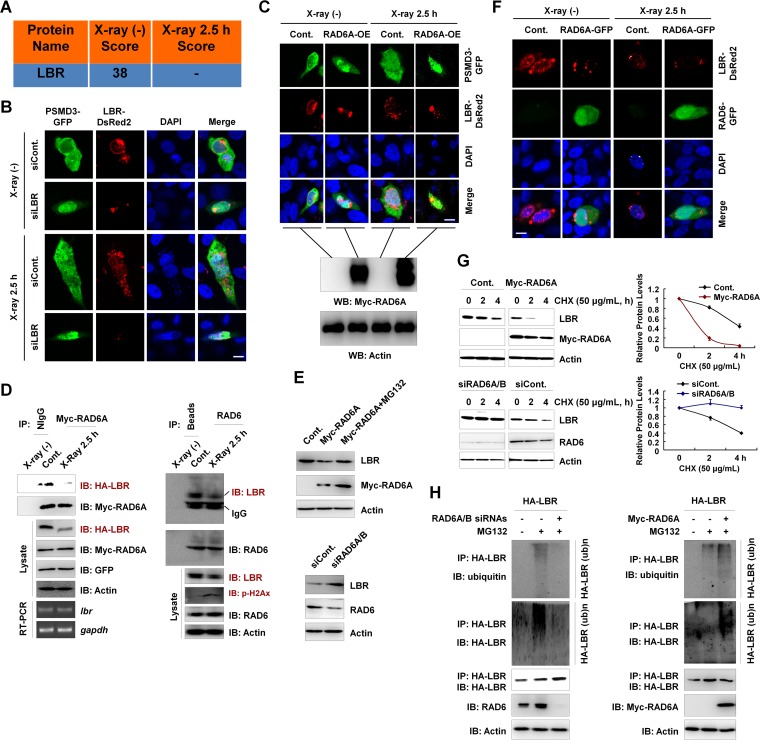
We observed elevated nuclear proteasome activity in response to X-ray irradiation, and this enhancement of nuclear proteasome activity likely depended on the existence of RAD6 (Fig. 3E). We next wondered how the proteasome nuclear translocation is regulated. In Schizosaccharomyces pombe, the RAD6 homolog Rhp6 regulates proteasome nuclear translocation through the Rhp6-Cut8 axis. Cut8 is a nuclear envelope protein that blocks the nuclear localization of proteasomes (23, 30, 31). Rhp6 interacts with Cut8 and promotes the ubiquitination and degradation of Cut8 via the ubiquitin-proteasome pathway, further contributing to the nuclear translocation of proteasomes (23). Interestingly, from data from our mass spectrometry analysis, we observed that the lamin B receptor (LBR), which functions as a nuclear envelope protein similar to Schizosaccharomyces pombe Cut8, is a candidate RAD6A binding protein under nonirradiated conditions (Fig. 5A). We next tested whether LBR controls the subcellular localization of proteasomes in human cells. LBR depletion resulted in the nuclear translocation of proteasomes, as indicated by the proteasome 26S subunit PSMD3, in both normal control (non-X-ray-irradiated) and X-ray-irradiated cells (Fig. 5B), indicating that LBR is likely a functional ortholog of Schizosaccharomyces pombe Cut8 in human cells. In addition, we also observed that X-ray irradiation also resulted in the weak nuclear translocation of proteasomes and a slight disruption of normal nuclear envelopes, as indicated by the LBR signals (Fig. 5B). Moreover, RAD6A overexpression also resulted in the significant nuclear translocation of proteasomes in both X-ray-irradiated and nonirradiated cells (Fig. 5C), suggesting that RAD6-mediated proteasome nuclear translocation is conserved in human cells and is likely achieved through controlling LBR protein levels.
FIG 5.
LBR regulates the nuclear localization of proteasomes, and the degradation of LBR is affected by RAD6. (A) LBR is a potential RAD6-interacting protein, as indicated in our mass spectrometry analysis in Fig. 1. The score refers to the obtained value analyzed by Mascot software on the basis of the original mass spectrum data. (B) Knockdown of LBR results in the nuclear translocation of proteasomes, as indicated by PSMD3 immunostaining. HEK293T cells were transfected with a GFP-tagged PSMD3 plasmid and a DsRed2-tagged LBR together with a control siRNA (siCont) or an LBR-specific siRNA (siLBR), as indicated, for 48 h. Cells were irradiated with X rays for 2.5 h or not irradiated. Cells were then subjected to immunofluorescence assays by confocal laser microscopy. DAPI (4′,6-diamidino-2-phenylindole) staining was used to indicate the cell nucleus. Bar, 10 μm. (C) RAD6 overexpression promotes the nuclear translocation of proteasomes. HEK293T cells stably expressing a Myc-tagged RAD6A plasmid were transfected with GFP-tagged PSMD3 to indicate the proteasomes and DsRed2-tagged LBR for 48 h. Cells were irradiated with X rays for 2.5 h or not irradiated. (Top) The cell nucleus is indicated by DAPI staining. Cells were then subjected to immunofluorescence assays by confocal laser microscopy. Bar, 10 μm. (Bottom) The results of validation of Myc-RAD6A overexpression by Western blotting (WB). (D) RAD6 interacts with LBR, and X-ray irradiation downregulates LBR protein levels. Myc-tagged RAD6A and HA-tagged LBR were cotransfected into HEK293T cells for 48 h. Cells were then subjected to X-ray irradiation (X-ray 2.5 h) or not irradiated (control) as indicated. Cell extracts were prepared and subjected to co-IP analyses with anti-Myc antibodies, and Western blot assays were performed with the indicated antibodies. Reverse transcription-PCR (RT-PCR) assays were also performed using cells treated similarly. The detected genes are indicated (left). gapdh, glyceraldehyde-3-phosphate dehydrogenase gene. For endogenous co-IP assays, HEK293T cells were harvested and subjected to co-IP assays with anti-RAD6 antibodies, followed by Western blot analyses with the indicated antibodies (right). (E) RAD6 controls LBR protein levels in a proteasome-dependent manner. HEK293T cells transfected with or without (control) a Myc-tagged RAD6A plasmid were treated with MG132 for 8 h or not treated, as indicated. (Top) Cell extracts were prepared and subjected to Western blot analyses with the indicated antibodies. (Bottom) Cells transfected with a control siRNA or RAD6A/B-specific siRNAs (siRAD6A/B) were lysed and subjected to Western blot assays with the indicated antibodies. (F) Immunostaining assays support the conclusion that RAD6 regulates LBR protein levels. HEK293T cells were transfected with an equal amount of DsRed2-tagged LBR together with or without (control) a GFP-tagged RAD6A plasmid for 48 h. Cells were then irradiated with X rays for 2.5 h or not. Cells were subjected to immunofluorescence assays by confocal laser microscopy. DAPI staining was used to indicate the cell nucleus. Bar, 10 μm. (G) RAD6 regulates the degradation of LBR. (Top) Cells transfected with an empty vector expressing Myc or Myc-tagged RAD6A were treated with 50 μg/ml cycloheximide (CHX) for the indicated times. (Left) Cells were then harvested and subjected to Western blot analyses with the indicated antibodies. (Right) The bands were quantified, and the bars indicate the standard deviations from three biological replicates. (Bottom) Cells transfected with a control siRNA or RAD6A/B-specific siRNAs were treated with 50 μg/ml CHX for the indicated times. (Left) Cells were then harvested and subjected to Western blot analyses with the indicated antibodies. (Right) The bands were quantified, and the bars indicate the standard deviations from three biological replicates. (H) RAD6 regulates the ubiquitination of LBR. (Left) HEK293T cells expressing HA-tagged LBR were transfected with a control siRNA (−) or RAD6A/B-specific siRNAs (+) for 48 h, and cells were treated with MG132 for 8 h or not treated, as indicated. Immunoprecipitation assays were performed under denaturing conditions with anti-HA antibodies, followed by Western blot analyses with the indicated antibodies. (Right) HEK293T cells were transfected with the HA-tagged LBR plasmid with or without a Myc-tagged RAD6A plasmid for 48 h, and cells were treated with MG132 for 8 h or not treated, as indicated. Immunoprecipitation assays were performed under denaturing conditions with anti-HA antibodies, followed by Western blot analyses with the indicated antibodies.
To prove this hypothesis, we first verified the protein-protein interaction between RAD6 and LBR under both normal control conditions and X-ray irradiation conditions. RAD6A interacted with LBR predominantly under the normal control conditions without X-ray irradiation, and the interaction was significantly abolished when cells were treated with X-ray irradiation (Fig. 5D, left). In addition, the results of our endogenous co-IP assays with anti-RAD6 antibodies also support this conclusion (Fig. 5D, right). Meanwhile, X-ray irradiation induced a significant downregulation of LBR (Fig. 5D, Lysate), suggesting a possible reason for the observed increase in nuclear proteasome activities after X-ray irradiation. Additionally, this decrease in LBR protein levels is likely not due to transcriptional regulation, as the LBR mRNA levels were not changed (Fig. 5D, left, RT-PCR [reverse transcription-PCR]). Because RAD6A interacts with LBR, we next examined the possible regulatory relationship between these two proteins. RAD6A overexpression decreased LBR protein levels, which was blocked by treatment with the proteasome inhibitor MG132, suggesting that RAD6A regulates the proteasome-mediated degradation of LBR (Fig. 5E, top). In contrast, the loss of RAD6 expression resulted in the accumulation of LBR protein levels, further supporting our hypothesis (Fig. 5E, bottom). Our immunofluorescence (IF) assays also indicated that RAD6A overexpression resulted in a significant decrease in the LBR protein levels and disruptions of the normal morphology of the LBR location (Fig. 5F). We next examined whether RAD6 regulates LBR protein degradation by a chase assay. RAD6 overexpression significantly promoted the LBR degradation rate (Fig. 5G, top), and RAD6 inhibition impaired LBR degradation (Fig. 5G, bottom). Lastly, we analyzed the effect of RAD6 on the ubiquitination of LBR. Consistently, the loss of RAD6 expression strikingly abolished the ubiquitination of LBR (Fig. 5H, left), while RAD6A overexpression strongly enhanced LBR ubiquitination (Fig. 5H, right).
Together, these results indicate that LBR is a key regulator of proteasome nuclear translocation in human cells and is downregulated in response to DNA damage. RAD6 regulates the LBR protein levels via the ubiquitin-proteasome pathway and then promotes the nuclear translocation of proteasomes.
LBR is degraded by proteasomes during the DNA damage response.
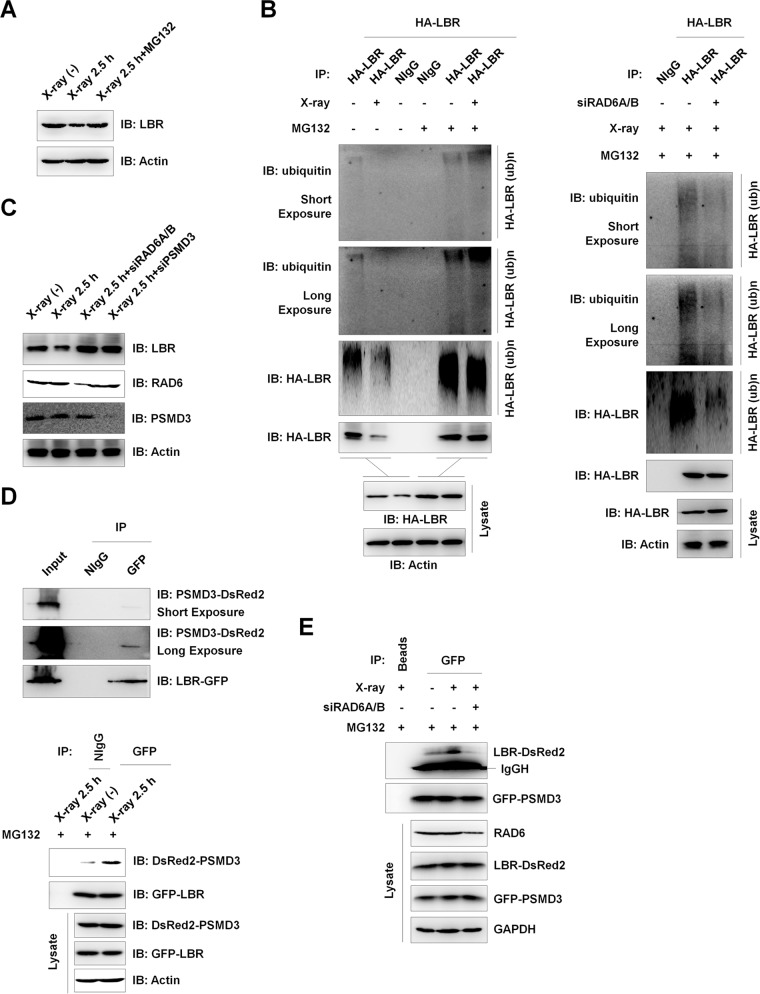
Because LBR protein levels are decreased after X-ray irradiation (Fig. 5D, Lysate), we wondered about the potential mechanisms of LBR regulation upon X-ray irradiation. We first examined whether the observed LBR protein level decrease was mediated by the proteasome-related pathway. The X-ray irradiation-induced decrease in LBR protein levels was blocked by treatment with MG132 (Fig. 6A). This observation suggested that the X-ray irradiation-induced downregulation of LBR likely occurred through the proteasome-mediated protein degradation process. We next analyzed the ubiquitination of LBR in the DNA damage response and found that LBR protein ubiquitination levels decreased significantly after X-ray irradiation without MG132 treatment (Fig. 6B, left, left two lanes). This was expected because the LBR protein is degraded after X-ray irradiation. However, when the cells were treated with MG132 to block the proteasome activities, we did not observe an increase in the level of LBR ubiquitination after X-ray irradiation. Instead, the LBR ubiquitination levels were similar between normal control cells and X-ray-irradiated cells, suggesting that X-ray irradiation does not alter the ubiquitination of LBR (Fig. 6B, left, right two lanes). Furthermore, when we knocked down RAD6 expression in HEK293T cells, LBR ubiquitination was also abolished under X-ray irradiation conditions, suggesting that there is an essential role for RAD6 in the control of LBR ubiquitination after X-ray irradiation (Fig. 6B, right). Because the interaction between RAD6 and LBR decreased significantly after X-ray irradiation (Fig. 5D), we propose that the LBR ubiquitination after X-ray irradiation is precatalyzed by RAD6 before X-ray irradiation. Consistently, the X-ray-induced downregulation of LBR protein levels was abolished when RAD6 or PSMD3 was depleted (Fig. 6C), further supporting the essential role of RAD6 and proteasomes in the control of LBR protein levels.
FIG 6.
DNA damage-induced downregulation of LBR protein levels occurs through the proteasome-mediated pathway and is regulated by RAD6 and PSMD3. (A) The X-ray-induced decrease in LBR depends on proteasome activity, suggesting that the observed LBR downregulation occurs through the proteasome-mediated protein degradation pathway. HEK293T cells treated with 25 μM MG132 for 8 h or not treated were subjected to X-ray irradiation or not irradiated, as indicated, and cells were recovered after another 2.5 h. Cell extracts were prepared and subjected to Western blot assays with the indicated antibodies. (B) X-ray irradiation does not alter the ubiquitination of LBR. HEK293T cells expressing HA-tagged LBR were treated with (+) MG132 for 8 h or not treated (−). Cells were subjected to X-ray irradiation or not irradiated as described above and recovered after another 2.5 h. Cells were harvested and subjected to immunoprecipitation assays under denaturing conditions with anti-HA antibodies. The precipitates were then used for Western blot analyses with the indicated antibodies. (C) The X-ray-induced degradation of LBR depends on the presence of RAD6 and PSMD3. HEK293T cells transfected with a control siRNA, RAD6A/B-specific siRNAs, or a PSMD3-specific siRNA (siPSMD3) were treated with X-ray irradiation or not irradiated, and cells were recovered after 2.5 h. Cell extracts were prepared and subjected to Western blot analyses with the indicated antibodies. (D) PSMD3 interacts with LBR, and their interaction is enhanced by X-ray irradiation. (Top two panels) HEK293T cells were cotransfected with a DsRed2-tagged PSMD3 plasmid and a GFP-tagged LBR plasmid for 48 h. Cell extracts were then prepared and subjected to co-IP assays with anti-GFP antibodies. Western blot analyses were performed with the indicated antibodies. (Bottom) HEK293T cells expressing GFP-LBR and DsRed2-PSMD3 were treated with MG132 for 8 h, and cells were then subjected to X-ray irradiation or not irradiated (control). The cells were recovered after another 2.5 h. Co-IP assays were employed with anti-GFP antibodies, and Western blot analyses were performed with the indicated antibodies. (E) RAD6 is essential for the interaction of PSMD3 and LBR under X-ray irradiation conditions. HEK293T cells expressing GFP-LBR and DsRed2-PSMD3 were transfected with a control siRNA (−) or RAD6A/B-specific siRNAs (+) for 48 h. Cells were then treated with MG132 for 8 h and were subjected to X-ray irradiation (+) or not irradiated (−). Cells were recovered after another 2.5 h. Cell extracts were prepared and subjected to co-IP assays with anti-GFP antibodies, followed by Western blot analyses with the indicated antibodies. IgGH, IgG heavy chain.
The Schizosaccharomyces pombe envelope protein Cut8 interacts with proteasomes both in vitro and in vivo (23). As our results also indicated that both PSMD3 and LBR are RAD6-interacting partners (Fig. 4A and 5A), we tested whether LBR interacts with proteasomes by using PSMD3 as a proteasome indicator. Indeed, LBR interacted with PSMD3 under nonirradiation conditions (Fig. 6D, top), and their interaction was enhanced after X-ray irradiation (Fig. 6D, bottom). In addition, the PSMD3-LBR interaction was abolished by RAD6 depletion, suggesting that RAD6 is essential for the PSMD3-LBR interaction (Fig. 6E).
Combined with the findings that the interaction of RAD6 with LBR was decreased (Fig. 5D) while the interaction of RAD6 with PSMD3 was enhanced after X-ray irradiation (Fig. 4B), we propose that the RAD6 protein released from LBR might recruit PSMD3 (proteasomes) to LBR by interacting with PSMD3 after X-ray irradiation.
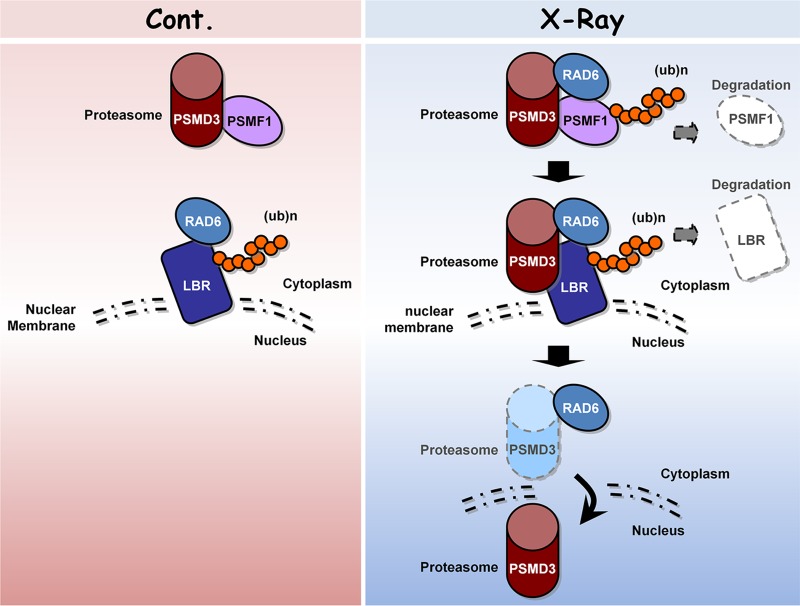
Taken together, this work provides valuable information about the interaction networks of RAD6 before and after DNA damage. This information will be helpful in more globally understanding the roles of RAD6 in the DNA damage response. Moreover, our interactome analysis also provides novel insights into new potential pathways that are employed by RAD6 in DNA damage repair. In addition, based on our proteomic analysis, we further showed that proteasome-related events likely play significant roles in the regulation of DNA damage repair. We found that DNA damage enhances proteasome activity and nuclear translocation, which are essential for the DNA damage response. The proteasome activity is regulated by RAD6 through controlling the ubiquitination and degradation of PSMF1, while the nuclear translocation of proteasomes is regulated by RAD6 via the control of the ubiquitination and degradation of LBR. When cells are exposed to X-ray irradiation, the degradation of PSMF1 and LBR by RAD6 is enhanced, further increasing proteasome activity and nuclear localization (Fig. 7).
FIG 7.
Working model. Under normal conditions, proteasomes are primarily localized in the cytoplasm and proteasome activities are inhibited by the inhibitory protein PSMF1. However, when cells encounter DNA damage stress, such as X-ray irradiation, the proteasome activity is enhanced and more proteasomes enter the nucleus. The proteasome activity is regulated by RAD6 through controlling the ubiquitination and degradation of PSMF1, while the nuclear translocation of proteasomes is also regulated by RAD6 via the control of LBR ubiquitination and degradation. When cells are exposed to X-ray irradiation, the degradation of PSMF1 and LBR by RAD6 is enhanced, further increasing proteasome activity and nuclear localization.
DISCUSSION
Efficient repair of damaged DNA is critical to maintain normal life processes in organisms. RAD6 is a key DNA damage repair regulator that controls both the error-prone and error-free DNA damage repair pathways (3, 4). In addition to the well-known RAD6-PCNA pathway, H2B monoubiquitination and HP1α are also involved in the RAD6-mediated DNA damage repair process, suggesting that multiple pathways are likely employed by RAD6 in the DNA damage response (10–12). Therefore, a global understanding of the RAD6-mediated DNA damage repair process is required. To this end, we performed an interactome analysis to dissect the dynamics of the RAD6 interaction networks in response to DNA damage. This proteomic analysis provides valuable insights into the novel pathways possibly involved in the RAD6-mediated DNA damage response. We found that the proteasome-related events are likely involved in the DNA damage response, as after DNA damage many RAD6 binding proteins are tightly related to proteasome regulation (Fig. 2). Moreover, proteasome activities are indeed essential for efficient HR and NHEJ repair regulated by RAD6 (Fig. 3B and C), supporting the conclusion that proteasome-related events are involved in RAD6-mediated DNA repair. These findings are consistent with previous reports that the ubiquitin-proteasome pathways participate in the regulation of DNA damage repair in both proteolytic and nonproteolytic manners (32–35). For instance, two proteolytic pathways, the Ubr1/Rad6-dependent N-end-rule pathway and the Ufd4/Ubc4-dependent ubiquitin fusion degradation (UFD) pathway, participate in the regulation of DNA damage repair in yeast by controlling the degradation of Mgt1 (33).
As proteasome activity and localization are both changed accordingly in response to DNA damage in a RAD6-dependent manner (Fig. 3D and E), we further determined the potential regulatory mechanisms. PSMF1 is a proteasome-inhibitory protein, and PSMF1 protein levels decreased after DNA damage (Fig. 4B and C), correlating with the increased proteasome activity after DNA damage. Moreover, X-ray irradiation induced an interaction between RAD6 and PSMF1 and further promoted the ubiquitination and degradation of PSMF1 (Fig. 4A, B, E, and F). Therefore, these findings suggest a possible mechanism employed by RAD6 to regulate proteasome activity in the DNA damage response and also suggest a novel role for RAD6 in the regulation of proteasome activity.
Our results showed that LBR is likely a critical regulator of proteasome nuclear translocation in the DNA damage response (Fig. 5B), and the degradation of LBR is also regulated by RAD6. In nonirradiated control cells, RAD6 interacts with and ubiquitinates LBR, further affecting the stability of LBR (Fig. 5D to H). However, due to the reduced interaction between LBR and proteasomes under the control conditions (Fig. 6D), the LBR protein is maintained at a relatively higher level. When cells encountered DNA damage stresses, the interaction between RAD6 and proteasomes was enhanced (Fig. 4B). This resulted in increased proteasome activity by promoting the degradation of PSMF1 (Fig. 4) and the recruitment of proteasomes to LBR (Fig. 6D and E), eventually leading to the degradation of LBR. Therefore, proteasomes can enter the nucleus and contribute to DNA damage repair (Fig. 7).
Similar to our findings that RAD6 regulates proteasome nuclear translocation in human cells, previous reports also suggested that the Schizosaccharomyces pombe homolog of RAD6, Rph6, regulates proteasome nuclear localization in Schizosaccharomyces pombe by controlling the degradation of the nuclear envelope protein Cut8 (23). Therefore, the role of RAD6 in the regulation of proteasome localization is conserved from yeast to human. No homolog of Schizosaccharomyces pombe Cut8 has been characterized in human cells, and our results suggest that the human LBR protein possesses a similar function in the control of proteasome localization and is also regulated by RAD6. Therefore, we propose that LBR is a functional homolog of Schizosaccharomyces pombe Cut8 in human cells.
MATERIALS AND METHODS
Cell culture and transfection.
Human embryonic kidney 293T (HEK293T) cells were cultured at 37°C in Dulbecco modified Eagle medium (DMEM; catalog number 11960-044; Gibco) supplemented with 10% fetal bovine serum and 1% penicillin and streptomycin (catalog number 15070-063; Gibco) in a 5% CO2 incubator. The transfection of constructs into cells was performed with the Lipofectamine 2000 reagent (catalog number 11668-019; Invitrogen) according to the manufacturer's standard protocol.
The two reporter cell lines used to analyze NHEJ and HR repair efficiencies, HCA2-I9a and HCA2-H15C, were grown in DMEM (catalog number 11960-044; Gibco) supplemented with 10% fetal bovine serum, 1× nonessential amino acids (catalog number 11140-050; Gibco), and 1% penicillin and streptomycin (catalog number 15070-063; Gibco) in an Hera240i incubator with 5% CO2 and 3% O2 at 37°C. The cells were transfected with different plasmids or small interfering RNAs (siRNAs) with a Lonza 4D electroporator using the DT-130 program.
Plasmid construction.
Plasmids pCMV-Myc, pCMV-HA, pEGFP-N1, and pDsRed2-N1 (catalog numbers 635689, 635690, 6085-1, and 632406, respectively; Clontech) expressing RAD6A, PSMF1, LBR, or PSMD3 were constructed by cloning the RAD6A, PSMF1, LBR, or PSMD3 PCR product into the pCMV-Myc, pCMV-HA, pEGFP-N1, and pDsRed2-N1 vectors, as indicated above and below.
Coimmunoprecipitation (co-IP).
Cells were transfected with different plasmids as indicated above and below. After 48 h, the cells were harvested, washed with ice-cold phosphate-buffered saline, resuspended in ATM lysis buffer (containing 100 mM Tris-HCl, pH 7.5, 150 mM NaCl, 0.2 mM EDTA, 20% glycerol, 0.4% NP-40, 2% Tween 20, and 0.2 mM phenylmethylsulfonyl fluoride [PMSF]), and sonicated on ice 10 times (for 3 s each time) with a 20% efficiency. The cell lysates were incubated with normal mouse IgG (as a negative control; catalog number sc-2025; Santa Cruz Biotechnology), anti-HA (Zhongshan Golden Bridge Company, People's Republic of China), or anti-GFP (Zhongshan Golden Bridge) antibody at 4°C overnight. Protein A/G-agarose beads (catalog number sc-2003; Santa Cruz Biotechnology) were subsequently added. The solution was incubated for another 3 h, followed by centrifugation to harvest the agarose beads after they had been washed 5 times with lysis buffer. The precipitated proteins were released by boiling in loading buffer and resolved via SDS-PAGE (12%). Immunoblot analyses were performed with the antibodies indicated in the figures.
Mass spectrometry analysis.
Mass spectrometry analysis was performed by PTM BioLabs, Inc. (Zhejiang Province, People's Republic of China). Briefly, for in-gel tryptic digestion, gel pieces were destained in 50 mM NH4HCO3 in 50% (vol/vol) acetonitrile (ACN) until they were clear. Gel pieces were dehydrated with 100 μl of 100% ACN for 5 min, the liquid was removed, and the gel pieces were rehydrated in 10 mM dithiothreitol and incubated at 5°C for 60 min. The gel pieces were again dehydrated in 100% ACN, the liquid was removed, and the gel pieces were rehydrated with 55 mM iodoacetamide. Samples were incubated at room temperature in the dark for 45 min. Gel pieces were washed with 50 mM NH4HCO3 and dehydrated with 100% ACN. Gel pieces were rehydrated with 10 ng/μl trypsin resuspended in 50 mM NH4HCO3 on ice for 1 h. Excess liquid was removed, and gel pieces were digested with trypsin at 37°C overnight. Peptides were extracted with 50% ACN–5% formic acid (FA), followed by 100% ACN. Peptides were dried to completion and resuspended in 2% ACN–0.1% FA.
The tryptic peptides were dissolved in 0.1% FA, and the solution was directly loaded onto a reversed-phase precolumn (Acclaim PepMap 100; Thermo Scientific). Peptide separation was performed using a reversed-phase analytical column (Acclaim PepMap rapid separation liquid chromatography [RSLC]; Thermo Scientific). The gradient was comprised of an increase from 5% to 40% solvent B (0.1% FA in 98% ACN) over 9 min, and the gradient then climbed to 80% solvent B in 3 min and was held at 80% solvent B for the last 3 min; all gradients were performed at a constant flow rate of 350 nl/min on an EASY-nLC 1000 ultraperformance liquid chromatography (UPLC) system. The resulting peptides were analyzed by use of a Q Exactive Plus hybrid quadrupole-Orbitrap mass spectrometer (Thermo Fisher Scientific).
The peptides were subjected to a nanospray ionization source followed by tandem mass spectrometry (MS/MS) in the Q Exactive Plus mass spectrometer (Thermo) coupled online to the UPLC. Intact peptides were detected in the Orbitrap mass spectrometer at a resolution of 70,000. Peptides were selected for MS/MS using a normalized collision energy (NCE) setting of 28; ion fragments were detected in the Orbitrap mass spectrometer at a resolution of 17,500. A data-dependent procedure that alternated between 1 MS scan followed by 20 MS/MS scans was applied for the top 20 precursor ions above a threshold ion count of 10,000 in the MS survey scan with a 15.0-s dynamic exclusion. The electrospray voltage applied was 2.0 kV. Automatic gain control (AGC) was used to prevent overfilling of the Orbitrap mass spectrometer; 5E4 ions were accumulated for the generation of MS/MS spectra. For the MS scans, the m/z scan range was 350 to 1,800.
The resulting MS/MS data were processed using the Mascot search engine (v.2.3.0). Tandem mass spectra were searched against those in the Swiss-Prot human proteome database. Trypsin/P was specified as the cleavage enzyme, and up to 2 missing cleavages were allowed. The mass error was set to 10 ppm for precursor ions and 0.02 Da for fragment ions. Carbamidomethyl on Cys was specified as a fixed modification, and oxidation on Met and acetylation on the protein N terminus were specified as variable modifications. The peptide ion score was set at ≥20.
Antibodies and Western blot analysis.
Antibodies against HA, Myc, DsRed2, and GFP were purchased from Zhongshan Golden Bridge. An antibody against RAD6 was purchased from Santa Cruz Biotechnology (catalog number sc-30078). Antibodies against PSMF1 and LBR were purchased from Proteintech (catalog numbers 12941-1-AP and 12398-1-AP, respectively). An antibody against PSMD3 was purchased from Santa Cruz Biotechnology (catalog number sc-107978). An antiubiquitin antibody was purchased from R&D (catalog number MAB701). All horseradish peroxidase (HRP)-conjugated secondary antibodies were purchased from Zhongshan Golden Bridge.
Cells were lysed in ATM lysis buffer (containing 100 mM Tris-HCl, pH 7.5, 150 mM NaCl, 0.2 mM EDTA, 20% glycerol, 0.4% NP-40, 2% Tween 20, and 0.2 mM PMSF). The protein concentration in the supernatant was measured with a bicinchoninic acid assay kit (catalog number 71285-3; Novagen). Then, the samples were loaded into a 15% gel to resolve the proteins. Different amounts of total protein were loaded in each experiment to facilitate the detection of different target proteins. After electrophoresis, the proteins were transferred to polyvinylidene difluoride membranes (catalog number 10600021; Amersham) and hybridized with primary antibodies at a dilution of 1:2,000. The HRP-labeled secondary antibodies (Zhongshan Golden Bridge) were applied at a dilution of 1:4,000. An enhanced chemiluminescence detection system (catalog number 345818; Calbiochem) was used to detect the signals on the membranes.
In vivo ubiquitination assay.
HEK293T cells expressing HA-tagged specific proteins were transfected with control siRNA, RAD6A/B-specific siRNAs, a plasmid expressing RAD6A, or an empty control plasmid. At 48 h posttransfection, an in vivo ubiquitination assay was performed under denaturing conditions. The cells were lysed with 100 μl of SDS lysis buffer containing 1% SDS, and the lysate was boiled for 15 min. The resulting lysate was centrifuged for 15 min at 12,000 rpm at 4°C. The supernatant was diluted to 0.1% SDS with 900 μl of ATM lysis buffer. The lysate was subsequently incubated with a normal mouse IgG antibody (catalog number sc-2025; Santa Cruz Biotechnology) or anti-HA antibody (catalog number 2616; Cell Signaling Technology) at 4°C overnight. Protein A/G-agarose beads were then added to precipitate the bound proteins. The ubiquitination levels of the different target proteins were detected in Western blot assays with antibodies against ubiquitin (catalog number MAB701; R&D).
Immunofluorescence and laser confocal microscopy.
Immunofluorescence staining was performed as described in our previous report (16). The primary antibody anti-LBR (catalog number 12398-1-AP; Proteintech) was used at a dilution of 1:50. The secondary antibody coupled to Texas Red (1:100) was purchased from the Zhongshan Golden Bridge Company, People's Republic of China. Images were photographed with a laser scanning confocal microscope (Leica) with a 100× oil immersion objective.
HR and NHEJ DNA repair efficiency analysis.
The HR and NHEJ DNA damage repair efficiency assays were performed as we previously described (12). Briefly, the HR reporter cassette consists of two mutated copies of GFP-Pem1. In the first copy of GFP-Pem1, the first GFP exon contains an insertion of two I-SceI recognition sites in an inverted orientation and a deletion of 22 nucleotides (Δ22), which ensures that GFP cannot be reconstituted by an NHEJ event. The second copy of GFP-Pem1 lacks a promoter, the first ATG, and the second exon of GFP. Upon induction of double-strand breaks (DSBs) by I-SceI transfection, gene conversion events reconstitute an active GFP gene. The NHEJ reporter cassette consists of a GFP gene under the control of a cytomegalovirus promoter with an engineered intron from the rat Pem1 gene. This intron is interrupted by an adenoviral exon (AD). The adenoviral exon is flanked by I-SceI recognition sites in an inverted orientation for the induction of DSBs. In this construct, the GFP gene is inactive. However, the induction of a DSB and successful NHEJ triggers the construct to express GFP.
Proteasome activity analysis.
The proteasome activity assays were performed with an Amplite fluorimetric proteasome 20S activity assay kit (product number: 13456; AAT Bioquest, CA) according to the manufacturer's instructions.
ACKNOWLEDGMENTS
This work was supported by the basic research program of the Ministry of Science and Technology of China (grant no. 2016YFA0100400, 2015CB964800, 2015CB856204, 2014CB964603, and 2014CB965001) and the National Natural Science Foundation of China (grant no. 91419304, 31330043, and 31271534).
We thank Zhiyong Mao's lab at Tongji University for help with the HR and NHEJ analyses.
We declare no conflicts of interest.
REFERENCES
- 1.Osborn AJ, Elledge SJ, Zou L. 2002. Checking on the fork: the DNA-replication stress-response pathway. Trends Cell Biol 12:509–516. doi: 10.1016/S0962-8924(02)02380-2. [DOI] [PubMed] [Google Scholar]
- 2.Roos WP, Thomas AD, Kaina B. 2016. DNA damage and the balance between survival and death in cancer biology. Nat Rev Cancer 16:20–33. doi: 10.1038/nrc.2015.2. [DOI] [PubMed] [Google Scholar]
- 3.Hoege C, Pfander B, Moldovan GL, Pyrowolakis G, Jentsch S. 2002. RAD6-dependent DNA repair is linked to modification of PCNA by ubiquitin and SUMO. Nature 419:135–141. doi: 10.1038/nature00991. [DOI] [PubMed] [Google Scholar]
- 4.Pfander B, Moldovan GL, Sacher M, Hoege C, Jentsch S. 2005. SUMO-modified PCNA recruits Srs2 to prevent recombination during S phase. Nature 436:428–433. [DOI] [PubMed] [Google Scholar]
- 5.Weissman AM. 2001. Themes and variations on ubiquitylation. Nat Rev Mol Cell Biol 2:169–178. doi: 10.1038/35056563. [DOI] [PubMed] [Google Scholar]
- 6.Ulrich HD, Jentsch S. 2000. Two RING finger proteins mediate cooperation between ubiquitin-conjugating enzymes in DNA repair. EMBO J 19:3388–3397. doi: 10.1093/emboj/19.13.3388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bailly V, Lamb J, Sung P, Prakash S, Prakash L. 1994. Specific complex formation between yeast RAD6 and RAD18 proteins: a potential mechanism for targeting RAD6 ubiquitin-conjugating activity to DNA damage sites. Genes Dev 8:811–820. doi: 10.1101/gad.8.7.811. [DOI] [PubMed] [Google Scholar]
- 8.Kim J, Hake SB, Roeder RG. 2005. The human homolog of yeast BRE1 functions as a transcriptional coactivator through direct activator interactions. Mol Cell 20:759–770. doi: 10.1016/j.molcel.2005.11.012. [DOI] [PubMed] [Google Scholar]
- 9.Kim J, Guermah M, McGinty RK, Lee JS, Tang Z, Milne TA, Shilatifard A, Muir TW, Roeder RG. 2009. RAD6-mediated transcription-coupled H2B ubiquitylation directly stimulates H3K4 methylation in human cells. Cell 137:459–471. doi: 10.1016/j.cell.2009.02.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nakamura K, Kato A, Kobayashi J, Yanagihara H, Sakamoto S, Oliveira DV, Shimada M, Tauchi H, Suzuki H, Tashiro S, Zou L, Komatsu K. 2011. Regulation of homologous recombination by RNF20-dependent H2B ubiquitination. Mol Cell 41:515–528. doi: 10.1016/j.molcel.2011.02.002. [DOI] [PubMed] [Google Scholar]
- 11.Moyal L, Lerenthal Y, Gana-Weisz M, Mass G, So S, Wang SY, Eppink B, Chung YM, Shalev G, Shema E, Shkedy D, Smorodinsky NI, van Vliet N, Kuster B, Mann M, Ciechanover A, Dahm-Daphi J, Kanaar R, Hu MC, Chen DJ, Oren M, Shiloh Y. 2011. Requirement of ATM-dependent monoubiquitylation of histone H2B for timely repair of DNA double-strand breaks. Mol Cell 41:529–542. doi: 10.1016/j.molcel.2011.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen S, Wang C, Sun L, Wang DL, Chen L, Huang Z, Yang Q, Gao J, Yang XB, Chang JF, Chen P, Lan L, Mao Z, Sun FL. 2015. RAD6 promotes homologous recombination repair by activating the autophagy-mediated degradation of heterochromatin protein HP1. Mol Cell Biol 35:406–416. doi: 10.1128/MCB.01044-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wood A, Krogan NJ, Dover J, Schneider J, Heidt J, Boateng MA, Dean K, Golshani A, Zhang Y, Greenblatt JF, Johnston M, Shilatifard A. 2003. Bre1, an E3 ubiquitin ligase required for recruitment and substrate selection of Rad6 at a promoter. Mol Cell 11:267–274. doi: 10.1016/S1097-2765(02)00802-X. [DOI] [PubMed] [Google Scholar]
- 14.Kim J, Roeder RG. 2009. Direct Bre1-Paf1 complex interactions and RING finger-independent Bre1-Rad6 interactions mediate histone H2B ubiquitylation in yeast. J Biol Chem 284:20582–20592. doi: 10.1074/jbc.M109.017442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kim J, Roeder RG. 2011. Nucleosomal H2B ubiquitylation with purified factors. Methods 54:331–338. doi: 10.1016/j.ymeth.2011.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen S, Wang DL, Liu Y, Zhao L, Sun FL. 2012. RAD6 regulates the dosage of p53 by a combination of transcriptional and posttranscriptional mechanisms. Mol Cell Biol 32:576–587. doi: 10.1128/MCB.05966-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lyakhovich A, Shekhar MP. 2003. Supramolecular complex formation between Rad6 and proteins of the p53 pathway during DNA damage-induced response. Mol Cell Biol 23:2463–2475. doi: 10.1128/MCB.23.7.2463-2475.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Read ML, Seed RI, Fong JC, Modasia B, Ryan GA, Watkins RJ, Gagliano T, Smith VE, Stratford AL, Kwan PK, Sharma N, Dixon OM, Watkinson JC, Boelaert K, Franklyn JA, Turnell AS, McCabe CJ. 2014. The PTTG1-binding factor (PBF/PTTG1IP) regulates p53 activity in thyroid cells. Endocrinology 155:1222–1234. doi: 10.1210/en.2013-1646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dover J, Schneider J, Tawiah-Boateng MA, Wood A, Dean K, Johnston M, Shilatifard A. 2002. Methylation of histone H3 by COMPASS requires ubiquitination of histone H2B by Rad6. J Biol Chem 277:28368–28371. doi: 10.1074/jbc.C200348200. [DOI] [PubMed] [Google Scholar]
- 20.Haddad DM, Vilain S, Vos M, Esposito G, Matta S, Kalscheuer VM, Craessaerts K, Leyssen M, Nascimento RM, Vianna-Morgante AM, De Strooper B, Van Esch H, Morais VA, Verstreken P. 2013. Mutations in the intellectual disability gene Ube2a cause neuronal dysfunction and impair parkin-dependent mitophagy. Mol Cell 50:831–843. doi: 10.1016/j.molcel.2013.04.012. [DOI] [PubMed] [Google Scholar]
- 21.Zimmermann C, Chymkowitch P, Eldholm V, Putnam CD, Lindvall JM, Omerzu M, Bjørås M, Kolodner RD, Enserink JM. 2011. A chemical-genetic screen to unravel the genetic network of CDC28/CDK1 links ubiquitin and Rad6-Bre1 to cell cycle progression. Proc Natl Acad Sci U S A 108:18748–18753. doi: 10.1073/pnas.1115885108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cai F, Chen P, Chen L, Biskup E, Liu Y, Chen PC, Chang JF, Jiang W, Jing Y, Chen Y, Jin H, Chen S. 2014. Human RAD6 promotes G1-S transition and cell proliferation through upregulation of cyclin D1 expression. PLoS One 9:e113727. doi: 10.1371/journal.pone.0113727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Takeda K, Yanagida M. 2005. Regulation of nuclear proteasome by Rhp6/Ubc2 through ubiquitination and destruction of the sensor and anchor Cut8. Cell 122:393–405. doi: 10.1016/j.cell.2005.05.023. [DOI] [PubMed] [Google Scholar]
- 24.Zaiss DM, Standera S, Holzhütter H, Kloetzel P, Sijts AJ. 1999. The proteasome inhibitor PI31 competes with PA28 for binding to 20S proteasomes. FEBS Lett 457:333–338. doi: 10.1016/S0014-5793(99)01072-8. [DOI] [PubMed] [Google Scholar]
- 25.Clemen CS, Marko M, Strucksberg KH, Behrens J, Wittig I, Gärtner L, Winter L, Chevessier F, Matthias J, Türk M, Tangavelou K, Schütz J, Arhzaouy K, Klopffleisch K, Hanisch FG, Rottbauer W, Blümcke I, Just S, Eichinger L, Hofmann A, Schröder R. 2015. VCP and PSMF1: antagonistic regulators of proteasome activity. Biochem Biophys Res Commun 463:1210–1217. doi: 10.1016/j.bbrc.2015.06.086. [DOI] [PubMed] [Google Scholar]
- 26.Sung P, Berleth E, Pickart C, Prakash S, Prakash L. 1991. Yeast RAD6 encoded ubiquitin conjugating enzyme mediates protein degradation dependent on the N-end-recognizing E3 enzyme. EMBO J 10:2187–2193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sancar G, Sancar C, Brügger B, Ha N, Sachsenheimer T, Gin E, Wdowik S, Lohmann I, Wieland F, Höfer T, Diernfellner A, Brunner M. 2011. A global circadian repressor controls antiphasic expression of metabolic genes in Neurospora. Mol Cell 44:687–697. doi: 10.1016/j.molcel.2011.10.019. [DOI] [PubMed] [Google Scholar]
- 28.Hong JH, Kaustov L, Coyaud E, Srikumar T, Wan J, Arrowsmith C, Raught B. 2015. KCMF1 (potassium channel modulatory factor 1) links RAD6 to UBR4 (ubiquitin N-recognin domain-containing E3 ligase 4) and lysosome-mediated degradation. Mol Cell Proteomics 14:674–685. doi: 10.1074/mcp.M114.042168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang C, Chang JF, Yan H, Wang DL, Liu Y, Jing Y, Zhang M, Men YL, Lu D, Yang XM, Chen S, Sun FL. 2015. A conserved RAD6-MDM2 ubiquitin ligase machinery targets histone chaperone ASF1A in tumorigenesis. Oncotarget 6:29599–29613. doi: 10.18632/oncotarget.5011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tatebe H, Yanagida M. 2000. Cut8, essential for anaphase, controls localization of 26S proteasome, facilitating destruction of cyclin and Cut2. Curr Biol 10:1329–1338. doi: 10.1016/S0960-9822(00)00773-9. [DOI] [PubMed] [Google Scholar]
- 31.Takeda K, Tonthat NK, Glover T, Xu W, Koonin EV, Yanagida M, Schumacher MA. 2011. Implications for proteasome nuclear localization revealed by the structure of the nuclear proteasome tether protein Cut8. Proc Natl Acad Sci U S A 108:16950–16955. doi: 10.1073/pnas.1103617108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schauber C, Chen L, Tongaonkar P, Vega I, Lambertson D, Potts W, Madura K. 1998. Rad23 links DNA repair to the ubiquitin/proteasome pathway. Nature 391:715–718. doi: 10.1038/35661. [DOI] [PubMed] [Google Scholar]
- 33.Hwang CS, Shemorry A, Varshavsky A. 2009. Two proteolytic pathways regulate DNA repair by cotargeting the Mgt1 alkylguanine transferase. Proc Natl Acad Sci U S A 106:2142–2147. doi: 10.1073/pnas.0812316106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Vlachostergios PJ, Patrikidou A, Daliani DD, Papandreou CN. 2009. The ubiquitin-proteasome system in cancer, a major player in DNA repair. Part 1: post-translational regulation. J Cell Mol Med 13:3006–3018. doi: 10.1111/j.1582-4934.2009.00824.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Vlachostergios PJ, Patrikidou A, Daliani DD, Papandreou CN. 2009. The ubiquitin-proteasome system in cancer, a major player in DNA repair. Part 2: transcriptional regulation. J Cell Mol Med 13:3019–3031. doi: 10.1111/j.1582-4934.2009.00825.x. [DOI] [PMC free article] [PubMed] [Google Scholar]