ABSTRACT
Mitochondrial life cycle and protein import are intricate cellular processes, which require precise coordination between the transport machineries of outer and inner mitochondrial membranes. Presequence translocase performs the indispensable function of translocating preproteins having N-terminal targeting sequences across the inner membrane. Tim23 forms the core of the voltage-gated import channel, while Tim17 is presumed to maintain the stoichiometry of the translocase. However, mechanistic insights into how Tim17 coordinates these regulatory events within the complex remained elusive. We demonstrate that Tim17 harbors conserved G/AXXXG/A motifs within its transmembrane regions and plays an imperative role in the translocase assembly through interaction with Tim23. Tandem motifs are highly essential, as most of the amino acid substitutions lead to nonviability due to the complete destabilization of the TIM23 channel. Importantly, Tim17 transmembrane regions regulate the dynamic assembly of translocase to form either the TIM23 (PAM)-complex or TIM23 (SORT)-complex by recruiting the presequence translocase-associated motor (PAM) machinery or Tim21, respectively. To a greater significance, tim17 mutants displayed mitochondrial DNA (mtDNA) instability, membrane potential loss, and defective import, resulting in organellar dysfunction. We conclude that the integrity of Tim17 transmembrane regions is critical for mitochondrial function and protein turnover.
KEYWORDS: mitochondria, presequence translocase, mtDNA stability, preprotein import, membrane potential
INTRODUCTION
The vast majority of mitochondrial proteins are encoded by nuclear genes and synthesized on cytosolic ribosomes with an amino-terminal positively charged cleavable signal sequence. The efficient import of mitochondrial proteins requires precise coordination between the translocases of the outer membrane (TOM complex) and the inner membrane (TIM23 complex) (1–9). After initial recognition and transport through TOM complex, these preproteins are actively sorted based on their destination. The preproteins directed toward the matrix compartment possess an N-terminal positively charged presequence and are imported into the matrix compartment with the aid of presequence translocase (TIM23 complex) (1–10). The core of presequence translocase consists of a membrane-bound channel component formed by Tim23 and Tim17 (11–15). Tim23 and Tim17 are phylogenetically related integral membrane proteins and have similar transmembrane topology. Tim23 contributes to the formation of voltage-gated protein-conducting pores, while the evidence suggests that Tim17 is majorly involved in regulating the channel activity (16–23). Other components such as Tim50 and Tim21 act as receptors and coordinate with the intermembrane space (IMS) domain of Tim23 to facilitate preprotein recognition and transfer from the TOM complex to presequence translocase (16, 17, 22, 24–32). Mgr2 has been recently identified as a member of active, preprotein-conducting translocases and functions as a gatekeeper to regulate the lateral release of preproteins into the inner membrane. In addition, Mgr2 also assists in Tim21-mediated coupling of Tim23 to the respiratory chain complexes III and IV (33–35).
The membrane potential (Δψm) across the inner membrane serves as the initial driving force for the preprotein import by presequence translocase (36). The complete translocation of preproteins into the matrix is facilitated by mitochondrial heat shock protein 70 (mtHsp70), which forms the core of the import motor and is recruited to the import channel via a membrane tether, Tim44 (37–41). The presequence translocase-associated motor (PAM) J-protein components, Pam18-Pam16 heterodimer, regulate the ATPase cycle of mtHsp70 together with the nucleotide exchange factor Mge1 in an ATP-dependent manner (42–48). The organization and recruitment of the PAM subcomplex to the core channel are mediated by additional nonessential components of presequence translocase, Pam17 and Tam41 (41, 49–52).
Due to the lack of high-resolution structural insights, the organization of TIM23 pores and the formation of protein-conducting channels together with Tim17 are largely unknown and constitute an intriguing aspect of the presequence translocase yet to be uncovered. A preliminary analysis using reconstituted proteoliposomes suggests that Tim23 forms a gated twin pore and that depletion of Tim17 or its truncation at the N terminus disrupts the architecture of the core channel (21, 23, 53). Intriguingly, impaired interaction of Tim17 with the TM2 region of Tim23 has been largely attributed to loss of Δψ leading to defective import (22, 54, 55). These observations collectively indicate that Tim17 plays an important role in modulating the voltage-dependent structural dynamics of the TIM23 channel. Moreover, the organization of the human TIM23 channel is highly intricate due to the presence of two Tim17 paralogs (Tim17A and Tim17B), which organize into two separate translocation channels (56). Recent reports indicate that upregulation of Tim17A expression is associated with the progression of breast cancer and could serve as a hallmark for the detection and prognosis of breast cancer. At the same time, knockdown experiments using small interference RNA (siRNA) indicated an oncogenic activity of TIMM17A in breast cancer, suggesting that it might represent a novel class of mitochondrial targets for tumor therapy (57, 58). Furthermore, overexpression of TIMM17A could rescue mitochondrial DNA (mtDNA) loss in human NT2 teratocarcinoma cells containing A3243G mutant mtDNA, which previously have been shown to lose mtDNA (59). This explicitly emphasizes the critical requirement of uncovering the role of Tim17 in maintenance of architecture of presequence translocase, thereby regulating and reprogramming the mitochondrial functions in cancer cells.
The G/AXXXG/A motifs are frequently present within the transmembrane regions and are known to favor helix-helix interactions in polytopic membrane proteins. Intriguingly, these motifs gained more attention as the three-dimensional structure of the transmembrane domain of glycophorin A could provide insight into how the glycine residues of the GXXXG motif stabilize helix-helix interactions and aid in homodimerization, thus underlining its central importance in the assembly process of membrane proteins (60–64). At the amino acid sequence level, both Tim17 and Tim23 contain a large number of G/AXXXG/A motifs within the transmembrane segments. Recent reports highlight that conditional mutants isolated within the G/AXXXG/A motifs of Tim23 transmembrane regions were found to impair Tim23 homodimerization as well as interactions with other components such as Tim17 (22, 65).
In the present report, we have delineated the importance of G/AXXXG/A motifs of Tim17 in the assembly of the core TIM23 channel. These motifs of Tim17 are responsible for the precise helix-helix packing, thus stabilizing the geometry of TIM23 pore through heterotopic interactions. Our biochemical and genetic analysis comprehensively provides evidence for the role of Tim17 in the recruitment of PAM subcomplex and Tim21 to the core channel forming Tim23. In addition, our report highlights the profound role of G/AXXXG/A motifs of Tim17 in the maintenance of inner membrane potential, mitochondrial integrity, and mtDNA stability, thereby signifying its importance in preserving the architecture of presequence translocase.
RESULTS
Conserved G/AXXXG/A motifs within the transmembrane regions of Tim17 are essential for its function.
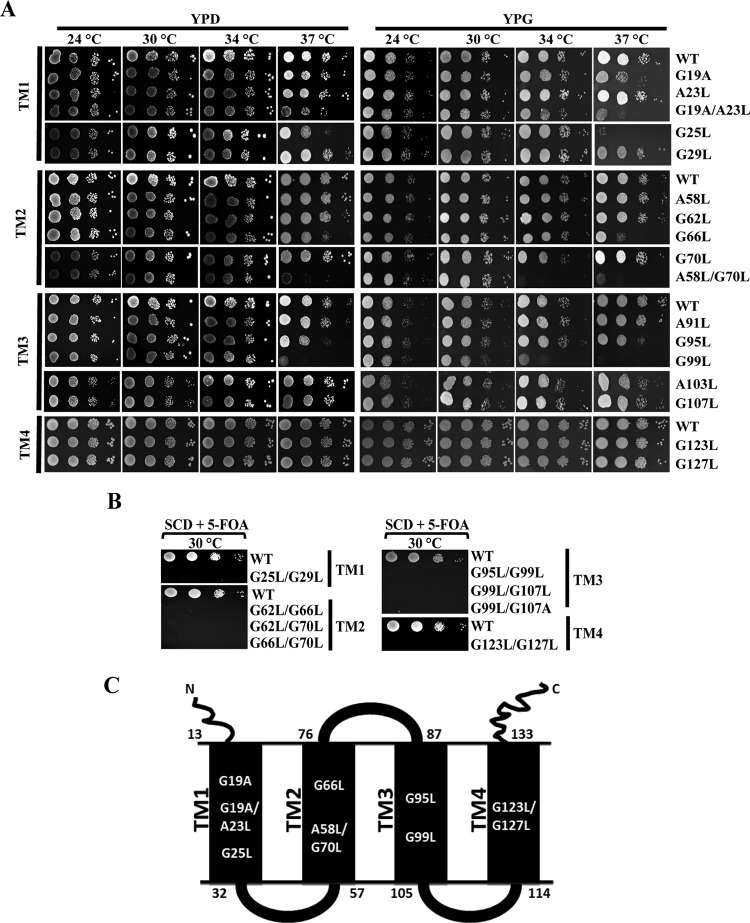
Tim17 protein (Tim17) is an essential subunit of the presequence translocase, sharing significant structural similarity with Tim23 and consisting of four predicted transmembrane regions (TM1, TM2, TM3, and TM4) with both its amino and carboxyl termini facing the IMS region (53, 66). Multiple-sequence alignment of Tim17 across species reveals the presence of several conserved G/AXXXG/A motifs in TM regions (see Fig. S1 in the supplemental material). To determine the importance of G/AXXXG/A motifs in Tim17 function, we have performed site-directed mutagenesis systematically to convert amino acids from glycine to either leucine or alanine, and similarly, alanines in these motifs were replaced with leucines. The mutant plasmids were transformed into a haploid Δtim17 strain containing a wild-type functional copy of the TIM17 gene on a URA3-based plasmid. The presence of the URA3 gene allowed the selection of cells that had lost the plasmid carrying the wild-type TIM17 gene on 5-fluoroorotic acid (5-FOA) medium. To test the growth phenotype, wild-type and mutant cells were spotted on either 1% yeast extract–2% peptone–2% glucose (YPD) or 1% yeast extract–2% peptone–3% glycerol (YPG) medium and incubated under permissive and nonpermissive temperature conditions. Three isolates with mutations from the TM1 region, namely, tim17G19A, tim17G19A/A23L, and tim17G25L, exhibited a temperature-sensitive (Ts) phenotype (Fig. 1A). Of these, the tim17G19A mutant displayed a Ts phenotype at 37°C in YPG medium only (Fig. 1A). At the same time, changing the glycine at position 19 to leucine (tim17G19L) caused the organism to display a lethal phenotype (data not shown). On the other hand, the tim17G19A/A23L and tim17G25L double mutant showed growth sensitivity in both YPD and YPG media at 37°C (Fig. 1A).
FIG 1.
Isolation of Ts mutants from different transmembrane regions of Tim17 protein. (A) Growth phenotype analysis. Wild-type (WT) and tim17 mutant strains isolated from transmembrane regions (TM1, TM2, TM3, and TM4) were allowed to grow until mid-log phase in liquid YPD medium at 30°C. Yeast cells corresponding to an A600 of optical density at 600 nm (OD600) of 0.5 were harvested, serially diluted, and spotted onto YPD or YPG medium. Plates were incubated at the indicated temperatures, and images were captured after 72 h. (B) Growth phenotypes of double tim17 mutants. Mutants from tandem G/AXXXG/A motifs were spotted onto medium supplemented with 5-fluoroorotic acid (5-FOA) and incubated at 30°C, and images were captured after 72 h. (C) Modular representation of Tim17 protein. The topology of different helices and loops and the positions of the amino acids spanning each of the segments of the Tim17 protein are highlighted. Amino acid positions of the Ts mutants from TM1, TM2, and TM3 and the lethal mutant from the TM4 region are indicated.
Similarly, two mutants with single or double amino acid substitutions in the TM2 region were isolated, namely, the tim17G66L and tim17A58L/G70L mutants, which showed temperature sensitivity at 37°C (Fig. 1A). Furthermore, the tim17A58L/G70L mutant displayed severe growth defect even at 34°C on YPG medium (Fig. 1A). In addition to that, two Ts mutants were isolated from the TM3 region, namely, the tim17G95L and tim17G99L mutants, which were found to be sensitive at 37°C on YPD/YPG media, while the tim17G99L mutant exhibited severe growth defects at 34°C (Fig. 1A). On the other hand, single-amino-acid substitutions in G/AXXXG/A motifs of the TM4 region did not yield any growth defects (Fig. 1A). Strikingly, tandem amino acid replacements in G/AXXXG/A motifs of every transmembrane region (TM1 to -4) exhibit a lethal phenotype (Fig. 1B). These mutants include those with tim17G25L/G29L (TM1), tim17G62L/G66L, tim17G62L/G70L, tim17G66L/G70L (TM2), tim17G95L/G99L, tim17G99L/G107L, tim17G99L/G107A (TM3), and tim17G123L/G127L (TM4) mutations, which exhibited nonviability in medium containing 5-FOA (Fig. 1B). In summary, mutations in G/AXXXG/A motifs from all the TM regions of Tim17 that are indicated in Fig. 1C resulted in either a Ts or a lethal phenotype, suggesting that all the transmembrane regions play an indispensable role in performing the essential functions of Tim17.
Tandem G/AXXXG/A motifs maintain the stability of Tim17.
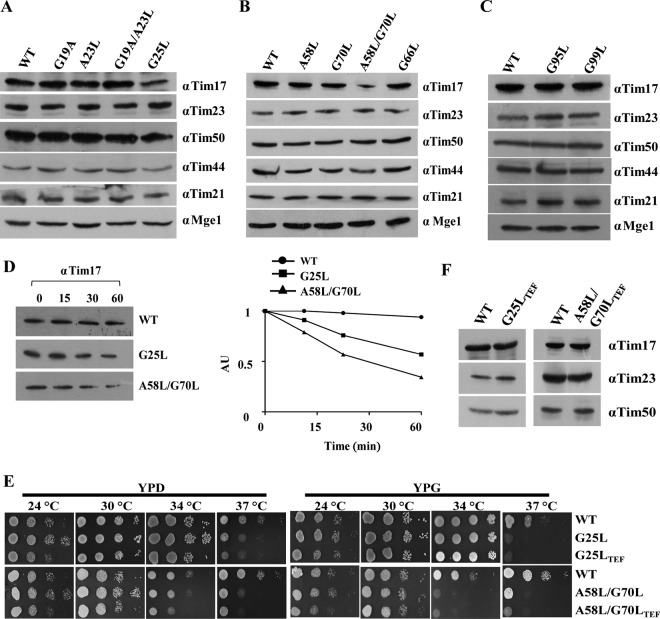
To determine the role of G/AXXXG/A motifs in the stability of Tim17, we measured the steady-state levels of mutant protein in the lysates. Most of the single-point mutations in G/AXXXG/A motifs were found to be stable and showed comparable Tim17 levels as well as other subunits of the presequence translocase, including Tim23, Tim50, Tim44, Tim21, and Mge1 (Fig. 2A to C). However, two mutants, namely, the tim17G25L (TM1) and tim17A58L/G70L (TM2) mutants, showed significant reduction in the protein levels compared to other subunits of presequence translocase, whose levels remained unaltered (Fig. 2A to C). We hypothesized that the reduction in steady-state protein levels associated with the tim17G25L (TM1) and tim17A58L/G70L (TM2) mutations might be a consequence of its reduced half-life. In order to test this hypothesis, we measured the relative abundance of proteins after inhibiting the protein translation using cycloheximide. Notably, the two mutants showed an enhanced rate of Tim17 degradation as a function of time, indicating that the decreased level of mutant protein is a consequence of compromised stability (Fig. 2D). To rule out the possibility that the growth defects associated with the Ts mutant were not due to reduced protein levels, we overexpressed the mutants under the control of the centromeric TEF promoter. Interestingly, as indicated in Fig. 2E, both tim17G25L and tim17A58L/G70L mutants failed to complement the growth defects at nonpermissive temperatures, even though the expression levels were equivalent to wild-type levels (Fig. 2F), suggesting that the Ts phenotype of mutants was indeed a consequence of its intrinsic functional defect.
FIG 2.
Measurement of steady-state protein levels and phenotypic analyses of transmembrane regions (TM1 to -3) mutants of Tim17. (A to C) Protein expression analysis. Steady-state protein levels of the wild type and tim17 mutants (from TM1, TM2, TM3) were analyzed by Western blotting using specific antibodies for different components of presequence translocase. (D) Analysis of protein half-lives. Wild-type and mutant cells were treated with cycloheximide (50 μg/ml) to inhibit the protein translation, and cells were collected at different time intervals. Lysates were prepared and analyzed by Western blotting (AU, arbitrary units). The amounts of different proteins were quantified by densitometry using ImageJ software. (E) The wild type and tim17 mutants (from TM1 and TM2) expressed under the control of the centromeric TEF promoter were serially diluted and spotted on the indicated media and incubated at different temperatures. (F) Protein overexpression is analyzed for tim17 mutants (TM1 and TM2) using desired specific antibodies by Western blotting.
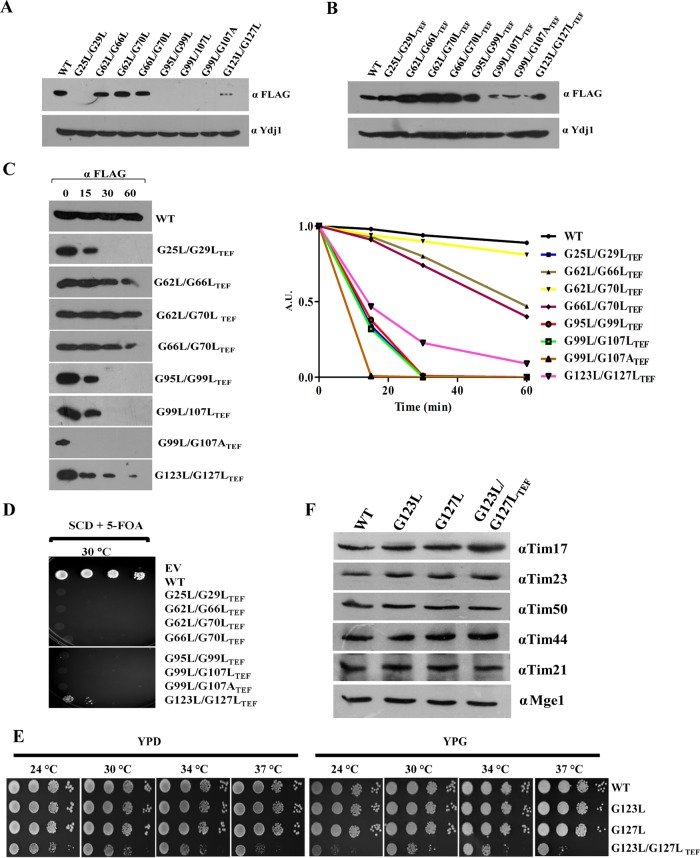
Similarly, our findings also highlight that the lethality associated with the double mutations in G/AXXXG/A motifs of TM1, TM3, and TM4 is due to a reduction in the protein levels as a result of the compromised stability of Tim17 (Fig. 3A to C). However, TM2 lethal mutants did not show much difference in the stability of the protein compared to the wild type (Fig. 3A to C). Further, we analyzed the growth defects of overexpressed mutant proteins by plasmid shuffling on 5-FOA (Fig. 3D). Surprisingly, only the tim17G123L/G127L-TEF strain was able to produce viable colonies on 5-FOA, while the other mutants were nonviable. To analyze the growth phenotype of the tim17G123L/G127L-TEF mutant, we subjected it to serial dilution assay on YPD and YPG. Remarkably, the tim17G123L/G127L-TEF mutant displayed a severe growth defect at all temperatures analyzed (Fig. 3E). However, the steady-state protein levels of different subunits of the presequence translocase in the tim17G123L/G127L-TEF mutant were found to be comparable with wild-type levels (Fig. 3F). In conclusion, our findings provide the first evidence to indicate that the tandem G/AXXXG/A motifs are critical for maintaining the stability of the transmembrane regions of Tim17.
FIG 3.
Analysis of steady-state proteins levels of lethal mutants. (A) C-terminal FLAG-tagged tim17 mutants were transformed into wild-type isolates harboring endogenous Tim17 (pRS-316 Tim17), and expression levels of mutant proteins were analyzed by Western blotting using anti-FLAG antibody. (B) Protein levels of tim17 mutants after overexpressing under the control of centromeric TEF promoter were analyzed as described earlier. (C) Analysis of protein half-life. Yeast cells expressing C-terminal FLAG-tagged wild-type and mutant Tim17 under an endogenous Tim17 (pRS316 Tim17) background were treated with cycloheximide (50 μg/ml) to inhibit the protein translation; cells were collected at different time intervals. Lysates were prepared and analyzed by Western blotting (AU, arbitrary units). The amounts of different proteins were quantified by densitometry using ImageJ software. (D) Spotting assay. Double mutants from tandem G/AXXXG/A motifs overexpressing Tim17 under the control of centromeric TEF promoter were spotted on medium supplemented with 5-fluoroorotic acid (5-FOA) and incubated at 30°C. Images were captured after 72 h. (E) Growth phenotype analysis. Yeast strains expressing the wild type, tim17G123L, tim17G127L (under the control of the endogenous promoter), and tim17G123L/G127L-TEF (under the control of the centromeric TEF promoter) from the TM4 segment were serially diluted, spotted onto the indicated media, and incubated at different temperatures. (F) Protein expression analysis of tim17 mutants. The steady-state protein levels of Tim17 and presequence translocase complex components in TM4 mutants were analyzed by immunodecoration with specific antibodies.
The integrity of Tim17 transmembrane regions is critical for preprotein sorting into matrix and inner mitochondrial membrane.
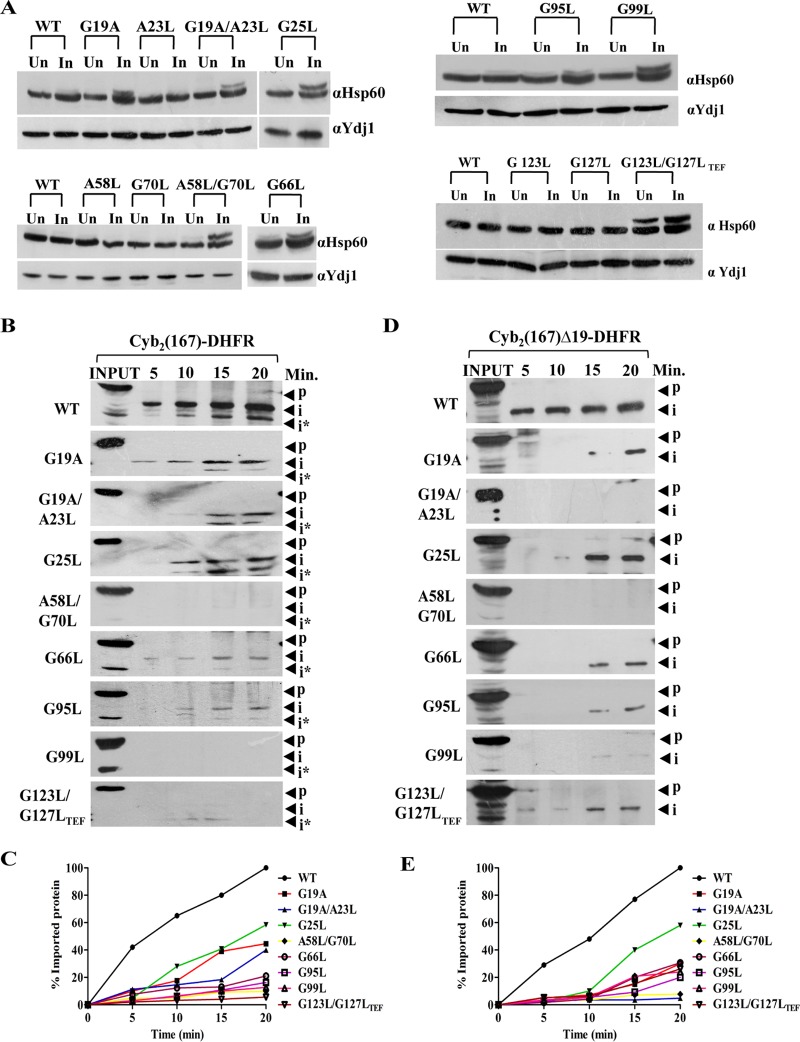
To evaluate the conditional growth phenotype of tim17 mutants, we first tested their ability to retain preprotein sorting into the mitochondrial matrix and inner membrane. To determine the matrix sorting, we subjected the mutants to in vivo precursor accumulation analysis using Hsp60 as a model protein, and the removal of N-terminal signal sequence by the matrix-processing peptidase was monitored. To induce phenotypic effects, the wild type and all Ts mutants were subjected to heat shock at 37°C and the accumulation of preproteins was monitored. In contrast to wild-type organisms, all the Ts mutants showed significant preprotein accumulation of Hsp60 precursor correlating with their observed growth phenotype (Fig. 4A).
FIG 4.
Critical role of Tim17 in regulating both matrix sorting and inner membrane sorting. (A) In vivo precursor accumulation assay. Wild-type and tim17 mutant cells were grown to early log phase and subjected to heat shock at 37°C for 4 h. Cell lysates were prepared and analyzed by Western blotting using Hsp60-specific antibodies; Un, uninduced; In, induced. (B) In vitro import kinetic analysis. Purified mitochondria from wild-type and different mutant yeast strains were subjected to heat shock at 37°C for 30 min to induce phenotype, followed by incubation with saturating amounts of purified cytb2 (1–167)-DHFR at 25°C. The reaction was terminated by the addition of valinomycin at different time intervals; aliquots were subjected to proteinase K treatment. The samples were subsequently analyzed by Western blotting using an anti-DHFR antibody; p, precursor; i and i*, intermediates. (C) The signal intensity of imported intermediate bands obtained from Western blots was quantified by densitometry using ImageJ software for cytb2 (1–167)-DHFR after normalizing the amounts of precursor imported in the wild type at 20 min as 100% import. (D and E) Mitochondria from wild-type and tim17 mutant cells were subjected to heat shock at 37°C for 30 min to induce phenotype followed by incubation with saturating amounts of purified cytb2 (1–167)Δ19-DHFR at 25°C. The import reaction was terminated by valinomycin at indicated time intervals, followed by treatment with proteinase K. The samples were subsequently analyzed by Western blotting using an anti-DHFR antibody; p, precursor; i, intermediate (D). The imported intermediate bands were quantified by densitometry using ImageJ software after normalizing the amounts of precursor imported in the wild type at 20 min as 100% import (E).
In order to gain insights into the role of G/AXXXG/A motifs of Tim17 in preprotein import to matrix and inner membrane of mitochondria, we performed in vitro import kinetics. We have utilized two model proteins. The first, cytb2 (1–167)-DHFR, comprised of the mouse dihydrofolate reductase (DHFR) fused to the N-terminal 167 amino acids of cytochrome b2, has an inner membrane sorting signal and thus localizes into the inner membrane of mitochondria. The second model protein, cytb2 (1–167)Δ19-DHFR, harbors a deletion of 19 amino acids of the inner membrane sorting signal and thereby localizes to the matrix. Mitochondria were isolated from yeast cells grown at permissive temperature and preincubated at 37°C to induce the mutant phenotype, followed by incubation with saturating amounts of two different precursor proteins in separate import reaction mixtures, and the import kinetics were monitored by Western blotting. In the case of inner membrane targeting, tim17A58L/G70L, tim17G99L, and tim17G123L/G127L-TEF mutants showed a severe defect in the import efficiency, while tim17G19A, tim17G19A/A23L, tim17G25L, tim17G66L, and tim17G95L mutants showed an intermediate defect consistent with their growth phenotype (Fig. 4B and C). On the other hand, tim17G19A, tim17G19A/A23L, tim17A58L/G70L, and tim17G99L mutants exhibited a significant defect in matrix targeting, while tim17G25L, tim17G66L, tim17G95L, and tim17G123L/G127LTEF mutants showed an intermediate import defect (Fig. 4D and E). Similarly, overexpression of tim17G25L and tim17A58L/G70L under the control of the TEF promoter also failed to rescue both in vivo and in vitro import defects (data not shown). In summary, we report that G/AXXXG/A motifs from transmembrane regions of Tim17 play a crucial role in both inner membrane sorting and matrix sorting via presequence translocase.
Tim17 transmembrane regions anchor PAM subcomplex and Tim21 to the core TIM23 channel.
In order to unravel the role of Tim17 in maintaining the architecture of presequence translocase, we initially subjected tim17 mutants to mitochondrial fractionation analysis by sonication and sodium carbonate extraction. All the mutant proteins were found to be partitioned into the pellet fraction, indicating that they were properly targeted and inserted into the inner membrane of mitochondria (data not shown).
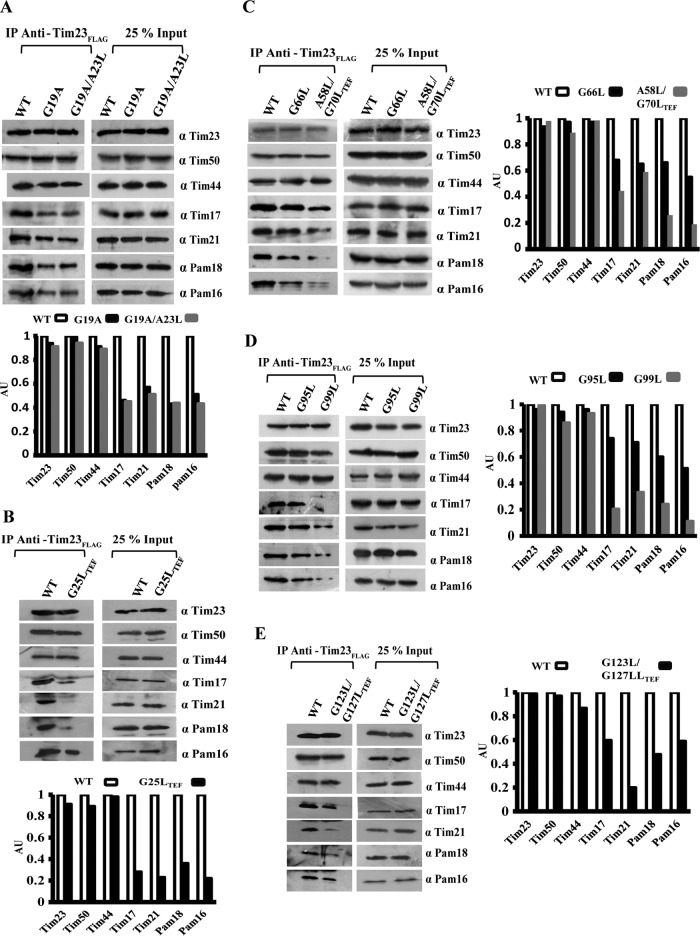
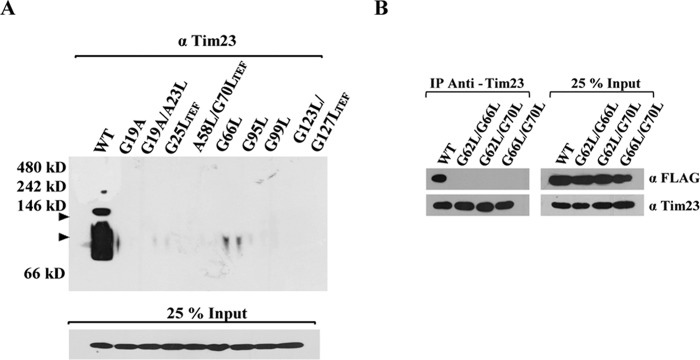
To address the influence of G/AXXXG/A mutation from different regions of Tim17 in maintaining the architecture of presequence translocase, the digitonin-lysed wild-type and mutant mitochondria were subjected to coimmunoprecipitation (co-IP) using anti-FLAG antibody directed against C-terminally FLAG-tagged Tim23. Our findings highlight that Ts mutants from the TM1 region, including tim17G19A, tim17G19A/A23L, and tim17G25L-TEF, showed a significant reduction in the copurification of Tim17 with Tim23 (Fig. 5A and B). At the same time, TM1 Ts mutants also showed impairment in the recruitment of the import motor-forming PAM subcomplex, including Pam18 and Pam16 (Fig. 5A and B). Furthermore, there was a compromised interaction of Tim21 with the core complex (Fig. 5A and B), while Tim50 and Tim44 were efficiently recovered together with Tim23. Similarly, all the Ts mutants from TM2, TM3, and TM4 showed impairment in the interaction of Tim17, Pam18, Pam16, and Tim21 with the core TIM23 channel irrespective of their mutations in these motifs across the full length of the protein (Fig. 5C to E). To further validate these observations, we analyzed the stability of TIM23 core complex in tim17 mutants using blue native PAGE (BN-PAGE). All the Ts mutants from different TM regions of Tim17 showed impaired formation of a distinct core complex with a molecular mass of 90 kDa corresponding to Tim23 and Tim17 (Fig. 6A, arrowheads). Besides, mutants also failed to assemble into higher-molecular-mass core complexes than those of the wild type, suggesting a compromised interaction of partner subunits with the core TIM23 channel (Fig. 6A, arrowheads). Likewise, our findings also highlight that the lethality of tim17G62L/G66L, tim17G62L/G70L, and tim17G66L/G70L (TM2) double mutants is attributable to the complete destabilization of the core channel due to the defective interaction of mutant Tim17 with Tim23 (Fig. 6B). Together, these observations provide direct evidence to highlight that the G/AXXXG/A motifs of Tim17 transmembrane regions play a pivotal role in maintaining the architecture of presequence translocase.
FIG 5.
All the TM regions of Tim17 are essential for Tim23 interaction. (A to E) Purified mitochondria from the wild type and tim17 mutants (TM1 to -4) were subjected to heat shock at 37°C for 30 min to induce phenotype. The mitochondria were lysed with digitonin-containing buffer, and the lysates were subjected to co-IP using anti-FLAG antibody. Samples were separated on SDS-PAGE gels and subjected to Western blot analysis using the indicated specific antibodies to detect the different components of presequence translocase. Twenty-five percent of the total sample served as loading control (Input) against 100% of immunoprecipitated product. The amounts of different immunoprecipitated proteins were quantified by densitometry using ImageJ software.
FIG 6.
The integrity of Tim17 transmembrane regions is important for maintaining the architecture of presequence translocase. (A) Analysis of stability of protein complexes using BN-PAGE. The digitonin-solubilized mitochondria from the wild type and tim17 mutants were subjected to BN-PAGE followed by immunoblotting against Tim23 antibodies. The positions of the complexes of the wild type are indicated by arrowheads. Twenty-five percent of the input sample was used as loading control. (B) Co-IP analysis. Tim17 and tim17 TM2 region lethal mutant proteins were C-terminally FLAG-tagged and transformed into a strain harboring endogenous Tim17 (pRS316-Tim17). Mitochondria were isolated and subjected to heat shock at 37°C for 30 min, followed by lysis in digitonin buffer. The lysates were subjected to co-IP using anti-Tim23 antibody. Samples were separated on SDS-PAGE gels and immunoblotted using anti-FLAG antibody. Twenty-five percent of the input was used as loading control.
Tim17 plays a critical role in the maintenance of mitochondrial DNA integrity.
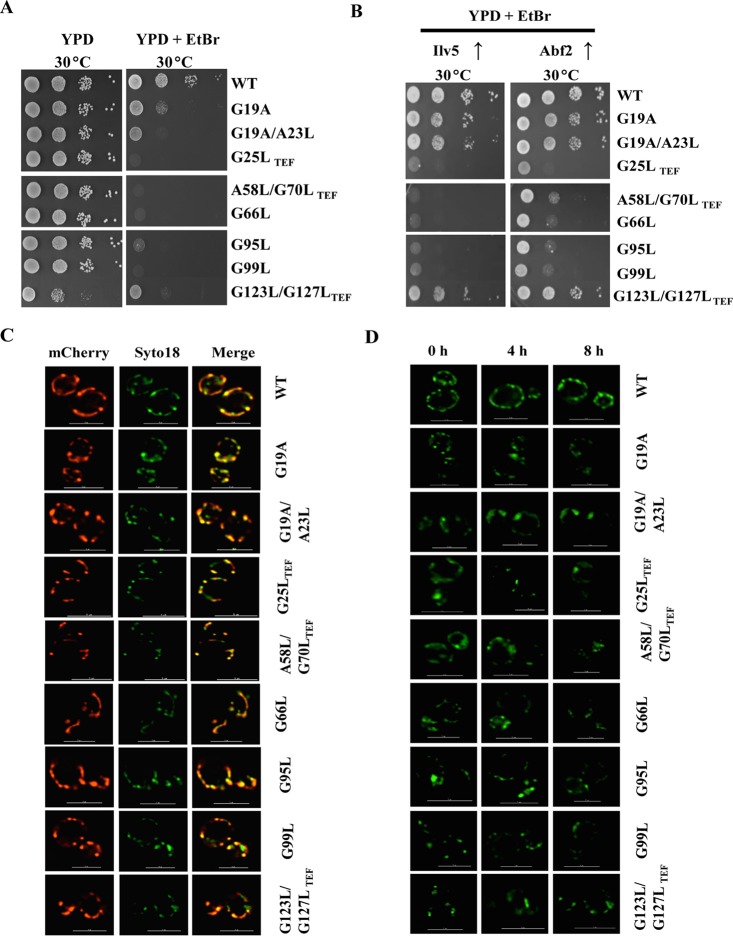
Mitochondrial DNA (mtDNA) is packaged into nucleoid structures having several mtDNA molecules bound to DNA binding proteins to form a nucleoprotein complex. Abf2 and Ilv5 are key mtDNA binding proteins that play a crucial role in the maintenance of mtDNA stability (67, 68). Interestingly, TIM17 is identified as a multicopy suppressor of mtDNA instability in Δilv5 cells, and its overexpression was able to rescue the mitochondrial DNA loss in cells lacking Abf2 (59). In order to unravel the role of Tim17 in the maintenance of mtDNA stability, we assessed the growth of tim17 mutants at a permissive temperature on YPD medium or YPD supplemented with ethidium bromide (EtBr), which promotes mtDNA loss to generate a petite-negative phenotype (ρ0). Strikingly, as indicated in Fig. 7A, all the tim17 mutants from transmembrane regions exhibited a petite-negative phenotype, as they failed to produce viable colonies on EtBr-supplemented YPD medium. Since overexpression of TIM17 stabilizes the mtDNA in Δilv5 and Δabf2 cells, we tested whether overexpression of ILV5 and ABF2 can suppress the mtDNA instability in tim17 mutants. Notably, the overexpression of ABF2 was able to completely rescue the petite-negative phenotype of tim17G19A, tim17G19A/A23L, and tim17G123L/G127L-TEF mutants, while the rescue was partial with the remaining tim17 mutants (Fig. 7B). Similarly, overexpression of ILV5 was also capable of suppressing the petite-negative phenotype of tim17G19A, tim17G19A/A23L, and tim17G123L/G127L-TEF mutants (Fig. 7B).
FIG 7.
Cells lacking functional Tim17 protein exhibit a petite-negative phenotype. (A and B) Growth phenotype analysis. Wild-type and mutant yeast cells were grown to mid-log phase at 30°C in liquid YPD medium. Equivalent amounts of cells from each strain were serially diluted and spotted onto YPD medium or YPD medium containing 40 μg/ml ethidium bromide. The plates were incubated at the permissive temperatures for 3 days, and images were captured after 72 h (A). Similarly, the wild type and tim17 conditional mutants were transformed with either ILV5 or ABF2 on a centromeric plasmid under the control of the TEF promoter. The transformed yeast strains were serially diluted and spotted onto YPD medium containing 40 μg/ml ethidium bromide and incubated at the permissive temperatures for 3 days. Images were captured after 72 h (B). (C and D) Quantification of the nuclei number in tim17 mutants. Yeast cells were transformed with mitochondrially targeted mCherry and allowed to grow to mid-log phase at 30°C. The cells were stained with 10 mM SYTO 18 for 20 min, followed by imaging analysis using a Delta Vision Elite fluorescence microscope (GE Healthcare), and the images were analyzed using SoftWoRx 6.1.3 software. The images from mCherry and SYTO 18 were superimposed (merge). The cells in all the panels were imaged at identical exposures to compare the fluorescence intensity levels. Bars, 5 μm. (C). Similarly, wild-type and the indicated tim17 mutant strains were grown to mid-log phase, followed by incubation at the restrictive temperature of 37°C for 0, 4, and 8 h. The mitochondrial nucleoid morphology was examined at each time interval by staining with SYTO 18 to a final concentration of 10 mM, followed by imaging analysis using a Delta Vision Elite fluorescence microscope (D). The cells in all the panels were subjected to identical exposures. Bars, 5 μm.
To investigate the effects of mutations in G/AXXXG/A motifs on mtDNA nucleoid structures, the wild type and tim17 mutants expressing a mitochondrially targeted sequence (MTS)-mCherry construct were grown to the mid-log phase under permissive conditions and stained with the mtDNA-specific dye SYTO18, and nucleoid morphology was analyzed by fluorescence imaging. As indicated in Fig. 7C, a colocalization of mitochondrial mCherry red with green SYTO18 dye, resulting in yellow or orange fluorescence, indicates the specific staining of mtDNA nucleoid structures. Under permissive conditions, most of the tim17 mutants displayed numbers of mtDNA nucleoid structures equivalent to the numbers of the wild type, with punctate morphology similar to that of the wild type (Fig. 7C, compare middle panels with SYTO18 labeling). A time course analysis was performed at restrictive temperature (37°C) to measure the stability of mtDNA nucleoid structures in tim17 mutants. At all the time points analyzed, wild-type isolates maintained many punctate nucleoids (25 ± 5, n = 20) with normal morphology. However, tim17 mutants showed fewer nucleoids (7 ± 3, n = 20) with coalescent nucleoid morphology at 4 and 8 h (Fig. 7D). Collectively, these findings indicate that mutations in G/AXXXG/A motifs of Tim17 result in aberrations in nucleoid quantity and morphology, signifying its importance in the maintenance of mtDNA stability.
Tim17 G/AXXXG/A motifs are important for maintaining inner membrane potential.
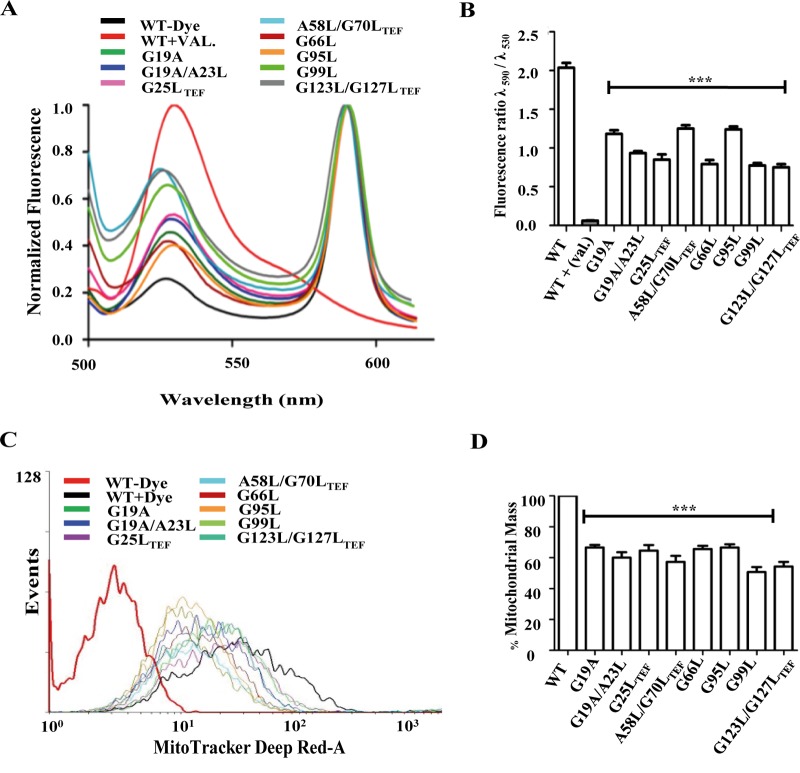
Translocation via presequence translocase is energetically driven by active membrane potential (Δψm) across the inner membrane. Reduced Δψm has been implicated in diminished rates of protein import across the inner membrane (22, 36, 53). To assess whether the mutations in G/AXXXG/A motifs of Tim17 impart loss in Δψm, we stained the cells with the polarity-sensitive JC-1 dye (69). In the presence of intact Δψm, the JC-1 dye is taken up by mitochondria to form an aggregate that gives a characteristic red fluorescence at 590 nm. On the other hand, dissipation in Δψm will result in the retention of dye outside the mitochondria, giving rise to a characteristic green fluorescence at 530 nm. The ratio of red to green fluorescence indicates the retention of Δψm across the inner membrane. Valinomycin, a potassium ionophore that disrupts the membrane polarity in isolated mitochondria, served as a positive control. Notably, compared to the wild type, all the tim17 mutants showed more than 35% reduction in the ratio of red to green fluorescence, indicating reduced Δψm across the inner membrane (Fig. 8A and B). To check whether the loss of Δψm leads to decrease in the organellar functionality, wild-type and mutant cells were stained with MitoTracker Deep Red-A dye, which is a membrane potential-dependent indicator of active mitochondrial mass (70–72). After inducing the phenotype by heat shock, the wild type and tim17 mutants were subjected to functional mass estimation using MitoTracker Deep Red-A fluorescence through flow-cytometric analysis (22). In agreement with our hypothesis, compared to the wild type, all the tim17 mutants exhibited a significant decrease in fluorescence, indicating that mutations in G/AXXXG/A motifs of Tim17 caused the reduction in functional mitochondria (Fig. 8C and D).
FIG 8.
The functional Tim17 protein is essential for maintaining the membrane polarity across the inner membrane. (A and B) Measurements of inner membrane polarity. Purified mitochondria from wild-type and tim17 mutant cells were stained with JC-1 dye for 15 min at room temperature. The fluorescence emission scan was recorded from 500 to 620 nm with an excitation of 490 nm (A). The mean fluorescence intensity obtained in the spectral analysis was quantified (B). *, P ≤ 0.05; **, P ≤ 0.01; ***, P ≤ 0.001. (C and D) Determination of functional mitochondrial mass. The indicated wild-type and tim17 mutant strains were grown to mid-log phase at 30°C, followed by treatment of heat shock at 37°C for 4 h. The cells were subjected to staining with MitoTracker Deep Red-A dye for 20 min and were analyzed by flow cytometer using the BD FACSCanto II analyzer (C). The mean fluorescence intensity obtained in FACS analysis was quantitated to estimate the relative total mass in the wild type and tim17 mutants (D). ***, P ≤ 0.001.
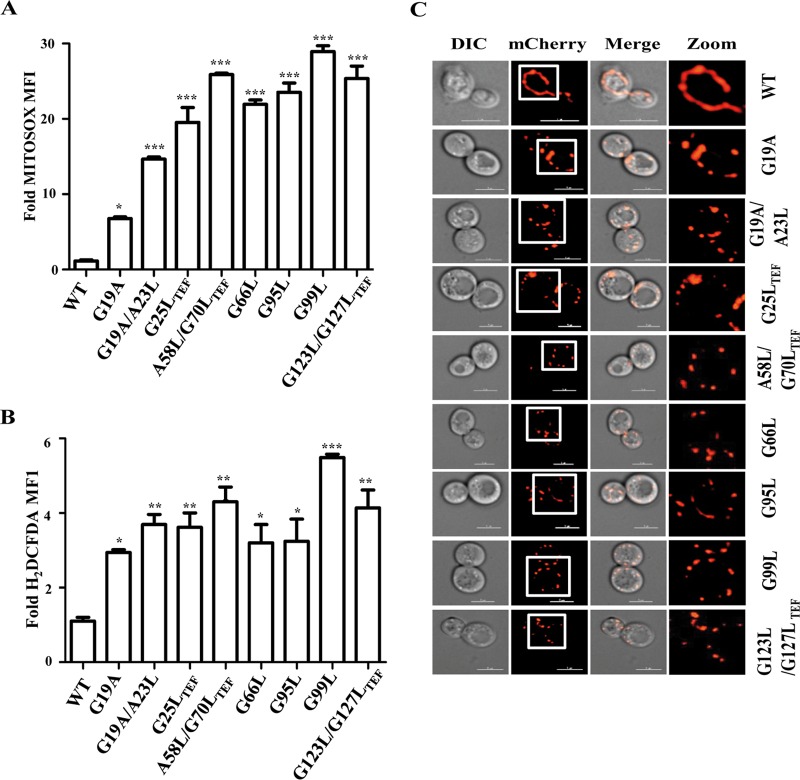
Defective import and loss of Δψm are often associated with the generation of reactive oxygen species (ROS) due to improper electron channeling across the electron transport chain (ETC) complexes leading to the formation of superoxide and hydroxyl radicals. Therefore, to assess the effects of mutations in tim17 on maintaining the free radical balance, we performed MitoSOX staining and analyzed mitochondrial superoxide levels by flow cytometry. Convincingly, all the tim17 mutants displayed a significant elevation in the mitochondrial superoxide levels compared to wild-type levels (Fig. 9A). This finding further explains the loss of functional mitochondria in tim17 mutants, as elevated superoxide levels are known to cause organellar dysfunction. Since organellar dysfunctions have shown to perturb the overall cellular redox balance in the cells, we next assessed the overall cellular ROS levels by 2′,7′-dichlorodihydrofluorescein diacetate (H2DCFDA) staining. As expected, all the tim17 mutants exhibited a significant increase in overall cellular ROS levels (Fig. 9B), indicating that Tim17 plays an important role in maintaining the cellular free radical balance.
FIG 9.
Analysis of ROS levels and mitochondrial morphology in tim17 mutants. (A) Measurement of mitochondrial superoxide levels by MitoSOX staining. WT and mutant yeast strains grown to mid-log phase were given heat shock at 37°C for 8 h followed by staining with MitoSOX and subsequently analyzed by flow cytometry. The relative fluorescence intensity is represented as the fold mean fluorescence intensity (MFI). *, P ≤ 0.05; **, P ≤ 0.01; ***, P ≤ 0.001. (B) Analysis of total cellular ROS by H2DCFDA staining. After growing to mid-log phase, WT and mutant yeast strains were given heat shock at 37°C for 8 h, followed by staining with H2DCFDA, and analyzed by flow cytometry. The relative fluorescence intensity is represented as the fold mean fluorescence intensity (MFI). *, P ≤ 0.05; **, P ≤ 0.01; ***, P ≤ 0.001. (C) Analysis of mitochondrial morphology. Wild-type and tim17 mutant strains expressing the MTS-mCherry construct were grown to mid-log phase at 30°C. The cells were subjected to heat shock at 37°C and incubated for 4 h to induce the mutant phenotype. Samples were collected and were mounted for imaging analysis using a Delta Vision Elite fluorescence microscope. The images were analyzed using SoftWoRx 6.1.3 software. The cells in all the panels were imaged at identical exposures to compare the fluorescence intensities. Bars, 5 μm.
Enhanced ROS production, mtDNA instability, and loss of Δψm are often implicated with alterations in mitochondrial morphology (73–76). So, we investigated the role of Tim17 in the maintenance of the organellar integrity; wild-type and all tim17 mutant strains were transformed with an MTS-mCherry construct, which decorates the mitochondria exclusively. After inducing the phenotype by heat shock, cells were subjected to fluorescence imaging to visualize the mitochondrial morphology. Strikingly, the loss of functional Tim17 led to mitochondrial fragmentation, as all the mutants showed small punctate structures while the wild type retained its tubular network of mitochondria (Fig. 9C, compare zoomed images on the right). In summary, our studies provide compelling evidence to highlight the role of Tim17 in the maintenance of mitochondrial integrity and biogenesis.
DISCUSSION
Tim17 contains conserved G/AXXXG/A signature motifs in different TM regions, which are often found in multispanning membrane proteins, including glycophorin A, the subunits of the mitochondrial ATP-synthase complex, and the amyloid precursor protein (61–65). These signature motifs are important for mediating homotypic as well as heterotypic interactions, thereby assisting cross talk between membrane proteins. Our previous analysis on Tim23 highlighted that these signature motifs are indispensable for maintaining the architecture of presequence translocase (22). In the current report, we elucidate the significance of G/AXXXG/A signature motifs in all TM regions that are critical for Tim17 in vivo functions. Our genetic analysis for the first time reveals that all the TM regions of Tim17 are indispensable for cell viability, unlike what is seen in Tim23, where TM3 and TM4 are dispensable for growth (22). Strikingly, our biochemical analysis using conditional mutants from all TM regions has revealed that recruitment of Tim17 to the core TIM23 channel is significantly impaired, thus highlighting the importance of these motifs in the maintenance of core TIM23 channel integrity. It has been previously reported that the interaction of Tim17 with Tim23 is essential for the proper organization of TIM23 complex (21). Since Tim23 exists as a dimer and can recruit two Tim17 proteins, destabilization of the core channel due to mutations in transmembrane regions of Tim17 might interfere in its helix-helix packing, with Tim23 leading to perturbations in helical periodicity. Although the tim17G19A mutation is a consensus in nature, it leads to the formation of an AXXXA motif (alanine at 19th and 23rd positions) which is known to maintain the thermostability of membrane proteins, but intermolecular hydrogen bonds are unlikely to be involved to the same extent as GXXXG motifs due to the increased interhelical distance caused by alanine compared to glycine (62). Additionally, most of the dual replacements of glycine residues in tandem G/AXXXG/A motifs were found to be lethal, and the lethality of these mutants was a consequence of the complete destabilization of the core channel. Moreover, these double mutants displayed reduced steady-state levels due to lower stability, indicating that the association of Tim17 with Tim23 is indispensable for the assembly of presequence translocase.
Mitochondrial protein translocation mediated by presequence translocase of inner membrane is dynamic in nature, as it can differentially assemble either into the TIM23 (PAM) complex or into the membrane-embedded TIM23 (SORT) complex in order to facilitate the preprotein import (17, 22, 77, 78). Importantly, all the G/AXXXG/A conditional mutants from different TM regions exhibited a significant decrease in the association of Pam18 and Pam16 to the core channel, indicating that Tim17 acts as an anchor for the recruitment of the “import motor” to assemble into the TIM23 (PAM) complex. On the other hand, a diminished copurification of Tim21 with Tim23 suggests that Tim17 also plays an indispensable role in the assembly of the membrane-embedded TIM23 (SORT) complex. Previous evidence highlights that Tim17 has been shown to play a key role in matrix sorting and membrane insertion of preproteins (17, 22, 53). In correlation with these observations, the conditional mutants of all TM regions showed a significant decrease in their ability to translocate inner membrane as well as the matrix sorted preproteins, thereby highlighting a dual role of Tim17 in protein translocation across the inner membrane, where it can recruit the PAM complex for targeting preproteins into the matrix and it can also anchor Tim21 to facilitate the membrane insertion of preproteins. Collectively, our analyses provide comprehensive evidence to highlight the role of G/AXXXG/A motifs from TM regions of Tim17 in regulating the dynamicity of presequence translocase and facilitating the protein import into the matrix and inner membrane sorting.
Voltage gating is an important event in protein translocation via presequence translocase, as it acts as the initial driving force for preprotein import. Recent reports highlight that the TM2 helix of Tim23 forms a core of the protein-conducting channel with one side facing the nonpolar environment and the opposite side facing the aqueous channel lumen. At the same time, the TM2 region of Tim23 undergoes structural changes in the absence of Δψm, leading to the channel opening toward the more polar environment (54, 55). Intriguingly, our previous study showed that the TM2 region of Tim23 plays an inevitable role in interacting with Tim17, which in turn helps in recruiting the PAM machinery. Further, our investigation also revealed that overexpression of Tim17 in TM2 mutants of Tim23 could suppress the Δψm defects to a significant level (22). In line with the above observations, G/AXXXG/A conditional mutants from all TM regions of Tim17 exhibited significant reduction in Δψm, thus underlining its indispensable role in maintaining inner membrane polarity. Presumably, the loss of membrane polarity due to mutation in G/AXXXG/A motifs can arise as a consequence of perturbed helix-helix packing between Tim23 and Tim17. A recent report highlights that the Tim17 disulfide bond is critical for protein translocation and channel gating (79). Additionally, mutants lacking the Tim17 disulfide bond showed larger Tim21-bound TIM23 complexes and depleted TIM23 core complex, suggesting “extensive remodelling of TIM23-complex” (79). On the other hand, our findings highlight that the mutations in G/AXXXG/A motifs result in complete destabilization of the TIM23 complex due to defective recruitment of Pam18, Pam16, and Tim21. This impaired recruitment of partner subunits subsequently affects the proper channeling of preproteins via the TIM23 complex, thus resulting in defective import. In summary, these results provide compelling evidence that Tim17 acts as a key regulator of channel gating by asserting voltage-coupled conformational dynamics of Tim23.
Proteins encoded by the mitochondrial genome are essential for its biogenesis. The mutations, deletions, or depletion of mitochondrial DNA (mtDNA) can lead to a plethora of inherited diseases, including mtDNA depletion syndrome (MDDS) (80–82). Previous evidence suggests that TIM17 is a multicopy suppressor of the mtDNA instability phenotypes in ilv5Δ and abf2Δ cells, thus highlighting its possible involvement in the maintenance of mtDNA stability (59). Our genetic and biochemical findings provide a direct evidence to highlight a role of Tim17 in the maintenance of mtDNA stability, as the G/AXXXG/A conditional mutants failed to grow on EtBr-supplemented medium, resulting into a petite-negative phenotype (Fig. 7A). Besides, mutants exhibit aberrant mitochondrial nucleoid structure and quantity, thus firmly establishing the role of Tim17 in the maintenance of mitochondrial genome and copy number. Earlier findings suggest that Tim54, a component of carrier translocase, plays an indirect role in maintaining the mtDNA stability by promoting the assembly of Yme1p (83). Similarly, we envision that the mtDNA instability in tim17 mutants could be a secondary effect due to the defective import of proteins required for stability and replication of mtDNA. Moreover, elevated mitochondrial oxidative stress is well known to cause mtDNA damage, leading to its fragmentation as well as accelerating the mitochondrial fission process. As a result, an accumulation of fragmented mitochondrial structures with coalescence morphology was observed with a low mtDNA copy number, thus promoting the formation of petite ([rho−]/[rho0]) phenotypes. Nevertheless, these insightful observations highlight the critical nature of G/AXXXG/A motifs of Tim17 in the dynamic organization of presequence translocase and mitochondrial genome maintenance.
In summary, our findings reveal previously elusive important functional attributes of Tim17 protein as a key regulator of Tim23 complex architecture, inner membrane polarity, mitochondrial morphology, and its genome maintenance. In humans, the altered expression of Tim17 paralogs was shown to be associated with mtDNA loss and cancer progression. Therefore, our results provide a fundamental platform to explore the role of Tim17 in the intricate organization of mammalian presequence translocase and its possible connection in the mitochondrial reprogramming pertinent to cancer progression.
MATERIALS AND METHODS
Genetic methods and yeast strains.
The yeast strains used in this study are listed in Table S1 in the supplemental material. The TIM17 gene was PCR amplified using genomic DNA from positions −500 to +500 and cloned into pRS314 vector. Plasmids encoding mutant tim17 were obtained by performing site-directed mutagenesis with high-fidelity Pfu-Turbo polymerase (Stratagene) using appropriate primers containing the required point mutations. The plasmid having the desired point mutation was transformed into a haploid Δtim17 strain (50) harboring a wild-type functional copy of the TIM17 gene on a URA3-based plasmid and selected on auxotrophic synthetic defined (SD) medium. The strains harboring only mutant tim17 were selected by plasmid shuffling in the presence of 5-FOA. The colonies were tested for the Ts phenotype by spotting on 1% yeast extract, 2% peptone, and 2% glucose (YPD) or 1% yeast extract, 2% peptone, and 3% glycerol (YPG) followed by incubation at different temperatures.
In vitro import assay.
In vitro import kinetics assays were performed with purified precursor proteins Cyb2 (1–167)-DHFR(His) and Cyb2 (1–167)Δ19-DHFR(His) as described previously (22). After inducing the phenotype at 37°C, wild-type (WT) and mutant mitochondria were incubated either with Cyb2 (1–167)-DHFR(His) or with Cyb2 (1–167)Δ19-DHFR(His) in import buffer (250 mM sucrose, 10 mM morpholinepropanesulfonic acid [MOPS]-KOH [pH 7.2], 80 mM KCl, 5 mM dithiothreitol, 5 mM MgCl2, 2 mM ATP, 2 mM NADH, 1% bovine serum albumin) for 5-, 10-, 15-, and 20-min intervals at 25°C. The import reaction was stopped by dissipating the membrane potential using 10 μg/ml of valinomycin (Sigma). Nonimported precursor proteins were removed by treatment with 0.1 mg/ml proteinase K (PK) for 20 min on ice, followed by addition of 1 mM phenylmethylsulfonyl fluoride (PMSF; US Biological) to inhibit proteinase K. The samples were separated on SDS-PAGE gels and processed for immunoblotting using specific antibodies. Image quantification was performed using ImageJ software.
Co-IP and BN-PAGE.
Purified wild-type and mutant mitochondria expressing FLAG-tagged Tim23 were subjected to heat shock for 30 min at 37°C to induce phenotype. Immunoprecipitation was performed using antibodies (Sigma) directed against FLAG tag. The mitochondrial lysates were prepared by resuspending 2 mg of mitochondria in the lysis buffer (20 mM MOPS-KOH [pH 7.4], 250 mM sucrose, 80 mM KCl, 5 mM EDTA, 1 mM PMSF) containing 1% digitonin for 45 min at 4°C. The cleared mitochondrial lysates were incubated with preequilibrated anti-FLAG-bound protein G-Sepharose beads for 1 h at 4°C. The beads were washed with the lysis buffer, and the samples were separated on SDS-polyacrylamide gels, followed by immunodecoration using specific antibodies.
For BN-PAGE analysis, mitochondria (200 μg) were first subjected to heat shock, followed by lysis in 90 μl of ice-cold digitonin buffer (1% digitonin [recrystallized], 20 mM Tris-HCl [pH 7.4], 0.1 mM EDTA, 50 mM NaCl, 10% glycerol, 1 mM PMSF) for 45 min at 4°C. The unsolubilized material was removed by centrifugation for 20 min at 50,000 × g. After addition of 10 μl of sample buffer (5% [wt/vol] Coomassie brilliant blue G-250, 100 mM bis-Tris [pH 7.0], 500 mM 6-aminocaproic acid), the supernatant was analyzed directly by BN-PAGE, followed by immunoblotting with specific antibodies.
Analysis of mitochondrial DNA loss and mitochondrial morphology.
To investigate the mitochondrial DNA instability, wild-type and mutant strains were grown to early log phase. Yeast cells corresponding to A600 of 0.5 were collected and subjected to 10-fold serial dilution. Each dilution was spotted on YPD or on YPD supplemented with 40 μg/ml of ethidium bromide. The plates were incubated at the permissive temperature of 30°C, and images were acquired at 72 h.
Visualization of mitochondria and analysis of mtDNA stability were performed as described previously (84–86). To visualize mitochondria, the yeast cells were transformed with pRS415 vector containing presequence of subunit 9 (pSU9) followed by mCherry fluorescent protein, which decorates the mitochondria exclusively. The cells were washed twice with 1× phosphate-buffered saline (PBS) and mounted with Antifade (Invitrogen) on agarose pads, and images were acquired with a Delta Vision Elite fluorescence microscope (GE Healthcare) using a 100× objective lens. Images were subsequently deconvolved and analyzed using SoftWoRx 6.1.3. software.
For analysis of mitochondrial DNA integrity, yeast cells expressing mitochondrially targeted mCherry fluorescent protein were grown to mid-log phase and incubated with 10 mM SYTO18 for 15 min at room temperature (25°C). Stained yeast cells were thoroughly washed and suspended in HEPES buffer and mounted with Antifade (Invitrogen) on agarose pads. Images were acquired with a Delta Vision Elite fluorescence microscope (GE Healthcare) using a 100× objective lens. The λex and λem used for SYTO18 were 468 nm and 533 nm, respectively, and the images were further analyzed using SoftWoRx 6.1.3. software.
Quantification of protein half-lives.
Wild-type and mutant yeast cells were grown in liquid YPD medium to mid-log phase at 30°C. Cycloheximide (50 μg/ml; US Biological) was added to the cultures to inhibit protein translation. Following the treatment, equal amounts of cells were collected at 0, 15, 30, and 60 min. Cell lysates were prepared and separated on SDS-polyacrylamide gels, followed by immunoblotting using the desired antibody. Data were quantified using ImageJ and analyzed using GraphPad Prism 5.
Fluorescence and flow-cytometric analysis.
To analyze membrane potential, 50 μg of mitochondria was first subjected to heat shock at 37°C, followed by incubation with JC-1 dye for 10 min at 25°C in the dark. Valinomycin (0.1 M) was added to wild-type mitochondria and used as a positive control for complete depolarization. The fluorescence measurement was performed using a JASCO spectrofluorometer (model 6300) at a fixed excitation wavelength of 490 nm, and the emission spectrum was recorded from 500 to 620 nm. The ratio of 590 to 530 nm was used as an indicator of membrane polarization.
For assessment of functional mitochondrial mass, wild-type and mutant strains were grown to early log phase and subjected to heat shock for 2 h. Cells were harvested, washed with PBS, and incubated with 10 mM MitoTracker Deep Red A dye (Invitrogen) in the dark for 15 min. Subsequently, cells were subjected for fluorescence-activated cell sorter (FACS) analysis using an excitation wavelength of 488 nm and an emission wavelength of 520 nm (Becton, Dickinson [BD] FACSCanto II flow cytometer). The data were analyzed using WinMDI, version 2.9, software.
Statistical analyses.
Statistical analyses were performed using GraphPad Prism 5.0 software. Error bars represent standard errors (SE) and are derived from three replicates. For significance testing by one-way analysis of variance, Dunnett's multiple-comparison posttest and Tukey's multiple-comparison tests were used to compare mutant against WT values and between columns, respectively. The amounts of different proteins in Fig. 2D, 3C, and 4C and E were quantified by densitometry using ImageJ software and illustrated in the form of a line graph.
Miscellaneous.
Orthologous Tim17 sequences from different species, including Saccharomyces cerevisiae, Neurospora crassa, Schizosaccharomyces pombe, Arabidopsis thaliana, and Homo sapiens, were retrieved from the NCBI database, and multiple-sequence alignment was performed using ClustalW2 program, which does a pairwise alignment followed by creation of a guide tree to carry out a multiple alignment. Isolation of mitochondria and precursor accumulation analysis (Hsp60) were performed as described in previously published protocols (22). Measurement of ROS was carried out as described earlier (84). The antisera used for immunodecoration against yeast-specific proteins, such as Hsp60, Mge1, Ydj1, Tim23, Tim17, Tim50, Tim44, Pam18, and Pam16 were raised in rabbits, as reported earlier (22). The Tim17 protein tagged with FLAG was detected using an anti-FLAG antibody (Sigma). All immunoblot analyses were performed using an enhanced chemiluminescence system (PerkinElmer) according to the manufacturer's instructions.
Supplementary Material
ACKNOWLEDGMENTS
We are grateful to Elizabeth A Craig, University of Wisconsin—Madison, for the Δtim17 haploid yeast strain and anti-Tim44 antibody. We thank the FACS Facility of the Indian Institute of Science, Bangalore, India. We thank Devanjan Sinha and Shubhi Srivastava for critical analysis of the manuscript.
This work was supported by a Swarnajayanthi fellowship from the Department of Science and Technology (grant no. DST/SJF/LSA-01/2011–2012), DBT-IISc partnership program (grant no. DBT/BF/PR/INS/2011-12/IISc), DST-FIST program (grant no. SR/FST/LSII-023/2009), and UGC-CAS SAP-II program (grant UGC LT. No. F. 5-2/2012. SAP-II) (to P.D). S.K.M. and A.O. acknowledge a research fellowship from University Grants Commission. G.P. and K.B. acknowledge a research fellowship from the Council of Scientific and Industrial Research.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/MCB.00491-16.
REFERENCES
- 1.Baker MJ, Frazier AE, Gulbis JM, Ryan MT. 2007. Mitochondrial protein-import machinery: correlating structure with function. Trends Cell Biol 17:456–464. doi: 10.1016/j.tcb.2007.07.010. [DOI] [PubMed] [Google Scholar]
- 2.Bauer MF, Hofmann S, Neupert W, Brunner M. 2000. Protein translocation into mitochondria: the role of TIM complexes. Trends Cell Biol 10:25–31. doi: 10.1016/S0962-8924(99)01684-0. [DOI] [PubMed] [Google Scholar]
- 3.Bolender N, Sickmann A, Wagner R, Meisinger C, Pfanner N. 2008. Multiple pathways for sorting mitochondrial precursor proteins. EMBO Rep 9:42–49. doi: 10.1038/sj.embor.7401126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chacinska A, Koehler CM, Milenkovic D, Lithgow T, Pfanner N. 2009. Importing mitochondrial proteins: machineries and mechanisms. Cell 138:628–644. doi: 10.1016/j.cell.2009.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Neupert W, Brunner M. 2002. The protein import motor of mitochondria. Nat Rev Mol Cell Biol 3:555–565. doi: 10.1038/nrm878. [DOI] [PubMed] [Google Scholar]
- 6.Neupert W, Herrmann JM. 2007. Translocation of proteins into mitochondria. Annu Rev Biochem 76:723–749. doi: 10.1146/annurev.biochem.76.052705.163409. [DOI] [PubMed] [Google Scholar]
- 7.Schmidt O, Pfanner N, Meisinger C. 2010. Mitochondrial protein import: from proteomics to functional mechanisms. Nat Rev Mol Cell Biol 11:655–667. doi: 10.1038/nrm2959. [DOI] [PubMed] [Google Scholar]
- 8.Truscott KN, Brandner K, Pfanner N. 2003. Mechanisms of protein import into mitochondria. Curr Biol 13:R326–R337. doi: 10.1016/S0960-9822(03)00239-2. [DOI] [PubMed] [Google Scholar]
- 9.van der Laan M, Hutu DP, Rehling P. 2010. On the mechanism of preprotein import by the mitochondrial presequence translocase. Biochim Biophys Acta 1803:732–739. doi: 10.1016/j.bbamcr.2010.01.013. [DOI] [PubMed] [Google Scholar]
- 10.Donzeau M, Kaldi K, Adam A, Paschen S, Wanner G, Guiard B, Bauer MF, Neupert W, Brunner M. 2000. Tim23 links the inner and outer mitochondrial membranes. Cell 101:401–412. doi: 10.1016/S0092-8674(00)80850-8. [DOI] [PubMed] [Google Scholar]
- 11.Bauer MF, Sirrenberg C, Neupert W, Brunner M. 1996. Role of Tim23 as voltage sensor and presequence receptor in protein import into mitochondria. Cell 87:33–41. doi: 10.1016/S0092-8674(00)81320-3. [DOI] [PubMed] [Google Scholar]
- 12.Dekker PJ, Keil P, Rassow J, Maarse AC, Pfanner N, Meijer M. 1993. Identification of MIM23, a putative component of the protein import machinery of the mitochondrial inner membrane. FEBS Lett 330:66–70. doi: 10.1016/0014-5793(93)80921-G. [DOI] [PubMed] [Google Scholar]
- 13.Emtage JL, Jensen RE. 1993. MAS6 encodes an essential inner membrane component of the yeast mitochondrial protein import pathway. J Cell Biol 122:1003–1012. doi: 10.1083/jcb.122.5.1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Maarse AC, Blom J, Keil P, Pfanner N, Meijer M. 1994. Identification of the essential yeast protein MIM17, an integral mitochondrial inner membrane protein involved in protein import. FEBS Lett 349:215–221. doi: 10.1016/0014-5793(94)00669-5. [DOI] [PubMed] [Google Scholar]
- 15.Ryan KR, Menold MM, Garrett S, Jensen RE. 1994. SMS1, a high-copy suppressor of the yeast mas6 mutant, encodes an essential inner membrane protein required for mitochondrial protein import. Mol Biol Cell 5:529–538. doi: 10.1091/mbc.5.5.529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Alder NN, Sutherland J, Buhring AI, Jensen RE, Johnson AE. 2008. Quaternary structure of the mitochondrial TIM23 complex reveals dynamic association between Tim23p and other subunits. Mol Biol Cell 19:159–170. doi: 10.1091/mbc.E07-07-0669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chacinska A, Lind M, Frazier AE, Dudek J, Meisinger C, Geissler A, Sickmann A, Meyer HE, Truscott KN, Guiard B, Pfanner N, Rehling P. 2005. Mitochondrial presequence translocase: switching between TOM tethering and motor recruitment involves Tim21 and Tim17. Cell 120:817–829. doi: 10.1016/j.cell.2005.01.011. [DOI] [PubMed] [Google Scholar]
- 18.Davis AJ, Ryan KR, Jensen RE. 1998. Tim23p contains separate and distinct signals for targeting to mitochondria and insertion into the inner membrane. Mol Biol Cell 9:2577–2593. doi: 10.1091/mbc.9.9.2577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kaldi K, Bauer MF, Sirrenberg C, Neupert W, Brunner M. 1998. Biogenesis of Tim23 and Tim17, integral components of the TIM machinery for matrix-targeted preproteins. EMBO J 17:1569–1576. doi: 10.1093/emboj/17.6.1569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lohret TA, Jensen RE, Kinnally KW. 1997. Tim23, a protein import component of the mitochondrial inner membrane, is required for normal activity of the multiple conductance channel, MCC. J Cell Biol 137:377–386. doi: 10.1083/jcb.137.2.377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Martinez-Caballero S, Grigoriev SM, Herrmann JM, Campo ML, Kinnally KW. 2007. Tim17p regulates the twin pore structure and voltage gating of the mitochondrial protein import complex TIM23. J Biol Chem 282:3584–3593. [DOI] [PubMed] [Google Scholar]
- 22.Pareek G, Krishnamoorthy V, D'Silva P. 2013. Molecular insights revealing interaction of Tim23 and channel subunits of presequence translocase. Mol Cell Biol 33:4641–4659. doi: 10.1128/MCB.00876-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Truscott KN, Kovermann P, Geissler A, Merlin A, Meijer M, Driessen AJ, Rassow J, Pfanner N, Wagner R. 2001. A presequence- and voltage-sensitive channel of the mitochondrial preprotein translocase formed by Tim23. Nat Struct Biol 8:1074–1082. doi: 10.1038/nsb726. [DOI] [PubMed] [Google Scholar]
- 24.Albrecht R, Rehling P, Chacinska A, Brix J, Cadamuro SA, Volkmer R, Guiard B, Pfanner N, Zeth K. 2006. The Tim21 binding domain connects the preprotein translocases of both mitochondrial membranes. EMBO Rep 7:1233–1238. doi: 10.1038/sj.embor.7400828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Geissler A, Chacinska A, Truscott KN, Wiedemann N, Brandner K, Sickmann A, Meyer HE, Meisinger C, Pfanner N, Rehling P. 2002. The mitochondrial presequence translocase: an essential role of Tim50 in directing preproteins to the import channel. Cell 111:507–518. doi: 10.1016/S0092-8674(02)01073-5. [DOI] [PubMed] [Google Scholar]
- 26.Gevorkyan-Airapetov L, Zohary K, Popov-Celeketic D, Mapa K, Hell K, Neupert W, Azem A, Mokranjac D. 2009. Interaction of Tim23 with Tim50 is essential for protein translocation by the mitochondrial TIM23 complex. J Biol Chem 284:4865–4872. doi: 10.1074/jbc.M807041200. [DOI] [PubMed] [Google Scholar]
- 27.Mokranjac D, Paschen SA, Kozany C, Prokisch H, Hoppins SC, Nargang FE, Neupert W, Hell K. 2003. Tim50, a novel component of the TIM23 preprotein translocase of mitochondria. EMBO J 22:816–825. doi: 10.1093/emboj/cdg090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mokranjac D, Popov-Celeketic D, Hell K, Neupert W. 2005. Role of Tim21 in mitochondrial translocation contact sites. J Biol Chem 280:23437–23440. doi: 10.1074/jbc.C500135200. [DOI] [PubMed] [Google Scholar]
- 29.Schulz C, Lytovchenko O, Melin J, Chacinska A, Guiard B, Neumann P, Ficner R, Jahn O, Schmidt B, Rehling P. 2011. Tim50's presequence receptor domain is essential for signal driven transport across the TIM23 complex. J Cell Biol 195:643–656. doi: 10.1083/jcb.201105098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tamura Y, Harada Y, Shiota T, Yamano K, Watanabe K, Yokota M, Yamamoto H, Sesaki H, Endo T. 2009. Tim23-Tim50 pair coordinates functions of translocators and motor proteins in mitochondrial protein import. J Cell Biol 184:129–141. doi: 10.1083/jcb.200808068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.van der Laan M, Wiedemann N, Mick DU, Guiard B, Rehling P, Pfanner N. 2006. A role for Tim21 in membrane-potential-dependent preprotein sorting in mitochondria. Curr Biol 16:2271–2276. doi: 10.1016/j.cub.2006.10.025. [DOI] [PubMed] [Google Scholar]
- 32.Yamamoto H, Esaki M, Kanamori T, Tamura Y, Nishikawa S, Endo T. 2002. Tim50 is a subunit of the TIM23 complex that links protein translocation across the outer and inner mitochondrial membranes. Cell 111:519–528. doi: 10.1016/S0092-8674(02)01053-X. [DOI] [PubMed] [Google Scholar]
- 33.Gebert M, Schrempp SG, Mehnert CS, Heisswolf AK, Oeljeklaus S, Ieva R, Bohnert M, von der Malsburg K, Wiese S, Kleinschroth T, Hunte C, Meyer HE, Haferkamp I, Guiard B, Warscheid B, Pfanner N, van der Laan M. 2012. Mgr2 promotes coupling of the mitochondrial presequence translocase to partner complexes. J Cell Biol 197:595–604. doi: 10.1083/jcb.201110047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ieva R, Heisswolf AK, Gebert M, Vogtle FN, Wollweber F, Mehnert CS, Oeljeklaus S, Warscheid B, Meisinger C, van der Laan M, Pfanner N. 2013. Mitochondrial inner membrane protease promotes assembly of presequence translocase by removing a carboxy-terminal targeting sequence. Nat Commun 4:2853. doi: 10.1038/ncomms3853. [DOI] [PubMed] [Google Scholar]
- 35.Ieva R, Schrempp SG, Opalinski L, Wollweber F, Hoss P, Heisswolf AK, Gebert M, Zhang Y, Guiard B, Rospert S, Becker T, Chacinska A, Pfanner N, van der Laan M. 2014. Mgr2 functions as lateral gatekeeper for preprotein sorting in the mitochondrial inner membrane. Mol Cell 56:641–652. doi: 10.1016/j.molcel.2014.10.010. [DOI] [PubMed] [Google Scholar]
- 36.Martin J, Mahlke K, Pfanner N. 1991. Role of an energized inner membrane in mitochondrial protein import. Delta psi drives the movement of presequences. J Biol Chem 266:18051–18057. [PubMed] [Google Scholar]
- 37.Berthold J, Bauer MF, Schneider HC, Klaus C, Dietmeier K, Neupert W, Brunner M. 1995. The MIM complex mediates preprotein translocation across the mitochondrial inner membrane and couples it to the mt-Hsp70/ATP driving system. Cell 81:1085–1093. doi: 10.1016/S0092-8674(05)80013-3. [DOI] [PubMed] [Google Scholar]
- 38.Liu Q, D'Silva P, Walter W, Marszalek J, Craig EA. 2003. Regulated cycling of mitochondrial Hsp70 at the protein import channel. Science 300:139–141. doi: 10.1126/science.1083379. [DOI] [PubMed] [Google Scholar]
- 39.Rassow J, Maarse AC, Krainer E, Kubrich M, Muller H, Meijer M, Craig EA, Pfanner N. 1994. Mitochondrial protein import: biochemical and genetic evidence for interaction of matrix hsp70 and the inner membrane protein MIM44. J Cell Biol 127:1547–1556. doi: 10.1083/jcb.127.6.1547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schneider HC, Berthold J, Bauer MF, Dietmeier K, Guiard B, Brunner M, Neupert W. 1994. Mitochondrial Hsp70/MIM44 complex facilitates protein import. Nature 371:768–774. doi: 10.1038/371768a0. [DOI] [PubMed] [Google Scholar]
- 41.Ting SY, Schilke BA, Hayashi M, Craig EA. 2014. Architecture of the TIM23 inner mitochondrial translocon and interactions with the matrix import motor. J Biol Chem 289:28689–28696. doi: 10.1074/jbc.M114.588152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.D'Silva PD, Schilke B, Walter W, Andrew A, Craig EA. 2003. J protein cochaperone of the mitochondrial inner membrane required for protein import into the mitochondrial matrix. Proc Natl Acad Sci U S A 100:13839–13844. doi: 10.1073/pnas.1936150100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.D'Silva PR, Schilke B, Walter W, Craig EA. 2005. Role of Pam16's degenerate J domain in protein import across the mitochondrial inner membrane. Proc Natl Acad Sci U S A 102:12419–12424. doi: 10.1073/pnas.0505969102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Frazier AE, Dudek J, Guiard B, Voos W, Li Y, Lind M, Meisinger C, Geissler A, Sickmann A, Meyer HE, Bilanchone V, Cumsky MG, Truscott KN, Pfanner N, Rehling P. 2004. Pam16 has an essential role in the mitochondrial protein import motor. Nat Struct Mol Biol 11:226–233. doi: 10.1038/nsmb735. [DOI] [PubMed] [Google Scholar]
- 45.Kozany C, Mokranjac D, Sichting M, Neupert W, Hell K. 2004. The J domain-related cochaperone Tim16 is a constituent of the mitochondrial TIM23 preprotein translocase. Nat Struct Mol Biol 11:234–241. doi: 10.1038/nsmb734. [DOI] [PubMed] [Google Scholar]
- 46.Li Y, Dudek J, Guiard B, Pfanner N, Rehling P, Voos W. 2004. The presequence translocase-associated protein import motor of mitochondria. Pam16 functions in an antagonistic manner to Pam18. J Biol Chem 279:38047–38054. [DOI] [PubMed] [Google Scholar]
- 47.Mokranjac D, Sichting M, Neupert W, Hell K. 2003. Tim14, a novel key component of the import motor of the TIM23 protein translocase of mitochondria. EMBO J 22:4945–4956. doi: 10.1093/emboj/cdg485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Truscott KN, Voos W, Frazier AE, Lind M, Li Y, Geissler A, Dudek J, Muller H, Sickmann A, Meyer HE, Meisinger C, Guiard B, Rehling P, Pfanner N. 2003. A J-protein is an essential subunit of the presequence translocase-associated protein import motor of mitochondria. J Cell Biol 163:707–713. doi: 10.1083/jcb.200308004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gallas MR, Dienhart MK, Stuart RA, Long RM. 2006. Characterization of Mmp37p, a Saccharomyces cerevisiae mitochondrial matrix protein with a role in mitochondrial protein import. Mol Biol Cell 17:4051–4062. doi: 10.1091/mbc.E06-04-0366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Schilke BA, Hayashi M, Craig EA. 2012. Genetic analysis of complex interactions among components of the mitochondrial import motor and translocon in Saccharomyces cerevisiae. Genetics 190:1341–1353. doi: 10.1534/genetics.112.138743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tamura Y, Harada Y, Yamano K, Watanabe K, Ishikawa D, Ohshima C, Nishikawa S, Yamamoto H, Endo T. 2006. Identification of Tam41 maintaining integrity of the TIM23 protein translocator complex in mitochondria. J Cell Biol 174:631–637. doi: 10.1083/jcb.200603087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.van der Laan M, Chacinska A, Lind M, Perschil I, Sickmann A, Meyer HE, Guiard B, Meisinger C, Pfanner N, Rehling P. 2005. Pam17 is required for architecture and translocation activity of the mitochondrial protein import motor. Mol Cell Biol 25:7449–7458. doi: 10.1128/MCB.25.17.7449-7458.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Meier S, Neupert W, Herrmann JM. 2005. Conserved N-terminal negative charges in the Tim17 subunit of the TIM23 translocase play a critical role in the import of preproteins into mitochondria. J Biol Chem 280:7777–7785. doi: 10.1074/jbc.M412158200. [DOI] [PubMed] [Google Scholar]
- 54.Alder NN, Jensen RE, Johnson AE. 2008. Fluorescence mapping of mitochondrial TIM23 complex reveals a water-facing, substrate-interacting helix surface. Cell 134:439–450. doi: 10.1016/j.cell.2008.06.007. [DOI] [PubMed] [Google Scholar]
- 55.Malhotra K, Sathappa M, Landin JS, Johnson AE, Alder NN. 2013. Structural changes in the mitochondrial Tim23 channel are coupled to the proton-motive force. Nat Struct Mol Biol 20:965–972. doi: 10.1038/nsmb.2613. [DOI] [PubMed] [Google Scholar]
- 56.Sinha D, Srivastava S, Krishna L, D'Silva P. 2014. Unraveling the intricate organization of mammalian mitochondrial presequence translocases: existence of multiple translocases for maintenance of mitochondrial function. Mol Cell Biol 34:1757–1775. doi: 10.1128/MCB.01527-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Salhab M, Patani N, Jiang W, Mokbel K. 2012. High TIMM17A expression is associated with adverse pathological and clinical outcomes in human breast cancer. Breast Cancer 19:153–160. doi: 10.1007/s12282-010-0228-3. [DOI] [PubMed] [Google Scholar]
- 58.Xu X, Qiao M, Zhang Y, Jiang Y, Wei P, Yao J, Gu B, Wang Y, Lu J, Wang Z, Tang Z, Sun Y, Wu W, Shi Q. 2010. Quantitative proteomics study of breast cancer cell lines isolated from a single patient: discovery of TIMM17A as a marker for breast cancer. Proteomics 10:1374–1390. doi: 10.1002/pmic.200900380. [DOI] [PubMed] [Google Scholar]
- 59.Iacovino M, Granycome C, Sembongi H, Bokori-Brown M, Butow RA, Holt IJ, Bateman JM. 2009. The conserved translocase Tim17 prevents mitochondrial DNA loss. Hum Mol Genet 18:65–74. doi: 10.1093/hmg/ddn313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Doura AK, Fleming KG. 2004. Complex interactions at the helix-helix interface stabilize the glycophorin A transmembrane dimer. J Mol Biol 343:1487–1497. doi: 10.1016/j.jmb.2004.09.011. [DOI] [PubMed] [Google Scholar]
- 61.Doura AK, Kobus FJ, Dubrovsky L, Hibbard E, Fleming KG. 2004. Sequence context modulates the stability of a GxxxG-mediated transmembrane helix-helix dimer. J Mol Biol 341:991–998. doi: 10.1016/j.jmb.2004.06.042. [DOI] [PubMed] [Google Scholar]
- 62.Kleiger G, Grothe R, Mallick P, Eisenberg D. 2002. GXXXG and AXXXA: common alpha-helical interaction motifs in proteins, particularly in extremophiles. Biochemistry 41:5990–5997. doi: 10.1021/bi0200763. [DOI] [PubMed] [Google Scholar]
- 63.McClain MS, Iwamoto H, Cao P, Vinion-Dubiel AD, Li Y, Szabo G, Shao Z, Cover TL. 2003. Essential role of a GXXXG motif for membrane channel formation by Helicobacter pylori vacuolating toxin. J Biol Chem 278:12101–12108. doi: 10.1074/jbc.M212595200. [DOI] [PubMed] [Google Scholar]
- 64.Melnyk RA, Kim S, Curran AR, Engelman DM, Bowie JU, Deber CM. 2004. The affinity of GXXXG motifs in transmembrane helix-helix interactions is modulated by long-range communication. J Biol Chem 279:16591–16597. doi: 10.1074/jbc.M313936200. [DOI] [PubMed] [Google Scholar]
- 65.Demishtein-Zohary K, Marom M, Neupert W, Mokranjac D, Azem A. 2015. GxxxG motifs hold the TIM23 complex together. FEBS J 282:2178–2186. doi: 10.1111/febs.13266. [DOI] [PubMed] [Google Scholar]
- 66.Wrobel L, Sokol AM, Chojnacka M, Chacinska A. 2016. The presence of disulfide bonds reveals an evolutionarily conserved mechanism involved in mitochondrial protein translocase assembly. Sci Rep 6:27484. doi: 10.1038/srep27484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Chen XJ, Butow RA. 2005. The organization and inheritance of the mitochondrial genome. Nat Rev Genet 6:815–825. doi: 10.1038/nrg1708. [DOI] [PubMed] [Google Scholar]
- 68.Spelbrink JN. 2010. Functional organization of mammalian mitochondrial DNA in nucleoids: history, recent developments, and future challenges. IUBMB Life 62:19–32. doi: 10.1002/iub.282. [DOI] [PubMed] [Google Scholar]
- 69.Cossarizza A, Salvioli S. 2001. Flow cytometric analysis of mitochondrial membrane potential using JC-1. Curr Protoc Cytom; Chapter 9:Unit 9.14. doi: 10.1002/0471142956.cy0914s13. [DOI] [PubMed] [Google Scholar]
- 70.Lugli E, Troiano L, Ferraresi R, Roat E, Prada N, Nasi M, Pinti M, Cooper EL, Cossarizza A. 2005. Characterization of cells with different mitochondrial membrane potential during apoptosis. Cytometry A 68:28–35. [DOI] [PubMed] [Google Scholar]
- 71.Wang C, Li Z, Lu Y, Du R, Katiyar S, Yang J, Fu M, Leader JE, Quong A, Novikoff PM, Pestell RG. 2006. Cyclin D1 repression of nuclear respiratory factor 1 integrates nuclear DNA synthesis and mitochondrial function. Proc Natl Acad Sci U S A 103:11567–11572. doi: 10.1073/pnas.0603363103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Zhou R, Yazdi AS, Menu P, Tschopp J. 2011. A role for mitochondria in NLRP3 inflammasome activation. Nature 469:221–225. doi: 10.1038/nature09663. [DOI] [PubMed] [Google Scholar]
- 73.Legros F, Lombes A, Frachon P, Rojo M. 2002. Mitochondrial fusion in human cells is efficient, requires the inner membrane potential, and is mediated by mitofusins. Mol Biol Cell 13:4343–4354. doi: 10.1091/mbc.E02-06-0330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Olichon A, Baricault L, Gas N, Guillou E, Valette A, Belenguer P, Lenaers G. 2003. Loss of OPA1 perturbates the mitochondrial inner membrane structure and integrity, leading to cytochrome c release and apoptosis. J Biol Chem 278:7743–7746. doi: 10.1074/jbc.C200677200. [DOI] [PubMed] [Google Scholar]
- 75.Osman C, Noriega TR, Okreglak V, Fung JC, Walter P. 2015. Integrity of the yeast mitochondrial genome, but not its distribution and inheritance, relies on mitochondrial fission and fusion. Proc Natl Acad Sci U S A 112:E947–E956. doi: 10.1073/pnas.1501737112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Wu S, Zhou F, Zhang Z, Xing D. 2011. Mitochondrial oxidative stress causes mitochondrial fragmentation via differential modulation of mitochondrial fission-fusion proteins. FEBS J 278:941–954. doi: 10.1111/j.1742-4658.2011.08010.x. [DOI] [PubMed] [Google Scholar]
- 77.Popov-Celeketic D, Mapa K, Neupert W, Mokranjac D. 2008. Active remodelling of the TIM23 complex during translocation of preproteins into mitochondria. EMBO J 27:1469–1480. doi: 10.1038/emboj.2008.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Wiedemann N, van der Laan M, Hutu DP, Rehling P, Pfanner N. 2007. Sorting switch of mitochondrial presequence translocase involves coupling of motor module to respiratory chain. J Cell Biol 179:1115–1122. doi: 10.1083/jcb.200709087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Ramesh A, Peleh V, Martinez-Caballero S, Wollweber F, Sommer F, van der Laan M, Schroda M, Alexander RT, Campo ML, Herrmann JM. 2016. A disulfide bond in the TIM23 complex is crucial for voltage gating and mitochondrial protein import. J Cell Biol 214:417–431. doi: 10.1083/jcb.201602074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Chatterjee A, Mambo E, Sidransky D. 2006. Mitochondrial DNA mutations in human cancer. Oncogene 25:4663–4674. doi: 10.1038/sj.onc.1209604. [DOI] [PubMed] [Google Scholar]
- 81.Dimmer KS, Jakobs S, Vogel F, Altmann K, Westermann B. 2005. Mdm31 and Mdm32 are inner membrane proteins required for maintenance of mitochondrial shape and stability of mitochondrial DNA nucleoids in yeast. J Cell Biol 168:103–115. doi: 10.1083/jcb.200410030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Taylor RW, Turnbull DM. 2005. Mitochondrial DNA mutations in human disease. Nat Rev Genet 6:389–402. doi: 10.1038/nrg1606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Hwang DK, Claypool SM, Leuenberger D, Tienson HL, Koehler CM. 2007. Tim54p connects inner membrane assembly and proteolytic pathways in the mitochondrion. J Cell Biol 178:1161–1175. doi: 10.1083/jcb.200706195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Bankapalli K, Saladi S, Awadia SS, Goswami AV, Samaddar M, D'Silva P. 2015. Robust glyoxalase activity of Hsp31, a ThiJ/DJ-1/PfpI family member protein, is critical for oxidative stress resistance in Saccharomyces cerevisiae. J Biol Chem 290:26491–26507. doi: 10.1074/jbc.M115.673624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Ingavale SS, Chang YC, Lee H, McClelland CM, Leong ML, Kwon-Chung KJ. 2008. Importance of mitochondria in survival of Cryptococcus neoformans under low oxygen conditions and tolerance to cobalt chloride. PLoS Pathog 4:e1000155. doi: 10.1371/journal.ppat.1000155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Kim JM, Yoshikawa H, Shirahige K. 2001. A member of the YER057c/yjgf/Uk114 family links isoleucine biosynthesis and intact mitochondria maintenance in Saccharomyces cerevisiae. Genes Cells 6:507–517. doi: 10.1046/j.1365-2443.2001.00443.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.