ABSTRACT
Construction of Listeria monocytogenes mutants by allelic exchange has been laborious and time-consuming due to lack of proficient selection markers for the final recombination event, that is, a marker conveying substance sensitivity to the bacteria bearing it, enabling the exclusion of merodiploids and selection for plasmid loss. In order to address this issue, we engineered a counterselection marker based on a mutated phenylalanyl-tRNA synthetase gene (pheS*). This mutation renders the phenylalanine-binding site of the enzyme more promiscuous and allows the binding of the toxic p-chloro-phenylalanine analog (p-Cl-phe) as a substrate. When pheS* is introduced into L. monocytogenes and highly expressed under control of a constitutively active promoter, the bacteria become sensitive to p-Cl-phe supplemented in the medium. This enabled us to utilize pheS* as a negative selection marker and generate a novel, efficient suicide vector for allelic exchange in L. monocytogenes. We used this vector to investigate the monocin genomic region in L. monocytogenes strain 10403S by constructing deletion mutants of the region. We have found this region to be active and to cause bacterial lysis upon mitomycin C treatment. The future applications of such an effective counterselection system, which does not require any background genomic alterations, are vast, as it can be modularly used in various selection systems (e.g., genetic screens). We expect this counterselection marker to be a valuable genetic tool in research on L. monocytogenes.
IMPORTANCE L. monocytogenes is an opportunistic intracellular pathogen and a widely studied model organism. An efficient counterselection marker is a long-standing need in Listeria research for improving the ability to design and perform various genetic manipulations and screening systems for different purposes. We report the construction and utilization of an efficient suicide vector for allelic exchange which can be conjugated, leaves no marker in the bacterial chromosome, and does not require the use of sometimes leaky inducible promoters. This highly efficient genome editing tool for L. monocytogenes will allow for rapid sequential mutagenesis, introduction of point mutations, and design of screening systems. We anticipate that it will be extensively used by the research community and yield novel insights into the diverse fields studied using this model organism.
KEYWORDS: counterselection, Listeria monocytogenes, allelic exchange, bacteriocins, monocin, mutagenesis, pheS
INTRODUCTION
Listeria monocytogenes is a Gram-positive, foodborne, facultative intracellular pathogen and the causative agent of listeriosis (1). Listeriosis can cause abortions during pregnancy in humans and may lead to encephalitis, septicemia, and death in immunosuppressed people, such as the elderly, HIV patients, and people after transplantation or chemotherapy. L. monocytogenes is being extensively studied as a model organism for intracellular pathogens via genetic manipulation (1–3). There are many genetic tools available for L. monocytogenes, including integrative plasmids, replicating plasmids, transposons, and temperature-sensitive plasmids, enabling various manipulations to be carried out (4). In order to genetically investigate the pathogenicity of this model organism in an unbiased manner, it is essential to construct clean mutations or genomic deletions of loci of interest. Generation of “scarless” deletion mutants, genomic point mutations, or genomic insertions in L. monocytogenes is performed by the standard allelic exchange method. When applying this method, a plasmid is site-specifically integrated into the bacterial chromosome by homologous recombination (first recombination event), and subsequently the plasmid is excised, again by homologous recombination (second recombination event), leaving the chromosome mutated. Selection for the first recombination event is fairly easily achieved, as the plasmids utilized harbor an antibiotic resistance gene and a temperature-sensitive origin of replication. Therefore, after introducing the plasmid into the bacteria, selection is attained by plating selective media at temperatures inhibiting plasmid replication (5–9). However, selection for the second recombination event is routinely executed by manually streaking colonies on plates with and without antibiotics and searching for bacteria regaining antibiotic sensitivity. This process is laborious, as only a small fraction of the bacteria go through the second recombination event to regain sensitivity after overnight culture growth. To enrich these bacteria, the population is grown for dozens of generations without antibiotics, which is highly time-consuming. A marker enabling the exclusion of merodiploids and selection for plasmid loss (a counterselection marker) can make the genomic editing process dramatically more efficient, and yet until now an efficient counterselection system for L. monocytogenes has not been available.
Some of the accepted counterselection approaches used in other bacteria, such as sacB and tetAR, necessitate features of Gram-negative bacteria and are therefore incompatible with L. monocytogenes (10). Recently, Abdelhamed et al. constructed and published a report of a counterselectable plasmid for L. monocytogenes which is based on the inducible transcription of secY antisense RNA (7). Notwithstanding, from our experience with inducible promoters in Listeria, leaky expression is always an issue and in this case may inflict a fitness cost to bacteria harboring the plasmid, even in the absence of an inducer. Moreover, this plasmid is 8,995 bp long, which may make inserting long fragments less efficient, and it cannot be conjugated, which necessitates the laborious preparation of competent L. monocytogenes cells from each parental strain. These constraints accumulate particularly when constructing strains with multiple genomic alterations sequentially.
In this study, we describe the construction of an efficient counterselection marker in L. monocytogenes, which is based on a mutated phenylalanyl-tRNA synthetase gene (pheS*) (11). This mutation renders the phenylalanine-binding site of the enzyme more promiscuous and allows the binding of the toxic p-chloro-phenylalanine (p-Cl-phe) analog as a substrate. Using this approach as a counterselection system we generated a novel, efficient suicide vector for allelic exchange in L. monocytogenes. As a proof of concept, two gene deletion mutations in the monocin (F-type bacteriocin) genomic region of the 10403S strain were generated by this method and are described below.
RESULTS
Construction and calibration of pheS* counterselection marker.
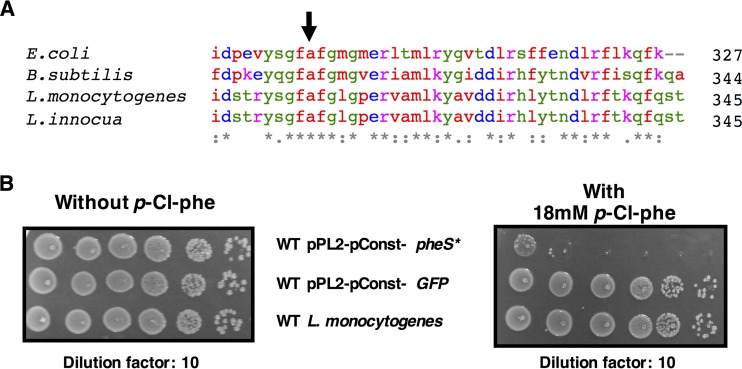
Mutating the conserved alanine corresponding to A294 in the PheS of Escherichia coli to glycine was previously shown to generate an effective counterselection system in E. coli, Enterococcus faecalis, Streptococcus mutans, and Thermus thermophilus (12–15). Using Clustal Omega, we aligned the amino acid sequences of PheS from E. coli to those of Listeria monocytogenes, Listeria innocua, and Bacillus subtilis and found the conserved alanine in all species (Fig. 1A) (16). We chose to clone the B. subtilis pheS gene and not the endogenous pheS of L. monocytogenes in order to avoid unwanted integration of the counterselection vector into the native L. monocytogenes pheS gene by homologous recombination during the first recombination event. When aligning the pheS DNA sequences from L. monocytogenes and B. subtilis using MUSCLE, it is evident that even though the total identity is high at 68.61%, the longest identity streak is only 18 bp long, and thus the chances for homologous recombination between the two genes are low (see Fig. S1 in the supplemental material) (17). Indeed, in our experience in working with this novel vector, no cases of such unwanted integration were encountered (so far, the system has been used to generate 20 different mutants). To this end, we cloned the B. subtilis pheS gene into the pPL2 integrative plasmid, replacing the corresponding codon GCA to GGA (A309G) to generate the pheS* mutation (for details, see Materials and Methods) (Fig. 1A and S1) (18). The mutated pheS* gene was expressed using a highly active constitutive promoter designated pConst (pPL2-pConst-pheS*). This promoter, consisting of the constitutive HyperSPO1 promoter fused to the full hly 5′ untranslated region (UTR), was constructed by Shen and Higgins and described in their work from 2005 (19). We next introduced pPL2-pConst-pheS* into L. monocytogenes, assessed the counterselection efficiency, and calibrated the p-Cl-phe concentration optimal for this system. By applying this expression system, we were able to restrict L. monocytogenes growth on plates containing 18 mM p-Cl-phe by ∼10−5-fold, as evident from the CFU ratio between the restricted and unrestricted bacteria. As a control we used the same vector expressing green fluorescent protein (GFP) instead of PheS* to show that the restriction does not arise from the vector or the highly active promoter itself, with no apparent growth restriction (Fig. 1B).
FIG 1.
Construction and calibration of the pheS* counterselection marker. (A) Amino acid multiple-sequence alignment of PheS protein C termini, achieved with Clustal Omega. The sequences of the indicated species were aligned, and the conserved alanine corresponding to A294 of E. coli was identified (arrow); on the right is the number of amino acids in the protein. (B) Growth of the indicated strains on BHI agar plates with or without 18 mM p-Cl-phe.
Construction of a novel suicide counterselection plasmid, pLR16-pheS*.
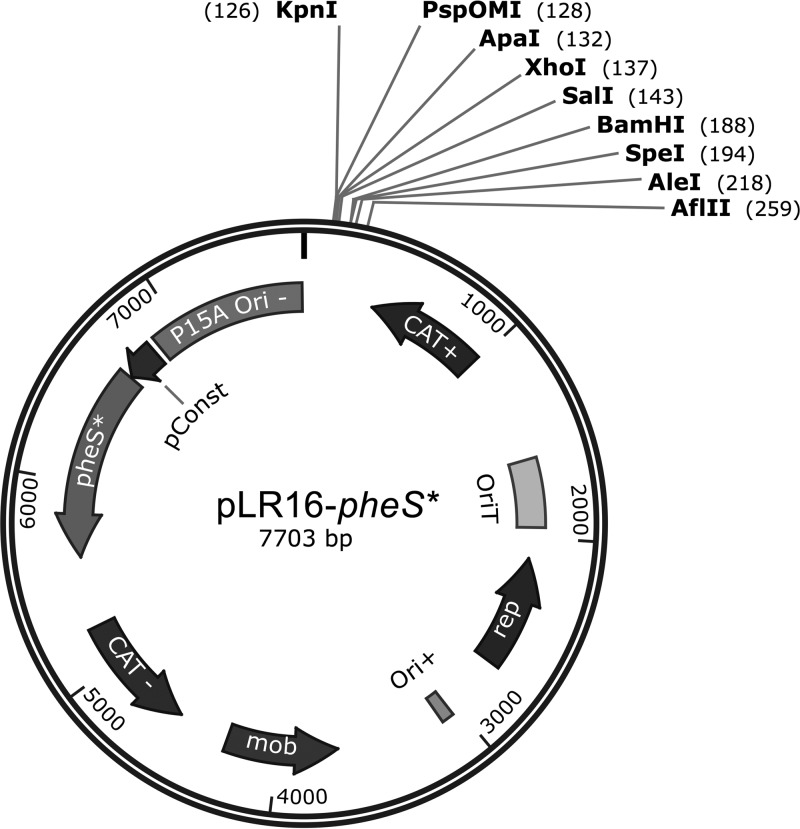
In order to use the pheS* counterselection marker for allelic exchange procedures, we generated a new plasmid, pLR16-pheS* (Fig. 2). This plasmid contains the temperature-sensitive Gram-positive origin of replication (Ori) first published as a part of pKSV7 (20). This Ori facilitates plasmid replication at lower temperatures (30°C) and does not allow replication at higher temperatures (41°C). As indicated above, this feature allows the integration of the plasmid into the target gene at higher temperatures as part of the first recombination event. The origin of transfer (OriT), chloramphenicol resistance genes, and multiple cloning site (MCS) used in pLR16-pheS* are all part of pPL2 and were previously published (18) (Fig. 2). During the construction of the plasmid, several cloning approaches were taken and their products assessed before reaching the operational plasmid. The full process is detailed in Materials and Methods and in Fig. S2 in the supplemental material, and the primers used are specified in Table 1.
FIG 2.
pLR16-pheS* annotated plasmid map. The plasmid layout of pLR16-pheS*, including the key functions annotated, is shown. Ori+ is a temperature-sensitive Gram-positive origin of replication first published as a part of pKSV7, and the origin of transfer (OriT), chloramphenicol resistance genes, and multiple cloning site (MCS) were published as a part of pPL2 (18, 35). The figure was generated using SnapGene software (GSL Biotech).
TABLE 1.
Primers used in this study
| Primer no. | Primer name | Sequence (5′→3′) |
|---|---|---|
| 1 | pConst F SacI | AAAAGAGCTCAATTTTGCAAAAAGTT |
| 2 | pConst R PstI | TTTCTGCAGCACTCTCCTTCTACATTTTTTAAC |
| 3 | pheS* SOE B.S. F | ATATCAGGGCTTCGGATTCG GAATGGGTGT TGA |
| 4 | pheS* SOE B.S. R | CCATTCCGAATCCGAAGCCC TGATATTCCT TCG |
| 5 | KpnI Phes BS R | TTTTGGTACCTTACGCCTGTTTAAACTGC |
| 6 | PstI Phes BS F | AAAACTGCAGATGGAAGAAAAGCTAAAACAGC |
| 7 | ZBM pGEX3 | CCGGGAGCTGCATGTGTCAGAGG |
| 8 | pPL2 rev | ACTATAGGGCGAATTGGAG |
| 9 | pPL2 Gibson F | CTATCAGCTGTCCCTCCTGTTCAGCTACTGACGGGGTGGTCCGGGAGCTGCATGT |
| 10 | pPL2 Gibson R | CCGCTAGCGCTGATGTCCGGCGGTGCTTTTGCCGTTACGCGAGCTCAATTTTGCAAAAAGTTG |
| 11 | pLR4 Gibson F | GCGTAACGGCAAAAGCACCGC |
| 12 | pLR4 Gibson R | ACCACCCCGTCAGTAGCT |
| 13 | Throw F | ACCTTACGCCTGTTTAAACT |
| 14 | Throw R | CGTTTCGGTGATGACGG |
| 15 | Δmonocin_A_SalI | AAAAGTCGACTTAAGGCTTGGGCAAATTTTG |
| 16 | Δmonocin_B | TGCGGAAGAGAAGTTAATTTGGTTGGATATCTCTCCAG |
| 17 | Δmonocin_C | AGAGATATCCAACCAAATTAACTTCTCTTCCGCAAAAC |
| 18 | Δmonocin_D_PspOMI | TTTTGGGCCCTGTTCAACCATTTGGTAACATTC |
| 19 | ΔftbQ-R_A_PspOMI | AAAGGGCCCGGCTTTATGAGTTCTTTTGATAAACAA |
| 20 | ΔftbQ-R_B | CGCTCCTTTTTGGAAAACCTCCTTTTTAGTTAC |
| 21 | ΔftbQ-R_C | GGAGGTTTTCCAAAAAGGAGCGGCC |
| 22 | ΔftbQ-R_D_SalI | AAAGTCGACCTATTTGGCGGCGGC |
| 23 | T3 | ATTAACCCTCACTAAAGGGA |
| 24 | pLR16 rev | CCAATAACTTAAGGGTAACTAGCC |
Construction of in-frame deletion mutants using pLR16-pheS*.
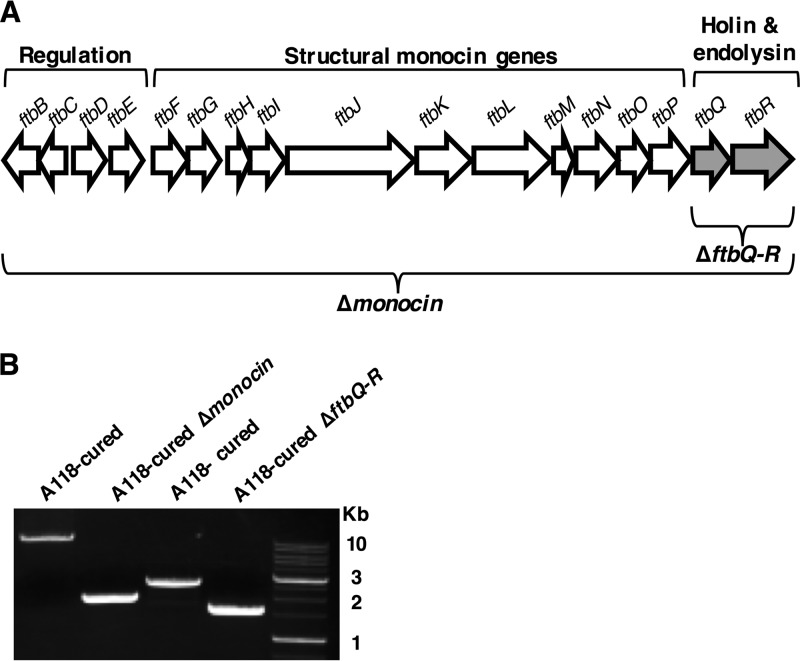
In order to validate the functionality of pLR16-pheS*, we constructed two in-frame deletion mutants of L. monocytogenes strain 10403S with mutations in the genomic region from LMRG_2362 to LMRG_2378. This region contains 17 genes of a phage origin that were shown, in other L. monocytogenes strains, to function as phage tail-like F-type bacteriocins, also named monocins (21, 22) (Fig. 3A). These monocins are produced upon induction of the SOS system, are released via bacterial lysis, and kill other Listeria strains (other than the producer strain), functioning as an altruistic mechanism to compete locally with neighboring bacteria. The first mutant we generated with mutations in this region had two genes deleted, LMRG_2377 and LMRG_2378 (1,132 bp, ΔftbQR mutant), which encode the monocin holin and endolysin, while the second had the complete monocin region, LMRG_2362 to LMRG_2378 deleted (10,727 bp, Δmonocin mutant). As part of our interest in the effects of phages and elements of phage origins on L. monocytogenes physiology and virulence (23, 24), we wanted to specifically assess the influence of the L. monocytogenes 10403S monocin on bacterial growth under SOS-inducing stress conditions. Since strain 10403S contains another prophage in its genome that is active, A118-like ϕ10403S, we constructed the deletion mutants on a 10403S strain with the A118 prophage cured (A118-cured strain). This allowed us to distinguish between effects caused by the monocin and the A118-like phage.
FIG 3.
Construction and validation of the Δmonocin and ΔftbQR mutants using pLR16-pheS*. (A) Schematic representation of the monocin gene region. The region is ~10.7 kbp long and consists of 17 genes (LMRG_2362 to LMRG_2378, here presented as ftbB to -R). Deletion of the whole region is designated Δmonocin, and deletion of the 1,132-bp long LMRG_2377 and LMRG_2378, encoding the holin and endolysin (marked in gray), is designated ΔftbQR. Annotation of gene groups is marked above the diagram; above the arrows, representing genes, are the gene names as designated by Lee et al. (21). (B) PCR verification of the deletion mutants. For the Δmonocin mutant, primers 15 and 18 (Table 1) were used, expecting ∼12,700-bp- and ∼2,000-bp-long products from A118 phage-cured L. monocytogenes and the Δmonocin mutant, respectively. For ΔftbQR, primers 19 and 22 (Table 1) were used, expecting ∼2,700-bp- and ∼1,600-bp-long products from A118-phage cured L. monocytogenes and ΔftbQR mutant, respectively. A 1-kb DNA ladder (GeneDireX) is shown on the right.
The deletions were carried out as specified in Materials and Methods. The pheS*-mediated selection for the second recombination event proved to be highly efficient, as about 1/100 of the bacteria grew on the selective p-Cl-phe plates, representing the typical plasmid loss rate of ∼1%. We verified the mutants using PCR and found DNA products of the expected size, indicating successful gene deletions (Fig. 3B).
The monocin gene region inhibits bacterial growth and causes lysis upon MC treatment.
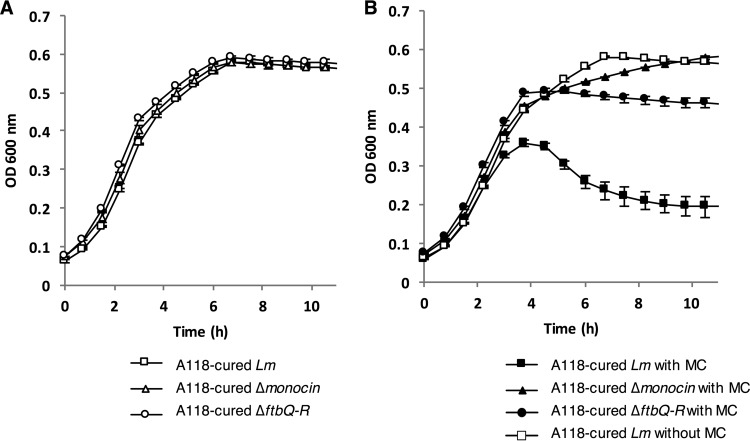
We next aimed to test whether the monocin of L. monocytogenes strain 10403S inhibits bacterial growth under SOS-inducing conditions and whether the monocin's holin and endolysin genes are involved. For induction of the SOS response we used mitomycin C (MC), a DNA-damaging agent that was shown before to activate monocin production (21). First, we tested the Δmonocin and ΔftbQR mutants' growth in rich brain heart infusion (BHI) medium at 30°C with no treatment and found them all to grow like the A118-cured parental strain (Fig. 4A). After treatment with MC and inspection of the optical density (OD) of the A118-cured strain, a sharp decrease was evident at 4 h posttreatment, representing bacterial lysis. To test whether this lysis is monocin mediated, we inspected the growth of the MC-treated Δmonocin mutant and found it to grow like untreated bacteria (A118-cured without MC treatment) with no detectable lysis. When inspecting the growth of the ΔftbQR mutant, we found no bacterial lysis, but at 4 h posttreatment growth was arrested, and the final OD was much lower than in untreated bacteria (Fig. 4B). Based on these observations, we concluded that the monocin region of L. monocytogenes strain 10403S is functional, imposes a significant fitness burden, and eventually causes bacterial lysis under conditions that induce severe DNA damage. Moreover, bacterial lysis is inflicted specifically by the holin and endolysin genes, carried at the end of the monocin gene region.
FIG 4.
Growth of L. monocytogenes with and without mitomycin C treatment. A118-cured L. monocytogenes (Lm) and Δmonocin mutant and ΔftbQR mutant bacteria were grown in BHI rich medium at 30°C without (A) and with (B) mitomycin C (MC) at 1.5 μg/ml. The experiment was performed in a 96-well format in a Synergy HT BioTek plate reader. Growth curves from one representative experiment are shown. Some error bars, representing the standard deviation for a triplicate sample, are hidden by the symbols.
DISCUSSION
In this work, we describe the construction of a novel and efficient suicide vector for allelic exchange in L. monocytogenes, pLR16-pheS*, employing the pheS*-mediated counterselection in this species. Prior to this system, many attempts to generate an efficient counterselection system in L. monocytogenes have been made by our laboratory and others, though they all failed for different reasons. Intrigued by the challenge, we cloned and tested multiple selection systems that are used in other bacteria, such as induction of streptomycin sensitivity (10), MazF toxin (25), and the upp marker (25), until we came across the PheS system, which worked.
Testing the induction of streptomycin sensitivity as an approach was appealing at the beginning, as strain 10403S is resistant to streptomycin and therefore no genomic manipulations were needed in order to apply the counterselection. We introduced an intact rpsL gene (conferring streptomycin sensitivity) from B. subtilis and from L. monocytogenes EGD-e on pPL2 plasmids into L. monocytogenes 10403S, but these clones did not show streptomycin sensitivity (data not shown). Nevertheless, even if this method had worked, it would be relevant only to Listeria strains naturally resistant to streptomycin and would therefore be a very limited tool. Another method considered was the utilization of the E. coli MazF toxin as a counterselection marker. This method is frequently applied in B. subtilis and necessitates the use of a highly expressing but tightly regulated inducible promoter. Unfortunately, there is no tight inducible system in Listeria that completely prevents leaky transcription. Leaky expression of the toxin was shown to provoke the rise of spontaneous MazF-resistant mutants, which dramatically decreases the efficiency of isolation of the desired mutants by generating “false-positive” counterselected colonies still harboring the inserted segment (25). Other methods, such as counterselection for upp, encoding uracil phosphoribosyltransferase that confers sensitivity to 5-fluorouracil, requires the deletion of upp in the parental strain, which may affect L. monocytogenes pathogenicity and skew the conclusions derived from the study of such mutants. The current system was a kind of last resort and was carefully adjusted by taking into account multiple considerations, some described below, that made it perfect for L. monocytogenes bacteria.
High expression of the B. subtilis PheS* and its functional similarity to the L. monocytogenes endogenous PheS protein are key for the effective restriction of bacterial growth. This is because the plasmid-encoded PheS* needs to compete with the native PheS for PheT-PheS complex formation (14, 26). Therefore, several promoters were tested, of which the pConst promoter demonstrated the best results. To assess possible problems arising from similarity and compatibility between PheS* and the bacteria, we initially mutated the endogenous L. monocytogenes pheS(A310G) to construct L. monocytogenes pheS* on the pPL2 plasmid, in addition to the B. subtilis pheS*. We then tested both constructs for growth restriction as described above. This L. monocytogenes-derived pheS* conveyed a slightly, yet not significantly, more severe growth restriction than B. subtilis pheS* (data not shown). This shows that even though these proteins are not identical (70% identity and 84% similarity), they have an extremely high functional resemblance (27). This is in contrast to the work published by Xie et al. in which they reported that they could not utilize pheS* markers from closely related bacteria in their Streptococcus mutans system (14). The authors managed to obtain effective counterselection only by using pheS* encoding the exact protein sequence as the pheS endogenous to their system, except for the mutated alanine. In order to avoid unwanted homologous recombination into the endogenous pheS during the first recombination event, the authors mutated a large section of pheS* to be riddled with silent mutations (14).
Other important parameters affecting pheS*-mediated toxicity are the potency of the substrate and the efficiency of the PheS*-mutated enzyme in using it. The potencies of p-Cl-phe from several manufacturers varied substantially; thus, p-Cl-phe concentration should be calibrated for maximal growth restriction when applying this system. It was recently reported by Miyazaki that mutating E. coli PheS T251A/A294G or T251S/A294G improves the pheS*-mediated toxicity (28). This threonine at position 251 is conserved in L. monocytogenes and B. subtilis, and an improved pheS* counterselection marker can be constructed and assessed, although we found ∼105 restriction to be satisfactory.
In the second recombination event there are two, theoretically equally probable, outcomes to plasmid loss: (i) reverting to wild type (WT) or (ii) manifesting the desired mutation. In our lab we used pLR16-pheS* to preform multiple gene deletions, in addition to the ones presented here, and many other genomic alterations, including insertions, substitutions, and point mutations. We found that roughly half of the colonies inspected after counterselection were WT and half were the desired mutants, agreeing with the theoretical probability. When all of the inspected bacteria are found to be parental revertants, one can conclude that the mutation attempted is toxic or highly unfavored. This may be concluded only when inspecting a sufficient number of colonies after the second recombination event. The efficiency of the counterselection on p-Cl-phe enables one to easily conclude toxicity and try to biologically solve the problem instead of technically repeating the procedure in vain.
We used pLR16-pheS* to investigate the monocin genomic region in L. monocytogenes strain 10403S by constructing two deletion mutations of the region. This genomic region was first mentioned in 1990 by Gohmann et al., where they identified a gene provoking an immune response in mice and named it lmaA (Listeria monocytogenes antigen A) (29). Later, lmaA and the three genes preceding it, lmaB, -C, and -D, were cloned. Furthermore, temperature-dependent differential expression and secretion of the proteins encoded by these genes were demonstrated (30). In a much more recent work, published in 2012, Hain et al. refer to the lma region in the EGD-e strain as a phage remnant and a monocin due to the fact that most of its open reading frames (ORFs) are related to phage genes, including a holin and endolysin module, while noticing that it lacks essential phage functions such as capsid structural genes (31). They show this region to be the only locus of phage origin conserved between Listeria lineages. This lma/monocin gene region/phage remnant locus is probably the conserved locus described to secrete bacteriolytic particles after UV induction in a work published in 1995 by Zink et al. (22). In that work, the authors characterized a holin/endolysin gene thought to be related to the monocin. Very recently, Lee at al. reported that this region from L. monocytogenes strain ATCC 35152 and Listeria innocua strain ATCC 33090 functions as an F-type-bacteriocin (21). The authors reported that this region, highly similar to the region in 10403S, produces phage tail-like bacteriocins with different killing spectra depending on the strain of origin. Furthermore, they cloned the complete monocin gene region from L. monocytogenes into B. subtilis, expressed it, and artificially altered its killing spectrum by means of genetic engineering. Due to the functionality of the region, the authors designated its genes ftbA to -R for F-type bacteriocins (Fig. 3).
So far, the monocin region of L. monocytogenes 10403S has not been studied. In our laboratory we study another phage of this strain, A118-like ϕ10403S, which resides as a prophage within the comK gene, affecting bacterial virulence (23). Studying this phage using in vitro systems, for example, inducing its lytic production by treatment with mitomycin C or UV irradiation, we came across the notion that we might induce the production of the monocin as well. To test whether the monocin region is active, meaning that it can lead to bacterial lysis upon these treatments, we generated two mutants with the complete monocin region and the holin and endolysin genes deleted in the background of A118-cured strain. The data clearly show that the monocin region is activated upon mitomycin C treatment and leads to robust bacterial lysis, which is dependent on the holin and endolysin genes. This is a demonstration of the activity of this region in strain 10403S, which should be taken into account in our study and others, especially those that are using 10403S as a vaccine platform (32).
Finally, the pheS* counterselection marker in L. monocytogenes is a potent genetic tool, as it can be applied in various genetic systems for different purposes, for example, systematic deletion and insertion of genes, generation of point mutations, curing of prophages, and designing selection systems for genetic screening. To the best of our knowledge, the pheS* counterselection system has not been used in B. subtilis or published. It is likely that such a system, utilizing the L. monocytogenes pheS* as constructed here, will work in B. subtilis and contribute to research in that important model organism as well (25). pLR16-pheS* itself is a highly efficient genome editing tool for L. monocytogenes, and we expect it to work for all Listeria strains and species. We anticipate that it will be extensively used by the research community and yield novel insights into the diverse fields studied using this model organism and perhaps other Gram-positive bacteria as well.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
L. monocytogenes strain 10403S was used as a WT strain, and L. monocytogenes 10403S cured of A118-like phage (A118-cured strain) (23) was used as the parental strain for all mutants generated in this work (Table 2). E. coli XL-1 Blue (Stratagene) was used for vector propagation. E. coli SM-10 was used for conjugative plasmid delivery to L. monocytogenes bacteria (33). L. monocytogenes strains were grown in brain heart infusion (BHI) (Merck) medium at 37°C or 30°C as specified, and E. coli strains were grown in Luria-Bertani (LB) (Acumedia) medium at 37°C. p-Chloro-phenylalanine (p-Cl-phe) (Acros Organics) was added to the BHI agar before autoclaving to help dissolve it. Immediately after autoclaving, the medium was vigorously stirred, cooled down to ∼55°C, and poured to plates. Antibiotics were used as specified: chloramphenicol (Cm), 10 μg/ml; mitomycin C (MC) (Sigma), 1.5 μg/ml; and streptomycin (Strep), 100 μg/ml. All restriction enzymes were purchased from New England BioLabs. We used Phusion polymerase for all cloning purposes and Taq polymerase for PCR ligation verifications of the different plasmids described in this paper.
TABLE 2.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Genotype | Reference or source |
|---|---|---|
| Escherichia coli strains | ||
| XL-1 Blue | recA1 endA1 gyrA96 thi-1 hsdR17 supE44 relA1 lac [F′ proAB lacIqZΔM15 Tn10 (Tetr)] | Stratagene |
| SM-10 | Conjugation donor; F− thi-1 thr-1 leuB6 recA tonA21 lacY1 supE44 (Muc+) λ− [RP4-2(Tc::Mu)] Kmr Tra+ | 33 |
| Listeria monocytogenes strains | ||
| 10403S | WT, Strepr | D. A. Portnoy, lab stock |
| A118 cured | 10403S ΔcomK phage | 23 |
| Δmonocin mutant | 10403S ΔcomK phage ΔLMRG_2362-2337 | This study |
| ΔftbQR mutant | 10403S ΔcomK phage ΔLMRG_2377-2378 | This study |
| WT + pPL2 pConst pheS* | 10403S harboring pPL2 integrative plasmid expressing a mutated B. subtilis pheS, p-Cl-pheS, Cmr | This study |
| WT + pPL2 pConst GFP | 10403S harboring pPL2 integrative plasmid expressing GFP | This study |
| Plasmids | ||
| pPL2 | E. coli/L. monocytogenes conjugative vector, integrative to tRNA-Arg gene, Cmr | 18 |
| pBHE261 | E. coli/L. monocytogenes conjugative vector, pKSV7 temp-sensitive Ori, Amp−r, Cm+r | 35 |
| pLR16-pheS* | E. coli/L. monocytogenes conjugative vector, pKSV7 temp-sensitive Ori, p-Cl-pheS, Cmr | This study |
| pLR16Δmonocin | pLR16 with upstream and downstream homology fragments inserted | This study |
| pLR16ΔftbQ-R | pLR16 with upstream and downstream homology fragments inserted | This study |
aAmpr, ampicillin resistance; Kmr, kanamycin resistance; Cmr, chloramphenicol resistance; p-Cl-Phes, p-chloro-phenylalanine sensitivity. −, Gram-negative resistance; +, Gram-positive resistance.
Construction of pheS* counterselection marker.
The pheS gene was amplified from the B. subtilis 168 genome and mutated by overlap extension PCR using primers 3 and 5 and primers 4 and 6. The PCR products were then used as templates for pheS* amplification using primers 5 and 6 (34) (Table 1). The mutated pheS* and pPL2 integrative plasmid (18) were digested using KpnI and PstI restriction enzymes, ligated using T4 ligase, transformed into E. coli XL-1 Blue competent bacteria, and selected on Cm. This pPL2-pheS* plasmid was digested using PstI and SacI restriction enzymes. pConst was amplified with primers 1 and 2, digested with the same enzymes, ligated upstream of pheS*, and transformed into XL-1 Blue. This pPL2-pConst-pheS* was transformed into E. coli SM-10 and conjugated into L. monocytogenes 10403S (33). All ligation products were validated by PCR and sequencing.
Construction of pLR16-pheS* counterselection vector.
An pKSV7oriT derivative vector (pBHE261), a kind gift from Peter Lauer (Aduro Biotech), and pPL2 were used as templates for pLR16-pheS* construction (18, 35). pPL2 was digested with XbaI, and digestion products were separated using a 1% agarose gel. The linearized 2,063-bp fragment containing the P15A Ori and the cat gene conveying Gram-negative Cm resistance was purified, circularized using T4 ligase, and transformed into XL-1 Blue cells to generate pLR1. The multiple cloning site (MCS) of pPL2 was amplified using primers 7 and 8 (Table 1), digested with HhaI and SacI, and ligated to pLR1 digested with AleI and SacI. The ligation product was transformed into XL-1 Blue to generate the 2,262-bp pLR2. pBHE261 and pLR2 were digested with AleI and SacI, and digestion products were separated using a 1% agarose gel. The 4,172 and 2,252-bp fragments, originating from pBHE261 and pLR2, respectively, were purified, ligated and transformed into XL-1 Blue to generate the 6,424-bp pLR4. The 4,172-bp fragment originating from pBHE261 contained the pKSV7 temperature-sensitive Ori+ for Gram-positive bacteria, the origin of transfer (OriT), a replication gene (rep), a gene necessary for conjugative mobilization (mob), and a gene conveying Cm resistance to Gram-positive bacteria (Cat+). pLR4 was linearized using PCR and primers 11 and 12, and the product was separated using a 1% agarose gel and purified. pConst-pheS* was amplified from pPL2-pConst-pheS* constructed in this study using primers 9 and 10 (Table 1), Gibson assembled with the linearized pLR4, and transformed into XL-1 Blue to generate the 7,812-bp pLR15-pheS* (36). pLR15-pheS* had a 109-bp region originating from pPL2 repeating twice. This region was deleted from the plasmid by linearizing it without the region by PCR using primers 13 and 14, circularizing using T4 ligase, and transforming into XL-1 Blue to generate the final 7,703-bp pLR16-pheS* plasmid (see Fig. S2 in the supplemental material). The final pLR16-pheS* was fully validated by sequencing.
Construction of deletion mutants using pLR16-pheS*.
In-frame deletion mutations of LMRG_2362 to LMRG_2378 (17 genes of the entire monocin region) (10,727 bp, Δmonocin mutant) and of LMRG_2377 and LMRG_2378, encoding the monocin holin and endolysin (1,132 bp, ΔftbQR mutant), were constructed using overlap extension PCR and primers 15 to 22 (Table 1) (34). Approximately 1,000-bp (Δmonocin mutant) or 800-bp (ΔftbQR mutant) fragments upstream and downstream from the deleted regions were separately amplified by PCR using primers A and B (i.e., those shown in Table 1 with _A and _B, respectively) and primers C and D (i.e., those shown in Table 1 with _C and _D, respectively). The products were separated using a 1% agarose gel, purified, and used as templates for the overlap extension PCR using primers A and D, generating fragments of ∼2,000 bp (Δmonocin mutant) or ∼1,600 bp (ΔftbQR mutant). The PCR products were separated using a 1% agarose gel. The purified product and pLR16-pheS* plasmid were digested with PspOMI and SalI, ligated using T4 ligase, and transformed into XL-1 Blue cells. Ligation products were first verified by PCR using primers 23 and 24 (Table 1) and then sequenced. Plasmids were transformed into SM-10 cells and then transferred to the L. monocytogenes 10403S A118-cured strain by conjugation by coincubation on BHI agar plates. Transconjugants were selected by incubation on BHI agar plates containing Strep and Cm at 37°C for 24 h. (It is important to note that if one wishes to use pLR16-pheS* to mutate a Strep-sensitive Listeria species or strain, cephamycin or Listeria selective agar should be applied instead of Strep.) Two L. monocytogenes transconjugants were inoculated into BHI broth supplemented with Cm and grown at 41°C with agitation overnight. Bacteria from this culture were spread on a BHI agar plate containing Cm at 41°C until large colonies were formed. These colonies represent bacteria with the plasmid fully integrated into their chromosomes, that is, after the first recombination event. One colony from each strain was inoculated in BHI broth and cultured at 30°C overnight, resulting in a mixed culture where typically about 1% of bacteria have lost the plasmid through the second recombination event. The culture was diluted 10−4, and 100 μl was plated on BHI agar supplemented with 18 mM p-Cl-phe and incubated at 37°C overnight. Single colonies that grew on the counterselection plates (without counterselection, there is a bacterial lawn) were PCR verified to contain the desired mutation using primers A and D. PCR-verified colonies were validated to be Cm sensitive and designated the Δmonocin and ΔftbQR mutants.
Growth curves.
Bacteria were grown overnight at 37°C with agitation in BHI broth, and then the culture was diluted 1:10 in BHI, incubated without agitation at 30°C to reach an OD at 600 nm (OD600) of ∼0.4, diluted to an OD600 of 0.15, and pipetted in triplicates into a 96-well plate with or without MC (1.5 μg/ml). The plates were incubated at 30°C using a Synergy HT BioTek plate reader, and the OD600 was measured every 15 min following 2 min of shaking.
Accession number(s).
The suicide-counterselection vector pLR16-pheS* was fully sequenced, and the sequence was submitted to GenBank under accession number KY286114.
Supplementary Material
ACKNOWLEDGMENTS
We thank Ilya Borovok for his helpful insights during this study and the Herskovits group members for testing the pheS* plasmid in their projects.
This work was funded by the European Research Council FP7 program, ERC starting grant for A.A.H. (PathoPhageHost).
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AEM.02927-16.
REFERENCES
- 1.Freitag NE, Port GC, Miner MD. 2009. Listeria monocytogenes—from saprophyte to intracellular pathogen. Nat Rev Microbiol 7:623–628. doi: 10.1038/nrmicro2171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hamon M, Bierne H, Cossart P. 2006. Listeria monocytogenes: a multifaceted model. Nat Rev Microbiol 4:423–434. doi: 10.1038/nrmicro1413. [DOI] [PubMed] [Google Scholar]
- 3.Drevets DA, Bronze MS. 2008. Listeria monocytogenes: epidemiology, human disease, and mechanisms of brain invasion. FEMS Immunol Med Microbiol 53:151–165. doi: 10.1111/j.1574-695X.2008.00404.x. [DOI] [PubMed] [Google Scholar]
- 4.Liu D. 2008. Handbook of Listeria monocytogenes. CRC Press, Boca Raton, FL. [Google Scholar]
- 5.Rychli K, Guinane CM, Daly K, Hill C, Cotter PD. 2014. Generation of nonpolar deletion mutants in Listeria monocytogenes using the “SOEing” method. Methods Mol Biol 1157:187–200. doi: 10.1007/978-1-4939-0703-8_16. [DOI] [PubMed] [Google Scholar]
- 6.Arnaud M, Chastanet A, Débarbouillé M. 2004. New vector for efficient allelic replacement in naturally nontransformable, low-GC-content, Gram-positive bacteria. Appl Environ Microbiol 70:6887–6891. doi: 10.1128/AEM.70.11.6887-6891.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Abdelhamed H, Lawrence ML, Karsi A. 2015. A novel suicide plasmid for efficient gene mutation in Listeria monocytogenes. Plasmid 81:1–8. doi: 10.1016/j.plasmid.2015.05.003. [DOI] [PubMed] [Google Scholar]
- 8.Behari J, Youngman P. 1998. Regulation of hly expression in Listeria monocytogenes by carbon sources and pH occurs through separate mechanisms mediated by PrfA. Infect Immun 66:3635–3642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li G, Kathariou S. 2003. An improved cloning vector for construction of gene replacements in Listeria monocytogenes. Appl Environ Microbiol 69:3020–3023. doi: 10.1128/AEM.69.5.3020-3023.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Reyrat J-M, Pelicic V, Gicquel B, Rappuoli R. 1998. Counterselectable markers: untapped tools for bacterial genetics and pathogenesis. Infect Immun 66:4011–4017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kast P, Hennecke H. 1991. Amino acid substrate specificity of Escherichia coli phenylalanyl-tRNA synthetase altered by distinct mutations. J Mol Biol 222:99–124. doi: 10.1016/0022-2836(91)90740-W. [DOI] [PubMed] [Google Scholar]
- 12.Kast P. 1994. pKSS—a second-generation general purpose cloning vector for efficient positive selection of recombinant clones. Gene 138:109–114. doi: 10.1016/0378-1119(94)90790-0. [DOI] [PubMed] [Google Scholar]
- 13.Kristich CJ, Chandler JR, Dunny GM. 2007. Development of a host-genotype-independent counterselectable marker and a high-frequency conjugative delivery system and their use in genetic analysis of Enterococcus faecalis. Plasmid 57:131–144. doi: 10.1016/j.plasmid.2006.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Xie Z, Okinaga T, Qi F, Zhang Z, Merritt J. 2011. Cloning-independent and counterselectable markerless mutagenesis system in Streptococcus mutans. Appl Environ Microbiol 77:8025–8033. doi: 10.1128/AEM.06362-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Carr JF, Danziger ME, Huang AL, Dahlberg AE, Gregory ST. 2015. Engineering the genome of Thermus thermophilus using a counterselectable marker. J Bacteriol 197:1135–1144. doi: 10.1128/JB.02384-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sievers F, Wilm A, Dineen D, Gibson TJ, Karplus K, Li W, Lopez R, McWilliam H, Remmert M, Söding J, Thompson JD, Higgins DG. 2011. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol Syst Biol 7:539. doi: 10.1038/msb.2011.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Edgar RC. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lauer P, Chow MYN, Loessner MJ, Portnoy DA, Calendar R. 2002. Construction, characterization, and use of two Listeria monocytogenes site-specific phage integration vectors. J Bacteriol 184:4177–4186. doi: 10.1128/JB.184.15.4177-4186.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shen A, Higgins DE. 2005. The 5′ untranslated region-mediated enhancement of intracellular listeriolysin O production is required for Listeria monocytogenes pathogenicity. Mol Microbiol 57:1460–1473. doi: 10.1111/j.1365-2958.2005.04780.x. [DOI] [PubMed] [Google Scholar]
- 20.Smith K, Youngman P. 1992. Use of a new integrational vector to investigate compartment-specific expression of the Bacillus subtilis spoIIM gene. Biochimie 74:705–711. doi: 10.1016/0300-9084(92)90143-3. [DOI] [PubMed] [Google Scholar]
- 21.Lee G, Chakraborty U, Gebhart D, Govoni GR, Zhou ZH, Scholl D. 2016. F-type bacteriocins of Listeria: a new class of phage tail-like structures reveals broad parallel co-evolution between tailed bacteriophages and high molecular weight bacteriocins. J Bacteriol 198:2784–2793. doi: 10.1128/JB.00489-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zink R, Loessner MJ, Scherer S. 1995. Charaterization of cryptic prophages (monocins) in Listeria and sequence analysis of a holin/endolysin gene. Microbiology 141:2577–2584. doi: 10.1099/13500872-141-10-2577. [DOI] [PubMed] [Google Scholar]
- 23.Rabinovich L, Sigal N, Borovok I, Nir-Paz R, Herskovits AA. 2012. Prophage excision activates Listeria competence genes that promote phagosomal escape and virulence. Cell 150:792–802. doi: 10.1016/j.cell.2012.06.036. [DOI] [PubMed] [Google Scholar]
- 24.Feiner R, Argov T, Rabinovich L, Sigal N, Borovok I, Herskovits AA. 2015. A new perspective on lysogeny: prophages as active regulatory switches of bacteria. Nat Rev Microbiol 13:641–650. doi: 10.1038/nrmicro3527. [DOI] [PubMed] [Google Scholar]
- 25.Dong H, Zhang D. 2014. Current development in genetic engineering strategies of Bacillus species. Microb Cell Fact 13:63. doi: 10.1186/1475-2859-13-63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mermershtain I, Finarov I, Klipcan L, Kessler N, Rozenberg H, Safro MG. 2011. Idiosyncrasy and identity in the prokaryotic phe-system: crystal structure of E. coli phenylalanyl-tRNA synthetase complexed with phenylalanine and AMP. Protein Sci 20:160–167. doi: 10.1002/pro.549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Miyazaki K. 2015. Molecular engineering of a PheS counterselection marker for improved operating efficiency in Escherichia coli. Biotechniques 58:86–88. doi: 10.2144/000114257. [DOI] [PubMed] [Google Scholar]
- 29.Göhmannn S, Wächter ML, Schiltz E, Goebel W, Chakraborty T. 1990. Characterization of a Listeria monocytogenes-specific protein capable of inducing delayed hypersensitivity in Listeria-immune mice. Mol Microbiol 4:1091–1099. doi: 10.1111/j.1365-2958.1990.tb00683.x. [DOI] [PubMed] [Google Scholar]
- 30.Schäferkordt S, Chakraborty T. 1997. Identification, cloning, and characterization of the Ima operon, whose gene products are unique to Listeria monocytogenes. J Bacteriol 179:2707–2716. doi: 10.1128/jb.179.8.2707-2716.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hain T, Ghai R, Billion A, Kuenne CT, Steinweg C, Izar B, Mohamed W, Mraheil MA, Domann E, Schaffrath S, Kärst U, Goesmann A, Oehm S, Pühler A, Merkl R, Vorwerk S, Glaser P, Garrido P, Rusniok C, Buchrieser C, Goebel W, Chakraborty T. 2012. Comparative genomics and transcriptomics of lineages I, II, and III strains of Listeria monocytogenes. BMC Genomics 13:144. doi: 10.1186/1471-2164-13-144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Brockstedt DG, Bahjat KS, Giedlin MA, Liu W, Leong M, Luckett W, Gao Y, Schnupf P, Kapadia D, Castro G, Lim JYH, Sampson-Johannes A, Herskovits AA, Stassinopoulos A, Bouwer HGA, Hearst JE, Portnoy DA, Cook DN, Dubensky TW. 2005. Killed but metabolically active microbes: a new vaccine paradigm for eliciting effector T-cell responses and protective immunity. Nat Med 11:853–860. doi: 10.1038/nm1276. [DOI] [PubMed] [Google Scholar]
- 33.Simon R, Priefer U, Pühler A. 1983. A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in Gram negative bacteria. Nat Biotechnol 1:784–791. doi: 10.1038/nbt1183-784. [DOI] [Google Scholar]
- 34.Horton RM, Cai Z, Ho SM, Pease LR. 1990. Gene splicing by overlap extension: tailor-made genes using the polymerase chain reaction. Biotechniques 8:528–535. [PubMed] [Google Scholar]
- 35.Lauer P, Hanson B, Lemmens EE, Liu W, Luckett WS, Leong ML, Allen HE, Skoble J, Bahjat KS, Freitag NE, Brockstedt DG, Dubensky TW. 2008. Constitutive activation of the PrfA regulon enhances the potency of vaccines based on live-attenuated and killed but metabolically active Listeria monocytogenes strains. Infect Immun 76:3742–3753. doi: 10.1128/IAI.00390-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gibson DG, Young L, Chuang R-Y, Venter JC, Hutchison CA, Smith HO. 2009. Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat Methods 6:343–345. doi: 10.1038/nmeth.1318. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.