ABSTRACT
The genetic diversity of bacterial populations nodulating Lupinus micranthus in five geographical sites from northern Tunisia was examined. Phylogenetic analyses of 50 isolates based on partial sequences of recA and gyrB grouped strains into seven clusters, five of which belong to the genus Bradyrhizobium (28 isolates), one to Phyllobacterium (2 isolates), and one, remarkably, to Microvirga (20 isolates). The largest Bradyrhizobium cluster (17 isolates) grouped with the B. lupini species, and the other five clusters were close to different recently defined Bradyrhizobium species. Isolates close to Microvirga were obtained from nodules of plants from four of the five sites sampled. We carried out an in-depth phylogenetic study with representatives of the seven clusters using sequences from housekeeping genes (rrs, recA, glnII, gyrB, and dnaK) and obtained consistent results. A phylogeny based on the sequence of the symbiotic gene nodC identified four groups, three formed by Bradyrhizobium isolates and one by the Microvirga and Phyllobacterium isolates. Symbiotic behaviors of the representative strains were tested, and some congruence between symbiovars and symbiotic performance was observed. These data indicate a remarkable diversity of L. micranthus root nodule symbionts in northern Tunisia, including strains from the Bradyrhizobiaceae, Methylobacteriaceae, and Phyllobacteriaceae families, in contrast with those of the rhizobial populations nodulating lupines in the Old World, including L. micranthus from other Mediterranean areas, which are nodulated mostly by Bradyrhizobium strains.
IMPORTANCE Lupinus micranthus is a legume broadly distributed in the Mediterranean region and plays an important role in soil fertility and vegetation coverage by fixing nitrogen and solubilizing phosphate in semiarid areas. Direct sowing to extend the distribution of this indigenous legume can contribute to the prevention of soil erosion in pre-Saharan lands of Tunisia. However, rhizobial populations associated with L. micranthus are poorly understood. In this context, the diversity of endosymbionts of this legume was investigated. Most Lupinus species are nodulated by Bradyrhizobium strains. This work showed that about half of the isolates from northern Tunisian soils were in fact Bradyrhizobium symbionts, but the other half were found unexpectedly to be bacteria within the genera Microvirga and Phyllobacterium. These unusual endosymbionts may have a great ecological relevance. Inoculation with the appropriate selected symbiotic bacterial partners will increase L. micranthus survival with consequent advantages for the environment in semiarid areas of Tunisia.
KEYWORDS: Bradyrhizobium, Lupinus micranthus, Microvirga, Phyllobacterium, nodulation
INTRODUCTION
Lupinus is the largest genus in the tribe Genisteae within the Fabaceae family, with around 275 species (1). Lupines are widely distributed in the Old and New Worlds, but only 15 species have been identified in the Mediterranean region (2–4). A few species from this genus have been cultivated in rotations with cereals for more than 2,000 years (5).
The endosymbiotic bacteria of cultivated lupines L. albus, L. angustifolius, and L. luteus belong mostly to Bradyrhizobium lineages (6–9). However, endosymbionts from most wild lupines are unknown or scarcely studied; among them are the Bradyrhizobium strains isolated from L. albescens native to Brazil (10) and strains from L. mariae-josephae from eastern Spain (11, 12). Also, Ochrobactrum and Microvirga strains have been isolated from L. honoratus in Argentina (13) and L. texensis in Texas (USA), respectively (14, 15).
L. micranthus is a wild lupine spread widely around the Mediterranean. Symbiotic and phylogenetic analyses of isolates from nodules of field-grown plants in Algeria and Spain recently showed that they all belong to the Bradyrhizobium genus (16). There are no reports addressing rhizobia nodulating L. micranthus in Tunisia, where four other Lupinus species have been described, namely L. albus, L. angustifolius, L. luteus, and L. cosentinii (17). This work shows an unexpected biodiversity among rhizobial isolates from L. micranthus in North Tunisia, including isolates belonging to Microvirga and Phyllobacterium genera.
The Microvirga genus comprises mainly soil bacteria, and currently, 13 species have been described (18–24). Of these species, only 4 are legume symbionts and were designated M. lotononidis and M. zambiensis, both isolated from Listia angolensis, M. vignae, nodulating Vigna unguiculata (21), and the already mentioned M. lupini, isolated from L. texensis (15).
The Phyllobacterium genus was originally described by D. H. Knösel (25) for bacteria isolated from leaf nodules of tropical ornamental plants. Thus far, this genus contains 10 species, six of which were isolated from root nodules of legumes and are designated as follows: Phyllobacterium sophorae, isolated from Sophora flavescens (26); P. trifolii, isolated from Trifolium pratense (27); P. leguminum, isolated from Argyrolobium uniflorum and Astragalus algerianus (28); P. loti, isolated from Lotus corniculatus (29); P. endophyticum, isolated from Phaseolus vulgaris (30); and P. ifriqiyense, isolated from Astragalus algerianus and Lathyrus numidicus (28). However, the capacities to induce effective nodulation on the original host plant were only demonstrated for P. trifolii and P. sophorae (26, 27). This diversity in symbiosis suggests that legume roots are the preferred habitats for species of the genus Phyllobacterium, more so than leaf nodules (31).
RESULTS
Isolation and diversity of L. micranthus-nodulating bacteria in northern Tunisia.
A total of 50 rhizobial isolates were obtained from root nodules collected in April 2015 from wild Lupinus micranthus growing in five different areas in North Tunisia (Table 1 and Fig. 1). These isolates nodulated and fixed nitrogen with L. micranthus plants inoculated under bacteriologically controlled conditions. Inoculated plants were dark green, clearly different from the yellowish, smaller noninoculated control plants, and produced 3 to 10 red nodules per plant at 30 days postinoculation. When the isolates were examined in free-living conditions, different phenotypic characteristics were observed on yeast extract mannitol agar (YMA) plates. Most of the isolates (i.e., 32) produced circular, convex, smooth, and mucilaginous colonies 2 to 3 mm in diameter at 28°C; half of them appeared after 3 to 4 days postinoculation and the other half after 6 to 7 days. Six isolates had extremely low growth rates and required more than 7 days to generate nonmucoid colonies.
TABLE 1.
Designations and geographical origins of L. micranthus endosymbionts in northern Tunisia
| Sites | Coordinates | Altitudea (m) | Soil pH | No. of isolates | Designation | Environment | No. of isolates (group)b |
|---|---|---|---|---|---|---|---|
| Borj Hfaiedh | 36°28′50.03″N 10°35′15.30″E | 106 | 8.5 | 11 | LmiB | Area with olive trees | 3 (II), 2 (V), 6 (VII) |
| El Alia | 37°11′23.26″N 10°00′19.83″E | 47 | 8.3 | 4 | LmiE | Area next to greenhouses | 4 (VII) |
| Hammamet | 36°23′38.98″N 10°36′57.47″E | 4 | 9.0 | 9 | LmiH | Semiurbanized area, next to the sea | 3 (IV), 6 (VII) |
| Mraissa | 36°45′13.00″N 10°33′14.20″E | 10 | 8.0 | 10 | LmiM | Area planted with cereals | 3 (I), 3 (III), 4 (VII) |
| Takelsa | 36°45′43.17″N 10°34′53.73″E | 51 | 7.3 | 16 | LmiT | Area planted with orange trees | 14 (I), 2 (VI) |
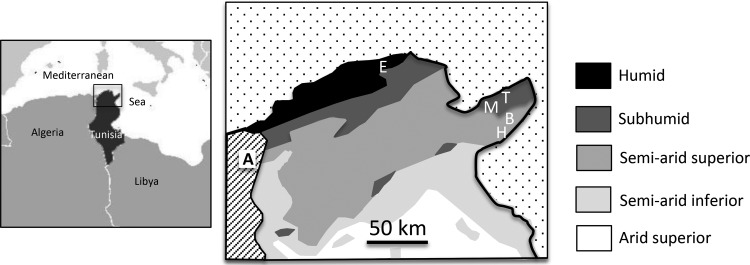
FIG 1.
Sites of plant sampling in northern Tunisia. Bioclimatic areas are indicated on the right. A, Algeria; E, El Alia; M, Mraissa; T, Takelsa; B, Borj Hfaiedh; H, Hammamet. Template maps were obtained from Wikipedia (left; https://upload.wikimedia.org/wikipedia/commons/a/af/Tunisia_Locator.png) and Tunisia's National Research Institute of Water, Forests and Rural Engineering (right; http://www.inrgref.agrinet.tn/an/?p=19).
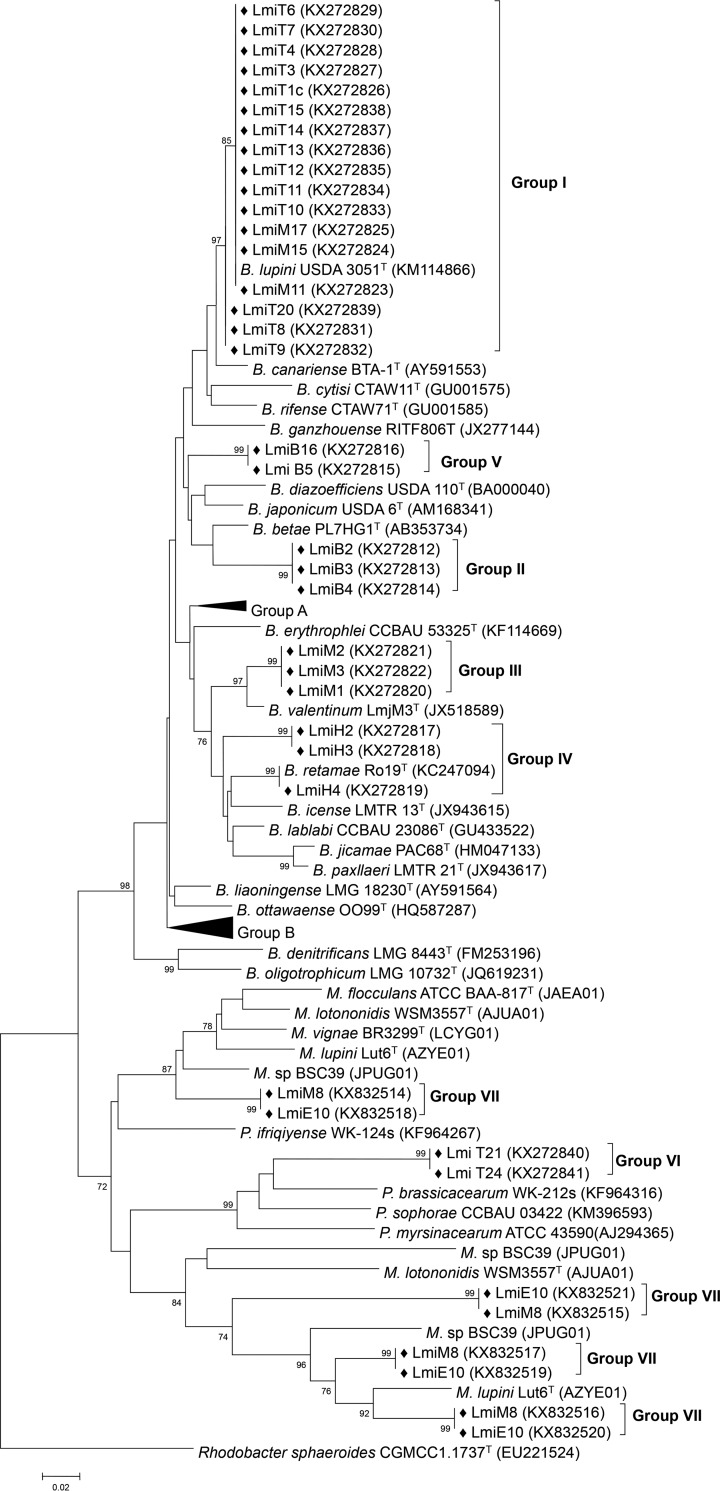
To evaluate the diversity of the isolates, an internal sequence of recA was obtained. Previous work in our group showed that the analysis of recA sequences is a very resolutive tool for estimating the similarities among rhizobial strains and predicting their lineages and genera (16, 33, 34). An analysis of recA sequences using BLAST enabled the clustering of the 50 L. micranthus-associated isolates into 7 groups (Fig. 2). Sequences from isolates in five groups (groups I, II, III, IV, and V) matched those of the genus Bradyrhizobium and were similar to B. lupini (17 isolates), B. betae (3 isolates), B. valentinum (3 isolates), B. retamae (3 isolates), and B. diazoefficiens (2 isolates) type strains, respectively. The 2 isolates in group VI matched type strains of species from the genus Phyllobacterium, and the last group (group VII, Fig. 2) contained 20 isolates that appeared similar to one another. Sanger chromatograms for recA sequences of these isolates displayed sequence ambiguities (up to 14 in 533 nucleotides [nt]). Preliminary draft genome sequences of two of these isolates (L. micranthus LmiM8 and LmiE10) suggested the existence of four nonidentical recA copies that might explain these results (Fig. 2). The location of group VII recA sequences in the recA phylogenetic tree predicts their membership in the Microvirga genus. The presence of several copies of recA in isolates LmiM8 and LmiE10 is in line with what has been described for M. lupini (two copies), M. lotononidis (two copies), and M. sp. BSC39 (three copies) (35–37). Therefore, a different housekeeping gene marker, gyrB, commonly used with Microvirga species (15, 21, 22), was selected for amplicon characterization. Sequencing of gyrB amplicons confirmed the affiliation of the 20 isolates in group VII to the Microvirga genus (Fig. 3).
FIG 2.
Neighbor-joining phylogenetic tree of Tunisian L. micranthus-associated (Lmi) isolates and reference strains based on recA (375 bp) sequences. Bootstrap values were calculated for 1,000 replications, and those greater than 70% are indicated at the internodes. Black diamonds (◆) indicate Tunisian Lmi isolates. Collapsed groups include group A: B. elkanii USDA76T, B. ferriligni CCBAU 51502T, B. pachyrhizi PAC48T, B. viridifuturi SEMIA 690T, B. embrapense CNPSo 2833T, and B. tropiciagri CNPSo 1112T; and group B: B. ingae BR 10250T, B. iriomotense EK05T, B. kavangense 14-3T, B. manausense BR 3351T, B. subterraneum 58 2-1T, B. yuanmingense CCBAU 10071T, B. vignae 7-2T, B. daqingense CCBAU 15774T, and B. huanghuaihaiense CCBAU 23303T. Group VII includes only two strains (LmiM8 and LmiE10) that contain 4 copies of recA (see the text for details). B., Bradyrhizobium; M., Microvirga; P., Phyllobacterium. Accession numbers from GenBank and whole-genome sequencing (NCBI) databases are shown in parentheses.
FIG 3.
Neighbor-joining phylogenetic tree of Tunisian L. micranthus-associated (Lmi) isolates and reference strains based on gyrB (623 bp) sequences. Bootstrap values were calculated for 1,000 replications, and those greater than 60% are indicated at the internodes. Tunisian Lmi isolates with identical gyrB sequences were collapsed in group VII: LmiM8, LmiE10, LmiH9, LmiE9, LmiH1, LmiB17, LmiB20, LmiB18, LmiB6, LmiB21, LmiB24, LmiE5, LmiE6, LmiH5, LmiH6, LmiH8, LmiH10, LmiM7, LmiM9, and LmiM10 (accession numbers KX784068 to KX784087). Other accession numbers from the GenBank database are shown in parentheses. B., Bradyrhizobium; M., Microvirga; Me., Methylobacterium.
The isolates included in Microvirga and Phyllobacterium genera had mean generation times of approximately 5 h in tryptone yeast (TY) medium in contrast with Bradyrhizobium isolates, which had a mean generation time of more than 9 h (Table 2). Microvirga colonies were pink in color after 8 days in YMA, as has been observed for other Microvirga strains, such as M. subterranea (20), M. lupini (15), and M. makkahensis (22). The growth rates of bradyrhizobial isolates from groups III and IV were similar to those of B. valentinum and B. retamae, respectively, with an extra-low growth rate phenotype on YMA plates.
TABLE 2.
Symbiotic characteristics and generation times of Tunisian Lupinus micranthus representative isolates
| Lmi isolate | Symbiovara | Groupb | Generation time (h)d | Nodulation phenotype with hostc: |
||||
|---|---|---|---|---|---|---|---|---|
| Lmi | Lan | Lmj | Vun | Mat | ||||
| T21 | A | VI | 5.5 | + | + | − | W | + |
| E10 | A | VII | 4.8 | + | + | W | W | + |
| M8 | A | VII | 4.6 | + | + | W | W | + |
| H4 | B | IV | 14.4 | + | + | + | + | + |
| M2 | B | III | 22.3 | + | W | + | + | + |
| B3 | C | II | 9.4 | + | − | − | + | + |
| T14 | genistearum | I | 13.1 | + | W | W | W | + |
| T3 | genistearum | I | 11.5 | + | + | W | W | + |
| B5 | genistearum | V | 16.4 | + | + | − | + | + |
As shown in Fig. 4.
Groups (I to VII) are defined in Fig. 2, 3, and 4. Phylogenetic similarities: group I, B. lupini; group II, B. betae; group III, B. valentinum; group IV, B. retamae; group V, B. diazoefficiens/B. rifense; group VI, Phyllobacterium sp.; and group VII, Microvirga sp.
Nodulation evaluated by color of nodules: +, red; W, white; −, no nodules. In all cases, plants with white nodules were smaller than plants with red nodules. Lmi, L. micranthus; Lan, L. angustifolius; Lmj, L. mariae-josephae; Vun, Vigna unguiculata; Mat, Macroptilium atropurpureum.
Generation times in TY medium at 28°C.
The L. micranthus nodule isolates studied were obtained from five locations in northern Tunisia, and their geographical origins are shown in Table 1 and Fig. 1. Among the seven groups identified, Bradyrhizobium isolates in groups II, III, and V were recovered from only one site, and the Phyllobacterium isolates were recovered only from nodules of plants collected in Takelsa. Moreover, in all but one of the sites, isolates belonged to at least two different groups. The exception was El Alia, where all 4 isolates were within the Microvirga group.
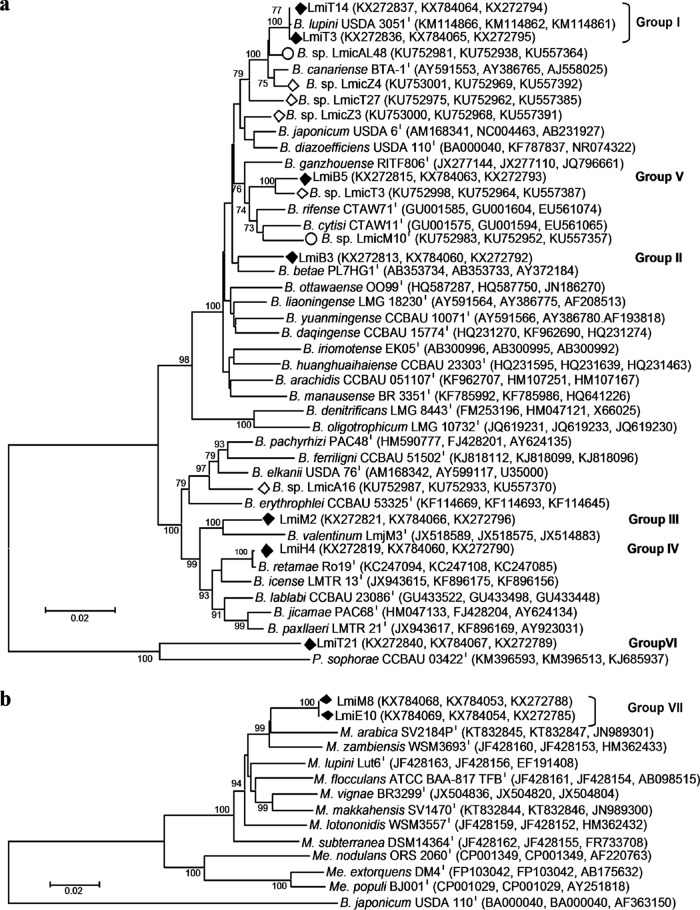
To perform a deeper multilocus phylogenetic analysis, 9 representatives of the seven clusters (with two from each of the two larger groups) were randomly selected. For Bradyrhizobium and Phyllobacterium isolates, rrs, recA, and glnII were chosen for analyses because they are considered robust markers for determining the phylogeny and taxonomy of rhizobia (38) (Fig. 4a). Of note, glnII seems to be absent from the genomes of Microvirga spp., since it does not appear in data banks or in any of the Microvirga genomes sequenced. Consistent with this, glnII did not appear in the draft genome sequences of the strains LmiM8 and LmiE10 obtained by our group (data not shown). Therefore, dnaK was used instead, and the phylogenetic tree for this group of strains was made with concatenated sequences from gyrB, dnaK, and rrs (Fig. 4b). These phylogenetic trees are in agreement with previous phylogenetic analyses, although group V appeared closer to B. rifense than to B. diazoefficiens, in contrast with the recA analysis. On the other hand, the representative Bradyrhizobium isolates were compared with L. micranthus-associated strains recently isolated in Algeria and Spain (16). Results shown in Fig. 4a indicate that the largest group present in soils from all three countries contains bacteria belonging to the B. lupini/B. canariense lineages. Group V contains isolates from Algeria and Tunisia, represented by isolates LmicT3 and LmiB5, respectively (Fig. 4a). Isolates from group IV were assigned to B. retamae species. The remaining groups of isolates identified in this study (group II, III, V, VI, and VII) may constitute different genospecies.
FIG 4.
Neighbor-joining phylogenetic trees of Tunisian L. micranthus-associated (Lmi) isolates and reference strains based on concatenated sequences. (a) Phylogeny based on recA (372 bp), glnII (451 bp), and rrs (1209 bp) sequences. (b) Phylogeny based on gyrB (621 bp), dnaK (741 bp), and rrs (1303 bp) sequences. Symbols: ◆, Tunisian Lmi isolates obtained in this work; ○, Lmic isolates from Spanish soils; and ♢, Lmic isolates from Algerian soils described in reference 16. Bootstrap values were calculated for 1,000 replications, and those greater than 70% are indicated at the internodes. Accession numbers from GenBank database are shown in parentheses. B., Bradyrhizobium; M., Microvirga; Me., Methylobacterium.
Phylogenetic analysis based on nodC sequence.
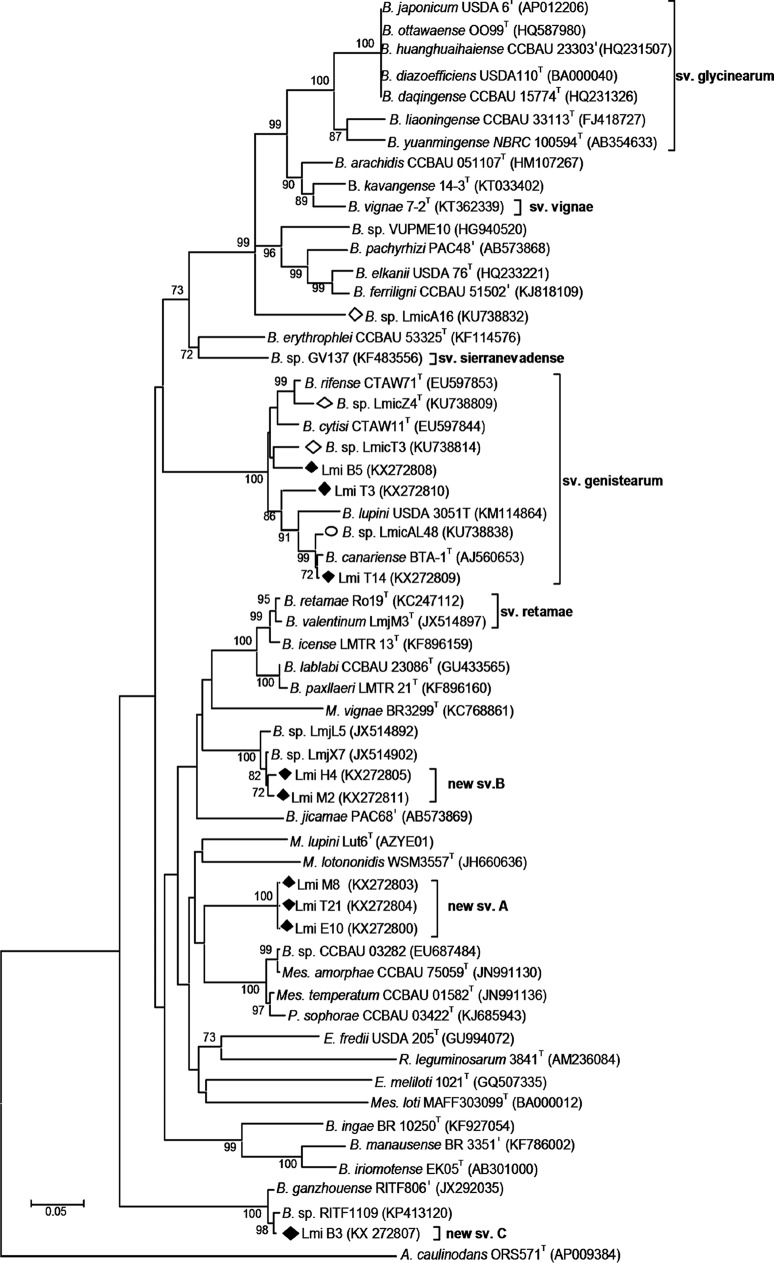
The nodC gene has been used to trace host range and symbiovars within the genus Bradyrhizobium (32, 39). An analysis of nodC sequences from representatives of the different groups of Tunisian L. micranthus-associated isolates suggests the existence of four different symbiovars (Fig. 5); three of them include Bradyrhizobium isolates and one includes Phyllobacterium and Microvirga-type isolates. Most of the representative isolates were symbiovar genistearum, which also encompasses strains able to nodulate legumes of the Genisteae tribe, such as the type strains of B. lupini, B. rifense, B. cytisi, and B. canariense. The two remaining symbiovars of Bradyrhizobium-type representative isolates may correspond to new symbiovars designated symbiovar B (LmiH4 and LmiM2), close to symbiovar retamae, and symbiovar C (LmiB3), which is distant from other Bradyrhizobium symbiovars, including symbiovar retamae nodulating Retama spp., and Lupinus mariae-josephae (11, 40) and symbiovar sierranevadense nodulating Genista versicolor (41). The fourth symbiovar, symbiovar A, comprised Microvirga and Phyllobacterium isolates with identical nodC sequences that define a new (100% bootstrap support) lineage with some similarity to nodC sequences from M. lupini and P. sophorae (Fig. 5).
FIG 5.
Neighbor-joining phylogenetic tree of Tunisian L. micranthus-associated (Lmi) isolates and reference strains based on nodC sequences. Symbols: ◆, Tunisian Lmi isolates obtained in this work; ○, Spanish Lmic isolates; and ♢, Algerian Lmic isolates described in reference 16. Bootstrap values were calculated for 1,000 replications, and those greater than 70% are indicated at the internodes. Accession numbers from the GenBank and whole-genome sequencing (NCBI) databases are shown in parentheses. B., Bradyrhizobium, E., Ensifer; M., Microvirga; Mes., Mesorhizobium; P., Phyllobacterium; R., Rhizobium; sv., symbiovar.
Plant nodulation tests of Tunisian L. micranthus-associated isolates.
We examined the symbiotic characteristics of representative isolates from the seven identified L. micranthus groups by cross-inoculation tests using Lupinus angustifolius, L. mariae-josephae, Vigna unguiculata, and Macroptilium atropurpureum as hosts (Table 2). No differences in the sizes of plants or appearances of the nodules were found when L. micranthus was inoculated with any of the representative isolates. However, clear differences in nodulation and symbiotic efficiency were observed when other hosts were inoculated with the same isolates. Isolate LmiB3 failed to nodulate L. angustifolius, and isolates LmiM2 and LmiT14 produced white nodules unable to fix nitrogen, whereas the remaining isolates produced healthy plants (Table 2). L. mariae-josephae plants were efficiently nodulated only by isolates of symbiovar B (Fig. 5), while isolates from other symbiovars induced no nodules or only white inefficient nodules. White inefficient nodules were also elicited in V. unguiculata by Microvirga and Phyllobacterium isolates grouped in symbiovar A and by Bradyrhizobium strains of symbiovar genistearum (group I), while red efficient nodules were elicited in this host by strains of Bradyrhizobium group V (symbiovar genistearum) and symbiovars B and C. Finally, M. atropurpureum plants were effectively nodulated by all L. micranthus-associated isolates (Table 2).
DISCUSSION
We performed a phylogenetic analysis of isolates from nodules of wild L. micranthus growing in five locations in northern Tunisia. The analysis showed a remarkable diversity of symbiotic bacteria, including isolates belonging to three genera, Bradyrhizobium, Microvirga, and Phyllobacterium, from three different families within the order Rhizobiales, namely Bradyrhizobiaceae, Methylobacteriaceae, and Phyllobacteriaceae, respectively. In previous studies, a clear predominance of Bradyrhizobium lineages was described for most Lupinus species (6, 7, 9, 12, 42). In lupine species from the Old World, B. canariense and B. japonicum are the two dominant rhizobial species in root nodules (8, 9). One exception is the B. valentinum species, a microsymbiont of L. mariae-josephae, a particular endemism of central-eastern Spain that grows in alkaline soils (11). In this study, most of the root nodule isolates (56%) were ascribed to the Bradyrhizobium genus, but the remaining 44% were, notably, fast-growing nonbradyrhizobia. This outcome contrasts with that from a recent study of L. micranthus symbiosis that identified all nodule isolates from Algeria and Spain soils as belonging to the genus Bradyrhizobium, mainly B. lupini/B. canariense (16). Similarly, most of the Tunisian bradyrhizobial isolates also belonged to the B. lupini/B. canariense lineage. A closer comparison of the five Tunisian Bradyrhizobium groups (groups I to V) with the five Bradyrhizobium groups from Algeria and the two groups from Spain (16) suggests the existence of specific phylogenetic groups for different geographical locations, as well as a greater diversity of these bradyrhizobia in northern Africa than in Spain (Fig. 6).
FIG 6.
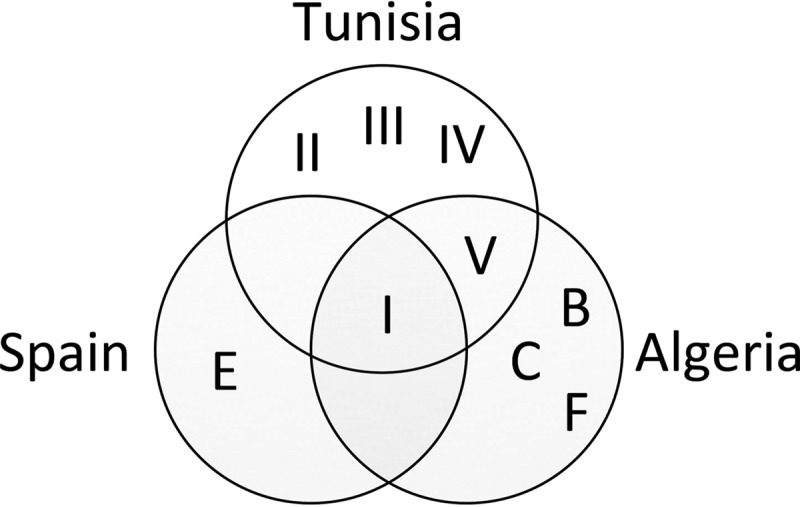
Venn diagram showing the Bradyrhizobium groups of L. micranthus-associated isolates shared by soils from different countries. Tunisian groups were defined in Fig. 2, and Spanish and Algerian groups were described by Bourebaba et al. (16). Group I in Tunisia corresponds to clusters A of Spain and Algeria, and group V corresponds to cluster D of Algeria.
Unexpectedly, a large fraction of the northern Tunisia L. micranthus root nodule isolates were fast-growing Rhizobiales from the Microvirga (20 isolates) and Phyllobacterium (2 isolates) genera. Although a large number of lupine root nodule isolates from around the world has been characterized (6, 7, 9, 10, 12, 16, 42), only one example of nodulation by Microvirga has been reported, namely M. lupini from the North American L. texensis species (14, 37). This appears to be a highly specific symbiosis, since no other Lupinus sp. has been reported that can form effective nitrogen-fixing nodules with M. lupini. Of the 13 Microvirga species currently described, only four are able to establish root nodule symbioses with different legumes, namely M. lupini with L. texensis in Texas-northeastern Mexico (15), M. zambiensis and M. lotononidis with Listia angolensis in Zambia and Angola, respectively (15), and M. vignae with Vigna unguiculata in Brazil (21). The limited and haphazard geographical distribution of Microvirga symbionts suggests that Tunisian Microvirga isolates from L. micranthus might have recently acquired their symbiotic capacity, perhaps by horizontal transfer from other symbionts, a phenomenon that has been evidenced in different systems (43–45). The Microvirga isolates from L. micranthus may constitute a new genospecies different from M. zambiensis/M. arabica, their closer symbiotic relatives (Fig. 4b). It should be emphasized that M. zambiensis fixes nitrogen in association with the South African legume Listia angolensis of the tribe Crotalarieae, sister to the tribe Genisteae, a large and diverse group that largely occurs in Africa (46).
A second remarkable result from this work is our finding that Phyllobacterium-related bacteria effectively nodulate lupines. Phyllobacterium strains have been identified in different environments (31), mostly as plant-associated bacteria (28, 30, 47, 48). To date, the Phyllobacterium genus contains 10 species, but only two, P. trifolii and P. sophorae, are able to induce effective nodules on their host legumes, Trifolium pratense and Sophora flavescens, respectively (26, 27). P. trifolii was also reported to induce white inefficient nodules in Lupinus albus (27). Strains of P. leguminum and P. ifriqiyense, isolated from nodules of Astragalus algerianus and Lathyrus numidicus, respectively, and growing in the infra-arid zone of southern Tunisia, were unable to induce effective nodules in any of the plants tested (28).
Soil pH can be an important factor determining the presence of different rhizobial species associated with one plant. Rodríguez-Echeverría et al. (49) studied the endosymbionts of Retama sphaerocarpa in acidic and alkaline soils. Like L. micranthus, R. sphaerocarpa is widely distributed in the Mediterranean and is nodulated by B. canariense only in soils with a pH below 7.0. In contrast, B. retamae appeared in nodules of plants growing in soils with a pH higher than 7.5. Similarly, in this work, most strains of the B. lupini/B. canariense lineage (group I) were isolated from neutral or mildly alkaline soils (Takelsa, pH 7.3, and Mraissa, pH 8.0), and Phyllobacterium strains (group VI) were identified only in Takelsa (Table 1). In contrast, Microvirga strains were found in all soils with a pH of 8 to 9 and were absent only in Takelsa. Thus, Microvirga strains appear to have a preference for alkaline soils, similar to that of the remaining Bradyrhizobium groups (II, III, IV and V) nodulating L. micranthus. In this respect, it is worth noting that groups III and IV are similar to B. valentinum and B. retamae, respectively, and these have been associated with alkaline soils (12, 34, 49). Overall, the available evidence suggests that symbiotic bacteria with certain genotypes are adapted to particular ecosystems where soil pH might play an important role. This assertion is consistent with the higher bradyrhizobial diversity observed in L. micranthus nodules from Tunisian (neutral or alkaline) soils versus Algerian and Spanish (acidic) soils (16).
The phylogeny of symbiotic nod genes has been frequently related to symbiotic performances of rhizobial strains (9, 42, 50, 51). Here, a large diversity of nodC sequences was observed within L. micranthus endosymbionts, which clustered into four symbiovars.
(i) Symbiovar genistearum was described by Vinuesa et al. (45) and includes many strains nodulating a large number of plants from the tribe Genisteae, including the genera Lupinus, Cytisus, Retama, Chamaecytisus, and Spartium, growing in Africa, Europe, and America (12, 33, 39, 40, 45). The majority of L. micranthus-associated isolates related to B. lupini (group I), and those similar to B. cytisi/B. rifense lineages (group V) belonged to this symbiovar. This is in line with the finding that most symbionts of Cytisus triflorus from Morocco and Algeria, as well as most isolates from L. micranthus from Algeria and Spain, belong to B. canariense/B. lupini and B. cytisi/B. rifense lineages and to symbiovar genistearum (16, 33, 52). Importantly, these microorganisms may be dominant in neutral or acidic soils.
(ii) Besides bradyrhizobial L. micranthus-associated isolates, symbiovar B included strains Bradyrhizobium sp. LmjX7 and Bradyrhizobium sp. LmjL5 isolated from nodules of L. mariae-josephae (34). Interestingly, L. mariae-josephae elicited only red effective nodules with strains of symbiovar B, while other symbiovars either did not produce nodules or produced white and inefficient nodules (Table 2). These results indicate that L. mariae-josephae and L. micranthus can be nodulated by a common Bradyrhizobium lineage.
(iii) Isolate LmiB3 appears to represent a new genospecies (close to B. betae) and a new symbiovar, far from other strains isolated from lupines or from other Genisteae legumes. In fact, the closest nodC sequences corresponded to Bradyrhizobium strains associated with Acacia melanoxylon grown in South China (53). At present, it is not clear why isolates from geographically distant plants as diverse as Lupinus and Acacia have nodC genes that are so similar.
(iv) Finally, the new symbiovar A, formed by Microvirga and Phyllobacterium strains, displayed similarities to Phyllobacterium strains isolated from root nodules of Sophora flavescens grown in Changzhi County in northern China (26) and with Mesorhizobium and Bradyrhizobium strains from root nodules of Caragana spp. from arid and semiarid alkaline deserts, also in northern China (54). Since Microvirga strains have also been found in arid soils from other areas (22), the capacity to proliferate in arid soils may be an important feature of the symbiosis between Microvirga and L. micranthus in the semiarid soils of Tunisia.
In conclusion, our study evidenced a wide biodiversity among the symbiotic bacteria isolated from L. micranthus nodules from northern Tunisia, including three genera, Bradyrhizobium and the unusual fast-growing Microvirga and Phyllobacterium. Our data and those from other sources suggest that the edaphoclimatic factors might be important for determining the occurrence of different L. micranthus microsymbionts. Given that L. micranthus is widely distributed in the Mediterranean basin, further studies from different edaphoclimatic areas might aid in gaining an understanding of the biology and specificity of this symbiosis.
MATERIALS AND METHODS
Isolation of bacterial strains from Lupinus micranthus nodules and culture conditions.
Endosymbiotic bacteria were isolated from field root nodules sampled from L. micranthus plants growing in five different locations in northern Tunisia separated by up to 120 km, namely Borj Hfaiedh, Hammamet, El Alia, Mraissa, and Takelsa (Table 1 and Fig. 1). Nodules were surface disinfected with 95% ethanol (1 min) and sodium hypochlorite (3 min) from a 1:4 dilution of commercial bleach. They were then rinsed 10 times with sterile distilled water and individually crushed on sterile plates. A loopful of the nodule suspension was streaked onto yeast extract mannitol agar (YMA) plates (55). Plates were incubated at 28°C until colonies were visualized, usually after 4 to 10 days. Single colonies were picked and checked for purity by repeated streaking on yeast extract mannitol medium and then maintained at 4°C. Isolates were maintained for long-term storage in yeast mannitol broth (YMB) containing 20% glycerol (vol/vol) at −80°C. The numbers, designations, and geographical origins of the isolates used in this study are listed in Table 1. All strains included in this study were checked for their ability to effectively renodulate L. micranthus.
Sizes and morphologies of colonies were assessed on YMA plates after 7 to 10 days of growth at 28°C. The generation times were determined by measuring the optical density at 600 nm (OD600) every 2 h in a 50-ml TY culture grown in a 250-ml Erlenmeyer flask incubated at 28°C with shaking (140 rpm). The starting OD600 for each of the cultures was 0.05.
Nodulation and cross-inoculation experiments.
The abilities of all purified isolates to renodulate their original hosts were demonstrated. L. micranthus, L. angustifolius, Vigna unguiculata, and Macroptilium atropurpureum seedlings were surface sterilized using 95% ethanol (1 min) and 25% sodium hypochlorite (3 min). L. mariae-josephae seeds required a scarification step by cutting the seed coat with a razor blade. Leonard jar units with 2 seedlings were inoculated with 1 ml of rhizobial suspension (109 cells ml−1). Plants were grown under bacteriologically controlled conditions in a greenhouse (16/8 h day/night at 25/23°C) and watered with sterile Jensen's liquid medium once per week for 3 to 8 weeks depending on the legume host. Uninoculated plants were included as negative controls.
DNA isolation and PCR amplification.
Total genomic DNA was isolated from a cell pellet from 2 ml of YMB culture by a simplified alkaline lysis procedure. The pellet was suspended in 20 μl of lysis solution (0.1 N NaOH and 0.5% SDS) and placed in hot water at 80 to 90°C for 15 min. The suspension was mixed with 100 μl of sterile distilled water and centrifuged at 13,000 × g for 10 min. Alternatively, DNA was isolated using DNeasy blood and tissue kit columns (Qiagen, Ltd.). PCR amplicons of genes coding for DNA recombination and repair protein RecA (recA), DNA gyrase subunit B (gyrB), 16S rRNA (rrs), glutamine synthetase 2 (glnII), chaperone protein DnaK (dnaK), and N-acetylglucosaminyltransferase (nodC) were obtained with primers and conditions previously described for recA, rrs, glnII, and nodC (12, 16) and for gyrB and dnaK (21). Amplicons were sequenced according to the procedures described by Sánchez-Cañizares et al. (12) and were used for phylogenetic analysis. Unincorporated primers and deoxynucleoside triphosphates (dNTPs) were removed from PCR products with the NucleoSpin Extract II (Macherey-Nagel) or, when needed, by gel electrophoresis followed by band purification with the same kit. Sequencing was performed at STABvida (Lisbon, Portugal) and sequences were edited and assembled with Geneious Pro 5.6.7 software (Biomatters, Ltd., Auckland, New Zealand).
Sequence and phylogenetic analysis.
Alignments of rrs sequences were performed using SINA service from the SILVA database (http://www.arb-silva.de/aligner) (56). Other sequences were aligned using CLUSTALW (57). The phylogenetic and molecular evolutionary analyses were carried out with MEGA 6.06 (58). The neighbor-joining statistical method and the Kimura two-parameter model (59) were used for all genes. Phylogenetic trees were subjected to 1,000 bootstrap replications, and preferred topologies were plotted.
Accession number(s).
GenBank accession numbers and whole-genome sequencing codes for the sequences obtained in this study are included in the corresponding trees in Fig. 2 (KX832514 to KX832521 and KX272812 to KX272841), Fig. 3 (KX784068 to KX784087), Fig. 4 (KX784060 to KX784067 and KX272785 to KX272796), and Fig. 5 (KX272800 to KX272811).
ACKNOWLEDGMENTS
We thank the reviewers for helpful comments. We also thank A. Bautista for excellent technical assistance.
This study was supported by the MINECO project BIO2013-43040 to J.P., MINECO CGL2011-26932 and MICROGEN CSD2009-00006 to J.I., and UPM AL16-PID-06 to L.R. A.M. acknowledges support from University of Gabès and Faculty of Sciences Tunis-University of El Manar Tunis for the Tunisian exchange and mobility fellowship.
REFERENCES
- 1.Cristofolini G. 1997. The biodiversity of the Leguminosae-Genisteae and its genesis. Lagascalia 19:121–128. [Google Scholar]
- 2.Ainouche AK, Bayer RJ. 1999. Phylogenetic relationships in Lupinus (Fabaceae: Papilionoideae) based on internal transcribed spacer sequences (ITS) of nuclear ribosomal DNA. Am J Bot 86:590–607. doi: 10.2307/2656820. [DOI] [PubMed] [Google Scholar]
- 3.Drummond CS, Eastwood RJ, Miotto STS, Hughes CE. 2012. Multiple continental radiations and correlates of diversification in Lupinus (Leguminosae): testing for key innovation with incomplete taxon sampling. Syst Biol 61:443–460. doi: 10.1093/sysbio/syr126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mahé F, Pascual H, Coriton O, Huteau V, Navarro Perris A, Misset M-T, Aınouche A. 2011. New data and phylogenetic placement of the enigmatic Old World lupin: Lupinus mariae-josephi H. Pascual. Genet Resour Crop Evol 58:101–114. doi: 10.1007/s10722-010-9580-6. [DOI] [Google Scholar]
- 5.Gladstones JS. 1988. Distribution, origin, taxonomy, history and importance, p 1–37. In Gladstones JS, Atkins CA, Hamblin J (ed), Lupins as crop plants: biology, production and utilization. CAB International, Wallingford, United Kingdom. [Google Scholar]
- 6.Barrera LL, Trujillo ME, Goodfellow M, García FJ, Hernández-Lucas I, Dávila G, van Berkum P, Martínez-Romero E. 1997. Biodiversity of bradyrhizobia nodulating Lupinus spp. Int J Syst Bacteriol 47:1086–1091. doi: 10.1099/00207713-47-4-1086. [DOI] [PubMed] [Google Scholar]
- 7.Jarabo-Lorenzo A, Pérez-Galdona R, Donate-Correa J, Rivas R, Velázquez E, Hernández M, Temprano F, Martínez-Molina E, Ruiz-Argüeso T, León-Barrios M. 2003. Genetic diversity of bradyrhizobial populations from diverse geographic origins that nodulate Lupinus spp. and Ornithopus spp. Syst Appl Microbiol 26:611–623. doi: 10.1078/072320203770865927. [DOI] [PubMed] [Google Scholar]
- 8.Stępkowski T, Żak M, Moulin L, Króliczak J, Golińska B, Narożna D, Safronova VI, Mądrzak CJ. 2011. Bradyrhizobium canariense and Bradyrhizobium japonicum are the two dominant rhizobium species in root nodules of lupin and serradella plants growing in Europe. Syst Appl Microbiol 34:368–375. doi: 10.1016/j.syapm.2011.03.002. [DOI] [PubMed] [Google Scholar]
- 9.Velázquez E, Valverde A, Rivas R, Gomis V, Peix A, Gantois I, Igual JM, León-Barrios M, Willems A, Mateos PF, Martínez-Molina E. 2010. Strains nodulating Lupinus albus on different continents belong to several new chromosomal and symbiotic lineages within Bradyrhizobium. Antonie Van Leeuwenhoek 97:363–376. doi: 10.1007/s10482-010-9415-7. [DOI] [PubMed] [Google Scholar]
- 10.Granada CE, Beneduzi A, Lisboa BB, Turchetto-Zolet AC, Vargas LK, Passaglia LM. 2015. Multilocus sequence analysis reveals taxonomic differences among Bradyrhizobium sp. symbionts of Lupinus albescens plants growing in arenized and non-arenized areas. Syst Appl Microbiol 38:323–329. doi: 10.1016/j.syapm.2015.03.009. [DOI] [PubMed] [Google Scholar]
- 11.Durán D, Rey L, Navarro A, Busquets A, Imperial J, Ruiz-Argüeso T. 2014. Bradyrhizobium valentinum sp. nov., isolated from effective nodules of Lupinus mariae-josephae, a lupine endemic of basic-lime soils in Eastern Spain. Syst Appl Microbiol 37:336–341. doi: 10.1016/j.syapm.2014.05.002. [DOI] [PubMed] [Google Scholar]
- 12.Sánchez-Cañizares C, Rey L, Durán D, Temprano F, Sánchez-Jiménez P, Navarro A, Polajnar M, Imperial J, Ruiz-Argüeso T. 2011. Endosymbiotic bacteria nodulating a new endemic lupine Lupinus mariae-josephi from alkaline soils in Eastern Spain represent a new lineage within the Bradyrhizobium genus. Syst Appl Microbiol 34:207–215. doi: 10.1016/j.syapm.2010.11.020. [DOI] [PubMed] [Google Scholar]
- 13.Trujillo ME, Willems A, Abril A, Planchuelo AM, Rivas R, Ludena D, Mateos PF, Martínez-Molina E, Velázquez E. 2005. Nodulation of Lupinus albus by strains of Ochrobactrum lupini sp. nov. Appl Environ Microbiol 71:1318–1327. doi: 10.1128/AEM.71.3.1318-1327.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Andam CP, Parker MA. 2007. Novel alphaproteobacterial root nodule symbiont associated with Lupinus texensis. Appl Environ Microbiol 73:5687–5691. doi: 10.1128/AEM.01413-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ardley JK, Parker MA, De Meyer SE, Trengove RD, O'Hara GW, Reeve WG, Yates RJ, Dilworth MJ, Willems A, Howieson JG. 2012. Microvirga lupini sp. nov., Microvirga lotononidis sp. nov. and Microvirga zambiensis sp. nov are alphaproteobacterial root-nodule bacteria that specifically nodulate and fix nitrogen with geographically and taxonomically separate legume hosts. Int J Syst Evol Microbiol 62:2579–2588. doi: 10.1099/ijs.0.035097-0. [DOI] [PubMed] [Google Scholar]
- 16.Bourebaba Y, Durán D, Boulila F, Ahnia H, Boulila A, Temprano F, Palacios JM, Imperial J, Ruiz-Argüeso T, Rey L. 2016. Diversity of Bradyrhizobium strains nodulating Lupinus micranthus on both sides of the western Mediterranean: Algeria and Spain. Syst Appl Microbiol 39:266–274. doi: 10.1016/j.syapm.2016.04.006. [DOI] [PubMed] [Google Scholar]
- 17.Pottier-Alapetite G. 1979. Flore de la Tunisie: angiospermes-dicotylédones, p 299–302. (In French.) Ministère de l'Enseignement supérieur et de la Recherche scientifique et ministère de l'Agriculture, Paris, France. [Google Scholar]
- 18.Amin A, Ahmed I, Habib N, Abbas S, Hasan F, Xiao M, Hozzein WN, Li WJ. 2016. Microvirga pakistanensis sp. nov., a novel bacterium isolated from desert soil of Cholistan, Pakistan. Arch Microbiol 198:933–939. doi: 10.1007/s00203-016-1251-3. [DOI] [PubMed] [Google Scholar]
- 19.Caputo A, Lagier JC, Azza S, Robert C, Mouelhi D, Fournier PE, Raoult D. 2016. Microvirga massiliensis sp. nov., the human commensal with the largest genome. Microbiologyopen 5:307–322. doi: 10.1002/mbo3.329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kanso S, Patel BK. 2003. Microvirga subterranea gen. nov., sp. nov., a moderate thermophile from a deep subsurface Australian thermal aquifer. Int J Syst Evol Microbiol 53:401–406. doi: 10.1099/ijs.0.02348-0. [DOI] [PubMed] [Google Scholar]
- 21.Radl V, Simões-Araújo JL, Leite J, Passos SR, Martins LMV, Xavier GR, Rumjanek NG, Baldani JI, Zilli JE. 2014. Microvirga vignae sp. nov., a root nodule symbiotic bacterium isolated from cowpea grown in semi-arid Brazil. Int J Syst Evol Microbiol 64:725–730. doi: 10.1099/ijs.0.053082-0. [DOI] [PubMed] [Google Scholar]
- 22.Veyisoglu A, Tatar D, Saygin H, Inan K, Cetin D, Guven K, Tuncer M, Sahin N. 2016. Microvirga makkahensis sp. nov., and Microvirga arabica sp. nov., isolated from sandy arid soil. Antonie Van Leeuwenhoek 109:287–296. doi: 10.1007/s10482-015-0631-z. [DOI] [PubMed] [Google Scholar]
- 23.Weon H-Y, Kwon S-W, Son J-A, Jo E-H, Kim S-J, Kim Y-S, Kim B-Y, Ka J-O. 2010. Description of Microvirga aerophila sp.nov. and Microvirga aerilata sp. nov., isolated from air, reclassification of Balneimonas flocculans Takeda et al. 2004 as Microvirga flocculans comb. nov. and emended description of the genus Microvirga. Int J Syst Evol Microbiol 60:2596–2600. doi: 10.1099/ijs.0.018770-0. [DOI] [PubMed] [Google Scholar]
- 24.Zhang J, Song F, Xin YH, Zhang J, Fang C. 2009. Microvirga guangxiensis sp. nov., a novel alpha proteobacterium from soil, and emended description of the genus Microvirga. Int J Syst Evol Microbiol 59:1997–2001. doi: 10.1099/ijs.0.007997-0. [DOI] [PubMed] [Google Scholar]
- 25.Knösel DH. 1962. Prufung von Bakterien auf Fahigkeit zur Sternbildung. Zentralbl Bakteriol Parasitenkd Infektionskr Hyg II Abt 116:79–100. [Google Scholar]
- 26.Jiao YS, Yan H, Ji ZJ, Liu YH, Sui XH, Zhang XX, Wang ET, Chen WX, Chen WF. 2015. Phyllobacterium sophorae sp. nov., a symbiotic bacterium isolated from root nodules of Sophora flavescens. Int J Syst Evol Microbiol 65:399–406. doi: 10.1099/ijs.0.067017-0. [DOI] [PubMed] [Google Scholar]
- 27.Valverde A, Velázquez E, Fernández-Santos F, Vizcaíno N, Rivas R, Mateos PF, Igual JM, Willems A. 2005. Phyllobacterium trifolii sp. nov., nodulating Trifolium and Lupinus in Spanish soils. Int J Syst Evol Microbiol 55:1985–1989. doi: 10.1099/ijs.0.63551-0. [DOI] [PubMed] [Google Scholar]
- 28.Mantelin S, Fischer-Le Saux M, Zakhia F, Béna G, Bonneau S, Jeder H, de Lajudie P, Cleyet-Marel JC. 2006. Emended description of the genus Phyllobacterium and description of four novel species associated with plant roots: Phyllobacterium bourgognense sp. nov., Phyllobacterium ifriqiyense sp. nov., Phyllobacterium leguminum sp. nov. and Phyllobacterium brassicacearum sp nov. Int J Syst Evol Microbiol 56:827–839. doi: 10.1099/ijs.0.63911-0. [DOI] [PubMed] [Google Scholar]
- 29.Sánchez M, Ramírez-Bahena MH, Peix A, Lorite M, Sanjuán J, Velázquez E, Monza J. 2014. Phyllobacterium loti sp. nov. isolated from nodules of Lotus corniculatus. Int J Syst Evol Microbiol 64:781–786. doi: 10.1099/ijs.0.052993-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Flores-Félix JD, Carro L, Velázquez E, Valverde A, Cerda-Castillo E, García-Fraile P, Rivas R. 2013. Phyllobacterium endophyticum sp. nov., isolated from nodules of Phaseolus vulgaris. Int J Syst Evol Microbiol 63:821–826. doi: 10.1099/ijs.0.038497-0. [DOI] [PubMed] [Google Scholar]
- 31.Mergaert J, Cnockaert MC, Swings J. 2002. Phyllobacterium myrsinacearum (subjective synonym Phyllobacterium rubiacearum) emend. Int J Syst Evol Microbiol 52:1821–1823. doi: 10.1099/00207713-52-5-1821. [DOI] [PubMed] [Google Scholar]
- 32.Rogel MA, Ormeño-Orrillo E, Martínez-Romero E. 2011. Symbiovars in rhizobia reflect bacterial adaptation to legumes. Syst Appl Microbiol 34:96–104. doi: 10.1016/j.syapm.2010.11.015. [DOI] [PubMed] [Google Scholar]
- 33.Ahnia H, Boulila F, Boulila A, Boucheffa K, Durán D, Bourebaba Y, Salmi A, Imperial J, Ruiz-Argüeso T, Rey L. 2014. Cytisus villosus from northeastern Algeria is nodulated by genetically diverse Bradyrhizobium strains. Antonie Van Leeuwenhoek 105:1121–1129. doi: 10.1007/s10482-014-0173-9. [DOI] [PubMed] [Google Scholar]
- 34.Durán D, Rey L, Sánchez-Cañizares C, Navarro A, Imperial J, Ruiz-Argüeso T. 2013. Genetic diversity of indigenous rhizobial symbionts of the Lupinus mariae-josephae endemism from alkaline-limed soils within its area of distribution in eastern Spain. Syst Appl Microbiol 36:128–136. doi: 10.1016/j.syapm.2012.10.008. [DOI] [PubMed] [Google Scholar]
- 35.Bailey AC, Kellom M, Poret-Peterson AT, Noonan K, Hartnett HE, Raymond J. 2014. Draft genome sequence of Microvirga sp. strain BSC39, isolated from biological soil crust of Moab, Utah. Genome Announc 2:e01197-14. doi: 10.1128/genomeA.01197-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Reeve W, Ardley J, Tian R, Meyer S, Terpolilli J, Melino V, Tiwari R, Yates R, O'Hara G, Howieson J, Ninawi M, Zhang X, Bruce D, Detter C, Tapia R, Han C, Wei CL, Huntemann M, Han J, Chen IM, Mavromatis K, Markowitz V, Szeto E, Ivanova N, Pagani I, Pati A, Goodwin L, Woyke T, Kyrpides N. 2013. Genome sequence of the Listia angolensis microsymbiont Microvirga lotononidis strain WSM3557T. Stand Genomic Sci 9:540–550. doi: 10.4056/sigs.4548266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Reeve W, Parker M, Tian R, Goodwin L, Teshima H, Tapia R, Han C, Han J, Liolios K, Huntemann M, Pati A, Woyke T, Mavromatis K, Markowitz V, Ivanova N, Kyrpides N. 2014. Genome sequence of Microvirga lupini strain LUT6(T), a novel Lupinus alphaproteobacterial microsymbiont from Texas. Stand Genomic Sci 9:1159–1167. doi: 10.4056/sigs.5249382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhang YM, Tian CF, Sui XH, Chen WF, Chen WX. 2012. Robust markers reflecting phylogeny and taxonomy of rhizobia. PLoS One 7:e44936. doi: 10.1371/journal.pone.0044936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Peix A, Ramírez-Bahena MH, Velázquez E, Bedmar EJ. 2015. Bacterial associations with legumes. CRC Crit Rev Plant Sci 34:17–42. doi: 10.1080/07352689.2014.897899. [DOI] [Google Scholar]
- 40.Guerrouj K, Ruíz-Díez B, Chahboune R, Ramírez-Bahena MH, Abdelmoumen H, Quinones MA, El Idrissi MM, Velázquez E, Fernández-Pascual M, Bedmar EJ, Peix A. 2013. Definition of a novel symbiovar (sv. retamae) within Bradyrhizobium retamae sp. nov., nodulating Retama sphaerocarpa and Retama monosperma. Syst Appl Microbiol 36:218–223. doi: 10.1016/j.syapm.2013.03.001. [DOI] [PubMed] [Google Scholar]
- 41.Cobo-Díaz JF, Martínez-Hidalgo P, Fernández-González AJ, Martínez-Molina E, Toro N, Velázquez E, Fernández-López M. 2014. The endemic Genista versicolor from Sierra Nevada National Park in Spain is nodulated by putative new Bradyrhizobium species and a novel symbiovar (sierranevadense). Syst Appl Microbiol 37:177–185. doi: 10.1016/j.syapm.2013.09.008. [DOI] [PubMed] [Google Scholar]
- 42.Stępkowski T, Hughes CE, Law IJ, Markiewicz Ł, Gurda D, Chlebicka A, Moulin L. 2007. Diversification of lupine Bradyrhizobium strains: evidence from nodulation gene trees. Appl Environ Microbiol 73:3254–3264. doi: 10.1128/AEM.02125-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Menna P, Hungria M. 2011. Phylogeny of nodulation and nitrogen-fixation genes in Bradyrhizobium: supporting evidence for the theory of monophyletic origin, and spread and maintenance by both horizontal and vertical transfer. Int J Syst Evol Microbiol 61:3052–3067. doi: 10.1099/ijs.0.028803-0. [DOI] [PubMed] [Google Scholar]
- 44.Moulin L, Béna G, Boivin-Masson C, Stępkowski T. 2004. Phylogenetic analyses of symbiotic nodulation genes support vertical and lateral gene co-transfer within the Bradyrhizobium genus. Mol Phylogenet Evol 30:720–732. doi: 10.1016/S1055-7903(03)00255-0. [DOI] [PubMed] [Google Scholar]
- 45.Vinuesa P, León-Barrios M, Silva C, Willems A, Jarabo-Lorenzo A, Pérez Galdona R, Werner D, Martínez-Romero E. 2005. Bradyrhizobium canariense sp. nov., an acid-tolerant endosymbiont that nodulates endemic genistoid legumes (Papilionoideae: Genisteae) from the Canary Islands, along with Bradyrhizobium japonicum bv. genistearum, Bradyrhizobium genospecies alpha and Bradyrhizobium genospecies beta. Int J Syst Evol Microbiol 55:569–575. doi: 10.1099/ijs.0.63292-0. [DOI] [PubMed] [Google Scholar]
- 46.Boatwright JS, Marianne MR, Wink M, Morozova T, van Wyk BE. 2008. Phylogenetic relationships of tribe Crotalarieae (Fabaceae) inferred from DNA sequences and morphology. Syst Bot 33:752–761. doi: 10.1600/036364408786500271. [DOI] [Google Scholar]
- 47.Bromfield ESP, Tambong JT, Cloutier S, Prévost D, Laguerre G, van Berkum P, Thi TVT, Assabgui R, Barran LR. 2010. Ensifer, Phyllobacterium and Rhizobium species occupy nodules of Medicago sativa (alfalfa) and Melilotus alba (sweet clover) grown at a Canadian site without a history of cultivation. Microbiology 156:505–520. doi: 10.1099/mic.0.034058-0. [DOI] [PubMed] [Google Scholar]
- 48.Rasolomampianina R, Bailly X, Fetiarison R, Rabevohitra R, Béna G, Ramaroson L, Raherimandimby M, Moulin L, De Lajudie P, Dreyfus B, Avarre JC. 2005. Nitrogen-fixing nodules from rose wood legume trees (Dalbergia spp.) endemic to Madagascar host seven different genera belonging to α- and β-Proteobacteria. Mol Ecol 14:4135–4146. doi: 10.1111/j.1365-294X.2005.02730.x. [DOI] [PubMed] [Google Scholar]
- 49.Rodríguez-Echeverría S, Moreno S, Bedmar EJ. 2014. Genetic diversity of root nodulating bacteria associated with Retama sphaerocarpa in sites with different soil and environmental conditions. Syst Appl Microbiol 37:305–310. doi: 10.1016/j.syapm.2013.11.003. [DOI] [PubMed] [Google Scholar]
- 50.Kalita M, Stępkowski T, Łotocka B, Małek W. 2006. Phylogeny of nodulation genes and symbiotic properties of Genista tinctoria bradyrhizobia. Arch Microbiol 186:87–97. doi: 10.1007/s00203-006-0124-6. [DOI] [PubMed] [Google Scholar]
- 51.Parker MA. 2012. Legumes select symbiosis island sequence variants in Bradyrhizobium. Mol Ecol 21:1769–1778. doi: 10.1111/j.1365-294X.2012.05497.x. [DOI] [PubMed] [Google Scholar]
- 52.Chahboune R, Barrijal S, Moreno S, Bedmar EJ. 2011. Characterization of Bradyrhizobium species isolated from root nodules of Cytisus villosus grown in Morocco. Syst Appl Microbiol 34:440–445. doi: 10.1016/j.syapm.2011.04.008. [DOI] [PubMed] [Google Scholar]
- 53.Lu JK, Dou YJ, Zhu YJ, Wang SK, Sui XH, Kang LH. 2014. Bradyrhizobium ganzhouense sp. nov., an effective symbiotic bacterium isolated from Acacia melanoxylon R. Br. nodules. Int J Syst Evol Microbiol 64:1900–1905. doi: 10.1099/ijs.0.056564-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Li M, Li Y, Chen WF, Sui XH, Li Y, Li Y, Wang ET, Chen WX. 2012. Genetic diversity, community structure and distribution of rhizobia in the root nodules of Caragana spp. from arid and semi-arid alkaline deserts, in the north of China. Syst Appl Microbiol 35:239–245. doi: 10.1016/j.syapm.2012.02.004. [DOI] [PubMed] [Google Scholar]
- 55.Vincent JM. 1970. A manual for the practical study of root nodule bacteria. International biological programme, number 15. Blackwell Scientific, Oxford, United Kingdom. [Google Scholar]
- 56.Pruesse E, Peplies J, Glockner FO. 2012. SINA: accurate high-throughput multiple sequence alignment of ribosomal RNA genes. Bioinformatics 28:1823–1829. doi: 10.1093/bioinformatics/bts252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Chenna R, Sugawara H, Koike T, Lopez R, Gibson TJ, Higgins DG, Thompson JD. 2003. Multiple sequence alignment with the Clustal series of programs. Nucleic Acids Res 31:3497–3500. doi: 10.1093/nar/gkg500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. 2013. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol Biol Evol 30:2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kimura M. 1980. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J Mol Evol 16:111–120. doi: 10.1007/BF01731581. [DOI] [PubMed] [Google Scholar]