ABSTRACT
This study developed RNA-based predictive models describing the survival of Vibrio parahaemolyticus in Eastern oysters (Crassostrea virginica) during storage at 0, 4, and 10°C. Postharvested oysters were inoculated with a cocktail of five V. parahaemolyticus strains and were then stored at 0, 4, and 10°C for 21 or 11 days. A real-time reverse transcription-PCR (RT-PCR) assay targeting expression of the tlh gene was used to evaluate the number of surviving V. parahaemolyticus cells, which was then used to establish primary molecular models (MMs). Before construction of the MMs, consistent expression levels of the tlh gene at 0, 4, and 10°C were confirmed, and this gene was used to monitor the survival of the total V. parahaemolyticus cells. In addition, the tdh and trh genes were used for monitoring the survival of virulent V. parahaemolyticus. Traditional models (TMs) were built based on data collected using a plate counting method. From the MMs, V. parahaemolyticus populations had decreased 0.493, 0.362, and 0.238 log10 CFU/g by the end of storage at 0, 4, and 10°C, respectively. Rates of reduction of V. parahaemolyticus shown in the TMs were 2.109, 1.579, and 0.894 log10 CFU/g for storage at 0, 4, and 10°C, respectively. Bacterial inactivation rates (IRs) estimated with the TMs (−0.245, −0.152, and −0.121 log10 CFU/day, respectively) were higher than those estimated with the MMs (−0.134, −0.0887, and −0.0732 log10 CFU/day, respectively) for storage at 0, 4, and 10°C. Higher viable V. parahaemolyticus numbers were predicted using the MMs than using the TMs. On the basis of this study, RNA-based predictive MMs are the more accurate and reliable models and can prevent false-negative results compared to TMs.
IMPORTANCE One important method for validating postharvest techniques and for monitoring the behavior of V. parahaemolyticus is to establish predictive models. Unfortunately, previous predictive models established based on plate counting methods or on DNA-based PCR can underestimate or overestimate the number of surviving cells. This study developed and validated RNA-based molecular predictive models to describe the survival of V. parahaemolyticus in oysters during low-temperature storage (0, 4, and 10°C). The RNA-based predictive models show the advantage of being able to count all of the culturable, nonculturable, and stressed cells. By using primers targeting the tlh gene and pathogenesis-associated genes (tdh and trh), real-time RT-PCR can evaluate the total surviving V. parahaemolyticus population as well as differentiate the pathogenic ones from the total population. Reliable and accurate predictive models are very important for conducting risk assessment and management of pathogens in food.
KEYWORDS: Vibrio parahaemolyticus, Eastern oyster, molecular predictive model, RNA, real-time RT-PCR, predictive models
INTRODUCTION
Vibrio parahaemolyticus is a Gram-negative, halophilic asporogenous and curved bacterium. It is a human pathogen that grows naturally in marine environments and can be isolated from various seafoods, including oysters, fish, and shrimp (1). Oysters are a nutrient-rich seafood popular worldwide. In the United States, oyster consumption increased in recent decades. Compared with consuming 10 to 12 pounds of seafood during the 1980s, the average American now eats approximately 16.5 pounds every year (2). Unlike most other foods, oysters are often consumed raw or uncooked (3).
Oysters feed by filtering large volumes of seawater. During this process, pathogenic microorganisms can accumulate and concentrate to levels up to 100 times greater than those present naturally in seawater (2, 4). The consumption of raw oysters has the potential to cause V. parahaemolyticus infection, with symptoms such as watery diarrhea, abdominal cramps, nausea, vomiting, and even septicemia (5, 6). The largest raw oyster-associated outbreak happened in the United States in 1998 and caused 416 illnesses in 13 states (2). In 2006, another outbreak of V. parahaemolyticus caused 177 illnesses in three states in the U.S. due to the consumption of raw oysters (7). In the U.S., most (about 72.8%) of the Vibrio infection cases occurred during summer months, when water temperatures were warmer (2). Other research also showed that higher densities of V. parahaemolyticus in harvested oysters were found in spring and summer months (7–9).
All V. parahaemolyticus isolates contain the thermolabile hemolysin (tlh) gene, a marker gene that has been used to qualitatively or quantitatively detect V. parahaemolyticus in different food systems (1). However, clinical illnesses caused by V. parahaemolyticus strains are more closely associated with the expression of the thermostable direct hemolysin (tdh) and tdh-related hemolysin (trh) genes. The prevalence of tdh gene-positive isolates ranges from 3% to 70% and the prevalence of the trh gene ranges from 17% to 60% in all V. parahaemolyticus isolates (4, 10).
One reason that V. parahaemolyticus has been a common problem in seafood is its ability to survive and persist over a range of temperatures. It was reported that the population of V. parahaemolyticus in oysters increased significantly when the temperature increased from 15°C to 30°C and decreased when temperatures were lower than 10°C (4, 11). To better ensure oyster safety and to control the levels of V. parahaemolyticus in harvested oysters, the National Shellfish Sanitation Program (NSSP) Guide requires harvested shellfish to be cooled to below 10°C (50°F) within 12, 18, 24, and 36 h after harvesting when the average monthly maximum air temperature is ≥27°C (80°F), between 15 and 27°C (60 to 80°F), between 10 and 15°C (50 to 60°F), and below 10°C (50°F), respectively (12).
An important method for validation of postharvest techniques and monitoring the behavior of V. parahaemolyticus is that of establishing predictive models. These models not only help with validation and monitoring but can also contribute information in a risk assessment. Through mathematical functions, primary predictive models are developed to describe population dynamics of pathogenic and spoilage bacteria under different environmental conditions (4). Based on the primary models, secondary predictive models are constructed to evaluate the effect of temperature on growth rates (GRs) or inactivation rates (IRs) of bacteria (11). Predictive models have been developed to describe the behavior of V. parahaemolyticus in slurries of inoculated oysters (Crassostrea gigas) over a range of temperatures from 10 to 30°C and in inoculated Pacific oysters (C. gigas) at temperatures from 4 to 30°C, as well as the behavior of naturally occurring V. parahaemolyticus in Eastern oysters (C. virginica) at temperatures ranging from 5 to 30°C (4, 7, 11).
Unfortunately, most of the current predictive models were constructed using plate counting methods. These methods are time-consuming and labor-intensive and do not culture the bacteria in a viable but a nonculturable (VBNC) or stressed state (13, 14). VBNC pathogenic bacteria are considered a threat to public health and food safety because they retain their viability and ability to express their virulence (15). Therefore, predictive traditional models (TMs) based on plate counting methods may underestimate the populations of bacteria (15, 16). With the development of molecular quantitative methods, predictive models based on real-time PCR and PCR-denaturing gradient gel electrophoresis (PCR-DGGE) methods have been reported to describe the survival and growth of Listeria monocytogenes and V. parahaemolyticus in food matrixes (13, 17). However, all of these molecular methods are based on DNA, which can be extracted from both live and dead bacteria (13, 18, 19).
Propidium monoazide (PMA)–real-time PCR based on DNA has been proposed to be a good way to avoid amplification of DNA isolated from dead cells (20). Unfortunately, PMA becomes increasingly toxic at higher concentrations and is very expensive; PMA–real-time PCR methods that include higher PMA concentrations are considered to be counterproductive and cost prohibitive (21). According to Nocker and Camper, PMA inhibits amplification of the DNA of the dead cells based on membrane integrity and cell wall penetration (22). Given this mechanism, Nocker et al. reported that the PMA method may not allow monitoring of the killing efficacy of UV treatment and other inactivation mechanisms (e.g., low-temperature storage) that do not directly impact cell membrane integrity. When membrane damage occurs as a secondary effect, little is known about the time span within which membranes become susceptible to PMA treatment (23). Thus, RNA, which can be extracted only from live bacteria, and RNA-based molecular methods serve as a more accurate way to quantitatively measure populations of surviving V. parahaemolyticus cells (24).
Pathogenic V. parahaemolyticus with the tdh or trh virulence gene has been recognized as a major factor causing human illnesses. Traditional plate counting methods (using, e.g., thiosulfate-citrate-bile salts-sucrose [TCBS] or TCBS agar) can detect the total V. parahaemolyticus population but cannot differentiate pathogenic V. parahaemolyticus from nonpathogenic V. parahaemolyticus. Real-time PCR has the advantage of being able to quantify the total V. parahaemolyticus population targeting the tlh gene and to differentiate and quantify the pathogenic V. parahaemolyticus population with the tdh or trh gene (25, 26).
To summarize, the objectives of this study were to establish and validate RNA-based predictive molecular models (MMs) using real-time reverse transcription-PCR (RT-PCR) to describe the survival of a V. parahaemolyticus cocktail in oysters during low-temperature storage.
RESULTS
tlh gene expression at 0, 4, and 10°C.
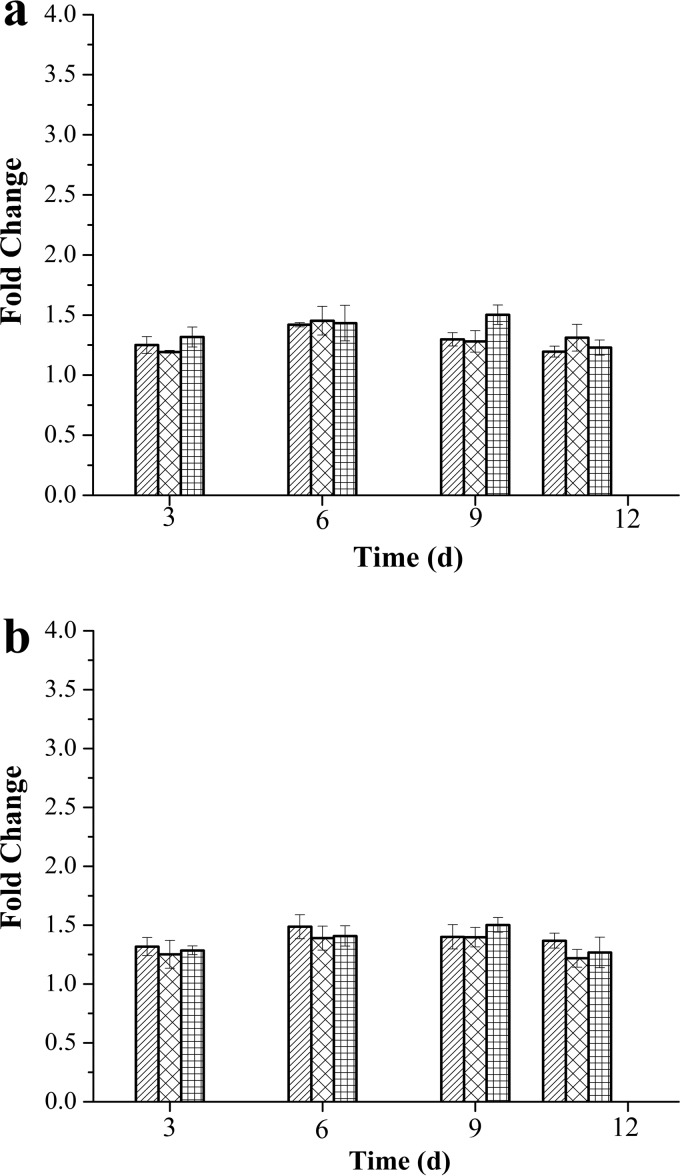
According to Meng et al., changes in the gene expression level that were less than 2 log2-fold were not considered to be significant (27). As shown in Fig. 1, the fold changes of tlh expression determined using pvsA and recA as reference genes showed no significant differences (P > 0.05) between different days (days 3, 6, 9, and 11) or different temperatures (0, 4, and 10°C). The transcription of the tlh gene was maintained at constant levels at different temperatures during storage.
FIG 1.
Fold change of tlh gene expression based on endogenous reference genes on days (d) 3, 6, 9, and 11 at 0, 4, and 10°C. (a) pvsA used as endogenous reference gene. (b) recA used as endogenous reference gene. Bars with diagonal lines represent fold change of tlh gene expression at 0°C; bars with diagonal crosshatching represent gene expression at 4°C; bars with squares represent gene expression at 10°C.
The primary models of V. parahaemolyticus in oysters.
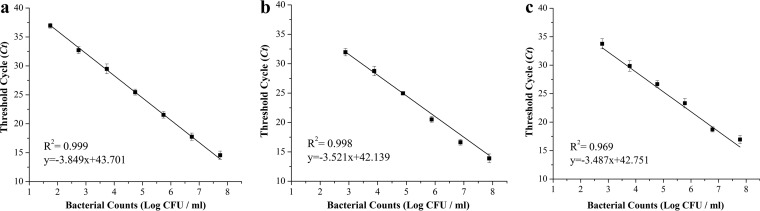
The final V. parahaemolyticus inoculation level in oysters was 5.449 ± 0.218 log10 CFU/g. By using the plate counting method and real-time RT-PCR, the populations of surviving V. parahaemolyticus in oysters were enumerated and calculated at different time intervals. For real-time RT-PCR, to convert the threshold cycle (CT) values to cell counts, standard curves based on the tlh, tdh, and trh genes were constructed (Fig. 2). The coefficient of determination (R2) values were 0.999, 0.998, and 0.969 and the equations were y = −3.849x + 43.701, y = −3.521x + 42.139, and y = −3.487x + 42.751 for the tlh, tdh, and trh genes, respectively.
FIG 2.
Quantification standard curves targeting tlh (a), tdh (b), and trh (c) genes in V. parahaemolyticus.
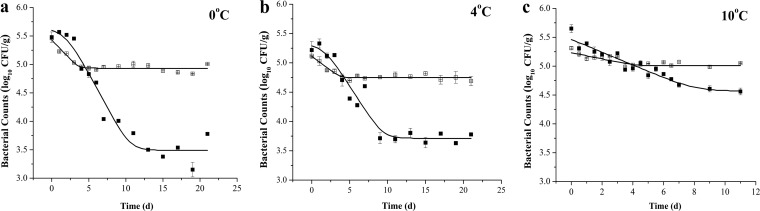
The survival data, obtained from the plate counting method and the real-time RT-PCR method at three storage temperatures, were then fitted by the Baranyi function (equation 3) (57) to obtain primary TMs and MMs (Fig. 3). The parameters of primary TMs and MMs are listed in Table 1. For primary TMs, the average R2 value for plate counting data fitted by the Baranyi function (equation 3) of three storage temperatures was 0.925. For storage at 0, 4, and 10°C, the population of V. parahaemolyticus decreased from the initial values (IVs) to the final values (FVs) with reductions of 2.109, 1.579, and 0.894 log10 CFU/g, respectively. The inactivation rates (IRs) showed an increasing trend as the temperature increased from 0 to 4 and 10°C. V. parahaemolyticus did not have lag times (LTs) when stored at 10°C. For primary MMs (Table 1), the average R2 value for molecular data fitted by the Baranyi function (equation 3) of the three storage temperatures was 0.859. The respective reductions from IVs to FVs were 0.493, 0.362, and 0.238 log10 CFU/g for 0, 4, and 10°C storage. The IRs also showed an increasing trend as temperature increased from 0 to 4 and 10°C storage. All of the LTs of the primary MMs of V. parahaemolyticus at 0, 4, and 10°C were zero.
FIG 3.
The primary MMs established based on the tlh gene and the primary TMs based on the plate counting method for V. parahaemolyticus after storage at 0 (a), 4 (b), and 10°C (c) for 21, 21, and 11 days, respectively. Filled squares represent plate count data; squares with boxes represent RT-PCR data; solid lines represent plate Baranyi fit data.
TABLE 1.
Inactivation parameters of the primary molecular and traditional models
| Parameter | Value(s)a |
||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0°C |
4°C |
10°C |
|||||||||||||
| R2 | Initial value (log10 CFU/g) | Final value (log10 CFU/g) | Lag time (day) | Inactivation rate (log10 CFU/day) | R2 | Initial value (log10 CFU/g) | Final value (log10 CFU/g) | Lag time (day) | Inactivation rate (log10 CFU/day) | R2 | Initial value (log10 CFU/g) | Final value (log10 CFU/g) | Lag time (day) | Inactivation rate (log10 CFU/day) | |
| Primary MMs | 0.881 | 5.428 ± 0.0477A | 4.935 ± 0.0177A | 0A | −0.134 ± 0.0263A | 0.891 | 5.114 ± 0.0335A | 4.752 ± 0.0125A | 0A | −0.0886 ± 0.0184A | 0.806 | 5.261 ± 0.0345A | 5.023 ± 0.0205A | 0A | −0.073 ± 0.0168A |
| Primary TMs | 0.943 | 5.601 ± 0.167A | 3.492 ± 0.0955B | 1.931 ± 0.528B | −0.245 ± 0.05B | 0.931 | 5.293 ± 0.138A | 3.714 ± 0.0718B | 1.605 ± 0.559B | −0.152 ± 0.0434B | 0.902 | 5.463 ± 0.0467A | 4.569 ± 0.0783B | 0A | −0.121 ± 0.0635B |
Different uppercase superscript roman letters indicate that a significant difference (P < 0.05) exists between the values determined for the parameters from the primary MMs and TMs.
The survival of V. parahaemolyticus with the tdh or trh virulence gene during cold storage.
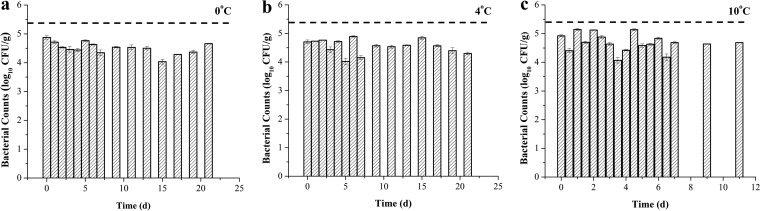
The survival behavior of pathogenic V. parahaemolyticus (tdh gene positive) in a V. parahaemolyticus cocktail in oysters was monitored during cold storage at 0, 4, and 10°C for 21, 21, and 11 days, respectively (Fig. 4). The initial populations of these V. parahaemolyticus were 4.876, 4.704, and 4.924 log10 CFU/g and the final populations were 4.663, 4.297, and 4.687 log10 CFU/g for storage at 0, 4, and 10°C, respectively. However, monitoring the survival of pathogenic V. parahaemolyticus containing the trh gene, no amplification was detected through real-time PCR after day 0. On day 0, the CT values for the trh gene were 26.737, 25.917, and 26.560 for 0, 4, and 10°C, respectively, representing 4.592, 4.827, and 4.643 log10 CFU/g.
FIG 4.
Survival of pathogenic V. parahaemolyticus (with the tdh virulence gene) in oysters during cold storage at 0°C (a), 4°C (b), and 10°C (c). The dotted line represents the initial total population of V. parahaemolyticus.
The secondary models of V. parahaemolyticus in oysters.
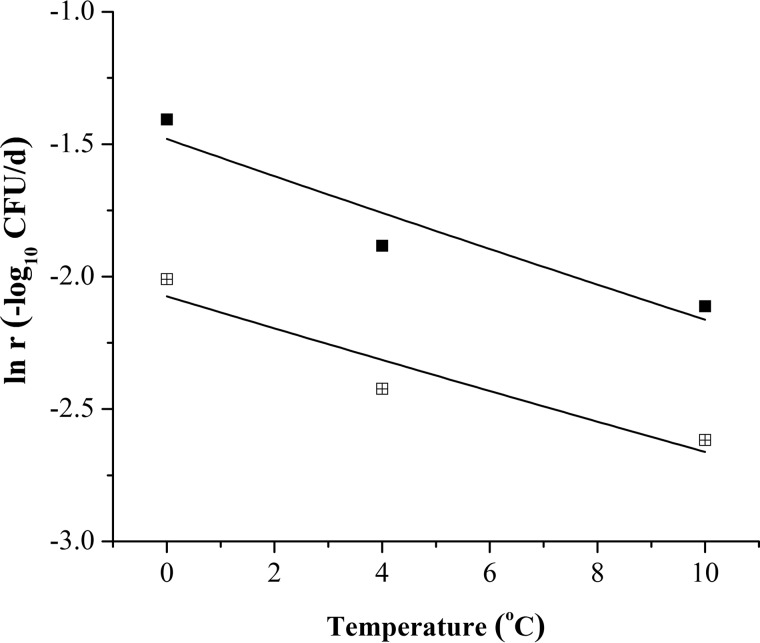
The secondary models were developed to describe the parameter changes of the primary models as a function of temperature. The IRs obtained from the primary TMs and MMs were used to produce the secondary TMs and MMs. The Arrhenius model (equation 4) (56) was utilized to describe the change of V. parahaemolyticus ln (−IRs) from 0 to 10°C (Fig. 5). For the secondary TMs, the coefficients A (collision factor) and Ea/R (activation energy/universal gas constant) were 9.156 × 10−10 and −5,280.115, respectively (equation 1). The R2 value of goodness of fit was 0.818 after Arrhenius function fitting. The values for Bf (bias factor [relative average deviation of predicted and observed values]) and Af (accuracy factor [spread of the results around the predicted values]) were 1.003 and 1.048, respectively.
| (1) |
For the secondary MMs, the A and Ea/R values were 7.503 × 10−9 and −4,543.456 (equation 2). The R2 value corresponding to the goodness of fit was 0.812 with Arrhenius equation fitting. The Bf and Af values were 1.001 and 1.031, respectively.
| (2) |
The secondary TM and MM were compared using one-way analysis of variance (ANOVA). Significant differences (P < 0.05) between them were found.
FIG 5.
Data from the secondary TM established based on IR data from the primary TMs are represented by filled squares. Data from the secondary MM established based on IR data from the primary MMs are represented by squares with boxes.
Validation of the secondary MMs in oysters.
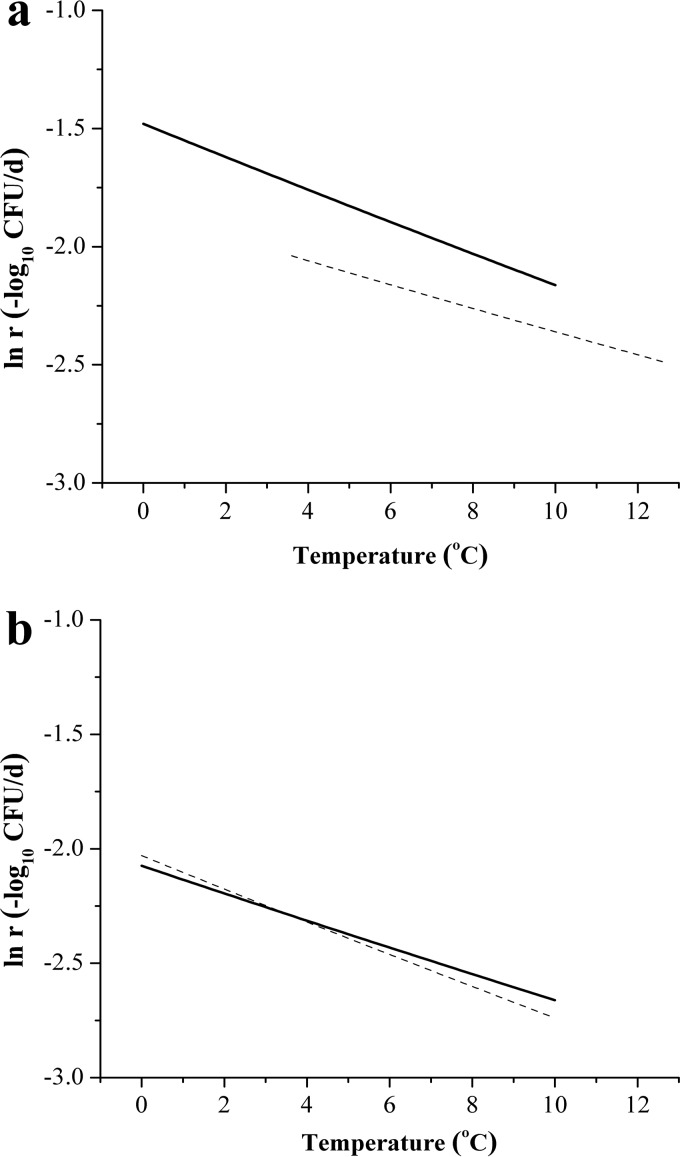
In order to validate the secondary models of V. parahaemolyticus in oysters, the secondary TM was compared with a previously reported secondary predictive model (4). The secondary model published by Fernandez-Piquer et al. described the effects of storage temperatures from 3.6 to 12.6°C on the survival of V. parahaemolyticus in inoculated Pacific oysters (C. gigas) (4). The secondary MM in oysters was compared with a secondary validation model built with oysters harvested in different months.
As shown in Fig. 6a, the secondary TM and the reported model had the same decreasing trend: as the temperature increased, the IRs increased. The Bf and Af values, calculated based on the comparison of IRs in the secondary TM and the reported model, were 0.887 and 1.127, respectively. The predicted IRs in the secondary TM were significant lower at every given temperature (P = 0.034 < 0.05) than the IRs predicted in the model reported by Fernandez-Piquer et al. (4). In Fig. 6b, the secondary MM and the validation model nearly overlapped. The Bf and Af values, calculated on the basis of the comparison of IRs in the secondary MM and validation model, were 1.004 and 1.015, respectively. No significant difference was seen between the IRs of the two models (P = 0.519 > 0.05). Differences between predictions of models were analyzed by one-way ANOVA, and the results indicated that the secondary MM in this study is a reliable predictor for IR changes as a result of temperature change.
FIG 6.
Validation of the secondary models. (a) Comparison of the secondary TM (solid line) in this study and the inactivation model reported by Fernandez-Piquer et al. (the secondary validation model [VM]) (4) (dashed line). (b) Comparison of the secondary MM (solid line) in this study and the validation model (the secondary VM) conducted in this study (dashed line).
DISCUSSION
The current study developed RNA-based predictive models to describe the survival of a V. parahaemolyticus cocktail in Eastern oysters (C. virginica) at low storage temperatures (0, 4, and 10°C). The predictive MMs showed great potential to predict V. parahaemolyticus activities in oysters during postharvest cold storage. During the past decade, predictive models have been established to describe the growth and inactivation of V. parahaemolyticus in different broth and seafood matrices (6, 11, 28, 29). In a recent report, the population of V. parahaemolyticus in a Korean oyster slurry decreased under conditions of storage at 10 and 15°C for 80 h and 70 h, respectively, with approximate IRs of −0.900 and −1.371 log10 CFU/day. Those authors also found that the V. parahaemolyticus numbers increased in nutrition broth in storage at 10 and 15°C (7). Both findings indicated that both the storage temperature and storage conditions such as nutrient concentrations impacted the survival of V. parahaemolyticus. According to Burnham et al., the minimum temperature for V. parahaemolyticus to grow in tryptic soy broth (TSB) was 5°C (30).
Traditionally, conventional plate counting methods have been used to collect data to construct predictive models (26, 31). For instance, TCBS agar is widely used to enumerate V. parahaemolyticus colonies and to monitor their growth and survival without enrichment (7). These methods do not count cells in VBNC and stressed states (32) and may underestimate bacterial levels (4, 13). In addition, the presence of background bacteria may negatively impact the enumeration efficiency of the plating media (17). Compared with traditional plate counting methods, molecular quantitative methods have the advantage of quantifying all of the viable bacteria, including cells in VBNC and stressed states (33, 34). In this study, V. parahaemolyticus RNA was extracted and the real-time RT-PCR method was used to construct predictive MMs. RNA is extracted only from live cells, which may give more accurate results when using real-time PCR (35). The tlh gene is used to establish the prediction models as it has been widely used as a species-specific marker for V. parahaemolyticus (36). In the genome of V. parahaemolyticus, the tlh gene has one copy, the tdh gene has two copies, and the trh gene has one copy (37). Before it was used to establish RNA-based models, tlh gene expression was evaluated first. As shown in this study, tlh gene expression is maintained at a constant level at different temperatures and can be used as a reliable target for monitoring survival of V. parahaemolyticus.
Three storage temperatures (0, 4, and 10°C) were tested in this study. According to Fernandez-Piquer et al. (4), V. parahaemolyticus was inactivated when the storage temperature of the oysters was below 12.6°C. V. parahaemolyticus stored at low temperature may enter into the viable but nonculturable (VBNC) or stressed state (38), and this may explain why V. parahaemolyticus was detected in winter months when water temperatures were below 10°C (8). Therefore, to better predict the survival or persistence of V. parahaemolyticus, the Baranyi model was utilized in this study to fit the collected data for establishing the primary TMs and MMs for V. parahaemolyticus in oysters during low-temperature storage (0, 4, and 10°C). The average R2 of the primary TMs was 0.925 and that of the primary MMs was 0.859, indicating that the Baranyi model was appropriate to fit data for the primary models in this study. The Baranyi model has been used to model the survival of high hydrostatic pressure (HHP)-treated Listeria innocua cells (39) and Enterobacter sakazakii in infant formula milk (40) in previous studies. To construct the secondary models, the Arrhenius model was used, as it is a common mathematical model to fit bacterial IR data. Fernandez-Piquer et al. applied the Arrhenius function to fit a secondary model of V. parahaemolyticus in oysters with an R2 value of 0.78 (4). In our study, the R2 values obtained from TM and MM were greater than 0.8, and Bf and Af were also within the satisfactory limit (1.0 < Bf < Af < 1.1), which indicated that the two secondary models have high goodness of fit (41).
According to the primary TMs, the total numbers of viable V. parahaemolyticus cells no longer changed after days 13.861, 13.219, and 10.123 at storage temperatures of 0, 4, and 10°C, respectively. However, according to the primary MMs, the numbers of viable V. parahaemolyticus cells did not change 5.881, 4.237, and 4.425 days after storage at the three respective temperatures (Fig. 3). The significant differences seen here indicate that many cells might have gradually entered the viable but nonculturable (VBNC) state or become stressed cells such that they did not grow on selective agar. Real-time RT-PCR is able to more sensitively and accurately quantify all of the viable cells (33) and provides a more accurate and reliable prediction than TMs.
A five-strain V. parahaemolyticus cocktail, including three nonpathogenic strains with the tlh gene, one pathogenic strain with the tdh gene, and one pathogenic strain with the trh gene, was used in this study. V. parahaemolyticus infections are mainly caused by tdh and trh genes (42–44). In reality, more-complicated V. parahaemolyticus strain mixtures and microfloras exist. Thus, models established by using cocktails made with both nonpathogenic and pathogenic strains mimic real contamination situations. The kinetic changes of the pathogenic V. parahaemolyticus strains in the cocktail were monitored by real-time RT-PCR. The population of pathogenic V. parahaemolyticus with the tdh gene did not decrease significantly, while the population of pathogenic V. parahaemolyticus with the trh gene could not be detected after day 0. This result showed that the V. parahaemolyticus strain with the trh gene had a poor survival capability during cold storage. This could also be the reason why the detection rate of pathogenic V. parahaemolyticus with the trh gene in the environment is low. Ward and Bej used a multiplexed real-time PCR to detect V. parahaemolyticus in oysters, with results that were 51% positive for the tlh gene and 12.1% for the tdh gene but negative for the trh gene (45). In the study conducted by Rizvi and Bej, their results showed that 58% of the 24 oysters tested were positive for the tlh gene, 21% for the tdh gene, and 0.7% for the trh gene (46). The prevalence of the trh gene was always the lowest among the three genes tested in those two studies.
A report by Burnham et al. showed that the levels of pathogenic (tdh-positive) V. parahaemolyticus decreased by 2.880 and 1.490 log10 CFU/ml in TSB during storage at 5°C and 8°C, respectively, for 10 days using the plate count method (30). According to Yang et al., pathogenic (tdh-positive) V. parahaemolyticus in salmon had reductions of 3.50, 2.50, and 1.00 log10 CFU/g during storage at 0°C, 3°C, and 9°C, respectively, for 10 days as shown using the plate count method (6). Although a lower reduction was observed for the tdh strain in our study, there are two potential reasons for the disparity. First of all, the methods used were different. This study used real-time RT-PCR to monitor the pathogenic strains in a cocktail, while the previous studies used plate counting methods. Second, previous studies were done using a single strain and not a cocktail. The interaction between pathogenic strains and nonpathogenic strains used in this study may have also impacted the final results. Regardless of these differences, all of the results and reports showed that pathogenic V. parahaemolyticus can survive for a long period of time in low-temperature storage.
The secondary TM and MM were established based on IRs generated from the primary TMs and MMs (Fig. 5). Due to the different methods (real-time RT-PCR versus TCBS analysis) used for enumerating surviving V. parahaemolyticus cells, the secondary MM is significantly different (P < 0.05) from the secondary TM. In order to validate the predictive models, the secondary TM was evaluated by comparing it to a reported secondary model of V. parahaemolyticus in Pacific oysters (4). The calculated Bf and Af values were not within the satisfactory limit (1.0 ≤ Bf ≤ Af ≤ 1.1), which suggests that the predicted IRs in the secondary TM are different from those of the reported model. In Fig. 6, the TM curve was significantly higher than that of the reported model. The IRs in the TM were significantly lower than those of the reported model at every given temperature (P < 0.05). Differences between the two studies may be due to the different oyster species, the bacterial strains used, bacterial strain variability, and/or the oyster host defenses. For the MM, oysters harvested in summer months were used to validate the model. The calculated Bf and Af values were within the satisfactory limits (1.0 ≤ Bf ≤ Af ≤ 1.1), which indicates that the predicted IRs in the secondary MM are close to those in the validation model in this study. The results from the secondary MM and the validation model were not significantly different (P > 0.05).
In summary, this study developed and validated RNA-based molecular predictive models to describe the survival of V. parahaemolyticus in oysters during low-temperature storage (0, 4, and 10°C). The primary MMs, as they can count all viable V. parahaemolyticus cells, give a more accurate evaluation of the surviving cells than the primary TMs, which were built based on plate counting methods. Among the V. parahaemolyticus strains studied, the level of pathogenic V. parahaemolyticus with the tdh gene showed no significant reduction (P > 0.05). These results reveal that it is important to have a more reliable and accurate predictive model system to better assist with V. parahaemolyticus risk assessment and management in seafood.
MATERIALS AND METHODS
Oysters.
One-year-old Eastern oysters (C. virginica) were harvested from Mobile Bay, AL, USA, and depurated under a flowthrough depuration system for 7 days at the Auburn University Marine Extension and Research Center (AUMERC) located at Dauphin Island, AL, USA. The oysters were then shipped overnight to the Food Microbiology laboratory at Auburn University, Auburn, AL. Upon arrival at the laboratory, oysters were washed to remove excess mud following protocols described by the American Public Health Association for the bacteriological examination of shellfish (47). Oysters were then stored at 0°C before inoculation.
Three uninoculated oysters from each shipment were randomly selected, and the presence of V. parahaemolyticus was checked by both a traditional plating method and real-time RT-PCR. The details of the protocols were the same as those of the procedures used for evaluating inoculated oysters, which are described below. Results from both the plate counting method and the real-time RT-PCR showed that the amounts of V. parahaemolyticus naturally present in the oysters (2 log10 CFU/g for the plate counting method and 2.74 log10 CFU/g for the real-time RT-PCR method) were below the limit of detection.
Bacterial strains.
A total of five V. parahaemolyticus strains (Table 2) were used in this study. Three strains were purchased from the American Type Culture Collection (ATCC) (Manassas, VA, USA), and the other two strains were isolates from previous oyster studies. A liquid V. parahaemolyticus cocktail was prepared by growing individual strains in 10 ml of tryptone soy broth (TSB) (BBL/Difco Laboratories, Sparks, MD, USA) supplemented with 3% NaCl at 37°C for 18 h. The overnight fresh cultures were washed by centrifugation (Eppendorf, Hauppauge, NY, USA) at 3,000 × g for 3 min and resuspended with phosphate-buffered saline (PBS; pH 7.4). The optical density at 600 nm (OD600) of each washed culture was adjusted to 1.7 ± 0.1 by using an Ultrospec 10-cell density meter (Amersham BioSciences, Piscataway, NJ, USA). To mix the cocktail, equal volumes of each washed culture were taken and mixed in a 15-ml Falcon sterile tube (VWR, Atlanta, GA, USA). The concentration of the V. parahaemolyticus cocktail was 1.12 × 108 CFU/ml.
TABLE 2.
Vibrio parahaemolyticus strains used in this study
Oyster inoculation and storage.
To inoculate the oysters, a 5-mm notch was drilled in the shell of each oyster approximately 50 mm from the hinge. After that, 100 μl of inoculum cocktail was injected into the muscle of the oyster using a sterile 1-ml syringe with a 23-gauge needle (Terumo, Somerset, NJ, USA) following the protocol described in previous reports (4, 51). The inoculated oysters were kept flat on the benchtop for 5 min to allow the liquid cultures to be absorbed by the oyster muscle before being stored at different temperatures.
Three storage temperatures and formats were chosen: on ice (∼0°C), in a 4°C refrigerator, and in a 10°C refrigerator. For the 0°C and 4°C storage conditions, three subsamples (three oysters/sample) of inoculated oysters were removed and analyzed once every day for the first week and then once every 2 days for two more weeks. For the 10°C storage condition, oysters were removed and analyzed every 12 h for the first week and then on days 9 and 11.
V. parahaemolyticus enumeration using the plate count method.
TCBS plates (BD, Sparks, MD, USA) were used to plate and enumerate the surviving V. parahaemolyticus bacteria. At each sampling point, three sets of three oysters were taken from each storage condition. The oysters were shucked, and the meat was removed from the shell. Approximately 25 g of oyster meat from each storage condition was then placed in a filtered Whirl-Pak bag (Nasco, Fort Atkinson, WI, USA) with 225 ml of PBS. The mixture was homogenized by the use of a Smasher laboratory blender (AES Chemunex, bioMérieux, France) at the normal speed for 2 min. The homogenized samples were diluted and plated. Colonies were enumerated after the plates were incubated at 37°C for 18 h. In addition, 1 ml of every homogenized sample was removed, transferred into a 15-ml Falcon sterile tube (VWR, Atlanta, GA, USA), and stored at −80°C with 2 ml of RNA Protect reagent (Qiagen, Hilden, Germany) for RNA extraction.
RNA extraction.
In order to quantify the number of V. parahaemolyticus bacteria containing tlh, tdh, and trh genes using real-time RT-PCR, standard curves for the three target genes were established. To extract the RNA for establishment of the tlh, tdh, and trh gene-based standard curves, 10-fold serial dilutions of the Vibrio parahaemolyticus cocktail, V. parahaemolyticus ATCC 43996, and V. parahaemolyticus ATCC 17082 were used, starting at cell densities of 5.5 × 107, 7.6 × 107, and 6 × 107 CFU/ml. RNA was extracted from 1 ml of each dilution using an RNeasy minikit (Qiagen, Valencia, CA, USA). During the RNA extraction, 80 μl of DNase I stock solution (Qiagen, Valencia, CA, USA) (a total of 30 units) was added to clean up any potential DNA contamination.
To extract RNA from inoculated oyster samples, 1 ml of each stored homogenized sample solution (from step 4) was used. The same RNeasy minikit was used to extract the RNA, and all of the RNA samples were stored immediately and maintained at −80°C until they were analyzed.
The concentration and purity of all RNA samples were analyzed using a NanoVue Plus spectrophotometer (GE Healthcare, Piscataway, NJ, USA). cDNA was generated by reverse transcription of 10 μl of RNA from each sample using a High Capacity cDNA reverse transcription kit (Applied Biosystems, Foster City, CA, USA) and then used for real-time PCR. The reverse transcription program consisted of 25°C for 10 min, 37°C for 120 min, and 85°C for 5 min.
Real-time PCR.
The primers used to target the tlh gene are F-tlh (5′-ACTCAACACAAGAAGAGATCGACAA-3′) and R-tlh (5′-GATGAGCGGTTGATGTCCAA-3′); the primers used for tdh are F-tdh (5′-TCCCTTTTCCTGCCCCC-3′) and R-tdh (5′-CGCTGCCATTGTATAGTCTTTATC-3′); and the primers used for trh are F-trh (5′-TTGCTTTCAGTTTGCTATTGGCT-3′) and R-trh (5′-TGTTTACCGTCATATAGGCGCTT-3′) (52). All of the primers were synthesized by Integrated DNA Technologies (Coralville, IA, USA).
SYBR green real-time PCR was carried out on an ABI 7500 system (Applied Biosystems, Foster City, CA, USA). Each reaction system consisted of 10 μl of PerfeCTa SYBR green SuperMix (Quanta Biosciences, Gaithersburg, MD, USA), 0.5 μl each of 10 μM forward and reverse primers, 6 μl of RNase-free water, and 3 μl of cDNA templates. Real-time PCR assays were conducted following a program with an initial denaturing period at 95°C for 10 min and then 40 cycles of 95°C for 15 s and 60°C for 1 min. Standard curves were built by constructing regression lines, with the x axis representing CT values and y axis representing log10 CFU per milliliter. The standard curves were used to quantitatively calculate the levels of surviving V. parahaemolyticus cells in inoculated oyster samples.
To monitor the survival of the pathogenic V. parahaemolyticus strains in the cocktail, real-time RT-PCR targeting the tdh and trh genes was conducted. The setup of PCRs was the same as described above.
Evaluation of tlh gene expression at low temperatures.
Real-time RT-PCR was used to monitor expression of the tlh gene in V. parahaemolyticus during storage at 0, 4, and 10°C. The relative quantification data of gene expression were calculated by a 2−ΔΔCT method. Using this method, the data were presented as the fold change in gene expression normalized to the endogenous reference gene and relative to that of the calibrator as follows: ΔΔCT = (CTtarget − CTreference)target − (CTtarget − CTreference)calibrator (53). Two housekeeping genes (pvsA and recA) were used as the endogenous reference genes for tlh gene expression monitoring. The primers for pvsA are F-pvsA (5′-CTCCTTCATCCAACACGAT-3′) and R-pvsA (5′-GGGCGAGATAATCCTTGT-3′) (54); the primers for recA are F-recA (5′-GCTAGTAGAAAAAGCGGGTG-3′) and R-recA (5′-GCAGGTGCTTCTGGTTGAG-3′) (58). The expression level of the tlh gene on day 0 was used as the calibrator.
RNA was extracted from inoculated oyster samples at 0, 4, and 10°C on days 0, 3, 6, 9, and 11 using an RNeasy minikit (Qiagen, Hilden, Germany). First, 10 μl of RNA from each sample was transcribed into cDNA using a High Capacity cDNA reverse transcription kit (Applied Biosystems, Foster City, CA, USA), and real-time PCR was then conducted. The reaction system was the same as described above. Triplicates were conducted.
Mathematical modeling.
The raw data obtained from the plate counting method and real-time RT-PCR method were transformed to a natural logarithm format. For the survival model, the Baranyi equation (see equation 3) was chosen to fit the data, and the calculations were performed using the DMfit tool of the Combase website (available at http://www.ifr.ac.uk/safety/dmfit/). The output data were then converted into the log10 format. The means and standard deviations were calculated. The fitted model based on the plate counting method was defined as the primary TM, and the model based on the real-time RT-PCR method was defined as the primary MM. Parameters of coefficient determination (R2), lag time (LT), minimum inactivation rate (IR), initial value (IV), and final value (FV) were obtained for the models. The Baranyi equation is as follows:
| (3) |
where y is the natural logarithm of the bacteria concentration at any given time (ln CFU per milliliter), y0 and yend are the initial value and the end value of y, A(t) is the equation governing the duration of the period (informally called the “shoulder”) preceding the log linear inactivation phase, t is time (day), m determines the smoothness of the transition from the exponential inactivation phase to the survival tail, μmin is the minimum value of the inactivation rate, ν is the rate at which the bacteria lose the ability to survive during the shoulder, and q0 is the initial physiological state of bacterial cells.
The Arrhenius equation (equation 4) was applied for the establishment of the secondary model to describe the IR of V. parahaemolyticus in oysters as a function of temperature. The equation is as follows:
| (4) |
where r is the inactivation rate (IR), T is the absolute temperature, Ea is the activation energy, R is the universal gas constant, and A is the collision factor. The equation was input into OriginPro 8.0 software (OriginLab Corporation, Northampton, MA, USA), which was then used to fit the IR data to obtain a plot of ln r against T and the values of ln A and Ea/R.
Evaluation and validation of models.
In order to evaluate the goodness of fit of the TMs and MMs, the coefficient of determination (R2), bias factor (Bf), and accuracy factor (Af) were calculated by using equations 5, 6, and 7 (17). The primary TMs were compared to the primary MMs through analysis of R2, LT, IR, IV, and FV values using one-way analysis of variance (ANOVA). To validate the MMs, the secondary MM was compared with a secondary validation model that was built by conducting a replication experiment with a new batch of harvested Eastern oysters using the same inoculation and storage processes. To validate the TMs, the fitted secondary TM of V. parahaemolyticus was compared with a model previously published by Fernandez-Piquer et al. (4). Differences between predicted IR data in the secondary models and validation models were evaluated by using one-way ANOVA, Bf, and Af as follows:
| (5) |
| (6) |
| (7) |
where n represents the number of observations and “predicted,” “observed,” and “mean” represent the predicted values, observed values, and average values, respectively.
In this study, R2 greater than 0.8 for the predictive models was considered representative of a good fitting performance (55). Bf and Af were calculated to evaluate the goodness of fit of the primary models and to validate the secondary models. Bf measures the relative average deviation of predicted and observed values. Af indicates the spread of the results around the predicted values. The 1.0 ≤ Bf ≤ Af ≤ 1.1 range was defined as a satisfactory limit (17).
Statistical analysis.
One-way ANOVA was applied using OriginPro 8.0 software (OriginLab Corporation, Northampton, MA, USA) to compare the different predictive models. P < 0.05 was regarded as representative of a statistically significant difference between models (4, 13, 29).
REFERENCES
- 1.Su YC, Liu C. 2007. Vibrio parahaemolyticus: a concern of seafood safety. Food Microbiol 24:549–558. doi: 10.1016/j.fm.2007.01.005. [DOI] [PubMed] [Google Scholar]
- 2.Iwamoto M, Ayers T, Mahon BE, Swerdlow DL. 2010. Epidemiology of seafood-associated infections in the United States. Clin Microbiol Rev 23:399–411. doi: 10.1128/CMR.00059-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ye M, Huang Y, Chen H. 2012. Inactivation of Vibrio parahaemolyticus and Vibrio vulnificus in oysters by high-hydrostatic pressure and mild heat. Food Microbiol 32:179–184. doi: 10.1016/j.fm.2012.05.009. [DOI] [PubMed] [Google Scholar]
- 4.Fernandez-Piquer J, Bowman JP, Ross T, Tamplin ML. 2011. Predictive models for the effect of storage temperature on Vibrio parahaemolyticus viability and counts of total viable bacteria in Pacific oysters (Crassostrea gigas). Appl Environ Microbiol 77:8687–8695. doi: 10.1128/AEM.05568-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.DaSilva L, Parveen S, DePaola A, Bowers J, Brohawn K, Tamplin ML. 2012. Development and validation of a predictive model for the growth of Vibrio vulnificus in postharvest shellstock oysters. Appl Environ Microbiol 78:1675–1681. doi: 10.1128/AEM.07304-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yang ZQ, Jiao XA, Li P, Pan ZM, Huang JL, Gu RX, Fang WM, Chao GX. 2009. Predictive model of Vibrio parahaemolyticus growth and survival on salmon meat as a function of temperature. Food Microbiol 26:606–614. doi: 10.1016/j.fm.2009.04.004. [DOI] [PubMed] [Google Scholar]
- 7.Yoon KS, Min KJ, Jung YJ, Kwon KY, Lee JK, Oh SW. 2008. A model of the effect of temperature on the growth of pathogenic and nonpathogenic Vibrio parahaemolyticus isolated from oysters in Korea. Food Microbiol 25:635–641. doi: 10.1016/j.fm.2008.04.007. [DOI] [PubMed] [Google Scholar]
- 8.Parveen S, Hettiarachchi KA, Bowers JC, Jones JL, Tamplin ML, McKay R, Beatty W, Brohawn K, DaSilva LV, DePaola A. 2008. Seasonal distribution of total and pathogenic Vibrio parahaemolyticus in Chesapeake Bay oysters and waters. Int J Food Microbiol 128:354–361. doi: 10.1016/j.ijfoodmicro.2008.09.019. [DOI] [PubMed] [Google Scholar]
- 9.Sobrinho PDSC, Destro MT, Franco BD, Landgraf M. 2014. A quantitative risk assessment model for Vibrio parahaemolyticus in raw oysters in Sao Paulo State, Brazil. Int J Food Microbiol 180:69–77. doi: 10.1016/j.ijfoodmicro.2014.04.008. [DOI] [PubMed] [Google Scholar]
- 10.Elexson N, Son R, Rukayadi Y, Zainazor TT, Ainy MN, Nakaguchi Y, Mitsuaki N. 2013. Biosafety of Vibrio parahaemolyticus biofilm from seafood using herbs and spices. J Life Med 1:71–82. doi: 10.14511/jlm.2013.010305. [DOI] [Google Scholar]
- 11.Parveen S, DaSilva L, DePaola A, Bowers J, White C, Munasinghe KA, Brohawn K, Mudoh M, Tamplin M. 2013. Development and validation of a predictive model for the growth of Vibrio parahaemolyticus in post-harvest shellstock oysters. Int J Food Microbiol 161:1–6. doi: 10.1016/j.ijfoodmicro.2012.11.010. [DOI] [PubMed] [Google Scholar]
- 12.Food and Drug Administration (FDA), Interstate Shellfish Sanitation Conference (ISSC). 2013. National Shellfish Sanitation Program (NSSP): guide for the control of molluscan shellfish. 2013 revision. FDA, Washington, DC. [Google Scholar]
- 13.Liao C, Peng ZY, Li JB, Cui XW, Zhang ZH, Malakar PK, Zhang WJ, Pan YJ, Zhao Y. 2015. Simultaneous construction of PCR-DGGE-based predictive models of Listeria monocytogenes and Vibrio parahaemolyticus on cooked shrimps. Lett Appl Microbiol 60:210–216. doi: 10.1111/lam.12376. [DOI] [PubMed] [Google Scholar]
- 14.Zhang ZH, Liu HQ, Lou Y, Xiao LL, Liao C, Malakar PK, Pan YJ, Zhao Y. 2015. Quantifying viable Vibrio parahaemolyticus and Listeria monocytogenes simultaneously in raw shrimp. Appl Microbiol Biotechnol 99:6451–6462. doi: 10.1007/s00253-015-6715-x. [DOI] [PubMed] [Google Scholar]
- 15.Ramamurthy T, Ghosh A, Pazhani GP, Shinoda S. 2014. Current perspectives on viable but non-culturable (VBNC) pathogenic bacteria. Front Public Health 2:103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Su CP, Jane WN, Wong HC. 2013. Changes of ultrastructure and stress tolerance of Vibrio parahaemolyticus upon entering viable but non-culturable state. Int J Food Microbiol 160:360–366. doi: 10.1016/j.ijfoodmicro.2012.11.012. [DOI] [PubMed] [Google Scholar]
- 17.Ye K, Wang H, Zhang X, Jiang Y, Xu X, Zhou G. 2013. Development and validation of a molecular predictive model to describe the growth of Listeria monocytogenes in vacuum-packaged chilled pork. Food Control 32:246–254. doi: 10.1016/j.foodcont.2012.11.017. [DOI] [Google Scholar]
- 18.Rudi K, Moen B, Dromtorp SM, Holck AL. 2005. Use of ethidium monoazide and PCR in combination for quantification of viable and dead cells in complex samples. Appl Environ Microbiol 71:1018–1024. doi: 10.1128/AEM.71.2.1018-1024.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wagner AO, Malin C, Knapp BA, Illmer P. 2008. Removal of free extracellular DNA from environmental samples by ethidium monoazide and propidium monoazide. Appl Environ Microbiol 74:2537–2539. doi: 10.1128/AEM.02288-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liu Y, Mustapha A. 2014. Detection of viable Escherichia coli O157: H7 in ground beef by propidium monoazide real-time PCR. Int J Food Microbiol 170:48–54. doi: 10.1016/j.ijfoodmicro.2013.10.026. [DOI] [PubMed] [Google Scholar]
- 21.Taylor MJ, Bentham RH, Ross KE. 2014. Limitations of using propidium monoazide with qPCR to discriminate between live and dead legionella in biofilm samples. Microbiol Insights 7:15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nocker A, Camper AK. 2009. Novel approaches toward preferential detection of viable cells using nucleic acid amplification techniques. FEMS Microbiol Lett 291:137–142. doi: 10.1111/j.1574-6968.2008.01429.x. [DOI] [PubMed] [Google Scholar]
- 23.Nocker A, Sossa-Fernandez P, Burr MD, Camper AK. 2007. Use of propidium monoazide for live/dead distinction in microbial ecology. Appl Environ Microbiol 73:5111–5117. doi: 10.1128/AEM.02987-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tse TPY. 2015. Virulence gene expression of Vibrio parahaemolyticus in the viable but nonculturable state. PhD dissertation California Polytechnic State University, San Luis Obispo, CA. [Google Scholar]
- 25.Postollec F, Falentin H, Pavan S, Combrisson J, Sohier D. 2011. Recent advances in quantitative PCR (qPCR) applications in food microbiology. Food Microbiol 28:848–861. doi: 10.1016/j.fm.2011.02.008. [DOI] [PubMed] [Google Scholar]
- 26.Tang X, Zhao Y, Sun X, Xie J, Pan Y, Malakar PK. 2015. Predictive model of Vibrio parahaemolyticus O3:K6 growth on cooked Litopenaeus vannamei. Ann Microbiol 65:487–493. doi: 10.1007/s13213-014-0884-1. [DOI] [Google Scholar]
- 27.Meng L, Alter T, Aho T, Huehn S. 2015. Gene expression profiles of Vibrio parahaemolyticus in viable but non-culturable state. FEMS Microbiol Ecol 91:fiv035. doi: 10.1093/femsec/fiv035. [DOI] [PubMed] [Google Scholar]
- 28.Boonyawantang A, Mahakarnchanakul W, Rachtanapun C, Boonsupthip W. 2012. Behavior of pathogenic Vibrio parahaemolyticus in prawn in response to temperature in laboratory and factory. Food Control 26:479–485. doi: 10.1016/j.foodcont.2012.02.009. [DOI] [Google Scholar]
- 29.Miles DW, Ross T, Olley J, McMeekin TA. 1997. Development and evaluation of a predictive model for the effect of temperature and water activity on the growth rate of Vibrio parahaemolyticus. Int J Food Microbiol 38:133–142. doi: 10.1016/S0168-1605(97)00100-1. [DOI] [PubMed] [Google Scholar]
- 30.Burnham VE, Janes ME, Jakus LA, Supan J, DePaola A, Bell J. 2009. Growth and survival differences of Vibrio vulnificus and Vibrio parahaemolyticus strains during cold storage. J Food Sci 74:M314–M318. doi: 10.1111/j.1750-3841.2009.01227.x. [DOI] [PubMed] [Google Scholar]
- 31.Lee YJ, Jung BS, Yoon HJ, Kim KT, Paik HD, Lee JY. 2014. Predictive model for the growth kinetics of Listeria monocytogenes in raw pork meat as a function of temperature. Food Control 44:16–21. doi: 10.1016/j.foodcont.2014.03.024. [DOI] [Google Scholar]
- 32.Willenburg E, Divol B. 2012. Quantitative PCR: an appropriate tool to detect viable but not culturable Brettanomyces bruxellensis in wine. Int J Food Microbiol 160:131–136. doi: 10.1016/j.ijfoodmicro.2012.09.012. [DOI] [PubMed] [Google Scholar]
- 33.Dinu LD, Bach S. 2013. Detection of viable but non-culturable Escherichia coli O157: H7 from vegetable samples using quantitative PCR with propidium monoazide and immunological assays. Food Control 31:268–273. doi: 10.1016/j.foodcont.2012.10.020. [DOI] [Google Scholar]
- 34.Mishra A, Taneja N, Sharma M. 2012. Viability kinetics, induction, resuscitation and quantitative real-time polymerase chain reaction analyses of viable but nonculturable Vibrio cholerae O1 in freshwater microcosm. J Appl Microbiol 112:945–953. doi: 10.1111/j.1365-2672.2012.05255.x. [DOI] [PubMed] [Google Scholar]
- 35.Derveaux S, Vandesompele J, Hellemans J. 2010. How to do successful gene expression analysis using real-time PCR. Methods 50:227–230. doi: 10.1016/j.ymeth.2009.11.001. [DOI] [PubMed] [Google Scholar]
- 36.López-Hernández KM, Pardío-Sedas VT, Lizárraga-Partida L, Williams JDJ, Martínez-Herrera D, Flores-Primo A, Rendón-Castro K. 2015. Environmental parameters influence on the dynamics of total and pathogenic Vibrio parahaemolyticus densities in Crassostrea virginica harvested from Mexico's Gulf coast. Mar Pollut Bull 91:317–329. doi: 10.1016/j.marpolbul.2014.11.015. [DOI] [PubMed] [Google Scholar]
- 37.Shalu OA, Pisanov RV, Monakhova EV. 2012. Efficiency of Vibrio parahaemolyticus tdh gene expression depends upon two point mutations in its promoter region. Russ J Genet 48:1177–1183. doi: 10.1134/S1022795412120125. [DOI] [PubMed] [Google Scholar]
- 38.Chen SY, Jane WN, Chen YS, Wong HC. 2009. Morphological changes of Vibrio parahaemolyticus under cold and starvation stresses. Int J Food Microbiol 129:157–165. doi: 10.1016/j.ijfoodmicro.2008.11.009. [DOI] [PubMed] [Google Scholar]
- 39.Saucedo-Reyes D, Marco-Celdrán A, Pina-Pérez MC, Rodrigo D, Martínez-López A. 2009. Modeling survival of high hydrostatic pressure treated stationary-and exponential-phase Listeria innocua cells. Innov Food Sci Emerg Technol 10:135–141. doi: 10.1016/j.ifset.2008.11.004. [DOI] [Google Scholar]
- 40.Pina Pérez MC, Rodrigo Aliaga D, Saucedo Reyes D, Martínez López A. 2007. Pressure inactivation kinetics of Enterobacter sakazakii in infant formula milk. J Food Prot 70:2281–2289. doi: 10.4315/0362-028X-70.10.2281. [DOI] [PubMed] [Google Scholar]
- 41.Kramer M. 2005. R2 statistics for mixed models, p 148–160. Proceedings of the Annual Conference on Applied Statistics in Agriculture. New Prairie Press at Kansas State University, Manhattan, KS. [Google Scholar]
- 42.Matsuda S, Kodama T, Okada N, Okayama K, Honda T, Iida T. 2010. Association of Vibrio parahaemolyticus thermostable direct hemolysin with lipid rafts is essential for cytotoxicity but not hemolytic activity. Infect Immun 78:603–610. doi: 10.1128/IAI.00946-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shimohata T, Takahashi A. 2010. Diarrhea induced by infection of Vibrio parahaemolyticus. J Med Invest 57:179–182. doi: 10.2152/jmi.57.179. [DOI] [PubMed] [Google Scholar]
- 44.Yanagihara I, Nakahira K, Yamane T, Kaieda S, Mayanagi K, Hamada D, Fukui T, Ohnishi K, Kajiyama S, Shimizu T, Sato M, Ikegami T, Ikeguchi M, Honda T, Hashimoto H. 2010. Structure and functional characterization of Vibrio parahaemolyticus thermostable direct hemolysin. J Biol Chem 285:16267–16274. doi: 10.1074/jbc.M109.074526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ward LN, Bej AK. 2006. Detection of Vibrio parahaemolyticus in shellfish by use of multiplexed real-time PCR with TaqMan fluorescent probes. Appl Environ Microbiol 72:2031–2042. doi: 10.1128/AEM.72.3.2031-2042.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rizvi AV, Bej AK. 2010. Multiplexed real-time PCR amplification of tlh, tdh and trh genes in Vibrio parahaemolyticus and its rapid detection in shellfish and Gulf of Mexico water. Antonie Van Leeuwenhoek 98:279–290. doi: 10.1007/s10482-010-9436-2. [DOI] [PubMed] [Google Scholar]
- 47.Hunt DA, Miescier J, Redman J, Salinger A, Lucas JP. 1984. Molluscan shellfish, fresh or fresh frozen oysters, mussels or clams, p 590–610. In Speck M. (ed), Compendium of methods for the microbiological examination of foods. American Public Health Association Inc., Washington, DC. [Google Scholar]
- 48.Fujino T. 1965. Taxonomic studies on the bacterial strains isolated from cases of “Shirasu” food poisoning (Pasteurella parahaemolytica) and related microorganisms. Biken J 8:63–71. [PubMed] [Google Scholar]
- 49.Barrow GI. 1974. Microbiological and other hazards from seafoods with special reference to Vibrio parahaemolyticus. Postgrad Med J 50:612–619. doi: 10.1136/pgmj.50.588.612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Colwell RR. 1970. Polyphasic taxonomy of the genus Vibrio: numerical taxonomy of Vibrio cholerae, Vibrio parahaemolyticus, and related Vibrio species. J Bacteriol 104:410–433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Garnier M, Labreuche Y, Garcia C, Robert M, Nicolas JL. 2007. Evidence for the involvement of pathogenic bacteria in summer mortalities of the Pacific oyster Crassostrea gigas. Microb Ecol 53:187–196. doi: 10.1007/s00248-006-9061-9. [DOI] [PubMed] [Google Scholar]
- 52.Nordstrom JL, Vickery MC, Blackstone GM, Murray SL, DePaola A. 2007. Development of a multiplex real-time PCR assay with an internal amplification control for the detection of total and pathogenic Vibrio parahaemolyticus bacteria in oysters. Appl Environ Microbiol 73:5840–5847. doi: 10.1128/AEM.00460-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Livak KJ, Schmittgen TD. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 54.Coutard F, Lozach S, Pommepuy M, Hervio-Heath D. 2007. Real-time reverse transcription-PCR for transcriptional expression analysis of virulence and housekeeping genes in viable but nonculturable Vibrio parahaemolyticus after recovery of culturability. Appl Environ Microbiol 73:5183–5189. doi: 10.1128/AEM.02776-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zou KH, Tuncali K, Silverman SG. 2003. Correlation and simple linear regression 1. Radiology 227:617–628. doi: 10.1148/radiol.2273011499. [DOI] [PubMed] [Google Scholar]
- 56.Arrhenius S. 1915. Quantitative laws in biological chemistry. G. Bell and Sons, Ltd., London, United Kingdom. [Google Scholar]
- 57.Baranyi J, Roberts TA. 1994. A dynamic approach to predicting bacterial growth in food. Int J Food Microbiol 23:277–294. doi: 10.1016/0168-1605(94)90157-0. [DOI] [PubMed] [Google Scholar]
- 58.Ma YJ, Sun XH, Xu XY, Zhao Y, Pan YJ, Hwang CA, Wu VC. 2015. Investigation of reference genes in Vibrio parahaemolyticus for gene expression analysis using quantitative RT-PCR. PLoS One 10:e0144362. doi: 10.1371/journal.pone.0144362. [DOI] [PMC free article] [PubMed] [Google Scholar]