Abstract
The complex allotetraploid genome is one of major challenges in cotton for repressing gene expression. Developing site-specific DNA mutation is the long-term dream for cotton breeding scientists. The clustered regularly interspaced short palindromic repeats/CRISPR-associated protein 9 (CRISPR/Cas9) system is emerging as a robust biotechnology for targeted-DNA mutation. In this study, two sgRNAs, GhMYB25-like-sgRNA1 and GhMYB25-like-sgRNA2, were designed in the identical genomic regions of GhMYB25-like A and GhMYB25-like D, which were encoded by cotton A subgenome and the D subgenome, respectively, was assembled to direct Cas9-mediated allotetraploid cotton genome editing. High proportion (14.2–21.4%) CRISPR/Cas9-induced specific truncation events, either from GhMYB25-like A DNA site or from GhMYB25-like D DNA site, were detected in 50% examined transgenic cotton through PCR amplification assay and sequencing analyses. Sequencing results also demonstrated that 100% and 98.8% mutation frequency were occurred on GhMYB25-like-sgRNA1 and GhMYB25-like-sgRNA2 target site respectively. The off-target effect was evaluated by sequencing two putative off-target sites, which have 3 and 1 mismatched nucleotides with GhMYB25-like-sgRNA1 and GhMYB25-like-sgRNA2, respectively; all the examined samples were not detected any off-target-caused mutation events. Thus, these results demonstrated that CRISPR/Cas9 is qualified for generating DNA level mutations on allotetraploid cotton genome with high-efficiency and high-specificity.
Cotton fiber and its derivative products play crucial roles in our daily life and the world economy, which had been estimated to directly determine the annual income of almost 100 million families from approximately 150 countries1. Its annually worldwide economic impact had been assessed at approximately US$500 billion1,2. The widely cultivated cotton cultivars are allotetraploid species, which consist of two set of subgenomes, “A subgenome” and “D subgenome”. Those two subgenomes cotton species reunited geographically by the transoceanic dispersal happened approximately 1–2 million years ago (MYA)3. This polyploidization confers many excellent properties on tetraploid cotton, including longer cotton fiber length and higher cotton fiber strength, which make it possible to cultivate modern spinnable cotton cultivars. However, the complex genome feature of allotetraploid cotton presents a new challenge for cotton genes functional analyses and the genetic improvement through transgenic approach. Previously, many cotton genes were identified to be implicated in cotton fibre development4,5,6,7,8,9,10,11,12,13,14,15,16, stress responsees17,18,19,20, and pathogen immune regulation through expressed sequence tag (EST)-based cDNA library coupled with conventional RNAi and gene overexpression strategies21,22,23,24. With the rapid development of high throughput deep sequencing technologies and bioinformatics, many diploid and allotetraploid cotton species were sequenced and assembled within recent years, including D-genome diploids cotton Gossypium raimondii (DD; 2n = 26)25,26, A-genome diploids Gossypium arboreum (AA; 2n = 26)27, ant the AtDt allotetraploid cotton species Gossypium hirsutum and Gossypium barbadense28,29,30. Those excellent contributions extremely facilitate gene identification of interest and subsequent vector construction for functional genes analyses and screening via ‘genotype-to-phenotype’ approach. Thus, the arsenal of cotton genomic manipulation urgently require to be updated to meet the demand for rapid and precise dissecting gene functional analyses.
Extensive studies have shown that high-frequent creating DNA double-strand breaks (DSBs) in desired nuclear DNA sites is a reliable approache to induce gene mutations31. DSBs can trigger two distinct endogenous DNA repair mechanisms, error-prone nonhomologous end joining (NHEJ) and homology-directed repair (HDR)32,33, respectively. For NHEJ-mediated repair, it simply bring the break ends together and rejoin them by DNA ligation without the guidance from homologous template. Therefore, NHEJ was considered as a “quick and dirty” approach to repair DSB-caused DNA breaks and frequently lead to indel mutations and loss of nucleotides at the repair sites33,34,35. In contrast to NHEJ, the completion of DHR depends on the homologous sequence as a guide template, which usually is its undamaged sister chromatid, and so produces more accurate repair than NHEJ34,35,36.
To artificially generate targeted DSBs in the genomic region of interest, several nucleases, including meganucleases (MN), zinc-finger nucleases (ZFN), transcription activator-like effector nucleases (TALEN), were engineered to catalyze site-specific cleavage through fusing a programmable, sequence-specific DNA-binding domain with a cleavage domain37,38,39,40,41,42,43,44.
The type II RNA-guided CRISPR/Cas9 system, which derived from the adaptive immunity mechanism of bacteria Streptococcus pyogenes, has been recently proven to be effective for targeted gene editing in a wildly range of organisms, including human cells, mice, zebrafish, bacteria, yeast, Arabidopsis thaliana, Nicotiana benthamiana, maize, wheat, populus, grape sorghum and rice36,45,46,47,48,49,50,51,52,53,54,55,56,57,58,59,60,61,62,63,64. The CRISPR/Cas9 system is composed of a Cas9 nuclease sequence and two noncoding RNA genes, a precursor CRISPR RNA (pre-crRNA) and a trans-activating crRNA (tracrRNA). By replacing those two RNA genes with an engineered single guide RNA (sgRNA), the sgRNA-Cas9 complex can specifically recognizes complementary DNA targets sequence that immediately upstream of a 5′-NGG or 5′-NAG protospacer adjacent motif (PAM) sequence through Watson-Crick base pairing, and then catalyzing a site-specific cleavage on the targeted DNA sequence 3–4 base pairs upstream of the PAM site. In this study, a set of CRISPR/Cas9 genome editing system was firstly proved to possess the feature of high-efficiency and high-specificity on allotetraploid cotton genome editing, which may extremely enhance cotton genomic study and application.
Results
Experimental design and Golden Gate assembly of sgRNAs
CRISPR/Cas9 technology is emerging as important genome manipulation techniques for precisely gene targeting and DNA editing. Given that the genome complexity of allotetraploid upland cotton, we sought to develop a high-efficient and time-saving CRISPR/Cas9 system for cotton research community. Based on optimizing maize-codon Cas9 protein and simplifying the assembly process of sgRNAs, Xing and colleagues (2014) validated the high efficiency and specificity of a set of CRISPR/Cas9 toolkit in model plant Arabidopsis45. Most importantly, the multiple-gene mutations could be transmitted to their progenies with the efficiency can reach up to 100%45.
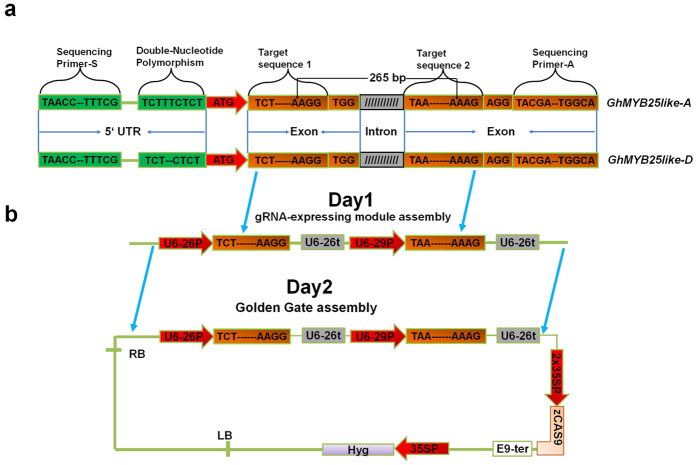
Based on screening bacterial artificial chromosomes (BACs) library, two GhMYB25-like cDNAs, referred to as GhMYB25-like A and GhMYB25-like D, were identified from upland cotton6. Although GhMYB25-like A and GhMYB25-like D are encoded by allotetraploid cotton A subgenome and the D subgenome respectively, they share a similar gene structure and highly conserved R2R3-binding domain6. Previous study show that GhMYB25-like play important roles in cotton fiber development6. Thus, GhMYB25-like A and GhMYB25-like D, which derived from A subgenome and the D subgenome, respectively, are optimum candidates for validating the effectiveness of CRISPR/Cas9 genome editing system. Through searching the DNA sequence of GhMYB25-like A and GhMYB25-like D, two 23-bp 5′-N20NGG-3′ types of genomic DNA sequence were chosen as target sites for designing CRISPR/Cas9 vectors (Fig. 1a).
Figure 1. Overview of CRISPR–Cas9-guided genome editing in GhMYB25-like A and GhMYB25-like D genes.
(a) Schematic description of selected DNA region of GhMYB25-like A and GhMYB25-like D. Two 23-bp 5′-N20NGG-3′ types of genomic DNA sequence are derived from the exons regions of GhMYB25-like A and GhMYB25-like D. The target sequences lying immediately upstream of the PAM sequence, TGG and AGG, were selected as guide sequence. The designed cleavage length is 265 bp. A pair of sequencing primer will be used for checking the effectiveness and efficiency of targeted DNA sites. And there are several single-nucleotide polymorphism and double-nucleotide polymorphism within sequencing region, which can be used to distinguish gene GhMYB25-like A and gene GhMYB25-like D. (b) Diagrams illustrating the procedure of vector construction. The upper panel is the premade gRNA-expressing modules. U6-26p and U6-29p are Arabidopsis U6 gene promoters, and U6-26t is Arabidopsis U6 gene terminators. The lower panel is the main functional component of this CRISPR/Cas9 binary vectors between the RB and LB. Both Cas9 protein and Hyg selectable markers are driven by 2X35 S promoter. ZCas9 and Hyg are Zea mays codon-optimized Cas9 sequence and hygromycin respectively.
By using PCR-based sgRNA(guide RNA) assembly system, two sgRNAs, GhMYB25-like-sgRNA1 and GhMYB25-like-sgRNA2, were rapidly introduced into sgRNA-expressing module with just one round of PCR reaction. As shown in Fig. 1b, the expressions of GhMYB25-like-sgRNA1 and GhMYB25-like-sgRNA2 were driven by Arabidopsis Pol III promoters, U6-26p and U6-29p, respectively. And each of GhMYB25-like-sgRNA1 and GhMYB25-like-sgRNA2 have their own terminators, U6-26t (Fig. 1b). The application of sgRNA-expressing module vectors facilitates the assembly process, and meanwhile guarantee the accuracy of those sgRNA expression cassettes. In my case, one day is sufficient to accomplish PCR amplification and PCR products purification. In the following step, the Type IIS restriction endonucleases (REases) BsaI, was employed to seamlessly integrate maize-codon optimized Cas9 and two GhMYB25-like sgRNA-expressing cassettes (Fig. 1b), which could be finished within 6 hours.
Gene transformation and evaluation of CRISPR/Cas9-mediated mutagenesis in cotton GhMYB25-like A and GhMYB25-like D
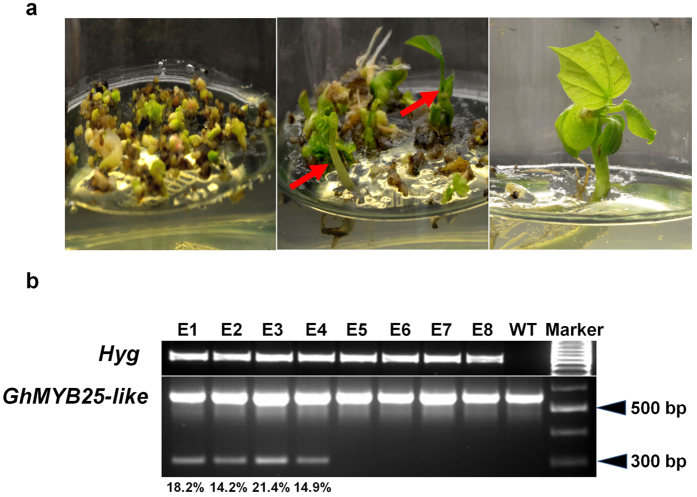
Agrobacterium tumefaciens (A. tumefaciens)-mediated cotton transformation and somatic embryogenesis were performed as described previously23,65,66. With several round of tissue subculture and antibiotic selection on selective medium, a lot of hygromycin-resistant cotyledon-stage embryos were generated from original explant (Left panel, Fig. 2a), excised cotton hypocotyl segments. Those antibiotic-resistant embryos continued to be cultured on hygromycin-containing medium and many plantlets were produced on selective medium (middle panel, Fig. 2a). Two plantlets, as shown by red arrow on Fig. 2a, from each independent transgenic event were sampled for DNA extraction and subsequent mutation analyses. To validate the exogenous T-DNA insertion in GhMYB25-like transgenic plantlets, DNAs extracted from twelve independent transgenic events were analyzed by PCR assay using gene specific primers for the hygromycin resistant gene. And eight DNA samples were detected the correct exogenous T-DNA insertion (Upper panel, Fig. 2b), which were referred to E1, E2, E3, E4, E5, E6, E6, E8, respectively.
Figure 2. Cotton transformation and identification of positive plantlets for mutation analyses.
(a) Regeneration of cotton GhMYB25-like transgenic plants. Sterilized cotton hypocotyl were excised into 5–8 mm segments for infecting with GhMYB25-like::CRISPR/Cas9-containing A. tumefaciens strain EHA105. Left panel is the hygromycin-resistant cotyledonary stage embryos generated from original hypocotyl segments, middle and right panel is part of regeneration plantlets. (b) Detection of CRISPR/Cas9-induced long DNA fragment deletions. The common PCR primers of GhMYB25-like A and GhMYB25-like D were synthesized for double-cleavage mutation analyses and genes cloning.
To evaluate the potential genomic DNA deletion occurred on the designed GhMYB25-like genomic regions, a pair of primers, covering the similar genomic area of GhMYB25-like A and GhMYB25-like D (Fig. 1a), was synthesized to detect the truncated cleavage product. As shown in lower panel of Fig. 2b, a specific smaller band, which below the main PCR product and the size is around 300 bp, was found in samples E1, E2, E3, E4, whereas only one main PCR band was detected in samples E5, E6, E7, E8. Based on our original design, the size of the main PCR product is 572 bp, and the designed cleavage length is 265 bp (Fig. 1a). Thus, our PCR results demonstrated that the precise cleavage events most probably occurred in our designed genomic regions of GhMYB25-like genes.
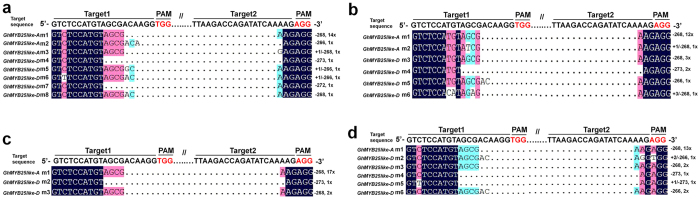
To further validate whether the changed PCR size was derived from CRISPR/Cas9-caused genomic truncation, we randomly picked 20 positive colonies generated from smaller PCR products of each transgenic cotton sample E1/E2/E3/E4 for sequencing analyses. Those sequencing results demonstrated that all 80 smaller sequences were the truncated versions of GhMYB25-like genomic sequences, either from GhMYB25-like A DNA site or from GhMYB25-like D DNA site (Fig. 3). To quantify the proportion of double cleavage, signal intensity of each band was measured by using ImageJ software ( https://imagej.nih.gov/ij/download.html). The cleavage DNA length was mostly concentrate on −268bp, which account for 87% and 92% in GhMYB25-like A DNA site of E1 and E2 samples, 100% in GhMYB25-like A DNA site of E1 and E2 samples, 67% in GhMYB25-like D DNA site of E3 sample (Table 1). Taken all together, these results indicated that this set of CRISPR/Cas9 genome editing system have the potential to efficiently generate long DNA fragment deletions on the selected genomic region.
Figure 3. Validation of CRISPR/Cas9-induced long DNA fragment deletions at both GhMYB25-like A and D subgenomic sites.
(a) to (d) Alignment of genomic sequences cloned from the truncated PCR products using E1, E2, E3, E4 DNA samples as templates.
Table 1. Percentage of different CRISPR/Cas9-caused truncated events.
| Sample | Gene | Rate of different nucleotide insertion (+) and deletion (−) (%) | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| −266 | −268 | −272 | −273 | +1/−273 | +1/−266 | +2/−266 | +1/−268 | +3/−268 | ||
| E1 | GhMYB25like-A | 7 | 87 | 7 | ||||||
| GhMYB25like-D | 20 | 20 | 20 | 40 | ||||||
| E2 | GhMYB25like-A | 92 | 8 | |||||||
| GhMYB25like-D | 14 | 43 | 29 | 14 | ||||||
| E3 | GhMYB25like-A | 100 | ||||||||
| GhMYB25like-D | 67 | 33 | ||||||||
| E4 | GhMYB25like-A | 100 | ||||||||
| GhMYB25like-D | 29 | 29 | 14 | 14 | 14 | |||||
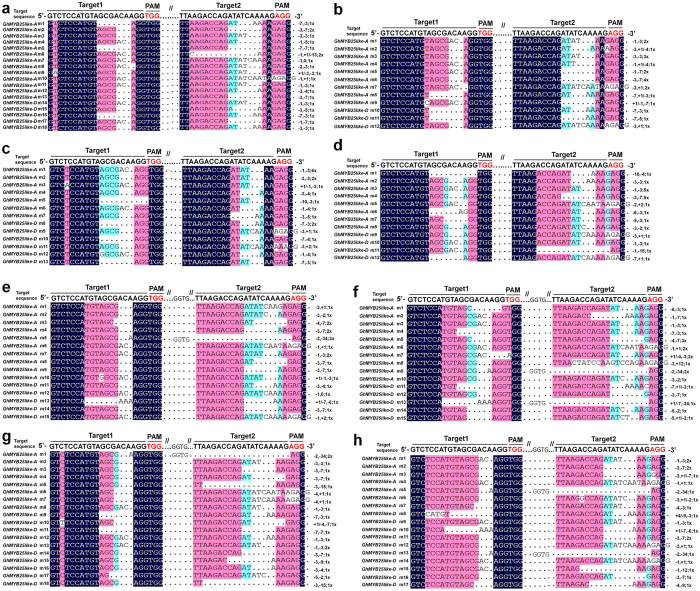
To investigate whether CRISPR/Cas9-mediated nucleotide insertion mutations and deletion events also precisely occurred in the main PCR products, 160 positive colonies, which cloned from the PCR products using E1, E2, E3, E4, E5, E6, E7, E8 DNA samples as templates, were randomly picked for sequencing analyses. In samples E1, E2, E3, E4, all the 159 examined Target1 and Target2 genomic sites precisely occurred genome editing events, except 1 DNA sites, which from E1 sample Target2, was not affected (Fig. 4). As shown in Table 2, most of the nucleotide insertion and deletion mutations were −1bp/−3bp/−7bp nucleotide deletion mutations and +1 bp insertion mutation.
Figure 4. Sequencing analyses and validation of CRISPR/Cas9-induced NHEJ mutations at both GhMYB25-like A and D subgenomic sites.
Sequence confirmation of nucleotide deletion and insertion mutations in cotton transgenic samples E1, E2, E3, E4, E5, E6, E7, and E8.
Table 2. Rate of CRISPR/Cas9-caused small nucleotide insertion (+) and deletion (−) events in cotton transgenic samples.
| Sample | Target sequence | Gene | Rate of different nucleotide insertion (+) and deletion (−) (%) | ||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | +1 | −1 | +1/−1 | +1/−2 | +1/−3 | +1/−4 | +1/−7 | −2 | ++2 | +2/−7 | −3 | −4 | +4/−9 | −5 | −6 | −7 | −9 | −10 | −12 | +12 | −15 | −34 | |||
| E1 | Target 1 | GhMYB25like-A | 40 | 7 | 7 | 33 | 13 | ||||||||||||||||||
| GhMYB25like-D | 40 | 40 | 20 | ||||||||||||||||||||||
| Target 2 | GhMYB25like-A | 7 | 7 | 33 | 7 | 7 | 27 | 13 | |||||||||||||||||
| GhMYB25like-D | 60 | 20 | 20 | ||||||||||||||||||||||
| E2 | Target 1 | GhMYB25like-A | 29 | 6 | 12 | 47 | 6 | ||||||||||||||||||
| GhMYB25like-D | 33 | 67 | |||||||||||||||||||||||
| Target 2 | GhMYB25like-A | 12 | 6 | 35 | 6 | 41 | |||||||||||||||||||
| GhMYB25like-D | 33 | 33 | 33 | ||||||||||||||||||||||
| E3 | Target 1 | GhMYB25like-A | 54 | 8 | 31 | 8 | |||||||||||||||||||
| GhMYB25like-D | 14 | 29 | 57 | ||||||||||||||||||||||
| Target 2 | GhMYB25like-A | 8 | 69 | 15 | 8 | ||||||||||||||||||||
| GhMYB25like-D | 14 | 14 | 43 | 29 | |||||||||||||||||||||
| E4 | Target 1 | GhMYB25like-A | 31 | 6 | 44 | 6 | 6 | 6 | |||||||||||||||||
| GhMYB25like-D | 50 | 25 | 25 | ||||||||||||||||||||||
| Target 2 | GhMYB25like-A | 6 | 6 | 50 | 6 | 31 | |||||||||||||||||||
| GhMYB25like-D | 50 | 25 | 25 | ||||||||||||||||||||||
| E5 | Target 1 | GhMYB25like-A | 20 | 7 | 20 | 40 | 13 | ||||||||||||||||||
| GhMYB25like-D | 40 | 40 | 20 | ||||||||||||||||||||||
| Target 2 | GhMYB25like-A | 13 | 7 | 20 | 40 | 20 | |||||||||||||||||||
| GhMYB25like-D | 20 | 20 | 20 | 20 | 20 | ||||||||||||||||||||
| E6 | Target 1 | GhMYB25like-A | 27 | 13 | 13 | 33 | 7 | 7 | |||||||||||||||||
| GhMYB25like-D | 20 | 40 | 40 | ||||||||||||||||||||||
| Target 2 | GhMYB25like-A | 13 | 7 | 33 | 27 | 7 | 13 | ||||||||||||||||||
| GhMYB25like-D | 20 | 20 | 20 | 20 | 20 | ||||||||||||||||||||
| E7 | Target 1 | GhMYB25like-A | 11 | 22 | 44 | 22 | |||||||||||||||||||
| GhMYB25like-D | 27 | 9 | 36 | 9 | 18 | ||||||||||||||||||||
| Target 2 | GhMYB25like-A | 22 | 22 | 22 | 11 | 22 | |||||||||||||||||||
| GhMYB25like-D | 9 | 36 | 9 | 27 | 9 | 9 | |||||||||||||||||||
| E8 | Target 1 | GhMYB25like-A | 33 | 11 | 44 | 11 | |||||||||||||||||||
| GhMYB25like-D | 27 | 9 | 9 | 36 | 9 | 9 | |||||||||||||||||||
| Target 2 | GhMYB25like-A | 11 | 11 | 11 | 33 | 22 | 11 | ||||||||||||||||||
| GhMYB25like-D | 18 | 18 | 9 | 27 | 9 | 9 | 9 | ||||||||||||||||||
Similarly, high proportion nucleotide insertion and deletion events were detected in samples E5, E6, E7, and E8 (Fig. 4). Except 1 DNA sites, which from E5 Target 2, still keep its wild type DNA sequence, all the rest of 159 examined genomic sites were detected nucleotide insertion or deletion mutation events (Fig. 4). As shown in Table 2, the nucleotide insertion and deletion mutations were mostly concentrate on −1bp/−2bp/−3bp/−7bp deletion mutations and +1 bp insertion mutation.
Thus, those results suggested that both GhMYB25-like-sgRNA1 and GhMYB25-like-sgRNA2 effectively and precisely guided cas9-mediated genome cleavage. Given that the high-efficient effect on both GhMYB25-like A and GhMYB25-like D genome sequence, this set of CRISPR/Cas9 genome editing system have the potential to generate DNA level knockout mutations on complex allotetraploid cotton genome.
Among the genome knockout transgenic events, mosaicism was observed in each transgenic event. Mosaicism sometimes may disturb later phenotypic analysis. Given that the double-cleavage DNA length was mostly on −268bp and the majority of small nucleotide mutations are −1bp/−3bp/−7bp nucleotide deletions, we infer that most but not all of the targeted genome editing events may occur in the transformed single cell stage. This can be eliminated during later stage of selection.
Off-target analyses
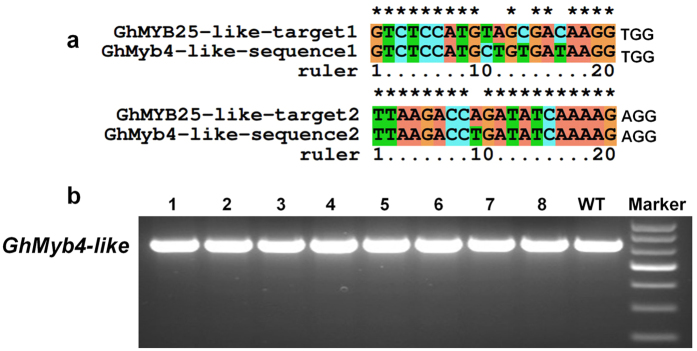
To evaluate the possibility of off-target effect, two putative off-target sequences, which derived from GhMYB4-like genomic sequence and have three and one mismatched nucleotides with GhMYB25-like-sgRNA1 and GhMYB25-like-sgRNA2, respectively (Fig. 5a), were employed to analyzing potential off-target events. Unlike the result in Fig. 2B, the truncate smaller band was not detected by PCR amplifying the predicted GhMYB4-like genomic region (Fig. 5b). To further exam the potential involvement of small nucleotide insertion and deletion mutations, PCR product amplified from DNA sample E3, which have the highest proportion (21.4%) of double cleavage and 100% small nucleotide deletion mutation (Table 1 and Fig. 3c), was cloned for sequencing analyses. As shown in Table 3, both of the putative off target sites, GhMYB4-like-sequence1 and GhMYB4-like-sequence2, were not detected any occurrence of mutation events. Those data suggested that this set of CRISPR/Cas9 genome editing system have high specificity.
Figure 5. Off-targets analyses of two potential off-target sites.
(a) Alignment of two putative off-target sites derived from GhMYB4-like genomic sequence. (b) Assessment of possible off-target-caused double-cleavage mutation.
Table 3. Mutation Analyses in two potential off-target sites.
| sgRNA name | Putative off-target sequence | No. of mismatched nucleotides | No. of examined events | No. of mismatched events |
|---|---|---|---|---|
| GhMYB25-like-sgRNA1 | GhMYB4-like-sequence 1 | 3 | 29 | 0 |
| GhMYB25-like-sgRNA2 | GhMYB4-like-sequence 2 | 1 | 29 | 0 |
Discussion
Cotton acts as one of the world’s major staple crop, contributing to approximately US$500 billion worldwide economic impact annually. Although the allotetraploid genome feature of upland cotton, which bring the challenges for cotton genetic improvement, cotton investigators never stop updating their biotechnology arsenal to more effectively and accurately dissect cotton genes functions, such as RNAi technology23, virus-induced gene silencing (VIGS) technology23,67, activation tagging technology68, constitutively or spatiotemporal gene expression technology69. With the revolutionary achievements in the cotton whole-genome sequencing and assembly, cotton research community urgent need a set of high-efficient and time-saving CRISPR/Cas9 system for cotton functional genome studies and the subsequent application.
To exam the qualification for effective CRISPR/Cas9-caused genomic editing in allotetraploid cotton genome, two sgRNAs (GhMYB25-like-sgRNA1 and GhMYB25-like-sgRNA2), designed in the identical genomic regions of A subgenome gene GhMYB25-like A and D subgenome gene GhMYB25-like D5,6, were employed to examine the efficacy of allotetraploid cotton genome editing. Through A. tumefaciens-mediated cotton transformation and antibiotic selection, eight independent positive transgenic plantlets samples, namely E1, E2, E3, E4, E5, E6, E6, E8 (Fig. 2B), were obtained for further analyzing the efficacy of the CRISPR/Cas9 system. Surprisingly, high proportion plausible cleavage products, which account for 18.2%, 14.2%, 21.4%, and 14.9% of the total PCR products produced from samples E1, E2, E3, and E4 DNA template (Fig. 2B), respectively, were detected directly through normal PCR amplification. Those potential double-cleavage events were further confirmed by sequencing analyses of those smaller PCR product, which showed that all of the 160 examined samples appeared to be derived from CRISPR/Cas9-triggered truncation events, either from truncated GhMYB25-like A genome region or from truncated GhMYB25-like D genome region (Table 1 and Fig. 3). The long genomic fragment deletions require high Cas9-sgRNAs-complex activity to ensure two designed cleavage sites be efficiently recognized and cleaved70,71. In addition, the accurate and high-efficient feature of this CRISPR/Cas9 system for long DNA fragment deletions provides the possibility to effectively replace undesired genomic area by introducing any desired engineered sequence fragment through endogenous HDR DNA repair mechanism72.
Current studies have demonstrated that most of DSBs are repaired by NHEJ-mediated repair mechanism and cause several nucleotide insertion mutations and deletion mutations48,49,51,71,72,73. Consistence with those discoveries, one main PCR product band, whose PCR sizes were very similar with their WT control lane, was detected in E1, E2, E3, E4, E5, E6, E7 and E8 DNA samples. However, sequencing analyses demonstrated that high proportion of nucleotide insertion and deletion occurred on predicted DNA cutting sites. Except two sites, which from E1 sample Target2 and E2 sample Target2, respectively, all the 318 examined sites exhibited mutations events (Fig. 4). Statistic data suggested that the majority of mutations detected in GhMYB25-like A and gene GhMYB25-like D genome were −1bp/−3bp/−7bp nucleotide deletions and +1 bp nucleotide insertion (Table 2). To sum up, these data demonstrated that this CRISPR/Cas9 genome editing system may be qualified for high-efficient generating DNA level mutations on allotetraploid cotton genome.
Off-target effect is a crucial factor for the application of CRISPR/Cas9 system. In our study, all the examined putative off-target sequences were completely match their original wild type genomic DNA sequence (Table 3), even though the second putative off-target sites only have 1 mismatched nucleotides with GhMYB25-like-sgRNA2 (Fig. 5). Several researches demonstrated that CRISPR/Cas9 system-caused off-target effect varies with different organisms and always very low in plants species49,74,75. Thus, this set of CRISPR/Cas9 system may have high-specificity in cotton genome editing.
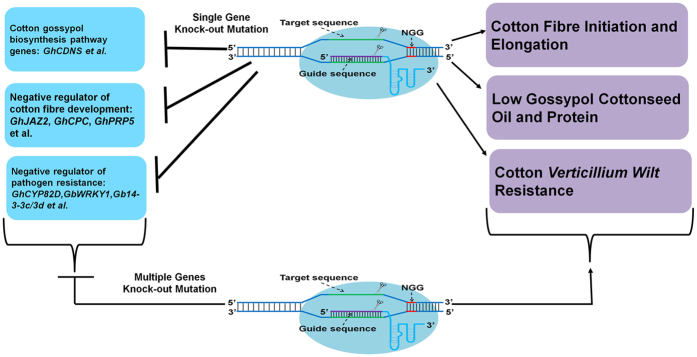
Cotton is the most major source of nature textile fibre, and its cottonseed is emerging as one of the most impotent renewable resources of plant oil and plant protein. In addition, cotton Verticillium wilt is the most destructive disease, which annually cause 250–310 million US dollars economic losses in China76. Given that the high-efficiency, high-specificity, and high proportion of germline transmission of this CRISPR/Cas9 system45, we may have the expectation that it have the potential to substitute current mainstream RNAi vectors for generating DNA level knockout mutation in cotton genome. As shown in (Fig. 6), many candidate genes regarding to cotton gossypol biosynthesis, negative regulator of cotton fibre development and Verticillium wilt resistance, can be selected as target genes for genetic improvement of cotton agronomic traits. Recently, CRISPR/Cas9 have been modified to generate site-specific transcription activation/repression and targeted DNA methylation/demethylation by fusing different engineered enzymes71,77,78. However, the key first step for those modifications is the efficient and specific guidance of those engineered enzymes. Thus, this set of CRISPR/Cas9 system probably also have the potential to modify for inducing site-specific methylation/demethylation and transcription activation/repression, which will further facilitate cotton gene functional researches and applications.
Figure 6. Prospect of the application of this set of CRISPR/Cas9 genome editing system.
Cotton fibre and cottonseed oil/protein are two major agronomic traits directly related to cotton economic value. Verticillium wilt is the main bottleneck hampering cotton fibre production and quality in cotton field. Through the different sgRNA-expression modules, single or multiple sgRNAs can be easily assembled for the specific or integrated improvement of cotton fibre quality, gossypol toxicity of cottonseed and Verticillium wilt resistance.
Methods and Materials
Plant Material and Growth
Cotton (Gossypium hirsutum L.) cv ‘YZ1’ were used in this study. The plants were grown in the East Carolina University Greenhouse or growth chambers with a 16-h-day/8-h-night cycle at 28 °C.
sgRNAs Design and Golden Gate Assembly of CRISPR/Cas9 System
To strict evaluate the efficacy and specificity of the CRISPR/Cas9 vectors, the sgRNAs design basically need to meet three standards: firstly, these sgRNAs target sites can be used to test the genome mutation efficacy on both G. hirsutum cotton A subgenome and D subgenome; secondly, several single-nucleotide polymorphism and double-nucleotide polymorphism near these two sgRNAs target sites can be used for distinguishing the identity of GhMYB25-like genes; thirdly, there are two highly similar sequences (have 1–3 mismatched nucleotides) with the designed sgRNAs can be used to estimate the off-target effect. Through comparing previously reported gene information and searching the NIBC cotton database, two sgRNAs, GhMYB25-like-sgRNA1 and GhMYB25-like-sgRNA2, which from GhMYB25-like A and GhMYB25-like D genomic sequences encoded by allotetraploid cotton A subgenome and the D subgenome respectively5,6, were designed for the assembly of sgRNA-expression module. The GhMYB25-like-sgRNA1 and GhMYB25-like-sgRNA2 expression module was assembled by direct PCR amplification using pCBC-DT1T2 template as described previously45. And then this PCR product was purified for the Golden Gate assembly of Cas9 expression module through the Type IIS restriction endonucleases (REases) BsaI reaction (R3535L, New England Biolabs).
Cotton Transformation
The assembled CRISPR/Cas9 vector was then transformed into Agrobacterium tumefaciens (strain EHA105). And the A. tumefaciens-mediated cotton G. hirsutum ‘YZ1′ was performed as previous reports65,66,79.
Genomic DNA Extraction and Mutations Analyses
The procedure of total genomic DNA isolation was followed the manual of Plant Genomic DNA Kit (TIANGEN, China). The specific primers designed from hygromycin resistance gene sequence were used for the identification of exogenous T-DNA insertion, and the primers sequence are Hyb-S2: TCGTTATGTTTATCGGCACTTTG, Hyb-A2: TATTGGGAATCCCCGAACAT. The common PCR primers of two GhMYB25-like genes were synthesized for double-cleavage mutation analyses and genes cloning, and the primers sequence are GhMYB25-like-S: TAACCAATTCTACCCACATTTTCG, GhMYB25-like-A: TGCCACTTTATCGGTTGTCGTA. PCR products were cloned by TA cloning reaction. And the positive colonies were randomly selected for sequencing (Genomics Core Facility, East Carolina University, NC). The sequences were analyzed and aligned through NCBI database and DNAman software.
Off-target analyses
We searched all potential off-target sites in cotton genome. Two putative off-target sites, namely GhMYB4-like sequence1 and GhMYB4-like sequence2, were only discovered from GhMyb4-like (LOC107894856) genome sequence, which had three or less mismatches. GhMYB4-like sequence1 and GhMYB4-like sequence2 have 3 and 1 mismatched nucleotides with the designed GhMYB25-like-sgRNA1 and GhMYB25-like-sgRNA2, respectively (Fig. 5a). To detect the potential double-cleavage event, a pair of primers, covering the putative GhMYB4-like cleavage genomic area, was designed for PCR amplification and sequencing analyses. The PCR primers are GhMyb4-like-S: TTCCCTTGCTTTCAACGCTC, GhMyb4-like-A: GTTTTAGGCTTCTGCGTCACG. The sequencing analyses are same as mention above.
Additional Information
How to cite this article: Li, C. et al. A high-efficiency CRISPR/Cas9 system for targeted mutagenesis in Cotton (Gossypium hirsutum L.). Sci. Rep. 7, 43902; doi: 10.1038/srep43902 (2017).
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Acknowledgments
The CRISPR-Cas9 vectors were kindly provided by Dr. Qi-jun Chen of China Agricultural University. This work is partially supported by the Cotton Incorporated.
Footnotes
The authors declare no competing financial interests.
Author Contributions C.L., T.U. and B.Z. designed the experiment. C.L. perform the experiment. C.L., T.U. and B.Z. did data analysis and wrote the manuscript. All authors reviewed the manuscript.
References
- Guan X., Song Q. & Chen Z. J. Polyploidy and small RNA regulation of cotton fiber development. Trends Plant Sci 19, 516–528 (2014). [DOI] [PubMed] [Google Scholar]
- Li C. & Zhang B. MicroRNAs in Control of Plant Development. J Cell Physiol 231, 303–313 (2016). [DOI] [PubMed] [Google Scholar]
- Wendel J. F. New World tetraploid cottons contain Old World cytoplasm. Proc Natl Acad Sci USA 86, 4132–4136 (1989). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tu L. L. et al. Genes expression analyses of sea-island cotton (Gossypium barbadense L.) during fiber development. Plant Cell Rep 26, 1309–1320 (2007). [DOI] [PubMed] [Google Scholar]
- Machado A., Wu Y., Yang Y., Llewellyn D. J. & Dennis E. S. The MYB transcription factor GhMYB25 regulates early fibre and trichome development. Plant J 59, 52–62 (2009). [DOI] [PubMed] [Google Scholar]
- Walford S. A., Wu Y., Llewellyn D. J. & Dennis E. S. GhMYB25-like: a key factor in early cotton fibre development. Plant J 65, 785–797 (2011). [DOI] [PubMed] [Google Scholar]
- Li H. B. et al. A cotton ascorbate peroxidase is involved in hydrogen peroxide homeostasis during fibre cell development. New Phytol 175, 462–471 (2007). [DOI] [PubMed] [Google Scholar]
- Tan J. et al. A genetic and metabolic analysis revealed that cotton fiber cell development was retarded by flavonoid naringenin. Plant Physiol 162, 86–95 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hao J. et al. GbTCP, a cotton TCP transcription factor, confers fibre elongation and root hair development by a complex regulating system. J Exp Bot 63, 6267–6281 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng F. et al. GbPDF1 is involved in cotton fiber initiation via the core cis-element HDZIP2ATATHB2. Plant Physiol 158, 890–904 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng Y. et al. GhCaM7-like, a calcium sensor gene, influences cotton fiber elongation and biomass production. Plant Physiol Biochem 109, 128–136 (2016). [DOI] [PubMed] [Google Scholar]
- Tang W. et al. The calcium sensor GhCaM7 promotes cotton fiber elongation by modulating reactive oxygen species (ROS) production. New Phytol 202, 509–520 (2014). [DOI] [PubMed] [Google Scholar]
- Li Y. et al. GbEXPATR, a species-specific expansin, enhances cotton fibre elongation through cell wall restructuring. Plant Biotechnol J 14, 951–963 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han J., Tan J., Tu L. & Zhang X. A peptide hormone gene, GhPSK promotes fibre elongation and contributes to longer and finer cotton fibre. Plant Biotechnol J 12, 861–871 (2014). [DOI] [PubMed] [Google Scholar]
- Wang L., Cook A., Patrick J. W., Chen X. Y. & Ruan Y. L. Silencing the vacuolar invertase gene GhVIN1 blocks cotton fiber initiation from the ovule epidermis, probably by suppressing a cohort of regulatory genes via sugar signaling. Plant J 78, 686–696 (2014). [DOI] [PubMed] [Google Scholar]
- Wan Q. et al. Small interfering RNAs from bidirectional transcripts of GhMML3_A12 regulate cotton fiber development. New Phytol 210, 1298–1310 (2016). [DOI] [PubMed] [Google Scholar]
- Yang X. et al. Expression profile analysis of genes involved in cell wall regeneration during protoplast culture in cotton by suppression subtractive hybridization and macroarray. J Exp Bot 59, 3661–3674 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Min L. et al. Cotton GhCKI disrupts normal male reproduction by delaying tapetum programmed cell death via inactivating starch synthase. Plant J(2013). [DOI] [PubMed] [Google Scholar]
- He X., Zhu L., Xu L., Guo W. & Zhang X. GhATAF1, a NAC transcription factor, confers abiotic and biotic stress responses by regulating phytohormonal signaling networks. Plant Cell Rep 35, 2167–2179 (2016). [DOI] [PubMed] [Google Scholar]
- Xu L. et al. Overexpression of GbWRKY1 positively regulates the Pi starvation response by alteration of auxin sensitivity in Arabidopsis. Plant Cell Rep(2012). [DOI] [PubMed] [Google Scholar]
- Guo W. et al. An ethylene response-related factor, GbERF1-like, from Gossypium barbadense improves resistance to Verticillium dahliae via activating lignin synthesis. Plant Mol Biol 91, 305–318 (2016). [DOI] [PubMed] [Google Scholar]
- Li Y. B. et al. The Thioredoxin GbNRX1 Plays a Crucial Role in Homeostasis of Apoplastic Reactive Oxygen Species in Response to Verticillium dahliae Infection in Cotton. Plant Physiol 170, 2392–2406 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C. et al. Cotton WRKY1 mediates the plant defense-to-development transition during infection of cotton by Verticillium dahliae by activating JASMONATE ZIM-DOMAIN1 expression. Plant Physiol 166, 2179–2194 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu L. et al. Differential Gene Expression in Cotton Defence Response to Verticillium dahliae by SSH. Journal of Phytopathology 159, 606–615 (2011). [Google Scholar]
- Wang K. et al. The draft genome of a diploid cotton Gossypium raimondii. Nat Genet 44, 1098–1103 (2012). [DOI] [PubMed] [Google Scholar]
- Paterson A. H. et al. Repeated polyploidization of Gossypium genomes and the evolution of spinnable cotton fibres. Nature 492, 423–427 (2012). [DOI] [PubMed] [Google Scholar]
- Li F. et al. Genome sequence of the cultivated cotton Gossypium arboreum. Nat Genet 46, 567–572 (2014). [DOI] [PubMed] [Google Scholar]
- Zhang T. et al. Sequencing of allotetraploid cotton (Gossypium hirsutum L. acc. TM-1) provides a resource for fiber improvement. Nat Biotechnol(2015). [DOI] [PubMed] [Google Scholar]
- Li F. et al. Genome sequence of cultivated Upland cotton (Gossypium hirsutum TM-1) provides insights into genome evolution. Nat Biotechnol(2015). [DOI] [PubMed] [Google Scholar]
- Yuan D. et al. The genome sequence of Sea-Island cotton (Gossypium barbadense) provides insights into the allopolyploidization and development of superior spinnable fibres. Scientific Reports 5, 17662 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson S. P. Sensing and repairing DNA double-strand breaks. Carcinogenesis 23, 687–696 (2002). [DOI] [PubMed] [Google Scholar]
- Sonoda E., Hochegger H., Saberi A., Taniguchi Y. & Takeda S. Differential usage of non-homologous end-joining and homologous recombination in double strand break repair. DNA Repair (Amst) 5, 1021–1029 (2006). [DOI] [PubMed] [Google Scholar]
- Johnson R. D. & Jasin M. Double-strand-break-induced homologous recombination in mammalian cells. Biochem Soc Trans 29, 196–201 (2001). [DOI] [PubMed] [Google Scholar]
- Lin Y., Lukacsovich T. & Waldman A. S. Multiple pathways for repair of DNA double-strand breaks in mammalian chromosomes. Mol Cell Biol 19, 8353–8360 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang F., Han M., Romanienko P. J. & Jasin M. Homology-directed repair is a major double-strand break repair pathway in mammalian cells. Proc Natl Acad Sci USA 95, 5172–5177 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang W., Yang B. & Weeks D. P. Efficient CRISPR/Cas9-Mediated Gene Editing in Arabidopsis thaliana and Inheritance of Modified Genes in the T2 and T3 Generations. Plos One 9, e99225 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao H. et al. Heritable targeted mutagenesis in maize using a designed endonuclease. Plant J 61, 176–187 (2010). [DOI] [PubMed] [Google Scholar]
- Puchta H. The repair of double-strand breaks in plants: mechanisms and consequences for genome evolution. J Exp Bot 56, 1–14 (2005). [DOI] [PubMed] [Google Scholar]
- Hermann M. et al. Evaluation of OPEN zinc finger nucleases for direct gene targeting of the ROSA26 locus in mouse embryos. Plos One 7, e41796 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sander J. D. et al. Selection-free zinc-finger-nuclease engineering by context-dependent assembly (CoDA). Nat Methods 8, 67–69 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sander J. D. et al. Predicting success of oligomerized pool engineering (OPEN) for zinc finger target site sequences. BMC Bioinformatics 11, 543 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cantos C., Francisco P., Trijatmiko K. R., Slamet-Loedin I. & Chadha-Mohanty P. K. Identification of “safe harbor” loci in indica rice genome by harnessing the property of zinc-finger nucleases to induce DNA damage and repair. Front Plant Sci 5, 302 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y. et al. Transcription activator-like effector nucleases enable efficient plant genome engineering. Plant Physiol 161, 20–27 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y. et al. Simultaneous editing of three homoeoalleles in hexaploid bread wheat confers heritable resistance to powdery mildew. Nat Biotechnol 32, 947–951 (2014). [DOI] [PubMed] [Google Scholar]
- Xing H. L. et al. A CRISPR/Cas9 toolkit for multiplex genome editing in plants. Bmc Plant Biol 14, 327 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang W. et al. Demonstration of CRISPR/Cas9/sgRNA-mediated targeted gene modification in Arabidopsis, tobacco, sorghum and rice. Nucleic Acids Res 41, e188 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng Z. et al. Efficient genome editing in plants using a CRISPR/Cas system. Cell Res 23, 1229–1232 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan D. et al. Efficient CRISPR/Cas9-mediated Targeted Mutagenesis in Populus in the First Generation. Sci Rep 5, 12217 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren C. et al. CRISPR/Cas9-mediated efficient targeted mutagenesis in Chardonnay (Vitis vinifera L.). Sci Rep 6, 32289 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J.-F. et al. Multiplex and homologous recombination-mediated genome editing in Arabidopsis and Nicotiana benthamiana using guide RNA and Cas9. Nat Biotech 31, 688–691 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nekrasov V., Staskawicz B., Weigel D., Jones J. D. & Kamoun S. Targeted mutagenesis in the model plant Nicotiana benthamiana using Cas9 RNA-guided endonuclease. Nat Biotechnol 31, 691–693 (2013). [DOI] [PubMed] [Google Scholar]
- Upadhyay S. K., Kumar J., Alok A. & Tuli R. RNA-guided genome editing for target gene mutations in wheat. G3 (Bethesda) 3, 2233–2238 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie K. & Yang Y. RNA-guided genome editing in plants using a CRISPR-Cas system. Mol Plant 6, 1975–1983 (2013). [DOI] [PubMed] [Google Scholar]
- Zhang H. et al. The CRISPR/Cas9 system produces specific and homozygous targeted gene editing in rice in one generation. Plant Biotechnol J 12, 797–807 (2014). [DOI] [PubMed] [Google Scholar]
- Mao Y. et al. Application of the CRISPR-Cas system for efficient genome engineering in plants. Mol Plant 6, 2008–2011 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miao J. et al. Targeted mutagenesis in rice using CRISPR-Cas system. Cell Res 23, 1233–1236 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang Z., Zhang K., Chen K. & Gao C. Targeted Mutagenesis in Zea mays Using TALENs and the CRISPR/Cas System. J Genet Genomics 41, 63–68 (2014). [DOI] [PubMed] [Google Scholar]
- Feng Z. et al. Multigeneration analysis reveals the inheritance, specificity, and patterns of CRISPR/Cas-induced gene modifications in Arabidopsis. Proc Natl Acad Sci USA 111, 4632–4637 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou H., Liu B., Weeks D. P., Spalding M. H. & Yang B. Large chromosomal deletions and heritable small genetic changes induced by CRISPR/Cas9 in rice. Nucleic Acids Res 42, 10903–10914 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie K., Zhang J. & Yang Y. Genome-wide prediction of highly specific guide RNA spacers for CRISPR-Cas9-mediated genome editing in model plants and major crops. Mol Plant 7, 923–926 (2014). [DOI] [PubMed] [Google Scholar]
- Cho S. W., Kim S., Kim J. M. & Kim J. S. Targeted genome engineering in human cells with the Cas9 RNA-guided endonuclease. Nat Biotechnol 31, 230–232 (2013). [DOI] [PubMed] [Google Scholar]
- Hwang W. Y. et al. Efficient genome editing in zebrafish using a CRISPR-Cas system. Nat Biotech 31, 227–229 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang W., Bikard D., Cox D., Zhang F. & Marraffini L. A. RNA-guided editing of bacterial genomes using CRISPR-Cas systems. Nat Biotech 31, 233–239 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li D. et al. Heritable gene targeting in the mouse and rat using a CRISPR-Cas system. Nat Biotech 31, 681–683 (2013). [DOI] [PubMed] [Google Scholar]
- Jin S. et al. Identification of a novel elite genotype for in vitro culture and genetic transformation of cotton. Biol Plantarum 50, 519–524 (2006). [Google Scholar]
- Firoozabady E. et al. Transformation of cotton (Gossypium hirsutum L.) by Agrobacterium tumefaciens and regeneration of transgenic plants. Plant Mol Biol 10 (1987). [DOI] [PubMed] [Google Scholar]
- Tuttle J. R., Idris A. M., Brown J. K., Haigler C. H. & Robertson D. Geminivirus-mediated gene silencing from Cotton leaf crumple virus is enhanced by low temperature in cotton. Plant Physiol 148, 41–50 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z. et al. PAG1, a cotton brassinosteroid catabolism gene, modulates fiber elongation. New Phytol 203, 437–448 (2014). [DOI] [PubMed] [Google Scholar]
- Zhang M. et al. Spatiotemporal manipulation of auxin biosynthesis in cotton ovule epidermal cells enhances fiber yield and quality. Nat Biotechnol 29, 453–458 (2011). [DOI] [PubMed] [Google Scholar]
- Zhou H., Liu B., Weeks D. P., Spalding M. H. & Yang B. Large chromosomal deletions and heritable small genetic changes induced by CRISPR/Cas9 in rice. Nucleic Acids Res 42 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowder L. G. et al. A CRISPR/Cas9 Toolbox for Multiplexed Plant Genome Editing and Transcriptional Regulation. Plant Physiol 169, 971–985 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shan Q., Wang Y., Li J. & Gao C. Genome editing in rice and wheat using the CRISPR/Cas system. Nat Protoc 9, 2395–2410 (2014). [DOI] [PubMed] [Google Scholar]
- Ma X. et al. A robustCRISPR/Cas9 system for convenient, high-efficiency multiplex genome editing in monocot and dicotplants. Mol Plant(2015). [DOI] [PubMed] [Google Scholar]
- Pattanayak V. et al. High-throughput profiling of off-target DNA cleavage reveals RNA-programmed Cas9 nuclease specificity. Nat Biotech 31, 839–843 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu Y. et al. High-frequency off-target mutagenesis induced by CRISPR-Cas nucleases in human cells. Nat Biotech 31, 822–826 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y. et al. Significant Improvement of Cotton Verticillium Wilt Resistance by Manipulating the Expression of Gastrodia Antifungal Proteins. Mol Plant 9, 1436–1439 (2016). [DOI] [PubMed] [Google Scholar]
- Vojta A. et al. Repurposing the CRISPR-Cas9 system for targeted DNA methylation. Nucleic Acids Res 44, 5615–5628 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu X. et al. A CRISPR-based approach for targeted DNA demethylation. Cell Discov 2, 16009 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang B. H., Feng R., Liu F. & Wang Q. L. High frequency somatic embryogenesis and plant regeneration of an elite Chinese cotton variety. Bot Bulletin Acad Sin 42, 7–16 (2001). [Google Scholar]