Abstract
The unfolded protein response (UPR) is an adaptive response to endoplasmic reticulum stress and the inositol-requiring enzyme 1α/X-box binding protein 1 (IRE1α/XBP1) pathway of the UPR is important in lipid metabolism. However, its role in bile acid metabolism remains unknown. We demonstrate that liver-specific Xbp1 knockout (LS-Xbp1−/−) mice had a 45% reduction in total bile acid pool. LS-Xbp1−/− mice had lower serum 7α-hydroxy-4-cholesten-3-one (C4) levels compared with Xbp1fl/fl mice, indicating reduced cholesterol 7α-hydroxylase (CYP7A1) synthetic activity. This occurred without reductions of hepatic CYP7A1 protein expression. Feeding LS-Xbp1−/− mice cholestyramine increased hepatic CYP7A1 protein expression to levels 2-fold and 8-fold greater than cholestyramine-fed and chow-fed Xbp1fl/fl mice, respectively. However, serum C4 levels remained unchanged and were lower than both groups of Xbp1fl/fl mice. In contrast, although feeding LS-Xbp1−/− mice cholesterol did not increase CYP7A1 expression, serum C4 levels increased significantly up to levels similar to chow-fed Xbp1fl/fl mice and the total bile acid pool normalized. In conclusion, loss of hepatic XBP1 decreased the bile acid pool and CYP7A1 synthetic activity. Cholesterol feeding, but not induction of CYP7A1 with cholestyramine, increased CYP7A1 synthetic activity and corrected the genotype-specific total bile acid pools. These data demonstrate a novel role of IRE1α/XBP1 regulating bile acid metabolism.
Keywords: cholesterol, cholesterol 7-alpha hydroxylase, liver, gene expression, endoplasmic reticulum, unfolded protein response, serum 7α-hydroxy-4-cholesten-3-one
Bile acids are amphipathic compounds that are synthesized in the liver from cholesterol. Bile acids facilitate hepatobiliary secretion and intestinal lipid absorption, regulate glucose and lipid metabolism in the liver, and regulate energy expenditure in the peripheral tissues (1, 2). Bile acids are synthesized from cholesterol through a classic (neutral) pathway and an alternative (acidic) pathway. These bile acid biosynthetic pathways involve a number of enzymes and the rate-limiting enzyme of the classic pathway is cholesterol 7α-hydroxylase (CYP7A1). Bile acids in the liver are secreted into bile, released into the intestine, reabsorbed in the ileum, and transported via the portal circulation back to the liver. Over 95% of the bile acid pool is conserved through this enterohepatic circulation, with the small amount of fecal and urinary bile acid loss compensated by de novo biosynthesis in the liver (2–4).
The unfolded protein response (UPR) is an adaptive cellular response to endoplasmic reticulum (ER) stress that maintains homeostasis by increasing protein processing capacity and attenuating protein translation. When the inositol-requiring enzyme 1α/X-box binding protein 1 (IRE1α/XBP1) pathway of the UPR is activated in the presence of ER stress, XBP1 mRNA undergoes unconventional splicing by phosphorylated IRE1α to remove a 26-nucleotide sequence, causing a translational frameshift and producing the transcriptionally active XBP1 spliced form. While this pathway of the UPR has been implicated in the pathogenesis of and as a protective response to liver injury (5–8), the IRE1α/XBP1 pathway is also important in hepatic lipid metabolism. In fact, liver-specific deletion of Xbp1 has been shown to reduce hepatic lipogenic gene expression, fatty acid synthesis, and VLDL secretion (9–11). Although bile acids are important hepatobiliary lipids that regulate metabolism, the role of XBP1 in bile acid metabolism remains unexplored.
MATERIALS AND METHODS
Materials
Cholestyramine resin and cholesterol were purchased from Sigma (St. Louis, MO). Antibodies against CYP7A1 and GAPDH were purchased from Proteintech (Rosemont, IL). GRP78 antibody was purchased from Cell Signaling Technology (Danvers, MA).
Animal use and treatment
Liver-specific Xbp1 knockout (LS-Xbp1−/−) mice were generated by breeding C57BL/6-Xbp1fl/fl mice (kindly provided by Dr. Laurie J. Glimcher, Harvard University, MA) with C57BL/6-albumin-Cre mice (Jackson Laboratory, ME) as previously described (6). LS-Xbp1−/− mice and control littermate Xbp1fl/fl mice were cohoused on a 14 h light/10 h dark cycle with free access to food and water. Male Xbp1fl/fl and LS-Xbp1−/− mice (8–10 weeks old) were randomly assigned to receive standard chow, chow supplemented with 2% (w/w) cholestyramine, or chow supplemented with 2% (w/w) cholesterol for 1 week. The mice were fasted for 4 h prior to euthanasia, blood was obtained using cardiac puncture, and the liver and ileum were removed and rinsed with ice-cold saline, sectioned, and snap-frozen in liquid nitrogen. For mice used in the total or organ-specific bile acid pool analysis experiments, the liver, gallbladder, and small intestine were collected from nonfasted mice and immediately minced in 100% methanol either separately for organ-specific bile acid analysis, or together for measurement of the total bile acid pool. In bile analysis experiments, bile was aspirated from the gallbladders of mice fasted for 4 h. All protocols and procedures were performed in conformity with the Public Health Service policy on the Humane Care and Use of Laboratory Animals and approved by the Northwestern University Institutional Animal Care and Use Committee guidelines.
Bile acid analysis
Total bile acid pool size, estimated as the total amount of bile acids circulating in the enterohepatic circulation, and bile acid contents and composition were measured by high-performance liquid chromatography as previously described (12). Samples were spiked with glycocholic acid as an internal standard to control for extraction efficiency. Individual bile acid species were identified by their characteristic retention times and by using bile acid standards. The fecal bile acid content was measured spectrophotometrically after a 72 h collection using a colorimetric assay kit according to the manufacturer’s instructions (GenWay Biotech, San Diego, CA) as previously described (12).
Serum biochemistries
Serum cholesterol was determined using an Infinity spectrophotometric assay according to the instructions of the manufacturer (Thermo Scientific). Serum 7α-hydroxy-4-cholesten-3-one (C4) measurement was performed at the Mayo Clinic Immunochemical Core Lab (Rochester, MN). Serum bile acids were measured colorimetrically.
RNA extraction and quantitative real-time PCR
Total RNA was extracted from frozen liver and ileum using TRIZOL reagent according to the manufacturer’s protocol (Invitrogen Life Technologies, Carlsbad, CA). One microgram of total RNA was reverse-transcribed to cDNA with the qScript cDNA synthesis kit (Quanta Bioscience, Gaithersburg, MD). Quantitative (q)PCR was then performed using Power SYBR® Green PCR Master Mix (Thermo Scientific, Waltham, MA) with the Applied Biosystems Prism 7300 sequence detection system (Applied Biosystems, Foster City, CA). Real-time data were collected for 40 cycles of 95°C, 10 s; 60°C, 1 min. Relative expression of the gene of interest was estimated by the ΔΔCt method using 18s as a reference gene. Samples were analyzed in duplicate, and experiments were repeated a minimum of three times. All primers were synthesized by Integrated DNA Technology (Coralville, IA). RNA-Seq, Gene Ontology (GO), and Kyoto Encyclopedia of Genes and Genomes (KEGG) analyses were performed as previously reported (6). The GEO accession number for the data set is GSE64824.
Preparation of whole liver homogenates and microsomal protein
Protein homogenates from frozen liver were isolated using T-Per protein extraction reagent (Thermo Scientific) containing protease inhibitor cocktails (EMD Millipore, Billerica, MA) and HaltTM phosphatase inhibitor (Thermo Scientific). Microsomal protein was isolated from liver homogenate via differential centrifugation. Livers were homogenized in microsome buffer (50 mM KH2PO4, 50 mM KCl, 100 mM sucrose, 30 mM EDTA, 50 mM NaCl, 2 mM DTT) and centrifuged at 8,600 g for 15 min. Supernatants were further centrifuged at 81,000 g for 1 h, washed, and resuspended in microsome buffer.
Western blotting
After protein quantification with Coomassie Plus protein assay reagent (Thermo Scientific), equal amounts of protein samples were subjected to immunoblotting for target proteins, and immunoreactive bands were visualized using Amersham ECL Western blotting detection reagents according to the manufacturer’s protocol (GE Healthcare, Piscataway, NJ). Densitometry was performed with ImageJ. CYP7A1 protein expression was normalized to GAPDH for liver homogenates. GRP78 was used to verify microsomal enrichment and protein loading. The results were expressed as a relative amount to chow-fed Xbp1fl/fl mice.
Free cholesterol assay
Hepatic lipids were extracted using a modified Folch method. Briefly, liver tissues were homogenized in 2 ml of chloroform:methanol (2:1). The homogenates were incubated at room temperature for 20 h, followed by centrifugation at 1,000 g for 5 min. Supernatants were washed with saline and solvents were removed by evaporation under a N2 stream. Dried lipids were resuspended in assay buffer, followed by free cholesterol measurement using a cholesterol quantitation kit (Sigma) according to manufacturer’s instructions.
Statistics
Data are shown as mean ± SEM. Comparison between two groups was performed using two-tailed Student’s t-test. Statistical significance was defined as P ≤ 0.05.
RESULTS
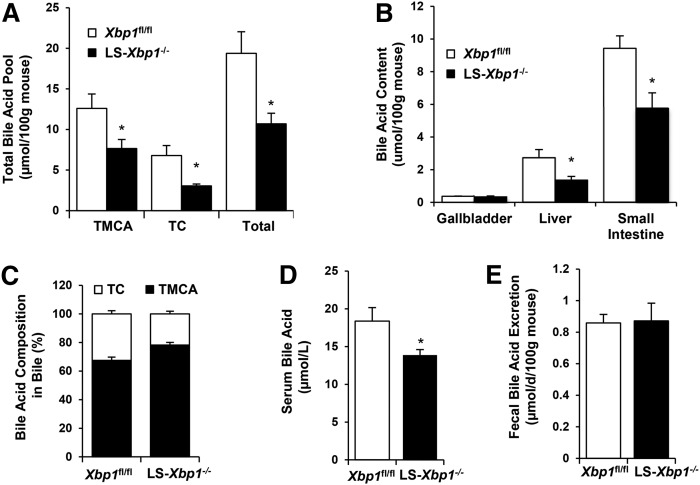
LS-Xbp1−/− mice have reduced hepatic fatty acid synthesis and altered lipoprotein metabolism (9, 10). Because bile acids are hepatobiliary lipids that regulate hepatic metabolism, we hypothesized that the IRE1α/XBP1 signaling pathway is also important in hepatic bile acid metabolism. Therefore, we initially examined the total bile acid pool and bile acid species in chow-fed LS-Xbp1−/− and Xbp1fl/fl mice. The total bile acid pool size was reduced by 45% in LS-Xbp1−/− mice, with total bile acids of 10.7 ± 1.3 μmol/100 g mouse and 19.4 ± 2.7 μmol/100 g mouse in LS-Xbp1−/− and Xbp1fl/fl mice, respectively (P < 0.05) (Fig. 1A). Tauromuricholic acid (TMCA) and taurocholic acid (TC) contents were also reduced by 40% and 56%, respectively (P < 0.05) in LS-Xbp1−/− mice. Bile acid contents were similarly reduced in the liver and small intestine in LS-Xbp1−/− mice, but not in the gallbladder (Fig. 1B). We also measured the bile acid composition of gallbladder bile. As shown in Fig. 1C, the bile in Xbp1fl/fl mice contained 67.5 ± 2.3% TMCA and 32.5 ± 2.3% TC; and the bile in LS-Xbp1−/− mice contained a slightly higher percentage of TMCA (78.2 ± 1.9%, P < 0.05) with a correspondingly lower percentage of TC (21.8 ± 1.9%, P < 0.05). Serum bile acid levels were also reduced in LS-Xbp1−/− mice, being 13.9 ± 0.7 μmol/l and 18.4 ± 1.8 μmol/l in LS-Xbp1−/− mice and Xbp1fl/fl mice, respectively (P < 0.05) (Fig. 1D). There were no differences in fecal bile acid output between the two genotypes (0.87 ± 0.11 μmol/day/100 g mouse vs. 0.86 ± 0.05 μmol/day/100 g in LS-Xbp1−/−and Xbp1fl/fl mice, respectively) (Fig. 1E). Urine bile acid excretion was less than 0.1% of the bile acid pool and also did not differ between the two genotypes. Therefore, enhanced bile acid excretion cannot account for the reduced bile acid pool in LS-Xbp1−/− mice.
Fig. 1.
Bile acid levels in LS-Xbp1−/− and Xbp1fl/fl mice. Total bile acid pool and bile acid species (A); bile acid content in gallbladder, liver, and small intestine (B); bile acid composition in bile (C); serum bile acid concentration (D); and fecal bile acid excretion (E) in LS-Xbp1−/− and Xbp1fl/fl mice (n = 3–5). The total bile acid pool, TMCA content, TC content, and serum bile acid level were reduced in LS-Xbp1−/− mice, while fecal bile acid excretion was unchanged. *P < 0.05 compared with Xbp1fl/fl mice.
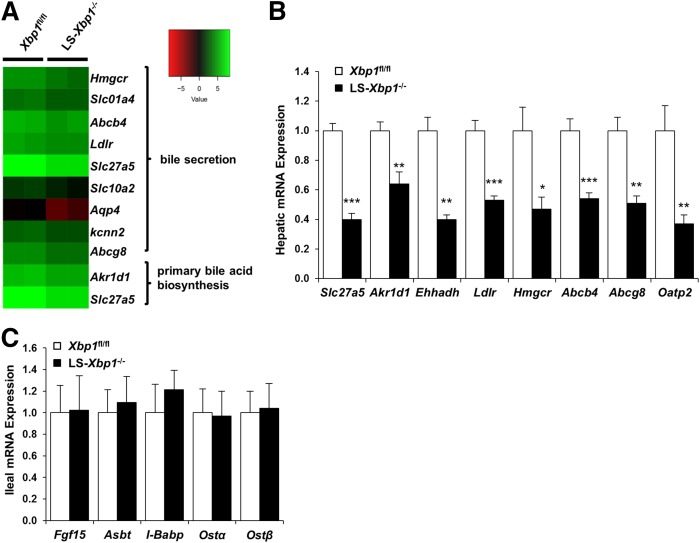
We next performed RNA-Seq on hepatic mRNA isolated from LS-Xbp1−/− and Xbp1fl/fl mice to identify bile acid metabolic genes that were altered in mice lacking hepatic Xbp1. KEGG pathway analysis of differentially expressed hepatic genes revealed that the primary bile acid biosynthesis and bile secretion pathways were downregulated in LS-Xbp1−/− mice (Fig. 2A). We subsequently performed qPCR on several of these genes to confirm the differences in hepatic gene expression (Fig. 2B). Because the ileum has an important role in bile acid metabolism, we analyzed ileal bile acid metabolic genes and determined that there were no differences between LS-Xbp1−/− and Xbp1fl/fl mice (Fig. 2C).
Fig. 2.
Hepatic and ileal gene expression of bile acid metabolism genes in LS-Xbp1−/− and Xbp1fl/fl mice. A: RNA-Seq and KEGG pathway analysis of bile acid metabolism genes. B: Hepatic bile acid metabolism gene expression in LS-Xbp1−/− and Xbp1fl/fl mice (n = 5). C: Ileal bile acid metabolism gene expression in LS-Xbp1−/− mice compared with Xbp1fl/fl mice (n = 4–5). Expression of multiple hepatic bile acid metabolism genes was reduced in LS-Xbp1−/− mice, while ileal gene expression was unchanged. *P < 0.05, **P < 0.01, ***P < 0.001 compared with Xbp1fl/fl mice.
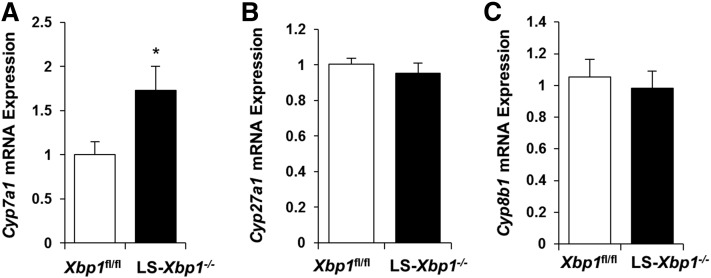
Because the total bile acid pool is reduced in LS-Xbp1−/− mice without changes in bile acid excretion, we next measured gene expression of the major bile acid synthetic genes, Cyp7a1, Cyp27a1, and Cyp8b1. Figure 3 demonstrates that hepatic gene expression of Cyp7a1 was higher in LS-Xbp1−/− mice compared with Xbp1fl/fl mice (P < 0.05), while gene expression of Cyp27a1 and Cyp8b1 was similar to the controls. Hepatic gene expression of bile acid synthetic pathway genes, Akr1d1 and Slc27a5, was reduced by 36% (P < 0.01) and 60% (P < 0.001), respectively (Fig. 2B). Hepatic Shp expression did not change in LS-Xbp1−/− mice compared with Xbp1fl/fl mice (1.30 ± 0.26 vs. 1.06 ± 0.13, respectively; n = 9).
Fig. 3.
Hepatic bile acid synthesis gene expression in LS-Xbp1−/− and Xbp1fl/fl mice. Hepatic gene expression of Cyp7a1 (A), Cyp27a1 (B), and Cyp8b1 (C) was measured in LS-Xbp1−/− and Xbp1fl/fl mice (n = 9). Cyp7a1 expression was increased in LS-Xbp1−/− mice, while expression of Cyp27a1 and Cyp8b1 was similar. *P < 0.05 compared with Xbp1fl/fl mice.
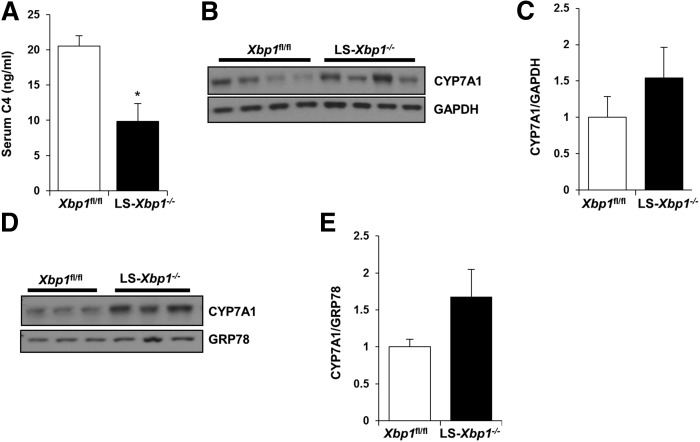
C4 is an intermediate in the classic pathway of bile acid synthesis and the measurement of serum C4 levels is an accurate method for detecting in vivo CYP7A1 synthetic activity (13–17). Figure 4A demonstrates that serum C4 levels were significantly lower in LS-Xbp1−/− mice, being 9.8 ± 2.6 ng/ml compared with 20.5 ± 1.5 ng/ml in Xbp1fl/fl mice (P < 0.05), indicating reduced CYP7A1 synthetic activity in LS-Xbp1−/− mice. Because the gene expression of Cyp7a1 was actually higher in LS-Xbp1−/− mice, we performed Western blotting to confirm CYP7A1 protein expression levels. CYP7A1 protein expression did not change in either the liver homogenate or the liver microsomal fraction from LS-Xbp1−/− mice (Fig. 4B–E). Therefore, the reduced hepatic CYP7A1 synthetic activity in LS-Xbp1−/− mice was not due to changes in CYP7A1 expression.
Fig. 4.
Hepatic CYP7A1 protein expression and serum C4 levels in LS-Xbp1−/− and Xbp1fl/fl mice. A: Serum C4 levels were measured to determine CYP7A1 synthetic activity in LS-Xbp1−/− and Xbp1fl/fl mice (n = 3–4). Representative Western blot (B) and densitometry quantification (C) of CYP7A1 protein expression in liver homogenates from LS-Xbp1−/− and Xbp1fl/fl mice (n = 4). GAPDH was used as a loading control. Representative Western blot (D) and densitometry quantification (E) of CYP7A1 protein expression in microsomes from LS-Xbp1−/− and Xbp1fl/fl mice (n = 3). GRP78 was used to confirm microsome enrichment and as a loading control. Although CYP7A1 protein expression in whole liver homogenates and microsomes was similar in both genotypes, CYP7A1 synthetic activity was reduced in LS-Xbp1−/− mice. *P < 0.05 compared with Xbp1fl/fl mice.
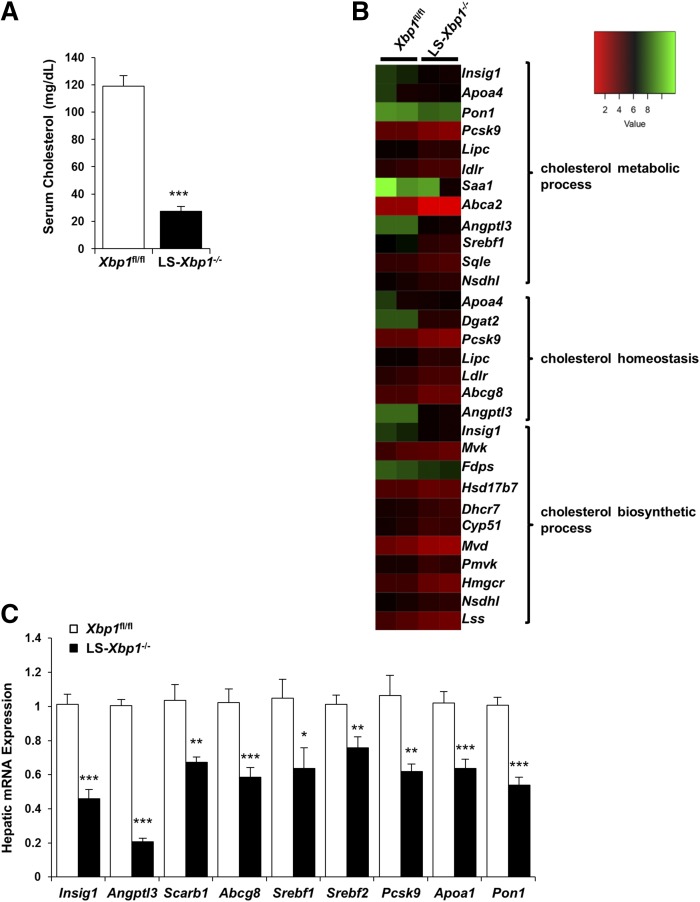
Cholesterol is the precursor for bile acid synthesis by CYP7A1 and serum cholesterol levels were reduced by 70% in LS-Xbp1−/− mice compared with Xbp1fl/fl mice (P < 0.001) (Fig. 5A). GO biological process analysis of differentially expressed hepatic genes identified by RNA-Seq revealed that the cholesterol metabolic process, cholesterol homeostasis, and cholesterol biosynthetic process pathways were downregulated in LS-Xbp1−/− mice (Fig. 5B). Figure 5C shows the qPCR measurement performed to confirm the RNA-Seq expression differences of some of these genes. Of note, there is no difference in the hepatic free cholesterol level between Xbp1fl/fl mice and LS-Xbp1−/− mice (2.33 ± 0.08 mg/g liver vs. 2.38 ± 0.11 mg/g liver, respectively; n = 5).
Fig. 5.
Serum cholesterol and hepatic gene expression of cholesterol metabolic genes in LS-Xbp1−/− and Xbp1fl/fl mice. A: Serum cholesterol was measured in LS-Xbp1−/− and Xbp1fl/fl mice (n = 4). B: RNA-Seq and GO pathway analysis of cholesterol metabolism genes. C: qPCR validation of hepatic cholesterol metabolic gene expression in LS-Xbp1−/− and Xbp1fl/fl mice (n = 9). Serum cholesterol and hepatic expression of several cholesterol metabolic genes were reduced in LS-Xbp1−/− mice. *P < 0.05, **P < 0.01, ***P < 0.001 compared with Xbp1fl/fl mice.
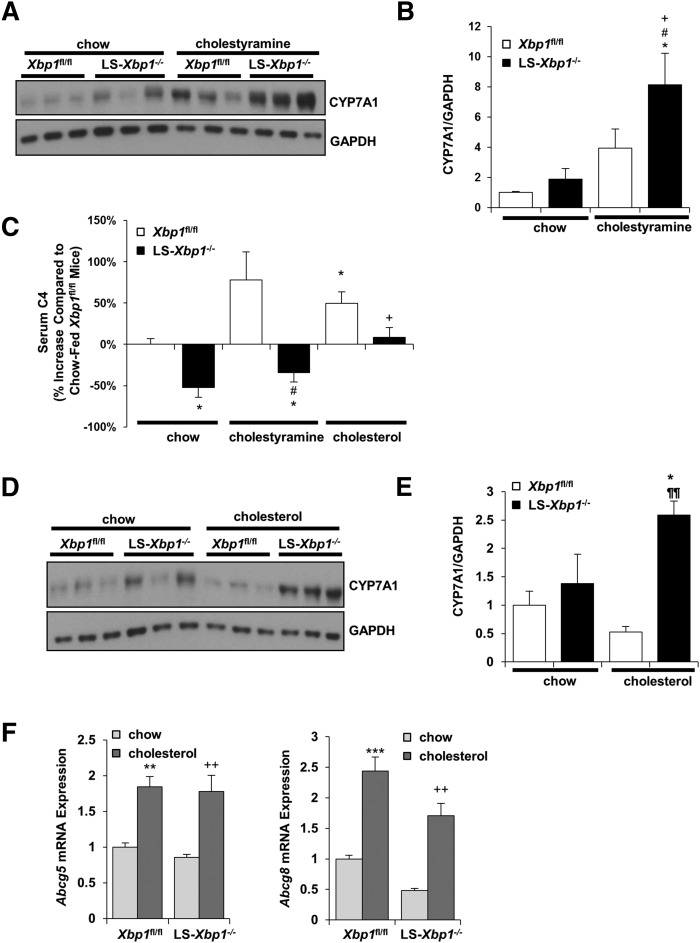
Cholestyramine is a bile acid binder that induces hepatic CYP7A1 expression and increases hepatic CYP7A1 activity in wild-type mice (18, 19). When LS-Xbp1−/− and Xbp1fl/fl mice were fed chow with 2% cholestyramine for 7 days, hepatic CYP7A1 protein expression increased in LS-Xbp1−/− mice, but not in Xbp1fl/fl mice, and the CYP7A1 protein expression levels were higher in LS-Xbp1−/− mice (Fig. 6A, B). Despite the increased hepatic CYP7A1 protein expression in LS-Xbp1−/− mice, serum C4 levels were 63% lower in cholestyramine-fed LS-Xbp1−/− mice compared with the cholestyramine-fed Xbp1fl/fl mice (P < 0.05) (Fig. 6C). In fact, although CYP7A1 protein expression was approximately 8-fold higher in the cholestyramine-fed LS-Xbp1−/− mice compared with the chow-fed Xbp1fl/fl mice, serum C4 levels still remained 34% lower than the baseline C4 levels in chow-fed Xbp1fl/fl mice (P < 0.05).
Fig. 6.
Serum C4 levels and hepatic CYP7A1 protein expression in LS-Xbp1−/− and Xbp1fl/fl mice fed chow, cholestyramine, or cholesterol. LS-Xbp1−/− and Xbp1fl/fl mice were fed chow, chow with cholestyramine (2% w/w), or chow with cholesterol (2% w/w) for 7 days. Western blot (A) and densitometry quantification (B) of hepatic CYP7A1 protein expression in chow-fed and cholestyramine-fed mice. GAPDH was used as a loading control. C: Changes in serum C4 levels in LS-Xbp1−/− and Xbp1fl/fl mice fed chow, cholestyramine, and cholesterol. Data are expressed as percent increase compared with chow-fed Xbp1fl/fl mice. Western blot (D) and densitometry quantification (E) of hepatic CYP7A1 protein expression in chow-fed mice and cholesterol-fed mice. GAPDH was used as a loading control. F: Hepatic gene expression of Abcg5 and Abcg8 was measured. *P < 0.05, **P < 0.01, ***P < 0.001 compared with chow-fed Xbp1fl/fl mice. +P < 0.05, ++P < 0.01 compared with chow-fed LS-Xbp1−/− mice. #P < 0.05 compared with cholestyramine-fed Xbp1fl/fl mice. ¶¶P < 0.01 compared with cholesterol-fed Xbp1fl/fl mice. n = 3–5.
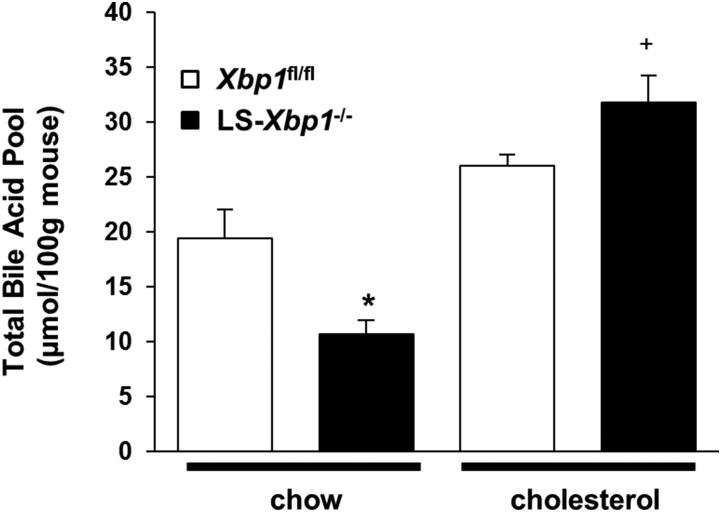
In contrast, when we treated LS-Xbp1−/− and Xbp1fl/fl mice with 2% cholesterol for 7 days, hepatic CYP7A1 protein expression did not change in either genotype of mice (Fig. 6D, E), even though the mRNA expression of the liver X receptor (LXR) target genes, Abcg5 and Abcg8, was upregulated in both genotypes (Fig. 6F). However, serum C4 levels increased in response to cholesterol feeding in both genotypes (Fig. 6C, P < 0.05). Although, hepatic CYP7A1 protein expression did not change in LS-Xbp1−/− mice, cholesterol feeding raised the serum C4 levels up to the baseline levels present in the chow-fed Xbp1fl/fl mice. Cholesterol feeding, but not induction of hepatic CYP7A1 protein levels with cholestyramine, increased the serum C4 levels of LS-Xbp1−/− mice. Consistent with these findings, cholesterol feeding increased the total bile acid pool size in LS-Xbp1−/− mice and corrected the genotypic differences in total bile acid pool size between Xbp1fl/fl mice and LS-Xbp1−/− mice (Fig. 7).
Fig. 7.
Total bile acid pool in Xbp1fl/fl and LS-Xbp1−/− mice after cholesterol feeding. LS-Xbp1−/− and Xbp1fl/fl mice were fed chow or chow with cholesterol (2% w/w) for 1 week. The total bile acid pool was measured. Cholesterol feeding corrected the total bile acid pool in LS-Xbp1−/− mice. *P < 0.05 compared with chow-fed Xbp1fl/fl mice (n = 4–5). +P < 0.05 compared with chow-fed LS-Xbp1−/− mice (n = 4–5).
DISCUSSION
Bile acids are synthesized in the liver and regulate metabolism in both normal physiology and pathophysiologic states. Cholestasis and other hepatic disorders can induce ER stress and activate the UPR (5, 20–22). Although UPR activation is a compensatory and protective response to ER stress, the hepatic IRE1α/XBP1 pathway of the UPR also regulates hepatic fatty acid and lipoprotein metabolism and secretion (9, 10, 23). Bile acids are biliary lipids that regulate many hepatic lipid metabolic and transport processes. Therefore, we investigated the role of the IRE1α/XBP1 pathway in bile acid metabolism.
We initially determined that liver-specific deletion of Xbp1 resulted in a reduced bile acid pool and decreased in vivo bile acid synthesis. Hepatic and ileal bile acid content was reduced, and TC and TMCA content in the bile acid pool was also lower in LS-Xbp1−/− mice than in Xbp1fl/fl littermate controls. Fecal bile acid excretion was similar in both genotypes, despite the different bile acid pool size. The bile acid cycling time and fecal loss per cycle are unknown in LS-Xbp1−/− mice, which could account for the observed fecal excretion. Fecal bile acid loss was low, and we cannot exclude the possibility that small differences in fecal loss of bile acid were not detected. Nonetheless, this does not detract from the significant findings of reduced serum C4 levels and decreased bile acid synthesis in LS-Xbp1−/− mice.
RNA-Seq (with confirmatory qPCR) and KEGG pathway analysis demonstrate that LS-Xbp1−/− mice had reduced expression of several hepatic genes involved in primary bile acid biosynthesis and bile secretion pathways. Many of these genes are regulated by the farnesoid X receptor, suggesting that their reduced expression may be either a primary effect of Xbp1 deletion or a secondary effect due to the reduced bile acid pool. Hepatic mRNA expression of the major bile acid synthetic enzymes, Cyp7a1, Cyp27a1, and Cyp8b1, was not reduced and, therefore, was not the primary cause of the reduced bile acid pool. In fact, Cyp7a1 gene expression increased, although protein expression was unchanged. Despite the changes in the bile acid pool, hepatic gene expression of Shp and ileal expression of Fgf15 and other ileal bile acid metabolic genes remained unchanged. Hepatic gene expression of the bile acid synthetic pathway genes, Slc27a5 and Akr1d1, was reduced in LS-Xbp1−/− mice. Although human mutations of these genes have been reported to be associated with rare cholestatic liver diseases (24–26), these enzymes are not believed to be rate-limiting for bile acid synthesis. Moreover, Slc27a5 and Akr1d1 are downstream to the production of C4 in the bile acid synthesis pathway. If the reduced bile acid synthesis was due to these enzymes, serum C4 levels should not be reduced and may in fact be elevated.
Serum C4 levels indicated that in vivo CYP7A1 synthetic activity was reduced in LS-Xbp1−/− mice, while Western blotting demonstrated that both hepatic and microsomal CYP7A1 protein levels were unchanged. CYP7A1 is not known to be posttranslationally regulated by phosphorylation and endogenous physiologic inhibitors for this enzyme have not been described. Our data on microsomal CYP7A1 protein expression also do not suggest that CYP7A1 protein in the ER was reduced. When CYP7A1 protein expression was induced by cholestyramine feeding in LS-Xbp1−/− mice, serum C4 levels did not increase and remained lower than those in Xbp1fl/fl mice, even though CYP7A1 protein levels were 8-fold higher than the CYP7A1 levels in chow-fed Xbp1fl/fl mice. Therefore, increasing CYP7A1 protein level by cholestyramine could not rescue CYP7A1 synthetic activity.
RNA-Seq data demonstrate that genes involved in cholesterol metabolism are downregulated in LS-Xbp1−/− mice relative to Xbp1fl/fl mice, which could cause reductions of cholesterol biosynthesis and intracellular trafficking. We observed decreases in serum cholesterol levels in the LS-Xbp1−/− mice. Therefore, we verified the hypocholesterolemia in this albumin-Cre-driven XBP1 deletion model and it is consistent with a previous report of hepatic XBP1-deficient mice using Mx1-Cre with poly (I:C) administration (9). Mice lacking hepatic XBP1 have nearly absent LDL levels and a lesser reduction of serum HDL (9, 10). Because cholesterol is the precursor for bile acid synthesis, alterations in hepatic cholesterol metabolism in LS-Xbp1−/− mice may affect bile acid synthesis and the bile acid pool.
Feeding rodents a diet supplemented with cholesterol increases the specific activity (picomoles per milligram protein per minute) of hepatic CYP7A1 and enriches hepatic microsomal cholesterol content (27, 28). When we fed LS-Xbp1−/− mice diets supplemented with 2% cholesterol for 1 week, hepatic CYP7A1 protein expression did not change; however, CYP7A1 synthetic activity increased. In fact, serum C4 levels in the cholesterol-fed LS-Xbp1−/− mice increased to the baseline levels present in chow-fed Xbp1fl/fl mice. Thus, cholesterol feeding rescued the diminished CYP7A1 activity in LS-Xbp1−/− mice, while the 8-fold increases of CYP7A1 protein induced by cholestyramine feeding had no effect.
Cholesterol is the substrate of CYP7A1 enzymatic activity. Gene expression of cholesterol biosynthesis and metabolic pathways are downregulated in LS-Xbp1−/− mice; and dietary feeding of cholesterol restores CYP7A1 activity and completely corrects the bile acid pool size phenotype. Therefore, the reduction in CYP7A1 activity in LS-Xbp1−/− mice may be due to decreased cholesterol availability to CYP7A1. Hepatic free cholesterol levels did not differ between Xbp1fl/fl and LS-Xbp1−/− mice, consistent with a previous report (9). However, the ER cholesterol content and the rate of cholesterol delivery to the ER in LS-Xbp1−/− mice are not known. In rodents, cholesterol feeding can increase hepatic CYP7A1 expression through LXR signaling (29–31). However, we did not observe increased expression of CYP7A1 after cholesterol feeding in Xbp1fl/fl mice, even though other LXR target genes, such as Abcg5 and Abcg8, were upregulated. Although this finding could be due to altered mouse genetics (wild-type vs. Xbp1fl/fl in C57BL/6 background strains), CYP7A1 expression in response to chronic cholesterol feeding may not always be increased under certain cholesterol feeding conditions (32).
This study demonstrates a novel role for hepatic XBP1 in the regulation of bile acid metabolism. Mice lacking hepatic Xbp1 have a reduced bile acid pool due to reduced CYP7A1 bile acid synthetic activity. In contrast to many studies on bile acid metabolism, the changes in hepatic CYP7A1 synthetic activity are not primarily due to changes of its level of expression, but are regulated by a cholesterol-responsive process. These data may have implications on hepatic and systemic lipid metabolism, as well as in the pathogenesis and treatment of cholestatic and fatty liver disorders.
Footnotes
Abbreviations:
- C4
- 7α-hydroxy-4-cholesten-3-one
- CYP7A1
- cholesterol 7α-hydroxylase
- ER
- endoplasmic reticulum
- GO
- Gene Ontology
- IRE1α
- inositol-requiring enzyme 1α KEGG, Kyoto Encyclopedia of Genes and Genomes
- LS-Xbp1−/−
- liver-specific X-box binding protein 1 knockout
- LXR
- liver X receptor
- TC
- taurocholic acid
- TMCA
- tauromuricholic acid
- UPR
- unfolded protein response
- XBP1
- X-box binding protein 1
This work was supported by National Institute of Diabetes and Digestive and Kidney Diseases Grant R01 DK093807, the George Lockerbie Liver Cancer Foundation, and the Max Goldenberg Foundation. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
REFERENCES
- 1.Thomas C., Pellicciari R., Pruzanski M., Auwerx J., and Schoonjans K.. 2008. Targeting bile-acid signalling for metabolic diseases. Nat. Rev. Drug Discov. 7: 678–693. [DOI] [PubMed] [Google Scholar]
- 2.Li T., and Chiang J. Y.. 2014. Bile acid signaling in metabolic disease and drug therapy. Pharmacol. Rev. 66: 948–983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Russell D. W. 2003. The enzymes, regulation, and genetics of bile acid synthesis. Annu. Rev. Biochem. 72: 137–174. [DOI] [PubMed] [Google Scholar]
- 4.Chiang J. Y. 2009. Bile acids: regulation of synthesis. J. Lipid Res. 50: 1955–1966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Malhi H., and Kaufman R. J.. 2011. Endoplasmic reticulum stress in liver disease. J. Hepatol. 54: 795–809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Liu X., Henkel A. S., LeCuyer B. E., Schipma M. J., Anderson K. A., and Green R. M.. 2015. Hepatocyte X-box binding protein 1 deficiency increases liver injury in mice fed a high-fat/sugar diet. Am. J. Physiol. Gastrointest. Liver Physiol. 309: G965–G974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lee J., and Ozcan U.. 2014. Unfolded protein response signaling and metabolic diseases. J. Biol. Chem. 289: 1203–1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hur K. Y., So J. S., Ruda V., Frank-Kamenetsky M., Fitzgerald K., Koteliansky V., Iwawaki T., Glimcher L. H., and Lee A. H.. 2012. IRE1alpha activation protects mice against acetaminophen-induced hepatotoxicity. J. Exp. Med. 209: 307–318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lee A. H., Scapa E. F., Cohen D. E., and Glimcher L. H.. 2008. Regulation of hepatic lipogenesis by the transcription factor XBP1. Science. 320: 1492–1496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.So J. S., Hur K. Y., Tarrio M., Ruda V., Frank-Kamenetsky M., Fitzgerald K., Koteliansky V., Lichtman A. H., Iwawaki T., Glimcher L. H., et al. . 2012. Silencing of lipid metabolism genes through IRE1alpha-mediated mRNA decay lowers plasma lipids in mice. Cell Metab. 16: 487–499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Glimcher L. H., and Lee A. H.. 2009. From sugar to fat: How the transcription factor XBP1 regulates hepatic lipogenesis. Ann. N. Y. Acad. Sci. 1173(Suppl 1): E2–E9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Figge A., Lammert F., Paigen B., Henkel A., Matern S., Korstanje R., Shneider B. L., Chen F., Stoltenberg E., Spatz K., et al. . 2004. Hepatic overexpression of murine Abcb11 increases hepatobiliary lipid secretion and reduces hepatic steatosis. J. Biol. Chem. 279: 2790–2799. [DOI] [PubMed] [Google Scholar]
- 13.Gälman C., Arvidsson I., Angelin B., and Rudling M.. 2003. Monitoring hepatic cholesterol 7alpha-hydroxylase activity by assay of the stable bile acid intermediate 7alpha-hydroxy-4-cholesten-3-one in peripheral blood. J. Lipid Res. 44: 859–866. [DOI] [PubMed] [Google Scholar]
- 14.Pattni S. S., Brydon W. G., Dew T., and Walters J. R.. 2012. Fibroblast growth factor 19 and 7alpha-hydroxy-4-cholesten-3-one in the diagnosis of patients with possible bile acid diarrhea. Clin. Transl. Gastroenterol. 3: e18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gälman C., Angelin B., and Rudling M.. 2005. Bile acid synthesis in humans has a rapid diurnal variation that is asynchronous with cholesterol synthesis. Gastroenterology. 129: 1445–1453. [DOI] [PubMed] [Google Scholar]
- 16.Steiner C., Othman A., Saely C. H., Rein P., Drexel H., von Eckardstein A., and Rentsch K. M.. 2011. Bile acid metabolites in serum: intraindividual variation and associations with coronary heart disease, metabolic syndrome and diabetes mellitus. PLoS One. 6: e25006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hashimoto M., Kobayashi K., Watanabe M., Kazuki Y., Takehara S., Inaba A., Nitta S., Senda N., Oshimura M., and Chiba K.. 2013. Knockout of mouse Cyp3a gene enhances synthesis of cholesterol and bile acid in the liver. J. Lipid Res. 54: 2060–2068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schwarz M., Russell D. W., Dietschy J. M., and Turley S. D.. 2001. Alternate pathways of bile acid synthesis in the cholesterol 7alpha-hydroxylase knockout mouse are not upregulated by either cholesterol or cholestyramine feeding. J. Lipid Res. 42: 1594–1603. [PubMed] [Google Scholar]
- 19.Chen J. Y., Levy-Wilson B., Goodart S., and Cooper A. D.. 2002. Mice expressing the human CYP7A1 gene in the mouse CYP7A1 knock-out background lack induction of CYP7A1 expression by cholesterol feeding and have increased hypercholesterolemia when fed a high fat diet. J. Biol. Chem. 277: 42588–42595. [DOI] [PubMed] [Google Scholar]
- 20.Tamaki N., Hatano E., Taura K., Tada M., Kodama Y., Nitta T., Iwaisako K., Seo S., Nakajima A., Ikai I., et al. . 2008. CHOP deficiency attenuates cholestasis-induced liver fibrosis by reduction of hepatocyte injury. Am. J. Physiol. Gastrointest. Liver Physiol. 294: G498–G505. [DOI] [PubMed] [Google Scholar]
- 21.Bochkis I. M., Rubins N. E., White P., Furth E. E., Friedman J. R., and Kaestner K. H.. 2008. Hepatocyte-specific ablation of Foxa2 alters bile acid homeostasis and results in endoplasmic reticulum stress. Nat. Med. 14: 828–836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sasaki M., Yoshimura-Miyakoshi M., Sato Y., and Nakanuma Y.. 2015. A possible involvement of endoplasmic reticulum stress in biliary epithelial autophagy and senescence in primary biliary cirrhosis. J. Gastroenterol. 50: 984–995. [DOI] [PubMed] [Google Scholar]
- 23.Wang S., Chen Z., Lam V., Han J., Hassler J., Finck B. N., Davidson N. O., and Kaufman R. J.. 2012. IRE1alpha-XBP1s induces PDI expression to increase MTP activity for hepatic VLDL assembly and lipid homeostasis. Cell Metab. 16: 473–486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Drury J. E., Mindnich R., and Penning T. M.. 2010. Characterization of disease-related 5beta-reductase (AKR1D1) mutations reveals their potential to cause bile acid deficiency. J. Biol. Chem. 285: 24529–24537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chong C. P., Mills P. B., McClean P., Gissen P., Bruce C., Stahlschmidt J., Knisely A. S., and Clayton P. T.. 2012. Bile acid-CoA ligase deficiency–a new inborn error of bile acid metabolism. J. Inherit. Metab. Dis. 35: 521–530. [DOI] [PubMed] [Google Scholar]
- 26.Setchell K. D., Heubi J. E., Shah S., Lavine J. E., Suskind D., Al-Edreesi M., Potter C., Russell D. W., O’Connell N. C., Wolfe B. , et al. 2013. Genetic defects in bile acid conjugation cause fat-soluble vitamin deficiency. Gastroenterology. 144: 945–955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tiemann M., Han Z., Soccio R., Bollineni J., Shefer S., Sehayek E., and Breslow J. L.. 2004. Cholesterol feeding of mice expressing cholesterol 7alpha-hydroxylase increases bile acid pool size despite decreased enzyme activity. Proc. Natl. Acad. Sci. USA. 101: 1846–1851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ren S., Marques D., Redford K., Hylemon P. B., Gil G., Vlahcevic Z. R., and Pandak W. M.. 2003. Regulation of oxysterol 7alpha-hydroxylase (CYP7B1) in the rat. Metabolism. 52: 636–642. [DOI] [PubMed] [Google Scholar]
- 29.Peet D. J., Turley S. D., Ma W., Janowski B. A., Lobaccaro J. M., Hammer R. E., and Mangelsdorf D. J.. 1998. Cholesterol and bile acid metabolism are impaired in mice lacking the nuclear oxysterol receptor LXR alpha. Cell. 93: 693–704. [DOI] [PubMed] [Google Scholar]
- 30.Chiang J. Y., Kimmel R., and Stroup D.. 2001. Regulation of cholesterol 7alpha-hydroxylase gene (CYP7A1) transcription by the liver orphan receptor (LXRalpha). Gene. 262: 257–265. [DOI] [PubMed] [Google Scholar]
- 31.Gupta S., Pandak W. M., and Hylemon P. B.. 2002. LXR alpha is the dominant regulator of CYP7A1 transcription. Biochem. Biophys. Res. Commun. 293: 338–343. [DOI] [PubMed] [Google Scholar]
- 32.Henkel A. S., Anderson K. A., Dewey A. M., Kavesh M. H., and Green R. M.. 2011. A chronic high-cholesterol diet paradoxically suppresses hepatic CYP7A1 expression in FVB/NJ mice. J. Lipid Res. 52: 289–298. [DOI] [PMC free article] [PubMed] [Google Scholar]