Abstract
Vitreoscilla filiformis is a Gram-negative bacterium isolated from spa waters and described for its beneficial effects on the skin. We characterized the detailed structure of its lipopolysaccharide (LPS) lipid A moiety, an active component of the bacterium that contributes to the observed skin activation properties. Two different batches differing in postculture cell recovery were tested. Chemical analyses and mass spectra, obtained before and after mild-alkali treatments, revealed that these lipids A share the common bisphosphorylated β-(1→6)-linked d-glucosamine disaccharide with hydroxydecanoic acid in an amide linkage. Short-chain FAs, hydroxydecanoic and dodecanoic acid, were found in a 2:1 ratio. The two lipid A structures differed by the relative amount of the hexa-acyl molecular species and phosphoethanolamine substitution of the phosphate groups. The two V. filiformis LPS batches induced variable interleukin-6 and TNF-α secretion by stimulated myelomonocytic THP-1 cells, without any difference in reactive oxygen species production or activation of caspase 3/7. Other different well-known highly purified LPS samples were characterized structurally and used as standards. The structural data obtained in this work explain the low inflammatory response observed for V. filiformis LPS and the previously demonstrated beneficial effects on the skin.
Keywords: cytokines, lipid biochemistry, lipopolysaccharide, mass spectrometry, skin, toll-like receptor, V. filiformis
Lipopolysaccharides (LPSs) are the main components of the outer-membrane of Gram-negative bacteria, representing about 106 molecules per bacterium (1). They are also major antigens displaying unique structures for each bacterium. This is a signature for which each small detail may exert important differences on LPS recognition by host cells and signal transduction (2). LPS does not necessarily induce a toxic response, so all LPSs are not endotoxins, but endotoxins are always composed of LPSs. LPSs are the first nonsecreted toxins found to interact with host innate immunity, generating cytokine production, immune cell recruitment, and specific host-cell responses leading, at higher doses, to multiple organ failure and septic shock (3). The smallest change in the structures of LPSs or their hydrophobic moiety, named lipid A, is directly linked to innate immune recognition and development of a specific host response. The interaction between LPS molecules and their innate receptor, the MD-2/TLR4 complex, is greatly affected by the fine details of LPS structures. First, LPS molecules are recognized by the LPS binding protein and presented as monomers to the soluble or membrane CD14 receptors. Then, the LPS is transferred to the MD-2/TLR4 complex (2, 4) allowing its dimerization of the receptor through ionic and hydrophobic interactions and activation of the cellular signaling pathways. Among these, the activation of the NFκB pathway directly induces rapid pro-inflammatory cytokine production. Secreted cytokines are ultimately central to the generation of efficient responses to pathogens and the activation of the adaptive immune system (5, 6).
The Neisseriaceae family members, including the Vitreoscilla genus, are obligate aerobic bacteria described for the first time in 1949 by E. G. Pringsheim (7) as colorless gliding filamentous organisms. Similar organisms were in fact described earlier by Cohn (8) in 1870. They are known as the first genus in which bacterial hemoglobin (VHb) was discovered (9).
After the discovery of the improvement of the atopic dermatitis skin syndrome by spa waters containing such nonpathogenic bacteria (10), interest was focused on the bacterial elements responsible for this beneficial effect.
During the last decades, biological activities have been found to be beneficial for the skin after application of Vitreoscilla filiformis biomass. These activities include: substance P antagonist (11), stimulation of skin anti-microbial peptide secretion, recovery of sunburn areas (12, 13), activation of MnSOD as an inducible free-radical scavenger in keratinocytes for protecting cells against the uncontrolled production of reactive oxygen species (ROS) (14), modulation of mouse and human cutaneous inflammatory responses inducing interleukin (IL)-10 production in dendritic cells (DCs), and priming of regulatory T cells (15). It was shown that the activation of two independent TLR pathways occurs following DC stimulation with a V. filiformis lysate (13, 15). First, the TLR2 pathway induces IL-10 secretion and activation of antioxidant responses. Second, the TLR4 pathway activation induces IL-12p70 production. In contrast to Escherichia coli LPSs, it was shown that V. filiformis LPSs stimulate anti-microbial peptide secretion by keratinocytes. DCs stimulated with V. filiformis LPS induce a Th1 polarization during T cell priming. This effect is not found with V. filiformis lysate stimulation. The role of V. filiformis lipid A, in contrast to E. coli lipid A, was found to promote a skin DC response through increased expression of costimulatory factors (B7-2) and phagocytosis (10, 16). Altogether, the effects of a V. filiformis lysate on the skin are dominated by TLR2 signaling and induction of tolerance (17), while V. filiformis lipid A acts through TLR4 signaling promoting pathogen clearance.
In dermatologic treatments including V. filiformis lysates, the structure of the biologically active part of the corresponding LPS had to be characterized. Herein, we present the chemical characterization of the lipids A of two closely related forms of LPS from V. filiformis and their TLR4-related myeloid response. The two LPS batches correspond to LPS extracted from a bacterial culture pellet, named V. filiformis (V.f)5-6, and LPS isolated from the whole biomass, including LPS from the pellet and LPS soluble in the growth medium, named V.f6+2 phosphoethanolamine (PEA) (V.f6+2PEA). We therefore stress that culture supernatants should always be tested for the presence of more soluble LPS molecules, which could be underestimated and lost while being part of LPS-relevant molecules for a given experiment.
We also demonstrate the importance of MS during the process of structural characterization of these molecules and for the control of LPS structures known to vary according to different bacterial growth conditions and showing discrepancies with structures described in the literature.
MATERIALS AND METHODS
Bacterial strains
V. filiformis (ATCC® 15551TM) was collected in two different manners as described in the European patent 2144995 A1 (18). The μ production was regulated at 0.12 H−1, the temperature at 26°C, and the pH at 7.00. The biomass was harvested by centrifugation (9,000 g at 4°C) and then stabilized by lyophilization.
Two batches were examined for lipid A characterization, the first batch corresponded to a first growth corresponding to the bacterial pellet of the whole biomass after centrifugation. The second corresponded to another growth in similar conditions, without pellet and supernatant separation before LPS extraction (i.e., the whole biomass).
E. coli J5 (ATCC® 43745TM) was cultured in lysogeny broth at 37°C overnight in our laboratory.
Bordetella pertussis 1414 and Neisseria meningitis bacteria were grown at the Institut Mérieux, Lyon, France and LPSs were extracted in the laboratory.
Reference LPS samples
Several purified LPS samples with already described structures were used as references.
The Haemophilus influenzae Eagan LPS sample was a generous gift from the National Research Center (Ottawa, Canada).
Rhodopseudomonas gelatinosa and Pseudomonas aeruginosa LPS were kindly provided by LPS-BioSciences (Orsay, France).
LPS and lipid A preparation
All LPS samples were repurified to obtain the necessary quality for optimal conditions of MS analysis and no contamination by other bacterial components required for biological use, as shown by UV spectrometry, SDS-PAGE, TLC, and amino acid analyses (19, 20).
The V. filiformis LPSs were extracted by the isobutyric acid/1 M ammonium hydroxide method (21). LPSs were purified by enzymatic treatments to remove DNA, RNA, and proteins, as already described (22). They were also extracted with a mixture of solvents to remove phospholipids and lipoproteins. Lipid A was prepared by mild detergent-facilitated hydrolysis of LPS and purified as before (23).
Alternatively, lipid A was obtained by direct hydrolysis of the lyophilized bacteria (24). Briefly, 10 mg of lyophilized bacteria were suspended in 400 μl of isobutyric acid and 1 M ammonium hydroxide (5:3, v:v), heated 2 h at 100°C with stirring, cooled to 4°C, and centrifuged. The supernatant was diluted with water (1:1, v:v) and lyophilized. The material obtained was then washed twice with 400 μl of methanol and centrifuged (2,000 g for 15 min). Finally, the insoluble lipid A was extracted once in a 100 to 200 μl mixture of chloroform:methanol:water (3:1.5:0.25, v:v:v).
Monophosphorylated lipids A were obtained by HCl 0.1 M hydrolysis of the lipid A for 10 min at 100°C, neutralization, and recovery by ultracentrifugation.
Identification of glycose absolute configurations
Lipids A (4 mg) were hydrolyzed with 0.5 ml of 4 M HCl at 100°C for 2 h. After cooling and extraction of FAs with chloroform, residual solutions were brought to neutrality by repeated evaporation under reduced pressure. After N-acetylation, the residue was treated with trifluoroacetic acid, R-(−)-2-butanol, per-acetylated, and analyzed by GC on a BP10 capillary column (Scientific Glass Engineering) using a program of 160°C (1 min) to 220°C, 5°C min−1 at 0.6 kPa (25).
Sequential liberation of ester-linked FAs by mild alkali treatment
This treatment was used to establish the lipid A acylation patterns (22). For the first-step liberation of primary ester-linked FAs, lipid A (200 μg) was suspended at 1 mg/ml in 28% ammonium hydroxide and stirred for 5 h at 50°C. The solutions were dried under a stream of nitrogen, the residues taken up in a mixture of chloroform:methanol:water (3:1.5:0.25, v:v) followed by MALDI-MS analysis. In this case, kinetics (15 min, 30 min, 2 h 30 min, and 5 h) were done to follow the complete process in parallel to the B. pertussis lipid A kinetics taken as a reference (26).
Chemical analyses
FAs were analyzed as in (27). Briefly, LPSs (1 mg) were submitted to strong acid treatment, 4 M HCl, 2 h at 100°C with 20:0 (20 μg) as internal standard, extraction with ethyl acetate, and esterification with diazomethane. GC-MS analysis was performed as before (28) using a Finnigan Mat 95S mass spectrometer.
The aqueous phase containing the free soluble compounds was lyophilized and hydrolyzed with 6 M HCl for 6 h at 95°C in order to remove the glucosamine (GlcN) phosphate group. Acid was removed under vacuum and the residue, recovered into a citrate buffer, was subjected to hexosamine, ethanolamine (EA), and PEA analysis.
The contents of these components were measured with a Hitachi L-8800 amino acid analyzer equipped with a 2620 MSC-PS column (ScienceTec, Les Ulis, France). The elution protocol recommended by the manufacturer for the separation of amino acids and hexosamine was used. Under these conditions, PEA, GlcN, and EA were eluted respectively at 3.5, 39.84, and 46.91 min.
TLC
TLC was done on glass HPTLC silica gel plates (Merck). Twenty micrograms of LPS were deposited at the origin of the HPTLC plate and chromatographed in a solvent mixture of isobutyric acid and 1 M ammonium hydroxide (5:3, v:v) (29). Products were visualized by charring (in an oven at 150°C for 5 min) after spraying with 10% sulfuric acid in ethanol.
SDS-polyacrylamide gel analysis of LPS
Fifteen percent polyacrylamide gel was used and 0.2 μg of LPS were loaded onto the 4% stacking gel. The LPS sample preparation, electrophoresis process, and the Tsai and Frasch silver nitrate coloration were performed as previously described (30, 31).
MALDI-MS analysis
MALDI-MS was done in the linear mode with delayed extraction using a PerSeptive Voyager STR (PE Biosystems, France) time-of-flight mass spectrometer and/or Shimadzu Axima Performance system. A suspension of lipid A (1 mg/ml) in chloroform:methanol:water (3:1.5:0.25, v:v:v) was desalted with a few grains of Dowex 50W-X8 (H+), 1 μl was deposited on the target mixed with 1 μl of the gentisic acid (2,5-dihydroxybenzoic acid) matrix suspended at 10 μg/μl in the same solvent or in 0.1 M aqueous citric acid, and dried (23). Analyte ions were desorbed from the matrix with pulses from a 337 nm nitrogen laser. Spectra were obtained in the negative-ion mode at 20 kV. When LPSs were analyzed directly, they were suspended in water and decationized in the same way, using the same matrix and conditions.
Cell culture
ATCC® TIB-202TM THP-1 cells were cultured in RPMI 1640 medium supplemented with 10% heat-inactivated (56°C for 30 min) fetal bovine serum (PAA Laboratory, GE Lifesciences), 100 IU/ml penicillin, and 100 μg/ml streptomycin under standard conditions.
LPS stimulations and cytokine production
For stimulations, 5 × 105 myelomonocytic cells (THP-1) were seeded in each well of a 12-well tissue culture plate (CELLSTAR®, Greinerbio-one, Gmbh, Germany) and stimulated with 0, 5, 10, 25, and 100 ng/ml LPS. THP-1 cells were stimulated for 6 or 30 h and TNF-α and IL-6 were measured in cell-free supernatants by ELISA kit (Ready-Set-Go!® human IL-6 and human TNF-α eBioscience, San Diego, CA). The OD of each well was read by using a microplate reader (96-well Maxisorp Nunc) at 450 nm (Multiskan EX; Thermo Fisher Scientific, Waltham, MA).
ROS production and caspase 3/7 activation
THP-1 cells (1 × 106) were seeded in 12-well tissue culture plates and stimulated for 1 or 24 h with or without LPS (100 ng/ml). Different controls were realized, N-acetyl-cysteine (10 mmol/l; HIDONAC, Zambon, France), a ROS taken as a production inhibitor, was incubated for 1 h before LPS stimulation. ROS induction was obtained with polymyristate-acetate (Sigma-Aldrich, St. Louis, MO) at 100 ng/ml incubation for 5 min just before labeling. Caspase 3/7 activation was controlled with polymyristate-acetate incubation at 100 ng/ml for 3 h before the 24 h-culture. Shortly, after stimulation, cells were incubated with mouse anti-human monoclonal antibody CD14 PE-Cy5.5® TuK4 (Invitrogen, Carlsbad, CA) for 15 min at room temperature. Cells were resuspended in PBS and labeled for either ROS production (CellRox Green reagent; Molecular Probes, Life Technologies, Carlsbad, CA) or caspase 3/7 activation (CellEvent Caspase3/7 Green detection reagent; Molecular Probes, Life Technologies) according to manufacturer’s instructions. Cell viability was measured with LIVE/DEAD fixable aqua dead cell stain (Molecular Probes, Life Technologies, Eugene, OR). Samples were analyzed with an Attune acoustic focusing cytometer (Thermo Fisher Scientific, Germantown, MD). Events (20,000–30,000) on CD14+ live cells were counted to measure intensity emission. Data were analyzed with the Attune Cytometer software v2.1. Data groups were compared two by two with the statistical Student’s t-test.
RESULTS
Total FA composition of the two forms of V. filiformis lipid A
Lipid A FA composition was found to correspond to 3-hydroxydecanoic acid [10:0(3-OH)] and dodecanoic acid (12:0) in the proportions 1.8:1 for the pellet sample and 1.6:1 for the whole biomass sample. Taking into account the heterogeneity of these lipids A and the tendency toward underestimation of the short-chain FAs, these proportions translated to 2 units of 10:0(3-OH) and 1 unit of 12:0 for the main molecular species.
The anomer of the GlcN-I glycosidic phosphate was determined to be α by the kinetics of phosphate release compared with the α- and β-P anomers of synthetic GlcNAc references. The glycose absolute configuration of D-GlcN was determined by GC-MS compared with l- and d-GlcN reference samples (32).
Molecular heterogeneity and distribution of the FAs between the two d-GlcN residues in lipid A from two V. filiformis bacterial samples
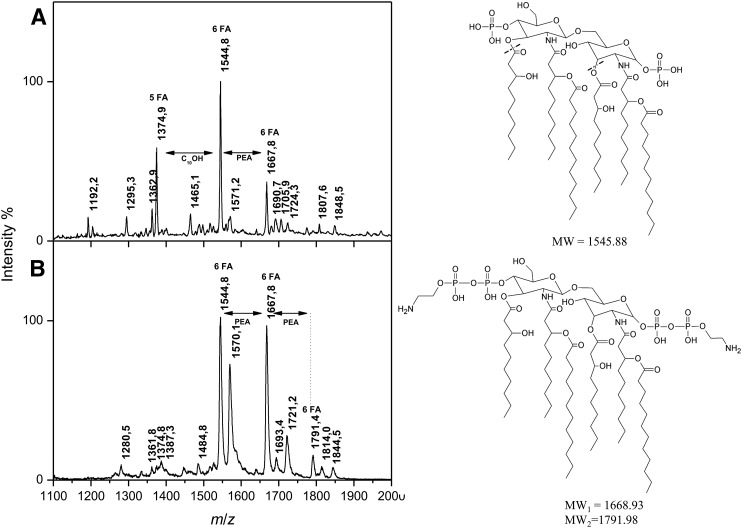
Negative-ion mass-spectra of lipid A isolated from the pellet and the whole biomass samples are shown in Fig. 1A, B, respectively. They share the major peak of deprotonated [M-H]− ions observed at m/z 1,544.8. Based on the overall chemical composition, this peak is attributed to a molecular species containing two GlcN, two phosphates, four hydroxydecanoic acids, and two dodecanoic acids (theoretical molecular weight (MW) = 1,545.88). Another common molecular ion observed at m/z 1667.8 carries an additional PEA residue (+123 u). While its abundance is relatively low for the pellet sample, it represents a major molecular species for the whole biomass sample. Interestingly, the latter also contains a molecular species with two PEA residues, which is observed at m/z 1,791.4. This means that both phosphate groups of the lipid A isolated from the whole biomass sample can be substituted, and that the level of PEA substitution varies from one batch to another. The pellet sample also displays more heterogeneity due to the presence of a penta-acyl molecular species observed at m/z 1,374.9 and containing three 10:0(3-OH) residues and two 12:0 residues. Other peaks observed in the mass-spectra correspond to dephosphorylated ions whose presence is most probably due to hydrolysis (corresponding molecular species are observed at −80 u, e.g., at m/z 1,295.3 and 1,465.1) or due to the MS fragmentation process (−98 u, e.g., m/z 1,570.1 and 1,693.4). In the light of the present data, we named the LPS from the pellet sample V.f5-6 to reflect the number of FAs in the two main molecular species, and the LPS from the biomass sample was named V.f6+2PEA to reflect the number of FAs in the main molecular species combined with PEA.
Fig. 1.
Negative-ion MALDI-TOF mass spectra of the lipid A moieties of the two V. filiformis LPS samples obtained after SDS-promoted pH 4.5 hydrolysis. A: V. filiformis lipid A isolated from the pellet sample (V.f5-6). B: V. filiformis lipid A isolated from the biomass sample (V.f6+2PEA).
The negative-ion mass-spectrum of monophosphorylated lipid A from V.f6+2PEA (data not shown) displays a major peak at m/z 1,465 and another one at m/z 1,587. These peaks correspond to monophosphoryl hexa-acyl lipid A produced by the loss of the glycosyl phosphate or EA pyrophosphate at the C1 position. In the molecular species observed at m/z 1,465, the C4′ position is substituted with phosphate, and the one observed at m/z 1,587 is substituted with EA pyrophosphate. This result validates the presence of two PEA residues in this lipid A.
A postsource-decay negative-mode experiment performed by MALDI-MS on the ion at m/z 1,668 (not shown) gave three main daughter-ion peaks appearing at m/z 1,570 (−98 u, phosphate) and at m/z 122.9 (PEA) and 98 (phosphate) in the low-mass region. The presence of EA and PEA was also confirmed after hydrolysis, and detection of the soluble released substituents was confirmed with the use of an amino acid analyzer after elution and comparison with standard molecules.
Kinetics of sequential liberation of ester-linked FAs from the V.f5-6 lipid A
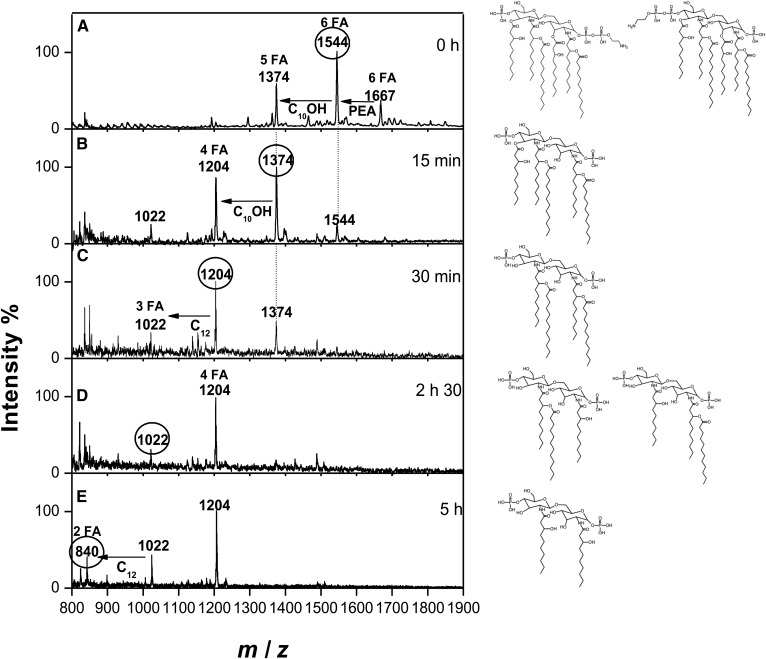
The kinetics of lipid A deacylation presented in Fig. 2 revealed, after 15 min of treatment, the almost complete release of the FA at C3, according to previous data (26). It corresponded to a 10:0(3-OH), according to the mass difference, which agrees with the FA composition previously determined. It was also linked to the concomitant increase of the penta-acyl molecular species at m/z 1,374, with the disappearance of the hexa-acylated molecular species at m/z 1,544. After 30 min, the peak at m/z 1,374 became a minor one and a molecular species at m/z 1,204 appeared, corresponding to a tetra-acyl molecular species. The loss of the two 10:0(3-OH) at C3 and C3′ was complete at 2 h 30 min and confirmed by GC/MS. The mass difference also corresponded to the loss of two 10:0(3-OH). This demonstrated that these 10:0(3-OH) were not carrying any other FA on their hydroxyl groups in the native structure. Therefore, the two 12:0 had to be substituting the two remaining 10:0(3-OH) present in amide linkage, as shown by MALDI-MS. The release of secondary 12:0 from the C2 and C2′ positions was much slower, and was not even completed after 5 h of treatment. A tri-acylated molecular species corresponding to a peak at m/z 1,022 increased progressively along the kinetics, it corresponded to the loss of a branched 12:0. The release of a second branched 12:0 led to a di-acyl molecular species at m/z 840. All data contributed to the design of a symmetrical FA distribution for the two V. filiformis lipid A structures. The FA distribution was also confirmed by fragmentation in the positive-ion mode giving a B1 fragment ion at m/z 764 (not shown) corresponding to GlcN-II, one phosphate residue, two 10:0(3-OH) residues, and one 12:0 residue (33, 34).
Fig. 2.
Monitoring of the FA release from the lipid A by negative-ion MALDI-TOF MS during alkaline hydrolysis and the corresponding structures. Example of V.f5-6 before hydrolysis (A), and 15 min (B), 30 min (C), 2 h 30 min (D), and 5 h (E) after hydrolysis. The structures presented in this figure correspond to the marked masses appearing in the spectra.
MALDI-MS analysis of LPS from different bacterial genera.
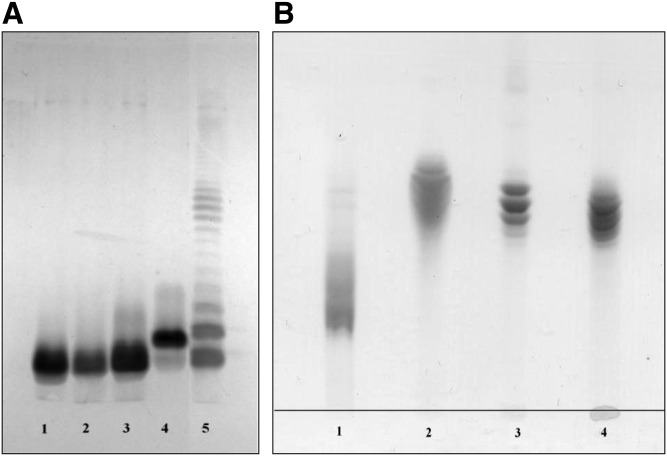
Bordetella, Escherichia, Haemophilus, Neisseria, Pseudomonas, and Rhodopseudomonas LPSs were highly purified for structural and biological comparison with the two V. filiformis LPSs. SDS-PAGE and TLC profiles of the reference samples used in this work are presented in Fig. 3. The predominant lipid A moiety molecular species of these LPS samples are presented in Fig. 4.
Fig. 3.
Analyses of purified LPS from the two V. filiformis samples compared with LPS standards by SDS-PAGE (A) and HPTLC (B). A: 1, E. coli J5 LPS; 2, V. filiformis V.f5-6 LPS; 3, V. filiformis V.f6+2PEA LPS; 4, B. pertussis 1414 LPS; 5, P. aeruginosa serotype O6 LPS. One-tenth of a microgram of each LPS was loaded per well for the SDS-PAGE electrophoresis. B: 1, B. pertussis 1414 LPS; 2, E. coli J5; 3, V. filiformis V.f5-6; 4, V. filiformis V.f6+2PEA. Twenty micrograms were deposited for HPTLC migration.
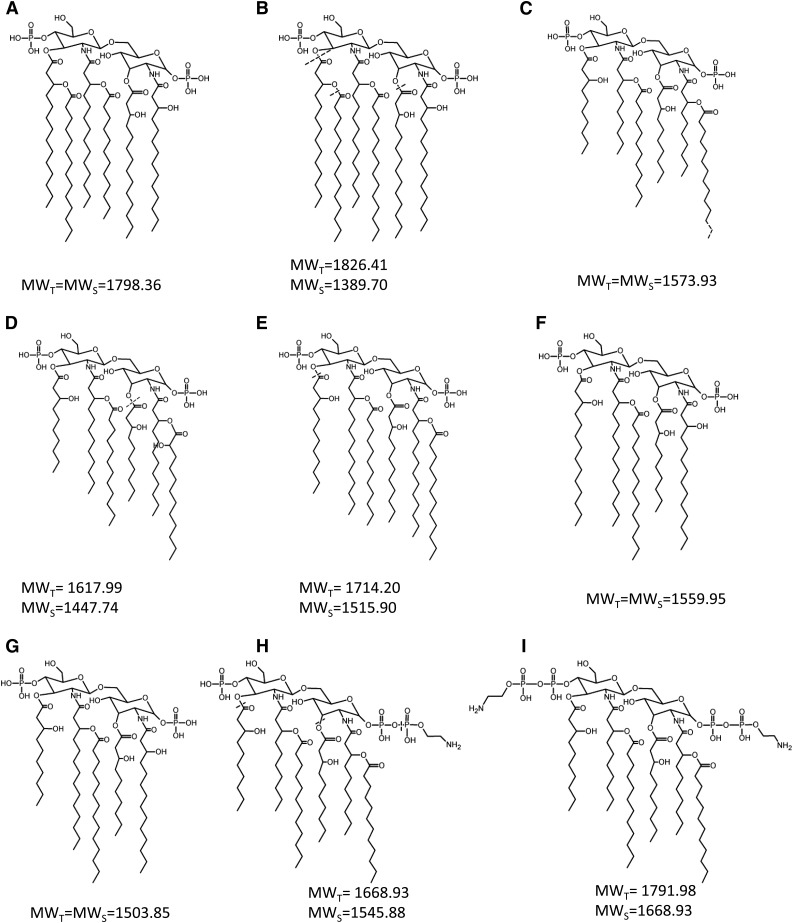
Fig. 4.
Lipid A structures of the major molecular species present in the purified LPS samples. The structures presented with the corresponding MWT given masses correspond to published data for all LPSs. The dashed lines indicate elements missing in the structures for each LPS sample used in this paper and corresponding to MWS. E. coli J5 (A), H. influenzae (B), R. gelatinosa (C), P. aeruginosa (D), N. meningitis (E), B. pertussis 1414 (F), B. pertussis 18-323 (G), V. filiformis V.f5-6 (H), V. filiformis V.f6+2PEA (I).
E. coli J5 lipid A structure is composed of the classical hexa-acylated molecular species (35). Two weaker signals corresponding to tetra- and penta-acyl molecular species were observed in this lipid A sample.
H. influenzae lipid A is usually not very heterogeneous, as described before (36), but the tested sample is composed of a major tetra-acyl molecular species followed by the penta-, tri-, and hexa-acyl molecular species with smaller intensity. Choi et al. (37) have already described the lipid A heterogeneity for this sample.
Frequently, Neisseria meningitidis lipid A structures differ from strain to strain by the presence of one or two PEA decorations on the GlcN (38). In our sample, the predominant LPS species possesses five FAs and does not display any PEA decoration.
B. pertussis 18-323 lipid A structure was shown, in our laboratory, to differ from the vaccine strain B. pertussis 1414 lipid A structure by the presence of a 10:0(3-OH) on the C3′ instead of a 14:0(3-OH) (39). The two detected major molecular species for B. pertussis 1414 are the tetra- and the penta-acylated forms, the latter lacking the 14:0(3-OH) at C3′.
P. aeruginosa lipid A structure has been described as composed of a bisphosphorylated GlcN disaccharide substituted by two amide-linked 12:0(3-OH), themselves substituted by one 12:0(2-OH) and one 12:0. The two other FAs on C3 and the C3′ are both 10:0(-3OH) (40). When we examined the strain at our disposal by MALDI-MS, the lipid A structure established according to previous studies was essentially composed of the penta-acyl molecular species, the 10:0(3-OH) at C3 being absent. Only small amounts of the hexa-acyl molecular species were detected.
The lipid A structure of the tested R. gelatinosa sample was found to be composed of six FAs, including four 10:0(3-OH), one 12:0, and one 12/14:0, as already described (41).
Comparison of IL-6 and TNF-α production after LPS stimulation
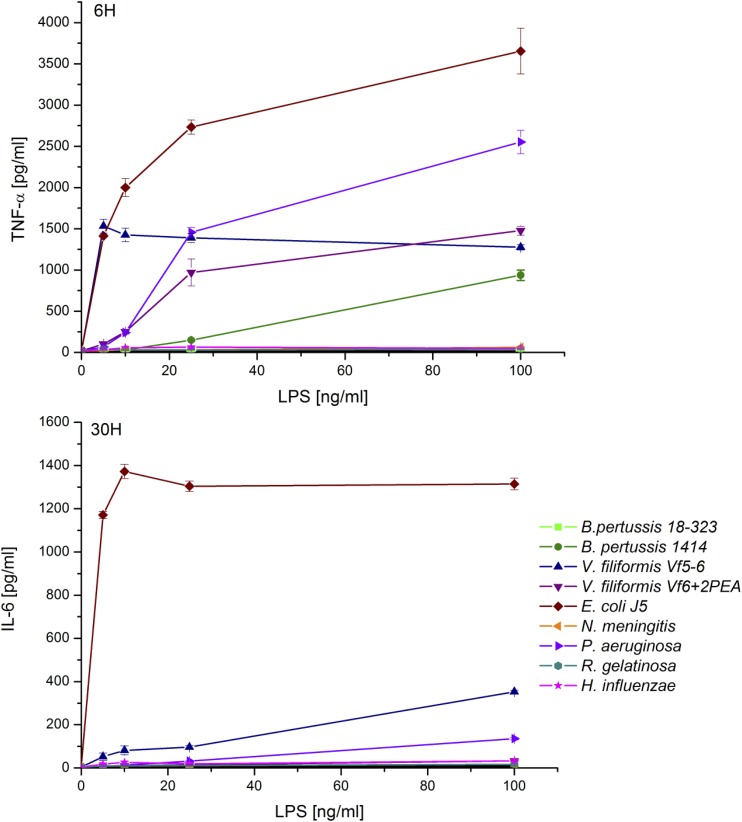
Although the pro-inflammatory response to stimulation by both V. filiformis LPS samples was lower than with E. coli J5 LPS, both batches showed differences in TNF-α and IL-6 responses (Fig. 5). Early cytokine production, as displayed by TNF-α, showed that V.f5-6 reaches a plateau at lower concentration (5 ng/ml) than V.f6+2PEA (25 ng/ml). In addition, late IL-6 production was measurable for V.f5 at 30 h, but not for V.f6+2PEA. In our experiment, E. coli J5 LPS showed the stronger IL-6 and TNF-α responses, whereas H. influenzae, B. pertussis 18-323, R. gelatinosa, and N. meningitidis LPS did not induce any cytokine production. B. pertussis 1414 and P. aeruginosa displayed an intermediate TNF-α and IL-6 response.
Fig. 5.
THP-1 cells pro-inflammatory response to LPS stimulation. TNF-α and IL-6 production following LPS dose response stimulation on THP-1 cells. The results presented on the figure are representative of three independent experiments, and were realized in triplicate. Results are expressed as mean ± SEM.
Caspase 3/7 activation and ROS production
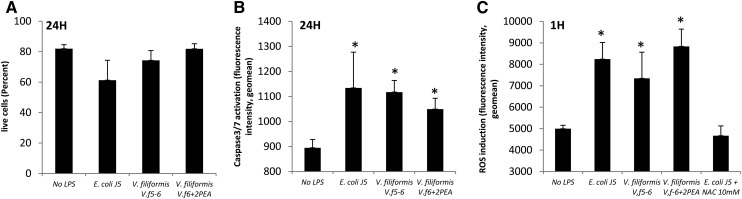
In order to better characterize the cellular response to V. filiformis LPS stimulation, ROS production and caspase 3/7 activation were compared with those of E. coli J5 LPS. No difference in cell viability was observed after E. coli J5 or both V. filiformis LPS stimulations (Fig. 6A). Caspase 3/7 activation was evaluated at 0, 1, 12, 24, and 48 h after stimulation and for the ROS induction at 0, 0.5, 1, 2, 12, 24, and 48 h (data not shown). Significant caspase 3/7 activation occurred after 12 h of LPS stimulation, reaching a plateau after 24 h. No difference in caspase 3/7 activation was found between E. coli J5 and V. filiformis LPS stimulations (Fig. 6B). ROS production started as early as 30 min, reaching a plateau after 1 h of LPS stimulation. No significant difference in ROS production was found after E. coli J5 or V. filiformis LPS stimulations (Fig. 6C).
Fig. 6.
ROS production and caspase 3/7 activation induced by LPS stimulation. Cell viability (A) and caspase 3/7 activation (B) were assessed after 24 h THP-1 stimulation. C: ROS production by THP-1 cells after 1 h stimulation. N-acetyl-l-cysteine was used to inhibit ROS production in E. coli J5 stimulation. *P < 0.05 compared to no LPS.
DISCUSSION
We describe here, for the first time, the structure of V. filiformis lipid A. This was important in order to understand the previously described remarkable skin activation capacities of this molecule in the bacterial lysate (42). We characterized two different batches, and found some differences. This structural variability in V. filiformis batches induces significant differences in some of the tested biological responses, stressing the necessity of controlling the fine structures in each LPS sample used for structure to activity relationship studies.
LPS and lipid A biosynthesis were remarkably well-described for E. coli by Raetz and Whitfield (43). The conserved process leads to the synthesis of whole LPS molecules. In the lipid A moiety, the “decorations” on the phosphate groups, or the heterogeneity in the number of FAs, are due to posttranslational modifications. The details of the structures are crucial for the recognition of the whole structure by the MD-2/TLR4 complex, hence for the induction of biological activities (4). The standard LPS samples presented in this work in comparison with the two V. filiformis LPSs displayed various examples of these modifications.
After characterization of the different standard LPS structures corresponding, as expected, to molecules with short-chain FAs, like 10:0(3-OH) and 12:0, we compared their cell activation capacities by TNF-α production, IL-6 production, and ROS induction; the cytotoxicity was illustrated by caspase 3/7 activation. We also compared these activations to those of a J5 E. coli LPS sample, a strong inflammatory inducer, involving all the characteristics for a good receptor recognition, and also selected for similarity in its molecular mass. Effectively, the size of the LPS molecular species is rarely taken into account in such comparisons. However, LPS molecular masses vary from 2 to 20 kDa, therefore it is obvious that their biologically active moieties (i.e., lipids A) correspond to almost 70% of deep rough-type LPS, 50% of LOS, and only 10–20% of long-chain smooth-type LPS. In addition, the LPS solubility and state of aggregation vary considerably from one type to the other.
We thus compared the production of two cytokines stimulated by the two Vitreoscilla LPS batches, with different known LPS samples originating from Bordetella, Escherichia, Haemophilus, Neisseria, Pseudomonas, and Rhodopseudomonas genera. All of them, except for Pseudomonas, corresponded to R-type LPS or LOS, as shown in Fig. 3. Observed differences resulted in the lack of one or two FAs or phosphate substitutions compared with published data; this was expected to induce differences in the MD-2/TLR4 activation and in cytokine production (2).
According to the structure and previously described biological data, we thus found that two of our samples, those isolated from Neisseria and Haemophilus, behaved as poor cytokine inducers; although they should not, according to their well-known described lipid A structure (37, 44). We then showed by MALDI-MS that these Neisseria and Haemophilus samples, usually described as displaying major hexa-acyl molecular species, were in the present case, respectively penta- and tri/tetra-acylated. The hypo-acylation of these samples explains their relative weak activity compared with E. coli J5 LPS. One of the best illustrated examples of structure to activity relationships, related to the low number and short length of FA carbon chains, is that of B. pertussis, displaying mixtures of tetra- and penta-acyl molecular lipid A species. This example has been extensively studied (20, 39, 45) and compared with standard LPS as well as to closely related structures differing by only two carbons in a single FA at C3′ or the presence of GlcN on both phosphate groups. Both B. pertussis lipids A were hypo-acylated with tetra- and penta-acyl molecular species. Interestingly, R. gelatinosa LPS did not induce any inflammatory responses, whereas it was previously described as being highly pyrogenic in rabbit and lethal for mice (46). R. gelatinosa LPS contains short-chain FAs: four 10:0(3-OH), one 12:0, and one 12/14:0. This type of lipid A structure, as shown with the many other examples of short-chain and hypo-acylated molecules, is not expected to induce high levels of inflammation (20). Several P. aeruginosa LPSs differing by their origin (biofilm, planktonic, cystic fibrosis, bronchiectasis) were studied by us and others (47, 48). In this work, P. aeruginosa forms of LPS induced an intermediate level of TNF-α and IL-6 secretions by THP-1 cells, although the penta-acyl lipid A form was predominant. This pentaacyl lipid A form was already described for inducing low levels of TNF-α in THP-1 with a good activation of TLR4 (49). When we compared the TNF-α and IL-6 activities of the two V. filiformis forms of LPS, we observed major differences: a lower TNF-α dose-response and no late cytokine production for V.f6+2PEA compared with V.f5-6. These results suggest that the addition of PEA significantly modifies cytokine production by host cells and formation of the LPS MD-2/TLR4 complex. Nevertheless, cytokine production is lower than that observed after E. coli LPS stimulation. Moderate cytokine production by both V. filiformis LPS forms is, however, not negligible, explaining their capacity to contribute to the effects described on skin and the induction of the innate immune response that is necessary for setting up the adaptive immune response.
In addition to cytokine production, the comparative effects of V. filiformis and E. coli LPS on myeloid ROS production, cell viability, and induction of apoptosis were tested. No significant difference was observed, in this case, between the different LPSs. ROS is a useful mechanism for phagocytes to eliminate pathogens by killing the intracellular bacteria and is an essential element in the activation of the innate immune response via TLR and NFκB (50). Caspase activation is known to regulate cell viability and apoptosis. This is associated with the known effect of V. filiformis LPS in inducing costimulatory effectors and the production of antimicrobial peptides (12, 13, 16) that promote efficient antimicrobial effects with a limited pro-inflammatory cytokine response.
In summary, we established the detailed structure of two batches of V. filiformis lipids A, one of the active contributors of the bacterial lysate used to treat atopic dermatitis. These structures, mostly composed of short-chain FAs and PEA, can substitute the phosphate groups. The two bacterial samples corresponded to two different biomass recovery methods, which, by itself, could explain not only the observed structural differences in the number of FAs, but also the presence of two PEA derivatives linked to the phosphate groups. By experience, we can anticipate that the PEA bis-substituted molecular species is more soluble than the nonsubstituted one, not only because PEA adds some natural solubility in water, but also because substituting the phosphate groups prevents LPS aggregation through bivalent cation capture by these phosphates. Therefore, it is not surprising that such molecular species could be solubilized and lost in the supernatant of the growth medium during the centrifugation process separating bacterial cells and solubilized free LPS molecules in the growth medium used in the usual LPS purification steps. In the other biomass batch, the non-PEA-substituted lipid A remained in the pellet. It is very interesting to give this additional example of the loss of natural LPS molecular species during LPS purification steps leading, in this particular example, to the biological differences observed between the two samples. Then, we compared their biological activities to standard LPS, whose structures were characterized in this work. Differences in cytokine production were observed between both V. filiformis LPSs compared with standard LPS. The moderate cytokine release matches with the described structures of both V. filiformis lipids A displaying short-chain FA. However, neither caspase 3/7 activation nor ROS production differences, compared with E. coli J5 LPS, appeared, confirming the maintained antimicrobial effects induced by V. filiformis LPS with limited pro-inflammatory cytokine production.
Acknowledgments
The authors thank Pr. Jean-Marc Cavaillon and Catherine Fitting for their help in preparing the cytokine experiments and Dr. Ian-Barry Holland for his kind help in editing the manuscript.
Footnotes
Abbreviations:
- DC
- dendritic cell
- EA
- ethanolamine
- GlcN
- glucosamine
- IL
- interleukin
- LPS
- lipopolysaccharide
- MW
- molecular weight
- PEA
- phosphoethanolamine
- ROS
- reactive oxygen species
- V.f
- Vitreoscilla filiformis
- 10:0(3-OH)
- 3-hydroxydecanoic acid
- 12:0
- dodecanoic acid
This work was supported by a Convention Industrielle de Formation par la Recherche (CIFRE) doctoral grant to A.B.
REFERENCES
- 1.Rietschel E. T., Kirikae T., Schade F. U., Mamat U., Schmidt G., Loppnow H., Ulmer A. J., Zähringer U., Seydel U., and Di Padova F.. 1994. Bacterial endotoxin: molecular relationships of structure to activity and function. FASEB J. 8: 217–225. [DOI] [PubMed] [Google Scholar]
- 2.Maeshima N., and Fernandez R. C.. 2013. Recognition of lipid A variants by the TLR4-MD-2 receptor complex. Front. Cell. Infect. Microbiol. 3: 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Martich G. D., Boujoukos J., and Suffredini F.. 1993. Response of man to endotoxin. Immunobiology. 187: 403–416. [DOI] [PubMed] [Google Scholar]
- 4.Park B. S., Song D. H., Kim H. M., Choi B-S., Lee H., and Lee J-O.. 2009. The structural basis of lipopolysaccharide recognition by the TLR4-MD-2 complex. Nature. 458: 1191–1195. [DOI] [PubMed] [Google Scholar]
- 5.Tissières P., and Pugin J.. 2009. The role of MD-2 in the opsonophagocytosis of Gram-negative bacteria. Curr. Opin. Infect. Dis. 22: 286–291. [DOI] [PubMed] [Google Scholar]
- 6.Drifte G., Dunn-Siegrist I., Tissières P., and Pugin J.. 2013. Innate immune functions of immature neutrophils in patients with sepsis and severe systemic inflammatory response syndrome. Crit. Care Med. 41: 820–832. [DOI] [PubMed] [Google Scholar]
- 7.Pringsheim E. G. 1951. The Vitreoscillaceae: A family of colourless, gliding, filamentous organisms. J. Gen. Microbiol. 5: 124–149. [DOI] [PubMed] [Google Scholar]
- 8.Cohn F. 1870. Über den Brunnenfaden (Crenothrix polyspora) mit Bemerkungen über die mikroskopische Analyse des Brunnenwassers. Beitr. z. Biol. d. Pflanz. 1: 108–131. [Google Scholar]
- 9.Webster D. A., and Hackett D. P.. 1966. The purification and properties of cytochrome o from Vitreoscilla. J. Biol. Chem. 241: 3308–3315. [PubMed] [Google Scholar]
- 10.Gueniche, A., and L. Breton, inventors. 2004. L’Oréal, assignee. Utilisation d’une fraction de lipopolysaccharides de bactérie filamenteuse non-fructifiante non-photosynthétique comme agent immunorégulateur cutanné. French patent FR2879461. 2004 Dec 17.
- 11.Pineau, N., R. Martin, L. Breton, and L. Aubert, inventors. 2001. L’Oréal, assignee. Cosmetic/pharmaceutical compositions comprising microorganism culture media. US patent US62422295(B1). 2001 Jun 5.
- 12.Mahe, Y. F., and R. Martin, inventors. 2008. L’Oréal, assignee. Use of a vitreoscilla filiformis lipopolysaccharide fraction as an agent for stimulating the synthesis of anti-microbial skin peptides. European patent EP1974720(A1). 2008 Oct 1.
- 13.Mahe Y. F., Perez M. J., Tacheau C., Fanchon C., Martin R., Rousset F., and Seite S.. 2013. A new Vitreoscilla filiformis extract grown on spa water-enriched medium activates endogenous cutaneous antioxidant and antimicrobial defenses through a potential Toll-like receptor 2/protein kinase C, zeta transduction pathway. Clin. Cosmet. Investig. Dermatol. 6: 191–196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mahé Y. F., Martin R., Aubert L., Billoni N., Collin C., Pruche F., Bastien P., Drost S. S., Lane A. T., and Meybeck A.. 2006. Induction of the skin endogenous protective mitochondrial MnSOD by Vitreoscilla filiformis extract. Int. J. Cosmet. Sci. 28: 277–287. [DOI] [PubMed] [Google Scholar]
- 15.Volz T., Skabytska Y., Guenova E., Chen K-M., Frick J-S., Kirschning C. J., Kaesler S., Röcken M., and Biedermann T.. 2014. Nonpathogenic bacteria alleviating atopic dermatitis inflammation induce IL-10-producing dendritic cells and regulatory Tr1 cells. J. Invest. Dermatol. 134: 96–104. [DOI] [PubMed] [Google Scholar]
- 16.Martin R. 2005. L’Oréal, assignee. Lipid A-type compound and composition containing it. US patent US2005/0118118 (A1). 2005 Jun 2. [Google Scholar]
- 17.Nakatsuji T., and Gallo R. L.. 2014. Dermatological therapy by topical application of non-pathogenic bacteria. J. Invest. Dermatol. 134: 11–14. [DOI] [PubMed] [Google Scholar]
- 18.Martin, R., and P. Hilaire. 2010. L’Oréal, assignee. Preparation of cosmetic active principles by culturing Vitreoscilla on thermal water and compositions comprising them. European patent EP2144995. 2010 Jan 20.
- 19.Tirsoaga A., Novikov A., Adib-Conquy M., Werts C., Fitting C., Cavaillon J. M., and Caroff M.. 2007. Simple method for repurification of endotoxins for biological use. Appl. Environ. Microbiol. 73: 1803–1808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Marr N., Novikov A., Hajjar A. M., Caroff M., and Fernandez R. C.. 2010. Variability in the lipooligosaccharide structure and endotoxicity among Bordetella pertussis strains. J. Infect. Dis. 202: 1897–1906. [DOI] [PubMed] [Google Scholar]
- 21.Caroff, M. 2004. Université Paris Sud, assignee. Novel method for isolating endotoxins. World Intellectual Property Organization patent WO/2004/062690 A1. 2004 Jul 29.
- 22.Tirsoaga A., El Hamidi A., Perry M., Caroff M., and Novikov A.. 2007. A rapid, small-scale procedure for the structural characterization of lipid A applied to Citrobacter and Bordetella strains: discovery of a new structural element. J. Lipid Res. 48: 2419–2427. [DOI] [PubMed] [Google Scholar]
- 23.Chafchaouni-Moussaoui I., Novikov A., Bhrada F., Perry M. B., Filali-Maltouf A., and Caroff M.. 2011. A new rapid and micro-scale hydrolysis, using triethylamine citrate, for lipopolysaccharide characterization by mass spectrometry. Rapid Commun. Mass Spectrom. 25: 2043–2048. [DOI] [PubMed] [Google Scholar]
- 24.El Hamidi A., Tirsoaga A., Novikov A., Hussein A., and Caroff M.. 2005. Microextraction of bacterial lipid A: easy and rapid method for mass spectrometric characterization. J. Lipid Res. 46: 1773–1778. [DOI] [PubMed] [Google Scholar]
- 25.El Hamidi A., Novikov A., Karibian D., Perry M. B., and Caroff M.. 2009. Structural characterization of Bordetella parapertussis lipid A. J. Lipid Res. 50: 854–859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Caroff M., and Novikov A.. 2011. Micromethods for lipid A isolation and structural characterization. In Microbial Toxins: Methods and Protocols. O. Holst, editor. Humana Press, Springer New York, Dordrecht, Heidelberg, London. 135–146. [DOI] [PubMed] [Google Scholar]
- 27.Wollenweber H. W., and Rietschel E. T.. 1990. Analysis of lipopolysaccharide (lipid A) fatty acids. J. Microbiol. Methods. 11: 195–211. [Google Scholar]
- 28.Basheer S. M., Guiso N., Tirsoaga A., Caroff M., and Novikov A.. 2011. Structural modifications occurring in lipid A of Bordetella bronchiseptica clinical isolates as demonstrated by matrix-assisted laser desorption/ionization time-of-flight mass spectrometry. Rapid Commun. Mass Spectrom. 25: 1075–1081. [DOI] [PubMed] [Google Scholar]
- 29.Caroff M. G., and Karibian D.. 1990. Several uses for isobutyric acid-ammonium hydroxide solvent in endotoxin analysis. Appl. Environ. Microbiol. 56: 1957–1959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Laemmli U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 227: 680–685. [DOI] [PubMed] [Google Scholar]
- 31.Tsai C. M., and Frasch C. E.. 1982. A sensitive silver stain for detecting lipopolysaccharides in polyacrylamide gels. Anal. Biochem. 119: 115–119. [DOI] [PubMed] [Google Scholar]
- 32.Caroff M., Tacken A., and Szabo L.. 1988. Detergent-accelerated hydrolysis of bacterial endotoxins and determination of the anomeric configuration of the glycosyl phosphate present in the “isolated lipid A” fragment of the Bordetella pertussis endotoxin. Carbohydr. Res. 175: 273–282. [DOI] [PubMed] [Google Scholar]
- 33.Karibian D., Brunelle A., Aussel L., and Caroff M.. 1999. 252Cf-plasma desorption mass spectrometry of unmodified lipid A: fragmentation patterns and localization of fatty acids. Rapid Commun. Mass Spectrom. 13: 2252–2259. [DOI] [PubMed] [Google Scholar]
- 34.Que N. L. S., Lin S., Cotter R. J., and Raetz C. R. H.. 2000. Purification and mass spectrometry of six lipid A species from the bacterial endosymbiont Rhizobium etli: demonstration of a conserved distal unit and a variable proximal portion. J. Biol. Chem. 275: 28006–28016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Heine H., Rietschel E. T., and Ulmer A. J.. 2001. The biology of endotoxin. Mol. Biotechnol. 19: 279–296. [DOI] [PubMed] [Google Scholar]
- 36.Karibian D., Deprun C., Szabo L., Le Beyec Y., and Caroff M.. 1991. 252Cf-plasma desorption mass spectrometry applied to the analysis of endotoxin lipid A preparations. Int. J. Mass Spectrom. Ion Process. 111: 273–286. [Google Scholar]
- 37.Choi J., Cox A. D., Li J., McCready W., and Ulanova M.. 2014. Activation of innate immune responses by Haemophilus influenzae lipooligosaccharide. Clin. Vaccine Immunol. 21: 769–776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cox A. D., Wright J. C., Li J., Hood D. W., Moxon E. R., and Richards J. C.. 2003. Phosphorylation of the lipid A region of meningococcal lipopolysaccharide: Identification of a family of transferases that add phosphoethanolamine to lipopolysaccharide. J. Bacteriol. 185: 3270–3277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shah N. R., Albitar-Nehme S., Kim E., Marr N., Novikov A., Caroff M., and Fernandez R. C.. 2013. Minor modifications to the phosphate groups and the C3′ acyl chain length of lipid A in two Bordetella pertussis strains, BP338 and 18-323, independently affect Toll-like receptor 4 protein activation. J. Biol. Chem. 288: 11751–11760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bhat R., Marx A., Galanos C., and Conrad R. S.. 1990. Structural studies of lipid a from Pseudomonas aeruginosa PAO1: occurrence of 4-amino-4-deoxyarabinose. J. Bacteriol. 172: 6631–6636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Masoud H., Lindner B., Weckesser J., and Mayer H.. 1990. The structure of the lipid A component of Rhodocyclus gelatinosus Dr2 lipopolysaccharide. Syst. Appl. Microbiol. 13: 227–233. [Google Scholar]
- 42.Gueniche A., Knaudt B., Schuck E., Volz T., Bastien P., Martin R., Röcken M., Breton L., and Biedermann T.. 2008. Effects of nonpathogenic gram-negative bacterium Vitreoscilla filiformis lysate on atopic dermatitis: A prospective, randomized, double-blind, placebo-controlled clinical study. Br. J. Dermatol. 159: 1357–1363. [DOI] [PubMed] [Google Scholar]
- 43.Raetz C. R. H., and Whitfield C.. 2002. Lipopolysaccharide endotoxins. Annu. Rev. Biochem. 71: 635–700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zughaier S. M., Tzeng Y., Zimmer S. M., Carlson R. W., Stephens D. S., and Datta A.. 2004. Neisseria meningitidis lipooligosaccharide structure-dependent activation of the macrophage CD14/Toll-like receptor 4 pathway. Infect. Immun. 72: 371–380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Marr N., Tirsoaga A., Blanot D., Fernandez R., and Caroff M.. 2008. Glucosamine found as a substituent of both phosphate groups in Bordetella lipid A backbones: role of a BvgAS-activated ArnT ortholog. J. Bacteriol. 190: 4281–4290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Galanos C., Roppel J., Weckesser J., Rietschel E. T., and Mayer H.. 1977. Biological activities of lipopolysaccharides and lipid A from Rhodospirillaceae. Infect. Immun. 16: 407–412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bedoux G., Vallee-Rehel K., Kooistra O., Zahringer U., and Haras D.. 2004. Lipid A components from Pseudomonas aeruginosa PAO1 (serotype O5) and mutant strains investigated by electrospray ionization ion-trap mass spectrometry. J. Mass Spectrom. 39: 505–513. [DOI] [PubMed] [Google Scholar]
- 48.Ciornei C. D., Novikov A., Beloin C., Fitting C., Caroff M., Ghigo J. M., Cavaillon J. M., and Adib-Conquy M.. 2010. Biofilm-forming Pseudomonas aeruginosa bacteria undergo lipopolysaccharide structural modifications and induce enhanced inflammatory cytokine response in human monocytes. Innate Immun. 16: 288–301. [DOI] [PubMed] [Google Scholar]
- 49.Hajjar A. M., Ernst R. K., Tsai J. H., Wilson C. B., and Miller S. I.. 2002. Human Toll-like receptor 4 recognizes host-specific LPS modifications. Nat. Immunol. 3: 354–359. [DOI] [PubMed] [Google Scholar]
- 50.Park H. S., Jung H. Y., Park E. Y., Kim J., Lee W. J., and Bae Y. S.. 2004. Cutting edge: direct interaction of TLR4 with NAD(P)H oxidase 4 isozyme is essential for lipopolysaccharide-induced production of reactive oxygen species and activation of NF-kappa B. J. Immunol. 173: 3589–3593. [DOI] [PubMed] [Google Scholar]