Abstract
Close contacts between organelles, often called membrane contact sites (MCSs), are regions where lipids are exchanged between organelles. Here, we identify a novel mechanism by which cells promote phospholipid exchange at MCSs. Previous studies have shown that phosphatidylserine (PS) synthase activity is highly enriched in portions of the endoplasmic reticulum (ER) in contact with mitochondria. The objective of this study was to determine whether this enrichment promotes PS transport out of the ER. We found that PS transport to mitochondria was more efficient when PS synthase was fused to a protein in the ER at ER-mitochondria contacts than when it was fused to a protein in all portions of the ER. Inefficient PS transport to mitochondria was corrected by increasing tethering between these organelles. PS transport to endosomes was similarly enhanced by PS production in regions of the ER in contact with endosomes. Together, these findings indicate that PS production at MCSs promotes PS transport out of the ER and suggest that phospholipid production at MCSs may be a general mechanism of channeling lipids to specific cellular compartments.
Keywords: endoplasmic reticulum, lipid transfer proteins, lipid biochemistry, mitochondria, phospholipids/trafficking, lipid transport, phosphatidylethanolamine, phosphatidylserine synthase
Membrane contact sites (MCSs) are regions where two organelles are closely apposed, often within 30 nm of one another. One function of these sites is to facilitate nonvesicular lipid exchange between organelles (1–3). This exchange can play roles in signaling pathways or can be a mechanism allowing intracellular lipid transport during stress conditions when vesicular trafficking is compromised, though it remains unclear how much nonvesicular lipid exchange occurs at most MCSs. Lipid exchange at some MCSs probably facilitates membrane biogenesis; this is particularly true of mitochondria (4) and chloroplasts (5), organelles that obtain most of the lipids needed for the biogenesis of their membranes from other organelles by nonvesicular transport that probably occurs at MCSs (6).
The mechanism of nonvesicular lipid exchange between organelles at MCSs is not well understood, but in most cases probably requires lipid transport proteins (LTPs) (7, 8). These proteins have domains that can bind lipid monomers in a hydrophobic pocket or cleft, which allows them to shuttle lipids between membranes (7). Some LTPs are enriched at MCSs, where they would need to diffuse only a short distance between organelles to facilitate lipid exchange.
The best-characterized MCSs are those between the endoplasmic reticulum (ER) and other organelles, particularly mitochondria (9–11) and the plasma membrane (12, 13). The lipid and protein composition of the ER at these MCSs probably differs from that of the rest of the ER (11, 14, 15). Interestingly, the specific activity of a number of enzymes required for glycerol phospholipid synthesis, which largely occurs in the ER, is higher in portions of the ER that form contacts with the plasma membrane (16) and mitochondria (15, 17) than in the rest of the ER, suggesting that this may be a common feature of MCSs formed by the ER. Whether the increase in specific activity is because the enzymes are more enriched at these sites or because the enzymes are more active there has not been determined.
ER that associates with mitochondria is often called mitochondria-associated membrane (MAM). One enzymatic activity that is enriched in MAM is phosphatidylserine (PS) synthase (17). This is true of both Saccharomyces cerevisiae (18) and mammalian cells (19), even though PS is synthesized by different mechanisms in these species, suggesting that enrichment of PS synthase in MAM plays an important role in PS metabolism (20). PS is transferred to mitochondria at ER-mitochondria contacts (21). This transport is thought to be nonvesicular, but the mechanism is not understood and the LTPs required have not been identified. Once PS is transported to mitochondria, it can be converted to phosphatidylethanolamine (PE) in mitochondria (21). In yeast, this conversion is performed by PS decarboxylase 1 (Psd1p) (22). Cells lacking Psd1p have substantially reduced levels of PE in mitochondria, suggesting that PS transport to mitochondria is important to maintain PE levels in mitochondria (8, 23). Yeast also has a second PS decarboxylase 2 (Psd2p) that resides in the endosomes (24, 25). It is not yet known whether PS synthase activity is also enriched in the portions of the ER that contact endosomes.
The fact that Psd1p and Psd2p are outside the ER, where most glycerol phospholipid synthesis occurs, has been used to investigate PS transport from the ER to mitochondria and endosomes (26). When cells are radiolabeled with [3H]serine, [3H]PS is synthesized in the ER by the sole PS synthase in yeast, Cho1p (27). The conversion of this radiolabeled PS to PE indicates that the PS has been transferred to either mitochondria or endosomes (28). The PE can then be transported back to the ER where it can be converted to phosphatidylcholine (PC; Fig. 1A) (29, 30).
Fig. 1.
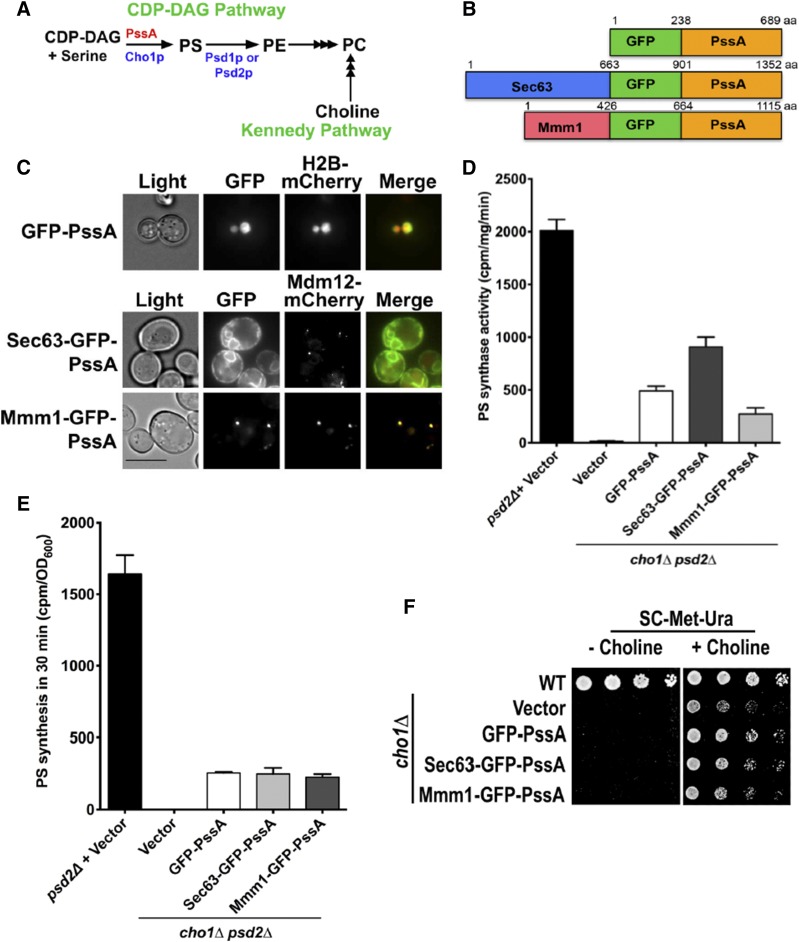
Localization and activity of PssA fusions used to study ER to mitochondria PS transport. A: Pathways of PC production in S. cerevisiae. Enzymes from yeast are in blue and red is used for E. coli. PC production by the Kennedy pathway requires exogenous choline. Multiple arrowheads indicate multiple steps. B: Diagram of PssA fusion proteins used to investigate PS transport to mitochondria. C: Growing cells expressing the indicated fusion proteins were visualized live. H2B-mCherry is a nuclear marker and Mdm12-mCherry is part of the ERMES complex. Scale bar, 5 μm. D: PS synthase activities of lysates from the indicated strains (mean ± SD, n = 3 independent experiments). E: Cells were labeled with [3H]serine and the amount of [3H]PS produced in 30 min was determined (mean ± SD, n = 3 independent experiments). F: The indicated strains were grown in SC with choline to mid-logarithmic growth phase, washed, and serial dilutions were spotted onto plates that either did or did not contain choline. The plates were incubated for 2 days at 30°C.
In this study, we investigated the function of the enrichment of PS synthase activity in the ER at ER-mitochondria contacts. We demonstrate that enrichment facilitates PS transport to mitochondria. We also show that PS synthesis at ER-endosome junctions similarly promotes PS transport from the ER to endosomes. These findings suggest that phospholipid production at MCSs promotes lipid exchange at these sites.
MATERIALS AND METHODS
Strains, plasmids, and growth media
The strains and plasmids used in this study are listed in supplemental Table S1. Yeast cells were grown in either YPD (1% yeast extract, 2% peptone, 2% glucose) or synthetic complete dextrose (SC), which contains 2% glucose, 0.67% yeast nitrogen base without amino acids, and amino acids from the appropriate dropout mix from US Biological Life Sciences. Synthetic complete glycerol (SG) media were the same as SC but with 3% glycerol instead of glucose. Where indicated, 2 mM choline were added to growth media.
Serial dilutions to assess growth
Cells were grown in synthetic complete dextrose to logarithmic growth phase, washed twice with sterile water, and adjusted to an OD600 of 1.0 in sterile water. Five-fold serial dilutions were prepared in water and spotted onto plates. The plates were incubated for 2 or 3 days at 30°C.
Isolation and purification of mitochondria
Isolation and purification of mitochondria were performed as described by Nunnari et al. (31). Briefly, cells were grown in SG with choline, harvested, and spheroplasts were prepared in 1.2 M sorbitol, 20 mM Tris (pH 7.4), and zymolyase 20T (1 mg/ml). Spheroplasts were suspended in ice-cold lysis buffer [0.6 M sorbitol, 20 mM HEPES (pH 7.4), 0.5 mM PMSF, and protease inhibitors] and homogenized with a dounce using a B-pestle. The cell homogenate was centrifuged twice for 5 min at 3,000 g to remove unlysed cells and debris. The resulting supernatant was centrifuged at 9,600 g for 10 min and the pellet containing crude mitochondria was resuspended in lysis buffer. To purify the mitochondria, crude mitochondria were loaded on OptiPrep (Axis-Shield) density gradient and the gradient was centrifuged at 64,000 g using an SW40 Ti rotor for 3 h at 4°C and the mitochondria were collected (31).
PS synthase activity assay
To prepare a cell-free protein extract from actively growing yeast, the cells were suspended in a lysis buffer [20 mM Tris (pH 7.4), 100 mM NaCl, and protease inhibitors] and lysed by agitation with glass beads in a Precellys24 homogenizer at 4°C. The PS synthase activity in 500 μg of cell extract protein was measured as described (32). The incorporation of [3H]serine into PS was measured for 15 min in the presence of 50 mM Tris-Cl (pH 8.0), 0.6 mM MnCl2, 0.25% Triton X-100, 0.2 mM CDP-DAG, and 0.5 mM [3H]serine (2 μCi). The assay was terminated by addition to 3 ml of CHCl3:methanol (1:2), lipids were extracted (33), and the CHCl3 phase was counted using a scintillation counter. When PS synthase activity was measured in the presence of phospholipids, the lipids (egg PE and brain PS; Avanti Polar Lipids) were added to the buffer and the solution was sonicated before beginning the assays. These assays were terminated after 20 min.
Steady state phospholipid analysis
Cells were grown in 50 ml SG with choline for at least four generations in the presence of 20 μCi [3H]acetate to an OD600 of 0.8–1.0. For whole cell analysis, cells were harvested and lysed with a Precellys24 homogenizer and lipids were extracted as described by Parks et al. (33). Phospholipids were separated by TLC as described by Vaden et al. (34). TLC plates were scanned on a RITA Star thin-layer analyzer (Raytest) to determine the counts per minute per phospholipid class and nanomoles of phospholipid were calculated using the formula 1,000 cpm/nmol lipid phosphate. This formula was determined by labeling WT cells grown for at least four generations in SG with choline containing 20 μCi [3H]acetate and measuring the amount of lipid phosphate per phospholipid class (35).
In vivo [3H]serine labeling
Cells were labeled with [3H]serine (American Radiolabeled Chemicals) as described by Achleitner et al. (28). Briefly, 2.5 OD600 units of cells from a saturated overnight culture were added to 25 ml of serine-free SC medium and incubated at 30°C. When the cells reached an OD600 of 0.4, 10 μg/ml myriocin (Sigma-Aldrich) was added, the cells were grown for 25 min and 10 μg/ml cerulenin was added to the medium. About 5 min later, 50 μCi [3H]serine was added to the medium and the cells were grown for an additional 30 min. After labeling, the cells were lysed in a Precellys24 homogenizer. Lipids were extracted as described by Parks et al. (33) and separated by high-pressure liquid chromatography (36, 37). The fractions containing PS, PE, and PC were collected and counted by liquid scintillation counter. The percent PS converted to PE was calculated using the formula: percent PS converted to PE = 100 × {cpm [3H]PE/(cpm [3H]PS + cpm [3H]PE)} .
Electron microscopy
Samples were prepared for electron microscopy (EM) as described (38), with some modifications. Briefly WT cells expressing empty vector or chiMERA were grown to mid-logarithmic growth phase, and 10 OD600 units of cells were harvested and incubated with 1 ml of fixative media [2.5% glutaraldehyde, 1.25% paraformaldehyde, and 40 mM potassium phosphate (pH 7.0)] for 30 min at room temperature. Cells were centrifuged, resuspended in 1 ml of fresh fixative medium and incubated on ice for 1 h. Cells were centrifuged and washed twice with 0.9% NaCl and once with water. Cells were incubated with a 2% solution of KMnO4 for 5 min at room temperature, centrifuged, and again resuspended with fresh solution of 2% KMnO4 for 45 min at room temperature for en-bloc staining. Cells were dehydrated using graded ethanol, and embedded using Spurr’s resin (Electron Microscopy Sciences). Semi- and ultrathin sections were obtained with a diamond knife (Diatome) on an ultra-microtome (Ultracut UCT; Leica-Microsystems), collected on formvar-coated 200 mesh copper grids (Electron Microscopy Sciences), post stained with uranyl acetate and lead citrate, and visualized with a Tecnai T12 TEM (FEI) operating at 120 kV. Pictures were recorded on a below-mounted Gatan 2k × 2k CCD camera. Brightness and contrast were adjusted to the entire images using Adobe Photoshop, version CC 2014.
Fluorescent microscopy
Cells were imaged live at room temperature by using an Olympus BX61 microscope, a UPlanApoX 100/1.35 lens, a QImaging Retiga EX camera, and iVision software (version 4.0.5).
Immunoblotting and antibodies
Proteins were extracted as described (39) from 2.0 OD600 units of cells, separated by 4–12% of SDS-PAGE gel (Invitrogen), and transferred to nitrocellulose membranes at 120 V for 2 h. The resulting membrane was analyzed using primary antibodies to GFP (1:1,000; Invitrogen) and anti-Pgk1 (1:1,000; Invitrogen). Proteins were visualized using IRDye secondary anti-mouse antibody (Li-COR Biosciences; 1:10,000). The blots were visualized with an Odyssey infrared imaging system (Li-COR Biosciences).
Statistical analysis
All the values reported in this paper represent the mean of three or more independent experiments. Student’s t-test (two-tailed, unpaired) was used to identify the statistical difference and differences were considered statistically significant when P < 0.05.
RESULTS
Escherichia coli PS synthase is active in S. cerevisiae
We wanted to determine whether the enrichment of PS synthase activity at ER-mitochondria contact sites increases PS transport to mitochondria. To address this, we targeted PS synthase to portions of the ER that are in close contact with mitochondria and determined whether this enhances PS transport from the ER to mitochondria. We reasoned that if PS production at contacts promotes PS transport to mitochondria, then targeting PS synthase to contact sites would result in greater PS transport to mitochondria than when PS synthase is localized all over the ER. For these experiments, we used E. coli PS synthase (PssA), a peripheral membrane protein (40) that has no homology to yeast PS synthase (Cho1p) (41) and that probably cannot interact with proteins that normally interact with Cho1p.
To target PssA to ER-mitochondria contacts, we fused it to a component of a complex called the ER-mitochondria encounter structure (ERMES), which tethers the ER to mitochondria in S. cerevisiae (42). ERMES forms a small number of puncta in cells, typically about one to ten per cell (43). We fused PssA to the ERMES component, Mmm1p, which resides in the ER, and to GFP for visualization. The resulting fusion, Mmm1-GFP-PssA (Fig. 1B), forms puncta that colocalize with a mCherry-tagged version of Mdm12p, another member of the ERMES complex (42) (Fig. 1C).
To localize PS synthase all over the ER, we fused PssA to Sec63p (44) and GFP. We confirmed that the resulting fusion, Sec63-GFP-PssA (Fig. 1B), was distributed throughout the ER (Fig. 1C). As an additional control, we also expressed GFP-PssA (Fig. 1B) in cells. Surprisingly, the fusion localized to the nucleus for unknown reasons; we confirmed that GFP-PssA was in the nucleus by colocalizing it with histone H2B fused to mCherry (Fig. 1C). We used immunoblotting to determine whether the fusions were expressed at similar levels and found that they were (supplemental Fig. S1).
Next, we determined whether the PssA fusion proteins were active. The fusions were expressed in cells lacking Cho1p (45) and the PS activity of whole cell lysates was measured. It should be noted that these strains also lacked Psd2p because this was necessary for the PS transport experiments described in the next section. All the PssA fusions were active, but had lower activity than endogenous PS synthase (Fig. 1D). We also wanted to estimate the relative rate of PS synthesis by the fusions in cells. To do that, we labeled cells with [3H]serine for 30 min and measured the total amount of PS produced. Because PS is converted to PE, the total amount of PS produced was taken to be the amount of radiolabeled PS and PE. We have previously found that when labeling cells with [3H]serine, it is necessary to treat them with the fatty acid synthase inhibitor, cerulenin, to prevent the radiolabel from being incorporated into PC via the one-carbon pathway (36). When labeling cells treated with cerulenin, very little radiolabeled PE is converted to PC (36). Therefore, total PS produced during a 30 min labeling is equal to the amount of radiolabeled PS and PE. We found that cells expressing the PssA fusions produce PS at similar rates, but at an ∼5-fold lower rate than cells expressing endogenous PS synthase (Fig. 1E).
We found that the low level of PS synthase activity in cells lacking Cho1p and expressing any of the PssA fusion was not sufficient for growth in conditions when most PE and PC must be derived from PS. Yeast cells lacking Cho1p can grow if they are supplemented with choline, which allows them to produce PC by the Kennedy pathway instead of from PS (46) (Fig. 1A). None of the PssA fusions allowed cells lacking Cho1p to grow in the absence of choline, indicating that although the PssA fusions produce PS in cells, it is not sufficient to support cell growth (Fig. 1F).
PS synthesis at ER-mitochondria contacts enhances PS transport to mitochondria
To estimate PS transport from the ER to mitochondria, we used a well-established assay that makes use of the fact that Psd1p, which converts PS to PE, is localized in the mitochondrial inner membrane (28, 47). The conversion of PS to PE by Psd1p can be used to estimate the rate of PS transport to mitochondria in cells that lack Psd2p, which is outside mitochondria (24). The PssA fusion proteins were expressed in cells that express Psd1p, but lack Cho1p and Psd2p. As a control, we used a strain that expresses Cho1p, but lacks Psd2p.
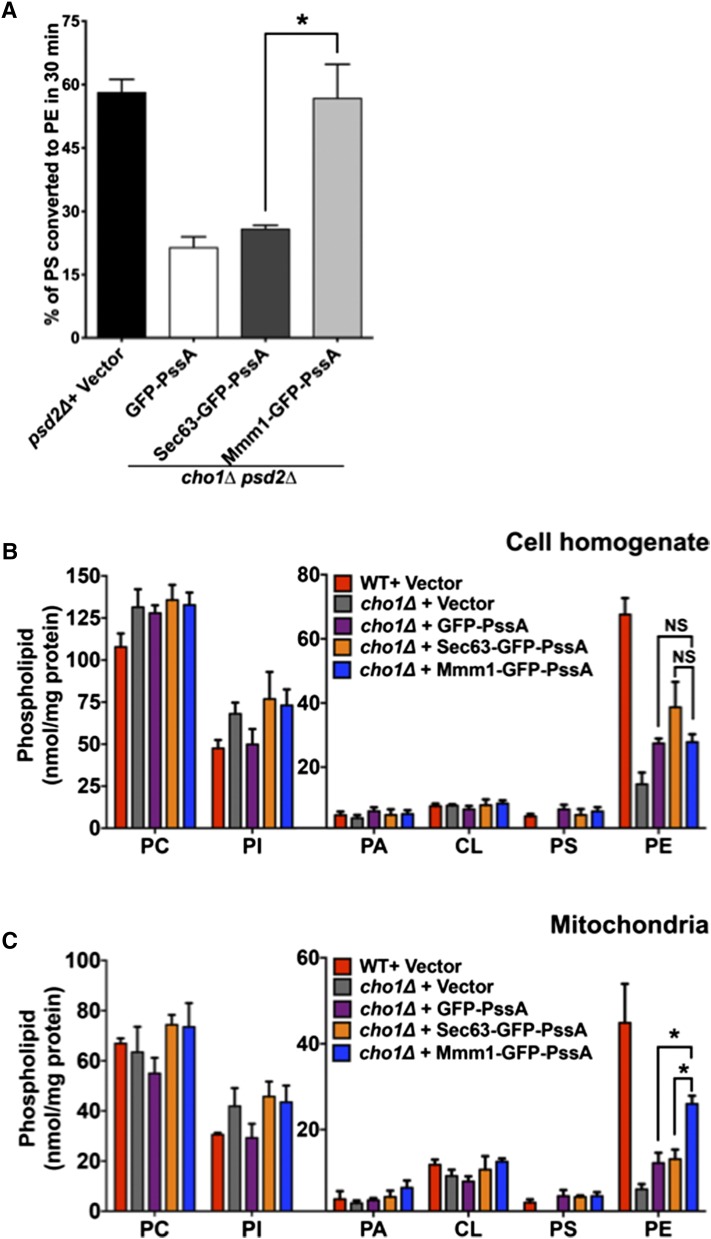
The strains were labeled for 30 min with [3H]serine and the percent of radiolabeled PS converted to PE was determined; we have previously shown that PS to PE conversion is linear during this time and that the percent of newly synthesized PS converted to PE in cells is independent of the amount of PS produced (36). In cells expressing endogenous PS synthase, ∼58% of PS was converted to PE (Fig. 2A). Only about 20% of PS was converted to PE in cells lacking Cho1p and expressing either GFP-PssA or Sec63-GFP-PssA. However, the percent of PS converted to PE increased to about 56% in cells expressing Mmm1-GFP-PssA. These results indicate that PS synthesis at ER-mitochondria contacts increases the rate of PS transport to mitochondria.
Fig. 2.
PS synthesis at ER-mitochondria contacts increases PS transport to mitochondria. A: The indicated strains were labeled with [3H]serine for 30 min and the percentage of [3H]PS converted to [3H]PE was determined (mean ± SD, n = 3 independent experiments). *P < 0.05, independent two-tailed t-test. Raw data used to calculate percent of [3H]PS converted to [3H]PE is in supplemental Fig. S2A, B. B, C: Steady state levels of phospholipids in whole cells (B) and mitochondria (C) from the indicated strains grown in SG with choline (mean ± SD, n = 3 independent experiments). *P < 0.05, independent two-tailed t-test. PI, phosphatidylinositol; PA, phosphatidic acid; CL, cardiolipin.
We wondered whether increased PS transport to mitochondria would alter the amount of PS and PE in mitochondria, inasmuch as most PE in mitochondria is derived from PS in mitochondria and not from PE synthesized outside mitochondria (8, 23). We measured phospholipid levels in whole cells and in purified mitochondria from cells expressing the PssA fusions. The cells were grown for at least four generations in a medium containing [3H]acetate, which labels all lipids. The relative amounts of PS, PE, PC, phosphatidylinositol, phosphatidic acid, and cardiolipin were determined. For these experiments, cells were grown in a medium containing glycerol, a nonfermentable carbon source, because previous studies have found that about 75% of mitochondrial PE is derived from PS when cells are grown on media with nonfermentable carbon sources; whereas in media with glucose, only about 50% of mitochondrial PE is derived from PS (23).
We found that there was no significant difference in the amount of PS and PE in whole cell lysate of the cells expressing any of the three PssA fusions (Fig. 2B). However, cells expressing Mmm1-GFP-PssA had significantly more PE in mitochondria than cells expressing either Sec63-GFP-PssA or GFP-PssA (Fig. 2C). The Mmm1-GFP-PssA-expressing cells did not have significantly more PS, but this is probably because most PS imported into mitochondria is converted into PE. Taken together, these findings indicate that PS production at ER-mitochondria contacts by Mmm1-GFP-PssA increases PS transfer to mitochondria and increases mitochondrial PE levels because most mitochondrial PE is derived from mitochondrial PS decarboxylation.
Increasing ER-mitochondria contacts enhances PS transport to mitochondria
Because PS production at ER-mitochondria contacts promotes PS transfer to mitochondria, we wondered whether increasing these contacts would increase the amount of PS transferred. If this was correct, increasing ER-mitochondria contacts would increase PS transport to mitochondria in cells expressing Sec63-GFP-PssA, because this fusion is all over the ER. In contrast, because Mmm1-GFP-PssA localizes to ER-mitochondria contacts, increasing these contacts in cells expressing this fusion would not be expected to increase PS transfer to mitochondria. We also predicted that increased ER-mitochondria contacts would not alter the rate of PS transport to mitochondria in cells expressing GFP-PssA because this protein is in the nucleus and should not become more enriched at ER-mitochondria contacts when they are increased because contacts are formed outside the nucleus.
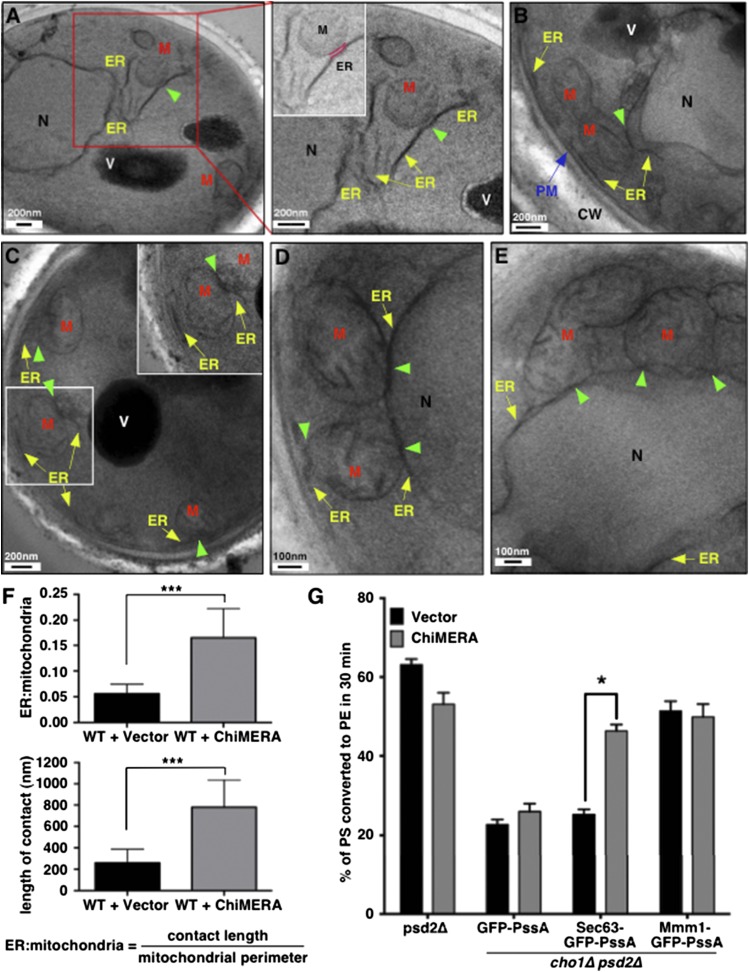
To increase contacts between the ER and mitochondria, we expressed an artificial ER-mitochondria tether, called ChiMERA (42), in the cells expressing the PssA fusions. ChiMERA has domains that insert into the ER membrane and the mitochondrial outer membrane, tethering the organelles. We used EM to quantify the increase in ER-mitochondria contacts when ChiMERA is expressed in cells. Contacts were quantified in two ways: the percent of the mitochondrial surface that has ER within 30 nm and the length of these contacts. In cells not expressing ChiMERA, about 5% of the surface of mitochondria had ER closely apposed and the average length of these contacts was about 250 nm (Fig. 3A, F). In cells expressing ChiMERA, ∼15% of the mitochondria surface had contacting ER and the length of these contacts was about 800 nm (Fig. 3B–F). Thus, ChiMERA causes about a 3-fold increase in the amount of contact between the ER and mitochondria.
Fig. 3.
Increasing ER-mitochondria contacts increases PS transport to mitochondria. A–E: WT cells expressing empty vector (A) or ChiMERA (B–E) were visualized by EM. Boxed regions are shown in higher magnification. ER-mitochondria contact is highlighted in pink in the inset of the right panel of (A). Yellow arrows denote ER membrane. Green arrowheads indicate contact sites between the ER and mitochondria. The blue arrow denotes plasma membrane. M, mitochondria; V, vacuole; N, nucleus; PM, plasma membrane; CW, cell wall. F: Quantification of length of contact between ER and mitochondria for experiments in (A–E). The top panel shows the average ER-mitochondria ratio and the lower panel shows the average length of contacts (mean ± SD, n = 15 individual cells). The ER-mitochondria ratio was determined by measuring the length of contacts between the ER and mitochondria divided by the length of mitochondrial perimeter in each image. ***P < 0.005, independent two-tailed t-test. G: The indicated strains were labeled with [3H]serine for 30 min and the percentage of [3H]PS converted to [3H]PE was determined (mean ± SD, n = 3 independent experiments). *P < 0.05, independent two-tailed t-test. Raw data used to calculate percent of [3H]PS converted to [3H]PE is in supplemental Fig. S2C, D.
We determined the percent of PS converted to PE in 30 min in cells expressing ChiMERA and the PssA fusions. As predicted, we found that the expression of ChiMERA markedly increased PS transport from ER to mitochondria in cells producing Sec63-GFP-PssA (Fig. 3G). In contrast, ChiMERA did not alter PS transport to mitochondria in cells expressing Mmm1-GFP-PssA or in cells expressing GFP-PssA. These findings provide further evidence that PS production at ER-mitochondria contacts promotes PS transport to mitochondria and indicate that increasing contact can increase transport when PS is synthesized all over the ER.
PS synthesis at ER-endosome contacts enhances PS transport to endosomes
Because PS production at ER-mitochondria contacts enhances PS transport to mitochondria, we wondered whether PS production at another contact would similarly increase PS transport out of the ER. We investigated PS transport from the ER to endosomes, which is thought to occur at contacts between these organelles (26). Studies of ER to endosome PS transport have made use of the localization of Psd2p in endosomes. In cells lacking Psd1p, the conversion of newly synthesized PS to PE indicates that the PS has been transferred from the ER, where it is synthesized, to endosomes containing Psd2p (24).
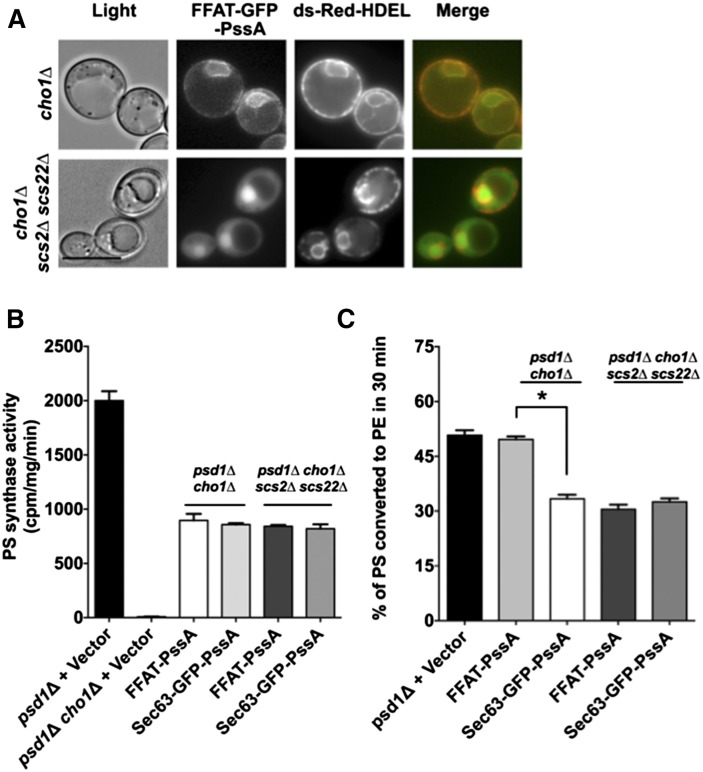
To determine whether PS production at ER-endosome contacts enhances PS transport to Psd2p-containing endosomes, we sought to target PssA to these contacts (just as we targeted PssA to ER-mitochondria contacts by fusing it to Mmm1p). ER-endosome contacts are not well-characterized, but it is thought that the protein Scs2p is enriched at these sites (48). This protein is the yeast homolog of vesicle-associated membrane protein-associated proteins (VAPs) in mammals, which are also thought to be enriched at some of the MCSs (10, 49, 50). VAPs and Scs2p bind proteins containing two phenylalanines in acidic tract (FFAT) motifs (51). Therefore, to promote the localization of PssA to ER-endosome contacts, we fused FFAT to GFP-PssA. We found that FFAT-GFP-PssA was all over the ER (Fig. 4A) and was expressed at levels similar to the other PssA fusion proteins (supplemental Fig. S1). However, in cells lacking Scs2p and the Scs2p homolog, Scs22p, the fusion was found largely in the nucleus (similarly to GFP-PssA; Fig. 1C), indicating that FFAT-GFP-PssA binds to Scs2p and Scs22p. It should be noted that even though Scs2p is thought to be enriched at some MCSs (48), GFP fusions to Scs2p are found all over the ER, presumably because Scs2p is not as enriched at contact sites as, for example, proteins in the ERMES complex (42).
Fig. 4.
PS synthesis at ER-endosome contacts increases PS transport to endosomes. A: Growing cells expressing FFAT-GFP-PssA and the ER marker, ds-Red-HDEL, were visualized live. Scale bar = 5 μm. B: PS synthase activities of lysates from the indicated strains (mean ± SD, n = 3 independent experiments). C: The indicated strains were labeled with [3H]serine for 30 min and the percentage of [3H]PS converted to [3H]PE was determined (mean ± SD, n = 3 independent experiments). *P < 0.05, independent two-tailed t-test. Raw data used to calculate percent of [3H]PS converted to [3H]PE is in supplemental Fig. S2E, F.
To determine whether enriching PS synthase at ER-endosome contacts promotes PS transport from ER to endosome, we expressed either FFAT-PssA or Sec63-GFP-PssA in cells lacking Cho1p and Psd1p (cho1Δ psd1Δ). As a control, these fusions were also expressed in cho1Δ psd1Δ cells that were also lacking Scs2p and Scs22p. We first determined the in vitro PS synthase activity of the cells expressing either FFAT-PssA or Sec63-GFP-PssA and found that they were similar (Fig. 4B). Next, we labeled these strains with [3H]serine for 30 min and measured the amount of PS converted to PE. We found that cho1Δ psd1Δ cells expressing FFAT-PssA converted significantly more radiolabeled PS to PE than those expressing Sec63-GFP-PssA (Fig. 4C). This difference was eliminated when Scs2p and Scs22p were lacking in these strains (Fig. 4C). Taken together, these findings suggest that enrichment of PS synthase at ER-endosome contacts promotes PS transport from the ER to endosomes.
Cho1p is inhibited by its product, PS
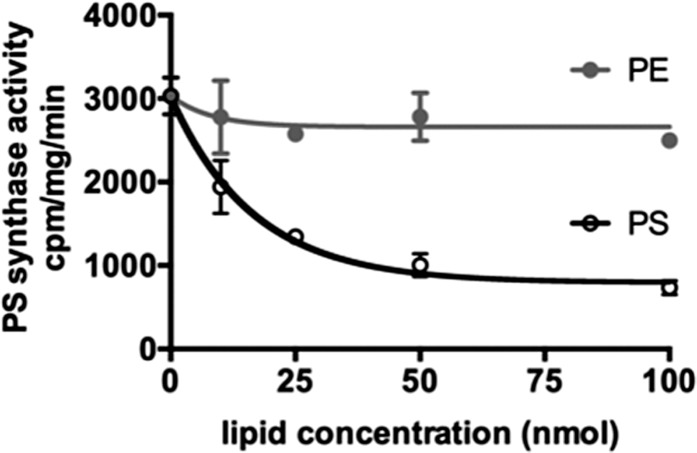
Our findings indicate that the enrichment of PS synthase activity in portions of the ER in contact with mitochondria increases PS transfer to mitochondria. How PS synthase activity is enriched at these contacts is not known. When Cho1-GFP is expressed in cells, it is found all over the ER (52), suggesting that Cho1p is either activated at contacts or inhibited in portions of the ER not in contact with mitochondria or other organelles. One way that PS synthase activity may be regulated is by its product, PS. It is known that PS inhibits mammalian PS synthases (53, 54) and mutations in the synthases that ablate inhibition by PS are known to cause disease (55). We wondered whether PS might also inhibit Cho1p. It was previously found that Cho1p is not inhibited by a number of phospholipids (56), but it was not known whether PS inhibited it. We found that PS reduces Cho1p activity in a concentration-dependent manner (Fig. 5). This finding suggests that Cho1p activity could be regulated by differences in PS concentration in the ER; as PS is transferred out of the ER at contact sites, the reduction in PS levels may trigger more PS synthesis.
Fig. 5.
PS inhibits Cho1p. PS synthase activities of lysates from WT W303 cells with the indicated amounts of added PE or PS (mean ± SD, n = 3 independent experiments).
DISCUSSION
In this study, we show that enrichment of PS synthase in regions of the ER that are in contact with mitochondria promotes the transfer of newly synthesized PS from the ER to mitochondria. Enrichment of PS synthase at ER-endosome contacts similarly promotes PS transport to endosomes. These findings suggest that a significant fraction of nonvesicular PS transport to mitochondria and endosomes occurs at MCSs and that PS made at MCSs is more readily transported out of the ER than PS made in other regions of the ER. This conclusion is consistent with a previous study that showed that PS transport from the ER to mitochondria slows significantly in cells in which ER-mitochondria tethering is reduced (47). Our findings are also consistent with work from Wu and Voelker (57) that showed that the in vitro rate of PS transfer from liposomes to acceptor membranes is dependent on the PS concentration in the liposomes. Thus, PS production at MCSs is a mechanism of channeling PS to specific organelles. Because a number of lipid synthetic enzymes are enriched in MAM (or are more active there) than in the rest of the ER (17), lipid synthesis in regions of the ER at MCSs may promote the transfer of a number of types of lipids out of the ER at these sites.
Why PS synthesized at MCSs is preferentially transported out of the ER remains an important question. It could be that PS diffusion in the ER is limited such that PS synthesized outside MCSs cannot readily reach these sites to be transported out of the ER. However, the only known diffusion barrier in the yeast ER is between mother cells and buds and there is no indication that diffusion barriers are present elsewhere in the ER (58–60). These studies examined the diffusion of proteins and not lipids, but it seems unlikely that lipid diffusion within the ER is impeded when protein diffusion is not. Another more plausible explanation for why PS produced at MCSs is more likely to be transported out of the ER is that PS synthesis is linked to PS transport. This possibility is supported by studies suggesting that newly synthesized PS is preferentially transported to mitochondria (18, 61). It may be that when PS is synthesized it can more readily desorb from the ER membrane than already existing PS. Once the PS has desorbed, it could be captured by LTPs and transferred to other organelles at MCSs (7). Our findings could also be explained if Cho1p in the ER can catalyze PS formation in membranes closely apposed to the ER. However, we have failed to find any evidence that Cho1p (or PssA fused to Sec63p) can generate PS in closely apposed membranes containing CDP-DAG (not shown). Therefore, we favor the model that newly synthesized PS can be more readily transported out of the ER than preexisting PS, perhaps because it can more readily desorb from the ER when it is synthesized.
It is not known how Cho1p is activated in portions of the ER at contact sites or inhibited outside these sites. We propose that Cho1p is inhibited when it is outside MAM or other MCSs. In this study, we found that Cho1p, like mammalian PS synthases (53, 54), is inhibited by its product, PS. This suggests a simple mechanism of activating Cho1p at MCSs; PS transport out of the ER at MCSs will stimulate Cho1p activity in these regions. Future studies will focus on determining whether other lipid synthases are regulated similarly to PS synthase and on understanding how lipid synthesis at MCSs promotes lipid transport out of the ER.
Supplementary Material
Acknowledgments
The authors thank Morgane Michaud and Orna Cohen-Fix for critically reading the manuscript.
Footnotes
Abbreviations:
- EM
- electron microscopy
- ER
- endoplasmic reticulum
- ERMES
- endoplasmic reticulum-mitochondria encounter structure
- FFAT
- two phenylalanines in an acidic tract
- LTP
- lipid transport protein
- MAM
- mitochondria-associated membrane
- MCS
- membrane contact site
- PC
- phosphatidylcholine
- PE
- phosphatidylethanolamine
- PS
- phosphatidylserine
- Psd1p
- phosphatidylserine decarboxylase 1
- Psd2p
- phosphatidylserine decarboxylase 2
- PssA
- Escherichia coli phosphatidylserine synthase
- SC
- synthetic complete dextrose
- SG
- synthetic complete glycerol
This work was supported by the Intramural Research Program of the National Institute of Diabetes and Digestive and Kidney Diseases. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
The online version of this article (available at http://www.jlr.org) contains a supplement.
REFERENCES
- 1.Elbaz Y., and Schuldiner M.. 2011. Staying in touch: the molecular era of organelle contact sites. Trends Biochem. Sci. 36: 616–623. [DOI] [PubMed] [Google Scholar]
- 2.Helle S. C., Kanfer G., Kolar K., Lang A., Michel A. H., and Kornmann B.. 2013. Organization and function of membrane contact sites. Biochim. Biophys. Acta. 1833: 2526–2541. [DOI] [PubMed] [Google Scholar]
- 3.Prinz W. A. 2014. Bridging the gap: membrane contact sites in signaling, metabolism, and organelle dynamics. J. Cell Biol. 205: 759–769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Horvath S. E., and Daum G.. 2013. Lipids of mitochondria. Prog. Lipid Res. 52: 590–614. [DOI] [PubMed] [Google Scholar]
- 5.Benning C. 2009. Mechanisms of lipid transport involved in organelle biogenesis in plant cells. Annu. Rev. Cell Dev. Biol. 25: 71–91. [DOI] [PubMed] [Google Scholar]
- 6.Voelker D. R. 2009. Genetic and biochemical analysis of non-vesicular lipid traffic. Annu. Rev. Biochem. 78: 827–856. [DOI] [PubMed] [Google Scholar]
- 7.Lev S. 2010. Non-vesicular lipid transport by lipid-transfer proteins and beyond. Nat. Rev. Mol. Cell Biol. 11: 739–750. [DOI] [PubMed] [Google Scholar]
- 8.Osman C., Voelker D. R., and Langer T.. 2011. Making heads or tails of phospholipids in mitochondria. J. Cell Biol. 192: 7–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lang A., John Peter A. T., and Kornmann B.. 2015. ER-mitochondria contact sites in yeast: beyond the myths of ERMES. Curr. Opin. Cell Biol. 35: 7–12. [DOI] [PubMed] [Google Scholar]
- 10.Phillips M. J., and Voeltz G. K.. 2016. Structure and function of ER membrane contact sites with other organelles. Nat. Rev. Mol. Cell Biol. 17: 69–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Vance J. E. 2014. MAM (mitochondria-associated membranes) in mammalian cells: lipids and beyond. Biochim. Biophys. Acta. 1841: 595–609. [DOI] [PubMed] [Google Scholar]
- 12.Gallo A., Vannier C., and Galli T.. 2016. Endoplasmic reticulum-plasma membrane associations: structures and functions. Annu. Rev. Cell Dev. Biol. 32: 279–301. [DOI] [PubMed] [Google Scholar]
- 13.Lebiedzinska M., Szabadkai G., Jones A. W., Duszynski J., and Wieckowski M. R.. 2009. Interactions between the endoplasmic reticulum, mitochondria, plasma membrane and other subcellular organelles. Int. J. Biochem. Cell Biol. 41: 1805–1816. [DOI] [PubMed] [Google Scholar]
- 14.Raturi A., and Simmen T.. 2013. Where the endoplasmic reticulum and the mitochondrion tie the knot: the mitochondria-associated membrane (MAM). Biochim. Biophys. Acta. 1833: 213–224. [DOI] [PubMed] [Google Scholar]
- 15.Flis V. V., and Daum G.. 2013. Lipid transport between the endoplasmic reticulum and mitochondria. Cold Spring Harb. Perspect. Biol. 5: a013235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pichler H., Gaigg B., Hrastnik C., Achleitner G., Kohlwein S. D., Zellnig G., Perktold A., and Daum G.. 2001. A subfraction of the yeast endoplasmic reticulum associates with the plasma membrane and has a high capacity to synthesize lipids. Eur. J. Biochem. 268: 2351–2361. [DOI] [PubMed] [Google Scholar]
- 17.Vance J. E. 1990. Phospholipid synthesis in a membrane fraction associated with mitochondria. J. Biol. Chem. 265: 7248–7256. [PubMed] [Google Scholar]
- 18.Gaigg B., Simbeni R., Hrastnik C., Paltauf F., and Daum G.. 1995. Characterization of a microsomal subfraction associated with mitochondria of the yeast, Saccharomyces cerevisiae. Involvement in synthesis and import of phospholipids into mitochondria. Biochim. Biophys. Acta. 1234: 214–220. [DOI] [PubMed] [Google Scholar]
- 19.Stone S. J., and Vance J. E.. 2000. Phosphatidylserine synthase-1 and -2 are localized to mitochondria-associated membranes. J. Biol. Chem. 275: 34534–34540. [DOI] [PubMed] [Google Scholar]
- 20.Vance J. E., and Tasseva G.. 2013. Formation and function of phosphatidylserine and phosphatidylethanolamine in mammalian cells. Biochim. Biophys. Acta. 1831: 543–554. [DOI] [PubMed] [Google Scholar]
- 21.Shiao Y. J., Lupo G., and Vance J. E.. 1995. Evidence that phosphatidylserine is imported into mitochondria via a mitochondria-associated membrane and that the majority of mitochondrial phosphatidylethanolamine is derived from decarboxylation of phosphatidylserine. J. Biol. Chem. 270: 11190–11198. [DOI] [PubMed] [Google Scholar]
- 22.Trotter P. J., Pedretti J., and Voelker D. R.. 1993. Phosphatidylserine decarboxylase from Saccharomyces cerevisiae. Isolation of mutants, cloning of the gene, and creation of a null allele. J. Biol. Chem. 268: 21416–21424. [PubMed] [Google Scholar]
- 23.Storey M. K., Clay K. L., Kutateladze T., Murphy R. C., Overduin M., and Voelker D. R.. 2001. Phosphatidylethanolamine has an essential role in Saccharomyces cerevisiae that is independent of its ability to form hexagonal phase structures. J. Biol. Chem. 276: 48539–48548. [DOI] [PubMed] [Google Scholar]
- 24.Trotter P. J., and Voelker D. R.. 1995. Identification of a non-mitochondrial phosphatidylserine decarboxylase activity (PSD2) in the yeast Saccharomyces cerevisiae. J. Biol. Chem. 270: 6062–6070. [DOI] [PubMed] [Google Scholar]
- 25.Gulshan K., Shahi P., and Moye-Rowley W. S.. 2010. Compartment-specific synthesis of phosphatidylethanolamine is required for normal heavy metal resistance. Mol. Biol. Cell. 21: 443–455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kannan M., Riekhof W. R., and Voelker D. R.. 2015. Transport of phosphatidylserine from the endoplasmic reticulum to the site of phosphatidylserine decarboxylase2 in yeast. Traffic. 16: 123–134. [DOI] [PubMed] [Google Scholar]
- 27.Yamashita S., and Nikawa J.. 1997. Phosphatidylserine synthase from yeast. Biochim. Biophys. Acta. 1348: 228–235. [DOI] [PubMed] [Google Scholar]
- 28.Achleitner G., Zweytick D., Trotter P. J., Voelker D. R., and Daum G.. 1995. Synthesis and intracellular transport of aminoglycerophospholipids in permeabilized cells of the yeast, Saccharomyces cerevisiae. J. Biol. Chem. 270: 29836–29842. [DOI] [PubMed] [Google Scholar]
- 29.Kuchler K., Daum G., and Paltauf F.. 1986. Subcellular and submitochondrial localization of phospholipid-synthesizing enzymes in Saccharomyces cerevisiae. J. Bacteriol. 165: 901–910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kodaki T., and Yamashita S.. 1987. Yeast phosphatidylethanolamine methylation pathway. Cloning and characterization of two distinct methyltransferase genes. J. Biol. Chem. 262: 15428–15435. [PubMed] [Google Scholar]
- 31.Nunnari J., Wong E. D., Meeusen S., and Wagner J. A.. 2002. Studying the behavior of mitochondria. Methods Enzymol. 351: 381–393. [DOI] [PubMed] [Google Scholar]
- 32.Choi H. S., Sreenivas A., Han G. S., and Carman G. M.. 2004. Regulation of phospholipid synthesis in the yeast cki1Delta eki1Delta mutant defective in the Kennedy pathway. The Cho1-encoded phosphatidylserine synthase is regulated by mRNA stability. J. Biol. Chem. 279: 12081–12087. [DOI] [PubMed] [Google Scholar]
- 33.Parks L. W., Bottema C. D., Rodriguez R. J., and Lewis T. A.. 1985. Yeast sterols: yeast mutants as tools for the study of sterol metabolism. Methods Enzymol. 111: 333–346. [DOI] [PubMed] [Google Scholar]
- 34.Vaden D. L., Gohil V. M., Gu Z., and Greenberg M. L.. 2005. Separation of yeast phospholipids using one-dimensional thin-layer chromatography. Anal. Biochem. 338: 162–164. [DOI] [PubMed] [Google Scholar]
- 35.Bartlett G. R. 1959. Colorimetric assay methods for free and phosphorylated glyceric acids. J. Biol. Chem. 234: 469–471. [PubMed] [Google Scholar]
- 36.Raychaudhuri S., and Prinz W. A.. 2008. Nonvesicular phospholipid transfer between peroxisomes and the endoplasmic reticulum. Proc. Natl. Acad. Sci. USA. 105: 15785–15790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang F., Zhao Y., and Wang P.. 2003. Separation and determination of phospholipids in biological samples by high-performance liquid chromatography. J. Chromatogr. Sci. 41: 142–144. [DOI] [PubMed] [Google Scholar]
- 38.Choudhary V., Ojha N., Golden A., and Prinz W. A.. 2015. A conserved family of proteins facilitates nascent lipid droplet budding from the ER. J. Cell Biol. 211: 261–271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Riezman H., Hase T., van Loon A. P., Grivell L. A., Suda K., and Schatz G.. 1983. Import of proteins into mitochondria: a 70 kilodalton outer membrane protein with a large carboxy-terminal deletion is still transported to the outer membrane. EMBO J. 2: 2161–2168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Carman G. M., and Dowhan W.. 1979. Phosphatidylserine synthase from Escherichia coli. The role of Triton X-100 in catalysis. J. Biol. Chem. 254: 8391–8397. [PubMed] [Google Scholar]
- 41.Okada M., Matsuzaki H., Shibuya I., and Matsumoto K.. 1994. Cloning, sequencing, and expression in Escherichia coli of the Bacillus subtilis gene for phosphatidylserine synthase. J. Bacteriol. 176: 7456–7461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kornmann B., Currie E., Collins S. R., Schuldiner M., Nunnari J., Weissman J. S., and Walter P.. 2009. An ER-mitochondria tethering complex revealed by a synthetic biology screen. Science. 325: 477–481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Voss C., Lahiri S., Young B. P., Loewen C. J., and Prinz W. A.. 2012. ER-shaping proteins facilitate lipid exchange between the ER and mitochondria in S. cerevisiae. J. Cell Sci. 125: 4791–4799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Prinz W. A., Grzyb L., Veenhuis M., Kahana J. A., Silver P. A., and Rapoport T. A.. 2000. Mutants affecting the structure of the cortical endoplasmic reticulum in Saccharomyces cerevisiae. J. Cell Biol. 150: 461–474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Letts V. A., Klig L. S., Bae-Lee M., Carman G. M., and Henry S. A.. 1983. Isolation of the yeast structural gene for the membrane-associated enzyme phosphatidylserine synthase. Proc. Natl. Acad. Sci. USA. 80: 7279–7283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Henry S. A., Kohlwein S. D., and Carman G. M.. 2012. Metabolism and regulation of glycerolipids in the yeast Saccharomyces cerevisiae. Genetics. 190: 317–349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lahiri S., Chao J. T., Tavassoli S., Wong A. K., Choudhary V., Young B. P., Loewen C. J., and Prinz W. A.. 2014. A conserved endoplasmic reticulum membrane protein complex (EMC) facilitates phospholipid transfer from the ER to mitochondria. PLoS Biol. 12: e1001969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Riekhof W. R., Wu W. I., Jones J. L., Nikrad M., Chan M. M., Loewen C. J., and Voelker D. R.. 2014. An assembly of proteins and lipid domains regulates transport of phosphatidylserine to phosphatidylserine decarboxylase 2 in Saccharomyces cerevisiae. J. Biol. Chem. 289: 5809–5819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wakana Y., Kotake R., Oyama N., Murate M., Kobayashi T., Arasaki K., Inoue H., and Tagaya M.. 2015. CARTS biogenesis requires VAP-lipid transfer protein complexes functioning at the endoplasmic reticulum-Golgi interface. Mol. Biol. Cell. 26: 4686–4699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Skehel P. A., Martin K. C., Kandel E. R., and Bartsch D.. 1995. A VAMP-binding protein from Aplysia required for neurotransmitter release. Science. 269: 1580–1583. [DOI] [PubMed] [Google Scholar]
- 51.Loewen C. J., and Levine T. P.. 2005. A highly conserved binding site in vesicle-associated membrane protein-associated protein (VAP) for the FFAT motif of lipid-binding proteins. J. Biol. Chem. 280: 14097–14104. [DOI] [PubMed] [Google Scholar]
- 52.Achleitner G., Gaigg B., Krasser A., Kainersdorfer E., Kohlwein S. D., Perktold A., Zellnig G., and Daum G.. 1999. Association between the endoplasmic reticulum and mitochondria of yeast facilitates interorganelle transport of phospholipids through membrane contact. Eur. J. Biochem. 264: 545–553. [DOI] [PubMed] [Google Scholar]
- 53.Kuge O., Hasegawa K., Saito K., and Nishijima M.. 1998. Control of phosphatidylserine biosynthesis through phosphatidylserine-mediated inhibition of phosphatidylserine synthase I in Chinese hamster ovary cells. Proc. Natl. Acad. Sci. USA. 95: 4199–4203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kuge O., Saito K., and Nishijima M.. 1999. Control of phosphatidylserine synthase II activity in Chinese hamster ovary cells. J. Biol. Chem. 274: 23844–23849. [DOI] [PubMed] [Google Scholar]
- 55.Sousa S. B., Jenkins D., Chanudet E., Tasseva G., Ishida M., Anderson G., Docker J., Ryten M., Sa J., Saraiva J. M., et al. 2014. Gain-of-function mutations in the phosphatidylserine synthase 1 (PTDSS1) gene cause Lenz-Majewski syndrome. Nat. Genet. 46: 70–76. [DOI] [PubMed] [Google Scholar]
- 56.Bae-Lee M., and Carman G. M.. 1990. Regulation of yeast phosphatidylserine synthase and phosphatidylinositol synthase activities by phospholipids in Triton X-100/phospholipid mixed micelles. J. Biol. Chem. 265: 7221–7226. [PubMed] [Google Scholar]
- 57.Wu W. I., and Voelker D. R.. 2004. Reconstitution of phosphatidylserine transport from chemically defined donor membranes to phosphatidylserine decarboxylase 2 implicates specific lipid domains in the process. J. Biol. Chem. 279: 6635–6642. [DOI] [PubMed] [Google Scholar]
- 58.Shcheprova Z., Baldi S., Frei S. B., Gonnet G., and Barral Y.. 2008. A mechanism for asymmetric segregation of age during yeast budding. Nature. 454: 728–734. [DOI] [PubMed] [Google Scholar]
- 59.Luedeke C., Frei S. B., Sbalzarini I., Schwarz H., Spang A., and Barral Y.. 2005. Septin-dependent compartmentalization of the endoplasmic reticulum during yeast polarized growth. J. Cell Biol. 169: 897–908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Chao J. T., Wong A. K., Tavassoli S., Young B. P., Chruscicki A., Fang N. N., Howe L. J., Mayor T., Foster L. J., and Loewen C. J.. 2014. Polarization of the endoplasmic reticulum by ER-septin tethering. Cell. 158: 620–632. [DOI] [PubMed] [Google Scholar]
- 61.Vance J. E. 1991. Newly made phosphatidylserine and phosphatidylethanolamine are preferentially translocated between rat liver mitochondria and endoplasmic reticulum. J. Biol. Chem. 266: 89–97. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.