Abstract
We determined for the first time the profiles of the nine most abundant unbound FFAs (FFAus) in human plasma. Profiles were determined for a standard reference plasma of pooled healthy adults for which the Lipid MAPSMAPS Consortium had determined the total FFA profiles. Measurements were performed by using 20 different acrylodan-labeled fatty acid binding protein mutants (probes), which have complementary specificities for the nine FFAs that comprise more than 96% of long-chain plasma FFA. The acrylodan fluorescence emission for each probe changes upon binding a FFAu. The plasma concentrations of each of the nine FFAus were determined by combining the measured fluorescence ratios of the 20 probes. The total molar FFAu concentration accounted for <10−5 of the total FFA concentration, and the mole fractions of the FFAu profiles were substantially different than the total FFA profiles. Myristic acid, for example, comprises 22% of the unbound versus 2.8% of the total. The most surprising difference is our finding of zero unbound cis-9-palmitoleic acid (POA), whereas the total POA was 7.2%. An unidentified plasma component appears to specifically prevent the release of POA. FFAus are the physiologically active FFAs, and plasma FFAu profiles may provide novel information about human health.
Keywords: lipidomics, fatty acid binding proteins, physical biochemistry, quantitation, albumin binding, fluorescence, metabolism
Large amounts of FFAs circulate in mammalian plasma to provide fuel for physiologic activity and substrates for more complex lipids and to participate in important signaling events (1, 2). FFAs are present in blood at concentrations from ∼100 µM to >1 mM, and almost all FFAs are bound to serum albumin. The pool of circulating FFAs is composed of as many as 40 distinct molecular species of FFAs. Approximately 30 of these are long-chain FFAs (14–26 carbons), of which 9 FFAs (Table 1) make up >96% of the total long-chain FFAs by molarity (3). The distribution of the different FFAs (the FFA profile) is expected to reflect the physiologic state, and changes in the profiles have been correlated with disease (4–8).
TABLE 1.
The 9 FFAs whose unbound plasma profiles were determined in this study
| Common Name | Abbreviation | Structuresa |
| Myristic | MA | 14:0 |
| Palmitic | PA | 16:0 |
| cis-9-palmitoleic | POA | 16:1 |
| Stearic | SA | 18:0 |
| Oleic | OA | 18:1 |
| Linoleic | LA | 18:2 |
| α-Linolenic | LNA | 18:3 (ω-3) |
| Arachidonic | AA | 20:4 |
| Eicoaspentaenoic 5,8,11,14,17 | EPA | 20:5 |
| Docosahexaenoic 4,7,10,13,16,19 | DHA | 22:6 |
These nine FFAs comprise 96% of the total FFA by molarity as determined by the Lipid MAPS Consortium (3).
Fatty acid chain length and double bond number.
Because most FFAs are bound to albumin, the total plasma FFA concentrations do not directly reflect the physiologically active FFA concentrations. The albumin-bound FFAs cannot enter cells, bind to proteins, or serve as substrates for FFA-utilizing enzymes. Only FFAs that are soluble monomers in the aqueous phase can be transported across membranes, bind to specific sites on proteins, and function in enzymatic reactions. The soluble FFAs (unbound FFA; FFAu) are a tiny fraction of the total FFAs (bound and unbound). In humans, the molar FFAu concentration is typically <10−5 of the total FFA concentration (9). As a consequence, measurements of total FFAs are insensitive to changes in FFAus.
Determination of the total FFA profile requires organic/aqueous phase separation of the plasma to extract the lipid components and separation of the FFA fraction from the other lipid species. The extracted FFAs are derivatized, chromatographed, and optimally detected by mass spectroscopy. Extraction by phase separation will alter the FFA profile from that of the plasma because different FFAs have different organic/aqueous partition coefficients. The distortion in the total FFA profile due to differences in partition can be addressed by using multiple FFA standards to account for these differences in partition (3).
Here, we describe a method to determine, for the first time, the concentrations of the different FFAus in human plasma. The method uses fluorescently labeled mutated fatty acid binding proteins (probes) that respond to the binding of FFAus by a change in the ratio of fluorescence (R ) at two different emission wavelengths, 550/460 nm. A set of 20 probes with complementary specificities for the nine FFAs was used to obtain the FFAu profiles. Each probe in the set was added directly and independently to a plasma sample, and the fluorescence ratio of each probe was measured at the same emission wavelengths.
We have applied this new method to determine the FFAu profile of the National Institute of Standards and Technology (NIST) standard reference material plasma (SRM) 1950. SRM 1950 is plasma pooled from 100 fasting (overnight) healthy adult donors, 50 females and 50 males, with an ethnic distribution representative of the population of the United States. Moreover, this sample’s total FFA profile has been determined by the Lipid MAPS Consortium (3). We find that the FFAu profile, expressed as mole fractions, is different from the Lipid MAPS Consortium profile of the total FFA. Some of the differences may be due to FFA-specific differences in albumin binding. However, not all the profile differences can be explained by differences in FFA-albumin binding affinities. Most surprisingly, although the mole fraction of cis-9-palmitoleate (POA) in the total FFA profile is 7.2% (3), the mole fraction of the unbound POA is 0. Here, we provide evidence that an unidentified plasma factor may specifically prevent the dissociation of POA from albumin, or a nonalbumin plasma component may bind POA with very high affinity; either mechanism would result in zero unbound POA.
MATERIALS AND METHODS
Materials
Buffer salts, HEPES Ultra, and the sodium salt of DHA were purchased from Sigma-Aldrich (St. Louis, MO). All other fatty acid sodium salts were obtained from NuChek Prep (Elysian, MN). Essentially fatty acid free human serum albumin (HSA) was purchased from SeraCare Life Sciences (Milford, MA). Samples and FFA/albumin complexes were diluted into the following buffer before measuring: 50 mM HEPES, 140 mM NaCl, 5 mM KCl, and 1 mM Na2HPO4 (pH 7.4).
Construction of FFAu-specific probes
We previously created FFAu probes from rat intestinal fatty acid binding protein (rI-FABP) mutants by labeling the mutants with acrylodan at lysine 27, position 27 being at a particularly favorable location near the ligand binding pocket (10). However, labeling reactions were frequently inefficient and somewhat nonspecific, producing unlabeled and multiply labeled products, as well as the desired single label at lysine 27. We have now produced probes using essentially the same method as in (10), except that acrylodan is reacted with a cysteine residue added near the binding pocket by substitution mutagenesis. Cysteine reacts with acrylodan much more efficiently and at a lower pH than lysine. For each promising probe from a screening library, the position of the cysteine relative to the binding pocket was optimized to maximize selectivity, binding affinity, and dynamic range. These cysteine-labeled probes are substantially more specific for individual FFAus and are more sensitive (larger signal) than the previous probes (10). Libraries of mutant fatty acid binding proteins (FABPs) were constructed by combinatorial mutagenesis of amino acid residues, primarily within the rI-FABP binding pocket (11, 12). All mutant FABPs have an E131D mutation plus a COOH-terminal Arg-Gly-six-histidine tag. Colonies picked from these libraries were grown, lysed, Ni-nitrilotriacetic acid (Ni-NTA) agarose affinity purified, labeled with acrylodan, and eluted in deep 96-well plates. Probes were screened for their response to the FFAus (shown in Table 1) in 384-well plates. The response was measured by the change in the ratio (R ) of the fluorescence intensities, excitation wavelength at 380 nm, and emission wavelengths at 460 and 550 nm. Probes with desirable specificity and sensitivity parameters were further improved by using the initially identified probe as a template to generate additional rounds of mutant FABP libraries.
Probe calibration
The rI-FABP mutants chosen during screening were grown to large scale and purified by using a combination of Ni-NTA affinity, ion-exchange, and gel-filtration chromatography. Purified protein was reacted with acrylodan, and the reaction product was purified by gel-filtration chromatography. Approximately 700 mg of purified protein was obtained from a 4 l Escherichia coli shake flask culture, and approximately 200 mg of purified probe could be produced from each protein preparation. Purified probes were calibrated by using equation 1 as described in ref. 10. Equation 1 relates, for each probe, the FFAu concentration for a single FFAu to the equilibrium dissociation constant (Kd), the ratio of fluorescence intensity at 460 nm in the absence of any FFAus to that at concentrations of the FFAus that saturate the response (Q), the R value at saturation (Rm), and R0, the R value at zero FFAus (10, 13).
| (Eq. 1) |
For each FFAu, a binding isotherm of the probe was determined, at 22°C, by measuring the increase in R with increasing FFAu concentrations. FFAu concentrations were clamped between 1 and 5,000 nM by using high concentrations of HSA complexed with FFA to strongly buffer the FFAu concentration. The complexes were prepared by titrating 600 µM HSA with FFA and using the previously described probe, ADIFAB2, to monitor the FFAu concentration during the titration (10). A nonlinear fit to the binding isotherm yielded the Kd, Q, and Rm values (10). Calibration parameters were determined for all 9 FFAs and 20 probes (supplemental Table S1).
Measurements of probe fluorescence in human plasma
Plasma FFAu profiles were determined from measurements made by using a suite of 20 different FFAu probes. These probes have complementary specificities for the nine FFAus of Table 1. Probe concentrations were 0.5 µM and at these concentrations did not significantly disturb the equilibrium FFAu concentrations in plasma samples diluted 50-fold in buffer (2% plasma).
Fluorescence was measured in 96-well plates by using a BMG PHERAstar plate reader (BMG LABTECH, Cary, NC) with a fixed filter module, capable of measuring two emission wavelengths simultaneously from each well of the 96-well plates, and all measurements were at 22 ± 3°C. The excitation filter of the module was centered at 380 nm and the emission filters at 460 and 550 nm; bandwidths for all filters were 10 nm.
Plasma samples and 600 µM fatty acid free HSA were each diluted 50-fold in measuring buffer (50 mM HEPES, 140 mM NaCl, 5 mM KCl, and 1 mM Na2HPO4 at pH 7.4) to final volumes of 150 μl in 96-well plates. Measurements of the fluorescence intensities at both emission wavelengths were performed first without added probe in the diluted samples to obtain sample and R o blank values. Probe was then added (one probe per well) to a final concentration in the wells of 0.5 µM, and the measurements of fluorescence were repeated. From the probe and blank measurements, the R values for each probe-sample combination were calculated as:
| (Eq. 2) |
where I(λ) and I(λ)blank are the probe and blank fluorescence intensities at wavelength . Measurements of all 96-well plates were repeated four times at ∼15 min intervals, where the time required for measurements of a 96-well plate was 60 s. From these measurements, the coefficient of variation (CV) for R, for the probes was ≤ 1.2% (supplemental Table S2).
A probe response (ΔR/Ro) is the change in the probe R value (ΔR = R − Ro) for a specific FFAu concentration, relative to R o. Rearranging equation 1, we obtained the following expression for ΔR/Ro:
| (Eq. 3) |
ΔR/Ro was calculated for each of the 20 probes of Table 2 and for the 9 FFAus of Table 1. The calculations used the calibration parameters (KdQ, Rm) (supplemental Table S1) and a concentration of 1 nM for each FFAu. The resulting probe response profiles for the 20 probes are shown in supplemental Table S3.
TABLE 2.
Sequences of the 20 probes used to determine FFAu profiles
| Probe | Mutations Relative to the Sequence of Fig. 1 |
| L2P22G6 | M18I G31Y A73G |
| L2P22G6K27C | M18I K27C G31Y A73G |
| L4BP4B9 | L72A A73W D74S |
| L10P7A4L30C | Y14L M18L K27A L30C G31Y L72A A73L Y117A |
| L11P7B3V26C | M21F V26C K27A L72A L78V L102V |
| L13EP16E11 | V26C K27A V49L L72A R106W Q115S |
| L19CP10C7 | −1G 1I Y14R M18L K27C S71I A73F Y117D |
| L36P15B7 | M18L I23V G31N F55R L72W A73T D74A R106A Q115E Y117V |
| L50BP4E2 | M18L M21F I23F K27C G31N L72T A73T D74A T76F |
| L50BP9D5 | M18L M21F I23L K27C G31N L72S A73T D74A T76I Y117N |
| L50AP31A2 | M18L I23L K27A L30C G31N L72S A73T D74A T76V |
| L61P8B12 | V8I Y14L M18L I23L K27Y L30C G31V S53I F55W L72G D74A L78V W82V |
| G91Y F93M L102V R106W Q115W Y117L | |
| L71BP33D1 | Y14W M18L M21I I23Y K27C G31N L38V L72W A73T D74F T76P R106A Q115E Y117V |
| L76P9E4 | Y14L M18Y I23T K27L L30C G31I L72G A73I D74A L78V W82P G91S F93M |
| A104F R106L Y117L Q115A | |
| L106P3B10 | −1G 1I M18L I23L K27C G31N L72T A73T D74A T76P L78I A104S Y117H F128Y |
| L118P4H5 | Y14W M18Q K27C L72A D74Q R106W |
| L119P3E5 | M18V K27C G31A L72A A73W |
| L127P3G2 | Y14W M18E M21V I23L K27C G31N L38V F55R L72W A73T D74A L78W R106A |
| Q115E Y117V | |
| L138P1H8N34C | −1G 1I M18L M21L I23Y N24C G31I F55V L72S A73S D74G T76V Y117H F128Y |
| L156P3F9V26C | Y14L M18L I23T V26C K27F G31A L72G D74A L78T W82V G91Y F93M L102V |
| R106W Q115W Y117L |
Determination of plasma FFAu profiles
FFAu profiles, the set of different FFAus in the sample (FFAi), were determined from the measured R values by using the following set of equations relating the measured R, the Ro values, the calibration parameters for each probe ( each FFAi,
| (Eq. 4) |
Equation 4 is for a single probe (j). Therefore, a set of 20 (j = 1–20) equations, 1 for each of the 20 different probes, was used to determine the profile for the n = 9, FFAi. The concentration of each FFAi in the sum of equation 4 multiplies a ratio that includes the calibration parameters ( for each for each probe j and for each of the nine FFAi. The set of 20 equations was solved simultaneously to determine the FFAu profiles [FFA1, FFA2,…..FFAN]. Because the probes are not absolutely specific (supplemental Table S3), multiple complementary probes were used so that the number (n = 20) of type 4 equations exceeded the number of FFAi in the profiles. The solution to this overdetermined system of equations was found by least-squares fitting, implemented in MLAB (Civilized Software, Silver Spring, MD).
An additional constraint on the solutions to equations 4 was used to identify outlier values arising from sample contamination and FFAi values resulting in saturating Rj values (Rj ≈ Rmj). The constraint is that the total FFAu ( must be the same for all probes. This follows from the definition of
| (Eq. 5) |
where is the mole fraction of each FFAi by definition .
Using equation 5, equation 4 can be solved for as,
| (Eq. 6) |
where for each probe j, the brackets <> represent weighted averages. The values were first calculated for each probe by using the αi obtained from the initial FFAu profile. The average value and SD of all probes was determined, and probes whose deviated by ≥2 SDs were identified. In most cases, the deviation could be corrected, or if saturation occurred, a new profile was determined with the outlier probe removed.
Artificial NIST
We created an “artificial NIST” plasma by combining FFA-HSA complexes of the nine FFAs in Table 1 using the ratios of the individual total FFA concentrations determined for the NIST 1950 SRM plasma by the Lipid MAPS Consortium (3). FFA profile measurements were performed by using artificial NIST diluted 50-fold in the buffer.
Estimates of FFAu profile uncertainties as a function of errors in R
The ability to determine FFAu profiles accurately from the measured R values depends on the uncertainties in the measured R values. To estimate what level of uncertainty will allow an accurate determination of the FFAu profiles, we used the mole fractions, the set of αi, determined from the measured NIST FFAu. These αi were used to generate R values for each probe by solving equation 6 for R j, where
| (Eq. 7) |
Starting sets of ideal R values (no errors) for 20 probes were calculated for each of three total FFAu concentrations: 0.5, 1.5, and 5.0 nM, a range that is typical for different donors. For each total FFAu concentration, a random, maximum error of 1% and 2%, was applied to 20 copies of the ideal 20-probe set of R values for each FFAu total. Twenty FFAu profiles at each total FFAu concentration were then determined from each nonideal set of R values by using MLAB. The CV for each FFAu in each FFAu profile was calculated from the set of the 20 “replicate” FFAu profiles (supplemental Table S4).
FFA-albumin binding constants
The HSA equilibrium dissociation constants (Kd), at 22°C, for the nine FFAus were determined from binding isotherms measured, as described previously for BSA (10). Measurements were done by titrating 600 µM fatty acid free HSA in HEPES buffer with the sodium salts of the fatty acids. The fatty acid salts were added slowly to reach concentrations from ∼90 to 3,300 µM (depending on the FFA) and incubated for 10 min, and the equilibrium FFAu concentrations were measured with ADIFAB2 (10). Scatchard analysis revealed a single class of between five and eight binding sites and Kd values ranging from ∼5 to 50 nM, depending on the FFA (supplemental Table S5). Supplemental Table S5 also shows that results for BSA, from our earlier study (10), are similar to those obtained in the present study for HSA.
RESULTS
Composition of the FFAu probes used to determine plasma FFAu profiles
The protein components of the probes used to determine FFAu profiles are mutants of a His-tagged rI-FABP with a Glu-131 to Asp substitution and a COOH-terminal affinity tag comprising Arg-132, Gly-133, and six histidines (Fig. 1). Site selection for mutagenesis (10) was guided by the X-ray crystallography and NMR structures for the wild-type protein (11, 12), as well as our previous studies of the thermodynamics of mutant FABPs (14, 15). The FFAu probes consisted of between 3 and 20 substitutional mutations of the wild-type protein, modified by the His tag addition (Fig. 1). Most of the mutations were within the FFA binding pocket of the wild-type protein, and in some cases a cysteine was introduced at positions on or near the protein’s α-helical region (Table 2). In the sequences presented in Table 2 and Fig. 1, amino acids are numbered from the first residue of the mature protein. The initiator methionine residue of the wild-type protein was removed by aminopeptidase activity, leaving an alanine as residue 1. Thus, counting backward and skipping zero, the initiator methionine is at residue position −1. Seventeen of the probes were labeled with acrylodan on a cysteine at positions 26, 27, or 30 (Table 2). The remaining three probes were labeled at lysine at position 27.
Fig. 1.
The amino acid sequence of the mature rI-FABP modified by His-tag addition at the carboxyl terminus. The residue positions are numbered starting with the first residue of the mature protein, which lacks its initiator methionine. FFAu probes are generated from this protein through substitution and insertion mutagenesis, followed by labeling the mutant proteins with acrylodan (Table 2).
Probe response profiles
The response profiles were calculated by using equation 3, a FFAu concentration of 1 nM for each of the nine FFAus, and the probe calibrations in supplemental Table S1. For each probe, the largest responses occurred for those FFAus for which the probe is most sensitive (supplemental Table S3). The 20 probes have complementary specificities for all nine FFAus, but no probe has absolute specificity for any single FFAu. Several probes have high degrees of specificity, for example; L11P783V26C for stearic acid (SA), L76P9E4 for oleic acid (OA), and L118P4H5 for DHA (supplemental Table S3). The probes also differ in the magnitude of ΔR/Ro (sensitivity), where, for example, ΔR/Ro for POA is 62% for L50BP9D5, 470% for L61P8B12, and 1,460% for L156P3F926C1.
Determination of the FFAu profiles
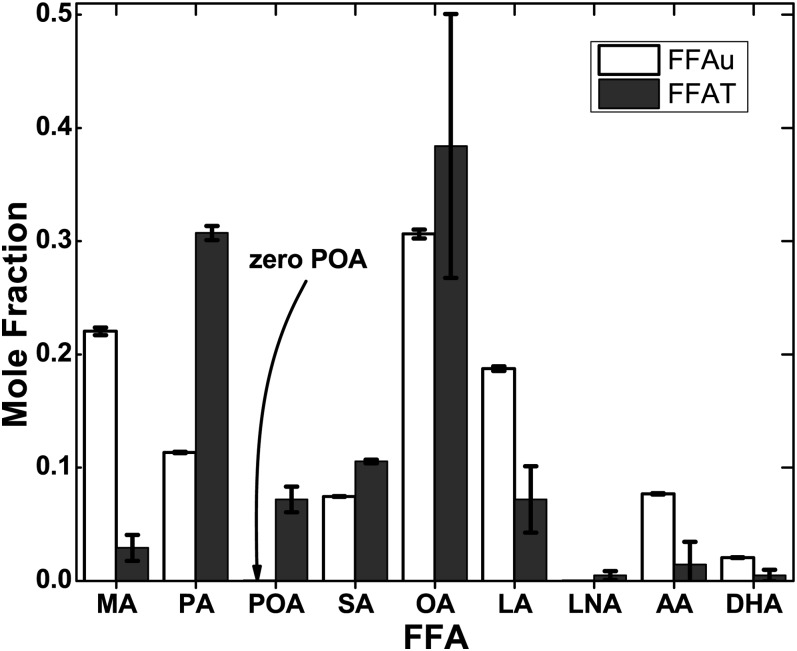
Measurements were carried out by using the NIST standard reference plasma (SRM 1950). R values were measured for each probe as defined by equation 2 and were repeated four times every 15 min. CVs for R values averaged over each duplicate at each of the four measurement times were ≤ 1.2% for all 20 probes (supplemental Table S2). FFAu profiles for the nine FFAus of Table 1 were determined from the measured R values of the 20 probes of Table 2. Profiles were solved individually for the separate measurements at different times (Table 3). There were no significant differences (all P > 0.05) by ANOVA among the profiles of Table 3. The total FFAu concentration ( was 1.6 nM, which is <10−5 of the total FFA concentration of 208 µM determined by the Lipid MMAPS Consortium (3). The profiles, as mole fractions, also revealed considerable differences between the FFAus and the total FFA profiles (Fig. 2). Although mole fractions of SA and OA were similar for the unbound and total profiles, the FFAu mole fractions for myristic acid (MA) (22%) was ∼10-fold larger than the total MA mole fraction (2.7%). The mole fraction of unbound palmitic acid (PA) was 30% of the total PA mole fraction, whereas unbound linoleic acid (LA), arachidonic acid (AA), and DHA mole fractions were more than 2.5-fold larger than their corresponding total FFA. The most extreme difference occurred for POA, where the mole fraction was ∼7% for the total FFA, but was zero for FFAu. Zero unbound POA is not specific to the NIST SRM 1950 sample. The concentration of unbound POA was found to be zero in >500 human donors, as well as in bovine, porcine, and rodent plasma (data not shown).
TABLE 3.
FFAu profiles of the NIST SRM 1950 human plasma
| Sample | Total FFAu (nM) | MA | PA | POA | SA | OA | LA | LNA | AA | DHA |
| NIST S1 Average | 1.6 | 0.222 | 0.108 | 0.000 | 0.074 | 0.303 | 0.196 | 0.000 | 0.076 | 0.021 |
| CV | 0.9% | 7.0% | 15.2% | NA | 2.1% | 2.7% | 6.7% | NA | 1.7% | 8.9% |
| NIST S2 Average | 1.6 | 0.219 | 0.118 | 0.000 | 0.075 | 0.311 | 0.179 | 0.000 | 0.077 | 0.021 |
| CV | 0.8% | 3.9% | 9.5% | NA | 1.4% | 1.9% | 5.2% | NA | 2.0% | 7.5% |
| NIST S3 Average | 1.6 | 0.221 | 0.115 | 0.000 | 0.075 | 0.305 | 0.187 | 0.000 | 0.078 | 0.020 |
| CV | 0.9% | 6.1% | 12.0% | NA | 1.5% | 2.1% | 6.6% | NA | 3.7% | 3.2% |
| NIST all average | 1.6 | 0.221 | 0.114 | 0.000 | 0.075 | 0.306 | 0.187 | 0.000 | 0.077 | 0.021 |
| CV | 1.4% | 5.3% | 11.8% | 1.7% | 2.3% | 6.9% | 2.6% | 6.6% | ||
| SD | 0.022 | 0.012 | 0.002 | NA | 0.001 | 0.013 | 0.007 | NA | 0.001 | 0.001 |
Three replicate NIST SRM 1950 samples, each in duplicate, were measured at four times, from 15 min after sample addition to the 96-well plates to 60 min later. The duplicate R values from each sample were averaged and used to determine FFAu profiles (mole fractions) separately for each of the four times and for the three samples. Averages and CVs for each sample’s profile and averages for all three samples are shown. The concentration for individual FFAu is mole fraction times the total FFAu. NA, not applicable.
Fig. 2.
FFAu profiles of NIST SRM 1950 plasma compared with the total FFA profiles of ref. 3. The results are shown as mole fractions of the nine FFAs normalized to unity. The total concentrations are 1.6 nM for the FFAus and 208 µM for the nine FFAs. Errors are SEM determined for FFAus from 12 measurements [3 replicate samples at 4 different times each (Table 3)]. The most striking difference between the total FFA and FFAu profiles is the absence of unbound POA.
FFAu profiling of artificial NIST
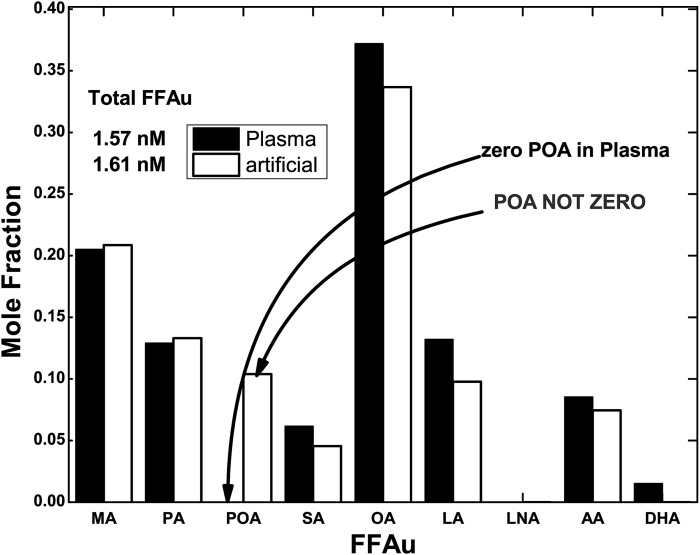
To assess whether the lack of POA detection is a methodologic problem, we prepared artificial NIST samples by combining fatty acid free HSA with a mixture of the nine total FFAs of Table 1, whose relative concentrations were those determined by the Lipid MAPS (3). The FFAu profiles of the artificial and actual NIST plasma were virtually identical, with the exception of POA, for which the mole fraction of the unbound POA was 12% for the artificial NIST sample, as compared with zero for the NIST plasma (Fig. 3). The failure to detect POA in plasma samples was therefore not a defect of the FFAu profile method.
Fig. 3.
FFAu profiles for NIST plasma (plasma) and artificial NIST (artificial). FFAu profiles for NIST plasma and artificial NIST plasma reveal almost identical profiles with the exception of POA, which is zero in the NIST plasma but 12% in the artificial NIST.
Estimates of uncertainties in the FFAu profiles
We estimated the level of uncertainty in the measured R values that would be needed to determine accurate FFAu profiles. The results show that the CVs of the individual FFAu components of the profile increase substantially with decreasing and with increasing CVs of the measured R values (supplemental Table S4). With R value CVs ≤ 1%, as in the present study, reliable FFAu levels (most CVs ≤ 50%) can be obtained for profiles with as low as 0.5 nM, which is about the lowest concentration observed in mammals. The results indicate that the limits of detection (±1 SD) are FFAu-dependent, and for the NIST samples with =1.6 nM yield an average of ∼7 pM for the nine FFAus of Table 1.
Potential mechanism for zero unbound POA in plasma
In an effort to understand the absence of unbound POA, we investigated the possibility that an unknown plasma component either prevents the dissociation of POA from albumin or itself binds POA with extremely high affinity. We determined the FFAu profiles of artificial NIST mixed with NIST plasma at artificial NIST to NIST plasma ratios from 9:1 to 1:1 (Table 4). The results indicate a monotonic reduction in unbound POA reaching a decrease of 73% at 1:1. In contrast to POA the other FFAus did not decrease, and most did not change [except linolenic acid (LNA); see Discussion)].
TABLE 4.
NIST plasma inhibits POA dissociation from artificial NIST
| Sample | Total FFAu (nM) | MA | PA | POA | SA | OA | LA | LNA | AA | DHA |
| Artificial NIST | 1.32 | 0.220 | 0.135 | 0.137 | 0.022 | 0.335 | 0.038 | 0.042 | 0.059 | 0.013 |
| 9:1 Artif:Plasma | 1.33 | 0.227 | 0.128 | 0.098 | 0.022 | 0.383 | 0.034 | 0.031 | 0.061 | 0.016 |
| 7:3 Artif:Plasma | 1.34 | 0.234 | 0.121 | 0.063 | 0.023 | 0.419 | 0.038 | 0.016 | 0.071 | 0.015 |
| 1:1 Artif:Plasma | 1.32 | 0.241 | 0.129 | 0.037 | 0.029 | 0.398 | 0.072 | 0.003 | 0.075 | 0.015 |
Artificial NIST, a mixture of nine FFAs and HSA, was titrated with NIST SRM 1950 plasma at ratios of artificial NIST to plasma from 9:1 to 1:1. FFAu profiles were determined from 100% artificial NIST and for each of the titration steps. Total FFAu and all FFAu mole fraction components were virtually unchanged at each step except for POA and LNA, which decreased by 73% and 93%, respectively.
DISCUSSION
FFAus are the physiologically active FFAs, but until now, their plasma profiles were unknown. Although FFAu profiles were previously determined in aqueous solutions of defined mixtures of ≤ 5 FFA with bovine albumin, it was unknown whether the method would be accurate in biological samples (7). Blood plasma contains many molecules that might interfere with the probe response, including ∼30 different FFAs that might interact with the FFAu probes that were available to Huber et al. (10). The ability to successfully profile blood samples is largely due to the discovery of probes with higher sensitivities and specificities than were available previously.
Validation of the FFAu profiles obtained by the FFAu probe method derives from in vivo and in vitro studies. FFAu profiles were determined in blood plasma of rats that were infused intravenously with oil emulsions of different compositions (16). The infusion was carried out in the presence of heparin, and the resulting hydrolysis of the oils generated oil-specific FFA. Comparing FFAu profiles determined before and after infusion demonstrated that the modulation of the plasma FFAu profile was consistent with the fatty acid compositions of the different oils. In addition, the study of Oh et al. used nicotinic acid to inhibit adipose lipolysis, which reduced by 4.5-fold (16). This reduction occurred for all FFAus, except AA, thereby increasing the unbound AA mole fraction from 10% to 50% of . The reduction of all FFAs except AA in rats was virtually identical with earlier studies of total FFA profiles of human subjects treated with nicotinic acid (8). We have also determined that addition of specific FFAus to plasma, as FFA:HSA complexes, yields FFAu profiles enriched in the added FFAus (data not shown). Additional evidence that the probes are highly specific for FFAus and are not affected by other plasma metabolites are the virtually identical FFAu profiles, except for POA, obtained in NIST SRM 1950 plasma and artificial NIST (Fig. 2).
An important feature of the FFAu profile measurements is that no alteration of the sample is required; measurements consist solely of adding each probe to a separate plasma sample and measuring the fluorescence ratio. In contrast, determination of total FFA profiles requires lipid extraction, derivatization, addition of standards, and GC/MS. The FFAu concentrations are more than 5 orders of magnitude smaller than total FFA, and their mole fractions are different from the total FFA profiles. As a consequence, changes in the physiological state due to disease and interactions of albumin with metabolites, hormones, and drugs detected by FFAu profiles may go undetected in the total FFA profile.
The FFAus are the physiologically active FFAs because FFAu monomers, but not albumin-bound FFA, react with other molecules. These interactions can be abolished by addition of albumin, which reduces FFAu without changing the total FFAs (10, 17–20). Moreover, physiologic reactions occur at the nanomolar concentrations of plasma FFAus, not the millimolar concentrations of total FFAs (10, 17). Determining the physiological effects of individual FFAs requires the use of nanomolar FFAu concentrations (Fig. 3 and Table 3). In vitro studies used FFA-albumin complexes to generate FFAu, presumed to be at physiologic concentrations. However, because FFAu concentrations increase exponentially with increasing ratios of FFA to albumin, especially above mole ratios of 4:1, reliable FFAu levels can be assured only by direct measurement of FFAu levels (21).
The FFAu profiles provide information about FFA metabolism that is not available from the total FFA profiles. This is because the FFAu profile also depends on: 1) the FFA-albumin binding constants, 2) interactions among the bound FFA, 3) interactions with drugs and other metabolites that can alter FFA binding to albumin, 4) metabolic alterations of albumin (22, 23), and 5) possibly albumin polymorphisms (24). The different albumin-binding characteristics of individual FFAs (supplemental Table S5) are consistent with the observed unbound and total profile differences for some FFAs. For example, the mole fractions of SA and OA are similar for the unbound and total profiles, possibly because SA and OA complexes with albumin have the smallest dissociation constants (4.3 and 4.7 nM, respectively). In contrast, the FFAu mole fraction for MA (22%), which has the largest albumin Kd value of the nine FFAs profiled (50 nM), is ∼10-fold larger than the total FFA mole fraction (2.7%). However, the correlation between total FFA, FFA-HSA Kd, and FFAus for other FFAs is less clear and may reflect additional interactions among bound FFAs.
The most striking difference between the observed FFAu and FFA profiles is the complete absence of unbound POA as compared with total POA. This is especially unexpected because the Kd for binding POA to HSA (27 nM) is about halfway between the Kd values of MA and PA. Moreover, the relatively large Kd for POA is also consistent with the unbound POA mole fraction in the artificial NIST profiles (12%) which is larger than the total mole fraction (7%). To begin to elucidate a mechanism for the lack of unbound POA, we considered the possibility that a factor in plasma binds specifically to albumin-bound POA and prevents its dissociation. The evidence for this is based on our measurements of FFAu profiles in artificial NIST that show a substantial decrease in unbound POA with increasing admixtures of NIST plasma (Table 4). If there were no inhibitory factor in NIST plasma, the unbound POA concentration would not change because FFA-HSA complexes are not affected significantly by dilution (21). Moreover, if all FFAs were freely exchangeable, the individual FFAu concentrations would be essentially the same at all steps in the titration as they are, except for POA (and possibly LNA) (Table 4). Similar to POA, the mole fraction of LNA in NIST plasma is zero, and its dissociation from albumin may also be blocked by a factor in NIST plasma. In contrast to POA, the mole fraction of total LNA is quite small (0.06%), and in other human blood samples, unbound LNA is occasionally nonzero. It is therefore unclear whether there is an inhibitory factor specific for LNA or whether its behavior reflects albumin-bound interactions with POA. It is also worth noting that only 35% of albumin has, on average, even a single bound FFA, and only 2.4% and 0.018% of albumin molecules on average have bound POA and LNA, respectively, suggesting a high affinity for the putative inhibitory factor. The results of Table 4 are also consistent with POA binding to an unknown serum factor with much higher affinity and a similar or greater concentration than HSA.
The surprising biochemical behavior of plasma POA likely has important physiologic ramifications. Studies suggest that POA has unique characteristics for regulating metabolism, insulin sensitivity, liver steatosis, and the risk of diabetes. In vitro studies indicate that PA reduces insulin sensitivity (25) through a process involving the activation of macrophages. This is essentially reversed by POA, which improves insulin sensitivity in muscle cells through an AMP-activated protein kinase mechanism (26, 27). Adipocyte FABP knockouts in mice resulted in increased circulating POA, improved insulin sensitivity, and reduced liver steatosis (28). In vivo studies of plasma POA levels and insulin sensitivity in humans have yielded conflicting outcomes (29, 30). Our results suggest that the unbound concentration of POA in plasma is tightly regulated and is not correlated with the plasma total POA concentration. Because unbound POA is biologically active, a correlation between plasma total POA FFA and clinical outcomes seems unlikely, unless a mechanism is activated that releases POA. Future studies will be needed to identify the factors influencing either stability of the POA-albumin complex or a nonalbumin molecule that binds POA with high affinity.
Supplementary Material
Acknowledgments
We thank Annie Tung-Chih Sun for preparation of mutants and Hiroto Kameyama for measurements and preliminary analysis of fluorescence.
Footnotes
Abbreviations:
- AA
- arachidonic acid
- CV
- coefficient of variation
- FABP
- fatty acid binding protein
- FFAu
- unbound FFA
- HSA
- human serum albumin
- LA
- linoleic acid
- LNA
- alpha-linolenic acid
- MA
- myristic acid
- NIST
- National Institute of Standards and Technology
- OA
- oleic acid
- PA
- palmitic acid
- POA
- palmitoleic acid
- probe
- acrylodan-labeled fatty acid binding mutant
- SA
- stearic acid
- SRM
- standard reference material
The online version of this article (available at http://www.jlr.org) contains a supplement.
REFERENCES
- 1.Nielsen S., Guo Z., Albu J. B., Klein S., O’Brien P. C., and Jensen M. D.. 2003. Energy expenditure, sex, and endogenous fuel availability in humans. J. Clin. Invest. 111: 981–988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Quehenberger O., and Dennis E. A.. 2011. The human plasma lipidome. N. Engl. J. Med. 365: 1812–1823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Quehenberger O., Armando A. M., Brown A. H., Milne S. B., Myers D. S., Merrill A. H., Bandyopadhyay S., Jones K. N., Kelly S., Shaner R. L., et al. 2010. Lipidomics reveals a remarkable diversity of lipids in human plasma. J. Lipid Res. 51: 3299–3305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yli-Jama P., Meyer H. E., Ringstad J., and Pedersen J. I.. 2002. Serum free fatty acid pattern and risk of myocardial infarction: a case- control study. J. Intern. Med. 251: 19–28. [DOI] [PubMed] [Google Scholar]
- 5.Villa P. M., Laivuori H., Kajantie E., and Kaaja R.. 2009. Free fatty acid profiles in preeclampsia. Prostaglandins Leukot. Essent. Fatty Acids. 81: 17–21. [DOI] [PubMed] [Google Scholar]
- 6.Lv W., and Yang T.. 2012. Identification of possible biomarkers for breast cancer from free fatty acid profiles determined by GC-MS and multivariate statistical analysis. Clin. Biochem. 45: 127–133. [DOI] [PubMed] [Google Scholar]
- 7.Tomita K., Teratani T., Yokoyama H., Suzuki T., Irie R., Ebinuma H., Saito H., Hokari R., Miura S., and Hibi T.. 2011. Plasma free myristic acid proportion is a predictor of nonalcoholic steatohepatitis. Dig. Dis. Sci. 56: 3045–3052. [DOI] [PubMed] [Google Scholar]
- 8.Hagenfeldt L., Wahren J., Pernow B., and Raf L.. 1972. Uptake of individual free fatty acids by skeletal muscle and liver in man. J. Clin. Invest. 51: 2324–2330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Huber A. H., Kampf J. P., Kwan T., Zhu B., Adams J. III, and Kleinfeld A. M.. 2014. Usefulness of serum unbound free fatty acid levels to predict death early in patients with ST-segment elevation myocardial infarction (from the Thrombolysis In Myocardial Infarction [TIMI] II Trial). Am. J. Cardiol. 113: 279–284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Huber A. H., Kampf J. P., Kwan T., Zhu B., and Kleinfeld A. M.. 2006. Fatty acid-specific fluorescent probes and their use in resolving mixtures of different unbound free fatty acids in equilibrium with albumin. Biochemistry. 45: 14263–14274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sacchettini J. C., Gordon J. I., and Banaszak L. J.. 1988. The structure of crystalline Escherichia coli-derived rat intestinal fatty acid binding protein at 2.5 A resolution. J. Biol. Chem. 263: 5815–5819. [PubMed] [Google Scholar]
- 12.Cistola D. P., Sacchettini J. C., Banaszak L. J., Walsh M. T., and Gordon J. I.. 1989. Fatty acid interactions with rat intestinal and liver fatty acid-binding proteins expressed in Escherichia coli. J. Biol. Chem. 264: 2700–2710. [PubMed] [Google Scholar]
- 13.Richieri G. V., Ogata R. T., and Kleinfeld A. M.. 1992. A fluorescently labeled intestinal fatty acid binding protein: Interactions with fatty acids and its use in monitoring free fatty acids. J. Biol. Chem. 267: 23495–23501. [PubMed] [Google Scholar]
- 14.Richieri G. V., Low P. J., Ogata R. T., and Kleinfeld A. M.. 1997. Mutants of rat intestinal fatty acid binding protein illustrate the critical role played by enthalpy-entropy compensation in ligand binding. J. Biol. Chem. 272: 16737–16740. [DOI] [PubMed] [Google Scholar]
- 15.Richieri G. V., Low P. J., Ogata R. T., and Kleinfeld A. M.. 1998. Thermodynamics of fatty acid binding to engineered mutants of the adipocyte and intestinal fatty acid binding proteins. J. Biol. Chem. 273: 7397–7405. [DOI] [PubMed] [Google Scholar]
- 16.Oh Y. T., Kim J., Kang I., and Youn J. H.. 2014. Regulation of hypothalamic-pituitary-adrenal axis by circulating free fatty acids in male Wistar rats: role of individual free fatty acids. Endocrinology. 155: 923–931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Richieri G. V., Ogata R. T., and Kleinfeld A. M.. 1994. Equilibrium constants for the binding of fatty acids with fatty acid-binding proteins from adipocyte, intestine, heart, and liver measured with the fluorescence probe ADIFAB. J. Biol. Chem. 269: 23918–23930. [PubMed] [Google Scholar]
- 18.Kleinfeld A. M., and Okada C.. 2005. Free fatty acid release from human breast cancer tissue inhibits cytotoxic T-lymphocyte-mediated killing. J. Lipid Res. 46: 1983–1990. [DOI] [PubMed] [Google Scholar]
- 19.Itoh Y., Kawamata Y., Harada M., Kobayashi M., Fujii R., Fukusumi S., Ogi K., Hosoya M., Tanaka Y., Uejima H., et al. 2003. Free fatty acids regulate insulin secretion from pancreatic beta cells through GPR40. Nature. 422: 173–176. [DOI] [PubMed] [Google Scholar]
- 20.Stulnig T. M., Berger M., Roden M., Stingl H., Raederstorff D., and Waldhausl W.. 2000. Elevated serum free fatty-acid concentrations inhibit T lymphocyte signaling. FASEB J. 14: 939–947. [DOI] [PubMed] [Google Scholar]
- 21.Richieri G. V., Anel A., and Kleinfeld A. M.. 1993. Interactions of long chain fatty acids and albumin: Determination of free fatty acid levels using the fluorescent probe ADIFAB. Biochemistry. 32: 7574–7580. [DOI] [PubMed] [Google Scholar]
- 22.Bhagavan N. V., Ha J. S., Park J. H., Honda S. A., Rios C. N., Sugiyama C., Fujitani G. K., Takeuchi I. K., and Ha C. E.. 2009. Utility of serum fatty acid concentrations as a marker for acute myocardial infarction and their potential role in the formation of ischemia-modified albumin: a pilot study. Clin. Chem. 55: 1588–1590. [DOI] [PubMed] [Google Scholar]
- 23.Lu J., Stewart A. J., Sadler P. J., Pinheiro T. J., and Blindauer C. A.. 2012. Allosteric inhibition of cobalt binding to albumin by fatty acids: implications for the detection of myocardial ischemia. J. Med. Chem. 55: 4425–4430. [DOI] [PubMed] [Google Scholar]
- 24.Peters T. 1996. Academic Press, San Diego, CA. 142–150. [Google Scholar]
- 25.Dimopoulos N., Watson M., Sakamoto K., and Hundal H. S.. 2006. Differential effects of palmitate and palmitoleate on insulin action and glucose utilization in rat L6 skeletal muscle cells. Biochem. J. 399: 473–481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Talbot N. A., Wheeler-Jones C. P., and Cleasby M. E.. 2014. Palmitoleic acid prevents palmitic acid-induced macrophage activation and consequent p38 MAPK-mediated skeletal muscle insulin resistance. Mol. Cell. Endocrinol. 393: 129–142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chan K. L., Pillon N. J., Sivaloganathan D. M., Costford S. R., Liu Z., Theret M., Chazaud B., and Klip A.. 2015. Palmitoleate reverses high fat-induced proinflammatory macrophage polarization via AMP-activated protein kinase (AMPK). J. Biol. Chem. 290: 16979–16988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cao H., Gerhold K., Mayers J. R., Wiest M. M., Watkins S. M., and Hotamisligil G. S.. 2008. Identification of a lipokine, a lipid hormone linking adipose tissue to systemic metabolism. Cell. 134: 933–944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Stefan N., Kantartzis K., Celebi N., Staiger H., Machann J., Schick F., Cegan A., Elcnerova M., Schleicher E., Fritsche A., et al. 2010. Circulating palmitoleate strongly and independently predicts insulin sensitivity in humans. Diabetes Care. 33: 405–407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fabbrini E., Magkos F., Su X., Abumrad N. A., Nejedly N., Coughlin C. C., Okunade A. L., Patterson B. W., and Klein S.. 2011. Insulin sensitivity is not associated with palmitoleate availability in obese humans. J. Lipid Res. 52: 808–812. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.