Abstract
The Metabolic Syndrome in Men (METSIM) study is a population-based study including 10,197 Finnish men examined in 2005–2010. The aim of the study is to investigate nongenetic and genetic factors associated with the risk of T2D and CVD, and with cardiovascular risk factors. The protocol includes a detailed phenotyping of the participants, an oral glucose tolerance test, fasting laboratory measurements including proton NMR measurements, mass spectometry metabolomics, adipose tissue biopsies from 1,400 participants, and a stool sample. In our ongoing follow-up study, we have, to date, reexamined 6,496 participants. Extensive genotyping and exome sequencing have been performed for essentially all METSIM participants, and >2,000 METSIM participants have been whole-genome sequenced. We have identified several nongenetic markers associated with the development of diabetes and cardiovascular events, and participated in several genetic association studies to identify gene variants associated with diabetes, hyperglycemia, and cardiovascular risk factors. The generation of a phenotype and genotype resource in the METSIM study allows us to proceed toward a “systems genetics” approach, which includes statistical methods to quantitate and integrate intermediate phenotypes, such as transcript, protein, or metabolite levels, to provide a global view of the molecular architecture of complex traits.
Keywords: type 2 diabetes, cardiovascular risk factors, metabolic disease, coronary artery disease, METSIM
Metabolic diseases and CVDs are caused by the effects and interactions of multiple genetic variants and lifestyle/environmental factors. Obesity, T2D, and coronary artery disease (CAD) are currently important causes of morbidity and mortality worldwide. Therefore, understanding the etiology and pathophysiology of these diseases is of great interest.
Genome-wide association studies (GWASs) have substantially increased our knowledge about the genetic basis of metabolic diseases and CVDs. Advances in next-generation sequencing techniques have made it possible to accumulate detailed genetic information at the level of the exome and genome. By contrast, the development in phenotyping has been considerably slower in recent years. Deep phenotyping of large randomly selected population-based studies is crucial for the success of genetic studies. Especially before the era of GWASs, population samples tended to be too small and poorly phenotyped with respect to clinical traits and laboratory measurements.
The Finnish population is a homogeneous isolate that has experienced a bottleneck in the 16th century in the settlement of Eastern Finland. A recent study demonstrated that Finns carry a significant enrichment of low frequency (1–5%) (1) and loss-of-function variants (frequency 0.5–5%) compared with other European and Asian populations (2). This offers advantages for studying rare variants in complex diseases. Distribution of common alleles in Finns, both synonymous and nonsynonymous, is unchanged by the bottleneck and, consequently, genetic association studies have reported similar associations of common variants with clinical and laboratory traits in Finns as in other European populations, as presented in this review. However, a strong influence of the bottleneck, particularly the proportional enrichment of rare deleterious variants, is also influenced by the unique history of the Finnish population and will not necessarily apply to all populations influenced by a bottleneck (2).
There are benefits to carrying out population-based epidemiological studies among Finns. The participation rate in these studies has been high (60–80%) during recent decades. Additionally, access to electronic nationwide health records (hospital discharge, mortality, cancer, and drug reimbursement registries) in Finland makes it possible to follow-up essentially all participants of any population-based study with respect to morbidity, mortality, and drug treatment of different diseases. Given this ideal setting to investigate nongenetic and genetic factors influencing the risk of different chronic diseases in Finns, we initiated the Metabolic Syndrome in Men (METSIM) study in 2005.
METSIM STUDY
Cross-sectional study
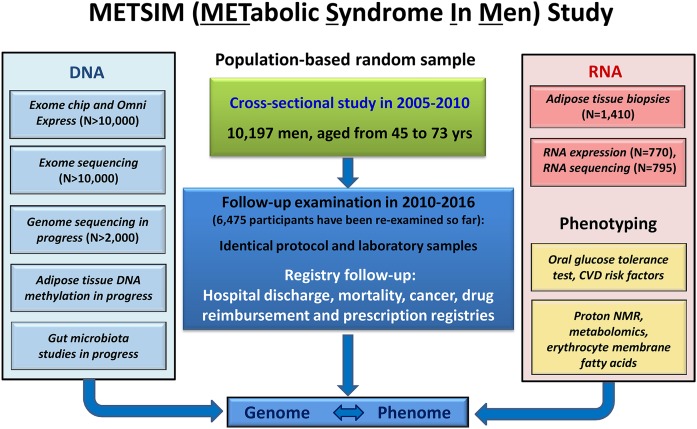
The METSIM study includes 10,197 men, aged from 45 to 73 years at entry, randomly selected from the population register of the Kuopio town, Eastern Finland, and examined in 2005–2010 (3). The aim of the study is to investigate nongenetic and genetic factors associated with T2D and CVD, and with cardiovascular risk factors in both cross-sectional and longitudinal settings. The METSIM study protocol includes, e.g., collection of data on drug treatment, CVD risk factors [smoking, exercise, diet, history of chronic diseases (including CAD, stroke, cardiac failure, medication, history of diabetes or early onset CAD in the family)], a questionnaire on the FINDRISC score (4), and measurement of height, weight, waist, hip, blood pressure, and fat percentage. Figure 1 presents the METSIM cross-sectional and follow-up study designs, including deep phenotyping and extensive DNA- and RNA-based genetic studies. Table 1 lists the diseases and traits investigated in the METSIM study, including nongenetic markers and genetics of the corresponding diseases/traits.
Fig. 1.
Description of the cross-sectional and longitudinal METSIM study. The cross-sectional study included 10,197 Finnish men, aged from 45 to 73 years. Phenotyping included several laboratory measurements in fasting, an OGTT, and proton NMR measurements from all participants. DNA-based genotyping includes OmniExpress for common and exome chip for low-frequency and rare variants, exome and genome sequencing, DNA methylation analysis in adipose tissue, and gut microbiome sequencing. Adipose tissue biopsies have been taken from 1,410 participants, RNA sequencing performed for 795 participants and RNA expression determined for 770 participants. The protocol of the follow-up study is identical to the cross-sectional study, and so far 6,496 individuals have participated in the follow-up. Additionally, all participants have registry follow-up allowing information on morbidity, mortality, and drug treatment to be obtained.
TABLE 1.
Diseases and traits investigated in the METSIM study
| Disease or Traita | Non-genetic Measurements | Genetic Association Studies |
| T2D (3) | Yes | Yes |
| Fasting or 2 h glucose | Yes | Yes |
| Glycated hemoglobin A1c | Yes | Yes |
| Insulin processing or insulin secretion | Yes | Yes |
| Insulin resistance | Yes | Yes |
| NASH | Yes | No |
| Height | Yes | Yes |
| BMI | Yes | Yes |
| Adiposity and fat distribution | Yes | Yes |
| Fat mass | Yes | Yes |
| Metabolic syndrome | Yes | Yes |
| Adiponectin | Yes | Yes |
| Dyslipidemia | Yes | Yes |
| Blood pressure | Yes | Yes |
| CAD (7, 8) | Yes | Yes |
| CVD (7, 8) | Yes | Yes |
A number in parentheses describes the reference where the criteria for different disease endpoints has been defined.
Laboratory studies include the following measurements obtained after a minimum of 10 h fasting: lipids, lipoproteins, apolipoproteins, adiponectin, bilirubin, alanine aminotransferase (ALT), bile acids, high sensitivity C-reactive protein, interleukin 1 receptor antagonist (IL-1RA), interleukin 1β, HbA1c, mass spectrometry metabolomics (Metabolon, Durham, NC), and proton NMR measurements (lipids and lipoproteins, amino acids, fatty acids of different lengths, and other low molecular weight metabolites). Additionally, an oral glucose tolerance test (OGTT) is performed to evaluate glucose tolerance (samples for glucose, insulin, proinsulin, and free fatty acids measured at 0, 30, and 120 minutes) (5). We also obtained subcutaneous adipose tissue samples from 1,410 participants. We assessed insulin sensitivity and insulin secretion using markers that we validated in a separate sample of 287 nondiabetic Finnish individuals from the region of Kuopio. We validated our insulin secretion markers against measures from the intravenous glucose tolerance test, and insulin sensitivity markers against the gold standard method to evaluate insulin sensitivity, the euglycemic-hyperinsulinemic clamp (6).
Follow-up study
The protocol of 5 year follow-up study is identical to that of the cross-sectional study. To date, we have reexamined 6,496 participants (participation rate 64%) and identified 693 new cases of T2D (a total of 2,106 incident and prevalent cases). The follow-up study will be finished in 2017. Diagnoses of myocardial infarction, stroke, and peripheral vascular disease (amputations) are verified from medical records against internationally accepted criteria for these diseases (7, 8). We also collect data from medical records on coronary angiograms, balloon angioplasty, by-pass coronary artery surgery, heart failure, and diabetic microvascular (retinopathy, nephropathy) and macrovascular complications. Participants signed a consent that allows us to use several Finnish registries: hospital discharge registry (includes all diagnoses of hospital admission), drug reimbursement and prescription registry (includes information on drug therapy for major chronic diseases, including diabetes, CAD, hypercholesterolemia, etc.), cancer registry, and mortality registry. Therefore, the coverage of the follow-up data of the participants is nearly 100% with respect to diagnoses of different diseases.
Genotyping, mRNA expression, and RNA sequencing
OmniExpress (Illumina) genotyping, exome chip (Illumina) genotyping genotyping, and exome sequencing have been performed for essentially all METSIM participants (9), and >2,000 METSIM participants have been whole-genome sequenced. Subcutaneous adipose tissue mRNA expression (Affymetrix) and RNA sequencing have been performed for 770 and 795 METSIM participants, respectively (10).
Ethics approval
The Ethics Committee of the University of Eastern Finland and Kuopio University Hospital approved the METSIM study, and this study was conducted in accordance with the Declaration of Helsinki. All study participants gave written informed consent.
Limitations of the METSIM study
The METSIM cohort includes only Finnish men. Therefore, the results reported in the METSIM participants need to be replicated in women and in other ethnic groups. The rationale to include only men was that in the age group from 45 to 73 years, the prevalence and incidence of T2D and CAD are higher in Finnish men than in Finnish women.
NON-GENETIC BIOMARKERS
T2D and hyperglycemia
A major interest in our studies based on the METSIM cohort has been to investigate the predictors for the development of hyperglycemia and the conversion to T2D. T2D is often preceded by a long period of prediabetes, characterized by insulin resistance and elevation of fasting (impaired fasting glucose) or 2 h glucose (impaired glucose tolerance) in an OGTT (11). Conversion to diabetes happens when the pancreas is no longer able to increase insulin secretion in a manner sufficient to compensate for insulin resistance in peripheral insulin-sensitive tissues. Characteristic findings of individuals with prediabetes and T2D are impaired insulin secretion and insulin resistance. Insulin resistance also plays a major role in the development of CVD (11).
We have identified several biomarkers as predictors for T2D and hyperglycemia (Table 2). We demonstrated that branch-chain amino acids (leucine, isoleucine) and aromatic amino acids (phenylalanine, tyrosine) were associated with increased risk of T2D and hyperglycemia, whereas glutamine was associated with decreased risk of T2D and hyperglycemia (12), confirming previous findings (13). Adjustment for insulin sensitivity abolished the significance of these associations, demonstrating for the first time in a longitudinal setting that insulin resistance is a major mechanism by which amino acids increase the risk of T2D (12). We also demonstrated for the first time that ketone bodies predict the conversion to T2D by impairing insulin secretion (14). Noncholesterol sterols (desmosterol, avenasterol) (15), glycerol, free fatty acids, and monounsaturated and saturated fatty acids predicted incident T2D (16). Palmitoleic acid increased and linoleic acid measured from plasma (17) or erythrocyte membranes (18) decreased the risk of T2D. High proinsulin level (19) and apolipoprotein/lipoprotein ratios (20) also predicted the conversion to T2D.
TABLE 2.
Association of different metabolites and laboratory measurements with T2D and hyperglycemia in the prospective METSIM study
| Marker | T2D | Hyperglycemia |
| Amino acids (11) | Alanine↑, leucine↑, isoleucine↑, tyrosine↑, phenylalanine↑, glutamine↓ | NA |
| Ketone bodies (13) | Acetoacetate↑, β-hydoxybutyrate↑ | Acetoacetate↑ |
| Non-cholesterol sterols (14) | Desmosterol↑, campesterol↓, avenasterol↑ | Desmosterol↑, campesterol↓, avenasterol↑ |
| Glycerol, FAs (proton NMR) (15) | Glycerol↑, free FAs↑, n-3 FAs↓, n-6 FAs↓, monounsaturated FAs↑, saturated n-7 and n-9 FAs↑ | Glycerol↑, free FFAs↑, n-6 FAs↓, monounsaturated FAs↑, saturated n-7 and n-9 FAs↑ |
| Plasma FAs in phospholipids, cholesteryl esters, and triglyceride fractions (16) | Dihomo-γ-linoleic acid↑ | Saturated FAs↑, palmitoleic acid↑, dihomo-γ-linolenic acid↑, linoleic acid↓ |
| Erythrocyte membrane FAs (17) | Palmitoleic acid↑, linoleic acid↓ | Palmitoleic acid↑, linoleic acid↓ |
| Proinsulin (18) | Fasting and glucose-stimulated proinsulin↑ | Fasting and glucose-stimulated proinsulin↑ |
| Lipoproteins and apolipoproteins (19) | ApoB/LDL cholesterol ratio↑, ApoA1/HDL cholesterol ratio↑ | ApoB/LDL cholesterol ratio↑, ApoA1/HDL cholesterol ratio↑ |
References are in parentheses. NA, not available.
Summary point.
Identification of biomarkers predicting the conversion to T2D is of great importance, especially for the prevention of diabetes. We, and others, have identified several nongenetic biomarkers (amino acids, ketone bodies, noncholesterol sterols, fatty acids) associated with the risk of T2D that improve the prediction for the conversion to diabetes beyond and above the classical risk factors for this disease (obesity, age, family history of diabetes, etc.). These findings need to be replicated in other ethnic groups and in women.
Nonalcoholic steatohepatitis
Non-alcoholic fatty liver disease is rapidly becoming the most common cause of liver disease, characterized by hepatic lipid accumulation contributing to insulin resistance, T2D, and hyperlipidemia. It can lead to nonalcoholic steatohepatitis (NASH) and liver cirrhosis, and ultimately to liver failure (21). Pathophysiological mechanisms leading to NASH have remained unclear, and noninvasive diagnosis of NASH is challenging. Therefore, finding simple laboratory markers for NASH is important. We hypothesized that increased IL-1RA serum levels in subjects with a high ALT level reflect increased inflammation in the liver. Indeed, we found that IL-1RA levels were associated with liver inflammation and ALT, independent of obesity, alcohol consumption, and insulin resistance in the METSIM study. Moreover, we found that liver expression of IL1RN, a gene encoding IL1-RA, correlated with liver steatosis and inflammation. Thus, increased IL1-RA levels in individuals with NASH could be directly linked to liver disease. Our findings suggest that IL-1RA is an important noninvasive inflammatory marker for NASH (22).
Dysregulation of the cholesterol synthesis pathway and accumulation of cholesterol in the liver play an important role in the pathogenesis of NASH. We measured serum and liver levels of three cholesterol precursor sterols (cholestenol, desmosterol, lathosterol) as serum surrogate markers of cholesterol synthesis in 110 obese individuals with detailed liver histology. We found that desmosterol in serum and liver associated with NASH. These results suggest that serum desmosterol is a marker of disturbed cholesterol metabolism in the liver and a potential biomarker for NASH (23).
Summary point.
NASH can be diagnosed accurately only by liver biopsy. Therefore, several laboratory measurements have been proposed as surrogate biomarkers, but no consensus about the best biomarker for NASH has been reached. IL-1RA and desmosterol as potential biomarkers for NASH need further validation studies.
GENETICS OF T2D, HYPERGLYCEMIA, INSULIN SECRETION, AND INSULIN RESISTANCE
T2D
GWASs.
The first replicated gene variant associated with T2D was the Pro12Ala amino acid substitution in the PPARG gene (24, 25). Interestingly, we demonstrated, using a Pro12Ala knock-in model, that, on chow diet, the Ala12Ala mice were leaner, had increased insulin sensitivity, and longer lifespans. High-fat feeding eliminated the beneficial effects of the Pro12Ala variant on adiposity and insulin sensitivity, demonstrating strong gene-environment interactions for this variant (26). Since 2007, GWASs and sequencing studies have provided new insights into the genetic basis of T2D, and currently about 90 loci for T2D and over 80 loci for glycemic traits have been published (27). The METSIM study has been an important European cohort in the identification and replication of several loci for T2D and for studies aiming to define causal mechanisms at the T2D susceptibility loci (28–34). However, these studies in aggregate explain only about 10–15% of the heritability of T2D.
Protective gene variants.
The METSIM study has been a part of three studies reporting protective gene variants for T2D. A study that combined sequence and genotype data on ∼150,000 individuals across five ancestry groups showed that two protein-truncating variants of SLC30A8 (p.Arg138* and p.Lys34Serfs*50) encoding zinc ZnT8 transport proteins are associated with T2D protection, suggesting that ZnT8 inhibition could be a therapeutic strategy in T2D prevention (35). Similarly, by whole-genome sequencing of 2,630 Icelanders, a low-frequency (1.5%) variant in intron 1 of CCND2 was found that reduced the risk of T2D by half (36). This finding was replicated in 29,956 individuals, including the participants of the METSIM cohort (37). A recent exome sequencing study reported that a low-frequency missense variant in the gene encoding glucagon-like peptide-1 receptor (GLP1R), the target of GLP1R agonists, was associated with decreased fasting glucose levels and the risk of T2D, consistent with GLP1R agonist therapies (38).
Low frequency variants.
The most recent study evaluated how much low-frequency variants (0.005 < minor allele frequency < 0.05) explain of the heritability of T2D. This study included whole-genome sequencing data from 2,657 European individuals with and without diabetes, and exome sequencing data from 12,940 individuals from five ancestry groups, including the METSIM cohort. This study did not support the notion that low-frequency variants explain a substantial portion of T2D heritability (39).
Hyperglycemia.
Several studies, including the METSIM cohort, have shown that common (40–43) and low-frequency/rare variants assayed by the exome array (44, 45) are associated with fasting or 2 h glucose concentrations (46, 47). Moreover, not all of 80 loci identified for hyperglycemia are risk loci for the conversion to T2D. The METSIM cohort has also been a part of the studies reporting common variants for glycated hemoglobin levels (48).
Summary point.
GWASs and exome and genome sequencing have identified close to 200 gene variants associated with T2D and hyperglycemia. Larger sample sizes of population studies will increase the number of gene variants even further, but genetic variants do not explain the epidemic of T2D observed today worldwide. Environmental and lifestyle factors and their interaction with genetic variants are the key to understanding the etiology and pathophysiology of T2D. Lifestyle factors also modify gene expression and methylation of DNA, which are likely to play an important role in the risk of T2D and other chronic diseases.
Insulin secretion and insulin resistance
Common variants.
Most of the common variants known to be associated with the risk of T2D affect insulin processing and insulin secretion and only a few affect insulin sensitivity (49–51). The METSIM cohort was important for this collaborative work given the large size of our cohort, as well as validated markers for insulin secretion and insulin sensitivity (52). Three studies, including the METSIM cohort, have searched for variants regulating insulin resistance (53–55). A common variant, rs11603334, was associated with T2D and proinsulin levels in the METSIM study. The T2D-risk allele increased transcriptional activity and was associated with increased ARAP1 expression in a small sample of human pancreatic islets, suggesting that increased ARAP1 expression may contribute to T2D susceptibility (56), although the T2D-risk allele at this locus is also associated with decreased STARD10 expression in larger studies of human pancreatic islets (57).
Low frequency and rare variants.
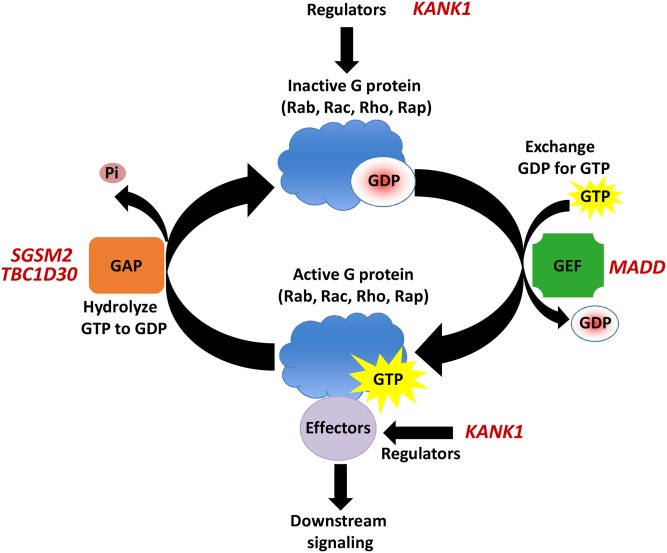
We were the first to report exome array results for insulin processing and secretion (9). We identified low-frequency coding variants associated with fasting proinsulin concentrations at the SGSM2 and MADD GWAS loci, and three new genes with low-frequency variants associated with fasting proinsulin or insulinogenic index (TBC1D30, KANK1, PAM) in 8,229 nondiabetic participants of the METSIM study. Of these genes, SGSM2, MADD, TBC1D30, and KANK1 regulate or function in G-protein signaling (Fig. 2). Our study demonstrated that, by exome array, it is possible to identify low-frequency variants that contribute to complex traits.
Fig. 2.
Roles of described genes in G protein signaling [small G protein signaling modulator 2 (SGSM2), MAP kinase activating death domain (MADD), TBC1 domain family member 30 (TBC1D30), and KN motif and ankyrin repeat domains 1 (KANK1)] have been shown to regulate or function in G protein signaling. GTP-binding proteins (G proteins) are characterized by their ability to bind and hydrolyze GTP and include members of Rab, Rac, Rho, Rap, and other families. G proteins are active when bound to GTP, but inactive when bound to GDP. Guanine nucleotide exchange factors (GEFs) catalyze the dissociation of GDP and the binding of GTP, thus promoting to active G protein state. When bound to GTP, G proteins remain active briefly and can activate G proteins by promoting GTP hydrolysis and a return to the inactive state. Adapted from (9).
Summary point.
Genetic studies have given important information about the etiology and pathophysiology of T2D. It is now clear that the most important pathophysiological mechanism leading to T2D is impaired insulin secretion, largely attributable to multiple gene variants regulating insulin secretion. By contrast, only a few gene variants regulating insulin sensitivity have been found, suggesting that insulin resistance is more likely to be an acquired trait (obesity, lack of exercise, unhealthy diet) than an inherited trait.
FINDING NEW FUNCTIONS FOR COMMON GENE VARIANTS ASSOCIATED WITH DIFFERENT CLINICAL AND LABORATORY MEASUREMENTS
In recent years, genetic studies have become much larger and technologies developed to identify low frequency and rare gene variants across the whole exome and genome have made it possible to identify causal variants for different diseases and traits. Understanding the biology behind genetic association is, however, the key for the understanding of the pathophysiology of different diseases. T2D is a good example of the problem of imprecise phenotypes because the pathophysiology of this disease involves several tissues, including pancreas, liver, skeletal muscle, fat, and brain. Phenotyping that goes beyond what is typically recorded in medical records or routine laboratory measurement is needed to dissect out mechanisms behind diseases of interest (58). Such “deep phenotyping” is possible in small clinical and family studies. The authors of a recent study performed detailed physiological (euglycemic clamp, insulin secretion) and in vitro characterization (insulin signaling in adipose tissue and skeletal muscle) in a limited number of carriers of the PTEN mutation (59). They were able to show that mutation carriers had decreased risk of T2D owing to enhanced insulin sensitivity (59).
Deep phenotyping is more challenging in large population-based studies for several reasons. First, deep phenotyping is expensive and time consuming. Second, large population-based cohorts have been collected several years ago when the significance of deep phenotyping in studies aiming to identify genes for complex diseases was not deemed practical. Third, deep phenotyping is successful only if study participants are willing to undertake time-consuming protocols (e.g., biopsies from tissues of interest, metabolic studies lasting for hours).
Deep phenotyping offers many advantages for genetic studies, and it could improve, for example, the classification of a disease of interest into the subtypes. In the majority of cases, T2D occurs especially in obese individuals who are typically insulin resistant, but this disease can also occur in normal weight individuals whose primary defect in glucose metabolism is impaired insulin secretion. Large sample sizes are needed to obtain statistically significant association of gene variants with T2D, a heterogenous disease defined by fasting and 2 h glucose concentrations. Deep phenotyping at the tissue, metabolite, and pathway levels related to the pathophysiology of T2D is likely to improve the understanding of the function of gene variants associated with T2D, and has the potential to reduce, to some extent, the sample sizes required to detect association. In the METSIM study, we have applied deep phenotyping, including proton NMR spectroscopy, to measure several metabolites for association studies with T2D risk gene variants. We found that the glucose-increasing alleles of rs780094 in glucokinase regulatory protein gene (GCKR) and rs174550 in fatty acid desaturase 1 (FADS1) were significantly associated with lipoprotein particles (5). The glucose-increasing allele of rs780094 of GCKR was additionally associated with decreased levels of the amino acids, alanine and isoleucine, and elevated levels of glutamine (12) and β-hydroxybutyrate (14). Thus, GCKR regulates not only glucose metabolism, but also lipoprotein and ketone body metabolism, and demonstrates that deep phenotyping offers a tool to identify new functions for the genes of interest.
Summary point
Phenotyping has been underdeveloped compared with genotyping in genetic studies. Recent studies applying proton NMR and mass spectrometry-based metabolomics, including the METSIM study, have shown the potential of deep phenotyping to identify new functions and pathways for the gene variants of interest. By contrast, proteomics, another essential tool for the understanding of the functions of cellular systems in human diseases, has not yet been widely applied because of many methodological challenges. New technological developments are urgently needed to advance the proteomics approach so that it may also be applied in genetic studies.
GENETICS OF ANTHROPOMETRIC TRAITS
Obesity and height
Obesity is heritable (60) and an important risk factor for T2D and CVD. The METSIM study has been a replication cohort for several GWASs aiming to identify common variants for height (61, 62) and BMI (63–66). The most recent meta-analysis included 339,224 individuals and identified 97 BMI-associated loci accounting for ∼2.7% of BMI variation. Genome-wide estimates suggested that common variants account for >20% of BMI variation (66).
Adiposity
The METSIM study has participated as a replication cohort for meta-analyses for adiposity and fat distribution (67–71). Interestingly, we found that the body-fat-decreasing allele near IRS1 was paradoxically associated with insulin resistance, dyslipidemia, and decreased adiponectin levels. We demonstrated, using the data from the METSIM study, that the variant near IRS1 is likely to have its primary effect on body fat percentage and that the association with decreased insulin sensitivity is partly mediated by changes in body fat percentage (69).
Summary point
Obesity and, especially, central obesity are important risk factors for T2D, CAD, hypertension, and certain cancers. Recent studies, including observations from the METSIM cohort, have shown that the gene variants associated with BMI are primarily expressed in the central nervous system, whereas gene variants associated with adiposity are primarily expressed in fat tissue (63–66). These findings from genetic studies have greatly contributed to better understanding of the etiology of obesity and adiposity.
GENETICS OF DYSLIPIDEMIA
Measurements
In the METSIM study, we have measured lipids (total cholesterol and triglycerides), lipoproteins (HDL, LDL, VLDL), and apolipoproteins (apoA1 and apoB). Additionally, we determined lipoprotein subclasses by proton NMR in native serum samples that provide quantitative molecular data on 14 lipoprotein subclasses and their lipid concentrations and composition (72).
GWASs
The METSIM study has been included in many GWAS meta-analyses identifying gene variants regulating LDL cholesterol, HDL cholesterol, and triglycerides. The first such study including the METSIM cohort reported 11 new variants for these traits, suggesting that the cumulative effect of several common variants contribute to polygenic dyslipidemia (73). Metabochip analysis, including trans-ethnic high-density genotyping, demonstrated the presence of allelic heterogeneity and the identification of population-specific variants (74) and the Global Lipids Genetics Consortium study reported the association of lipid and lipoprotein loci with metabolic and cardiovascular traits, including coronary heart disease and blood pressure (75). The most recent of these studies identified associations of lipid and lipoprotein levels with 62 new loci, bringing the total number of lipid-associated loci to 157 (75). All but one of the loci included protein-coding genes. Of the 62 new loci, 38 included genes whose role in the regulation of blood lipid levels has support from previous literature or databases, but for 24 loci such evidence has not been documented.
Functional studies
It has been challenging to convert GWAS data into mechanistic insights. The METSIM study data has been helpful in generating functional information for some GWAS variants, e.g., a variant rs7575840 in the APOB gene region associated with LDL cholesterol level. Adipose tissue RNA expression analysis indicated that rs7575840 alters the expression of APOB and a regional noncoding RNA (BU630349) (76). Another study examined the functional regulatory effects of 25 noncoding variants at the GALNT2 locus associated with HDL cholesterol in the METSIM cohort. This study demonstrated that at least two noncoding variants play key roles in influencing GALNT2 expression (77). We identified a gene locus for high serum triglycerides in Mexicans on chromosome 18q11.2 harboring a regulatory gene variant (rs17259126), and demonstrated that it disrupts normal hepatocyte nuclear factor 4α binding and decreases the expression of the TMEM241 gene in 795 adipose RNAs from the METSIM cohort. Our findings suggest that decreased transcript levels of TMEM241 contribute to increased triglyceride levels in Mexicans (78).
Summary point
Lipid-associated loci have been shown to associate strongly with CAD, T2D, BMI, and blood pressure (75). Therefore, genetic studies have greatly contributed not only to the understanding of the pathways that modify lipid and lipoprotein levels in humans, but also to the understanding of the links between lipids and lipoproteins and other chronic diseases. These studies may also facilitate the design of new therapies for cardiovascular and metabolic diseases. More mechanistic studies are needed, especially for those 24 new gene variants that have recently been associated with lipids and lipoproteins, having no support from previous literature or databases (75).
GENETICS OF BLOOD PRESSURE AND OTHER CARDIOVASCULAR RISK FACTORS
Blood pressure
Elevated blood pressure is a heritable trait and an important risk factor for CVD. The METSIM study participated in the first GWAS meta-analysis for elevated blood pressure including a total of 34,433 subjects of European ancestry in which eight common variants were found to be associated with systolic or diastolic blood pressure (79). A second GWAS meta-analysis including 200,000 individuals of European descent identified sixteen new loci (80). A genetic risk score based on 29 genome-wide significant variants was associated with hypertension, stroke, and CAD (80). Only three of the loci associated with blood pressure in Europeans were replicated in African Americans (81). A recent study including 192,000 individuals identified 30 new blood pressure or hypertension-associated genetic regions. This study also identified three rare missense variants in RBM47, COL21A1, and RRAS with larger effects than common variants (82).
Metabolic syndrome
We conducted a GWAS on the metabolic syndrome and its component traits in four Finnish cohorts consisting of 2,637 cases and 7,927 controls, both free of diabetes. A previously known lipid locus, the APOA1/C3/A4/A5 gene cluster region, was associated with the metabolic syndrome in all four study samples. However, we did not find evidence for a common genetic basis for clustering of metabolic syndrome traits (83). We have also identified a variant in SIRT3 that is suggestive of a genetic association with the metabolic syndrome (84).
Adiponectin
Circulating levels of adiponectin, a hormone produced predominantly by adipocytes, are highly heritable. The METSIM study has contributed to a GWAS meta-analysis of 39,883 individuals of European ancestry where eight novel loci were identified for adiponectin level. The genetic risk score, including several common risk variants for adiponectin level, showed an association with T2D, increased triglycerides, obesity markers, fasting insulin, and insulin resistance (85). However, a Mendelian randomization study did not support the conclusion that the association of adiponectin levels with insulin resistance and T2D is causal (86).
Summary point
Several new gene variants have recently been associated with blood pressure. More importantly, these studies have proved the causal link between elevated blood pressure and stroke and CAD, given the fact that the genetic risk score for blood pressure was associated directly with CVD endpoints. Our study has not found evidence for a common genetic basis of the metabolic syndrome (83), and similarly there is no causal link between adiponectin and insulin resistance and T2D.
GENETICS OF CAD
CAD is a major disease causing morbidity and mortality worldwide. Therefore, it is important to understand the genetic basis of CAD. The METSIM study participated in a GWAS meta-analysis of 63,746 CAD cases and 130,681 controls that identified new loci for CAD. Fifteen loci reached genome-wide significance and all 46 putative susceptibility loci explained approximately 10.6% of CAD heritability. The four most significant pathways mapping to these networks were linked to lipid metabolism and inflammation, underscoring the causal role of these pathways in the genetics of CAD (87). Importantly, this study showed that both LDL cholesterol and triglyceride levels, but not HDL cholesterol levels, were causally related to the risk of CAD (87).
The METSIM cohort was involved in another study investigating specifically the possibility that increased triglyceride levels are causally associated with CAD. Using 185 common variants mapped for plasma lipids, the study demonstrated that total triglyceride levels correlated with the magnitude of its effect on CAD risk, taking into account the effects on LDL cholesterol and/or HDL cholesterol levels. This study suggests that triglyceride-rich lipoproteins can causally influence the risk for CAD (88).
Summary point
Accumulating evidence from genetic studies demonstrates that the levels of LDL cholesterol and triglycerides are causally associated with the risk of CAD, whereas this causality is missing for HDL cholesterol level. These results have had important implications for the treatment of dyslipidemia.
TRANSCRIPTOMICS AND MicroRNA
Transcriptomics and RNA sequencing
The GWAS approach has been highly successful in the identification of common genetic variation contributing to normal and pathological traits. However, the mechanistic steps between genetic variation and different traits often remain poorly understood (89). Only a limited number of large-scale human population studies are available where extensive genotyping, phenotyping, and transcriptomics have been performed to obtain information on the functionality of the gene variants.
The first large study including samples from peripheral blood (N = 1,002) and subcutaneous fat (N = 673) was published from the Icelandic population. The study showed that more than 50% of all gene expression traits in adipose tissue strongly correlated with clinical traits related to obesity, compared with only about 10% in blood (90). Heritability was a highly significant contributor to variation in gene expression, and there was an approximately 50% overlap of genetic signals between the two tissues (90). Given the fact that adipose tissue is very important for metabolic diseases and CVDs, we collected subcutaneous abdominal adipose tissue biopsies from 1,410 randomly selected METSIM participants who did not have chronic diseases or chronic medication. Expression profiling using the Affymetrix U219 microarray expression quantitative trait locus (eQTL) mapping was performed for 770 METSIM individuals with both genotype and expression data available. We provided evidence for >100 loci for which eQTLs are coincident with GWAS loci, suggesting that these genes may be involved in human metabolic traits (91).
We also compared the results with those in 100 diverse commercially available inbred strains of mice, called the Hybrid Mouse Diversity Panel (HMDP), and found consistent associations between the traits and the expression of 25 genes in humans and mice (91). The HDMP is a collection of approximately 100 well-characterized inbred strains of mice that can be used to analyze the genetic and environmental factors underlying complex traits (92, 93). There are several benefits of the HMDP. It makes it possible to control environmental factors, guarantees the access for global molecular phenotyping, and makes it possible to integrate separate studies. All published data of the HMDP are available (92).
In another study, we sequenced 600 adipose tissue RNA samples from METSIM participants (Illumina TrueSeq RNA Prep kit and the Illumina Hiseq 2000 platform) and implemented a new method to identify genes whose expression is significantly associated with complex traits in individuals without directly measured expression levels. This method integrates gene expression measurements with summary association statistics from large-scale GWASs to identify genes whose cis-regulated expression is associated with complex traits. We identified 69 new genes significantly associated with obesity-related traits (BMI, lipids, and height). Many of these genes were also associated with relevant phenotypes in the HMDP (10).
MicroRNA
MicroRNAs (miRNAs) are small noncoding RNAs that regulate gene expression and determine how genetic variants affect different phenotypes. Information about genetic factors contributing to miRNA expression is limited; therefore, we examined variation of miRNA expression in adipose tissue in 200 METSIM participants. We reliably quantified 356 miRNA species expressed in human adipose tissue using genome-wide expression arrays and next-generation sequencing. Genetic variation of miRNA expression was substantially less than that of mRNAs. Twenty-four miRNAs were significantly associated with the traits of the metabolic syndrome (94).
Summary point
Adipose tissue gene expression studies have been important to reveal eQTLs that contribute to the understanding of the function of the genes. However, adipose tissue gene expression studies also have limitations. Gene expression in a single tissue may not be relevant for the mechanisms of some of the cardiometabolic traits, and the colocalization of an eQTL with a disease locus may be coincidental. Additionally, adipose tissue includes several cell types affecting gene expression profile, and adipose tissue gene expression is also influenced by gender, diet, and other environmental factors (91).
FUTURE PLANS
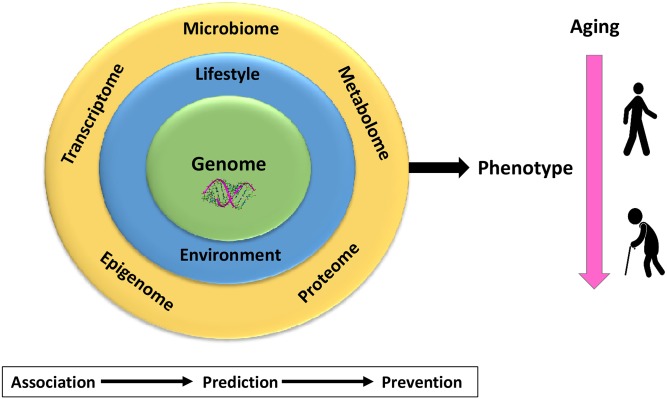
Genome sequencing
Extensive genotyping and exome sequencing have been performed for essentially all participants of the METSIM study. Genome sequencing has been performed for >2,000 METSIM study participants with and without CVD events, and continues in 2017–2018. (Fig. 3)
Fig. 3.
A roadmap for the genome and phenome analyses in the METSIM study. The whole-genome sequencing and extensive phenotyping are ongoing. Phenotyping includes metabolomics, proteomics, epigenomics, adipose tissue transciptomics, and microbiome analysis. The interaction between the genome and phenome will be extensively investigated as well as the interaction between the genome and lifestyle/environmental factors and aging. Prediction models will be developed using the Mendelian randomization approach by using genetic variants to estimate the causal contribution of a given risk marker to the risk of a given disease (especially T2D and CAD).
Prediction models
The METSIM follow-up study and registry follow-up allow continuous monitoring of the incidence of several diseases, including T2D and CAD, that makes it possible to develop genetic and nongenetic prediction models for T2D, cardiovascular complications, and cardiovascular risk factors. However, observational studies do not prove causality of a given risk factor. Therefore, we will apply Mendelian randomization methods by using genetic variants to estimate the causal contribution of a given risk marker to the risk of a given disease (95). This information is crucial for the planning of prevention of complex diseases, including T2D and CVD.
Omics
Recent major advances in omics technologies (metabolomics, proteomics, etc.) have enabled high-throughput analysis of a variety of molecular processes. We will continue our deep phenotyping by applying these technologies, as well as epigenomics and microbiota studies aiming to associate these phenotypes with common, low frequency, and rare gene variants. Integrative analyses across multiple omics platforms are important as we investigate the etiologies of complex diseases.
Metabolome and proteome
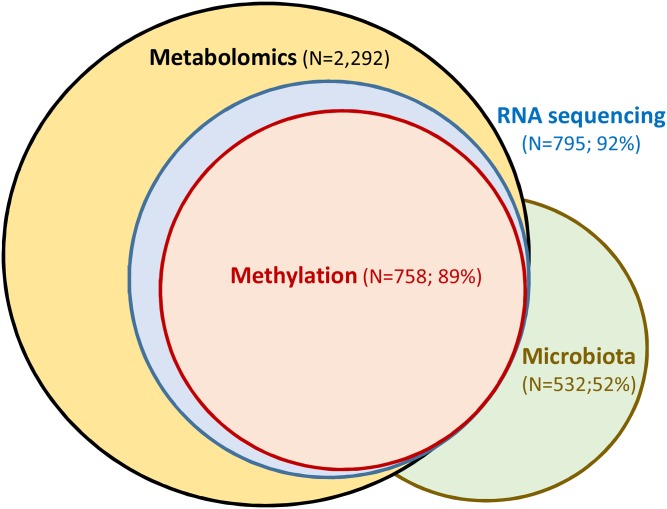
Proton NMR and mass spectrometry allow high throughput analysis of multiple metabolites. In the METSIM study, we have proton NMR data for >10,000 participants and mass spectrometry-based metabolomics results currently for 2,292 participants. These data make it possible to correlate metabolite concentrations with clinical traits and adipose tissue gene expression data, and to identify gene variants that regulate metabolite concentrations. Proteomics allows studies of the entire set of proteins, the proteome, by mass spectrometry and other methods. Recent advances in mass spectrometry and computational analysis of the results have made this technology an attractive method to characterize the proteome (96). As shown in Fig. 4, we have RNA sequencing (N = 795) and adipose tissue methylation data (N = 758) especially from those METSIM study participants who also have mass spectrometry-based metabolomics data (N = 2,292). Additionally, we have so far collected stool samples from 532 METSIM participants for gut microbiota sequencing and about 50% of them also have metabolomics data.
Fig. 4.
The number of the METSIM participants having data on adipose tissue RNA sequencing (N = 795), adipose tissue methylation (N = 758), and gut microbiota (N = 532) in relation to mass spectrometry-based metabolomics (N = 2,292). About 90% of participants who have RNA sequencing and methylation results and about 50% of participants who have microbiota analyses also have metabolomics data.
Microbiome
Diet plays an important role in obesity, metabolic syndrome, and other metabolic diseases, especially in T2D. Recently, the gut microbiota has become a focus of interest because it is at the intersection of diet and metabolic health. Both animal models and human studies have accumulated data to show that the gut microbiota mediates the effects of the diet on the host metabolic status (97). In 2017–2018, we aim to collect additional stool samples from about 500 METSIM participants for microbiota studies.
Epigenetics
Methylation of DNA cytosine bases plays an important role in the regulation of gene expression. DNA methylation is variable among human populations, in part heritable, and controlled by genes both in cis and in trans (98). To examine the methylation pattern in the METSIM cohort, we have constructed reduced representation bisulfite sequencing libraries from adipose tissue biopsies. The sequences obtained from bisulfite sequencing libraries are enriched in genes and CpG islands, and cover approximately two million CpGs out of the 30 million CpGs in the human genome. The Illumina HiSeq platform is used for sequencing of the libraries. We aim to sequence about 800 adipose tissue DNA samples in 2017–2018.
CONCLUSIONS
The METSIM study provides a resource for nongenetic and genetic studies of metabolic diseases and CVDs. It is a randomly selected large population-based sample of men (N = 10,197) from Eastern Finland having unique genetic background and extensive genetic data (OmniExpress chip, exome chip, and exome sequence in nearly all, genome sequence in many). We have performed deep phenotyping of this cohort and are currently expanding the phenotype further to include data on microbiota, RNA sequencing, adipose tissue methylation, and mass spectrometry for metabolites and proteins. Our findings, so far, suggest that we are able to identify new nongenetic biomarkers for T2D, insulin secretion, and insulin sensitivity. Additionally we have been able to find new low-frequency and rare variants in the METSIM study alone and in collaboration with others, and we have reported their association with proinsulin levels and insulin secretion. The METSIM study data have been useful in several meta-analyses to identify new common variants for T2D, hyperglycemia, anthropometric traits, and cardiovascular risk factors.
The METSIM study is a part of the Accelerated Medicines Partnership Type 2 Diabetes (AMP T2D) project supported by the Foundation for the National Institutes of Health (99). This project is designed to create a knowledge portal by building a database of DNA sequence, functional genomic and epigenomic, and clinical data from T2D studies with cardiac and renal complications. A broad range of genetic and phenotype data from the METSIM study will soon be available from the database of genotypes and phenotypes (dbGaP). The generation of a phenotype and genotype resource in the METSIM study allows us and other investigators to proceed toward a “systems genetics” approach (100), which includes statistical methods to quantitate and integrate intermediate phenotypes, such as transcript, protein, or metabolite levels to provide a global view of the molecular architecture of complex traits.
Footnotes
Abbreviations:
- ALT
- alanine aminotransferase
- CAD
- coronary artery disease
- eQTL
- expression quantitative trait locus
- GCKR
- glucokinase regulatory protein gene
- GLP1R
- glucagon-like peptide-1 receptor
- GWAS
- genome-wide association study
- HMDP
- Hybrid Mouse Diversity Panel
- IL-1RA
- interleukin 1 receptor antagonist
- METSIM
- Metabolic Syndrome in Men
- miRNA
- microRNA
- NASH
- nonalcoholic steatohepatitis
- OGTT
- oral glucose tolerance test
The studies included in this work were funded by grants from the National Institutes of Health, the Academy of Finland, the University of Eastern Finland, the Juselius Foundation, the Finnish Diabetes Research Foundation, and the Finnish Foundation for Cardiovascular Research. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
REFERENCES
- 1.Lim E. T., Wurtz P., Havulinna A. S., Palta P., Tukiainen T., Rehnstrom K., Esko T., Magi R., Inouye M., Lappalainen T., et al. 2014. Distribution and medical impact of loss-of-function variants in the Finnish founder population. PLoS Genet. 10: e1004494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lek M., Karczewski K. J., Minikel E. V., Samocha K. E., Banks E., Fennell T., O’Donnell-Luria A. H., Ware J. S., Hill A. J., Cummings B. B., et al. 2016. Analysis of protein-coding genetic variation in 60,706 humans. Nature. 536: 285–291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Stancáková A., Javorský M., Kuulasmaa T., Haffner S. M., Kuusisto J., and Laakso M.. 2009. Changes in insulin sensitivity and insulin release in relation to glycemia and glucose tolerance in 6,414 Finnish men. Diabetes. 58: 1212–1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lindström J., and Tuomilehto J.. 2003. The diabetes risk score: a practical tool to predict type 2 diabetes risk. Diabetes Care. 26: 725–731. [DOI] [PubMed] [Google Scholar]
- 5.Stančáková A., Paananen J., Soininen P., Kangas A. J., Bonnycastle L. L., Morken M. A., Collins F. S., Jackson A. U., Boehnke M. L., Kuusisto J., et al. 2011. Effects of 34 risk loci for type 2 diabetes or hyperglycemia on lipoprotein subclasses and their composition in 6,580 nondiabetic Finnish men. Diabetes. 60: 1608–1616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Laakso M., Zilinskaite J., Hansen T., Boesgaard T. W., Vanttinen M., Stancakova A., Jansson P. A., Pellme F., Holst J. J., Kuulasmaa T., et al. EUGENE2 Consortium . 2008. Insulin sensitivity, insulin release and glucagon-like peptide-1 levels in persons with impaired fasting glucose and/or impaired glucose tolerance in the EUGENE2 study. Diabetologia. 51: 502–511. [DOI] [PubMed] [Google Scholar]
- 7.Thygesen K., Alpert J. S., Jaffe A. S., Simoons M. L., Chaitman B. R., and White H. D.; Writing Group on behalf of the Joint ESC/ACCF/AHA/WHF Task Force for the Universal Definition of Myocardial Infarction . 2012. Third universal definition of myocardial infarction. Glob. Heart. 7: 275–295. [DOI] [PubMed] [Google Scholar]
- 8.European Stroke Initiative Executive Committee; EUSI Writing Committee, Olsen T. S., Langhorne P., Diener H. C., Hennerici M., Ferro J., Sivenius J., Wahlgren N. G., and Bath P.. 2003. European Stroke Initiative recommendations for stroke management-update 2003. Cerebrovasc. Dis. 16: 311–337. [DOI] [PubMed] [Google Scholar]
- 9.Huyghe J. R., Jackson A. U., Fogarty M. P., Buchkovich M. L., Stancakova A., Stringham H. M., Sim X., Yang L., Fuchsberger C., Cederberg H., et al. 2013. Exome array analysis identifies new loci and low-frequency variants influencing insulin processing and secretion. Nat. Genet. 45: 197–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gusev A., Ko A., Shi H., Bhatia G., Chung W., Penninx B. W., Jansen R., de Geus E. J., Boomsma D. I., Wright F. A., et al. 2016. Integrative approaches for large-scale transcriptome-wide association studies. Nat. Genet. 48: 245–252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Laakso M., and Kuusisto J.. 2014. Insulin resistance and hyperglycaemia in cardiovascular disease development. Nat. Rev. Endocrinol. 10: 293–302. [DOI] [PubMed] [Google Scholar]
- 12.Stancáková A., Civelek M., Saleem N. K., Soininen P., Kangas A. J., Cederberg H., Paananen J., Pihlajamaki J., Bonnycastle L. L., Morken M. A., et al. 2012. Hyperglycemia and a common variant of GCKR are associated with the levels of eight amino acids in 9,369 Finnish men. Diabetes. 61: 1895–1902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang T. J., Larson M. G., Vasan R. S., Cheng S., Rhee E. P., McCabe E., Lewis G. D., Fox C. S., Jacques P. F., Fernandez C., et al. 2011. Metabolite profiles and the risk of developing diabetes. Nat. Med. 17: 448–453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mahendran Y., Vangipurapu J., Cederberg H., Stancakova A., Pihlajamaki J., Soininen P., Kangas A. J., Paananen J., Civelek M., Saleem N. K., et al. 2013. Association of ketone body levels with hyperglycemia and type 2 diabetes in 9,398 Finnish men. Diabetes. 62: 3618–3626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cederberg H., Gylling H., Miettinen T. A., Paananen J., Vangipurapu J., Pihlajamaki J., Kuulasmaa T., Stancakova A., Smith U., Kuusisto J., et al. 2013. Non-cholesterol sterol levels predict hyperglycemia and conversion to type 2 diabetes in Finnish men. PLoS One. 8: e67406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mahendran Y., Cederberg H., Vangipurapu J., Kangas A. J., Soininen P., Kuusisto J., Uusitupa M., Ala-Korpela M., and Laakso M.. 2013. Glycerol and fatty acids in serum predict the development of hyperglycemia and type 2 diabetes in Finnish men. Diabetes Care. 36: 3732–3738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lankinen M. A., Stancakova A., Uusitupa M., Agren J., Pihlajamaki J., Kuusisto J., Schwab U., and Laakso M.. 2015. Plasma fatty acids as predictors of glycaemia and type 2 diabetes. Diabetologia. 58: 2533–2544. [DOI] [PubMed] [Google Scholar]
- 18.Mahendran Y., Agren J., Uusitupa M., Cederberg H., Vangipurapu J., Stancakova A., Schwab U., Kuusisto J., and Laakso M.. 2014. Association of erythrocyte membrane fatty acids with changes in glycemia and risk of type 2 diabetes. Am. J. Clin. Nutr. 99: 79–85. [DOI] [PubMed] [Google Scholar]
- 19.Vangipurapu J., Stancakova A., Kuulasmaa T., Kuusisto J., and Laakso M.. 2015. Both fasting and glucose-stimulated proinsulin levels predict hyperglycemia and incident type 2 diabetes: a population-based study of 9,396 Finnish men. PLoS One. 10: e0124028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fizelova M., Miilunpohja M., Kangas A. J., Soininen P., Kuusisto J., Ala-Korpela M., Laakso M., and Stancakova A.. 2015. Associations of multiple lipoprotein and apolipoprotein measures with worsening of glycemia and incident type 2 diabetes in 6607 non-diabetic Finnish men. Atherosclerosis. 240: 272–277. [DOI] [PubMed] [Google Scholar]
- 21.Lallukka S., and Yki-Jarvinen H.. 2016. Non-alcoholic fatty liver disease and risk of type 2 diabetes. Best Pract. Res. Clin. Endocrinol. Metab. 30: 385–395. [DOI] [PubMed] [Google Scholar]
- 22.Pihlajamäki J., Kuulasmaa T., Kaminska D., Simonen M., Karja V., Gronlund S., Kakela P., Paakkonen M., Kainulainen S., Punnonen K., et al. 2012. Serum interleukin 1 receptor antagonist as an independent marker of non-alcoholic steatohepatitis in humans. J. Hepatol. 56: 663–670. [DOI] [PubMed] [Google Scholar]
- 23.Simonen M., Mannisto V., Leppanen J., Kaminska D., Karja V., Venesmaa S., Kakela P., Kuusisto J., Gylling H., Laakso M., et al. 2013. Desmosterol in human nonalcoholic steatohepatitis. Hepatology. 58: 976–982. [DOI] [PubMed] [Google Scholar]
- 24.Deeb S. S., Fajas L., Nemoto M., Pihlajamaki J., Mykkanen L., Kuusisto J., Laakso M., Fujimoto W., and Auwerx J.. 1998. A Pro12Ala substitution in PPARgamma2 associated with decreased receptor activity, lower body mass index and improved insulin sensitivity. Nat. Genet. 20: 284–287. [DOI] [PubMed] [Google Scholar]
- 25.Altshuler D., Hirschhorn J. N., Klannemark M., Lindgren C. M., Vohl M. C., Nemesh J., Lane C. R., Schaffner S. F., Bolk S., Brewer C., et al. 2000. The common PPARgamma Pro12Ala polymorphism is associated with decreased risk of type 2 diabetes. Nat. Genet. 26: 76–80. [DOI] [PubMed] [Google Scholar]
- 26.Heikkinen S., Argmann C., Feige J. N., Koutnikova H., Champy M. F., Dali-Youcef N., Schadt E. E., Laakso M., and Auwerx J.. 2009. The Pro12Ala PPARgamma2 variant determines metabolism at the gene-environment interface. Cell Metab. 9: 88–98. [DOI] [PubMed] [Google Scholar]
- 27.Mohlke K. L., and Boehnke M.. 2015. Recent advances in understanding the genetic architecture of type 2 diabetes. Hum. Mol. Genet. 24: R85–R92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zeggini E., Scott L. J., Saxena R., Voight B. F., Marchini J. L., Hu T., de Bakker P. I., Abecasis G. R., Almgren P., Andersen G., et al. 2008. Meta-analysis of genome-wide association data and large-scale replication identifies additional susceptibility loci for type 2 diabetes. Nat. Genet. 40: 638–645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lyssenko V., Nagorny C. L., Erdos M. R., Wierup N., Jonsson A., Spegel P., Bugliani M., Saxena R., Fex M., Pulizzi N., et al. 2009. Common variant in MTNR1B associated with increased risk of type 2 diabetes and impaired early insulin secretion. Nat. Genet. 41: 82–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Voight B. F., Scott L. J., Steinthorsdottir V., Morris A. P., Dina C., Welch R. P., Zeggini E., Huth C., Aulchenko Y. S., Thorleifsson G., et al. 2010. Twelve type 2 diabetes susceptibility loci identified through large-scale association analysis. Nat. Genet. 42: 579–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Morris A. P., Voight B. F., Teslovich T. M., Ferreira T., Segre A. V., Steinthorsdottir V., Strawbridge R. J., Khan H., Grallert H., Mahajan A., et al. 2012. Large-scale association analysis provides insights into the genetic architecture and pathophysiology of type 2 diabetes. Nat. Genet. 44: 981–990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.DIAbetes Genetics Replication And Meta-analysis (DIAGRAM) Consortium; Asian Genetic Epidemiology Network Type 2 Diabetes (AGEN-T2D) Consortium; South Asian Type 2 Diabetes (SAT2D) Consortium; Mexican American Type 2 Diabetes (MAT2D) Consortium; Type 2 Diabetes Genetic Exploration by Nex-generation sequencing in muylti-Ethnic Samples (T2D-GENES) Consortium, Mahajan A., Go M. J., Zhang W., Below J. E., Gaulton K. J., et al. 2014. Genome-wide trans-ancestry meta-analysis provides insight into the genetic architecture of type 2 diabetes susceptibility. Nat. Genet. 46: 234–244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Prokopenko I., Poon W., Magi R., Prasad B. R., Salehi S. A., Almgren P., Osmark P., Bouatia-Naji N., Wierup N., Fall T., et al. 2014. A central role for GRB10 in regulation of islet function in man. PLoS Genet. 10: e1004235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gaulton K. J., Ferreira T., Lee Y., Raimondo A., Magi R., Reschen M. E., Mahajan A., Locke A., Rayner N. W., Robertson N., et al. 2015. Genetic fine mapping and genomic annotation defines causal mechanisms at type 2 diabetes susceptibility loci. Nat. Genet. 47: 1415–1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Flannick J., Thorleifsson G., Beer N. L., Jacobs S. B., Grarup N., Burtt N. P., Mahajan A., Fuchsberger C., Atzmon G., Benediktsson R., et al. 2014. Loss-of-function mutations in SLC30A8 protect against type 2 diabetes. Nat. Genet. 46: 357–363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Steinthorsdottir V., Thorleifsson G., Sulem P., Helgason H., Grarup N., Sigurdsson A., Helgadottir H. T., Johannsdottir H., Magnusson O. T., Gudjonsson S. A., et al. 2014. Identification of low-frequency and rare sequence variants associated with elevated or reduced risk of type 2 diabetes. Nat. Genet. 46: 294–298. [DOI] [PubMed] [Google Scholar]
- 37.Yaghootkar H., Stancakova A., Freathy R. M., Vangipurapu J., Weedon M. N., Xie W., Wood A. R., Ferrannini E., Mari A., Ring S. M., et al. 2015. Association analysis of 29,956 individuals confirms that a low-frequency variant at CCND2 halves the risk of type 2 diabetes by enhancing insulin secretion. Diabetes. 64: 2279–2285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Scott R. A., Freitag D. F., Li L., Chu A. Y., Surendran P., Young R., Grarup N., Stancakova A., Chen Y., Varga T. V., et al. 2016. A genomic approach to therapeutic target validation identifies a glucose-lowering GLP1R variant protective for coronary heart disease. Sci. Transl. Med. 8: 341ra76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fuchsberger C., Flannick J., Teslovich T. M., Mahajan A., Agarwala V., Gaulton K. J., Ma C., Fontanillas P., Moutsianas L., McCarthy D. J., et al. 2016. The genetic architecture of type 2 diabetes. Nature. 536: 41–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chen W. M., Erdos M. R., Jackson A. U., Saxena R., Sanna S., Silver K. D., Timpson N. J., Hansen T., Orru M., Grazia Piras M., et al. 2008. Variations in the G6PC2/ABCB11 genomic region are associated with fasting glucose levels. J. Clin. Invest. 118: 2620–2628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Prokopenko I., Langenberg C., Florez J. C., Saxena R., Soranzo N., Thorleifsson G., Loos R. J., Manning A. K., Jackson A. U., Aulchenko Y., et al. 2009. Variants in MTNR1B influence fasting glucose levels. Nat. Genet. 41: 77–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Li X., Shu Y. H., Xiang A. H., Trigo E., Kuusisto J., Hartiala J., Swift A. J., Kawakubo M., Stringham H. M., Bonnycastle L. L., et al. 2009. Additive effects of genetic variation in GCK and G6PC2 on insulin secretion and fasting glucose. Diabetes. 58: 2946–2953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dupuis J., Langenberg C., Prokopenko I., Saxena R., Soranzo N., Jackson A. U., Wheeler E., Glazer N. L., Bouatia-Naji N., Gloyn A. L., et al. 2010. New genetic loci implicated in fasting glucose homeostasis and their impact on type 2 diabetes risk. Nat. Genet. 42: 105–116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wessel J., Chu A. Y., Willems S. M., Wang S., Yaghootkar H., Brody J. A., Dauriz M., Hivert M. F., Raghavan S., Lipovich L., et al. 2015. Low-frequency and rare exome chip variants associate with fasting glucose and type 2 diabetes susceptibility. Nat. Commun. 6: 5897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mahajan A., Sim X., Ng H. J., Manning A., Rivas M. A., Highland H. M., Locke A. E., Grarup N., Im H. K., Cingolani P., et al. 2015. Identification and functional characterization of G6PC2 coding variants influencing glycemic traits define an effector transcript at the G6PC2-ABCB11 locus. PLoS Genet. 11: e1004876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Saxena R., Hivert M. F., Langenberg C., Tanaka T., Pankow J. S., Vollenweider P., Lyssenko V., Bouatia-Naji N., Dupuis J., Jackson A. U., et al. 2010. Genetic variation in GIPR influences the glucose and insulin responses to an oral glucose challenge. Nat. Genet. 42: 142–148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Scott R. A., Chu A. Y., Grarup N., Manning A. K., Hivert M. F., Shungin D., Tonjes A., Yesupriya A., Barnes D., Bouatia-Naji N., et al. 2012. No interactions between previously associated 2-hour glucose gene variants and physical activity or BMI on 2-hour glucose levels. Diabetes. 61: 1291–1296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Soranzo N., Sanna S., Wheeler E., Gieger C., Radke D., Dupuis J., Bouatia-Naji N., Langenberg C., Prokopenko I., Stolerman E., et al. 2010. Common variants at 10 genomic loci influence hemoglobin A(1)(C) levels via glycemic and nonglycemic pathways. Diabetes. 59: 3229–3239. [Erratum. 2011. Diabetes 60: 1050–1051.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ingelsson E., Langenberg C., Hivert M. F., Prokopenko I., Lyssenko V., Dupuis J., Magi R., Sharp S., Jackson A. U., Assimes T. L., et al. 2010. Detailed physiologic characterization reveals diverse mechanisms for novel genetic Loci regulating glucose and insulin metabolism in humans. Diabetes. 59: 1266–1275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Koeck T., Olsson A. H., Nitert M. D., Sharoyko V. V., Ladenvall C., Kotova O., Reiling E., Ronn T., Parikh H., Taneera J., et al. 2011. A common variant in TFB1M is associated with reduced insulin secretion and increased future risk of type 2 diabetes. Cell Metab. 13: 80–91. [DOI] [PubMed] [Google Scholar]
- 51.Dimas A. S., Lagou V., Barker A., Knowles J. W., Magi R., Hivert M. F., Benazzo A., Rybin D., Jackson A. U., Stringham H. M., et al. 2014. Impact of type 2 diabetes susceptibility variants on quantitative glycemic traits reveals mechanistic heterogeneity. Diabetes. 63: 2158–2171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Stancáková A., Kuulasmaa T., Paananen J., Jackson A. U., Bonnycastle L. L., Collins F. S., Boehnke M., Kuusisto J., and Laakso M.. 2009. Association of 18 confirmed susceptibility loci for type 2 diabetes with indices of insulin release, proinsulin conversion, and insulin sensitivity in 5,327 nondiabetic Finnish men. Diabetes. 58: 2129–2136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Manning A. K., Hivert M. F., Scott R. A., Grimsby J. L., Bouatia-Naji N., Chen H., Rybin D., Liu C. T., Bielak L. F., Prokopenko I., et al. 2012. A genome-wide approach accounting for body mass index identifies genetic variants influencing fasting glycemic traits and insulin resistance. Nat. Genet. 44: 659–669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Scott R. A., Lagou V., Welch R. P., Wheeler E., Montasser M. E., Luan J., Magi R., Strawbridge R. J., Rehnberg E., Gustafsson S., et al. 2012. Large-scale association analyses identify new loci influencing glycemic traits and provide insight into the underlying biological pathways. Nat. Genet. 44: 991–1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Walford G. A., Gustafsson S., Rybin D., Stancakova A., Chen H., Liu C. T., Hong J., Jensen R. A., Rice K., Morris A. P., et al. 2016. Genome-wide association study of the modified Stumvoll Insulin Sensitivity Index identifies BCL2 and FAM19A2 as novel insulin sensitivity loci. Diabetes. 65: 3200–3211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kulzer J. R., Stitzel M. L., Morken M. A., Huyghe J. R., Fuchsberger C., Kuusisto J., Laakso M., Boehnke M., Collins F. S., and Mohlke K. L.. 2014. A common functional regulatory variant at a type 2 diabetes locus upregulates ARAP1 expression in the pancreatic beta cell. Am. J. Hum. Genet. 94: 186–197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.van de Bunt M., Manning Fox J. E., Dai X., Barrett A., Grey C., Li L., Bennett A. J., Johnson P. R., Rajotte R. V., Gaulton K. J., et al. 2015. Transcript expression data from human islets links regulatory signals from genome-wide association studies for type 2 diabetes and glycemic traits to their downstream effectors. PLoS Genet. 11: e1005694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Delude C. M. 2015. Deep phenotyping: the details of disease. Nature. 527: S14–S15. [DOI] [PubMed] [Google Scholar]
- 59.Pal A., Barber T. M., Van de Bunt M., Rudge S. A., Zhang Q., Lachlan K. L., Cooper N. S., Linden H., Levy J. C., Wakelam M. J., et al. 2012. PTEN mutations as a cause of constitutive insulin sensitivity and obesity. N. Engl. J. Med. 367: 1002–1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Farooqi I. S., and O’Rahilly S.. 2007. Genetic factors in human obesity. Obes. Rev. 8(Suppl 1): 37–40. [DOI] [PubMed] [Google Scholar]
- 61.Wood A. R., Esko T., Yang J., Vedantam S., Pers T. H., Gustafsson S., Chu A. Y., Estrada K., Luan J., Kutalik Z., et al. 2014. Defining the role of common variation in the genomic and biological architecture of adult human height. Nat. Genet. 46: 1173–1186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Joshi P. K., Esko T., Mattsson H., Eklund N., Gandin I., Nutile T., Jackson A. U., Schurmann C., Smith A. V., Zhang W., et al. 2015. Directional dominance on stature and cognition in diverse human populations. Nature. 523: 459–462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Willer C. J., Speliotes E. K., Loos R. J., Li S., Lindgren C. M., Heid I. M., Berndt S. I., Elliott A. L., Jackson A. U., Lamina C., et al. 2009. Six new loci associated with body mass index highlight a neuronal influence on body weight regulation. Nat. Genet. 41: 25–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Speliotes E. K., Willer C. J., Berndt S. I., Monda K. L., Thorleifsson G., Jackson A. U., Lango Allen H., Lindgren C. M., Luan J., Magi R., et al. 2010. Association analyses of 249,796 individuals reveal 18 new loci associated with body mass index. Nat. Genet. 42: 937–948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Berndt S. I., Gustafsson S., Magi R., Ganna A., Wheeler E., Feitosa M. F., Justice A. E., Monda K. L., Croteau-Chonka D. C., Day F. R., et al. 2013. Genome-wide meta-analysis identifies 11 new loci for anthropometric traits and provides insights into genetic architecture. Nat. Genet. 45: 501–512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Locke A. E., Kahali B., Berndt S. I., Justice A. E., Pers T. H., Day F. R., Powell C., Vedantam S., Buchkovich M. L., Yang J., et al. 2015. Genetic studies of body mass index yield new insights for obesity biology. Nature. 518: 197–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Lindgren C. M., Heid I. M., Randall J. C., Lamina C., Steinthorsdottir V., Qi L., Speliotes E. K., Thorleifsson G., Willer C. J., Herrera B. M., et al. 2009. Genome-wide association scan meta-analysis identifies three Loci influencing adiposity and fat distribution. PLoS Genet. 5: e1000508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Heid I. M., Jackson A. U., Randall J. C., Winkler T. W., Qi L., Steinthorsdottir V., Thorleifsson G., Zillikens M. C., Speliotes E. K., Magi R., et al. 2010. Meta-analysis identifies 13 new loci associated with waist-hip ratio and reveals sexual dimorphism in the genetic basis of fat distribution. Nat. Genet. 42: 949–960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kilpeläinen T. O., Zillikens M. C., Stancakova A., Finucane F. M., Ried J. S., Langenberg C., Zhang W., Beckmann J. S., Luan J., Vandenput L., et al. 2011. Genetic variation near IRS1 associates with reduced adiposity and an impaired metabolic profile. Nat. Genet. 43: 753–760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Shungin D., Winkler T. W., Croteau-Chonka D. C., Ferreira T., Locke A. E., Magi R., Strawbridge R. J., Pers T. H., Fischer K., Justice A. E., et al. 2015. New genetic loci link adipose and insulin biology to body fat distribution. Nature. 518: 187–196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Lu Y., Day F. R., Gustafsson S., Buchkovich M. L., Na J., Bataille V., Cousminer D. L., Dastani Z., Drong A. W., Esko T., et al. 2016. New loci for body fat percentage reveal link between adiposity and cardiometabolic disease risk. Nat. Commun. 7: 10495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Soininen P., Kangas A. J., Wurtz P., Suna T., and Ala-Korpela M.. 2015. Quantitative serum nuclear magnetic resonance metabolomics in cardiovascular epidemiology and genetics. Circ Cardiovasc Genet. 8: 192–206. [DOI] [PubMed] [Google Scholar]
- 73.Kathiresan S., Willer C. J., Peloso G. M., Demissie S., Musunuru K., Schadt E. E., Kaplan L., Bennett D., Li Y., Tanaka T., et al. 2009. Common variants at 30 loci contribute to polygenic dyslipidemia. Nat. Genet. 41: 56–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Wu Y., Waite L. L., Jackson A. U., Sheu W. H., Buyske S., Absher D., Arnett D. K., Boerwinkle E., Bonnycastle L. L., Carty C. L., et al. 2013. Trans-ethnic fine-mapping of lipid loci identifies population-specific signals and allelic heterogeneity that increases the trait variance explained. PLoS Genet. 9: e1003379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Global Lipids Genetics Consortium, Willer C. J., Schmidt E. M., Sengupta S., Peloso G. M., Gustafsson S., Kanoni S., Ganna A., Chen J., Buchkovich M. L., et al. 2013. Discovery and refinement of loci associated with lipid levels. Nat. Genet. 45: 1274–1283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Haas B. E., Weissglas-Volkov D., Aguilar-Salinas C. A., Nikkola E., Vergnes L., Cruz-Bautista I., Riba L., Stancakova A., Kuusisto J., Soininen P., et al. 2011. Evidence of how rs7575840 influences apolipoprotein B-containing lipid particles. Arterioscler. Thromb. Vasc. Biol. 31: 1201–1207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Roman T. S., Marvelle A. F., Fogarty M. P., Vadlamudi S., Gonzalez A. J., Buchkovich M. L., Huyghe J. R., Fuchsberger C., Jackson A. U., Wu Y., et al. 2015. Multiple hepatic regulatory variants at the GALNT2 GWAS locus associated with high-density lipoprotein cholesterol. Am. J. Hum. Genet. 97: 801–815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Rodríguez A., Gonzalez L., Ko A., Alvarez M., Miao Z., Bhagat Y., Nikkola E., Cruz-Bautista I., Arellano-Campos O., Munoz-Hernandez L. L., et al. 2016. Molecular characterization of the lipid genome-wide association study signal on chromosome 18q11.2 implicates HNF4A-mediated regulation of the TMEM241 gene. Arterioscler. Thromb. Vasc. Biol. 36: 1350–1355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Newton-Cheh C., Johnson T., Gateva V., Tobin M. D., Bochud M., Coin L., Najjar S. S., Zhao J. H., Heath S. C., Eyheramendy S., et al. 2009. Genome-wide association study identifies eight loci associated with blood pressure. Nat. Genet. 41: 666–676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.International Consortium for Blood Pressure Genome-Wide Association Studies, Ehret G. B., Munroe P. B., Rice K. M., Bochud M., Johnson A. D., Chasman D. I., Smith A. V., Tobin M. D., Verwoert G. C., et al. 2011. Genetic variants in novel pathways influence blood pressure and cardiovascular disease risk. Nature. 478: 103–109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Fox E. R., Young J. H., Li Y., Dreisbach A. W., Keating B. J., Musani S. K., Liu K., Morrison A. C., Ganesh S., Kutlar A., et al. 2011. Association of genetic variation with systolic and diastolic blood pressure among African Americans: the Candidate Gene Association Resource study. Hum. Mol. Genet. 20: 2273–2284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Surendran P., Drenos F., Young R., Warren H., Cook J. P., Manning A. K., Grarup N., Sim X., Barnes D. R., Witkowska K., et al. 2016. Trans-ancestry meta-analyses identify novel rare and common variants associated with blood pressure and hypertension. Nat. Genet. 48: 1151–1161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Kristiansson K., Perola M., Tikkanen E., Kettunen J., Surakka I., Havulinna A. S., Stancakova A., Barnes C., Widen E., Kajantie E., et al. 2012. Genome-wide screen for metabolic syndrome susceptibility Loci reveals strong lipid gene contribution but no evidence for common genetic basis for clustering of metabolic syndrome traits. Circ Cardiovasc Genet. 5: 242–249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Hirschey M. D., Shimazu T., Jing E., Grueter C. A., Collins A. M., Aouizerat B., Stancakova A., Goetzman E., Lam M. M., Schwer B., et al. 2011. SIRT3 deficiency and mitochondrial protein hyperacetylation accelerate the development of the metabolic syndrome. Mol. Cell. 44: 177–190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Dastani Z., Hivert M. F., Timpson N., Perry J. R., Yuan X., Scott R. A., Henneman P., Heid I. M., Kizer J. R., Lyytikainen L. P., et al. 2012. Novel loci for adiponectin levels and their influence on type 2 diabetes and metabolic traits: a multi-ethnic meta-analysis of 45,891 individuals. PLoS Genet. 8: e1002607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Yaghootkar H., Lamina C., Scott R. A., Dastani Z., Hivert M. F., Warren L. L., Stancakova A., Buxbaum S. G., Lyytikainen L. P., Henneman P., et al. 2013. Mendelian randomization studies do not support a causal role for reduced circulating adiponectin levels in insulin resistance and type 2 diabetes. Diabetes. 62: 3589–3598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.CARDIoGRAMplusC4D Consortium, P. Deloukas, S. Kanoni, C. Willenborg, M. Farrall, T. L. Assimes, J. R. Thompson, E. Ingelsson, D. Saleheen, J. Erdmann, et al. 2013. Large-scale association analysis identifies new risk loci for coronary artery disease. Nat. Genet. 45: 25–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Do R., Willer C. J., Schmidt E. M., Sengupta S., Gao C., Peloso G. M., Gustafsson S., Kanoni S., Ganna A., Chen J., et al. 2013. Common variants associated with plasma triglycerides and risk for coronary artery disease. Nat. Genet. 45: 1345–1352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Edwards S. L., Beesley J., French J. D., and Dunning A. M.. 2013. Beyond GWASs: illuminating the dark road from association to function. Am. J. Hum. Genet. 93: 779–797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Emilsson V., Thorleifsson G., Zhang B., Leonardson A. S., Zink F., Zhu J., Carlson S., Helgason A., Walters G. B., Gunnarsdottir S., et al. 2008. Genetics of gene expression and its effect on disease. Nature. 452: 423–428. [DOI] [PubMed] [Google Scholar]
- 91.Civelek M., Wu Y., Pan C., Raulerson C. K., Ko A., He A., Tilford C., Saleem N. K., Stančáková A., Scott L. J., et al. 2017. Genetic regulation of adipose gene expression and cardio-metabolic traits. Am. J. Hum. Genet. In press . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Lusis A. J., Seldin M. M., Allayee H., Bennett B. J., Civelek M., Davis R. C., Eskin E., Farber C. R., Hui S., Mehrabian M., et al. 2016. The Hybrid Mouse Diversity Panel: a resource for systems genetics analyses of metabolic and cardiovascular traits. J. Lipid Res. 57: 925–942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Rau C. D., Civelek M., Pan C., and Lusis A. J.. 2017. A suite of tools for biologists that improve accessibility and visualization of large systems genetics datasets: applications to the Hybrid Mouse Diversity Panel. Methods Mol. Biol. 1488: 153–188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Civelek M., Hagopian R., Pan C., Che N., Yang W. P., Kayne P. S., Saleem N. K., Cederberg H., Kuusisto J., Gargalovic P. S., et al. 2013. Genetic regulation of human adipose microRNA expression and its consequences for metabolic traits. Hum. Mol. Genet. 22: 3023–3037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Davey Smith G., and Hemani G.. 2014. Mendelian randomization: genetic anchors for causal inference in epidemiological studies. Hum. Mol. Genet. 23: R89–R98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Walther T. C., and Mann M.. 2010. Mass spectrometry-based proteomics in cell biology. J. Cell Biol. 190: 491–500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Sonnenburg J. L., and Backhed F.. 2016. Diet-microbiota interactions as moderators of human metabolism. Nature. 535: 56–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Bell J. T., Pai A. A., Pickrell J. K., Gaffney D. J., Pique-Regi R., Degner J. F., Gilad Y., and Pritchard J. K.. 2011. DNA methylation patterns associate with genetic and gene expression variation in HapMap cell lines. Genome Biol. 12: R10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.American Diabetes Association. Genetics portal for type 2 diabetes debuts. 2015. Accessed June 7, 2015, at http://www.diabetesdispatchextra.org/genetics-portal-for-type-2-diabetes-debuts.
- 100.Civelek M., and Lusis A. J.. 2014. Systems genetics approaches to understand complex traits. Nat. Rev. Genet. 15: 34–48. [DOI] [PMC free article] [PubMed] [Google Scholar]