Abstract
Chemoreception is essential for survival. Some chemicals signal the presence of nutrients or toxins, others the proximity of mating partners, competitors, or predators. Chemical signal transduction has therefore been studied in multiple organisms. In Drosophila species, a number of odorant receptor genes and various other types of chemoreceptors were found. Three main gene families encode for membrane receptors and one for globular proteins that shuttle compounds with different degrees of affinity and specificity towards receptors. By sequencing the genome of Drosophila nigrosparsa, a habitat specialist restricted to montane/alpine environment, and combining genomics and structural biology techniques, we characterised odorant, gustatory, ionotropic receptors and odorant binding proteins, annotating 189 loci and modelling the protein structure of two ionotropic receptors and one odorant binding protein. We hypothesise that the D. nigrosparsa genome experienced gene loss and various evolutionary pressures (diversifying positive selection, relaxation, and pseudogenisation), as well as structural modification in the geometry and electrostatic potential of the two ionotropic receptor binding sites. We discuss possible trajectories in chemosensory adaptation processes, possibly enhancing compound affinity and mediating the evolution of more specialized food, and a fine-tuned mechanism of adaptation.
Chemosensory systems allow animals to orient themselves in a sea of chemical compounds. Some of these chemicals signal the presence of nutrients or toxins, others the proximity of mating partners, competitors, or predators. Signal intensity can be crucial: The quantity of sugar in a food source reflects its nutrient value, just like bitter compounds may reflect its toxicity, and the same stimuli can be attractive at low but aversive at high concentrations1,2. Being able to identify and quantify chemicals is crucial to survival and adaptation1,2.
Chemical signal transduction has been well studied and is generally similar in multiple organisms across the tree of life. From nematodes to crustaceans, insects, and vertebrates, chemical compounds are detected through interactions with specific receptors on the dendrites of both olfactory sensory neurons and gustatory receptor neurons3. The activation of these receptors induces a messenger cascade leading to ion channel activation and receptor neuron depolarization4. So far, four large gene families were discovered to mediate chemical responses from the environment, and further kinds of chemoreceptors likely await discovery5. Three of these gene families encode for membrane receptors (odorant, gustatory, and ionotropic receptors), while a fourth encodes for odorant binding proteins, globular proteins secreted by sensory neuron accessory cells1.
The three-dimensional arrangement of the insect odorant receptor (OR) family is still under debate since no clear structural information is available yet. Seven putative transmembrane helices (7-TMHs) were predicted1,6. In insect ORs, the amino-terminus is located toward the cytoplasm, and a ligand-gated ion channel function was inferred, which implies a signal transduction mechanism independent from G proteins6,7. Therefore, the odour response spectrum, the termination kinetics, and the level of spontaneous activity are hypothesized to be directly dependent on the receptor itself4,8, mediating both excitatory and inhibitory responses in the same cell9. The functional unit of the OR family consists of homomeric and heteromeric structures6 that involve the interaction of single odorant receptors with a co-receptor protein, as shown for Or22a and Orco8,10,11.
Compared with ORs, gustatory receptors (GRs) are a more divergent gene family and are generally more related to sweet, bitter, umami, and salt concentration taste12,13. They are just weakly related to ORs, with which they share the putative seven helices. Knowledge about GRs’ three-dimensional structure and their mechanisms of signal transduction is very poor1,14.
The ionotropic receptors (IRs) are related to ionotropic glutamate receptors (iGluRs), which mediate the excitatory transmission and are key to synaptic plasticity and to learning and memory processes15. Most IRs have two conserved domains: An extracellular 1st half ligand-binding domain (S1) (Lig_chan-Glu_bd; Pfam: PF10613) at the amino-terminus followed by the ligand-gated ion channel domain (Lig_chan; Pfam: PF00060), which is composed of three transmembrane helices (TMHs), an ion channel pore (P), and the 2nd half of the ligand-binding domain (S2). A third domain, the amino terminal domain (ATD), present in all iGluRs, can be present, depending on the gene and species, and does not share a strong homology signal with the iGluR sub-family16. Unlike other receptors, IRs confer response to many organic acids and amines, some of which play important behavioural roles, such as the induction of male courtship by food-related odour17.
The fourth gene family, the odorant binding proteins (OBPs), is a quite divergent family. It encodes for small globular and soluble proteins, which bind hydrophilic odorant compounds with different degrees of affinity and specificity to shuttle them to the underlying receptors, in form monomers and/or homodimers18. OBPs appear to be involved in odorant recognition, binding compounds, and in Drosophila are generally subdivided in six sub-families based on structural features, functional information, and phylogenetic relationships: Classic, Minus-C, Plus-C, Dimer, ABPII, and CRLBP18,19.
Drosophila species are saprophagous insects that feed and breed on a wide range of fermenting plant materials including fruits, flowers, slime fluxes, decaying bark, leaves, and stems, and on fungi20,21. Because of this, Drosophila species provide an excellent system to study how changes in the chemosensory repertoire are associated with behaviour and ecology, both in terms of food preference and sexual behaviour. Because the composition of volatiles drastically changes during food ripening and fermentation, they can potentially provide different cues to different species. Among the distinctive ecological niches occupied by the multitude of Drosophila species, D. nigrosparsa is a habitat specialist, restricted to the European montane/alpine zone with a maximum density at about 2000 m above sea level, and most abundant at the timber line22. Its life history starts being revealed, and its preferred oviposition substrate are fungi23. Given the harsh conditions of the habitat, the fly likely evolved particular adaptations to inhabit the alpine ecosystem.
Here, as a first step to understand the genetic basis of the species’ ecological adaptation and life history, we analysed the genetic and the molecular architecture of the majority of genes related to chemosensation, aiming to identify signatures of evolutionary pressures (positive, purifying, and relaxed selection). For a wider view of the adaptation process for D. nigrosparsa, we combined genomics and structural biology. In doing so, after sequencing the D. nigrosparsa genome, and using transcriptomic data, we annotated the four main chemosensory gene families (CGFs), analysed their repertoire, and inferred structural changes related to the ligand binding domain of proteins under selection.
Results
CGF gene annotation
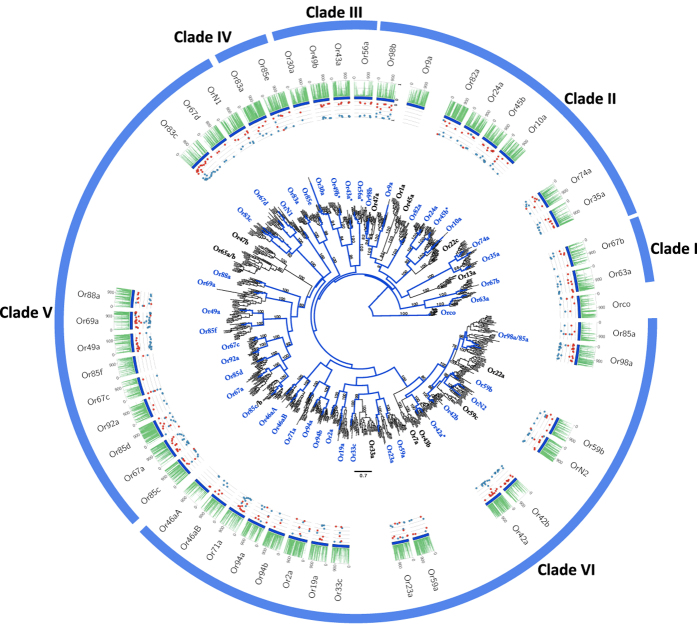
To characterize adaptation in the CGF gene repertoire in D. nigrosparsa, we sequenced its genome (SI Appendix 1) and annotated CGF genes by mapping previously annotated Drosophila CGF protein sequences against the de novo D. nigrosparsa genome assembly. The analysis for ORs resulted in 55 candidate OR loci (SI Appendix 1, Fig. 1, Table S1), grouped in 45 of the 58 clusters obtained from the phylogenetic analysis of all Drosophila OR loci. The number of OR clusters did not differ from those in other Drosophila subgenus species and S. flava (minimum 43 in D. grimshawi; maximum 47 in D. mojavensis). Here, we define two terms: in-paralogs and out-paralogs. In-paralogs are paralogs in a given lineage produced by duplications after the split, which gave rise to that lineage and therefore represent lineage-specific gene expansions. Out-paralogs are paralogs in a lineage that evolved by gene duplications before emergence of the lineage24. This distinction is key to identifying lineage-specific gene expansions and adaptations. In nine clusters, we recovered paralogous genes, in seven of them only out-paralogs (DnigOrN2, DnigOr42, DnigOr66d, DnigOr30a DnigOr98b, DnigOr10a, DnigOr74a), in one only in-paralogs (DnigOr9a), and in another cluster one out-paralog and two in-paralogs (DnigOr83c, Fig. 1). Protein domain predictors found the 7tm_6 Pfam domain in all loci, and a prediction from one to seven TMHs (Figure S2a). Because ORs bear 7-TMHs, for this study, we considered as putatively functional proteins those having: (i) a minimum of 6-TMHs, and ii) a Pfam domain coverage ≥70%25. A total of 33 loci were assigned as functional, while the remaining nine were classified as putative pseudogenes. We found that the distribution of TMHs predicted for the receptors in clade V, encompassing Or67d to Or85c (Fig. 1), was biased towards 6-TMHs instead of 7-TMHs. Namely, 89% of sequences were predicted with 6-TMHs, compared with 17% in the other clades. Looking at the domain sequence identities of this cluster, we observed a significantly lower value in clade V (average sequence identity of 22%) than in all other clades (average sequence identity of 25%; p-values < 0.002, one-tailed Wilcoxon rank-sum test; Figure S2b).
Figure 1. Phylogenetic relationships of the candidate D. nigrosparsa Odorant Receptors (ORs).
Subdivided into six clades, the phylogeny describes D. nigrosparsa loci with respect to 13 other Drosophila species. The maximum likelihood tree was rooted by the OR co-receptor orthologs. Bootstrap support ≥70 for branches is indicated. Branches coloured in blue correspond to the lineages leading to D. nigrosparsa loci. Asterisks after the OR name indicate genes under positive selection. The histogram represents highly conserved elements (PhastCons) of each alignment. The PhastCons index ranges between 0 and 1 and can be interpreted as the probability that each base is a conserved element, based on the assumptions of the model and the maximum-likelihood parameter estimates. The dot plots report the distribution of synonymous (α; blue) and non-synonymous (β; red) substitution rates over sites inferred by the MEME model, which ranges between 0 and 1 (posterior probability >0.95).
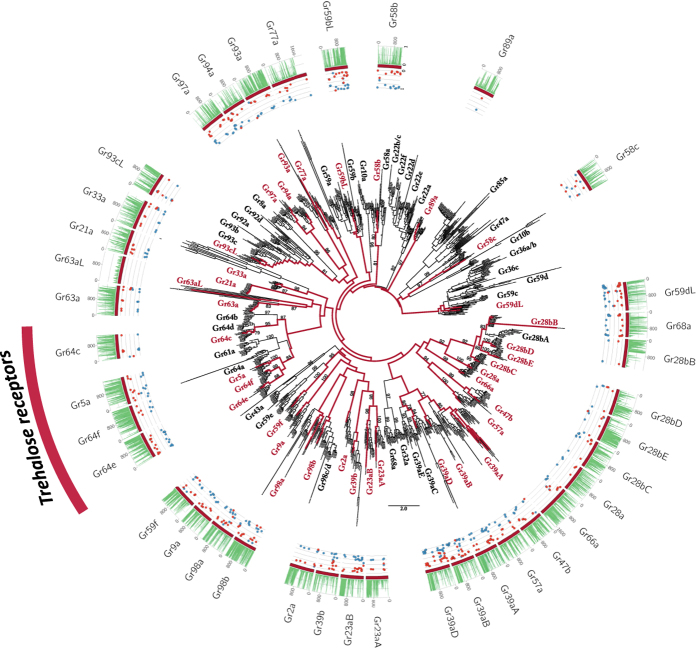
Forty-seven candidate loci for the GR gene family were annotated. All genes and transcripts were aligned to build the GR phylogenetic family tree. Of the 72 clusters, D. nigrosparsa GR loci grouped into 37, lacking between eight and 12 loci. Paralogs were found in five clusters, of which three contained just out-paralogs (DnigGr59dL, DnigGr93cL, and DnigGr98a), one only in-paralogs (DnigGr58b), and one three out- and two in-paralogs (SI Appendix 1, Fig. 2, Table S2). Protein domain predictors found the 7tm_7 Pfam domain in 43 loci and the Trehalose_recp domain in four, and the protein topology predictions found between four and eight TMHs (Figure S2c). Applying the same criteria as for ORs (see above); 31 loci were assigned as functional and six as putative pseudogenes.
Figure 2. Phylogenetic relationships of the candidate D. nigrosparsa Gustatory Receptors (GRs).
The phylogeny describes D. nigrosparsa loci with respect to 12 other Drosophila species. The sugar-related genes are highlighted as trehalose receptors. The maximum likelihood tree was midpoint rooted. Bootstrap support ≥70 for branches is indicated. Red branches correspond to the lineages leading to D. nigrosparsa loci. Asterisks after the GR name indicate genes under positive selection. Histogram and dot plots as in Fig. 1.
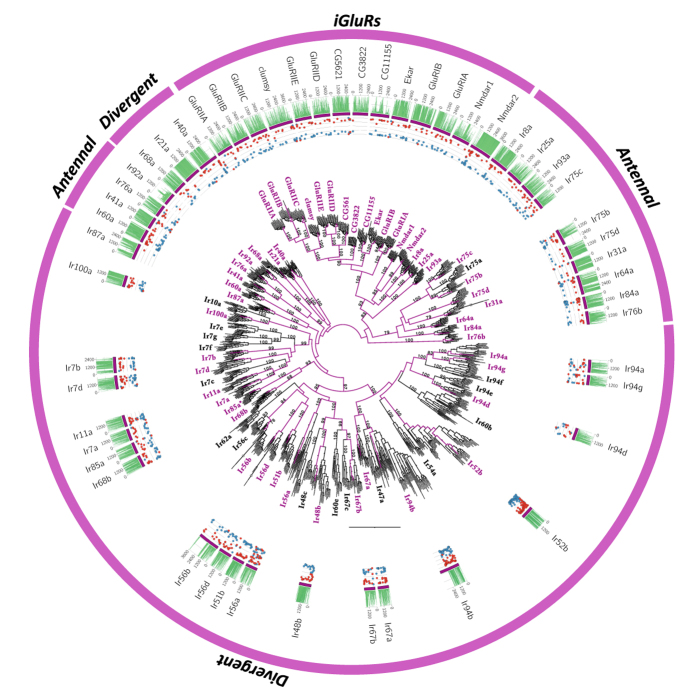
About ionotropic receptors, we annotated both odour-specific IRs and iGluRs and found a total of 54 loci, clustering in 53 of the 67 clusters (SI Appendix 1, Fig. 3, Table S3). All 14 members of Kainate, AMPA, and NMDA receptors5 were found. We recovered all iGluRs, 17 of the 18 antennal and 21 of the ~30 divergent IRs. Only in-paralogs were found in one antennal (Ir75b) and in one divergent IR (Ir87a). Of the 39 loci, 26 (67%) were recovered with a complete gene model, of which only one was assigned as putative pseudogene (the divergent IR DnigIr94d). Protein domain predictors found Lig_chan-Glu_bd and Lig_chan domains, with a different degree of divergence among subfamilies. The iGluRs were recovered with sequence identities of 40% and 49% for Lig_chan-Glu_bd and Lig_chan domains, respectively. In contrast, between the other two IR subfamilies antennal and divergent divergence increased significantly. In detail, similarity of the Lig_chan-Glu_bd domain decreased from 24% in antennal to 18% in divergent and similarity of the Lig_chan domain decreased from 26% in antennal to 16% in divergent (p-adjusted <0.05, Wilcoxon rank-sum test; Figure S2d).
Figure 3. Phylogenetic relationships of the candidate D. nigrosparsa Ionotropic Receptors (IRs).
Phylogenetic relationships of putative Ionotropic (IRs) and Ionotropic Glutamate receptors (iGluRs) in D. nigrosparsa and 12 other Drosophila species. IRs are subdivided into antennal and divergent. The maximum likelihood tree was midpoint rooted. Bootstrap support ≥70 for branches is indicated. Purple branches correspond to the lineages leading to D. nigrosparsa loci. Asterisks after the IR name indicate genes under positive selection. Histogram and dot plots as in Fig. 1.
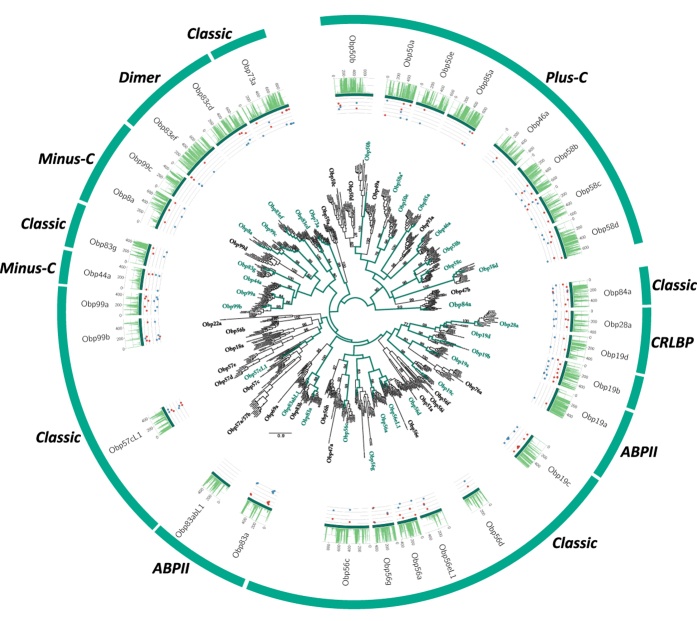
Thirty-two candidate OBP loci were identified, aligned and grouped in 31 of the 57 Drosophila clusters (SI Appendix 1, Fig. 4, Table S4), 30 assigned as putative full-length OBPs, and between 10 and 11 loci were assigned as missing. Only one in-paralog was found, the Plus-C DnigObp58b, and based on conserved cysteines, one locus (DnigObp19b) was annotated as putative pseudogene, bearing only four of the six conserved cysteines. Protein domain predictors found the PBP_GOBP Pfam domain in all loci.
Figure 4. Phylogenetic relationships of the candidate D. nigrosparsa Odorant Binding Proteins (OBPs).
The gene family tree describes phylogenetic relationships of the candidate D. nigrosparsa OBPs with the 12 other Drosophila species, subdivided according to the classic cysteine criteria. The maximum likelihood tree was midpoint rooted. Bootstrap support ≥70 for branches is indicated. Green branches correspond to the lineages leading to D. nigrosparsa loci. Asterisks after the OBP name indicate genes under positive selection. Histogram and dot plots as in Fig. 1.
Evolutionary forces on D. nigrosparsa chemosensory genes and their structural rearrangements
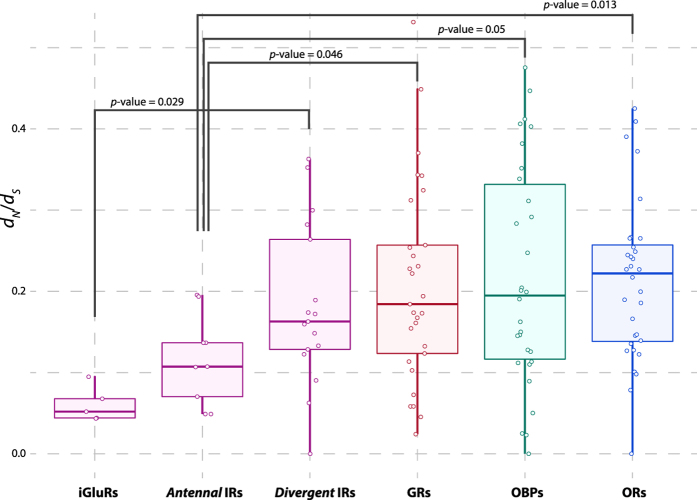
After the CGF annotation, episodic diversifying selection was tested only on full-length loci. Putative signals of selection were recorded in five of 28 OR loci tested (18%; DnigOr42a, DnigOr49ba, DnigOr43a, DnigOr56a, DnigOr45a), in one of 24 GR loci tested (4%; DnigGr64e), in two of 21 odorant-related IRs tested (9%; antennal DnigIr84a. divergent DnigIr85a), and in one of the 29 OBP loci tested (3%; Plus-C DnigObp50a). The distribution of the mean ω (dN/dS) across the D. nigrosparsa branches indicated that the iGluRs and antennal IRs are evolving under significantly different selective pressures. The iGluRs (medians: ωiGluR = 0.06) were found to be under stronger purifying selection than the IRs (ωDivergent = 0.17; p-value = 0.028) and the antennal IRs (ωAntennal = 0.12) under stronger purifying selection than the other three gene families (ωOrs = 0.23, p-value = 0.013; ωGrs = 0.19, p-value = 0.046; ωObps = 0.20, p-value = 0.051; one-tailed Wilcoxon rank-sum test) (Fig. 5). Although the ORs showed a higher rate of positive selection, the medians from ORs, GRs, and OBPs were statistically indistinguishable from each other (p-value > 0.05; one-tailed Wilcoxon rank-sum test). Given the generally stronger purifying selection in antennal IRs than the other gene families, we searched for a putative signal of relaxation amongst the full-length antennal IRs in D. nigrosparsa, and detected a significant relaxation rate (k < 1) in DnigIr21a (ω = 0.10; k = 0.78) and DnigIr93a (ω = 0.14; k = 0.67) (p-adjusted < 0.05) (Figure S3).
Figure 5. Evolutionary pressure in the D. nigrosparsa chemosensory gene families and sub-families.
Boxplots depicting distribution and median (horizontal line) of ω (dN/dS) rates in each gene family and sub-family for D. nigrosparsa. Distributions were significantly different in four comparisons (Wilcoxon rank-sum tests).
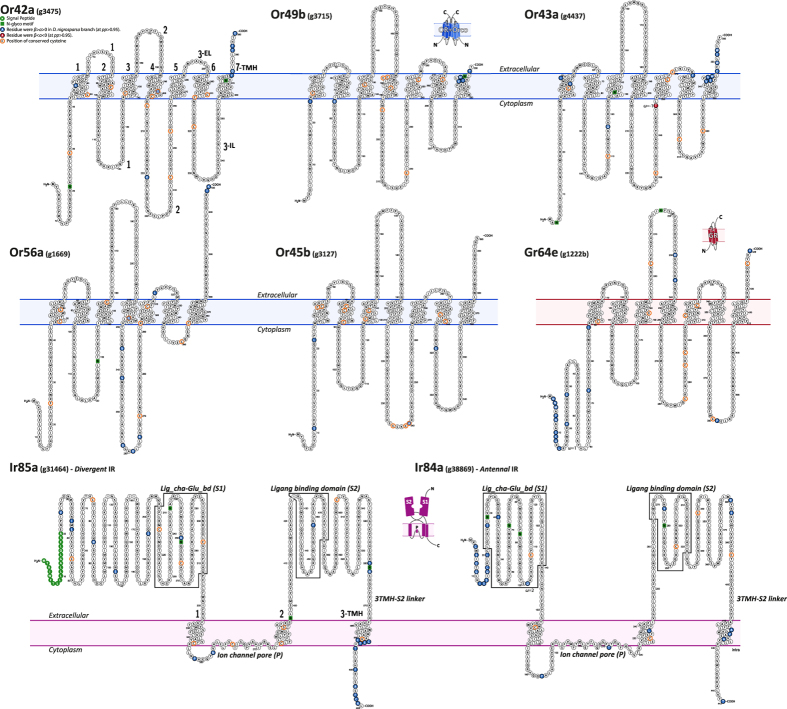
Given the absence of suitable OR/GR templates in Protein Data Bank, prediction of three-dimensional models would not be reliable6. Therefore, we focused on the prediction of the receptor topology organization. Drosophila nigrosparsa ORs had a general topology organization of 7-TMHs connected by three intracellular (ILs) and three extracellular loops (ELs) of variable lengths6. These receptors also included two variable regions at the N-terminus and C-terminus that protrude toward the cytosolic and extracellular compartments (Fig. 6)6. There are three critical regions for the functionality of these receptors: the N-terminus, the EL2, and IL3/TMH7, which have strong evolutionary constraints6,26. Performing site-to-site selection tests (MEME27) on loci under putative episodic positive selection, we aimed to identify codons for which non-synonymous (β) substitution rates are significantly higher than synonymous (α) substitution rates and zero (β > α > 0). The aim was to identify D. nigrosparsa branch-specific mutations with possible implications for protein structure functionality. We observed that a great portion, 48% (50/105) of all modifications, occurred on the amino and carboxyl terminus in all proteins, corresponding to the more variable and disordered protein regions28. The remaining 52% occurred on TMHs, ILs, and ELs. To better characterize the receptor structural rearrangements, we focused our analysis on highly conserved regions between D. nigrosparsa protein sequences and their orthologs29,30. Eight point mutations were isolated on ORs, which were grouped according to their structural localization: DnigOr42aC196S, (i.e. C196S represents a cysteine substituted by a serine in position 196), and DnigOr56aC224G in TMH4; DnigOr42aL221K, DnigOr43aT276F, DnigOr56aT258T, DnigOr56aE261R, and DnigOr45bC240A in IL2; and DnigOr45bT319E in IL3. Two were identified in GR64e: DnigGr64eL230W in EL2 and DnigGr64eD381P in IL3 (Figs 6, S4).
Figure 6. Protein topology of D. nigrosparsa receptors under positive selection.
The predicted topologies of odorant, gustatory, and ionotropic receptors, showing distribution of non-synonymous substitutions (β > α > 0) on conserved residues (see methods, Figure S4) between D. nigrosparsa and the 12 Drosophila species (blue circles) and conserved cysteines (orange circles) along protein domains. Seven trans-membrane domains were inferred for ORs and GRs, three for IRs.
On the two IR receptors under positive selection (Fig. 7a,b), the site-to-site test helped us identify candidate structural changes. A total of 45 codons for the two loci were identified, most of them in variable and/or disordered regions, such as the C-terminus and the divergent ATD (i.e., ANF_receptor, Figure S2d)31, involved in channel assembly and/or co-factor binding32. Only nine mutations were located in conserved regions (Figure S4): three occurred in the Lig_cha-Glu_bd (S1) domain (DnigIr85aH260N, DnigIr85aM401I, DnigIr84aG101Q), three (DnigIr85aH560G, DnigIr85aS562N, DnigIr84aQ380R) in the TMH3-S2 linker, the region connecting S2 with TMH3, one (DnigIr84aA221T) in the ion channel pore (P), one (DnigIr84aM235L) in TMH2, and one (DnigIr84aF276Y) in S2 (Figs 6 and 7).
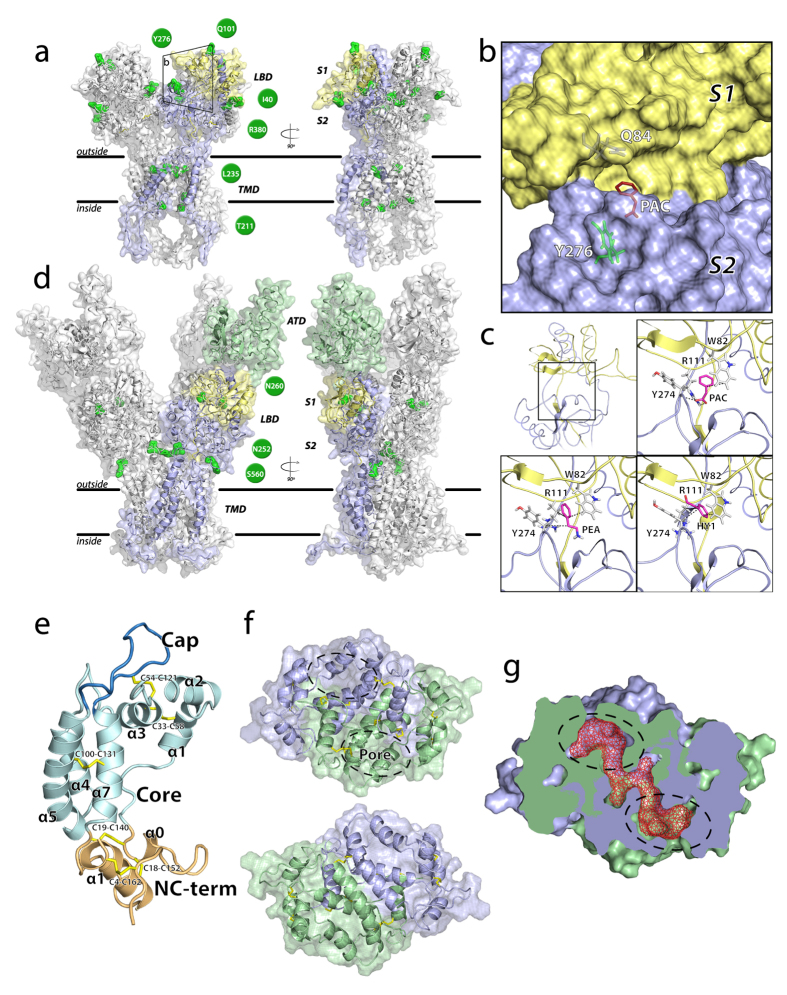
Figure 7. Three-dimensional protein structure prediction of DnigIr84a, DnigIr85a, and DnigObp50a.
Protein models of DnigIr84a (a) and DnigIr85a (d) homotetramers. Only one chain was colour-coded according to its protein domains (yellow: Lig_chan-Glu_bd, Pfam PF10613; lilac: Lig_chan, Pfam PF00060; green: ATD, Pfam PF01094; TMD: transmembrane domain). The ligand binding domain (LBD) is subdivided in two halves, S1 and S2. Residues for which β > α > 0 in conserved regions (Figure S4) are depicted in green, as well as position and residue, which are shown in green circles. (b) Binding pocket of DnigIr84a. Its pore differs from D. melanogaster by two residues, Q84 and Y276. (c) The ligand-binding site of DnigIr84a and the three best pose for PAC, HY1, and PEA. (e) Schematic representation of the DnigObp50a and its three domains, the Core (pale green), the Cap (blue), and the NC-term domain (ochre). Sequence position of cysteines forming disulfide bonds are shown (yellow), together with α-helices. (f) Schematic representation of DnigObp50a three-dimensional domain-swapped dimer (lilac and green). Two views are shown, one from the face of the dimer formed by the swapped NC-term domain (up) and one from the face formed by the α5 and α5′ helices (bottom). A tunnel with pores on each side runs through the dimer interface. (g) Surface representation of the DnigObp50a swapped dimer (slice). The tunnel is represented by red mesh.
The homology between IRs and iGluRs allowed us to predict reliable three-dimensional structure models important to infer possible structural role of mutated residues in the binding site. We analysed the predicted structure of Ir84a and characterized it in terms of cavities and binding pockets. Two large cavities were located in the ion channel domain and in the region of Ir84a corresponding to the binding site of the iGluRs33,34, constituted by 28 residues. Using as reference DmelIr84a, we identified six mutated residues (21%; DnigIr84aK81T, DnigIr84aE84Q, DnigIr84aD87P, DnigIr84aR101Q, DnigIr84aL114P, DnigIr84aF276Y) and a proline deletion immediately after DnigIr84aY274 (Fig. 7). All amino acids forming the putative binding site cavity were used as active residues (i.e., residues likely to be involved in binding) in three docking simulations, each using a specific small compound as ligand: phenylethylamine (PEA), phenylacetic acid (PAC), and phenylacetaldehyde (HY1)17. Complexes predicted by the HADDOCK algorithm were grouped in clusters according to the fraction of common contacts (fcc). PEA and PAC had higher average HADDOCK scores of the most representative cluster for the three ligands than HY1 (89%, 72%, and 23% for the refined complexes of HY1, PEA, and PAC, respectively). The PAC molecule had the best pose among all the ligands in terms of HADDOCK scores. Best scoring complexes for the DnigIr84a-ligand PEA and PAC suggested that three residues in the binding site (DnigIr84aW82, DnigIr84aR111, DnigIr84aY274) might play a pivotal role in the binding of the three ligands. In details, these residues contacted the aromatic compounds in more than 90% of the proposed docking solutions. However, in more than 60% of the PEA poses, residue DnigIr84aQ84 established a hydrogen bond with the amino group of the PEA. Three other contacts were found in more than 90% of the predicted HY1 complexes: DnigIr84aR111, DnigIr84aS273, and DnigIr84aR324, which mainly established hydrophobic interactions.
A single OBP was detected with a signature of positive selection, DnigObp50a. While the translated putative protein sequence showed a good phylogenetic signal with other Drosophila orthologs (bs = 98), the locus was topologically basal and quite distant from other loci (Fig. 4), with a very low sequence similarity with respect to the loci from other species (between 47% and 62% with D. erecta and D. mojavensis, respectively). To understand the putative biological functionality, we built a three-dimensional model of DnigObp50a, and revealed a compact domain-swapped dimer structure (Fig. 7e,f). Each 193-amino-acid long monomer contained seven α-helices (α1-5, α7, α8 plus α0). The secondary structure element α6 was the only helix predicted with a random coil structure by HHpred35. The model had good structure similarity with the template (RMS: 2.644), and the monomer had the typical three-partition domain organization: a central Core domain, resembling the classical OBP fold (α1-7, and a loop); a flanking NC-tem domain, made by the two terminal domains linked by two disulfide bridges (α0 and α8); and a Cap domain made by the coil region and a long protruding loop. In contrast to the template, for which two channels are described, we found only a single L-shaped tunnel (Fig. 7g), bearing two pores formed by the equal contribution of the two monomers. As in the crystal structure, the inner cavity is located at the interface between α7 and α7′, which also represents one of the most conserved regions in the alignment. The conformation of the NC-term domain, which does not interact with the internal part of the Core domain, was oriented toward the solvent shaping a putative ligand binding site as already observed in other paralogs19,36.
Putative odorant receptive landscape in D. nigrosparsa
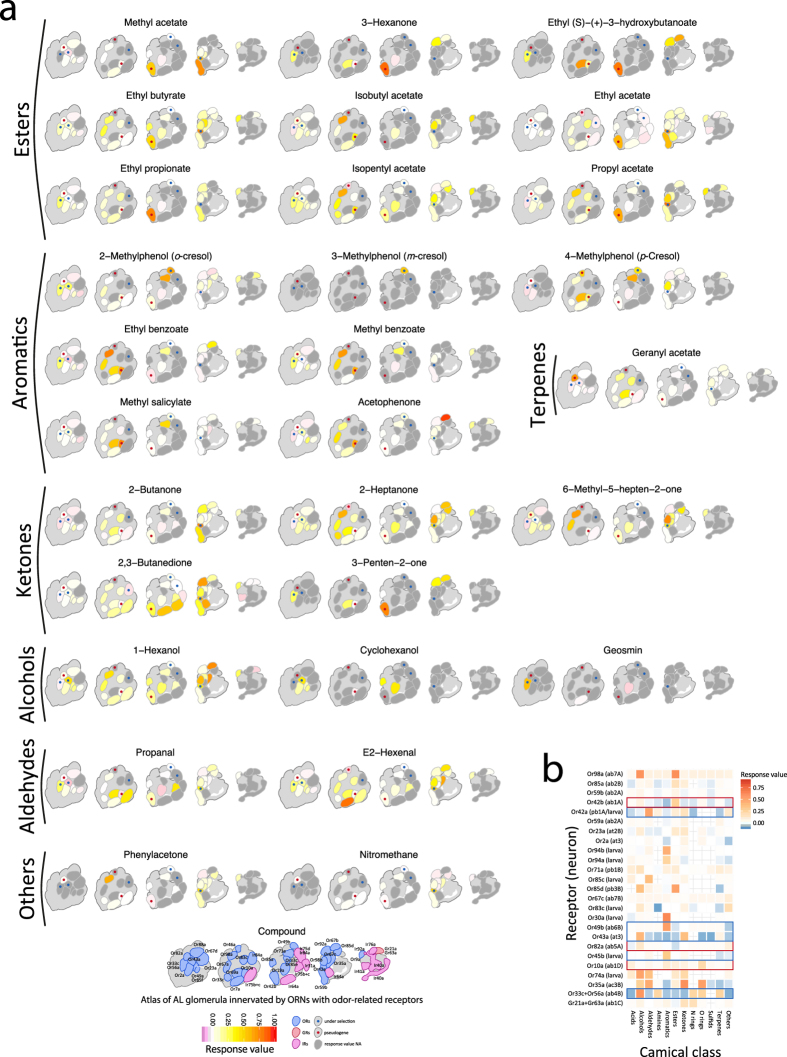
Once we had genetically characterized the putative chemosensory toolkit of D. nigrosparsa, we asked how this could reflect changes or modifications in the chemosensory receptive landscape of the species. Starting by looking at the available data of D. melanogaster’s olfactome and using the neural data from the Database of Odorant Receptors (DoOR v2.0.0)37,38, we mapped all available odorant interactions using the set of D. nigrosparsa receptors we characterized. Projecting knowledge on the peripheral and central aspects of odour detection of D. melanogaster on D. nigrosparsa may introduce errors, but lacking other options. We thus cautiously used the results of this approach to gain a first overview of the odorant reception landscape. Thereby, we showed that D. nigrosparsa possesses the full set of receptors capable of detecting a wide range of chemical classes (response value threshold ≥0.25) (Fig. 8). Focusing on odorant-related receptors under diversifying selection, we noted that Or42a, located both in the basiconic sensilla (palp) of the adult and in the dorsal organ dome in the larva of D. melanogaster39, might show a wide range of affinity to ~20 compounds including ketones, esters, aldehydes, alcohols, and others, all related to fruit scents and fermentation activity14,39. Interestingly, DnigOr67c had a significantly higher conservation level and was well affine to a wide range of chemical classes including its primary ligand, ethyl lactate, generated from malolactic fermentation and signalling a hospitable environment14. Or56a had a more specific pathway, a very narrow and conserved spectrum of action in Drosophila40, and is related to the detection and a subsequent repulsion response to geosmin41.
Figure 8. Odorant receptor-to-glomeruli projection map of response values to compounds and their chemical classes.
In flies, like in other insects and vertebrates, odorants bind to odorant receptors of dendrites of bipolar olfactory receptor neurons (ORNs). Each ORN expresses one or very few receptors and sends its axons to make connections with the second order neurons in the glomeruli of the antennal lobes (ALs) of the brain. This figure shows (a) the heatmap response level of glomeruli of the AL connected with ORN expressing specific receptors with the more specific chemical compounds (response value ≥0.75) to the positively selected receptors as in D. melanogaster, whenever data are available; (b) the heatmap of response level of receptors with the different chemical classes. Blue and red rectangles represent pseudogenes and receptors under selection, respectively.
Discussion
Until today, 26 genomes of drosophilid species have been published. Of them, only five (excluding S. flava) belong to the Drosophila subgenus, as does D. nigrosparsa. Sequenced genome sizes range from 121 Mbp in D. busckii to ~200 Mbp in D. mojavensis (194 Mbp), D. grimshawi (200 Mbp), and D. virilis (206 Mbp), to 254 Mbp in D. albomicans. Generally, eukaryote genome size reflects the genomic content in repeated sequences42. The Drosophila radiation is not an exception; transposable elements (TE) accumulation is a major factor of genome size variation, and both TE accumulation and genome size are fully explained by the phylogeny43. According to the CGF phylogenies now available, D. nigrosparsa’s phylogenetic position is within the Drosophila subgenus. When considering just OR-based phylogenies, it is most closely related to D. grimshawi and S. flava. This is in line with phylogenetic reconstructions using the complete mtDNA and single-copy ortholog genes (Cicconardi et al. submitted). Given that our size estimation for the D. nigrosparsa genome of 221.5 Mbp is consistent with those for other drosophilid species (~200 Mbp), we believe we have sequenced the great majority of the genome. This is also supported by the almost full list (94.5%) of BUSCO ortholog genes annotated and the rather high RNA-seq mappability of ~84%44.
The genome of D. nigrosparsa allowed us to begin the investigation of the unique characteristics and adaptations of this mountain fly. Here, we studied the main four CGFs, annotating 189 loci and recording an uneven proportion of presence/absence across CGFs. Indeed, while the set of ORs, iGluRs, and antennal IRs looks quite comprehensive, many genes belonging to GRs, OBPs, and divergent IRs seem to be missing. In interpreting the lack of these loci, we tend to reject a bias in assembling the genome as causal. Rather, it may represent gene loss. As shown in previous studies, gene turnover rate tends to vary across chemosensory gene families; ORs, OBPs, and antennal IRs have the least dynamic rates, with lower death rate (δ) values across different models (average δOR = 0.0017, δOBP = 0.0011, δAnt IR = 0.0003), while GRs and divergent IRs have higher dynamic rates and about 2-fold higher death rates (average δGR = 0.0023; δDiv IR = 0.0026)45. Specialist species such as D. sechellia16,45, exclusively depending on Morinda citrifolia as a host plant, and D. erecta, depending on Pandanus candelabrum, have actually lost several GRs45. Although in our study this hypothesis was not fully validated, it is plausible that, especially for GRs and divergent IRs, a number of genes could have been lost through pseudogenisation during D. nigrosparsa’s early adaptation and rapidly deleted from the genome16,46,47. Unfortunately, it is very difficult to validate this theory, and strong evidence of profound gene loss in D. nigrosparsa is lacking.
Stronger signs of adaptation could, instead, be found in several genes bearing signs of evolutionary pressures. We found that nine of the 11 full-length CGFs are under putative diversifying positive selection (five ORs, one GR, one antennal IR, one divergent IR, one OBP), two antennal IRs bear a putative signal of relaxation, and in six loci putative pseudogenisation occurred (three single-copy ORs, two out-paralogous ORs, and one divergent IR). Here, based on the functional roles of some of these genes, we present hypotheses on how D. nigrosparsa might have adapted. Or49b and Or42a are known to play an important role in the attraction to aromatic and alcoholic compounds related to food14,48, while Or56a is pivotal in the detection of geosmin, a volatile toxic compound produced by a number of fungi49, bacteria50, and cyanobacteria51, which activates a conserved pathway among all Drosophila species, triggering a single class of sensory neurons and targeting the DA2 glomerulus (Fig. 8)40. In D. nigrosparsa, Or56a and Or42a lost two cysteines, highly conserved across their orthologues, both at TMH4 (Fig. 6). Cysteines play a key role in structurally related receptors by the formation of disulfide bonds29,52. Thus, the loss of disulfide bridges, particularly when found in combination with other surrounding cysteines (i.e., DnigOr42aC196S surrounded by: DnigOr42aC139, DnigOr42aC203, DnigOr42aC243, DnigOr42aC254), might have an impact on the receptors’ folding in the cytoplasmic membrane29,53, and/or mediate affinity to compounds54. This might allow a higher degree of the receptor’s flexibility. We also found that the residues of intracellular loops (IL2 and IL3) evolve at a high rate. In these loops, the chemical/physical change due to a single amino-acid mutation might influence intracellular recruitment of other proteins once the receptor is bound to a ligand, influencing the signal transduction mechanism and modulating the receptor’s ability to recognize molecular partners29,53,55. These changes could affect the ion channel formation and functioning once the heterodimer is formed6. Drosophila nigrosparsa is a mushroom breeder (with more than 93% of eggs deposited on them in experiments) but not specialised on a single fungal taxon23. Modifications in DnigOr56a receptor may thus possibly reflect important adaptation towards different types of toxic compounds possibly present in fungi, and their recognition in non-suitable fungi species might inhibit positive chemotaxis, avoiding the oviposition40. Given that fungi are known to produce various volatile substances such as 1-octen-3-ol (called mushroom alcohol)56, DnigOr56a receptor modifications may alternatively be key to identifying suitable oviposition substrate using substances produced by fungi. This scenario would be in line with findings for Drosophila specialised on yeast57, such as D. melanogaster, D. mojavensis, and D. suzukii58,59.
We also found signs of diversifying positive selection in odorant-related IRs, specifically in one antennal (DnigIr84a) and one divergent (DnigIr85a) IR. In D. melanogaster, Ir84a effectively binds specific aromatic compounds, like PAC and PEA, commonly present on food sources, and the activation of the Ir84a pathway may promote male courtship in the presence of food, complementing the functions of pheromone receptors in regulating mate choice, thus explaining the widespread Drosophila behaviour to mate predominantly on their food17. Given the availability of crystal structures of phylogenetically and functionally related iGluRs33,34,60,61, it was possible to investigate the structural role of the modifications in these two D. nigrosparsa IRs. By predicting the three-dimensional homotetramer models for both receptors, and considering the strong purifying selection acting on antennal IRs (Fig. 5), it is likely that, among others, DnigIr84a successfully contributes to the adaptation to an environment where food is scarce or available only for a short time62. To support this hypothesis, we studied the binding site cavity and its surrounding and found that 21% of the residues are different between DnigIr84a and DmelIr84a, and two residues (DnigIr84aE84 and DnigIr84aF276) outline the cavity opening. This might cause difference in the compound accessibility in the pocket between the two proteins in ligand binding kinetics and affinity. The close proximity of the DmelIr84a insertion to the cavity entering might also underlie differences in ligand entering and recognition.
Overall, the results obtained by the data-driven docking are in accordance with the experimental data on binding affinity available for D. melanogaster reported from in vivo experiments17. The most representative contacts identified by the docking simulations do not involve the aforementioned residues, with the exception of the hydrogen bond between DnigIr84aE84 and PEA. These findings point to a more indirect effect caused by differences in the geometry and electrostatic potential of the two binding sites, underlying a fine-tuning adaptation mechanism for this particular class of receptors.
GRs are together with some other IRs involved in the gustatory response. In insects, the taste receptor system not only covers a wide ligand spectrum of sugars, bitter substances, and salts but also includes reception of pheromone and somatosensory stimulants63. In characterizing D. nigrosparsa GRs, we found duplication events, and putative loss-of-function in three other receptors. In Drosophila, the response to sugar is typically mediated by classes of sensory neurons mediated by eight GRs, namely Gr5a, Gr61a, and members of the Gr64a–f cluster64,65. In D. nigrosparsa, we were not able to annotate four of them (Gr61a, Gr64a, Gr64b, Gr64d), but one of them, DnigGr64e, specifically related to low-sugar stimuli and linked to glycerol, a by-product of yeast fermentation present in fermented fruit, was detected as the only GR bearing a signature of positive diversifying selection. In this receptor, we found modifications occurring on EL2, a protein region possibly involved in the binding site6. Both the possible loss-of-function, and the positive selection in this important receptor cluster may represent a process of adaptation towards low sugar content food.
Odorant and gustatory receptors are located in dendrite membranes embedded in lymph in contact with the air through pores in the sensilla’s cuticle1,66. Chemicals need to enter the aqueous lymph to interact with the odorant receptors, and because OBPs contain hydrophobic binding pockets, they play a key role in solubilizing hydrophobic odours1,67,68. Among them, only Obp50a, secreted by auxiliary cells of the antennal trichoid sensilla69, was found with a signature of diversifying selection, but no information is available about its compound affinity. By reconstructing the DnigObp50a three-dimensional structure, we made plausible all signs of putative functionality suggested by conserved features such as the disulfide bonds and the α helix configuration and specific characteristics such as the configuration of the channel pore, very different from other Drosophila orthologues. Although more evidence is needed, we suggest that this protein represents a possible additional adaptation/innovation of this alpine species.
In conclusion, insect chemosensory systems have emerged as prominent models in neuroscience. The investigation of their functions and mechanisms has revealed surprising answers to fundamental questions of how animals detect and process chemical stimuli and adapt to the environment. Using our comparative-genomics and structural-proteomics approach, we have described putative signatures of adaptation in the four CGFs in a habitat-specialist fly. We have detected a stronger adaptation of ORs compared with GRs, IRs, and OBPs. Like the majority of Drosophilidae, D. nigrosparsa also has the chemosensory arsenal for the exploration of fermentation-related food source, and it is possible to hypothesize that mutations on several receptors might enhance their affinity to compounds, mediating the evolution of more specialized food preferences in both adults and larvae.
Methods
Sequencing and genome assembly
The genome of D. nigrosparsa was sequenced from 150 pooled individuals of the iso-female line Iso12 kept at the University of Innsbruck, approximately 50 generations after its establishment. Six gDNA libraries were constructed, three with a short insert size (nominal size < 500 bp), consisting of one overlapped paired-end library (Ovl) of 100–300 nucleotide (nt) read length and two standard paired-end libraries of 100 and 130 nt lengths, and three mate-pair libraries with longer insert size (nominal sizes 2 kbp, 5 kbp, 10 kbp), outsourced at IGA Technology Services (http://www.igatechnology.com). The Ovl library was sequenced on Illumina MiSeq, all others on Illumina HiSeq2500. Raw reads were quality filtered using Trimmomatic v.0.3270 with a sliding window equal to 10% of the read length and a quality score of 10 without trimming, deduplicated with FastUniq71, and error corrected with SOAPec72 (kmer = 17). The estimated raw base error ratio was very low at 0.0010 to 0.0003.
ALLPATHS-LG73 was used to assemble the genome, which was done in two iterations. In the first iteration, all reads were used to run a first draft genome (PLOIDITY = 2), using the Blobology pipeline74 to filter out possible contaminants. In brief, the pipeline consists of a first BLASTn v.2.2.29+ search of contigs against the whole nucleotide database of GenBank, retaining taxonomic information from the best hit, followed by mapping of genomic read libraries against contigs. By plotting each contig’s GC content versus coverage, possible contaminant contigs from organisms hosted by the target species were detected. All reads mapping to contaminant contigs were filtered out from each library and all remaining ones used to run the second iteration to assemble the genome. Genome completeness was assessed searching the 2675 arthropod orthologous genes from the Benchmarking Universal Single-Copy Orthologs (BUSCO)75 on a provisional complete genome annotation. Annotation of the genome was performed mapping all D. melanogaster protein sequences and RNA reads onto the genome and used as hints for the BRAKER pipeline76.
Gene family datasets and identification of CGFs in Drosophila nigrosparsa
The dataset for all gene families from the Drosophila 12 genomes consortium77 was obtained from Almeida et al.45. For each gene family, both nt and amino acid (aa) sequences were used, together with the assignment of orthology. For ORs, both nt and aa sequences from Scaptomyza flava78 were added to the study.
To identify and characterize chemosensory genes in D. nigrosparsa, aa sequences from all available Drosophilidae species were used as queries in a tBLASTn v.2.2.29+ search on the D. nigrosparsa assembly with an e-value cutoff of 1e−5 78. From all hits, genomic regions of 15 kbp were extracted from the assembly and used as input file for Augustus v.3.2.179 (–genemodel = partial –species = fly). The ab initio transcriptome of D. nigrosparsa was assembled using TopHat80 and Cufflinks v.2.2.181, using the available RNA-seq library of D. nigrosparsa82. Each gene was then manually annotated taking care of tBLASTn and Augustus results, together with the ab initio transcriptome and spliced reads. Drosophila nigrosparsa sequences that did not align to the entire length of orthologous Drosophila sequences were labelled incomplete only when gaps where present; sequences with nonsense mutations were annotated as putative pseudogenes. Gene annotation on the D. nigrosparsa genome was confined to the four gene families for this study.
Phylogenies of chemosensory gene families and orthology assignment
We inferred the evolutionary relationships of the ORs, GRs, IRs, and OBPs annotated from the D. nigrosparsa genome, and 12 (13 for ORs) Drosophilidae species using aa sequences. Pseudogenes and incomplete genes were included in the analysis. For each CGF, aa sequences were aligned using ClustalW v.1.2.1 EBI web server83 (settings: mbed: true; mbediteration: true; iterations: 5; gtiterations: 5; hmmiterations: 5), and the phylogeny was inferred using maximum likelihood (ML) search as implemented in RAxML v.8.1.2084 (Rapid BS search with MRE-based bootstopping criterion “–f a”), using the best-fit amino acid substitution model inferred with ProtTest v.3.4.185. For OR phylogeny, the Odorant receptor coreceptor (Orco/Or83b) was used as outgroup78,86. Gene orthology was subsequently assigned based on the phylogenetic tree.
To assign orthology, count the actual number of genes, evaluate functional completeness in each of the four protein families, and evaluate the presence or absence of genes in the genomes of D. nigrosparsa and the other species, we used the gene family phylogeny, manually annotated clusters of closely related genes, and adopted the classification by Sonnhammera & Kooninb24,87. Protein domains were searched using the HHpred server35, using both Pfam databases available, pfamA_29.0 and pfam_03Feb16.
Gene nomenclature
Among the four datasets, only the IR family required a major curation given its lack of orthologous assignment (see previous section) and correspondence between aa and nt sequences. We manually curated the conformity and annotation by searching between annotated nt and aa sequences using BLAT. All D. nigrosparsa genes were named according to their orthology with D. melanogaster.
Diversifying positive selection analysis
To assess roles of diversifying positive selection on the evolution of CGFs, signatures of selection were searched for each putative full-length gene of the four protein families. CDSs were aligned using MACSE v.1.01b88, which implements a pairwise CDS alignment method that detects CDS preserving codon structure. From each gene alignment, gaps and ambiguously aligned sites were removed with Gblocks v.0.91b89 (settings: −t = c −b1 = $b −b2 = $b −b3 = 1 −b4 = 6 −b5 = h; where $b is the number of sequences divided by two plus one) (Cicconardi et al. submitted). The alignment was afterwards used to infer a de novo gene tree with an ML search performed with FastTree v.2.1.8 SSE390. Signatures of diversifying selection were inferred using the adaptive branch-site random effects likelihood (aBSREL91) algorithm as implemented in the HYPHY batch language92, testing only branches leading to D. nigrosparsa genes. Using the Benjamini and Hochberg method93, p-values were adjusted and a threshold of 0.05 used.
We performed the RELAX94 test to search for a putative signal of relaxation amongst the full-length antennal IRs in D. nigrosparsa, using as reference all branches. All p-values were adjusted for multiple comparisons. In brief, RELAX tests the hypothesis of evolutionary rate relaxation in selected branches of a phylogenetic tree compared with reference branches. A k value is computed to evaluate whether selective strength was relaxed (k < 1) or intensified (k > 1).
To detect site-wise positive selection, the mixed effects model of evolution (MEME)27 was used as implemented in HYPHY, searching for the best substitution model also implemented in HYPHY. The analysis was performed in an explorative manner, gaining insight into the proportion of negative and positive pressure for all genes (plots in Figs 1, 2, 3 and 4) using as threshold a posterior probability >0.95.
In an explorative analysis, highly conserved elements (HCEs) were also searched for each gene using the PhastCons algorithm95. This was done running PhyloFit (PHAST package v.1.396) to estimate a neutral phylogenetic model (also considered as the nonconserved model in PhastCons), using as topology the one previously computed as the guide tree, followed by PhastCons (PHAST package) to estimate conservation for each nucleotide position of all aligned genes.
Protein topology, modelling, and docking
To predict receptor transmembrane topology for all ORs, GRs, and IRs, the TOPCONS webserver v.2.097 was adopted. Protein topology analyses and modelling were performed only for putative full-length proteins for which signs of positive selection were found. We were not able to predict the three-dimensional structure of ORs and GRs because no experimental structures with a satisfying amino acid sequence identity were available. We used the Protter webserver98 together with the HMMTOP webserver99 to draw the expected 7-TMHs topology.
Modeller v.9.16100 was adopted to predict three-dimensional structures of IRs and OBP. Two templates were used for DnigIr85a (pdb codes: 4u2p,4uqq), and another one for both DnigIr84a (pdb code: 4u2p) and DnigObp50a (pdb code: 4ij7). The homotetramers for IRs and the homodimer for DnigObp50a were obtained superimposing our models to AMPA subtype iGluR (pdb code: 3KG2).
The ligand-receptor complexes of DnigIr84a bound to three of its known ligands: PEA, HY1, and PAC17 were predicted using the HADDOCK v.2.2 algorithm101. The algorithm options were set as recommended for small molecule docking in Sennhauser et al.102, with final docking poses set to 500. Complexes were grouped in clusters according to the fraction of common contacts (fcc). We used the 3 V tool103 to calculate cavities on the three-dimensional model of Ir84a and highlight the residues shaping the putative binding site cavity. To analyze the resulting docking poses, we calculated all contacts under 4.0 Å distance between DnigIr84a residues and the small molecules using the R package Bio3d v.2.2-4104. Specific interactions for the most representative contacts were calculated using Ligplus105.
Additional Information
How to cite this article: Cicconardi, F. et al. Chemosensory adaptations of the mountain fly Drosophila nigrosparsa (Insecta: Diptera) through genomics’ and structural biology’s lenses. Sci. Rep. 7, 43770; doi: 10.1038/srep43770 (2017).
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Material
Acknowledgments
This work was supported by the Austrian Federal Ministry of Science, Research and Economy, BMWFW, as part of the Infrastrukturprogramm of the Focal Point Scientific Computing at the University of Innsbruck. F.C. was supported by the Austrian Science Fund (P23949 and P23409). The computational results presented were achieved using Vienna Scientific Cluster 3 and Mach Scientific Cluster. We thank Michael Fink for help with using Mach Scientific Cluster, Magdalena Tratter for help in the wet lab, and Kurt LaButti and Christian Schlötterer for their helpfulness and readiness to discuss.
Footnotes
The authors declare no competing financial interests.
Author Contributions F.C., F.M.S. and B.C.S. designed the study. W.A. and F.M.S. performed breeding and prepared samples for DNA-seq analysis. F.C. performed the genomic bioinformatics analysis. D.D., P.P.O., and F.C. performed the protein modeling. P.P.O. performed the docking analysis. All authors contributed to the biological interpretation of the results, wrote, read, and approved the final manuscript.
References
- Joseph R. M. & Carlson J. R. Drosophila chemoreceptors: a molecular interface between the chemical world and the brain. Trends Genet. 31, 1–13 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Depetris-Chauvin A., Galagovsky D. & Grosjean Y. Chemicals and chemoreceptors: ecologically relevant signals driving behavior in Drosophila. Front. Ecol. Evol. 3, 41 (2015). [Google Scholar]
- Hansson B. S. & Stensmyr M. C. Evolution of insect olfaction. Neuron 72, 698–711 (2011). [DOI] [PubMed] [Google Scholar]
- Wicher D. et al. Drosophila odorant receptors are both ligand-gated and cyclic-nucleotide-activated cation channels. Nature 452, 1007–1012 (2008). [DOI] [PubMed] [Google Scholar]
- Benton R., Vannice K. S., Gomez-Diaz C. & Vosshall L. B. Variant ionotropic glutamate receptors as chemosensory receptors in Drosophila. Cell 136, 149–162 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hopf T. A. et al. Amino acid coevolution reveals three-dimensional structure and functional domains of insect odorant receptors. Nat Commun 6, 6077 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smart R. et al. Drosophila odorant receptors are novel seven transmembrane domain proteins that can signal independently of heterotrimeric G proteins. Insect Biochem. Mol. Biol. 38, 770–780 (2008). [DOI] [PubMed] [Google Scholar]
- Benton R., Sachse S., Michnick S. W. & Vosshall L. B. Atypical membrane topology and heteromeric function of Drosophila odorant receptors in vivo. PLoS Biol. 4, 240–257 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hallem E. A. & Carlson J. R. Coding of odors by a receptor repertoire. Cell 125, 143–160 (2006). [DOI] [PubMed] [Google Scholar]
- Larsson M. C. et al. Or83b encodes a broadly expressed odorant receptor essential for Drosophila olfaction. Neuron 43, 703–714 (2004). [DOI] [PubMed] [Google Scholar]
- German P. F., van der Poel S., Carraher C., Kralicek A. V. & Newcomb R. D. Insights into subunit interactions within the insect olfactory receptor complex using FRET. Insect Biochem. Mol. Biol. 43, 138–145 (2013). [DOI] [PubMed] [Google Scholar]
- Robertson H. M., Warr C. G. & Carlson J. R. Molecular evolution of the insect chemoreceptor gene superfamily in Drosophila melanogaster. Proc. Natl. Acad. Sci. USA 100 Suppl, 14537–14542 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freeman E. G., Wisotsky Z. & Dahanukar A. Detection of sweet tastants by a conserved group of insect gustatory receptors. Proc. Natl. Acad. Sci. USA. 111, 1598–1603 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mansourian S. & Stensmyr M. C. The chemical ecology of the fly. Curr. Opin. Neurobiol 34, 95–102 (2015). [DOI] [PubMed] [Google Scholar]
- Yelshanskaya M. V., Li M. & Sobolevsky A. I. Structure of an agonist-bound ionotropic glutamate receptor. Science 345, 1070–1074 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Croset V. et al. Ancient protostome origin of chemosensory ionotropic glutamate receptors and the evolution of insect taste and olfaction. PLoS Genet. 6, e1001064 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grosjean Y. et al. An olfactory receptor for food-derived odours promotes male courtship in Drosophila. Nature 478, 236–240 (2011). [DOI] [PubMed] [Google Scholar]
- Leal W. S. Odorant reception in insects: Roles of receptors, binding proteins, and degrading enzymes. Annu. Rev. Entomol 58, 373–391 (2013). [DOI] [PubMed] [Google Scholar]
- Vieira F. G. & Rozas J. Comparative genomics of the odorant-binding and chemosensory protein gene families across the arthropoda: Origin and evolutionary history of the chemosensory system. Genome Biol. Evol 3, 476–490 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markow T. A. & O’Grady P. Reproductive ecology of Drosophila. Funct. Ecol 22, 747–759 (2008). [Google Scholar]
- Carson H. L. The ecology of Drosophila breeding sites. Harold L. Lyon Arbor. Lect. No. 2, 1–27 (1971). [Google Scholar]
- Bächli G. Results from a Survey of the Biodiversity of Diptera (Insecta) in the Stilfserjoch National Park (Italy), vol. 1, Drosophilidae (Studia Dipterololica, 16, 2008).
- Kinzner M.-C. et al. Oviposition Substrate of the Mountain Fly Drosophila nigrosparsa (Diptera: Drosophilidae). PLoS One 11, e0165743 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonnhammera E. L. & Kooninb E. V. Orthology, paralogy and proposed classification for paralog subtypes. Trends Genet 18, 619–620 (2002). [DOI] [PubMed] [Google Scholar]
- Leoni G., Le Pera L., Ferrè F., Raimondo D. & Tramontano A. Coding potential of the products of alternative splicing in human. Genome Biol. 12, R9 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hopf T. A. et al. Three-dimensional structures of membrane proteins from genomic sequencing. Cell 149, 1607–1621 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murrell B. et al. Detecting individual sites subject to episodic diversifying selection. PLoS Genet. 8, e1002764 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Echave J., Spielman S. J. & Wilke C. O. Causes of evolutionary rate variation among protein sites. Nat. Rev. Genet. 17, 109–121 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wheatley M. et al. Lifting the lid on GPCRs: The role of extracellular loops. Br. J. Pharmacol. 165, 1688–1703 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venkatakrishnan A. J. et al. Molecular signatures of G-protein-coupled receptors. Nature 494, 185–194 (2013). [DOI] [PubMed] [Google Scholar]
- Rytz R., Croset V. & Benton R. Ionotropic receptors (IRs): chemosensory ionotropic glutamate receptors in Drosophila and beyond. Insect Biochem. Mol. Biol. 43, 888–897 (2013). [DOI] [PubMed] [Google Scholar]
- Jin R. et al. Crystal structure and association behaviour of the GluR2 amino-terminal domain. EMBO J. 28, 1812–1823 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alberstein R., Grey R., Zimmet A., Simmons D. K. & Mayer M. L. Glycine activated ion channel subunits encoded by ctenophore glutamate receptor genes. Proc. Natl. Acad. Sci. 112, E6048–E6057 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armstrong N. & Gouaux E. Mechanisms for activation and antagonism of an AMPA-sensitive glutamate receptor: crystal structures of the GluR2 ligand binding core. Neuron 28, 165–181 (2000). [DOI] [PubMed] [Google Scholar]
- Söding J., Biegert A. & Lupas A. N. The HHpred interactive server for protein homology detection and structure prediction. Nucleic Acids Res. 33, 244–248 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsitsanou K. E. et al. Crystal and solution studies of the ‘plus-C’ odorant-binding protein 48 from Anopheles gambiae: Control of binding specificity through three-dimensional domain swapping. J. Biol. Chem. 288, 33427–33438 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galizia C. G., Münch D., Strauch M., Nissler A. & Ma S. Integrating heterogeneous odor response data into a common response model: A DoOR to the complete olfactome. Chem. Senses 35, 551–563 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muench D. & Galizia C. G. DoOR 2.0 - Comprehensive mapping of Drosophila melanogaster odorant responses. Sci. Rep. 6, 1–14 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vosshall L. B. & Stocker R. F. Molecular architecture of smell and taste in Drosophila. Annu. Rev. Neurosci. 30, 505–533 (2007). [DOI] [PubMed] [Google Scholar]
- Stensmyr M. C. et al. A conserved dedicated olfactory circuit for detecting harmful microbes in Drosophila. Cell 151, 1345–1357 (2012). [DOI] [PubMed] [Google Scholar]
- Becher P. G., Bengtsson M., Hansson B. S. & Witzgall P. Flying the fly: Long-range flight behavior of Drosophila melanogaster to attractive odors. J. Chem. Ecol. 36, 599–607 (2010). [DOI] [PubMed] [Google Scholar]
- Biémont C. & Vieira C. Junk DNA as an evolutionary force. Nature 443, 521–524 (2006). [DOI] [PubMed] [Google Scholar]
- Sessegolo C. et al. Strong phylogenetic inertia on genome size and transposable element content among 26 species of flies. Biol. Lett. 12, 521–524 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobin A. et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Almeida F. C., Sánchez-Gracia A., Campos J. L. & Rozas J. Family size evolution in Drosophila chemosensory gene families: a comparative analysis with a critical appraisal of methods. Genome Biol. Evol. 6, 1669–1682 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison P. M., Milburn D., Zhang Z., Bertone P. & Gerstein M. Identification of pseudogenes in the Drosophila melanogaster genome. Nucleic Acids Res. 31, 1033–1037 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrov D. a., Chao Y. C., Stephenson E. C. & Hartl D. L. Pseudogene evolution in Drosophila suggests a high rate of DNA loss. Mol. Biol. Evol. 15, 1562–1567 (1998). [DOI] [PubMed] [Google Scholar]
- Kapsetaki S.-E. et al. The bacterial metabolite 2-aminoacetophenone promotes association of pathogenic bacteria with flies. Nat. Commun. 5, 4401 (2014). [DOI] [PubMed] [Google Scholar]
- Saadoun I., Schrader K. K. & Blevins W. T. Identification of geosmin as a volatile metabolite of Anabaena sp. J. Basic Microbiol. 41, 51–55 (2001). [DOI] [PubMed] [Google Scholar]
- Gerber N. N. & Lechevalier H. A. Geosmin, an earthly-smelling substance isolated from actinomycetes. Appl. Microbiol. 13, 935–938 (1965). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jüttner F. & Watson S. B. Biochemical and ecological control of geosmin and 2-methylisoborneol in source waters. Applied and Environmental Microbiology 73, 4395–4406 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner R. M. et al. Mutational analysis of cysteine residues of the insect odorant co-receptor (Orco) from Drosophila melanogaster reveals differential effects on agonist- and odorant-tuning receptor-dependent activation. J. Biol. Chem. 289, 31837–31845 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagerström M. C. & Schiöth H. B. Structural diversity of G protein-coupled receptors and significance for drug discovery. Nat. Rev. Drug Discov. 7, 339–357 (2008). [DOI] [PubMed] [Google Scholar]
- Schiedel A. C. et al. The four cysteine residues in the second extracellular loop of the human adenosine A2B receptor: Role in ligand binding and receptor function. Biochem. Pharmacol. 82, 389–399 (2011). [DOI] [PubMed] [Google Scholar]
- Shukla A. K. et al. Visualization of arrestin recruitment by a G-protein-coupled receptor. Nature 512, 218–222 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morawicki R. O. & Beelman R. B. Study of the biosynthesis of 1-octen-3-ol using a crude homogenate of Agaricus bisporus in a bioreactor. J. Food Sci. 73, 1–5 (2008). [DOI] [PubMed] [Google Scholar]
- Davis T. S., Crippen T. L., Hofstetter R. W. & Tomberlin J. K. Microbial volatile emissions as insect semiochemicals. J. Chem. Ecol. 39, 840–859 (2013). [DOI] [PubMed] [Google Scholar]
- Scheidler N. H., Liu C., Hamby K. A., Zalom F. G. & Syed Z. Volatile codes: Correlation of olfactory signals and reception in Drosophila-yeast chemical communication. Sci. Rep. 5, 14059 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hickner P. V. et al. The making of a pest: Insights from the evolution of chemosensory receptor families in a pestiferous and invasive fly, Drosophila suzukii. BMC Genomics 17, 648 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyerson J. R. et al. Structural mechanism of glutamate receptor activation and desensitization. Nature 514, 328–334 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dürr K. L. et al. Structure and dynamics of AMPA receptor GluA2 in resting, pre-open, and desensitized states. Cell 158, 778–792 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagy L. & Grabherr G. The biology of alpine habitats. Oxford Univ. Press 58, 7250–7 (2009). [Google Scholar]
- Isono Kumio & Hiromi M. Molecular and cellular designs of insect taste receptor system. Front. Cell. Neurosci. 4, 20 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahanukar A., Lei Y. T., Kwon J. Y. & Carlson J. R. Two Gr genes underlie sugar reception in Drosophila. Neuron 56, 503–516 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wisotsky Z., Medina A., Freeman E. & Dahanukar A. Evolutionary differences in food preference rely on Gr64e, a receptor for glycerol. Nat. Neurosci. 14, 1534–1541 (2011). [DOI] [PubMed] [Google Scholar]
- Sánchez-Gracia A., Vieira F. G. & Rozas J. Molecular evolution of the major chemosensory gene families in insects. Heredity (Edinb). 103, 208–216 (2009). [DOI] [PubMed] [Google Scholar]
- Pelosi P., Iovinella I., Felicioli A. & Dani F. R. Soluble proteins of chemical communication: An overview across arthropods. Front. Physiol. 5, 1–13 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou S., Mackay T. F. C. & Anholt R. R. H. Tuning the chemosensory window. Fly (Austin). 4, 230–235 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin F., Boto T., Gomez-Diaz C. & Alcorta E. Elements of olfactory reception in adult Drosophila melanogaster. Anat. Rec. 296, 1477–1488 (2013). [DOI] [PubMed] [Google Scholar]
- Bolger A. M., Lohse M. & Usadel B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu H. et al. FastUniq: A fast de novo duplicates removal tool for paired short reads. PLoS One 7, 1–6 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo R. et al. SOAPdenovo2: an empirically improved memory-efficient short-read de novo assembler. Gigascience 1, 18 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gnerre S. et al. High-quality draft assemblies of mammalian genomes from massively parallel sequence data. Proc. Natl. Acad. Sci. USA 108, 1513–1518 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S., Jones M., Koutsovoulos G., Clarke M. & Blaxter M. Blobology: exploring raw genome data for contaminants, symbionts and parasites using taxon-annotated GC-coverage plots. Front. Genet 4, 237 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simão F. A., Waterhouse R. M., Ioannidis P., Kriventseva E. V. & Zdobnov E. M. BUSCO: Assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics 31, 3210–3212 (2015). [DOI] [PubMed] [Google Scholar]
- Hoff K. J., Lange S., Lomsadze A., Borodovsky M. & Stanke M. BRAKER1: Unsupervised RNA-Seq-based genome annotation with GeneMark-ET and AUGUSTUS. Bioinformatics 32, 767–769 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark A. G. et al. Evolution of genes and genomes on the Drosophila phylogeny. Nature 450, 203–218 (2007). [DOI] [PubMed] [Google Scholar]
- Goldman-Huertas B. et al. Evolution of herbivory in Drosophilidae linked to loss of behaviors, antennal responses, odorant receptors, and ancestral diet. Proc. Natl. Acad. Sci. USA 112, 3026–3031 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanke M., Schöffmann O., Morgenstern B. & Waack S. Gene prediction in eukaryotes with a generalized hidden Markov model that uses hints from external sources. BMC Bioinformatics 7, 62 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D. et al. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol 14, R36 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C. et al. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat. Protoc 7, 562–578 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arthofer W. et al. Genomic Resources Notes Accepted 1 August 2014–30 September 2014. Mol. Ecol. Resour 15, 228–229 (2014). [DOI] [PubMed] [Google Scholar]
- Larkin M. a. et al. Clustal W and Clustal X version 2.0. Bioinformatics 23, 2947–2948 (2007). [DOI] [PubMed] [Google Scholar]
- Stamatakis A. RAxML version 8: A tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30, 1312–1313 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darriba D., Taboada G. L. & Posada D. ProtTest 3: fast selection of best-fit models of protein evolution. Bioinformatics 27, 1–4 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox A. N., Pitts R. J., Robertson H. M., Carlson J. R. & Zwiebel L. J. Candidate odorant receptors from the malaria vector mosquito Anopheles gambiae and evidence of down-regulation in response to blood feeding. Proc. Natl. Acad. Sci. USA 98, 14693–14697 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonnhammer E. L. L. & Östlund G. InParanoid 8: Orthology analysis between 273 proteomes, mostly eukaryotic. Nucleic Acids Res 43, D234–D239 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ranwez V., Harispe S., Delsuc F. & Douzery E. J. P. MACSE: Multiple alignment of coding SEquences accounting for frameshifts and stop codons. PLoS One 6, e22594 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castresana J. Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol. Biol. Evol 17, 540–552 (2000). [DOI] [PubMed] [Google Scholar]
- Price M. N., Dehal P. S. & Arkin A. P. FastTree 2 - Approximately maximum-likelihood trees for large alignments. PLoS One 5, e9490 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith M. D. et al. Less is more: an adaptive branch-site random effects model for efficient detection of episodic diversifying selection. Mol. Biol. Evol 32, 1342–1353 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosakovsky Pond S. L., Frost S. D. W. & Muse S. V. HyPhy: Hypothesis testing using phylogenies. Bioinformatics 21, 676–679 (2005). [DOI] [PubMed] [Google Scholar]
- Benjamini Y. & Hochberg Y. Controlling the false discovery rate: A practical and powerful approach to multiple testing. J. R. Stat. Soc. Ser. B Methodol 57, 289–300 (1995). [Google Scholar]
- Wertheim J. O., Murrell B., Smith M. D., Kosakovsky Pond S. L. & Scheffler K. RELAX: Detecting relaxed selection in a phylogenetic framework. Mol. Biol. Evol 32, 1–13 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siepel A. et al. Evolutionarily conserved elements in vertebrate. insect. worm. and yeast genomes. Genome Res 15, 1034–1050 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubisz M. J., Pollard K. S. & Siepel A. PHAST and RPHAST: phylogenetic analysis with space/time models. Brief. Bioinform 12, 41–51 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsirigos K. D., Peters C., Shu N., Käll L. & Elofsson A. The TOPCONS web server for consensus prediction of membrane protein topology and signal peptides. Nucleic Acids Res 43, W401–407 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Omasits U., Ahrens C. H., Müller S. & Wollscheid B. Protter: Interactive protein feature visualization and integration with experimental proteomic data. Bioinformatics 30, 884–886 (2014). [DOI] [PubMed] [Google Scholar]
- Tusnády G. E. & Simon I. The HMMTOP transmembrane topology prediction server. Bioinformatics 17, 849–850 (2001). [DOI] [PubMed] [Google Scholar]
- Webb B. & Sali A. Protein structure modeling with MODELLER. Methods Mol. Biol 1137, 1–15 (2014). [DOI] [PubMed] [Google Scholar]
- Dominguez C., Boelens R. & Bonvin A. HADDOCK: a protein− protein docking approach based on biochemical or biophysical Information. J. Am. Chem. Soc 125, 1731–1737 (2003). [DOI] [PubMed] [Google Scholar]
- Sennhauser G., Amstutz P., Briand C., Storchenegger O. & Grütter M. G. Drug export pathway of multidrug exporter AcrB revealed by DARPin inhibitors. PLoS Biol 5, 0106–0113 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voss N. R. & Gerstein M. 3V: Cavity, channel and cleft volume calculator and extractor Nucleic Acids Res. 38, 555–562 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant B. J., Rodrigues A. P. C., ElSawy K. M., McCammon J. A. & Caves L. S. D. Bio3d: An R package for the comparative analysis of protein structures. Bioinformatics 22, 2695–2696 (2006). [DOI] [PubMed] [Google Scholar]
- Laskowski R. A. & Swindells M. B. LigPlot+: Multiple ligand-protein interaction diagrams for drug discovery. J. Chem. Inf. Model 51, 2778–2786 (2011). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.