Abstract
Background
Verticillium dahliae is a phytopathogenic fungal pathogen that causes vascular wilt diseases responsible for considerable decreases in cotton yields. The complex mechanism underlying cotton resistance to Verticillium wilt remains uncharacterized. Identifying an endogenous resistance gene may be useful for controlling this disease.
Results
We cloned the ribosomal protein L18 (GaRPL18) gene, which mediates resistance to Verticillium wilt, from a wilt-resistant cotton species (Gossypium arboreum). We then characterized the function of this gene in cotton and Arabidopsis thaliana plants. GaRPL18 encodes a 60S ribosomal protein subunit important for intracellular protein biosynthesis. However, previous studies revealed that some ribosomal proteins are also inhibitory toward oncogenesis and congenital diseases in humans and play a role in plant disease defense. Here, we observed that V. dahliae infections induce GaRPL18 expression. Furthermore, we determined that the GaRPL18 expression pattern is consistent with the disease resistance level of different cotton varieties. GaRPL18 expression is upregulated by salicylic acid (SA) treatments, suggesting the involvement of GaRPL18 in the SA signal transduction pathway. Virus-induced gene silencing technology was used to determine whether the GaRPL18 expression level influences cotton disease resistance. Wilt-resistant cotton species in which GaRPL18 was silenced became more susceptible to V. dahliae than the control plants because of a significant decrease in the abundance of immune-related molecules. We also transformed A. thaliana ecotype Columbia (Col-0) plants with GaRPL18 according to the floral dip method. The plants overexpressing GaRPL18 were more resistant to V. dahliae infections than the wild-type Col-0 plants. The enhanced resistance of transgenic A. thaliana plants to V. dahliae is likely mediated by the SA pathway.
Conclusion
Our findings provide new insights into the role of GaRPL18, indicating that it plays a crucial role in resistance to cotton “cancer”, also known as Verticillium wilt, mainly regulated by an SA-related signaling pathway mechanism.
Electronic supplementary material
The online version of this article (doi:10.1186/s12870-017-1007-5) contains supplementary material, which is available to authorized users.
Keywords: Cotton, Verticillium wilt, Resistance gene, Ribosomal protein, GaRPL18, Salicylic acid
Background
Verticillium dahliae Kleb. is a destructive phytopathogenic fungus that causes wilt diseases on more than 400 plant species, including cotton (Gossypium arboreum) [1, 2]. Verticillium dahliae infects cotton by penetrating the roots. It then spreads across the root cortex and invades the xylem vessels where it forms the conidia responsible for the colonization of vascular tissues and functional impairment. This results in several symptoms, including wilting, discoloration, necrosis, and defoliation [3–6]. Cotton fiber quality and annual yields decrease as a result of Verticillium wilt induced by V. dahliae, and a severe outbreak can lead to yield losses of more than 30% [7, 8]. In China, more than 40% of the cotton-growing area is threatened by Verticillium wilt, potentially causing considerable decreases in cotton production and serious economic losses each year. Furthermore, the fungus can survive for long periods in the soil even without a host, making Verticillium wilt difficult to control using practical and effective chemical treatments [9, 10]. Numerous methods are used to reduce the incidence of Verticillium wilt, such as the application of tillage, soil solarization, soil amendments, and biological controls. However, these are not always efficient or effective [11, 12]. Soil fumigation, which is by far the most effective treatment for inhibiting the propagation of Verticillium species, is costly and can have lethal effects on human health and the environment [7, 13]. The identification and isolation of disease-responsive candidate genes, along with the development of disease-resistant transgenic cotton cultivars, are essential for managing Verticillium wilt [14–16].
The ribosomal protein (RP) has complex structures that differ in prokaryotes and eukaryotes. The eukaryotic ribosome is composed of two unequal subunits (60S and 40S), four ribosomal RNAs (rRNAs), and 82 different RPs. The small ribosomal subunit is composed of a single 18S rRNA and approximately 33 proteins, while the large subunit comprises 28S/25S, 5.8S, and 5S rRNAs, as well as approximately 49 proteins [17–19]. The ribosome is a highly conserved protein that is essential for cellular activities. Although its main function is to synthesize proteins, recent in-depth studies have revealed that it is also important for cell growth, division, and development, and gene regulation [20–22]. Recently, a study has shown that overexpression of the N-terminal 99 amino acids of ribosomal protein L3 confers resistance to pokeweed antiviral protein and the Fusarium mycotoxin deoxynivalenol in tobacco [23]. Another study has shown that ribosomal protein L12 and ribosomal protein L19 are important in nonhost disease resistance in N. benthamiana and A. thaliana. In addition, these genes also play a minor role in basal resistance against virulent pathogens [24]. In particular, a recent study examining ribosomal protein S14 (RPS14) and cancer concluded that this protein can specifically interact with murine double minute 2 (MDM2) to inhibit the degradation of p53 by MDM2 ubiquitin, thereby promoting p53 activity. In gastric and colorectal cancer cells the cell cycle is arrested and tumor cell growth is inhibited in the presence of abundant RPS14 [25]. Another study revealed that ribosomal protein L4 can also regulate the MDM2–p53 loop to regulate p53 activity [26]. Although these studies suggest that RPs are important for disease resistance, they did not include cotton species. Therefore, a more thorough characterization of the function of the cotton RP may be useful for the breeding of Verticillium wilt-resistant cotton varieties.
Under natural conditions, plants frequently encounter diverse potential pathogens. Plants are constantly evolving to cope with these biotic stresses. For example, plants have evolved an immune system that includes constitutive and inducible defense systems that offer protection from potentially dangerous pathogens [27, 28]. Plants also produce several endogenous signaling molecules that help regulate plant defense responses, including jasmonic acid (JA), salicylic acid (SA), and ethylene (ET), all of which are involved in complex signal transduction networks. These biochemical molecules function cooperatively or antagonistically to increase plant resistance to different pathogens [29–32]. Our study revealed that cotton ribosomal protein L18 (GaRPL18) expression levels can be upregulated by accumulated SA, suggesting that RPs can mediate cotton resistance to Verticillium wilt through the SA signaling pathway. While SA is crucial for plant defenses and acquired systemic resistance, it is predominantly involved in the former [33, 34]. Increased SA levels in plant pathogen-challenged tissues and applications of exogenous SA induce the expression of pathogenesis-related (PR) genes, thereby enhancing resistance to invading pathogens [35, 36]. The activation of plant immune responses is also associated with increases in the production of reactive oxygen intermediates and nitric oxide (NO) levels [37]. While the signal transduction networks underlying all plant response mechanisms are complex, crosstalk between the different signaling molecules and networks provides plants with a powerful means of regulating immune responses [38, 39].
In this study, we focused on determining whether GaRPL18 is important for cotton resistance to Verticillium wilt caused by V. dahliae. Our objective was also to identify the signaling pathway associated with cotton defense responses. To verify the expression of GaRPL18, we harvested G. arboreum ‘HuNanChangDeTieZiMian’ samples at different time points after treatments with V. dahliae, methyl jasmonate (MeJA), SA, or ET. We observed that the GaRPL18 expression level increased significantly following V. dahliae and SA treatments. Moreover, we used virus-induced gene silencing (VIGS) technology and transgenic Arabidopsis thaliana lines overexpressing GaRPL18 to functionally characterize GaRPL18 in cotton. Complementary physiology and molecular experiments confirmed that GaRPL18 significantly contributes to cotton resistance against the fungal wilt pathogen V. dahliae via a mechanism related to the SA signaling pathway. Our findings provide insights into the molecular features and functions of a cotton RP gene related to increased resistance to Verticillium wilt.
Methods
Plant sources and growth conditions
Seeds of G. arboreum ‘HuNanChangDeTieZiMian’ (resistant) and ‘NaShangQuXiaoHua’ (susceptible) were obtained from the Institute of Cotton Research of the Chinese Academy of Agricultural Sciences. The GaRPL18 overexpression vector (i.e., 35S::GaRPL18) was inserted into wild-type (WT) A. thaliana Columbia ecotype (Col-0) plant. The transgenic plants transformed with 35S::GaRPL18 were grown in a greenhouse at 22 °C and 60% relative humidity under a 16-h light/8-h dark photoperiod. The seeds from different G. arboreum cultivars were incubated in another greenhouse at 25 °C and 80% relative humidity under a 16-h light/8-h dark photoperiod.
Culturing of Verticillium dahliae and inoculation of plants
An antagonistic defoliating Verticillium dahliae isolate (Vd07038) was grown on potato dextrose agar medium at 25 °C for 6 days. Colonies were then cultured in Czapek’s medium [3% (w/v) sucrose, 0.2% (w/v) NaNO3, 0.05% (w/v) MgSO4 · 7H2O, 0.05% (w/v) KCl, 0.002% (w/v) FeSO4 · 7H2O, and 0.131% (w/v) KH2PO4] for 5–7 days at 25 °C with shaking (150 rpm). For V. dahliae treatments, 10 ml conidial suspensions (107 conidia/ml in sterile distilled water) were applied to the bottom of pots containing seedlings. Similarly, A. thaliana plants were grown for 20 days before a 10-ml conidial suspension was injected into the soil using a sterile needle. Control plants were inoculated with an equal volume of sterile distilled water. For in vitro treatments, seedlings were inoculated with V. dahliae, and the extent of stunting was determined using a previously described method [40]. Seedlings were inoculated with a 2-μl conidial suspension (5 × 103 conidia/ml) 2 weeks after germinating.
cDNA cloning and construction of the GaRPL18 overexpression construct
Total RNA was extracted from V. dahliae-resistant G. arboreum ‘HuNanChangDeTieZiMian’ plants using the RNAprep Pure Plant Kit (Tiangen, Beijing, China). The purified RNA was used as a template to prepare cDNA with the PrimeScript™ II 1st Strand cDNA Synthesis Kit (Takara, Dalian, China). The 450-bp full-length GaRPL18 coding sequence with XbaI and AscI linkers was cloned using the primers GaRPL18-F and GaRPL18-R (Table 1). For overexpression studies, the 35S::GaRPL18 vector was constructed by digesting the GaRPL18 coding sequence with XbaI and AscI (BioLabs). The digested sequence was then inserted into a modified pCAMBIA3300 (Cambia) plant binary vector containing the glufosinate (Basta) resistance gene. This vector was used to transform Agrobacterium tumefaciens strain GV3101 using a freeze–thaw method.
Table 1.
primers used in the research
| Primer name | Forward and reverse primers(5’-3’) |
|---|---|
| GaRPL18-F | TCTAGAATGAAGCTTTGGGCCACCAA |
| GaRPL18-R | GGCGCGCCCATAAACAAGTTGGGTTT |
| VGaRPL18-F | ACTAGTATGAAGCTTTGGGCCACCAA |
| VGaRPL18-R | GGCGCGCCCATAAACAAGTTGGGTTT |
| QGaRPL18-F | AATGAAGTCCGTGCCAAATCCAAG |
| QGaRPL18-R | CGGAGCCAAATGCCGTAGTTCTTTC |
| Gahistone3-F | AAGACTGATTTGCGTTTCCA |
| Gahistone3-R | GCGCAAAGGTTGGTGTCTTC |
| AtUBQ10-F | AACTTTGGTTTGTGTTTTGG |
| AtUBQ10-R | TCGACTTGTCATTAGAAAGAAAGAGATAA |
| V-QPCR-F | AACAACAGTCCGATGGATAATTC |
| V-QPCR-R | GTACCGGGCTCGAGATCG |
| PAL-F | TGGTGGCTGAGTTTAGGAAA |
| PAL-R | TGAGTGAGGCAATGTGTGA |
| 4CL-F | ATTCAAAAGGGAGATGCC |
| 4CL-R | GAGAAGGGCAAAGCAACA |
| Basic chitinase-F | CTTAGCCCAAACTTCCCA |
| Basic chitinase-R | TACATTGAGTCCACCGAGAC |
| β-1, 3-glucanase-F | CACAGGTGCTGAAGTTGGT |
| β-1, 3-glucanase-R | CGATGGAGGGAAAGATGA |
| Cadinene synthase-F | TAACAACAATGATGCCGAGAA |
| Cadinene synthase-R | ATGGTCCAAAGATGCTACTGC |
| AtORA59-F | TCATTTGACCAATCCTTCCTTT |
| AtORA59-R | CCGTTTCCRCACRCCTCTGTAT |
| AtPDFl.2-F | ACCCTTATCTTCGCTGCTCTTG |
| AtPDFl.2-R | ATGTCCCACTTGGCTTCTCG |
| AtVSP2-F | CTTTCACTTCTCTTGCTCTTGGC |
| AtVSP2-R | GCAGTTGGCGTAGTTGATGGA |
| AtNPRl-F | GGCTTGCGGAGAAGACGAC |
| AtNPRl-R | ACGACGATGAGAGAGTTTACGG |
| AtPRl-F | GCTACGCAGAACAACTAAGAGGC |
| AtPRl-R | CCAGACAAGTCACCGCTACC |
| AtPR3-F | GAGACACCGCCACGAGGAA |
| AtPR3-R | TTGCTTGAAACAGTAGCCCCAT |
| AtAC02-F | TGTTCCTCCTCTCAACCACTC |
| AtAC02-R | CCGACATCCTGTTTCCTTCT |
| AtEIN3-F | TCAAGGCTTTGTTTATGGGATTA |
| AtEIN3-R | GCAAGGTATGAGGAGTCGGTC |
| AtERFl-F | GAGAATGACCAATAAGAAGACGAA |
| AtERFl-R | CTCCCAAATCCTCAAAGACAAC |
F Forward primer, R Reverse primer
Bioinformatics analysis
We used the National Center for Biotechnology Information online BLAST tool to analyze the GaRPL18 sequence (https://blast.ncbi.nlm.nih.gov/Blast.cgi). The Gene Structure Display Server 2.0 was then used to analyze gene structures. We also used the ExPASy online tool (http://web.expasy.org/compute_pi/) to predict the isoelectric point and molecular weight. An image of the 3D structure was developed with the PyMOL program (http://www.pymol.org/).
Generation of the virus-induced gene silencing construct and pathogen inoculation
We used Cotton leaf crumple virus (CLCrV)-based vectors (i.e., pCLCrVA and pCLCrVB) for VIGS, with CLCrV:CHLI (encoding magnesium chelatase subunit I) as a positive control [41, 42]. The GaRPL18 fragment was amplified by polymerase chain reaction (PCR) using ‘HuNanChangDeTieZiMian’ cDNA and the VGaRPL18-F/VGaRPL18-R primers (Table 1). The PCR product was digested with SpeI and AscI (BioLabs) and inserted into the pCLCrVA vector. The constructs (i.e., pCLCrVA-GaRPL18, pCLCrVA, and pCLCrVB) were then used to transform A. tumefaciens strain GV3101. The cotyledons of 7-day-old Verticillium wilt-resistant cotton seedlings were then injected with equal amounts of CLCrV vectors. After a 24-h incubation in darkness, the cotton seedlings were transferred to the greenhouse and inoculated with V. dahliae,10 days after the vector infiltration.
Arabidopsis thaliana transformation and molecular analysis
Agrobacterium tumefaciens strain GV3101 containing the GaRPL18 overexpression vector was used to transform Arabidopsis Col-0 via the floral dip method [43]. The T0 transgenic seeds were then spread evenly over soil in a pot. After 1 week, seedlings were sprayed with 0.1% Basta to select positive transformants. The false-positive seedlings turned yellow before dying. Transgenic seeds of the T1 generation were selected on plates of Murashige and Skoog (MS) medium containing 0.1% Basta. After a few days, lines with segregation ratios of approximately 3:1 (i.e., Basta resistant: Basta sensitive) were used to generate T2 lines. The transgenic seeds of the T2 generation were also selected on MS medium containing Basta to identify T3 homozygous lines. The T3 lines with the transgene and the correct segregation ratio were detected based on quantitative reverse transcription (qRT)-PCR analysis of GaRPL18 expression. Only stable homozygous T4 lines exhibiting high GaRPL18 expression levels were chosen for further functional analyses.
Quantitative reverse transcription polymerase chain reaction
We extracted total RNA from the roots and leaves of cotton plants as well as from the leaves of GaRPL18-overexpressing A. thaliana and WT plants using the RNAprep Pure Plant Kit. The RNA was used to prepare cDNA with the PrimeScript™ RT Reagent Kit with gDNA Eraser (Perfect Real Time; Takara). The Gahistone3 (Cotton_A_11188) and ubiquitin10 (accession: At4g05320) genes were used as internal controls for cotton and A. thaliana, respectively. We designed all qRT-PCR primers with the Primer Premier 6.0 program (Table 1). Diluted cDNA was used as the template for the qRT-PCR, which was conducted with SYBR® Premix Ex Taq™ (Tli RNaseH Plus; Takara) and an ABI 7900 qRT-PCR System (Applied Biosystems, CA, USA). The three-step method involved the following PCR conditions: 40 cycles of 95 °C for 30 s, 95 °C for 5 s, and 60 °C for 30 s. We analyzed the dissociation curves for each reaction and used the 2−ΔΔCT method [44] to calculate the expression level of each target gene. All reactions were conducted with at least three biological replicates.
Quantification of Verticillium dahliae colonization
We used a previously described qRT-PCR approach to detect and quantify V. dahliae colonization. The qRT-PCR analysis with the V-QPCR-F and V-QPCR-R primer pair (Table 1) was completed as previously described [45].
Methyl jasmonate, salicylic acid, and ethylene treatments
Plants were treated with 1 mM MeJA, SA, or ET solutions. Cotton or A. thaliana seedlings were grown in pots incubated in a greenhouse. They were sprayed with different solutions at the foliar stage. Control plants were treated with water at the same pH.
Measurements of free salicylic acid, nitric oxide, H2O2, and catalase levels
The abundance of the immune system-related molecules SA, NO, H2O2, and catalase (CAT) was monitored using different methods. The free SA content was determined via the Rigol L3000 high performance liquid chromatography system (Beijing, China) as previously described [46]. We ground leaf samples in liquid nitrogen for subsequent measurements of NO, H2O2, and CAT levels using a Quantitative Assay Kit (Nanjing Jiancheng, Beijing, China).
Cell death assay
Plant cell death was visualized with trypan blue staining as previously described with several modifications [47]. Leaves were soaked in trypan blue dye (1 g phenol, 1 mg trypan blue, 1 ml lactic acid, and 1 ml glycerol dissolved in 1 ml sterile distilled water) and then stained by boiling. After cooling to room temperature, samples were decolorized with a chloral hydrate solution (2.5 g/ml).
Verticillium dahliae recovery assay
To determine the effects of a V. dahliae infection on cotton and A. thaliana plants, we analyzed stem fragments from the first stem node. The cotton and A. thaliana stems were 4.5 cm and 3 cm long, respectively. The stems were cleaned according to a previously described method [48], and then sliced into six parts. The stem fragments were placed on potato dextrose agar in plates, which were incubated at 25 °C. Plant susceptibility to infection was defined according to the number of stem sections from which the fungus grew.
Analysis of the disease index, stunting, and chlorosis
For cotton plants, the disease index (DI) was calculated according to the following formula: DI = [(∑disease grades × number of infected plants)/(total checked plants × 4)] × 100%. Seedlings were classified into five grades (i.e., grade 0, 1, 2, 3, and 4) according to the symptoms on the cotyledons and true leaves [49, 50]. The disease severity for A. thaliana plants was graded on a 0–5 scale, and the DI was calculated with the following formula as previously described [40]: DI = [(∑disease grades × number of infected plants)/(total checked plants × 5)] × 100%. The DI represents a comprehensive and objective measure of plant health, with high DI values corresponding to serious infections. The extent of stunting was rated on a 0–3 scale (0 = no stunting; 1 = moderate reduction in leaf area; 2 = considerable decrease in leaf area; and 3 = considerable decrease in leaf area, leaf number, and stem length). Leaf chlorosis was rated on a 0–4 scale (0 = no symptoms; 1 = up to 25% chlorotic leaves; 2 = up to 50% chlorotic leaves; 3 = up to 75% chlorotic leaves; and 4 = up to 100% chlorotic leaves) [40].
Results
Analysis of GaRPL18 structure and expression patterns in cotton plants treated with Verticillium dahliae or hormones
Based on the results of an unpublished transcriptomics analysis of disease responses in G. arboreum plants, we analyzed GaRPL18 fragment sequences. ‘HuNanChangDeTieZiMian’ and ‘NaShangQuXiaoHua’ plants are resistant and susceptible to Verticillium wilt, respectively. We cloned the 1211-bp full-length GaRPL18 sequence using the GaRPL18-F and GaRPL18-R primers (Table 1) along with ‘HuNanChangDeTieZiMian’ cDNA. GaRPL18 was mapped to chromosome Ca3. It was localized to the complementary strand of the reference genome between positions 22987558 bp and 22988768 bp. The Gene Structure Display Server 2.0 program was used to analyze gene structures. Exons and introns are displayed as colored boxes and green lines, respectively (Fig. 1a). GaRPL18 contains two introns as well as three exons that are 49, 64, and 450 bp long (from right to left). The gene consists of a 450-bp opening reading frame encoding 149 amino acids (Fig. 1a). GaRPL18 also contains a conserved ribosomal_L18ae domain sequence (indicated with a blue box in Fig. 1a). The theoretical isoelectric point and molecular weight of the encoded protein were calculated as 10.46 and 17.9KDa, respectively, according to the ExPASy online tool. The 3D structure revealed that the ribosomal_L18ae domain (indicated in blue in Fig. 1b) is largely composed of beta turns and alpha helices. Because the GaRPL18 function is unknown, especially in V. dahliae-infected cotton plants, we examined GaRPL18 expression patterns in samples harvested from Verticillium wilt-resistant and -susceptible cotton lines at different time points after inoculations. The GaRPL18 expression levels were stable over the duration of the experiment in the control plants treated with water (Fig. 1c, d). In contrast, GaRPL18 expression levels fluctuated in the susceptible cotton plants. Additionally, in the Verticillium wilt-resistant cotton plants treated with V. dahliae, GaRPL18 expression was considerably upregulated at 6 h after inoculation, and peaked after 12 h, in both roots and leaves. To identify the signal pathway associated with GaRPL18, we examined the GaRPL18 expression patterns in hormone-treated ‘HuNanChangDeTieZiMian’ plants. We observed that GaRPL18 expression was differentially affected by the plant hormones. GaRPL18 expression was rapidly induced, reaching a peak level at 6 h after treatment, following the application of exogenous SA, but remained at baseline levels following MeJA or ET treatments (Fig. 1e). These results suggest that GaRPL18 enhances the resistance of cotton to V. dahliae, and affects the SA-mediated signaling pathway.
Fig. 1.
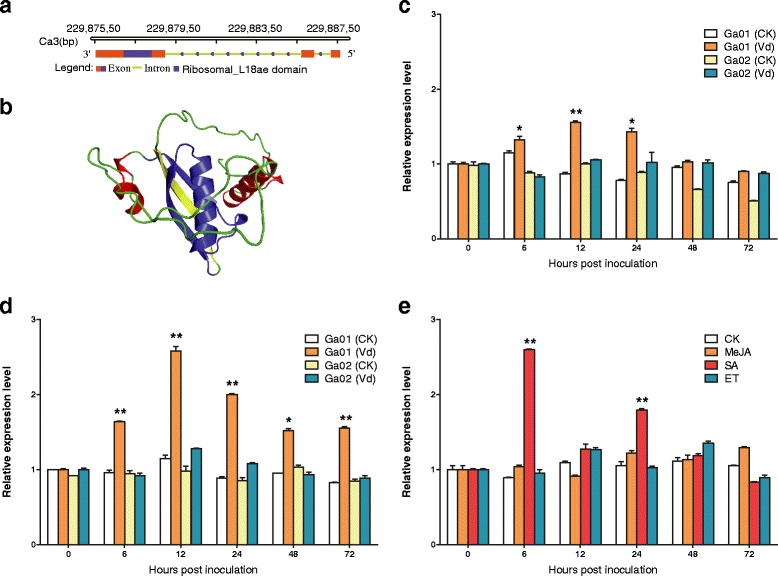
Structural analysis and expression patterns of GaRPL18 under Verticillium dahliae stress or hormone treatment in cotton. a Gene structure analysis. b Protein 3D structure. c,d After seeding for 3 weeks, the cotton seedlings of ‘HuNanChangDeTieZiMian’ (resistant, Ga01) and ‘NaShangQuXiaoHua’ (susceptible, Ga02) were inoculated with V. dahliae spores or an equal sterile distilled water (Mock control). The sample of c is the leaf, while d is root. e The leaves of resistant cotton (‘HuNanChangDeTieZiMian’) were treated by exogenous hormones or an equal sterile distilled water. The Mock and treated roots and leaves were harvested at 0, 6, 12, 24, 48, and 72 hpi, and expression levels were determined by qRT-PCR using Gahiston3 as the internal reference gene. Expression levels of Mock control was normalized to ‘1’, error bars represent the standard deviation of three biological replicates. Asterisks indicate statistically significant differences, as determined by Student’s t-test (*P <0.05; **P <0.001)
Interactions between GaRPL18 and PR genes
To further clarify the effects of GaRPL18 on Verticillium wilt resistance in cotton plants, we monitored the expression levels of PR genes at different time points after wilt-resistant cotton plants were inoculated with V. dahliae. Phenylpropanoid metabolism is critical for cotton defense responses, and involves the core genes phenylalanine ammonia lyase (PAL; Cotton_A_00465) and 4-coumarate: CoA ligase (4CL; Cotton_A_02864) [51]. In leaf tissue, the expression levels of the GaRPL18-related PAL and 4CL genes were highest at 12 h, while the expression of 4CL in the roots peaked after 48 h. Additionally, PAL expression levels continued to increase in inoculated plants. The basic chitinase (Cotton_A_36866) and β-1, 3-glucanase (Cotton_A_36866) genes are important for plant disease resistance because the encoded enzymes can decompose the cell walls of pathogens, thereby protecting plants from disease [27]. In leaves, the expression levels of GaRPL18-related basic chitinase and β-1,3-glucanase genes peaked after 48 h. In contrast, the highest basic chitinase and β-1,3-glucanase expression levels in the roots were observed after 6 h and 24 h, respectively. Cadinene synthase (Cotton_A_34493) is a key gene involved in synthesizing terpenoids, which are critical phytoalexins in cotton [52]. The cadinene synthase expression level rapidly increased in the leaves and roots of inoculated plants, peaking after 48 h (Fig. 2).
Fig. 2.
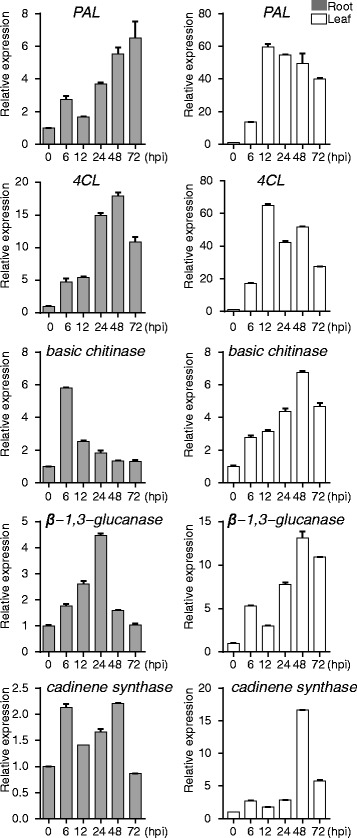
Interactions between GaRPL18 and other pathogenesis related (PR) genes. Time course in the transcription of resistant cotton defense-related pathogenesis related (PR) genes from different time points after inoculation. Total RNAs were extracted from 3-week-old cotton plants after treatment with V. dahliae, and infected roots and leaves were harvested at 0, 6, 12, 24, 48, and 72 hpi. The expression levels of PR gene were determined by qRT-PCR using Gahiston3 as the internal reference gene. Error bars represent the standard deviation of three biological replicates. Asterisks indicate statistically significant differences, as determined by Student’s t-test (*P <0.05; **P <0.001). The left part is the result of root (gray), the right part is the result of leaf (white)
Silencing of GaRPL18 in cotton considerably weakened resistance to Verticillium dahliae
To clarify the function of GaRPL18 in cotton responses against V. dahliae, we used CLCrV-based VIGS to generate GaRPL18-knockdown plants. Two weeks after ‘HuNanChangDeTieZiMian’ plants underwent an Agrobacterium tumefaciens infiltration, we used qRT-PCR to assess the gene silencing efficiency. The abundance of GaRPL18 transcripts was significantly lower in CLCrV:GaRPL18 plants than in the controls (Fig. 3c), indicating that GaRPL18 was effectively silenced in these plants. Additionally, following V. dahliae inoculations, there were no obvious disease symptoms on the tissues of the vector control plants, while necrotic, yellowish, stunted, and wilting leaves were observed on CLCrV:GaRPL18 plants (Fig. 3a). Using a recovery assay, we examined the extent of the V. dahliae colonization in the infected stems of treated plants. This analysis was used to determine the virulence of the fungus for each plant. There were more fungal colonies in CLCrV:GaRPL18 plants than in CLCrV control plants (Fig. 3b). We used DI values to confirm that the resistance of cotton plants to V. dahliae was compromised in the absence of GaRPL18 expression. The DI values were significantly higher for CLCrV:GaRPL18 plants than for the control plants at 20 and 25 days post-inoculation (dpi) (Fig. 3d).
Fig. 3.
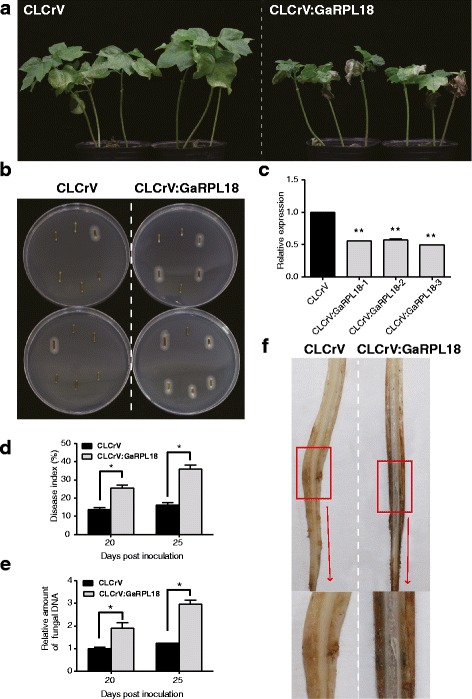
Ribosomal protein GaRPL18 is involved in the resistance of cotton to V. dahliae. a Disease symptoms induced with V. dahliae strain on the leaves of empty-vector control plants and CLCrV:GaRPL18 plants at 25 dpi. b 25-dpi stem sections were plated on PDA medium, incubating at 25 °C. Photos were taken at 7 day after plating. c Transcript levels of GaRPL18 in leaves of empty-vector control plants and CLCrV:GaRPL18 plants, which were determined by qRT-PCR using Gahiston3 as the internal reference gene. Value of CLCrV was normalized to ‘1’. d Assessment of Disease Index (DI) at 20 dpi and 25 dpi. Error bars represent the standard deviation of three biological replicates (n ≥ 30). e The levels of Verticillium dahliae biomass by qRT-PCR on the leaves of empty-vector control plants and CLCrV:GaRPL18 plants at 20 dpi and 25 dpi. Error bars represent the standard deviation of three biological replicates. Asterisks indicate statistically significant differences, as determined by Student’s t-test (*P <0.05; **P <0.001). f The infected CLCrV:GaRPL18 plants showed vascular browning in xylem at 25 dpi
Using qRT-PCR, we quantified V. dahliae colonization levels. The expression levels of V. dahliae genes indicated that CLCrV:GaRPL18 plants were significantly more sensitive to V. dahliae than the controls (Fig. 3e). Moreover, we observed that the xylem of CLCrV:GaRPL18 plants exhibited greater vascular browning than the controls (Fig. 3f). An additional examination of cotton cell morphology in inoculated plants revealed that the vascular bundle cells of CLCrV:GaRPL18 plants were longer and larger than those of control plants (Fig. 4a). This was likely because V. dahliae mycelia may have more easily penetrated CLCrV:GaRPL18 plant than the control. To visually contrast the differences in the leaves of plants inoculated with V. dahliae, we used trypan blue to stain dead cells. The trypan blue staining area was larger and the color was more intense in the CLCrV:GaRPL18 leaves, particularly in the veins, than in the control leaves (Fig. 4b).
Fig. 4.
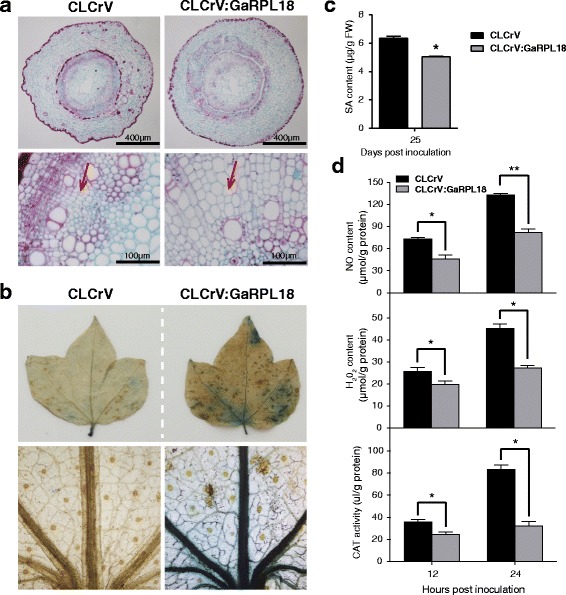
Effect of silencing of GaRPL18 in cotton under V. dahliae stress. a The observation of cell morphology of the stems of CLCrV and CLCrV:GaRPL18 cotton plants at 25 dpi. b Trypan blue staining was used to visualize cell death of the leaves of CLCrV and CLCrV:GaRPL18 cotton plants at 25 dpi. c Measuring the SA content of CLCrV and CLCrV:GaRPL18 cotton plants at 25 dpi via HPLC. Bars represent standard error from three biological replicates. Asterisks indicate statistically significant differences (Student’s t-test; *P <0.05). d Leaves of CLCrV and CLCrV:GaRPL18 cotton plants were harvested at 12 hpi and 24 hpi to measure the contents of NO, H2O2 and CAT. Error bars represent the standard deviation of three biological replicates. Asterisks indicate statistically significant differences when compared with control (Student’s t-test; *P <0.05; **P <0.001)
Salicylic acid, nitric oxide, H2O2, and catalase levels decreased in GaRPL18-silenced cotton plants upon Verticillium dahliae infection
To further evaluate the role of GaRPL18 and its relationship with the SA pathway in cotton defense responses to V. dahliae infection, we analyzed the SA content and the abundance of several other immune-responsive compounds (i.e., NO, H2O2, and CAT) in V. dahliae-treated plants. All compounds were present in lower amounts in inoculated CLCrV:GaRPL18 plants than in CLCrV control plants (Fig. 4c, d). These results indicate that silencing GaRPL18 in wilt-resistant cotton plants inhibits V. dahliae-induced production of SA, NO, H2O2, and CAT, further confirming that GaRPL18 is closely related to the SA signaling pathway.
Transgenic Arabidopsis thaliana seeds overexpressing GaRPL18 increased in size and weight, while seedlings became more resistant to Verticillium dahliae
Arabidopsis thaliana ecotype Col-0 was transformed to generate transgenic lines overexpressing GaRPL18 under the control of the 35S promoter. We then used a variety of methods to select homozygous GaRPL18-overexpressing transgenic lines, including a 0.1% Basta treatment, PCR, and qRT-PCR (Additional file 1: Figure S1a–e). We observed that the seeds of stable GaRPL18-overexpressing T4 plants were larger than those of WT plants (Additional file 1: Figure S1f). Analyses of seed weight per 1,000 mature dried seeds, seed length, seed width, and the length: width ratio revealed that the values were higher in GaRPL18-overexpressing transgenic plants than in WT controls, although this difference was not significant for the length: width ratio (Additional file 1: Figure S1g–i). We then used an in vitro technique to assess the resistance of A. thaliana seedlings to V. dahliae. The GaRPL18-overexpressing transgenic plants were compared with the WT plants daily. As expected, the transgenic A. thaliana plants were more resistant to Verticillium wilt than the WT plants. All WT plants were dead within 14 days of being inoculated with V. dahliae, while the transgenic plants were still alive (Fig. 5a). In WT plants, wilting was observed as early as 5 dpi, and the plants rapidly became yellow and exhibited stunted growth. At 14 dpi, the WT leaves were observed to be more stunted (grade 3) than the transgenic leaves (grade 2). Additionally, chlorosis was more severe in WT plants (grade 4) than in transgenic plants (grade 2) (Fig. 5b, c). This further confirms that the transgenic A. thaliana plants overexpressing GaRPL18 were more resistant to Verticillium wilt than the WT plants.
Fig. 5.
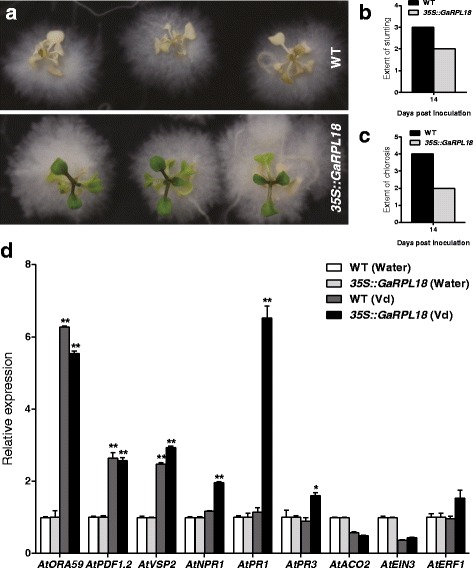
Effects of overexpression of GaGRPL18 in A. thaliana seedings and the effects of signaling pathway-related marker genes under V. dahliae stress. a Disease symptoms of in vitro grown seedings. 2-week-old plantlets were treatment with V. dahliae. Photos were taken at 14 days after inoculated. b Extent of stunting in WT and 35S::GaRPL18 plants at 14 dpi. c Extent of leaf chlorosis in WT and 35S::GaRPL18 plants at 14 dpi. WT : wild-type. d Expression patterns of JA, SA and ET signaling pathway-related marker genes in WT and GaRPL18 transgenic A. thaliana under water or V. dahliae challenge for 24 h. The expression levels was monitored by qRT-PCR using AtUBQ10 as an internal control gene. Expression levels of untreated control was normalized to ‘1’. Error bars represent the standard deviation of three biological replicates. Asterisks indicate statistically significant differences, as determined by Student’s t-test (*P <0.05; **P <0.001). WT: wild-type
Effects of GaRPL18 overexpression on the salicylic acid signaling pathway in Verticillium dahliae-treated transgenic Arabidopsis thaliana plants
We examined the expression level of signaling pathway-related genes, including those involved in JA biosynthesis [i.e., octadecanoid-responsive Arabidopsis AP2/ERF domain protein 59 (AtORA59), plant defensin 1.2 (AtPDF1.2), and vegetative storage protein 2 (AtVSP2)], SA biosynthesis [i.e., non-expressor of pathogenesis-related genes 1 (AtNPR1), pathogenesis-related gene 1 (AtPR1), and pathogenesis-related gene 3 (AtPR3)], and ET biosynthesis [i.e., amino cyclopropane carboxylate oxidase 2 (AtACO2), ethylene insensitive 3 (AtEIN3), and ethylene response factor 1 (AtERF1)]. As shown in Fig. 5D, over-expression of GaRPL18 in the Col-0 background did not affect expression of genes related to the JA, SA and ET pathways; these marker genes had similar expression levels in WT and over-expression plants without V. dahliae stress. In addition, the expression levels of the JA- and ET-related marker genes were similar overall between the WT controls and GaRPL18- overexpressing transgenic A. thaliana plants at 24 h post-inoculation (hpi). However, the SA-related genes were expressed more highly in the rosette leaves of transgenic plants than in the WT leaves at 24 h post-inoculation. The expression levels in the WT leaves were unchanged (Fig. 5d). Thus, GaRPL18 overexpression had little effect on the signal transduction pathways regulating JA and ET biosynthesis, but had a significant effect on the SA biosynthesis signal transduction pathway.
Application of exogenous salicylic acid increases the resistance of transgenic Arabidopsis thaliana plants to Verticillium dahliae
We examined the resistance of GaRPL18-overexpressing transgenic A. thaliana plants to V. dahliae infection. When T4 lines were inoculated with V. dahliae, the transgenic plants were noticeably more resistant to the fungus than the WT plants. We then pretreated the rosette leaves of WT and transgenic A. thaliana seedlings with 1 mM SA for 24 h. The leaves were then inoculated with V. dahliae and treated daily with supplemental exogenous SA for 4 days. The transgenic A. thaliana plants were considerably more resistant to V. dahliae than the WT plants following the SA treatments (Fig. 6a). Additionally, we used the recovery assay to examine the colonization of infected WT and transgenic stems by V. dahliae. The number of colonies differed significantly between the WT, transgenic, and SA-treated transgenic plants. Overall, the transgenic plants grew better than the WT plants, with the SA-treated transgenic plants exhibiting the greatest resistance to V. dahliae. There were no differences between the WT and SA-treated WT plants (Fig. 6b). This is further evidence of the relationship between GaRPL18 and SA. To more clearly observe the effects of V. dahliae on rosette leaves, we removed the rosette leaves from plants at 15 dpi. Consistent with our previous results, the SA treatment enhanced the resistance of transgenic A. thaliana rosette leaves to V. dahliae, but had little effect on WT leaves (Fig. 6c).
Fig. 6.
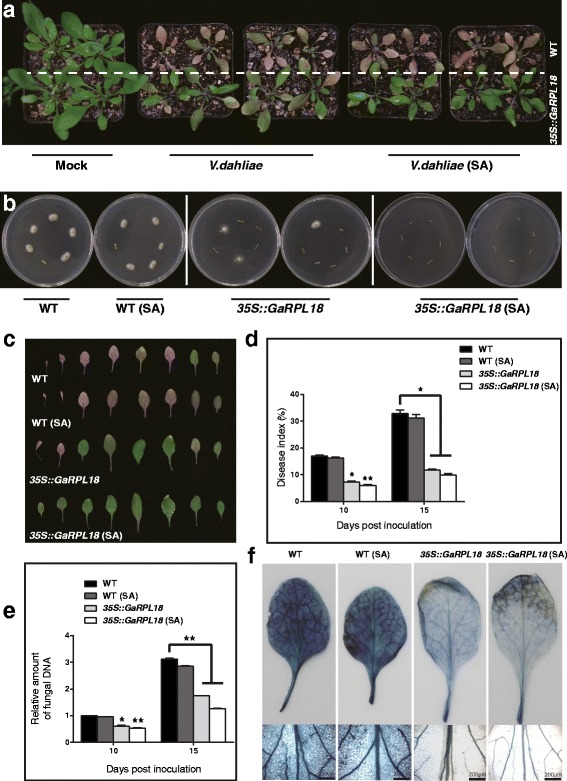
Effects of -overexpression of GaRPL18 and exogenous SA in defense response to V. dahliae in A. thaliana. a Disease symptoms induced with V. dahliae strain on the rosette leaves of WT, GaRPL18 overexpressed plants and exogenous SA-treated plants. 3-week-old seedings were inoculated with V. dahliae, and pretreated transgenic A. thaliana and wild-type seedlings rosette leaves with 1 mM SA for 24 h before inoculation with V. dahliae. Then, supplementing exogenous SA every day until four days. The control plants were sprayed with water. Photos were taken at 15 dpi. b 15-dpi stem sections were plated on PDA medium, incubating at 25 °C. Photographed at 7 day after plating. c The observation of A. thaliana rosette leaves. d Assessment of Disease Index (DI) at 10 dpi and 15 dpi. Error bars represent the standard deviation of three biological replicates (n ≥ 30). Asterisks indicate statistically significant differences when compared with control (Student’s t-test; *P <0.05; **P <0.001). e Quantification the biomass of V. dahliae by qRT-PCR using AtUBQ10 as an internal control gene on the rosette leaves of WT, GaRPL18 overexpressed plants and SA-treated plants at 10 dpi and 15dpi. Error bars represent the standard deviation of three biological replicates. Asterisks indicate statistically significant differences, as determined by Student’s t-test (*P <0.05; **P <0.001). f Trypan blue staining was used to visualize cell death of the leaves of A. thaliana at 15 dpi
We calculated the DI values and quantified the V. dahliae colonization using qRT-PCR for WT plants, SA-treated WT plants, transgenic plants, and SA-treated transgenic plants. The DI value for WT plants was significantly higher than that of transgenic plants, including those treated with SA (Fig. 6d). Analyses of the expression level of fungal genes revealed that SA-treated transgenic plants were the most disease-resistant, while WT plants were the most susceptible (Fig. 6e). The trypan blue staining results were similar for WT and SA-treated WT plants. We observed dark blue veins and large stained mesophyll areas. The veins of transgenic leaves were light blue, and the mesophyll was not extensively stained. These observations are consistent with the greater disease resistance of SA-treated transgenic plants (Fig. 6f).
Discussion
Verticillium wilt is one of the most lethal fungal diseases of plants and significantly reduces the quality and annual yields of cotton. Therefore, clarifying the molecular mechanism underlying cotton resistance to V. dahliae is essential for developing new wilt-resistant cotton varieties. Previous studies involving V. dahliae infections revealed that this pathogen attacks susceptible plants by germinating on the roots. It then employs various mechanisms to move through the root cortex into the plant vascular system, allowing for further colonization of the plant. When the fungus has completely infected the plant, it produces microsclerotial spores that remain stable and dormant in the soil until they are exposed to a new host [53]. To breed new cotton varieties resistant to Verticillium wilt, more research is required to identify resistance-related genes. Technological improvements have facilitated the mining and functional analysis of genes. There are some reports that describe the application of genes encoding proteins associated with resistance to V. dahliae to develop transgenic A. thaliana plants capable of inhibiting disease development. These genes include those encoding glutathione S-transferase [54, 55], rate-limiting enzymes [56], and transcriptional regulators [57]. However, efforts involving gene mining have been insufficient, and the cotton-producing regions of China are still affected by annual outbreaks of Verticillium wilt. The lack of known wilt-resistance genes in cotton makes the identification and functional characterization of V. dahliae resistance-related genes essential for the development of new wilt-resistant cotton varieties. In this study, we confirmed that GaRPL18, which encodes an RP, influences resistance to Verticillium wilt.
The function and characteristics of RPs during protein synthesis have been thoroughly researched, and recent in-depth studies confirmed that RPs also have important roles in other processes [20–22]. In particular, some studies have shown PRs play an important role in plant disease resistance [23, 24]. Recent cancer studies concluded that the RPs can regulate the expression of key genes in the MDM2–p53 regulatory loop, thereby promoting p53 activity and suppressing tumor growth [25, 26, 58–60]. Additionally, the RP has other effects related to disease resistance. For example, mutations to RPS19 are associated with a congenital erythroblastopenia with a decreased abundance or lack of erythroid precursors [61]. However, little is known about the function and characteristics of RP-encoding genes during cotton resistance to V. dahliae. In this study, we determined that the RP mediates the resistance of cotton plants to V. dahliae.
To confirm the role of GaRPL18 in cotton pathogen defense responses, we cloned the gene using cDNA produced from RNA extracted from wilt-resistant cotton plants. We also analyzed the GaRPL18 expression patterns in different cotton varieties treated with V. dahliae. The GaRPL18 expression level was rapidly and considerably upregulated in infected wilt-resistant cotton plants, but was relatively unchanged in the wilt-susceptible cotton plants. We then examined the expression of PR genes in wilt-resistant cotton plants. We confirmed that cotton resistance to V. dahliae results from interactions among various disease resistance systems that function together to resist pathogen invasions. Using VIGS technology, we determined that the silencing of GaRPL18 in wilt-resistant cotton plants significantly decreased resistance to V. dahliae. Additionally, the NO, H2O2, and CAT contents were lower in cotton plants in which GaRPL18 was silenced than in vector-control plants. An invasion by V. dahliae induces a spike in NO content, which is important for disease resistance in plants [62]. An earlier study concluded that the H2O2 content is significantly higher in disease-resistant tomato plants than in control plants, and that CAT plays an important role in early defense responses [63]. Our results indicate that NO, H2O2, and CAT affect the GaRPL18- related responses to V. dahliae in cotton plants. These observations confirm that GaRPL18 is important for cotton resistance to V. dahliae, which is consistent with the anti-carcinogenic effects reported for other RPs. We used transgenic A. thaliana plants to further analyze V. dahliae disease resistance traits based on DI values, V. dahliae colonization, trypan blue staining, and recovery assays. Moreover, while the rosette leaves of WT plants gradually exhibited disease symptoms with increasing fungal growth and spread, almost no disease symptoms were observed on the rosette leaves of GaRPL18-overexpressing transgenic plants. This implies that the fungus was inhibited in the roots of the transgenic plants, and prevented from spreading into the rosette leaves. Additionally, in vitro analyses of the wilt-resistance of GaRPL18-overexpressing A. thaliana plants further confirmed the role of GaRPL18 in plant defense responses to V. dahliae. This suggests that the accumulation of GaRPL18 considerably impedes pathogen colonization, even at the seedling stage. In summary, we confirmed that GaRPL18-overexpressing transgenic plants were more resistant to V. dahliae than WT plants, and that the extent of V. dahliae colonization was significantly lower in transgenic plants than in WT plants.
We also observed that GaRPL18 expression was rapidly upregulated by SA, reaching peak levels 6 h after the application of the hormone. In contrast, MeJA and ET treatments had minimal effects on GaRPL18 expression. Therefore, we hypothesized that GaRPL18 may be associated with the SA signaling pathway. This was verified by a significant decrease in SA levels in V. dahliae-infected CLCrV:GaRPL18 plants. The activation of SA signaling in stressed plants can stimulate the expression of downstream disease-resistance genes to provide protection from pathogens [64, 65]. There are many genes related to the SA signaling pathway, including NPR1, PR1, PR3, WRKYs, and AtGSTF6 [66–69]. To verify these results, the JA-, SA- and ET-related marker gene expression levels were examined in V. dahliae-infected transgenic A. thaliana plants overexpressing GaRPL18. The upregulated expression of SA signaling pathway-related genes (i.e., AtNPR1, AtPR1, and AtPR3) and SA content confirmed our hypothesis. To further clarify the link between GaRPL18 and SA signaling, we treated the transgenic and WT plants with exogenous SA, and observed that the hormone significantly increased the resistance of transgenic plants to V. dahliae. These findings imply that GaRPL18, working in concert with the SA signaling pathway, has a strongly antimicrobial effect on V. dahliae. Therefore, GaRPL18 may be useful for breeding Verticillium wilt-resistant cotton varieties.
Conclusions
To the best of our knowledge, this study is the first to examine the RP function related to cotton resistance to V. dahliae. We used VIGS technology to confirm that GaRPL18 is important for the resistance of cotton plants to V. dahliae infections. Our data also suggest that breeding new cultivars that overexpress GaRPL18 may be an effective way to control Verticillium wilt of cotton plants. Finally, this study revealed that SA is an important factor related to the cotton defense response system, and that the mechanism of GaRPL18-associated V. dahliae resistance is related to the SA signaling pathway.
Acknowledgements
We thank the members of Professor Zhu’s groups (Institute of Cotton Research, Chinese Academy of Agricultural Sciences) for providing the V. dahliae isolate and their technical assistance.
Funding
This work was supported by The Major Program of Joint Funds (Sinkiang) of the National Natural Science Foundation of China (Grant No. U1303282) and the Zhengzhou Science and Technology Program (Grant No. 153PXXCY180). The funding bodies were not involved in designing the study, the collection, analysis, and interpretation of data, or the preparation of the manuscript.
Availability of data and materials
All data generated or analyzed during this study are included in this published article and its supplementary information files.
Authors’ contributions
Conceived and designed the experiments: QG, ZY, SH, and FL. Conducted the experiments: QG and ZY. Analyzed the data: QG and ZY. Contributed reagents/materials/analysis tools: QW, HIB, JZ, YZ, and EC. Wrote the manuscript: QG. All authors have read and approved the current version of the manuscript.
Competing interests
The authors declare that there are no competing interests.
Consent for publication
Not applicable.
Ethics approval and consent to participate
Not applicable.
Abbreviations
- 4CL
4-coumarate: CoA ligase
- ACO2
Amino cyclopropane carboxylate oxidase 2
- CAT
Catalase
- CLCrV
Cotton leaf crumple virus
- DI
Disease index
- EIN3
Ethylene insensitive 3
- ERF1
Ethylene response factor 1
- ET
Ethylene
- JA
Jasmonic acid
- MDM2
Murine double minute 2
- MeJA
Methyl jasmonate
- NO
Nitric oxide
- NPR1
Nonexpressor of pathogenesis-related gene 1
- ORA59
Octadecanoid-responsive Arabidopsis AP2/ERF domain protein 59
- PAL
Phenylalanine ammonia lyase
- PDF1.2
Plant defensin 1.2
- PR
Pathogenesis related
- RP
Ribosomal protein
- SA
Salicylic acid
- VIGS
Virus-induced gene silencing
- VSP2
Vegetative storage protein 2
- WT
Wild-type
Additional file
Screening process of stable transgenic T4 lines and the effects of over-expressing GaRPL18 in A. thaliana. a, b, c The process of product homozygous T3 lines. d Detection of positive lines. e The expression levels of GaRPL18 were determined by qRT-PCR using AtUBQ10 as an internal control gene. Error bars represent the standard deviation of three biological replicates. f The observation of seeds of GaRPL18-Transgenic A. thaliana and WT. g Seed weight per 1,000 mature dried seeds. h Seed length and seed width. i The ratio of length to width. (PDF 9473 kb)
Contributor Information
Qian Gong, Email: 15201609648@163.com.
Zhaoen Yang, Email: yangzhaoen0925@126.com.
Xiaoqian Wang, Email: aywxiaoqian@163.com.
Hamama Islam Butt, Email: hambutt80@hotmail.com.
Eryong Chen, Email: lovelycheneryong@163.com.
Shoupu He, Email: zephyr0911@126.com.
Chaojun Zhang, Email: Zcj1999@yeah.net.
Xueyan Zhang, Email: zhangxueyan_caas@126.com.
Fuguang Li, Phone: 86-372-2562256, Email: aylifug@163.com.
References
- 1.Carrero-Carrón I, Trapero-Casas JL, Olivares-García C, Monte E, Hermosa R, Jiménez-Díaz RM. Trichoderma asperellum is effective for biocontrol of Verticillium wilt in olive caused by the defoliating pathotype of Verticillium dahliae. Crop Prot. 2016;88:45–52. doi: 10.1016/j.cropro.2016.05.009. [DOI] [Google Scholar]
- 2.Wei F, Passey T, Xu X. Effects of individual and combined use of bio-fumigation-derived products on the viability of Verticillium dahliae microsclerotia in soil. Crop Prot. 2016;79:170–176. doi: 10.1016/j.cropro.2015.09.008. [DOI] [Google Scholar]
- 3.Gerik J HO. Study of field-grown cotton roots infected with Verticillium dahliae using an immunoenzymatic staining technique. Phytopathol Ecol Epidemiol. 1988;78(9):1174–78.
- 4.Jiménez-Díaz RM, Cirulli M, Bubici G, del Mar Jiménez-Gasco M, Antoniou PP, Tjamos EC. Verticillium wilt, a major threat to olive production: current status and future prospects for its management. Plant Dis. 2012;96(3):304–329. doi: 10.1094/PDIS-06-11-0496. [DOI] [PubMed] [Google Scholar]
- 5.Xu L, Zhu L, Tu L, Guo X, Long L, Sun L, Gao W, Zhang X. Differential gene expression in cotton defence response to Verticillium dahliae by SSH. J Phytopathol. 2011;159(9):606–615. doi: 10.1111/j.1439-0434.2011.01813.x. [DOI] [Google Scholar]
- 6.Cheng H-Q, Han L-B, Yang C-L, Wu X-M, Zhong N-Q, Wu J-H, Wang F-X, Wang H-Y, Xia G-X. The cotton MYB108 forms a positive feedback regulation loop with CML11 and participates in the defense response against Verticillium dahliae infection. J Exp Bot. 2016;67(6):1935–1950. doi: 10.1093/jxb/erw016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fradin EF, Thomma BP. Physiology and molecular aspects of Verticillium wilt diseases caused by V. Dahliae and V. Albo-atrum. Mol Plant Pathol. 2006;7(2):71–86. doi: 10.1111/j.1364-3703.2006.00323.x. [DOI] [PubMed] [Google Scholar]
- 8.Cai HX Y, Mo J, Sun Q, Yang J, Liu J. Molecular research and genetic engineering of resistance to Verticillium wilt in cotton: a review. Afr J Biotechnol. 2009;8(25):7363–7372. [Google Scholar]
- 9.Zhang W, Zhang H, Qi F, Jian G. Generation of transcriptome profiling and gene functional analysis in Gossypium hirsutum upon Verticillium dahliae infection. Biochem Biophys Res Commun. 2016;473(4):879–885. doi: 10.1016/j.bbrc.2016.03.143. [DOI] [PubMed] [Google Scholar]
- 10.Han Q, Wu F, Wang X, Qi H, Shi L, Ren A, Liu Q, Zhao M, Tang C. The bacterial lipopeptide iturins induce Verticillium dahliae cell death by affecting fungal signalling pathways and mediate plant defence responses involved in pathogen-associated molecular pattern-triggered immunity. Environ Microbiol. 2015;17(4):1166–1188. doi: 10.1111/1462-2920.12538. [DOI] [PubMed] [Google Scholar]
- 11.Liu S-S, Rao A, Bradleigh Vinson S. Biological control in china: past, present and future – an introduction to this special issue. Biol Control. 2014;68:1–5. doi: 10.1016/j.biocontrol.2013.05.005. [DOI] [Google Scholar]
- 12.Zeng H, Chen R, Luo X, Tian J. Isolation and anti-Verticillium dahliae activity from Bacillus axarquiensis TUBP1 protein. Process Biochem. 2016;51(10):1691–98.
- 13.Zhang J, Fang H, Zhou H, Sanogo S, Ma Z. Genetics, breeding, and marker-assisted selection for Verticillium wilt resistance in cotton. Crop Sci. 2014;54(4):1289. doi: 10.2135/cropsci2013.08.0550. [DOI] [Google Scholar]
- 14.Miao W, Wang X, Li M, Song C, Wang Y, Hu D, Wang J. Genetic transformation of cotton with a harpin-encoding gene hpaXoo confers an enhanced defense response against different pathogens through a priming mechanism. BMC Plant Biol. 2010;10:67. doi: 10.1186/1471-2229-10-67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Parkhi V, Kumar V, Campbell LM, Bell AA, Shah J, Rathore KS. Resistance against various fungal pathogens and reniform nematode in transgenic cotton plants expressing Arabidopsis NPR1. Transgenic Res. 2010;19(6):959–975. doi: 10.1007/s11248-010-9374-9. [DOI] [PubMed] [Google Scholar]
- 16.Li F, Shen H, Wang M, Fan K, Bibi N, Ni M, Yuan S, Wang X. A synthetic antimicrobial peptide BTD-S expressed in Arabidopsis thaliana confers enhanced resistance to Verticillium dahliae. Mol Gen Genet MGG. 2016;291(4):1647–1661. doi: 10.1007/s00438-016-1209-9. [DOI] [PubMed] [Google Scholar]
- 17.Tollervey JVD. Ribosome synthesis IN saccharomyces cerevisiae. Annu Rev Genet. 1999;33:261–311. doi: 10.1146/annurev.genet.33.1.261. [DOI] [PubMed] [Google Scholar]
- 18.Belknap JEGWR. Isolation of a ubiquitin-ribosomal protein gene (ubi3) from potato and expression of its promoter in transgenic plants. Plant Mol Biol. 1994;24:119–127. doi: 10.1007/BF00040579. [DOI] [PubMed] [Google Scholar]
- 19.Perry RP. Balanced production of ribosomal proteins. Gene. 2007;401(1-2):1–3. doi: 10.1016/j.gene.2007.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Barakat A, Szick-Miranda K, Chang IF, Guyot R, Blanc G, Cooke R, Delseny M, Bailey-Serres J. The organization of cytoplasmic ribosomal protein genes in the Arabidopsis genome. Plant Physiol. 2001;127(2):398–415. doi: 10.1104/pp.010265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rogalski M, Schottler MA, Thiele W, Schulze WX, Bock R. Rpl33, a nonessential plastid-encoded ribosomal protein in tobacco, is required under cold stress conditions. Plant Cell. 2008;20(8):2221–2237. doi: 10.1105/tpc.108.060392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Petibon C, Parenteau J, Catala M, Elela SA. Introns regulate the production of ribosomal proteins by modulating splicing of duplicated ribosomal protein genes. Nucleic Acids Res. 2016;44(8):3878–3891. doi: 10.1093/nar/gkw140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tumer RDNE. Expression of a truncated form of ribosomal protein L3 confers resistance to pokeweed antiviral protein and the Fusarium mycotoxin deoxynivalenol. Mol Plant-Microbe Interact. 2005;18(8):762–770. doi: 10.1094/MPMI-18-0762. [DOI] [PubMed] [Google Scholar]
- 24.Nagaraj S, Senthil-Kumar M, Ramu VS, Wang K, Mysore KS. Plant ribosomal proteins, RPL12 and RPL19, play a role in nonhost disease resistance against bacterial pathogens. Front Plant Sci. 2015;6:1192. doi: 10.3389/fpls.2015.01192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhou X, Hao Q, Liao J, Zhang Q, Lu H. Ribosomal protein S14 unties the MDM2–p53 loop upon ribosomal stress. Oncogene. 2012;32(3):388–396. doi: 10.1038/onc.2012.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.He X, Li Y, Dai M-S, Sun X-X. Ribosomal protein L4 is a novel regulator of the MDM2-p53 loop. Oncotarget. 2016;7(13):16217–226. [DOI] [PMC free article] [PubMed]
- 27.Bu B, Qiu D, Zeng H, Guo L, Yuan J, Yang X. A fungal protein elicitor PevD1 induces Verticillium wilt resistance in cotton. Plant Cell Rep. 2014;33(3):461–470. doi: 10.1007/s00299-013-1546-7. [DOI] [PubMed] [Google Scholar]
- 28.Eva Häffner PK, Richard S, Anna T, Elke D. ERECTA, salicylic acid, abscisic acid, and jasmonic acid modulate quantitative disease resistance of Arabidopsis thaliana to Verticillium longisporum. BMC Plant Biol. 2014;14(85):1471–2229. doi: 10.1186/1471-2229-14-85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lorenzo O, Solano R. Molecular players regulating the jasmonate signalling network. Curr Opin Plant Biol. 2005;8(5):532–540. doi: 10.1016/j.pbi.2005.07.003. [DOI] [PubMed] [Google Scholar]
- 30.Glazebrook J. Contrasting mechanisms of defense against biotrophic and necrotrophic pathogens. Annu Rev Phytopathol. 2005;43:205–227. doi: 10.1146/annurev.phyto.43.040204.135923. [DOI] [PubMed] [Google Scholar]
- 31.Spoel SH, Johnson JS, Dong X. Regulation of tradeoffs between plant defenses against pathogens with different lifestyles. Proc Natl Acad Sci U S A. 2007;104(47):18842–18847. doi: 10.1073/pnas.0708139104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Balbi V, Devoto A. Jasmonate signalling network in Arabidopsis thaliana: crucial regulatory nodes and new physiological scenarios. New Phytol. 2008;177(2):301–318. doi: 10.1111/j.1469-8137.2007.02292.x. [DOI] [PubMed] [Google Scholar]
- 33.Grant M, Lamb C. Systemic immunity. Curr Opin Plant Biol. 2006;9(4):414–420. doi: 10.1016/j.pbi.2006.05.013. [DOI] [PubMed] [Google Scholar]
- 34.Sang-Wook Park EK, Dhirendra K, Stephen M, Klessig DF. Methyl salicylate is a critical mobile signal for plant systemic acquired resistance. Science. 2008;318(5):113–116. doi: 10.1126/science.1147113. [DOI] [PubMed] [Google Scholar]
- 35.Sappl PG, Oa-Sn L, Singh KB, Harvey Millar A. Proteomic analysis of glutathione S-transferases of Arabidopsis thaliana reveals differential salicylic acid-induced expression of the plant-specific phi and tau classes. Plant Mol Biol. 2004;54:205–219. doi: 10.1023/B:PLAN.0000028786.57439.b3. [DOI] [PubMed] [Google Scholar]
- 36.Bari R, Jones JD. Role of plant hormones in plant defence responses. Plant Mol Biol. 2009;69(4):473–488. doi: 10.1007/s11103-008-9435-0. [DOI] [PubMed] [Google Scholar]
- 37.Caarls L, Pieterse CM, Van Wees SC. How salicylic acid takes transcriptional control over jasmonic acid signaling. Front Plant Sci. 2015;6:170. doi: 10.3389/fpls.2015.00170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fradin EF, Zhang Z, Juarez Ayala JC, Castroverde CD, Nazar RN, Robb J, Liu CM, Thomma BP. Genetic dissection of Verticillium wilt resistance mediated by tomato Ve1. Plant Physiol. 2009;150(1):320–332. doi: 10.1104/pp.109.136762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Li A, Zhang R, Pan L, Tang L, Zhao G, Zhu M, Chu J, Sun X, Wei B, Zhang X, et al. Transcriptome analysis of H2O2-treated wheat seedlings reveals a H2O2-responsive fatty acid desaturase gene participating in powdery mildew resistance. PLoS One. 2011;6(12):e28810. doi: 10.1371/journal.pone.0028810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Veronese P, Narasimhan ML, Stevenson RA, Zhu J-K, Weller SC, Subbarao KV, Bressan RA. Identification of a locus controllingVerticilliumdisease symptom response inArabidopsis thaliana. Plant J. 2003;35(5):574–587. doi: 10.1046/j.1365-313X.2003.01830.x. [DOI] [PubMed] [Google Scholar]
- 41.Tuttle JR, Idris AM, Brown JK, Haigler CH, Robertson D. Geminivirus-mediated gene silencing from Cotton leaf crumple virus is enhanced by low temperature in cotton. Plant Physiol. 2008;148(1):41–50. doi: 10.1104/pp.108.123869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gu Z, Huang C, Li F, Zhou X. A versatile system for functional analysis of genes and microRNAs in cotton. Plant Biotechnol J. 2014;12(5):638–649. doi: 10.1111/pbi.12169. [DOI] [PubMed] [Google Scholar]
- 43.Clough SJBA. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998;16(6):735–743. doi: 10.1046/j.1365-313x.1998.00343.x. [DOI] [PubMed] [Google Scholar]
- 44.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25(4):402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 45.Atallah ZK, Bae J, Jansky SH, Rouse DI, Stevenson WR. Multiplex real-time quantitative PCR to detect and quantify Verticillium dahliae colonization in potato lines that differ in response to Verticillium wilt. Phytopathology. 2007;97(7):865–872. doi: 10.1094/PHYTO-97-7-0865. [DOI] [PubMed] [Google Scholar]
- 46.Julia Dewdney TLR, Mary C. Wildermuth: three unique mutants of Arabidopsis identify eds loci required for limiting growth of a biotrophic fungal pathogen. Plant J. 2000;24(2):205–218. doi: 10.1046/j.1365-313x.2000.00870.x. [DOI] [PubMed] [Google Scholar]
- 47.Choi DS, Hwang BK. Proteomics and functional analyses of pepper abscisic acid-responsive 1 (ABR1), which is involved in cell death and defense signaling. Plant Cell. 2011;23(2):823–842. doi: 10.1105/tpc.110.082081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhang Y, Wang X, Yang S, Chi J, Zhang G, Ma Z. Cloning and characterization of a Verticillium wilt resistance gene from Gossypium barbadense and functional analysis in Arabidopsis thaliana. Plant Cell Rep. 2011;30(11):2085–2096. doi: 10.1007/s00299-011-1115-x. [DOI] [PubMed] [Google Scholar]
- 49.Wang YQ, Chen DJ, Wang DM, Huang QS, Yao ZP, Liu FJ, Wei XW, Li RJ. Over-expression of Gastrodia anti-fungal protein enhances Verticillium wilt resistance in coloured cotton. Plant Breed. 2004;123:454–459. doi: 10.1111/j.1439-0523.2004.01005.x. [DOI] [Google Scholar]
- 50.Zhang B, Yang Y, Chen T, Yu W, Liu T, Li H, Fan X, Ren Y, Shen D, Liu L, et al. Island cotton Gbve1 gene encoding a receptor-like protein confers resistance to both defoliating and non-defoliating isolates of Verticillium dahliae. PLoS One. 2012;7(12):e51091. doi: 10.1371/journal.pone.0051091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Xu L, Zhu L, Tu L, Liu L, Yuan D, Jin L, Long L, Zhang X. Lignin metabolism has a central role in the resistance of cotton to the wilt fungus Verticillium dahliae as revealed by RNA-Seq-dependent transcriptional analysis and histochemistry. J Exp Bot. 2011;62(15):5607–5621. doi: 10.1093/jxb/err245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ma D, Hu Y, Yang C, Liu B, Fang L, Wan Q, Liang W, Mei G, Wang L, Wang H, et al. Genetic basis for glandular trichome formation in cotton. Nat Commun. 2016;7:10456. doi: 10.1038/ncomms10456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Klimes A, Dobinson KF, Thomma BP, Klosterman SJ. Genomics spurs rapid advances in our understanding of the biology of vascular wilt pathogens in the genus Verticillium. Annu Rev Phytopathol. 2015;53:181–198. doi: 10.1146/annurev-phyto-080614-120224. [DOI] [PubMed] [Google Scholar]
- 54.Nutricati E, Miceli A, Blando F, De Bellis L. Characterization of two Arabidopsis thaliana glutathione S-transferases. Plant Cell Rep. 2006;25(9):997–1005. doi: 10.1007/s00299-006-0146-1. [DOI] [PubMed] [Google Scholar]
- 55.Wisser RJ, Kolkman JM, Patzoldt ME, Holland JB, Yu J, Krakowsky M, Nelson RJ, Balint-Kurti PJ. Multivariate analysis of maize disease resistances suggests a pleiotropic genetic basis and implicates a GST gene. Proc Natl Acad Sci U S A. 2011;108(18):7339–7344. doi: 10.1073/pnas.1011739108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Mo HJ, Sun YX, Zhu XL, Wang XF, Zhang Y, Yang J, Yan GJ, Ma ZY. Cotton S-adenosylmethionine decarboxylase-mediated spermine biosynthesis is required for salicylic acid- and leucine-correlated signaling in the defense response to Verticillium dahliae. Planta. 2016;243(4):1023–1039. doi: 10.1007/s00425-015-2463-5. [DOI] [PubMed] [Google Scholar]
- 57.Meng X, Li F, Liu C, Zhang C, Wu Z, Chen Y. Isolation and characterization of an ERF transcription factor gene from cotton (Gossypium barbadense L.) Plant Mol Biol Report. 2009;28(1):176–183. doi: 10.1007/s11105-009-0136-x. [DOI] [Google Scholar]
- 58.Bai D, Zhang J, Xiao W, Zheng X. Regulation of the HDM2-p53 pathway by ribosomal protein L6 in response to ribosomal stress. Nucleic Acids Res. 2014;42(3):1799–1811. doi: 10.1093/nar/gkt971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zhu Y, Poyurovsky MV, Li Y, Biderman L, Stahl J, Jacq X, Prives C. Ribosomal protein S7 is both a regulator and a substrate of MDM2. Mol Cell. 2009;35(3):316–326. doi: 10.1016/j.molcel.2009.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Chen J, Guo K, Kastan MB. Interactions of nucleolin and ribosomal protein L26 (RPL26) in translational control of human p53 mRNA. J Biol Chem. 2012;287(20):16467–16476. doi: 10.1074/jbc.M112.349274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Natalia Draptchinskaia PG, Björn Andersson, Monica Pettersson, Thiébaut-Noël Willig,, Irma Dianzani SB, Gil Tchernia, Joakim Klar1, Hans Matsson, Dimitri Tentler,, Narla Mohandas BCND. The gene encoding ribosomal protein S19 is mutated in Diamond-Blackfan anaemia. nature genetics. 1999;21(2):169–75. [DOI] [PubMed]
- 62.Fu-Mei Shi Y-ZL. Verticillium dahliae toxins-induced nitric oxide production in Arabidopsis is major dependent on nitrate reductase. BMC Rep. 2008;41(1):79–85. [DOI] [PubMed]
- 63.Gayoso C, Pomar F, Novo-Uzal E, Merino F, de Ilarduya OM. The Ve-mediated resistance response of the tomato to Verticillium dahliae involves H2O2, peroxidase and lignins and drives PAL gene expression. BMC Plant Biol. 2010;10:232. doi: 10.1186/1471-2229-10-232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Pieterse CM, Leon-Reyes A, Van der Ent S, Van Wees SC. Networking by small-molecule hormones in plant immunity. Nat Chem Biol. 2009;5(5):308–316. doi: 10.1038/nchembio.164. [DOI] [PubMed] [Google Scholar]
- 65.Zhang Y, Wang XF, Ding ZG, Ma Q, Zhang GR, Zhang SL, Li ZK, Wu LQ, Zhang GY, Ma ZY. Transcriptome profiling of Gossypium barbadense inoculated with Verticillium dahliae provides a resource for cotton improvement. BMC Genomics. 2013;14(637):2–18. doi: 10.1186/1471-2164-14-637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Dubreuil-Maurizi C, Vitecek J, Marty L, Branciard L, Frettinger P, Wendehenne D, Meyer AJ, Mauch F, Poinssot B. Glutathione deficiency of the Arabidopsis mutant pad2-1 affects oxidative stress-related events, defense gene expression, and the hypersensitive response. Plant Physiol. 2011;157(4):2000–2012. doi: 10.1104/pp.111.182667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Herrera-Vasquez A, Salinas P, Holuigue L. Salicylic acid and reactive oxygen species interplay in the transcriptional control of defense genes expression. Front Plant Sci. 2015;6:171. doi: 10.3389/fpls.2015.00171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Manohar M, Tian M, Moreau M, Park SW, Choi HW, Fei Z, Friso G, Asif M, Manosalva P, von Dahl CC, et al. Identification of multiple salicylic acid-binding proteins using two high throughput screens. Front Plant Sci. 2014;5:777. doi: 10.3389/fpls.2014.00777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ying X-B, Dong L, Zhu H, Duan C-G, Du Q-S, Lv D-Q, Fang Y-Y, Garcia JA, Fang R-X, Guo H-S. RNA-dependent RNA polymerase 1 fromNicotiana tabacumSuppresses RNA silencing and enhances viral infection inNicotiana benthamiana. Plant Cell. 2010;22(4):1358–1372. doi: 10.1105/tpc.109.072058. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data generated or analyzed during this study are included in this published article and its supplementary information files.


