Abstract
HECT E3 ubiquitin ligases (~28 known) are associated with many phenotypes in eukaryotes, and are important drug targets. However, assays used to screen for small molecule inhibitors of HECT E3s are complex and require ATP, Ub, E1, E2, and HECT E3 enzymes, producing three covalent thioester enzyme intermediates E1~Ub, E2~Ub, and HECT E3~Ub (~ indicates thioester bond), and mixtures of polyubiquitin chains. To reduce the complexity of the assay, we developed a novel class of fluorescent probes UbFluor that act as mechanistically relevant pseudosubstrates of HECT E3s. These probes undergo a direct transthiolation reaction with the catalytic cysteine of HECT E3s, producing the catalytically active HECT E3~Ub thioester accompanied by fluorophore release. Thus, a fluorescence polarization assay can continuously monitor UbFluor consumption by HECT E3s, and changes in UbFluor consumption rendered by biochemical point mutations or small molecule modulation of HECT E3 activity.
Keywords: ubiquitin, HECT ligase, fluorescence polarization, thioester
INTRODUCTION
Throughout the human body, over 800 ubiquitin enzymes control the timely degradation and activity of proteins involved in all aspects of eukaryotic biology (Ciechanover, 2013; Varshavsky, 2012). Homologous to E6AP Carboxyl Terminus (HECT)-type ubiquitin ligases (~28 known in humans) harbor a catalytic cysteine and form an obligatory HECT E3~Ub thioester enzyme intermediate prior to ubiquitin transfer onto lysine residues of protein substrates (Huibregtse et al., 1995; Scheffner et al., 1995). Typical HECT E3 ligase is defined by a ~350 amino acid C-terminal catalytic HECT domain. Genetic inactivation and mutations to HECT E3s cause a wide range of abnormal and disease-related phenotypes in animals and humans, which highlights their significance in eukaryotic biology (Scheffner and Kumar, 2014). Some HECT E3s such as HERC2 are also involved in coloration of human eyes, skin, and hair (Wilde et al., 2014). Furthermore, HECT E3 ligases are also hijacked by viruses that redirect HECT substrate specificity such as when E6AP ligase targets p53 in HPV18 and HPV16 positive cervical cancer cells (Scheffner et al., 1993a).
Given the significance of HECT E3s in eukaryotic and human biology, two challenges need to be addressed: a) downstream substrates of HECT E3s need to be identified to elucidate HECT-regulated signaling pathways (Persaud and Rotin, 2011), and b) selective chemical probes for HECT E3 ligases are needed to validate these enzymes as drug targets and to link HECT E3 enzyme function and cell phenotype (Kathman et al., 2015; Zhang et al., 2016). Both of these challenges can be addressed by developing selective small molecule inhibitors of HECT E3s. To meet the first challenge, quantitative proteomic methods can identify direct downstream substrates of HECT E3s upon their inhibition with small molecules (Kim et al., 2011; Ordureau et al., 2015).
One challenge associated with the discovery of HECT E3 ligase inhibitors is the complexity of enzymatic assays that require ATP, Ub, E1, E2, and HECT E3 enzymes. The reaction is further complicated by the formation of three covalent intermediates E1~Ub, E2~Ub, and HECT E3~Ub, and potential mixtures of autoubiquitinated HECT E3s and free polyubiquitin chains of varying lengths (Ronchi and Haas, 2012; Scheffner et al., 1995). Altogether, this complexity makes enzymatic assays to study HECT E3 biochemistry or high-throughput screens operationally difficult. Furthermore, when high-throughput screening is performed, false positives may arise due to off-target inhibition of E1 or E2 enzymes, their respective thioesters, or from interaction with protein-based reagents that are often added to quantify reaction products.
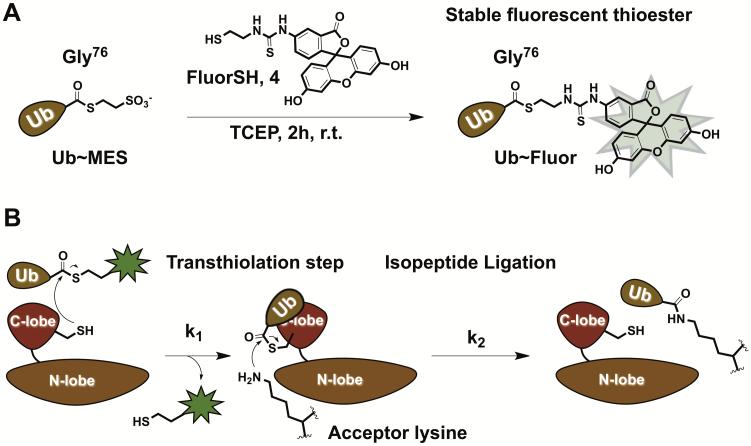
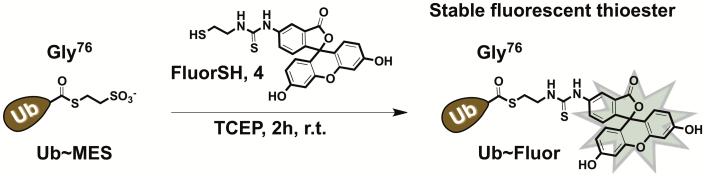
To overcome these challenges, we developed a novel class of fluorescent ubiquitin probes UbFluor, which in turn are based on the Bypassing System (Krist et al., 2016; Park et al., 2015). UbFluor is a thioester conjugate between the C-terminal Gly76 of Ub and fluorescein thiol (Figure 1A) (Krist et al., 2016). This conjugate can be easily prepared from the known Ub~MES thioester upon its transthiolation with fluorescent thiol (Figure 1A). UbFluor can undergo a transthiolation reaction with the catalytic cysteine of HECT E3, liberating the fluorophore and forming the catalytically active HECT E3~Ub thioester (k1 step). Kinetics of the fluorophore release from UbFluor can be monitored in real-time using fluorescence polarization (FP) methods. Subsequent to transthiolation, ubiquitin can be discharged from the HECT E3~Ub thioester onto an acceptor lysine. Acceptor lysine can be derived from UbFluor itself (to form Ub-UbFluor as a reaction product), exogenously added lysine, or a lysine residue of the substrate or of the ligase itself (k2 step, Figure 1B).
Figure 1.
A) Synthesis of UbFluor from Ub~MES. B) UbFluor reacts with HECT E3 ligase through transthiolation (k1) to produce HECT E3~Ub thioester and liberates FluorSH. Clearance of HECT E3~Ub can be accomplished through isopeptide bond ligation. If there are defects in transthiolation or isopeptide ligation steps, consumption of UbFluor will be inhibited under MT reaction conditions, i.e. 10 to 20-fold excess of UbFluor over enzyme.
Following discharge of ubiquitin, HECT E3 can react with a second molecule of UbFluor. Thus, if the isopeptide ligation step is hindered by a point mutation or small molecule, inactive HECT E3~Ub thioester accumulates and the consumption of UbFluor is inhibited. This effect can be observed under multiple turnover (MT) reaction conditions with an excess of UbFluor (10 to 20-fold excess relative to the enzyme). The inhibition of UbFluor consumption under these conditions can also be inhibited if the first transthiolation step (k1) is defective. It is possible to differentiate between these two possibilities by conducting the reaction either under single-turnover conditions (ST, 5 to 20-fold excess of HECT E3 over UbFluor) or under MT conditions (10 to 20-fold excess of UbFluor over the enzyme). Under ST, HECT E3 ligase has the opportunity to react with only one molecule of UbFluor. In this case, the consumption of UbFluor will reflect the transthiolation rate of UbFluor by HECT E3. If UbFluor consumption is inhibited under ST conditions upon biochemical point mutations or in the presence of small molecules, transthiolation (k1) between HECT E3 and UbFluor is likely defected. MT conditions, on the other hand, require both transthiolation (k1) and isopeptide ligation (k2) steps to proceed efficiently. If the consumption of UbFluor is inhibited under MT conditions, but not under ST, HECT E3 may have an isopeptide ligation defect.
To characterize the interaction between UbFluor and HECT domain, we measured the Michaelis-Menten parameters of a MT reaction with the HECT domain of the yeast ligase Rsp5. Measuring the consumption of UbFluor (25 – 500 μM) by Rsp5 HECT (1 μM), we determined an apparent kcat of 0.094 ± 0.010 s−1 and an apparent KM of 791.9 ± 121 μM. Experiments with large amounts of UbFluor (500 μM) are not amenable to an efficient analysis of many mutants, or for quantifying the impact of small molecules on catalysis. Therefore, we routinely run UbFluor assays under pseudo first-order reaction conditions to obtain an apparent bimolecular rate constant kobs (M−1s−1) where reaction rate = kobs[enzyme][UbFluor]. This approximation is valid for concentrations of UbFluor that are far below KM (~800 μM). Thus, we can calculate kobs under ST (5 μM ligase with 0.25, 0.50, 0.75, and 1.0 μM UbFluor) or MT (1 μM ligase with 10, 12.5, 15, and 20 μM UbFluor) conditions. Comparing a set of HECT domain mutants with kobs values determined in UbFluor assays incorporates measurements of at least four UbFluor concentrations into one value.
To determine kinetics from a UbFluor FP assay, polarization values must be converted to units of concentration. At both high and low UbFluor concentrations, we observed a linear relationship between polarization and the ratio of uncleaved to cleaved UbFluor. Therefore, polarization values registered during an assay can easily be converted to concentration units according to linear transformations.
Unique advantages of the developed method include the ability to measure the catalytic activity of the isolated HECT domain (MW ~45 kDa) that can be easily expressed in E. coli. While typical full length HECT E3s such as Nedd4-1 have a molecular weight of ~100 kDa and can be expressed in E. coli, other HECT E3s such as ARF-BP1/MULE/HUWE1 have much larger molecular weights around ~500 kDa and are difficult to express in E. coli or insect cells. In this and similar cases, one can study the truncated version of the HUWE1 catalytic HECT domain with UbFluor. Thus, just as kinase inhibitors target the kinase ATP active site (Knight and Shokat, 2005), it could be possible to develop HECT E3 ligase inhibitors that target the catalytic HECT domain (active site) and display useful pharmacological properties. Importantly, since UbFluor lacks E2 enzyme, this assay has limited potential to discover inhibitors that disrupt the binding of E2 enzyme to the catalytic HECT domain. However, we have shown that despite lacking E2 enzyme, UbFluor and its non-fluorescent analogues are largely processed by a native HECT E3 mechanism based on analysis of UbFluor reactions with HECT E3 mutants (Kamadurai et al., 2013; Krist et al., 2016).
This protocol describes a) the synthesis of UbFluor (Basic Protocol 1), and b) procedures to conduct enzymatic assays for HECT E3s, and c) data analysis.
STRATEGIC PLANNING
FP assays can be conducted in 384-well plates with typical reaction volumes of ~20 μL. Reagent dispensing can be automated, and many measurements can be made in a short period of time. Typically, 1-4 enzyme mutants can be analyzed in one day. Importantly, because it lacks E2 enzyme, the ubiquitination reaction by HECT domain is slower with UbFluor than in the native cascade. Typical native ubiquitination reactions proceed on the second timescale, while reactions with UbFluor occur on the minute timescale. Since UbFluor is a thioester, free thiols such as dithiothreitol (DTT) or β-mercaptoethanol (BME) must not be used in enzyme solutions or reaction buffers, because they will cause a non-specific consumption of UbFluor.
BASIC PROTOCOL 1
E1 ENZYME UBA1 EXPRESSION
This protocol describes the expression of mouse ubiquitin-activating E1 enzyme (UBA1) from E. coli and its subsequent purification. This enzyme is used in the chemoenzymatic synthesis of Ub~MES, which is later converted to the final probe UbFluor. If a lab is not equipped to express protein, UBA1 enzyme can be obtained from a commercial vendor. However, when large amounts of UbFluor are needed, e.g., for high-throughput screening assays, it might be more affordable to produce E1 enzyme in-house than to buy it from commercial vendors.
Materials
REAGENTS
E. coli BL21 (DE3) cells (Fisher Scientific)
E1 enzyme UBA1 plasmid for expression in E. coli, (pET-28b mE1) available from addgene (plasmid #32534)
Kanamycin sulfate (Aldrich)
LB Agar (Fisher Scientific)
LB broth (Fisher Scientific)
0.5 M Isopropyl β-D-1-thiogalactopyranoside in water
Resuspension buffer: 50 mM Tris pH 8.0, 150 mM NaCl, 0.1% Triton X-100, 1 mM DTT, with Complete Protease EDTA-free Inhibitor Cocktail (Roche) (see recipe)
Complete Protease EDTA-free Inhibitor Cocktail (Roche)
Ni-NTA agarose (Qiagen)
Ni-NTA wash buffer: 50 mM Na2HPO4/NaH2PO4 pH 8.0, 150 mM NaCl (see recipe)
Ni-NTA elution buffer: 50 mM Na2HPO4/NaH2PO4 pH 8.0, 150 mM NaCl, 300 mM imidazole (see recipe)
E1 storage buffer: 20 mM HEPES pH 8.0, 100 mM NaCl, 1 mM DTT (see recipe)
EQUIPMENT AND MATERIALS
Lab benchtop scale
Petri Dishes (Fisher Scientific)
250 mL baffled Erlenmeyer flask (Fisher Scientific)
Incubator/shaker that can reach 37 °C
2 L baffled Erlenmeyer flask (Fisher Scientific)
Absorbance spectrophotometer (to measure optical density of E. coli solution)
Polycarbonate centrifuge bottles (500 mL volume and 40 mL volume sizes)
High-speed centrifuge for E1 expression steps (Thermo, Sorvall RC 6+ or comparable)
Centrifuge rotor for pelleting cells in 500 mL bottles (PTI/Thermo Scientific F10S-6x500y centrifuge rotor or comparable)
Centrifuge rotor for clearing lysate (Thermo Scientific F21S-8x50y centrifuge rotor or comparable)
Sonicator/Sonic dismembrator (Fisher scientific FB505 or comparable)
0.45 μm syringe filter (Fisher Scientific)
Econo Column (10 cm × 2.5 cm; BioRad)
Rocking Platform (BioRad)
Centrifuge for 50 mL conical tubes (Eppendorf 5430 R or equivalent)
FPLC system (Akta)
HiLoad Superdex 200 (GE Healthcare)
Amicon 30 kDa MWCO spin filters
−80 °C freezer
Procedure for E1 enzyme UBE1 expression
1.) Transform E. coli BL21 (DE3) cells with a pET-mE1 plasmid using 15 μg/mL kanamycin for selection on agar plates.
2.) Once transformed BL21 colonies are obtained on agar plates, inoculate an overnight culture (50 mL LB broth with 15 μg/mL kanamycin in a 250 mL baffled flask) with a colony and culture at 37 °C, with shaking at 225 rpm overnight.
3.) Add the overnight culture to 1 L LB broth with with 15 μg/mL kanamycin in a 2 L baffled flask. Set to rock at 37 °C, 225 rpm. Measure the optical density at 600 nm (OD) every hour.
4.) When the OD reaches ~0.6, add 1 mL of 0.5 M IPTG solution (final IPTG concentration 0.5 mM) to the flask and set to 16 °C, 225 rpm for 20 hours.
-
5.) After 20 hours transfer the culture to 500 mL bottles appropriate for centrifugation. Centrifuge at 6000 g, for 20 minutes at 4 °C. Decant the supernatant and collect the cell pellets.
Cell pellets can be stored at −80 °C for several months.
6.) Resuspend the cell pellets in resuspension buffer: 50 mM Tris pH 8.0, 150 mM NaCl, 0.1% Triton X-100, 1 mM DTT with protease inhibitor (Complete Protease EDTA-free Inhibitor Cocktail, Roche) (see recipe). From a 1L expression, we typically resuspend cells with 20 mL resuspension buffer.
7.) Distribute resuspended cells to 40 mL centrifuge bottles and then lyse cells by sonication.
8.) Centrifuge the cell lysate at 18,000 g for 40 minutes at 4 °C. Collect supernatant.
9.) Pass the cleared cell lysate through a 0.45 μm syringe filter to eliminate any remaining cellular debris.
10.) Prepare a column (10 cm × 2.5 cm BioRad Econo column) for protein purification. Rinse the column with DI water and clamp it upright in a cold room (4 °C). Add Ni-NTA agarose (2 mL slurry) and drain the storage buffer. Wash the Ni-NTA resin with 3 × 20 mL of cold Ni-NTA wash buffer: 50 mM Na2HPO4/NaH2PO4, 150 mM NaCl, to equilibrate the resin with this buffer.
11.) Add lysate to the equilibrated Ni-NTA agarose, close the column, and then incubate for 2 hours at 4 °C with gentle rocking using a rocking platform. This is to ensure a complete saturation of the beads to maximize the protein yield.
Alternatively, lysate can be added to the column and eluted with a slow flow rate (~1 mL/min). Running the subsequent flow-through over the Ni-NTA agarose again may improve protein yield.
12.) Elute unbound proteins from the column.
13.) Wash the resin with Ni-NTA wash buffer using gravity flow (5 × 15 mL with a ~1 mL/min flow rate).
14.) Elute E1 enzyme with cold Ni-NTA elution buffer: 50 mM Na2HPO4/NaH2PO4 pH 8.0, 150 mM NaCl, 300 mM imidazole (3 × 3 mL).
15.) Purify E1 enzyme through a HiLoad Superdex 200 FPLC column equilibrated with E1 storage buffer. A typical flow rate is 0.5 mL/min.
16.) Combine fractions containing E1 enzyme and concentrate to ~ 2 mL with Amicon 30 kDa MWCO spin filters. The typical concentration of E1 enzyme is ~10 μM, or 1.2 mg/mL. E1 enzyme activity can be tested using previously reported methods (An and Statsyuk, 2013).
BASIC PROTOCOL 2
SYNTHESIS OF UbFluor
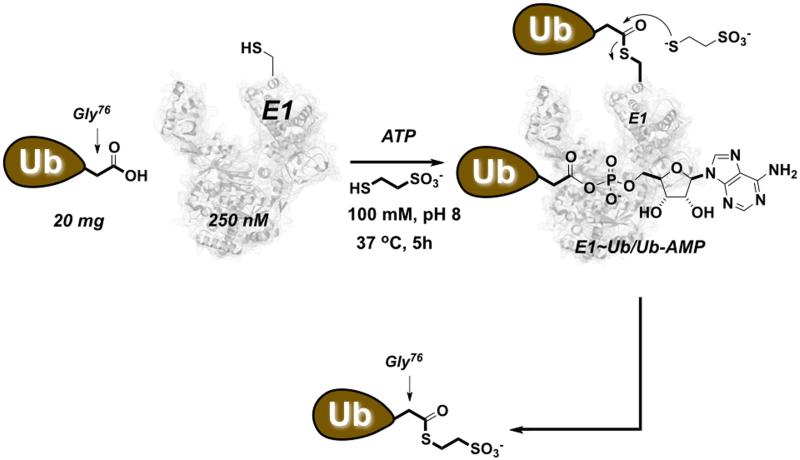
Synthesis of UbFluor requires commercially available ATP and ubiquitin, ubiquitin-activating E1 enzyme (E1), and 2-mercaptoethanesulfonic acid sodium salt. The synthesis also requires the fluorescent thiol FluorSH that can be synthesized in three chemical steps. These steps are based on very simple and robust chemistry which does not require advanced chemical synthesis skills. If the available laboratory space is not suitable for chemical synthesis, such work can easily be performed by properly trained personnel at any chemistry department equipped with fume hoods. UbFluor synthesis involves the intermediate thioester Ub~MES, which can be prepared by a chemoenzymatic approach with ATP, E1, ubiquitin, and 2-mercaptoethanesulfonic acid sodium salt (Figure 2) (El Oualid et al., 2010).
Figure 2.
Chemoenzymatic synthesis of Ub~MES. Ubiquitin is incubated with E1 enzyme in the presence of ATP and 100 mM 2-mercaptoethanesulfonic acid sodium salt (MESNa). The initially formed E1~Ub thioester reacts with MESNa, producing Ub~MES and regenerated E1 enzyme.
Upon reaction of Ub~MES with an excess of FluorSH, 2-mercaptoethanesulfonate in Ub~MES is replaced with FluorSH via transthiolation to produce the desired UbFluor (Figure 1A) (Krist et al., 2016). We found this final step to be sensitive to FluorSH purity such that FluorSH should be first purified with HPLC and lyophilized. If HPLC purification is not performed, then the subsequent conjugation step is not reproducible.
Materials
REAGENTS
500 mM Na2HPO4/NaH2PO4 pH 8.0 pH 8.0 (see recipe)
Adenosine triphosphate disodium salt hydrate (ATP, Aldrich)
2-mercaptoethanesulfonic acid sodium salt (MESNa, Aldrich)
Magnesium chloride
Ubiquitin from bovine erythrocytes (Aldrich)
Storage Buffer A: 25 mM NaCl, 12.5 mM HEPES pH 6.7 (see recipe)
Cysteamine hydrochloride
Trifluoroacetic acid (TFA)
Methylene chloride
Trityl chloride
1 M NaOH aqueous solution
Saturated sodium chloride aqueous solution
Magnesium sulfate (anhydrous)
Diethyl ether/n-pentane (1:4 ratio)
Dimethylformamide (DMF)
Fluorescein isothiocyanate (Aldrich, isomer 1)
N,N-diisopropylethylamine
50 mM HEPES pH 6.5 (see recipe)
Silica gel, standard grade (Sorbent technologies, 60 Å, 40-63 μm particle size or comparable)
Ethyl acetate
Sand
Methanol
Trifluoroacetic acid
Triethylsilane
Acetonitrile/water (3:7) with 0.1 % TFA (see recipe)
DMSO/H2O (1:1) (see recipe)
Sodium bicarbonate
1 M HEPES pH 7.5 (see recipe)
Tris(2-carboxyethyl)phosphine hydrochloride (Aldrich)
Guanidine hydrochloride
Storage Buffer B: 250 mM NaCl, 12.5 mM HEPES pH 6.0 (see recipe)
1 × PBS with 20 mM BME (see recipe)
MATERIALS AND EQUIPMENT
Lab benchtop scale
50 mL Falcon conical tubes
Incubator that can reach 37 °C
Amicon 3 kDa MWCO spin filters
Centrifuge for 50 mL conical tubes (Eppendorf 5430 R or equivalent)
FPLC system (Akta)
HiLoad Superdex 75 FPLC column (GE Healthcare)
Nanodrop 2000 Spectrophotometer (Thermo Scientific) or equivalent
Chemistry fume hood equipped with nitrogen and vacuum sources
Stir bar with standard stir plate
Glass round-bottomed flasks (single 24/40 neck, 100 mL and 250 mL sizes) (single 14/20 neck, 25 mL size)
Glass separatory funnels (30 mL and 250 mL sizes)
Glass Erlenmeyer flasks (50 mL and 250 mL sizes)
Glass cone funnel for 24/40 joint
Whatman filter paper (70 mm)
Fritted filter funnel, 150 mL
High-vacuum pump (Welch DuoSeal, 1×10−4 torr)
Glass chromatography column (250 mL, 1” diameter)
Glass elbow joint for N2(g) on glass chromatography column
Tygon tubing for N2(g)
Rotary evaporator
20 mL glass scintillation vial
Aluminum foil
Standard lab vortex mixer (up to 3000 rpm)
Symphony ultrasonic cleaner (VWR) or equivalent sonication bath
HPLC system
HPLC column (Restek Pinnacle DB C18)
Lyophilizer
LCTOF mass spectrometer for intact protein analysis
HiTrap Desalting columns (5 mL size, GE Healthcare)
−80 °C freezer
Procedure for Ub~MES synthesis
-
1.) In a 50 mL conical tube, combine the following in the given order of addition: 500 mM Na2HPO4/NaH2PO4 pH 8.0 (2.4 mL; 50 mM final concentration)
E1 enzyme UBE1 (volume adjusted so final concentration is 0.25 μM) – from previous protocol
ATP (129 mg, 10 mM final concentration)
MESNa (384 mg, 100 mM final concentration)
magnesium chloride (234 μL of 1 M solution, 10 mM final concentration),
ubiquitin (20 mg, 100 μM final concentration).
Add double distilled water to a final total volume of 23.4 mL.
2.) Cap the 50 mL conical tube and then gently invert the tube a few times to dissolve and mix the contents.
3.) Place the tube in a 37 °C incubator without agitation for 5 hours.
4.) Concentrate the reaction solution (23.4 mL) to ~2 mL with an Amicon 3 kDa MWCO spin filter.
5.) Purify Ub~MES through a HiLoad Superdex 75 FPLC column equilibrated with Storage Buffer A.
6.) Combine fractions of Ub~MES and concentrate to ~ 2 mL with Amicon 3 kDa MWCO spin filters. Typical concentrations range from 400-1200 μM, as determined by nanodrop spectrophotometer. Prepare a stock solution of Ub~MES (700 μM in Storage Buffer A) for subsequent conjugation to FluorSH.
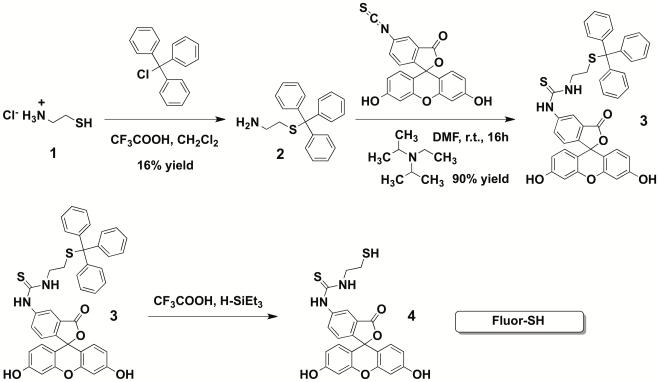
Procedure for synthesis of fluorescein thiol (FluorSH; Figure 3)
Figure 3.
Synthesis of FluorSH, 4
CAUTION: Perform these steps in a fume hood.
Procedure for synthesis of trityl-protected cysteamine, 2
7.) Combine cysteamine hydrochloride (1 g, 8.8 mmol), trifluoroacetic acid (1.3 mL, 1.9 g, 17.0 mmol) and methylene chloride (30 mL) with a stir bar in a 100 mL round bottomed flask. Then add trityl chloride (2.4 g, 8.8 mmol) and allow the reaction to stir for 16 hours at room temperature.
8.) After 16 hours, pour the reaction solution to a glass separatory funnel, and add 1 M NaOH solution (20 mL). Close the funnel and then shake it vigorously for a few minutes (relieve the pressure as necessary) so that the aqueous and organic layers thoroughly mix. Clamp the funnel upright so that the layers separate after several minutes.
9.) Drain the funnel and collect the phases separately (bottom layer = organic, top layer = aqueous). Add the organic phase back to the funnel with additional methylene chloride (50 mL).
10.) Add saturated sodium chloride solution (50 mL) to the separatory funnel, and shake the flask as described above. Allow layers to separate. Collect the organic phase (bottom layer) in an Erlenmeyer flask (250 mL size), avoiding passage of any water to the flask.
11.) Add 1-2 g magnesium sulfate (anhydrous) to the Erlenmeyer flask and swirl it with the organic phase to soak up trace amounts of water.
12.) Place a glass funnel with Whatman filter paper in the top of a round bottomed flask (250 mL size). Slowly add the contents of the Erlenmeyer flask to the funnel in order to filter off magnesium sulfate; only clear organic solution should enter the round-bottomed flask.
13.) Add a stir bar to the flask, place the flask over a stir plate, and begin stirring vigorously. Slowly add ether/n-pentane solution (1:4 ratio of ether:pentane) until no additional precipitate forms.
14.) Filter off the crystalline white solid with a fritted filter funnel fit to the round-bottomed flask and connected to house (hood) vacuum.
15.) Add the solid to a clean round-bottomed flask and connect to a high-vacuum pump (oil pump) for several hours to remove trace solvent.
Procedure for synthesis of trityl-protected fluorescein thiol, 3
16.) Combine trityl-protected cysteamine (from previous step) (0.33 g, 1.0 mmol), DMF (5 mL), and fluorescein isothiocyanate (0.40 g, 1.0 mmol) with a stir bar in a round-bottomed flask (25 mL size). Then add N,N-diisopropylethylamine (346 μL, 2.0 mmol) dropwise and allow the mixture to stir at room temperature for 16 hours.
17.) After 16 hours, reduce the volume of the reaction solution by stirring the solution vigorously under a strong stream of nitrogen gas for 3-5 hours. The volume of the solution should now be 1-2 mL.
18.) Add the concentrated reaction solution to 48 mL of aqueous 50 mM HEPES pH 6.5 in a 50 mL conical tube.
19.) Centrifuge the precipitated product at 4000 g for 10 minutes at room temperature. Decant the supernatant to leave a solid pellet at the bottom of the tube.
20.) Prepare a silica gel plug: make a slurry of silica gel (3-4 g) in ethyl acetate (~50 mL). Mix this slurry well and then pour to a glass chromatography column (250 mL, 1” diameter). After the silica has settled to the bottom of the column, add a small amount of sand to the top of the silica. At this point, the silica layer should be 4-5 cm thick and the sand layer should be 1 cm thick. Use Tygon tubing to connect a vented nitrogen outlet in the hood to the glass elbow joint. Use this joint to lightly add nitrogen pressure to the glass column to slowly elute ethyl acetate (flash chromatography).
Do not flash ethyl acetate past the silica gel layer. Always keep a layer of ethyl acetate over the silica gel.
21.) Dissolve the pellet in methanol (2-3 mL). Add this solution onto the sand layer in the glass chromatography tube. Open the column stopcock so that the methanol solution moves onto the silica gel. Then slowly add ethyl acetate (50 mL) to the column. Begin collecting the solvent that elutes from the column. Use nitrogen pressure to elute solution from the column at a faster rate.
22.) Add the ethyl acetate/methanol solution (8% MeOH, 50 mL) to the top of the column and elute with nitrogen pressure.
-
23.) Combine all eluted organics in a round-bottomed flask (250 mL size).
Use a rotary evaporator (“rotovap”) to remove solvent (down to ~50 torr with a 30 °C water bath). An orange powder should precipitate. Place the flask under a ‘high’ vacuum to remove trace amounts of solvent.
Procedure for deprotection of trityl-protected fluorescein thiol to provide FluorSH, 4
24.) In a 20 mL glass scintillation vial, combine trityl-protected Fluor (from previous step) (100 mg, 0.14 mmol) with trifluoroacetic acid (1 mL, 1.5 g, 13.0 mmol) and water (25 μL). Agitate the vial to dissolve the fluorophore.
25.) To the dissolved fluorophore, add triethylsilane (25 μL, 18.2 mg, 0.16 mmol. Then cap the vial, cover it with aluminum foil to protect from light, and set to shake at a medium rate on a standard lab vortex mixer (~1000 rpm) for two hours at room temperature.
26.) After two hours, pour the reaction solution into diethyl ether (50 mL in a 50 mL conical tube) to precipitate the product.
27.) Centrifuge the precipitated product at 4000 g, for 10 minutes at 4 °C. Decant the supernatant to leave a solid pellet at the bottom of the tube.
28.) Dissolve the pellet from step #4 in acetonitrile/water (3:7) with 0.1% TFA (typical volume is 2 mL) adding a few drops of triethylamine to assist dissolution of the solid.
29.) Purify FluorSH with a gradient HPLC method (ramp from 30% to 95% acetonitrile in water, 0.1% TFA, over 30 minutes; column: Restek Pinnacle DB C18). We found that purification of FluorSH using HPLC is essential to the reproducibility of the subsequent conjugation step to produce UbFluor.
-
30.) Combine fractions of the major peak (typical elution time 12 minutes) and lyophilize overnight to obtain an orange/red powder.
Lyophilized UbFluor can be stored at −20 °C in the conical vial until the synthesis of UbFluor (below). We recommend to fill the vial with nitrogen to prevent sulfur oxidation, and to protect it from light by wrapping it with aluminum foil.
Procedure for synthesis of UbFluor ( Figure 4 )
Figure 4.
Synthesis of UbFluor.
-
31.) Dissolve lyophilized FluorSH (from step 31) in a few milliliters of methanol to completely dissolve it. Remove 5-10 μL of solution for mass analysis by LCTOF. Transfer this solution to a tared 20 mL glass scintillation vial and remove methanol with a rotary evaporator. Remove trace methanol with a nitrogen stream for 30 minutes in a chemistry fume hood.
Collection of FluorSH with methanol ensures complete recovery of the fine FluorSH powder, which may otherwise stick to the sides of containers after lyophilization.
-
32.) Once methanol has been removed, determine the net mass of FluorSH.
We typically obtain 20-30 mg lyophilized FluorSH from 100 mg of trityl-protected fluorescein thiol.
-
33.) Dissolve FluorSH to 50 mM in DMSO/H2O (1:1), adding a minimal amount of saturated aqueous NaHCO3 (~30 μL saturated NaHCO3 solution to 20 mg FluorSH). This step will produce a stock solution of FluorSH which can be used for conjugation with Ub~MES.
After adding NaHCO3, the solution should change from yellow to red. It may be necessary to briefly sonicate the solution using an ultrasonic cleaner bath in order to fully dissolve FluorSH.
-
34.) In each of 4 × 2 mL microcentrifuge tubes, combine the following in the given order of addition:
1 M HEPES pH 7.5 (111 μL, 100 mM final concentration)
100 mM TCEP (100 μL, 9 mM final concentration)
6 M guanidinium chloride (300 μL, 1.62 M final concentration)
50 mM FluorSH (300 μL, 13.5 mM final concentration) – from step 34
700 μM Ub~MES (300 μL, 189 μM final concentration) – from step 6
The final solution volume is 1111 μL.
After setting up the reactions, leftover FluorSH solution can be stored at −20 °C. If the Ub~MES solution is more concentrated than 700 μM, use a proportionately lower volume to maintain 189 μM final concentration and add double distilled water to make the final volume 1111 μL.
35.) Close the reaction tubes, cover in aluminum foil, and set to lightly vortex using a standard lab vortexer for 2 hours at room temperature.
36.) Add Storage Buffer B (389 μL) to each tube to bring the total volume to 1500 μL.
-
37.) Purify each reaction through a HiTrap Desalting column. Follow manufacturer’s instructions.
Each HiTrap Desalting Column can be reused. We wash the columns with five or more column volumes of 1 M NaCl, 5 mM DTT, 50 mM Tris pH 7.5, and then with five or more column volumes of pure water to remove any remaining bound FluorSH and other leftover reaction components.
-
38.) Further purify UbFluor through a HiLoad Superdex 75 FPLC column equilibrated with Storage Buffer B.
We observe UbFluor to elute around 120 mL elution volume, while unmodified ubiquitin elutes around 90 mL elution volume.
39.) Combine fractions of UbFluor and concentrate to ~ 2 mL with Amicon 3 kDa MWCO spin filters.
40.) Analyze the final product with SDS-PAGE/fluorescence scanning to ensure the absence of free fluorophore. Run intact protein mass spectrometry to verify UbFluor purity MW: 9013 m/z.
41.) Aliquot UbFluor, snap freeze in liquid nitrogen, and store at −80 °C.
Procedure for determining UbFluor concentration
42.) Prepare a solution containing 50 μM FluorSH and 50 μM ubiquitin in 1 × PBS with 20 mM BME. Use the 1 × PBS with 20 mM BME buffer to make two-fold serial dilutions to achieve final concentrations of FluorSH and ubiquitin: 25, 20, 15, 12.5, 10, 7.5, 5, 2.5 μM.
43.) Use a Nanodrop 2000 (Thermo) or equivalent spectrophotometer to measure the emission intensity of this dilution series to obtain a standard curve (emission at 512 nm).
44.) Dilute a sample of UbFluor (3 μL) with 1 × PBS, 20 mM BME (27 μL), gently mix with pipette, and then incubate at room temperature for 5 minutes. Under these conditions BME cleaves fluorescein from Ub.
-
45.) Measure fluorescence intensity with the spectrometer, and use the generated standard curve to calculate the concentration of UbFluor.
For 3 μL UbFluor mixed with 27 μL buffer, the calculated concentration should be multipled by 10 to obtain the concentration of the stock UbFluor solution.
BASIC PROTOCOL 3
FLUORESCENCE POLARIZATION ASSAY WITH UBFLUOR
Fluorescence polarization (FP) is a widely employed method to monitor enzyme activity in response to biochemical point mutations or small molecule inhibitors (Levine et al., 1997; Liang et al., 2014). Because it is a ratiometric technique, the signal is independent of fluorophore concentration. Furthermore, fluorescence intensity variations due to the presence of impurities produce minor interference with the assay. When conducting Michaelis-Menten experiments we needed to use large concentrations of UbFluor (>400 μM), and observed self-quenching of the fluorophore (Zhuang et al., 2000). However, we verified that the FP signal was not affected at these high concentrations, and were able to conduct kinetic measurements in the quenching regime up to 500 μM UbFluor (Krist et al., 2016). We have found that the FP assay is robust and performs very well with a range of HECT E3 ligases including Nedd4-1, WWP1, E6AP, ITCH, Nedd4-2 and others. We would like to highlight the following feature of this assay: typically, the consumption of UbFluor under ST reaction conditions (5 to 20-fold excess of HECT E3 over UbFluor) is complete after about 40 minutes. MT reaction conditions (10 to 20-fold excess of UbFluor over HECT E3) can require up to 6h for 50% consumption of UbFluor.
Furthermore, we have found that different HECT E3s display different activity in assays with UbFluor under MT reaction conditions. For example, 100 nM WWP1 HECT is able to process UbFluor, while Nedd4-1 HECT domain only consumes UbFluor when the Nedd4-1 concentration is equal to or greater than 500 nM. To increase the activity of HECT domain, we found it useful to express N-terminally extended versions of the catalytic HECT domain. In this case, the enzyme is also more prone to autoubiquitination, which can increase the rate of UbFluor consumption under MT conditions.
As a cautionary note, full length HECT E3 ligases may exist in an autoinhibitory conformation and could have very low activity in UbFluor assays (Mari et al.). In this case, such autoinhibition can either be destabilized with point mutations, or by truncation (deletion) of one of the interacting domains. For example, in Nedd4-1 HECT E3, the N-terminal C2 domain occludes the C-terminal HECT domain. This conformation inhibits transthiolation of Nedd4-1 with E2~Ub thioesters, such that deletion of the C2 domain in Nedd4-1 activates the enzyme toward transthiolation.
Finally, please be aware that stock solutions of commercially available enzymes may contain thiols such as DTT and BME that can cleave UbFluor and affect polarization measurements. Any enzyme solution containing thiol must be desalted twice or purified via FPLC to completely remove thiol, as these are not tolerated even at concentrations as low as 1 μM. However, we find that in general, UbFluor tolerates free amines such as lysine at concentrations up to at least 100 mM for at least 30 minutes.
Materials
REAGENTS
UbFluor, from Basic Protocol 2
HECT ligase (either the catalytic domain or full length enzyme, available from Boston Biochem and obtainable from E.coli expression)
240 μM Tween-20
24.4 mM TCEP
UbFluor Assay Buffer: 1.5 M NaCl, 500 mM HEPES pH 7.5 (see recipe)
EQUIPMENT AND MATERIALS
1.5 mL microcentrifuge tubes
Centrifuge (for 1.5 mL microcentrifuge tubes, Thermo Legend Micro 21R or equivalent)
384-well plate (low volume, low binding, Corning #3820)
Centrifuge for 384-well plates (Jouan RC1022 or comparable)
Synergy 4 (BioTek) fluorescence plate reader running Gen5 software (BioTek).
Procedure for FP assay with UbFluor
-
1.) Except for the HECT ligase, combine all components of a given reaction (total volume: 25 μL) in a 1.5 mL microcentrifuge tube.
Our reactions typically contain the following components at the given final concentrations: 150 mM NaCl, 6 μM Tween-20, 0.5 mM TCEP, 50 mM HEPES pH 7.5 (Use the corresponding stock solutions given in the Recipes section) with 0.25 – 20 μM UbFluor.
-
2.) Add the required amount of HECT ligase to the side of each microcentrifuge tube, making sure the ligase does not touch the UbFluor solution at the bottom of the tube.
Our reactions typically contain 0.5 – 5 μM HECT ligase.
3.) Centrifuge all reaction tubes at 10,000 g for seven seconds to start the reactions.
4.) Transfer each reaction to a well in a 384-well plate (load 20 μL), pipetting each solution five times, while loading to fully mix.
Depending on how the reactions are set up, it may be necessary to use a fresh pipette tip for each tube.
-
5.) Once all reactions have been loaded to the plate, centrifuge the plate at 1,500 g for 8 seconds, then place plate in Synergy 4 plate reader and begin FP readings.
The dead time between spinning HECT ligase into solution and FP measurements is typically ~2 minutes. This can be expected when measuring 4 – 8 reactions during a given reading. The plate centrifuge should be as close to the plate reader as possible to minimize dead time.
For ST experiments, 5 μM HECT ligase is treated with 0.25, 0.50, 0.75, or 1.0 μM UbFluor and analyzed every 5 – 15 seconds by the plate reader. For MT assays, 1 μM HECT ligase is treated with 10, 12.5, 15, or 20 μM UbFluor and analyzed every 5 – 20 seconds by the plate reader.
It is also important to note that in MT reactions, isopeptide ligation occurs. The enzyme products in this case could be di-UbFluor described previously, polyubiquitin chains of varied length, and autoubiquitinated HECT E3. It is thus recommended to analyze MT reactions at given time points with western blots using ubiquitin antibodies and in-gel fluorescence scanning to confirm that isopeptide ligation took place. We found that some HECT E3s such as WWP1 and ITCH autoubiquitinate in the presence of UbFluor, and some such as Rsp5 and Nedd4-1 autoubiquitinate poorly. This trend correlates with the observed production of free polyubiquitin chains – HECT domains of WWP1 and ITCH more actively produce polyubiquitin chains than Rsp5 and Nedd4-1 HECT domains (Krist et al., 2016).
As an orthogonal assay, it may be possible to monitor the consumption of UbFluor by HECT E3 and formation of HECT E3~Ub thioester using SDS-PAGE with in-gel fluorescence scanning or Coomassie staining under non-reducing conditions. Since BME and DTT cleave UbFluor thioester, do not use them in the Laemmli loading buffer. Typical dynamic range of the FP assay is ~100 mP.
ALTERNATE PROTOCOL 1
FLUORESCENCE POLARIZATION ASSAY WITH UBFLUOR FOR FAST REACTIONS – ALTERNATE TO BASIC PROTOCOL 3
This alternate protocol for FP assays with UbFluor allows the measurement of initial rates of UbFluor reactions that occur very rapidly due to the enzyme choice, reagent concentrations, or presence of activating reagents or proteins. This protocol is appropriate for reactions which are completed on timescales as short as 3 minutes. Typically, reactions requiring this protocol are single-turnover reactions using a HECT E3 that is activated due to a point mutation. Since this protocol measures one reaction at a time, multiple repetitions of steps 4-9 are required for monitoring the concentration dependence of these reactions and obtaining kcat/KM values.
Additional Materials
None
Procedure for FP assay with UbFluor for fast reactions
-
1.) Except for the HECT ligase, combine all components of a given reaction (total volume: 25 μL) in 1.5 mL microcentrifuge tubes.
Our reactions typically contain the following components at the given final concentrations: 150 mM NaCl, 6 μM Tween-20, 0.5 mM TCEP, 50 mM HEPES pH 7.5 (Use the corresponding stock solutions given in the Reagents and Solutions section) with 0.25 – 20 μM UbFluor.
2.) Transfer the contents of each 1.5 mL tube (all reaction components except HECT ligase) in the reaction series to consecutive wells in a 384-well plate.
3.) Centrifuge the plate at 1,500 g for 10 seconds.
-
4.) Prepare the Synergy 4 plate reader Gen5 software for FP readings of the first well in the reaction series (or the subsequent individual well as the protocol is repeated for multiple reaction conditions).
For rapid reactions, the plate reader should be set to make FP readings every 2-5 seconds.
5.) Collect and prepare all solutions and pipettors prior to steps 6-9.
-
6.) Add the required amount of HECT ligase stock solution to the first well in the reaction series, pipetting up and down three times to ensure complete injection of the solution into the reaction mixture. This starts the reaction; timing of the reaction should begin here.
Our reactions typically contain 0.5 – 5 μM HECT ligase final concentration.
-
7.) Using a separate pipettor set to 15 μL, pipet the reaction mixture 5 times to fully mix the solution in the well, being careful not to introduce air bubbles into the mixture.
This step can be omitted if the HECT stock solution volume added in step 6 is large enough to fully mix the reaction. However, the typical HECT ligase stock solution concentration is >5 times the final HECT concentration, necessitating a small addition volume which is insufficient for complete mixing.
-
8.) Place plate in Synergy 4 plate reader and begin FP readings.
The dead time between adding HECT ligase into solution and FP measurements is typically 10-45 seconds.
9.) Repeat steps 4-9 for each prepared reaction mixture until all reactions have been completed.
BASIC PROTOCOL 4
DATA ANALYSIS FOR UBFLUOR FP ASSAY
Before running any assays with enzyme, it is important to determine the baseline polarizations obtained with your plate reader for ‘intact’ vs ‘cleaved’ UbFluor. This will allow conversion of raw polarization data to concentration units of consumed UbFluor. Under ST conditions (0.25 – 1.00 μM UbFluor), we observe maximal polarization values to be ~155 mP. Under MT conditions (10 – 20 μM UbFluor), we observe maximal polarization values to be ~138 mP. Under single and MT conditions, cleaved UbFluor provides polarization values ~30 mP. For a given batch of UbFluor, the ‘intact’ UbFluor polarization values typically do not vary by more than 2-3 mP. We recommend using a single batch of UbFluor and HECT E3 for any experiments involving direct comparisons in order to minimize the effects of potential batch-to-batch variation.
Additional Materials
None.
Procedure for data analysis of UbFluor FP assay
-
1.) To determine the baseline polarization of ‘intact’ UbFluor, run a polarization experiment according to the method described above in Basic Protocol 3: Fluorescence polarization (FP) assay without enzyme or with catalytically dead enzyme. Observe the polarization over 30 minutes. The average of these polarization values can be considered the ‘intact’ polarization level.
Under MT reaction conditions, we always observe an initial slight decrease in UbFluor polarization signal, even in the absence of enzyme.
2.) To determine the baseline polarization of ‘cleaved’ UbFluor run a polarization experiment according to the method described above in Basic Protocol 3: Fluorescence polarization (FP) assay with the addition of 1 mM BME. Observe the polarization over the course of 30 minutes. The average of these polarization values can be considered the ‘cleaved’ polarization level. In-gel fluorescence scanning should also be used to confirm a complete cleavage of UbFluor in the presence of 1 mM BME to ensure that the observed polarization corresponds to cleaved UbFluor product.
-
3.) To convert raw polarization values to units of UbFluor concentration, use the determined ‘intact’ and ‘cleaved’ values to build linear transformations for a given concentration of UbFluor. Polarization can be the independent variable (x), and UbFluor concentration can be the dependent variable (y). Each concentration of UbFluor should utilize a different equation for data transformation.
Example: For 10 μM UbFluor, plot (138 mP, 10 μM UbFluor) and (30 mP, 0 μM UbFluor). Calculate the equation of a linear fit for these two points. Use this equation to convert raw polarization values to units of UbFluor concentration.
4.) Under pseudo-first order reaction conditions ([UbFluor]<<800 μM, KM) the observed rate is linear with respect to enzyme and UbFluor concentrations, so the rate = kobs [Enzyme][UbFluor]. Plotting UbFluor concentrations (independent variable) vs the rate of UbFluor consumption (dependent variable) produces a linear curve for both single and MT reactions. The slope of this line is kobs [Enzyme]. By dividing this value by enzyme concentration the value of kobs can be obtained.
REAGENTS AND SOLUTIONS
Use ddH2O from Thermo Scientific Nanopure or equivalent purification system
Resuspension buffer: 50 mM Tris pH 8.0, 150 mM NaCl, 0.1% Triton X-100, 1 mM DTT with protease inhibitor (Complete Protease EDTA-free Inhibitor Cocktail, Roche).
50 mM Tris base
150 mM NaCl
Adjust pH to 8.0 by dropwise addition of 1 M HCl
0.1 % Triton X-100 (v/v)
1 mM DTT
1 tablet Complete Protease EDTA-free Inhibitor Cocktail, Roche
Use within one day.
Ni-NTA wash buffer: 50 mM Na2HPO4/NaH2PO4 pH 8.0, 150 mM NaCl
0.47 M Na2HPO4, sodium phosphate dibasic
0.034 M NaH2PO4, sodium phosphate monobasic
150 mM NaCl
Adjust pH to 8.0 by dropwise addition of 1 M NaOH
Store for up to 1 month; check pH to ensure quality.
Ni-NTA elution buffer: 50 mM Na2HPO4/NaH2PO4 pH 8.0, 150 mM NaCl, 300 mM imidazole
0.47 M Na2HPO4, sodium phosphate dibasic
0.034 M NaH2PO4, sodium phosphate monobasic
150 mM NaCl
Adjust pH to 8.0 by dropwise addition of 1 M NaOH
300 mM imidazole
Use within 1 day. May prepare from Ni-NTA wash buffer (see recipe, above), adding imidazole on day of use.
E1 storage buffer: 20 mM HEPES pH 8.0, 100 mM NaCl, 1 mM DTT
20 mM HEPES
100 mM NaCl
Adjust pH to 8.0 by dropwise addition of 1 M NaOH
1 mM DTT
Use within 1 day. May eliminate DTT, store up to 1 month at room temperature in a dark place, and add DTT to needed portion.
500 mM NaH2PO4/Na2HPO4 pH 8.0
0.47 M Na2HPO4, sodium phosphate dibasic
0.034 M NaH2PO4, sodium phosphate monobasic
Adjust pH to 8.0 by dropwise addition of 1 M NaOH
Store at room temperature for up to 1 month
Storage Buffer A: 25 mM NaCl, 12.5 mM HEPES pH 6.7
25 mM NaCl
12.5 mM HEPES
Adjust pH to 7.5 by dropwise addition of 1 M NaOH
Store at room temperature for up to 1 month in a dark place; check pH to ensure quality.
50 mM HEPES pH 6.5
50 mM HEPES
Adjust pH to 6.5 by dropwise addition of 1 M NaOH
Store at room temperature for up to 1 month in a dark place; check pH to ensure quality.
Acetonitrile/water (3:7) with 0.1 % TFA
30% v/v acetonitrile
0.1% v/v TFA
Store indefinitely at room temperature.
DMSO/H2O 1:1
50% DMSO in water (v/v)
Use within 1 day; store solutions containing this mixture at −20°C
1 M HEPES pH 7.5
1 M HEPES
Adjust pH to 7.5 by addition of solid NaOH pellets and/or dropwise addition of 1 M NaOH
Store at room temperature for up to 1 month in a dark place; check pH to ensure quality.
Storage Buffer B: 250 mM NaCl, 12.5 mM HEPES pH 6.0
250 mM NaCl
12.5 mM HEPES
Adjust pH to 6.0 by dropwise addition of 1 M NaOH
Store at room temperature for up to 1 month in a dark place; check pH to ensure quality.
1 × PBS with 20 mM BME
137 mM NaCl
2.7 mM KCl
10 mM Na2HPO4
1.8 mM KH2PO4
Adjust pH to 7.4 if necessary
20 mM BME
Use within 1 day; may eliminate BME, store up to 1 month at room temperature in a dark place, and add BME to needed portion.
UbFluor Assay Buffer: 1.5 M NaCl, 500 mM HEPES pH 7.5, 10x
1.5 M NaCl
500 mM HEPES
Adjust pH to 7.5 by addition of solid NaOH pellets and/or dropwise addition of 1 M NaOH.
Store at room temperature for up to 1 month in a dark place; check pH to ensure quality.
COMMENTARY
Background Information
Protein ubiquitination is an important posttranslational modification that regulates protein stability, activity, and localization. Approximately 800 ubiquitin enzymes regulate the ubiquitination state of some ~19,000 substrates, and represent a vast and unexplored area of the druggable genome (Kim et al., 2011). The drug discovery potential of the ubiquitin system is comparable to that of three other families: kinases (~500 known), proteases (~600 known), and GPCRs (~800 known). One unique feature of the ubiquitin cascades is their inherent complexity since ubiquitin conjugation requires three enzymes: E1, E2, and E3s. The situation is further complicated because ~300 Cullin-RING E3s are multi-subunit ligases (Bennett et al.). HECT E3 ligases are the simplest and most well characterized E3 ligases that harbor a catalytic cysteine that accepts ubiquitin from an E2~Ub thioester (Scheffner et al., 1995). The resulting HECT E3~Ub thioester can ligate ubiquitin onto the lysine of a protein substrate. Recent studies suggest that genetic inactivation of HECT E3s is associated with either abnormal phenotypes in mice or disease related phenotypes in humans (Table 1).
Table 1.
Connection between genetic inactivation of HECT E3s and mouse/human phenotypes.
| HECT E3 | Mouse Phenotype | Human Phenotype |
|---|---|---|
|
Gene: UBE3A
Protein: E6AP |
Deletion of maternal copy of UBE3A impairs synapse development and plasticity. Angelman syndrome phenotypes: motor dysfunction, inducible seizures, impaired long term potentiation (Jiang et al., 1998; Miura et al., 2002). |
Angelman Syndrome: Deletion or inactivating missense mutation of maternal UBE3A allele (chromosome 15q11-q13) (Albrecht et al., 1997; Jiang et al., 1999; Kishino et al., 1997). Autism: duplication or triplication of maternally inherited chromosome 15q11- q13 (Veenstra-VanderWeele et al., 1999). Missense mutation of T485A, that abrogates inhibitory phosphorylation of UBE3A by protein kinase A (Yi et al., 2015). Cervical cancer: E6AP (UBE3A) is highjacked by a high risk HPV 16 and 18 viruses that produce E6 protein. E6 protein binds E6AP ligase and p53, promoting polyubiquitination and proteasomal degradation of p53(Scheffner et al., 1993b). |
| ITCH |
ITCH−/− mice: skin scratching, severe immune disease resulting in death at 4-6 months. Alveolar proteinosis and interstitial inflammation in lungs, lymphoid hyperplasia (Hustad et al., 1995; Perry et al., 1998). Protection from diet induced obesity (Marino et al., 2014). |
Homozygous truncating mutations in ITCH in humans are associated with organomegaly, failure to thrive, developmental delay, dysmorphic features, and autoimmune inflammatory cell infiltration of the lungs, liver, and gut (Lohr et al., 2010). |
| Nedd4-1 |
Nedd4-1−/− mice: death at birth, body weight 65% less than that of wild type (Cao et al., 2008). Nedd4-1 +/− mice: fertile and viable, protection from diet induced obesity. Glucose tolerant, reduced insulin sensitivity. Less lipid accumulation and accelerated adipose lipolysis. Decreased long-term spatial memory (Li et al., 2015). Mechanism: positive regulator of Insulin and IGF-1 growth pathway. Inactivates PTEN that dephosphorylates tyrosine of IRS-1 (Shi et al., 2014). |
N/A |
|
Nedd4-2,
Nedd4L |
Nedd4-2−/− mice: Develop normally, size is similar to wt mice. Lungs have collapsed alveolar spaces and fail to inflate to a functional level. Turn blue upon birth, short gasping breath, die 10-60 min after revival. Increased ENaC expression in lungs, kidneys, which leads to premature lung fluid clearance, and a failure to inflate the lungs causing respiratory distress and perinatal lethality. Nedd4-2−/− survivors (4.2%) die after 22 days. Lungs are infiltrated with inflammatory cells such as macrophages and neutrophils (Boase et al., 2011). Nedd4-2-/+ mice: increased blood pressure at normal salt diet (0.3% Na+) 109.4 mm Hg vs 87.1 mm Hg in wild type (Boase et al., 2011). |
Lyddle Syndrome, chronic hypertension. Mutations in PY motif of ENaC that prevent its binding to WW domains of Nedd4-2 and ubiquitination (Schild et al., 1996). |
|
ARF-BP1
MULE HUWE1 |
ARF-BP1−/− mice: embryonic lethality, hemorrhage in the abdominal region by E14.5. Activation of p53 and caspase cleavage (Kon et al., 2012). Mice with ARF-BP1−/− knock out in pancreatic β-cells: loss of pancreatic β-cells with age, hyperglycemia and insulin deficiency. Diabetic conditions are rescued with the concomitant deletion of p53 in pancreatic β-cells (Kon et al., 2012). Known substrate: p53 |
Missense mutations of HUWE1 are associated with the phenotypic severity in a case of familial idiopathic intellectual disability (Isrie et al., 2013). Copy-number gains of HUWE1 are associated with nonsyndromic intellectual disability (Froyen et al., 2012). Duplication or mutations of HUWE1 is associated with mental retardation (Froyen et al., 2008). Mutation of HUWE1 was found in hepatocellular carcinoma patient (Liu et al., 2012). |
Given the involvement of HECT E3s in biology, significant efforts are directed toward identifying substrates of HECT E3s to uncover biological pathways they regulate (Persaud et al., 2009), and to develop chemical probes that target these E3s to validate them as drug targets (Cao et al., 2014; Peter et al., 2014). Importantly, small molecule probes for HECT E3s can be used to identify substrates of E3 ligases using quantitative proteomics methods (Kim et al., 2011). Pharmacological inhibition of an E3 ligase should cause a decrease in the ubiquitination of direct downstream substrates of that E3. These changes can be detected upon immunoprecipitation of ubiquitinated peptides with GGK-antibodies upon trypsin digestion of cell lysates. This strategy was used to identify protein substrates of Cullin-RING E3 ligases using an inhibitor of Culling-RING E3 MLN4924 (Kim et al., 2011).
Thus, the discovery and development of small molecule probes for HECT E3s will serve dual purposes: a) small molecule probes can be coupled with quantitative proteomics to identify protein substrates of HECT E3s, and b) these small molecules can be used as tool compounds to validate HECT E3s as drug targets.
One challenge associated with the discovery of chemical probes of HECT E3s is the complexity of enzymatic assays, which translate into difficulties during high-throughput screening, and subsequent SAR and dose-response studies. A typical enzymatic reaction requires E1, E2, HECT E3 enzymes, ubiquitin, and ATP. Furthermore, during the reaction three covalent enzyme intermediates are produced: E1~Ub, E2~Ub, and E3~Ub thioesters. Some assays measure the quantity of polyubiquitin chains with Tandem Ubiquitin Binding Entities (TUBEs). Overall, at least six reagents must be used in the assay, which create substantial operational complexity and add cost during large scale high-throughput screening or SAR studies. Furthermore, false positive hits may arise due to off-target inhibition of E1 and E2 enzymes or their respective thioesters. Such an assay is also not convenient for academic laboratories, especially in cases when many alanine mutants of HECT E3s need to be tested (Kamadurai et al., 2013). Western blot quantitation can be complicated and may require optimization by adjusting the concentrations of ATP, Ub, E1, E2, and HECT E3 enzymes. Thus it is clear that a simple two-component assay to measure HECT E3 ligase activity upon biochemical point mutations, or incubation with small molecules would be advantageous.
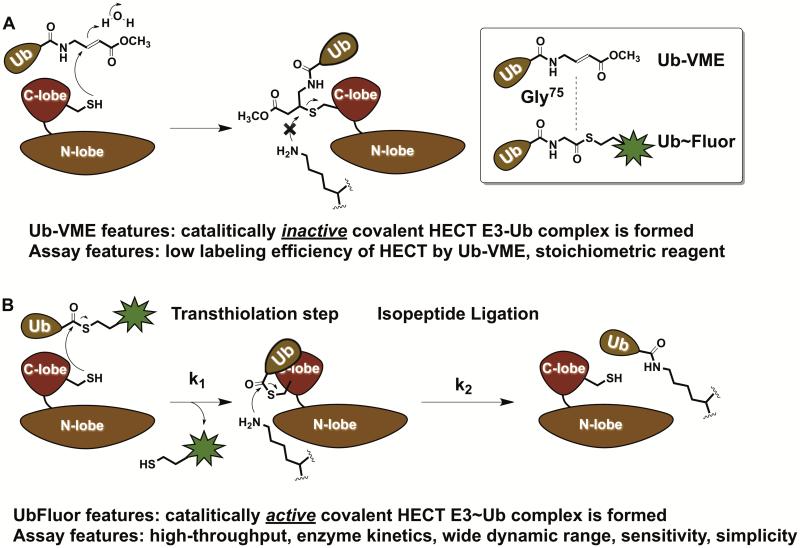
One approach to develop such assays relies on Ubiquitin-vinylmethyl ester (Ub-VME) activity-based probes and their analogues. Ub-VME were originally developed as activity-based probes for deubiquitinating enzymes (DUBs) in vitro and in cell lysates (Borodovsky et al., 2002). These probes are suicide inhibitors that bind deubiquitinating enzymes and react covalently and irreversibly with the catalytic cysteine. These probes were successfully used to covalently label DUBs in cell lysates leading to the discovery of previously unknown DUBs. Since HECT E3s have a catalytic cysteine, two known Ub binding sites, and form covalent HECT E3~Ub thioester intermediates, it is rational to propose that Ub-VME probes can be used to label the catalytic cysteine of HECT E3s. Accordingly, it is possible to screen for small molecule inhibitors that prevent Ub-VME labeling of HECT catalytic cysteines in simple competition assays. Indeed, it was shown that this strategy can be used to covalently label HECT E3s in cell lysates and in vitro (Love et al., 2009). However, Ub-VME and its analogues are stoichiometric suicide inhibitors and cannot be used in functional enzymatic assays.
To develop more relevant probes for HECT E3s we developed the fluorescent C-terminal ubiquitin thioesters UbFluor (Krist et al., 2016). Chemically, these probes are similar to Ub-VME yet fundamentally different. Ub-VME is a suicide inhibitor of DUBs and HECT E3s, and upon labeling the catalytic cysteine form a catalytically inactive complex. UbFluor and its non-fluorescent analogue Ub~MES, on the other hand, undergo a transthiolation reaction with the catalytic cysteine of HECT E3s and form a catalytically active HECT E3~Ub thioester, which can ligate ubiquitin onto the lysine of a protein substrate (Figure 5). In its essence, this is the first example of ATP-, E1-, and E2-independent protein ubiquitination reactions in vitro (Park et al., 2015), and recently an E1- and E2-independent ubiquitination reaction was discovered in L. Pneumophila (Qiu et al., 2016). As of today, technicians in other academic laboratories and core facilities have been producing UbFluor, and are using UbFluor in biochemical assays and high-throughput screens to discover chemical probes of HECT E3 ligases. We envision that the simplicity and robustness of these assays should facilitate rapid training of high-throughput screening personnel in academic and industrial laboratories. It is important to note that besides HECT E3s (~28 known), there are ~14 Ring-Between-Ring E3s that also bear a catalytic cysteine (Dove et al., 2016), and ~30 bacterial E3 ligases that are known to form the covalent E3~Ub thioester intermediates (Hicks and Galan, 2010). Most likely, UbFluor can be used to monitor the enzymatic activity of and screen for small molecule inhibitors of these E3 ligases as well. Further applications of UbFluor probes in enzymatic studies and high-throughput screening assays for HECT E3s and other E3s will be reported elsewhere.
Figure 5.
Fundamental differences between the previously described electrophilic Ub-VME probes and novel C-terminal fluorescent UbFluor probes. A) Ub-VME is a suicide inhibitor of HECT E3 ligases and thus is a stoichiometric reagent, which does not allow enzyme kinetic studies. B) Fluorescent C-terminal ubiquitin thioesters UbFluor undergo an efficient transthiolation reaction with the catalytic cysteine of HECT E3, generating the catalytically active HECT E3~Ub thioester that can transfer the ubiquitin onto the acceptor lysine. UbFluor enables enzymatic assays, and detects defects in both transthiolation (k1) and isopeptide ligation (k2) steps. UbFluor-based assays are robust, simple, fast, and amenable to high-throughput screening (384-well plates). Only two components are required (UbFluor and HECT E3), which stand in sharp contrast to commercially available assays for HECT E3s that require at least 8 reagents– ATP, Ub, E1, E2, HECT E3, and three detection reagents based on TUBEs to quantify the amount of polyubiquitin chains.
Critical Parameters
Since UbFluor is a thioester, assays must be strictly run in the absence of free thiol such as DTT or BME, which will cleave the thioester between Ub and fluorescein. UbFluor is stable in the presence of another reducing reagent, TCEP. Thus, when there is a need to use reducing agent with UbFluor, TCEP should be the reagent of choice. UbFluor is generally stable in the presence of 100 mM of lysine. In addition, caution should be exercised when small molecules are screened against HECT E3 ligase using UbFluor: if such small molecules have thiol or amine groups they may cleave UbFluor even in the absence of HECT E3.
Some small molecules and biological reagents can also cause an increase in fluorescence polarization over time. We observed that bovine serum albumin increases polarization of UbFluor in a time-dependent manner. Fluorescent small molecules may also disrupt fluorescence polarization readings; thus, such molecules should be excluded from use in UbFluor assays as modulators or labeling agents.
Troubleshooting
Issue: FluorSH does not dissolve in conjugation solution containing Ub~MES.
Possible solutions:
-Make sure to HPLC-purify FluorSH.
- Add more saturated NaHCO3 or DMSO (about 10 μL of either)
Issue: The blank (no enzyme sample) demonstrates a slight drop in polarization over the first 10-20 minutes of reaction.
Possible solution:
−This is a common phenomenon that we have observed and is typically more pronounced under MT conditions than ST conditions. Using a higher concentration of enzyme may help if reactions are difficult to distinguish from the blank.
Issue: More than 90% of UbFluor is consumed before the FP measurement begins.
Possible solution:
-Use a lower concentration of ligase in the reaction or utilize the Alternate Protocol 1 ‘Fast’ protocol.
Anticipated Results
Basic Protocol 1 will provide sufficient UBE1 enzyme that can be stored and used to chemoenzymatically produce Ub~MES. Basic Protocol 2 will produce sufficient quantities of FluorSH than can be stored and used when needed for conjugation to Ub~MES to produce UbFluor. Basic Protocol 2 will also produce the required amounts of UbFluor for fluorescence polarization studies. Basic Protocol 3 will establish proper reaction conditions to measure the progress of the enzymatic reaction between HECT E3 and UbFluor, and establish a framework for enzyme kinetic studies. Furthermore, the developed conditions can be used to establish HTS assays to screen for chemical probes for HECT E3s. Basic protocol 4 will make the user familiar with data analysis and interpretation. Our typical rates under ST reaction conditions are 0.03 μM/min (5 μM HECT E3, 0.25 – 1 μM UbFluor) and under MT reaction conditions are 0.15 μM/min (0.5 μM HECT E3, 5-20 μM UbFluor) (Figure 6-7).
Figure 6.
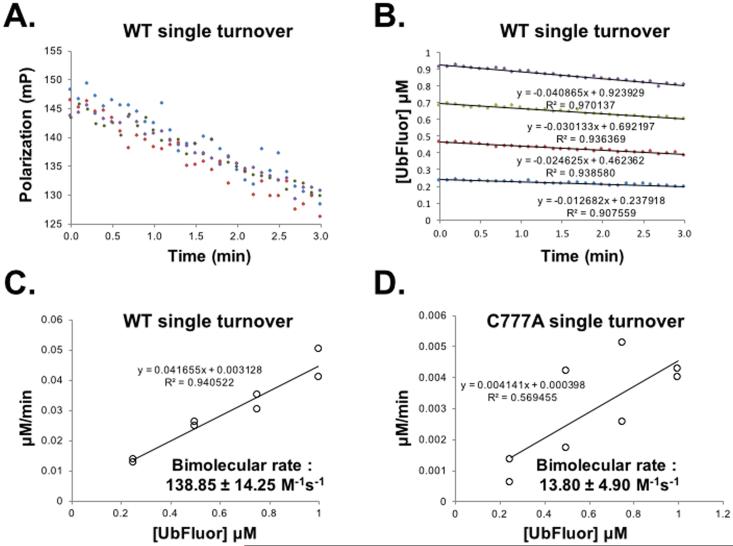
A bimolecular rate constant can be obtained from ST conditions. (A) Raw polarization data from a ST experiment with WT Rsp5 HECT: Rsp5 HECT (5 μM) with UbFluor (0.25 (blue), 0.50 (red), 0.75 (green), 1.00 μM (purple). Measurements were taken every 6 seconds for 3 minutes, at which point 20% of available UbFluor had been consumed. (B) Polarization values from (A) are converted to concentrations of UbFluor according to Table S1. (C) Slopes from the lines in (B) and from a replicate experiment performed at the same enzyme and UbFluor concentrations are plotted against UbFluor concentrations. The resulting slope of the line (kobs = 0.041655 min-1) is converted into the bimolecular rate by dividing by enzyme concentration and 60 sec/min. (D) The same procedure described for (A – C) is performed for the catalytically inactive mutant Rsp5 HECT C777A. For plots in (C) and (D), error of bimolecular rate was calculated using the “linest” function in Microsoft Excel to specify the error in the slope. The R2 values for linear trendlines in panel A are > 0.90. Data in panel A appear more scattered than in panel B because of the y-axis scale difference. Measurement of UbFluor consumption at each of four UbFluor concentrations was performed twice (8 total measurements). All data points are shown in (C) and (D). Reproduced from (Krist et al., 2016).
Figure 7.
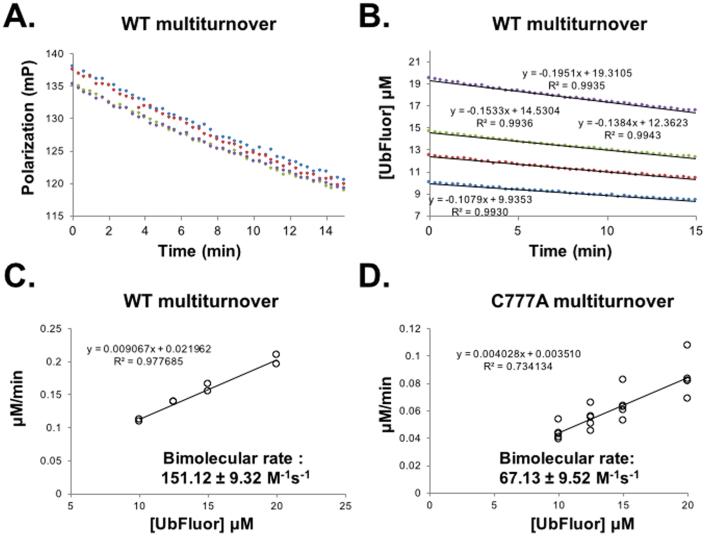
A bimolecular rate constant can be obtained from MT conditions. (A – C are also given as main text Figure 5). (A) Raw polarization data from a MT experiment with WT Rsp5 HECT: Rsp5 HECT (1 μM) with UbFluor (10 (blue), 12.5 (red), 15 (green), 20 μM (purple)). Measurements were taken every 20 seconds for 15 minutes. (B) Polarization values from (A) are converted to concentrations of UbFluor according to Table S1. (C) Slopes from the lines in (B) and from a replicate experiment performed at the same enzyme and UbFluor concentrations are plotted against UbFluor concentrations. The resulting slope of the line (kobs = 0.009067 min-1) is converted into the bimolecular rate by dividing by enzyme concentration and 60 sec/min. (D) The same procedure described for (A – C) is performed for the catalytically inactive mutant Rsp5 HECT C777A. For plots in (C) and (D), error of bimolecular rate was calculated using the “linest” function in Microsoft Excel to specify the error in the slope. Measurement of UbFluor consumption at each of four UbFluor concentrations was performed twice for WT (8 total measurements), and was performed 5 times for C777A (20 total measurements). All data points are shown in (C) and (D). Reproduced from (Krist et al., 2016)
Time Considerations
For typical experiments with HECT ligase, assays are run in the presence of upstream E1 and E2 enzymes, ATP and ubiquitin. Assembling these components and maintaining laboratory stocks can be time consuming and expensive. UbFluor, on the other hand, allows a simple, two-component reaction that can be monitored by fluorescence polarization, obviating the need for more labor intensive SDS-PAGE gels.
Preparation of UBE1 enzyme should require 3-5 days, and the subsequent preparation of Ub~MES requires one day for synthesis and FPLC size exclusion purification. Conjugation of FluorSH to Ub~MES should require 1-2 days for the reaction and subsequent purification. A skilled chemist can complete the synthesis of FluorSH in 7-10 days.
30-60 minutes should be allowed to prepare a set of UbFluor reactions for FP measurement. Data analysis can require several hours to days depending on the size of the dataset.
ACKNOWLEDGEMENT
(mandatory for NIH, optional for all others): Funding from Northwestern University and the CLP Cornew Innovation Award is greatly acknowledged. A. V. S. is a Pew Scholar in the Biomedical Sciences, supported by the Pew Charitable Trusts. D. T. K. has been supported by National Institute of General Medical Sciences Training Grant 5T32GM008382. D. T. K. is supported by an NU Nicholson Fellowship. Research reported in this publication was supported by NIGMS of the National Institutes of Health under award number R01GM987654 to A.V.S. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
LITERATURE CITED
- Albrecht U, Sutcliffe JS, Cattanach BM, Beechey CV, Armstrong D, Eichele G, Beaudet AL. Imprinted expression of the murine Angelman syndrome gene, Ube3a, in hippocampal and Purkinje neurons. Nat. Genet. 1997;17:75–78. doi: 10.1038/ng0997-75. [DOI] [PubMed] [Google Scholar]
- An H, Statsyuk AV. Development of Activity-Based Probes for Ubiquitin and Ubiquitin-like Protein Signaling Pathways. Journal of the American Chemical Society. 2013;135:16948–16962. doi: 10.1021/ja4099643. [DOI] [PubMed] [Google Scholar]
- Bennett EJ, Rush J, Gygi SP, Harper JW. Dynamics of Cullin-RING Ubiquitin Ligase Network Revealed by Systematic Quantitative Proteomics. Cell. 143:951–965. doi: 10.1016/j.cell.2010.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boase NA, Rychkov GY, Townley SL, Dinudom A, Candi E, Voss AK, Tsoutsman T, Semsarian C, Melino G, Koentgen F, Cook DI, Kumar S. Respiratory distress and perinatal lethality in Nedd4-2-deficient mice. Nat. Commun. 2011;2:287. doi: 10.1038/ncomms1284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borodovsky A, Ovaa H, Kolli N, Gan-Erdene T, Wilkinson KD, Ploegh HL, Kessler BM. Chemistry-based functional proteomics reveals novel members of the deubiquitinating enzyme family. Chem. Biol. 2002;9:1149–1159. doi: 10.1016/s1074-5521(02)00248-x. [DOI] [PubMed] [Google Scholar]
- Cao XR, Lill NL, Boase N, Shi PP, Croucher DR, Shan H, Qu J, Sweezer EM, Place T, Kirby PA, Daly RJ, Kumar S, Yang B. Nedd4 controls animal growth by regulating IGF-1 signaling. Sci. Signal. 2008;1:ra5. doi: 10.1126/scisignal.1160940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao Y, Wang C, Zhang X, Xing G, Lu K, Gu Y, He F, Zhang L. Selective small molecule compounds increase BMP-2 responsiveness by inhibiting Smurf1-mediated Smad1/5 degradation. Sci. Rep. 2014;4:4965. doi: 10.1038/srep04965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciechanover A. Intracellular protein degradation: from a vague idea through the lysosome and the ubiquitin-proteasome system and onto human diseases and drug targeting. Bioorg. Med. Chem. 2013;21:3400–3410. doi: 10.1016/j.bmc.2013.01.056. [DOI] [PubMed] [Google Scholar]
- Dove KK, Stieglitz B, Duncan ED, Rittinger K, Klevit RE. Molecular insights into RBR E3 ligase ubiquitin transfer mechanisms. EMBO Rep. 2016 doi: 10.15252/embr.201642641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El Oualid F, Merkx R, Ekkebus R, Hameed DS, Smit JJ, de Jong A, Hilkmann H, Sixma TK, Ovaa H. Chemical Synthesis of Ubiquitin, Ubiquitin-Based Probes, and Diubiquitin. Angew. Chem. Intl. Ed. 2010;49:10149–10153. doi: 10.1002/anie.201005995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Froyen G, Belet S, Martinez F, Santos-Reboucas CB, Declercq M, Verbeeck J, Donckers L, Berland S, Mayo S, Rosello M, Pimentel MM, Fintelman-Rodrigues N, Hovland R, Rodrigues dos Santos S, Raymond FL, Bose T, Corbett MA, Sheffield L, van Ravenswaaij-Arts CM, Dijkhuizen T, Coutton C, Satre V, Siu V, Marynen P. Copy-number gains of HUWE1 due to replication- and recombination-based rearrangements. Am. J. Hum. Genet. 2012;91:252–264. doi: 10.1016/j.ajhg.2012.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Froyen G, Corbett M, Vandewalle J, Jarvela I, Lawrence O, Meldrum C, Bauters M, Govaerts K, Vandeleur L, Van Esch H, Chelly J, Sanlaville D, van Bokhoven H, Ropers HH, Laumonnier F, Ranieri E, Schwartz CE, Abidi F, Tarpey PS, Futreal PA, Whibley A, Raymond FL, Stratton MR, Fryns JP, Scott R, Peippo M, Sipponen M, Partington M, Mowat D, Field M, Hackett A, Marynen P, Turner G, Gecz J. Submicroscopic duplications of the hydroxysteroid dehydrogenase HSD17B10 and the E3 ubiquitin ligase HUWE1 are associated with mental retardation. Am. J. Hum. Genet. 2008;82:432–443. doi: 10.1016/j.ajhg.2007.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hicks SW, Galan JE. Hijacking the host ubiquitin pathway: structural strategies of bacterial E3 ubiquitin ligases. Curr. Opin. Microbiol. 2010;13:41–46. doi: 10.1016/j.mib.2009.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huibregtse J, Scheffner M, Beaudenon S, Howley P. A family of proteins structurally and functionally related to the E6-AP ubiquitin-protein ligase. Proc. Natl. Acad. Sci. U.S.A. 1995;92:2563–2567. doi: 10.1073/pnas.92.7.2563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hustad CM, Perry WL, Siracusa LD, Rasberry JC, Cobb L, Cattanach BM, Kovatch R, Copeland NG, Jenkins NA. Molecular Genetic Characterization of Six Recessive Viable Alleles of the Mouse agouti Locus. Genetics. 1995;140:255–265. doi: 10.1093/genetics/140.1.255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isrie M, Kalscheuer VM, Holvoet M, Fieremans N, Van Esch H, Devriendt K. HUWE1 mutation explains phenotypic severity in a case of familial idiopathic intellectual disability. Eur. J. Med. Genet. 2013;56:379–382. doi: 10.1016/j.ejmg.2013.05.005. [DOI] [PubMed] [Google Scholar]
- Jiang Y.-h., Armstrong D, Albrecht U, Atkins CM, Noebels JL, Eichele G, Sweatt JD, Beaudet AL. Mutation of the Angelman Ubiquitin Ligase in Mice Causes Increased Cytoplasmic p53 and Deficits of Contextual Learning and Long-Term Potentiation. Neuron. 1998;21:799–811. doi: 10.1016/s0896-6273(00)80596-6. [DOI] [PubMed] [Google Scholar]
- Jiang Y.-h., Lev-Lehman E, Bressler J, Tsai T-F, Beaudet AL. Genetics of Angelman Syndrome. Am. J. Hum. Gen. 1999;65:1–6. doi: 10.1086/302473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamadurai HB, Qiu Y, Deng A, Harrison JS, MacDonald C, Actis M, Rodrigues P, Miller DJ, Souphron J, Lewis SM, Kurinov I, Fujii N, Hammel M, Piper R, Kuhlman B, Schulman BA. Mechanism of ubiquitin ligation and lysine prioritization by a HECT E3. eLife. 2013;2:e00828. doi: 10.7554/eLife.00828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kathman SG, Span I, Smith AT, Xu Z, Zhan J, Rosenzweig AC, Statsyuk AV. A Small Molecule That Switches a Ubiquitin Ligase From a Processive to a Distributive Enzymatic Mechanism. J. Am. Chem. Soc. 2015;137:12442–12445. doi: 10.1021/jacs.5b06839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim W, Bennett EJ, Huttlin EL, Guo A, Li J, Possemato A, Sowa ME, Rad R, Rush J, Comb MJ, Harper JW, Gygi SP. Systematic and quantitative assessment of the ubiquitin-modified proteome. Mol. Cell. 2011;44:325–340. doi: 10.1016/j.molcel.2011.08.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kishino T, Lalande M, Wagstaff J. UBE3A/E6-AP mutations cause Angelman syndrome. Nat. Genet. 1997;15:70–73. doi: 10.1038/ng0197-70. [DOI] [PubMed] [Google Scholar]
- Knight ZA, Shokat KM. Features of selective kinase inhibitors. Chem. Biol. 2005;12:621–637. doi: 10.1016/j.chembiol.2005.04.011. [DOI] [PubMed] [Google Scholar]
- Kon N, Zhong J, Qiang L, Accili D, Gu W. Inactivation of arf-bp1 induces p53 activation and diabetic phenotypes in mice. J. Biol. Chem. 2012;287:5102–5111. doi: 10.1074/jbc.M111.322867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krist DT, Park S, Boneh GH, Rice SE, Statsyuk AV. UbFluor: a mechanism-based probe for HECT E3 ligases. Chem. Sci. 2016;7:5587–5595. doi: 10.1039/c6sc01167e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine LM, Michener ML, Toth MV, Holwerda BC. Measurement of Specific Protease Activity Utilizing Fluorescence Polarization. Anal. Biochem. 1997;247:83–88. doi: 10.1006/abio.1997.2047. [DOI] [PubMed] [Google Scholar]
- Li JJ, Ferry RJ, Jr., Diao S, Xue B, Bahouth SW, Liao FF. Nedd4 haploinsufficient mice display moderate insulin resistance, enhanced lipolysis, and protection against high-fat diet-induced obesity. Endocrinology. 2015;156:1283–1291. doi: 10.1210/en.2014-1909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang Q, Dexheimer TS, Zhang P, Rosenthal AS, Villamil MA, You C, Zhang Q, Chen J, Ott CA, Sun H, Luci DK, Yuan B, Simeonov A, Jadhav A, Xiao H, Wang Y, Maloney DJ, Zhuang Z. A selective USP1-UAF1 inhibitor links deubiquitination to DNA damage responses. Nat. Chem. Biol. 2014;10:298–304. doi: 10.1038/nchembio.1455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu YX, Zhang SF, Ji YH, Guo SJ, Wang GF, Zhang GW. Whole-exome sequencing identifies mutated PCK2 and HUWE1 associated with carcinoma cell proliferation in a hepatocellular carcinoma patient. Oncol. Lett. 2012;4:847–851. doi: 10.3892/ol.2012.825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohr NJ, Molleston JP, Strauss KA, Torres-Martinez W, Sherman EA, Squires RH, Rider NL, Chikwava KR, Cummings OW, Morton DH, Puffenberger EG. Human ITCH E3 ubiquitin ligase deficiency causes syndromic multisystem autoimmune disease. Am. J. Hum. Genet. 2010;86:447–453. doi: 10.1016/j.ajhg.2010.01.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love KR, Pandya RK, Spooner E, Ploegh HL. Ubiquitin C-terminal electrophiles are activity-based probes for identification and mechanistic study of ubiquitin conjugating machinery. ACS Chem. Biol. 2009;4:275–287. doi: 10.1021/cb9000348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mari S, Ruetalo N, Maspero E, Stoffregen Mira C, Pasqualato S, Polo S, Wiesner S. Structural and Functional Framework for the Autoinhibition of Nedd4-Family Ubiquitin Ligases. Structure. 22:1639–1649. doi: 10.1016/j.str.2014.09.006. [DOI] [PubMed] [Google Scholar]
- Marino A, Menghini R, Fabrizi M, Casagrande V, Mavilio M, Stoehr R, Candi E, Mauriello A, Moreno-Navarrete JM, Gomez-Serrano M, Peral B, Melino G, Lauro R, Fernandez Real JM, Federici M. ITCH deficiency protects from diet-induced obesity. Diabetes. 2014;63:550–561. doi: 10.2337/db13-0802. [DOI] [PubMed] [Google Scholar]
- Miura K, Kishino T, Li E, Webber H, Dikkes P, Holmes GL, Wagstaff J. Neurobehavioral and Electroencephalographic Abnormalities in Ube3aMaternal-Deficient Mice. Neurobiol. Dis. 2002;9:149–159. doi: 10.1006/nbdi.2001.0463. [DOI] [PubMed] [Google Scholar]
- Ordureau A, Munch C, Harper JW. Quantifying ubiquitin signaling. Mol. Cell. 2015;58:660–676. doi: 10.1016/j.molcel.2015.02.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park S, Krist DT, Statsyuk AV. Protein ubiquitination and formation of polyubiquitin chains without ATP, E1 and E2 enzymes. Chem. Sci. 2015;6:1770–1779. doi: 10.1039/c4sc02340d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perry WL, Hustad CM, Swing DA, O'Sullivan TN, Jenkins NA, Copeland NG. The itchy locus encodes a novel ubiquitin protein ligase that is disrupted in a18H mice. Nat. Genet. 1998;18:143–146. doi: 10.1038/ng0298-143. [DOI] [PubMed] [Google Scholar]
- Persaud A, Alberts P, Amsen EM, Xiong X, Wasmuth J, Saadon Z, Fladd C, Parkinson J, Rotin D. Comparison of substrate specificity of the ubiquitin ligases Nedd4 and Nedd4-2 using proteome arrays. Mol. Syst. Biol. 2009;5:333. doi: 10.1038/msb.2009.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Persaud A, Rotin D. Use of proteome arrays to globally identify substrates for E3 ubiquitin ligases. In: Castrillo J, Oliver S, editors. Methods in Molecular Biology. Vol. 759. Springer, Inc.; New York, NY, USA: 2011. pp. 215–224. [DOI] [PubMed] [Google Scholar]
- Peter S, Bultinck J, Myant K, Jaenicke LA, Walz S, Muller J, Gmachl M, Treu M, Boehmelt G, Ade CP, Schmitz W, Wiegering A, Otto C, Popov N, Sansom O, Kraut N, Eilers M. Tumor cell-specific inhibition of MYC function using small molecule inhibitors of the HUWE1 ubiquitin ligase. EMBO Mol. Med. 2014;6:1525–1541. doi: 10.15252/emmm.201403927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu J, Sheedlo MJ, Yu K, Tan Y, Nakayasu ES, Das C, Liu X, Luo ZQ. Ubiquitination independent of E1 and E2 enzymes by bacterial effectors. Nature. 2016;533:120–124. doi: 10.1038/nature17657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronchi VP, Haas AL. Measuring rates of ubiquitin chain formation as a functional readout of ligase activity. Methods in molecular biology (Clifton, N.J.) 2012;832:197–218. doi: 10.1007/978-1-61779-474-2_14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheffner M, Huibregtse J, Vierstra R, PM H. The HPV-16 E6 and E6-AP complex functions as a ubiquitin-protein ligase in the ubiquitination of p53. Cell. 1993a;75:495–505. doi: 10.1016/0092-8674(93)90384-3. [DOI] [PubMed] [Google Scholar]
- Scheffner M, Huibregtse JM, Vierstra RD, Howley PM. The HPV-16 E6 and E6-AP complex functions as a ubiquitin-protein ligase in the ubiquitination of p53. Cell. 1993b;75:495–505. doi: 10.1016/0092-8674(93)90384-3. [DOI] [PubMed] [Google Scholar]
- Scheffner M, Kumar S. Mammalian HECT ubiquitin-protein ligases: biological and pathophysiological aspects. Biochim. Biophys. Acta. 2014;1843:61–74. doi: 10.1016/j.bbamcr.2013.03.024. [DOI] [PubMed] [Google Scholar]
- Scheffner M, Nuber U, Huibregtse J. Protein ubiquitination involving an E1-E2-E3 enzyme ubiquitin thioester cascade. Nature. 1995;373:81–83. doi: 10.1038/373081a0. [DOI] [PubMed] [Google Scholar]
- Schild L, Lu Y, Gautschi I, Schneeberger E, Lifton RP, Rossier BC. Identification of a PY motif in the epithelial Na channel subunits as a target sequence for mutations causing channel activation found in Liddle syndrome. Embo J. 1996;15:2381–2387. [PMC free article] [PubMed] [Google Scholar]
- Shi Y, Wang J, Chandarlapaty S, Cross J, Thompson C, Rosen N, Jiang X. PTEN is a protein tyrosine phosphatase for IRS1. Nat. Struct. Mol. Biol. 2014;21:522–527. doi: 10.1038/nsmb.2828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varshavsky A. The ubiquitin system, an immense realm. Annu. Rev. Biochem. 2012;81:167–176. doi: 10.1146/annurev-biochem-051910-094049. [DOI] [PubMed] [Google Scholar]
- Veenstra-VanderWeele J, Gonen D, Leventhal BL, Cook EH., Jr. Mutation screening of the UBE3A/E6-AP gene in autistic disorder. Mol. Psychiatry. 1999;4:64–67. doi: 10.1038/sj.mp.4000472. [DOI] [PubMed] [Google Scholar]
- Wilde S, Timpson A, Kirsanow K, Kaiser E, Kayser M, Unterländer M, Hollfelder N, Potekhina I, Schier W, Thomas M, Burger J. Direct evidence for positive selection of skin, hair, and eye pigmentation in Europeans during the last 5,000 y. Proc. Natl. Acad. Sci. U.S.A. 2014;111:4832–4837. doi: 10.1073/pnas.1316513111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi JJ, Berrios J, Newbern JM, Snider WD, Philpot BD, Hahn KM, Zylka MJ. An Autism-Linked Mutation Disables Phosphorylation Control of UBE3A. Cell. 2015;162:795–807. doi: 10.1016/j.cell.2015.06.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W, Wu KP, Sartori MA, Kamadurai HB, Ordureau A, Jiang C, Mercredi PY, Murchie R, Hu J, Persaud A, Mukherjee M, Li N, Doye A, Walker JR, Sheng Y, Hao Z, Li Y, Brown KR, Lemichez E, Chen J, Tong Y, Harper JW, Moffat J, Rotin D, Schulman BA, Sidhu SS. System-Wide Modulation of HECT E3 Ligases with Selective Ubiquitin Variant Probes. Mol Cell. 2016;62:121–136. doi: 10.1016/j.molcel.2016.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhuang X, Ha T, Kim HD, Centner T, Labeit S, Chu S. Fluorescence quenching: A tool for single-molecule protein-folding study. Proc. Nat. Acad. Sci. U.S.A. 2000;97:14241–14244. doi: 10.1073/pnas.97.26.14241. [DOI] [PMC free article] [PubMed] [Google Scholar]