Abstract
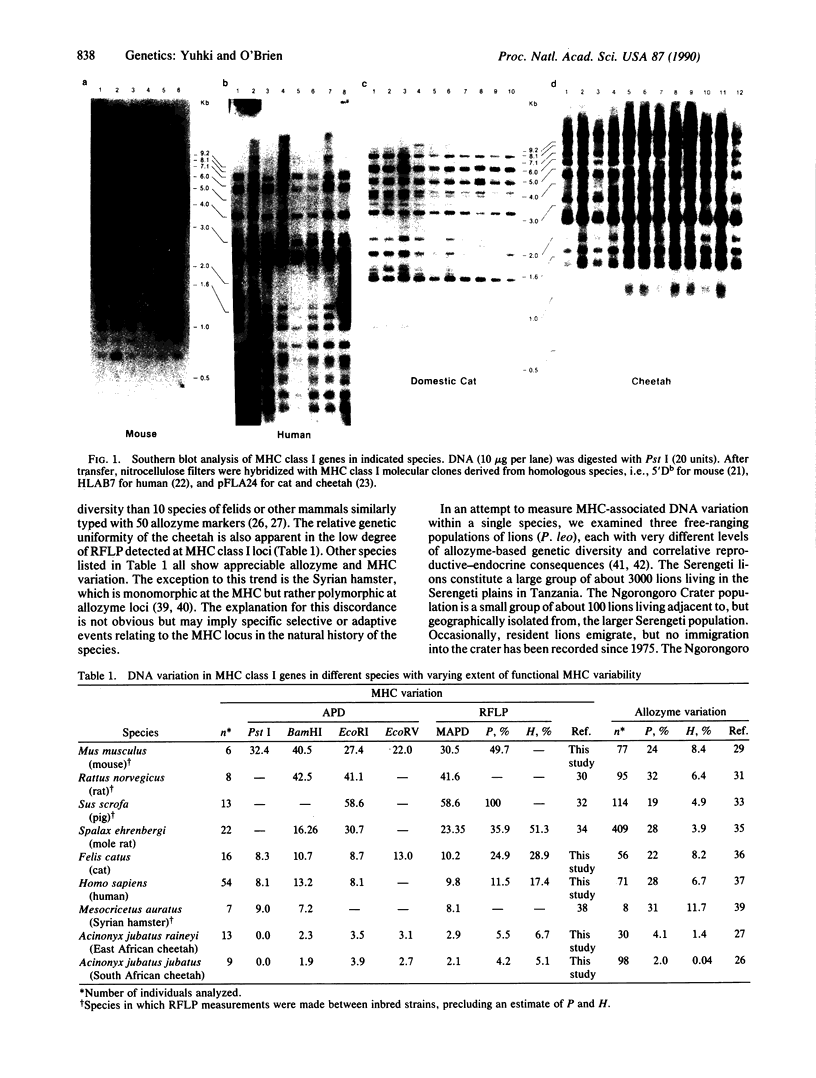
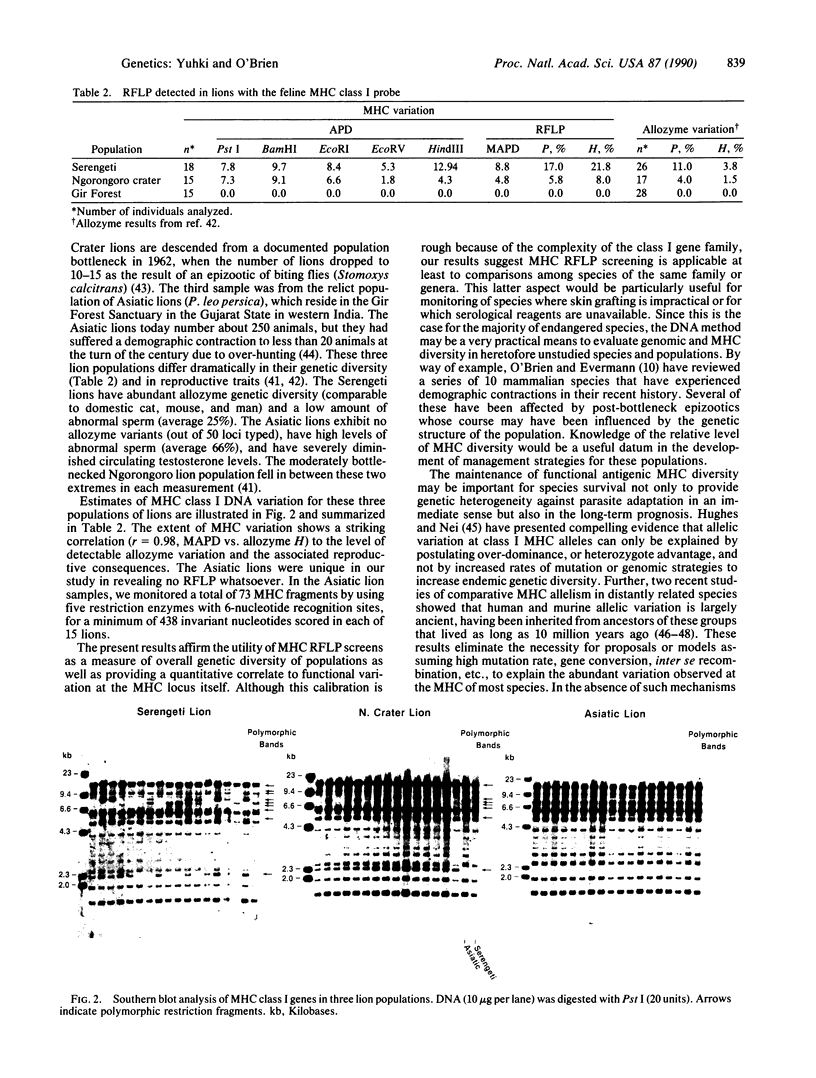
The major histocompatibility complex (MHC) is a multigene complex of tightly linked homologous genes that encode cell surface antigens that play a key role in immune regulation and response to foreign antigens. In most species, MHC gene products display extreme antigenic polymorphism, and their variability has been interpreted to reflect an adaptive strategy for accommodating rapidly evolving infectious agents that periodically afflict natural populations. Determination of the extent of MHC variation has been limited to populations in which skin grafting is feasible or for which serological reagents have been developed. We present here a quantitative analysis of restriction fragment length polymorphism of MHC class I genes in several mammalian species (cats, rodents, humans) known to have very different levels of genetic diversity based on functional MHC assays and on allozyme surveys. When homologous class I probes were employed, a notable concordance was observed between the extent of MHC restriction fragment variation and functional MHC variation detected by skin grafts or genome-wide diversity estimated by allozyme screens. These results confirm the genetically depauperate character of the African cheetah, Acinonyx jubatus, and the Asiatic lion, Panthera leo persica; further, they support the use of class I MHC molecular reagents in estimating the extent and character of genetic diversity in natural populations.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bjorkman P. J., Saper M. A., Samraoui B., Bennett W. S., Strominger J. L., Wiley D. C. The foreign antigen binding site and T cell recognition regions of class I histocompatibility antigens. Nature. 1987 Oct 8;329(6139):512–518. doi: 10.1038/329512a0. [DOI] [PubMed] [Google Scholar]
- Chardon P., Vaiman M., Kirszenbaum M., Geffrotin C., Renard C., Cohen D. Restriction fragment length polymorphism of the major histocompatibility complex of the pig. Immunogenetics. 1985;21(2):161–171. doi: 10.1007/BF00364868. [DOI] [PubMed] [Google Scholar]
- Csaikl F. Electrophoretic comparison of Syrian and Chinese hamster species. Heredity (Edinb) 1984 Feb;52(Pt 1):141–144. doi: 10.1038/hdy.1984.14. [DOI] [PubMed] [Google Scholar]
- Doherty P. C., Zinkernagel R. M. A biological role for the major histocompatibility antigens. Lancet. 1975 Jun 28;1(7922):1406–1409. doi: 10.1016/s0140-6736(75)92610-0. [DOI] [PubMed] [Google Scholar]
- Eriksson K., Halkka O., Lokki J., Saura A. Enzyme polymorphism in feral, outbred and inbred rats (Rattus norvegicus). Heredity (Edinb) 1976 Dec;37(3):341–349. doi: 10.1038/hdy.1976.98. [DOI] [PubMed] [Google Scholar]
- Figueroa F., Günther E., Klein J. MHC polymorphism pre-dating speciation. Nature. 1988 Sep 15;335(6187):265–267. doi: 10.1038/335265a0. [DOI] [PubMed] [Google Scholar]
- Flavell R. A., Allen H., Burkly L. C., Sherman D. H., Waneck G. L., Widera G. Molecular biology of the H-2 histocompatibility complex. Science. 1986 Jul 25;233(4762):437–443. doi: 10.1126/science.3726537. [DOI] [PubMed] [Google Scholar]
- Good M. F., Berzofsky J. A., Maloy W. L., Hayashi Y., Fujii N., Hockmeyer W. T., Miller L. H. Genetic control of the immune response in mice to a Plasmodium falciparum sporozoite vaccine. Widespread nonresponsiveness to single malaria T epitope in highly repetitive vaccine. J Exp Med. 1986 Aug 1;164(2):655–660. doi: 10.1084/jem.164.2.655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris H., Hopkinson D. A. Average heterozygosity per locus in man: an estimate based on the incidence of enzyme polymorphisms. Ann Hum Genet. 1972 Jul;36(1):9–20. doi: 10.1111/j.1469-1809.1972.tb00578.x. [DOI] [PubMed] [Google Scholar]
- Hughes A. L., Nei M. Pattern of nucleotide substitution at major histocompatibility complex class I loci reveals overdominant selection. Nature. 1988 Sep 8;335(6186):167–170. doi: 10.1038/335167a0. [DOI] [PubMed] [Google Scholar]
- Lawlor D. A., Ward F. E., Ennis P. D., Jackson A. P., Parham P. HLA-A and B polymorphisms predate the divergence of humans and chimpanzees. Nature. 1988 Sep 15;335(6187):268–271. doi: 10.1038/335268a0. [DOI] [PubMed] [Google Scholar]
- Mann D. L., Popovic M., Sarin P., Murray C., Reitz M. S., Strong D. M., Haynes B. F., Gallo R. C., Blattner W. A. Cell lines producing human T-cell lymphoma virus show altered HLA expression. Nature. 1983 Sep 1;305(5929):58–60. doi: 10.1038/305058a0. [DOI] [PubMed] [Google Scholar]
- Mayer W. E., Jonker M., Klein D., Ivanyi P., van Seventer G., Klein J. Nucleotide sequences of chimpanzee MHC class I alleles: evidence for trans-species mode of evolution. EMBO J. 1988 Sep;7(9):2765–2774. doi: 10.1002/j.1460-2075.1988.tb03131.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGuire K. L., Duncan W. R., Tucker P. W. Syrian hamster DNA shows limited polymorphism at class I-like loci. Immunogenetics. 1985;22(3):257–268. doi: 10.1007/BF00404485. [DOI] [PubMed] [Google Scholar]
- Nevo E., Shaw C. R. Genetic variation in a subterranean mammal, Spalax ehrenbergi. Biochem Genet. 1972 Dec;7(3):235–241. doi: 10.1007/BF00484821. [DOI] [PubMed] [Google Scholar]
- Nishimoto H., Kikutani H., Yamamura K., Kishimoto T. Prevention of autoimmune insulitis by expression of I-E molecules in NOD mice. 1987 Jul 30-Aug 5Nature. 328(6129):432–434. doi: 10.1038/328432a0. [DOI] [PubMed] [Google Scholar]
- Nizetić D., Figueroa F., Nevo E., Klein J. Major histocompatibility complex of the mole-rat. II. Restriction fragment polymorphism. Immunogenetics. 1985;22(1):55–67. doi: 10.1007/BF00430594. [DOI] [PubMed] [Google Scholar]
- O'Brien S. J., Roelke M. E., Marker L., Newman A., Winkler C. A., Meltzer D., Colly L., Evermann J. F., Bush M., Wildt D. E. Genetic basis for species vulnerability in the cheetah. Science. 1985 Mar 22;227(4693):1428–1434. doi: 10.1126/science.2983425. [DOI] [PubMed] [Google Scholar]
- O'Brien S. J., Wildt D. E., Bush M., Caro T. M., FitzGibbon C., Aggundey I., Leakey R. E. East African cheetahs: evidence for two population bottlenecks? Proc Natl Acad Sci U S A. 1987 Jan;84(2):508–511. doi: 10.1073/pnas.84.2.508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'brien S. J., Wildt D. E., Goldman D., Merril C. R., Bush M. The cheetah is depauperate in genetic variation. Science. 1983 Jul 29;221(4609):459–462. doi: 10.1126/science.221.4609.459. [DOI] [PubMed] [Google Scholar]
- Palmer M., Wettstein P. J., Frelinger J. A. Evidence for extensive polymorphism of class I genes in the rat major histocompatibility complex (RT1). Proc Natl Acad Sci U S A. 1983 Dec;80(24):7616–7620. doi: 10.1073/pnas.80.24.7616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ralls K., Brugger K., Ballou J. Inbreeding and juvenile mortality in small populations of ungulates. Science. 1979 Nov 30;206(4422):1101–1103. doi: 10.1126/science.493997. [DOI] [PubMed] [Google Scholar]
- Rice M. C., Gardner M. B., O'Brien S. J. Genetic diversity in leukemia-prone feral house mice infected with murine leukemia virus. Biochem Genet. 1980 Oct;18(9-10):915–928. doi: 10.1007/BF00500124. [DOI] [PubMed] [Google Scholar]
- Schrier P. I., Bernards R., Vaessen R. T., Houweling A., van der Eb A. J. Expression of class I major histocompatibility antigens switched off by highly oncogenic adenovirus 12 in transformed rat cells. 1983 Oct 27-Nov 2Nature. 305(5937):771–775. doi: 10.1038/305771a0. [DOI] [PubMed] [Google Scholar]
- Schwartz R. H. Immune response (Ir) genes of the murine major histocompatibility complex. Adv Immunol. 1986;38:31–201. doi: 10.1016/s0065-2776(08)60006-1. [DOI] [PubMed] [Google Scholar]
- Sood A. K., Pereira D., Weissman S. M. Isolation and partial nucleotide sequence of a cDNA clone for human histocompatibility antigen HLA-B by use of an oligodeoxynucleotide primer. Proc Natl Acad Sci U S A. 1981 Jan;78(1):616–620. doi: 10.1073/pnas.78.1.616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Streilein J. W., Duncan W. R. On the anomalous nature of the major histocompatibility complex in Syrian hamsters, Hm-1. Transplant Proc. 1983 Jun;15(2):1540–1545. [PubMed] [Google Scholar]
- Wildt D. E., O'Brien S. J., Howard J. G., Caro T. M., Roelke M. E., Brown J. L., Bush M. Similarity in ejaculate-endocrine characteristics in captive versus free-ranging cheetahs of two subspecies. Biol Reprod. 1987 Mar;36(2):351–360. doi: 10.1095/biolreprod36.2.351. [DOI] [PubMed] [Google Scholar]
- Winkler C., Schultz A., Cevario S., O'Brien S. Genetic characterization of FLA, the cat major histocompatibility complex. Proc Natl Acad Sci U S A. 1989 Feb;86(3):943–947. doi: 10.1073/pnas.86.3.943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuhki N., Heidecker G. F., O'Brien S. J. Characterization of MHC cDNA clones in the domestic cat. Diversity and evolution of class I genes. J Immunol. 1989 May 15;142(10):3676–3682. [PubMed] [Google Scholar]
- Yuhki N., O'Brien S. J. Molecular characterization and genetic mapping of class I and class II MHC genes of the domestic cat. Immunogenetics. 1988;27(6):414–425. doi: 10.1007/BF00364427. [DOI] [PubMed] [Google Scholar]
- Zinkernagel R. M., Doherty P. C. MHC-restricted cytotoxic T cells: studies on the biological role of polymorphic major transplantation antigens determining T-cell restriction-specificity, function, and responsiveness. Adv Immunol. 1979;27:51–177. doi: 10.1016/s0065-2776(08)60262-x. [DOI] [PubMed] [Google Scholar]