Abstract
Auxiliary metabolic genes (AMG) are commonly found in the genomes of phages that infect cyanobacteria and increase the fitness of the cyanophage. AMGs are often homologs of host genes, and also typically related to photosynthesis. For example, the ΦcpeT gene in the cyanophage P-HM1 encodes a putative phycobiliprotein lyase related to cyanobacterial T-type lyases, which facilitate attachment of linear tetrapyrrole chromophores to Cys-155 of phycobiliprotein β-subunits, suggesting that ΦCpeT may also help assemble light-harvesting phycobiliproteins during infection. To investigate this possibility, we structurally and biochemically characterized recombinant ΦCpeT. The solved crystal structure of ΦCpeT at 1.8-Å resolution revealed that the protein adopts a similar fold as the cyanobacterial T-type lyase CpcT from Nostoc sp. PCC7120 but overall is more compact and smaller. ΦCpeT specifically binds phycoerythrobilin (PEB) in vitro leading to a tight complex that can also be formed in Escherichia coli when it is co-expressed with genes encoding PEB biosynthesis (i.e. ho1 and pebS). The formed ΦCpeT·PEB complex was very stable as the chromophore was not lost during chromatography and displayed a strong red fluorescence with a fluorescence quantum yield of ΦF = 0.3. This complex was not directly able to transfer PEB to the host phycobiliprotein β-subunit. However, it could assist the host lyase CpeS in its function by providing a pool of readily available PEB, a feature that might be important for fast phycobiliprotein assembly during phage infection.
Keywords: bacteriophage, microbiology, photosynthetic pigment, protein crystallization, viral protein
Introduction
Bacteriophages are the most abundant entities on our planet and outnumber by far the quantity of their bacterial host cells (1, 2). During the last decade a number of cyanophages, viruses that infect cyanobacteria, have been isolated and their genomes have been sequenced (3–5). Many of the phage-encoded genes are homologs of their host and often encode functional proteins that are important for production of phage progeny (replication, transcription, and nucleotide metabolism). In addition, auxiliary metabolic genes (AMG)3 involved in photosynthesis have been identified in a number of cyanophage genomes and it was shown that they increase phage fitness during infection (6). One example is the cyanophage-encoded gene psbA encoding the core reaction center protein D1, which was shown to be expressed during infection with the amount of phage D1 protein increasing at the same time (7). In addition to the core reaction center encoding genes, several cyanophage genomes encode genes for enzymes/proteins involved in light-harvesting (4, 6, 8). Among them members of the family of ferredoxin-dependent bilin reductases, enzymes involved in the biosynthesis of the light-harvesting pigments phycocyanobilin (PCB) and phycoerythrobilin (PEB) have been identified. Although the PCB-producing enzyme PcyA resembles its host counterpart, the phage copy of a PEB biosynthesis gene encodes a bifunctional protein combining the activities of two cyanobacterial host genes (8). This new phycoerythrobilin synthase (PebS) efficiently produces PEB from a biliverdin precursor and is thus far solely found in genomes of cyanophages, often together with genes encoding a heme oxygenase (enzyme that converts heme to biliverdin, the substrate of PebS). Although it was shown that the genes are expressed during infection their function still remains elusive (8). However, it has been speculated that their expression also contributes to phage fitness by enhancing the light-harvesting capacity. In this regard, the cyanophage S-PM2 was shown to induce increased synthesis of the light-harvesting phycobiliprotein phycoerythrin (PE) in Synechococcus sp. WH7803 during infection (9). Inspection of the genome of this phage revealed that it lacks genes involved in pigment biosynthesis but contains one gene related to light-harvesting, cpeT. cpeT encodes a putative phycobiliprotein lyase (4), a protein likely to be involved in supporting the attachment of PEB to the apo-phycobiliprotein. Thereby, the lyase provides a scaffold for the pigment to ensure binding in the right conformation to facilitate correct stereospecific ligation to a conserved cysteine residue within the phycobiliprotein. CpeT belongs to the so-called T-type lyases and phycocyanobilin-specific homologs of CpeT (i.e. CpcT) were shown to specifically serve the position equivalent to cysteine 153 in the β-subunit of phycocyanin (PC) (10). Therefore, it is postulated that in analogy CpeT lyases serve the βCys-153 of PE. CpeT homologs can be found in several other sequenced cyanophage genomes including the Prochlorococcus infecting myovirus P-HM1 (11).
Within this study we present the first structural and biochemical characterization of a putative CpeT phycobiliprotein lyase from a virus. P-HM1 infects the high-light adapted Prochlorococcus marinus ecotype MED4, which only possesses a much degenerated form of a phycobiliprotein, the PE β-subunit (i.e. CpeB) (12). Our study confirmed the ability of recombinant P-HM1CpeT (ΦCpeT) to bind PEB and its precursor 15,16-dihydrobiliverdin forming a stable complex. Interestingly, the CpeT·PEB complex displayed a strong red fluorescence, a feature that was also confirmed with other phage-encoded CpeT homologs. The crystal structure with data to 1.8 Å provides additional insight into these features.
Results
Overall Structure of ΦCpeT
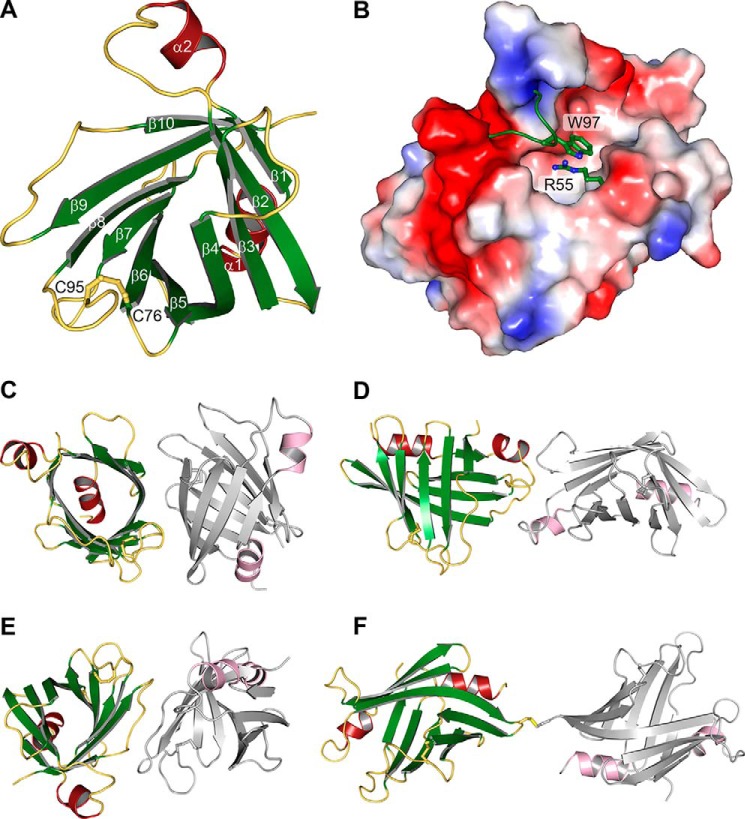
To obtain insights into the structure-function relationship of AMGs in cyanophages we solved the crystal structure of recombinant ΦCpeT (Fig. 1) from Prochlorococcus phage P-HM1 at 1.8-Å resolution (PDB code 5HI8; Table 1). ΦCpeT is the first biochemically characterized phycobiliprotein (PBP) lyase from a cyanophage. ΦCpeT adopts a typical barrel-like fold with 10 β-strands (Fig. 2). This barrel is formed by two sheets that lie on top of each other and are rotated by 90 degrees. They are connected by the kinked β-strand 4. This composition leads to a wide opening of the barrel, leaving a large cleft on its “side.” Closure of the two sheets is accomplished on one hand by the central N-terminal α-helix (the barrel bottom) and on the other hand by several loop regions between the strands. The opening of the barrel is even further extended because the loop between strands 1 and 2 is bent toward the outside of the barrel, stabilized by the structural element of α-helix 2. Another striking feature is the unusual composition of the loop between β-strand 7 and strand 8 (loop7–8). It is very long and the first four amino acid residues form a bulb to the exterior that is not stabilized by an interaction with other parts of the structure. However, an intramolecular cysteine bridge between Cys-76 and Cys-95 keeps it in place and stabilizes it.
FIGURE 1.
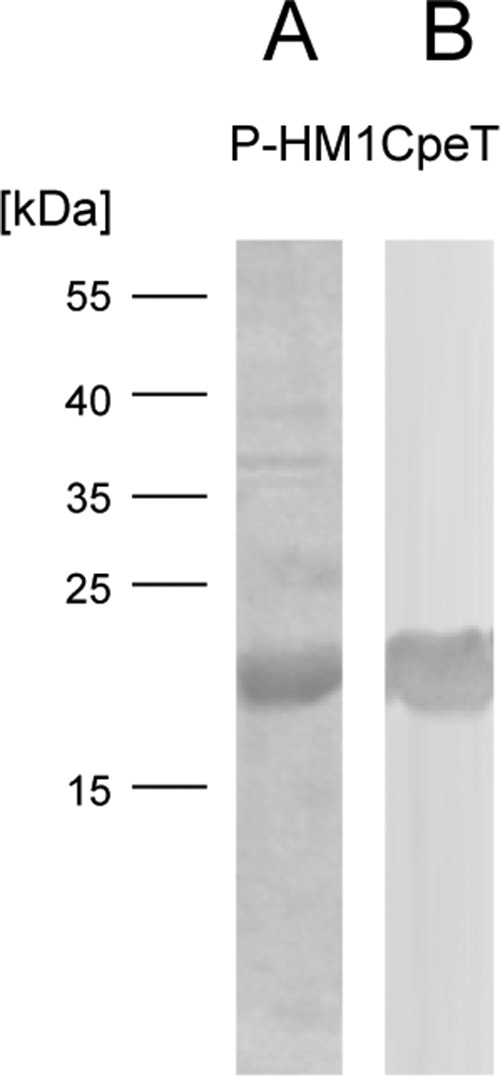
Purification of ΦCpeT using affinity chromatography. A, SDS-PAGE analysis of purified StrepII-P-HM1CpeT protein (19 kDa). B, Western blot transfer and immunological detection via StrepII-tag antibody.
TABLE 1.
Crystallographic data
| Low resolution dataset | High resolution dataset | |
|---|---|---|
| Crystallographic data | ||
| Space group | C 1 2 1 | C 1 2 1 |
| Cell dimensions | ||
| a, b, c (Å) | 62.67, 63.90, 93.48 | 63.29, 61.68, 93.34 |
| α, β, γ (degree) | 90, 109.56, 90 | 90, 109.78, 90 |
| Wavelength (Å) | 0.979 | 1.00 |
| Resolution (Å) | 50–2.5 (2.72–2.5)a | 50–1.80 (1.85–1.80) |
| CC1/2 | 99.4 (66.5) | 99.1 (70.4) |
| I/σ(I) | 6.63 (1.02) | 7.37 (1.18) |
| Rmeas (%) | 22.6 (184.7) | 10.0 (112.3) |
| Completeness (%) | 99.6 (99.1) | 97.9 (95.7) |
| Redundancy | 5.5 | 3.5 |
| Refinement | ||
| Rwork/Rfree (%) | 20.6/22.8 | |
| No. of reflections used | 60,319 | |
| No. of atoms | ||
| Protein | 4,497 | |
| Magnesium | 2 | |
| Acetate | 7 | |
| Water | 77 | |
| Average B-factor (Å2) | 35.27 | |
| Bond lengths root mean square deviations (Å) | 0.012 | |
| Bond angles root mean square deviations (°) | 1.341 | |
a Values in parentheses represent the highest resolution shell.
FIGURE 2.
Crystal structure of ΦCpeT. A, schematic representation of the β-barrel structure with its intramolecular disulfide bridge. B, electrostatic surface of the potential binding pocket of ΦCpeT. C–F, crystallographic interfaces as predicted by the PISA server.
The Crystallographic Arrangement Suggests a Monomer
The structure showed lowest R-factors in space group C2, despite the fact that scaling programs XDS and POINTLESS suggested space group F222. Hence, the asymmetric unit possesses two molecules.
According to the PISA server, all contacts show only small interaction surfaces. Two interaction sites exhibit buried areas of 604 and 532 Å2, respectively (Fig. 2, C and E). The next smaller contact area buries only 419 Å2 (Fig. 2D). Calculated, theoretical ΔG values are 1.4 and −2.0 kcal/mol, and are hence below the threshold for a reasonable interaction in solution. One crystal contact that is not recognized by PISA is stabilized by an intermolecular disulfide bridge, between residues Cys-65 of different molecules (Fig. 2F). Due to the fact that there is no additional residue contributing to this interaction, it seems likely that this covalent bond can only be formed in crystals and is irrelevant for the behavior of the protein in solution.
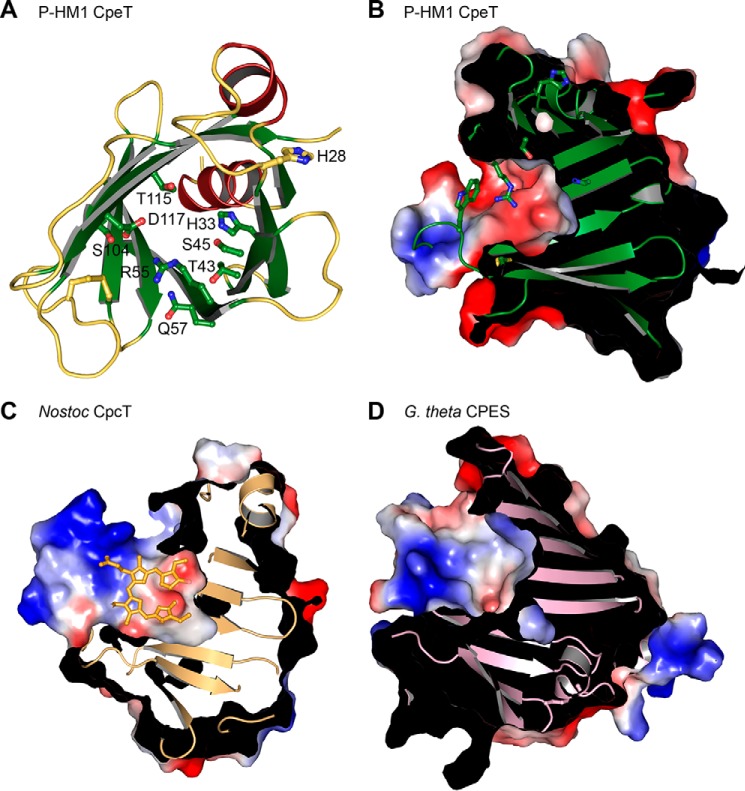
The Central Cavity Is Closed
The bottom of the central cavity is formed by the N-terminal helix. The deeper half is exclusively composed of hydrophobic residues, the majority of which are phenylalanines. A single hydrophilic residue, His-33, is located in the middle of the pocket. All other polar amino acids are positioned at the top rim of the opening and point into the upper part of the cavity. These include Ser-45, Gln-57, Ser-104, Thr-115, Asp-117, and Arg-55 (Fig. 3A). Arg-55 points into the center of the cavity, thereby strongly reducing its accessible volume. Compared with its surrounding and the majority of the structure, the guanidine group has a significantly higher B value of ∼70 Å2 (average B value: 35 Å2), indicating flexibility. The extended loop7–8 creates a lid on top of the cavity, by placing the side chain of Trp-97 centrally onto the opening. Loop7–8, and particularly the side chain of Trp-97 is highly flexible with an average B factor of 91 Å2.
FIGURE 3.
Binding pockets of PBP lyases. A, schematic representation of ΦCpeT indicating all hydrophilic residues that are located on the inner side. B, electrostatic surface potential of the binding pocket of ΦCpeT. C, surface of Nostoc sp. CpcT with bound PCB (light orange). D, binding pocket of the S-type PBP lyase CPES from G. theta (PDB code 4TQ2).
ΦCpeT Specifically Binds Bilins with Reduced 15,16-Double Bond
Cyanobacterial PBP lyases are chaperon-like enzymes that ensure the correct and regiospecific attachment of phycobilins to specific cysteine residues in the PBP. To address whether cyanophage encoded PBP lyases resemble their host counterpart, recombinant ΦCpeT was tested for its ability to bind various phycobilins. In their free form, phycobilins adopt a cyclic, helical porphyrin-like conformation (Z,Z,Z,s,s,s) which is visible as a broad absorption peak in the longer wavelength region (13). Upon binding to ΦCpeT, PEB displayed a significant change in its absorption properties. Binding resulted in an absorbance shift from 540 to 607 nm, concomitant with a strong increase in the extinction coefficient (from ϵ540 nm = 21.763 mm−1 cm−1 to ϵ607nm 55.247 mm−1 cm−1) indicating a change in bilin conformation to a more stretched, possibly protonated form (Fig. 4A). Addition of 15,16-DHBV, the biosynthetic precursor of PEB also resulted in a change in absorption but significantly weaker with new absorption peaks appearing at 560 and 605 nm (Fig. 4B). In contrast, addition of the related phycobilin PCB to ΦCpeT did not have a significant influence on the absorption properties (Fig. 4C) suggesting a strong preference of ΦCpeT for bilins with reduced 15,16-double bond.
FIGURE 4.
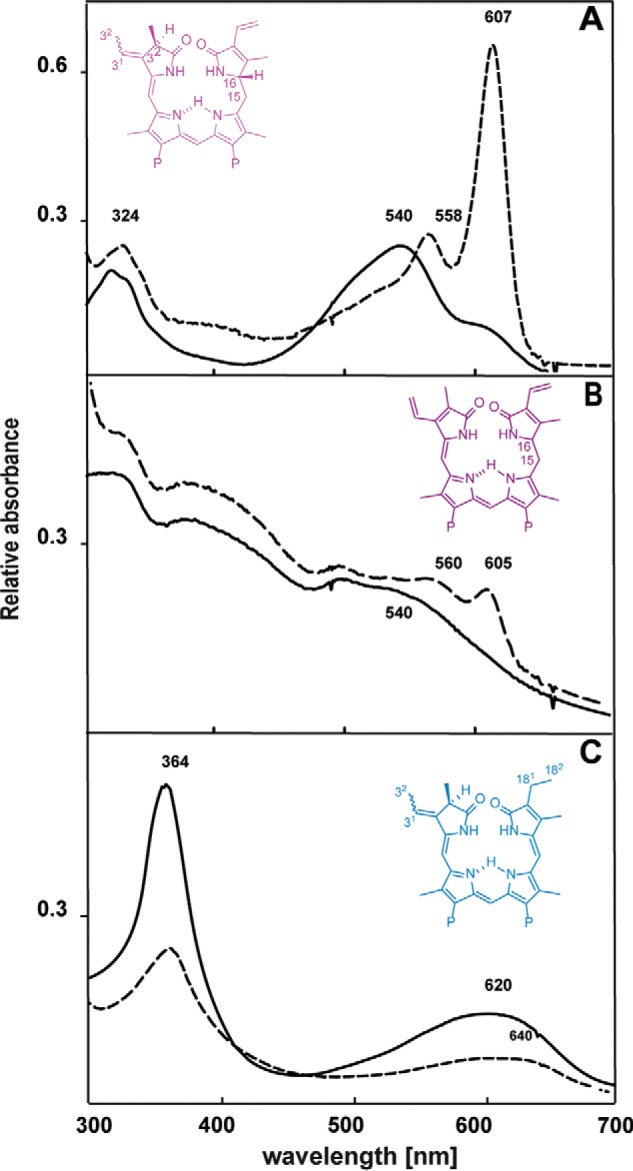
Bilin binding to ΦCpeT. Absorption spectra of free bilins in comparison to absorption spectra after addition of equimolar amounts ΦCpeT (10 μm), solid lines correspond to free bilin spectra, dashed lines reflect the ΦCpeT·bilin samples PEB (A), 15,16-DHBV (B), and PCB (C). The structures of the bilin is shown as insets. For PEB and PCB the waved bond stands for the two possible isomers (i.e. 3(Z) and 3(E)), respectively.
The ΦCpeT·PEB Is Highly Stable and Displays a Strong Red Fluorescence
The in vitro assembled ΦCpeT·PEB complex forms very rapidly with the chromophore being noncovalently bound to the lyase (data not shown). Interestingly and in contrast to other PEB-specific lyases (i.e. CpeS/CPES) (14, 15), the formed complex displayed strong red fluorescence (Fig. 5). Excitation at 540 nm resulted in a red fluorescence emission at 617 nm with a fluorescence quantum yield of ΦF = 0.3.
FIGURE 5.
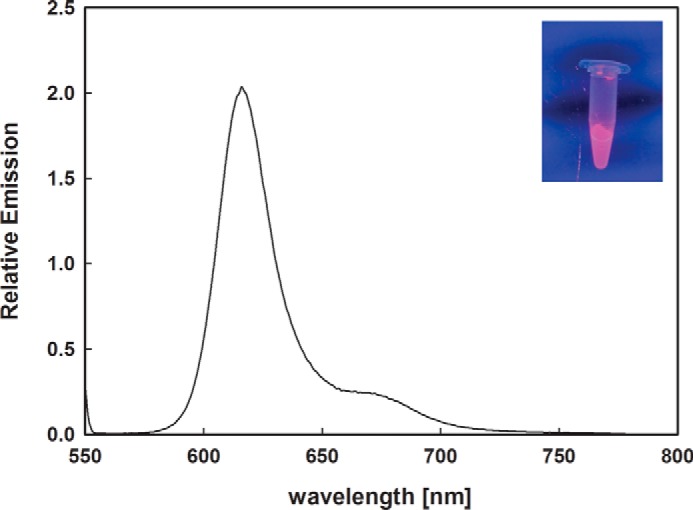
Fluorescence spectroscopy of the ΦCpeT·PEB complex. The ΦCpeT·PEB complex shows a strong fluorescence emission at 617 nm after excitation with 540 nm. Inset shows the complex under UV light (340 nm).
The ΦCpeT·PEB Complex Can Be Formed and Detected in Escherichia coli
Due to this reasonable fluorescence quantum yield of ∼30% we further investigated whether ΦCpeT might be suitable as a new fluorescent marker, similar to those recently published (16, 17). Therefore, we co-expressed ΦCpeT together with the genes for the biosynthesis of PEB (8) in E. coli to see whether the complex can also be formed in vivo. Co-expression of all required genes in E. coli resulted in dark blue cells. Subsequent purification of ΦCpeT yielded an intensely blue dyed protein indicating that the complex had been formed in E. coli and was stable enough to sustain the affinity chromatography. Bound PEB was verified via UV-visible spectroscopy and HPLC analysis. The absorption spectrum of the in vivo complex isolated from E. coli was nearly identical to the one assembled in vitro. The bound bilin from the in vivo complex was identified as the 3(Z)-isomer of PEB (Fig. 6).
FIGURE 6.
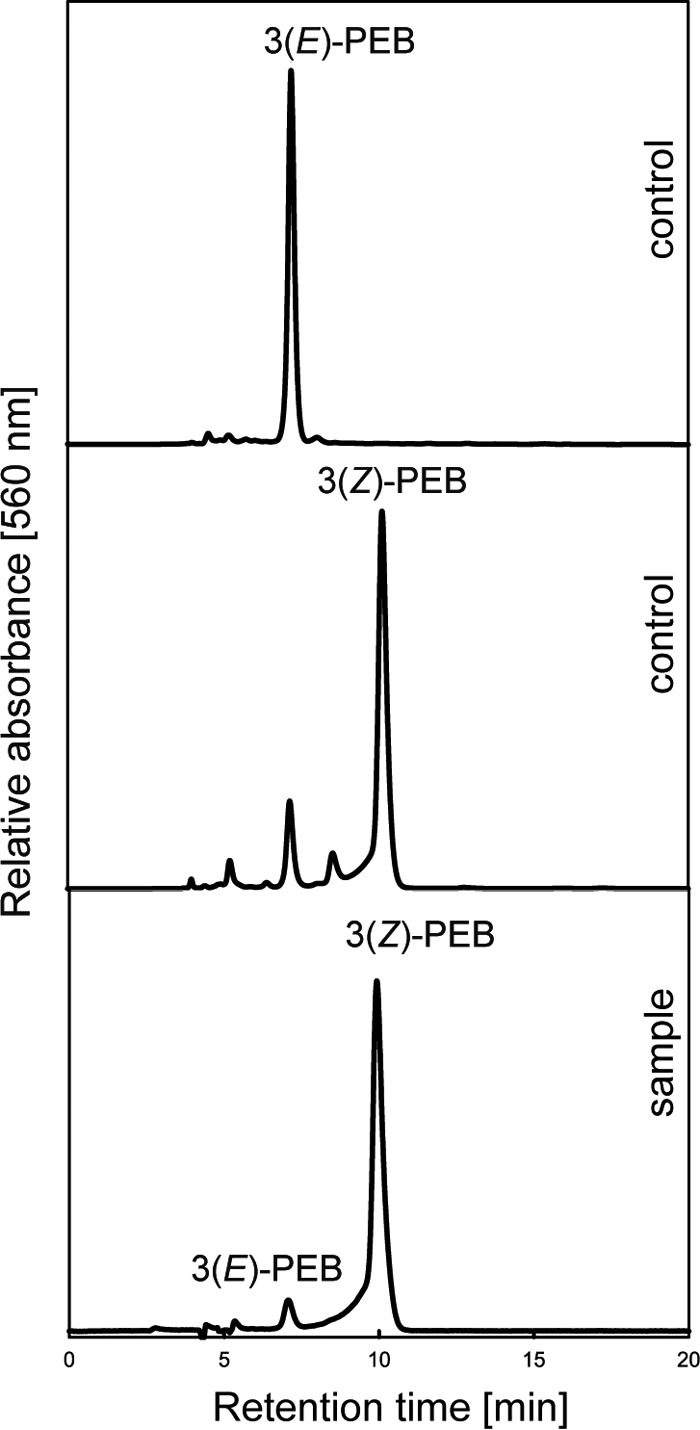
HPLC analysis of extracted bilin from ΦCpeT after co-expression with PEB-biosynthesis genes in E. coli using a C18 reverse-phase Luna C18 column (Phenomenex). Bilins were detected at 560 nm with subsequent whole spectrum analysis of elution peaks.
Co-expression experiments with the bilins 15,16-DHBV and PCB resulted in only minor pigmentation of the E. coli cells with a purified ΦCpeT having no bilin bound (data not shown). This, furthermore, strengthens the observation that ΦCpeT has a strong preference for PEB.
Interestingly, the strong red fluorescence of the in vivo formed ΦCpeT·PEB complex was able to label E. coli cells as monitored by fluorescence microscopy (Fig. 7). Therefore, ΦCpeT could be a suitable candidate for a new fluorescence reporter.
FIGURE 7.
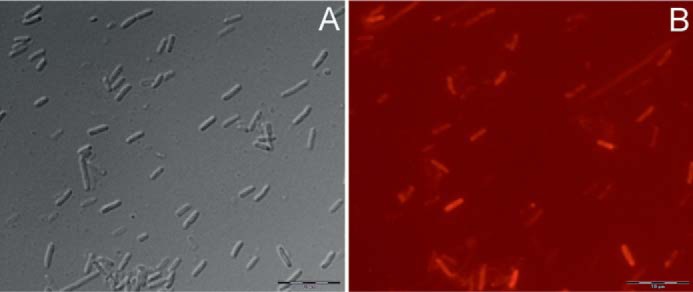
Fluorescence microscopy of E. coli cells expressing ΦCpeT and PEB biosynthesis genes employing an Olympus BX51 epifluorescence microscope. A, brightfield image; B, fluorescence image taken in Texas Red fluorescence mode. Bars represent 10 μm.
To finally test whether the observed fluorescence is a feature only found with CpeT from cyanophage P-HM1 we cloned and tested several other cyanophage-encoded CpeTs (Table 2) in a co-expression experiment. All proteins displayed the same features yielding a stable complex with PEB in E. coli and displaying strong red fluorescence with small variations in the emission wavelength (Table 3).
TABLE 2.
Plasmid-based expression constructs employed in this study
| pASK_PHM1cpeT | pASK-IBA45+ derivate containing cyanophage P-HM1 cpeT, AmpR |
| pASK_Syn1cpeT | pASK-IBA45+ derivate containing cyanophage Syn1 cpeT, AmpR |
| pASK_Syn9cpeT | pASK-IBA45+ derivate containing cyanophage Syn9 cpeT, AmpR |
| pASK_Syn19cpeT | pASK-IBA45+ derivate containing cyanophage Syn19 cpeT, AmpR |
| pASK_SPM2cpeT | pASK-IBA45+ derivate containing cyanophage S-PM2 cpeT, AmpR |
| pASK_RSM4cpeT | pASK-IBA45+ derivate containing cyanophage R-SM41 cpeT, AmpR |
| pColaDuet1MED4cpeB | pCOLADuet1 derivative, containing P. marinus MED4 cpeB, KanR |
TABLE 3.
Absorption und fluorescence emission properties of phage CpeT:PEB complexes
| Origin of ΦcpeT | Absorption maximum of CpeT·PEB complex | Fluorescence emission maximum of CpeT·PEB complex (λex = 595 nm) |
|---|---|---|
| P-HM1 | 607 | 616 |
| Syn1 | 604 | 613 |
| Syn9 | 608 | 622 |
| Syn19 | 603 | 618 |
| S-PM2 | 597 | 614 |
| P-RSM4 | 607 | 618 |
ΦCpeT Does Not Transfer PEB to Host CpeB but Is Able to Assist the Host Lyase CpeS
Supposing the phage-encoded PBP lyase might adopt a function during infection we tested whether the PEB-loaded ΦCpeT is able to transfer the chromophore to the host apo-PBP. Cyanophage P-HM1 was isolated from the host P. marinus MED4. P. marinus MED4 is special as it is a high light-adapted strain possessing only a much degenerated β-subunit of phycoerythrin (CpeB). This subunit possesses only one chromophore binding site for PEB at position Cys-82, which is in vitro served by the S-type lyase CpeS (15, 18). ΦCpeT from a coexpression experiment and fully loaded with PEB was incubated with purified PmCpeB (15) and the fluorescence emission was followed over time. The initial emission at 617 nm is derived from the ΦCpeT·PEB complex (see above) (Fig. 8). Over time, no change in fluorescence emission is observed indicating that PEB remained bound to ΦCpeT. We then asked whether the presence of the host lyase CpeS would have an influence on the transfer potential. Interestingly enough, further addition of unloaded host CpeS changed the emission properties to those resembling holo-CpeB (λem = 569 nm (15)) suggesting a transfer of PEB from ΦCpeT to CpeB likely assisted by CpeS (Fig. 8).
FIGURE 8.
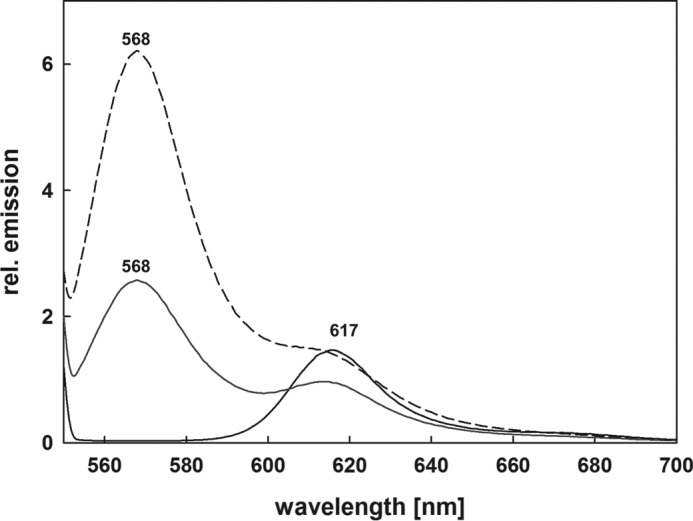
Bilin transfer assays employing ΦCpeT and PmCpeB in combination with PmCpeS. The solid spectrum shows fluorescence emission of the ΦCpeT·PEB complex incubated with purified recombinant PmCpeB. The obtained fluorescence emission is solely due the ΦCpeT·PEB complex and no transfer to PmCpeB is observed; addition of purified recombinant host PBP lyase CpeS after 15 min is shown as dotted spectrum (···). After 30 min incubation the spectrum shown in dashes is observed (- - -). The additional fluorescence emission at 568 nm is due to the assembled PmCpeB (15). See text for details.
Discussion
T-type lyases are the least characterized of all PBP lyases. CpcT lyases specifically support the addition of PCB to βCys-155 of either phycocyanin or phycoerythrocyanin (10, 19, 20). However, paralogs of these lyases can also be found in PE containing cyanobacteria that contain PEB in their β-subunits. Therefore it has been hypothesized that CpeT lyases are involved in the specific attachment of PEB to βCys-155 (21). With ΦCpeT we present the first structural and biochemical characterization of a PEB-specific T-type lyase.
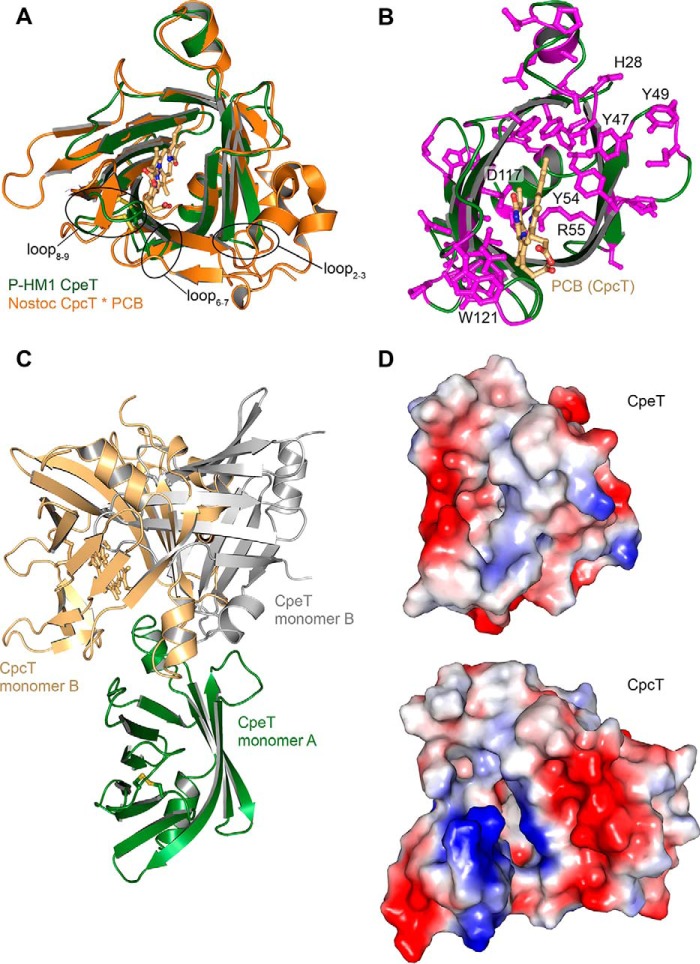
Small Cyanophage T-lyase Retains Structural Features of Large Cyanobacterial Counterpart
The cyanobacterial PBP lyase CpcT from Nostoc sp. PCC 7120 is thus far the only T-type lyase with known structure (20). This lyase specifically binds PCB and facilitates the attachment to Cys-155 of PC β-subunits (CpcB). Comparison with CpcT shows that ΦCpeT is smaller, which might be expected for a viral protein (Fig. 9, A and D). Hence, to retain the overall structure, the phage lyase shows partially significant differences. In CpcT the loop between strands 6 and 7 forms a big extension that folds back to the outside of the β-barrel. In contrast, the corresponding loop in ΦCpeT is very short and only contains three amino acid residues. However, in both structures this loop leads to an extended opening of the barrel, which is stabilized by the intramolecular disulfide bridge. Another difference is the loop between strands 8 and 9, which harbors the second cysteine of the disulfide bridge (Cys-95 in ΦCpeT) as well as the constricting Trp-97.
FIGURE 9.
Structural comparison of T-type lyases. A, overlay of ΦCpeT (green) and Nostoc sp. CpcT (orange) (PDB code 4O4S). Differently sized loops are indicated. B, schematic representation, showing ΦCpeT (green) and Nostoc CpcT between conserved residues (magenta). PCB ligand of CpcT is shown in light orange. C, overlay of the CpcT dimer (only one monomer shown in orange) and the closest ΦCpeT dimer (green and gray). D, electrostatic surfaces of Nostoc sp. CpcT and ΦCpeT.
Dimer Formation of ΦCpeT Is Different to a Cyanobacterial PBP Lyase
ΦCpeT elutes as a dimer in size exclusion chromatography (data not shown) and the crystal substructure also suggests protein dimerization; however, PISA scores are not high enough to predict interaction. For CpcT, Zhou et al. (20) claim that dimer formation is mandatory for protein function and binding of PCB. Helix α2 builds a plug that protrudes into the other monomer, contributing to ligand binding. This dimer does not exist in ΦCpeT (Fig. 9C). Although one of the crystal contacts in ΦCpeT resembles the dimer of CpcT, one of the subunits is tilted by a few degrees; impeding a functional role as in CpcT.
Binding of PEB Resembles S-type PBP Lyases
ΦCpeT was solved in the apo state without bound ligand. Although, ΦCpeT and PEB form such a strong and stable complex, any attempts to soak or co-crystallize it with its substrate PEB failed. Therefore, it can thus far only be speculated and inferred from spectroscopic data how the bilin is bound.
Our biochemical data indicate that ΦCpeT has a strong preference for bilins with reduced 15,16-double bond (i.e. 15,16-DHBV and 3(E/Z)-PEB). Isolation of the ligand from the co-expression experiment in E. coli revealed that the bound ligand of ΦCpeT is 3(Z)-PEB, which is in agreement with PEB isolated from the eukaryotic CPES lyase from Guillardia theta (14). It is, however, in contrast to our previous suggestion that the (E/Z)-configuration of the lyase-bound bilin determines the final configuration at the newly formed stereocenter at C31 of the bound phycobilin (R versus S) (14). Following that hypothesis, the preferred isomer for T-type lyases should be the 3(E)-isomer as the newly formed stereocenter at C31 of PEB at Cys-155 is in the S-configuration, whereas in most other positions it is in the R-configuration (22). Future biochemical analysis of the transfer reaction to other PBP and with cyanobacterial T-type lyases will finally have to prove or disapprove this hypothesis.
Common to all PEB-specific PBP lyases is a strong increase in the extinction coefficient of PEB concomitant with an absorbance shift to longer wavelength. Absorption of free PEB changes upon addition of PmCpeS and GtCPES to 590 and 599 nm, respectively, whereas addition of ΦCpeT shifts the absorption even further to 607 nm. This behavior has previously been attributed to conformational changes of PEB upon binding to the lyase to a more stretched conformation and possibly also protonation (14, 15). This binding behavior is substantially different to Nostoc CpcT, where the soaked ligand (i.e. PCB) adopts a Z,Z,Z,s,s,s geometry in an M-helical conformation when bound in the pocket (20). Hence, ΦCpeT, despite sharing a similar fold with the Nostoc T-type lyase, likely binds the ligand similar to the PEB-specific S-type lyases (14, 15).
Interestingly, all residues proposed to play a major role in chromophore ligation in CpcT are conserved in ΦCpeT. The central Arg-55 (Fig. 3A), located at the same position as Arg-66 in Nostoc CpcT, also points into the pocket. Because of the absence of a ligand, ΦCpeT Arg-55 has a very high B-factor and protrudes further into the pocket. Regardless of the difference in bilin binding, this Arg is likely to play a major role in ligand binding or even ligand detection. The other two proposed catalytic residues Tyr-66 and Asp-163 in CpcT are conserved and similarly positioned in ΦCpeT (Tyr-54, Asp-117).
In Nostoc CpcT, Gln-55 forms an H-bond with the A-ring carbonyl oxygen. This residue is Ser-45 in ΦCpeT and will be too far away, if PEB would adopt the same cyclic conformation in ΦCpeT as PCB in CpcT. CpcT Arg-68, interacting with the propionyl of ring C, is replaced by Gln-57 in ΦCpeT. All remaining hydrophilic residues that point into the binding pocket are conserved between both structures (Fig. 3A).
The PCB Nostoc CpcT structure cannot easily explain how PCB could be transferred in such a ring-like conformation, where pyrrole rings A and B point toward the pocket bottom and render it inaccessible from the solvent. Hence, an extended, more linear conformation is likely as this would enable the A-ring to point toward the opening of the pocket.
Activity of P-HM1 CpeT
Interestingly, ΦCpeT has a highly conserved ring of tyrosine residues, which are pointing into the solvent (Fig. 9B). They are located at the rim of the opening on a flexible loop. We propose that those residues are involved in interaction with the receiving protein and might play a role in PEB transfer.
Hydrophilic residues within the binding pocket are rare, very different than the arrangement in S-type lyases such as CPES (14). Apart from a few hydroxyl groups from serine and threonine residues, only four hydrophilic residues are present (Fig. 3A). The conserved Arg-55 and Asp-113, which are proposed to play a role in catalytic activity (20), as well as Tyr-54 are positioned at the same locations as in Nostoc CpcT. As binding modes of ligands seem to be different between CpcT (circular) and ΦCpeT (extended) it remains curious why all important residues are conserved and in similar positions. One explanation would be that the PCB-bound Nostoc CpcT structure represents a pre-catalytic state, possibly associated with the chaperone function of lyases, upon which the ligand undergoes a structural rearrangement to a linear form. Spectroscopically, this can be observed by a shift in peak wavelength for PEB, but not for PCB. This would explain how the deeply buried PCB A-ring in CpcT could be attached to the PBP Cys residue.
The intramolecular disulfide bridge is crucial for protein function (Fig. 2A). It is debated whether the interaction supports overall structural rigidity or whether the cysteines serve a further function in catalysis (20, 23). Judging by the structure, the disulfide bridge seems to be important for positioning of several loops. This includes loop7–8 harboring the pocket-lid Trp-97. For PEB to bind to the inner pocket of ΦCpeT, this loop and Trp-97 have to undergo considerable structural changes. This might affect the disulfide bridge and potentially catalysis.
Comparing binding pockets between the circularly binding CpcT and the linearly binding ΦCpeT and CPES does not reveal explanations for the difference. Both ΦCpeT and CPES have rather short binding pockets (Fig. 3), whereas CpcT has an extended, positively charged pocket, which is barely occupied by the circular ligand.
Auxiliary Metabolic Genes and Their Function during Infection
Many of the cyanophage-encoded AMGs are involved in phycobilin metabolism. Not only genes involved in phycobilin biosynthesis are encoded in high numbers in phage genomes but also genes assisting in the attachment of the bilin to the PBP. Recent transcriptome data revealed that all these genes are transcribed during infection suggesting a functional role during infection (24). Interestingly, thus far no genes encoding PBP subunits have been identified in cyanophage genomes. The cpeT gene investigated in this study originates from the P. marinus MED4 infecting phage P-HM1 but homologs can also be found in other cyanophage genomes (i.e. Syn1, Syn9, Syn19, S-PM2, and R-SM4). Inspection of the genome of the phage revealed that this phage is one of the rare examples (5/74) that encode bilin biosynthesis genes (i.e. ho1 and pebS) together with cpeT. Therefore, the phage has all the genetic information for the formation of a ΦCpeT·PEB complex during infection. This somehow could constitute a pigment pool for the delivery of PEB to the host PBP lyase CpeS. As the host PBP CpeB has only one chromophore binding site at Cys-82 (15), it is not too surprising that a T-type lyase cannot transfer its pigment as these lyases are usually specific for the βCys-155 position of the PBP (which is absent in Pm MED4 CpeB). Therefore we postulate an assisting/regulatory function of ΦCpeT during infection. Regulatory function of T-type lyases have been suggested earlier because the CpcT-like lyase CpcT2 of Nostoc sp. PCC7120 was also shown to be inactive in vitro (10). This, however, still does not rule out a PBP lyase function if the host bacterium possesses a PE β-subunit with all conserved cysteine residues for bilin binding, including Cys-155.
Independent studies, furthermore, revealed that infection of Synechococcus sp. WH7803 with the cpeT-containing cyanophage S-PM2 led to a slight increase in PE content as well as induced transcription of the host mpeAB genes (encoding PBP α- and β-subunits) (9). In this context, ΦCpeT was shown to bind upstream of mpeAB additionally supporting a regulatory function (6).
Experimental Procedures
All chemicals were obtained in laboratory grade or better. pASK-IBA45+ and Steptactin®-Sepharose were obtained from IBA GmbH (Göttingen, Germany). HPLC-grade acetone, acetonitrile, and formic acid were obtained from Mallinckrodt Baker (Griesheim, Germany) and Sep-Pak cartridges from Waters (Milford, MA).
Construction of cpeT Expression Vectors
Cyanophage P-HM1_cpeT (PHM1_028) was amplified via PCR from a synthetic gene template (MWG Eurofins Operon) using the following primer: 5′-gcgaattctatgatagataaattttgt-3′ and 5′-cgctcgagttatacattctttttgaag-3. The primer included sequences for restriction enzyme recognition sites (underlined; EcoRI and XhoI) for cloning into a similarly cut pASK-IBA45+ to yield pPHM1cpeT. With a similar strategy, the cpeT genes from the cyanophages Syn1, Syn9, Syn19, S-PM2, and P-RSM4 were cloned (Table 2). Details for the cloning can be obtained upon request. All plasmids were verified by sequencing (MWG-Biotech AG, Ebersberg, Germany).
Production and Purification of Recombinant Cyanophage CpeT
An overnight culture of E. coli BL21(DE3) carrying pPHM1cpeT or any other cpeT construct was diluted 1:100 in fresh LB medium supplemented with sorbitol (100 mm), betaine (2.5 mm), and ampicillin (100 μg/ml). The culture was incubated at 37 °C with shaking (120 rpm) until an optical density (A578 nm) of 0.7 was reached. After a cooling step to 17 °C, the production of P-HM1_CpeT was induced with 20 ng/ml of anhydrotetracycline. The culture was subsequently incubated overnight at 17 °C (∼16 h). Cells were harvested by centrifugation (Sorvall RCplus, SLA3000, 4,000 rpm, 10 min) and stored at −20 °C until further use or immediately resuspended in lysis buffer (50 mm Tris-HCl, pH 7.5, 100 mm NaCl, 5% glycerol). Cells were disrupted by two passages through a Constant Systems Cell Disruptor at 40,000 lb/in2. The cell debris was separated by centrifugation (Sorvall RC5, SS34, 19,000 rpm, 1 h) and the supernatant loaded onto a Streptactin®-Sepharose column. Purification was performed following the manufacturer's instructions. The buffer system used for purification was based on sodium phosphate buffer (60 mm sodium phosphate, 100 mm NaCl, pH 7.5). Purified protein was dialyzed against assay buffer (60 mm sodium phosphate, 300 mm NaCl, pH 7.5) and concentrated using Vivaspin 6 concentrator devices (molecular mass cut off 10,000 Da; Sartorius Stedim Biotech GmbH, Göttingen, Germany). Concentration of proteins was determined using a calculated molar extinction coefficient at 280 nm (protcalc.sourceforge.net).
SDS-PAGE and Western Blot
Protein samples were analyzed using 12.5 or 15% SDS-PAGE and immunological detection of the Strep-tag was done according to the manufacturer's manual (IBA GmbH).
Co-production of P-HM1_CpeT and PEB Biosynthesis
For the overproduction of the CpeT·PEB complex in E. coli the vectors pTDho1pebS (8) and pPHM1cpeT (or any other cpeT construct) were co-transformed into E. coli BL21(DE3). The overnight culture (16 h) was diluted 1:100 into fresh LB medium containing ampicillin (100 μg/ml) and chloramphenicol (33 μg/ml). After incubation at 37 °C with shaking until an optical density (A578 nm) of 0.7, the cultures were cooled down and overexpression the PEB biosynthesis genes was induced by the addition of 0.5 mm isopropyl β-thiogalactopyranoside. After incubation for 1 h at 17 °C, P-HM1cpeT expression was induced by adding anhydrotetracycline (20 ng/ml). The cells were further incubated overnight at 17 °C (16 h), harvested by centrifugation (Sorvall RCplus, SLA3000, 4.000 rpm, 10 min), and stored at −20 °C until further use. Purification of the CpeT·PEB complex was performed as described for the apo-CpeT.
High Performance Liquid Chromatography
To identify the nature of the bound PEB isomer within CpeT after co-expression, the bound pigment was extracted from the lyase and analyzed via HPLC analysis as described before (14). The purified CpeT·PEB complex was mixed 10-fold with 0.1% TFA to denature the protein. After a brief centrifugation step, the supernatant containing the bilin was loaded on a preconditioned Sep-Pak C18 column, washed by 6 ml of 0.1% TFA, and eluted with acetonitrile. Eluted samples were directly loaded onto a Phenomenex Luna C18 reversed phase column and analyzed as described before using an Agilent Technologies 1100 series system (14).
Preparation of Bilins
BV IXα was obtained from Frontier Scientific (Logan, UT). The preparation of DHBV was performed under anaerobic conditions as described before (26, 27). 3(E)-PCB was isolated from Spirulina platensis (28), 3(E)- and 3(Z)-PEB was isolated from Porphyridium purpureum or produced enzymatically employing recombinant PebS (8). Bilins were diluted in a small amount of DMSO immediately before use. The concentration was determined in MeOH/HCl (2.5% for BV IX α and PCB, 5% for DHBV and PEB) employing the following extinction coefficients: ϵ571 nm = 46.9 mm−1 cm−1 (3(E)-PEB) (29) and ϵ685nm = 37.15 mm−1 cm−1 (3(E)-PCB) (30). As there are no reported extinction coefficients for 3(Z)-PEB and DHBV the ϵ of 3(E)-PEBs was used (15).
Bilin Binding Assay
Lyase bilin binding assays were performed as described before (14, 15). Briefly, equimolar amounts (10 μm) of bilin and CpeT were mixed in assay buffer (60 mm sodium phosphate, 100 mm NaCl, pH 7.5) and absorption was immediately measured using an Agilent Technologies 8453 UV-visible diode array spectrophotometer. The absorption spectra were compared with those of the free bilins (diluted in DMSO, filled up with assay buffer).
Fluorescence Spectroscopy
Fluorescence emission measurements were performed in sodium phosphate buffer (60 mm, 100 mm NaCl, pH 7.5). The excitation wavelength corresponded to the maximal absorption of the bilin. Spectra were taken in an Aminco-Bowman 2 FA-256 Series fluorimeter.
Fluorescence Microscopy
Fluorescence microscopy of E. colicells expressing ΦCpeT in combination with the PEB chromophore biosynthesis were performed on a Olympus BX51 epifluorescence microscope (Olympus) using a U-UCD8 condenser and a UPlanSApo 100XO objective. Images were taken using a CC12 digital color camera and cell imaging software. Fluorescence signals were detected using the TXRed fluorescent mode with a U-LH100HGAPO burner and a U-RFL-T power supply.
Crystallization, Data Collection, and Structure Determination
The protein was crystallized at 10 mg/ml. Crystals grew to 10–30 μm in 0.1 m glycine, 0.05 m magnesium acetate, 32% polyethylene glycol 400, pH 9.5 (1:1 μl), within 3 days. At maximum size they were immediately collected and flash frozen in liquid nitrogen, using no further cryo additive. A dataset was taken, using the PILATUS 6M detector at X10SA beamline at the SLS (Villingen, Switzerland) and processed using the XDS program suite, followed by scaling using XSCALE and XDSCONV (31). The structure was initially solved by molecular replacement using the program Phaser in space group F222 as suggested by XDS and POINTLESS (32). A model containing the β-sheet of Nostoc CpcT (Protein Data Bank code 4O4O) after deleting all loop regions was used successfully. Arp/Warp (33) in the atom update mode was used to improve electron density map by overfitting, which allowed further manual building using Coot (34), followed by phenix.refine (35) refinement. A significant improvement in model quality was observed by changing the space group to C2 with two molecules per asymmetric unit. Using a new high resolution dataset in phenix.refine and extensive manual building further improved the model. Final refinement by phenix.refine included NCS, TLS refinement, ADP weight, and stereochemistry weight optimization and refinement of individual B-factors. Final coordinates have been deposited in the PDB database under accession number 5HI8.
Author Contributions
R. G. and J. S. crystallized the protein, R. G. collected data, solved the structure, and wrote the paper, J. S., J. H., A. H., N. R., and J. W. performed biochemical experiments, E. H. helped with structure determination, and N. F. D. designed research, analyzed data, and wrote the paper.
Acknowledgments
We appreciate the assistance of Thorben Dammeyer, Thilo Lerari, Carina Niedenführ, Janina Pauls, and Christian Scholte in the initial phase of this project. We thank Penny Chisholm for providing phage lysates and Barbara Klein for providing a Porphyridium start culture. Crystallographic experiments were performed on beamline X10SA at the Suisse Light Source (Villigen, Switzerland) and beamlines at the European Synchrotron Radiation Facility (Grenoble, France). We thank all local contacts and our colleagues from the MPI of Molecular Physiology, Dortmund, for help during data collection.
This work was supported by grants from the Deutsche Forschungsgemeinschaft and the European Union within the Ziel2.NRW Science-to-Business program (to N. F. D.). The authors declare that they have no conflicts of interest with the contents of this article.
The atomic coordinates and structure factors (code 5HI8) have been deposited in the Protein Data Bank (http://wwpdb.org/).
- AMG
- auxiliary metabolic gene
- PCB
- phycocyanobilin
- PEB
- phycoerythrobilin
- PebS
- phycoerythrobilin synthase
- PE
- phycoerythrin
- PBP
- phycobiliprotein
- DHBV
- 15,16-dihydrobiliverdin
- PDB
- Protein Data Bank.
References
- 1. Fuhrman J. A. (1999) Marine viruses and their biogeochemical and ecological effects. Nature 399, 541–548 [DOI] [PubMed] [Google Scholar]
- 2. Suttle C. A. (2005) Viruses in the sea. Nature 437, 356–361 [DOI] [PubMed] [Google Scholar]
- 3. Sullivan M. B., Coleman M. L., Weigele P., Rohwer F., and Chisholm S. W. (2005) Three Prochlorococcus cyanophage genomes: signature features and ecological interpretations. PLos Biol. 3, e144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Mann N. H., Clokie M. R., Millard A., Cook A., Wilson W. H., Wheatley P. J., Letarov A., and Krisch H. M. (2005) The genome of S-PM2, a “photosynthetic” T4-type bacteriophage that infects marine Synechococcus strains. J. Bacteriol. 187, 3188–3200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Weigele P. R., Pope W. H., Pedulla M. L., Houtz J. M., Smith A. L., Conway J. F., King J., Hatfull G. F., Lawrence J. G., and Hendrix R. W. (2007) Genomic and structural analysis of Syn9, a cyanophage infecting marine Prochlorococcus and Synechococcus. Environ. Microbiol. 9, 1675–1695 [DOI] [PubMed] [Google Scholar]
- 6. Puxty R. J., Millard A. D., Evans D. J., and Scanlan D. J. (2015) Shedding new light on viral photosynthesis. Photosynth. Res. 126, 71–97 [DOI] [PubMed] [Google Scholar]
- 7. Lindell D., Sullivan M. B., Johnson Z. I., Tolonen A. C., Rohwer F., and Chisholm S. W. (2004) Transfer of photosynthesis genes to and from Prochlorococcus viruses. Proc. Natl. Acad. Sci. U.S.A. 101, 11013–11018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Dammeyer T., Bagby S. C., Sullivan M. B., Chisholm S. W., and Frankenberg-Dinkel N. (2008) Efficient phage-mediated pigment biosynthesis in oceanic cyanobacteria. Curr. Biol. 18, 442–448 [DOI] [PubMed] [Google Scholar]
- 9. Shan J., Jia Y., Clokie M. R., and Mann N. H. (2008) Infection by the “photosynthetic” phage S-PM2 induces increased synthesis of phycoerythrin in Synechococcus sp. WH7803. FEMS Microbiol. Lett. 283, 154–161 [DOI] [PubMed] [Google Scholar]
- 10. Zhao K. H., Zhang J., Tu J. M., Böhm S., Plöscher M., Eichacker L., Bubenzer C., Scheer H., Wang X., and Zhou M. (2007) Lyase activities of CpcS- and CpcT-like proteins from Nostoc PCC7120 and sequential reconstitution of binding sites of phycoerythrocyanin and phycocyanin β-subunits. J. Biol. Chem. 282, 34093–34103 [DOI] [PubMed] [Google Scholar]
- 11. Sullivan M. B., Huang K. H., Ignacio-Espinoza J. C., Berlin A. M., Kelly L., Weigele P. R., DeFrancesco A. S., Kern S. E., Thompson L. R., Young S., Yandava C., Fu R., Krastins B., Chase M., Sarracino D., et al. (2010) Genomic analysis of oceanic cyanobacterial myoviruses compared with T4-like myoviruses from diverse hosts and environments. Environ. Microbiol. 12, 3035–3056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Hess W. R., Partensky F., van der Staay G. W., Garcia-Fernandez J. M., Börner T., and Vaulot D. (1996) Coexistence of phycoerythrin and a chlorophyll a/b antenna in a marine prokaryote. Proc. Natl. Acad. Sci. U.S.A. 93, 11126–11130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Krois D., and Lehner H. (1993) Helically fixed chiral bilirubins and biliverdins: a new insight into the conformational, associative and dynamic features of linear tetrapyrrols. J. Chem. Soc. Perkin Trans. 2, 1351–1360 [Google Scholar]
- 14. Overkamp K. E., Gasper R., Kock K., Herrmann C., Hofmann E., and Frankenberg-Dinkel N. (2014) Insights into the biosynthesis and assembly of cryptophycean phycobiliproteins. J. Biol. Chem. 289, 26691–26707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Wiethaus J., Busch A. W., Kock K., Leichert L. I., Herrmann C., and Frankenberg-Dinkel N. (2010) CpeS is a lyase specific for attachment of 3Z-PEB to Cys-82 of β-phycoerythrin from Prochlorococcus marinus MED4. J. Biol. Chem. 285, 37561–37569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Rodriguez E. A., Tran G. N., Gross L. A., Crisp J. L., Shu X., Lin J. Y., and Tsien R. Y. (2016) A far-red fluorescent protein evolved from a cyanobacterial phycobiliprotein. Nat. Methods 13, 763–769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Shu X., Royant A., Lin M. Z., Aguilera T. A., Lev-Ram V., Steinbach P. A., and Tsien R. Y. (2009) Mammalian expression of infrared fluorescent proteins engineered from a bacterial phytochrome. Science 324, 804–807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Steglich C., Frankenberg-Dinkel N., Penno S., and Hess W. R. (2005) A green light-absorbing phycoerythrin is present in the high-light-adapted marine cyanobacterium Prochlorococcus sp. MED4. Environ. Microbiol. 7, 1611–1618 [DOI] [PubMed] [Google Scholar]
- 19. Shen G., Saunée N. A., Williams S. R., Gallo E. F., Schluchter W. M., and Bryant D. A. (2006) Identification and characterization of a new class of bilin lyase: the cpcT gene encodes a bilin lyase responsible for attachment of phycocyanobilin to Cys-153 on the β-subunit of phycocyanin in Synechococcus sp. PCC 7002. J. Biol. Chem. 281, 17768–17778 [DOI] [PubMed] [Google Scholar]
- 20. Zhou W., Ding W.-L., Zeng X.-L., Dong L.-L., Zhao B., Zhou M., Scheer H., Zhao K.-H., and Yang X. (2014) Structure and mechanism of the phycobiliprotein lyase CpcT. J. Biol. Chem. 289, 26677–26689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Overkamp K. E., and Frankenberg-Dinkel N. (2014) Phycobiliproteins, Biosynthesis, Assembly and Applications. in The Porphyrin Handbook (Ferreira G., Kadish K. M., Smith K. M., and Guilard R., eds) pp. 187–226, World Scientific Publishing Company, Singapore [Google Scholar]
- 22. Ficner R., Lobeck K., Schmidt G., and Huber R. (1992) Isolation, crystallization, crystal structure analysis and refinement of B-phycoerythrin from the red alga Porphyridium sordidum at 2.2-Å resolution. J. Mol. Biol. 228, 935–950 [DOI] [PubMed] [Google Scholar]
- 23. Zhang J., Sun Y. F., Zhao K. H., and Zhou M. (2012) Identification of amino acid residues essential to the activity of lyase CpcT1 from Nostoc sp. PCC7120. Gene 511, 88–95 [DOI] [PubMed] [Google Scholar]
- 24. Doron S., Fedida A., Hernández-Prieto M. A., Sabehi G., Karunker I., Stazic D., Feingersch R., Steglich C., Futschik M., Lindell D., and Sorek R. (2016) Transcriptome dynamics of a broad host-range cyanophage and its hosts. ISME J. 10, 1437–1455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Terry M. J. (2002) Biosynthesis and analysis of bilins. in Heme, Chlorophyll, and Bilins, Methods and Protocols (Smith G. A., and Witty M., eds) pp. 273–291, Humana Press, Totowa, NJ [Google Scholar]
- 26. Busch A. W., Reijerse E. J., Lubitz W., Frankenberg-Dinkel N., and Hofmann E. (2011) Structural and mechanistic insight into the ferredoxin-mediated two-electron reduction of bilins. Biochem. J. 439, 257–264 [DOI] [PubMed] [Google Scholar]
- 27. Tu S. L., Gunn A., Toney M. D., Britt R. D., and Lagarias J. C. (2004) Biliverdin reduction by cyanobacterial phycocyanobilin:ferredoxin oxidoreductase (PcyA) proceeds via linear tetrapyrrole radical intermediates. J. Am. Chem. Soc. 126, 8682–8693 [DOI] [PubMed] [Google Scholar]
- 28. Terry M. J., Wahleithner J. A., and Lagarias J. C. (1993) Biosynthesis of the plant photoreceptor phytochrome. Arch. Biochem. Biophys. 306, 1–15 [DOI] [PubMed] [Google Scholar]
- 29. Gossauer A., and Klahr E. (1979) Synthesen von Gallenfarbstoffen, VIII. Total synthese des racem: Phycoerythrobilin-dimethylesters. Chem. Ber. 112, 2243–2255 [Google Scholar]
- 30. Weller J. P., and Gossauer A. (1980) Synthesen von Gallenfarbstoffen, X. Synthese und Photoisomerisierung des racem: Phytochromobilin-dimethylesters. Chem. Ber. 113, 1603–1611 [Google Scholar]
- 31. Kabsch W. (2010) XDS. Acta Crystallogr. D Biol. Crystallogr. 66, 125–132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Evans P. (2006) Scaling and assessment of data quality. Acta Crystallogr. D Biol. Crystallogr. 62, 72–82 [DOI] [PubMed] [Google Scholar]
- 33. Langer G., Cohen S. X., Lamzin V. S., and Perrakis A. (2008) Automated macromolecular model building for X-ray crystallography using ARP/wARP version 7. Nat. Protocols 3, 1171–1179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Emsley P., and Cowtan K. (2004) Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 [DOI] [PubMed] [Google Scholar]
- 35. Adams P. D., Afonine P. V., Bunkóczi G., Chen V. B., Davis I. W., Echols N., Headd J. J., Hung L.-W., Kapral G. J., Grosse-Kunstleve R. W., McCoy A. J., Moriarty N. W., Oeffner R., Read R. J., Richardson D. C., et al. (2010) PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66, 213–221 [DOI] [PMC free article] [PubMed] [Google Scholar]