Abstract
Deregulated expression of N-acetylgalactosaminyltransferases (GALNTs), which is responsible for the initial step of mucin-type O-glycosylation, could produce abnormal truncated O-glycans and thereby exert pivotal functions during malignant transformation. GALNT4 is one of the few isoforms preferring to catalyze partial GalNAc-glycosylated substrates and modify the sites not utilized by other known GALNTs. This study aims to evaluate the impact of GALNT4 expression on malignant transformation of hepatocellular carcinoma (HCC). Immunohistochemistry and in situ hybridization analysis were performed to assess GALNT4 and miR-9 level in clinical specimens, respectively. GALNT4 expression is markedly repressed in primary HCC tissues, and reduced expression of GALNT4 is significantly associated with adverse survival of patients with HCC. Functional investigations demonstrate that repressed GALNT4 could promote migration, invasion, anoikis resistance, and stemness of HCC cells in vitro as well as tumor growth in vivo. The wild-type GALNT4 could modify O-linked glycosylation on EGFR and thus modulate the activity of EGFR. A luciferase activity assay further identified microRNA-9 (miR-9) as the crucial specific arbitrator for GALNT4 expression in HCC cells. Furthermore, restoring GALNT4 expression attenuates miR-9-mediated oncogenic functions. Kaplan-Meier survival analysis indicates that the miR-9/GALNT4 expression signature yields promising prognostic significance to refine the risk stratification of patients with HCC. In conclusion, this study establishes the miR-9/GALNT4 axis as a potential adverse prognostic factor and therapeutic target for HCC patients.
Keywords: anoikis, glycobiology, glycosyltransferase, hepatocellular carcinoma, microRNA (miRNA), migration, post-transcriptional regulation, GALNT4, Prognostic biomarker, microRNA-9
Introduction
Hepatocellular carcinoma (HCC)4 affects more than half a million individuals annually and is the third leading cause of cancer-related death worldwide (1). Although hepatic resection, chemotherapy, and liver transplantation are widely used to improve outcomes of patients with HCC, the mortality rate remains high, which is largely attributable to the high rate of tumor recurrence and distant metastasis after surgery (2). Revealing the molecular mechanisms during hepatocarcinogenesis is therefore urgent to the development of early diagnosis and novel therapeutic strategies for HCC.
In mammals, there are two major types of glycosylation, termed N-linked and O-linked. The most common type of the latter is mucin-type O-glycosylation, which henceforth will be referred to simply as O-glycosylation. The initiating step of O-glycosylation is catalyzed by a large family of up to 20 different UDP-GalNAc:polypeptide N-acetylgalactosaminyltransferases (GALNTs), in which N-acetylgalactosamine (GalNAc) is transferred to the hydroxyl groups of serine and threonine amino acid residues (3, 4). Now it is clear that all but one of the 20 isoforms of humans are composed of two globular domains containing a catalytic domain and a ricin-like lectin carbohydrate binding domain. The study of Gerken et al. (5) revealed an inspiring discovery that the lectin domain could direct glycopeptide substrate glycosylation in an N- or C-terminal direction, which is probably attributable to the binding of the lectin domain to an existing glycan on the glycopeptide. This might contribute to the preference of specific substrates and/or specific sites on the same substrates of different isoforms. Over the years, several GALNTs have been determined to be necessary for, or associated with, malignant transformation and progression of cancers. For instance, GALNT3 might promote the growth and survival of pancreatic cancer cells and be identified as a potential prognostic factor for lung cancer and renal cell carcinoma (6–8). GALNT6 might contribute to mammary carcinogenesis and be determined as an independent prognosticator for pancreatic cancer (9–11). GALNT14 might modulate apoptotic signaling of cancer cells through modifying the proapoptotic receptors (12). The vital roles of GALNT1 and GALNT2 in HCC have been well elaborated (13, 14). GALNT4 is one of the isoforms preferring to catalyze partial GalNAc-glycosylated substrates and modify the sites not utilized by other known GALNTs (15, 16). It has been determined that mutation of the α-subdomain of GALNT4 (D459H) could eliminate its lectin activity, which abolishes its ability to glycosylate the specific substrates (16). To date, however, the expression pattern and functional significance of GALNT4 in HCC have not been elucidated.
In this study, we first analyzed the expression of GALNT4 by qRT-PCR and immunohistochemical analysis in clinical specimens and their correlations with clinicopathologic characteristics and clinical outcome of patients with HCC. We further explored the functions exerted by GALNT4 in HCC cells in vitro and in vivo. The main specific miRNA targeting GALNT4 in HCC cells was identified. Importantly, the clinical significance of miRNA/GALNT4 expression signature for patients with HCC was evaluated.
Results
Decreased GALNT4 Expression Correlates with Poor Prognosis in Patients with HCC
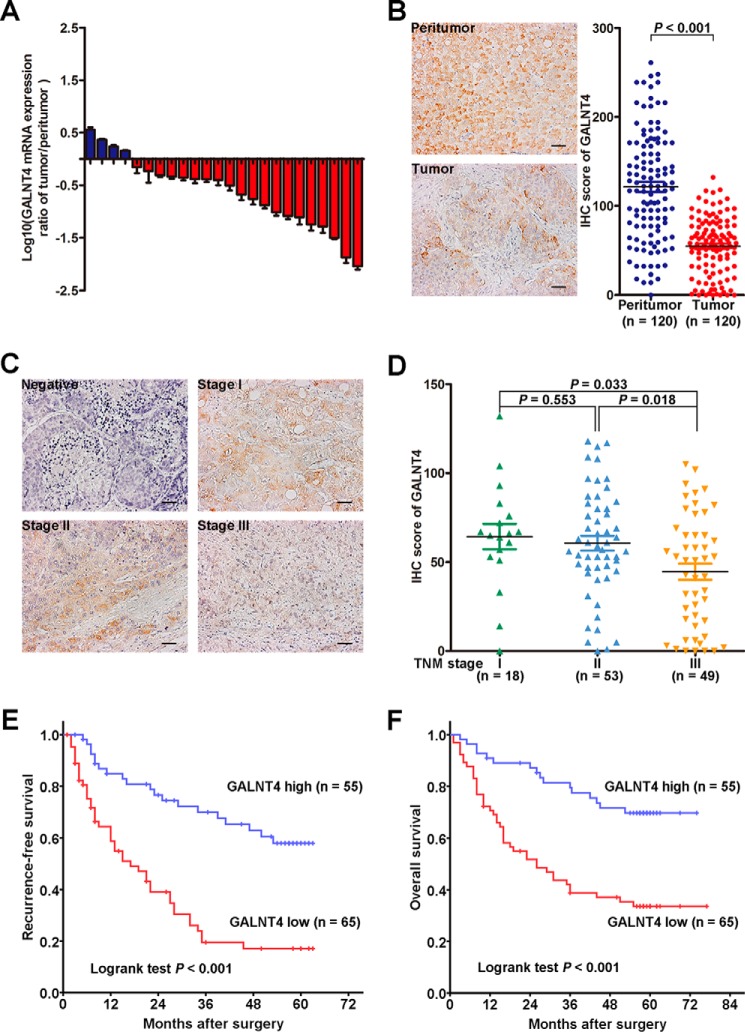
To evaluate the expression of GALNT4 in HCC, the mRNA expression of GALNT4 was detected in 24 pairs of primary HCC tumors and their corresponding peritumor tissues by qRT-PCR. Down-regulation of GALNT4 was determined in 20 of 24 of HCC tumors (Fig. 1A). Moreover, GALNT4 expression was further assessed in an additional 120 pairs of primary HCC tumors and their corresponding peritumor tissues by immunohistochemistry (IHC). Consistently, we observed that GALNT4 expression was significantly decreased in tumors (p < 0.001; Fig. 1B). Of note, tumors with advanced stages (TNM stage III) presented a markedly decreased level of GALNT4 compared with those with early stages, TNM stage I and II (p = 0.033 and p = 0.018, respectively; Fig. 1, C and D). For further analysis, patients were dichotomized into a “GALNT4 high” group and “GALNT4 low” group. Kaplan-Meier survival analysis revealed that patients with low GALNT4 expression suffered from earlier recurrence and poorer overall survival (log rank test, p < 0.001 and p < 0.001, respectively; Fig. 1, E and F).
FIGURE 1.
Decreased GALNT4 expression correlates with poor prognosis in patients with HCC. A, GALNT4 mRNA was detected in 24 pairs of primary HCC tumor and corresponding peritumor tissues by qRT-PCR. B, representative IHC images of GALNT4 expression in tumor and peritumor tissues of HCC patients (left) and scatter plots for IHC scores of GALNT4 expression in tumor and matched peritumor tissues of patients with HCC (right). Scale bar, 50 μm (original magnification, ×200). C, representative IHC images of GALNT4 expression in each TNM stage. Scale bar, 50 μm (original magnification, ×200). D, scatter plots for corresponding evaluated IHC scores of GALNT4 in each TNM stage. E and F, Kaplan-Meier survival analysis of HCC patients for RFS and OS according to the expression of GALNT4 (high expression subgroup, n = 55; low expression subgroup, n = 65). p value was calculated by log rank test.
The association between GALNT4 expression and clinicopathologic features of HCC was further investigated. As presented in Table 1, lower level of GALNT4 correlated with larger tumor size (p = 0.005), poorer tumor differentiation (p = 0.016), higher rates of vascular invasion (p = 0.017), higher Barcelona Clinic liver cancer stage (p = 0.014), and TNM stage (p = 0.026).
TABLE 1.
Correlation between GALNT4 expression and patient characteristics
Boldface values indicate p < 0.05 (t test for continuous variables and χ2 test for categorical variables). HBsAg, hepatitis B surface antigen; ALT, alanine aminotransferase; AFP, α-fetoprotein; BCLC, Barcelona Clinic liver cancer staging.
| Characteristic | Patients (n = 120) |
GALNT4 expression |
p | ||
|---|---|---|---|---|---|
| Number | % | High (n = 55) | Low (n = 65) | ||
| Age (years)a | 120 | 100.0 | 51.9 ± 10.8 | 50.6 ± 12.2 | 0.538 |
| Gender | 0.369 | ||||
| Female | 17 | 14.2 | 10 | 7 | |
| Male | 103 | 85.8 | 45 | 58 | |
| Liver cirrhosis | 0.962 | ||||
| Absent | 106 | 88.3 | 48 | 58 | |
| Present | 14 | 11.7 | 7 | 7 | |
| Child-Pugh | 0.834 | ||||
| A | 114 | 95.0 | 52 | 62 | |
| B | 6 | 5.0 | 3 | 3 | |
| HBsAg | 0.835 | ||||
| Negative | 15 | 12.5 | 6 | 9 | |
| Positive | 105 | 87.5 | 49 | 56 | |
| ALT (IU) | 0.399 | ||||
| ≤40 | 55 | 45.8 | 28 | 27 | |
| >40 | 65 | 54.2 | 27 | 38 | |
| AFP (ng/ml) | 0.225 | ||||
| ≤20 | 38 | 31.7 | 21 | 17 | |
| >20 | 82 | 68.3 | 34 | 48 | |
| Tumor size (cm) | 0.005 | ||||
| ≤5 | 72 | 60.0 | 41 | 31 | |
| >5 | 48 | 40.0 | 14 | 34 | |
| Tumor number | 0.342 | ||||
| Single | 97 | 80.8 | 47 | 50 | |
| Multiple | 23 | 19.2 | 8 | 15 | |
| Tumor encapsulation | 0.426 | ||||
| Complete | 64 | 53.3 | 32 | 32 | |
| None | 56 | 46.7 | 23 | 33 | |
| Tumor differentiation | 0.016 | ||||
| 1 + 2 | 84 | 70.0 | 45 | 39 | |
| 3 + 4 | 36 | 30.0 | 10 | 26 | |
| Vascular invasion | 0.017 | ||||
| Absent | 61 | 50.8 | 35 | 26 | |
| Present | 59 | 49.2 | 20 | 39 | |
| BCLC stage | 0.014 | ||||
| 0 + A | 65 | 54.2 | 37 | 28 | |
| B + C | 55 | 45.8 | 18 | 37 | |
| TNM stage | 0.026 | ||||
| I + II | 71 | 59.2 | 39 | 32 | |
| III | 49 | 40.8 | 16 | 33 | |
a The results of continuous variables are expressed as mean ± S.D.
Cox proportional hazards analysis identified GALNT4 expression as an independent favor prognostic factor for recurrence-free survival (RFS) (hazard ratio, 0.317; 95% confidence interval, 0.182–0.553; p < 0.001) and overall survival (OS) (hazard ratio, 0.415; 95% confidence interval, 0.226–0.761; p = 0.005) (Tables 2 and 3).
TABLE 2.
Univariate Cox regression analyses of recurrence-free survival and overall survival after surgery in 120 HCCs
Boldface values indicate p < 0.05. p values are from Cox regression analysis. CI, confidence interval; HR, hazard ratio, HBsAg, hepatitis B surface antigen; ALT, alanine aminotransferase; AFP, α-fetoprotein; BCLC, Barcelona Clinic liver cancer staging.
| Characteristics | RFS |
OS |
||
|---|---|---|---|---|
| HR (95% CI) | p | HR (95% CI) | p | |
| Age | 1.002 (0.980–1.024) | 0.892 | 0.996 (0.974–1.019) | 0.719 |
| Gender (male versus female) | 2.083 (0.952–4.557) | 0.068 | 2.346 (0.942–5.848) | 0.069 |
| Liver cirrhosis (present versus absent) | 0.933 (0.446–1.951) | 0.855 | 1.230 (0.531–2.851) | 0.631 |
| Child-Pugh (B versus A) | 1.233 (0.450–3.381) | 0.686 | 2.620 (1.126–6.097) | 0.026 |
| HBsAg (positive versus negative) | 0.690 (0.362–1.318) | 0.264 | 1.388 (0.599–3.217) | 0.448 |
| ALT, IU (>40 versus ≤40) | 1.195 (0.732–1.951) | 0.478 | 1.446 (0.857–2.441) | 0.169 |
| AFP, ng/ml (>20 versus ≤20) | 1.069 (0.636–1.798) | 0.802 | 2.024 (1.074–3.814) | 0.030 |
| Tumor size, cm (>5 versus ≤5) | 2.766 (1.677–4.561) | <0.001 | 4.321 (2.518–7.416) | <0.001 |
| Tumor number (multiple versus single) | 1.010 (0.540–1.887) | 0.976 | 1.043 (0.554–1.963) | 0.897 |
| Tumor encapsulation (none versus complete) | 1.716 (1.050–2.804) | 0.032 | 1.724 (1.029–2.889) | 0.040 |
| Tumor differentiation (3 + 4 versus 1 + 2) | 1.400 (0.829–2.365) | 0.211 | 1.611 (0.944–2.750) | 0.082 |
| Vascular invasion (present versus absent) | 2.030 (1.237–3.332) | 0.005 | 2.599 (1.520–4.442) | <0.001 |
| BCLC stage (B + C versus 0 + A) | 2.927 (1.770–4.837) | <0.001 | 4.720 (2.673–8.333) | <0.001 |
| TNM stage (III versus I + II) | 1.730 (1.055–2.836) | 0.031 | 4.124 (2.394–7.104) | <0.001 |
| GALNT4 (high versus low) | 0.286 (0.167–0.489) | <0.001 | 0.313 (0.176–0.557) | <0.001 |
TABLE 3.
Multivariate Cox regression analyses of recurrence-free survival and overall survival after surgery in 120 HCCs
Boldface values indicate p < 0.05. p values are from Cox regression analysis. NA, not applicable; CI, confidence interval; HR, hazard ratio; AFP, α-fetoprotein; BCLC, Barcelona Clinic liver cancer staging.
| Characteristics | RFS |
OS |
||
|---|---|---|---|---|
| HR (95% CI) | p | HR (95% CI) | p | |
| Child-Pugh (B versus A) | NA | NA | 1.530 (0.600–3.902) | 0.376 |
| AFP, ng/ml (>20 versus ≤20) | NA | NA | 1.388 (0.724–2.659) | 0.326 |
| Tumor size, cm (>5 versus ≤5) | 0.781 (0.271–2.254) | 0.650 | 0.642 (0.219–1.886) | 0.423 |
| Tumor encapsulation (none versus complete) | 1.298 (0.751–2.244) | 0.352 | 1.027 (0.577–1.826) | 0.929 |
| Vascular invasion (present versus absent) | 1.369 (0.804–2.330) | 0.250 | 1.640 (0.903–2.977) | 0.106 |
| BCLC stage (B + C versus 0 + A) | 3.220 (1.109–9.355) | 0.033 | 4.362 (1.444–13.178) | 0.009 |
| TNM stage (III versus I + II) | 1.037 (0.568–1.895) | 0.905 | 2.334 (1.241–4.388) | 0.009 |
| GALNT4 (high versus low) | 0.317 (0.182–0.553) | <0.001 | 0.415 (0.226–0.761) | 0.005 |
GALNT4 Abrogates the Tumorigenicity of HCC Cells in Vivo
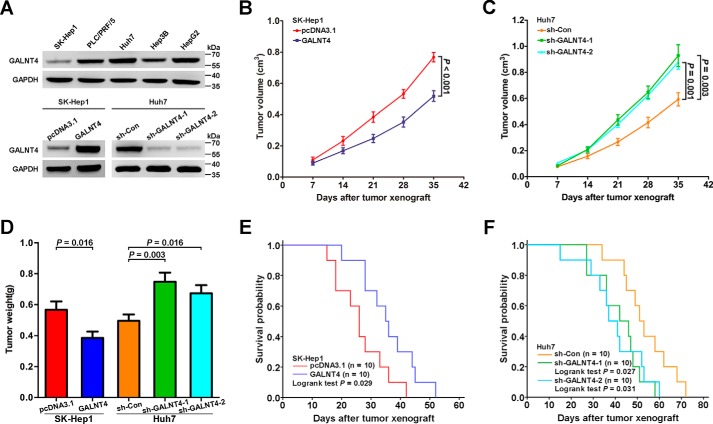
To further illustrate the role performed by GALNT4 during hepatocarcinogenesis observed on clinical specimens, we conducted in vivo experiments using the nude mice model. We first detected GALNT4 protein level in five HCC cell lines (Fig. 2A). Then we stably overexpressed GALNT4 in SK-Hep1 cells (which possessed the lowest level of GALNT4 expression) and stably knocked down GALNT4 in Huh7 cells (which possessed the highest level of GALNT4 expression) for further studies. Two short hairpin RNAs (shRNAs) were applied to generate two knockdown clones. Stable overexpression and knockdown of GALNT4 were confirmed by Western blotting (Fig. 2A).
FIGURE 2.
GALNT4 suppresses hepatoma cells growth in vivo. A, GALNT4 protein level in five HCC cell lines was determined by Western blotting (top). Western blot analysis to detect GALNT4 expression in SK-Hep1 stably transfected with empty vector or pcDNA3.1-GALNT4, respectively, and in Huh7 stably transfected with control shRNA or two specific shRNAs against GALNT4, respectively (bottom). GAPDH was used as the endogenous control. Shown are tumor growth curves (B and C) and quantification of weight of the tumors (D) that developed in nude mice subcutaneously injected with aforementioned stably transfected cells as described in A (n = 10 in each group). E and F, Kaplan-Meier survival analysis was conducted for the aforementioned xenograft mice (n = 10 in each group); p value was calculated by log rank test. Error bars, S.E.
To examine the effect of GALNT4 on tumor growth in vivo, nude mice were subcutaneously injected with the indicated cells. The results showed that GALNT4-expressing tumors had a significantly decreased growth rate when compared with a control group (p < 0.001; Fig. 2B). Consistent with the smaller tumor volume, GALNT4-expressing tumors had less mass than the control group (p = 0.016; Fig. 2D). Accordingly, tumor formation in nude mice with GALNT4-silencing Huh7 was markedly augmented compared with control group (tumor growth rate, p = 0.003 (shRNA-1) and p = 0.001 (shRNA-2) (Fig. 2C); tumor weight, p = 0.003 (shRNA-1) and p = 0.016 (shRNA-2) (Fig. 2D)). Furthermore, Kaplan-Meier survival curves revealed that mice receiving GALNT4-expressing SK-Hep1 had a significantly longer life span than mice injected with pcDNA3.1 (p = 0.029; Fig. 2E). On the other hand, mice inoculated GALNT4-silencing Huh7 had a worse survival than the control group (p = 0.027 (shRNA-1) and p = 0.031 (shRNA-2); Fig. 2F).
Silence of GALNT4 Promotes Malignant Phenotype of HCC Cells in Vitro
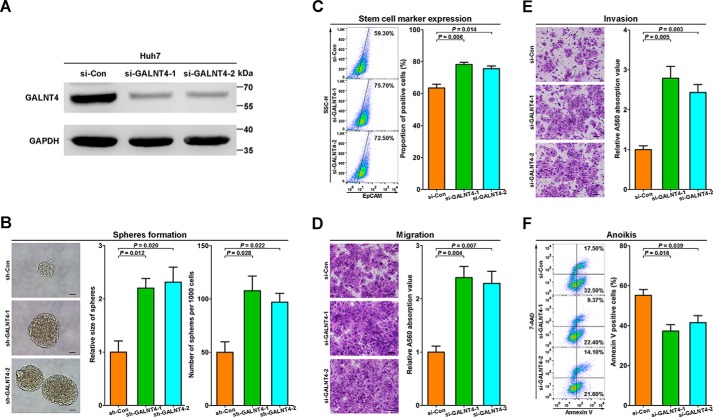
Our in vivo results in turn prompted us to elucidate the functional significance of GALNT4 in HCC cells. Then we transiently knocked down GALNT4 in Huh7 cells by two siRNAs for further studies. The knockdown of GALNT4 was confirmed by Western blotting (Fig. 3A).
FIGURE 3.
Knockdown of GALNT4 promotes malignant phenotype of HCC cells in vitro. A, the down-regulation of GALNT4 in Huh7 after being transfected with indicated constructs was confirmed by Western blotting. GAPDH was used as the endogenous control. B, representative micrograph of tumor spheres formed by stably transfected cells after 7 days of culture and corresponding quantification of sphere size and numbers. Sphere size is determined by its diameter. Scale bar, 50 μm. C, expression of EpCAM was measured by FACS after 48 h of sphere-forming culture. D, representative images of migration assay and corresponding quantification determined by A560 absorbance. Scale bar, 50 μm. E, representative images of invasion assay and corresponding quantification determined by A560 absorbance. Scale bar, 50 μm. F, representative flow cytometry of anoikis assays and corresponding quantification. Positive populations were analyzed by Annexin V/7-AAD staining. Considering that the sphere-forming assay needs to last for 7 days, stably knocked down cells were used in this part. Transiently transfected cells were used in other analysis. All data are from three independent experiments. Error bars, S.E.
Because the TMA analysis indicated that GALNT4 expression was significantly associated with tumor differentiation (Table 1), the effects of GALNT4 on stemness of HCC cells were further investigated. Considering that the sphere-forming assay needs to last for 7 days, stably knocked down cells were used in this part. The assay showed that GALNT4-silencing Huh7 formed dramatically more and larger spheres compared with control cells (p = 0.012 (shRNA-1) and p = 0.020 (shRNA-2) for size, p = 0.028 (shRNA-1) and p = 0.022 (shRNA-2) for numbers; Fig. 3B). Furthermore, the membrane expression of the well documented stem cell marker, EpCAM, on sphere-derived cells (48 h) was detected by flow cytometry analysis. Down-regulation of GALNT4 in Huh7 significantly promoted EpCAM expression (p = 0.006 (siRNA-1) and p = 0.014 (siRNA-2); Fig. 3C).
Given that GALNT4 expression was inversely correlated with TNM stage and vascular invasion, as summarized in Table 1, we speculated that GALNT4 might also be involved in the migration and invasiveness of HCC cells. As shown in Fig. 3D, migration assays showed that silencing of GALNT4 by siRNA dramatically promotes cell migration of Huh7 (p = 0.004 (siRNA-1) and p = 0.007 (siRNA-2)). Similarly, the Matrigel invasion assay also determined that knockdown of GALNT4 enhanced cell invasion of Huh7 (p = 0.005 (siRNA-1) and p = 0.003 (siRNA-2); Fig. 3E).
Besides the capabilities of migration and invasion, anoikis resistance also plays a critical role in tumor metastasis for its faculty to maintain anchorage-independent survival and growth of cancer cells during local dissemination and distant colonization. To investigate whether GALNT4 could modulate anoikis resistance of HCC cells, an in vitro anoikis assay was performed in Huh7 transfected with indicated constructs. Prepared cells were analyzed by flow cytometry. As presented in Fig. 3F, GALNT4 silencing significantly alleviated cell anoikis (p = 0.016 (siRNA-1) and p = 0.039 (siRNA-2)) during detached culture.
GALNT4 Attenuates Cellular Migration, Invasion, and Stemness and Induces Anoikis of HCC Cells in Vitro, but the Glycosyltransferase-dead Mutant Failed to Be Competent to Exert These Effects
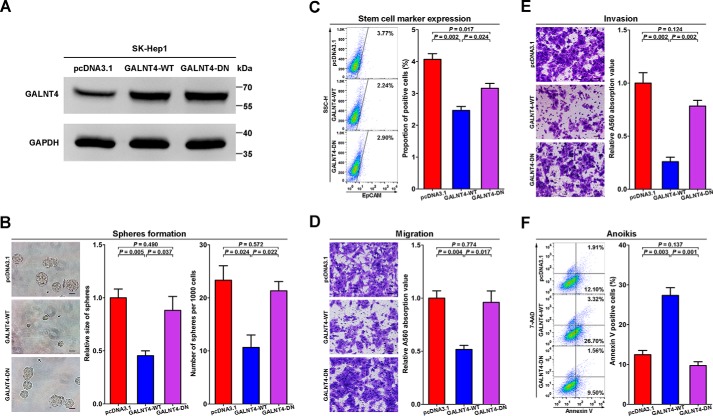
Next, we overexpressed wild-type GALNT4 (GALNT4-WT) and glycosyltransferase-dead mutant (GALNT4-DN) in SK-Hep1 cells, respectively. The overexpression of GALNT4-WT and GALNT4-DN was confirmed by Western blotting (Fig. 4A).
FIGURE 4.
GALNT4 attenuates cellular migration, invasion, and stemness and induces anoikis of HCC cells in vitro, but the glycosyltransferase-dead mutant failed to be competent to exert these effects. A, overexpression of GALNT4-WT and GALNT4-DN in SK-Hep1 were confirmed by Western blot. B, representative micrograph of tumor spheres formed by stably transfected cells after 7 days of culture and corresponding quantification of sphere size and numbers. Sphere size is determined by its diameter. Scale bar, 50 μm. C, expression of EpCAM was measured by FACS after 48 h of sphere-forming culture. D, representative images of migration assay and corresponding quantification determined by A560 absorbance. Scale bar, 50 μm. E, representative images of invasion assay and corresponding quantification determined by A560 absorbance. Scale bar, 50 μm. F, representative flow cytometry of anoikis assays and corresponding quantification. Positive populations were analyzed by Annexin V/7-AAD staining. Considering that the sphere-forming assay needs to last for 7 days, stably overexpressed cells were used in this part. Transiently transfected cells were used in other analysis. All data are from three independent experiments. Error bars, S.E.
Considering that a sphere-forming assay needs to last for 7 days, stably overexpressed cells were used in this part. The sphere-forming assay showed that GALNT4-WT-expressing SK-Hep1 formed much fewer and smaller spheres compared with control cells (p = 0.005 for size, p = 0.024 for numbers; Fig. 4B), whereas GALNT4-DN had no effect on size or number of spheres.
Furthermore, reintroduction of GALNT4-WT markedly suppressed the expression of EpCAM (p = 0.002; Fig. 4C) on SK-Hep1 after 48 h of sphere-forming culture. Although GALNT4-DN also inhibited the expression of EpCAM (p = 0.017; Fig. 4C), the expression of EpCAM on GALNT4-DN-expressing SK-Hep1 was significantly higher than on GALNT4-WT-expressing cells (p = 0.024; Fig. 4C).
As shown in Fig. 4D, migration assays showed that ectopic expression of GALNT4-WT but not GALNT4-DN strongly impaired the cell motility of SK-Hep1 compared with control cells (p = 0.004). Similarly, the Matrigel invasion assay also determined that GALNT4-WT markedly inhibited cell invasion (p = 0.002; Fig. 4E), whereas GALNT4-DN did not. Furthermore, in vitro anoikis analysis indicated that GALNT4-WT but not GALNT4-DN dramatically induced cell anoikis (p = 0.003; Fig. 4F) during detached culture.
GALNT4 Expression Is Directly Regulated by miR-9
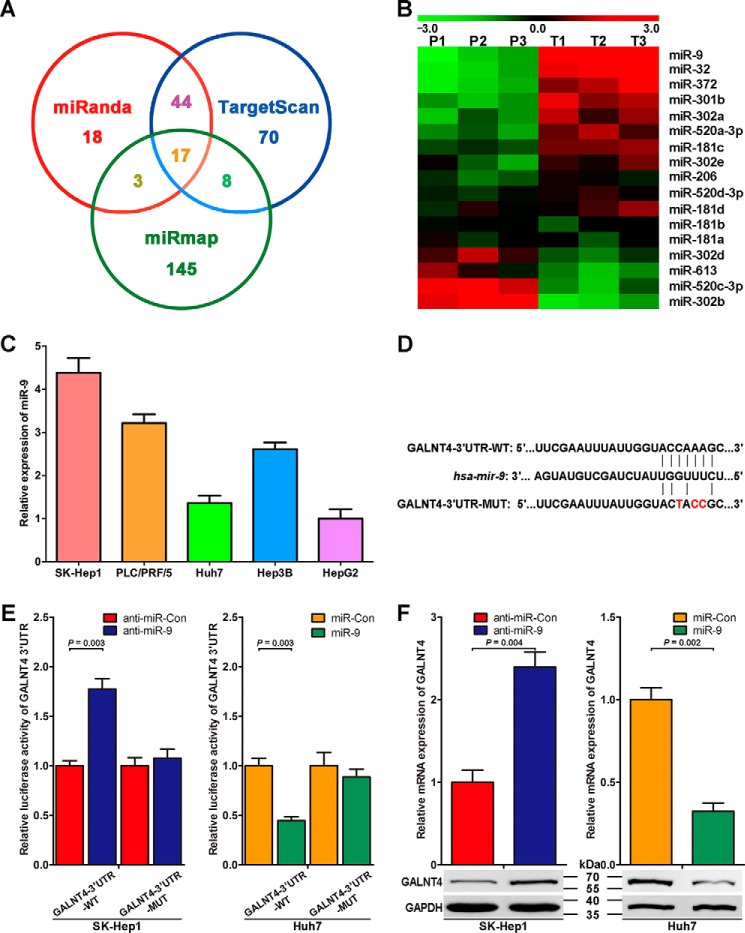
Because microRNAs are deeply involved in the modulation of gene expression at the post-transcription level, we asked whether certain miRNAs are responsible for the down-regulation of GALNT4 in HCC cells. To identify specific miRNAs targeting GALNT4, we used three miRNA target-predicting algorithms (TargetScan (release 6.2), miRanda, and miRmap) to screen out 17 potential miRNAs that overlapped among three data sets (Fig. 5A). qRT-PCR analysis of these 17 miRNAs in a panel of peritumor and tumor tissues determined that miR-9 expression is up-regulated with the greatest -fold changes in tumor compared with peritumor tissues (Fig. 5B). Thus, the expression of miR-9 in the aforementioned five HCC cell lines was detected by qRT-PCR, which presents inverse correlation trends with the protein level of GALNT4 in these cell lines (Fig. 5C).
FIGURE 5.
GALNT4 expression is directly repressed by miR-9. A, Venn diagrams showing the number of microRNAs identified to target GALNT4 as predicted by three algorithms (TargetScan, miRanda, and miRmap). B, heat map showing the average normalized relative expression levels of 17 miRNAs screened out in A in three pairs of tumor and matched peritumor tissues randomly selected from clinical specimens of Fig. 1A. U6 small nuclear RNA was used as internal control. P and T, peritumor and tumor tissues, respectively. C, miR-9 expression in five HCC cell lines was determined by qRT-PCR. U6 small nuclear RNA was used as the internal control. Data are from three independent experiments. D, sequences of miR-9 and the potential miR-9 binding sites at the 3′-UTR of GALNT4 mRNA. Also shown are nucleotides mutated in GALNT4-3′-UTR mutant. E, Dual-LuciferaseTM assays showing the effects of miR-9 mimic, miR-9 antisense, and their control oligonucleotides on wild-type 3′-UTR or mutant 3′-UTR, respectively. F, qRT-PCR and Western blotting analysis confirming that miR-9 mimic and miR-9 antisense could regulate GALNT4 expression. GAPDH serves as an internal control. All data are from three independent experiments. Error bars, S.E.
To determine whether GALNT4 is a direct target gene of miR-9, we cloned the 3′-UTR fragment of GALNT4 containing the conserved sequences of binding sites for miR-9 into a Dual-LuciferaseTM reporter construct (Fig. 5D, WT). We next generated mutations in the binding sites to abrogate miR-9-GALNT4 3′-UTR interaction (Fig. 5D, MUT). As shown in Fig. 5E, inhibition of endogenous miR-9 expression by anti-miR-9 significantly increased the luciferase activity of the wild-type reporter (p = 0.003). Importantly, overexpression of the miR-9 mimic markedly suppressed the luciferase activity of the wild-type reporter (p = 0.003; Fig. 5E). In contrast, both anti-miR-9 and miR-9 mimic had a minimal effect on the mutant reporter construct. qRT-PCR and Western blotting were then conducted to determine the effect of miR-9 on the regulation of GALNT4 expression in vitro. As presented in Fig. 5F, ectopic expression of anti-miR-9 dramatically increased GALNT4 mRNA level (p = 0.004), whereas transfection of miR-9 mimic significantly reduced GALNT4 mRNA level (p = 0.002). Corresponding to the change of GALNT4 mRNA level, GALNT4 protein expression also could be elevated by anti-miR-9 and suppressed by miR-9 mimic (Fig. 5F, bottom).
GALNT4 Attenuates miR-9-induced Migration, Invasion, Anoikis Resistance, and Stemness of HCC Cells
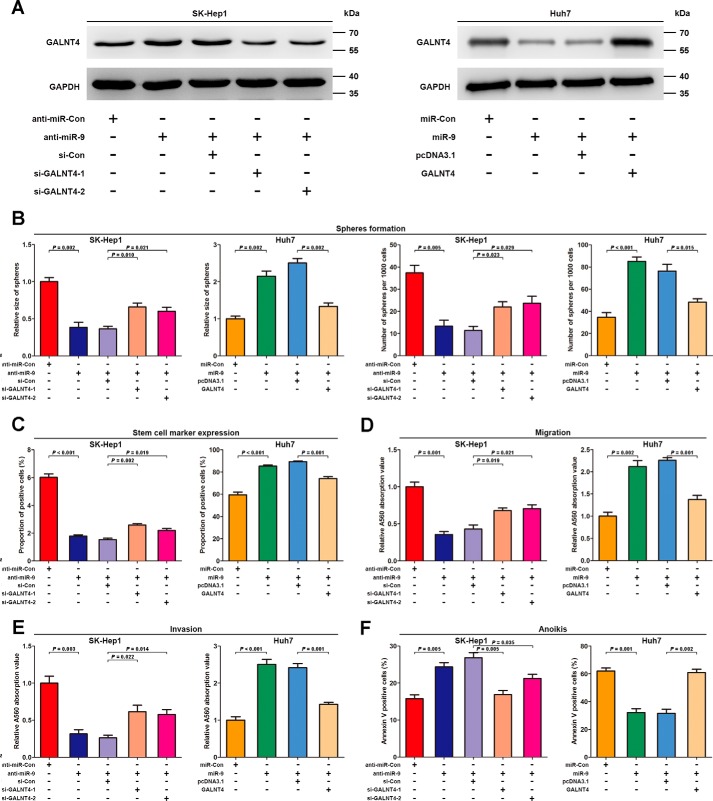
Having recently demonstrated that GALNT4 expression is directly repressed by miR-9, we further examined the role of GALNT4 in miR-9-induced phenotype of HCC cells. Co-transfection of GALNT4 siRNAs or pcDNA3.1-GALNT4 vector without its 3′-UTR reversed the effects of anti-miR-9 or miR-9 mimic on GALNT4 protein level, respectively (Fig. 6A). Performing a series of functional studies, we found that suppression of endogenous miR-9 expression by anti-miR-9 strongly inhibited sphere-forming ability (size, p = 0.002; number, p = 0.005 (Fig. 6B)), EpCAM expression (p < 0.001; Fig. 6C), migration (p = 0.001; Fig. 6D), invasion (p = 0.003; Fig. 6E), and anchorage-independent survival (p = 0.005; Fig. 6F) of SK-Hep1. In contrast, ectopic expression of miR-9 significantly promoted sphere-forming ability (size, p = 0.002; number, p < 0.001 (Fig. 6B)), EpCAM expression (p < 0.001; Fig. 6C), migration (p = 0.002; Fig. 6D), invasion (p < 0.001; Fig. 6E), and anchorage-independent survival (p = 0.001; Fig. 6F) of Huh7. Furthermore, these documented effects of anti-miR-9 or miR-9 mimic could be partially abolished via co-transfection of GALNT4 siRNAs or pcDNA3.1-GALNT4, respectively (Fig. 6).
FIGURE 6.
GALNT4 attenuates miR-9-induced migration, invasion, anoikis resistance, and stemness phenotype in HCC cells. SK-Hep1 transiently transfected with anti-miR-Con, anti-miR-9, or co-transfected anti-miR-9 with non-targeting control siRNA or GALNT4 siRNAs, respectively; Huh7 transiently transfected with miR-Con, miR-9 mimic, or co-transfected miR-9 mimic with pcDNA3.1 or pcDNA3.1-GALNT4, respectively. A, Western blotting analysis for SK-Hep1 and Huh7 transiently transfected with the indicated plasmids, respectively. GAPDH serves as an internal control. Quantification of sphere formation analysis (B), expression of EpCAM on sphere-derived cells (48 h) (C), migration assay (D), invasion assay (E), and anoikis assay (F) were documented when these analysis were performed in the indicated populations of each cell lines mentioned above. Sphere size is determined by its diameter. All data are from three independent experiments. Error bars, S.E.
GALNT4 Modulates the Activity of EGFR via O-Linked Glycosylation
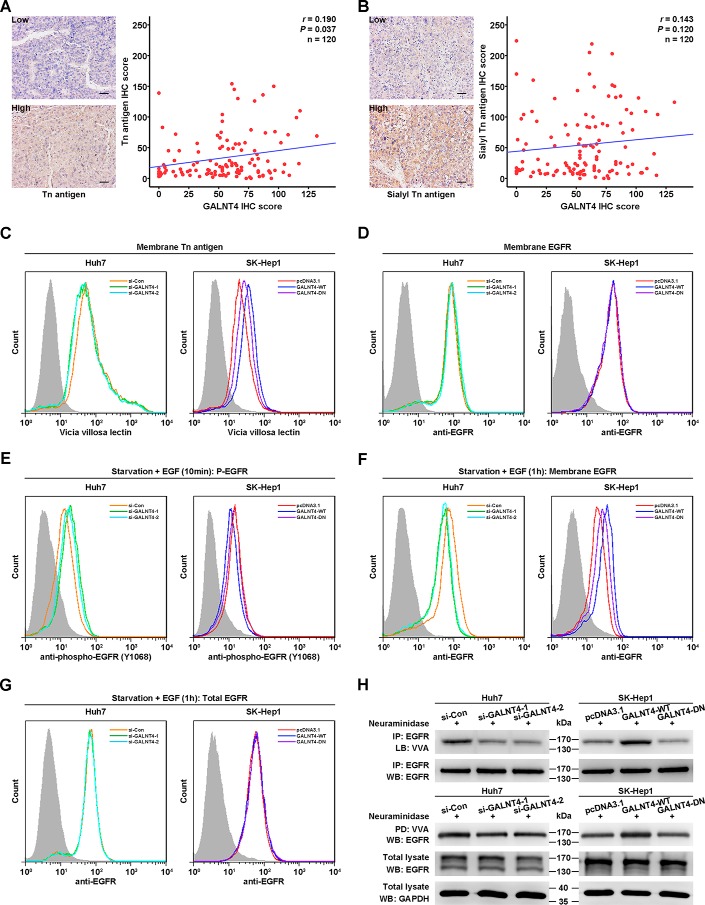
According to the above results indicating that the glycosyltransferase-dead mutant (GALNT4-DN) failed to exert the functions of the wild-type GALNT4, we speculated that GALNT4 might exert functions depending on its glycosyltransferase activity in HCC cells. We first detected the expression level of the products of GALNTs, Tn antigen and its derivative sialyl-Tn antigen, in clinical specimens by IHC analysis and evaluated whether GALNT4 level is associated with the level of these two structures. As presented in Fig. 7A, GALNT4 level was positively correlated with Tn antigen level in HCC specimens when the values for each patient were plotted (r = 0.190, p = 0.037). However, the level of sialyl-Tn antigen was not significantly associated with GALNT4 level (r = 0.143, p = 0.120) (Fig. 7B).
FIGURE 7.
GALNT4 modulates the activity of EGFR via O-linked glycosylation. A, representative images of IHC staining with Tn antigen in tumor tissues from the same set of specimens as in Fig. 1 (left). Scale bar, 50 μm (original magnification, ×200). The correlation between Tn antigen and GALNT4 expression was assessed by Spearman correlation test (right). B, representative images of IHC staining with sialyl-Tn antigen in tumor tissues from the same set of specimens as in Fig. 1 (left). Scale bar, 50 μm (original magnification, ×200). The correlation between sialyl-Tn antigen and GALNT4 expression was assessed by Spearman correlation test (right). C, flow cytometry analysis of membrane Tn antigen level on transfected Huh7 cells (left) and SK-Hep1 cells (right) by fluorescein-conjugated VVA. D, flow cytometry analysis of membrane EGFR level on transfected Huh7 cells (left) and SK-Hep1 cells (right) by PE-conjugated EGFR. E, flow cytometry analysis of phospho-EGFR (Tyr-1068) level on transfected Huh7 cells (left) and SK-Hep1 cells (right) by fluorescein-conjugated phospho-EGFR. Cells were serum-starved overnight and then treated with 100 ng/ml EGF at 37 °C for 10 min. F, flow cytometry analysis of membrane EGFR level on transfected Huh7 cells (left) and SK-Hep1 cells (right) by PE-conjugated EGFR. Cells were serum-starved overnight and then treated with 100 ng/ml EGF at 37 °C for 1 h. G, flow cytometry analysis of total EGFR level on transfected Huh7 cells (left) and SK-Hep1 cells (right) by PE-conjugated EGFR. Cells were serum-starved overnight and then treated with cycloheximide and 100 ng/ml EGF at 37 °C for 1 h. H, Huh7 cells and SK-Hep1 cells were transiently transfected with indicated plasmids. Cell lysates treated with neuraminidase were immunoprecipitated (IP) by anti-EGFR antibody and then lectin-blotted (LB) by VVA (top). Cell lysates treated with neuraminidase were pulled down (PD) by agarose-bound VVA and then immunoblotted (WB) by anti-EGFR antibody (bottom). Huh7 cells were transiently transfected with siRNAs specifically targeting GALNT4, and SK-Hep1 cells were transiently transfected with GALNT4-WT and GALNT4-DN.
To further confirm that GALNT4 has the ability to alter the level of Tn antigen in HCC cells, we detected the level of membrane Tn antigen in HCC cells transfected with the indicated plasmids by flow cytometry. Knockdown of GALNT4 can decrease the level of membrane Tn antigen, whereas overexpression of the GALNT4-WT but not GALNT4-DN can increase the Tn antigen level (Fig. 7C), which demonstrated that GALNT4 indeed exerts its glycosyltransferase activity in HCC cells.
Because previous studies have well documented that GALNT1, GALNT2, and GALNT10 can modify the O-glycosylation of epidermal growth factor receptor (EGFR) in HCC cells (13, 14, 17), we next aimed at distinguishing GALNT4 from these GALNTs or establishing similarities for the activity of EGFR in HCC cells. Our data showed that the membrane EGFR level was not affected by GALNT4 (Fig. 7D). Then we evaluated the phosphorylation of EGFR in HCC cells serum-starved overnight and stimulated by EGF for 10 min. As presented in Fig. 7E, knockdown of GALNT4 led the increase of EGFR (Tyr-1068) phosphorylation, whereas overexpression of GALNT-WT but not GALNT-DN decreased the level of P-EGFR (Tyr-1068).
Furthermore, after serum starvation overnight and stimulation by EGF for 1 h, the membrane EGFR of GALNT4-silencing cells was decreased, whereas overexpression of GALNT4-WT but not GALNT4-DN increased the surface EGFR level (Fig. 7F), because EGF-induced endocytosis leads to two different fates of EGFR: return to the cell surface by recycling or delivery to lysosomes for degradation. However, the total EGFR level was not affected by GALNT4 (Fig. 7G). Thus, GALNT4 might only inhibit EGF-induced endocytosis without affecting its degradation rate, similar to the effect of GALNT2 (14).
Because the activity of EGFR could be modulated by GALNT4, we then aimed to determine whether GALNT4 could modify the glycosylation of EGFR. First, the cell lysates were immunoprecipitated by anti-EGFR antibody. The enriched EGFR was treated by neuraminidase and then analyzed by lectin blotting using biotinylated Vicia villosa lectin (VVA). As showed in Fig. 7H, knockdown of GALNT4 decreased the Tn structure on EGFR, whereas GALNT4-WT but not GALNT4-DN increased Tn structure. In addition, EGFR could be pulled down by agarose-bound VVA after the cell lysates were treated by neuraminidase. Knockdown of GALNT4 decreased the amount of EGFR by VVA pull-down. And more EGFR could be pulled down in the GALNT4-WT-overexpressing cells but not GALNT4-DN-overexpressing cells (Fig. 7H). These results demonstrated that GALNT4 could also modify the O-linked glycosylation and regulate the activity of EGFR.
Inverse Correlated Expression Pattern of miR-9 and GALNT4 Contributes to Refine the Risk Stratification of Patients with HCC
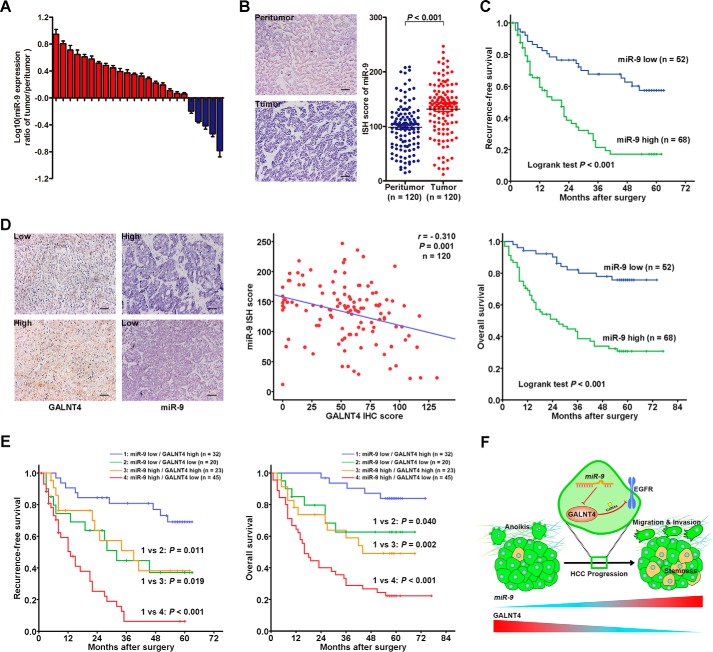
We next evaluated the expression of miR-9 in the same set of clinical specimens by qRT-PCR and in situ hybridization (ISH) analysis. Up-regulation of miR-9 was determined in 19 of 24 HCC tumors by qRT-PCR (Fig. 8A). ISH analysis also indicated that miR-9 expression was significantly up-regulated in tumors compared with their adjacent peritumor tissues (p < 0.001; Fig. 8B). In addition, patients with a high level of miR-9 presented earlier recurrence (p < 0.001; Fig. 8C) and shorter life span (p < 0.001; Fig. 8C) when compared with the miR-9 low expression subgroup. In line with the aforementioned results in vitro, GALNT4 protein level was inversely correlated with miR-9 level in HCC specimens when the values for each patient were plotted (r = −0.310, p = 0.001; Fig. 8D). Furthermore, subgroup analysis by Kaplan-Meier methods indicated that patients with a miR-9 high and GALNT4 low expression signature had the worst clinical outcomes, whereas patients with a miR-9 low and GALNT4 high expression signature had the best clinical outcomes (Fig. 8E).
FIGURE 8.
Inverse correlated expression pattern of miR-9 and GALNT4 contributes to refine the risk stratification of patients with HCC. A, miR-9 expression was detected in 24 pairs of primary HCC tumors and corresponding peritumor tissues by qRT-PCR. Analysis was performed in triplicate and normalized to the internal control, U6 small nuclear RNA. The y axis depicts log10(-fold change of miR-9 expression). Tissue specimens were collected from the same set of patients as in Fig. 1A. All data are from three independent experiments. B, representative ISH images of miR-9 expression in tumor and matched peritumor tissues of HCC patients from the same set of specimens as in Fig. 1 (left) and scatter plots for ISH scores of miR-9 expression in tumor and matched peritumor tissues. Scale bar, 50 μm (original magnification, ×200). C, Kaplan-Meier survival analysis of HCC patients for RFS and OS according to the expression of miR-9 (high expression subgroup, n = 68; low expression subgroup, n = 52). p value was calculated by log rank test. D, representative images of ISH staining with miR-9 and IHC staining with GALNT4 in tumor tissues (left). Scale bar, 50 μm. The correlation between miR-9 and GALNT4 expression was assessed by Spearman correlation test (right). E, subgroup analysis of HCC cases according to the expression profile of miR-9 and GALNT4 by the Kaplan-Meier method. p value was calculated by log rank test. F, proposed model of the role of the miR-9/GALNT4/EGFR axis for migration, invasion, anoikis resistance, and stemness properties in HCC cells during HCC progression.
Discussion
Deregulated expression of glycosyltransferases is a common feature of many tumor entities, including HCC. In this study, oncogenic miR-9-mediated down-regulation of GALNT4 was frequently presented in primary HCC tissues and associated with poor prognosis. Our work demonstrated that miR-9-mediated GALNT4 expression plays a central role in modulating extensive malignant behaviors of HCC cells, depending on its glycosyltransferase activity, and the miR-9/GALNT4 expression signature shows potential prognostic significance for patients with HCC (Fig. 8F).
The product of the initiating step of mucin-type O-glycosylation catalyzed by GALNTs is Tn antigen, which can be utilized to produce sialyl-Tn antigen, T antigen, and sialyl-T antigen by following enzymatic steps (18). Aberrant expression of these truncated O-glycans is functionally important, although it is only starting to be understood how this is linked to malignant characteristics of cancer cells (19). In line with the dual effects of truncated O-glycans, GALNTs can act as oncogenic promoters or tumor suppressors, probably depending on the different context of specific cell or cancer types. For instance, repressed expression of GALNT7 enhances invasion and immunosuppression in melanoma, whereas it suppresses growth and invasion of cervical cancer cells (20, 21). In the current study, we found that the expression of GALNT4 is frequently down-regulated in tumor tissues compared with the peritumor tissues of patients with HCC. In general, HCC is associated in most cases with chronic liver diseases, such as chronic hepatitis and cirrhosis (22). Just as the HCC cells sprout and transform into malignant tumor tissues, the peritumor tissues also undergo precancerous but not malignant changes. Of course, they are also difficult to regard as very normal tissues. Thus, the IHC staining-positive cells in peritumor tissues are liver cells possessing precancerous properties but not transformed malignant cells.
Further exploration demonstrated that ectopic expression of GALNT4 could inhibit migration and invasion of HCC cells. As the essential prerequisite for distant metastasis, the capacity of anoikis resistance was extremely suppressed by GALNT4.
The gaining of stemness by HCC cells has been recognized as a capability to generate cellular populations possessing strong metastatic potential from a small group of initiating cells and contributing to disease recurrence and progression (23). This study substantiated that GALNT4 could suppress stemness of HCC cells. Collectively, our results indicate the repressive effect of GALNT4 in self-renewal and propagation of stem cell-like HCC cells, which might further hinder distant colonization during tumor metastasis.
Accumulating evidence suggests that miRNAs play a crucial role in tumorigenesis and metastasis of HCC (24). Meanwhile, there are several examples of miRNA-mediated GALNT expression involving in malignant biological processes of corresponding tumors (17, 20, 21). In this study, we screened out miR-9 as the most probable miRNA directly targeting GALNT4 in HCC cells. Moreover, the up-regulation of miR-9 in primary HCC tissues was determined, and its promotion effect on migration, invasion, anoikis resistance, and stemness was also revealed. These findings on miR-9 are partially coherent with the results of other groups (25, 26). Indeed, miR-9 is associated with breast cancer stem cell phenotype and could increase the ability of HCC cells to form spheres (27, 28).
Previous studies have revealed that both GALNT1 and GALNT2 play pivotal roles in HCC tissues and cells (13, 14). Both of them could modify O-glycosylation of EGFR but exert opposite effects on the activity and stabilization of EGFR, which indicates that different ppGalNAc T isoforms, including GALNT4, might have different specific substrates and/or prefer modifying sites. Thus, GALNT1, -2, and -4 might all contribute to the alteration of GalNAc transferase activity and the production of total Tn antigen in HCC cells, but they also have their own unique or partially overlapping substrate panels. We have searched the TargetScan database and found that all of these three isoforms have miR-9 regulatory elements. This demonstrated that these isoforms might be subject to common regulatory mechanisms and exert synergistic effects. As presented in our current work, GALNT4 also could modify O-glycosylation of EGFR and then inhibit its activation and endocytosis in HCC cells. Similar effects of GALNT2 have been elucidated, which further confirms the synergy.
Experimental Procedures
Cell Lines and Clinical Samples
Human hepatoma cell lines Huh7, PLC/PRF/5, HepG2, SK-Hep1, and Hep3B were acquired from the Shanghai Cell Bank of the Chinese Academy of Sciences (Shanghai, China), where they were authenticated by DNA fingerprinting analysis using STR markers and characterized by mycoplasma detection. A total of 144 consecutive patients with HCC who underwent partial hepatectomy at Nantong Tumor Hospital (Nantong University, Jiangsu, China) were enrolled in this study. Clinical specimens were collected from 2003 to 2005. Pairs of HCC tumors and their matched peritumor tissues of 120 patients were exploited to construct the tissue microarray. The remaining 24 pairs of tumor tissues and adjacent peritumor tissues were used to isolate RNAs. Ethics approval was obtained from the research medical ethics committee of Fudan University, and informed consent was obtained from each patient.
Plasmid Construction
The wild-type and mutant 3′-UTR fragments of GALNT4 mRNA containing the predicted miR-9 binding sites were amplified and cloned in pGL3 control vector, respectively. The wild-type GALNT4 expression plasmid was kindly provided by Dr. Hisashi Narimatsu (National Institute of Advanced Industrial Science and Technology, Tsukuba, Japan). Glycosyltransferase-dead mutant GALNT4 (GALNT4-DN) was amplified by PCR and constructed into the pcDNA3.1. The D459H mutant has been confirmed to abolish the ability to glycosylate the specific substrates of GALNT4 (16). The PCR primer sets used are presented in Table 4.
TABLE 4.
Primers used for plasmid construction and qRT-PCR
| Primer name | Sequence (5′–3′) |
|---|---|
| GALNT4-3′-UTR-WT | |
| Forward | TTTCTAGATTTGCCTCGTAAAATGAA |
| Reverse | CATCTAGAAAAAGTTTGATTTGCCAT |
| GALNT4-3′-UTR-MUTa | |
| Forward | GAATTTATTGGTACTACCGCTCTCTT |
| Reverse | AGCCAAAGAGAGCGGTAGTACCAATA |
| GALNT4-DNb | |
| Forward 1c | AAGGTACCCTTCGAAGGAGATAGAAC |
| Reverse 1 | TGTTGTCAGGAGAATTATAATGTAAACA |
| Forward 2 | TCTCGTCTGAATGTTTACATTATAATTC |
| Reverse 2c | AACTCGAGGTCCTATTTCTCAAAACT |
| GAPDH | |
| Forward | GTCAAGGCTGAGAACGGGAA |
| Reverse | AAATGAGCCCCAGCCTTCTC |
| GALNT4 | |
| Forward | CTGGCGTTTTTAACAGTGGC |
| Reverse | ATCCTCGTTGAGCTGGAGTT |
| hsa-miR-32 | |
| Forward | TATTGCACATTACTAAGTTGCA |
| hsa-miR-181a | |
| Forward | AACATTCAACGCTGTCGGTGAGT |
| hsa-miR-181b | |
| Forward | AACATTCATTGCTGTCGGTGGGT |
| hsa-miR-181c | |
| Forward | AACATTCAACCTGTCGGTGAGT |
| hsa-miR-181d | |
| Forward | AACATTCATTGTTGTCGGTGGGT |
| hsa-miR-372 | |
| Forward | AAAGTGCTGCGACATTTGAGCGT |
| hsa-miR-613 | |
| Forward | AGGAATGTTCCTTCTTTGCC |
| hsa-miR-206 | |
| Forward | TGGAATGTAAGGAAGTGTGTGG |
| hsa-miR-301b | |
| Forward | CAGTGCAATGATATTGTCAAAGC |
| hsa-miR-9 | |
| Forward | TCTTTGGTTATCTAGCTGTATGA |
| hsa-miR-302a | |
| Forward | TAAGTGCTTCCATGTTTTGGTGA |
| hsa-miR-302b | |
| Forward | TAAGTGCTTCCATGTTTTAGTAG |
| hsa-miR-302d | |
| Forward | TAAGTGCTTCCATGTTTGAGTGT |
| hsa-miR-302e | |
| Forward | TAAGTGCTTCCATGCTT |
| hsa-miR-520a-3p | |
| Forward | AAAGTGCTTCCCTTTGGACTGT |
| hsa-miR-520c-3p | |
| Forward | AAAGTGCTTCCTTTTAGAGGGT |
| hsa-miR-520d-3p | |
| Forward | AAAGTGCTTCTCTTTGGTGGGT |
| U6snRNA | |
| Forward | CTCGCTTCGGCAGCACA |
| Reverse | AACGCTTCACGAATTTGCGT |
a MUT, mutant.
b DN, inactive form.
c Primers specifically bind to the linker sequence of the wild-type GALNT4 plasmid.
Transfection and RNA Interference
Transient and stable transfections were performed as described previously (29). pcDNA3.1-GALNT4 and its control, pcDNA3.1 vector, were used in transient and stable overexpression experiments. Duplex siRNA against GALNT4 and non-targeting control siRNA (OriGene, Technologies Inc., Rockville, MD) were used to transiently knock down endogenous GALNT4 expression. shRNA against GALNT4 and non-targeting control shRNA (OriGene Technologies) were used to stably knock down endogenous GALNT4 expression. miR-9 mimic or antisense (anti-miR-9) oligonucleotide with their corresponding scrambled sequence (miR-Con and anti-Con, respectively) (Invitrogen) were transfected into HCC cells to force or repress the expression of miR-9. Transfection was conducted using FuGENE HD transfection reagent (Roche Applied Science) according to the manufacturer's instructions.
RNA Extraction and qRT-PCR
Total RNA was isolated from clinical samples and cultured cell lines using TRIzol reagent (Invitrogen) according to the manufacturer's protocol. qRT-PCR analysis was performed as described in our previous study (29). The expression of 17 miRNAs in Fig. 5B was measured by NCodeTM miRNA first-strand cDNA synthesis and qRT-PCR kits (Invitrogen). Furthermore, the expression of miR-9 was validated in cell lines and clinical specimens by TaqMan miRNA assays (Applied Biosystems, Foster City, CA). GAPDH and U6 small nuclear RNAs were used as internal control when determining the expression of GALNT4 and miRNAs, respectively. The PCR primer sets used are presented in Table 4.
Western Blotting and Lectin Blotting
Western blotting was performed as described previously (29). Rabbit anti-GALNT4 polyclonal antibody (catalogue no. 12897-1-AP, Proteintech Group, Inc., Chicago, IL) was applied to detect the protein level of GALNT4. Mouse anti-EGFR monoclonal antibody (Santa Cruz Biotechnology, Inc., Dallas, TX) and mouse anti-GAPDH monoclonal antibody (Santa Cruz Biotechnology) were used to evaluate the protein level of EGFR and GAPDH, respectively. For lectin blotting, the PVDF membrane was first blocked by Carbo-FreeTM blocking solution (Vector Laboratories, Burlingame, CA) for 30 min at room temperature. Then the membrane was incubated with biotinylated VVA (Vector Laboratories) at 10 μg/ml for 30 min at room temperature and detected by VECTASTAIN ABC reagents (Vector Laboratories).
Lectin Pull-down and EGFR Immunoprecipitation
For lectin pull-down, neuraminidase (New England Biolabs, Ipswich, MA) was used to remove sialic acid, and cell lysates were incubated with agarose-bound VVA (Vector Laboratories) at 4 °C for 4 h followed by washing with lysis buffer three times. Immunoprecipitation was conducted with an immunoprecipitation kit (Roche Applied Science) according to the manufacturer's instructions. Briefly, cell lysates were incubated with 3 μg/ml mouse anti-EGFR monoclonal antibody (Santa Cruz Biotechnology) at 4 °C for 1 h and then added to 50 μl of Protein G-agarose suspension and incubated overnight at 4 °C with a rotator. The precipitated EGFR after neuraminidase treatment was then subjected to Western blotting.
In Silico Prediction for MicroRNAs Targeting GALNT4-3′-UTR
Three algorithms were used to predict potential miRNAs targeting 3′-UTR of GALNT4: TargetScan (release 6.2) confined to conserved miRNAs among vertebrates, miRanda with the threshold of mirSVR score sum of −0.1, and miRmap confined to ΔG open = 70, probability exact = 10, conservation phylop = 70, and miRmap score = 10.
Migration and Matrigel Invasion Assay
A cell migration assay was performed in a 24-well Boyden chamber with 8-μm pore size polycarbonate membrane (Corning Inc.) as described previously (30). Briefly, 3 × 104 cells transfected with indicated plasmids in 250 μl of DMEM containing 2% FBS were seeded into the upper chamber, and 500 μl of DMEM containing 10% FBS were added into the lower well for 24 h. An invasion assay was done by the same procedure, except that the membrane was coated with 40 μg of Matrigel (BD Biosciences) to form a matrix barrier and cultured for 48 h. The cells that had migrated and invaded were stained by cell stain (Millipore, Billerica, MA) and washed by extraction buffer (Millipore, Billerica, MA). The absorbance was assessed by a universal microplate reader (BIO-TEK Instruments, Minneapolis, MN).
In Vitro Anoikis Assay
In vitro anoikis analysis was performed as described previously (31). Briefly, HCC cells transfected with the indicated plasmids were seeded into poly-HEMA-coated 6-well tissue culture plates (Corning) with DMEM containing 2% FBS. Cells were collected to detect anoikis by flow cytometry.
Sphere Culture
The transfected cells were cultured in suspension in serum-free DMEM/F-12 (Invitrogen) containing B27 (1:50; Invitrogen), 20 ng/ml EGF (Sigma-Aldrich), and 20 ng/ml basic fibroblast growth factor (Invitrogen) in poly-HEMA-coated 6-well tissue culture plates (Corning) for 7 days. Spheres larger than 50 μm were counted.
Activation and Endocytosis of EGFR
To evaluate the effect of GALNT4 on phosphorylation of EGFR, the transfected cells were serum-starved overnight and then cultured with 100 ng/ml EGF (Sigma-Aldrich) at 37 °C for 10 min. The cells were washed by cold PBS and fixed to conduct the flow cytometry. To assess the alteration of membrane and total EGFR amount after EGF stimulation, the transfected cells were serum-starved overnight and then cultured with 100 ng/ml EGF at 37 °C for 1 h. The cells were washed by cold PBS and prepared for flow cytometry.
Flow Cytometry
To perform the flow cytometric analysis, cells were dissociated into single-cell populations and incubated with PE-conjugated EpCAM antibody (BD Biosciences), Annexin V and 7-AAD reagent (BD Biosciences), fluorescein-conjugated VVA (Vector Laboratories), fluorescein-conjugated anti-phospho-EGFR (Tyr-1068) (R&D Systems, Minneapolis, MN), and PE-conjugated EGFR antibody (Abcam, Cambridge, MA) at 4 °C for 30 min, respectively. For phospho-EGFR and total EGFR staining, cells were pretreated with the BD Cytofix/CytopermTM Fixation/Permeabilization Solution Kit (BD Biosciences) according to the manufacturer's instructions. Data were acquired by FACSCalibur (BD Biosciences) and analyzed by FlowJo software (TreeStar, Ashland, OR).
Tumor Xenograft Experiment
Four-week-old male athymic nude mice (Foxn1nu/nu, BALB/c background) were purchased from the Shanghai Laboratory Animal Center (Chinese Academy of Sciences, Shanghai, China). Tumor xenograft experiments in nude mice were carried out as described previously (29). Briefly, 1 × 107 viable cells stably transfected with pcDNA-3.1-GALNT4, shRNA against GALNT4, or the corresponding control vectors were subcutaneously injected into the flank on each side of the nude mice. Ten mice were randomly enrolled in each group. Tumor volume, tumor weight, and survival of mice were recorded. Tumor volumes were calculated using the formula, volume = (L × W2)/2, where L is length at the widest point of the tumor and W is the maximum width perpendicular to L. All animal procedures were approved by the animal care and use committee of Fudan University.
Luciferase Activity Assay
The luciferase activity assay was conducted with the Dual-LuciferaseTM reporter assay system (Promega, Madison, WI) according to the manufacturer's protocol. Activity of the co-expressed Renilla luciferase was used to normalize the transfection efficiency.
ISH and IHC
ISH and IHC on formalin-fixed and paraffin-embedded sections were performed as described previously (17). Probes targeting miR-9 or scrambled miRNA (50 nm; digoxigenin-labeled locked nucleic acid probes) (Exiqon, Vedbaek, Denmark) were used in ISH analysis. Rabbit anti-GALNT4 polyclonal antibody (Proteintech Group, Inc., Chicago, IL), biotinylated VVA (Vector Laboratories), and anti-sialyl-Tn antibody (Abcam) were used in IHC analysis. For ISH analysis, specimens were pretreated with proteinase K for 5 min. Specimens of both ISH and IHC analysis were incubated with PBS containing 3% H2O2 at 37 °C for 30 min. The staining intensity of specimens in both ISH and IHC analysis was evaluated by two independent pathologists blinded to the clinical outcome and clinicopathological data of patients. A semiquantitative H-score, which ranged from 0 to 300, was calculated by multiplying the staining intensity (0, negative; 1, weak; 2, moderate; 3, strong) by the distribution area proportion (0–100%) at each intensity for each sample.
Statistical Analysis
Data were presented as mean ± S.E. Patients were dichotomized into low and high expression subgroups by a “minimum p value” approach conducted by X-tile (32) according to miR-9 ISH scores and GALNT4 IHC scores, respectively. Statistical analyses involved Student's t test, χ2 test, log rank test, and Spearman correlation test. Data were analyzed using X-tile software version 3.6.1 (Yale University, New Haven, CT), IBM SPSS Statistics version 21.0 (IBM Corp., Armonk, NY), MedCalc software version 11.4.2.0 (MedCalc, Mariakerke, Belgium), and GraphPad Prism version 5 (GraphPad Software, La Jolla, CA). All statistical tests were two-tailed, and differences were considered significant at a level of <0.05.
Author Contributions
Y. L. performed acquisition, analysis, and interpretation of data; statistical analysis; and drafting of the manuscript. H. L., L. Y., Q. W., W. L., Q. F., W. Z., and H. Z. contributed technical and material support. J. X. and J. G. contributed the study concept and design, analyzed and interpreted data, drafted the manuscript, obtained funding, and supervised the study. All authors read and approved the final manuscript.
This work was supported by National Key Projects for Infectious Diseases of China Grants 2012ZX10002012-007 and 2016ZX10002018-008; National Natural Science Foundation of China Grants 31100629, 31270863, 81401988, 81471621, 81472227, 31570803, and 81572352; and Program for New Century Excellent Talents in University Grant NCET-13-0146. The authors declare that they have no conflicts of interest with the contents of this article.
- HCC
- hepatocellular carcinoma
- GALNT
- N-acetylgalactosaminyltransferase
- miR-9
- microRNA-9
- RFS
- recurrence-free survival
- OS
- overall survival
- qRT-PCR
- quantitative RT-PCR
- IHC
- immunohistochemistry
- EGFR
- EGF receptor
- VVA
- V. villosa lectin
- ISH
- in situ hybridization
- PE
- phycoerythrin
- 7-AAD
- 7-aminoactinomycin D.
References
- 1. El-Serag H. B., and Rudolph K. L. (2007) Hepatocellular carcinoma: epidemiology and molecular carcinogenesis. Gastroenterology 132, 2557–2576 [DOI] [PubMed] [Google Scholar]
- 2. Llovet J. M., Burroughs A., and Bruix J. (2003) Hepatocellular carcinoma. Lancet 362, 1907–1917 [DOI] [PubMed] [Google Scholar]
- 3. Clausen H., and Bennett E. P. (1996) A family of UDP-GalNAc: polypeptide N-acetylgalactosaminyl-transferases control the initiation of mucin-type O-linked glycosylation. Glycobiology 6, 635–646 [DOI] [PubMed] [Google Scholar]
- 4. Ten Hagen K. G., Fritz T. A., and Tabak L. A. (2003) All in the family: the UDP-GalNAc:polypeptide N-acetylgalactosaminyltransferases. Glycobiology 13, 1R–16R [DOI] [PubMed] [Google Scholar]
- 5. Gerken T. A., Revoredo L., Thome J. J., Tabak L. A., Vester-Christensen M. B., Clausen H., Gahlay G. K., Jarvis D. L., Johnson R. W., Moniz H. A., and Moremen K. (2013) The lectin domain of the polypeptide GalNAc transferase family of glycosyltransferases (ppGalNAc Ts) acts as a switch directing glycopeptide substrate glycosylation in an N- or C-terminal direction, further controlling mucin type O-glycosylation. J. Biol. Chem. 288, 19900–19914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Taniuchi K., Cerny R. L., Tanouchi A., Kohno K., Kotani N., Honke K., Saibara T., and Hollingsworth M. A. (2011) Overexpression of GalNAc-transferase GalNAc-T3 promotes pancreatic cancer cell growth. Oncogene 30, 4843–4854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Gu C., Oyama T., Osaki T., Li J., Takenoyama M., Izumi H., Sugio K., Kohno K., and Yasumoto K. (2004) Low expression of polypeptide GalNAc N-acetylgalactosaminyl transferase-3 in lung adenocarcinoma: impact on poor prognosis and early recurrence. Br. J. Cancer 90, 436–442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Kitada S., Yamada S., Kuma A., Ouchi S., Tasaki T., Nabeshima A., Noguchi H., Wang K. Y., Shimajiri S., Nakano R., Izumi H., Kohno K., Matsumoto T., and Sasaguri Y. (2013) Polypeptide N-acetylgalactosaminyl transferase 3 independently predicts high-grade tumours and poor prognosis in patients with renal cell carcinomas. Br. J. Cancer 109, 472–481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Park J. H., Katagiri T., Chung S., Kijima K., and Nakamura Y. (2011) Polypeptide N-acetylgalactosaminyltransferase 6 disrupts mammary acinar morphogenesis through O-glycosylation of fibronectin. Neoplasia 13, 320–326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Park J. H., Nishidate T., Kijima K., Ohashi T., Takegawa K., Fujikane T., Hirata K., Nakamura Y., and Katagiri T. (2010) Critical roles of mucin 1 glycosylation by transactivated polypeptide N-acetylgalactosaminyltransferase 6 in mammary carcinogenesis. Cancer Res. 70, 2759–2769 [DOI] [PubMed] [Google Scholar]
- 11. Li Z., Yamada S., Inenaga S., Imamura T., Wu Y., Wang K. Y., Shimajiri S., Nakano R., Izumi H., Kohno K., and Sasaguri Y. (2011) Polypeptide N-acetylgalactosaminyltransferase 6 expression in pancreatic cancer is an independent prognostic factor indicating better overall survival. Br. J. Cancer 104, 1882–1889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Wagner K. W., Punnoose E. A., Januario T., Lawrence D. A., Pitti R. M., Lancaster K., Lee D., von Goetz M., Yee S. F., Totpal K., Huw L., Katta V., Cavet G., Hymowitz S. G., Amler L., and Ashkenazi A. (2007) Death-receptor O-glycosylation controls tumor-cell sensitivity to the proapoptotic ligand Apo2L/TRAIL. Nat. Med. 13, 1070–1077 [DOI] [PubMed] [Google Scholar]
- 13. Huang M. J., Hu R. H., Chou C. H., Hsu C. L., Liu Y. W., Huang J., Hung J. S., Lai I. R., Juan H. F., Yu S. L., Wu Y. M., and Huang M. C. (2015) Knockdown of GALNT1 suppresses malignant phenotype of hepatocellular carcinoma by suppressing EGFR signaling. Oncotarget 6, 5650–5665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Wu Y. M., Liu C. H., Hu R. H., Huang M. J., Lee J. J., Chen C. H., Huang J., Lai H. S., Lee P. H., Hsu W. M., Huang H. C., and Huang M. C. (2011) Mucin glycosylating enzyme GALNT2 regulates the malignant character of hepatocellular carcinoma by modifying the EGF receptor. Cancer Res. 71, 7270–7279 [DOI] [PubMed] [Google Scholar]
- 15. Bennett E. P., Hassan H., Mandel U., Mirgorodskaya E., Roepstorff P., Burchell J., Taylor-Papadimitriou J., Hollingsworth M. A., Merkx G., van Kessel A. G., Eiberg H., Steffensen R., and Clausen H. (1998) Cloning of a human UDP-N-acetyl-α-d-galactosamine:polypeptide N-acetylgalactosaminyltransferase that complements other GalNAc-transferases in complete O-glycosylation of the MUC1 tandem repeat. J. Biol. Chem. 273, 30472–30481 [DOI] [PubMed] [Google Scholar]
- 16. Hassan H., Reis C. A., Bennett E. P., Mirgorodskaya E., Roepstorff P., Hollingsworth M. A., Burchell J., Taylor-Papadimitriou J., and Clausen H. (2000) The lectin domain of UDP-N-acetyl-d-galactosamine:polypeptide N-acetylgalactosaminyltransferase-T4 directs its glycopeptide specificities. J. Biol. Chem. 275, 38197–38205 [DOI] [PubMed] [Google Scholar]
- 17. Wu Q., Liu H. O., Liu Y. D., Liu W. S., Pan D., Zhang W. J., Yang L., Fu Q., Xu J. J., and Gu J. X. (2015) Decreased expression of hepatocyte nuclear factor 4α (Hnf4α)/microRNA-122 (miR-122) axis in hepatitis B virus-associated hepatocellular carcinoma enhances potential oncogenic GALNT10 protein activity. J. Biol. Chem. 290, 1170–1185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Tarp M. A., and Clausen H. (2008) Mucin-type O-glycosylation and its potential use in drug and vaccine development. Biochim. Biophys. Acta 1780, 546–563 [DOI] [PubMed] [Google Scholar]
- 19. Brockhausen I. (1999) Pathways of O-glycan biosynthesis in cancer cells. Biochim. Biophys. Acta 1473, 67–95 [DOI] [PubMed] [Google Scholar]
- 20. Gaziel-Sovran A., Segura M. F., Di Micco R., Collins M. K., Hanniford D., Vega-Saenz de Miera E., Rakus J. F., Dankert J. F., Shang S., Kerbel R. S., Bhardwaj N., Shao Y., Darvishian F., Zavadil J., Erlebacher A., et al. (2011) miR-30b/30d regulation of GalNAc transferases enhances invasion and immunosuppression during metastasis. Cancer Cell 20, 104–118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Peng R. Q., Wan H. Y., Li H. F., Liu M., Li X., and Tang H. (2012) MicroRNA-214 suppresses growth and invasiveness of cervical cancer cells by targeting UDP-N-acetyl-α-d-galactosamine:polypeptide N-acetylgalactosaminyltransferase 7. J. Biol. Chem. 287, 14301–14309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Farazi P. A., and DePinho R. A. (2006) Hepatocellular carcinoma pathogenesis: from genes to environment. Nat. Rev. Cancer 6, 674–687 [DOI] [PubMed] [Google Scholar]
- 23. Rountree C. B., Mishra L., and Willenbring H. (2012) Stem cells in liver diseases and cancer: recent advances on the path to new therapies. Hepatology 55, 298–306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Yang N., Ekanem N. R., Sakyi C. A., and Ray S. D. (2015) Hepatocellular carcinoma and microRNA: new perspectives on therapeutics and diagnostics. Adv. Drug Deliv. Rev. 81, 62–74 [DOI] [PubMed] [Google Scholar]
- 25. Wang Y., Lee A. T., Ma J. Z., Wang J., Ren J., Yang Y., Tantoso E., Li K. B., Ooi L. L., Tan P., and Lee C. G. (2008) Profiling microRNA expression in hepatocellular carcinoma reveals microRNA-224 up-regulation and apoptosis inhibitor-5 as a microRNA-224-specific target. J. Biol. Chem. 283, 13205–13215 [DOI] [PubMed] [Google Scholar]
- 26. Sun Z., Han Q., Zhou N., Wang S., Lu S., Bai C., and Zhao R. C. (2013) MicroRNA-9 enhances migration and invasion through KLF17 in hepatocellular carcinoma. Mol. Oncol. 7, 884–894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Gwak J. M., Kim H. J., Kim E. J., Chung Y. R., Yun S., Seo A. N., Lee H. J., and Park S. Y. (2014) MicroRNA-9 is associated with epithelial-mesenchymal transition, breast cancer stem cell phenotype, and tumor progression in breast cancer. Breast Cancer Res. Treat. 147, 39–49 [DOI] [PubMed] [Google Scholar]
- 28. Drakaki A., Hatziapostolou M., Polytarchou C., Vorvis C., Poultsides G. A., Souglakos J., Georgoulias V., and Iliopoulos D. (2015) Functional microRNA high throughput screening reveals miR-9 as a central regulator of liver oncogenesis by affecting the PPARA-CDH1 pathway. BMC Cancer 15, 542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Xu J., Yun X., Jiang J., Wei Y., Wu Y., Zhang W., Liu Y., Wang W., Wen Y., and Gu J. (2010) Hepatitis B virus X protein blunts senescence-like growth arrest of human hepatocellular carcinoma by reducing Notch1 cleavage. Hepatology 52, 142–154 [DOI] [PubMed] [Google Scholar]
- 30. Liu H., Xu L., He H., Zhu Y., Liu J., Wang S., Chen L., Wu Q., Xu J., and Gu J. (2012) Hepatitis B virus X protein promotes hepatoma cell invasion and metastasis by stabilizing Snail protein. Cancer Sci. 103, 2072–2081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Xu J., Liu H., Chen L., Wang S., Zhou L., Yun X., Sun L., Wen Y., and Gu J. (2012) Hepatitis B virus X protein confers resistance of hepatoma cells to anoikis by up-regulating and activating p21-activated kinase 1. Gastroenterology 143, 199–212.e4 [DOI] [PubMed] [Google Scholar]
- 32. Camp R. L., Dolled-Filhart M., and Rimm D. L. (2004) X-tile: a new bio-informatics tool for biomarker assessment and outcome-based cut-point optimization. Clin. Cancer Res. 10, 7252–7259 [DOI] [PubMed] [Google Scholar]