Abstract
The mitochondrial outer membrane (OM) contains single and multiple membrane-spanning proteins that need to contain signals that ensure correct targeting and insertion into the OM. The biogenesis of such proteins has so far essentially only been studied in yeast and related organisms. Here we show that POMP10, an OM protein of the early diverging protozoan Trypanosoma brucei, is signal-anchored. Transgenic cells expressing variants of POMP10 fused to GFP demonstrate that the N-terminal membrane-spanning domain flanked by a few positively charged or neutral residues is both necessary and sufficient for mitochondrial targeting. Carbonate extraction experiments indicate that although the presence of neutral instead of positively charged residues did not interfere with POMP10 localization, it weakened its interaction with the OM. Expression of GFP-tagged POMP10 in inducible RNAi cell lines shows that its mitochondrial localization depends on pATOM36 but does not require Sam50 or ATOM40, the trypanosomal analogue of the Tom40 import pore. pATOM36 is a kinetoplastid-specific OM protein that has previously been implicated in the assembly of OM proteins and in mitochondrial DNA inheritance. In summary, our results show that although the features of the targeting signal in signal-anchored proteins are widely conserved, the protein machinery that mediates their biogenesis is not.
Keywords: evolution, membrane biogenesis, mitochondrial transport, outer membrane, Trypanosoma brucei
Introduction
Mitochondria perform many important functions and are essential for all eukaryotes (1). Whereas a small number of proteins are synthesized in the organelle, >95% of the mitochondrial proteome is nuclear-encoded, synthesized in the cytosol, and subsequently imported into mitochondria (2–4). Mitochondria consist of four compartments: the outer and the inner membrane that surround the soluble intermembrane space (IMS)2 and the matrix, respectively. Thus, nuclear-encoded proteins not only need to be targeted to mitochondria but also sorted to their correct intra-mitochondrial destination. The mitochondrial outer membrane (OM) is of special interest. It builds the interface between the organelle and the cytosol and forms a barrier across which all communication between the organelle and its surroundings must occur (5–7). Mitochondrial OM proteins mediate apoptosis, fission, fusion and interaction with other organelles as well as transport of metabolites and precursor proteins. Whereas mitochondrial protein import in general has been extensively studied, we still have large gaps in the understanding of the biogenesis of mitochondrial OM proteins (8–11).
Integral OM proteins are either anchored by a transmembrane β-barrel or by single or multiple α-helices. The single membrane-spanning proteins can be further categorized into N-terminal (signal)-anchored, internally anchored or C-terminal (tail)-anchored proteins. OM proteins generally contain internal targeting signals. For β-barrel proteins the OM targeting signal appears to be a dedicated β-hairpin motif (12), whereas for single membrane-spanning proteins the targeting signal is confined to the transmembrane domain and positive residues in its flanking regions (13–17).
Biogenesis of α-helically anchored OM proteins involves a number of import pathways, many of which are still poorly characterized. β-Barrel proteins are first imported into the IMS by the translocase of the OM (TOM), where they bind to the small translocase of the inner membrane (TIM) chaperones that hand them over to the sorting and assembly machinery (SAM) by which they are integrated into the OM (2, 18). The insertion of some of the single membrane-spanning OM proteins depends exclusively on TOM subunits. Tom22 on the other hand requires the TOM receptors but subsequently is inserted into the OM by SAM and Mdm10 (19). Finally, insertion of some OM proteins is mediated by a protein complex consisting of Mim1 and Mim2 (20–22), termed mitochondrial import complex. The exact role of the mitochondrial import complex and its mode of action is still unknown (23). Moreover, it has recently been shown that a few proteins may be able to insert into the OM independently of protein factors and that at least in one case this is facilitated by specific lipids (24, 25).
Mapping the variation of the import systems in different eukaryotes provides insight into the evolution of the mitochondrial protein import systems, which is key to understanding how the endosymbiotic ancestor of mitochondria converted into an organelle (26–29). However, most of what we know about mitochondrial protein import stems from work done in the yeast Saccharomyces cerevisiae and a few related organisms.
Here we have studied the biogenesis of POMP10 (present in the outer mitochondrial membrane proteome 10) (7), a putative signal-anchored mitochondrial OM protein in the parasitic protozoan Trypanosoma brucei, which is essentially unrelated to yeast and one of the most early diverging eukaryotes (30, 31). T. brucei has a mitochondrion that in the insect stage of the parasite's life cycle is capable of oxidative phosphorylation. In our study we have identified the features of the import signal of POMP10. Moreover, we show that its import is independent of both Sam50 and the trypanosomal functional analog of Tom40, termed archaic translocase of the mitochondrial OM 40 (ATOM40) (32–34). However, using RNAi-mediated ablation it could be demonstrated that the biogenesis of POMP10 does require the trypanosomatid-specific OM protein termed peripheral ATOM36 (pATOM36) that is loosely associated with the ATOM complex (35). pATOM36 has recently been shown to mediate assembly of the ATOM complex as well as mitochondrial DNA inheritance (36).
Results
POMP10 Is a Signal-anchored Mitochondrial OM Protein
POMP10 is an abundant mitochondrial OM protein that is conserved in all kinetoplastids and which in T. brucei has a predicted molecular mass of 62.2 kDa. It does not have either conserved domains or orthologues outside the kinetoplastids. RNAi-mediated ablation of POMP10 does not affect growth or mitochondrial morphology (Fig. 1); its function therefore remains unknown. Most prediction programs indicate that POMP10 has a single transmembrane domain at its very N terminus (Table 1) that on both sides is flanked by positively charged amino acids (7). Bioinformatic analysis suggests it to be a signal-anchored protein, which would expose its soluble C-terminal domain to the cytosol. To experimentally confirm the predicted topology, we produced a transgenic cell line expressing a full-length POMP10 variant whose C terminus is fused to GFP. Fig. 2A shows that the POMP10-GFP fusion is expressed and that during cell fractionation it behaves like the mitochondrial marker proteins. This was confirmed by immunofluorescence (IF) analysis, which yields a staining pattern that is identical to the mitochondrial OM protein ATOM40 (see Fig. 4A). Protease protection assays using mitochondria isolated under isotonic conditions show that GFP-tagged POMP10 is protease-sensitive in intact mitochondria like the previously characterized protein import receptor ATOM69 (32). The IMS-localized TbTim9 and the matrix protein mtHsp70, however, were only digested after the membranes were dissolved by Triton (Fig. 2B).
FIGURE 1.
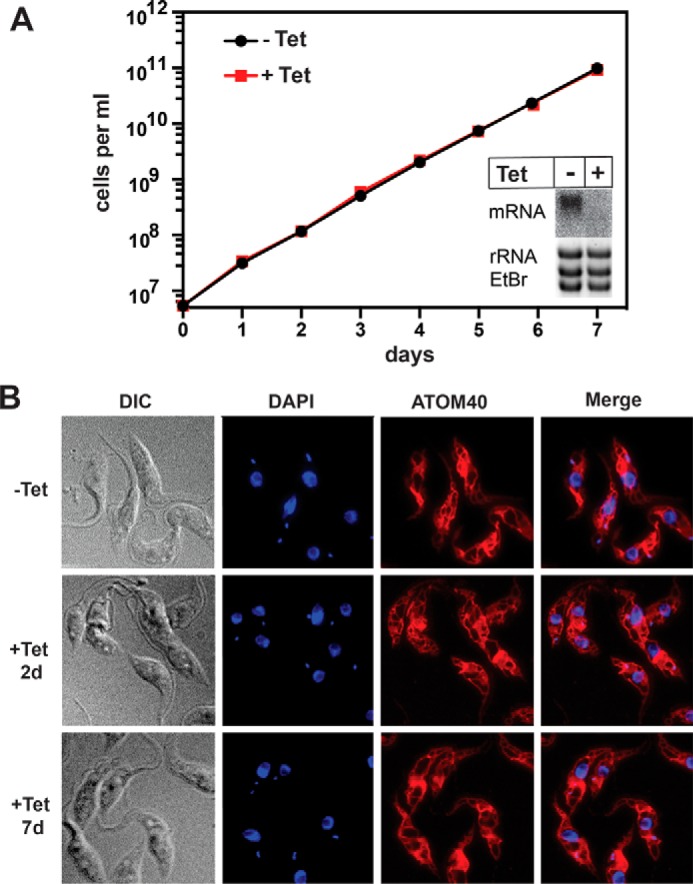
RNAi-mediated ablation of POMP10 did not affect growth or mitochondrial morphology. A, growth curve of uninduced tetracycline (−Tet) and induced (+Tet) POMP10-RNAi cell line. Inset, Northern blot of the POMP10 mRNA after 2 days of RNAi induction. Ethidium bromide-stained gel showing the rRNAs served as a loading control. B, immunofluorescence analysis of induced (2 d and 7 d) and uninduced RNAi cells. Cells were stained with ATOM40 antiserum as a mitochondrial marker (red) and 4′,6-diamidin-2-phenylindol (DAPI) to highlight nuclear and mitochondrial DNA (blue). DIC, differential interference contrast.
TABLE 1.
Prediction of transmembrane domains using the indicated prediction tools
| Name of server | Position of predicted transmembrane domains | Number of predicted transmembrane domains |
|---|---|---|
| DAS | 12–18, 230–233 | 2 |
| HMMTOP | 8–25 | 1 |
| MEMSAT | 8–25 | 1 |
| PRED-TMR | 8–25 | 1 |
| SPLIT | 227–248 | 1 |
| TMHMM | 7–25 | 1 |
| TOPCONS | 7–25 | 1 |
FIGURE 2.
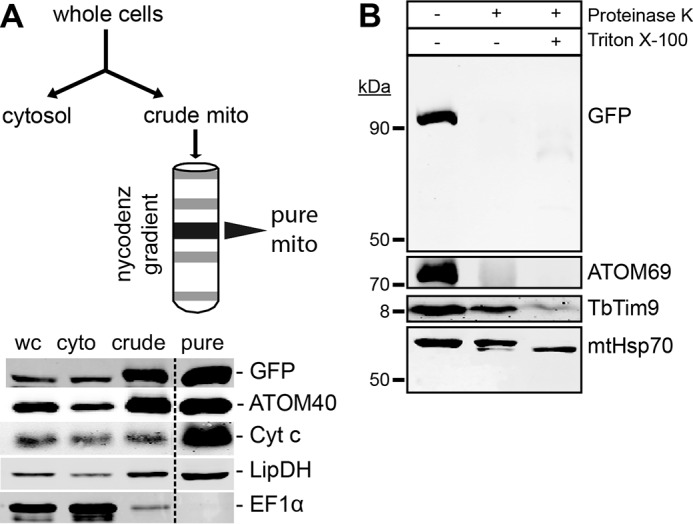
POMP10 is a signal-anchored mitochondrial OM protein. A, upper panel, outline of the mitochondrial purification scheme. Lower panel, immunoblot analysis of the indicated fractions of a mitochondrial purification from the cell line expressing full-length POMP10 that is C-terminally tagged with GFP (POMP10-GFP). The fractions (5 μg of each) depicted are whole cells (wc), cytosol (cyto), crude mitochondria (crude), and pure mitochondria (pure). The samples were separated by SDS-PAGE, blotted, and probed for GFP, the mitochondrial markers ATOM40, cytochrome c (Cyt c), and LipDH and the cytosolic marker EF1α. B, immunoblots of a protease protection assay using gradient-purified structurally intact mitochondria isolated from cell lines expressing POMP10-GFP analyzed by anti-GFP antiserum. The additions of proteinase K and of Triton X-100 are indicated. The OM protein ATOM69 served as a control OM protein. The IMS protein Tim9 and the matrix protein mtHsp70 served as controls for OM integrity. In the presence of Triton X-100 mtHsp70 was degraded to a protease-resistant fragment.
FIGURE 4.
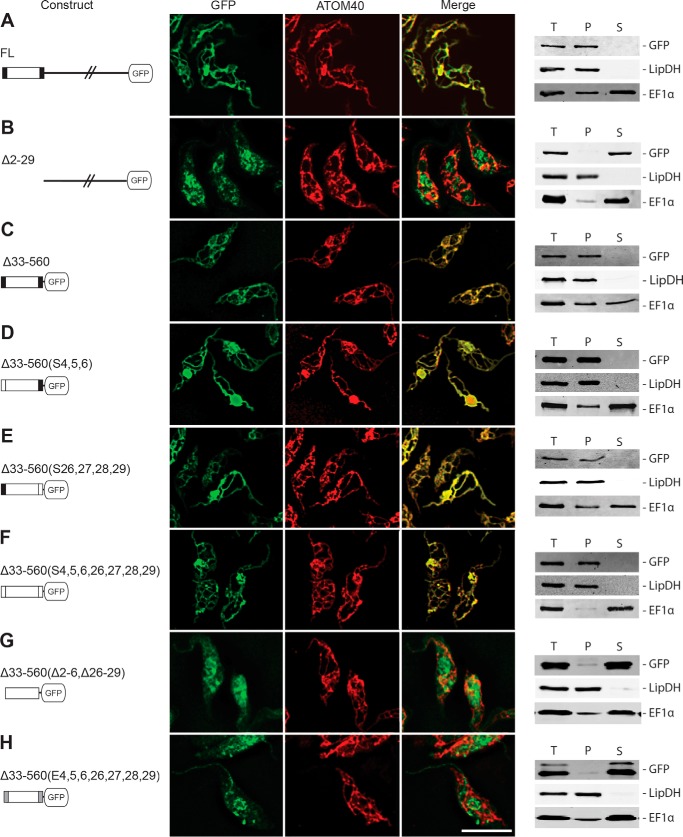
Characterization of the targeting signal in POMP10. IF analysis of the indicated POMP10-GFP fusion proteins (A–H, left columns) in procyclic forms of transgenic T. brucei (A–H, middle column). Samples were probed for GFP and the mitochondrial marker ATOM40, respectively. An overlap of both signals is also shown (Merge). Bar, 10 μm. Crude digitonin-based cell fractionation analysis (A–H, right column) of cell lines expressing the indicated POMP10-GFP fusion proteins. Total cellular extract (T), crude mitochondrial fraction (P), and digitonin-extracted cytosolic fraction (S) and shown. LipDH and EF1α served as mitochondrial and cytosolic markers, respectively. FL, full-length.
This indicates that the C-terminal soluble domain of the protein is exposed to the cytosol and thus establishes POMP10 as a signal-anchored mitochondrial OM protein. On the other hand, the small TIM chaperone TIM9, which serves as a soluble marker of the IMS, is resistant to the treatment, illustrating that the purified mitochondria that were used in the assay have an intact OM. Only when the membrane barrier is destroyed by the addition of detergent does TIM9 become susceptible to the treatment and become degraded.
Transgenic Cell Lines Expressing POMP10-GFP Fusions
To identify the signal that directs POMP10 to the OM, we produced a series of transgenic cell lines that allow tetracycline-inducible expression of a number of POMP10 variants, all of which were fused to GFP at their C terminus. The variants include a deletion of the membrane-spanning domain as well as a series of constructs in which the soluble domain was replaced by GFP. The differences between the latter concerns point mutations that replace the positively charged amino acids of either the cytosolic or the IMS-exposed membrane-flanking regions with neutral or negatively charged amino acids, respectively. Moreover, in one variant all positively charged residues were deleted (Fig. 3A). The immunoblot analysis of whole cell extracts from all transgenic cell lines using an anti-GFP antiserum shows that all POMP10-GFP fusions are expressed at comparable levels (Fig. 3B).
FIGURE 3.
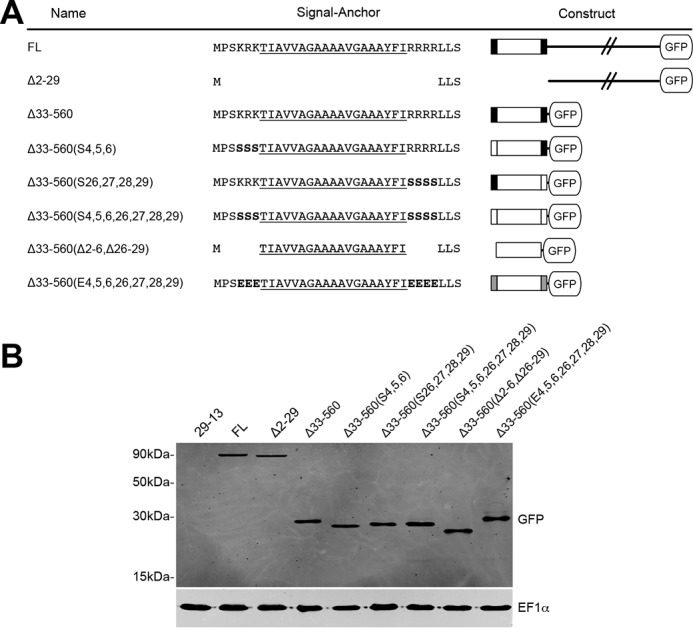
Transgenic cell lines expressing POMP10-GFP fusions. A, FL, full-length GFP fusion. For the other fusion proteins the extent of the deletions (Δ) as well as the positions that were replaced with either serine (S) or glutamate (E) are indicated on the left. Amino acid sequences of the signal-anchor domains in the indicated POMP10-GFP fusion proteins are shown in the middle. The predicted membrane-spanning domain is underlined. The serine and glutamate residues that were used to replace the positively charged flanking amino acids are indicated in bold. The symbols used for the different GFP constructs are shown on the right: black line, cytosolic domain; box, signal-anchor domain; large white box, predicted membrane-spanning domain; small black boxes, positively charged flanking sequence(s); small white boxes, uncharged flanking sequence(s); small gray boxes, negatively charged flanking sequences. B, immunoblot analysis of total cellular extracts from equal cell numbers of the parental strain T. brucei 29-13 and the indicated transgenic cell lines expressing the various POMP10-GFP fusion proteins were probed for GFP and EF1α, which serves as a loading control.
The Transmembrane Domain of POMP10 Contains the Import Signal
The localization of the different POMP10-GFP fusions was analyzed by IF as well as by isolation of crude mitochondrial fractions using digitonin extraction (37). Moreover, to test whether the mitochondrially localized fusion proteins are stably inserted into the OM, we performed carbonate extraction at pH 10.8 and 11.5, respectively. This treatment disrupts protein-protein interactions but should not interfere with protein-lipid interactions. Insolubility after carbonate extraction at high pH is used as an operational definition for integral membrane proteins (38).
Fig. 4A shows that the full-length POMP10-GFP fusion is correctly localized to mitochondria and behaves as an integral membrane protein (Fig. 5A). However, if the N-terminal transmembrane domain is deleted, the POMP10-GFP fusion becomes soluble at low digitonin concentration, and it accumulates outside mitochondria in a punctate-like pattern as evidenced by IF (Fig. 4B). However, the complementary fusion protein in which the transmembrane domain, including the positively charged flanking residues, was directly fused to the GFP, on the other hand, was correctly localized and inserted into the OM (Fig. 4C and Fig. 5B).
FIGURE 5.
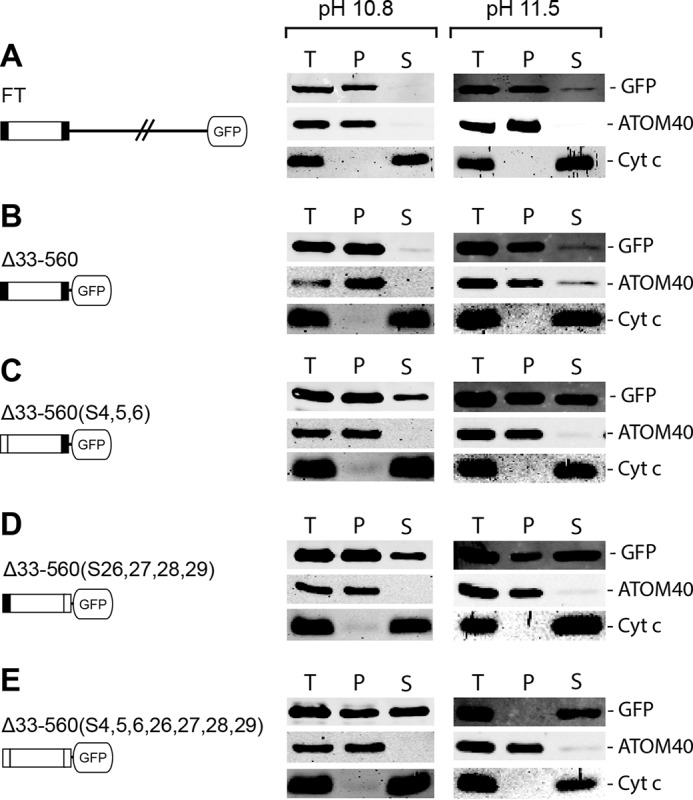
Carbonate extractions at high pH of membrane-associated POMP10-GFP fusion proteins. Immunoblots of equal cells equivalents of total (T), pellet (P), and supernatant (S) fractions of a carbonate-extracted crude mitochondrial pellet obtained by digitonin extraction of cells expressing the indicated POMP10-GFP variants (A–E). The extraction was performed at both pH 10.8 and pH 11.5, and immunoblots were analyzed by anti-GFP. ATOM40 and Cyt c served as markers representing an integral and peripheral membrane protein, respectively.
The Import Signal of POMP10 Does Not Depend on Positively Charged Amino Acids
Further GFP variants that were tested either had the three N-terminal positively charged amino acids that flank the transmembrane domain (Fig. 4D) or the four positively charged lysines that flank the region C-terminally (Fig. 4E) or both (Fig. 4F) replaced by serines. All these POMP10-GFP fusions are still mitochondrially localized as evidenced by IF, which results in a staining pattern that is congruent with the mitochondrial marker ATOM40. Moreover, like the full-length POMP10, all these variants are recovered in a crude mitochondrial fraction obtained by digitonin extraction (Fig. 4, D–F, right panel). However, the carbonate extraction experiments in Fig. 5, C–E, show that replacing the positive charges by neutral ones on either side of the membrane-spanning domain causes a fraction of each of the POMP10-GFP fusions to be released into the supernatant. The soluble fraction became larger at higher pH and/or if both sides of the membrane-spanning domain carried neutral residues (Fig. 5E), indicating that in the absence of positively charged flanking amino acids the transmembrane domain, although still targeted to the mitochondrial OM, is less efficiently inserted into or anchored in the lipid bilayer.
Overexpression of most of the truncated POMP10-GFP constructs (Fig. 4, D–F) that are targeted to mitochondria altered the morphology of the mitochondrial network as evidenced by enlarged sections that stain positive for mitochondrial marker proteins. All of these cell lines also grew more slowly, albeit to different extents, when expressing GFP fusion proteins (Fig. 6). This suggests that the altered mitochondrial morphology affects the overall fitness of the cells. However, the mechanism by which overexpression of the POMP10 membrane-spanning domain and its mutant derivatives when fused to GFP affects mitochondrial morphology remains to be elucidated.
FIGURE 6.
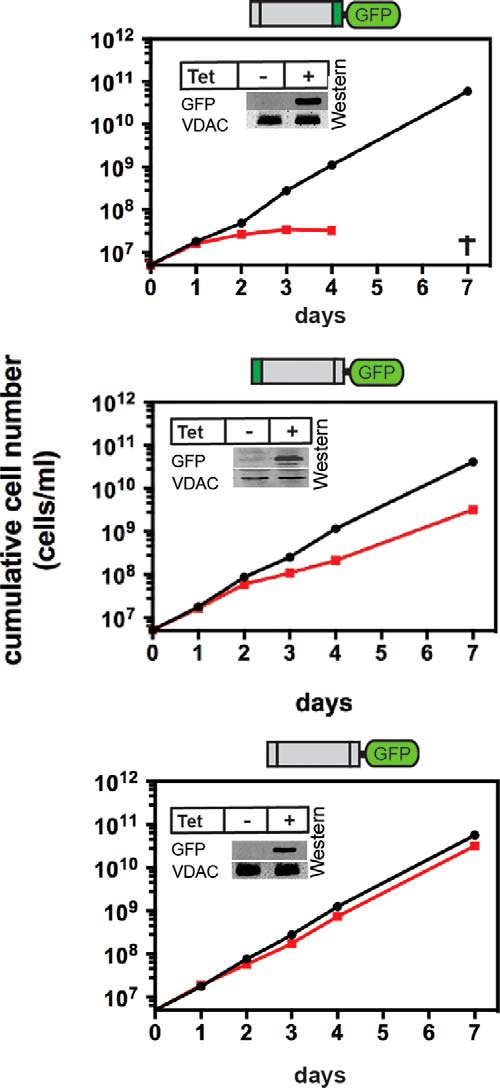
Expression of some of the truncated POMP10-GFP fusions interferes with growth. Growth curves of uninduced (black line) and induced (red line) cell lines expressing the indicated recombinant POMP10-GFP fusion proteins. All analyzed cell lines show an altered mitochondrial morphology (see Fig. 4, D–F). Insets, immunoblots confirming expression of the GFP fusion proteins. VDAC serves as loading control. Tet, tetracycline.
Negatively Charged Flanking Sequences Abolish the POMP10 Import Signal
Although the membrane-spanning domain of POMP10-GFP is still targeted to mitochondria if the flanking positively charged amino acids are replaced by neutral amino acids (Fig. 4F), targeting was abolished if the transmembrane flanking regions were lost or if the positive residues were replaced by negatively charged glutamates (Fig. 4, G and H). IF analysis shows that the resulting POMP10-GFP fusions appear in part to be localized to discrete structures that do not overlap with the mitochondrial marker ATOM40. Moreover, the protein is released into the supernatant at low concentrations of digitonin.
In Vivo Biogenesis of POMP10 Depends on pATOM36
Having identified the signal that targets POMP10 to the mitochondrial OM, we wanted to investigate which protein factors might mediate its biogenesis. The mitochondrial OM of T. brucei contains three protein complexes involved in mitochondrial protein import. The most important one is the ATOM complex, the pore-forming component of which is the β-barrel protein ATOM40 (32–34). As a functional analogue of the TOM complex, it is required for import of essentially all mitochondrial proteins. Insertion of β-barrel protein from the IMS side into the OM requires the SAM complex, the pore-forming component of which is the β-barrel protein SAM50 (39). Finally, there is the trypanosomatid-specific protein pATOM36 that dynamically associates with the ATOM complex (35). Recent work has shown that pATOM36 mediates the biogenesis of a number of OM proteins including subunits of the ATOM complex (36). To test the involvement of the three protein complexes in the targeting and import of POMP10, we expressed the full-length version of the protein fused to GFP in tetracycline-inducible ATOM40, Sam50, and pATOM36 RNAi cell lines, respectively.
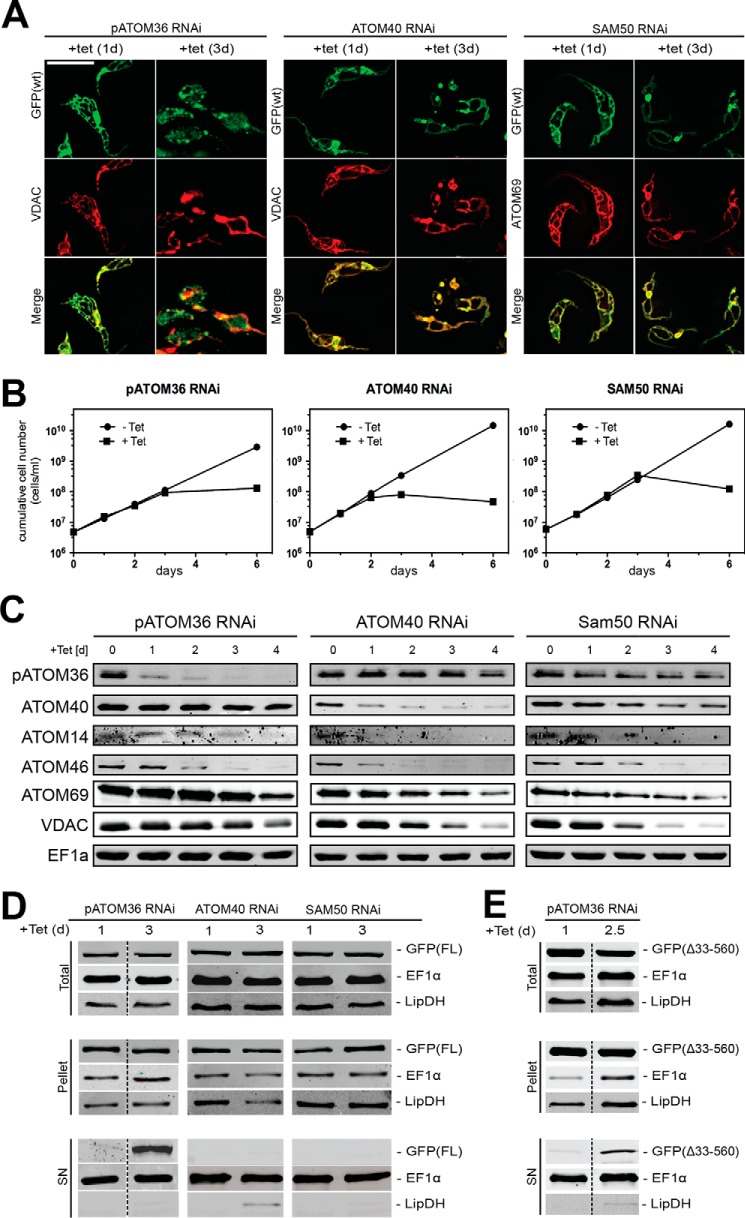
Using IF the localization of the GFP-tagged fusion protein was analyzed in the three RNAi cell lines. Cells that had been induced with tetracycline for 1 day served as a control as the GFP-tagged fusion protein was already expressed, but the RNAi effects were not yet visible. At this early time point of induction the staining pattern of the POMP10-GFP fusion protein coincided in all three cell lines with the mitochondrial marker (Fig. 7A). After 3 days of tetracycline induction all cell lines showed a growth arrest (Fig. 7B), which has been described before (33, 35, 39). Moreover, as expected, if mitochondrial protein import was abolished, the morphology of mitochondria was drastically altered (Fig. 7A) (40). However, in the ATOM40 and the Sam50 RNAi cell lines, despite the aberrant morphology of the organelles, the localization pattern of the POMP10-GFP fusion was essentially identical to the mitochondrial markers. This indicates that ablation of neither of the two proteins affects POMP10 biogenesis. In contrast, ablation of pATOM36 caused accumulation of POMP10-GFP outside mitochondria as evidenced by the diffuse and punctate signal that only partially overlapped with the mitochondrial marker (Fig. 7A).
FIGURE 7.
In vivo biogenesis of POMP10 depends on pATOM36. A, IF analysis of full-length POMP10 that is C-terminally tagged with GFP (GFP(FL): green) and mitochondrial marker proteins (VDAC and ATOM69: red) in tetracycline (tet)-inducible pATOM36 (left panels), ATOM40 (middle panels), and SAM50 (right panels) RNAi cell lines. Time of induction in days (d) is indicated at the top. Bar, 10 μm. B, growth curves of tetracycline-inducible pATOM36 (left panels), ATOM40 (middle panels), and SAM50 (right panels) RNAi cell lines that co-express full-length POMP10-GFP and that were used in A. C, equal cell equivalents of the same RNAi cell lines shown in A (left panel, pATOM36; middle panel, ATOM40; right panel, SAM50) were analyzed for the presence or absence of pATOM36, ATOM40, ATOM14, ATOM46, ATOM69, VDAC, and cytosolic EF1a using immunoblots. Time of RNAi induction is indicated at the top of each panel. D, crude digitonin-based cell fractionation analysis of the same cell lines shown in A and B. Total cellular extract (top panels), crude mitochondrial fraction (pellet, middle panels), and digitonin-extracted cytosolic fraction (SN, bottom panels) were probed for POMP10-GFP (GFP(FL)). LipDH and EF1α served as the mitochondrial and cytosolic markers, respectively. E, same as D, but a tetracycline-inducible pATOM36 RNAi cell line that co-expresses the POMP10 fusion protein in which the transmembrane domain including the positively charged flanking residues was directly fused to the GFP was analyzed.
Fig. 7C shows immunoblots for the three RNAi cell lines. RNAi directed against pATOM36 led to a rapid ablation of the protein. There is a decrease of ATOM14 and ATOM46, whose biogenesis has previously been shown to depend on pATOM36 (36). The levels of ATOM40, ATOM69, and VDAC, however, remained constant, except for a slight drop that was observed after 4 days of induction (Fig. 7C, left panel). As expected, ATOM40 were early and efficiently down-regulated in the ATOM40-RNAi cell line. The same was seen for the ATOM complex subunits ATOM14 and ATOM46. Moreover, we saw down-regulation of ATOM69 and VDAC. The level of pATOM36, however, remained constant throughout induction and only slightly decreased at the latest time point (Fig. 7C, middle panel). RNAi of Sam50 finally led to ablation of the two β-barrel proteins VDAC and ATOM40. As a consequence of the latter, a decline of ATOM14 and ATOM46 and, to a lesser extent ATOM69, was also observed. The level of pATOM36, on the other hand, remained essentially constant (Fig. 7C, right panel). Finally, elongation factor 1α (EF1α), which serves as a cytosolic control, was not affected in any of the three cell lines.
In summary, these results illustrate that pATOM36-RNAi has the most restricted effects on the tested proteins. Most importantly the ATOM complex subunits were much less reduced than when ATOM40 was ablated. Yet it is in pATOM36-RNAi cell line that cytosolic accumulation of POMP10-GFP was observed. This confirmed that pATOM36, but not the ATOM complex or Sam50, is required for the correct localization of POMP10.
It could be expected that in the absence a functional mitochondrial OM insertion machinery the POMP10-GFP might be directed to the ER. Fig. 8 shows that this is not the case, as in the absence of pATOM36 only very little of the staining corresponding to the POMP10-GFP fusion protein overlapped with the ER.
FIGURE 8.
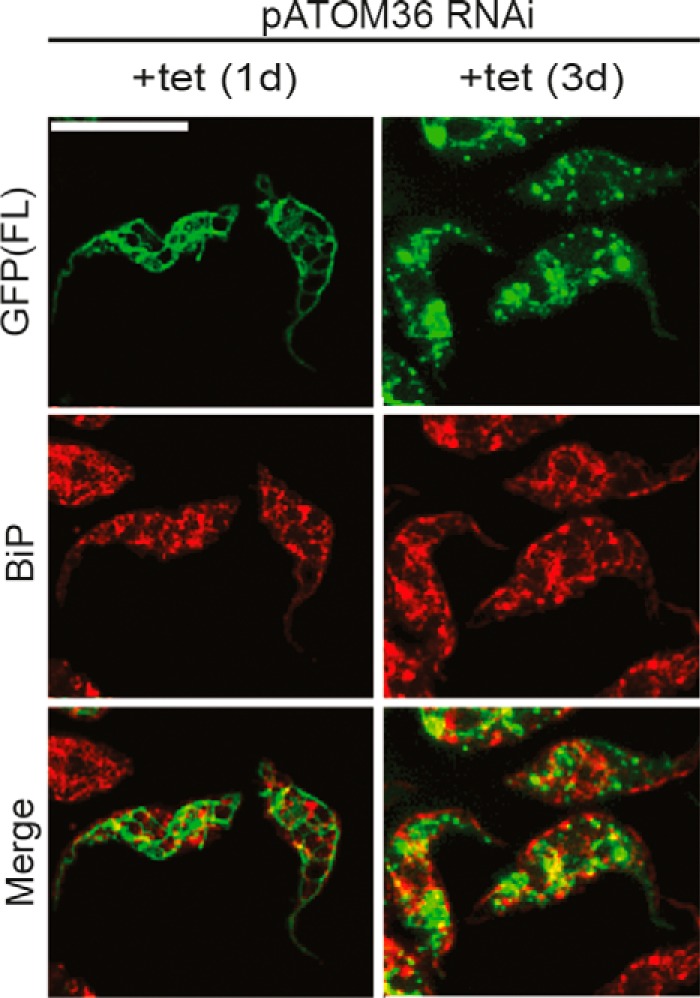
Ablation of pATOM36 did not cause mislocalization to the ER. IF analysis of full-length POMP10 that is C-terminally tagged with GFP (POMP(FL): green) and the ER marker protein BiP (red) in the tetracycline-inducible pATOM36 RNAi cell line. Time of induction in days (d) is indicated at the top. Bar, 10 μm.
All the above-mentioned RNAi cell lines expressing full-length POMP10-GFP were also analyzed by extraction with low concentrations of digitonin. In Fig. 7D the resulting total, mitochondria-enriched pellet, and supernatant fractions were analyzed on immunoblots probed for the POMP10-GFP fusion protein as well as for EF1α and lipoamide dehydrogenase (LipDH) that serve as cytosolic and mitochondrial markers, respectively. Whereas in cell lines ablated for ATOM40 and SAM50 the POMP10-GFP fusion protein was quantitatively recovered in the mitochondria-enriched pellet, a fraction of the protein was released into the supernatant in the absence of pATOM36. This indicates that the mitochondrial localization of POMP10 depends on pATOM36. The same was observed not only for the full-length POMP10 fusion protein but also if only the membrane-spanning domain was fused to GFP (ΔGFP(33–560)) (Fig. 7E).
In Vitro Biogenesis of POMP10 Depended on pATOM36
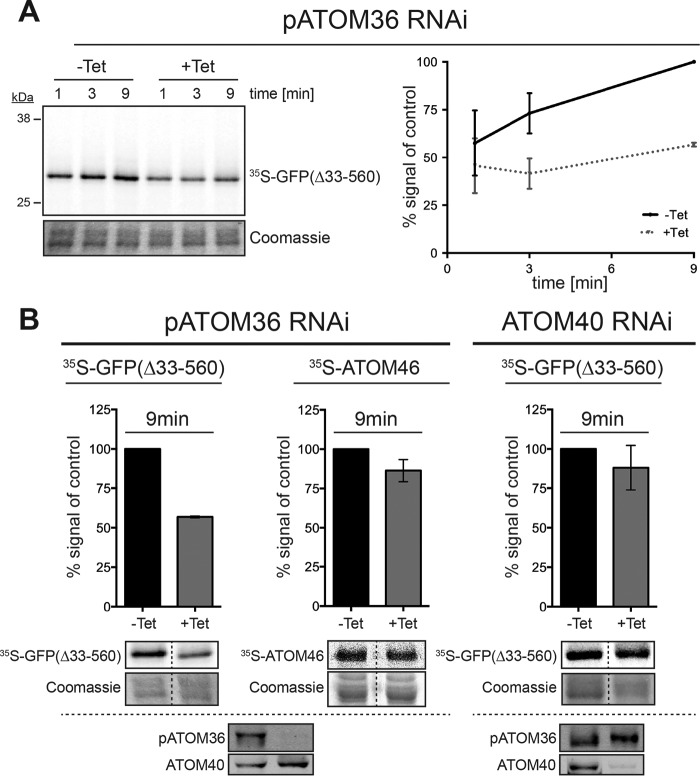
To confirm that pATOM36 plays an important role in the biogenesis of POMP10, we performed in vitro insertion experiments. To that end we used the POMP10 variant in which the membrane-spanning domain was directly fused to GFP (GFP(Δ33–560)) and whose in vivo biogenesis was shown to depend on pATOM36 (Fig. 7D). Thus, the substrate was in vitro translated in the presence of [35S]methionine using reticulocyte lysate and subsequently incubated with mitochondria isolated from either uninduced cells or from cells ablated for pATOM36. To measure insertion of the protein into the mitochondrial OM, the reisolated organelles were extracted with sodium carbonate at pH 11.5, and the resulting pellets were analyzed by SDS/PAGE analysis (36). Fig. 9A shows a time-dependent increase of the radioactive substrate protein in the carbonate-insoluble pellet fractions of organelles that stem from uninduced cells, indicating the substrate is inserted into the OM. However, in mitochondria isolated from pATOM36-ablated cells much less insertion was observed. Thus, as observed in vivo, pATOM36 facilitates the in vitro insertion of POMP10-GFP(Δ33–560) into the mitochondrial OM. Interestingly, another pATOM36 substrate, ATOM46, behaved differently in the same assay (Fig. 9B, middle panel). As described previously in the case of ATOM46, pATOM36 mediated the integration of the protein into the ATOM complex rather than its insertion into the membrane (36). Fig. 9B, right panel, finally shows that membrane insertion of POMP10-GFP(Δ33–560) was not affected in mitochondria ablated for ATOM40 even though in these organelles the levels of all ATOM subunits were lower than in the induced pATOM36 cell line (Fig. 7C) (32, 36).
FIGURE 9.
In vitro biogenesis of POMP10 depends on pATOM36. A, left panel, in vitro insertion assay of the [35S]Met-labeled POMP10-GFP(Δ33–560) using mitochondria isolated from the uninduced tetracycline (−Tet) and induced (+Tet, 2 days) procyclic pATOM36-RNAi cell line. Incubation times are indicated at the top. The pellet fraction of an alkaline carbonate extraction was separated by SDS-PAGE and analyzed by autoradiography. Lower panel, the section of the Coomassie-stained gel served as loading controls. Graph on the right, quantification of triplicate experiments shown on the left. Standard errors are indicated. B, graph of triplicate in vitro insertion assays (9-min time point) using isolated mitochondria of uninduced and induced pATOM36 and ATOM40 RNAi cell lines, respectively. The tested substrates are indicated at the top. Standard errors are indicated. The SDS-gels representing typical experiments including their corresponding loading controls are indicated below the graphs. Bottom panels, immunoblot confirming the specific down-regulation of pATOM36 and ATOM40 in the respective RNAi cell lines.
Discussion
In a recent study we purified the OM of T. brucei and show by a combination of protein abundance profiling and label-free quantitative mass spectrometry that its proteome consists of 82 different proteins of which 42 contain one or more putative transmembrane domains (7). Among them seven proteins are bioinformatically predicted to be signal-anchored. In the present study we selected one of these proteins, termed POMP10, to investigate its biogenesis pathway.
Using a protease protection assay we confirmed that POMP10 is indeed a signal-anchored protein. In a first series of experiments we showed that the targeting signal of POMP10 for the OM is confined to the predicted membrane-spanning domain and a few flanking residues. We then demonstrated that the positively charged flanking residues can be replaced by neutral ones without affecting mitochondrial localization. However, if the positively charged flanking residues are replaced by negatively charged ones or deleted, targeting of the protein was abolished.
The targeting sequence of signal-anchored proteins has previously been analyzed in yeast, humans, and plants. In all three systems the signal was confined to a membrane-spanning domain of “moderate” hydrophobicity, which however did not show sequence similarity between different signal-anchored proteins of the same or different species (13, 14). Moreover, whereas for human and plant signal-anchored proteins a C-terminal positively charged flanking region was an additional requirement for OM targeting (13, 41, 42), this was not the case for the corresponding proteins of yeast, as their positively charged residues could be replaced by neutral ones without affecting the targeting (14, 15). Although in the latter case the protein was still correctly targeted, its integration into the OM membrane was weakened.
The targeting signal of trypanosomal POMP10 conforms to this picture. As in the other systems, it consists of a transmembrane domain with moderate hydrophobicity that includes N-terminal and C-terminal flanking regions containing positively charged residues. However, as in yeast but unlike in plants and humans, these residues are not essential for mitochondrial targeting but do influence the strength of the interaction with the OM.
Yeast and humans belong to the eukaryotic supergroup of the Opisthokont and plants to the supergroup of the Archaeplastida, which represent two of the six main branches of the eukaryotic phylogenetic tree (43). T. brucei is a member of the supergroup of the Excavata, which are essentially unrelated to Archaeplastida and Opisthokont. The fact that the general features of the targeting signal for signal-anchored OM proteins are similar in all three systems suggests that they may be conserved in all eukaryotes.
Although the targeting signal of signal-anchored mitochondrial OM proteins has been investigated in fungi, mammals, and plants our knowledge of the components required for the biogenesis of these proteins is restricted to the two fungal species S. cerevisiae and Neurospora crassa. It has been shown in yeast that neither the SAM complex nor the import receptors Tom20 and Tom70, both of which are themselves signal-anchored proteins, are required for the biogenesis of signal-anchored OM proteins. Moreover, it has been demonstrated that the pore protein Tom40 can be blocked without affecting the import of signal-anchored proteins, although Tom20 still requires Tom40 for its biogenesis (9–11). Finally, a protein complex consisting of Mim1 and Mim2 has been characterized that is involved in import and assembly of at least some signal-anchored OM proteins (22, 23, 44, 45). Mim1 and Mim2 were also shown to be involved in the insertion and/or assembly of some tail-anchored as well as multimembrane-spanning proteins. However, the mechanism by which Mim1 and Mim2 exert their function remains to be investigated.
In trypanosomes we showed that membrane insertion in vivo and in vitro of the signal-anchored POMP10 did not require ATOM40 or Sam50, the pore-forming components of the ATOM and the SAM complexes. However, biogenesis of POMP10 did depend on pATOM36 a trypanosomatid-specific OM protein. A recent study has shown that pATOM36 has a dual localization. Similar to ATOM40 it is localized all over the OM; however, in contrast to ATOM40, it is also enriched at the tripartite attachment complex (TAC), the structure that links the mitochondrial DNA to the basal body of the flagellum. The dual localization of pATOM36 reflects its dual function in (i) the assembly of OM proteins, including most subunits of the ATOM complex (32) and (ii) the segregation of the replicated mitochondrial genome (35, 36).
Ablation of pATOM36 resulted in the accumulation of a fraction of POMP10 outside mitochondria in vivo and a much reduced membrane insertion in vitro, suggesting that the protein is required for targeting and/or insertion of the protein into the OM. This is different from the previously analyzed substrates of pATOM36 such as ATOM complex subunits, which in the absence of the pATOM36 were still inserted into the membrane but not assembled into high molecular weight complexes (36).
It is tempting to speculate that the kinetoplastid-specific pATOM36 might be a functional analogue of fungal Mim1 and -2, which were shown to be involved in the insertion and/or assembly of some signal-anchored OM proteins in yeast (22, 23, 44, 45). Thus, although the targeting signals for signal-anchored proteins appear to be conserved in all eukaryotes, this is not the case for the factors mediating their biogenesis. There is no sequence similarity between pATOM36 and Mim1/Mim2, and the molecular masses of the two sets of proteins are also very different: pATOM36, 36 kDa; Mim1, 13 kDa; Mim2, 11 kDa. The only recognizable motif that is shared between the two groups of proteins are two GXXXG(A) motifs in their predicted transmembrane domains. However this motif is quite frequent in transmembrane domains.
In summary our results show the existence of evolutionary distinct biogenesis factors for signal-anchored proteins in the different eukaryotic supergroups. Moreover, the fact that Mim1 and Mim2 are absent from mammals, which as the fungi belong to the Opisthokont, indicates that there might be different biogenesis factors even within the same supergroup. Identification of biogenesis factors for signal-anchored OM proteins in different phylogenetic groups is important, as it may allow the identification of shared traits between them that will help to define the fundamental biochemical features mandatory for their function.
Experimental Procedures
Production of Transgenic Cell Lines
Transgenic procyclic cell lines are based on T. brucei 29-13 (46). Cells were grown at 27 °C in SDM79 supplemented with 10% (v/v) FCS. All plasmids are based on the pLew100 expression vector in which the phleomycin resistance gene has been replaced by a puromycin resistance gene (47) and in which the ORF coding for enhanced GFP was inserted. PCR amplicons corresponding to the different variants of POMP10 (Tb927.11.13180) were cloned into this enhanced GFP expression vector to allow expression of C-terminally tagged variants. Cells were grown to mid-exponential phase and transfected with the corresponding linearized plasmid (48) followed by selection in medium containing puromycin. DNA constructs were verified by sequencing. RNAi constructs targeting ATOM40 (33), SAM50 (49), and pATOM36 (35) have been described elsewhere.
Immunoblotting
Protein samples were separated by SDS-PAGE and transferred onto nitrocellulose membranes (GE Healthcare). Membranes were blocked in 5% (w/v) milk in PBS-Tween (137 mm CaCl2, 2.7 mm KCl, 10 mm Na2HPO4, 1.8 mm KH2PO4, 0.1% (v/v) Tween 20, pH 7.4) and incubated overnight with the appropriate antibody at 4 °C. Monoclonal primary antibodies used were: mouse anti-GFP (Roche Applied Science, dilution 1:1,000) and mouse anti-EF1α (Merck Millipore, dilution 1:10,000). Polyclonal primary antibodies produced in rabbits were: anti-VDAC (dilution 1:1,000) (50), anti-ATOM40 (dilution 1:1,000) (32), anti-Cyt c (dilution 1:1,000) (7), and anti-TIM9 (1:20) (7) and rabbit anti-LipDH (1:10,000) (provided by L. Krauth-Siegel, Heidelberg University). Detection of the antibodies was done with an Odyssey Infrared imaging system using IRDye secondary antibodies (Li-COR Biosciences, dilution 1:20,000).
Immunofluorescence
The cells were collected, washed in PBS, and subsequently resuspended in ice-cold buffer. All further steps were performed in a wet chamber. Cells were fixed for 10 min in freshly made 4% paraformaldehyde in PBS, pH 7.2, and rinsed with ice-cold PBS. Cells were incubated in PBS containing 0.2% Triton X-100 for 5 min. Primary antibodies were rabbit anti-VDAC (1:100), rabbit anti-ATOM40 (1:1000), rabbit anti-ATOM69 (1:3) (32), and rabbit anti-BIP (1:5000). Cy3-conjugated goat anti-rabbit (dilution 1:500) was used as a secondary antibody. Cells were rinsed in ice-cold buffer, post-fixed in cold methanol, and mounted with VectaShield containing DAPI (Vector Laboratories). Fluorescence images were taken with Leica fluorescence microscope (Leica Microsystems). Images stacks were recorded and deconvoluted using LAS X software from Leica (Leica Microsystems).
Digitonin Extractions
Cell membranes were lysed by resuspension of 108 cells in SoTe buffer (20 mm Tris-HCL, pH 7.5, 0.6 m sorbitol, and 2 mm EDTA) containing 0.015% (w/v) digitonin followed by differential centrifugation. This yielded a mitochondria-enriched pellet and a fraction enriched for cytosolic proteins (37). All samples were analyzed by immunoblot experiments.
Carbonate Extractions
Mitochondria-enriched fractions were resuspended in 160 μl of 100 mm Na2CO3, pH 11.5 or 10.8. Thereafter, 80 μl were removed and mixed with 20 μl of 5× SDS loading buffer and boiled to serve as the “total” sample. The remaining 80 μl were incubated on ice for 10 min and centrifuged (100,000 × g, 4 °C, 10 min). The supernatant was transferred to a new tube, and 20 μl of 5× SDS loading buffer was added before boiling. The pellet was resuspended in 80 μl of 100 mm Na2CO3, and 20 μl of 5× SDS loading buffer was added before boiling. All samples were analyzed by SDS-PAGE.
Protease Protection Assay
Isotonically isolated mitochondria (25 μg) from cells (51, 52) expressing POMP10-GFP were resuspended in 20 mm Tris-HCl, pH 7.2, 15 mm KH2PO4, 20 mm MgSO4, 0.6 m sorbitol in a total volume of 50 μl in the presence of proteinase K (10 mg/ml) containing or not 0.5% (v/v) Triton-X100 followed by incubation on ice for 15 min. Reactions were stopped by adding phenylmethylsulfonyl fluoride at 5 mm, and mitochondria were centrifuged (6800 × g, 4 °C), resuspended in SDS-loading buffer, and boiled.
In Vitro Insertion of Proteins into the Mitochondrial OM
The membrane insertion assay was done exactly as described in Käser et al. 36). [35S]Met-labeled POMP10-GFP(Δ33–560) and ATOM46 were synthesized using the TNT T7 Quick for PCR in vitro translation kit (Promega). For the coupled transcription and translation, gel-eluted PCR-fragments were used that consisted of the T7 RNA polymerase promoter fused to the complete ORF of the corresponding substrates.
Author Contributions
J. B. designed and performed the research in Figs. 1–8. S. K. designed and performed the research Figs. 7C and 9. J. B. and S. K. analyzed the results and prepared the figures. J. M. planned and analyzed the experiments. A. S. analyzed the data and wrote the paper.
This work was supported by Swiss National Science Foundation Grant 138355 (to A. S.) and in part by the National Center of Competence in Research (NCCR) “RNA & Disease”. The authors declare that they have no conflicts of interest with the contents of this article.
- IMS
- soluble intermembrane space
- OM
- outer membrane
- TOM
- translocase of the OM
- TIM
- translocase of the inner membrane
- SAM
- sorting and assembly machinery
- ATOM40
- archaic translocase of the mitochondrial OM 40
- pATOM36
- peripheral ATOM36
- IF
- immunofluorescence
- LipDH
- lipoamide dehydrogenase
- VDAC
- voltage-dependent anion channel protein
- EF1α
- elongation factor 1α
- Cyt c
- cytochrome c
- ER
- endoplasmic reticulum.
References
- 1. Friedman J. R., and Nunnari J. (2014) Mitochondrial form and function. Nature 505, 335–343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Chacinska A., Koehler C. M., Milenkovic D., Lithgow T., and Pfanner N. (2009) Importing mitochondrial proteins: machineries and mechanisms. Cell 138, 628–644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Schulz C., Schendzielorz A., and Rehling P. (2015) Unlocking the presequence import pathway. Trends Cell Biol. 25, 265–275 [DOI] [PubMed] [Google Scholar]
- 4. Neupert W., and Herrmann J. M. (2007) Translocation of proteins into mitochondria. Annu. Rev. Biochem. 76, 723–749 [DOI] [PubMed] [Google Scholar]
- 5. Zahedi R. P., Sickmann A., Boehm A. M., Winkler C., Zufall N., Schönfisch B., Guiard B., Pfanner N., and Meisinger C. (2006) Proteomic analysis of the yeast mitochondrial outer membrane reveals accumulation of a subclass of preproteins. Mol. Biol. Cell 17, 1436–1450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Schmitt S., Prokisch H., Schlunck T., Camp D. G. 2nd, Ahting U., Waizenegger T., Scharfe C., Meitinger T., Imhof A., Neupert W., Oefner P. J., and Rapaport D. (2006) Proteome analysis of mitochondrial outer membrane from Neurospora crassa. Proteomics 6, 72–80 [DOI] [PubMed] [Google Scholar]
- 7. Niemann M., Wiese S., Mani J., Chanfon A., Jackson C., Meisinger C., Warscheid B., and Schneider A. (2013) Mitochondrial outer membrane proteome of Trypanosoma brucei reveals novel factors required to maintain mitochondrial morphology. Mol. Cell. Proteomics 12, 515–528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Shore G. C., McBride H. M., Millar D. G., Steenaart N. A., and Nguyen M. (1995) Import and insertion of proteins into the mitochondrial outer membrane. Eur. J. Biochem. 227, 9–18 [DOI] [PubMed] [Google Scholar]
- 9. Becker T., Vögtle F. N., Stojanovski D., and Meisinger C. (2008) Sorting and assembly of mitochondrial outer membrane proteins. Biochim. Biophys. Acta 1777, 557–563 [DOI] [PubMed] [Google Scholar]
- 10. Dukanovic J., and Rapaport D. (2011) Multiple pathways in the integration of proteins into the mitochondrial outer membrane. Biochim. Biophys. Acta 1808, 971–980 [DOI] [PubMed] [Google Scholar]
- 11. Walther D. M., and Rapaport D. (2009) Biogenesis of mitochondrial outer membrane proteins. Biochim. Biophys. Acta 1793, 42–51 [DOI] [PubMed] [Google Scholar]
- 12. Jores T., Klinger A., Groß L. E., Kawano S., Flinner N., Duchardt-Ferner E., Wöhnert J., Kalbacher H., Endo T., Schleiff E., and Rapaport D. (2016) Characterization of the targeting signal in mitochondrial β-barrel proteins. Nat. Commun. 7, 12036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Kanaji S., Iwahashi J., Kida Y., Sakaguchi M., and Mihara K. (2000) Characterization of the signal that directs Tom20 to the mitochondrial outer membrane. J. Cell Biol. 151, 277–288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Waizenegger T., Stan T., Neupert W., and Rapaport D. (2003) Signal-anchor domains of proteins of the outer membrane of mitochondria: structural and functional characteristics. J. Biol. Chem. 278, 42064–42071 [DOI] [PubMed] [Google Scholar]
- 15. McBride H. M., Millar D. G., Li J. M., and Shore G. C. (1992) A signal-anchor sequence selective for the mitochondrial outer membrane. J. Cell Biol. 119, 1451–1457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Wattenberg B., and Lithgow T. (2001) Targeting of C-terminal (tail)-anchored proteins: understanding how cytoplasmic activities are anchored to intracellular membranes. Traffic 2, 66–71 [DOI] [PubMed] [Google Scholar]
- 17. Borgese N., Brambillasca S., and Colombo S. (2007) How tails guide tail-anchored proteins to their destinations. Curr. Opin. Cell Biol. 19, 368–375 [DOI] [PubMed] [Google Scholar]
- 18. Ulrich T., and Rapaport D. (2015) Biogenesis of β-barrel proteins in evolutionary context. Int. J. Med. Microbiol. 305, 259–264 [DOI] [PubMed] [Google Scholar]
- 19. Thornton N., Stroud D. A., Milenkovic D., Guiard B., Pfanner N., and Becker T. (2010) Two modular forms of the mitochondrial sorting and assembly machinery are involved in biogenesis of alpha-helical outer membrane proteins. J. Mol. Biol. 396, 540–549 [DOI] [PubMed] [Google Scholar]
- 20. Waizenegger T., Schmitt S., Zivkovic J., Neupert W., and Rapaport D. (2005) Mim1, a protein required for the assembly of the TOM complex of mitochondria. EMBO Rep. 6, 57–62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Becker T., Pfannschmidt S., Guiard B., Stojanovski D., Milenkovic D., Kutik S., Pfanner N., Meisinger C., and Wiedemann N. (2008) Biogenesis of the mitochondrial TOM complex: Mim1 promotes insertion and assembly of signal-anchored receptors. J. Biol. Chem. 283, 120–127 [DOI] [PubMed] [Google Scholar]
- 22. Dimmer K. S., Papić D., Schumann B., Sperl D., Krumpe K., Walther D. M., and Rapaport D. (2012) A crucial role for Mim2 in the biogenesis of mitochondrial outer membrane proteins. J. Cell Sci. 125, 3464–3473 [DOI] [PubMed] [Google Scholar]
- 23. Stefan Dimmer K., and Rapaport D. (2010) The enigmatic role of Mim1 in mitochondrial biogenesis. Eur. J. Cell Biol. 89, 212–215 [DOI] [PubMed] [Google Scholar]
- 24. Vögtle F. N., Keller M., Taskin A. A., Horvath S. E., Guan X. L., Prinz C., Opalińska M., Zorzin C., van der Laan M., Wenk M. R., Schubert R., Wiedemann N., Holzer M., and Meisinger C. (2015) The fusogenic lipid phosphatidic acid promotes the biogenesis of mitochondrial outer membrane protein Ugo1. J. Cell Biol. 210, 951–960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Sauerwald J., Jores T., Eisenberg-Bord M., Chuartzman S. G., Schuldiner M., and Rapaport D. (2015) Genome-wide screens in Saccharomyces cerevisiae highlight a role for cardiolipin in biogenesis of mitochondrial outer membrane multispan proteins. Mol. Cell. Biol. 35, 3200–3211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Hewitt V., Alcock F., and Lithgow T. (2011) Minor modifications and major adaptations: the evolution of molecular machines driving mitochondrial protein import. Biochim. Biophys. Acta 1808, 947–954 [DOI] [PubMed] [Google Scholar]
- 27. Lithgow T., and Schneider A. (2010) Evolution of macromolecular import pathways in mitochondria, hydrogenosomes and mitosomes. Philos. Trans. R. Soc. Lond. B. Biol. Sci. 365, 799–817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Mani J., Meisinger C., and Schneider A. (2016) Peeping at TOMs-diverse entry gates to mitochondria provide insights into the evolution of eukaryotes. Mol. Biol. Evol. 33, 337–351 [DOI] [PubMed] [Google Scholar]
- 29. Eckers E., Cyrklaff M., Simpson L., and Deponte M. (2012) Mitochondrial protein import pathways are functionally conserved among eukaryotes despite compositional diversity of the import machineries. Biol. Chem. 393, 513–524 [DOI] [PubMed] [Google Scholar]
- 30. Cavalier-Smith T. (2010) Kingdoms Protozoa and Chromista and the eozoan root of the eukaryotic tree. Biol. Lett. 6, 342–345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. He D., Fiz-Palacios O., Fu C. J., Fehling J., Tsai C. C., and Baldauf S. L. (2014) An alternative root for the eukaryote tree of life. Curr. Biol. 24, 465–470 [DOI] [PubMed] [Google Scholar]
- 32. Mani J., Desy S., Niemann M., Chanfon A., Oeljeklaus S., Pusnik M., Schmidt O., Gerbeth C., Meisinger C., Warscheid B., and Schneider A. (2015) Novel mitochondrial protein import receptors in Kinetoplastids reveal convergent evolution over large phylogenetic distances. Nat. Commun. 6, 6646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Pusnik M., Schmidt O., Perry A. J., Oeljeklaus S., Niemann M., Warscheid B., Lithgow T., Meisinger C., and Schneider A. (2011) Mitochondrial preprotein translocase of trypanosomatids has a bacterial origin. Curr. Biol. 21, 1738–1743 [DOI] [PubMed] [Google Scholar]
- 34. Harsman A., Niemann M., Pusnik M., Schmidt O., Burmann B. M., Hiller S., Meisinger C., Schneider A., and Wagner R. (2012) Bacterial origin of a mitochondrial outer membrane protein translocase: new perspectives from comparative single channel electrophysiology. J. Biol. Chem. 287, 31437–31445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Pusnik M., Mani J., Schmidt O., Niemann M., Oeljeklaus S., Schnarwiler F., Warscheid B., Lithgow T., Meisinger C., and Schneider A. (2012) An essential novel component of the non-canonical mitochondrial outer membrane protein import system of trypanosomatids. Mol. Biol. Cell 23, 3420–3428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Käser S., Oeljeklaus S., Týč J., Vaughan S., Warscheid B., and Schneider A. (2016) Outer membrane protein functions as integrator of protein import and DNA inheritance in mitochondria. Proc. Natl. Acad. Sci. U.S.A. 113, E4467–E4475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Schneider A., Bouzaidi-Tiali N., Chanez A.-L., and Bulliard L. (2007) ATP production in isolated mitochondria of procyclic Trypanosoma brucei. Methods Mol. Biol. 372, 379–387 [DOI] [PubMed] [Google Scholar]
- 38. Fujiki Y., Hubbard A. L., Fowler S., and Lazarow P. B. (1982) Isolation of intracellular membranes by means of sodium carbonate treatment: application to endoplasmic reticulum. J. Cell Biol. 93, 97–102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Sharma S., Singha U. K., and Chaudhuri M. (2010) Role of Tob55 on mitochondrial protein biogenesis in Trypanosoma brucei. Mol. Biochem. Parasitol. 174, 89–100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Stojanovski D., Rissler M., Pfanner N., and Meisinger C. (2006) Mitochondrial morphology and protein import: a tight connection? Biochim. Biophys. Acta 1763, 414–421 [DOI] [PubMed] [Google Scholar]
- 41. Lee J., Lee H., Kim J., Lee S., Kim D. H., Kim S., and Hwang I. (2011) Both the hydrophobicity and a positively charged region flanking the C-terminal region of the transmembrane domain of signal-anchored proteins play critical roles in determining their targeting specificity to the endoplasmic reticulum or endosymbiotic organelles in Arabidopsis cells. Plant Cell 23, 1588–1607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Suzuki H., Maeda M., and Mihara K. (2002) Characterization of rat TOM70 as a receptor of the preprotein translocase of the mitochondrial outer membrane. J. Cell Sci. 115, 1895–1905 [DOI] [PubMed] [Google Scholar]
- 43. Burki F. (2014) The eukaryotic tree of life from a global phylogenomic perspective. Cold Spring Harb. Perspect. Biol. 6, a016147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Becker T., Wenz L. S., Krüger V., Lehmann W., Müller J. M., Goroncy L., Zufall N., Lithgow T., Guiard B., Chacinska A., Wagner R., Meisinger C., and Pfanner N. (2011) The mitochondrial import protein Mim1 promotes biogenesis of multispanning outer membrane proteins. J. Cell Biol. 194, 387–395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Lueder F., and Lithgow T. (2009) The three domains of the mitochondrial outer membrane protein Mim1 have discrete functions in assembly of the TOM complex. FEBS Lett. 583, 1475–1480 [DOI] [PubMed] [Google Scholar]
- 46. Wirtz E., Leal S., Ochatt C., and Cross G. A. (1999) A tightly regulated inducible expression system for conditional gene knock-outs and dominant-negative genetics in Trypanosoma brucei. Mol. Biochem. Parasitol. 99, 89–101 [DOI] [PubMed] [Google Scholar]
- 47. Bochud-Allemann N., and Schneider A. (2002) Mitochondrial substrate level phosphorylation is essential for growth of procyclic Trypanosoma brucei. J. Biol. Chem. 277, 32849–32854 [DOI] [PubMed] [Google Scholar]
- 48. McCulloch R., Vassella E., Burton P., Boshart M., and Barry J. D. (2004) Transformation of monomorphic and pleomorphic Trypanosoma brucei. Methods Mol. Biol. 262, 53–86 [DOI] [PubMed] [Google Scholar]
- 49. Tschopp F., Charrière F., and Schneider A. (2011) In vivo study in Trypanosoma brucei links mitochondrial transfer RNA import to mitochondrial protein import. EMBO Rep. 12, 825–832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Pusnik M., Charrière F., Mäser P., Waller R. F., Dagley M. J., Lithgow T., and Schneider A. (2009) The single mitochondrial porin of Trypanosoma brucei is the main metabolite transporter in the outer mitochondrial membrane. Mol. Biol. Evol. 26, 671–680 [DOI] [PubMed] [Google Scholar]
- 51. Schneider A., Charrière F., Pusnik M., and Horn E. K. (2007) Isolation of mitochondria from procyclic Trypanosoma brucei. Methods Mol. Biol. 372, 67–80 [DOI] [PubMed] [Google Scholar]
- 52. Hauser R., Pypaert M., Häusler T., Horn E. K., and Schneider A. (1996) In vitro import of proteins into mitochondria of Trypanosoma brucei and Leishmania tarentolae. J. Cell Sci. 109, 517–523 [DOI] [PubMed] [Google Scholar]