Abstract
trans-Aconitic acid (TAA) is an isomer of cis-aconitic acid (CAA), an intermediate of the tricarboxylic acid cycle that is synthesized by aconitase. Although TAA production has been detected in bacteria and plants for many years and is known to be a potent inhibitor of aconitase, its biosynthetic origins and the physiological relevance of its activity have remained unclear. We have serendipitously uncovered key information relevant to both of these questions. Specifically, in a search for novel nematicidal factors from Bacillus thuringiensis, a significant nematode pathogen harboring many protein virulence factors, we discovered a high yielding component that showed activity against the plant-parasitic nematode Meloidogyne incognita and surprisingly identified it as TAA. Comparison with CAA, which displayed a much weaker nematicidal effect, suggested that TAA is specifically synthesized by B. thuringiensis as a virulence factor. Analysis of mutants deficient in plasmids that were anticipated to encode virulence factors allowed us to isolate a TAA biosynthesis-related (tbr) operon consisting of two genes, tbrA and tbrB. We expressed the corresponding proteins, TbrA and TbrB, and characterized them as an aconitate isomerase and TAA transporter, respectively. Bioinformatics analysis of the TAA biosynthetic gene cluster revealed the association of the TAA genes with transposable elements relevant for horizontal gene transfer as well as a distribution across B. cereus bacteria and other B. thuringiensis strains, suggesting a general role for TAA in the interactions of B. cereus group bacteria with nematode hosts in the soil environment. This study reveals new bioactivity for TAA and the TAA biosynthetic pathway, improving our understanding of virulence factors employed by B. thuringiensis pathogenesis and providing potential implications for nematode management applications.
Keywords: bacterial genetics, bacterial toxin, biosynthesis, small molecule, tricarboxylic acid cycle (TCA cycle) (Krebs cycle), Bacillus thuringiensis, aconitate isomerase, membrane transporter, nematicidal factor, trans-aconitic acid
Introduction
Bacillus thuringiensis is an important entomopathogen that belongs to the Bacillus cereus group along with the human opportunistic pathogen B. cereus and mammalian etiological agent of anthrax Bacillus anthracis (1). Recent, research has classified B. thuringiensis as a bacterial pathogen of alternative nematode hosts (2–4), which may help to explain the complex ecology of B. thuringiensis that was previously thought to have a sole insect host (2, 5, 6). This finding further contextualizes already established interactions between B. thuringiensis and nematodes (1–4, 7), including free-living and parasitic species. The relationship between nematodes and B. thuringiensis is of great importance, because it not only gives new insights into the evolution and ecology of this important environmental microorganism (2, 3, 7) but also provides promising resources or strategies for nematode management.
B. thuringiensis bacterium is capable of undergoing a complete life cycle involving infection, germination, and reproduction stages inside Caenorhabditis elegans (1, 8). During the processes, a variety of virulence factors in nematicidal strains exhibit toxicity and cause the eventual death of the nematode host. Many important nematicidal factors of B. thuringiensis have been identified. Crystallized proteins, such as Cry5, Cry6, Cry13, Cry14, Cry21, and Cry55, are the predominant nematicidal toxins (8–15), and the application of Cry6A (16) or truncated Cry5B (17) in transgenic plants conferred significantly improved resistance to plant-parasitic nematodes. Meanwhile, chitinases (18), metalloproteinases (1, 4), lantibiotics (19), and a two-domain Nel protein (20) of B. thuringiensis are also active against nematodes. In addition to the most studied protein or peptide toxins, B. thuringiensis also secretes small active compounds to intoxicate nematodes. One identified compound in B. thuringiensis is thuringiensin, a 701-Da secondary metabolite that is highly synthesized and secreted by a 12-kb acyl carrier protein-dependent gene cluster (21). Thuringiensin displays excellent nematicidal activity against both free-living and plant-parasitic nematodes (22, 23). However, such small molecules are rarely reported in B. thuringiensis, leaving a gap in our understanding of the contributions of small molecular compounds to B. thuringiensis pathogenesis.
trans-Aconitic acid (TAA)4 is a small unsaturated tricarboxylic acid that is a natural isomer of cis-aconitic acid (CAA) in the tricarboxylic acid (TCA) cycle (24). Although it is a strong inhibitor of aconitase in the TCA cycle (25, 26), TAA is highly produced by Pseudomonas bacteria (27, 28) and sugar-containing plants (29–31). It has been speculated that TAA can neutralize alkaline compounds absorbed by roots (32) and act as an antifeedant against the rice pest brown planthopper (33). However, these identified biological roles only partially explain the significance of naturally accumulated TAA, suggesting additional unrevealed biofunctions for TAA. Two TAA biosynthetic pathways have long been hypothesized: aconitate isomerase-mediated biosynthesis from a cis-aconitic acid substrate in both microbes (27, 28) and plants (29, 30), and a citric acid dehydratase-mediated synthesis reaction from citric acid substrate specific to maize (31). Although TAA is closely related to the central cellular metabolism of the TCA cycle, no exact genes have been identified as responsible for the in vivo formation of TAA.
In this study, we discovered that a thuringiensin-producing strain, B. thuringiensis CT-43 (34, 35), also highly produces the unusual cellular metabolite TAA. Bioassays conducted on nematodes revealed an unexpected lethal activity for TAA, suggesting that it is a novel nematicidal factor of the B. thuringiensis pathogen. Using genetic and biochemical techniques, we determined the TAA biosynthetic gene operon consisting of tbrA and tbrB genes in strain CT-43 as well as the isomerism and transportation processes mediated by aconitate isomerase TbrA and TAA transporter TbrB in the TAA biosynthesis of B. thuringiensis. Further, the distribution of the tbr operon across the B. cereus and B. thuringiensis strains indicated a general role for TAA in the interactions of bacteria with nematodes in soil environments.
Results
CT-A Is a Nematicidal Compound of B. thuringiensis CT-43
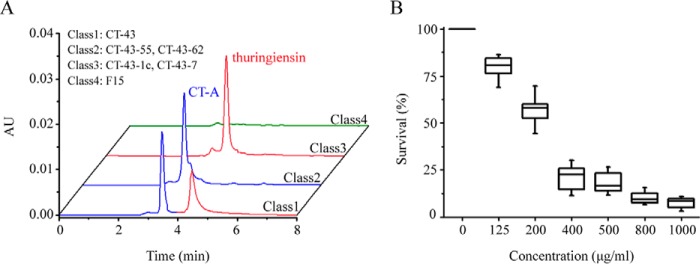
B. thuringiensis CT-43 (non-flagellum and previously classified as B. thuringiensis subsp. chinensis) (35) harbors 10 native plasmids carrying diverse toxic genes against nematode and insect targets (34). Thuringiensin is a nematicidal factor of strain CT-43, with a gene located on the pCT127 plasmid (21, 22). HPLC analysis of the culture supernatants of strain CT-43 and its plasmid-deficient mutants (35) showed different production profiles for thuringiensin and an undetermined high yielding compound, named CT-A. According to the production pattern, we divided these strains into four classes (Fig. 1A). To identify whether the highly produced CT-A was nematicidal, similar to thuringiensin, we prepared a highly purified sample of CT-A (Fig. 2A) from the culture supernatant of the thuringiensin-deficient mutant strain CT-43-55 in class 2 (Fig. 1A) by HPLC and conducted a bioassay on second-stage juveniles (J2s) of the root knot nematode Meloidogyne incognita (36), one of the most damaging plant-parasitic nematodes to global agriculture (37). As a result, the survival of M. incognita decreased with an increase in CT-A concentration (Fig. 1B). In particular, 44.1% of the population died when subjected to a low concentration (200 μg/ml); a moderate level of 400 μg/ml caused >78% mortality, and when CT-A level increased to 1,000 μg/ml, 92.1% of the nematodes died. The concentration at which 50% of M. incognita die (LC50) after 72 h was calculated as 235.5 μg/ml using probit analysis (Fig. 1B). These results suggested that secreted CT-A was a nematicidal component of the B. thuringiensis CT-43.
FIGURE 1.
The CT-A component prepared from B. thuringiensis CT-43-55 in class 2 showed nematicidal activity on the plant-parasitic nematode M. incognita. A, classification of strain CT-43 and its plasmid-deficient mutants according to the production profile of CT-A (blue peak) and thuringiensin (red peak). The plasmid-deficient mutant CT-43-55 in class 2 is preferred for CT-A preparation over strains that produce thuringiensin. B, bioassay of a highly purified CT-A sample extracted from B. thuringiensis CT-43-55 on the J2s of the plant-parasitic root knot nematode M. incognita. The bioassay included three biological repeats, and treatment at each TAA concentration included three technological repeats. After 72 h, total and living J2s were counted and used to calculate the LC50 values by probit analysis (IBM SPSS software). Error bars, S.D. AU, absorbance units.
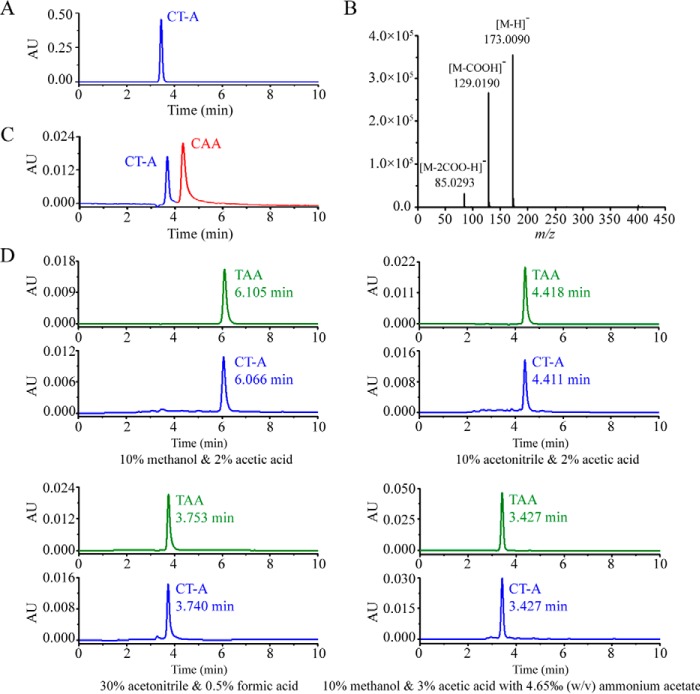
FIGURE 2.
Structure identification of CT-A molecule as trans-aconitic acid. A, highly purified CT-A sample prepared from strain CT-43-55 by HPLC. B, Q-TOF-MS analysis of highly purified CT-A. The m/z 173.0090 ion represents the mass of the [M − H]− ion of CT-A. Analysis of the 173.0090 ion with the METLIN database revealed that CT-A was aconitic acid (AA). The m/z signals at 129.0190 and 85.0293 indicated the decarboxylation of one and two carboxyl groups from the [M − H]− ion, respectively. C, HPLC analysis of a mixed sample of CT-A extract from strain CT-43-55 and a CAA commercial standard showed two separated peaks. D, HPLC analysis of retention times of CT-A extract from strain CT-43-55 with a trans-aconitic acid (TAA) commercial standard under four different mobile phase conditions. The retention times and compositions of each mobile phase are provided. AU, absorbance units.
The Nematicide CT-A Is trans-Aconitic Acid
To determine the structure of CT-A, the highly purified CT-A sample (Fig. 2A) was subjected to LC-Q-TOF-MS. The m/z 173.0090 ion representing the mass of the [M − H]− ion of the CT-A molecule is shown in Fig. 2B. Matching the signal of 173.0090 with METLIN, a metabolite mass spectral database (38), indicated that CT-A was aconitic acid (C6H6O6, 174.11 Da), a known unsaturated tricarboxylic acid with two natural isomers: CAA and TAA (Fig. 3A). CAA and TAA commercial standards were analyzed by HPLC to determine whether CT-A is CAA or TAA, according to specific retention times. The CT-A peak was distinct from the CAA peak (Fig. 2C). However, the retention times of CT-A were consistent with those of the TAA standard under four tested mobile phase conditions (Fig. 2D), indicating that CT-A is TAA. NMR spectroscopy and IR spectroscopy revealing the internal structure of the CT-A molecule further confirmed this identification (data not shown).
FIGURE 3.
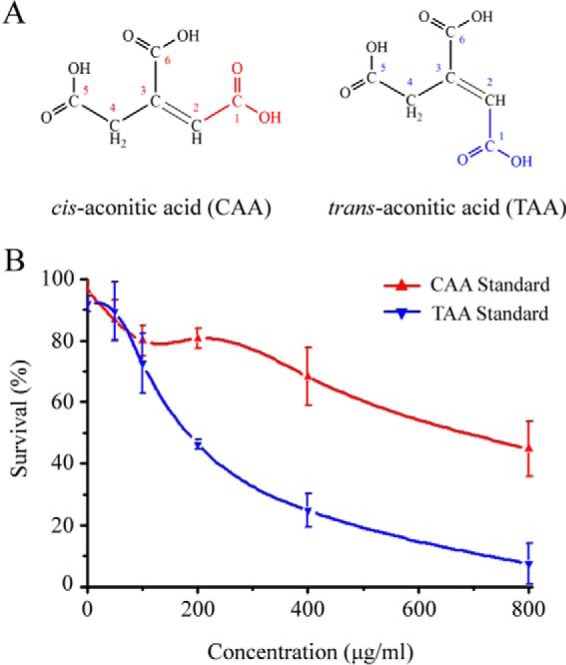
TAA displayed much stronger nematicidal activity than CAA on J2s of M. incognita. A, chemical structural formulas of CAA and TAA isomers. The conformational differences between the 1-carboxyl groups in CAA and TAA molecules are highlighted in red and blue, respectively. B, bioassays of CAA and TAA standards on J2s of M. incognita. Each bioassay included three biological repeats, and treatment for each concentration contained three technological repeats. After 72 h, the total and living numbers of J2s in CAA and TAA bioassays were recorded and used to calculate the LC50 values by probit analysis (IBM SPSS software). Data are means ± S.D. (error bars).
To verify the nematicidal activity of the TAA molecule, we examined the effect of a commercial TAA standard on M. incognita. Nematode survival decreased as the TAA standard concentration increased, and the LC50 value of the TAA standard on M. incognita after 72 h was calculated as 226.3 μg/ml (Fig. 3B, blue line), which was consistent with that of the highly purified CT-A (Fig. 1B). These results demonstrated that TAA was a nematicide. CAA, whose pH value was similar to TAA (data not shown), showed a significantly higher LC50 value of 912.1 μg/ml (Fig. 3B, red line), ruling out the possibility that TAA kills nematodes mainly through its acidity. Overall, these results confirmed that the highly produced CT-A in B. thuringiensis CT-43 was TAA, a TCA cycle-related metabolite that displays a novel nematicidal bioactivity.
The Operon Consisting of tbrA and tbrB Genes Is Responsible for TAA Biosynthesis in B. thuringiensis CT-43
Before CT-A was identified as TAA, we presumed that it was another nematicidal secondary metabolite of B. thuringiensis CT-43 other than thuringiensis and began to isolate its biosynthesis genes within the CT-43 genome. Given that most of the toxins encoding genes in B. thuringiensis are plasmid-borne (21, 39), we first investigated the relationship of CT-A formation with plasmids present in various plasmid-deficient mutants of strain CT-43 to preliminarily locate CT-A biosynthesis genes (Table 1). As shown by mutants CT-43-55 and CT-43-62 in class 2, the deficiency of plasmid pCT127 affected the production of thuringiensin but not CT-A; in CT-43-1c (class 3), deficiency of only the pCT281 plasmid abolished CT-A production, and CT-43-7 (class 3) or F15 (class 4) whose pCT281 plasmid was absent also produced no CT-A. Together, these data indicated that CT-A biosynthesis-related genes were located on the biggest plasmid, pCT281 (281,231 bp; 276 genes), of strain CT-43. Because bioinformatics analysis of the pCT281 sequence with antiSMASH, a microbial secondary metabolite biosynthetic gene cluster database (40), showed no cluster targets, we adopted a full sequence-wide deletion strategy covering any possible compound biosynthesis-related genes on pCT281 (Table 2) and successfully deleted one acetyltransferase, one N-hydroxyarylamine O-acetyltransferase, and 11 hypothetical protein-encoding genes on pCT281 (Table 2) through a temperature-sensitive (TS) replicon-mediated homologous recombination method (21). No genes were CT-A biosynthesis-related, because CT-A production was not affected in these mutants (data not shown). However, both efficiencies of transformation in the wild strain CT-43 and homologous recombination via temperature stress on the TS replicon were confirmed to be low. We failed to target CT-A biosynthesis genes by this deletion strategy until CT-A was identified as the cellular metabolite TAA.
TABLE 1.
Native plasmid distribution and CT-A (TAA) production in B. thuringiensis CT-43 and its plasmid-deficient mutants
Plus and minus signs indicate the presence and absence of relevant plasmid, respectively.
| Plasmid | Size | Class 1: CT-43 (CT-A) | Class 2 |
Class 3 |
Class 4: F15 (no CT-A) | ||
|---|---|---|---|---|---|---|---|
| CT-43-55 (CT-A) | CT-43-62 (CT-A) | CT-43-1c (no CT-A) | CT-43-7 (no CT-A) | ||||
| bp | |||||||
| pCT281 | 281,231 | + | + | + | − | − | − |
| pCT127 | 127,885 | + | − | − | + | + | − |
| pCT83 | 83,590 | + | + | + | + | + | + |
| pCT72 | 72,074 | + | + | + | + | + | + |
| pCT51 | 51,488 | + | + | + | + | − | + |
| pCT14 | 14,860 | + | + | + | + | + | + |
| pCT9547 | 9,547 | + | + | + | + | + | + |
| pCT8513 | 8,513 | + | + | + | + | + | + |
| pCT8252 | 8,252 | + | + | + | + | + | + |
| pCT6880 | 6,880 | + | + | + | + | + | + |
TABLE 2.
Design of full sequence-wide gene deletions in plasmid pCT281 of B. thuringiensis CT-43
The full sequence of plasmid pCT281 (∼281 kb) was divided into 28 regions (every 10 kb from 0 to 281 kb). Primer locations in each region defined the deleted sequence range, within which certain genes that were functionally unidentified or predicted to be biosynthesis-related were targeted and described according to the National Center for Biotechnology Information (NCBI) database. The primer locations in blue and red represent the completed constructions of gene-knockout recombinant vectors and the further completed transformations of relevant vectors into strain CT-43, respectively. The targeted genes in green were successfully deleted genes.
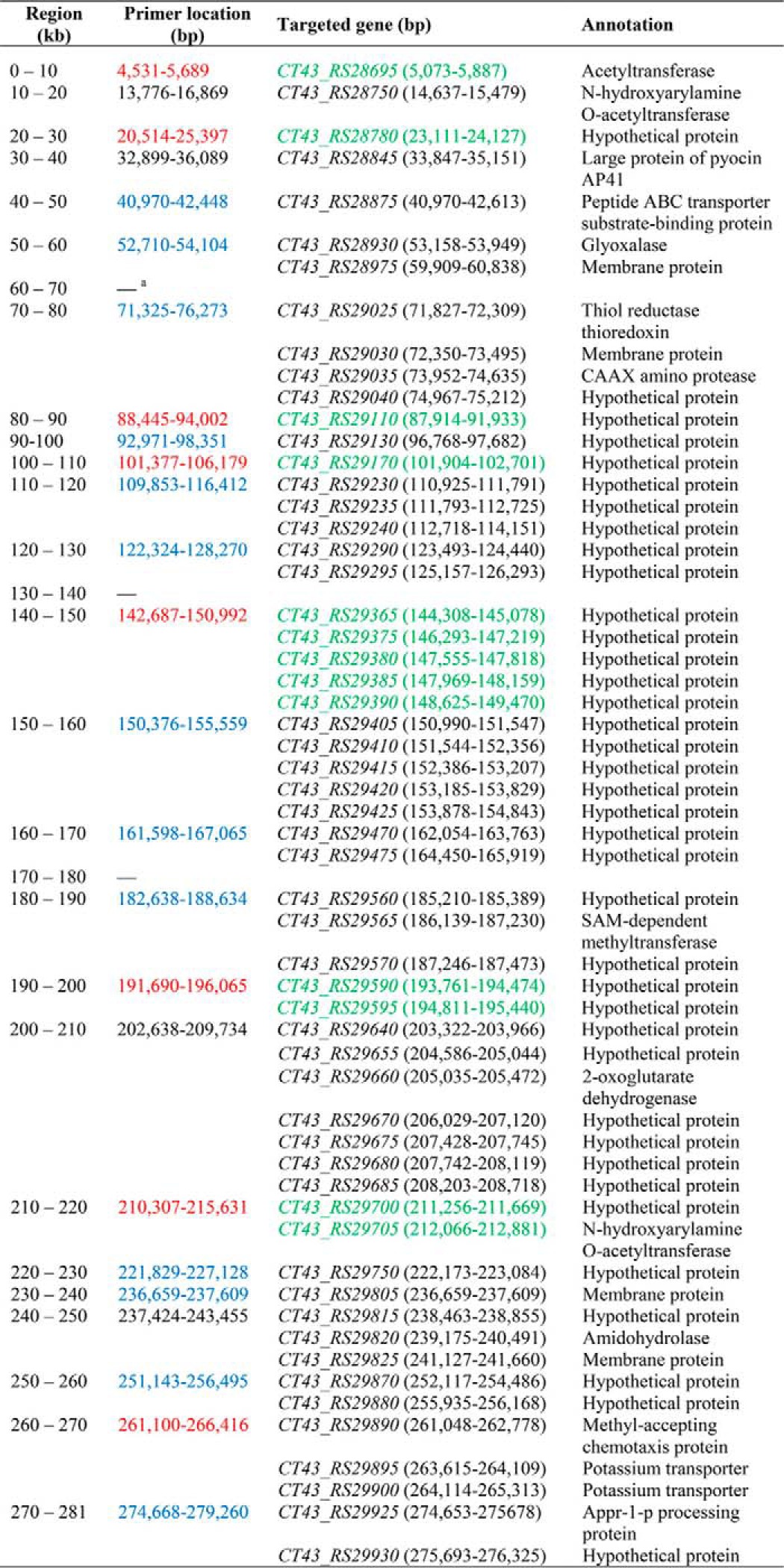
a No genes were targeted in this region.
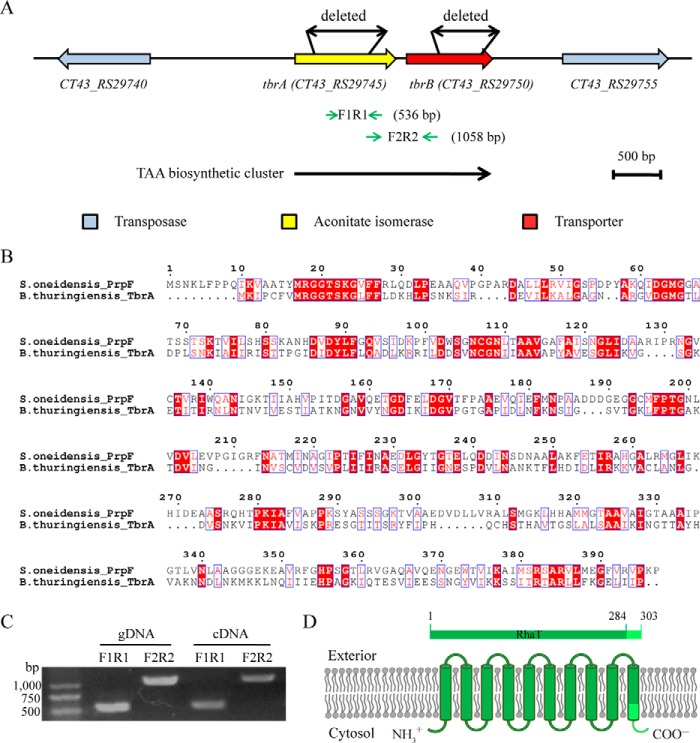
Although known for almost a century (25, 41), TAA biosynthesis genes and processes remain unclear in vivo. As mentioned above, aconitate isomerase has long been proposed to be responsible for TAA biosynthesis in microbes by driving the interconversion between CAA and TAA (27, 28). However, no genes of aconitate isomerase have been genetically identified or can be referenced in the B. thuringiensis bacterium. Thus, we analyzed all of the genes in pCT281 using CD-Search (42) to identify isomerous function-related genes. Fortunately, we found the CT43_RS29745 (1,074 bp) gene, which was the sole target of pCT281 (Fig. 4A) and showed sequence homology to the reported isomerase 2-methyl aconitate cis/trans-isomerase PrpF (43). The CT43_RS29745 gene encodes a hypothetical protein whose sequence shared 27% identity and 97% coverage with that of PrpF (397 aa) in Shewanella oneidensis MR-1 (Fig. 4B) (43). Additionally, PrpF was speculated to catalyze the interconversion of 2-methyl CAA and 2-methyl TAA in the 2-methylcitric acid cycle (43, 44). Based on these findings, we proposed that CT43_RS29745 is a gene encoding aconitate isomerase that is responsible for TAA biosynthesis in B. thuringiensis CT-43 and named it TAA biosynthesis-related gene A (tbrA). Meanwhile, another hypothetical protein-encoding gene, tbrB (CT43_RS29750, 912 bp), which was located 111 bp downstream of the tbrA and constituted an operon with tbrA (Fig. 4, A and C), also attracted our attention. Bioinformatics analysis of the TbrB protein showed an l-rhamnose-H+ transporter (RhaT) domain (45, 46) and 10 transmembrane helices (Fig. 4D), indicating TbrB as a potential TAA transporter.
FIGURE 4.
Bioinformatics analysis of putative TAA biosynthesis-related (tbr) genes tbrA and tbrB in B. thuringiensis CT-43. A, gene organization of the tbr operon region on plasmid pCT281. Two transposase genes are shown in blue; putative aconitate isomerase gene tbrA and predicted membrane protein gene tbrB are shown in yellow and red, respectively. Double-headed black arrows indicate the deleted regions within tbrA and tbrB ORFs in gene deletion experiments. Two pairs of inverted green arrows indicate the amplifying regions in the operon analysis experiment of tbrA and tbrB genes, and the sizes of the amplification products F1R1 and F2R2 are provided. B, sequence alignment of TbrA protein of B. thuringiensis CT-43 with PrpF protein from S. oneidensis MR-1. The alignment was performed using ClustalX and highlighted using ESPript version 3.0. Identical residues are shown in red, and residues with similar side chains are boxed. C, operon analysis of tbrA and tbrB genes in B. thuringiensis CT-43 by reverse transcription PCR. gDNA, the genomic DNA of strain CT-43. D, schematic of the putative transmembrane protein TbrB. Ten transmembrane helices of TbrB are shown in green. The l-rhamnose-H+ transporter (RhaT) domain that locates at the amino terminus of TbrB protein (aa 1–284) is indicated in dark green, and the rest of the carboxyl-terminal sequence (aa 285–303) is shown in light green.
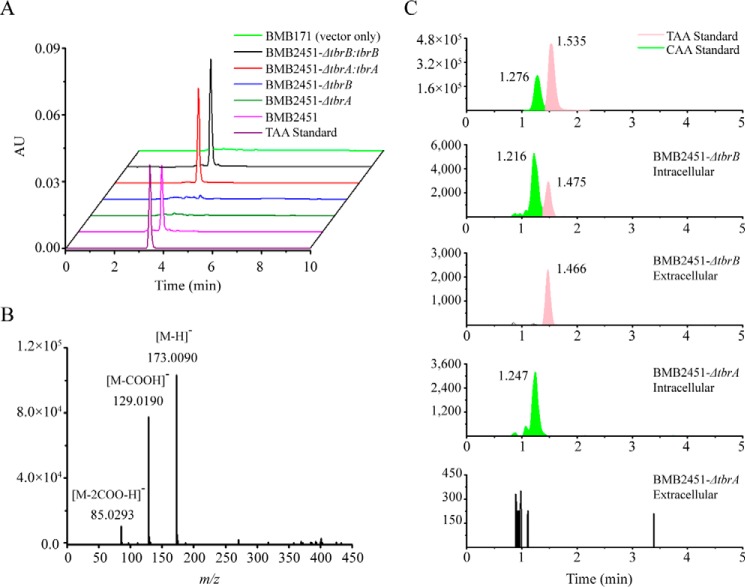
To test whether the operon comprising the tbrA and tbrB genes is responsible for TAA biosynthesis, we performed heterologous expression of the operon carried by the pHT304 vector (47) in B. thuringiensis BMB171 (48), an acrystalliferous B. thuringiensis mutant producing no TAA. This resulted in a recombinant termed BMB2451. HPLC and Q-TOF-MS analysis of the culture supernatant of BMB2451 confirmed TAA production (Fig. 5, A (pink line) and B). Subsequent gene deletions of tbrA and tbrB in the recombinant BMB2451 generated the recombinants BMB2451-ΔtbrA and BMB2451-ΔtbrB, respectively. TAA production was not observed in either supernatant by HPLC analysis (Fig. 5A, dark green and blue lines). When either tbrA or tbrB was complemented via another compatible vector pEMB0603 (39) in BMB2451-ΔtbrA:tbrA or BMB2451-ΔtbrB:tbrB, TAA production was restored (Fig. 5A, red and black lines). Together, these results genetically verified that the operon consisting of tbrA and tbrB was the TAA biosynthetic gene cluster of B. thuringiensis CT-43.
FIGURE 5.
Genetic verification of the TAA biosynthetic gene cluster. A, HPLC analysis of TAA in the culture supernatants of BMB2451 (pink line), BMB2451-ΔtbrA (dark green line), BMB2451-ΔtbrB (blue line), BMB2451-ΔtbrA:tbrA (red line), and BMB2451-ΔtbrB:tbrB (black line) recombinant strains. TAA standard (purple line) and the supernatant extract of BMB171 with empty vector pHT304 (light green line) were used as positive and negative controls, respectively. B, Q-TOF-MS confirmation of TAA production by heterologous expression of the tbr operon in BMB2451. C, Q-TOF-MS analysis of TAA presence in intracellular and extracellular fractions of BMB2451-ΔtbrA and BMB2451-ΔtbrB mutants. The m/z 173.0090 ion indicating the mass of [M − H]− patterned aconitic acid was used to extract a TAA signal from the total ion chromatograms (TIC). Retention times of extracted CAA (green peak) and TAA (pink peak) signals are provided. AU, absorbance units.
TbrA Protein Acts as Aconitate Isomerase in the Formation of TAA in B. thuringiensis
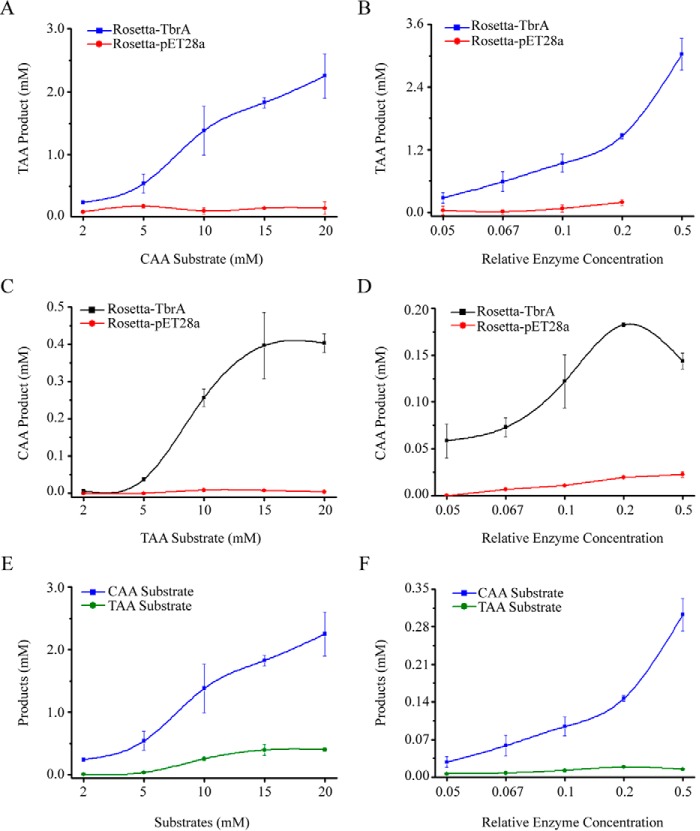
To verify the aconitate isomerase activity of TbrA protein, a TbrA-inducible Escherichia coli recombinant Rosetta-TbrA was constructed with the expression vector pET28a. After induction, the cell-free extracts of Rosetta-TbrA and Rosetta-pET28a (with empty vector only) were used for in vitro catalytic assays. First, we used CAA as a substrate to test TAA formation. Using a constant level of Rosetta-TbrA cell-free extract and a CAA concentration gradient from 2 to 20 mm, TAA formation was found to increase as CAA concentration increased. TAA formation was not detected in the Rosetta-pET28a cell-free extract (Fig. 6A). Meanwhile, when the CAA level was constant (10 mm), increasing the content of Rosetta-TbrA cell-free extract also resulted in increasing TAA formation. Rosetta-pET28a cell-free extract showed no enzymatic activity (Fig. 6B). These results demonstrated that TbrA could catalyze CAA into TAA. Next, we used TAA as a substrate to test CAA formation. As shown, CAA was detected in cell-free extracts of Rosetta-TbrA but not Rosetta-pET28a (Fig. 6, C and D), which demonstrated that TbrA catalyzes a reversible reaction from CAA to TAA, and the equilibrium favors isomerization into TAA (Fig. 6, E and F). Moreover, tbrA deletion abolished in vivo TAA formation in mutant BMB2451-ΔtbrA, as observed by Q-TOF-MS analysis (Fig. 5C). Together, these results demonstrated that TbrA has aconitate isomerase activity that mediates TAA formation in B. thuringiensis.
FIGURE 6.
Enzymatic assays of aconitate isomerase activity of TbrA protein. Cell-free extracts of induced Rosetta-TbrA and Rosetta-pET28a (control) recombinants were used for in vitro catalytic reactions. When CAA was used as substrate (A and B), TAA formation was tested by HPLC with varying CAA substrate concentrations of 2, 5, 10, 15, and 20 mm (A) and different relative enzyme concentrations of 0.05, 0.067, 0.1, 0.2, and 0.5 (B). Similar assays were performed when TAA was treated as substrate (C and D). The efficiencies of using CAA and TAA substrates to synthesize TAA and CAA products by TbrA protein were compared with different conditions of substrate (E) and enzyme (F) concentrations. Data are means ± S.D. (error bars).
TbrB Is a Membrane Transporter of TAA in B. thuringiensis
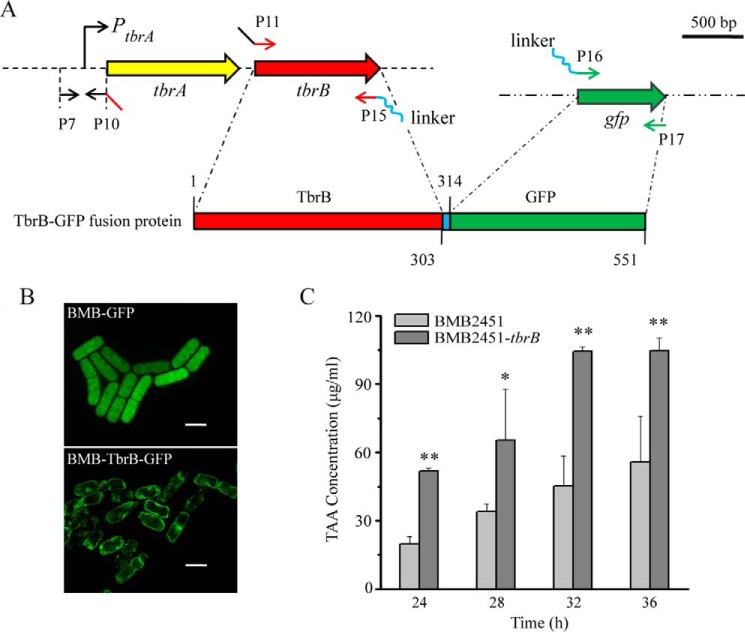
To determine the subcellular localization of TbrB, a putative membrane protein, we generated a fusion gene construct by connecting tbrB ORF to the green fluorescent protein (gfp) gene under the control of the tbrA promoter region (Fig. 7A). This construct was then transformed into BMB171, resulting in recombinant BMB-TbrB-GFP expressing a TbrB-GFP fusion protein. As shown in Fig. 7B, an obvious green fluorescent signal indicating TbrB-GFP localization appeared specifically at the cell membrane of BMB-TbrB-GFP. In the control, GFP was distributed uniformly inside the cells of BMB-GFP. This result demonstrated that TbrB is a membrane protein of B. thuringiensis.
FIGURE 7.
Characterization of TbrB as a TAA transporting membrane protein. A, construction schematic of TbrB-GFP fusion protein. SOE-PCR was performed to connect the promoter region of tbrA (PtbrA), the tbrB ORF (red), a 10-aa peptide linker-encoding sequence (blue), and the gfp ORF (green). Primers are highlighted in different colors to indicate individual overlapped regions. B, fluorescence image of BMB-TbrB-GFP showed membrane location of TbrB-GFP protein. BMB-GFP cells were used as control. Scale bar, 2 μm. C, TAA production was substantially promoted in recombinant BMB2451-tbrB by introducing additional copies of the tbrB gene into BMB2451 (**, p < 0.01; *, p < 0.05; one-way ANOVA followed by Tukey's honest significant difference test). All data are means ± S.D. (error bars).
Q-TOF-MS analysis of the intracellular contents of strain BMB2451-ΔtbrB revealed the presence of TAA (Fig. 5C), indicating active TAA formation despite tbrB function in vivo. However, TAA presence in the extracellular fraction of BMB2451-ΔtbrB was significantly reduced compared with BMB2451, where tbrB was functional (Fig. 5A). Furthermore, tbrB copy number was increased in recombinant BMB2451-tbrB, which was constructed based on the BMB2451 strain. This strain showed a substantially higher level of TAA in the culture supernatant than BMB2451 (Fig. 7C), in agreement with reports that the introduction of additional transporter genes into metabolite-producing bacteria could significantly promote product yield (49). Together, these results demonstrated a TAA membrane transporter role for the TbrB protein of B. thuringiensis.
The Biological Processes of TAA Biosynthesis in B. thuringiensis
TAA biosynthesis through an isomerous pathway in B. thuringiensis is proposed in Fig. 8. First, the substrate of aconitate isomerase, CAA, is formed as an intermediate through citric acid dehydration by aconitase (ACO) in the TCA cycle. Instead of being hydrated to isocitric acid by further action of ACO, a portion of CAA is isomerized into TAA by the TbrA protein. Although the isomerization is reversible, TbrA isomerase equilibrium favors TAA. The membrane-anchored TbrB protein transports TAA out of cells, resulting in extracellular accumulation of TAA.
FIGURE 8.
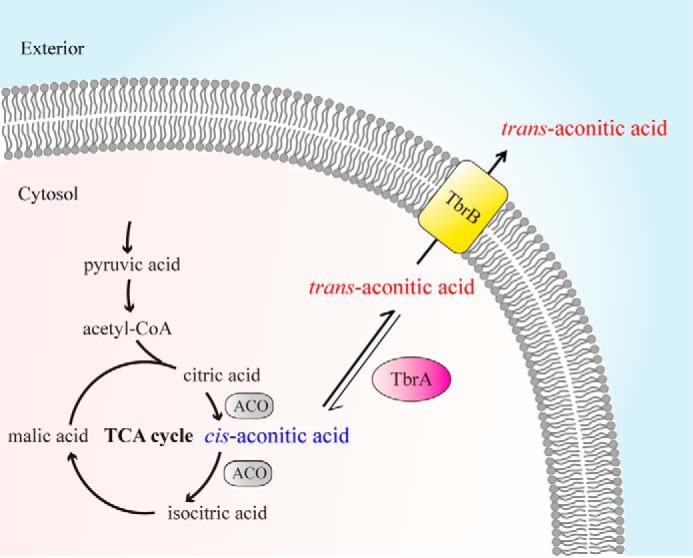
Model of the TAA biosynthetic pathway in B. thuringiensis. The TCA cycle synthesizes CAA through citric acid dehydration by ACO (gray). Instead of further hydration to isocitric acid by ACO, a portion of CAA is isomerized into TAA by aconitate isomerase TbrA (pink), which catalyzes the reversible reaction from CAA to TAA but favors the equilibrium for TAA, as indicated by the thicker black arrow. Intracellular TAA is then transported outside cells by the membrane-anchored TbrB protein (yellow), resulting in extracellular accumulation of TAA.
Distribution of TAA Biosynthetic Gene Cluster in the B. cereus Group
To determine whether other B. cereus group bacteria have the potential to produce TAA, we investigated the presence of the identified TAA genes in the B. cereus group. As shown in Table 3, a 2,097-bp sequence comprising a 1,074-bp tbrA ORF sequence, an 111-bp intergenic region, and a 912-bp tbrB ORF sequence (Fig. 4A) is found in B. thuringiensis and B. cereus bacteria. All targets are located on plasmids except for strains whose genomes were incomplete. In addition, two transposase genes were found at both ends of the TAA gene cluster (Fig. 4A), indicating possible acquisition of the TAA biosynthetic gene cluster by B. cereus and B. thuringiensis bacteria through horizontal gene transfer.
TABLE 3.
B. cereus group bacteria with TAA biosynthetic gene cluster
| Strain | Location | Identitya |
|---|---|---|
| % | ||
| B. thuringiensis | ||
| CT-43 | pCT281 | |
| YBT-1520 | pBMB293 | 100 |
| HD-1 | pBMB299 | 100 |
| HD-29 | pBMB267 | 100 |
| IS5056 | pIS56–285 | 100 |
| T01-328 | —b | 100 |
| Leapi01 | — | 100 |
| YC-10 | pYC-1 | 100 |
| Serovar mexicanensis | — | 100 |
| Serovar tolworthi | pKK2 | 100 |
| B. cereus | ||
| ISP2954 | — | 100 |
| BMG1.7 | — | 100 |
| HuB13-1 | — | 100 |
a Identity between TAA biosynthetic gene cluster of strain CT-43 and predicted genes in other B. cereus group bacteria.
b —, location undetermined.
Discussion
Despite its relationship to central metabolism, TAA biosynthesis and biofunction have remained unclear since its discovery almost a century ago. In this study, we discovered a novel biological role of TAA (also named CT-A before the structure identification; Fig. 2) as a nematicidal molecule of the B. thuringiensis bacterium and revealed how this small metabolite is biosynthesized.
TAA is an unusual cellular metabolite, and thus the fact that B. thuringiensis synthesizes and secretes large amounts of TAA suggests a specific purpose (e.g. use of TAA as a nematicidal factor). TAA is not necessary for basic biological metabolisms and is known to strongly inhibit the activity of aconitase (25). When intracellular TAA accumulates, the cell must clear it through methylation (50) or transport to avoid inhibition and maintain the normal operation of the TCA cycle. Thus, TAA biosynthesis does not appear to be necessary for central metabolism. With the newly identified nematicidal activity of TAA, we can now reasonably answer why B. thuringiensis produces high levels of TAA and reveal it as a novel virulence factor of this nematode pathogen. CAA, which is an essential cellular metabolite, has a pH similar to that of its isomer TAA but displayed a much weaker nematicidal effect (Fig. 3B). This further suggests that TAA is a specifically produced molecule whose trans-conformation contributes to its nematicidal activity.
We noticed similar effects contributed by the specific trans-structure of TAA. For example, in vitro, TAA inhibited the growth and transformation of Leishmania donovani, the etiological agent of kala-azar, whereas CAA had no effect (51, 52). In barnyard grass, a high endogenous TAA level was revealed to function as an antifeedant of rice brown planthopper, and CAA was determined to be non-functional (33). In addition, TAA has an anti-edematogenic effect (53) that may be due to its unexpected specific inhibitory effect on human phosphodiesterase 7 (PDE7) (54), an inflammatory disease-associated enzyme. Based on these reports, we conclude that TAA is a multifunctional natural product despite its simple molecular structure.
To our knowledge, this is the first report demonstrating the cellular TAA biosynthetic pathway in microorganisms. In 1961, Rao and Altekar (27) proposed the existence of a new enzyme catalyzing the interconversion of aconitates and named it aconitate isomerase. In 1971, Klinman and Rose prepared an aconitate isomerase sample from the total protein of Pseudomonas putida cells and determined its kinetic properties (28) and reaction mechanism (55). Since then, Pseudomonas has been the only reported TAA-metabolizing bacteria; few studies have focused on identification of relevant DNA determinants or characterization of the TAA biosynthesis pathway. Here, through genetic and biochemical methods, we determined that the hypothetical protein-encoding gene tbrA is an aconitate isomerase gene that is responsible for intracellular TAA formation in the B. thuringiensis bacterium. For extracellular accumulation, the tbrB gene encoding a transporter containing a 10-transmembrane helix is required (Fig. 8). According to Klinman and Rose (28), the molecular mass of the putative aconitate isomerase from P. putida was 78 ± 10 kDa, which is significantly larger than the 38.13-kDa mass of TbrA in B. thuringiensis. This finding indicates that although they drive the same type of reactions, aconitate isomerases from these two bacteria differ in sequence, suggesting possible species-specific origins. PrpF was proposed to work with an aconitase-like (AcnD) enzyme in the 2-methylcitric acid cycle and to isomerize 2-methyl CAA and 2-methyl TAA (43). Because 2-methyl aconitic acids are not commercially available, the isomerase activity of PrpF protein is unverified. Characterization of the aconitate isomerase function of TbrA, the homologous protein of PrpF, could support the putative 2-methyl aconitate cis/trans-isomerase role of PrpF protein.
B. thuringiensis and B. cereus are B. cereus group bacteria that inhabit diverse environments, including soil, freshwater, invertebrates, and insectivorous mammals (56). Compared with B. subtilis, a plant-associated bacterium, B. thuringiensis and B. cereus strains contain more nitrogen metabolism-associated genes than those in carbon metabolism, indicating that their hosts are likely to be animals (57, 58). Nematodes are the most abundant soil metazoans. Most nematodes in soil are bacteria feeders, and generally 90–99% of nematode habitats have high microbial activity (59). Thus, a soil environment represents a suitable place for the interactions of bacteria and nematodes, where diverse virulence factors are produced and operate. The distribution of the TAA biosynthetic gene cluster in B. thuringiensis and B. cereus (Table 3) suggests the potential to produce high levels of nematicidal TAA in these strains. The transposase genes at both ends of the cluster enable the mobility of the TAA biosynthetic genes. These findings indicate that TAA may contribute to bacteria toxicity, synergistically with other active elements, such as Cry toxins or metalloproteinases, in soil bacteria-nematode interactions. Although the nematicidal mechanism of TAA is unclear, we hypothesized it to be metabolism-associated. TAA inhibition of aconitase would limit isocitric acid production in the TCA cycle (26, 52, 60). The operation of a glyoxylate shunt, an essential pathway for the survival of parasites during infection (49, 61), may thus be affected in the J2s of M. incognita, where the glyoxylate shunt is highly expressed and dependent (62). This is because isocitric acid is a substrate of the first enzyme, isocitrate lyase, in the glyoxylate shunt (49). Combined with the above-mentioned inhibitory effect of TAA on parasite Leishmania spp., these facts raise interesting scientific issues, such as whether the glyoxylate shunt can be significantly influenced by TAA or whether TAA has broad activity in glyoxylate shunt-dependent parasites. Although TAA has long been used as an industrial material, these established bioactivities for TAA have potential in promoting its future biological usage in agricultural pest management.
Experimental Procedures
Bacterial Strains, Plasmids, and Culture Conditions
The bacterial strains and plasmids used in the present study are listed in Table 4. All E. coli and B. thuringiensis strains were cultured in Luria-Bertani (LB) medium at 37 and 28 °C, respectively. When appropriate, antibiotics were added at the following final concentrations: 100 μg/ml ampicillin, 25 μg/ml erythromycin and chloramphenicol, and 50 μg/ml kanamycin.
TABLE 4.
Bacterial strains and plasmids used in this study
| Strain and plasmid | Description | Source or reference |
|---|---|---|
| DH5α | E. coli cloning host | Ref. 63 |
| Rosetta-TbrA | E. coli Rosetta expressing the TbrA protein | This work |
| Rosetta-pET28a | E. coli Rosetta with empty vector pET28a | This work |
| CT-43 | B. thuringiensis, no flagellum; TAA and thuringiensin producer | Refs. 34 and 35 |
| CT-43-55 | Strain CT-43 derivative; TAA producer | Ref. 35 |
| CT-43-62 | Strain CT-43 derivative; TAA producer | Ref. 35 |
| CT-43-7 | Strain CT-43 derivative; thuringiensin producer | Ref. 35 |
| CT-43-1c | Strain CT-43 derivative; thuringiensin producer | Ref. 35 |
| F15 | Strain CT-43 derivative; no TAA and thuringiensin production | This work |
| BMB171 | Acrystalliferous mutant of B. thuringiensis | Ref. 48 |
| BMB2451 | BMB171 with plasmid pBMB2451 | This work |
| BMB2451-ΔtbrA | BMB171 with plasmid pBMB2451-ΔtbrA | This work |
| BMB2451-ΔtbrB | BMB171 with plasmid pBMB2451-ΔtbrB | This work |
| BMB2451-ΔtbrA:tbrA | BMB171 with plasmids pBMB2451-ΔtbrA and pEMB0603-tbrA | This work |
| BMB2451-ΔtbrB:tbrB | BMB171 with plasmids pBMB2451-ΔtbrB and pEMB0603-tbrB | This work |
| BMB-TbrB-GFP | BMB171 with plasmid pBMB-tbrB-gfp | This work |
| BMB-GFP | BMB171 with plasmid pBMB-gfp | This work |
| BMB2451-tbrB | BMB2451 with plasmid pEMB0603-tbrB | This work |
| pHT304 | E. coli–B. thuringiensis shuttle vector | Ref. 47 |
| pHT304-TS | pHT304 derivative with a temperature-sensitive Bacillus replicon | Ref. 21 |
| pMD19-T | E. coli cloning vector | Takara |
| pET28a | E. coli expression vector | Takara |
| pET28a-tbrA | TbrA protein expressing plasmid | This work |
| pEMB0603 | E. coli-B. thuringiensis shuttle BAC vector | Ref. 39 |
| pEMB0603-tbrA | pEMB0603 with intact tbrA gene | This work |
| pEMB0603-tbrB | pEMB0603 with tbrB ORF fused with tbrA gene promoter | This work |
| pBMB2451 | pHT304 carrying tbrA and tbrB genes | This work |
| pBMB2451-ΔtbrA | tbrA gene deleted pBMB2451 derivative | This work |
| pBMB2451-ΔtbrB | tbrB gene deleted pBMB2451 derivative | This work |
| pBMB-tbrB-gfp | pHT304 with tbrB-gfp fusion gene | This work |
| pBMB-gfp | pHT304 with gfp gene | This work |
CT-A (TAA) Extraction, Purification, and Detection
The extraction procedure for crude CT-A (TAA) was identical to that for thuringiensin (21). Samples were stored at 4 °C until use. For CT-A (TAA) purification, sufficient crude sample was first prepared from the CT-43-55 strain over other strains that produce thuringiensin. Purification was carried out using two rounds of preparation with an HPLC system consisting of a Waters 1525 pump, a Waters 2489 UV-visible detector, and a Waters SunFireTM prep C18 OBDTM column (150 × 19 mm, 5 μm; Waters Corp.). In the first round, crude CT-A (TAA) was dissolved in deionized water and carried by a mobile phase containing 10% methanol and 2% acetic acid. The CT-A (TAA) peak was collected and concentrated under reduced pressure using rotary evaporators. Procedures in the second round were similar to the first round, except for the mobile phase (30% acetonitrile and 0.05% formic acid). Finally, the sample was dissolved in deionized water and freeze-dried into powder. The purity of the resulting CT-A (TAA) met the requirements for structure identification, as indicated by a single peak displayed in six HPLC mobile phases (data not shown).
To detect CT-A (TAA) or CAA using HPLC, a similar system was adopted as described above except for the TC-C18 column (250 × 4.6 mm, 5 μm; Agilent). Isocratic elution of a 10-μl sample volume was delivered at a flow rate of 1.0 ml/min and monitored at 260 nm for 10 min. To analyze the sample with LC-MS, a high resolution LC-Q-TOF-MS system was used, which was composed of an Agilent 1260 LC device attached to a dual-source electrospray ionization ion source equipped with a G6540A Q-TOF-MS system (Agilent). Samples were analyzed in negative ion mode using diode array detection at 260 nm. Calibration was delivered using standard references with masses of 112.9855 and 1,033.9881 Da. The quadrupole was set to pass ions from m/z 50 to 1,500. Data were analyzed by Agilent MassHunter qualitative analysis software version B.05.00. CAA (Sigma-Aldrich) and TAA (Tokyo Chemical Industry) commercial standards were purchased and used for analysis.
Gene Deletion in B. thuringiensis CT-43
Genes in plasmid pCT281 that were considered CT-A biosynthesis-related are listed in Table 2. Generally, the target fragment harboring candidate genes on pCT281 was first amplified from the CT-43 genome and cloned into the pMD19-T vector (Takara, Dalian, China). By digestion with appropriate restriction enzymes, the central fragment of the inserted DNA was removed and replaced by a digested spectinomycin resistance gene (spcr) fragment. The inserted DNA on pMD19-T was cloned into a pHT304-TS vector (21) and electrophoretically introduced to strain CT-43. After culturing at 42 °C, the bacteria were simultaneously incubated on spcr and ermr solid agar plates to screen for mutants that were resistant to spectinomycin but not erythromycin. Finally, the in vivo gene mutation was verified by sequencing.
Identification and Characterization of TAA Biosynthetic Genes
PCR primers used in this study are listed in Table 5. For heterologous expression, a 2,451-bp PCR fragment harboring tbrA and tbrB genes was cloned into a 6.5-kb E. coli-B. thuringiensis shuttle vector pHT304 (47) to yield the pBMB2451 plasmid, which was then transferred into the B. thuringiensis host BMB171 (48) to generate the recombinant strain BMB2451. To delete the tbrA gene, the inserted 2,451-bp DNA fragment on pBMB2451 was transferred to the 2.7-kb pMD19-T vector at BamHI and SphI sites. Using this recombinant plasmid as template, reverse PCR was performed to generate a DNA product containing an incomplete tbrA ORF, the intact tbrB ORF, and the vector sequence, which was then digested at the XbaI site introduced by the reverse primers and subjected to self-ligation. The shortened DNA fragment on vector pMD19-T was transferred back to pHT304, resulting in the tbrA-deleted plasmid pBMB2451-ΔtbrA. Transforming the plasmid pBMB2451-ΔtbrA into the BMB171 host generated the mutant BMB2451-ΔtbrA. Similar operations were applied to construct thetbrB-deleted mutant BMB2451-ΔtbrB. For tbrA gene complementation, a compatible plasmid (39), recombinant pEMB0603-tbrA carrying a 1,868-bp PCR product containing the intact tbrA gene, was introduced into BMB2451-ΔtbrA, resulting in the complemented strain BMB2451-ΔtbrA:tbrA. To complement the tbrB gene, splicing overlap extension PCR (SOE-PCR) was performed to fuse the promoter region of tbrA with tbrB ORF, generating a 1,407-bp product that was inserted into pEMB0603. The generated pEMB0603-tbrB was introduced into BMB2451-ΔtbrB, resulting in the complemented strain BMB2451-ΔtbrB:tbrB.
TABLE 5.
Primers used in this study
Restriction sites and a 10-aa peptide linker-encoding sequence in P15 and P16 primers are underlined.
| Primer and description | Sequence (5′–3′) |
|---|---|
| For amplifying tbr operon | |
| P1 | TATGGATCCGGATACCAAGTTGTAGGAGGTG |
| P2 | GCCGCATGCGCTAATAGGCTTATTGCTTC |
| For deleting tbrA gene | |
| P3 | TATTCTAGATCCTGGTGTAGTGGAAATTCG |
| P4 | GCCTCTAGAGTCATCCCTAAGATTGCGGT |
| For deleting tbrB gene | |
| P5 | TATTCTAGAAGGGACTAAGAATAGAAAAGG |
| P6 | GCCTCTAGAAGAAGGGCAGCAAGTTCA |
| For complementing tbrA gene | |
| P7 | TATGGATCCCCGTGAAGCGTCTCATCCTAG |
| P8 | GCCAAGCTTCTGCTCTTGTCGCACGTCTCG |
| For complementing tbrB gene | |
| P9 | TATGGATCCCCGTGAAGCGTCTCATCCTAG |
| P10 | GTCGCTTTTTTAAGATTATCCATTTATTAGCATCTCCTTTTTATG |
| P11 | CATAAAAAGGAGATGCTAATAAATGGATAATCTTAAAAAAGCGAC |
| P12 | GCCGCATGCGCTAATAGGCTTATTGCTTC |
| For analyzing tbr operon | |
| F1 | GATTAAAGTCGGGTCAGG |
| R1 | CTGCTGATAATGCGAGCG |
| F2 | TCGCTCGCATTATCAGCAG |
| R2 | TGTCGCACGTCTCGTCTTT |
| For amplifying tbrA ORF | |
| P13 | GGAATTCCATATGATGAAAATACCTTGTTTTGTT |
| P14 | GCCCTCGAGAGGTATTATTAATTCGCCTTT |
| For constructing tbrB-gfp fusion gene (also P7, P10, and P11) | |
| P15 | GCCACCTCCGCCTGAACCGCCTCCACCTGATGAACTTGCTGCCCTTCTTTG |
| P16 | TCAGGTGGAGGCGGTTCAGGCGGAGGTGGCATGAGTAAAGGAGAAG |
| P17 | ACATGCATGCTTATTTGTATAGTTCA |
| For identifying pCT6880 of strain CT-43 | |
| P6880-1 | GCTAACAGCATTAGGTGTGC |
| P6880-2 | GCATATGTGGTGTCGCTTC |
| For identifying pCT8252 of strain CT-43 | |
| P8252-1 | CGATTGAAGAGACGTGTAG |
| P8252-2 | CAGAGGATCTCAATCCTAAG |
| For identifying pCT8513 of strain CT-43 | |
| P8513-1 | CGCAACGATGATGGAAGCA |
| P8513-2 | CATTCTACCTTCAGCATCACC |
| For identifying pCT9547 of strain CT-43 | |
| P9547-1 | GGAATGAGATGGTCGAATC |
| P9547-2 | GCATGTAGAGAACGTACAGC |
| For identifying pCT14 of strain CT-43 | |
| P14-1 | TCGTCAGCATTCATTTGAGC |
| P14-2 | GGGTTATCCGTTATATCCTG |
| For identifying pCT51 of strain CT-43 | |
| P51-1 | CACTGGAATGGTGATGTAGA |
| P51-2 | GTATCAGGCATCTTCTGCAC |
| For identifying pCT72 of strain CT-43 | |
| P72-1 | ATCGGTACAACTGGTTCAGG |
| P72-2 | GATGTTGCGGTATGCTAATC |
| For identifying pCT83 of strain CT-43 | |
| P83-1 | GAAGACGGTAATGGATGAAG |
| P83-2 | GCTGCTATACCAATAGACG |
| For identifying pCT127 of strain CT-43 | |
| P127-1 | CAACGAATGTAGTGAACGG |
| P127-2 | CGTCGCTAGGGTAACTATAG |
| For identifying pCT281 of strain CT-43 | |
| P281-1 | GGAGAGTCTGGATTCGT |
| P281-2 | GCGATGTCACCTATCGTTG |
In the operon analysis of tbrA and tbrB genes, the total RNA of strain CT-43 was prepared and reverse-transcribed into cDNA. RT-PCR was carried out with a primer pair designed to span the interval region between the tbrA ORF and tbrB ORF (Fig. 4A). To increase the copy number of the tbrB gene based on BMB2451, a recombinant BMB2451-tbrB was constructed by introducing the plasmid pEMB0603-tbrB into BMB2451. Supernatants of BMB2451 and BMB2451-tbrB cultures were sampled at different time points and subjected to TAA extraction as described above. The TAA level was quantified by HPLC and calibrated using the optical density of the cell culture at 600 nm (A600). All of the PCR products amplified in this study were verified by sequencing.
Enzymatic Assay of Aconitate Isomerase Activity of TbrA Protein
The gene tbrA was amplified from the genomic DNA of B. thuringiensis CT-43 and cloned into NdeI and XhoI sites of the expression vector pET28a to generate a recombinant plasmid pET28a-tbrA, which was then transformed into E. coli Rosetta to yield a recombinant Rosetta-TbrA for TbrA-inducible expression. The Rosetta-TbrA strain was cultured in 5 ml of LB medium with kanamycin and chloramphenicol at 37 °C for 4 h and transferred into 100 ml of LB medium with an appropriate amount of antibiotics in a ratio of 1:100 and cultured for 3 h, followed by the addition of 0.1 mm isopropyl-β-d-thiogalactoside (final concentration). The TbrA protein was induced at 28 °C for 6 h. Cells were harvested and resuspended in precooled 50 mm Tris-HCl buffer (pH 8.0) with 10% glycerol; the cell-free supernatant of the Rosetta-TbrA strain was collected by centrifugation after high pressure shaking at 4 °C. To test the aconitate isomerase activity of the cell-free extracts of the Rosetta-TbrA strain, CAA and TAA standards (adjusted with 5 m NaOH to pH 7.0) were used as substrates. Two control reactions, one lacking the substrate component and one lacking the cell-free extract component (enzyme), were also performed to calibrate the original presence of TAA and CAA in the cell-free extracts and the non-enzymatic isomerization between the two isomers. The reaction was conducted in a 500-μl test volume at 37 °C for 30 min and terminated by the addition of 20 μl of 6 m HCl. The reaction products were analyzed by HPLC in a mobile phase containing 10% methanol and 0.1% formic acid. The recombinant Rosetta-pET28a harboring the empty vector pET28a was treated with the same induction and culture operations as the negative control strain throughout the enzymatic assays.
Fluorescence Microscopy
SOE-PCR was performed to construct the tbrB-gfp fusion gene that comprised the promoter region of tbrA, the tbrB ORF, a 10-aa peptide linker-encoding sequence, and the gfp ORF (Fig. 7A). The fusion gene was introduced into host BMB171 by pHT304 vector to generate the recombinant BMB-TbrB-GFP. The bacteria was grown in LB medium at 28 °C for 24 h, and 1 ml of the cell culture was centrifuged, washed with 1× PBS buffer (pH 7.4), and resuspended in 0.5 ml of PBS. The strain BMB171-GFP, which constitutively expresses GFP protein, was used as a control and subjected to the same treatments. Cells were visualized and photographed using a Nikon structured illumination superresolution microscope (N-SIM). Images were processed using NIS-Elements advanced research microscope imaging software.
Nematode Bioassay
The root knot nematodes M. incognita were maintained on tomato roots (Solanum lycopersicum L., cv. Rutgers). Sixty days after nematode inoculation, egg masses of M. incognita were collected and sterilized with sodium hypochlorite and hatched in sterilized deionized water into J2s at 20 °C for 3 days. In the bioassay performed in 96-well plates, each well contained 20–40 J2s in a 100-μl test volume. The nematicidal effects of CT-A (TAA) extracts and CAA and TAA commercial standards were tested. Sterilized deionized water was used as a control. Treatments and bioassays were performed in triplicate. After 72 h, both total and living nematode numbers were recorded.
Bioinformatics Analysis
The antiSMASH database (version 3.0.5) and the batch Conserved Domain Search service (CD-Search) were used to identify secondary metabolite biosynthetic gene clusters and isomerase genes on plasmid pCT281, respectively. The protein sequence alignment of TbrA from B. thuringiensis CT-43 and PrpF from S. oneidensis MR-1 was performed using ClustalX and highlighted using ESPript version 3.0. The transmembrane helix prediction of the TbrB protein was conducted using the TMHMM Server version 2.0. Protein BLAST and nucleotide BLAST were used to analyze the distribution of the TAA biosynthetic gene cluster among all genome sequences of the B. cereus group strains in GenBankTM.
Statistical Analysis
One-way ANOVA with Tukey's honest significant difference test was performed to identify statistically significant differences in TAA production by B. thuringiensis strains. In nematode bioassays, the LC50 values were calculated by probit analysis. IBM SPSS (Statistical Package for the Social Sciences) software version 20.0 was used for these analyses.
Author Contributions
C. D., S. C., and M. S. designed the research. C. D., S. C., X. S., and X. N. performed the experiments and analyzed the data. C. D. wrote the paper. J. Z. and M. S. revised the paper. M. S., D. P., L. R., and Y. D. provided suggestions. All authors reviewed the results and approved the final version of the manuscript.
Acknowledgment
We thank Prof. Q. J. Kang (Shanghai Jiao Tong University) for suggestions on structure identification.
This work was supported by the China 948 Program of the Ministry of Agriculture (Grant 2016-X21), National Natural Science Foundation of China Grants 31670085 and 31171901, and National Key Research and Development Program of China Grant 2017YFC204521201. The authors declare that they have no conflicts of interest with the contents of this article.
- TAA
- trans-aconitic acid
- CAA
- cis-aconitic acid
- TCA
- tricarboxylic acid
- TS
- temperature-sensitive
- J2s
- second-stage juveniles
- ACO
- aconitase
- SOE-PCR
- splicing overlap extension PCR
- ANOVA
- analysis of variance
- aa
- amino acid(s).
References
- 1. Peng D., Lin J., Huang Q., Zheng W., Liu G., Zheng J., Zhu L., and Sun M. (2016) A novel metalloproteinase virulence factor is involved in Bacillus thuringiensis pathogenesis in nematodes and insects. Environ. Microbiol. 18, 846–862 [DOI] [PubMed] [Google Scholar]
- 2. Ruan L., Crickmore N., Peng D., and Sun M. (2015) Are nematodes a missing link in the confounded ecology of the entomopathogen Bacillus thuringiensis? Trends Microbiol. 23, 341–346 [DOI] [PubMed] [Google Scholar]
- 3. Zheng J., Peng D., Chen L., Liu H., Chen F., Xu M., Ju S., Ruan L., and Sun M. (2016) The Ditylenchus destructor genome provides new insights into the evolution of plant parasitic nematodes. Proc. Biol. Sci. 10.1098/rspb.2016.0942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Luo X., Chen L., Huang Q., Zheng J., Zhou W., Peng D., Ruan L., and Sun M. (2013) Bacillus thuringiensis metalloproteinase Bmp1 functions as a nematicidal virulence factor. Appl. Environ. Microbiol. 79, 460–468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Sanahuja G., Banakar R., Twyman R. M., Capell T., and Christou P. (2011) Bacillus thuringiensis: a century of research, development and commercial applications. Plant Biotechnol. J. 9, 283–300 [DOI] [PubMed] [Google Scholar]
- 6. Bravo A., Likitvivatanavong S., Gill S. S., and Soberón M. (2011) Bacillus thuringiensis: a story of a successful bioinsecticide. Insect Biochem. Mol. Biol. 41, 423–431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Zheng Z., Zheng J., Zhang Z., Peng D., and Sun M. (2016) Nematicidal spore-forming Bacilli share similar virulence factors and mechanisms. Sci. Rep. 6, 31341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Luo H., Xiong J., Zhou Q., Xia L., and Yu Z. (2013) The effects of Bacillus thuringiensis Cry6A on the survival, growth, reproduction, locomotion, and behavioral response of Caenorhabditis elegans. Appl. Microbiol. Biotechnol. 97, 10135–10142 [DOI] [PubMed] [Google Scholar]
- 9. Hu Y., Platzer E. G., Bellier A., and Aroian R. V. (2010) Discovery of a highly synergistic anthelmintic combination that shows mutual hypersusceptibility. Proc. Natl. Acad. Sci. U.S.A. 107, 5955–5960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Iatsenko I., Nikolov A., and Sommer R. J. (2014) Identification of distinct Bacillus thuringiensis 4A4 nematicidal factors using the model nematodes Pristionchus pacificus and Caenorhabditis elegans. Toxins 6, 2050–2063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Iatsenko I., Boichenko I., and Sommer R. J. (2014) Bacillus thuringiensis DB27 produces two novel protoxins, Cry21Fa1 and Cry21Ha1, which act synergistically against nematodes. Appl. Environ. Microbiol. 80, 3266–3275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Lenane I. J., Bagnall N. H., Josh P. F., Pearson R. D., Akhurst R. J., and Kotze A. C. (2008) A pair of adjacent genes, cry5Ad and orf2–5Ad, encode the typical N- and C-terminal regions of a Cry5Aδ-endotoxin as two separate proteins in Bacillus thuringiensis strain L366. FEMS Microbiol. Lett. 278, 115–120 [DOI] [PubMed] [Google Scholar]
- 13. Liu Y., Ye W., Zheng J., Fang L., Peng D., Ruan L., and Sun M. (2014) High-quality draft genome sequence of nematocidal Bacillus thuringiensis Sbt003. Stand. Genomic Sci. 9, 624–631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Salehi Jouzani G., Seifinejad A., Saeedizadeh A., Nazarian A., Yousefloo M., Soheilivand S., Mousivand M., Jahangiri R., Yazdani M., Amiri R. M., and Akbari S. (2008) Molecular detection of nematicidal crystalliferous Bacillus thuringiensis strains of Iran and evaluation of their toxicity on free-living and plant-parasitic nematodes. Can. J. Microbiol. 54, 812–822 [DOI] [PubMed] [Google Scholar]
- 15. Wei J. Z., Hale K., Carta L., Platzer E., Wong C., Fang S. C., and Aroian R. V. (2003) Bacillus thuringiensis crystal proteins that target nematodes. Proc. Natl. Acad. Sci. U.S.A. 100, 2760–2765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Li X. Q., Wei J. Z., Tan A., and Aroian R. V. (2007) Resistance to root-knot nematode in tomato roots expressing a nematicidal Bacillus thuringiensis crystal protein. Plant Biotechnol. J. 5, 455–464 [DOI] [PubMed] [Google Scholar]
- 17. Li X. Q., Tan A., Voegtline M., Bekele S., Chen C. S., and Aroian R. V. (2008) Expression of Cry5B protein from Bacillus thuringiensis in plant roots confers resistance to root-knot nematode. Biol. Control 47, 97–102 [Google Scholar]
- 18. Zhang L., Yu J., Xie Y., Lin H., Huang Z., Xu L., Gelbic I., and Guan X. (2014) Biological activity of Bacillus thuringiensis (Bacillales: Bacillaceae) Chitinase against Caenorhabditis elegans (Rhabditida: Rhabditidae). J. Econ. Entomol. 107, 551–558 [DOI] [PubMed] [Google Scholar]
- 19. Garsin D. A., Sifri C. D., Mylonakis E., Qin X., Singh K. V., Murray B. E., Calderwood S. B., and Ausubel F. M. (2001) A simple model host for identifying Gram-positive virulence factors. Proc. Natl. Acad. Sci. U.S.A. 98, 10892–10897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Ruan L., Wang H., Cai G., Peng D., Zhou H., Zheng J., Zhu L., Wang X., Yu H., Li S., Geng C., and Sun M. (2015) A two-domain protein triggers heat shock pathway and necrosis pathway both in model plant and nematode. Environ. Microbiol. 17, 4547–4565 [DOI] [PubMed] [Google Scholar]
- 21. Liu X. Y., Ruan L. F., Hu Z. F., Peng D. H., Cao S. Y., Yu Z. N., Liu Y., Zheng J. S., and Sun M. (2010) Genome-wide screening reveals the genetic determinants of an antibiotic insecticide in Bacillus thuringiensis. J. Biol. Chem. 285, 39191–39200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Noel G. R. (1990) Evaluation of thuringiensin for control of Heterodera glycines on soybean. J. Nematol. 22, 763–766 [PMC free article] [PubMed] [Google Scholar]
- 23. Liu X., Ruan L., Peng D., Li L., Sun M., and Yu Z. (2014) Thuringiensin: a thermostable secondary metabolite from Bacillus thuringiensis with insecticidal activity against a wide range of insects. Toxins 6, 2229–2238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Altekar W. W., and Rao M. R. (1963) Microbiological dissimilation of tricarballylate and trans-aconitate. J. Bacteriol. 85, 604–613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Saffran M., and Prado J. L. (1949) Inhibition of aconitase by trans-aconitate. J. Biol. Chem. 180, 1301–1309 [PubMed] [Google Scholar]
- 26. Sugimoto T., Kato T., and Park E. Y. (2014) Functional analysis of cis-aconitate decarboxylase and trans-aconitate metabolism in riboflavin-producing filamentous Ashbya gossypii. J. Biosci. Bioeng. 117, 563–568 [DOI] [PubMed] [Google Scholar]
- 27. Rao M. R., and Altekar W. W. (1961) Aconitate isomerase. Biochem. Biophys. Res. Commun. 4, 101–105 [DOI] [PubMed] [Google Scholar]
- 28. Klinman J. P., and Rose I. A. (1971) Purification and kinetic properties of aconitate isomerase from Pseudomonas putida. Biochemistry 10, 2253–2259 [DOI] [PubMed] [Google Scholar]
- 29. Thompson J. F., Schaefer S. C., and Madison J. T. (1990) Determination of aconitate isomerase in plants. Anal. Biochem. 184, 39–47 [DOI] [PubMed] [Google Scholar]
- 30. Thompson J. F., Schaefer S. C., and Madison J. T. (1997) Role of aconitate isomerase in trans-aconitate accumulation in plants. J. Agric. Food Chem. 45, 3684–3688 [Google Scholar]
- 31. Brauer D., and Teel M. R. (1981) Metabolism of trans-aconitic acid in maize: I. purification of two molecular forms of citrate dehydrase. Plant Physiol. 68, 1406–1408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Miller R. E., and Cantor S. M. (1951) Aconitic acid, a by-product in the manufacture of sugar. Adv. Carbohydr. Chem. 6, 231–249 [DOI] [PubMed] [Google Scholar]
- 33. Kim M., Koh H. S., Obata T., Fukami H., and Ishii S. (1976) Isolation and identification of trans-aconitate as the antifeedant in barnyard grass against the brown planthopper Nilaparvata Lugens (Stål) (Homoptera: Delphacidae). Appl. Ent. Zool. 11, 53–57 [Google Scholar]
- 34. He J., Wang J., Yin W., Shao X., Zheng H., Li M., Zhao Y., Sun M., Wang S., and Yu Z. (2011) Complete genome sequence of Bacillus thuringiensis subsp. chinensis strain CT-43. J. Bacteriol. 193, 3407–3408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Sun M., and Yu Z. (1996) Characterization of insecticidal crystal proteins of Bacillus thuringiensis subsp. chinensis CT-43. Acta Microbiologica Sinica 36, 303–306 [PubMed] [Google Scholar]
- 36. Abad P., Gouzy J., Aury J. M., Castagnone-Sereno P., Danchin E. G., Deleury E., Perfus-Barbeoch L., Anthouard V., Artiguenave F., Blok V. C., Caillaud M. C., Coutinho P. M., Dasilva C., De Luca F., Deau F., et al. (2008) Genome sequence of the metazoan plant-parasitic nematode Meloidogyne incognita. Nat. Biotechnol. 26, 909–915 [DOI] [PubMed] [Google Scholar]
- 37. Jones J. T., Haegeman A., Danchin E. G., Gaur H. S., Helder J., Jones M. G., Kikuchi T., Manzanilla-López R., Palomares-Rius J. E., Wesemael W. M., and Perry R. N. (2013) Top 10 plant-parasitic nematodes in molecular plant pathology. Mol. Plant Pathol. 14, 946–961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Smith C. A., O'Maille G., Want E. J., Qin C., Trauger S. A., Brandon T. R., Custodio D. E., Abagyan R., and Siuzdak G. (2005) METLIN, a metabolite mass spectral database. Ther. Drug Monit. 27, 747–751 [DOI] [PubMed] [Google Scholar]
- 39. Luo Y., Ruan L. F., Zhao C. M., Wang C. X., Peng D. H., and Sun M. (2011) Validation of the intact zwittermicin A biosynthetic gene cluster and discovery of a complementary resistance mechanism in Bacillus thuringiensis. Antimicrob. Agents Chemother. 55, 4161–4169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Blin K., Medema M. H., Kottmann R., Lee S. Y., and Weber T. (2017) The antiSMASH database, a comprehensive database of microbial secondary metabolite biosynthetic gene clusters. Nucleic Acids Res. 45, D555–D559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Bernheim F. (1928) The specificity of the dehydrases. Biochem. J. 22, 1178–1192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Marchler-Bauer A., and Bryant S. H. (2004) CD-Search: protein domain annotations on the fly. Nucleic Acids Res. 32, W327–W331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Grimek T. L., and Escalante-Semerena J. C. (2004) The acnD Genes of Shewenella oneidensis and Vibrio cholerae encode a new Fe/S-dependent 2-methylcitrate dehydratase enzyme that requires prpF Function in vivo. J. Bacteriol. 186, 454–462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Garvey G. S., Rocco C. J., Escalante-Semerena J. C., and Rayment I. (2007) The three-dimensional crystal structure of the PrpF protein of Shewanella oneidensis complexed with trans-aconitate: insights into its biological function. Protein Sci. 16, 1274–1284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Tate C. G., Muiry J. A., and Henderson P. J. (1992) Mapping, cloning, expression, and sequencing of the rhaT gene, which encodes a novel L-rhamnose-H+ transport protein in Salmonella typhimurium and Escherichia coli. J. Biol. Chem. 267, 6923–6932 [PubMed] [Google Scholar]
- 46. Tate C. G., and Henderson P. J. (1993) Membrane topology of the l-rhamnose-H+ transport protein (RhaT) from Enterobacteria. J. Biol. Chem. 268, 26850–26857 [PubMed] [Google Scholar]
- 47. Arantes O., and Lereclus D. (1991) Construction of cloning vectors for Bacillus thuringiensis. Gene 108, 115–119 [DOI] [PubMed] [Google Scholar]
- 48. He J., Shao X., Zheng H., Li M., Wang J., Zhang Q., Li L., Liu Z., Sun M., Wang S., and Yu Z. (2010) Complete genome sequence of Bacillus thuringiensis mutant strain BMB171. J. Bacteriol. 192, 4074–4075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Cordes T., Michelucci A., and Hiller K. (2015) Itaconic acid: the surprising role of an industrial compound as a mammalian antimicrobial metabolite. Annu. Rev. Nutr. 35, 451–473 [DOI] [PubMed] [Google Scholar]
- 50. Cai H., and Clarke S. (1999) A novel methyltransferase catalyzes the methyl esterification of trans-aconitate in Escherichia coli. J. Biol. Chem. 274, 13470–13479 [DOI] [PubMed] [Google Scholar]
- 51. Misra S., Sanyal T., Sarkar D., Bhattacharya P. K., and Ghosh D. K. (1989) Evaluation of antileishmanial activity of trans-aconitic acid. Biochem. Med. Metab. Biol. 42, 171–178 [DOI] [PubMed] [Google Scholar]
- 52. Kar S., Kar K., Bhattacharya P. K., and Ghosh D. K. (1993) Experimental visceral leishmaniasis: role of trans-aconitic acid in combined chemotherapy. Antimicrob. Agents Chemother. 37, 2459–2465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Garcia Ede F., de Oliveira M. A., Godin A. M., Ferreira W. C., Bastos L. F., Coelho Mde M., and Braga F. C. (2010) Antiedematogenic activity and phytochemical composition of preparations from Echinodorus grandiflorus leaves. Phytomedicine 18, 80–86 [DOI] [PubMed] [Google Scholar]
- 54. Chen S. T., Lee I. S., and Chen Y. C. (April 6, 2015) Trans-aconitic acid compounds and uses thereof for inhibiting phosphodiesterase 7. U. S. Patent 20150104531
- 55. Klinman J. P., and Rose I. A. (1971) Mechanism of the aconitate isomerase reaction. Biochemistry 10, 2259–2266 [DOI] [PubMed] [Google Scholar]
- 56. Raymond B., Johnston P. R., Nielsen-LeRoux C., Lereclus D., and Crickmore N. (2010) Bacillus thuringiensis: an impotent pathogen? Trends Microbiol. 18, 189–194 [DOI] [PubMed] [Google Scholar]
- 57. Zhu L., Peng D., Wang Y., Ye W., Zheng J., Zhao C., Han D., Geng C., Ruan L., He J., Yu Z., and Sun M. (2015) Genomic and transcriptomic insights into the efficient entomopathogenicity of Bacillus thuringiensis. Sci. Rep. 5, 14129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Ivanova N., Sorokin A., Anderson I., Galleron N., Candelon B., Kapatral V., Bhattacharyya A., Reznik G., Mikhailova N., Lapidus A., Chu L., Mazur M., Goltsman E., Larsen N., D'Souza M., et al. (2003) Genome sequence of Bacillus cereus and comparative analysis with Bacillus anthracis. Nature 423, 87–91 [DOI] [PubMed] [Google Scholar]
- 59. Zhou J., Li X., Jiang Y., Wu Y., Chen J., Hu F., and Li H. (2011) Combined effects of bacterial-feeding nematodes and prometryne on the soil microbial activity. J. Hazard. Mater. 192, 1243–1249 [DOI] [PubMed] [Google Scholar]
- 60. Middendorf P. J., and Dusenbery D. B. (1993) Fluoroacetic acid is a potent and specific inhibitor of reproduction in the nematode Caenorhabditis elegans. J. Nematol. 25, 573–577 [PMC free article] [PubMed] [Google Scholar]
- 61. McKinney J. D., Höner zu Bentrup K., Muñoz-Elías E. J., Miczak A., Chen B., Chan W. T., Swenson D., Sacchettini J. C., Jacobs W. R. Jr., and Russell D. G. (2000) Persistence of Mycobacterium tuberculosis in macrophages and mice requires the glyoxylate shunt enzyme isocitrate lyase. Nature 406, 735–738 [DOI] [PubMed] [Google Scholar]
- 62. Lourenço-Tessutti I. T., Souza Junior J. D., Martins-de-Sa D., Viana A. A., Carneiro R. M., Togawa R. C., de Almeida-Engler J., Batista J. A., Silva M. C., Fragoso R. R., and Grossi-de-Sa M. F. (2015) Knock-down of heat-shock protein 90 and isocitrate lyase gene expression reduced root-knot nematode reproduction. Phytopathology 105, 628–637 [DOI] [PubMed] [Google Scholar]
- 63. Hanahan D. (1983) Studies on transformation of Escherichia coli with plasmids. J. Mol. Biol. 166, 557–580 [DOI] [PubMed] [Google Scholar]