Abstract
Bone metastases remain as a serious health concern because of limited therapeutic options. Here, we report that crosstalk between ROR1-HER3 and the Hippo-YAP pathway promotes breast cancer bone metastasis in a long noncoding RNA-dependent fashion. Mechanistically, the orphan receptor tyrosine kinase ROR1 phosphorylates HER3 at a previously unidentified site Tyr1307, upon neuregulin stimulation, independently of other ErbB family members. p-HER3 Tyr1307 recruits the LLGL2-MAYA-NSUN6 RNA-protein complex to methylate Hippo/MST1 at Lys59. This methylation leads to MST1 inactivation and activation of YAP target genes in tumor cells, which elicits osteoclast differentiation and bone metastasis. Furthermore, increased ROR1, p-HER3 Tyr1307 and MAYA levels correlate with tumor metastasis and unfavorable outcomes. Our data provide insights into the mechanistic regulation and linkage of the ROR1-HER3 and Hippo-YAP pathway in cancer-specific context, and also imply valuable therapeutic targets for bone metastasis and possible therapy-resistant tumors.
Keywords: Bone metastasis, Breast cancer, Orphan receptor tyrosine kinase, ROR1, Hippo-YAP pathway, long noncoding RNA, MAYA, CTGF, MST1, Non-histone protein methylation, Locked nucleic acid
Introduction
Bone metastasis continues to be a severe health concern although significant progress has been made. When bone metastatic lesions develop, bone residential tumor cells produce a cohort of osteolytic factors1. These osteolytic factors, including connective tissue growth factor (CTGF), trigger differentiation of osteoclast precursors derived from monocytes2. CTGF is one of the signature genes that are significantly upregulated in patients with relapse to the bone compared to patients with relapse elsewhere based on gene expression analysis3.
The Hippo-YAP pathway controls cell proliferation, tumorigenesis, chemoresistance, and metastasis in human tumors4–6. The core Hippo cassette comprises MST1/2 (Hippo), LATS1/2 (Large Tumor Suppressor kinase1/2), and YAP (yes-associated protein) signaling cascade7,8. MST1/2 is a protein serine/threonine kinase that negatively regulates cell growth, and mutation of the Hippo genes leads to organ overgrowth9,10 In mammals, MST1/2 phosphorylates LATS1/2, which further phosphorylates YAP for cytoplasmic sequestration and degradation11. When MST1/2 kinase activity is inhibited, by an unknown mechanism, YAP accumulates in the nucleus for transcriptional activation12.
The receptor tyrosine kinase (RTKs)-like orphan receptors (RORs), like all other RTKs13, possesses extracellular ligand-binding domain, yet their ligands, cellular effects, and downstream signaling pathways are largely unknown14. The two ROR family members, ROR1 and ROR2, were originally found to be involved in skeletal, cardiorespiratory, and neurological development15,16. Rising evidence has indicated that both ROR proteins are highly expressed in multiple human cancer types, including leukemia, ovarian cancer, and breast cancer17–22. Although ROR1 treated as a pseudokinase originally based on the observation that ROR1 was unable to be autophosphorylated23,24, some studies have also demonstrated the moderate autocatalytic kinase activity of ROR1 in vitro25–27. Mechanisms of the enzymatic activity, substrates, and downstream signaling pathway of ROR1 are still elusive.
Cytoplasmic long noncoding RNAs (lncRNAs) are involved in regulating mRNA stabilization and transport as well as microRNA sponging28–31. It has also been suggested that cytoplasmic lncRNAs are important mediators of intracellular signaling pathways. For example, lnc-DC modulates the phosphorylation status of STAT332, NKILA regulates IkB phosphorylation and degradation33, and LINK-A regulates HIF1α signaling34.
Here, we demonstrate that neuregulin-1 (NRG1) triggers the heterodimerization of ROR1 and HER3, leading to HER3 phosphorylation by ROR1 at Tyr1307. Phosphorylated HER3 recruits the adaptor protein LLGL2, lncRNA MAYA (MST1/2-Antagonizing for YAP Activation), and methyltransferase NSUN6, which methylates MST1 at Lys59. The methylation abolishes MST1 kinase activity and activates YAP and target genes. These events lead to cancer cell-induced osteoclast differentiation and bone resorption. Our studies identify the crosstalk between the ROR1/HER3-LLGL2-MAYA-NSUN6 signaling axis and the Hippo-YAP pathway, and suggest a promising therapeutic strategy for bone metastatic patients.
RESULTS
ROR1 promotes the colonization and growth of breast cancer cells within the bone
Enhanced ROR1level has been observed in many blood and solid malignancies. To access its function and regulation under tumor-specific context, we first surveyed the genetic alterations of ROR1 gene. In sarcoma, ovarian, pancreatic, breast and lung cancer types, the ROR1 locus is amplified in 2–6% of patients (Fig. 1a). In breast cancer, ROR1 locus is amplified in triple-negative breast cancer (TNBC) subtype compared to the status of other biomarkers (Supplementary Fig. 1a). ROR1 expression is significantly upregulated in TNBC compared to non-TNBC subtypes (Fig. 1b and Supplementary Fig. 1b). Immunohistochemical (IHC) staining of breast cancer tissue microarrays (TMAs) revealed that high levels of ROR1 correlated with the TNBC subtype and poor survival (Fig. 1c, d and Supplementary Table 1). Furthermore, ROR1 expression is more correlated with non-small cell lung carcinoma (NSCLC) than with small cell lung carcinoma (Supplementary Fig. 1c). To enable functional characterization of ROR1, we generated ROR1 knockout (KO) breast cancer cell lines using CRISPR/Cas9 technology (Supplementary Fig. 1d, e). Two individual clones of ROR1 KO breast cancer cell line exhibited reduced proliferation (Supplementary Fig. 1f, g). ROR1 KO also inhibited migration and invasion of breast cancer cells and overexpression of wild-type (WT) ROR1, but not the kinase dead K506A mutant27, rescued the migration and invasion of ROR1 KO cells (Supplementary Fig. 1h–k).
Figure 1. ROR1 promotes the colonization and growth of breast cancer cells within the bone.
(a) Distribution of alteration frequency of ROR1 in multiple cancer types. (b) Box plot comparing ROR1 expression in TNBC, ERPR−/HER2+, ERPR+/HER2− and ERPR+/HER2+ breast cancer subtypes (n=119, 30, 482, 80 tumors respectively, one-way ANOVA). The boxes show the median±1 quartile, with whiskers extending to the most extreme data point within 1.5 interquartile ranges from the box boundaries. (c) Immunohistochemical (IHC) detection of ROR1 in TNBC, ERPR−/HER2+, ERPR+/HER2− and ERPR+/HER2+ breast cancer subtypes (n=23, 72, 17 and 30 tumors respectively; median, one-way ANOVA). (d) Survival analysis of ROR1 low and high breast cancer patients (top, scale bars: 40 μm; bottom, n=58 and 92 patients respectively, log rank test). (e) Heatmap representing color-coded expression levels of 57 YAP1 target genes in parental and ROR1 KO cells. (f) Immunoblotting (IB) detection of indicated proteins in MDA-MB-231 or BoM-1833 cells. (g) CTGF ELISA assay in parental and ROR1 KO BoM-1833 cells. (h and i) Osteoclast differentiation assays in the presence of M-CSF only, M-CSF+RANKL, combined M-CSF+RANKL and conditioned media (CM) from ROR1 KO BoM-1833 cells or KO cells transfected with indicated plasmids (h, scale bars: 200 μm; i, quantification). (j) Quantification of osteoclast differentiation in the presence of M-CSF only, M-CSF+RANKL, combined M-CSF+RANKL and conditioned media (CM) from ROR1 KO BoM-1833 cells or CM from ROR1 KO BoM-1833 cells KO supplemented with PBS or recombinant CTGF (50 ng ml−1). (k–m) Representative BLI images (k), BLI signal quantification (l, n=10 mice per group) and metastatic tumor numbers (m) of mice intracardially injected with parental or ROR1 KO BoM-1833 cells at week 5. For e, g, i, j, l and m, mean ± s.e.m. were derived from n=3 independent experiments (n.s., p>0.05, *p<0.05, **p<0.01 and ***p<0.001, two-tailed paired Student’s t-test). Unprocessed original scans of all blots with size marker for f are shown in Supplementary Fig. 9. Statistics source data for d, h–i and k–l are in Supplementary Table 8.
It has recently been reported that ROR1 mediates alternative Wnt signaling to activate the YAP pathway for organ development35, which prompts us to examine the expression of YAP1-regulated genes36–39 in ROR1 KO cells. Significantly, the expression of 37 out of the 57 YAP1-regulated genes was downregulated by depletion of ROR1 (Fig. 1e). Activation of YAP has been shown to promote tumor progression and metastasis40,41. Furthermore, YAP1-regulated genes including CTGF may play important roles in promoting osteolysis during bone metastatic lesion development42. Consistently, we found hyper-activation of YAP pathway and elevated expression of CTGF in bone metastatic BoM-1833 cells3 compared to parental MDA-MB-231 cells. Interestingly, the ROR1 protein level is upregulated in BoM-1833 cells, and ROR1 KO reduced CTGF production (Fig. 1f, g). Then we determined the role of ROR1 in cancer cell-induced osteoclast differentiation. We found that the differentiation of osteoclast precursors to mature osteoclasts was stimulated with conditioned media (CM) from parental but not ROR1 KO BoM-1833 cells, and this impaired osteoclast differentiation was rescued by CM from ROR1 KO cells expressing WT ROR1 but not K506A mutant (Fig. 1h, i). Interestingly, CM from ROR1 KO BoM-1833 cells supplemented with recombinant CTGF rescued osteoclast differentiation (Supplementary Fig. 1l and Fig. 1j).
Next, we aimed to determine the role of ROR1 in breast cancer bone metastasis. BoM-1833 cells, containing a stably-expressed firefly luciferase reporter, were inoculated to nude mice via intra-cardiac injection, and metastatic progression was monitored weekly by Bioluminescence imaging (BLI). The tumor burden of mouse limbs was greatly reduced by ROR1 KO (Fig. 1k, l). The bone metastatic lesion number was also decreased when ROR1 was knocked out (Fig. 1m). Therefore, these data suggest the important role of ROR1 in promoting the colonization and growth of breast cancer cells within the bone through regulation of Hippo-YAP pathway.
ROR1 phosphorylates HER3 at Tyr1307 in an ErbB-independent manner
To identify the potential substrates or regulators of ROR1, we immunoprecipitated endogenous ROR1 and identified ROR1-binding proteins by mass spectrometry (MS). Interestingly, we found that HER3 was the top protein that binds ROR1 (Supplementary Fig. 2a and Supplementary Table 2). The ROR1-associated HER3 harbored a previously unknown phosphorylation site of Tyr1307 (Fig. 2a and Supplementary Table 2). Hence, we generated phosphorylation-specific antibodies targeting p-HER3 (Tyr1307) (Supplementary Fig. 2b) for further cellular and tissue studies.
Figure 2. ROR1-dependent phosphorylation of HER3 at Tyr1307 correlates with breast cancer clinical parameters.
(a) Annotated MS/MS spectrum assigned to the HER3 peptide AFQGPGHQAPHVH[p]YAR, at 926.924 Da. Data acquired from analysis of the tryptic digest by high-sensitivity LC-MS/MS on an Orbitrap Elite high-resolution mass spectrometer. (b) Immunoprecipitation (IP) and IB detection of indicated proteins in MDA-MB-231 cells with NRG1 treatment. (c and d) IP followed by IB detection of HER3 phosphorylation and HER3-ROR1 interaction in MDA-MB-231cells without transfection (c), and pre-treated with Dacomitinib (PF299, 100 nM) (d) followed by NRG1 treatment. (e) IB detection of indicated proteins in cells transfected with indicated siRNAs followed by NRG1 treatment. (f and g) IP and IB detection of indicated proteins in MDA-MB-231 cells transfected with indicated siRNAs (f) or 32D cells transfected with indicated expression vectors (g) followed by NRG1 stimulation. (h and i) In vitro kinase assay was performed using His6-HER3 intracellular domain (ICD) WT or Y1307F mutant and FLAG-tagged ROR1 WT or extracellular domain (ECD) (h) or His6-FL HER3, GST-tagged WT ROR1 ICD or K506A mutant (i). (j) IP and IB detection of indicated proteins in MDA-MB-231 cells transfected with indicated plasmids followed by NRG1 treatment. (k) IHC staining intensity of p-HER3 (Tyr1307) in TNBC, ERPR-/HER2+, ERPR+/HER2- and ERPR+/HER2+ breast cancer subtypes (n=23, 72 17 and 30 patients respectively, median, one-way ANOVA). (l) Kaplan-Meier survival analysis of p-HER3 (Tyr1307) low and high breast cancer patients by IHC staining (top, scale bars: 40 μm; bottom, n=43 and 107 patients respectively, log rank test). (m) Correlation analysis showing the positive correlation of staining intensity of ROR1 (Fig. 1c) with that of p-HER3 (Tyr1307) within the TNBC subgroup. Fisher’s exact test was used (n=23 patients, ***p<0.001; R2=correlation coefficient). Unprocessed original scans of all blots with size marker are shown in Supplementary Fig. 9. Statistics source data for l are in Supplementary Table 8.
It has been well-established that HER3 forms heterodimers with EGFR and HER2 in response to NRG1 and plays important roles in a variety of cancer types43,44. We hypothesized that ROR1 and HER3 might form heterodimers, and found that HER3 associated with EGFR, HER4 and ROR1 upon NRG1 stimulation (Fig. 2b). NRG1 triggered the phosphorylation at previously known HER3 tyrosine sites (Tyr1197, Tyr1222, Tyr1289, and Tyr1328)45 as well as at a previously unknown Tyr1307 site (Fig. 2c). Although the pan-EGFR inhibitor Dacomitinib (PF299804) abolished HER3 phosphorylation at those 4 tyrosine sites, neither tyrosine phosphorylation of Tyr1307 nor the ROR1-HER3 interaction was affected by this inhibitor (Fig. 2d). In contrast, ROR1 knockdown abolished phosphorylation of HER3 at Tyr1307 but not at the EGFR-dependent phosphorylation sites (Fig. 2e).
Next, we found that knockdown of EGFR showed minimal effects on ROR1-HER3 interaction or the status of p-HER3 (Tyr1307) (Fig. 2f). NRG1 stimulation triggered interaction between exogenous ROR1 and HER3, as well as p-HER3 (Tyr1307) in ERBB-null 32D cells (Fig. 2g), suggesting a possibility that ROR1 may directly phosphorylate HER3. In vitro kinase assay showed that full-length (FL) ROR1 but not extracellular domain (ECD) phosphorylated the HER3 intracellular domain (ICD) at Tyr1307 site (Fig. 2h). Consistently, bacterially-expressed ICD of WT ROR1, but not K506A mutant phosphorylates HER3 at Tyr1307 in vitro (Fig. 2i). In breast cancer cells, expression of ROR1 K506A mutant abolished Tyr1307 phosphorylation of HER3 upon NRG1 stimulation (Fig. 2j).
It has been reported that ROR1 is tyrosine phosphorylated by c-MET and SRC at the proline-rich domain and the kinase domain, respectively46. Our data indicated that neither the tyrosine phosphorylation of ROR1 nor the ROR1-SRC/MET interaction was affected by NRG1 stimulation (Supplementary Fig. 2c). Although ROR1 tyrosine phosphorylation was abolished by SRC inhibitor, Saracatinib (Sara.), the NRG1-induced p-HER3 (Tyr1307) was not significantly affected (Supplementary Fig. 2d). In 32D cells, ROR1 exhibited undetectable tyrosine phosphorylation and association with SRC (Supplementary Fig. 2e), suggesting that the NRG1-triggered ROR1-HER3 pathway is independent of EGFR-SRC-MET signaling in breast cancer cells.
High levels of HER3 expression correlate with unfavorable outcomes of TNBC patients47,48 To ascertain whether p-HER3 (Tyr1307) promotes proliferation and invasion of breast cancer cells, we knocked out HER3 and found that HER3 KO impaired cell proliferation and mobility (Supplementary Fig. 2f–k). p-HER3 (Tyr1307) is highly increased in TNBC compared to other breast cancer subtypes (Fig. 2k). Further, the p-HER3 (Tyr1307) correlates with breast cancer patient outcomes (Fig. 2l). Interestingly, the level of ROR1 in TNBC strongly correlates with p-HER3 Tyr1307 status (R2 = 0.839) (Fig. 2m). Furthermore, the p-HER3 (Tyr1307) is elevated in lung adenocarcinomas compared to normal lung tissues (Supplementary Fig. 2l); lung adenocarcinomas at metastatic stage (TnN>0M≥0) showed an increased p-HER3 (Tyr1307) level compared to non-metastatic stage (TnN0M0) (Supplementary Fig. 2l). High level of p-HER3 (Tyr1307) also correlates with unfavorable outcomes for lung adenocarcinoma patients (Supplementary Fig. 2m).
Crosstalk between ROR1-HER3 and Hippo-YAP pathway
Phosphorylation of a receptor tyrosine kinase triggers the recruitment of proteins that contain an SH2 domain to mediate downstream signaling49. SH2 domain containing protein BCAR3 (breast cancer anti-estrogen resistance 3) identified by MS (see Supplementary Fig. 2a) could be recruited to p-HER3 (Tyr1307). To test this, we mutated HER3 Tyr1307 to Phe (Y1307F) and Tyr1197/Tyr1222/Tyr1289/Tyr1328 to Phe (4Y-F). We found that the expression of the HER3 Y1307F, but not 4Y-F mutant abolished HER3-BCAR3 interaction upon NRG1 stimulation (Fig. 3a). Consistently, the deletion of the SH2 domain of BCAR3 abolished HER3-BCAR3 interaction but showed no effect on p-HER3 (Tyr1307) (Fig. 3b). Next, we synthesized biotinylated-HER3 peptides harboring either phosphorylated- or unphosphorylated- Tyr1197, Tyr1222, Tyr1289, Tyr1328, and Tyr1307 residues with their corresponding flanking amino acid sequences. Streptavidin pull-down showed that the HER3 peptide harboring p-Tyr1307, but not other p-Tyr residues, strongly interacts with BCAR3 (Fig. 3c).
Figure 3. Crosstalk between ROR1-HER3 and the Hippo-YAP pathway.
(a) IP followed by IB detection of ROR1-HER3 interaction, p-HER3 (Tyr1307) phosphorylation, and p-HER3-BCAR3 interaction in MDA-MB-231 cells transfected with indicated plasmids followed by NRG1 treatment. (b) IP followed by IB detection of indicated proteins in MDA-MB-231 cells transfected with indicated plasmids followed by NRG1 treatment. (c) Peptide pull-down assay using phosphorylated or unphosphorylated peptides harboring indicated amino acid with corresponding flanking sequence were subjected to IB detection using indicated antibodies. (d) In vitro pull-down assay was performed by incubating BCAR3 with indicated recombinant proteins. BCAR3-associated proteins were detected by IB using indicated antibodies. (e) In vitro phosphorylation assay was performed using FLAG-tagged WT ROR1 or extracellular domain (ECD) and GST-tagged LLGL2. (f) IP and IB detection of indicated proteins in cells transfected with indicated siRNAs followed by NRG1 stimulation. (g) GST pull-down assay was performed by incubating either unphosphorylated (No-P)- or Tyr499 phosphorylated (P)-LLGL2 with indicated recombinant proteins. The associated proteins were detected by IB using indicated antibodies. Positive interactions are shown in red. (h) IB detection of indicated proteins in MDA-MB-231 cells treated with NRG1 according to indicated time course. Unprocessed original scans of all blots with size marker are shown in Supplementary Fig. 9.
We further investigated the downstream signaling pathways of BCAR3. Recombinant protein pull-down assay indicated that BCAR3 directly binds LLGL2 (lethal giant larvae homolog 2) but not other proteins identified by MS (Fig. 3d). In Drosophila, the adaptor protein LLGL2 plays a critical role in mediating the Hippo-YAP pathway that triggered by cell-cell junctions50. Our MS confirmed the associations of ROR1, HER3, and BCAR3 with LLGL2 (Supplementary Fig. 3a). Furthermore, a panel of Hippo complex proteins [STK4 (MST1), SAV1, MOB1, and RASSF1 (Ras association domain family member 1)] were observed to bind both ROR1 and LLGL2 (Supplementary Fig. 3a and Supplementary Table 3). MS also revealed the LLGL2 phosphorylation at Tyr499, thus a site-specific antibody against p-LLGL2 (Tyr499) was generated (Supplementary Fig. 3b, c). ROR1 was the most plausible candidate for LLGL2 (Tyr499) phosphorylation in the context of this specific signaling cascade, and in vitro kinase assay confirmed this hypothesis (Fig. 3e). Consistently, overexpression of the kinase-dead mutant of ROR1 abolished phosphorylation of LLGL2 at Tyr499 in NRG1-treated cells (Supplementary Fig. 3d). Knockdown of BCAR3 eliminated the interaction between HER3 and LLGL2 as well as p-LLGL2 (Tyr499) (Fig. 3f). GST-LLGL2 pull-down showed that both nonphosphorylated (No-P-) and phosphorylated (P-) LLGL2 binds BCAR3, but only P-LLGL2 (Tyr499) binds MST1 (Fig. 3g).
A previously unknown di-methylation modification of MST1 at Lys59 [MST1 (Lys59me2)] was also identified from MS and confirmed by methylation-specific antibodies (Supplementary Fig. 3e, Supplementary Table 3 and Supplementary Fig. 3f). Lysophosphatidic acid (LPA) has been shown to trigger YAP1 activation via G-protein-coupled receptor signaling51. Our data showed that NRG1, but not LPA, triggered MST1 (Lys59me2), and MST1 methylation status negatively correlates with MST1 (Thr183) phosphorylation and YAP1 (Ser127) phosphorylation (Fig. 3h and Supplementary Fig. 3g).
LncRNA MAYA is involved in YAP activation
We found a putative protein methyltransferase, NSUN6, in both ROR1 and LLGL2 pull-down experiments (see Supplementary Fig. 2a and 3a). NSUN6 is an RNA-binding ribosomal protein that can mediate RNA methylation52. Therefore, we hypothesized that lncRNA molecules could associate with NSUN6 to regulate MST1 methylation. To identify the lncRNAs that might be involved in the YAP signaling pathway, we transfected the human Lincode® siRNA library into MCF-7 cells that were engineered with a TEAD-driven luciferase reporter, and subsequently the relative YAP1 activity was determined. We set up a threshold at log2 ≤ -3 (red line) to pinpoint 40 lncRNAs that were potentially required for YAP1-dependent transcription (Fig. 4a and Supplementary Table 4).
Figure 4. LncRNA MAYA is required for activation of YAP.
(a) Screening of human Lincode® siRNA library in MCF-7 cells. (b) Box plots comparing MAYA expression in matched (top) and unmatched (bottom) breast tumors and normal tissues (top, n=105 patients for each group; bottom, n=837 and 105 patients respectively, Wilcoxon test). (c) MAYA expression in breast tumor or normal tissue (NBT). Left, RNAScope®, scale bars: 40 μm; right, quantification of training (n=54 tumors and 60 NBTs) and validation (n=180 tumors and 30 NBTs) sets based on 2 independent experiments. (d) MAYA expression in non-metastatic (TnN0M0) and metastatic (TnN>0M≥0) breast tumors (n=38 and 62 tumors respectively). (e) Kaplan-Meier survival analysis of MAYA low and high breast cancer patients (n=57 and 103 patients respectively, log rank test). (f) ChIP-qPCR detection of YAP1 occupancy on the promoter of YAP1 target genes in NRG1-treated cells. (g) RT-qPCR detection of indicated genes in cells transfected with indicated siRNAs followed by NRG1 treatment. (h) Box plot comparing the expression level of 29 YAP1 target genes in MDA-MB-231 cells transfected with indicated siRNAs (mean ± s.e.m. were derived from n=3 independent experiments, ***p<0.001, Wilcoxon test). (i) IB detection of indicated proteins in MAYA knockdown MDA-MB-231 cells. (j) Quantification of CTGF in BoM-1833 cells harboring indicated shRNAs. (k and l) TRAP staining (k, scale bars: 200 μm) and quantification (l) of osteoclast differentiation upon treatment with M-CSF, M-CSF+RANKL or combined M-CSF+RANKL and CM from indicated siRNA-transfected BoM-1833 cells supplemented with CTGF (50 ng ml−1) or CCN3 (50 ng ml−1). (m) Measurement of helical peptides released from human bone plates incubated with primary osteoclasts treated as in k. For b and h, the boxes show the median±1 quartile, with whiskers extending to the most extreme data point within 1.5 interquartile ranges from the box boundaries. For d, f, g, j, l and m, mean ± s.e.m. were derived from n=3 independent experiments (n.s., p>0.05, *p<0.05 and **p<0.01, two-tailed paired Student’s t-test). Unprocessed original scans of all blots with size marker are shown in Supplementary Fig. 9. Statistics source data for c and k–l are in Supplementary Table 8.
We then picked up the top 5 lncRNAs for RNA pull-down experiments to identify whether they bind the Hippo-YAP pathway proteins. Interestingly, LOC645249 (MNX1-AS1) binds LLGL2, MST1, SAV1, MOB1, NSUN6 and RASSF1, suggesting that it may mediate the YAP pathway activation; thus we renamed this lncRNA as MST1/2-Antagonizing lncRNA for YAP Activation (MAYA) (Supplementary Fig. 4a and Supplementary Table 5). We performed a 5′- and 3′-Rapid amplification of cDNA ends (RACE) assay to determine the entire sequence of MAYA, confirming that the sequence detected in MDA-MB-231 cells is identical to that archived in RefSeq database (Supplementary Fig. 4b). The expression pattern and splicing variation of MAYA in human tissues and breast cancer cells were validated by Northern blot (Supplementary Fig. 4c, d).
The Cancer Genome Atlas (TCGA) database mining revealed that MAYA expression is upregulated in breast cancer compared to normal tissue in both matched and unmatched sample pools (Fig. 4b). In addition, RNAscope® analysis confirmed that 71% and 62% of breast cancer tissues exhibited positive MAYA staining in training and validation study respectively (Fig. 4c). Furthermore, MAYA expression in breast cancer is associated with advanced stage (TnN>0M≥0) (p=0.049) (Fig. 4d) and unfavorable patient outcomes (p=0.0085) (Fig. 4e). As revealed by RNAscope® 2.0 analysis, MAYA was upregulated in the majority of human solid tumors (Supplementary Fig. 4e). High levels of MAYA also correlate with lung adenocarcinoma, metastatic stage (TnN>0M≥0) and poor outcomes (Supplementary Fig. 4f, g).
We next test whether MAYA is required for YAP1 activation in this specific signaling context, finding that MAYA depletion significantly impaired the occupancy of YAP1 on promoter of its target genes (Fig. 4f) and expression of these genes under NRG1 stimulation (Fig. 4g, h). The phosphorylation of MST1 (Thr183), MOB1 (Thr35), LATS1 (Thr1079), and YAP (Ser127) were all increased upon MAYA knockdown (Fig. 4i). Accordingly, MAYA knockdown significantly reduced CTGF secretion in BoM-1833 cells (Fig. 4j), and consequently impaired cancer cell-induced osteoclast differentiation and bone resorption, which could be partially rescued by introduction of recombinant CTGF (Fig. 4k–m). Notably, MAYA is not involved in LPA or cell density-dependent YAP1-target gene regulation (Supplementary Fig. 4h, i).
LncRNA-mediated, NSUN6-dependent methylation inhibits the kinase activity of MST1
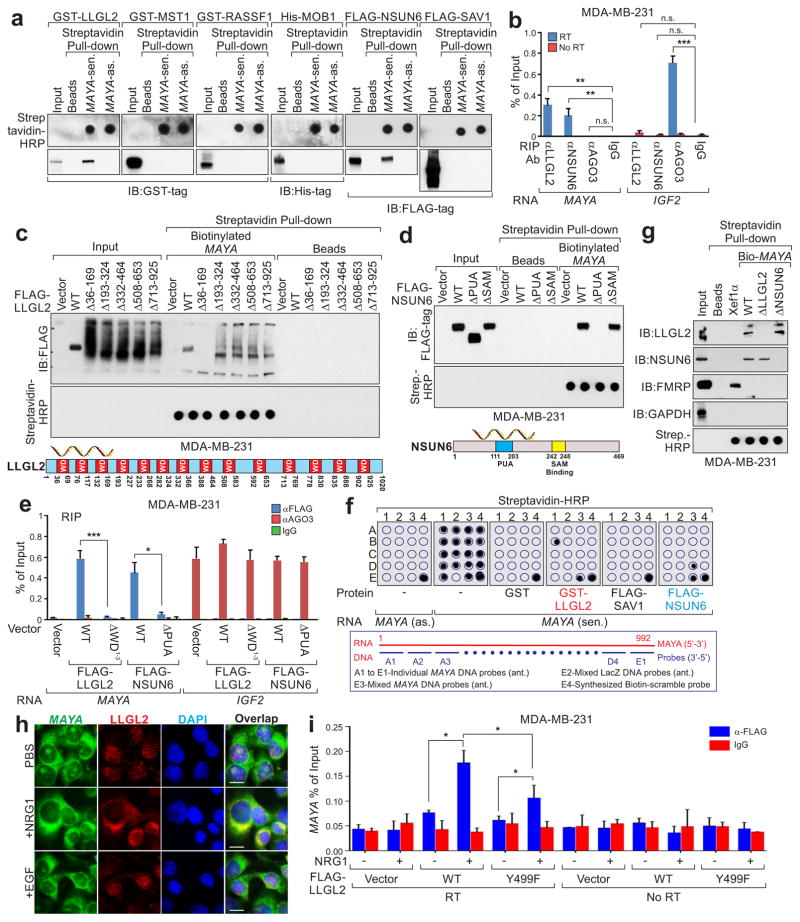
RNA pull-down in cell lysates showed that only the sense transcript of MAYA binds a panel of Hippo pathway proteins (Supplementary Fig. 5a, b). However, in vitro RNA pull-down with bacterially-expressed recombinant proteins revealed that only LLGL2 and NSUN6 directly bind MAYA (Fig. 5a), and the interactions were further confirmed by RNA immunoprecipitation (RIP) assay (Fig. 5b). LLGL2 contains 14 WD40 repeats that act as scaffolds to interact with RNA molecules for cellular functions50,53. To map the RNA-binding region of LLGL2, we generated a series of LLGL2 deletion mutants, finding that removal of the first three WD40 domains (ΔWD1–3) of LLGL2 (Δ36–169) abolished the MAYA-LLGL2 interaction (Fig. 5c). Similarly, PUA domain of NSUN6 (Δ111–203) is required for MAYA-NSUN6 interaction (Fig. 5d). The necessity of the WD40 domains 1–3 of LLGL2 and the PUA domain of NSUN6 for MAYA binding were confirmed in vivo by RIP assay (Fig. 5e).
Figure 5. Characterization of MAYA-LLGL2-NSUN6 associations.
(a) In vitro transcribed MAYA sense (sen.) or antisense (as.) transcripts were incubated with indicated recombinant proteins for in vitro streptavidin RNA pull-down assay, followed by IB detection using indicated antibodies. (b) RNA immunoprecipitation (RIP) assays were performed using indicated antibodies in MDA-MB-231 cells. (c and d) Top panel: FLAG-tagged WT or deletion mutants of LLGL2 (c) and NSUN6 (d) expressed in MDA-MB-231 cells were subjected to streptavidin RNA pull-down assay with biotinylated MAYA, followed by IB using anti-FLAG antibody. The presence of RNA transcripts was detected using streptavidin-HRP by dot-blot assay. (e) RIP assay using indicated antibodies in MDA-MB-231 cells transfected with the indicated plasmids. (f) In vitro RNA-protein binding followed by dot-blot assays using biotinylated MAYA sense (sen.) or anti-sense (as.) transcripts with recombinant proteins as indicated. Bottom, annotation of DNA probes targeting MAYA applied on the blot. (g) Streptavidin pull-down assay was performed using biotinylated FL, truncated MAYA, and Xef1α transcript and cell lysates extracted from MDA-MB-231 cells followed by IB using indicated antibodies. The presence of RNA transcripts was detected using streptavidin-HRP by dot-blot assay. (h) Immuno-RNA FISH detection using FISH probes targeting MAYA and antibody targeting LLGL2 in MDA-MB-231 cells with indicated treatment (scale bars: 20 μm). (i) RIP assay using anti-FLAG antibody in MDA-MB-231 cells transfected with the indicated plasmids followed by NRG1 treatment. For b, e, and i, mean ± s.e.m. were derived from n=3 independent experiments (n.s., p>0.05, *p<0.05, **p<0.01 and ***p<0.001, two-tailed paired Student’s t-test). Unprocessed original scans of all IB blots with size marker are shown in Supplementary Fig. 9. For a, c, d, f and g, unprocessed original scans of dot blots are also shown in Supplementary Fig. 9. Raw images of 3 independent experiments for h are in Supplementary Table 8.
To characterize the specific RNA sequences required for these interactions, we performed an in vitro RNA pull-down followed by a dot-blot assay as previously described34,53. We found that sequence B1 (nt. 241–300) of MAYA is essential for LLGL2 binding, and D3 (nt. 841–900) is responsible for NSUN6 binding (Fig. 5f), which was further confirmed by RNA pull-down assay with the corresponding MAYA mutants (Supplementary Fig. 5c and Fig. 5g). Furthermore, RNA electrophoretic mobility shift assay (REMSA) indicated that sequence nt. 251–290 and nt. 851–890 of MAYA directly binds LLGL2 and NSUN6, respectively (Supplementary Fig. 5d). Both RNA fluorescence in situ hybridization (FISH) and cell fractionation assay indicated that MAYA is localized to the cytosol (Fig. 5h and Supplementary Fig. 5e). Immuno-RNA FISH and RIP assays suggested that MAYA co-localized with LLGL2 upon NRG1 stimulation, which was impaired in the presence of LLGL2 Y499F mutant (Fig. 5h, i).
In the presence of FL MAYA, but not ΔLLGL2 or ΔNSUN6 deletion transcripts, bacterially-expressed LLGL2 could pull-down NSUN6 in vitro (Fig. 6a). These experiments suggested that LLGL2-MAYA-NSUN6 complex might be responsible for MST1 methylation on Lys59. To test this, we performed in vitro protein methylation assay and observed that only NSUN6 was able to methylate MST1 in the presence of SAM (S-adenosyl methionine) (Fig. 6b). Lys59 of MST1, in charge of ATP-binding, is critical for the kinase activity and autophosphorylation of MST154. We found that mutation of MST1 Lys59 to Ala or knockdown of NSUN6 impaired NRG1-triggered MST1 methylation at Lys59 (Fig. 6c, d). Consistently, knockout of LLGL2 or knockdown of MAYA abolished MST1’s methylation, leading to hyperphosphorylation of MST1 at Thr183 (Fig. 6e, f).
Figure 6. LncRNA-mediated, NSUN6-dependent methylation inhibits the kinase activity of MST1.
(a) Top, schematic diagram showing the direct interactions of MAYA with LLGL2 and NSUN6. Bottom, GST pull-down assay was performed by incubating GST-LLGL2 with NSUN6 in the presence of FL or domain deletion MAYA transcripts. LLGL2-NSUN6 interaction was detected by IB. (b) In vitro methylation assay was performed using recombinant FLAG-NSUN6 and indicated recombinant proteins with or without [3H]-SAM followed by autoradiography or IB detection with indicated antibody. (c–e) IB detection of MST1 Lys59 methylation in MDA-MB-231 cells transfected with indicated plasmids (c), siRNAs (d) or in LLGL2 KO cells (e) followed by NRG1 treatment. (f) IP and IB detection of indicated proteins in MDA-MB-231 cells transfected with indicated siRNAs followed by NRG1 treatment. (g) IP followed by IB detection of p-LLGL2 (Tyr499)- and NSUN6-dependent MST1 Lys59 methylation in MDA-MB-231 cells transfected with indicated plasmids followed by NRG1 treatment. (h) Left, graphic illustration of NSUN6 domain deletion mutants; right, IB detection of MST1 Lys59 methylation and Thr183 phosphorylation in cells transfected with indicated plasmids followed by NRG1 treatment. (i) In vitro methylation assay followed by kinase assay using GST-MST1 and FLAG-NSUN6 (WT and ΔSAM mutant) in the presence of [32P]-ATP and/or SAM. MST1 was purified by GST pull-down and detected by autoradiography or IB. (j) Heatmap representing color-coded fold change of NRG1-induced 57 YAP1 target genes determined by RT-qPCR in parental, ROR1 KO, LLGL2 KO, NSUN6 KO, and HER3 KO MDA-MB-231 cells. 21 genes were found to be commonly downregulated under all the KO conditions (n=3 independent experiments, *p<0.05, two-tailed paired student t-test). Unprocessed original scans of all blots with size marker are shown in Supplementary Fig. 9.
We further investigated the regulatory roles of LLGL2 and NSUN6 in modulating MST1 methylation. We found that both LLGL2 Y499F and ΔWD1–3 deletion mutants abolished ligand-induced MST1 methylation (Fig. 6g). In addition, unphosphorylated LLGL2 (Y499F) still associated with NSUN6, but failed to recruit MST1 (Fig. 6g) while ΔWD1–3 LLGL2 mutant could recruit MST1, but failed to associate with NSUN6 (Fig. 6g). Similarly, expression of both the NSUN6 ΔSAM (Δ242–248) and the ΔPUA (Δ111–203) mutants impaired MST1 methylation in vivo (Fig. 6h). To validate the hypothesis that methylation at Lys59 abolishes the kinase activity and autophosphorylation of MST1, we performed an in vitro methylation followed by kinase assay. In the presence of SAM, methylated MST1 failed to be phosphorylated at Thr183, but the unmethylated MST1 was autophosphorylated at Thr183 in the presence of ATP (Fig. 6i). At the cellular level, LLGL2 KO but not NSUN6 KO, decreased cell proliferation (Supplementary Fig. 6a–d). However, both LLGL2 and NSUN6 KO reduced cell mobility (Supplementary Fig. 6e–h). Among 57 YAP1-regulated genes examined, NRG1-induced expression of 25, 37, 40 and 29 genes was significantly downregulated by ROR1 KO, LLGL2 KO, NSUN6 KO and HER3 KO, respectively, and 21 genes were commonly reduced (Fig. 6j). Thus, we have identified the crosstalk between the ROR1/HER3-LLGL2-MAYA-NSUN6 signaling axis and the Hippo pathway.
To further demonstrate the mechanistic linkage, we expressed WT ROR1 and the K506A mutant in ROR1 KO cells for rescue experiments. We found that the expression of WT ROR1, but not the K506A mutant restored the NRG1-triggered phenotypes (Fig. 7a). Furthermore, we performed rescue experiments using HER3 Y1307E and LLGL2 Y499E phosphorylation-mimic mutants, respectively. The HER3 Y1307E mediated the HER3-BCAR3 interaction and recruitment of LLGL2 to phosphorylated HER3 (Fig. 7b and Supplementary Fig. 7a), and expression of the LLGL2 Y499E mutant in ROR1 KO cells rescued MST1 methylation and YAP1 hypophosphorylation (Supplementary Fig. 7a and Fig. 7c), and activation of YAP1 target genes in the absence of NRG1 (Fig. 7d and Supplementary Fig. 7b, c).
Figure 7. The ROR1/HER3-LLGL2/MAYA/NSUN6 signaling axis regulates YAP activity.
(a–c) IP and IB detection of indicated proteins in ROR1 KO cells transfected with indicated plasmids followed by NRG1 treatment. (d) RT-qPCR detection of CTGF expression in ROR1 KO cells transfected with indicated expression vectors followed by NRG1 stimulation. (e and f) IB detection of indicated proteins (e) and RT-qPCR detection of CTGF expression (f) in MDA-MB-231 cells transfected with LNAs against MAYA followed by overexpression of indicated plasmids and NRG1 treatment (100 ng ml−1 for 1 hr). (g) Quantification of osteoclast differentiation in the presence of M-CSF only, M-CSF+RANKL, or combined M-CSF+RANKL and CM from scramble (Scr) or MAYA LNA-transfected BoM-1833 cells rescued with indicated plasmids. (h) Measurement of helical peptides released from human bone plates incubated with primary osteoclasts treated as in g. For d, f, g and h, mean ± s.e.m. were derived from n=3 independent experiments (n.s. p>0.05, *p<0.05 and **p<0.01, two-tailed paired Student’s t-test). Unprocessed original scans of all blots with size marker are shown in Supplementary Fig. 9.
Next, we knocked down MAYA by locked nucleic acid (LNA) and rescued the expression of MAYA using LNA-resistant FL MAYA or ΔLLGL2 and ΔNSUN6 mutants, respectively (Supplementary Fig. 7d, e). MAYA knockdown in both breast and lung cancer cells showed dramatic effects on NRG1-dependent MST1 (Lys59) methylation, but minimal effects on phosphorylations of HER3 (Tyr1307) and LLGL2 (Tyr499) (Fig. 7e and Supplementary Fig. 7f). The NRG1-induced expression of YAP1-regulated genes was attenuated in MAYA knockdown cells but rescued by expressing FL MAYA, but not ΔLLGL2 and ΔNSUN6 deletion mutants in these cells (Fig. 7f and Supplementary Fig. 7g, h). As a control, a NRG1-induced gene ETS255, was not affected by MAYA knockdown (Supplementary Fig. 7i). Furthermore, CM from MAYA knockdown cells failed to enhance osteoclast differentiation (Fig. 7g and Supplementary Fig. 7j) and bone resorption by osteoclast (Fig. 7h). Notably, these effects were rescued by CM from MAYA knockdown cells expressing FL MAYA, but not ΔLLGL2 and ΔNSUN6 deletion mutants (Fig. 7g, h and Supplementary Fig. 7j).
MAYA serves as a promising therapeutic target for bone metastasis
We examined the expression level of MAYA in MDA-MB-231 parental and organ specific metastatic derivatives, including LM2 (lung)56, BoM-1833 (bone)56, BRN (brain)57, and a brain metastatic cell line derived from BT474 and its parental58. We found that MAYA was highly expressed in BoM-1833 cells, suggesting the potential role of MAYA in bone metastasis from breast cancer (Supplementary Fig. 8a). In BoM-1833 cells, MAYA knockdown showed minimal effects on NRG1-dependent p-HER3 (Tyr1307) and p-LLGL2 (Tyr449), but dramatic effects on MST1 (Lys59me2) and NRG1-triggered hypophosphorylation of MST1, LATS1, YAP1, as well as the expression of CTGF (Fig. 8a, b). Consistently, exogenous expression of FL MAYA, but not ΔLLGL2 or ΔNSUN6 mutants in MAYA knockdown BoM-1833 cells rescued these phenotypes (Fig. 8b). Inducible knockdown of MAYA by shRNAs inhibited proliferation, migration and invasion of BoM-1833 cells (Supplementary Fig. 8b–e).
Figure 8. MAYA serves as a promising therapeutic target for bone metastasis.
(a) IB detection of indicated proteins in BoM-1833 cells transfected with MAYA or control siRNAs. (b) IB detection of indicated proteins in BoM-1833 cells transfected with LNAs against MAYA followed by overexpression of indicated plasmids and NRG1 treatment. (c–f) Representative BLI images (c), bone colonization (d), femur μCT (e), and specimen CT and histological images (f, scale bars: 200 μm) of nude mice with intracardiac injection of BoM-1833 cells harboring indicated shRNAs. Yellow arrows indicate tumor lesions in femur (e, n=5 mice per group). (g–j) Quantification of bone parameters from representative specimen CT scans in f. BV/TV, bone volume/tissue volume ratio; Tb. th., trabecular thickness; Tb. sp., trabecular separation; Ct. th., cortical bone thickness. (k) Quantification of TRAP+ osteoclasts of histological sections from mouse femur in f. (l–o) Schematic diagram of LNA treatment time course (l, top) and femur μCT (l, bottom), representative BLI images (m), bone colonization (n), and RT-qPCR detection of MAYA expression from bone metastatic lesions (o) of mice with intracardiac injection of BoM-1833 cells followed by intravenous LNA administration (n=5 mice per group). Yellow arrows indicate tumor lesions in the femur (l). For d, g–k, n and o, mean ± s.e.m. were derived from n=3 independent experiments (n.s., p>0.05, *p<0.05 and **p<0.01, two-tailed paired Student’s t-test). Unprocessed original scans of all blots with size marker are shown in Supplementary Fig. 9. Statistics source data for c–d, f–k and m–n can be found in Supplementary Table 8.
We then investigated the role of MAYA in breast cancer bone metastasis using the intra-cardiac injection experimental metastasis mouse model. Compared to control shRNA (Ctl), inducible knockdown of MAYA significantly decreased hind-limb tumor burden (Fig. 8c, d). Micro computed tomography (Micro-CT) imaging revealed that mice inoculated with MAYA knockdown cells exhibited decreased bone lesions (Fig. 8e). Quantitative real-time RT-PCR analysis of bone tumor samples from mice inoculated with MAYA knockdown cells revealed a substantial decrease of MAYA expression (Supplementary Fig. 8f). Specimen CT revealed that MAYA knockdown increased trabecular bone mass in the femur (Fig. 8f). Quantification analyses confirmed a noteworthy enhancement of bone volume, trabecular thickness, and decreased trabecular spacing, but insubstantial differences in cortical bone thickness when MAYA was depleted (Fig. 8g–j). Histological TRAP staining indicated that MAYA knockdown impaired the number of osteoclasts relative to bone surface area (Fig. 8k). Therefore, these data suggest the importance of MAYA in promoting breast cancer metastasis to the bone.
Next, we examined the potential therapeutic value of MAYA LNAs using the same mouse model. After intra-cardiac inoculation of BoM-1833 cells, nude mice were intravenously injected with scramble (Scr) LNA or LNA targeting MAYA (15 mg/kg, 3 times per week) for 3 weeks. BLI imaging showed that the bone tumor burden was greatly reduced in mice treated with MAYA LNA (Fig. 8l–n). The expression level of MAYA from mice bone metastatic lesions was significantly reduced upon MAYA LNA treatment (Fig. 8o). We further explored MAYA’s tumor promoting and bone metastasis function in lung cancer using A549-Luc cells and observed similar effects (Supplementary Fig. 8g–l).
DISCUSSION
Recent studies have revealed that the activity of YAP/TAZ transcriptional co-activators and LATS1 kinase could be regulated by nutritional stress59, GPCR signaling51, WNT signaling35, and the mevalonate pathway60. However, the modulation of the Hippo kinase MST1/2 is not yet understood. Our study found that NRG1-bound ROR1/HER3 heterodimer recruits adaptor LLGL2 and methyltransferase NSUN6 in an lncRNA dependent fashion. The LLGL2-MAYA-NSUN6 module methylates MST1 at Lys59, which abolishes MST1’s kinase activity and consequently leads to hypophosphorylation of LATS1 and YAP. YAP is activated and accumulates in the nuclear to stimulate target genes expression involved in tumor cell proliferation and bone metastasis (Supplementary Fig. 8m).
Orphan receptor tyrosine kinase ROR1 transduces cellular signaling through both kinase-dependent and -independent mechanisms. ROR1 associates with EGFR, HER3, SRC or c-MET in response to EGF or Wnt stimulation to promote cancer cell proliferation, survival, and invasion27,46. Our data indicate that in TNBC cells, ROR1 forms heterodimer with HER3 to phosphorylate HER3 and LLGL2 upon NRG1 stimulation. It is possible that ROR1 dynamically forms hetero-dimers, or -trimers with other (receptor) tyrosine kinases in the presence of different ligands. ROR1 is subjected to tyrosine phosphorylation by SRC and MET within the kinase domain and proline-rich domain, respectively46. Although our data indicate that NRG1 exhibited minimal effect on the tyrosine phosphorylation of ROR1, it is possible that other ligands or signals may induce these modifications and regulate the kinase activity of ROR1. The downstream signaling pathways triggered by ROR1 might vary depending on the cell/tissue types. Previous studies suggested that ROR1 phosphorylates SRC to activate AKT pathway20,27. Our findings demonstrated the crosstalk between ROR1-HER3 and the Hippo-YAP pathway mediated by lncRNAs under cancer-specific context. Nuclear ErbB family members have been shown to modulate gene expression as transcriptional co-activator61. For example, nuclear ErbB4 has been suggested to associate with YAP and to regulate expression of YAP target genes62. It is possible that truncated nuclear-HER3 was translocated into the nucleus to trigger YAP transcriptional program.
The role of MAYA in mediating the LLGL2-NSUN6 interaction is an intriguing paradigm. The complicated secondary structure of long noncoding RNA molecules provides multiple protein binding sites, which may scaffold the RNA-protein complex formation34,63. Given uncharacteristic RNA-binding domains recently identified, such as WD40, SH2 and SH3 domains64, we anticipate that lncRNA-dependent regulatory mechanisms are widely applicable in signal transduction and epigenetic modifications. Although we show that growth factor enhances the association between LLGL2, MAYA, and NSUN6 to methylate MST1, it’s worthy to investigate whether or not the LLGL2-MAYA-NSUN6 complex assembly is regulated by other ligands.
For TNBC and NSCLC patients, anti-EGFR targeted therapies have been one of the primary treatments. However, a key challenge lies in how to overcome acquired resistance to tyrosine kinase inhibitors (TKIs). Recently, EGFR mutations have been linked to bone metastasis65. The highly upregulated ROR1 expression and YAP pathway activation in bone metastatic cells signified the potential therapeutic values of combined targeting ROR1 and YAP pathway against bone metastasis. Indeed, ROR1 has been implicated in EGFR-resistance in NSCLC patients66. Our observation that breast cancer patients with strong p-HER3 Tyr1307 staining exhibited unfavorable progression-free survival outcomes compared to patients with low HER3 staining suggest that p-HER3 Tyr1307 and MAYA may serve as predictive biomarkers to forecast bone metastasis and refer to commence targeted anti-EGFR therapy. Further, targeting ROR1 using monoclonal antibodies or small molecule inhibitors may prove to be a promising therapeutic option for leukemia, TNBC, and NSCLC patients. Finally, our study demonstrates the effectiveness of LNA oligonucleotide-based inhibition of breast cancer bone metastasis, which could be further developed as a valuable therapeutic strategy either alone or in combination with EGFR inhibitors, to overcome resistance to EGFR-targeted therapies.
METHODS
Clinical samples
Fresh frozen breast carcinomas and their adjacent normal tissues were purchased from Asterand Bioscience. Breast cancer, lung cancer, and multiple cancer type tissue microarrays were purchased from US Biomax, Biochain, and US BioLab. The fresh frozen primary tumor and normal breast tissues (Yixin Bre-01 cohort) were obtained from Yixing People’s Hospital in China. The study protocol was approved by the Institutional Review Board of Nanjing Medical University (Nanjing, China). All tissue samples were collected in compliance with informed consent policy. Clinical tissue information for all data presented in Figs 1c, d, 2k-m, 4c-e and Supplementary Figs 2l, m, and 4e-g is summarized in Supplementary Table 1.
Cell culture, treatment, transfection, electroporation, and lentiviral transduction
Human breast cancer cells MDA-MB-231 (ATCC), MCF-7 (ATCC), MDA-MB-231-BRN, BT474 and BT474-BRN (kindly provided by Dr. Dihua Yu at M.D. Anderson Cancer Center) were maintained in Dulbecco’s Modified Eagle’s Medium (DMEM)/F12 with 5% Fetal Bovine Serum (FBS), and 1% penicillin/streptomycin (pen/strep). The TEAD Reporter containing MCF-7 (BPS Bioscience) was cultured according to vendor’s instructions. MDA-MB-231-LM2 and MDA231-BoM-1833 were kindly provided by Dr. Xiang Zhang (Baylor College of Medicine) and maintained in DMEM (Life technologies) with 10% FBS and 1% pen/strep. Human non-small cell lung cancer cells A549 and A549-Luc luciferase expressing cells (Caliper Life Sciences) were maintained in RPMI 1640 with 10% FBS and 1% pen/strep. Mouse lymphoblast cell line 32D Clone 3 (ATCC® CRL11346™) was maintained in RPMI 1640 with 4.5 g/L glucose, 10% FBS and 10% mouse Interleukin-3 culture supplement. All cell lines were authenticated by autosomal STR profiles provided by the MDACC Characterized Cell Line core. MDA-MB-231, MCF-7, MDA-MB-231-BRN, BT474 and BT474-BRN, MDA-MB-231-LM2 and MDA231-BoM-1833 cell lines were tested negative for mycoplasma by the MDACC Characterized Cell Line core. The TEAD Reporter containing MCF-7 cell line has been screened using the PCR-based VenorGeM Mycoplasma Detection kit to confirm the absence of mycoplasma by BPS Bioscience. A549 and A549-Luc luciferase expressing cells are confirmed to be pathogen free by the IMPACT Profile I (PCR) at the University of Missouri Research Animal Diagnostic and Investigative Laboratory as indicated by vendor. 32D cell line was assayed for mycoplasma, by the Hoechst stain, PCR and the standard culture test by ATCC. None of the cell lines used was found in the database of commonly misidentified cell lines that are maintained by ICLAC and NCBI Biosample.
For treatment, cells were serum starved overnight-24 hours followed by treatment with growth factors (PeproTech), EGF (10 ng/ml), NRG1 (100 ng/ml) or LPA (1 μM, Cayman Chemical) for 30 minutes or 2 hours. Under specified conditions, cell were pre-treated with pan-EGFR inhibitor Dacomitinib (PF299804, PF299) at 100 nM, c-SRC inhibitor Saracatinib (AZD0530) and c-MET inhibitor JNJ-38877605 (Selleckchem) at 5 μM for 2 hrs.
siRNA or LNA transfections were performed using DharmaFECT4 (GE Healthcare). Plasmids transfections were performed using Lipofectamine3000 (Life Technologies). Electroporation of 32D cells with DNA plasmids was performed using Amaxa™ 4D-Nucleofector™ System. Lentiviruses were produced in HEK293T cells with ViraPower™ Lentiviral Expression System (Life Technologies) and used to transduce target cells.
Plasmids and constructs
The FL LLGL2, MST1, NSUN6, BCAR3, HER3, and ROR1 mammalian expression vectors were obtained from Origene Technologies. Bacterial expression vectors for His6-HER3 (WT and mutants) and HER3 ICD were constructed by subcloning the corresponding gene sequences into pET-DEST42 vectors using the Gateway system (Life Technologies). GST-tagged LLGL2 (WT and mutants) were constructed into the pGEX-5X-1 backbone (GE Healthcare). Mammalian expression vector for FL MAYA and mutants were constructed by subcloning the gene sequences into pBabe backbone (Addgene). To generate LNA#3-resistant MAYA mammalian expression vectors used in the rescue experiments, LNA#3 targeting sequences GAG CCT TTG CAA AGA G were mutated to GAG CCT TTC GTA AGA G. All mutants were generated using QuikChange™ Lightning Site-Directed Mutagenesis Kit (Agilent Technologies).
Antibodies
The antibodies used in this study are summarized in Supplementary Table 6. The specificity of antibodies was confirmed by peptide competition assay with unphosphorylated/unmethylated peptide, phosphorylated/methylated peptide, or corresponding scrambled phosphorylated/methylated peptide (modified residue was exchanged position with its adjacent residue).
Generation of knockout cell lines
CRISPR/Cas9 KO double nickase plasmids of ROR1 (h, sc-401841-NIC), HER3 (h, sc-400146-NIC), LLGL2 (h, sc-404667-NIC) and NSUN6 (h, sc-415118-NIC) were used to generate stable knockout cell lines of MDA-MB-231 and BoM-1833 according to manufacturer’s instructions. Either individual or pooled single cell clones with efficient target gene KO (as indicated in figures) were used for downstream functional experiment to minimize the clonal variation.
siRNA, shRNA, and LNA™
Lincode siRNA library (G-301005-01), Lincode® SMARTpool® siRNA targeting MAYA (R-036866), Lincode® non-targeting control siRNAs (D-001320), and ON-TARGETplus SMARTpool® siRNA targeting ROR1 (L-003171), BCAR3 (L-011469), LLGL2 (L-019812), HER3 (L-003127), EGFR (L-003114), NSUN6 (L-018822), and DOT1L (L-014900) from GE Healthcare Dharmacon were used in this study. The knockdown efficiency and specificity of all siRNAs were validated either by RT-qPCR or immunoblotting. The shRNAs targeting MAYA were designed using the shERWOOD algorithm and cloned into UltramiR scaffold in pZIP lentiviral vector (transOMIC technologies). For in vivo inducible knockdown of MAYA in xenograft experiments, shRNA sequences were constructed into pZIP-TRE3GS lentiviral vector (transOMIC technologies). LNAs targeting MAYA or a scrambled sequence were designed and synthesized from Exiqon. Supplementary Table 7 contains detailed sequence information of siRNAs, shRNAs and LNAs.
Protein recombination and purification
GST-LLGL2, GST-MST1, BCAR3 and GST-RASSF1 were obtained from Abnova. FLAG-NSUN6, FLAG-SAV1, FLAG-MST1, FLAG-ROR1 (WT and extracellular domain), and FLAG-HER3 were obtained from Origene Technologies. His6-MOB1 was purchased from Novoprotein Inc. GST-ROR1 was obtained from SignalChem. His6-HER3 intracellular domain (ICD) and WT His6-MST1 and corresponding mutants were expressed in E.coli strain BL21-CodonPlus (DE3)-RIPL (Agilent Technologies) and purified using HisPur™ Cobalt Spin Columns (Life Technologies). Domain deletion or point mutants of GST-ROR1 ICD and GST-LLGL2 were purified using Pierce™ GST Spin Purification Kit (Life Technologies). Biotinylated HER3 and phospho-HER3 peptides were synthesized from Elim Biopharmaceuticals Inc.
Cell lysis, fractionation, immunoprecipitation, and immunoblotting
Cells lysis preparation, immunoprecipitation and immunoblotting were performed as previously described34, 53. For cell fractionation, cytoplasmic and nuclear fractions were separated using cytoplasmic and nuclear protein/RNA extraction kit (Thermo-Fisher Scientific) following manufacturer’s instructions.
Human CTGF ELISA
The human CTGF levels in the conditioned media of cultured cells were quantitatively determined in triplicate by OmniKine™ ELISA kit (Assay Biotechnology).
In vitro osteoclast differentiation
Human osteoclasts were obtained from Lonza and differentiated in vitro according to vendor’s instructions. Conditioned media collected from the KO cells or cells transfected with indicated LNAs/plasmids after 72 hours were added to osteoclast cells at day 3 of differentiation in a 6-well plate. In rescue experiments, recombinant CTGF or CCN3 (PeproTech) was supplemented in conditioned media as indicated. Cells were TRAP stained on day 7 using Acid Phosphatase, Leukocyte (TRAP) kit (Sigma-Aldrich), and TRAP+-multinucleated cells were quantified as mature osteoclasts. Quantitative TRAP assay was performed using TRACP & ALP assay kit (Takara). For osteoclast resorption function analyses, bone marrow osteoclast differentiation was conducted in OsteoAssay bone plates (Lonza) and osteoclast activity was determined by quantifying the type I collagen helical peptide α1 (I) 620–633 released from bone into culture medium using MicroVue Bone Helical Peptide EIA assay (Quidel).
Tumor bone metastasis analysis
All animal experiments were performed in accordance with protocol approved by the Institutional Animal Care and Use Committee of MD Anderson. Animals arrived in our facility were randomly put into cages with five mice each. They were implanted with respective tumor cells in the unit of cages, which were randomly selected. The animal experiment was set up to use 5–10 mice per group to detect a 2-fold difference with power of 80% and at the significance level of 0.05 by a two-sided test for significant studies (RaoSoft Inc. sample size calculator). The luciferase-labelled BoM-1833 or A549-Luc cells (2 × 105) in 50 μl 1× PBS were intracardially injected into the left ventricle of nu/nu, female 4–6-week-old nude mice using a 100-μl Hamilton Microliter™ syringe. To induce MAYA shRNA expression, mice were fed with 1 mg/ml of tetracycline in drinking water containing 2% sucrose.
To assess the effect of MAYA knockdown with LNA™, mice were intravenously injected with in vivo-grade LNAs against MAYA (Exiqon) in PBS (15 mg/kg) three times per week for three weeks after BoM-1833 cells injection. Bone metastases were quantified by BLI every week using an IVIS Spectrum Xenogen Imaging System (Caliper Life Sciences). The osteolytic metastatic lesions on the femurs and tibiae were visualized using the micro-CT imaging. The hind limbs or other organs were removed and fixed for histological analysis. For the structural analysis of trabecular and cortical bones, mouse femurs and tibias were fixed in 70% ethanol and scanned at 7 μm resolution using the eXplore Locus micro-computed tomography instrument (μCT, GE Healthcare) at the Small Animal Imaging Facility of MDACC. The images were reconstructed and trabecular bone parameters were calculated using the eXplore Utilities software (GE Healthcare). For all animals producing respective tumors, they were randomly divided into groups. No animals were excluded from the analysis. The investigators were not blinded to allocation during experiments and outcome assessment.
RNAi screening
Dharmacon Lincode® SMARTpool® siRNA Library-Human NR LncRNA RefSeq v54 (#G-301000) (GE Dharmacon) was used. 1×104 TEAD Reporter-MCF-7 cells were plated in 96-well plates 24 hours prior to transfection. The cells were transfected with either the 15 pmol Lincode® siRNA library or siRNA control (GE Healthcare Dharmacon). siGLO green transfection indicator was included to show that over 80% of the cells were transfected using this method. Cells were lysed 72 hours after transfection and assayed for luciferase activity using the Dual-Glo Luciferase reporter system (Promega) and activities were monitored by Synergy H4 Hybrid Multi-Mode Microplate Reader (BioTek). The protein concentrations of diluted cell lysates (1:100) were measured in parallel using Bio-Rad protein assay kit (Bio-rad), which was used to normalize the luciferase activity. The firefly luciferase activity for each 96-well plate was subtracted with background, normalized by protein concentration, and further normalized by control siRNA as fold changes. Log2 of relative fold changes were plotted. The hits with log2 ≤ -3 were selected as top candidates.
RNA biology assays
The in vitro transcription, RNA pull-down/mass spectrometry analysis, In vitro RNA-protein binding assay, In vitro RNA pull-down coupled with dot-blot assay and RT-qPCR were performed as previously described34, 53. RNA electrophoretic mobility shift assay (REMSA) was performed as previously described34, 53 using bacterial recombinant LLGL2 and NSUN6 with synthesized RNA oligonucleotides corresponding to nt. 241–300 and nt. 841–900 (GE Healthcare Dharmacon). 5′- and 3′-rapid amplification of cDNA ends (RACE) was performed using SMARTer® RACE 5′/3′ Kit (Clontech). MAYA RNA expression was detected using the NorthernMax® Kit (Ambion) with biotin-labelled LNA probes (Exiqon) in cells and using premade human tissue Northern blot (Zyagen). Supplementary Table 7 contains detailed sequence information of primers and probes.
RNAScope® assay, immuno-RNA FISH and histological analysis/microscopy
Detection of MAYA expression using RNAScope® probe (designed by Advanced Cell Diagnostics) was performed on breast or lung cancer cell lines and tissue microarrays with RNAScope® 2.0 High Definition Assay kit according to the manufacturer’s instructions (Advanced Cell Diagnostics). The images were visualized with Zeiss Axioskop2 plus Microscope, and the slides were scanned on the Automated Cellular Image System III (ACIS III, Dako, Denmark) for quantification by digital image analysis.
Immuno-RNA FISH was performed as previously described34. In brief, the specimen slides from RNA FISH was blocked with blocking buffer (1×PBS/5% goat serum/0.3% Triton™ X-100) for 1 hr at room temperature followed by incubation with diluted primary antibody in dilution buffer (1×PBS/1% BSA/0.3% Triton™ X-100) for 2 hrs at room temperature, then incubate with fluorochrome-conjugated secondary antibody for 1 hr at room temperature in the dark, the slides were rinsed and mounted for visualization.
Immunohistochemistry (IHC) staining was performed as previously described34, 53. For Von Kossa staining, slides were stained in 1% silver nitrate followed by wash, then incubated in formaldehyde, sodium carbonate solution to develop the stain followed by wash, then placed in 5% sodium thiosulfate solution and rinsed. The slides were counterstained in van Gieson stain. For tartrate-resistant acid phosphatase (TRAP) staining, slides were incubated in acetate buffer of sodium acetate and tartaric aid, pH 5.0 at 37°C for 20 min; then the substrate naphthol AS-MX phosphate was added, followed by the color developer, fast red TR hemi (zinc chloride) salt at 37°C until color development was observed in TRAP positive cells. The slides were washed and counterstained in Harris’s hematoxylin. Images were acquired with Zeiss Axioskop2 plus microscope and analyzed with ProgRes Capture Pro software. The slides were scanned on the Automated Cellular Image System III (Dako, Agilent) for quantification by digital image analysis.
Image analysis and quantification for RNAScope® assay and immunohistochemistry
A total score of MAYA expression was calculated from both the percentage of positive cells and intensity. High and low expression was defined using the mean score of all samples as a cutoff point. Spearman rank correlation was used for statistical analyses of the correlation between each marker and clinical stage. The RNAScope® and IHC staining were categorized into five grades 0, 1+, 2+, 3+ and 4+, according to the following criteria: 0) no staining or less than 5% tumor cells in each field (3 fields) examined; 1+) 5%-10% tumor cells have staining in each field (3 fields) examined; 2+) 10%–25% tumor cells have staining in each field (3 fields) examined; 3+) 25%–50% tumor cells have staining in each field (3 fields) examined; 4+) 50%–100% tumor cells have staining in each field (3 fields) examined. If the staining score was ≥2+, we considered this case to be MAYA positive. The positive staining percentages for MAYA over total case number were averaged and shown. For quantification analysis of RNAScope® signal and IHC staining, the staining density for each tissue sample were determined by Image-Pro plus 6.0 (Media Cybernetics) and calculated based on the average staining intensity and the percentage of positively stained cells.
In vitro kinase assay
WT or mutant substrate proteins were incubated with 50 μl of in vitro kinase assay buffer I (SignalChem) containing 100 μM ATP (cold reaction) or 10 μCi [γ-32P] ATP and indicated protein kinase for 1 hour at 30°C. To recover the phosphorylated GST-tagged protein, Pierce™ Glutathione Magnetic Beads (Millipore) were added into reactions and the bound proteins were washed and eluted according to vendor’s instructions. The eluted proteins were dialyzed against the appropriate buffers for downstream assays or separated by SDS-PAGE and detected by Coomassie Blue staining, autoradiography, or immunoblotting.
In vitro methylation assay
Purified recombinant proteins were incubated (1 hour, 30 °C) with 1 μg of recombinant FLAG-NSUN6 in 30 μl of methylation buffer [50 mM HEPES pH 8.0, 0.01% (v/v) NP-40, 10 mM NaCl, 1 mM DTT, and 1 mM PMSF] supplemented with 2 μl of S-adenosyl-L-[methyl-[3H]methionine ([3H]-SAM, Perkin Elmer; for radioactive methylation) or 20 nmol of S-adenosyl-L-methionine sulfate p-toluenesulfonate (SAMe-PTS, Sigma-Aldrich; for nonradioactive methylation). SDS loading buffer was added to methylation reactions and boiled followed by separation on a 4–12% SDS-PAGE gel. The resulting protein bands were visualized by Coomassie blue staining, immunoblotting, or autoradiography using EN3HANCE™ spray (Perkin Elmer).
ChIP and RIP assays, cell proliferation assay, migration and invasion assays
Cell fixation and chromatin preparation were performed using truChIP Chromatin Shearing Kits on Covaris M220 focused Ultrasonicator (Covaris). The downstream procedure for ChIP was conducted as previously described53. RIP assay, cell proliferation, migration and invasion assays were performed as previously described34.
Computational analysis of TCGA RNA-Seq data
We downloaded the breast cancer RNA-seq BAM files from UCSC Cancer Genomics Hub (CGHub, https://cghub.ucsc.edu/). Quantification and statistical analysis of MAYA expression were performed as previously described34.
Statistics and reproducibility
The experiment was set up to use 3–5 samples/repeats per experiment/group/condition to detect a 2-fold difference with power of 80% and at the significance level of 0.05 by a two-sided test for significant studies. All experiments including IP/IB and Immuno-FISH were carried out with three biological replicates. Panels in Figs 1d, h–i, j, k–l, 2l, 4c, k–l, 5h, 8c–d, f–k, m–n and Supplementary Figs 1i–l, 2i–l, 6e–h, 7j, 8d, e, i, j and k–l shows a representative image of three independent experiments. Analyses of relative gene expression were determined using the 2-ΔΔCt method with GAPDH or B2M as the internal reference genes. The relative quantities of ChIP samples were normalized by individual inputs, respectively. Results are reported as mean ± standard error of the mean (S.E.M.) of at least three independent experiments. Each exact n value is indicated in the corresponding figure legend or in the figure. Statistical analysis was performed using GraphPad Prism 7 software. Comparisons were analyzed by two tailed paired Student’s t-test, Wilcoxon test or one-way ANOVA test (n.s., p>0.05, *p<0.05, **p<0.01, and ***p<0.001), as indicated in individual figures. Fisher’s exact test was implemented for statistical analyses of the correlation between markers and clinical parameters. For survival analysis, the expression of MAYA or phosphorylation density of indicated proteins was treated as a binary variant and divided into ‘high’ and ‘low’ level. Kaplan-Meier survival curves were compared using the log rank test. The experiments were not randomized. The investigators were not blinded to allocation during experiments and outcome assessment.
Data availability
The breast cancer RNA-seq data used to analyze MAYA expression were derived from the TCGA Research Network: http://cancergenome.nih.gov/, and the breast cancer RNA-seq BAM files were downloaded from the UCSC Cancer Genomics Hub (CGHub, https://cghub.ucsc.edu/). Source data for all mass spectrometry experiments (Figs. 2a, and Supplementary Figs. 2a, 3a, 3b, 3e, 4a) have been provided as Supplementary Table 2, 3 and 5. Statistics source data for Figs 1d, h–i, j, k–l, 2l, 4c, k–l, 5h, 8c–d, f–k, m–n, and Supplementary Figs 1i–l, 2i–l, 6e–h, 7j, 8d, e, i, j and k–l have been provided in Supplementary Table 8. All other data supporting the findings of this study are available from the corresponding author on reasonable request.
Supplementary Material
Acknowledgments
We are grateful to Dr. Joan Massague and Dr. Xiang Zhang for providing the MDA-MB-231 LM2 and BoM-1833 cell lines and to Dr. Dihua Yu for providing the MDA-MB-231-BRN and BT474-BRN cells. We thank Mr. D. Aten for assistance with figure presentation. This work was supported by National Institutes of Health Pathway to Independence Award (R00CA166527) and Cancer Prevention Research Institute of Texas First-time Faculty Recruitment Award (R1218) grants to L.Q.Y. and National Institutes of Health Pathway to Independence Award (R00DK094981) to C.R.L.
Footnotes
Author contributions
C.L.L. and S.Y.W. devised and performed most experiments. K.L., S.J. and G.G. helped with mouse intracardiac injections. A.F.L., Z.X. and Q.S.H. helped with biochemistry studies. D.H.H. performed mass spectrometry analysis. Clinical specimens were ascertained and processed by J.W.Z. and Y.Z. The histological staining and corresponding analysis were performed by K.L. P.K.P. assisted with manuscript drafting. J.Y., L.H. Z.Y.C and H.L. performed bioinformatics analysis. S.X.Z and M.C.H. contributed to discussion and data interpretation. L.Q.Y. and C.R.L. initiated and supervised the project and wrote the paper with input from all authors.
References
- 1.Guise TA, et al. Basic mechanisms responsible for osteolytic and osteoblastic bone metastases. Clin Cancer Res. 2006;12:6213s–6216s. doi: 10.1158/1078-0432.CCR-06-1007. [DOI] [PubMed] [Google Scholar]
- 2.Waning DL, Guise TA. Molecular mechanisms of bone metastasis and associated muscle weakness. Clin Cancer Res. 2014;20:3071–3077. doi: 10.1158/1078-0432.CCR-13-1590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kang Y, et al. A multigenic program mediating breast cancer metastasis to bone. Cancer Cell. 2003;3:537–549. doi: 10.1016/s1535-6108(03)00132-6. [DOI] [PubMed] [Google Scholar]
- 4.Pan D. The hippo signaling pathway in development and cancer. Developmental cell. 2010;19:491–505. doi: 10.1016/j.devcel.2010.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Harvey KF, Zhang X, Thomas DM. The Hippo pathway and human cancer. Nat Rev Cancer. 2013;13:246–257. doi: 10.1038/nrc3458. [DOI] [PubMed] [Google Scholar]
- 6.Meng Z, Moroishi T, Guan KL. Mechanisms of Hippo pathway regulation. Genes Dev. 2016;30:1–17. doi: 10.1101/gad.274027.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yu FX, Guan KL. The Hippo pathway: regulators and regulations. Genes Dev. 2013;27:355–371. doi: 10.1101/gad.210773.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hansen CG, Moroishi T, Guan KL. YAP and TAZ: a nexus for Hippo signaling and beyond. Trends Cell Biol. 2015;25:499–513. doi: 10.1016/j.tcb.2015.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhao B, Tumaneng K, Guan KL. The Hippo pathway in organ size control, tissue regeneration and stem cell self-renewal. Nat Cell Biol. 2011;13:877–883. doi: 10.1038/ncb2303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Thompson BJ, Sahai E. MST kinases in development and disease. J Cell Biol. 2015;210:871–882. doi: 10.1083/jcb.201507005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Halder G, Dupont S, Piccolo S. Transduction of mechanical and cytoskeletal cues by YAP and TAZ. Nat Rev Mol Cell Biol. 2012;13:591–600. doi: 10.1038/nrm3416. [DOI] [PubMed] [Google Scholar]
- 12.Piccolo S, Dupont S, Cordenonsi M. The biology of YAP/TAZ: hippo signaling and beyond. Physiol Rev. 2014;94:1287–1312. doi: 10.1152/physrev.00005.2014. [DOI] [PubMed] [Google Scholar]
- 13.Bergeron JJ, Di Guglielmo GM, Dahan S, Dominguez M, Posner BI. Spatial and Temporal Regulation of Receptor Tyrosine Kinase Activation and Intracellular Signal Transduction. Annu Rev Biochem. 2016;85:573–597. doi: 10.1146/annurev-biochem-060815-014659. [DOI] [PubMed] [Google Scholar]
- 14.Green JL, Kuntz SG, Sternberg PW. Ror receptor tyrosine kinases: orphans no more. Trends Cell Biol. 2008;18:536–544. doi: 10.1016/j.tcb.2008.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Forrester WC. The Ror receptor tyrosine kinase family. Cellular and molecular life sciences : CMLS. 2002;59:83–96. doi: 10.1007/s00018-002-8407-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yoda A, Oishi I, Minami Y. Expression and function of the Ror-family receptor tyrosine kinases during development: lessons from genetic analyses of nematodes, mice, and humans. Journal of receptor and signal transduction research. 2003;23:1–15. doi: 10.1081/RRS-120018757. [DOI] [PubMed] [Google Scholar]
- 17.Baskar S, et al. Unique cell surface expression of receptor tyrosine kinase ROR1 in human B-cell chronic lymphocytic leukemia. Clin Cancer Res. 2008;14:396–404. doi: 10.1158/1078-0432.CCR-07-1823. [DOI] [PubMed] [Google Scholar]
- 18.Daneshmanesh AH, et al. Orphan receptor tyrosine kinases ROR1 and ROR2 in hematological malignancies. Leuk Lymphoma. 2013;54:843–850. doi: 10.3109/10428194.2012.731599. [DOI] [PubMed] [Google Scholar]
- 19.Zhang S, et al. ROR1 is expressed in human breast cancer and associated with enhanced tumor-cell growth. PLoS One. 2012;7:e31127. doi: 10.1371/journal.pone.0031127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang S, et al. The onco-embryonic antigen ROR1 is expressed by a variety of human cancers. Am J Pathol. 2012;181:1903–1910. doi: 10.1016/j.ajpath.2012.08.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Borcherding N, Kusner D, Liu GH, Zhang W. ROR1, an embryonic protein with an emerging role in cancer biology. Protein & cell. 2014;5:496–502. doi: 10.1007/s13238-014-0059-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chien HP, et al. Expression of ROR1 has prognostic significance in triple negative breast cancer. Virchows Arch. 2016;468:589–595. doi: 10.1007/s00428-016-1911-3. [DOI] [PubMed] [Google Scholar]
- 23.Gentile A, Lazzari L, Benvenuti S, Trusolino L, Comoglio PM. Ror1 is a pseudokinase that is crucial for Met-driven tumorigenesis. Cancer Res. 2011;71:3132–3141. doi: 10.1158/0008-5472.CAN-10-2662. [DOI] [PubMed] [Google Scholar]
- 24.Bicocca VT, et al. Crosstalk between ROR1 and the Pre-B cell receptor promotes survival of t(1;19) acute lymphoblastic leukemia. Cancer Cell. 2012;22:656–667. doi: 10.1016/j.ccr.2012.08.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Masiakowski P, Carroll RD. A novel family of cell surface receptors with tyrosine kinase-like domain. J Biol Chem. 1992;267:26181–26190. [PubMed] [Google Scholar]
- 26.Oishi I, et al. Spatio-temporally regulated expression of receptor tyrosine kinases, mRor1, mRor2, during mouse development: implications in development and function of the nervous system. Genes to cells : devoted to molecular & cellular mechanisms. 1999;4:41–56. doi: 10.1046/j.1365-2443.1999.00234.x. [DOI] [PubMed] [Google Scholar]
- 27.Yamaguchi T, et al. NKX2-1/TITF1/TTF-1-Induced ROR1 is required to sustain EGFR survival signaling in lung adenocarcinoma. Cancer Cell. 2012;21:348–361. doi: 10.1016/j.ccr.2012.02.008. [DOI] [PubMed] [Google Scholar]
- 28.Rashid F, Shah A, Shan G. Long Non-coding RNAs in the Cytoplasm. Genomics, proteomics & bioinformatics. 2016;14:73–80. doi: 10.1016/j.gpb.2016.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Willingham AT, et al. A strategy for probing the function of noncoding RNAs finds a repressor of NFAT. Science. 2005;309:1570–1573. doi: 10.1126/science.1115901. [DOI] [PubMed] [Google Scholar]
- 30.Yoon JH, Abdelmohsen K, Gorospe M. Posttranscriptional gene regulation by long noncoding RNA. J Mol Biol. 2013;425:3723–3730. doi: 10.1016/j.jmb.2012.11.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tichon A, et al. A conserved abundant cytoplasmic long noncoding RNA modulates repression by Pumilio proteins in human cells. Nat Commun. 2016;7:12209. doi: 10.1038/ncomms12209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang P, et al. The STAT3-binding long noncoding RNA lnc-DC controls human dendritic cell differentiation. Science. 2014;344:310–313. doi: 10.1126/science.1251456. [DOI] [PubMed] [Google Scholar]
- 33.Liu B, et al. A cytoplasmic NF-kappaB interacting long noncoding RNA blocks IkappaB phosphorylation and suppresses breast cancer metastasis. Cancer Cell. 2015;27:370–381. doi: 10.1016/j.ccell.2015.02.004. [DOI] [PubMed] [Google Scholar]
- 34.Lin A, et al. The LINK-A lncRNA activates normoxic HIF1alpha signalling in triple-negative breast cancer. Nat Cell Biol. 2016;18:213–224. doi: 10.1038/ncb3295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Park HW, et al. Alternative Wnt Signaling Activates YAP/TAZ. Cell. 2015;162:780–794. doi: 10.1016/j.cell.2015.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mohseni M, et al. A genetic screen identifies an LKB1-MARK signalling axis controlling the Hippo-YAP pathway. Nat Cell Biol. 2014;16:108–117. doi: 10.1038/ncb2884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cordenonsi M, et al. The Hippo transducer TAZ confers cancer stem cell-related traits on breast cancer cells. Cell. 2011;147:759–772. doi: 10.1016/j.cell.2011.09.048. [DOI] [PubMed] [Google Scholar]
- 38.Zanconato F, et al. Genome-wide association between YAP/TAZ/TEAD and AP-1 at enhancers drives oncogenic growth. Nat Cell Biol. 2015;17:1218–1227. doi: 10.1038/ncb3216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhang J, et al. YAP-dependent induction of amphiregulin identifies a non-cell-autonomous component of the Hippo pathway. Nat Cell Biol. 2009;11:1444–1450. doi: 10.1038/ncb1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gomez M, Gomez V, Hergovich A. The Hippo pathway in disease and therapy: cancer and beyond. Clinical and translational medicine. 2014;3:22. doi: 10.1186/2001-1326-3-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Moroishi T, Hansen CG, Guan KL. The emerging roles of YAP and TAZ in cancer. Nat Rev Cancer. 2015;15:73–79. doi: 10.1038/nrc3876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shimo T, et al. Pathogenic role of connective tissue growth factor (CTGF/CCN2) in osteolytic metastasis of breast cancer. J Bone Miner Res. 2006;21:1045–1059. doi: 10.1359/jbmr.060416. [DOI] [PubMed] [Google Scholar]
- 43.Engelman JA, et al. MET amplification leads to gefitinib resistance in lung cancer by activating ERBB3 signaling. Science. 2007;316:1039–1043. doi: 10.1126/science.1141478. [DOI] [PubMed] [Google Scholar]
- 44.Sergina NV, et al. Escape from HER-family tyrosine kinase inhibitor therapy by the kinase-inactive HER3. Nature. 2007;445:437–441. doi: 10.1038/nature05474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hellyer NJ, Kim MS, Koland JG. Heregulin-dependent activation of phosphoinositide 3-kinase and Akt via the ErbB2/ErbB3 co-receptor. J Biol Chem. 2001;276:42153–42161. doi: 10.1074/jbc.M102079200. [DOI] [PubMed] [Google Scholar]
- 46.Gentile A, Lazzari L, Benvenuti S, Trusolino L, Comoglio PM. The ROR1 pseudokinase diversifies signaling outputs in MET-addicted cancer cells. Int J Cancer. 2014;135:2305–2316. doi: 10.1002/ijc.28879. [DOI] [PubMed] [Google Scholar]
- 47.Tao JJ, et al. Antagonism of EGFR and HER3 enhances the response to inhibitors of the PI3K-Akt pathway in triple-negative breast cancer. Science signaling. 2014;7:ra29. doi: 10.1126/scisignal.2005125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bae SY, et al. HER3 status by immunohistochemistry is correlated with poor prognosis in hormone receptor-negative breast cancer patients. Breast cancer research and treatment. 2013 doi: 10.1007/s10549-013-2570-6. [DOI] [PubMed] [Google Scholar]
- 49.Yaffe MB. Phosphotyrosine-binding domains in signal transduction. Nat Rev Mol Cell Biol. 2002;3:177–186. doi: 10.1038/nrm759. [DOI] [PubMed] [Google Scholar]
- 50.Johnson R, Halder G. The two faces of Hippo: targeting the Hippo pathway for regenerative medicine and cancer treatment. Nature reviews. Drug discovery. 2014;13:63–79. doi: 10.1038/nrd4161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yu FX, et al. Regulation of the Hippo-YAP pathway by G-protein-coupled receptor signaling. Cell. 2012;150:780–791. doi: 10.1016/j.cell.2012.06.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Haag S, et al. NSUN6 is a human RNA methyltransferase that catalyzes formation of m5C72 in specific tRNAs. RNA. 2015;21:1532–1543. doi: 10.1261/rna.051524.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Xing Z, et al. lncRNA Directs Cooperative Epigenetic Regulation Downstream of Chemokine Signals. Cell. 2014;159:1110–1125. doi: 10.1016/j.cell.2014.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Glantschnig H, Rodan GA, Reszka AA. Mapping of MST1 kinase sites of phosphorylation. Activation and autophosphorylation. J Biol Chem. 2002;277:42987–42996. doi: 10.1074/jbc.M208538200. [DOI] [PubMed] [Google Scholar]
- 55.Sapru MK. Neuregulin-1 regulates expression of the Ets-2 transcription factor. Life Sci. 2001;69:2663–2674. doi: 10.1016/s0024-3205(01)01343-1. [DOI] [PubMed] [Google Scholar]
- 56.Minn AJ, et al. Genes that mediate breast cancer metastasis to lung. Nature. 2005;436:518–524. doi: 10.1038/nature03799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhang C, Yu D. Microenvironment determinants of brain metastasis. Cell & bioscience. 2011;1:8. doi: 10.1186/2045-3701-1-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zhang S, et al. SRC family kinases as novel therapeutic targets to treat breast cancer brain metastases. Cancer Res. 2013;73:5764–5774. doi: 10.1158/0008-5472.CAN-12-1803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wang W, et al. AMPK modulates Hippo pathway activity to regulate energy homeostasis. Nat Cell Biol. 2015;17:490–499. doi: 10.1038/ncb3113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sorrentino G, et al. Metabolic control of YAP and TAZ by the mevalonate pathway. Nat Cell Biol. 2014;16:357–366. doi: 10.1038/ncb2936. [DOI] [PubMed] [Google Scholar]
- 61.Lin SY, et al. Nuclear localization of EGF receptor and its potential new role as a transcription factor. Nat Cell Biol. 2001;3:802–808. doi: 10.1038/ncb0901-802. [DOI] [PubMed] [Google Scholar]
- 62.Haskins JW, Nguyen DX, Stern DF. Neuregulin 1-activated ERBB4 interacts with YAP to induce Hippo pathway target genes and promote cell migration. Science signaling. 2014;7:ra116. doi: 10.1126/scisignal.2005770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Xue Z, et al. A G-Rich Motif in the lncRNA Braveheart Interacts with a Zinc-Finger Transcription Factor to Specify the Cardiovascular Lineage. Mol Cell. 2016;64:37–50. doi: 10.1016/j.molcel.2016.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Castello A, et al. Insights into RNA biology from an atlas of mammalian mRNA-binding proteins. Cell. 2012;149:1393–1406. doi: 10.1016/j.cell.2012.04.031. [DOI] [PubMed] [Google Scholar]
- 65.Bittner N, et al. Bone Metastases and the EGFR and KRAS Mutation Status in Lung Adenocarcinoma--The Results of Three Year Retrospective Analysis. Pathology oncology research : POR. 2015;21:1217–1221. doi: 10.1007/s12253-015-9955-2. [DOI] [PubMed] [Google Scholar]
- 66.Karachaliou N, et al. ROR1 as a novel therapeutic target for EGFR-mutant non-small-cell lung cancer patients with the EGFR T790M mutation. Translational lung cancer research. 2014;3:122–130. doi: 10.3978/j.issn.2218-6751.2014.03.02. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The breast cancer RNA-seq data used to analyze MAYA expression were derived from the TCGA Research Network: http://cancergenome.nih.gov/, and the breast cancer RNA-seq BAM files were downloaded from the UCSC Cancer Genomics Hub (CGHub, https://cghub.ucsc.edu/). Source data for all mass spectrometry experiments (Figs. 2a, and Supplementary Figs. 2a, 3a, 3b, 3e, 4a) have been provided as Supplementary Table 2, 3 and 5. Statistics source data for Figs 1d, h–i, j, k–l, 2l, 4c, k–l, 5h, 8c–d, f–k, m–n, and Supplementary Figs 1i–l, 2i–l, 6e–h, 7j, 8d, e, i, j and k–l have been provided in Supplementary Table 8. All other data supporting the findings of this study are available from the corresponding author on reasonable request.