Abstract
Nocardiosis caused by Nocardia seriolae is one of the major threats in the aquaculture of Seriola species (yellowtail; S. quinqueradiata, amberjack; S. dumerili and kingfish; S. lalandi) in Japan. Here, we report the complete nucleotide genome sequence of N. seriolae UTF1, isolated from a cultured yellowtail. The genome is a circular chromosome of 8,121,733 bp with a G+C content of 68.1% that encodes 7,697 predicted proteins. In the N. seriolae UTF1 predicted genes, we found orthologs of virulence factors of pathogenic mycobacteria and human clinical Nocardia isolates involved in host cell invasion, modulation of phagocyte function and survival inside the macrophages. The virulence factor candidates provide an essential basis for understanding their pathogenic mechanisms at the molecular level by the fish nocardiosis research community in future studies. We also found many potential antibiotic resistance genes on the N. seriolae UTF1 chromosome. Comparative analysis with the four existing complete genomes, N. farcinica IFM 10152, N. brasiliensis HUJEG-1 and N. cyriacigeorgica GUH-2 and N. nova SH22a, revealed that 2,745 orthologous genes were present in all five Nocardia genomes (core genes) and 1,982 genes were unique to N. seriolae UTF1. In particular, the N. seriolae UTF1 genome contains a greater number of mobile elements and genes of unknown function that comprise the differences in structure and gene content from the other Nocardia genomes. In addition, a lot of the N. seriolae UTF1-specific genes were assigned to the ABC transport system. Because of limited resources in ocean environments, these N. seriolae UTF1 specific ABC transporters might facilitate adaptation strategies essential for marine environment survival. Thus, the availability of the complete N. seriolae UTF1 genome sequence will provide a valuable resource for comparative genomic studies of N. seriolae isolates, as well as provide new insights into the ecological and functional diversity of the genus Nocardia.
Introduction
Members of the genus Nocardia are Gram-positive, non-motile and aerobic actinomycetes, belonging to the family Nocardiaceae. This genus contains more than 90 recognized species and are widely distributed in both aquatic and terrestrial habitats [1]. Many species of this genus are known as the causative agent of nocardiosis in humans and a variety of animals, which cause various clinical diseases and high mortality rates in some cases [2, 3]. In aquatic environments, four species of Nocardia, N. asteroides, N. seriolae, N. salmonicida and N. crassostreae, have also been found in diseased aquatic animals [4].
In Japan, members of the genus Seriola, including yellowtail (S. quinqueradiata), amberjack (S. dumerili), and kingfish (S. lalandi), are the most produced and economically important aquaculture fish species. Nocardiosis, caused by N. seriolae [5] (initially reported as N. kampachi [6]), is one of the most serious economic threats in the Seriola aquaculture. N. seriolae also infects other fish species including both marine and freshwater fishes and is found in other Asian countries [7]. To date, only two antibiotics, sulfamonomethoxine and sulfisozole sodium, are licensed for treatment of N. seriolae infections in Japan [8, 9]. Although these antibiotics are valuable for the control of nocardiosis, there are some concerns about the emergence of antibiotic-resistant strains and environmental impacts. Vaccination is thought to be another effective strategy for control of nocardiosis. However, the intracellular parasitic nature of N. seriolae makes development of vaccines for the disease difficult [10].
Complete genome sequences of pathogenic bacteria provide a powerful tool for understanding their biology, including mechanisms of bacterial pathogenicity and their drug-resistant properties, as well as for the development of new genetic and molecular approaches for disease control strategies [11]. So far, four Nocardia species have been fully sequenced, including three agents of human nocardiosis, N. farcinica IFM 10152 [12], N. brasiliensis HUJEG-1 [13] and N. cyriacigeorgica GUH-2 [14], and a rubber and gutta-percha-degrading strain, N. nova SH22a isolated from a root of Couma macrocarpa [15]. Although two draft sequences of N. seriolae isolates, ZJ0503 [16] and N-2927 [17] have been reported recently, these draft genome sequences consist of a large number of contigs, 319 contigs in 315 scaffolds for ZJ0503 [16] and 339 large contigs (>500bp) for N-2927 [17]. Therefore a complete genome sequence of N. seriolae is essential for a robust annotation, overall genome organization and comparative genomics of this species [18].
In this study, using Single Molecule, Real-Time (SMRT) DNA sequencing [19, 20], the complete genome nucleotide sequence of N. seriolae UTF1 isolated from a yellowtail that succumbed to nocardiosis in Japan was determined and annotated. We explored the virulence factors and antibiotic resistance gene candidates in the N. seriolae UTF1 genome. In addition, to investigate genomic diversity, the N. seriolae UTF1 genome sequence were compared with the four existing complete genomes of Nocardia. This is, to the best of our knowledge, the first report of the complete genome of the fish pathogenic Nocardia species. This genomic information will provide a reference genome data set of N. seriolae that could provide a basis for understanding the ecological and functional diversity of the genus Nocardia by comparative studies in future studies.
Materials and methods
Genome sequencing and assembly
N. seriolae UTF1 was originally isolated from a cultured yellowtail (Seriola quinqueradiata) in 2008 (Miyazaki Prefecture, Japan). This isolate was cultured in Brain Heart Infusion Broth (Difco, Sparks, MD, USA) at 25°C for 5 days under constant shaking with 150 rpm. After treatment with lysozyme, genomic DNA was extracted using the Maxwell Cell DNA Purification Kit (Promega, Madison, WI, USA). The nucleotide sequence of the N. seriolae UTF1 was determined by the Pacific Biosciences (PacBio) RS sequencing platform (Pacific Biosciences, Inc., CA, USA) at Tomy Digital Biology Co., Ltd (Tokyo, Japan). Briefly, genomic DNA (7 μg) was sheared using the g-TUBE (Covaris Inc., MA, USA) and a library was prepared using a DNA Template Prep Kit v2.0 (Pacific Biosciences) by the manufacturer’s instructions. The library was run in four Single Molecule, Real-Time (SMRT) cells on a PacBio RS sequencer (Pacific Biosciences) using P4C2 chemistry and a 120 minute data collection mode. The PacBio RS platform generated 227,796 sequence reads (mean read length: 4,756 bp, N50 read length: 7,364 bp) with 1,083,416,572 bp, providing a 133-fold sequencing coverage of the genome (Table 1). De novo assembly of sequence reads was performed using a SMRT Analysis v2.2.0 software package (Pacific Biosciences) and resulted in two contigs with lengths of 6,093,704 bp and 2,035,833 bp.
Table 1. Sequencing statistics of the N. seriolae UTF1 genome.
| Number of Reads | 227,796 |
| N50 Read Length (bp) | 7,364 |
| Mean Read Length (bp) | 4,756 |
| Number of Bases | 1,083,416,572 |
| Average Reference Coverage | 113.01 |
| Number of contigs | 2 |
For the genome sequence finishing, gaps between the two contigs were amplified by a long-range PCR method. The both gap-flanking sequences encoded ribosomal RNA (rRNA) operons, and therefore the PCR primer sets were designed for the outside of the region of rRNA operons. Using the gap-flanking PCR primer sets shown in S1 Table, long-range PCR was conducted using Phusion High-Fidelity PCR master mix with HF buffer (Thermo Fisher Scientific, Waltham, MA) following the manufacturer’s protocol. The long-range PCR amplification was performed using 50 ng of extracted total genomic DNA of N. seriolae UTF1 with an initial denaturation step of 30 s at 98°C and then a two-step PCR procedure (35 cycles of 98°C for 10 s and 72°C for 3 min), and 10 min of final extension at 72°C. The long-range PCR successfully amplified approximately 7–8 kb fragments correspond to the both gap regions and the two PCR products were purified with QIAquick PCR Purification Kit (QIAGEN, Hilden, Germany). The purified long-range PCR products (100 ng) were shotgun-sequenced using the Ion PGM platform (Thermo Fisher Scientific) and the sequence reads were assembled using V-GAP [21]. These two assembled sequences aligned to the two contigs, and consequently the complete nucleotide sequence of N. seriolae UTF1 comprising a circular chromosome was determined. The final sequence was submitted to DDBJ under accession number AP017900.
Genome annotation
The complete genome sequence of N. seriolae UTF1was annotated using the Rapid Annotations using Subsystems Technology (RAST) server v2.0 with SEED data [22] and using BLASTP [23] against the NCBI RefSeq protein data [24] (E value threshold of 1E-5). Functional categories of the predicted genes of N. seriolae UTF1 and four other Nocardia spp. genes were assigned with the Clusters of Orthologous Groups of proteins (COGs) database [25] using COGsoft [26] and with the Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway database using KEGG Orthology And Links Annotation (BlastKOALA) program [27]. Virulence factors of N. seriolae UTF1 were predicted by BLASTP against the Virulence Factor Database (VFDB) set A (a core dataset that covers genes associated with experimentally verified VFs) [28] and the virulence genes data set of N. farcinica IFM 10152 at the Nocardia farcinica Genome Project Page (http://nocardia.nih.go.jp/), using a cut off E-value of 1E−5. These BLAST results were then filtered using the criteria of query coverage per HSP (qcovhsp) greater than 80% and sequence similarity greater than 50%. The organization of the mce operon structures was identified based on the annotation result from the RAST server and visualized using Easyfig version 2.1 [29]. Genes for antibiotic resistance were estimated by Antibiotic Resistance Genes Database (ARDB) [30] with default parameters.
Comparative genomics
To Identify and characterize the gap regions in the previous reported N. seriolae draft genome sequences, the contig sequences of N. seriolae ZJ0503 (GenBank: NZ_JNCT01000000, 319 contigs) and N-2927 (GenBank: NZ_BAWD02000000, 339 contigs) were aligned to the complete sequence of N. seriolae UTF1 genome using MUMmer version 3.22 [31]. The uncovered region sequences in the comparison with ZJ0503 and with N-2927 were subjected to BLASTX search against the NCBI RefSeq database (E value threshold of 1E−5).
Four available complete genome sequences of the genus Nocardia, N. farcinica IFM 10152 (GenBank: AP006618), N. brasiliensis HUJEG-1 (GenBank: CP003876), N. cyriacigeorgica GUH-2 (GenBank: FO082843) and N. nova SH22a (GenBank: CP006850), were used for the comparative genomic analysis with N. seriolae UTF1. For visualization of circular genome comparisons, the BLASTN-based ring image was generated by BLAST Ring Image Generator (BRIG) version 0.95 [32], with N. seriolae UTF1 as a reference. Dot plots of complete nucleotide sequences were generated by MUMmer version 3.22 and the mummerplot script and the Unix program gnuplot [31]. Average Nucleotide Identity (ANI) and Amino Acid Identity (AAI) were calculated using the ANI calculator and AAI calculator, respectively (default settings) [33]. The orthologous and species-specific genes were identified using OrthoMCL [34]. Comparisons of functional profiling of the N. seriolae UTF1 and four other Nocardia spp. complete genomes were carried out by the method by Verma et al. (2014) [35] with some modification. The top 50 SEED subsystems [36, 37] from the RAST analysis and the KEGG modules assigned by BlastKOALA were clustered hierarchically by the abundance of gene content for each categories among the Nocardia genomes using Cluster 3.0 software [38]. The results were visualized by Java Treeview version 1.1.6r4 [39].
Results and discussion
Assembly and general genomic features
The relatively large genome size (6–10 Mbp) with high GC contents (approximately 70%) of genus Nocardia [40] makes the completion of their genome sequences difficult. Because of the genomic complexity, draft assemblies of N. seriolae from an Illumina MiSeq platform [16] or a Roche 454-GS Junior System combined with the Illumina reads (NCBI Sequence Read Archive, DRX020602) [8, 17] consist of a large number of contigs. It should be noted that our initial draft sequence assembly of the N. seriolae UTF1 genomes using a 454 GS-FLX+ System resulted in a total of 134 scaffolds comprising 365 contigs (unpublished data). The PacBio RS platform, a third-generation sequencing technology and based on single-molecule real-time (SMRT) sequencing, can achieve unbiased GC coverage with extremely long reads [19, 20]. This platform has been employed successfully for sequencing in complex bacterial genomes such as those with extremely high GC content genomes [41] and with multiple chromosomes containing more repetitive sequences [42]. In this study, we determined the complete genome nucleotide sequence of N. seriolae UTF1 using the PacBio RS.
Our de novo assembly with a 133-fold genome coverage of PacBio RS long-reads (mean read length: 4,756 bp, N50 read length: 7,364 bp) produced two large contigs with lengths of 6,093,704 bp and 2,035,833 bp (Table 1). The flanking sequences at both sides of the two assembled contigs encoded ribosomal RNA genes. Since the length of multiple copies of rRNA operons (16S-23S-5S rRNA) are approximately 5 kb, the current sequence reads with an average of 4,756 bp may have not been able to fully cover these rRNA operon regions. It should be noted that the newest PacBio RS II sequencer generates an average read length of 10–15 kb [43], and therefore the new sequencer is assumed to be able to assemble around these rRNA operon regions. After closing the gaps with a long-range PCR method, the complete nucleotide sequence of N. seriolae UTF1 comprising a circular chromosome of 8,121,733 bp with a G+C content of 68.1% was determined (Fig 1 and Table 2). The complete genome of N. seriolae UTF1 contains 7,697 predicted coding DNA sequences (CDSs) with an average length of 909 bp, 4 rRNA operons, and 62 transfer RNA (tRNA) sequences (Table 2). The genome sizes vary among the fully sequenced Nocardia genomes that range from 6,021,225 bp for N. farcinica IFM 10152 to 9,436,348 bp for N. brasiliensis HUJEG-1, while the numbers of CDSs also vary among them that range from 5,491 for N. farcinica IFM 10152 to 8,414 for N. brasiliensis HUJEG-1 (Table 2). On the other hand, the N. seriolae UTF1 genome has four rRNA operons and 62 tRNAs, while three rRNA operons and 49–53 tRNAs have been found in the other four fully sequenced Nocardia genomes (Table 2).
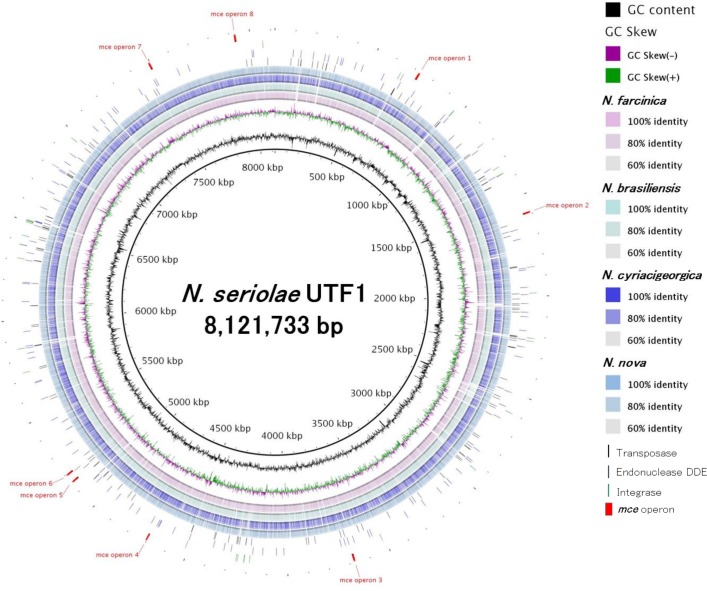
Fig 1. Comparative genomic map of N. seriolae UTF1 genome and the other four Nocardia complete genomes.
The BLASTN-based ring image was generated by BLAST Ring Image Generator (BRIG) version 0.95 [32]. The innermost two rings show GC content (black) and GC skew (purple/green). The remaining rings (rings 3–6) represent a BLASTN comparison with complete genome of N. farcinica IFM 10152 (magenta), N. brasiliensis HUJEG-1(cyan), N. cyriacigeorgica GUH-2 (blue) and N. nova SH22a (pale blue). Bars indicate the position of mobile-element related genes in the N. seriolae UTF1 genome such as transposases (black), endonuclease DDE (blue) and integrase (green). The outermost red boxes highlight the 8 mce operons found in the N. seriolae UTF1 genome.
Table 2. Comparison of genomic features of N. seriolae UTF1 and the other four Nacardia spp. complete genomes.
| Species | host | Accesion number | Size (bp) | GC% | CDS | rRNA | tRNA | Reference |
|---|---|---|---|---|---|---|---|---|
| N. seriolae UTF1 | Seriola quinqueradiata | AP017900 | 8,121,733 | 68.1 | 7,697 | 4 | 62 | This study |
| N. farcinica IFM 10152 | Homo sapiens | AP006618 | 6,021,225 | 70.8 | 5,674 | 3 | 53 | [12] |
| N. brasiliensis HUJEG-1 | Homo sapiens | CP003876 | 9,436,348 | 68.0 | 8,414 | 3 | 51 | [13] |
| N. cyriacigeorgica GUH-2 | Homo sapiens | FO082843 | 6,194,645 | 68.4 | 5,491 | 3 | 49 | [14] |
| N. nova SH22a | A root of Couma macrocarpa | CP006850 | 8,348,532 | 67.8 | 7,583 | 3 | 49 | [15] |
The genome sequences of N. seriolae UTF1 and the previously reported N. seriolae isolates were quite similar. The N. seriolae UTF1 genome showed 99.99 and 99.95% ANI with ZJ0503 [16] and N-2927 [17], respectively. It should be noted that the ANI between N-2927 and U-1 [8], the most recently reported draft genome of N. seriolae isolate, was 100%. Therefore, we did not use the U-1 genome for further comparisons and analysis. To clarify the uncovered regions (gaps) in the previously reported N. seriolae genome sequences, the contig sequences of N. seriolae ZJ0503 [16] (319 contigs) and N-2927 [17] (339 contigs) were aligned to the complete sequence of N. seriolae UTF1 genome. As a result, 300 uncovered regions (average length: 1,373 bp) were detected in the draft genome of ZJ0503, while 297 uncovered regions (average length: 1,502 bp) were detected in the draft genome of N-2927. These gap sequences were subjected to BLASTX searches against the NCBI RefSeq database (E value threshold of 1E−5) and as a result, 294 (98.0%) for ZJ0503 and 290 (97.6%) for N-2927 had significant BLAST hits. The BLAST results revealed that most assigned genes (76.9% for ZJ0503 and 73.1% for N-2927) were mobile element-related, such as for transposase, endonuclease DDE and integrase (S1 Fig). The comparative genomic map of N. seriolae UTF1 with ZJ0503 and N-2927 also showed that these mobile element-related genes are interspersed across the N. seriolae UTF1 genome and coincides with the gap regions of ZJ0503 and N-2927 (S2 Fig). Since the nucleotide length of these genes was more than 1kb, these repeat sequences have a significant influence during de novo assembly when relatively short reads (several hundred bp) from MiSeq and 454-GS Junior are used [8, 16, 17]. In the present study, the PacBio long-reads (mean read length: 4,756 bp, N50 read length: 7,364 bp) could cover over these repeated sequences and achieve completion of the N. seriolae UTF1 genome.
Overview of N. seriolae UTF1 virulence factors
Like causal agents of human nocardiosis, N. seriolae is considered to be an intracellular pathogen that invades and grows within host cells, even including phagocytes [44, 45]. The intracellular nature of this bacteria makes it difficult to control disease. As a first step toward understanding the virulence factors and pathogenic properties of N. seriolae UTF1, we conducted BLASTP searches of N. seriolae UTF1 CDSs against the Virulence Factor Database (VFDB) [28] and a well-annotated virulence genes data set of N. farcinica (http://nocardia.nih.go.jp/) (E-value < 1E-5, sequence length overlap > 80% and sequence similarity > 50%). The VFDB search identified 173 CDSs as candidate virulence factors (S2 Table). In addition, almost all of N. farcinica putative virulence genes were found in the N. seriolae UTF1 genome (Table 3).
Table 3. Virulence factor candidates in the N. seriolae UTF1 by comparison with the virulence genes data set of N. farcinica.
| Category | N. seriorae UTF1 ORF ID | N. farcinica gene ID* | Gene* | Description* | %Identity | %Similarity | E value |
|---|---|---|---|---|---|---|---|
| Cell wall proteins | ORF-145 | nfa1810 | fbpA | mycolyltransferase | 70.9 | 79.9 | 5.00E-159 |
| ORF-7590 | nfa1820 | fbpB | mycolyltransferase | 56.4 | 75.9 | 7.00E-67 | |
| ORF-150 | nfa1830 | fbpC | mycolyltransferase | 57.7 | 73.0 | 2.00E-37 | |
| Metal importers | ORF-5098 | nfa37790 | ideR | transcriptional regulator | 66.0 | 79.4 | 4.00E-84 |
| ORF-950 | nfa7630 | nbtA | thioesterase | 85.7 | 90.2 | 0 | |
| ORF-951 | nfa7640 | nbtB | polyketide synthase | 86.4 | 90.0 | 5.00E-60 | |
| ORF-952 | nfa7650 | nbtC | polyketide synthase | 87.3 | 93.0 | 1.00E-143 | |
| ORF-953 | nfa7660 | nbtD | non-ribosomal peptide synthetase | 88.5 | 93.8 | 0 | |
| ORF-954 | nfa7670 | nbtE | non-ribosomal peptide synthetase | 91.6 | 96.2 | 4.00E-161 | |
| ORF-750 | nfa7680 | nbtF | non-ribosomal peptide synthetase | 93.0 | 97.1 | 2.00E-108 | |
| ORF-948 | nfa7610 | nbtG | lysine-N-oxygenase | 84.7 | 93.1 | 0 | |
| ORF-747 | nfa6190 | nbtS | salicylate synthase | 81.4 | 88.5 | 4.00E-126 | |
| ORF-749 | nfa6200 | nbtT | salicylate-AMP ligase | 82.5 | 91.3 | 4.00E-126 | |
| Oxidative and nitrosative stresses | ORF-5118 | nfa37890 | ahpC | alkylhydroperoxide reductase | 68.0 | 79.0 | 5.00E-149 |
| ORF-5119 | nfa37900 | ahpD | alkylhydroperoxidase | 68.8 | 79.3 | 0 | |
| ORF-325 | nfa55390 | katC | catalase | 79.0 | 87.8 | 0 | |
| ORF-4212 | nfa29500 | katG | catalase-peroxidase | 63.8 | 77.8 | 1.00E-169 | |
| ORF-6237 | nfa45490 | narG | nitrate reductase alpha subunit | 71.3 | 84.8 | 0 | |
| ORF-6238 | nfa45500 | narH | nitrate reductase beta subunit | 72.4 | 83.3 | 1.00E-78 | |
| ORF-6240 | nfa45520 | narI | nitrate reductase gamma subunit | 75.3 | 84.6 | 0 | |
| ORF-6239 | nfa45510 | narJ | nitrate reductase delta subunit | 74.1 | 83.8 | 1.00E-157 | |
| ORF-6265 | nfa45610 | nirB | nitrite reductase (NAD(P)H) subunit | 76.9 | 86.9 | 0 | |
| ORF-6264 | nfa45600 | nirD | nitrite reductase (NAD(P)H) subunit | 75.6 | 84.0 | 1.00E-173 | |
| ORF-5117 | nfa37880 | oxyR | hydrogen peroxide sensing transcriptional regulator | 66.9 | 78.6 | 2.00E-165 | |
| ORF-418 | nfa52980 | sodC | superoxide dismutase | 78.2 | 86.4 | 1.00E-140 | |
| ORF-68 | nfa1210 | sodF | superoxide dismutase | 40.5 | 57.5 | 3.00E-112 | |
| Penetration into mammalian cells | ORF-4357 | nfa34810 | inv | invasin | 65.6 | 80.4 | 2.00E-119 |
| Phagosome arresting | ORF-5937 | nfa13510 | ndk | nucleoside diphosphate kinase | 44.8 | 57.6 | 2.00E-45 |
| ORF-5498 | nfa16310 | ptpA | protein-tyrosine phosphatase | 48.7 | 61.4 | 2.00E-88 | |
| ORF-2644 | nfa18680 | ptpB | protein-tyrosine phosphatase | 60.2 | 70.7 | 0 | |
| other | ORF-2873 | nfa19960 | tlyA | putative cytotoxin/hemolysin | 77.4 | 83.2 | 6.00E-135 |
* according to the Nocardia farcinica Genome Project Page (http://nocardia.nih.go.jp/).
Mammalian cell entry (Mce)-family proteins, virulence factors of Mycobacterium tuberculosis (the class Actinobacteria), have the ability to enter into mammalian cells and survive inside the macrophage [46]. The genome of M. tuberculosis contains four mce operons which comprise eight genes per operon in identical manner (two yrbE genes, A and B; six mce genes, A, B, C, D, E and F) [47, 48]. Mce proteins are found in diverse Actinobacteria including Nocardia spp. [49]. Six copies of mce operons have been found in the three human clinical isolates (N. farcinica IFM 10152 [12], N. brasiliensis HUJEG-1 [13] and N. cyriacigeorgica GUH-2 [14]), whereas 14 mce operons have been found in N. nova SH22, which was isolated from a plant root and has the ability of rubber and gutta-percha degradation [15]. Recently, Carrillo-González et al. (2016) demonstrated the importance of mce proteins for Nocardia pathogenesis from whole-genome comparison of an attenuated N. brasiliensis HUJEG-1 and the parental strain [50]. In the N. seriolae UTF1 genome, we found eight complete mce loci with nucleotide length of 6,814 bp to 8,964 bp (Figs 1 and 2). It should be noted that the locus mce3 has two extra genes (endonuclease DDE and orf3406) between mce3E and mce3F (Fig 2). However, the influence of these two extra genes to the function of the mce3 operon is unclear. Amino acid sequence similarities of N. seriolae UTF1 Mce1 proteins with the other four Nocardia species, and two Actinobacteria, Rhodococcus equi and M. tuberculosis are shown in S3 Table. The Mce1C protein is highly conserved among Mce1 proteins (87.8–90.7% similarities with Nocardia spp., 82.0% similarity with R. equi and 63.8% similarity with M. tuberculosis), while the Mce1E protein exhibit a relatively lower sequence homology (75.4–80.0% similarities with Nocardia spp., 75.3% similarity with R. equi and 63.3% similarity with M. tuberculosis). Invasin also plays a role in attachment and penetration into host cells by several bacterial species [51, 52], Nocardia species also possess an invasin gene [12, 13] and the N. seriolae UTF1 ORF-4357 is very similar to N. farcinica IFM 10152 invasin (80.4% similarity) (Table 3). Since Nocardia species are facultative intracellular pathogens, they most likely use this protein for entry into host cells. Further studies to determine the function of Nocardia invasin involved in the host cell entry are required.
Fig 2. The organization of 8 mce operons in N. seriolae UTF1 genome.
The figure was generated using Easyfig 2.1 [29]. Arrows represent yrbE genes, mce genes and two additional genes, DDE (endonuclease DDE) and orf3406 (hypothetical protein) in mce3. Values indicate the number of base positions. Asterisks (mce6* and mce7*) indicate reverse complement orientation.
Nocardia species can modulate phagocyte function and can grow in macrophages. It has been reported that catalase and superoxide dismutase (SOD) impair the ability of the oxidative killing mechanisms of phagocytes [53]. In the N. seriolae UTF1 genome, two catalases (katC, ORF-325; katG, ORF-4212) and two sod (sodC, ORF-418; sodF, ORF-68) genes are present (Table 3). We also found the N. seriolae UTF1 CDSs that are homologous to the N. farcinica IFM 10152 nitrate reductase genes, narG (84.8%), narH (83.3%), narI (84.6%), narJ (83.8%), nirB (86.9%) and nirD (84.0%) (Table 3), which may contribute to survival under low-oxygen conditions in stimulated macrophages [54, 55]. Recently, it has been reported that the attenuated N. brasiliensis HUJEG-1 lost a catalase, SOD and several nitrate reductase genes, suggesting that these genes may be associated to Nocardia spp. pathogenesis [50]. In addition to the above oxidative and nitrosative stress-related genes, alkylhydroperoxidases, AhpC and AhpD, have a crucial role for antioxidant defense in mycobacterial species [56, 57], particularly when the KatG catalase-peroxidase activity is depressed [58]. In the AhpC/AhpD antioxidant defense system, OxyR acts as a positive regulator for their expression [59]. The N. seriolae UTF1 genome contains these genes similar to N. farcinica IFM 10152 with a protein homology of ahpC (79.0%), ahpD (79.3%) and oxyR (78.6%) (Table 3).
A phagosome, a cellular compartment, is essential for intracellular killing and digesting of pathogenic microorganisms [60]. Nucleoside diphosphate kinase (Ndk) and protein tyrosine phosphatase A (PtpA) arrest macrophage phagosomal maturation for the intracellular survival and persistence of pathogenic mycobacteria [61]. The N. seriolae UTF1 ORF-5937 and ORF-5498 are homologous to N. farcinica IFM 10152 ndk (57.6%) and ptpA (61.4%), respectively (Table 3). Since iron is in very low concentration and in an insoluble state within macrophages, efficient iron-acquisition systems are required for pathogenic bacteria to survive [61]. Putative N. seriolae UTF1 metal importer related genes (ideR and nbtA–G, S, T) were identified (Table 3), and may contribute to the abilities of N. seriolae to survive in fish tissues including within macrophages.
Other virulence factor candidates were also found in the N. seriolae UTF1 genome. Antigen 85 (Ag85) complex of M. tuberculosis is a family of fibronectin binding proteins (Fbp) that plays an essential role in the pathogenesis of tuberculosis [62]. The Ag85 complex consists of three proteins (Ag85A, Ag85B and Ag85C: encoded by the genes fbpA, fbpB and fbpC) that possess mycolyltransferase activity involved in the final stages of mycobacterial cell wall assembly [63]. The N. seriolae UTF1 genome was found to have at least seven putative fbp genes: three fbpA (ORF-144, ORF-145 and ORF-147), two fbpB (ORF-146 and ORF-7590) and two fbpC (ORF-148 and ORF-150). Among these seven fbp gene candidates, protein sequences of ORF-145 (fbpA), ORF-7590 (fbpB) and ORF-150 (fbpC) were most similar to N. farcinica IFM 10152 fbpA (79.9%), fbpB (75.9%) and fbpC (73.0%) (Table 3). TlyA proteins of the pathogenic bacteria has a virulent hemolytic ability [64, 65], and the N. seriolae UTF1 ORF-2873 encodes a TlyA, with a homology of 83.2% to the N. farcinica IFM 10152 (Table 3). In contrast to the PtpA (mentioned above), PtpB is considered to be nonessential for the phagosome arresting function of M. tuberculosis [61]. On the other hand, Zhoua et al. (2010) reported that M. tuberculosis PtpB depresses the innate immune responses by inhibiting the signaling pathway involved in interleukin-6 (IL-6) production and promoting host cell survival by activating the Akt pathway for their survival in macrophages [66]. The N. seriolae UTF1 genome contains a ptpB gene (ORF-2644), with a similar protein in N. farcinica IFM 10152 (70.7%) (Table 3).
Overall, the whole-genome analysis of N. seriolae UTF1 reveals that the genome contains known virulence genes of mycobacteria and human clinical Nocardia isolates for host cell invasion, modulation of phagocyte function and surviving inside the macrophages. Therefore, the presence of these virulence genes in the N. seriolae UTF1 may explain their ability to survive intracellularly and within macrophages. The virulence gene set of N. seriolae UTF1 we present provides the material basis for further study of their pathogenic mechanisms at the molecular level. In addition, the complete genome sequence of N. seriolae UTF1 can be utilized for comparing the genomes between pathogenic and non (less)-pathogenic isolates to more fully resolve the genes responsible for N. seriolae pathogenesis in future studies.
Potential antibiotic resistance genes of N. seriolae UTF1
In general, Nocardia spp. are naturally resistant to many antibiotics and most β-lactams [7, 67–69]. The N. seriolae UTF1 encodes at least 14 β-lactamases, while the number of β-lactamase genes is one for N. farcinica IFM 10152 [12], 29 for N. brasiliensis HUJEG-1 [13] and 12 for N. cyriacigeorgica GUH-2 [14]. In addition, as found in the draft genome of N. seriolae genomes [8, 16, 17], the N. seriolae UTF1 genome has one vancomycin and two fluoroquinolones resistant gene candidates according to the RAST annotation. To explore more antibiotic resistant genes in the N. seriolae UTF1 chromosome, all their CDSs were compared with the Antibiotic Resistance Genes Database (ARDB) [30]. This analysis identified 20 CDSs as candidate antibiotic resistant genes that were classified into 16 antibiotic resistance gene types (Table 4). The presence of these antibiotic resistance genes of the N. seriolae UTF1 may, in part, explain the difficulty in treating diseases caused by N. seriolae.
Table 4. Potential antibiotic resistance genes of N. seriolae UTF1 according to the Antibiotic Resistance Genes Database (ARDB).
| Resistance gene type | Antibiotic resistance | Description | UTF1 ORF ID | Best Hit Accession | E-Value | Score |
|---|---|---|---|---|---|---|
| aac(6’)-Ic | Amikacin, dibekacin, isepamicin, netilmicin, sisomicin, tobramycin | Aminoglycoside N-acetyltransferase, which modifies aminoglycosides by acetylation. | ORF-1348 | AAA26549 | 6.00E-32 | 130 |
| bacA | Bacitracin | Undecaprenyl pyrophosphate phosphatase, which consists in the sequestration of Undecaprenyl pyrophosphate. | ORF-4188 | CAL13705 | 8.00E-23 | 101 |
| bl2a_iii2 | Penicillin | Class A beta-lactamase. This enzyme breaks the beta-lactam antibiotic ring open and deactivates the molecule's antibacterial properites. | ORF-6138 | YP_089932 | 8.00E-65 | 241 |
| carA | Lincosamide, macrolide, streptogramin b | ABC transporter system, Macrolide-Lincosamide-Streptogramin B efflux pump. | ORF-4637 | AAC32027 | 2.00E-10 | 58 |
| catbB1 | Chloramphenicol | Group B chloramphenicol acetyltransferase, which can inactivate chloramphenicol. Also referred to as xenobiotic acetyltransferase. | ORF-1707 | AAK88712 | 3.00E-06 | 45 |
| dfrA26 | Trimethoprim | Group A drug-insensitive dihydrofolate reductase, which can not be inhibited by trimethoprim. | ORF-6939 | CAL48457 | 8.00E-29 | 120 |
| macB | Macrolide | Resistance-nodulation-cell division transporter system. Multidrug resistance efflux pump. Macrolide-specific efflux system. | ORF-1987 | YP_001453760 | 9.00E-54 | 204 |
| ORF-4813 | YP_001571041 | 1.00E-46 | 180 | |||
| ORF-6113 | YP_001334578 | 5.00E-47 | 181 | |||
| ORF-6376 | YP_001453760 | 2.00E-49 | 189 | |||
| mfpA | Fluoroquinolone | Pentapeptide repeat family, which protects DNA gyrase from the inhibition of quinolones. | ORF-3773 | ABL05132 | 6.00E-08 | 52 |
| otr(B) | Tetracycline | Major facilitator superfamily transporter, tetracycline efflux pump. | ORF-1750 | AAD04032 | 1.00E-105 | 375 |
| srmB | Lincosamide, macrolide, streptogramin b | ABC transporter system, Macrolide-Lincosamide-Streptogramin B efflux pump. | ORF-4948 | CAA45050 | 9.00E-68 | 250 |
| tcmA | Tetracenomycin c | Major facilitator superfamily transporter. Resistance to tetracenomycin C by an active tetracenomycin C efflux system which is probably energized by transmembrane electrochemical gradients. | ORF-4285 | AAA67509 | 3.00E-05 | 40 |
| tcr3 | Tetracycline | Major facilitator superfamily transporter, tetracycline efflux pump. | ORF-6293 | BAA07390 | 8.00E-99 | 355 |
| vanRB | Vancomycin | VanB type vancomycin resistance operon genes, which can synthesize peptidoglycan with modified C-terminal D-Ala-D-Ala to D-alanine—D-lactate. | ORF-5538 | ABB53368 | 1.00E-13 | 70 |
| ORF-7016 | ABB53368 | 3.00E-16 | 79 | |||
| vanRC | Vancomycin | VanC type vancomycin resistance operon genes, which can synthesize peptidoglycan with modified C-terminal D-Ala-D-Ala to D-alanine—D-serine. | ORF-2855 | AAY67971 | 3.00E-31 | 129 |
| vatB | Streptogramin a | Virginiamycin A acetyltransferase, which can inactivate the target drug. | ORF-4237 | YP_001038094 | 9.00E-10 | 57 |
| vatC | Streptogramin a | Virginiamycin A acetyltransferase, which can inactivate the target drug. | ORF-3078 | AAG21695 | 6.00E-49 | 187 |
It has been reported that N. seriolae isolates could be divided into two phenotypic groups using α-glucosidase (α-glu) activity (α-glu-positive or -negative) [7, 68]. These two groups showed different oxytetracycline (OTC: a tetracyclinic antibiotics)/erythromycin (Em: a macrolide antibiotic) susceptibility profiles. Most of the α-glu-positive isolates were OTC-resistant and Em-sensitive, while most of α-glu-negative isolates were OTC-sensitive and Em-resistant [69,70]. N. seriolae UTF1 was α-glu-positive isolate which exhibited resistance to OTC and sensitivity to Em. Ismail et al. 2011 [69] found that OTC-resistant strains of N. seriolae possess tet(K) and/or tet(L) gene(s), while the Em-resistant strains possessed mef(A) and msr(D) genes. The tet(K) and tet(L) genes are generally found on small transmissible plasmids [71,72], while mef(A) and msr(D) genes are encoded in chromosomes of Gram-positive bacteria and associated with conjugative transposons [73]. Despite obtaining a 1.1 Gbp (133-fold genome coverage) of PacBio RS long-reads in this study and a 247 Mbp (30-fold genome coverage) of 454 reads (unpublished data), we could not find any plasmids or tet(K) and tet(L) gene sequences. The absence of these genes might have been caused by plasmid elimination during culture propagation. On another front, the N. seriolae UTF1 chromosome was found to have two tetracycline resistance gene candidates, otr(B) (ORF-1750) and tcr3 (ORF-6293) (Table 4), both found in Streptomyces spp. (Actinobacteria) [71], suggesting that these two genes might enhance some degree of resistance to OTC in N. seriolae.
As expected, mef(A) and msr(D) genes were not found in the N. seriolae UTF1 chromosome. However, the results from ARDB includes three candidates for macrolide efflux pump genes: carA (ORF-4637), macB (ORF-1987, ORF-4813, ORF-6113 and ORF-6376) and srmB (ORF-4948) (Table 4). Since N. seriolae UTF1 is an Em-sensitive isolate, these three candidates are not likely to be involved in Em-resistance of N. seriolae UTF1. In particular, MacB requires TolC and MacA for its function in E. coli [74,75], but the N. seriolae UTF1 genome lacks both genes. Thus, further information on the antibiotic profile of more N. seriolae isolates, as well as their genomic sequences are needed for an accurate view of their antibiotic resistance genes, and the complete genome sequence of the N. seriolae UTF1 can be used as a reference for these surveys in future studies.
Genome comparison with fully sequenced genomes of the genus Nocardia
Four available complete genome sequences of the genus Nocardia; N. farcinica IFM 10152, N. brasiliensis HUJEG-1, N. cyriacigeorgica GUH-2 and N. nova SH22a, were used for comparison of the genomic structure with the N. seriolae UTF1. From the comparative genomic map (Fig 1), we found that there were no large-scale variations among the genomes, but a considerable number of non-homologous regions are scattered around the N. seriolae UTF1 genome. Most of these non-homologous regions were linked to mobile element-related genes (transposase, endonuclease DDE and integrase) (Fig 1), suggesting they serve as plastic and variable regions for the N. seriolae UTF1 genome.
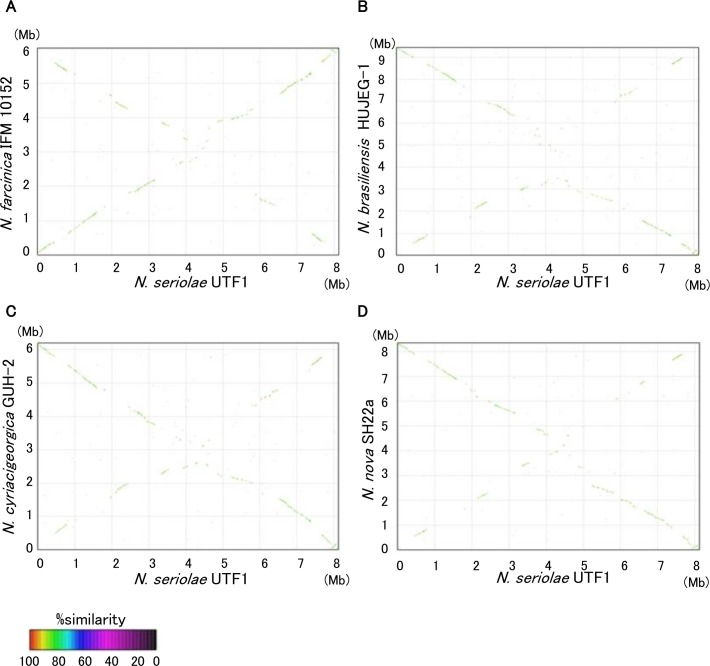
The whole-genome alignments of N. seriolae UTF1 with the other four Nocardia displayed an X-shaped distribution across the origin of replication (Fig 3), which is explained by the fork replication theory [76]. By contrast, the alignments of the three agents of human nocardiosis; N. farcinica IFM 10152, N. brasiliensis HUJEG-1 and N. cyriacigeorgica GUH-2, showed only a few visible symmetric inversions and a diagonal line with a slope of approximately 1 (S3 Fig). On the other hand, the alignments of N. nova SH22a and the three human clinical isolates were arranged in an X-shaped distribution as in the case of N. seriolae UTF1. Thus, the degree of symmetrical inversions indicates that the genetic distance among the three human clinical isolates are close to each other, while N. seriolae UTF1 and N. nova SH22a are genetically distant from these species.
Fig 3.
Dot plot comparisons of N. seriolae UTF1 genome against N. farcinica IFM 10152 (A), N. brasiliensis HUJEG-1 (B), N. cyriacigeorgica GUH-2 (C) and N. nova SH22a (D). Nucleotide-based alignments were performed with MUMmer version 3.22 and dot plots were generated by the mummerplot script and the Unix program gnuplot [31].
The ANI and AAI based pairwise comparisons between each genome are shown in Table 5. Typically, the ANI values between genomes of the same bacterial species show above 95%, while the values below 75% are too divergent to be compared based on this measurement [33]. For the latter case, AAI provides a much more robust resolution. AAI cut-offs for genus and species boundary have been estimated to be 55–60% and 85–90%, respectively [33]. The ANI and AAI between N. seriolae UTF1 and the other four Nocardia genomes ranged between 79.21–79.88% and 68.96–69.62%, respectively. Like the results of whole-genome alignments, the three human clinical isolates showed higher ANI and AAI values (81.19–81.61% ANI and 73.61–74.99% AAI) than the comparisons with N. nova SH22a (79.43–79.89% ANI and AAI (68.06–69.82%) and with N. seriolae UTF1 (described above). On the other hand, the ANI and AAI between N. seriolae UTF1 and N. nova SH22a had relatively low values, that displayed 79.63% ANI and 69.17% AAI, respectively. Overall, the ANI and AAI values indicate that the three human clinical isolates are a genetically similar group and N. seriolae UTF1, an agent of fish nocardiosis, is genetically distant from these species as well as N. nova SH22, an environmental isolate.
Table 5. Average nucleotide identity (ANI, upper grids) and average amino acid identity (AAI, lower grids) values (in percent) calculated between Nocardia genomes.
| N. seriolae UTF1 | N. farcinica IFM 10152 | N. brasiliensis HUJEG-1 | N. cyriacigeorgica GUH-2 | N. nova SH22a | |
|---|---|---|---|---|---|
| N. seriolae UTF1 | - | 79.88 | 79.21 | 79.88 | 79.63 |
| N. farcinica IFM 10152 | 69.17 | - | 81.36 | 81.61 | 79.77 |
| N. brasiliensis HUJEG-1 | 68.96 | 73.61 | - | 81.19 | 79.43 |
| N. cyriacigeorgica GUH-2 | 69.62 | 74.99 | 73.68 | - | 79.89 |
| N. nova SH22a | 69.17 | 69.21 | 68.06 | 69.82 | - |
We also focused on similarity of the functional profiling among the five complete Nocardia genomes. Fig 4 shows a dendrogram constructed based on the top 50 subsystems from the RAST analysis. Interestingly, although a distant genetic relationship was observed between N. seriolae UTF1 and the three agents of human nocardiosis through whole genome comparisons, the analysis revealed that the functional repertoire of N. seriolae UTF1 is closer to the three agents of human nocardiosis than N. nova SH22a (Fig 4). Similar results were obtained with the dataset of overall subsystems (S4 Fig) and the KEGG modules assigned by BlastKOALA (S5 Fig). These findings suggest that the closer functional relationship between the agents of the human and fish nocardiosis may be associated with their adaptations to infected animal hosts and pathogenic properties.
Fig 4. Functional profiling of the five Nocardia complete genomes.
Heat map shows the abundance of the top 50 subsystems [36, 37] enriched in the five Nocardia genomes. The color scale indicates the abundance of gene content for each category.
In summary, when the five complete genomes of Nocardia are compared, we found that N. seriolae UTF1 is genetically distant from the three agents of human nocardiosis, but is similar to the functional repertoire of them. Further research studies are required to more fully resolve the phylogenetic relationship of Nocardia spp. [40] and the differentially enriched pathways according to their habitat and lifestyle, when a greater number of completely sequenced and thoroughly annotated clinical and environmental Nocardia isolates become available.
Finding functional features in N. seriolae UTF1 genome
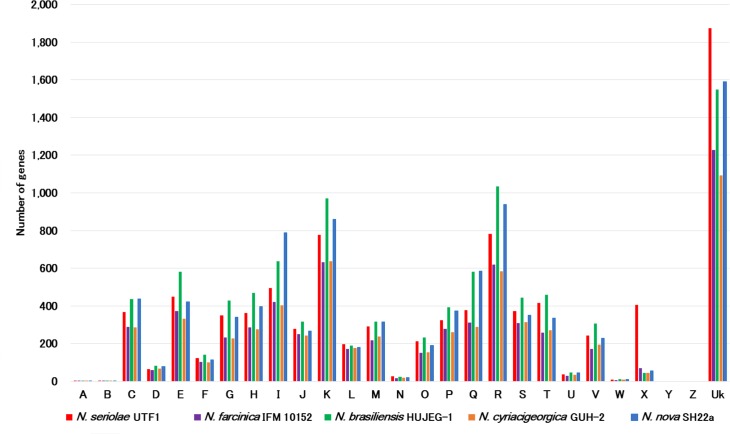
The complete sequence of the N. seriolae UTF1 genome has been determined for the first time as a marine Nocardia species, and therefore it is interesting to investigate the characteristic features of its genome. According to the COG classifications, the distribution of COG categories is mostly similar among Nocardia genomes (Fig 5). In the N. seriolae UTF1, the number of genes for the category ‘Mobilome: prophages, transposons (X)’ and ‘Unknown’ (not assignable to COG categories) is higher than that in the other four Nocardia genomes (Fig 5). Notably, genes related to ‘Mobilome: prophages, transposons (X)’ are quite abundant in the N. seriolae UTF1. The N. seriolae UTF1 genome contains 406 genes in this category, compared to only 71 in N. farcinica IFM 10152, 44 in N. brasiliensis HUJEG-1, 45 in N. cyriacigeorgica GUH-2 and 56 in N. nova SH22a (S4 Table). According to the comparative genomic map in Fig 1, these mobile element genes are interspersed throughout the N. seriolae UTF1genome, and their sequences correspond to the variable regions (Fig 1). Overall, the abundance of the genes related to mobile element proteins and unknown function in the N. seriolae UTF1 genome can partially explain the divergence of their genome structure and gene content from the other Nocardia genomes (Figs 1 and 3).
Fig 5. Comparison of COG distribution of N. seriolae UTF1 and the other four Nocardia genomes.
COG definitions are described as follows: A, RNA processing and modification; B, Chromatin structure and dynamics; C, Energy production and conversion; D, Cell cycle control, cell division, chromosome partitioning; E, Amino acid transport and metabolism; F, Nucleotide transport and metabolism; G, Carbohydrate transport and metabolism; H, Coenzyme transport and metabolism; I, Lipid transport and metabolism; J, Translation, ribosomal structure and biogenesis; K, Transcription; L, Replication, recombination and repair; M, Cell wall/membrane/envelope biogenesis; N, Cell motility; O, Posttranslational modification, protein turnover, chaperones; P, Inorganic ion transport and metabolism; Q, Secondary metabolites biosynthesis, transport and catabolism; R, General function prediction only; S, Function unknown; T, Signal transduction mechanisms; U, Intracellular trafficking, secretion, and vesicular transport; V, Defense mechanisms; W, Extracellular structures; X, Mobilome: prophages, transposons; Y, Nuclear structure; Z, Cytoskeleton. ‘Uk’ indicate unknown (unassigned genes). The number of ORFs with each COG category are listed in S4 Table.
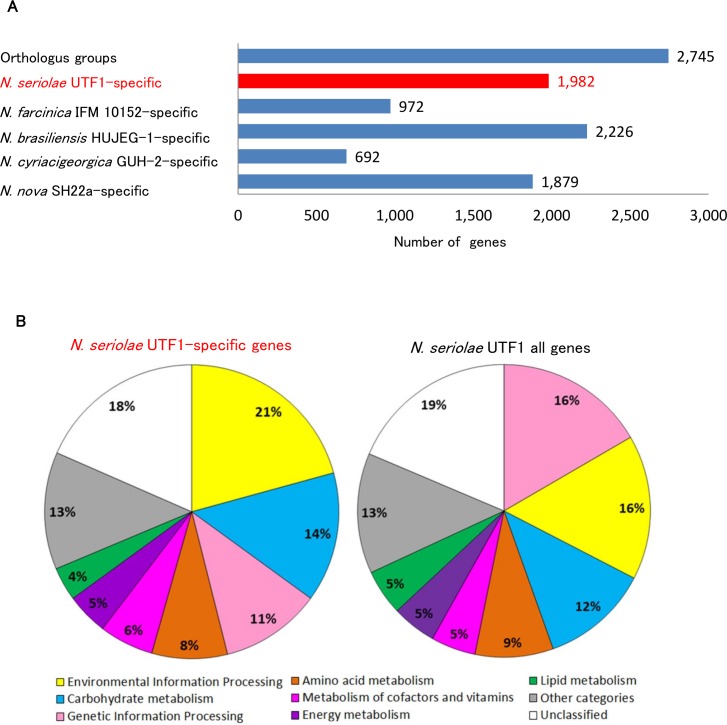
To further characterize the genomic features of N. seriolae UTF1, we focus on the functional characterization of the N. seriolae UTF1-specific genes. As based on the OrthoMCL clustering, 2,745 orthologous genes were presented in all 5 Nocardia genomes and 1,982 genes were unique to N. seriolae UTF1 (Fig 6A). Of the 1,982 N. seriolae UTF1-specific genes, 217 genes (10.9%) were annotated in the KEGG database. The proportion of KEGG categories of the N. seriolae UTF1-specific genes shows some differences compared to those of all N. seriolae UTF1 genes (Fig 6B). The 'Environmental Information Processing' category (21%) are the most abundant in the N. seriolae UTF1-specific genes, followed by ‘Carbohydrate metabolism’ (14%) (Fig 6B). Focusing on the abundant KEGG modules, 14 genes are assigned in the ABC transport system (Module ID: M00254 and M00258) (Table 6). Because of limited resources in ocean environments, marine bacteria require various efficient transport systems to capture essential nutrients [77]. Therefore, the N. seriolae UTF1-specific transport systems-related genes identified in the present study may have a role in adaptation to the marine environment. In addition, most of the N. seriolae UTF1-specific genes (89.1%) were not assignable to KEGG. Further study on such transport systems-related genes and to the genes of unknown function will provide unique insight into their adaptation to fish hosts in the aquatic environment.
Fig 6.
Identification of N. seriolae UTF1-specific genes (A) and their functional annotations (B). The orthologous and species-specific genes were identified using OrthoMCL [34] (A). The protein sequences of 1,982 N. seriolae UTF1-specific genes were functionally annotated with metabolic information from the Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway database using KEGG Orthology And Links Annotation (BlastKOALA) program [27] (B).
Table 6. Functional classification of unique genes of N. seriolae UTF1 by KEGG modules.
| KEGG Module | Module ID | Description | Number of genes |
|---|---|---|---|
| Environmental information processing | |||
| ABC-2 type and other transport systems | M00254 | ABC-2 type transport system | 12 |
| M00258 | Putative ABC transport system | 2 | |
| Mineral and organic ion transport system | M00188 | NitT/TauT family transport system | 1 |
| M00190 | Iron(III) transport system | 2 | |
| M00299 | Spermidine/putrescine transport system | 4 | |
| Phosphotransferase system (PTS) | M00273 | PTS system, fructose-specific II component | 3 |
| Phosphate and amino acid transport system | M00233 | Glutamate transport system | 1 |
| Two-component regulatory system | M00452 | CusS-CusR (copper tolerance) two-component regulatory system | 2 |
| M00475 | BarA-UvrY (central carbon metabolism) two-component regulatory system | 1 | |
| Bacterial secretion system | M00335 | Sec (secretion) system | 1 |
| Drug efflux transporter/pump | M00713 | Fluoroquinolone resistance, efflux pump LfrA | 1 |
| Drug resistance | M00742 | Aminoglycoside resistance, protease FtsH | 1 |
| M00743 | Aminoglycoside resistance, protease HtpX | 1 | |
| M00727 | Cationic antimicrobial peptide (CAMP) resistance, N-acetylmuramoyl-L-alanine amidase AmiA and AmiC | 1 | |
| M00745 | Imipenem resistance, repression of porin OprD | 2 | |
| M00714 | Multidrug resistance, efflux pump QacA | 1 | |
| Energy metabolism | |||
| ATP synthesis | M00159 | V-type ATPase, prokaryotes | 5 |
| M00149 | Succinate dehydrogenase, prokaryotes | 4 | |
| M00151 | Cytochrome bc1 complex respiratory unit | 1 | |
| Nitrogen metabolism | M00530 | Dissimilatory nitrate reduction, nitrate = > ammonia | 1 |
| Methane metabolism | M00345 | Formaldehyde assimilation, ribulose monophosphate pathway | 1 |
| Carbohydrate and lipid metabolism | |||
| Fatty acid metabolism | M00083 | Fatty acid biosynthesis, elongation | 3 |
| M00082 | Fatty acid biosynthesis, initiation | 1 | |
| Central carbohydrate metabolism | M00307 | Pyruvate oxidation, pyruvate = > acetyl-CoA | 3 |
| Nucleotide and amino acid metabolism | |||
| Serine and threonine metabolism | M00555 | Betaine biosynthesis, choline = > betaine | 1 |
| Cysteine and methionine metabolism | M00021 | Cysteine biosynthesis, serine = > cysteine | 2 |
| Other amino acid metabolism | M00027 | GABA (gamma-Aminobutyrate) shunt | 1 |
| Genetic information processing | |||
| Ribosome | M00178 | Ribosome, bacteria | 1 |
Conclusions
The complete nucleotide sequence of N. seriolae UTF1 consists of a circular chromosome of 8,121,733 bp with a G+C content of 68.1% and 7,697 predicted CDSs. The genome possesses known bacterial virulence genes that have functions in host cell invasion, modulation of phagocyte function and for survival within macrophages. The detected candidate virulence factors provide a novel resource for further study of their pathogenic mechanisms at the molecular level in the fish nocardiosis research community. We also found many antibiotic resistance genes on the N. seriolae UTF1 chromosome, suggesting natural resistance of this bacteria to many drugs. Our comparative analysis with the four existing complete Nocardia spp. genomes revealed that the N. seriolae UTF1 genome structure and gene content differs from the other Nocardia genomes due to a large amount of mobile element genes. In addition, there are homologs of many transporters among the N. seriolae UTF1-specific genes allowing us to speculate on their role in adaptation in the marine environment. Thus, we expect that the availability of the complete genome of N. seriolae UTF1 can be used as the reference sequence not only for N. seriolae isolates, but also for the comparative genomic studies of genus Nocardia as an example of a marine fish pathogen to provide insights into the ecological and functional diversity of this genus [1] in the near future.
Supporting information
The contig sequences of N. seriolae ZJ0503 (GenBank: NZ_JNCT01000000, 319 contigs) and N-2927 (GenBank: NZ_BAWD02000000, 339 contigs) were aligned to the complete sequence of N. seriolae UTF1 genome. Three hundred (average length: 1373 bp) and 297 (average length: 1502 bp) uncovered regions were detected in the comparison with ZJ0503 and with N-2927, respectively. These gap sequences were subjected to BLASTX search against the NCBI RefSeq database (E value threshold of 1E−5). As a result, 294 (98.0%) for ZJ0503 and 290 (97.6%) for N-2927 have significant BLAST hit. Values represent number of genes with the best BLAST hit for ZJ0503 (red) and N-2927 (blue).
(PDF)
The BLASTN-based ring image was generated by BLAST Ring Image Generator (BRIG) version 0.95 [32]. The innermost two rings show GC content (black) and GC skew (purple). The second and third innermost rings represent a BLASTN comparison with the draft genomes of ZJ0503 (blue) and N-2927 (red), respectively. Bars indicate the position of mobile-element related genes in the N. seriolae UTF1 genome such as transposases (black), endonuclease DDE (blue) and integrase (green).
(PDF)
Nucleotide-based alignments were performed with MUMmer version 3.22 and dot plots were generated by the mummerplot script and the Unix program gnuplot [31].
(PDF)
(TIF)
(TIF)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
Acknowledgments
We would like to thank Dr. Kristian von Schalburg (Dept. of Molecular Biology and Biochemistry, S.F.U., Vancouver, B.C., Canada) for his help in preparing the manuscript.
Data Availability
All relevant data are within the paper and its Supporting Information files. The complete genome sequence of the Nocardia seriolae UTF1 was submitted to DDBJ under the accession number AP017900.
Funding Statement
This study was supported by a Grant-in-Aid (Marine Metagenomics for Monitoring the Coastal Microbiota) from the Ministry of Agriculture, Forestry and Fisheries of Japan (There are no grant number and the URLs.). (MK, T. Kobayashi, AF, SN, YS, T. Kamaishi, YN and MY received the funding).
References
- 1.Luo Q, Hiessl S, Steinbuchel A. Functional diversity of Nocardia in metabolism. Environ Microbiol. 2014;16(1):29–48. 10.1111/1462-2920.12221 [DOI] [PubMed] [Google Scholar]
- 2.Kandi V. Human Nocardia Infections: A review of pulmonary nocardiosis. Cureus. 2015;7(8):e304 10.7759/cureus.304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lerner PI. Nocardiosis. Clin Infect Dis. 1996;22(6):891–903. [DOI] [PubMed] [Google Scholar]
- 4.Labrie L, Ng J, Tan Z, Komar C, Ho E, Grisez L. Nocardial infections in fish: an emerging problem in both freshwater and marine aquaculture systems in Asia In: Bondad-Reantaso M, Mohan C, Crumlish M, Subasinghe R, editors. Diseases in Asian aquaculture VI. Manila, Republic of the Philippines: Fish health section, Asian. Fisheries Society; 2008. p. 297–312. [Google Scholar]
- 5.Kudo T, Hatai K, Seino A. Nocardia seriolae sp. nov. Causing Nocardiosis of Cultured Fish. Int J Syst Evol Microbiol. 1988;38(2):173–8. [Google Scholar]
- 6.Kariya T, Kubota S, Nakamura Y, Kira K. Nocardial infection in cultured yellowtail Seriola quinqueradiata and S. purpurscens. Bacteriological study-I. Fish Pathol. 1968;3:16–23 (in Japanese). [Google Scholar]
- 7.Shimahara Y, Huang Y-F, Tsai M-A, Wang P-C, Yoshida T, Lee J-L, et al. Genotypic and phenotypic analysis of fish pathogen, Nocardia seriolae, isolated in Taiwan. Aquaculture. 2009;294(3–4):165–71. [Google Scholar]
- 8.Imajoh M, Sukeda M, Shimizu M, Yamane J, Ohnishi K, Oshima S. Draft genome sequence of erythromycin- and oxytetracycline-sensitive Nocardia seriolae strain U-1 (NBRC 110359). Genome Announc. 2016;4(1). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ismail TF, Nakamura A, Nakanishi K, Minami T, Murase T, Yanagi S, et al. Modified resazurin microtiter assay for in vitro and in vivo assessment of sulfamonomethoxine activity against the fish pathogen Nocardia seriolae. Fish Sci. 2012;78(2):351–7. [Google Scholar]
- 10.Nayak SK, Shibasaki Y, Nakanishi T. Immune responses to live and inactivated Nocardia seriolae and protective effect of recombinant interferon gamma (rIFN gamma) against nocardiosis in ginbuna crucian carp, Carassius auratus langsdorfii. Fish Shellfish Immunol. 2014;39(2):354–64. 10.1016/j.fsi.2014.05.015 [DOI] [PubMed] [Google Scholar]
- 11.Strauss EJ, Falkow S. Microbial pathogenesis: genomics and beyond. Science. 1997;276(5313):707–12. [DOI] [PubMed] [Google Scholar]
- 12.Ishikawa J, Yamashita A, Mikami Y, Hoshino Y, Kurita H, Hotta K, et al. The complete genomic sequence of Nocardia farcinica IFM 10152. Proc Natl Acad Sci U S A. 2004;101(41):14925–30. 10.1073/pnas.0406410101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vera-Cabrera L, Ortiz-Lopez R, Elizondo-Gonzalez R, Ocampo-Candiani J. Complete genome sequence analysis of Nocardia brasiliensis HUJEG-1 reveals a saprobic lifestyle and the genes needed for human pathogenesis. PLoS One. 2013;8(6):e65425 10.1371/journal.pone.0065425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zoropogui A, Pujic P, Normand P, Barbe V, Belli P, Graindorge A, et al. The Nocardia cyriacigeorgica GUH-2 genome shows ongoing adaptation of an environmental Actinobacteria to a pathogen's lifestyle. BMC Genomics. 2013;14:286 10.1186/1471-2164-14-286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Luo Q, Hiessl S, Poehlein A, Daniel R, Steinbuchel A. Insights into the microbial degradation of rubber and gutta-percha by analysis of the complete genome of Nocardia nova SH22a. Appl Environ Microbiol. 2014;80(13):3895–907. 10.1128/AEM.00473-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Xia L, Cai J, Wang B, Huang Y, Jian J, Lu Y. Draft genome sequence of Nocardia seriolae ZJ0503, a fish pathogen isolated from Trachinotus ovatus in China. Genome Announc. 2015;3(1). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Imajoh M, Fukumoto Y, Yamane J, Sukeda M, Shimizu M, Ohnishi K, et al. Draft genome sequence of Nocardia seriolae strain N-2927 (NBRC 110360), isolated as the causal agent of nocardiosis of yellowtail (Seriola quinqueradiata) in Kochi prefecture, Japan. Genome Announc. 2015;3(2). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fraser CM, Eisen JA, Nelson KE, Paulsen IT, Salzberg SL. The value of complete microbial genome sequencing (you get what you pay for). J Bacteriol. 2002;184(23):6403–5. 10.1128/JB.184.23.6403-6405.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chin CS, Alexander DH, Marks P, Klammer AA, Drake J, Heiner C, et al. Nonhybrid, finished microbial genome assemblies from long-read SMRT sequencing data. Nature methods. 2013;10(6):563–9. 10.1038/nmeth.2474 [DOI] [PubMed] [Google Scholar]
- 20.Eid J, Fehr A, Gray J, Luong K, Lyle J, Otto G, et al. Real-time DNA sequencing from single polymerase molecules. Science. 2009;323(5910):133–8. 10.1126/science.1162986 [DOI] [PubMed] [Google Scholar]
- 21.Nakamura Y, Yasuike M, Nishiki I, Iwasaki Y, Fujiwara A, Kawato Y, et al. V-GAP: Viral genome assembly pipeline. Gene. 2016;576(2 Pt 1):676–80 [DOI] [PubMed] [Google Scholar]
- 22.Overbeek R, Olson R, Pusch GD, Olsen GJ, Davis JJ, Disz T, et al. The SEED and the Rapid Annotation of microbial genomes using Subsystems Technology (RAST). Nucleic Acids Res. 2014;42(Database issue):D206–14. 10.1093/nar/gkt1226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215(3):403–10. 10.1016/S0022-2836(05)80360-2 [DOI] [PubMed] [Google Scholar]
- 24.Pruitt KD, Tatusova T, Maglott DR. NCBI reference sequences (RefSeq): a curated non-redundant sequence database of genomes, transcripts and proteins. Nucleic Acids Res. 2007;35(Database issue):D61–5. 10.1093/nar/gkl842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Galperin MY, Makarova KS, Wolf YI, Koonin EV. Expanded microbial genome coverage and improved protein family annotation in the COG database. Nucleic Acids Res. 2015;43(Database issue):D261–9. 10.1093/nar/gku1223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kristensen DM, Kannan L, Coleman MK, Wolf YI, Sorokin A, Koonin EV, et al. A low-polynomial algorithm for assembling clusters of orthologous groups from intergenomic symmetric best matches. Bioinformatics. 2010;26(12):1481–7. 10.1093/bioinformatics/btq229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kanehisa M, Sato Y, Morishima K. BlastKOALA and GhostKOALA: KEGG Tools for functional characterization of genome and metagenome sequences. J Mol Biol. 2016;428(4):726–31. 10.1016/j.jmb.2015.11.006 [DOI] [PubMed] [Google Scholar]
- 28.Chen L, Zheng D, Liu B, Yang J, Jin Q. VFDB 2016: hierarchical and refined dataset for big data analysis-10 years on. Nucleic Acids Res. 2016;44(D1):D694–7. 10.1093/nar/gkv1239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sullivan MJ, Petty NK, Beatson SA. Easyfig: a genome comparison visualizer.Bioinformatics (Oxford, England). 2011;27(7):1009–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liu B, Pop M. ARDB—Antibiotic Resistance Genes Database. Nucleic Acids Res. 2009;37(Database issue):D443–7. 10.1093/nar/gkn656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kurtz S, Phillippy A, Delcher AL, Smoot M, Shumway M, Antonescu C, et al. Versatile and open software for comparing large genomes. Genome Biol. 2004;5(2):R12 10.1186/gb-2004-5-2-r12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Alikhan NF, Petty NK, Ben Zakour NL, Beatson SA. BLAST Ring Image Generator (BRIG): simple prokaryote genome comparisons. BMC Genomics. 2011;12:402 10.1186/1471-2164-12-402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rodriguez-R L, Konstantinidis K. Bypassing cultivation to identify bacterial species. Microbe Magazine.
- 34.Li L, Stoeckert CJ Jr., Roos DS. OrthoMCL: identification of ortholog groups for eukaryotic genomes. Genome Res. 2003;13(9):2178–89. 10.1101/gr.1224503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Verma H, Kumar R, Oldach P, Sangwan N, Khurana JP, Gilbert JA, et al. Comparative genomic analysis of nine Sphingobium strains: insights into their evolution and hexachlorocyclohexane (HCH) degradation pathways. BMC Genomics. 2014;15:1014 10.1186/1471-2164-15-1014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.DeJongh M, Formsma K, Boillot P, Gould J, Rycenga M, Best A. Toward the automated generation of genome-scale metabolic networks in the SEED. BMC Bioinformatics. 2007;8:139 10.1186/1471-2105-8-139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Overbeek R, Begley T, Butler RM, Choudhuri JV, Chuang HY, Cohoon M, et al. The subsystems approach to genome annotation and its use in the project to annotate 1000 genomes. Nucleic Acids Res. 2005;33(17):5691–702. 10.1093/nar/gki866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.de Hoon MJ, Imoto S, Nolan J, Miyano S. Open source clustering software. Bioinformatics. 2004;20(9):1453–4. 10.1093/bioinformatics/bth078 [DOI] [PubMed] [Google Scholar]
- 39.Saldanha AJ. Java Treeview—extensible visualization of microarray data. Bioinformatics. 2004;20(17):3246–8. 10.1093/bioinformatics/bth349 [DOI] [PubMed] [Google Scholar]
- 40.Tamura T, Matsuzawa T, Oji S, Ichikawa N, Hosoyama A, Katsumata H, et al. A genome sequence-based approach to taxonomy of the genus Nocardia. Antonie van Leeuwenhoek. 2012;102(3):481–91. 10.1007/s10482-012-9780-5 [DOI] [PubMed] [Google Scholar]
- 41.Shin SC, Ahn do H, Kim SJ, Lee H, Oh TJ, Lee JE, et al. Advantages of single-molecule real-time sequencing in high-GC content genomes. PLoS One. 2013;8(7):e68824 10.1371/journal.pone.0068824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Miyamoto M, Motooka D, Gotoh K, Imai T, Yoshitake K, Goto N, et al. Performance comparison of second- and third-generation sequencers using a bacterial genome with two chromosomes. BMC Genomics. 2014;15:699 10.1186/1471-2164-15-699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Stadermann KB, Weisshaar B, Holtgräwe D. SMRT sequencing only de novo assembly of the sugar beet (Beta vulgaris) chloroplast genome. BMC Bioinformatics. 2015;16(1):1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Itano T, Nakaoka N, Kawakami H, Kono T, Sakai M. Comparison of sensitivity between yellowtail Seriola quinqueradiata and red sea bream Pagrus major to Nocardia seriolae. Fish Pathol. 2006;41(4):135–9. [Google Scholar]
- 45.Kusuda R, Kimura Y, Hamaguchi M. Changes in Peripheral and Peritoneal Leucocytes in Yellowtail Seriola quinqueradiata Immunized with Nocardia kampachi. Nihon Suisan Gakkai-shi 1989;55(7):1183–8 (in Japanese with English abstract). [Google Scholar]
- 46.Arruda S, Bomfim G, Knights R, Huima-Byron T, Riley LW. Cloning of an M. tuberculosis DNA fragment associated with entry and survival inside cells. Science. 1993;261(5127):1454–7. [DOI] [PubMed] [Google Scholar]
- 47.Cole ST, Brosch R, Parkhill J, Garnier T, Churcher C, Harris D, et al. Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature. 1998;393(6685):537–44. 10.1038/31159 [DOI] [PubMed] [Google Scholar]
- 48.Zhang F, Xie JP. Mammalian cell entry gene family of Mycobacterium tuberculosis. Mol Cell Biochem. 2011;352(1–2):1–10. 10.1007/s11010-011-0733-5 [DOI] [PubMed] [Google Scholar]
- 49.Casali N, Riley LW. A phylogenomic analysis of the Actinomycetales mce operons. BMC Genomics. 2007;8:60 10.1186/1471-2164-8-60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gonzalez-Carrillo C, Millan-Sauceda C, Lozano-Garza HG, Ortiz-Lopez R, Elizondo-Gonzalez R, Welsh O, et al. Genomic changes associated with the loss of Nocardia brasiliensis virulence in mice after 200 in vitro passages. Infect Immun. 2016;84(9):2595–606. 10.1128/IAI.00329-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Liu H, Semino-Mora C, Dubois A. Mechanism of H. pylori intracellular entry: an in vitro study. Front Cell Infect Microbiol. 2012;2:13 10.3389/fcimb.2012.00013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Seo KS, Kim JW, Park JY, Viall AK, Minnich SS, Rohde HN, et al. Role of a new intimin/invasin-like protein in Yersinia pestis virulence. Infect Immun. 2012;80(10):3559–69. 10.1128/IAI.00294-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Beaman BL, Black CM, Doughty F, Beaman L. Role of superoxide dismutase and catalase as determinants of pathogenicity of Nocardia asteroides: importance in resistance to microbicidal activities of human polymorphonuclear neutrophils. Infect Immun. 1985;47(1):135–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Akhtar S, Khan A, Sohaskey CD, Jagannath C, Sarkar D. Nitrite reductase NirBD is induced and plays an important role during in vitro dormancy of Mycobacterium tuberculosis. J Bacteriol. 2013;195(20):4592–9. 10.1128/JB.00698-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Khan A, Sarkar D. Nitrate reduction pathways in mycobacteria and their implications during latency. Microbiology. 2012;158(Pt 2):301–7. 10.1099/mic.0.054759-0 [DOI] [PubMed] [Google Scholar]
- 56.Koshkin A, Knudsen GM, Ortiz De Montellano PR. Intermolecular interactions in the AhpC/AhpD antioxidant defense system of Mycobacterium tuberculosis. Arch Biochem Biophys. 2004;427(1):41–7. 10.1016/j.abb.2004.04.017 [DOI] [PubMed] [Google Scholar]
- 57.Koshkin A, Nunn CM, Djordjevic S, Ortiz de Montellano PR. The mechanism of Mycobacterium tuberculosis alkylhydroperoxidase AhpD as defined by mutagenesis, crystallography, and kinetics. J Biol Chem. 2003;278(32):29502–8. 10.1074/jbc.M303747200 [DOI] [PubMed] [Google Scholar]
- 58.Sherman DR, Mdluli K, Hickey MJ, Arain TM, Morris SL, Barry CE 3rd, et al. Compensatory ahpC gene expression in isoniazid-resistant Mycobacterium tuberculosis. Science. 1996;272(5268):1641–3. [DOI] [PubMed] [Google Scholar]
- 59.Hahn JS, Oh SY, Roe JH. Role of OxyR as a peroxide-sensing positive regulator in Streptomyces coelicolor A3(2). J Bacteriol. 2002;184(19):5214–22. 10.1128/JB.184.19.5214-5222.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zimmerli S, Majeed M, Gustavsson M, Stendahl O, Sanan DA, Ernst JD. Phagosome-lysosome fusion is a calcium-independent event in macrophages. J Cell Biol. 1996;132(1–2):49–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Forrellad MA, Klepp LI, Gioffre A, Sabio y Garcia J, Morbidoni HR, de la Paz Santangelo M, et al. Virulence factors of the Mycobacterium tuberculosis complex. Virulence. 2013;4(1):3–66. 10.4161/viru.22329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wiker HG, Harboe M. The antigen 85 complex: a major secretion product of Mycobacterium tuberculosis. Microbiol Rev. 1992;56(4):648–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Belisle JT, Vissa VD, Sievert T, Takayama K, Brennan PJ, Besra GS. Role of the major antigen of Mycobacterium tuberculosis in cell wall biogenesis. Science. 1997;276(5317):1420–2. [DOI] [PubMed] [Google Scholar]
- 64.Martino MC, Stabler RA, Zhang ZW, Farthing MJ, Wren BW, Dorrell N. Helicobacter pylori pore-forming cytolysin orthologue TlyA possesses in vitro hemolytic activity and has a role in colonization of the gastric mucosa. Infect Immun. 2001;69(3):1697–703. 10.1128/IAI.69.3.1697-1703.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Rahman A, Srivastava SS, Sneh A, Ahmed N, Krishnasastry MV. Molecular characterization of tlyA gene product, Rv1694 of Mycobacterium tuberculosis: a non-conventional hemolysin and a ribosomal RNA methyl transferase. BMC Biochemistry. 2010;11:35 10.1186/1471-2091-11-35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zhou B, He Y, Zhang X, Xu J, Luo Y, Wang Y, et al. Targeting mycobacterium protein tyrosine phosphatase B for antituberculosis agents. Proc Natl Acad Sci U S A. 2010;107(10):4573–8. 10.1073/pnas.0909133107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Hashemi-Shahraki A, Heidarieh P, Bostanabad SZ, Hashemzadeh M, Feizabadi MM, Schraufnagel D, et al. Genetic diversity and antimicrobial susceptibility of Nocardia species among patients with nocardiosis. Sci Rep. 2015;5:17862 10.1038/srep17862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Shimahara Y, Nakamura A, Nomoto R, Itami T, Chen SC, Yoshida T. Genetic and phenotypic comparison of Nocardia seriolae isolated from fish in Japan. J Fish Dis. 2008;31(7):481–8. 10.1111/j.1365-2761.2008.00920.x [DOI] [PubMed] [Google Scholar]
- 69.Ismail TF, Takeshita A, Umeda N, Itami T, Yoshida T. Application of α-glucosidase activity and drug susceptibility tests to epidemiological studies on the fish pathogen Nocardia seriolae. Fish Sci. 2011;77(1):113–8. [Google Scholar]
- 70.Ismail TF, Takeshita A, Umeda N, Itami T, Yoshida T. The Use of Chromogenic media for α-glucosidase determination and presumptive drug susceptibility profiles in the fish pathogen Nocardia seriolae. Fish Pathol. 2011;46(2):62–4. [Google Scholar]
- 71.Chopra I, Roberts M. Tetracycline antibiotics: mode of action, applications, molecular biology, and epidemiology of bacterial resistance. Microbiol Mol Biol Rev. 2001;65(2):232–60. 10.1128/MMBR.65.2.232-260.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Roberts MC. Update on acquired tetracycline resistance genes. FEMS Microbiol Lett. 2005;245(2):195–203. 10.1016/j.femsle.2005.02.034 [DOI] [PubMed] [Google Scholar]
- 73.Varaldo PE, Montanari MP, Giovanetti E. Genetic elements responsible for erythromycin resistance in streptococci. Antimicrob Agents Chemother. 2009;53(2):343–53. 10.1128/AAC.00781-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Kobayashi N, Nishino K, Yamaguchi A. Novel macrolide-specific ABC-type efflux transporter in Escherichia coli. J Bacteriol. 2001;183(19):5639–44. 10.1128/JB.183.19.5639-5644.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Tikhonova EB, Devroy VK, Lau SY, Zgurskaya HI. Reconstitution of the Escherichia coli macrolide transporter: the periplasmic membrane fusion protein MacA stimulates the ATPase activity of MacB. Mol Microbiol. 2007;63(3):895–910. 10.1111/j.1365-2958.2006.05549.x [DOI] [PubMed] [Google Scholar]
- 76.Tillier ER, Collins RA. Genome rearrangement by replication-directed translocation. Nat Genet. 2000;26(2):195–7. 10.1038/79918 [DOI] [PubMed] [Google Scholar]
- 77.Tang K, Jiao N, Liu K, Zhang Y, Li S. Distribution and functions of TonB-dependent transporters in marine bacteria and environments: implications for dissolved organic matter utilization. PLoS One. 2012;7(7):e41204 10.1371/journal.pone.0041204 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The contig sequences of N. seriolae ZJ0503 (GenBank: NZ_JNCT01000000, 319 contigs) and N-2927 (GenBank: NZ_BAWD02000000, 339 contigs) were aligned to the complete sequence of N. seriolae UTF1 genome. Three hundred (average length: 1373 bp) and 297 (average length: 1502 bp) uncovered regions were detected in the comparison with ZJ0503 and with N-2927, respectively. These gap sequences were subjected to BLASTX search against the NCBI RefSeq database (E value threshold of 1E−5). As a result, 294 (98.0%) for ZJ0503 and 290 (97.6%) for N-2927 have significant BLAST hit. Values represent number of genes with the best BLAST hit for ZJ0503 (red) and N-2927 (blue).
(PDF)
The BLASTN-based ring image was generated by BLAST Ring Image Generator (BRIG) version 0.95 [32]. The innermost two rings show GC content (black) and GC skew (purple). The second and third innermost rings represent a BLASTN comparison with the draft genomes of ZJ0503 (blue) and N-2927 (red), respectively. Bars indicate the position of mobile-element related genes in the N. seriolae UTF1 genome such as transposases (black), endonuclease DDE (blue) and integrase (green).
(PDF)
Nucleotide-based alignments were performed with MUMmer version 3.22 and dot plots were generated by the mummerplot script and the Unix program gnuplot [31].
(PDF)
(TIF)
(TIF)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files. The complete genome sequence of the Nocardia seriolae UTF1 was submitted to DDBJ under the accession number AP017900.