Abstract
Background
The unfolded protein response (UPR) refers to intracellular stress signaling pathways that protect cells from the stress caused by accumulation of unfolded or misfolded proteins in the endoplasmic reticulum (ER). The UPR signaling is crucially involved in the initiation and progression of a variety of human diseases by modulating transcriptional and translational programs of the stressed cells. In this study, we analyzed the gene expression signatures of primary stress sensors and major mediators of UPR pathways in a variety of tissues/organs of human and murine species.
Methods
We first analyzed protein sequence similarities of major UPR transducers and mediators of human and murine species, and then examined their gene expression profiles in 26 human and mouse common tissues based on the microarray datasets of public domains. The differential expression patterns of the UPR genes in human diseases were delineated. The involvements of the UPR genes in mouse pathology were also analyzed with mouse gene knockout models.
Results
The results indicated that expression patterns and pathophysiologic involvements of the major UPR stress sensors and mediators significantly differ in 26 common tissues/organs of human and murine species. Gene expression profiles suggest that the IRE1α/XBP1-mediated UPR pathway is induced in secretory and metabolic tissues or organs. While deletion of the UPR trans-activator XBP1 leads to pathological phenotypes in mice, alteration in XBP1 is less associated with human disease conditions.
Conclusions
Expression signatures of the major UPR genes differ among tissues or organs and among human and mouse species. The differential induction of the UPR pathways reflects the pathophysiologic differences of tissues or organs. The difference in UPR induction between human and mouse suggests the limitation of using animal models to study human pathophysiology or drugology associated with environmental stress.
Keywords: Animal models, endoplasmic reticulum stress, environmental stress, transcriptional signature, unfolded protein response
INTRODUCTION
Unfolded protein response (UPR) refers to intracellular stress signaling pathways that are originated from the endoplasmic reticulum (ER) in response to accumulation of unfolded or misfolded proteins in the ER lumen.[1,2] The UPR is conserved in all the known eukaryotic species, ranging from yeast, worm, to mammals. The UPR pathways are mediated through three ubiquitously expressed transducers, namely, inositol-requiring I (IREI), PKR-like ER kinase/pancreatic eIF2α kinase (PERK), and activating transcription factor 6 (ATF6).[2,3] When cells encounter ER stress, PERK is activated to phosphorylate translation initiation factor eIF2α, leading to translational suppression and subsequent reduction in translocation of newly synthesized proteins into the ER. Upon activation of the UPR, IREIα is also activated to splice the mRNA encoding X-box-binding protein 1 (XBP1). Spliced XbpI mRNA encodes a potent bZIP transcription factor that activates expression of a number of ER chaperones and enzymes to promote protein folding, secretion of correctly folded proteins, and degradation of misfolded proteins. Under ER stress conditions, the UPR transducer ATF6 is also activated to function as a transcription factor that plays partially redundant roles of XBP1 in facilitating protein folding and secretion as well as degradation of misfolded proteins.[4,5] In principal, through three pathways, the UPR is activated to reduce the amount of new proteins translocated into the ER lumen, to increase degradation of misfolded proteins, and to bolster ER protein folding and secretion capacities. However, when ER stress gets prolonged or the adaptive UPR responses are not sufficient to resolve the accumulation of unfolded or misfolded proteins, the UPR signaling will initiate cell death programs to eliminate the stressed cells. Typically, ER stress-induced programmed cell death is mediated by PERK/eIF2α UPR pathway.[2,3] Under chronic or severe ER stress, PERK-mediated phosphorylation of eIF2α leads to translation of some selective mRNAs while it causes attenuation of protein translation in general. In mammals, phosphorylated eIF2α can mediate translation of ATF4 which induces expression of a pro-apoptotic factor CHOP/GADDI53, leading to ER stress-induced apoptosis. In addition, under stress condition, ATF4 can induce expression of the growth arrest and DNA damage-inducible protein GADD34.[6,7] GADD34 interacts with the catalytic subunit of type I protein serine/threonine phosphatase to dephosphorylate eIF2α, allowing most protein synthesis to resume. Thus, induction of GADD34 under ER stress conditions provides a negative feedback regulation in the PERK/eIF2α UPR pathway.
Recent discoveries in the mechanisms and roles of physiologic UPR signaling, coupled with the studies on genetically engineer animal models, have led to significant expansion in the scope and consequence of the UPR.[8] A variety of pathophysiologic stimuli, environmental stress, and even lifestyles can directly or indirectly induce ER stress and activate the same UPR pathways induced by biochemical or pharmacological drugs. It has been demonstrated that the IREIα/XBP1-mediated UPR pathway is required for normal differentiation of plasma cells as well as for function and survival of dendritic cells.[9–11] The PERK-mediated UPR pathway is a key regulator of energy metabolism and is required for pancreatic β cells function and survival.[12–15] The UPR is crucial for many specialized cell types, such as macrophages, pancreatic β cells, and neural oligodendrocytes, to make survival or death decision under stress conditions.[8] Indeed, disruption or hyperactivation of the UPR signaling is associated with a variety of systemic diseases, such as metabolic disease, cardiovascular disease, neurodegenerative disease, and cancer.
Because UPR signaling is crucial to cell differentiation, function, and survival, we asked whether expression profiles of the major UPR genes can indicate states of the pathophysiology of specialized tissues or organisms. Here, we analyzed the expression profiles of major UPR genes in human and mouse tissues as well as in human diseases based on the databases of public domains. Our analyses suggest that the expression signatures of the UPR genes differ among tissues and species. The UPR gene expression profiles reflect the functional differences of tissues or organs that are associated with human diseases.
METHODS
Microarray-based gene expression data analysis
Microarray-based gene expression data for 26 human and mouse common tissues were extracted from the microarray datasets of BioGPS (www.biogps.org). Fold changes of expression levels of the major UPR genes in human or mouse tissues were determined by normalizing to expression levels of the genes in the cerebellum which were defined as 1. Clustered heat map of gene expression in different tissues or organs was drawn based on the fold changes of gene expression.
Analysis of gene expression patterns associated with human diseases
Expression profiles of major UPR genes in human diseases were extracted from the European Bioinformatics Institute of European Molecular Biology Laboratory (EMBL-EBI, http://www.ebi.ac.uk/). Up- or down-regulation of UPR gene expression in human diseases was determined by comparing the expression levels of the genes in the tissues associated with the particular human diseases to that in the normal human tissues.
Determination of the correlation between gene knockout/dysfunction and animal phenotypes
Phenotypes of animal models with knockout or dysfunctional UPR genes were obtained from the Mouse Genome Informatics database (MGI, http://www.informatics.jax.org/) and the literature search of PubMed database (www.ncbi.pubmed).
RESULTS
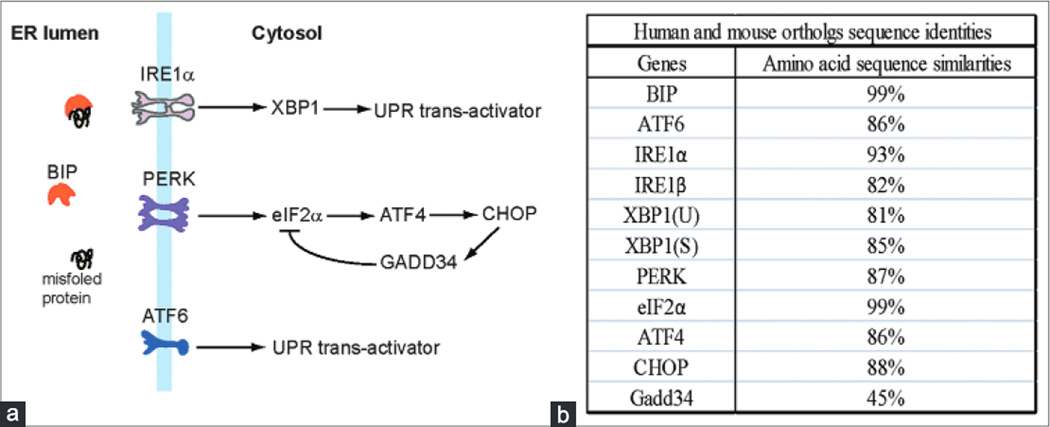
The major UPR genes are highly conserved in human and mouse, but their expression profiles vary significantly among tissues. We examined homologies of the major transducers and mediators of the UPR pathways in human and mouse species. These UPR transducers and mediators include (I) ER chaperone GRP78/BIP (BIP), (2) primary UPR transducer IREIα and its homolog IREIβ, (3) UPR transactivator XBP1 (the target of IREIα), (4) UPR transducer ATF6, (5) UPR transducer PERK, (6) translation initiation factor eIFα (the target of PERK), and (7) UPR mediators of the PERK/eIF2α pathways including ATF4, CHOP, and GADD34 [Figure Ia]. The amino acid sequences of the major UPR transducers and mediators, except GADD34, are highly conserved in human and mouse, with orthologous sequence identities of over 80% [Figure Ib; Supplementary Figure I]. However, GADD34, a nontypical UPR-associated protein factor that modulates the PERK/eIFα UPR pathway by dephosphorylating eIF2α,[6,7] displays only 45% sequence similarity between human and mouse species.
Figure I.
(a) The unfolded protein response branches and major unfolded protein response mediators. (b) The amino acid sequence similarities of the major unfolded protein response genes. The protein sequences were analyzed based on the database from NCBI (http://blast.ncbi.nlm.nih.gov/) [Supplementary Figure I]
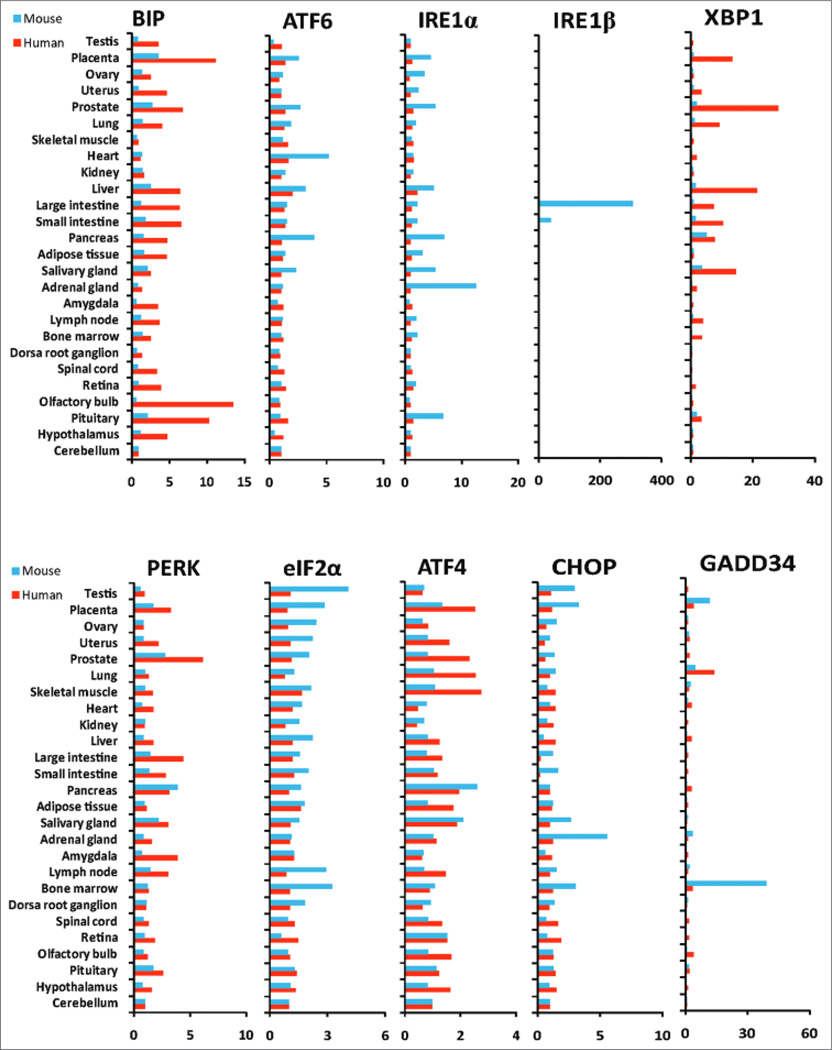
Next, we analyzed expression signatures of the major UPR genes in specialized human and mouse tissues or organs based on the gene expression microarray datasets from BioGPS (www.biogps.org). For this analysis, we defined the mRNA levels transcribed from the UPR genes in the cerebellum tissue as the baseline (1-fold). Fold changes of the mRNA levels in other tissues or organs were determined by normalizing to the mRNA levels in the cerebellum. Because the UPR signaling is associated with cellular physiology,[2] highly proliferative tissues or organs, such as intestines, reproductive organs, and glands, display higher expression levels of the UPR target genes BIP and XBP1, compared to the tissues or organs containing permanent cells, such as skeletal muscle and cerebellum [Figure 2 and Supplementary Table I]. For this reason, we selected cerebellum as the control tissue to normalize expression fold changes of the UPR genes in human or mouse tissues. In human, expression of the BIP mRNA is highly induced in specialized metabolic, inflammatory, and secretory tissues or organs, such as those of the reproductive system, gastrointestinal organs, immune system, and neuronal tissues, compared to that in the cerebellum [Figure 2]. The UPR transactivator XBP1 is also highly expressed in the reproductive tissues, gastrointestinal organs, and immune system, but not in the neuronal tissues. Different from the expression profiles of BIP and XBP1, expression levels of the other major UPR genes, including ATF6, IREIα, IREIβ, PERK, eIF2α, ATF4, and CHOP, in most metabolic and secretory tissues or organs are similar to those in the cerebellum [Figure 2]. Induction of both BIP and XBP1, the downstream targets of the UPR transducer IREIα, implicates elevation of the IREIα/XBP1-mediated UPR pathway.[2,3] However, the expression profiles of the IREIα gene are opposite to those of XBP1 in the human tissues [Figure 2]. This inconsistency may be due to the reverse correlation between IREIα protein activation and IREIα mRNA levels as it has been proven that the activated IREIα can decrease its own mRNA.[16] Therefore, the reduced levels of the IREIα mRNA are consistent with activation of the IREIα/XBP1-mediated UPR pathway. In addition, expression levels of the genes involved in the PERK/eIF2α-mediated UPR pathway, including PERK, eIF2α, ATF4, and CHOP, in these metabolic and secretory tissues are similar to or even lower than those in cerebellum under the physiological condition [Figure 2]. Since the PERK/eIF2α pathway leads to protein translational attenuation and ER stress-induced apoptosis,[2] the relatively low expression profiles of the genes involved in the PERK/eIF2α pathway suggest that stress-induced protein translation attenuation and apoptosis programs are not prevalent in the specialized tissues or organs whose primary functions are associated with protein secretion and metabolism under physiological conditions.
Figure 2.
Comparison of the major unfolded protein response gene expression profiles in human and mouse tissues. Microarray gene expression data for human and mouse tissues were extracted from BioGPS (www.biogps.org). Fold changes of gene expression levels in human (red bars) or mouse (blue bars) tissues were determined by normalized to expression levels of the genes in cerebellum (which were defined as I). A full matrix of normalized expression scores were given in Supplementary Table I
In mouse tissues, expression profiles of the major UPR genes also vary significantly [Figure 2 and Supplementary Table I]. Different from the gene expression signatures in the human tissues, the expression levels of the BIP and XbpI genes in most metabolic, inflammatory, and secretory organs or tissues are not as high as that in the cerebellum. Interestingly, expression of the eIF2α gene is more induced in most of the reproductive tissues, immune organs, and neuronal tissues, compared to that in the cerebellum [Figure 2]. Because only the phosphorylated eIF2α, but not the total eIF2α, is the substrate of the UPR transducer PERK,[2,8] increased expression of the eIF2α gene alone does not necessarily indicate the elevation of the PERK/eIF2α UPR pathway. Instead, the increased expression of the eIF2α genes may be correlated with elevated protein translation in these tissues or organs.
Expression signatures of the major UPR genes in human and mouse significantly differ. We compared the expression signatures of the major UPR genes in human and mouse tissues. Compared to the mouse tissues, human reproductive tissues, gastrointestinal organs, immune system, and neuronal tissues display higher expression levels of the genes encoding BIP and XBP1 [Figure 2]. Correlated with increased expression of BIP and XBP1, the expression levels of IREIα are decreased in the human tissues, consistent with the fact that activated IREIα can decrease the IREIα mRNA.[16] These profiles suggest that the basal induction of the IREIα/XBP1-mediated UPR signaling in human tissues is higher than that in mouse tissues. Given the roles of IREIα/XBP1 UPR signaling in protein secretion, metabolism, and homeostasis maintenance,[2,8] higher induction of the IREIα/XBP1-mediated UPR in human tissues suggest that the human tissues may possess more robust physiological programs and stress-adaption capability, compared to the mouse tissues. Another interesting observation is that IREIα, the homolog of IREIα, is more induced in mouse large and small intestines, compared to that in the cerebellum [Figure 2]. It has been reported that IREIβ is involved in mucin secretion in goblet cells,[17] intestinal lipid absorption,[18] and chylomicron production.[19] Induction of the IREIα gene in mouse intestine tissues suggests that mice may utilize IREIβ-mediated signaling to possess unique capabilities in food digestion and metabolism.
We also observed higher induction of the UPR transducer PERK and its downstream substrate ATF4 in most human tissues examined, compared to those in mice [Figure 2]. However, expression profile of the other substrates of PERK, including eIF2α, CHOP, and GADD34, are not consistent with those of PERK and ATF4. This may be explained by the fact that eIF2α, CHOP, and GADD34 are not solely UPR targets. Although PERK-mediated UPR signaling leads to phosphorylation of eIF2α protein, it does not regulate expression of the eIF2α gene. Expression of CHOP and GADD34 can be regulated by many other signals in addition to UPR signaling.[20,21] The low induction profile of CHOP in human tissues suggests that PERK-mediated UPR signaling may not necessarily lead to stress-induced apoptosis under physiologic conditions.
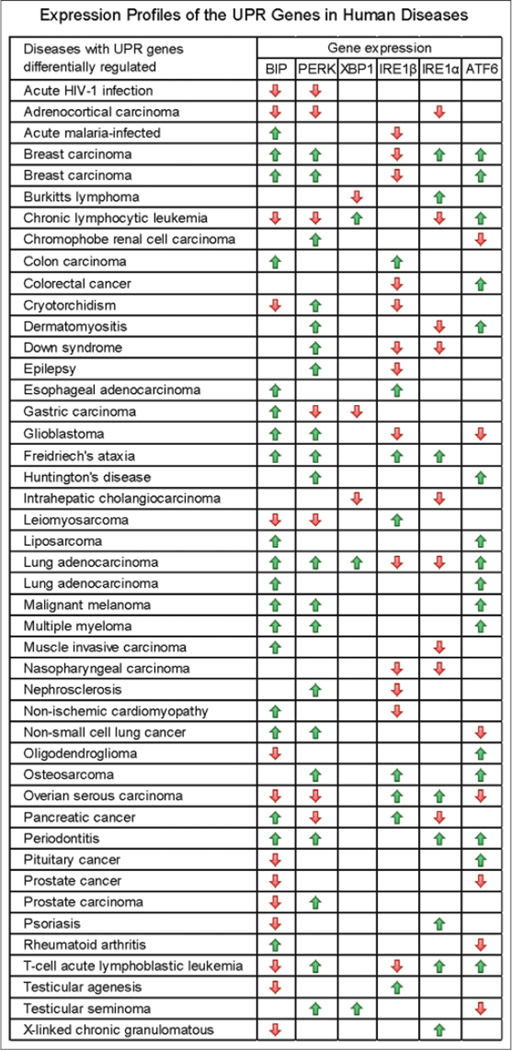
Expression profiles of the UPR genes vary under different human disease conditions. We analyzed expression signatures of the major UPR genes, including BIP, PERK, XBP1, IREIα, IREIβ, and ATF6, in human diseases based on the database from the EMBL-EBI (http://www.ebi.ac.uk/). Altered expression of multiple UPR transducers or mediators was found in 45 human disease conditions, ranging from cancers, infectious diseases, to neuronal diseases [Figure 3 and Supplementary Table 2]. Expression of the BIP, PERK, IREIα, IREIβ, and ATF6 genes is frequently modulated under human disease conditions. Surprisingly, altered expression of the UPR transactivator XBP1 is only associated with a small number of human diseases [Figure 3]. Note that expression of IREIβ, an IREIα homolog with unknown function in human, appears to be frequently modulated in human diseases.
Figure 3.
Expression profiles of the major unfolded protein response genes in human diseases. Data were extracted and analyzed based on the database from European Bioinformatics Institute of European Molecular Biology Laboratory (http://www.ebi.ac.uk/). The green or red arrows indicate up- or down-regulation of the genes in human disease tissues. The up- or down-regulations were determined by comparing the expression levels of the genes in the disease tissues to those in normal tissues. The t-test statistics was shown in Supplementary Table 2
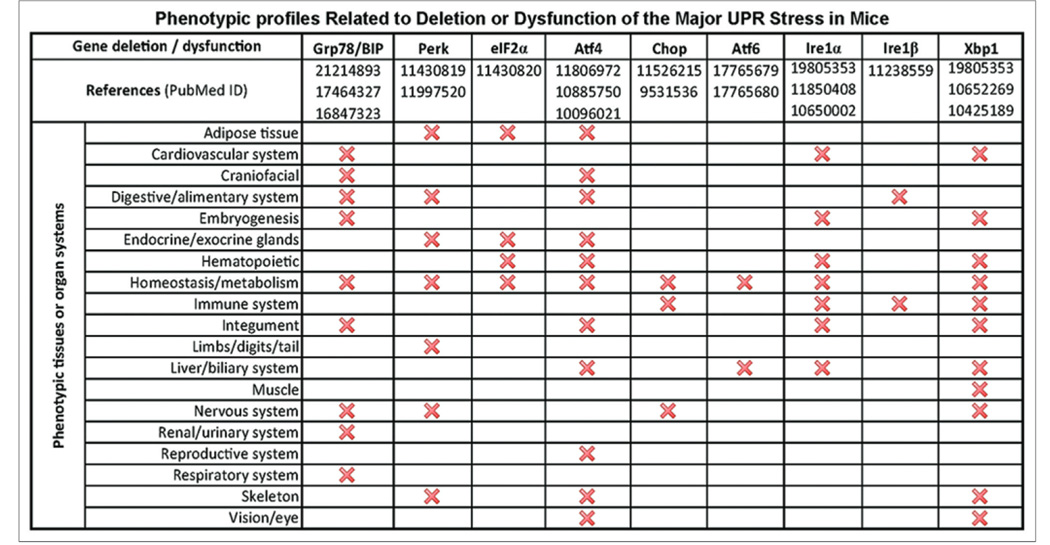
Deletion of the UPR gene significantly affects mouse pathophysiology. To gain insights into the impact of the UPR genes in mouse pathophysiology, we examined the phenotypic profiles related to deletion or dysfunction of the UPR genes in mice. Based on the MGI database (http://www.informatics.jax.org/) and the published literature from PubMed (www.ncbi.pubmed), we generated a profile of phenotypes related to deletion or dysfunction of the UPR genes in mice [Figure 4]. Deletion or dysfunction of the BIP, PERK, or IREIα gene leads to pathological phenotypes in many mice physiological systems. However, distinct from the UPR profiles in human diseases [Figure 3], dysfunction of XBP1 or ATF4 results in pathologic phenotypes in a variety of physiological systems in mice [Figure 4], suggesting that XBP1 and ATF4 are crucial to mouse pathophysiology.
Figure 4.
Profiles of pathological phenotypes of mouse models with unfolded protein response gene deletion or dysfunction. The animal phenotype information was summarized based on the animal database from Mouse Genome Informatics (http://www.informatics.jax.org/) and the literature of PubMed database (www.ncbi.pubmed)
DISCUSSION
The differential expression signatures of the UPR genes in normal human and mouse tissues or disease models have important implications in the understanding of human and mouse physiology. Our analyses indicate: (I) The major UPR genes are highly conserved but their expression profiles vary significantly among the tissues of human and mouse species; (2) relatively high induction of the IREIα/XBP1-mediated UPR branch, but not the PERK-mediated UPR branch, is observed in secretory and metabolic organs or tissues; (3) the expression signatures of the major UPR genes in human tissues are different from that in mouse tissues; and (4) the involvements of the UPR genes in human diseases are different from that in mouse pathophysiology. In particular, XBP1 and ATF4 are crucially involved in mouse pathophysiology, but not much in human diseases.
Based on the gene expression profiles of the ER chaperone BIP, the UPR transducer IREIα, and the UPR transactivator XBP1, it is apparent that the secretory and metabolic tissues or organs, such as those in the reproductive, gastrointestinal, immune, and neuronal systems, have high basal levels of the IREIα/XBP1-mediated UPR signaling [Figure 2]. This is consistent with the roles of the IREIα/XBP1 UPR signaling in facilitating protein secretion, metabolism, and homeostasis maintenance under physiological “stress” conditions.[2,8] Importantly, the degree of induction of the IREIα/XBP1 signaling in human tissues are likely higher than those in mouse tissues [Figure 2], implicating that human tissues may possess more robust adaptation programs in dealing with physiological demands and stress challenges. Interestingly, induction of IREIβ, a homolog of IREIα, is evidenced in mouse small and large intestines [Figure 2]. Because it has been reported that IREIβ is involved in mucin secretion and lipid transport in mouse digestive system,[17–19] it is possible that IREIβ-mediated stress signaling may provide a molecular basis for mice to achieve their uniqueness in food uptake, digestion, and energy metabolism.
Another important observation from this study is the different profiles for the involvement of the UPR genes in human disease and mouse pathology [Figures 3 and 4], XBP1, a UPR transactivator highly induced in human tissues under the physiologic condition [Figure 2], is associated with only a few types of human diseases [Figure 3]. Instead, ATF6, a UPR transducer that displays partial functional redundancy with XBP1,[4,5] is more relevant to the occurrence of human diseases. It is possible that ATF6 may not only compensate XBP1 dysregulation but also play additional indispensable roles in human. In contrast, deletion of XBP1 is critically involved in mouse pathology [Figure 4]. These observations suggest that induction profiles and pathophysiologic involvements of the UPR genes in human and mouse are significantly different. This finding is important because it confirms the limitation of using animal models to study human pathophysiology or drugology, particularly for those that are associated with environmental stress conditions. Given the fact that animal models have been widely used as platforms to study human diseases and to test therapeutic drugs, this study is informative to the research communities of biomedicine and public health.
Supplementary Material
Acknowledgments
Financial support and sponsorship
Portions of this work were supported by the National Institutes of Health Grants DK090313 and ES017829, American Heart Association Grants 09GRNT2280479, and Department of Defense Breast Cancer Program grant BC095179PI to KZ.
Footnotes
Conflicts of interest
There are no conflicts of interest.
REFERENCES
- 1.Kaufman RJ. Stress signaling from the lumen of the endoplasmic reticulum: Coordination of gene transcriptional and translational controls. Genes Dev. 1999;13:1211–1233. doi: 10.1101/gad.13.10.1211. [DOI] [PubMed] [Google Scholar]
- 2.Ron D, Walter P. Signal integration in the endoplasmic reticulum unfolded protein response. Nat Rev Mol Cell Biol. 2007;8:519–529. doi: 10.1038/nrm2199. [DOI] [PubMed] [Google Scholar]
- 3.Zhang K, Kaufman RJ. Signaling the unfolded protein response from the endoplasmic reticulum. J Biol Chem. 2004;279:25935–25938. doi: 10.1074/jbc.R400008200. [DOI] [PubMed] [Google Scholar]
- 4.Okada T, Yoshida H, Akazawa R, Negishi M, Mori K. Distinct roles of activating transcription factor 6 (ATF6) and double-stranded RNA-activated protein kinase-like endoplasmic reticulum kinase (PERK) in transcription during the mammalian unfolded protein response. Biochem J. 2002;366(Pt 2):585–594. doi: 10.1042/BJ20020391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yamamoto K, Sato T, Matsui T, Sato M, Okada T, Yoshida H, et al. Transcriptional induction of mammalian ER quality control proteins is mediated by single or combined action of ATF6alpha and XBP1. Dev Cell. 2007;13:365–376. doi: 10.1016/j.devcel.2007.07.018. [DOI] [PubMed] [Google Scholar]
- 6.Ma Y, Hendershot LM. Delineation of a negative feedback regulatory loop that controls protein translation during endoplasmic reticulum stress. J Biol Chem. 2003;278:34864–34873. doi: 10.1074/jbc.M301107200. [DOI] [PubMed] [Google Scholar]
- 7.Novoa I, Zeng H, Harding HP, Ron D. Feedback inhibition of the unfolded protein response by GADD34-mediated dephosphorylation of eIF2alpha. J Cell Biol. 2001;153:1011–1022. doi: 10.1083/jcb.153.5.1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhang K, Kaufman RJ. From endoplasmic-reticulum stress to the inflammatory response. Nature. 2008;454:455–462. doi: 10.1038/nature07203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhang K, Wong HN, Song B, Miller CN, Scheuner D, Kaufman RJ. The unfolded protein response sensor IREIalpha is required at 2 distinct steps in B cell lymphopoiesis. J Clin Invest. 2005;115:268–281. doi: 10.1172/JCI21848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Reimold AM, Iwakoshi NN, Manis J, Vallabhajosyula P, Szomolanyi-Tsuda E, Gravallese EM, et al. Plasma cell differentiation requires the transcription factor XBP-I. Nature. 2001;412:300–307. doi: 10.1038/35085509. [DOI] [PubMed] [Google Scholar]
- 11.Iwakoshi NN, Pypaert M, Glimcher LH. The transcription factor XBP-I is essential for the development and survival of dendritic cells. J Exp Med. 2007;204:2267–2275. doi: 10.1084/jem.20070525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Harding HP, Zeng H, Zhang Y, Jungries R, Chung P, Plesken H, et al. Diabetes mellitus and exocrine pancreatic dysfunction in perk−/− mice reveals a role for translational control in secretory cell survival. Mol Cell. 2001;7:1153–1163. doi: 10.1016/s1097-2765(01)00264-7. [DOI] [PubMed] [Google Scholar]
- 13.Oyadomari S, Harding HP, Zhang Y, Oyadomari M, Ron D. Dephosphorylation of translation initiation factor 2alpha enhances glucose tolerance and attenuates hepatosteatosis in mice. Cell Metab. 2008;7:520–532. doi: 10.1016/j.cmet.2008.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Scheuner D, Song B, McEwen E, Liu C, Laybutt R, Gillespie P, et al. Translational control is required for the unfolded protein response and in vivo glucose homeostasis. Mol Cell. 2001;7:1165–1176. doi: 10.1016/s1097-2765(01)00265-9. [DOI] [PubMed] [Google Scholar]
- 15.Scheuner D, Vander Mierde D, Song B, Flamez D, Creemers JW, Tsukamoto K, et al. Control of mRNA translation preserves endoplasmic reticulum function in beta cells and maintains glucose homeostasis. Nat Med. 2005;11:757–764. doi: 10.1038/nm1259. [DOI] [PubMed] [Google Scholar]
- 16.Tirasophon W, Lee K, Callaghan B, Welihinda A, Kaufman RJ. The endoribonuclease activity of mammalian IREI autoregulates its mRNA and is required for the unfolded protein response. Genes Dev. 2000;14:2725–2736. doi: 10.1101/gad.839400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tsuru A, Fujimoto N, Takahashi S, Saito M, Nakamura D, Iwano M, et al. Negative feedback by IREIβ optimizes mucin production in goblet cells. Proc Natl Acad Sci U S A. 2013;110:2864–2869. doi: 10.1073/pnas.1212484110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Iqbal J, Queiroz J, Li Y, Jiang XC, Ron D, Hussain MM. Increased intestinal lipid absorption caused by IreIβ deficiency contributes to hyperlipidemia and atherosclerosis in apolipoprotein E-deficient mice. Circ Res. 2012;110:1575–1584. doi: 10.1161/CIRCRESAHA.112.264283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Iqbal J, Dai K, Seimon T, Jungreis R, Oyadomari M, Kuriakose G, et al. IREIbeta inhibits chylomicron production by selectively degrading MTP mRNA. Cell Metab. 2008;7:445–455. doi: 10.1016/j.cmet.2008.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ron D, Habener JF. CHOP, a novel developmentally regulated nuclear protein that dimerizes with transcription factors C/EBP and LAP and functions as a dominant-negative inhibitor of gene transcription. Genes Dev. 1992;6:439–453. doi: 10.1101/gad.6.3.439. [DOI] [PubMed] [Google Scholar]
- 21.Liebermann DA, Hoffman B. Myeloid differentiation (MyD)/growth arrest DNA damage (GADD) genes in tumor suppression, immunity and inflammation. Leukemia. 2002;16:527–541. doi: 10.1038/sj.leu.2402477. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.