Abstract
Phenol- and naphthalene-degrading indigenous Pseudomonas pseudoalcaligenes strain C70 has great potential for the bioremediation of polluted areas. It harbours two chromosomally located catechol meta pathways, one of which is structurally and phylogenetically very similar to the Pseudomonas sp. CF600 dmp operon and the other to the P. stutzeri AN10 nah lower operon. The key enzymes of the catechol meta pathway, catechol 2,3-dioxygenase (C23O) from strain C70, PheB and NahH, have an amino acid identity of 85%. The metabolic and regulatory phenotypes of the wild-type and the mutant strain C70ΔpheB lacking pheB were evaluated. qRT-PCR data showed that in C70, the expression of pheB- and nahH-encoded C23O was induced by phenol and salicylate, respectively. We demonstrate that strain C70 is more effective in the degradation of phenol and salicylate, especially at higher substrate concentrations, when these compounds are present as a mixture; i.e., when both pathways are expressed. Moreover, NahH is able to substitute for the deleted PheB in phenol degradation when salicylate is also present in the growth medium. The appearance of a yellow intermediate 2-hydroxymuconic semialdehyde was followed by the accumulation of catechol in salicylate-containing growth medium, and lower expression levels and specific activities of the C23O of the sal operon were detected. However, the excretion of the toxic intermediate catechol to the growth medium was avoided when the growth medium was supplemented with phenol, seemingly due to the contribution of the second meta pathway encoded by the phe genes.
Introduction
In nature, aromatics are an extensively distributed class of organic compounds. Xenobiotic aromatic pollutants are usually the major concern because of their environmental persistence and toxicity to living organisms. Despite that, microorganisms possess biochemical pathways enabling them to use these compounds as sole carbon and energy sources [1–4].
Biodegradation pathways of aromatic compounds converge towards intermediates such as catechol, protocatechuate, gentisate and hydroquinone before ring cleavage occurs [5]. In aerobic degradation pathways, catechol and its substituted derivatives are channelled towards either an ortho-cleavage pathway by catechol 1,2-dioxygenase (C12O) or a meta-cleavage pathway by catechol 2,3-dioxygenase (C23O) [3, 5].
Extradiol dioxygenases (C23Os) are more versatile than the intradiol enzymes (C12Os), cleaving very different substrates and occurring in a wider variety of pathways [6]. It is vital for bacteria to avoid the accumulation of catechol during the degradation of aromatic compounds because it can repress the synthesis of the catechol dioxygenase systems [7–11]. For example, a high inhibitory effect of this metabolite was confirmed in Pseudomonas putida F1, where catechol concentrations higher than 0.09 mM significantly decreased both benzene- and catechol-associated respiration of the strain [11]. To avoid the accumulation of toxic intermediates, bacteria have developed multiple strategies: the expression of C23O-reactivating ferredoxins, the recruitment of inactivation-resistance C23Os, gene duplications that increase catechol consumption or the balanced induction of enzymes involved in the production and degradation of catechol [12, 13].
Several research groups have found that bacteria possess multiple C12Os and/or C23Os [14–20]. Gene duplication seems to be a widespread phenomenon amongst TOL plasmids and gives them a selective advantage in the natural environment over those with only single copies [16]. P. aeruginosa JI104, which can utilize different aromatic compounds, has three C23Os. These isoenzymes can coexist as a result of gene duplication and further divergence due to the accumulation of mutations in the duplicated genes that are free of selective pressures [21]. Possessing multiple catechol dioxygenases helps keep the intracellular concentration of catechol or it derivatives low, probably preventing its toxicity [20].
In the natural environment, bacteria are mostly exposed to mixtures of aromatic compounds, and their survival and distribution depends on the capability and versatility of bacterial metabolism [9, 22]. In the Baltic Sea, bacterioplankton is influenced by coastal industries and marine traffic [23], both of which can cause pollution containing various compounds. We have shown previously that bacteria isolated from this environment possess IncP-9 family plasmids [24] and can degrade phenol, benzoate, m-toluate, salicylate, naphthalene [25] and toluene [26]. Vedler et al. [25] also showed that 3 out of 38 strains degrading phenol via the catechol meta pathway have two to three phylogenetically different C23O genes. One of these strains, Pseudomonas pseudoalcaligenes C70, is able to grow on naphthalene in addition to phenol, but not on benzoate or toluate, and was selected for future investigation in this study. Whole genome sequencing revealed the occurrence of three genetically different sequences of the 16S rRNA gene, one most similar to P. pseudoalcaligenes and the other two to P. mendocina. Despite this finding, strain C70 was classified as a P. pseudoalcaligenes based on the sequence of the rpoB gene and its phenotypic characteristics [25].
The aim of the present research was to identify the physiological role of C23Os in P. pseudoalcaligenes strain C70 and to evaluate their potential to degrade a mixture of phenol and salicylate. We would like to note that bacteria endowed with functional lower nah and phe (dmp-like) pathways have never been described and published. We characterized the expression and functioning of the redundant C23O genes (pheB and nahH) by generating the C70ΔpheB strain and by analysing the effect of this mutation on cell growth and the effective consumption of a mixture of salicylate and phenol.
Materials and methods
Bacterial strains, plasmids and culture conditions
The studied Pseudomonas pseudoalcaligenes strain C70 [24, 25] (CELMS, University of Tartu, Estonia) and the other strains and plasmids are shown in S1 Table. Pure cultures were stored in 30% glycerol at -80°C. The C70 strain was incubated on agar plates with minimal medium containing M9 salts [27] and trace elements [28] supplemented with phenol (PHE) (1.3 mM), salicylate (SAL) (1.3 mM) or R2A medium (Difco, USA) at 30°C. Strain C70ΔpheB was grown on LB medium or naphthalene vapour with kanamycin (Km, 50 μg ml-1) at 30°C. Escherichia coli strain DH5α containing the pTZ57R/T plasmid (Thermo Fisher Scientific, USA) was grown on LB medium with ampicillin (15 μg ml-1) at 37°C. Batch cultivation of cells was performed in 200 ml Erlenmeyer flasks containing 50 ml minimal medium supplemented with 3 mM PHE and SAL as a single substrate and as a mixture at 30°C on a rotary shaker. Growth was followed spectrophotometrically at 580 nm.
Sequencing and assembly of the draft genome of strain C70
The total DNA from the bacterial strain was isolated using the QIAGEN Plasmid Mega Kit (QIAGEN, Netherlands) that enriches the plasmid DNA over the chromosomal DNA. The isolated DNA was fragmented, samples for whole genome sequencing were prepared using the Illumina TruSeq® DNA PCR-Free LT Sample Preparation Kit (Illumina, USA) and were sequenced by default paired-end read protocols (2x100 bp) using an Illumina HiSeq2500 (Illumina, USA).
The whole genome was assembled using the Velvet version 1.2.09 software [29] as described previously [26]. The total length of the assembled contigs was 4,561,427 bp (576-fold sequencing coverage) in 506 contigs. The contig N50 was 549,228 bp with the longest scaffold (1,109,403 bp).
Gaps in nah lower and phe operons were sequenced using the 3730xl DNA Analyzer (Applied Biosystems, Thermo Fisher Scientific, USA) using the BigDye® Terminator v3.1 Cycle Sequencing Kit (Applied Biosystems, Thermo Fisher Scientific, USA) and protocols provided by the manufacturer.
The nucleotide sequences of the nah lower and phe operons of the strain C70 were deposited in GenBank under accession numbers KU695544 and KU695543, respectively.
DNA analysis
The GenBank database search used the NCBI ORF Finder and BLAST programs. The deduced amino acid sequences of C23Os of strain C70 were aligned with C23Os of the reference strains from the GenBank database using BioEdit version 7.0.5.3 [30]. Program MEGA6 was applied to construct the phylogenetic tree. Next to the branches is the percentage of replicate trees in which the associated taxa clustered together in the bootstrap test (1000 replicates) [31]. The tree is drawn to scale, with branch lengths in the same units as those of the evolutionary distances used to infer the phylogenetic tree. The JTT matrix-based method was used for computing the evolutionary distances that are presented as the units of the number of amino acid substitutions per site [31].
qRT-PCR
Total RNA for quantification of mRNA levels of nahH and pheB genes was isolated from exponential phase cells of P. pseudoalcaligenes C70 using the NucleoSpin RNA II kit (Macherey-Nagel, USA). Cells were grown on R2A medium or R2A and 2.5 mM SAL, R2A and 2.5 mM PHE or R2A and the mixture of PHE and SAL (both 2.5 mM) media. DNase I (Thermo Fisher Scientific, USA) was used to treat the RNA samples. A NanoDrop ND-1000 Spectrophotometer (Thermo Fisher Scientific, USA) was used to assess the RNA concentrations and purity. The SuperScript III Platinum SYBR green one-step qRT-PCR kit (Invitrogen, Thermo Fisher Scientific, USA) was used according to the manufacturer’s protocol (in a total reaction volume of 10 μl) for the quantitative reverse transcription-PCR (qRT-PCR) assay performed on the Rotor-Gene Q system (QIAGEN). A 10 ng portion of the total RNA was used for each reaction. The nahH and pheB genes were amplified using the primers C70aF and C70aR, and C70bF and C70bR, and the polA gene was amplified using the primers polAXhoylev and polAXhoall (S2 Table). The PCRs were performed using the following program: 50°C for 3 min and 95°C for 5 min, followed by 40 cycles of denaturation for 15 s, annealing at 58°C for 30 s, extension at 72°C for 20 s and 40°C for 1 min. At the end of the run, melting curve analyses were performed by increasing the temperature from 72 to 95°C and 3 s at 0.35°C with continuous fluorescence recording. The Rotor-GeneQ software version 2.02 (QIAGEN) was used to analyse the raw data, and mRNA amounts were calculated using the LinRegPCR software version 2013.0 [32]. Data from three separate qRT-PCR experiments performed on three independently extracted RNAs were averaged and normalized against polA levels.
Enzyme activity assay
Crude extracts were prepared and the activities of C23O and the protein concentrations were measured as described in [33] and [34]. The accumulation of 2-hydroxymuconic semialdehyde (HMS) was monitored by measuring the absorbance at 375 nm (ɛ = 14.7 mM l-1 cm-1).
Mutagenesis of the putative C23O genes
The bacterial strains and plasmids used and constructed in the nahH and pheB mutagenesis experiment are described in S1 Table. To disrupt the nahH and pheB genes in P. pseudoalcaligenes strain C70, a 1.8 kb nahH-containing DNA region and a 1.8 kb pheB-containing DNA region were amplified by PCR from genomic DNA with the corresponding pairs of primers (S2 Table) C70a1F –C70a2R and C70b1F –C70b2R. The amplified PCR products were cloned into the pTZ57R/T vector (Thermo Fisher Scientific, USA), resulting in pTZ57R/C70nahH and pTZ57R/C70pheB constructs. Then, the nahH region was excised from pTZ57R/C70nahH with the AatII and NotI enzymes, and pheB was excised from pTZ57R/C70pheB using Eco130I. Approximately 1 kb sized DNA fragments of both genes, nahH and pheB, were deleted and replaced with the Gmr and Kmr gene, respectively, which were earlier amplified by PCR from the pBK-miniTn7-ΩGm using the primers GmY and GmA (S2 Table) and from pUTmini-Tn5 Km2 with primer KmSac (S2 Table) and afterwards cleaved with Ecl136II. Before ligation of the Gmr and Kmr gene fragments into the open constructs, the AatII, NotI and Eco130I ends of the constructs were blunt-ended. From the pTZ57RΔC70nahH::gm and pTZ57RΔC70pheB::km constructs, the sequences ΔC70nahH::gm and ΔC70pheB::km were excised with KpnI-PaeI, respectively, and inserted into the plasmid vector pGP704 to obtain the constructs pGP704ΔC70nahH::gm and pGP704ΔC70pheB::km. These constructs were transferred from E. coli CC118λpir (S1 Table) into the P. pseudoalcaligenes strain C70 by conjugation using the helper plasmid pRK2013 (S1 Table). Transconjugants were chosen on agar plates supplemented with kanamycin (50 μg ml-1) and naphthalene vapour or gentamycin (10 μg ml-1) and PHE; benzylpenicillin (1,500 μg ml-1). Transconjugants were confirmed with PCR analysis using the primers KmOc (S2 Table) and C70b1F and GmA and C70a1F, and with nahH and pheB inner primers C70aF and C70aR; C70bF and C70bR (S2 Table). Conjugation experiments were carried out in triplicate.
Measurement of phenol and salicylate tolerance using respiration analysis
Oxygen-consumption rates (μ) of the bacterial cells pre-grown on 1.3 mM PHE and SAL were determined using the manometric respiratory system OxiTop® Control sensor (WTW, Germany). Tests were performed in two replicates in 250 ml bottles containing 100 ml mineral medium [27] with different PHE (0.7–15 mM) and SAL (0.7–8 mM) concentrations at 30°C for up to five days. The initial cell concentration in the bottles was 106 CFU ml-1 (wt C70 and C70ΔpheB). Oxygen consumption was measured based on the pressure drop in the bottles [35], and calculations of the specific oxygen-consumption rate and parameters of inhibition of the specific oxygen-consumption rate by PHE and SAL as determined using the Luong equation were performed as described by [36].
Chemical analyses
All samples taken from growth experiments for chemical analysis were centrifuged at 12,000 x g for 1 min at 4°C, and the supernatants were stored at -20°C until analysis. Analysis of the substrates and metabolites were performed using an Agilent Technologies (USA) 1200 Series high-performance liquid chromatography (HPLC) system with a Quaternary pump, an Autosampler and a 5-channel variable wavelength UV-Vis detector. The separation of compounds was achieved using an Agilent ZORBAX Eclipse XDB-C18 (4.6 x 250 mm i.d., 5 μm) analytical column and guard column (12.5 x 4.6 mm i.d.) of the same material (Agilent Technologies, USA). Elution was carried out at a flow rate of 0.80 ml min-1 within 15 minutes with a mobile phase gradient as follows: 0 min B (methanol)/C (standard buffer of 0.1% formic acid and 1 mM ammonium acetate, pH ~2.8) 45/55, 9 min 75/25, 11 min 45/55, and 13 min 45/55. The separation of the mixtures (injection volume 20 μl) was carried out at room temperature and the UV-Vis detector was set at 274 nm. The combined standard uncertainties of the results were between 2.1% and 3.4%. The preparation of the sample, instrumental measurements (both samples and calibration standards) and integration of the peaks are the main contributions to the uncertainties [37].
Results and discussion
Analysis of the genes of the phenol and naphthalene (lower) catabolic pathways
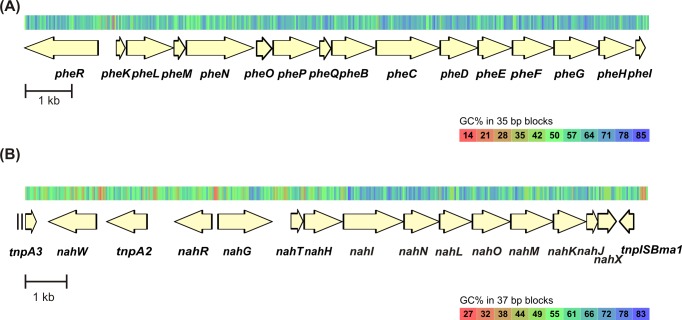
Pseudomonas pseudoalcaligenes strain C70 has two catechol meta pathways for the degradation of phenol and naphthalene (Fig 1). One of them is a part of the phenol degradation operon pheRKLMNOPQBCDEFGHI, which is structurally similar to the dmp operon of Pseudomonas sp. CF600. This operon is regulated by the XylR family-type regulatory gene pheR [38]. The other catechol meta route is a naphthalene lower pathway encoded by the classical nah operon (nahGTHINLOMKJX, also known as the sal operon) as described in P. putida G7 pNAH7 [39]. These enzymes convert salicylate over catechol to pyruvate and acetyl-CoA. This nah operon is regulated by the LysR family-type regulatory gene nahR [39]. The analysis of the sal operon structure in P. pseudoalcaligenes strain C70 revealed its high identity level with the sal operon described in P. stutzeri AN10 (CCUG 29243) [40]. Both strains have a second salicylate 1-hydroxylase gene (named as nahW in P. stutzeri AN10), in addition to nahG described from strain NAH7, and they share 100% amino acid identity. The strains NAH7, C70 and AN10 have the nahX gene with unknown function in the sal operon [41].
Fig 1. Genetic organization of the phe and sal operons of Pseudomonas pseudoalcaligenes C70.
(A) The genes of the phe operon—pheR, transcriptional regulator: pheKLMNOP, components of phenol hydroxylase; pheQ, ferredoxin-like gene; pheB, catechol 2,3-dioxygenase; pheC, 2-hydroxymuconic semialdehyde dehydrogenase; pheD, 2-hydroxymuconic semialdehyde hydrolase; pheE, 2-oxopent-4-dienoate hydratase; pheF, acetaldehyde dehydrogenase; pheG, 4-hydroxy-2-oxovalerate aldolase; pheH, 4-oxalocrotonate decarboxylase; pheI, 4-oxalocrotonate isomerase. (B) The genes of the sal operon—tnpA, transposase-like gene: nahW, salicylate 1-hydroxylase; nahR, regulatory gene; nahG, salicylate 1-hydroxylase; nahT, chloroplast ferredoxin-like protein coding gene; nahH, catechol 2,3-dioxygenase; nahI, hydroxymuconic semialdehyde dehydrogenase; nahN, hydroxymuconic semialdehyde hydrolase; nahL, 2-oxopent-4-enoate hydratase; nahO, acetaldehyde dehydrogenase; nahM, 2-oxo-4-hydroxypentanoate aldolase; nahK, 4-oxalocrotonate decarboxylase; nahJ, 4-oxalocrotonate isomerase; nahX, hypothetical protein coding gene; tnpISBma1, transposase-like gene.
As the nah lower operon is located between two transposable elements in C70, we can hypothesize that this operon was probably acquired from an outside source through horizontal gene transfer. Operons that encode aromatic degradation enzymes located on plasmids or chromosomes are frequently carried on transposons or flanked by insertion elements, providing their transferability [42]. Amino acid similarities of the different aromatic pathway enzymes and the diverging organization of catabolic genes suggest that several different gene clusters (e.g., meta pathways) may be combined in modules to which other peripheral genes may be added. Furthermore, it shows that many DNA rearrangements have occurred during the evolution of different pathways. This is also supported by the finding that the exposure of a community to a severe environment induces gene transfer, recombination and transposition events, thus conferring a higher survival capacity [5].
The strain C70 was shown to contain one large plasmid; therefore, the total DNA for whole genome sequencing was isolated using a method that enriches plasmid DNA over the chromosomal DNA. This led to some assembled contigs with at least 10-fold higher coverage, suggesting that these contigs are most likely of plasmid origin. However, all the genes necessary for phenol and naphthalene degradation were found on contigs with lower coverage, indicating that the phe and sal operon are located in the chromosome. In the plasmid, we did not find any aromatic catabolic genes. The nahH gene has been considered a plasmid encoded in the genus Pseudomonas, but in most of the naphthalene-degrading Pseudomonas stutzeri strains studied to date, this gene is chromosomally encoded [43]. The analysis of sequences did not reveal any catechol 1,2-dioxygenase or gentisate 1,2-dioxygenase encoding genes that might also be involved in the degradation of PHE and SAL. Because there were few data suggesting the coexistence of two catechol meta pathways in the same strain, we focused on the characterization of the key enzymes C23Os.
Phylogeny of redundant catechol 2,3-dioxygenases of strain C70
For phylogenetic analysis, a neighbour-joining tree based on deduced amino acid sequences of C23Os (PheB and NahH, 307 aa) of C70 was constructed together with C23O proteins accessible in GenBank (S1 Fig). Our analysis indicated that PheB from the phe operon in strain C70 was closest to the PhhB from P. putida P35X, with an amino acid identity of 95% (Cluster II in S1 Fig). As expected, NahH from strain C70 clustered together with the other C23O genes determined by nah lower operons (Cluster I, S1 Fig) and it was closest to chromosomally encoded NahH from P. stutzeri AN10, with an amino acid identity of 99.7%. NahH from C70 differed from that of AN10 by two bases, resulting in a tyrosine (Tyr) at position 218 instead of histidine (His). Two C23Os of strain C70, PheB and NahH, shared an amino acid identity of 85%. They also differed at position 218; NahH had a Tyr in this position, but PheB had phenylalanine (Phe). The catalytic properties of C23Os can change drastically even due to the change of a single amino acid [44]. Pseudomonads (for example, strain 1YB2) that have C23Os with a Tyr at amino acid position 218 are found in contaminated environments with high and low concentrations of benzene and toluene, which is compatible with a low turnover number and a high affinity of C23O for catechol. However, a C23O carrying His at position 218 (as in the case of the Pseudomonas sp. strain 1YdBTEX2) could be isolated only from a site with high concentrations of the same compounds, which is in accordance with a high turnover number and a low affinity for catechol. This suggests that in response to pollutant concentrations, the enzymes were positively selected in the environment [44, 45].
Our studied strain was isolated from Baltic Sea water taken from the southern area of the Finnish gulf, near a high sea traffic and port area; therefore, the water may be slightly contaminated with aromatic compounds. P. stutzeri AN10 was isolated from polluted marine sediments of the West Mediterranean Sea [46].
Construction of mutant strains and testing their tolerance to higher substrate concentrations
To establish the role of the redundant catechol meta pathways in strain C70, the knockout mutants of the two genes pheB and nahH, encoding the key enzymes of both pathways, C23O, were constructed. While the mutant strain C70ΔpheB (with the pheB gene disrupted with Kmr gene) was successfully obtained, we were unable to construct C70ΔnahH, although we attempted to use the alternative method of Martinez-Garcia and de Lorenzo [47].
The higher concentrations of aromatic compounds and their metabolites in the growth medium can be toxic to bacteria. For example, the growth of Escherichia coli and Burkholderia cepacia were inhibited in LB by the addition of 5 mM SAL [48] and on phenanthrene by SAL at concentrations >1.25 mM [49], respectively. The tolerance of wt C70 and C70ΔpheB strains to the high concentration of PHE and SAL was tested by determination of the inhibition of the oxygen consumption rate (Fig 2).
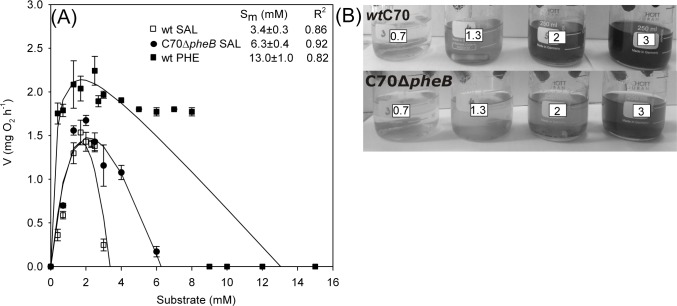
Fig 2. Determination of maximum inhibitory concentrations of phenol and salicylate, and accumulation of intermediates in salicylate-containing growth medium.
(A) The phenol (PHE)- and salicylate (SAL)-dependent oxygen consumption rate curves of wt and C70ΔpheB strains for determination of the maximum inhibitory concentrations of phenol and salicylate, Sm, using the Luong model. Error bars indicate the standard deviation. (B) Accumulation of dark brown metabolic intermediates of SAL catabolism with different SAL concentrations (0.7, 1.3, 2.0 and 3.0 mM) in wt and C70ΔpheB strains.
The C70ΔpheB strain tolerated higher SAL concentrations than the wt. The maximum inhibitory concentrations of SAL (at which no oxygen consumption was observed, Sm, using the Luong model) were 6.3±0.4 mM for C70ΔpheB and 3.4±0.3 mM for the wt strain. During growth on higher concentrations of SAL, the formation of dark pigments was observed at concentrations of 2 mM and 3 mM in the wt and C70ΔpheB strains, respectively (Fig 2B). These pigments are the result of the polymeric aggregates of quinonic intermediates, spontaneously formed in solution after the oxidation of catechol, and this dark brown pigment is toxic even for bacteria able to degrade catechol [20, 50]. A highly inhibitory effect of accumulating catechol to benzene-associated respiration has been shown in Pseudomonas putida F1 [11]. The maximum inhibitory concentration (Sm) of PHE in the wt strain was 13.0±1.0 mM and the accumulation of insoluble dark pigments in the growth medium was not determined. Juang and Tsai [10] have also shown that the inhibitory effect of SAL on the growth of Pseudomonas putida is higher than that of PHE.
Based on these data, we can conclude that the maximum inhibitory concentration of SAL in the strain possessing two catechol meta pathways and the C70ΔpheB strain, with a disrupted pheB gene and an intact nahH gene, are different. At the same time, the SAL concentrations where the values of the maximum specific oxygen-consumption rate were the highest are quite similar. In addition, the wt strain can tolerate higher PHE concentrations than SAL concentrations.
Effect of inducers on the expression and activity of two catechol 2,3-dioxygenase genes
The induction experiments were carried out to determine the effect of PHE and SAL as inducers on the expression of C23Os in wt and C70ΔpheB. The bacterial cells were grown to mid-log phase on R2A or on a mixture of R2A and SAL (2.5 mM) or/and PHE (2.5 mM), after which the corresponding mRNA was quantified by quantitative reverse transcription-PCR. The expression levels of the pheB and nahH genes are presented as relative to the housekeeping reference gene polA in Fig 3A. The results revealed that the expression of C23O was achieved by the induction of pheB by PHE and nahH by SAL in wt C70. In C70ΔpheB, the expression level of the nahH gene induced by SAL remained at the same level as in the wt strain; in the case of growth on PHE, the expression level of the nahH gene remained at the basal level (Fig 3A). As expected, the disrupted pheB gene was not expressed in C70ΔpheB. In addition, it seems that NahH is induced by SAL, although there is some basal level of expression without SAL, whereas PheB is inducible only by PHE (Fig 3A).
Fig 3. Expression levels of nahH and pheB and specific activities of C23O in strains C70 and C70ΔpheB on different growth substrates.
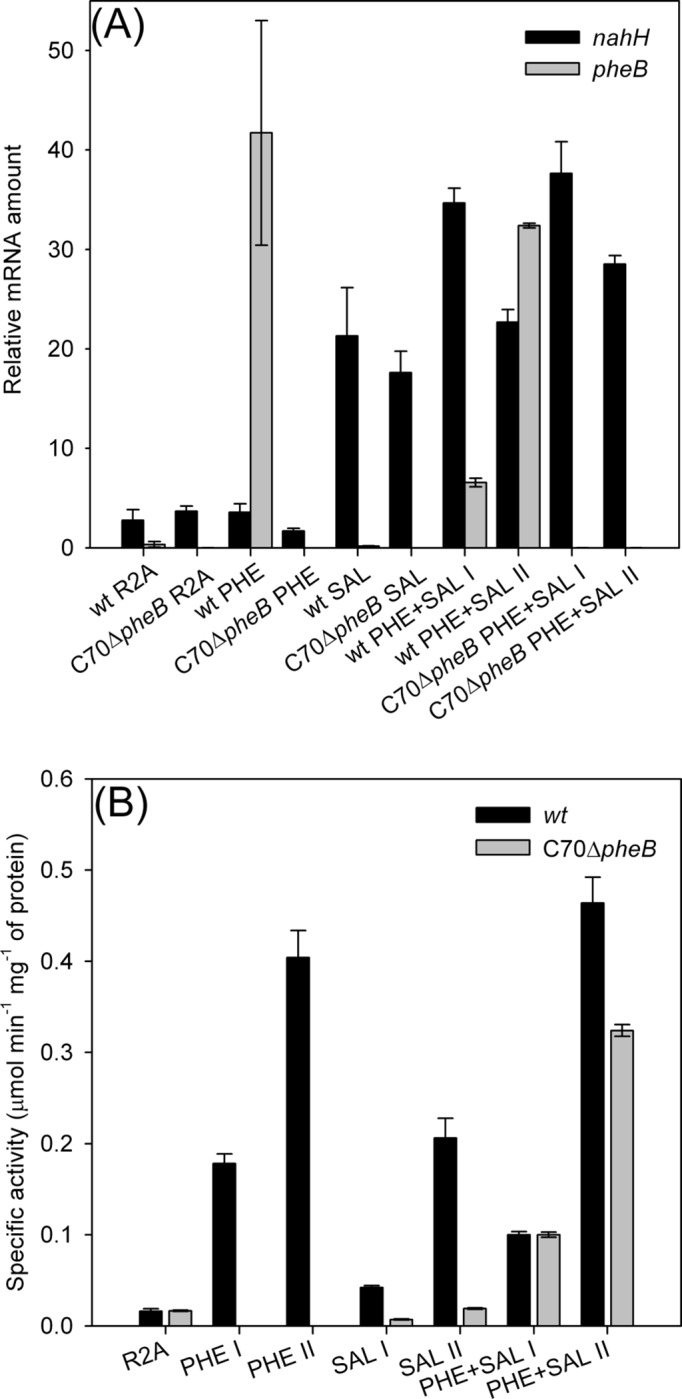
(A) For qRT-PCR analysis of the mRNA transcripts, the cells were grown on R2A medium, R2A and phenol (PHE, 2.5 mM), R2A and salicylate (SAL, 2.5 mM), or R2A and the mixture of SAL and PHE. The relative expression ratios (y-axis) were calculated as relative values of the reference gene polA expression level of the respective C70 strain. (B) Specific activities of C23O of the wt and C70ΔpheB strains were determined from cells grown on minimal medium supplemented with phenol, salicylate or the mixture of them (all substrates 3 mM). Samples were taken from the middle (I or no specification) or the late-exponential (II) growth phase.
Specific activity values of C23O determined in the wt strain grown on minimal medium containing PHE or SAL (Fig 3B) supported the results obtained in the qRT-PCR experiments (Fig 3A) that the activity of C23O is lower in cells grown on SAL than on PHE. At the same time, in C70ΔpheB grown in SAL, the specific activities were lower than that of the wt strain under the same conditions. The finding that specific activities are higher in the late-exponential than in the middle-exponential growth phase are in accordance with results obtained by Hugouvieux-Cotte-Pattat et al. [51] during the study of the induction of the TOL catabolic operons.
Since the expression levels and specific activities of C23Os in wt and C70ΔpheB strains on single substrates were different, experiments were conducted to clarify the degradation of SAL and PHE (both as a mixture and single substrate) by the studied strains.
Growth on single substrates: Identification of accumulation of intermediates
As mentioned above, the accumulation of dark brown pigments was observed when strains were grown on SAL (3 mM) (Fig 2B). In addition, the accumulation of a yellow compound was observed in all experiments. Furthermore, the biomass yield of wt and C70ΔpheB grown on SAL (Fig 4A and 4C) was lower than that of the wt strain grown on PHE (Fig 4B). To determine which intermediates accumulate in the growth media, HPLC and spectrophotometric analysis was conducted.
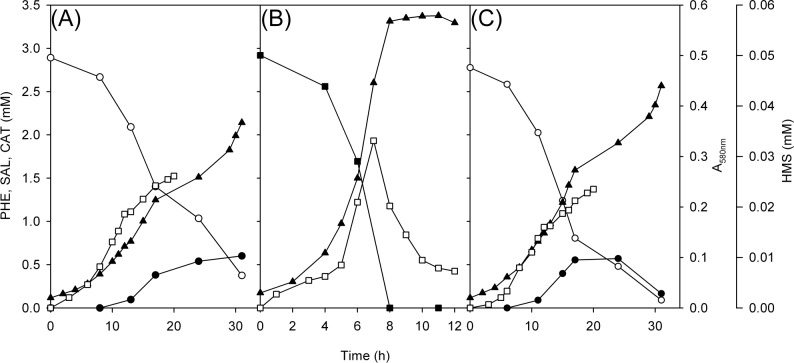
Fig 4. Growth, degradation of phenol and salicylate as single substrates and the accumulation of intermediates in strains C70 and C70ΔpheB.
Growth (▲, measured as absorbance at 580 nm), degradation of phenol (■) and salicylate (○) (both 3 mM, as single substrates), and accumulation of catechol (●) and 2-hydroxymuconic semialdehyde (HMS, □) in strains C70 (A, B) and C70ΔpheB (C). Concentrations of the substrates and intermediates are the averages of triplicate determinations; the combined standard uncertainties of the results were between 2.1% and 3.4%.
Accumulation of 2-hydroxymuconic semialdehyde (HMS) was observed when strains were grown on SAL and PHE (Fig 4). However, catechol (CAT) was determined in the growth medium only during the growth of the wt and C70ΔpheB strains on SAL (Fig 4A and 4C) after the appearance of HMS. Based on these results, we hypothesized that NahH is more sensitive to the accumulation of HMS than PheB, and therefore, CAT accumulates when strains are grown on SAL. Consequently, a lower growth yield on SAL than on PHE was determined. These assumptions are supported by higher specific activity values (both in the middle and at the end of the logarithmic growth phase, Fig 3B) and expression levels of C23O in the wt strain grown on PHE rather than on SAL (Fig 3A). Although in the case of the growth of the C70ΔpheB strain on SAL a reversible accumulation of catechol was observed (Fig 4C), we cannot conclude whether it was consumed by the bacteria or if the concentration was diminished due to autoxidation. The formation of dark brown pigments after ~15 h of incubation on SAL caused an increase in optical density at 580 nm that interfered with the determination of biomass concentration at that wavelength.
The assumption that catechols have an inhibitory effect on C23Os has been confirmed by several groups [11, 50, 52], but the role of HMS in the degradation of aromatic compounds has received little attention. Muñoz et al. [11] reported that the accumulation of catechol has a highly inhibitory effect on benzene- and catechol-associated respiration. They also suggested that the accumulation of the yellow intermediate had an effect, but they did not investigate it further. While modelling the growth of Pseudomonas sp. NCIMB 9688 on benzene, Monero et al. [53] concluded that HMS is a competitive inhibitor and its accumulation within the cell has an effect on the kinetic control of the meta pathway by the enzyme responsible for HMS degradation. The formed HMS can be degraded in two branches of the meta pathway by 2-hydroxymuconic semialdehyde dehydrogenase (HMSD) or hydrolase (HMSH), and the primary substrates control the whole set of enzymes [3, 54, 55]. However, experiments in mutant strains have shown that HMS is preferentially degraded by HMSD [38, 55]. Our experiments indicated that the accumulation of HMS influences the expression levels and specific activities of the C23O of the sal operon when strains are cultivated on SAL as a single substrate. In the case of strain C70, the accumulation of catechol is observed after the appearance of HMS in the growth medium, resulting in an inhibition of C23O and in turn the inability to degrade all the catechol produced by two salicylate 1-hydroxylases.
Catechol degradation intermediates are also able to inactivate the dioxygenases responsible for their cleavage [12, 52, 56]. Cerdan et al. [57] have shown that sensitivity of different C23Os to catechols can vary. They reported that NahH from P. putida G7 has a higher affinity constant and apparent rate constant of enzyme inactivation for catechol and 3-methylcatechol than XylE from P. putida KT2440. To avoid the accumulation of toxic intermediates, bacteria have evolved different strategies. For example, a mutant strain of P. putida F1 gained the ability to degrade styrene due to a single base pair mutation in toluene dioxygenase that resulted in attenuated activity of the enzyme and decreased the rate of 3-vinylcatechol production compared to the wt strain [52]. Another possibility to avoid catechol toxicity is to increase the rate of catechol-consuming reactions through duplication of catechol dioxygenase genes. For example, Cupriavidus pinatubonensis JMP134 (pJP4) (formerly Ralstonia eutropha JMP134) requires multiple copies of chlorocatechol 1,2-dioxygenase for efficient turnover of 3-chlorocatechol and growth on 3-chlorobenzoic acid [56]. Therefore, the ability to grow on aromatic compounds depends on a delicate balance between catechol-producing and catechol-consuming reactions.
However, in the case of our strain, simply having two copies of the C23O gene in the genome does not have such an effect when grown on a single substrate. During the growth of the wt strain on SAL, the accumulation of catechol is still detected because only one gene coding for C23O is expressed.
Growth on a mixture of substrates: Constitutively expressed nahH compensates for the function of disrupted pheB
The strain C70ΔpheB is not able to grow on minimal medium containing PHE as a single substrate. However, we were curious about what would occur when grown on a mixture of PHE and SAL. The growth yield of wt and C70ΔpheB strains grown on the mixture of substrates was at the same level, although there was a delay in the growth of the C70ΔpheB strain compared with the wt strain (Fig 5).
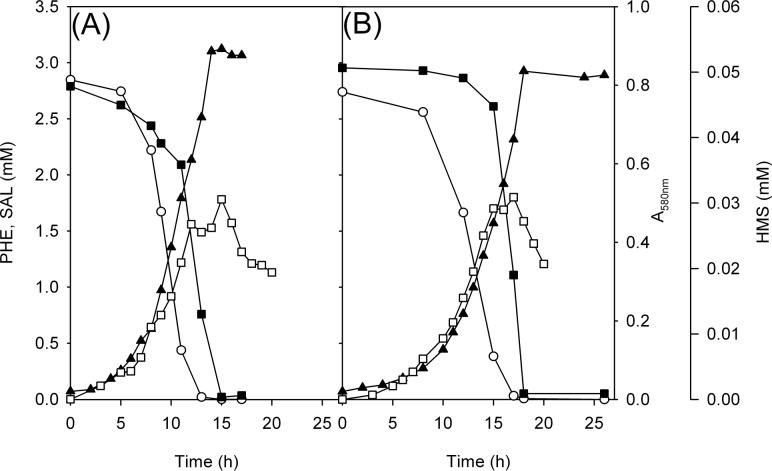
Fig 5. Growth and degradation of a mixture of phenol and salicylate and accumulation of intermediates in strains C70 and C70ΔpheB.
Growth (▲, measured as absorbance at 580 nm), degradation of phenol (■) and salicylate (○) (both 3 mM), and accumulation of 2-hydroxymuconic semialdehyde (HMS, □) in strains C70 (A) and C70ΔpheB (B). The concentrations of the substrates are the averages of triplicate determinations; the combined standard uncertainties of the results were between 2.1% and 3.4%.
HPLC analysis of the growth medium of both strains revealed that SAL was degraded before PHE, despite the fact that this substrate supports less efficient growth than PHE. This is probably due to the presence of two salicylate hydroxylase genes. Additionally, the accumulation of catechol was not detected (Fig 5). qRT-PCR analysis of the expression of nahH and pheB genes from wt and C70ΔpheB growing on R2A with a mixture of substrates showed that the nahH gene was expressed at a high level in the middle-exponential growth phase, whereas the pheB gene expression in the wt was higher in the late-exponential growth phase (Fig 3A). These data are in accordance with the results of the determination of specific activities of C23O, where higher values were also observed in the late-exponential phase (Fig 3B). The expression level of the nahH in C70ΔpheB grown on a mixture of PHE and SAL is almost at the same level through the exponential growth phase, although higher C23O activity values were obtained from late-exponential growth phase cells (Fig 3). Based on these results, we suggest that nahH can substitute/complement the pheB gene while strains are grown in a mixture of substrates, allowing the C70ΔpheB to use PHE as the growth substrate (Fig 5). Furthermore, as catechol does not accumulate in wt or C70ΔpheB in conditions where the meta pathway encoded by the phe genes is also expressed, the enzymes of this pathway downstream from C23O probably also contribute to the degradation of CAT and HMS produced from SAL.
Results obtained from the laboratory experiments with single strains and single substrates are usually not applicable in real pollution situations, as often the mixture of compounds is exposed to nature and the degradation of a particular component can be affected by other factors such as formed intermediates [7–9, 58]. In the case of strain C70, we found that this strain benefits from the two redundant catechol meta pathways, resulting in more efficient biodegradation of SAL only when PHE is also present in the growth medium and both pathways are expressed.
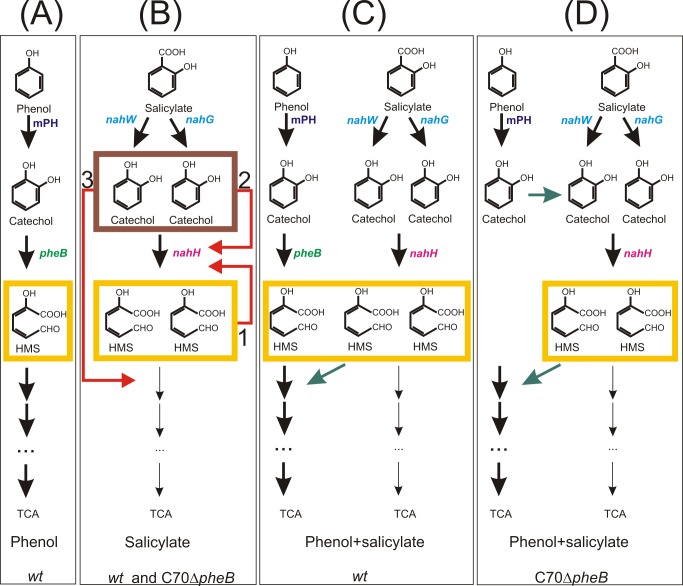
To sum up, we demonstrate that P. pseudoalcaligenes strain C70, harbouring two catechol meta pathways, encoded by two redundant catabolic operons, phe and nah, is more effective in the degradation of PHE and SAL, especially at higher substrate concentrations, when these compounds are present as a mixture; i.e., when both pathways are expressed. Induction experiments showed that NahH can substitute/complement the deleted PheB in PHE degradation when strains are cultivated in the mixture of substrates (Fig 6D). The accumulation of yellow HMS was observed in all substrates while the toxic intermediate catechol was detected in the growth medium only during the degradation of SAL by wt and C70ΔpheB strains (Fig 6).
Fig 6. Role of accumulation of intermediates in the catabolism of phenol and salicylate as single or mixed substrates in C70 and C70ΔpheB.
The figure sketches the degradation pathways and genes (mPH—multicomponent phenol hydroxylase; pheB and nahH—C23O; nahW and nahG—salicylate 1-hydroxylase) involved in phenol and salicylate catabolism as single (A, B) and mixed substrates (C, D). The supposed negative effect of accumulating intermediates 2-hydroxymuconic semialdehyde (HMS, yellow box) and catechol (brown box) to the functioning of involved catabolic genes are indicated with red arrows (with numbers 1, 2 and 3). Green arrows point to convergence or divergence of degradation of intermediates.
The accumulation of catechol inhibited the activity of C23O and probably also the HMS degrading enzyme (Fig 6B). In the case of the substrate mixture, catechol was not excreted into the growth medium, probably due to the contribution of the second meta pathway encoded by the phe genes (Fig 6C and 6D). SAL was degraded before PHE by both strains, probably as the result of bearing two salicylate hydroxylase genes. Therefore, we suppose that possessing two catechol degradation meta pathways gives an advantage to strain C70 in the environment, as contaminated ecosystems typically contain heterogeneous mixtures of organic compounds. Further research will be conducted to determine the substrate specificity of C23Os, the role of two redundant salicylate hydroxylase genes in strain C70 and the mechanism by which the PHE degradation pathway mitigates catechol toxicity during the degradation of SAL from the mixture of the substrates.
Supporting information
The phylogenetic tree is based on deduced amino acid sequences (307 aa) of the catechol 2,3-dioxygenases of strain C70 and reference strains from GenBank. The percentage of replicate trees in which the associated taxa clustered together in the bootstrap test (1000 replicates) is shown next to the branches.
(TIF)
(DOC)
(DOCX)
Acknowledgments
Isolated strains are deposited in the Collection of Environmental and Laboratory Microbial Strains (CELMS; financed by the Estonian Ministry of Education and Research), the public catalogue of which is available on the Estonian Electronic Microbial dataBase (EEMB) website http://eemb.ut.ee. We are grateful to Sofia Tšepelevitš for assistance with the HPLC analysis of the samples. This manuscript was edited for English language by American Journal Experts (AJE).
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
This research was supported by the Institute of Molecular and Cell Biology, University of Tartu; by the research grants SF0180026s08 (Prof. Ain Heinaru), IUT20-19, and CELMS from the Estonian Ministry of Education and Research, and by the Estonian Science Foundation grant 7827. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Widdel F, Rabus R. Anaerobic biodegradation of saturated and aromatic hydrocarbons. Curr Opin Biotechnol. 2001;12: 259–76. [DOI] [PubMed] [Google Scholar]
- 2.Lovley DR. Cleaning up with genomics: Applying molecular biology to bioremediation. Nat Rev Microbiol. 2003;1: 35–44. 10.1038/nrmicro731 [DOI] [PubMed] [Google Scholar]
- 3.Díaz E, Jimenez JI, Nogales J. Aerobic degradation of aromatic compounds. Curr Opin Biotechnol. 2013;24: 431–42. 10.1016/j.copbio.2012.10.010 [DOI] [PubMed] [Google Scholar]
- 4.Coronado E, Roggo C, Johnson DR, van der Meer JR. Genome-wide analysis of salicylate and dibenzofuran metabolism in Sphingomonas wittichii RW1. Front Microbiol. 2012;3: 300 10.3389/fmicb.2012.00300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.van der Meer JR, de Vos WM, Harayama S, Zehnder AJ. Molecular mechanisms of genetic adaption to xenobiotic compounds. Microbiol Rev. 1992;56: 677–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Vaillancourt FH, Bolin JT, Eltis LD. The ins and outs of ring-cleaving dioxygenases. Crit Rev Biochem Mol Biol. 2006;41: 241–67. 10.1080/10409230600817422 [DOI] [PubMed] [Google Scholar]
- 7.Saez PB, Rittmann BE. Biodegradation kinetics of a mixture containing a primary substrate (phenol) and an inhibitory co-metabolite (4-chlorophenol). Biodegradation. 1993;4: 3–21. [DOI] [PubMed] [Google Scholar]
- 8.Reardon KF, Mosteller DC, Bull Rogers JD. Biodegradation kinetics of benzene, toluene, and phenol as single and mixed substrates for Pseudomonas putida F1. Biotechnol Bioeng. 2000;69: 385–400. [DOI] [PubMed] [Google Scholar]
- 9.Heinaru E, Viggor S, Vedler E, Truu J, Merimaa M, Heinaru A. Reversible accumulation of p-hydroxybenzoate and catechol determines the sequential decomposition of phenolic compounds in mixed substrate cultivations in pseudomonads. FEMS Microbiol Ecol. 2001;37: 79–89. [Google Scholar]
- 10.Juang RS, Tsai SY. Growth kinetics of Pseudomonas putida in the biodegradation of single and mixed phenol and sodium salicylate. Biochem Eng J. 2006;31: 133–40. [DOI] [PubMed] [Google Scholar]
- 11.Muñoz R, Díaz LF, Bordel S, Villaverde S. Inhibitory effects of catechol accumulation on benzene biodegradation in Pseudomonas putida F1 cultures. Chemosphere. 2007;68: 244–52. 10.1016/j.chemosphere.2007.01.016 [DOI] [PubMed] [Google Scholar]
- 12.George KW, Hay AG. Bacterial strategies for growth on aromatic compounds. Adv Appl Microbiol. 2011;74: 1–33. 10.1016/B978-0-12-387022-3.00005-7 [DOI] [PubMed] [Google Scholar]
- 13.George KW, Hay A. Less is more: reduced catechol production permits Pseudomonas putida F1 to grow on styrene. Microbiology-Sgm. 2012;158: 2781–8. [DOI] [PubMed] [Google Scholar]
- 14.Aoki K, Konohana T, Shinke R, Nishira H. Two catechol 1,2-dioxygenases from aniline-assimilating bacterium, Frateuria species ANA-18. Agric Biol Chem. 1984;48: 2097–104. [Google Scholar]
- 15.Keil H, Lebens MR, Williams PA. TOL plasmid pWW15 contains two nonhomologous, independently regulated catechol 2,3-oxygenase genes. J Bacteriol. 1985;163: 248–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Odonnell KJ, Williams PA. Duplication of both xyl catabolic operons on TOL plasmid pww15. J Gen Microbiol. 1991;137: 2831–8. 10.1099/00221287-137-12-2831 [DOI] [PubMed] [Google Scholar]
- 17.Kim SI, Leem SH, Choi JS, Chung YH, Kim S, Park YM, et al. Cloning and characterization of two catA genes in Acinetobacter lwoffii K24. J Bacteriol. 1997;179: 5226–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Murakami S, Okuno T, Matsumura E, Takenaka S, Shinke R, Aoki K. Cloning of a gene encoding hydroxyquinol 1,2-dioxygenase that catalyzes both intradiol and extradiol ring cleavage of catechol. Biosci Biotechnol Biochem. 1999;63: 859–65. 10.1271/bbb.63.859 [DOI] [PubMed] [Google Scholar]
- 19.Pessione E, Giuffrida MG, Mazzoli R, Caposio P, Landolfo S, Conti A, et al. The catechol 1,2 dioxygenase system of Acinetobacter radioresistens: isoenzymes, inductors and gene localisation. Biological Chemistry. 2001;382: 1253–61. 10.1515/BC.2001.156 [DOI] [PubMed] [Google Scholar]
- 20.Jimenez JI, Perez-Pantoja D, Chavarria M, Diaz E, de Lorenzo V. A second chromosomal copy of the catA gene endows Pseudomonas putida mt-2 with an enzymatic safety valve for excess of catechol. Environ Microbiol. 2014;16: 1767–78. 10.1111/1462-2920.12361 [DOI] [PubMed] [Google Scholar]
- 21.Kitayama A, Achioku T, Yanagawa T, Kanou K, Kikuchi M, Ueda H, et al. Cloning and characterization of extradiol aromatic ring-cleavage dioxygenases of Pseudomonas aeruginosa JI104. J Ferment Bioeng. 1996;82: 217–23. [Google Scholar]
- 22.Choi KY, Zylstra GJ, Kim E. Benzoate catabolite repression of the phthalate degradation pathway in Rhodococcus sp. strain DK17. Appl Environ Microbiol. 2007;73: 1370–4. 10.1128/AEM.02379-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.HELCOM. Available from: http://www.helcom.fi/.
- 24.Jutkina J, Heinaru E, Vedler E, Juhanson J, Heinaru A. Occurrence of plasmids in the aromatic degrading bacterioplankton of the Baltic Sea. Genes. 2011;2: 853–68. 10.3390/genes2040853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vedler E, Heinaru E, Jutkina J, Viggor S, Koressaar T, Remm M, et al. Limnobacter spp. as newly detected phenol-degraders among Baltic Sea surface water bacteria characterised by comparative analysis of catabolic genes. Syst Appl Microbiol. 2013;36: 525–32. 10.1016/j.syapm.2013.07.004 [DOI] [PubMed] [Google Scholar]
- 26.Heinaru E, Naanuri E, Grunbach M, Jõesaar M, Heinaru A. Functional redundancy in phenol and toluene degradation in Pseudomonas stutzeri strains isolated from the Baltic Sea. Gene. 2016;589: 90–8. 10.1016/j.gene.2016.05.022 [DOI] [PubMed] [Google Scholar]
- 27.Adams MH. Bacteriophages. New York: Interscience Publishers; 1959. [Google Scholar]
- 28.Bauchop T, Elsden SR. The growth of micro-organisms in relation to their energy supply. J Gen Microbiol. 1960;23: 457–69. 10.1099/00221287-23-3-457 [DOI] [PubMed] [Google Scholar]
- 29.Zerbino DR, Birney E. Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Res. 2008;18: 821–9. 10.1101/gr.074492.107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hall T. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/ NT. Nucleic Acids Symp Ser. 1999;41: 95–8. [Google Scholar]
- 31.Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6: Molecular Evolutionary Genetics Analysis Version 6.0. Mol Biol Evol. 2013;30: 2725–9. 10.1093/molbev/mst197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ruijter JM, Ramakers C, Hoogaars WM, Karlen Y, Bakker O, van den Hoff MJ, et al. Amplification efficiency: linking baseline and bias in the analysis of quantitative PCR data. Nucleic Acids Res. 2009;37: e45 10.1093/nar/gkp045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hegeman GD. Synthesis of the enzymes of the mandelate pathway by Pseudomonas putida. I. Synthesis of enzymes by the wild type. J Bacteriol. 1966;91: 1140–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bradford MM. A rapid sensitive method for the quantification of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72: 248–54. [DOI] [PubMed] [Google Scholar]
- 35.Juhanson J, Truu J, Heinaru E, Heinaru A. Survival and catabolic performance of introduced Pseudomonas strains during phytoremediation and bioaugmentation field experiment. FEMS Microbiol Ecol. 2009;70: 446–55. 10.1111/j.1574-6941.2009.00754.x [DOI] [PubMed] [Google Scholar]
- 36.Jõesaar M, Heinaru E, Viggor S, Vedler E, Heinaru A. Diversity of the transcriptional regulation of the pch gene cluster in two indigenous p-cresol-degradative strains of Pseudomonas fluorescens. FEMS Microbiol Ecol. 2010;72: 464–75. 10.1111/j.1574-6941.2010.00858.x [DOI] [PubMed] [Google Scholar]
- 37.Ellison SLR, Rösslein M, Williams A. Eurachem/CITAC Guide, Quantifying uncertainty in analytical measurement, 2nd ed. Berlin: Eurachem; 2000. [Google Scholar]
- 38.Powlowski J, Shingler V. Genetic and biochemistry of phenol degradation by Pseudomonas sp. CF600. Biodegradation. 1994;5: 219–36. [DOI] [PubMed] [Google Scholar]
- 39.Yen KM, Gunsalus IC. Plasmid gene organization: naphthalene/salicylate oxidation. Proc Natl Acad Sci U S A. 1982;79: 874–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bosch R, Garcia-Valdes E, Moore ER. Complete nucleotide sequence and evolutionary significance of a chromosomally encoded naphthalene-degradation lower pathway from Pseudomonas stutzeri AN10. Gene. 2000;245: 65–74. [DOI] [PubMed] [Google Scholar]
- 41.Sota M, Yano H, Ono A, Miyazaki R, Ishii H, Genka H, et al. Genomic and functional analysis of the IncP-9 naphthalene-catabolic plasmid NAH7 and its transposon Tn4655 suggests catabolic gene spread by a tyrosine recombinase. J Bacteriol. 2006;188: 4057–67. 10.1128/JB.00185-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Williams PA, Sayers JR. The evolution of pathways for aromatic hydrocarbon oxidation in Pseudomonas. Biodegradation. 1994;5: 195–217. [DOI] [PubMed] [Google Scholar]
- 43.Rossello-Mora RA, Lalucat J, Garcia-Valdes E. Comparative biochemical and genetic analysis of naphthalene degradation among Pseudomonas stutzeri strains. Appl Environ Microbiol. 1994;60: 966–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Junca H, Plumeier I, Hecht HJ, Pieper DH. Difference in kinetic behaviour of catechol 2,3-dioxygenase variants from a polluted environment. Microbiology-Sgm. 2004;150: 4181–7. [DOI] [PubMed] [Google Scholar]
- 45.Suenaga H, Mizuta S, Miyazaki K. The molecular basis for adaptive evolution in novel extradiol dioxygenases retrieved from the metagenome. FEMS Microbiol Ecol. 2009;69: 472–80. 10.1111/j.1574-6941.2009.00719.x [DOI] [PubMed] [Google Scholar]
- 46.Garcia-Valdes E, Cozar E, Rotger R, Lalucat J, Ursing J. New naphthalene-degrading marine Pseudomonas strains. Appl Environ Microbiol. 1988;54: 2478–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Martinez-Garcia E, de Lorenzo V. Engineering multiple genomic deletions in Gram-negative bacteria: analysis of the multi-resistant antibiotic profile of Pseudomonas putida KT2440. Environ Microbiol. 2011;13: 2702–16. 10.1111/j.1462-2920.2011.02538.x [DOI] [PubMed] [Google Scholar]
- 48.Pomposiello PJ, Bennik MHJ, Demple B. Genome-wide transcriptional profiling of the Escherichia coli responses to superoxide stress and sodium salicylate. J Bacteriol. 2001;183: 3890–902. 10.1128/JB.183.13.3890-3902.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lee DS, Lee MW, Woo SH, Park JM. Effects of salicylate and glucose on biodegradation of phenanthrene by Burkholderia cepacia PM07. J Microbiol Biotechn. 2005;15: 859–65. [Google Scholar]
- 50.Park W, Jeon CO, Cadillo H, DeRito C, Madsen EL. Survival of naphthalene-degrading Pseudomonas putida NCIB 9816–4 in naphthalene-amended soils: toxicity of naphthalene and its metabolites. Appl Microbiol Biotechnol. 2004;64: 429–35. 10.1007/s00253-003-1420-6 [DOI] [PubMed] [Google Scholar]
- 51.Hugouvieux-Cotte-Pattat N, Köhler T, Rekik M, Harayama S. Growth-phase-dependent expression of the Pseudomonas putida TOL plasmid pWW0 catabolic genes. J Bacteriol. 1990;172: 6651–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.George KW, Kagle J, Junker L, Risen A, Hay AG. Growth of Pseudomonas putida F1 on styrene requires increased catechol-2,3-dioxygenase activity, not a new hydrolase. Microbiology-Sgm. 2011;157: 89–98. [DOI] [PubMed] [Google Scholar]
- 53.Monero A, Lanza L, Zilli M, Sene L, Converti A. Batch kinetics of Pseudomonas sp. growth on benzene. Modeling of product and substrate inhibitions. Biotechnol Prog. 2003;19: 676–9. 10.1021/bp025764s [DOI] [PubMed] [Google Scholar]
- 54.Sala-Trepat JM, Murray K, Williams PA. The metabolic divergence in the meta cleavage of catechols by Pseudomonas putida NCIB 10015. Physiological significance and evolutionary implications. Eur J Biochem. 1972;28: 347–56. [DOI] [PubMed] [Google Scholar]
- 55.Bertini L, Cafaro V, Proietti S, Caporale C, Capasso P, Caruso C, et al. Deepening TOL and TOU catabolic pathways of Pseudomonas sp. OX1: cloning, sequencing and characterization of the lower pathways. Biochimie. 2013;95: 241–50. 10.1016/j.biochi.2012.09.018 [DOI] [PubMed] [Google Scholar]
- 56.Perez-Pantoja D, Ledger T, Pieper DH, Gonzalez B. Efficient turnover of chlorocatechols is essential for growth of Ralstonia eutropha JMP134(pJP4) in 3-chlorobenzoic acid. J Bacteriol. 2003;185: 1534–42. 10.1128/JB.185.5.1534-1542.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Cerdan P, Rekik M, S. H. Substrate specificity differences between two catechol 2,3-dioxygenases encoded by the TOL and NAH plasmids from Pseudomonas putida. Eur J Biochem. 1995;229: 113–8. [DOI] [PubMed] [Google Scholar]
- 58.Smith MR, Ewing M, Ratledge C. The interactions of various aromatic substrates degraded by Pseudomonas sp. NCIB 10643: synergistic inhibition of growth by two compounds that serve as growth substrates Appl Microbiol Biotechnol. 1991;34: 536–8. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The phylogenetic tree is based on deduced amino acid sequences (307 aa) of the catechol 2,3-dioxygenases of strain C70 and reference strains from GenBank. The percentage of replicate trees in which the associated taxa clustered together in the bootstrap test (1000 replicates) is shown next to the branches.
(TIF)
(DOC)
(DOCX)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.