Structural and dynamic properties of a full-length bacteriophytochrome reveal details of the molecular regulation mechanism.
Keywords: phytochrome, allostery, sensor, effector, diguanylate cyclase, c-di-GMP, HDX-MS, dynamics, SAXS
Abstract
Nature has evolved an astonishingly modular architecture of covalently linked protein domains with diverse functionalities to enable complex cellular networks that are critical for cell survival. The coupling of sensory modules with enzymatic effectors allows direct allosteric regulation of cellular signaling molecules in response to diverse stimuli. We present molecular details of red light–sensing bacteriophytochromes linked to cyclic dimeric guanosine monophosphate–producing diguanylyl cyclases. Elucidation of the first crystal structure of a full-length phytochrome with its enzymatic effector, in combination with the characterization of light-induced changes in conformational dynamics, reveals how allosteric light regulation is fine-tuned by the architecture and composition of the coiled-coil sensor-effector linker and also the central helical spine. We anticipate that consideration of molecular principles of sensor-effector coupling, going beyond the length of the characteristic linker, and the appreciation of dynamically driven allostery will open up new directions for the design of novel red light–regulated optogenetic tools.
INTRODUCTION
During evolution, nature has developed a remarkably modular architecture of covalently linked protein domains. Combining individual building blocks from an array of diverse functionalities enabled organisms to develop complex cellular networks that are critical for homeostasis. The frequently observed coupling of sensory modules with enzymatic effectors enables direct allosteric regulation of, for example, second messenger levels in response to diverse stimuli. Recently, light-regulated enzymes have attracted special attention because of their potential for optogenetic applications (1). However, naturally occurring sensor-effector couples are limited, and our current understanding of molecular mechanisms underlying their modularity prevents the rational design of efficient, novel sensor-effector combinations. Optogenetic platforms based on near-infrared light photoreceptors have recently attracted special attention because of their potential for deep tissue stimulation in mammalian systems (2, 3). Most of these optogenetic tools are based on natural, noncovalent interactions of plant phytochromes with transcription factors, which can be adapted to a variety of cellular targets [reviewed by Tischer and Weiner (1)]. In contrast, the design of novel tools for direct allosteric control of enzymatic activity appears more complex because of a limited understanding of molecular mechanisms involved in signal transduction between sensor and effector domains. Nevertheless, bacterial phytochromes have been successfully used for the generation of artificial red light–regulated adenylyl cyclases (4) and phosphodiesterases (5). However, establishing tools for direct manipulation of enzymatic activities still requires substantial screening efforts, and concepts learned from successful designs are not easily transferable even to closely related systems. Especially the lack of structural information for natural or artificial full-length phytochromes precludes the functional interpretation of linker elements that covalently tether sensor to effector.
To overcome this limitation, we focused on naturally occurring systems that allow red light regulation of the bacterial second messenger bis-(3′-5′)-cyclic dimeric guanosine monophosphate (c-di-GMP) (6) by linking canonical bacteriophytochromes (7) with GGDEF domains (8) featuring diguanylyl cyclase (DGC) activity [phytochrome-activated diguanylyl cyclase (PadC)]. Although related systems have been characterized previously (9–11), we concentrated on natural variants that feature a defined PAS-GAF-PHY-GGDEF domain architecture (Fig. 1A) and initially characterized three constructs belonging to subgroups with characteristic linker length variations between the PHY and GGDEF domains. The systematic differences of one or two heptad repeats support the relevance of a coiled-coil structure preceding the DGC (fig. S1). This structural element is also observed in other sensor DGCs (12, 13) and is functionally relevant for GGDEF regulation (14, 15). However, molecular details of the structural changes induced by the reversible photoswitching of the phytochrome sensor between its dark-state conformation (Pr) and its red light–activated Pfr form with respect to long-range signaling to effector domains are not well understood.
Fig. 1. Overview of IsPadC.
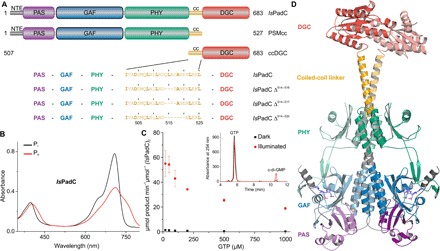
(A) Schematic representation of IsPadC constructs characterized in this study. Individual domains are colored in dark gray, violet, blue, green, orange, and red for the N-terminal extension (NTE), Per-ARNT-Sim (PAS), cGMP phosphodiesterase–adenylyl cyclase–FhlA (GAF), phytochrome-associated (PHY), coiled-coil (cc), and DGC domains, respectively. Coiled-coil truncations are denoted as, for example, IsPadC Δ514–520 for the variant deleted in one heptad including residues 514 to 520. (B) Spectral characteristics of dark-adapted (Pr state) IsPadC in comparison to the Pfr state obtained after red light illumination. (C) Kinetic characterization of GTP to c-di-GMP conversion. The characteristic GTP concentration dependence of initial rates of c-di-GMP formation is indicative of substrate inhibition, which is observed for both light- and dark-state activities. Product formation was quantified in triplicate for several reaction times, and the SD of individual points contributed to the error estimation of the linear fit that is used to calculate the initial rate of product formation. The SE of the estimate from the linear regression is shown as an error bar for each GTP concentration. The inset shows representative traces of the high-performance liquid chromatography (HPLC) assay used for quantifying product formation. (D) Overall structure of IsPadC with individual domains in cartoon representation colored according to (A), with one protomer highlighted in pale colors. The biliverdin cofactor, in slate color, and its attachment site Cys17 from the NTE are shown as stick models.
Here, we describe the first full-length crystal structure of a bacteriophytochrome in its Pr state linked to its functional, enzymatic output, revealing a parallel dimeric arrangement of the sensor and effector domains. This structure provides a foundation for increasing our understanding of the fine-tuned coupling mechanism of phytochrome sensors with various effector domains. The characterization of in-solution conformational dynamics substantiates the involvement of previously proposed functionally relevant structural elements observed in phytochromes (10, 16, 17) and highlights their dynamic interplay with the coiled-coil sensor-effector linker region. Our results demonstrate how allosteric light regulation of enzymatic effectors is fine-tuned by the architecture and composition of the coiled-coil linker and by the central helical spine of phytochromes without direct interaction of the sensory module. We anticipate that consideration of molecular principles of sensor-effector coupling, going beyond the length of the characteristic linker, will open up new directions for the design of novel red light–regulated optogenetic tools.
RESULTS
Structure of a full-length red light–activated DGC
We characterized three naturally occurring PadC homologs with different linker length between their PHY and DGC domains. All constructs feature a characteristic bacteriophytochrome photocycle (Fig. 1B, Table 1, and fig. S2, A and B) and light-regulated conversion of two molecules of guanosine triphosphate (GTP) to c-di-GMP (Fig. 1C and fig. S2, C and D). Because PadC from Idiomarina sp. A28L (IsPadC) showed the highest dynamic range of c-di-GMP formation, we focused on this construct for a more detailed biophysical characterization.
Table 1. Comparison of PadC kinetics of substrate conversion and dark-state recoveries.
| Dark-state recovery at 700 nm* | Comparison of initial rates† at 200 μM GTP (μmol product min−1 μmol−1 enzyme2) | ||||||
| Construct | τ1 (s) | Relative A1 (%) | τ2 (s) | Relative A2 (%) | Dark state | Light state | Fold activation |
| IsPadC | 30 ± 1 | 46 ± 2 | 118 ± 5 | 54 ± 1 | 0.8 ± 0.1 | 34.2 ± 0.4 | 43× |
| MaPadC | 24.9 ± 0.4 | 50.0 ± 0.5 | 458 ± 56 | 50 ± 3 | 2 ± 0.2 | 15.4 ± 0.7 | 8× |
| TsPadC | 18.9 ± 0.2 | 80 ± 1 | 71 ± 3 | 20 ± 1 | 53 ± 6 | 42 ± 6 | −1.2× |
| IsPadC Δ514–516 | 13.5 ± 0.2 | 40.0 ± 0.3 | 308 ± 12 | 60 ± 1 | 11 ± 1 | 10 ± 2 | — |
| IsPadC Δ514–517 | 13.1 ± 0.7 | 11.5 ± 0.3 | 115 ± 1 | 89 ± 2 | 25 ± 7 | 49 ± 3 | 2× |
| IsPadC Δ514–520 | 11.9 ± 0.3 | 86 ± 1 | 393 ± 9 | 14.3 ± 0.1 | 27.2 ± 0.6 | 36 ± 4 | 1.3× |
| IsPadC Δ442–477::SG | n.d.‡ | n.d. | — | — | 48 ± 6 | 30 ± 1 | −1.6× |
| IsPadC PSMcc | 19 ± 1 | 23 ± 1 | 148 ± 2 | 77.0 ± 0.3 | — | — | — |
| IsPadC ccDGC | — | — | — | — | 0.2 ± 0.1 | −4× (to IsPadC) | |
*Dark-state recoveries of the constructs were fit to a second-order exponential decay. After red light illumination of 1 min, changes in absorption at 700 nm were followed over 5 min, with automatic sampling every 5 s and an integration time of 0.01 s. The contribution of each phase in the dark recovery process is represented as relative amplitude. The SE of the estimate from the nonlinear curve fit corresponding to y = A1*exp(−x/τ1) + A2*exp(−x/τ2) + y0 was used as error indicator.
†Comparison of product formation between the various constructs was performed for initial reaction rates at 50 μM GTP. Initial rates are quantified from experimental triplicates for three time points, and the SD of individual points contributed to the error estimation of the linear fit that is used to calculate the initial rate of product formation. The SE of the estimate from the linear regression is used as error indicator.
‡The recovery of this construct could not be determined because it appears to be locked in the Pfr-enriched state after red light illumination and does not significantly change its spectrum during a 24-hour incubation at room temperature in the dark.
We crystallized full-length IsPadC under dark conditions and also obtained a GTP-bound complex from soaking experiments. In addition, a truncated version of IsPadC corresponding to the photosensory module with the coiled-coil linker (PSMcc) was crystallized. The structure of full-length IsPadC was determined from selenium single-wavelength anomalous dispersion data to 3.0 Å resolution (table S1). Two molecules of IsPadC are present in the asymmetric unit, revealing a parallel dimer architecture throughout the protein with a central twofold symmetry axis that is perturbed by a subtle kink of the GGDEF dimer (Fig. 1D). Almost identical parallel phytochrome dimers are observed in both dimeric species present in the higher-resolution PSMcc structure (fig. S3). Notably, the N-terminal extension, providing the covalent attachment site for the biliverdin cofactor and extending from the characteristic figure-of-eight knot (18), interacts with parts of the hairpin/tongue region that folds back from the PHY domain to the cofactor attachment site in the GAF domain, similar to observations in Cph1 (19). The β-hairpin character of the tongue region and the conformation of the biliverdin cofactor including functionally important surrounding residues (20) (fig. S3B) show that IsPadC has been crystallized in its dark-adapted Pr state (fig. S4). The overall PSM monomer closely resembles that of previously published phytochrome structures [reviewed by Anders and Essen (7)], but the PSM dimer features a tighter association of the PHY domains because of their direct extension of the terminal PHY-α5 helices with the coiled-coil linker element. Largely polar and charged residues line the dimer interface along the central helical spine, connecting the GAF and PHY domains, as well as α5 of PHY. Starting from residue Leu501, a characteristic coiled-coil signature is observed (fig. S1), which contains 3.5 heptad repeats that are directly extended by conserved residues of the characteristic turn preceding α0 of GGDEF domains. The dimeric arrangement of the DGCs positions the substrate binding half active sites at the interface of the GGDEF domains, which closely resembles the overall architecture of a GTPαS-bound DGC dimer reported previously (21). Individual GGDEF domains superpose very well, and also the dimeric arrangement that is essential for positioning GTP for the condensation reaction is similar (fig. S5A). In line with this observation, the GTP-soaked IsPadC structure shows only moderate changes that are essentially restricted to a more pronounced bending of the coiled-coil linker coupled to a subtle dimer rearrangement of the GGDEF domains. A more detailed description and comparison of structural features of the three crystal structures is presented in the Supplementary Materials. Considering the additional interdomain contacts formed upon GTP binding in the dark-state IsPadC dimer (fig. S5B), this provides a structural rationale for the characteristic substrate inhibition observed for phytochrome-linked DGCs (Fig. 1C and fig. S2, C and D). Substrate inhibition might serve as an additional regulation mechanism for c-di-GMP synthesis, the central bacterial second messenger involved in lifestyle transitions (6), because GTP levels strongly correlate with growth conditions. However, the in vivo relevance of this observation remains to be shown.
Conformational dynamics of red light signal transduction
Because the mode of DGC regulation observed in IsPadC extends current concepts of activation by dimerization or product inhibition by domain immobilization (6, 15) and rather resembles recent observation for a constitutively dimeric, Zn-regulated GGDEF domain (21), we set out to characterize the in-solution conformational dynamics of dark-state and light-activated conformations using hydrogen-deuterium exchange coupled to mass spectrometry (HDX-MS). This method provides information on the conformational dynamics in various functionally relevant states (22) and hence has proven powerful in addressing signaling mechanisms in light-activated proteins (23). We performed HDX-MS of IsPadC under dark and constant red light conditions and, to address the functional role of the coiled-coil linker, compared the results to experiments with the PSMcc construct (figs. S6 and S7). Figure 2 illustrates the overall effect of illumination on IsPadC and highlights the importance of the central helical spine of the phytochrome dimer that interacts with the coiled-coil sensor-effector linker. Only slowly exchanging amides are affected by illumination in these regions, indicating that light induces no change in α-helical structure but rather increases the conformational dynamics. Closer inspection of the region surrounding the biliverdin cofactor identifies changes in conformational dynamics of structural elements that include critical residues shown to be affected by the 15Z to 15E isomerization of the biliverdin cofactor (Fig. 2B) (16, 20). Structural rearrangements of residues surrounding the conserved DIP motif (including Asp199) eventually enable the structural reorganization of the tongue region (24). On the basis of the HDX data, the tongue region of IsPadC constitutes a moderately dynamic system that, upon illumination, increases its conformational entropy, as indicated by its rapid amide hydrogen exchange kinetics. This contrasts with observations of stable helical structures in bathy phytochromes (16) and the long-lived Pfr state of Deinococcus radiodurans phytochrome (20, 24). However, considering that the linker region shows an increase in conformational dynamics upon illumination, the loss of a stable Pr-state tongue conformation enables the structural plasticity of the PHY domains and the central helical spine to perturb the coiled-coil region (Fig. 2C and movies S1 and S2). A closer inspection of the coiled-coil composition indicates that two different helical registers can be populated by the linker element (Fig. 3). Register 1, as observed in the dark-state crystal structure, differs from register 2 only by a subtle rearrangement within the heptad units (Fig. 3, C and D). Such a rotation places the strictly conserved Asn residues (Asn512 and Asn519; fig. S1) in the central a position of the heptad repeats and could thereby explain their functional relevance. A similar hydrogen bond–stabilized coiled-coil architecture is also observed in the prototypic GCN4 leucine zipper, and strikingly, an artificial superactive GCN4-GGDEF fusion (14) features the highly conserved DXLT motif of GGDEF domains, positioned directly after the coiled-coil, in exactly the same relative orientation to the heptad architecture proposed for register 2 of IsPadC (Fig. 3, D and F). To test the involvement of coiled-coil rearrangements in regulating DGC activity, we generated variants of IsPadC that are stabilized in either register 1 or register 2 by substituting the corresponding destabilizing residues (boxed residues in Fig. 3, C and D) by either leucine or valine residues, respectively. In vivo screening of c-di-GMP production revealed that the IsPadC D504L A518L variant (stabilized in register 1) can no longer be activated by red light illumination (Fig. 3E). This contrasts with observations of the variant stabilized in register 2 (IsPadC S505V A526V) that is active under both dark and red light conditions. Apparently, regulation of DGC activity is enabled by a fine-tuned balancing of the relative population of the two registers, which directly influences the positioning of functionally important residues of the GGDEF domain. In wild-type IsPadC and under dark conditions, the transition between the two registers is inhibited by the stable tongue conformation that restricts the dynamics of the central helical spine; but upon illumination, this inhibitory mechanism is released and DGC activity is increased by a more frequent population of the rotated “active” register. This conclusion is supported by an IsPadC variant deleted in the tongue region that features a substantially increased dark-state activity (Table 1 and fig. S8). Interestingly, illumination of this construct still modulates the enzymatic activity and results in a reduction of DGC activity, indicating that a complex interplay of the conformational dynamics of the tongue region and the central helical spine is required for the regulation of effector domains. In this respect, it should be noted that the lack of hydrophobic packing for significant parts of the PAS-GAF dimer interface and the central helical spine is responsible for the various dimeric arrangements of PSM structures reported (7). The characteristic structural rearrangements of the dimer interface from a pivot point localized to the central helical spine indicate that the plasticity of this region (Fig. 4) plays an important functional role in providing the conformational flexibility to adapt to different output modules with potentially different regulatory mechanisms (4, 5, 20).
Fig. 2. Light-induced changes in conformational dynamics of full-length IsPadC observed by HDX-MS.
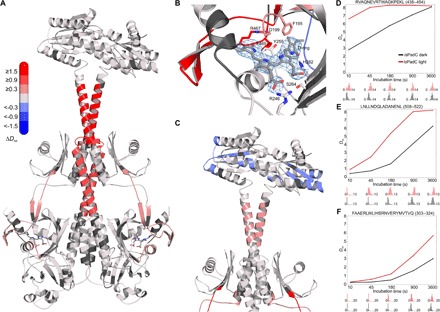
(A) IsPadC structure colored according to changes in relative deuterium incorporation (ΔDrel) between light-state and dark-adapted IsPadC after 15 min of deuteration (Drel of IsPadClight − Drel of IsPadCdark). The scale bar in the top left corner indicates the changes in ΔDrel, with blue corresponding to reduced deuterium incorporation and red reflecting increased exchange of amide protons upon red light illumination. The biliverdin cofactor is shown as gray stick model. (B) Close-up view of the biliverdin-binding pocket and the PHY tongue region. The 2Fo – Fc electron density map contoured at 1σ around the cofactor is shown as a light blue mesh. Important residues are shown as stick models, and the coloration corresponds to ΔDrel of the 45-s exchange time point. Residues 200 to 207 were removed for clarity. However, the coloration of residues Phe195 and Asp199 reflect the changes in Drel of the entire region 191 to 207. This region shows a higher deuterium incorporation of peptides at the interface of the PHY tongue and the biliverdin D-ring in the light state, highlighting the importance of the conformational dynamics of this region upon isomerization of the biliverdin cofactor. (C) Close-up view of the coiled-coil linker and the DGC domain colored according to ΔDrel after 10 s of deuteration. Several structural elements of the GGDEF domain, including substrate binding elements, show a reduction in conformational dynamics upon illumination. (D to F) Deuterium uptake curves of selected IsPadC peptides, with Drel plotted against the deuteration time for light- and dark-state HDX-MS experiments. The lower parts show software-estimated abundance distributions of individual deuterated species on a scale from undeuterated to all exchangeable amides deuterated. Deuteration plots for a peptide corresponding to the PHY tongue element (D), the PSM-DGC linker (E), and the internal helical spine linking the GAF and PHY domains (F). Drel values are shown as the mean of three independent measurements, and error bars correspond to the SD. An overview of all analyzed peptides is provided in figs. S6 and S7.
Fig. 3. Coiled-coil architecture of the sensor-effector linker.
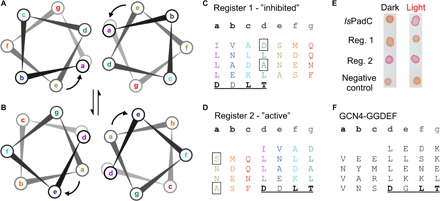
(A) Heptad register observed in the crystal structure of IsPadC (register 1). (B) A rotation of heptad positions e to a within the coiled-coil populates the “register 2” architecture. (C and D) Heptad units of the sensor-effector linker in registers 1 and 2, respectively, rainbow-colored according to the heptad repeats of register 1. Coiled-coil destabilizing residues are boxed. The highly conserved DXLT motif of GGDEF domains is underlined. (E) The assignment of active and inhibited states to registers 2 and 1, respectively, is confirmed by the observed DGC activity in a cell-based screening system. Wild-type IsPadC shows the expected increase in DGC activity upon red light illumination, as seen by the red coloration of the cells. In contrast, an IsPadC variant stabilizing register 1 can no longer be activated upon illumination, whereas the register 2 stabilizing variant is constitutively active. (F) For comparison, the heptad units of a superactive, artificial GCN4-GGDEF fusion (14) are also shown.
Fig. 4. Structural plasticity of phytochrome dimerization.
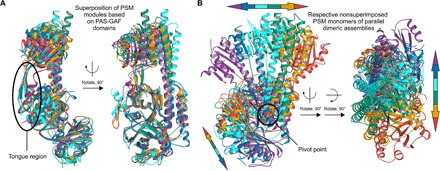
(A) Superposition of PSM modules from different published phytochrome structures featuring a parallel dimeric assembly to IsPadC (cyan) revealed almost identical arrangements of the PAS-GAF bidomain [root mean square deviation (RMSD) of 1.5, 1.2, 1.0, 0.9, and 0.9 Å for Protein Data Bank (PDB) 4OUR_B (blue) (60), 3G6O_A (violet) (16), 4Q0J_A (orange) (61), 5AKP_B (green) (26), and 5C5K_B (red) (20), respectively]. Irrespective of the β-hairpin or α-helical character of the tongue region, the PHY domains cluster in similar positions relative to the PAS-GAF domains. However, a characteristic flexibility of the central helix connecting the GAF and PHY domains is apparent, which ultimately results in altered positioning of the terminal PHY domain helices that link to the respective output modules. (B) The structural plasticity of the central helical spine and, consequently, the overall parallel dimeric phytochrome assembly are even more pronounced when comparing the nonsuperposed monomers. In this case, monomers of PSM modules have been aligned to the PAS-GAF-PHY monomer of IsPadC chain A, and the respective other monomers are displayed (RMSD of 3.2, 2.3, 2.2, 2.2, and 1.2 Å for PDB 3G6O_A, 4Q0J_A, 5AKP_B, 5C5K_B, and 4OUR_B, respectively). Again, no clustering of the PHY domain orientation with respect to the Pr- or Pfr-state character of the tongue can be observed. However, the structural differences among various phytochrome dimers are nonrandom and occur along a specific trajectory that corresponds to rotation at the dimer interface. The two extremes of this rotation correspond to structures obtained for Pfr-state crystals (violet and red). In contrast, the only parallel phytochrome structures with adjacent C-terminal domains (green and cyan) cluster in the middle of the overall trajectory. Although this suggests that the more pronounced dimer rotation of other PSM assemblies might be due to missing interactions of their output modules and linker regions, the characteristic structural transition reflecting the plasticity of the PHY domain dimerization is very likely functionally relevant for phytochromes in general.
Importance of the sensor-effector linker length and composition
The conformational dynamics of the central helical spine and the coiled-coil linker are finely tuned to achieve their function in the full-length protein. In the PSMcc construct, HDX-MS analyses revealed significantly increased dynamics of the central helical regions, and hence, the composition of both the helical spine and the linker appears to have evolved to stabilize the weak dimer interfaces (PAS-GAF, PHY, and GGDEF) at their ends in the dark state (movies S3 and S4). On the basis of the characterization of truncation variants within the heptad repeats (Table 1 and fig. S8), it is apparent that not only the linker length but also the composition plays an important role. The IsPadC Δ514–517 and Δ514–520 deletions should allow almost identical arrangements of the effector; however, their reduced coiled-coil character and the impact of the deletions on the relative stabilities of the two coiled-coil registers observed for wild-type IsPadC (Fig. 3) prevent the dark-state inhibition observed for the wild-type construct. Therefore, both truncations have a reduced dynamic range of light-regulated c-di-GMP formation. The additionally characterized IsPadC Δ514–516 construct that apparently results in a change of the relative positioning of sensor and effector preferentially produces the linear pppGpG intermediate observed in c-di-GMP biosynthesis (25). The ccDGC construct alone (compare Fig. 1) also catalyzes intermediate formation with lower efficiency (Table 1), suggesting that the native coiled-coil linker sequence alone is not sufficient to properly stabilize the GGDEF domains in the productive coiled-coil register. The benefit of functional regulation and prevention of futile GTP degradation has apparently led to the evolutionary generation of sensor-GGDEF couples with strong conservation of defined linker length. The central position of GTP in primary metabolism and the essential regulation of the open half active sites of GGDEF domains might be reasons for this systematic conservation in sensory GGDEF systems (12, 13). PadC from Thioalkalivibrio sp. ALMg3 (TsPadC) features an inversion of the signal, resulting in subtle light-inhibited DGC activity with an identical linker length to the IsPadC Δ514–520 variant but with different composition. Similarly, MaPadC, having a seven-residue-longer linker with a charged heptad insertion, shows changes in the kinetic profile of DGC activity with less pronounced substrate inhibition. Apparently, the linker elements have been fine-tuned for their requirements not only in length but also in their amino acid composition, which likely coevolved together with other functionally relevant structural elements.
Signal transduction requires the conformational coupling of multiple structural elements
In the context of phytochrome signaling, our HDX-MS data substantiate the involvement of previously proposed functionally relevant structural elements observed in phytochromes (10, 16, 17) and support the hypothesis that observed differences in the PSM dimer architectures, and the plasticity of the central PHY domain dimerization (Fig. 4), are functionally relevant. Whether the overall assembly is stabilized by an α-helical or a β-hairpin conformation of the tongue is irrelevant for the proposed model of functional phytochrome regulation (Fig. 5A) and rather reflects the evolutionary adaptation to respond to different light qualities or to modulate the dynamic range of various output modules. This is supported by the observation of increased dynamics in the Pr state of bathy phytochromes upon far-red light illumination (26), which resembles observations upon red light irradiation for all three PadC constructs characterized in this study (fig. S9), irrespective of whether they are light-activated or light-inhibited. In addition, solution-scattering studies with full-length IsPadC show an increased conformational flexibility between the PHY and GGDEF domains relative to the crystal structure (Fig. 5B and fig. S10). Light activation induces no pronounced, immediate structural rearrangements but a slow and reversible oligomerization process with a rate constant of 0.2 min−1 derived from the time-dependent increase of the small-angle x-ray scattering (SAXS)–derived molecular mass (fig. S10C). Only moderate light-induced structural changes have also recently been assigned to full-length D. radiodurans phytochrome (27). Therefore, the appreciation of a fine-tuned allosteric regulation mechanism following the violin model (Fig. 5) (28) will be important to appreciate general aspects of phytochrome signaling.
Fig. 5. Schematic model of signal integration pathways in phytochrome-linked enzymatic effectors and in-solution structure of IsPadC.
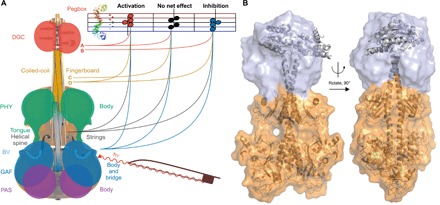
(A) The characteristic structure of IsPadC and regulatory properties of closely related homologs suggest a model of signal transduction corresponding to the violin model (28). Instead of a linear cascade of structural changes resulting in enzyme activation, the conformational dynamics of the whole system define the population of functionally relevant states, leading to either activation or inactivation with similar overall architectures (37). In the case of the phytochrome-violin, the pegbox corresponds to the effector domain, whose activity is tuned by the sensory module. Hitting the right chords on the enzymatic activity clef for stimulating GTP turnover is more complex than a specific actuation of the fingerboard, which would correspond to, for example, variation of its length (yellow lines and linkers C and D), and additionally depends on properties of the strings (gray), the shape of the violin body (blue), and the effector-pegbox (red; for example, two different DGC constructs A and B). The characteristic structural changes observed for various phytochrome structures (Fig. 4), which had also been interpreted as specific light-induced rearrangements (20, 24), reflect the structural plasticity of phytochrome dimerization. The latter, in turn, enables the modulation of the conformational dynamics of the overall system and thereby allows complex, evolutionary fine-tuning of the body of phytochrome-violin to optimize the output functionality as required by each system. The absence of characteristic structural changes, such as a defined rotation or a separation of the coiled-coil linker, enables the realization of systems featuring activation or inactivation within the same molecular architecture. (B) SAXS-based structural model for the IsPadC dimer in its dark-adapted Pr state. The surface represents the conformational space sampled by the GGDEF domains in the seven best structures according to the fit between the experimental and back-calculated SAXS data (compare fig. S10). Structures are aligned to the PAS-GAF-PHY domains.
DISCUSSION
The consideration of a complex interplay of multiple functionally relevant structural regions helps in generalizing and understanding previously made mechanistic proposals for long-range signal transduction in phytochromes. The importance of the tongue element and the central helical spine has been identified early on during the characterization of truncated phytochrome variants (10, 16, 17, 29, 30). However, in the absence of the corresponding output modules, the structural changes induced by illumination might not always be representative of the full-length proteins, as observed recently for D. radiodurans phytochrome (24, 27). For dimeric output modules, such as histidine kinases or DGCs, the functional coupling with the coiled-coil sensor-effector linker and the extended dimerization interface has important regulatory consequences for long-range signal transduction. In this respect, the similarities in overall architecture, for example, to sensors of blue light linked to histidine kinases (31) or adenylyl cyclases (32) raise the question of common principles in sensor-effector coupling. Although this is one of the central questions in understanding the astonishing modularity of naturally evolved sensor-effector systems, the divergence of postulated mechanisms even within individual sensory modules currently prevents a unifying answer. However, appreciation of a regulatory mechanism in line with the violin model (28) allows the conclusion that multiple output mechanisms can be realized within the same overall architecture. For example, removal of the tongue region in IsPadC still allows regulation of the effector module via the central helical spine, a signaling route used by evolutionary-related cyanobacteriochrome systems (33). Along this line, even antiparallel dimers or phytochrome monomers might be functionally relevant in different systems; for example, the binding of monomeric Rhodopseudomonas palustris phytochrome 1 to PpsR dimers, in analogy to blue light photoreceptors, shows interesting parallels to molecular principles identified for the AppA-PpsR system (34, 35). Consequently, even molecular details of long-range signal transduction in parallel dimeric phytochrome systems might not be conserved in every detail. Although a precise register switching and a characteristic length of the coiled-coil appear important for the regulation of GGDEF domains, the requirements for functional regulation of other enzymatic functionalities might feature different molecular characteristics such as regulated unfolding (36) or large-scale rotational mechanisms (20, 27).
As far as phytochrome engineering for direct allosteric regulation of effector domains is concerned, our results show that the natural selection of linker length does not necessarily correlate with a high dynamic range that would be desirable for optogenetic tools. Therefore, an improved understanding of the effect of linker length and composition variations on phytochrome signaling and how this eventually translates into molecular mechanisms of regulation for various effector domains will be required to enable a more rational design approach. Especially the observation of signal inversion for closely related systems suggests that an ensemble of differently populated functionally relevant states with differing energies needs to be considered (37). In this respect, the observation of two helical registers for phytochrome-linked DGCs and the modulation of their relative population upon activation provide an interesting perspective for interpreting successful and unsuccessful rationally designed sensor-effector fusions. Similar regulatory effects of transitions between different coiled-coil registers have been observed for mediating cargo binding to dynein by BicD (38) as well as modulating interactions of the cell cycle regulator Nek2 (39). In the context of long-range signaling, another interesting parallel is the rotary mechanism proposed for transmembrane signaling and its integration by HAMP domains (40). However, considering the dynamic properties of these coiled-coil structures, it will be interesting to see whether the major contribution to regulation is the rotary forces induced by the register switch or rather the conformational dynamics during the continuous transition between energetically similar coiled-coil registers. The latter mechanism has recently been shown to be critical for functional protein-protein interactions in the case of streptococcal M proteins (41). In the context of phytochrome-regulated DGCs, the involvement of energetically similar coiled-coil registers could also provide a rationale for the evolutionary adaptation of the dynamic range of light activation by altering the coiled-coil composition as well as by modifying structural elements that are functionally coupled to the sensor-effector linker.
In summary, we show that phytochrome regulation depends on the molecular characteristics of the central helical spine, the tongue region, and the coiled-coil linker region. We provide initial insights into the relative contributions of various sensor-effector elements to the energetic coupling of individual domains (42) that will eventually allow a better understanding of molecular mechanisms involved in phytochrome signaling. The appreciation of dynamically driven allostery (43) and the possibility to both activate and inhibit enzymatic activity in similar systems allow the conclusion that the rational design of sensor-effector couples needs to consider functional aspects of both sensor and effector domains that are inherently linked to their conformational dynamics.
MATERIALS AND METHODS
Protein expression and purification
The coding sequences of PadC homologs from Idiomarina sp. A28L, Marinimicrobium agarilyticum, and Thioalkalivibrio sp. ALMg3 (sequence accession numbers WP_007419415, WP_027329460, and WP_026331574, respectively) corresponding to IsPadC, MaPadC, and TsPadC were synthesized with codon optimization for expression in Escherichia coli (GeneArt, Life Technologies). Gene cassettes were cloned into the pETM-11 vector by restriction ligation using the Nco I and Not I sites for expression with a tobacco etch virus (TEV)–cleavable polyhistidine tag. Truncation variants of IsPadC were produced by site-directed mutagenesis following the protocol described by Liu and Naismith (44) (primers used in this work are listed in table S2A).
PadC variants were produced as (His)6-tagged holoproteins in complex with the biliverdin chromophore in E. coli BL21 (DE3) by coexpression of heme oxygenase (HO-1) from Synechocystis sp. PCC6803. For this purpose, we replaced the kanamycin cassette of pT7-ho1 (9) with the chloramphenicol resistance marker of pACYC184. Cells grown to mid-log phase at 37°C in LB medium supplemented with 0.3% glucose and 8.3 mM MgCl2 were shifted to 16°C for induction with 0.25 mM isopropyl-β-d-thiogalactopyranoside (IPTG) for ~15 hours in the presence of δ-aminolevulinic acid (10 mg liter−1). After harvesting, cells were disrupted in lysis buffer (compare table S2B) by combining lysozyme (100 μg ml−1) and sonication (2 × 5 min, 100 W, continuous mode; Labsonic L, Sartorius). Lysates were clarified by ultracentrifugation (206,000g) and affinity-purified on a Ni2+-Sepharose matrix (Ni Sepharose 6 Fast Flow, GE Healthcare) by gravity flow in a Protino column (Macherey-Nagel). After sample loading, proteins were washed with lysis buffer supplemented with 40 mM imidazole and finally eluted with 250 mM imidazole containing lysis buffer. Eluted fractions were then subjected to overnight dialysis (table S2B) in the presence of TEV protease at a ratio of ~1:16 for TEV/substrate. The histidine-tagged TEV protease and the cleaved histidine tag were removed by reloading the dialyzed sample onto the Ni2+ column and collecting the flow through. After concentration (Amicon Ultra-15, Merck Millipore), PadC samples were purified by size-exclusion chromatography on a 16/60 Superdex 200 prep grade column (GE Healthcare) with buffers according to table S2B. The purified proteins were concentrated, aliquoted, flash-frozen in liquid nitrogen, and conserved at −80°C until needed.
Selenomethionine-substituted IsPadC was expressed in E. coli BL21 (DE3) coexpressing heme oxygenase using modified minimal growth medium containing increased concentrations of certain amino acids for metabolic inhibition of methionine biosynthesis (45) and purified following the procedure described above.
The IsPadC ccDGC construct was initially purified in its c-di-GMP–bound form. To obtain product-free protein preparations, samples were incubated with the phosphodiesterase RocR and repurified by a third Ni column step to remove His-tagged RocR and by gel filtration to remove the pGpG product from the phosphodiesterase reaction. Purification of RocR was performed as previously described by Rao et al. (46) using the affinity purification materials described above.
Cell-based DGC assay
DGC activity was screened by adaptation of a previously described protocol (47). E. coli BL21 (DE3) cells containing the pT7-ho1 helper plasmid and transformed with pETM-11–based PadC expression plasmids were grown at 30°C in YESCA medium [casamino acids (10 mg ml−1) and yeast extract (1.5 mg ml−1)] supplemented with MgSO4 (0.05 mg ml−1), FeSO4 (0.5 μg ml−1), kanamycin (30 μg ml−1), and chloramphenicol (34 μg ml−1) to an absorbance of 0.5 at 600 nm. Each culture was then induced with 0.25 mM IPTG and δ-aminolevulinic acid (10 mg liter−1) for 4 hours at 16°C. Afterward, 3 μl of the induced culture was spotted on YESCA agar plates containing kanamycin (30 μg ml−1), chloramphenicol (34 μg ml−1), and Congo red (0.01 mg ml−1) and incubated at 20°C for 16 hours in the dark or under constant red light illumination (75 μW cm−2 at 630 nm). As a negative control, we used a pETM-11 AppA construct (35) that does not show any DGC activity.
Spectroscopic characterization
Ultraviolet (UV)–visible absorption spectra were acquired with a Specord 200 Plus spectrophotometer (Analytik Jena) using protein samples diluted in their corresponding storage buffers (compare table S2B). Dark-adapted Pr-state absorption spectra were measured from samples equilibrated under nonactinic conditions and minimizing the contact time with measuring light. Pfr-enriched spectra were recorded immediately after red light irradiation [1.8 mW cm−2 at 630 nm; Luminea light-emitting diode (LED)] using a charge-coupled device (CCD)–based Flame spectrometer (Ocean Optics) with low-intensity measuring light (mercury-xenon lamp). All spectra and time course experiments were measured at room temperature.
Pr-state recovery kinetics were followed at 700 and 750 nm after 1 min of red light illumination. Integration times and time intervals of the time scans were adjusted for the individual variants to minimize the effect of the actinic measuring lights. Reported time constants of dark-state recovery differ by less than 10% between 700- and 750-nm recordings.
Crystallization and structure elucidation
Dark-adapted IsPadC was crystallized at 289 K using a hanging-drop vapor diffusion setup. Drops (2 μl) containing equal volumes of protein solution at 7 mg/ml and reservoir solution [0.1 M bis-tris (pH 5.5), 0.1 M ammonium acetate, 17% (w/v) polyethylene glycol 10,000] were equilibrated against 500 μl of reservoir solution. Plate-like elongated crystals appeared after overnight incubation under dark conditions and reached final dimensions within 5 days. Pronounced drop-to-drop variability in crystallization behavior was observed, and some initially clear drops produced differently shaped crystals during prolonged incubation (~1 month). Structure elucidation revealed that these crystals correspond to a proteolytic degradation product corresponding to the IsPadC PSMcc fragment. Crystals were harvested from 2-μl drops equilibrated against 35 μl of reservoir solution in a sitting-drop vapor diffusion setup as described for full-length IsPadC.
Crystals of selenomethionine-labeled IsPadC were obtained following the same protocol as for native IsPadC in the presence of 1 mM dithioerythritol and were collected after 15 days of crystal growth. Crystals were harvested under low-intensity green light conditions (520 ± 20 nm LED) by transferring to a cryoprotectant solution (reservoir solution containing 25% glycerol) and subsequent flash-freezing in liquid nitrogen. GTP soaks of full-length IsPadC crystals were performed by a 5-min incubation in the cryoprotectant solution supplemented with 10 mM GTP (Roth) under nonactinic light conditions. Diffraction data for native and Se-Met IsPadC were collected at beamlines ID29-10 and ID23-1, respectively, of the European Synchrotron Radiation Facility (ESRF). Data were processed using the XDS program package (48), and Se-Met data sets collected at multiple positions were merged for successful phasing by selenium single-wavelength anomalous dispersion (table S1).
The crystal structure of IsPadC was solved by a combination of molecular replacement and single-wavelength anomalous dispersion phasing. Because of the moderate quality of the initial anomalous electron density map obtained from the 3 Å Se-Met data set with PHENIX AutoSol (49), individual search models of the PAS-GAF, PHY, and GGDEF domains were generated using PHENIX Sculptor (50) on the basis of the IsPadC sequence and the structures of the PAS-GAF module from R. palustris bacteriophytochrome RpA3015 (PDB 4S21) (30), the structure of the PHY domain of bacteriophytochrome RpBphP3 from R. palustris (PDB 4R70) (30), and the structure of the GGDEF domain of WpsR from Pseudomonas aeruginosa (PDB 3I5B) (14), respectively. Two molecules of each search model were successfully placed in the asymmetric unit using PHENIX Phaser (51). The initial model of a PAS-GAF-PHY-GGDEF dimer was subsequently used for rigid-body fitting in the anomalous density map and refined in several rounds of maximum likelihood least-squares refinement of models modified with Coot using σA-weighted 2mFo − DFc and Fo − Fc electron density maps. Refinement included experimental phase information using a phased maximum likelihood target (MLHL). In addition, torsion noncrystallographic symmetry (NCS) restraints and secondary structure restraints were included together with Translation/Libration/Screw (TLS) groups for the individual domains of the protein. During the final rounds of refinement, reference model restraints from the higher-resolution PSMcc structure were included.
The crystal structure of IsPadC PSMcc fragment was solved by molecular replacement using PHENIX Phaser with an intermediate model of the PAS-GAF-PHY fragment from the full-length structure of IsPadC as search model. Refinement included an initial simulated annealing (torsion) step followed by several rounds of maximum likelihood least-squares refinement of models modified with Coot, as described above. TLS and secondary structure restraints were applied during the refinement.
The crystal structure of IsPadC soaked with GTP was solved by molecular replacement using PHENIX Phaser with models of the dimeric IsPadC PSM containing the coiled-coil linker and the dimer of the IsPadC DGC. Refinement included an initial rigid-body fit of individual domains because the linker element revealed a more pronounced kink in the soak crystal, followed by several rounds of maximum likelihood least-squares refinement of models modified with Coot, as described above. TLS, NCS, and secondary restraints were applied during the refinement.
Kinetic characterization of PadC constructs
To monitor the conversion of GTP to c-di-GMP, we used an HPLC-based assay adapted from Enomoto et al. (11). Briefly, appropriate amounts of enzyme were incubated with various GTP concentrations at 20°C in reaction buffer containing 50 mM Hepes (pH 7.0), 150 mM NaCl, and 50 mM MgCl2. Reactions were stopped at different time points by heat denaturation at 95°C for 1 min. Samples were cleared by centrifugation, and the supernatant was subjected to HPLC analysis using a reversed-phase column (SunFire C18 4.6 mm × 100 mm, Waters) equilibrated in 10 mM triethylammonium formate (pH 6.0). Substrate and product were separated using a linear, 23-min gradient from 2 to 20% MeOH at a flow rate of 1 ml min−1. To remove inhibitory components from the GTP stock (Roth), it was initially purified by HPLC using a similar protocol. GTP (25 mg) was dissolved in 10 mM Hepes (pH 7.0) and purified in several runs over a preparative HPLC column (SunFire C18 10 mm × 100 mm, Waters) equilibrated in 10 mM triethylammonium formate (pH 6.0) using a MeOH gradient from 10 to 100%. Purified GTP fractions were then lyophilized and resuspended in 10 mM Hepes (pH 7.0) with final adjustment to a pH between 7 and 8. For the following red light–activated reactions, samples were preilluminated (0.7 mW cm−2 at 630 nm, Luminea LED) for 1 min before starting the reaction and maintained under constant illumination during the course of the reaction. For quantification of c-di-GMP formation, all samples were corrected for the amount of product formed during the heat inactivation step. The identity of the c-di-GMP peak was confirmed by MS, and the linear intermediate was identified by collecting representative fractions and verifying subsequent conversion to c-di-GMP by wild-type IsPadC. All activities were normalized to the dimer concentrations of the respective constructs.
Hydrogen-deuterium exchange coupled to mass spectrometry
Deuterium-labeling experiments were performed to address the effect of illumination on the conformational dynamics of full-length IsPadC and functional fragments thereof. Proteins were diluted to a final concentration of 200 μM in their storage buffers (compare table S2B) under nonactinic light conditions. Aliquots (2.5 μl) were preequilibrated at 20°C for 1 min under dark conditions or with red light illumination (0.7 mW cm−2 at 630 nm, Luminea LED), and the deuterium-labeling reaction started by 20-fold dilution in D2O containing 10 mM Hepes (pD 7.0), 150 mM NaCl, and 10 mM MgCl2. Aliquots (5 μl) were removed after 10 s, 45 s, 3 min, 15 min, and 60 min and quenched with 50 μl of ice-cold 200 mM ammonium formic acid (pH 2.5). The quenched labeling reaction (50 μl) was injected into a cooled HPLC setup, as described previously (52). Briefly, deuterated samples were digested on an immobilized pepsin column (Poroszyme, Applied Biosystems) operated at 10°C. Resulting peptides were desalted on a 2-cm C18 guard column (DiscoveryBIO C18, Sigma) and separated during a 7-min acetonitrile gradient (15 to 50%) in the presence of 0.6% (v/v) formic acid on a reversed-phase column (XR ODS 75 mm × 3 mm, 2.2 μM; Shimadzu). Peptides were then infused into a maXis electrospray ionization ultrahigh-resolution time-of-flight (UHR-TOF) mass spectrometer (Bruker). Deuterium incorporation was analyzed and quantified using the Hexicon 2 software package (http://hx2.mpimf-heidelberg.mpg.de) (53).
Limited proteolysis
Trypsin digestion patterns of all three PadC homologs were obtained by following the proteolytic degradation over 60 min in the dark or with constant red light illumination (0.7 mW cm−2 at 630 nm, Luminea LED) at 20°C. A 1:100 (w/w) ratio of protease to enzyme was used for IsPadC, 1:750 (w/w) ratio for MaPadC, and 1:1000 (w/w) ratio for TsPadC with PadC concentrations varying between 6 and 8 μM. Tryptic digests were performed in reaction buffer containing 10 mM tris-HCl (pH 8.0), 500 mM NaCl, and 10 mM MgCl2. PadC (5 μg) was quenched per time point upon mixing with SDS sample buffer [4× stock: 10% (w/v) glycerol, 0.6% (w/v) tris-HCl (pH 6.8), 2% (w/v) SDS, 0.02% (w/v) bromophenol blue, 1.5% (w/v) dithiothreitol] and heating at 95°C for 5 min. All samples were loaded onto a 12% SDS–polyacrylamide gel for electrophoretic separation of proteolytic fragments.
Small-angle x-ray scattering
SAXS data for solutions of IsPadC in the light and dark states were recorded on an in-house SAXS instrument (SAXSess mc2, Anton Paar) equipped with a Kratky camera, a sealed x-ray tube source, and a two-dimensional Princeton Instruments PI•SCX:4300 (Roper Scientific) CCD detector. For dark- and light-state measurements, the protein solutions were either kept in the dark or exposed to red light (1.8 mW cm−2 at 630 nm, Luminea LED) for 90 min before the measurements. The scattering patterns were measured with a 180-min exposure time (1080 frames, 10 s each) for several solute concentrations ranging from 0.8 to 13.0 mg/ml (fig. S10). Radiation damage was excluded on the basis of a comparison of individual frames of the 180-min exposures, where no changes were detected. A range of momentum transfer of 0.012 Å−1 < s < 0.63 Å−1 was covered [s = 4π sin(θ)/λ, where 2θ is the scattering angle and λ = 1.542 Å is the x-ray wavelength].
All SAXS data were analyzed with the package ATSAS (version 2.5). The data were processed with the SAXSQuant software (version 3.9) and desmeared using the programs GNOM and GIFT (54, 55). The forward scattering I(0), the radius of gyration Rg, the maximum dimension Dmax, and the interatomic distance distribution functions [P(R)] were computed with the program GNOM (54). The masses of the solutes were evaluated by comparison of the forward scattering intensity with that of a human serum albumin reference solution (molecular mass, 69 kDa) and using Porod’s law. To generate ab initio shape models, a total of 50 models were calculated using the program DAMMIF (56) and aligned and averaged using the program DAMCLUST. C2 symmetry was defined. The ab initio shape models were aligned with the crystal structure of IsPadC using the program SUPCOMB (57). The structures of IsPadC were modeled using the program CORAL (58). Input was the crystal structure of IsPadC determined here and SAXS data. The orientation of the DGC domains was kept flexible during the calculations, and no dimerization interface was restrained to account for dynamics. A total of 50 structures were calculated, and the best structures based on the fit to the experimental data were selected to prepare the figures.
Supplementary Material
Acknowledgments
We thank M. Gomelsky, K. Gruber, and P. Macheroux for critical reading of the manuscript as well as stimulating discussions and E. Zenzmaier for technical support. We are grateful to I. Schlichting for providing access to the maXis UHR-TOF mass spectrometer and the research infrastructure at the Max Planck Institute for Medical Research in Heidelberg (Germany) to perform the HDX-MS measurements. We appreciate improvements to the Hexicon 2 software package by R. Lindner. We thank the ESRF staff of beamlines ID23-1, ID29, and ID30A-3 for excellent support and D. von Stetten for help with the microspectrophotometer setup (59). The helper plasmid pT7-ho1 for coexpression of heme oxygenase was provided by M. Gomelsky (University of Wyoming, Laramie) and is derived from a pCB1 vector generated by G. A. Gambetta and J. C. Lagarias (University of California, Davis). The expression vector for the phosphodiesterase RocR cloned into pET26 was provided by Z.-X. Liang (Nanyang Technological University, Singapore). Funding: The project has been initiated with a startup financing grant by the Graz University of Technology (F-AF5-648-01) to A.W. G.G. and S.E. are supported by the Austrian Science Fund through the PhD programme “DK Molecular Enzymology” (W901). C.G. gratefully acknowledges the postdoctoral fellowship program of the Helmholtz Zentrum München. T.M. is supported by the Bavarian Ministry of Sciences, Research and the Arts (Bavarian Molecular Biosystems Research Network), the Deutsche Forschungsgemeinschaft (Emmy Noether program MA 5703/1-1), and the Austrian Science Fund (FWF): P28854. The authors gratefully acknowledge support from NAWI Graz. A.W. acknowledges funding from the Austrian Science Fund (FWF): P27124. Author contributions: G.G. cloned, expressed, purified, and characterized PadC variants. G.G. also performed limited proteolysis experiments and crystallized IsPadC variants. G.G. and A.W. solved the crystal structures and performed HDX-MS experiments. S.E. conducted the kinetic characterization of all DGC constructs. Solution-scattering experiments were performed by C.G. and T.M. U.V. cloned, expressed, purified, and characterized the tongue deletion variant of IsPadC. The manuscript was written by G.G. and A.W. with all authors’ input. Competing interests: The authors declare that they have no competing interests. Data and materials availability: Atomic coordinates and structure factors for the reported crystal structure have been deposited to the Protein Data Bank under accession numbers 5LLW, 5LLX, and 5LLY. All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. Additional data related to this paper may be requested from A.W. (andreas.winkler@tugraz.at).
SUPPLEMENTARY MATERIALS
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/3/2/e1602498/DC1
Supplementary Results
fig. S1. Multiple sequence alignment of PadC homologs generated with Jalview (64).
fig. S2. Spectroscopic and kinetic characterization of TsPadC and MaPadC.
fig. S3. Characterization of the IsPadC PSMcc variant.
fig. S4. IsPadC crystal and spectral characteristics of dark-state crystallized IsPadC.
fig. S5. Effect of substrate binding on the overall architecture of IsPadC.
fig. S6. Summary of HDX experiments.
fig. S7. Individual deuterium incorporation plots of all evaluated peptides.
fig. S8. Spectroscopic and kinetic characterization of IsPadC deletion variants.
fig. S9. Time course of tryptic digests of IsPadC, MaPadC, and TsPadC under dark and light conditions.
fig. S10. Details of SAXS measurements.
table S1. Data collection, phasing, and refinement statistics.
table S2. Overview of oligonucleotides and buffers.
movie S1. Changes in conformational dynamics upon red light illumination of IsPadC.
movie S2. Changes in conformational dynamics upon red light illumination of the IsPadC PSMcc variant.
movie S3. The influence of effector deletion on conformational dynamics of the dark-state IsPadC PSMcc assembly.
movie S4. The influence of effector deletion on conformational dynamics of the light-state IsPadC PSMcc assembly.
REFERENCES AND NOTES
- 1.Tischer D., Weiner O. D., Illuminating cell signalling with optogenetic tools. Nat. Rev. Mol. Cell Biol. 15, 551–558 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pathak G. P., Vrana J. D., Tucker C. L., Optogenetic control of cell function using engineered photoreceptors. Biol. Cell 105, 59–72 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Piatkevich K. D., Subach F. V., Verkhusha V. V., Engineering of bacterial phytochromes for near-infrared imaging, sensing, and light-control in mammals. Chem. Soc. Rev. 42, 3441–3452 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ryu M.-H., Kang I.-H., Nelson M. D., Jensen T. M., Lyuksyutova A. I., Siltberg-Liberles J., Raizen D. M., Gomelsky M., Engineering adenylate cyclases regulated by near-infrared window light. Proc. Natl. Acad. Sci. U.S.A. 111, 10167–10172 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gasser C., Taiber S., Yeh C. M., Wittig C. H., Hegemann P., Ryu S., Wunder F., Möglich A., Engineering of a red-light–activated human cAMP/cGMP-specific phosphodiesterase. Proc. Natl. Acad. Sci. U.S.A. 111, 8803–8808 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Römling U., Galperin M. Y., Gomelsky M., Cyclic di-GMP: The first 25 years of a universal bacterial second messenger. Microbiol. Mol. Biol. Rev. 77, 1–52 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Anders K., Essen L.-O., The family of phytochrome-like photoreceptors: Diverse, complex and multi-colored, but very useful. Curr. Opin. Struct. Biol. 35, 7–16 (2015). [DOI] [PubMed] [Google Scholar]
- 8.Ryjenkov D. A., Tarutina M., Moskvin O. V., Gomelsky M., Cyclic diguanylate is a ubiquitous signaling molecule in bacteria: Insights into biochemistry of the GGDEF protein domain. J. Bacteriol. 187, 1792–1798 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tarutina M., Ryjenkov D. A., Gomelsky M., An unorthodox bacteriophytochrome from Rhodobacter sphaeroides involved in turnover of the second messenger c-di-GMP. J. Biol. Chem. 281, 34751–34758 (2006). [DOI] [PubMed] [Google Scholar]
- 10.Anders K., Daminelli-Widany G., Mroginski M. A., von Stetten D., Essen L.-O., Structure of the cyanobacterial phytochrome 2 photosensor implies a tryptophan switch for phytochrome signaling. J. Biol. Chem. 288, 35714–35725 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Enomoto G., Nomura R., Shimada T., Win N.-N., Narikawa R., Ikeuchi M., Cyanobacteriochrome SesA is a diguanylate cyclase that induces cell aggregation in Thermosynechococcus. J. Biol. Chem. 289, 24801–24809 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Glantz S. T., Carpenter E. J., Melkonian M., Gardner K. H., Boyden E. S., Ka-Shu Wong G., Chow B. Y., Functional and topological diversity of LOV domain photoreceptors. Proc. Natl. Acad. Sci. U.S.A. 113, E1442–E1451 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Möglich A., Ayers R. A., Moffat K., Structure and signaling mechanism of Per-ARNT-Sim domains. Structure 17, 1282–1294 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.De N., Navarro M. V. A. S., Raghavan R. V., Sondermann H., Determinants for the activation and autoinhibition of the diguanylate cyclase response regulator WspR. J. Mol. Biol. 393, 619–633 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schirmer T., Jenal U., Structural and mechanistic determinants of c-di-GMP signalling. Nat. Rev. Microbiol. 7, 724–735 (2009). [DOI] [PubMed] [Google Scholar]
- 16.Yang X., Kuk J., Moffat K., Conformational differences between the Pfr and Pr states in Pseudomonas aeruginosa bacteriophytochrome. Proc. Natl. Acad. Sci. U.S.A. 106, 15639–15644 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Burgie E. S., Vierstra R. D., Phytochromes: An atomic perspective on photoactivation and signaling. Plant Cell 26, 4568–4583 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wagner J. R., Brunzelle J. S., Forest K. T., Vierstra R. D., A light-sensing knot revealed by the structure of the chromophore-binding domain of phytochrome. Nature 438, 325–331 (2005). [DOI] [PubMed] [Google Scholar]
- 19.Essen L.-O., Mailliet J., Hughes J., The structure of a complete phytochrome sensory module in the Pr ground state. Proc. Natl. Acad. Sci. U.S.A. 105, 14709–14714 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Burgie E. S., Zhang J., Vierstra R. D., Crystal structure of Deinococcus phytochrome in the photoactivated state reveals a cascade of structural rearrangements during photoconversion. Structure 24, 448–457 (2016). [DOI] [PubMed] [Google Scholar]
- 21.Zähringer F., Lacanna E., Jenal U., Schirmer T., Boehm A., Structure and signaling mechanism of a zinc-sensory diguanylate cyclase. Structure 21, 1149–1157 (2013). [DOI] [PubMed] [Google Scholar]
- 22.Konermann L., Pan J., Liu Y.-H., Hydrogen exchange mass spectrometry for studying protein structure and dynamics. Chem. Soc. Rev. 40, 1224–1234 (2011). [DOI] [PubMed] [Google Scholar]
- 23.Lindner R., Heintz U., Winkler A., Applications of hydrogen deuterium exchange (HDX) for the characterization of conformational dynamics in light-activated photoreceptors. Front. Mol. Biosci. 2, 33 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Takala H., Björling A., Berntsson O., Lehtivuori H., Niebling S., Hoernke M., Kosheleva I., Henning R., Menzel A., Ihalainen J. A., Westenhoff S., Signal amplification and transduction in phytochrome photosensors. Nature 509, 245–248 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ross P., Weinhouse H., Aloni Y., Michaeli D., Weinberger-Ohana P., Mayer R., Braun S., de Vroom E., van der Marel G. A., van Boom J. H., Benziman M., Regulation of cellulose synthesis in Acetobacter xylinum by cyclic diguanylic acid. Nature 325, 279–281 (1987). [DOI] [PubMed] [Google Scholar]
- 26.Otero L. H., Klinke S., Rinaldi J., Velázquez-Escobar F., Mroginski M. A., Fernández López M., Malamud F., Vojnov A. A., Hildebrandt P., Goldbaum F. A., Bonomi H. R., Structure of the full-length bacteriophytochrome from the plant pathogen Xanthomonas campestris provides clues to its long-range signaling mechanism. J. Mol. Biol. 428, 3702–3720 (2016). [DOI] [PubMed] [Google Scholar]
- 27.Björling A., Berntsson O., Lehtivuori H., Takala H., Hughes A. J., Panman M., Hoernke M., Niebling S., Henry L., Henning R., Kosheleva I., Chukharev V., Tkachenko N. V., Menzel A., Newby G., Khakhulin D., Wulff M., Ihalainen J. A., Westenhoff S., Structural photoactivation of a full-length bacterial phytochrome. Sci. Adv. 2, e1600920 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kornev A. P., Taylor S. S., Dynamics-driven allostery in protein kinases. Trends Biochem. Sci. 40, 628–647 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yang X., Kuk J., Moffat K., Crystal structure of Pseudomonas aeruginosa bacteriophytochrome: Photoconversion and signal transduction. Proc. Natl. Acad. Sci. U.S.A. 105, 14715–14720 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yang X., Stojković E. A., Ozarowski W. B., Kuk J., Davydova E., Moffat K., Light signaling mechanism of two tandem bacteriophytochromes. Structure 23, 1179–1189 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Diensthuber R. P., Bommer M., Gleichmann T., Möglich A., Full-length structure of a sensor histidine kinase pinpoints coaxial coiled coils as signal transducers and modulators. Structure 21, 1127–1136 (2013). [DOI] [PubMed] [Google Scholar]
- 32.Ohki M., Sugiyama K., Kawai F., Tanaka H., Nihei Y., Unzai S., Takebe M., Matsunaga S., Adachi S.-i., Shibayama N., Zhou Z., Koyama R., Ikegaya Y., Takahashi T., Tame J. R. H., Iseki M., Park S.-Y., Structural insight into photoactivation of an adenylate cyclase from a photosynthetic cyanobacterium. Proc. Natl. Acad. Sci. U.S.A. 113, 6659–6664 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cornilescu C. C., Cornilescu G., Burgie E. S., Markley J. L., Ulijasz A. T., Vierstra R. D., Dynamic structural changes underpin photoconversion of a blue/green cyanobacteriochrome between its dark and photoactivated states. J. Biol. Chem. 289, 3055–3065 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bellini D., Papiz M. Z., Structure of a bacteriophytochrome and light-stimulated protomer swapping with a gene repressor. Structure 20, 1436–1446 (2012). [DOI] [PubMed] [Google Scholar]
- 35.Winkler A., Heintz U., Lindner R., Reinstein J., Shoeman R. L., Schlichting I., A ternary AppA–PpsR–DNA complex mediates light regulation of photosynthesis-related gene expression. Nat. Struct. Mol. Biol. 20, 859–867 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schultz J. E., Natarajan J., Regulated unfolding: A basic principle of intraprotein signaling in modular proteins. Trends Biochem. Sci. 38, 538–545 (2013). [DOI] [PubMed] [Google Scholar]
- 37.Motlagh H. N., Wrabl J. O., Li J., Hilser V. J., The ensemble nature of allostery. Nature 508, 331–339 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liu Y., Salter H. K., Holding A. N., Johnson C. M., Stephens E., Lukavsky P. J., Walshaw J., Bullock S. L., Bicaudal-D uses a parallel, homodimeric coiled coil with heterotypic registry to coordinate recruitment of cargos to dynein. Genes Dev. 27, 1233–1246 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Croasdale R., Ivins F. J., Muskett F., Daviter T., Scott D. J., Hardy T., Smerdon S. J., Fry A. M., Pfuhl M., An undecided coiled coil: The leucine zipper of nek2 kinase exhibits atypical conformational exchange dynamics. J. Biol. Chem. 286, 27537–27547 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hulko M., Berndt F., Gruber M., Linder J. U., Truffault V., Schultz A., Martin J., Schultz J. E., Lupas A. N., Coles M., The HAMP domain structure implies helix rotation in transmembrane signaling. Cell 126, 929–940 (2006). [DOI] [PubMed] [Google Scholar]
- 41.Stewart C. M., Buffalo C. Z., Valderrama J. A., Henningham A., Cole J. N., Nizet V., Ghosh P., Coiled-coil destabilizing residues in the group A Streptococcus M1 protein are required for functional interaction. Proc. Natl. Acad. Sci. U.S.A. 113, 9515–9520 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Motlagh H. N., Hilser V. J., Agonism/antagonism switching in allosteric ensembles. Proc. Natl. Acad. Sci. U.S.A. 109, 4134–4139 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Guo J., Zhou H.-X., Protein allostery and conformational dynamics. Chem. Rev. 116, 6503–6515 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Liu H., Naismith J. H., An efficient one-step site-directed deletion, insertion, single and multiple-site plasmid mutagenesis protocol. BMC Biotechnol. 8, 91 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Van Duyne G. D., Standaert R. F., Karplus P. A., Schreiber S. L., Clardy J., Atomic structures of the human immunophilin FKBP-12 complexes with FK506 and rapamycin. J. Mol. Biol. 229, 105–124 (1993). [DOI] [PubMed] [Google Scholar]
- 46.Rao F., Yang Y., Qi Y., Liang Z.-X., Catalytic mechanism of cyclic di-GMP-specific phosphodiesterase: A study of the EAL domain-containing RocR from Pseudomonas aeruginosa. J. Bacteriol. 190, 3622–3631 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Antoniani D., Bocci P., Maciag A., Raffaelli N., Landini P., Monitoring of diguanylate cyclase activity and of cyclic-di-GMP biosynthesis by whole-cell assays suitable for high-throughput screening of biofilm inhibitors. Appl. Microbiol. Biotechnol. 85, 1095–1104 (2010). [DOI] [PubMed] [Google Scholar]
- 48.Kabsch W., XDS. Acta Crystallogr. D Biol. Crystallogr. D66, 125–132 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Adams P. D., Afonine P. V., Bunkóczi G., Chen V. B., Davis I. W., Echols N., Headd J. J., Hung L.-W., Kapral G. J., Grosse-Kunstleve R. W., McCoy A. J., Moriarty N. W., Oeffner R., Read R. J., Richardson D. C., Richardson J. S., Terwilliger T. C., Zwart P. H., PHENIX: A comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. D66, 213–221 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bunkóczi G., Read R. J., Improvement of molecular-replacement models with Sculptor. Acta Crystallogr. D Biol. Crystallogr. D67, 303–312 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.McCoy A. J., Grosse-Kunstleve R. W., Adams P. D., Winn M. D., Storoni L. C., Read R. J., Phaser crystallographic software. J. Appl. Cryst. 40, 658–674 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Winkler A., Barends T. R. M., Udvarhelyi A., Lenherr-Frey D., Lomb L., Menzel A., Schlichting I., Structural details of light activation of the LOV2-based photoswitch PA-Rac1. ACS Chem. Biol. 10, 502–509 (2015). [DOI] [PubMed] [Google Scholar]
- 53.Lindner R., Lou X., Reinstein J., Shoeman R. L., Hamprecht F. A., Winkler A., Hexicon 2: Automated processing of hydrogen-deuterium exchange mass spectrometry data with improved deuteration distribution estimation. J. Am. Soc. Mass Spectrom. 25, 1018–1028 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Svergun D. I., Determination of the regularization parameter in indirect-transform methods using perceptual criteria. J. Appl. Cryst. 25, 495–503 (1992). [Google Scholar]
- 55.Bergmann A., Fritz G., Glatter O., Solving the generalized indirect Fourier transformation (GIFT) by Boltzmann simplex simulated annealing (BSSA). J. Appl. Cryst. 33, 1212–1216 (2000). [Google Scholar]
- 56.Franke D., Svergun D. I., DAMMIF, a program for rapid ab-initio shape determination in small-angle scattering. J. Appl. Cryst. 42, 342–346 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kozin M. B., Svergun D. I., Automated matching of high- and low-resolution structural models. J. Appl. Cryst. 34, 33–41 (2001). [Google Scholar]
- 58.Petoukhov M. V., Franke D., Shkumatov A. V., Tria G., Kikhney A. G., Gajda M., Gorba C., Mertens H. D. T., Konarev P. V., Svergun D. I., New developments in the ATSAS program package for small-angle scattering data analysis. J. Appl. Cryst. 45, 342–350 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.McGeehan J., Ravelli R. B. G., Murray J. W., Owen R. L., Cipriani F., McSweeney S., Weik M., Garman E. F., Colouring cryo-cooled crystals: Online microspectrophotometry. J. Synchrotron Rad. 16, 163–172 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Burgie E. S., Bussell A. N., Walker J. M., Dubiel K., Vierstra R. D., Crystal structure of the photosensing module from a red/far-red light-absorbing plant phytochrome. Proc. Natl. Acad. Sci. U.S.A. 111, 10179–10184 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Burgie E. S., Wang T., Bussell A. N., Walker J. M., Li H., Vierstra R. D., Crystallographic and electron microscopic analyses of a bacterial phytochrome reveal local and global rearrangements during photoconversion. J. Biol. Chem. 289, 24573–24587 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Krissinel E., Henrick K., Inference of macromolecular assemblies from crystalline state. J. Mol. Biol. 372, 774–797 (2007). [DOI] [PubMed] [Google Scholar]
- 63.Tarnawski M., Barends T. R. M., Schlichting I., Structural analysis of an oxygen-regulated diguanylate cyclase. Acta Crystallogr. D Biol. Crystallogr. D71, 2158–2177 (2015). [DOI] [PubMed] [Google Scholar]
- 64.Waterhouse A. M., Procter J. B., Martin D. M. A., Clamp M., Barton G. J., Jalview Version 2—A multiple sequence alignment editor and analysis workbench. Bioinformatics 25, 1189–1191 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Takala H., Lehtivuori H., Hammarén H., Hytönen V. P., Ihalainen J. A., Connection between absorption properties and conformational changes in Deinococcus radiodurans phytochrome. Biochemistry 53, 7076–7085 (2014). [DOI] [PubMed] [Google Scholar]
- 66.Svergun D., Barberato C., Koch M. H. J., CRYSOL—A program to evaluate x-ray solution scattering of biological macromolecules from atomic coordinates. J. Appl. Cryst. 28, 768–773 (1995). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/3/2/e1602498/DC1
Supplementary Results
fig. S1. Multiple sequence alignment of PadC homologs generated with Jalview (64).
fig. S2. Spectroscopic and kinetic characterization of TsPadC and MaPadC.
fig. S3. Characterization of the IsPadC PSMcc variant.
fig. S4. IsPadC crystal and spectral characteristics of dark-state crystallized IsPadC.
fig. S5. Effect of substrate binding on the overall architecture of IsPadC.
fig. S6. Summary of HDX experiments.
fig. S7. Individual deuterium incorporation plots of all evaluated peptides.
fig. S8. Spectroscopic and kinetic characterization of IsPadC deletion variants.
fig. S9. Time course of tryptic digests of IsPadC, MaPadC, and TsPadC under dark and light conditions.
fig. S10. Details of SAXS measurements.
table S1. Data collection, phasing, and refinement statistics.
table S2. Overview of oligonucleotides and buffers.
movie S1. Changes in conformational dynamics upon red light illumination of IsPadC.
movie S2. Changes in conformational dynamics upon red light illumination of the IsPadC PSMcc variant.
movie S3. The influence of effector deletion on conformational dynamics of the dark-state IsPadC PSMcc assembly.
movie S4. The influence of effector deletion on conformational dynamics of the light-state IsPadC PSMcc assembly.


