Abstract
Multiprotein complexes transduce cellular signals through extensive interaction networks, but the ability to analyze these networks in cells from small clinical biopsies is limited. To address this, we applied an adaptable multiplex matrix system to physiologically relevant signaling protein complexes isolated from a cell line or from human patient samples. Focusing on the proximal T cell receptor (TCR) signalosome, we assessed 210 pairs of PiSCES (proteins in shared complexes detected by exposed surface epitopes). Upon stimulation of Jurkat cells with superantigen-loaded antigen-presenting cells, this system produced high-dimensional data that enabled visualization of network activity. A comprehensive analysis platform generated PiSCES biosignatures by applying unsupervised hierarchical clustering, principal component analysis, an adaptive nonparametric with empirical cutoff analysis, and weighted correlation network analysis. We generated PiSCES biosignatures from 4-mm skin punch biopsies from control patients or patients with the autoimmune skin disease alopecia areata. This analysis distinguished disease patients from the controls, detected enhanced basal TCR signaling in the autoimmune patients, and identified a potential signaling network signature that may be indicative of disease. Thus, generation of PiSCES biosignatures represents an approach that can provide information about the activity of protein signaling networks in samples including low-abundance primary cells from clinical biopsies.
INTRODUCTION
Cells perceive and respond to their environment by engaging receptors and transmitting intracellular messages through signal transduction cascades. This process is largely controlled by networks of proteins that bind to each other, dissociate, and advance signal progression along biochemical pathways (1). Signalosomes are formed when proteins acting as network hubs orchestrate interactions with other protein nodes to simultaneously control activation of various signaling pathways (2, 3). It is this modular, conditional interconnectivity between proteins and pathways that is largely responsible for providing the logic circuits required for signal transmission, synthesizing instructions for discrete cellular responses from multiple signaling inputs (4–6). However, despite its biological importance, the empirical assessment of signaling protein complexes at the network level is restricted by technological limitations, especially in the case of small clinical samples that provide small amounts of biomaterial for assessment. Most current protein-protein interaction methodologies are designed to work with copious amounts of biomaterial to generate maps of possible or potential intermolecular connections. In contrast, the ability to measure network activity under different physiologic conditions could (i) validate protein-protein associations predicted by literature and database archives and (ii) define signature combinations and the relative quantities of co-associated proteins related with specific biologic signals or pathologic states (7, 8). Thus, the generation of new approaches to improving accessibility to physiologic molecular networks is of particular interest, with high potential to accelerate our understanding of human signal transduction relevant to diagnosis, pharmacology, and medicine (9–11).
Here, we present a high-sensitivity, multiplex, microsphere-based matrix analysis to assess network protein complexes in an extensive signalosome, applicable to both human transformed cells and small primary patient biopsies. Physiologic network activity was visualized in a biosignature that simultaneously measured specific combinations and relative quantities of PiSCES (proteins in shared complexes detected by exposed surface epitopes). We focused on the human T cell antigen receptor (TCR) signalosome, applying data from Jurkat cells stimulated by superantigen-bearing, antigen-presenting cells (APCs) to optimize the experimental and analytical workflow. Subsequently, PiSCES analysis was applied to small primary clinical biopsies from control donors or patients with the autoimmune skin disease alopecia areata. We showed that network signatures (i) distinguished patients from control groups, (ii) detected autoreactive T cell signaling, and (iii) generated a hypothesis regarding a disease-associated network signature. Thus, we present PiSCES analysis as a scalable multiplex approach that could potentially be applied to any network of interactive proteins that may be of interest in signal transduction and disease pathogenesis.
RESULTS
Multiplex signaling protein complexes from superantigen-stimulated Jurkat cells
To study the physiologic network activity of protein complexes in human T cells, we developed a scalable, multiplex, microsphere-based approach for the analysis of 20 cellular proteins in a pairwise combination matrix, resulting in 210 distinct observations of PiSCES. On the basis of previous knowledge and current interactome data for the TCR signalosome (Fig. 1) (12, 13), we assembled a panel of immunoprecipitation antibodies covalently coupled to distinct microsphere classes, which are defined by the proportion of two dyes within the polystyrene latex material base. Multiplex immunoprecipitation was achieved when physiologic proteins were captured from cell lysates onto the microsphere panel and co-associated proteins were detected with fluorochrome-labeled antibodies (Fig. 2A, fig. S1, and table S1). Data were analyzed by flow cytometry on a Bio-Plex 200 instrument with a customized setup that was optimized for the analysis of protein complexes (Fig. 2B and fig. S2). The lower plate carrier unit of the instrument was placed in a commercial sandwich prep refrigerator to maintain protein complexes at 4°C before data acquisition. The upper flow cytometer portion, which needed to be maintained at room temperature, resided on an insulated acrylic divider placed on top of the refrigerator, whereas a bored hole in the acrylic enabled the sample injection port needle of the flow cytometer to access samples from the refrigerated plate carrier. Thus situated, immunoprecipitation beads bearing captured protein complexes were kept cold throughout the data acquisition process. To examine strong TCR signals in a human system, we stimulated Jurkat cells (a human T cell leukemia cell line) for 5 min with staphylococcal enterotoxin E (SEE) superantigen presented by Raji cells, which were used as APCs. Protein complexes were captured on the multiplex immunoprecipitation bead panel, and distinct microsphere classes were identified (Fig. 2C), together with specific multiprotein co-associations that were measured (individual examples, Fig. 2D). Upon initial network analysis, unsupervised hierarchical clustering distinguished unstimulated from stimulated data sets (Fig. 2E), as did principal component analysis (PCA) (Fig. 2F).
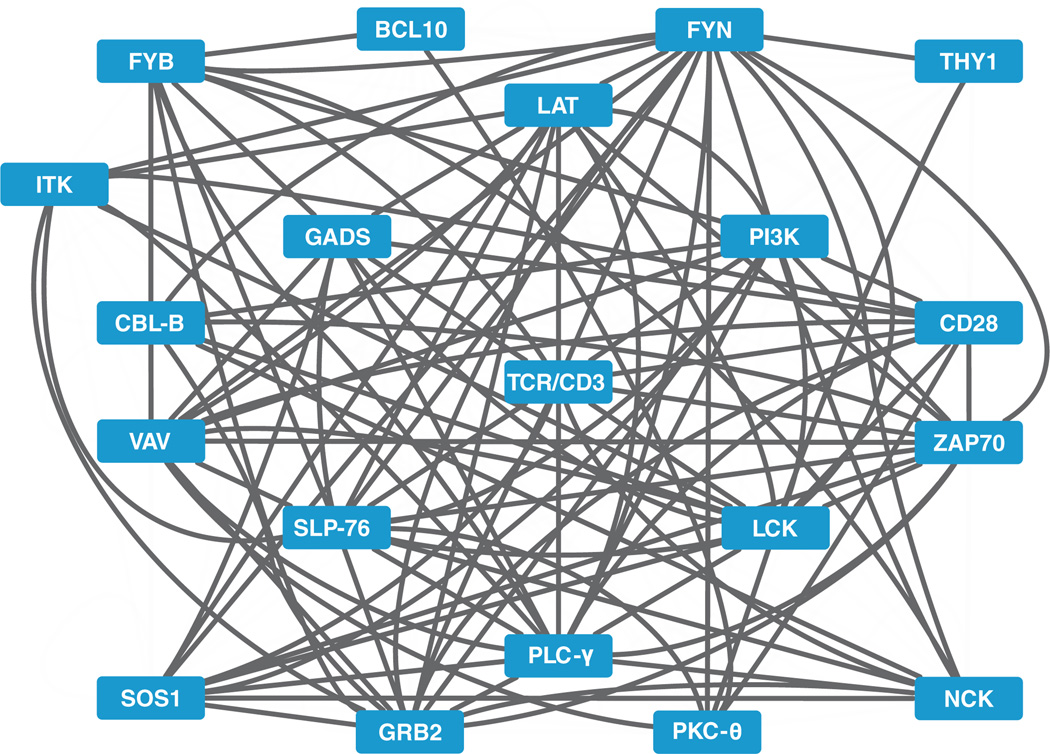
Fig. 1. A protein-protein interaction web consisting of the proximal TCR signalosome.
Visualization of many indirect and direct interactions among 20 proteins that mediate proximal TCR signaling, according to published reports or the Protein Interaction Network Analysis (PINA) database (43). Note that not all of the possible interactions among the collection are depicted, and this visualization provides no information regarding the activity of protein complexes under specific physiologic conditions.
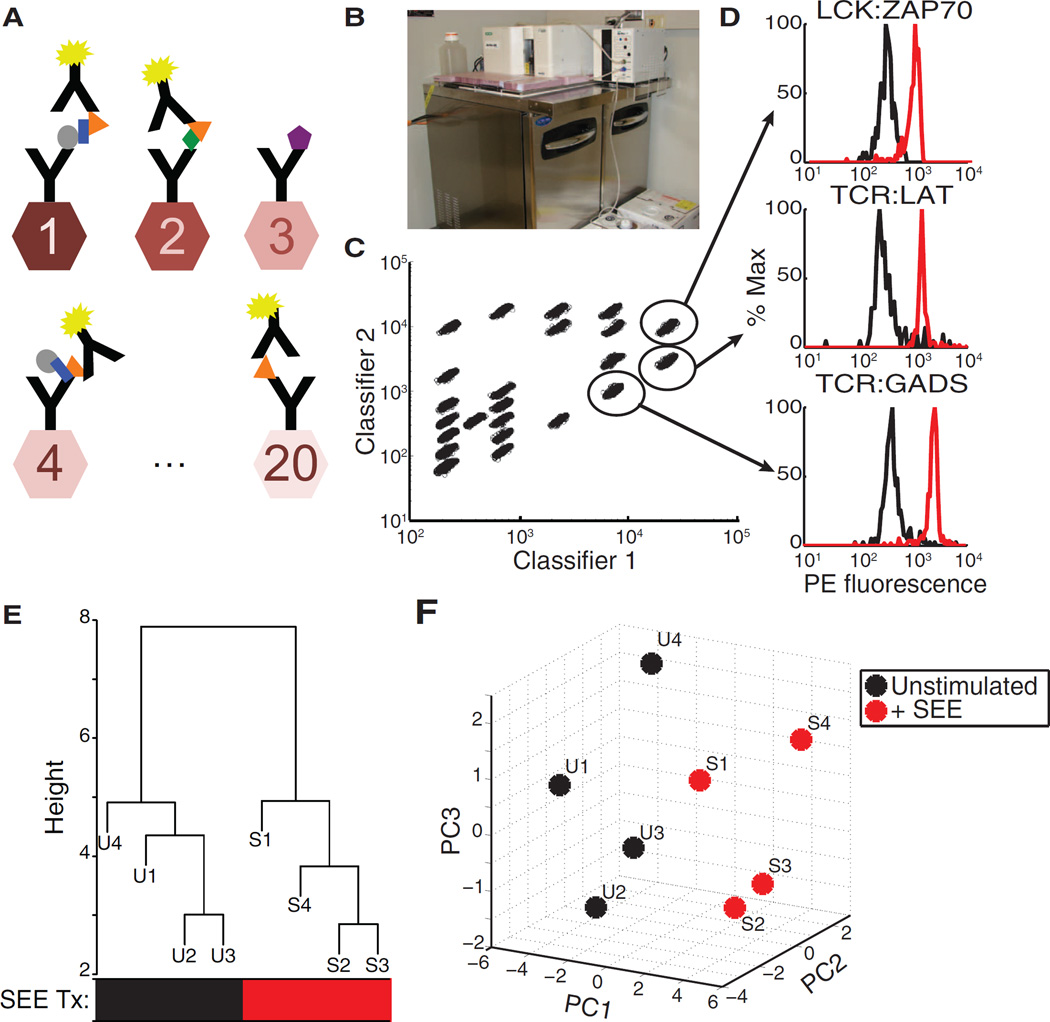
Fig. 2. PiSCES workflow for network profiling of TCR signalosome activation.
(A) Protein complexes immunoprecipitated on microsphere color classes coupled to capture antibodies were detected with a fluorophore-coupled detection (probe) antibody. (B) A Bio-Plex 200 instrument with a customized setup was used to refrigerate samples at 4°C through the data acquisition process. An insulated plexiglass divider enabled the upper flow cytometer portion to remain at room temperature, whereas the lower plate carrier and samples were maintained at 4°C. See fig. S2 for further details. (C) Bead classes were identified by the ratio of two classifier dyes. (D) PE fluorescence readings of example protein-protein associations that increased in intensity upon 5-min stimulation of Jurkat cells with SEE-loaded Raji cells. Black trace, unstimulated; red trace, stimulated. (E) Hierarchical clustering of PiSCES network median fluorescence intensity (MFI) values distinguished SEE-stimulated [S, red treatment (Tx) group] from unstimulated (U, black Tx group) conditions. Data from four independent experiments (labeled 1, 2, 3, and 4) were included in the analysis. P = 0.028, Fisher’s exact test. (F) PCAof log2 MFI from the four experiments (labeled 1, 2, 3, and 4) also distinguished SEE-stimulated from unstimulated conditions, displayinga stimulation axis that was largely dominated by principal component 1 (PC1).
Biostatistical evaluation for PiSCES network analysis
The effectiveness of various statistical approaches was evaluated for PiSCES measurements (figs. S3 and S4). We evaluated nonparametric Kolmogorov-Smirnov (KS) and Wilcoxon rank-sum (WRS) tests, as well as parametric methods based on linear regression (LR) or the Student’s t test (Tm). LR was modeled after the general approach of a method that was previously described for multiplex microbead data (eq. S1) (14). To assess the false-positive rate (FPR) and the true-positive rate (TPR) of each of the four statistical approaches, we used bootstrapping (resampling) from the empirical distributions of each PiSCES measurement in the SEE-stimulated Jurkat data set described earlier (Fig. 2). Each experiment generated two empirical distributions, one from each duplicate. For each duplicate, resampling with 500 iterations was performed to generate a distribution (fig. S3A, black trace). This process was repeated to generate a comparator distribution (fig. S3A, blue trace). Because these distributions (fig. S3A, compare the black and blue traces) were generated identically from the same data set, any differences observed represented false positives. Thus, the P values from these 500 iterations generated the anticipated FPR or type I error. Next, we artificially shifted the comparator distribution by a fixed percentage to evaluate TPR (TPR = 1 – β, where β = type II error; fig. S3B, black versus blue traces). The ability of the different statistical tests to detect shifts in PiSCES measurements was visualized by comparing the relative TPR across a range of FPRs (fig. S3C). Additionally, the FPRs and TPRs from the different statistical tests were compared with an α cutoff = 0.05 (fig. S3, D to F). On the basis of these comparisons, we selected the nonparametric KS statistical test to analyze PiSCES data sets.
To enable incorporation of interexperimental replicates, we defined a method to identify high-confidence, statistically significant PiSCES that were consistently identified as hits in at least 70% of experiments, which we refer to as adaptive nonparametric with empirical cutoff (ANC) (see Materials and Methods and eqs. S2 and S3). To estimate statistical power, we applied the α cutoff from the SEE stimulation experiments to the simulated distribution shifts (fig. S3, A to C) over a 1.1- to 1.5-fold range and found the power to be adequate (fig. S3G); for example, for a consistent shift of 1.2-fold, we predicted that there would be at least 90% power in 90% of the PiSCES measurements. To select an optimal number of experiments appropriate for ANC analysis, we plotted the number of consistent hits observed as the number of experimental replicates increased. With the SEE-stimulated Jurkat cell data set, we found that three independent experiments were sufficient to reach an apparent plateau (fig. S3H), and we considered this experimental number to be a minimum that would be applied in the present work.
We also used weighted correlation network analysis (WCNA) (15) to evaluate PiSCES data, reasoning that many protein-protein interactions involved in signalosome formation would change in a correlated manner. Application of WCNA generated a network heat map that revealed distinct modules that were highly correlated (fig. S4). Each module was summarized by an “eigenvector” that was based on the overall behavior of the module, and each PiSCES measurement was assigned a module membership (MM) value that represented the correlation with this eigenvector, as previously described in detail (15). We found that one set of PiSCES measurements, arbitrarily tagged as the turquoise module, was statistically significantly correlated with stimulation by SEE (fig. S4, A and B).
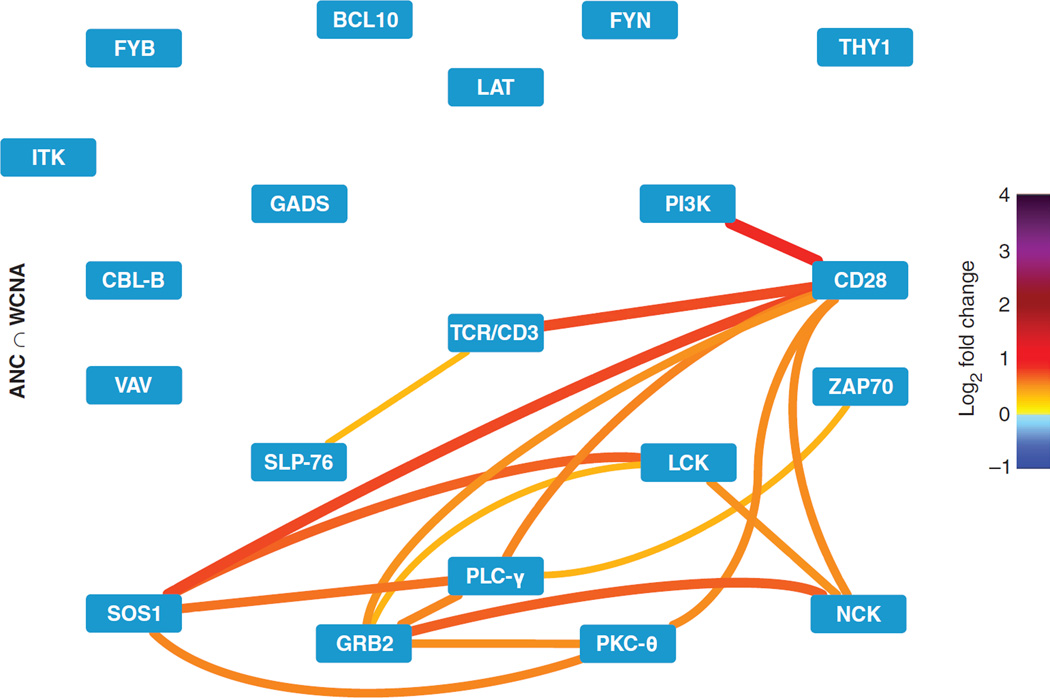
Upon comparison of the ANC and WCNA results, we found that these methods coincided in most of the statistically significant stimulation-induced changes that were identified (Fig. 3, A to C). PiSCES differences that were significant in WCNA, but not in ANC, typically showed very small fold changes (1.08 ± 0.008), whereas those that were significant in ANC, but not in WCNA, generally displayed a small positive correlation with the turquoise module, but missed the inclusion cutoff (MM > 0.5; inclusion cutoff, 0.7) (see Materials and Methods) (15), and displayed small fold changes < 1.5 (Fig. 3, C and D). These results suggest that PiSCES measurements whose statistical significance was discrepant between the two analyses likely included differences that (i) were trending toward (but did not reach) statistical significance, (ii) had higher variance that reduced sensitivity, or (iii) were false positives (Fig. 3, C and D). Therefore, we visualized the network of hits common to both analyses as a high-confidence group of coregulated PiSCES that responded to stimulation by SEE (Fig. 4). As expected upon cross-linking, the TCR-CD3 complex displayed a loop in the visualization, indicating that multiple copies of TCR-CD3 appeared in shared complexes. TCR-CD3 also formed shared complexes with lymphocyte-specific protein tyrosine kinase (LCK), ζ chain–associated protein kinase 70 (ZAP70), linker for activation of T cells (LAT), Src homology 2 (SH2) domain–containing leukocyte protein of 76 kD (SLP–76), growth factor receptor–bound protein 2 (GRB2)–related adaptor downstream of Shc (GADS), phospholipase C–γ (PLC-γ), and others, and many interactions among these proteins were also observed. We conclude that TCR engagement by superantigen produced a clear PiSCES signaling profile, which identified specifically induced protein co-associations that participated in TCR signalosome activity.
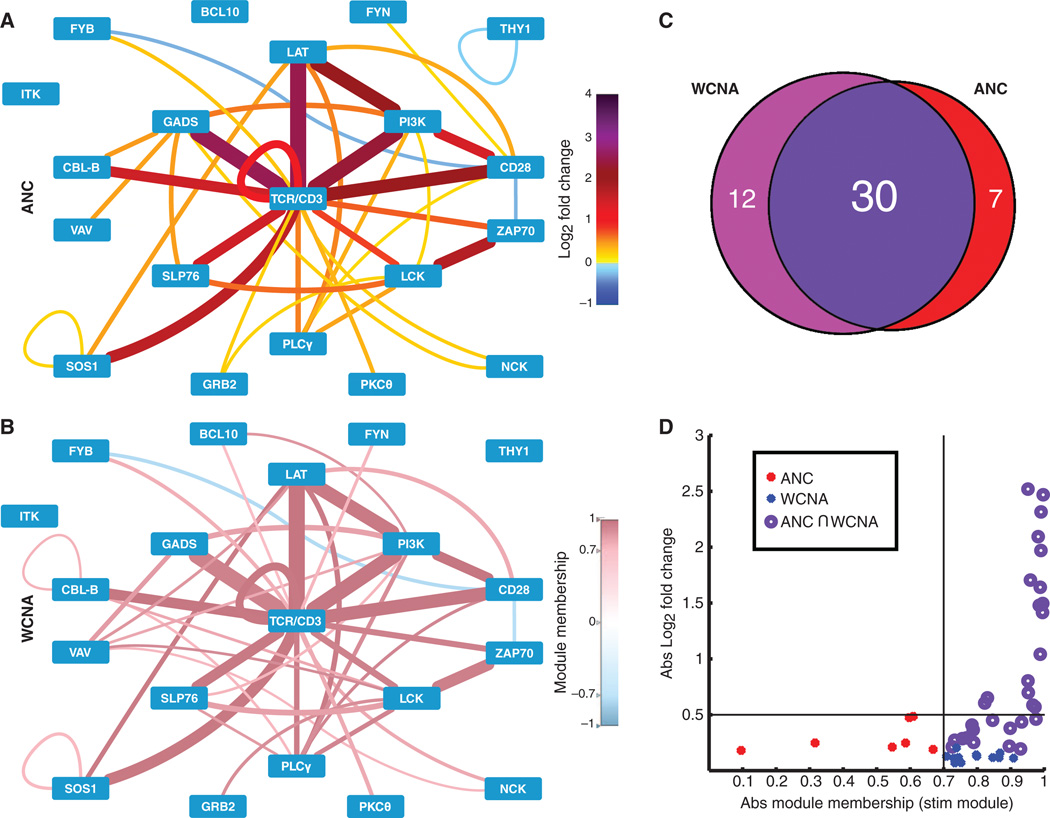
Fig. 3. Comparison of ANC and WCNA analyses.
(A) ANC analysis of four independent SEE stimulation experiments identified PiSCES that exhibited statistically significant changes in three of four experiments for the SEE data set. Edge color and thickness correspond to mean log2 fold change (color legend on the right). (B) WCNA analysis of the MFI values from the same SEE data set. PiSCES with MM > 0.7 in the turquoise stimulation network (see fig. S4 and Materials and Methods) are visualized in the node-edge diagram. Edge thickness corresponds to mean log2 fold change, whereas edge color corresponds to MM (color legend on the right). (C) Venn diagram of the overlap between both methods of analysis. Because most hits coincided between the two analyses, we concluded that the two analyses produced compatible results. (D) Hits identified by ANC (red), WCNA (blue), or both (dark purple) are graphed by the absolute value (Abs) of mean log2 fold change and the absolute value of MM in the stimulation (stim) (turquoise) module.
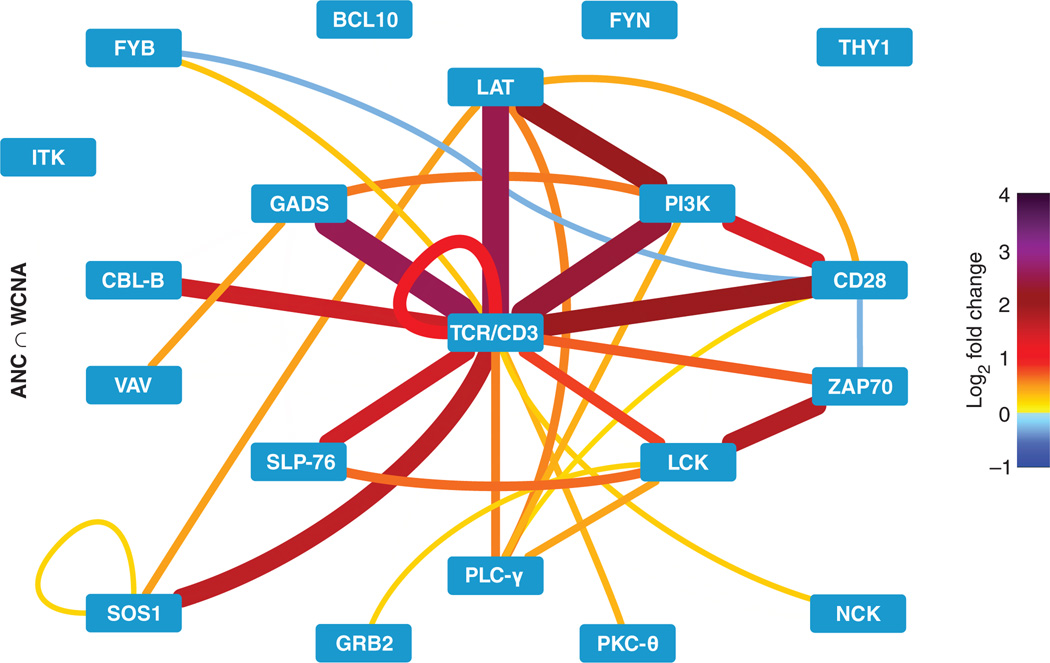
Fig. 4. High-confidence PiSCES network signature of SEE-induced proximal TCR signaling.
Shared hits identified by both ANC and WCNA analyses (ANC ∩ WCNA) are displayed for the PiSCES data set introduced in Figs. 2 and 3, in which Jurkat cells were either left unstimulated or stimulated for 5 min with SEE-loaded Raji cells. Edge color and thickness correspond to the mean log2 fold change (color legend on the right) from four independent experiments.
PiSCES analysis of CD28 costimulation
To isolate stimulation of the T cell costimulatory receptor CD28 as a variable and visualize its potential contribution to PiSCES networks, Jurkat cells were stimulated for 5 min with SEE-loaded Raji cells in the presence of either cytotoxic T lymphocyte–associated protein 4 (CTLA4)–immunoglobulin (Ig) or nonspecific human IgG. CTLA4-Ig binds to the CD28 ligands B7.1 and B7.2 and blocks them from binding to CD28 (16), whereas control IgG does not, enabling physiological engagement of CD28 on the Jurkat cells by the B7 ligands on the Raji cells. Normalizing the CD28-permissive condition to the CD28-blocked condition, network visualization showed a signature with CD28 as a central player that was induced to form shared complexes with phosphoinositide 3-kinase, PLC-γ, protein kinaseC–θ, GRB2, son of sevenless homolog 1 (SOS1), TCR, and other proteins, and additional protein pairings in these pathways were also observed (Fig. 5) (17). We conclude that PiSCES analysis can reveal network signatures of signaling protein complexes that are indicative of co-stimulatory pathway activity.
Fig. 5. High-confidence PiSCES network signature of CD28 costimulation.
Network activity profile is displayed for Jurkat cells stimulated by SEE-loaded Raji cells in the presence of physiological CD28 engagement (control human IgG–treated) normalized to the CD28-blocked condition (CTLA4-Ig–treated). Shared hits identified by both ANC and WCNA analyses (ANC ∩ WCNA) are displayed. Edge color and thickness correspond to mean log2 fold change (color legend on the right) from three independent experiments.
PiSCES analysis of T cells from biopsies of patients with the autoimmune condition alopecia areata
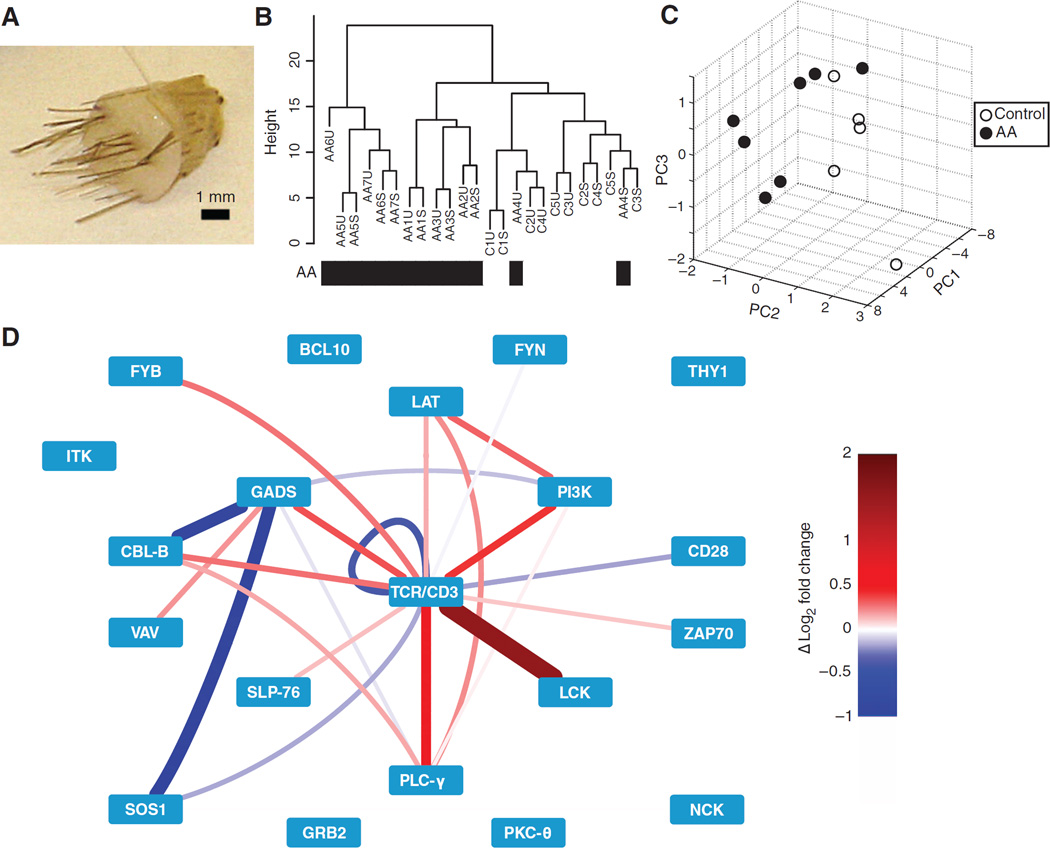
To seek insight into protein signaling network activity in human disease, we applied PiSCES analysis to the few primary T cells obtained from 4-mm skin punch biopsies of the scalp, donated by control or alopecia areata patients (Fig. 6A and table S2). Alopecia areata is an autoimmune disorder involving T cell–associated hair follicle pathology; however, no autoreactive TCRs, confirmed T cell autoantigens, or TCR signaling activities have yet been described for the human disease (18, 19). Experiments were performed by stimulating half of the biopsy-isolated T cells with plate-bound anti-CD3 and anti-CD28 antibodies for 5 min, whereas the other half of the cells were processed in parallel without stimulation. In addition, frozen aliquots of Jurkat cells were processed in each experiment, serving for intra-assay normalization and interassay comparison. Visualization of the stimulation-induced PiSCES network suggested overall agreement between control and alopecia areata samples (fig. S5), with many of the same protein complexes being formed during signaling. Despite this similarity, unsupervised hierarchical clustering (Fig. 6B) and PCA (Fig. 6C) revealed a separation between control patients and all but one alopecia areata patient, suggesting that the underlying PiSCES measurements contained patterns that distinguished the disease condition at a network level. Consistent with autoreactive TCR involvement in human alopecia areata, a statistically significant enrichment in TCR-responsive basal protein complexes in alopecia areata patients over controls was observed, comparing average normalized protein complex intensities in the absence of exogenous stimulation (Fig. 6D). This was determined by a resampling-based test, which showed that the higher percentage of protein pairs whose intensities were greater in alopecia areata samples than in controls (alopecia areata > control) from among the set of stimulation-inducible PiSCES (defined in fig. S5) represented a statistically significant increase over the percentage of protein pairs with alopecia areata > control in randomly resampled sets from the full PiSCES network of measurements. We conclude that combining unsupervised hierarchical clustering, PCA, ANC, and WCNA produced a clear PiSCES biosignature that distinguished the control from the disease condition. Furthermore, in exogenously stimulated cells, WCNA analysis identified a PiSCES module that was statistically significantly correlated with alopecia areata status (Fig. 7). The module revealed a subnetwork of potential interest, in which, among other observations, the balance of activity between GADS and GRB2 in the TCR signalosome may favor GADS in cells from alopecia areata patients, but may favorGRB2 in cells from control patients (Fig. 7). We propose that new hypotheses can be generated when PiSCES analysis is applied to physiologic protein networks involved in human signal transduction and disease.
Fig. 6. PiSCES network analysis of clinical biopsies distinguishes the alopecia areata patient group from the control patient group and suggests enhanced basal TCR signaling network activity in alopecia areata.
(A) A 4-mm scalp punch biopsy from which T cells were isolated. (B) Hierarchical clustering of the normalized fold changes in MFI predominately separates the seven alopecia areata patient samples from the five control patient samples. S, exogenously stimulated; U, unstimulated. P = 0.015, extension of Fisher’s exact test. (C) PCA of basal (not exogenously stimulated) PiSCES activity among all stimulation-responsive protein complexes (identified in fig. S5) separates alopecia areata patient samples from control patient samples. (D) Difference in the basal activity of stimulation-responsive network PiSCES in the alopecia areata patient group as compared to the control patient group (red indicates greater activity in the alopecia areata cells; blue indicates greater activity in the control patient cells) suggests that more stimulation-responsive PiSCES were basally increased in relative abundance in the alopecia areata patient group versus the control patient group. P = 0.039 with control-clustered patient AA4 removed; P = 0.115 with patient AA4 included, using a resampling-based test.
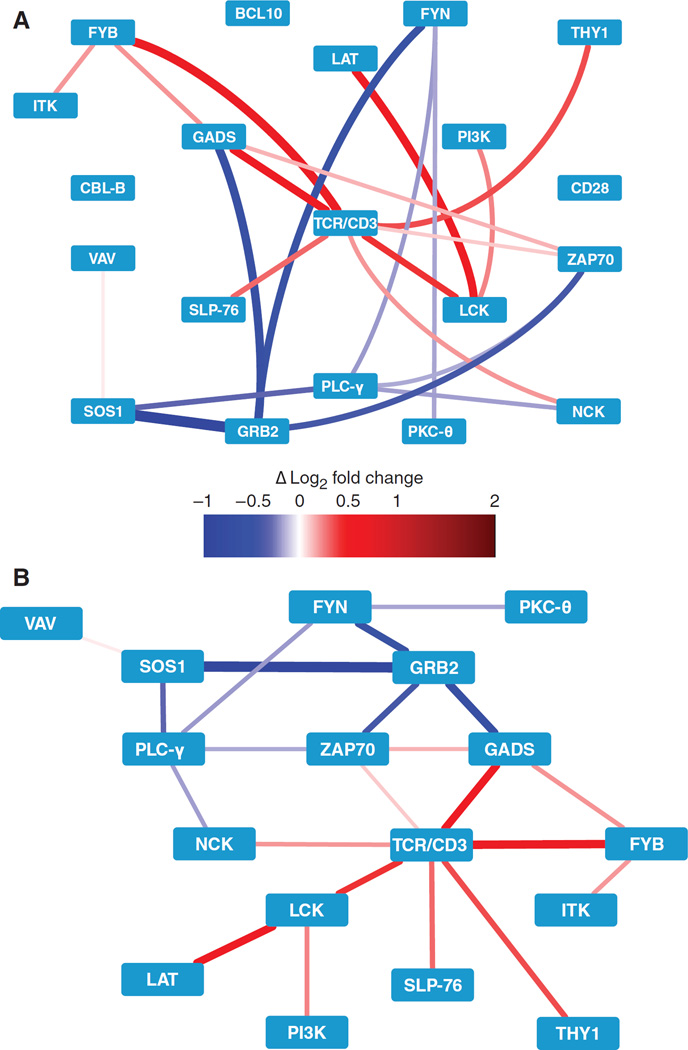
Fig. 7. PiSCES network activity suggests a disease-associated signaling profile in T cells from alopecia areata patients.
(A) Using PiSCES data from patient and control T cells that were exogenously stimulated for 5 min with plate-bound anti-CD3 and anti-CD28 antibodies, WCNA analysis of normalized MFI fold changes identified a PiSCES subnetwork module that distinguished the alopecia areata patient group from the control patient group. Red, enhanced in the alopecia areata condition; blue, enhanced in the control condition. (B) An alternative view of the subnetwork module, with the nodes positioned on the basis of the activity in alopecia areata patient samples (n = 7) versus control patient samples (n = 5).
DISCUSSION
Various proteomic, genetic, and other techniques are now in use to discover and document proteins that can bind to each other to compose an interactome. In the case of the TCR signalosome, years of previous work, together with the ongoing acquisition of data, reveal a highly interactive protein network (Fig. 1) that is thought to be capable of manifesting different activities to transduce distinct biological signals, depending on the strength of the antigen encountered (12, 13,20, 21). However, note that the complicated, so-called “hairball” (22) visualization (Fig. 1) conveys a map of protein co-associations that are deemed possible (because they have been reported), without containing information about the actual protein interaction activity that occurs to transmit a specific biological signal. For this reason, there remains an outstanding need for the generation of new approaches that could harvest known information about the interactome and apply it to data acquisition from human cells. In the case of primary patients, cells may originate from low-abundance clinical samples; thus, the applicability of genetic engineering or protein tagging can be quite limited. Our multiplex, microsphere matrix–based PiSCES analysis (Fig. 2) is designed to contribute toward overcoming these obstacles. PiSCES analysis enables the measurement of defined subnetworks of proteins from the interactome, validating their participation in physiologic signaling contexts and revealing how their specificity and activity compose signals in transformed and primary patient T cells.
Although singleplex and multiplex immunoprecipitation detected by flow cytometry can be highly sensitive down to the femtomole to attomole analyte range (23–25), application of this technology to multiprotein complexes in signal transduction required the investigation of biostatistical analysis techniques. The present investigation involved the comparison of several statistical approaches using both empirical and computer-simulated experimentation to select an applicable statistical approach for PiSCES analysis. Statistical evaluation of PiSCES signatures across the high-dimensional matrix of measurements was achieved through an analysis termed ANC, based on KS statistics and empirically determined α-cutoff criteria. ANC maintained a fixed type I error (0.05) adjusted for multiple comparisons, while identifying changes in PiSCES that were consistently statistically significant across experiments (fig. S3). Additionally, we observed by WCNA that many stimulation-inducible protein-protein associations changed in a correlated manner (fig. S4). Hits that were common to both ANC and WCNA provided a high-confidence, stimulation-responsive network profile (Figs. 3, 4, and 5). In summary, the protein-pair matrix data of PiSCES were able to achieve network-level visualization and analysis of the activity of a finite subset of signaling protein complexes from the interactome centered around the human TCR signalosome.
Constructed and optimized with data from experiments with SEE-stimulated Jurkat cells, PiSCES analysis proved capable of assessing activity from small samples from clinical patients. PiSCES biosignatures (unsupervised hierarchical clustering, PCA, ANC, and WCNA) distinguished alopecia areata patient groups from control groups and were suggestive of basal TCR signaling activity in lesional alopecia areata T cells (Fig. 6). Furthermore, upon exogenous stimulation, amodule within the TCR signalosome network was identified, generating a new hypothesis with specific ramifications (Fig. 7). The SH2 domains of GADS and GRB2 compete for binding to the phosphorylated amino acid residues Tyr171 and Tyr191 of LAT [GRB2 can also bind to Tyr226 (26)]; in T cells from alopecia areata lesions, the GADS:GRB2 balance may favor relatively more GADS activity in the signalosome than what is observed in T cells from control patient biopsies, in which the balance may favor GRB2. Additional observations in the data set were consistent with this model. WhereasGRB2recruits SOS1 (alopecia areata < control) to LAT, leading to the activation of the Ras–mitogen-activated protein kinase (MAPK) pathway, GADS recruits SLP-76 (alopecia areata > control), which in turn recruits FYB (alternatively called ADAP or SLAP130; alopecia areata > control), which can link to cytoskeletal remodeling and integrin signaling (27, 28). Together, these observations suggest that a subnetwork involving the balance between GADS and GRB2may play a role in alopecia areata. We propose that the unique visualization of network protein signaling activity by PiSCES has the potential to generate insights into human signal transduction and generate hypotheses regarding disease pathways.
The PiSCES approach provides access to physiologic protein networks, although it is subject to certain limitations that are also shared by other leading protein interaction methodologies. First, because it depends on antibody binding to exposed surface epitopes of multiprotein complexes under native (nondenatured) conditions, PiSCES is not expected to detect every protein-protein interaction that may be present. For example, a specific protein could be bound by interacting partners in a way that makes it inaccessible to immunoprecipitation or detection (probe) antibodies. This limitation applies to some extent to all methods that bind proteins to capture them before subsequently analyzing their interacting partners. Second, PiSCES data provide a network signature, but this signature does not define the identities of all of the proteins that compose complexes. For example, although TCR:ZAP70 and TCR:LAT are both protein pairs found in shared complexes upon stimulation, PiSCES data do not reveal that the doubly phosphorylated immunoreceptor tyrosine-based activation motifs of CD3 bind to the tandem SH2 domains of ZAP70 and that the same ZAP70 molecules bind through their catalytic domain to the phosphotyrosines of LAT, resulting in detection of TCR in shared complexes with LAT. Because PiSCES data lack this information, the data do not refute the possibility that the TCR and LAT might appear in shared complexes through a molecular pathway that is different from the simple one just described.
Despite this limitation, PiSCES data can validate the presence and relative quantities of physiologic proteins in shared complexes isolated from clinical samples—information that is otherwise generally unobtainable. Other methods that perform vast surveys of protein-protein interactions also lack information on the precise constituents of specific multiprotein complexes, which is one of the current major limitations in the interactome field (29–33). There are two notable consequences of these limitations as they affect PiSCES signatures: (i) quantification is not absolute but is relative between two assessed conditions under which each measurement can be influenced by the underlying activity of heterogeneous complexes, and (ii) like all other methods for analyzing protein-protein interactions, PiSCES data reflect the characteristics of a dominant assay; that is, whereas positive data support the hypothesis that two proteins join shared complexes, the failure to detect two proteins together does not disprove the hypothesis that they may interact with each other. Overall, PiSCES analysis takes a step forward in providing a network biosignature that visualizes the physiologic activity of signaling protein complexes from human tissue samples, although further progress is needed to overcome other technological barriers that currently limit the amount of information obtainable from the human clinical interactome.
The PiSCES approach is potentially adaptable to any defined network of interacting proteins and is scalable up to a maximum of 500 distinct microsphere classes in current use. The strategies outlined here, which involve reagent screening, data evaluation, and network profiling, could be applied to generate a multiprotein matrix analysis system for any human interaction network relevant to biomedicine. Our interest in genetically nonengineered primary patient samples drove us to build the system with antibodies as the capture and detection reagents, a strategy that supplied access to many quality control–ready candidates. Given that multinational efforts are aimed at generating monoclonal antibodies specific for every open reading frame of the human proteome, even more extensive antibody collections are likely to become available in the future (34, 35). Moreover, many other non–antibody-based protein-binding reagents are compatible with multiplex microsphere chemistry and are predicted to function well within the PiSCES analysis framework. Finally, it is anticipated that PiSCES analysis could potentially contribute to composite signatures generated in conjunction with other high-dimensional data (including cellular mass cytometry, multiplex cytokine and phosphoflow analysis, and genome-wide expression analysis) to further our understanding of immune-mediated diseases (36). In conclusion, we present PiSCES analysis as a broadly applicable strategy that is useful for identifying biosignatures of multiprotein complex activity in network signal transduction and disease pathways.
MATERIALS AND METHODS
Cell culture
Jurkat E6-1 cells [American Type Culture Collection (ATCC) TIB-152] and JRT3-T3.5 cells (ATCC TIB-153) were purchased from ATCC. Jurkat cell derivatives lacking specific proteins (37), which were used for antibody screening, and Raji cells, which were used as APCs, were provided by K. Hedin (Mayo Clinic). All cells were cultured in RPMI medium (Life Technologies) with 10% Cosmic Calf serum (HyClone), 2 mM l-glutamine (Life Technologies), penicillin (100 U/ml; Life Technologies), and streptomycin (100 µg/ml; Life Technologies) and maintained in an incubator at 37°C with 5% CO2.
Cell stimulation and lysate preparation
For SEE stimulation, Raji cells (APCs) were incubated with SEE (5 µg/ml; Toxin Technologies) for 2 hours in serum-free RPMI at 37°C. For CD28-blocking experiments, the Raji cells were also incubated with either human CTLA4-Ig fusion protein (3.33 µg/ml; Bio X Cell) or nonspecific human IgG (3.33 µg/ml; Jackson ImmunoResearch). The Raji cells were pelleted at 300g for 5 min, the supernatant was discarded, and the cells were fixed by resuspension in 0.05% gluteraldehyde in phosphate-buffered saline (PBS), as previously described (38). After 30 s, fixation was halted by the addition of PBS and 200mMglycine, and the fixed cells were washed three times in PBS and counted with a hemocytometer. Jurkat cells, which had been placed in fresh medium (0.35 × 106 cells/ml) the previous evening, were counted and pelleted. Then, 15 × 106 Jurkat cells were resuspended in 200 µl of ice-cold PBS, mixed with 20 × 106 Raji cells in an equal volume of ice-cold PBS, and centrifuged for 5 min at 300g at 4°C to facilitate cell-cell conjugation. The supernatant was discarded, and stimulation was commenced as the Jurkat cell–Raji cell pellet was placed in a water bath for 5 min at 37°C before being flash-frozen in liquid nitrogen. Control, unstimulated cells were mixed with nonloaded, fixed Raji cells in parallel. Frozen pellets were used immediately or were stored briefly at −80°C before being lysed in lysis buffer [150 mM NaCl, 50 mM tris (pH 7.4), 1% digitonin (High Purity, Millipore), 1× Halt protease/phosphatase inhibitors (Pierce), 10 mM NaF, 2 mM sodium orthovanadate, and 10 mM iodoacetamide].
Skin-biopsy T cells and lysate preparation
All procedures involving human subjects were approved by the Mayo Clinic Institutional Review Board (protocol #12-004980). Cases were identified on the basis of established clinical criteria for alopecia areata. Punch biopsies (4 mm) were taken from alopecia areata patients from perilesional areas with active hair loss but still contained hair. Specimens were immediately transported to the laboratory on ice. Biopsy samples from control patients were taken from normal-appearance scalp skin adjacent to skin lesions (not due to alopecia areata) that were removed during routine dermatological procedures. T cells were isolated by the punch biopsy crawl-out method as described previously (39, 40). Stimulation plates were prepared by adding the anti-CD3 clone OKT3 (10 µg/ml, purified in-house from hybridoma supernatant) and anti-CD28 (2 µg/ml, clone CD28.2, BioLegend) in PBS to each well of a six-well plate, incubating the wells overnight at 4°C, and washing them three times in PBS. Half of the harvested T cell preparation was added to a well coated with anti-CD3 and anti-CD28 antibodies, whereas the other half was added to an uncoated well (both on ice). Cells in the plate were centrifuged at 300g for 5 min at 4°C, the supernatant was aspirated and discarded, and the plate was floated in a water bath for 5 min at 37°C to activate signaling. The plate was then immediately placed on ice, and 200 µl of ice-cold lysis buffer was added to each well.
Antibody screening and coupling to microspheres
We first screened three to five antibodies per target for their ability to capture and detect proteins from postnuclear cell lysates. Using a collection of cell lines that had or lacked each target protein, we empirically confirmed the binding and specificity of each antibody pair in uniplex, microsphere, immunoprecipitation-based capture followed by flow cytometry analysis (table S1 and fig. S1). Each selected immunoprecipitation antibody was covalently coupled to a distinct bead class of Luminex MagPlex COOH beads (Bio-Rad), according to the manufacturer’s recommended amine coupling protocol. Each detection (probe) antibody was purchased in R-phycoerythrin (R-PE)–conjugated form or was biotinylated with Sulfo-NHS-Biotin (Thermo Scientific). Bead coupling efficiency was checked by staining beads with antibodies specific for the appropriate antibody host species and class, with fluorescence intensity acquired on an Accuri C5 flow cytometer (BD Biosciences).
Multiplex capture and detection of protein complexes
In the TCR signalosome, 20 signaling proteins were targeted in a minimum 20 × 20 matrix of capture and detection antibodies (table S1). This generated a minimum of 400 protein co-association measurements, including 210 total unique protein co-associations: [(20 capture × 20 probe) – 20 homotypic combinations)/2] + 20 homotypic combinations = 210. Some experiments included capture and detection antibody combinations targeting additional proteins, but this study focuses only on 20 targets that were assessed in all experiments. Because the multiplex microbead approach is scalable, additional capture and detection reagents and microbead classes can be used as desired for the addition of new targets to the collection. A master mix containing equal numbers of each antibody-coupled Luminex bead class was prepared and distributed into postnuclear cell lysate samples in duplicate. All steps were performed at 4°C or on ice, and other technical practices were followed as described previously (23, 24, 41). Protein complexes were immunoprecipitated from samples overnight, washed twice in Fly-P buffer [50mMtris (pH 7.4), 100 mM NaCl, 1% bovine serum albumin, and 0.02% sodium azide], and distributed into as many wells of a 96-well plate as there were probes. Detection antibodies were added and incubated for 1 hour, with gentle agitation at 500 rpm in a cold room. A Bio-Plex Pro II magnetic plate washer was used to wash microbeads and captured complexes three times in Fly-P buffer. Wells containing biotinylated detection antibodies were then incubated for 30 min with streptavidin-PE. After three washes in the plate washer, the microbeads were resuspended in 125 µl of Fly-P buffer, and fluorescence data were acquired on a Bio-Plex 200 instrument that was calibrated according to the manufacturer’s recommendations and run on the “high RP target” setting. Data files were exported in both Microsoft Excel and XML formats for further processing.
Data preprocessing and inclusion criteria
XML output files were parsed to acquire the raw data for use in MATLAB, R statistical package, Cytoscape, and other analysis and visualization platforms. For each well from a data acquisition plate, data were processed to (i) eliminate doublets on the basis of the doublet discriminator intensity (>5000 and <25,000 arbitrary units; Bio-Plex 200), (ii) identify specific bead classes within the bead regions used, and (iii) pair individual bead PE fluo-rescence measurements with their corresponding bead regions. This processing generated a distribution of PE intensity values for each pairwise protein PiSCES measurement.
Statistical tests evaluated for application to PiSCES analysis
The KS and WRS nonparametric tests were evaluated with merged distributions from two intraexperimental duplicates, where each duplicate provided fluorescence intensity measurements from ~100 microspheres. These tests were compared against two parametric methods based on LR or Tm. The data subjected to LR analysis were the fluorescence intensities from ~100 measurements per intraexperimental duplicate, having been modeled after the general approach of a method that was previously described for multiplex microbead data (eq. S1) (14). In contrast, Tm used MFI as a summary representation of each distribution, resulting in a statistical test that used two central values per intraexperimental duplicate. Model assumptions of these statistical tests differ, with KS being sensitive to changes in distribution features including variance, WRS emphasizing distribution median, LR emphasizing distribution mean, and Tm emphasizing population central value.
Unsupervised hierarchical clustering and PCA
For hierarchical clustering, filtered MFI intensities (exclusion criterion, MFI < 100) or filtered normalized fold-change values (exclusion criterion, 0.9 < fold change < 1.1) were clustered using the average method with a Euclidean distance matrix in R statistical package (flashClust) (42). PCA was performed using the built-in function in MATLAB. Unless otherwise specified, the input variables for PCA included either log2 MFI or log2 fold-change values for each of the PiSCES measurements performed, where each stimulation condition in one experiment constituted an observation.
ANC analysis
Differences in specific protein-pair PiSCES measurements were considered hits if they were consistently identified as statistically significant in at least 70% of experiments with the KS test. The α-cutoff value required per experiment to determine statistical significance was calculated to maintain an overall type I error of 0.05 (adjusted for multiple hypothesis testing with Bonferroni correction). The full description of the α-cutoff calculation (defined as ακ) is provided in eq. S2, and an example is provided in eq. S3. Further adjustment to the cutoff for statistical significance was imposed to account for technical error as follows. We first calculated the P values for all PiSCES measurements within intraexperimental duplicate probe wells. If a medianP value for duplicate wells was <0.05 (Bonferroni-corrected), then those duplicate wells were excluded from the analysis because of presumed manual error. For the remaining measurements across all related conditions and experiments, a combined empirical distribution of intraexperimental duplicate P values was used to account for technical error by identifying the P value at the percentile defined by ακ. Linear interpolation using the five nearest points to this target percentile (defined by ακ) was used to precisely mark this P value cutoff. As a result, the ακ-associated P value cutoff was customized for the empirical technical error in an experimental set and was used to define statistical significance.
Weighted correlation network analysis
The WGCNA package for R (15) was used to perform WCNA. Microsoft Excel output files were used to average the two duplicate sample MFIs for each protein-pair PiSCES measurement. Either MFI or fold-change values between conditions were used as the inputs for WCNA. Data were then (i) filtered to remove weakly detected or weakly changed PiSCES (<100MFI or 0.9 < fold change < 1.1, respectively) and (ii) subjected to log2 transformation. Soft thresholding using a power adjacency function was selected, and the power (exponent) value was determined by identifying the value that resulted in the best approximation of scale-free topology. The minimum module size was set to 5, and closely related modules were merged when the dissimilarity value (MEDissThres) was <0.25. Closely related PiSCES were assigned to modules, and eigenvectors, which are vectors describing the first principal component summarizing the general behavior for each module, were calculated. Each module was assigned an arbitrary identification color. Correlations between experimental traits (for example, “SEE stimulation,” ‘”AA status,” etc.) and module eigenvectors were visualized with the “Module-trait relationship” function. Modules with P < 0.05 for a given trait were considered to be statistically significantly correlated with that trait. PiSCES belonging to modules of interest were defined as those with MM > 0.7 and P < 0.05. PiSCES with MM < 0.7 were excluded even if the associated module inclusion P value was <0.05.
Data visualization
PiSCES analysis by hierarchical clustering, PCA, ANC, and WCNA used complete unedited data sets from all experiments, unless otherwise specified. In contrast, node-edge diagrams only visualized subsets of data and were generated with the publicly free open network resource Cytoscape. For protein pairs that had multiple measurements targeting different epitope combinations, the measurement with the greatest mean log2 fold-change value was selected for node-edge diagram visualizations.
Other statistics and analysis
Box-and-whiskers plots (boxplots) display the median as a central value, with box edges representing the 25th and 75th percentiles. Maximum whisker lengths were q3 + 1.5(q3 − q1) and q1 – 1.5(q3 − q1), where q1 and q3 are the values of the first and third quartiles, respectively. For the resampling-based statistical test in Fig. 6D, the set of stimulation-responsive PiSCES (defined in fig. S5) were compared against 50,000 randomly selected sets from the full network of measurements to determine whether there was a statistically significant enrichment for PiSCES whose mean normalized intensity was increased in alopecia areata samples, compared to that in control samples. Unsupervised hierarchical clustering was performed to generate the trees in Figs. 2E and 6B. To assess the statistical significance of the clusters in Fig. 2E, we performed Fisher’s exact test. For Fig. 6B, unsupervised hierarchical clustering was performed, and the tree structure was generated with data from unstimulated and stimulated cells for each patient. Because the alopecia areata patients predominately separated into two of three tree branch clusters, an extension of Fisher’s exact test was performed to calculate the statistical significance of observing no control patients in these two groups. Note that each patient’s unstimulated cell data tended to cluster closely with the same patient’s stimulated cell data, likely indicating sample dependency between these two conditions. As a result, we performed the test by treating each patient, not each sample, as the independent variable.
Supplementary Material
Acknowledgments
We thank our volunteer patients; J. Kidd for control skin sample acquisition; B. Alarcón, K. Hedin, L. Pease, and J. Regueiro for kind provision of cell lines; and R. Stiles for antibody purification.
Funding: This work was supported by NIH grants R01GM103841 (to A.G.S., J.E.E.-P., D.G., and C.N.), MH102244 (to S.E.P.S.), and T32AI7425 (to S.C.N. and B.K.R.); Mayo Graduate School (to S.C.N. and B.K.R.); and the philanthropic partnership of B. Fineman with Mayo Clinic (to A.G.S.).
Footnotes
SUPPLEMENTARY MATERIALS
www.sciencesignaling.org/cgi/content/full/9/439/rs7/DC1
Equations
Fig. S1. Screening strategy for multiplex panel antibodies, using SLP-76 as an example.
Fig. S2. Instrument setup for optimal analysis of protein complexes.
Fig. S3. Development and evaluation of ANC analysis.
Fig. S4. Application and evaluation of WCNA analysis for the SEE-stimulated and un-stimulated Jurkat cell data.
Fig. S5. Stimulation-induced PiSCES network is similar between control and alopecia areata patient groups.
Table S1. Validated antibody pairs used to identify each target in Jurkat cells.
Table S2. Phenotypic characteristics of alopecia areata patients and controls and their T cell populations.
Author contributions: S.E.P.S., S.C.N., B.K.R., D.G., C.N., and A.G.S. designed experiments; S.E.P.S., S.C.N., B.K.R., and T.R.D. performed experiments; G.F.S., C.N.W., and R.R.T. diagnosed and recruited patient donors, identified sites of active lesions, and acquired biopsies; S.E.P.S., S.C.N., J.P.S., J.E.E.-P., C.N., and A.G.S. analyzed and interpreted data; S.E.P.S., S.C.N., J.E.E.-P., C.N., and A.G.S. wrote the manuscript; A.G.S. conceived the project; and all authors contributed creative intellectual input and revised manuscript drafts.
Competing interests: The authors declare that they have no competing interests.
REFERENCES AND NOTES
- 1.Pawson T. Specificity in signal transduction: From phosphotyrosine-SH2 domain interactions to complex cellular systems. Cell. 2004;116:191–203. doi: 10.1016/s0092-8674(03)01077-8. [DOI] [PubMed] [Google Scholar]
- 2.Papin JA, Hunter T, Palsson BO, Subramaniam S. Reconstruction of cellular signalling networks and analysis of their properties. Nat. Rev. Mol. Cell Biol. 2005;6:99–111. doi: 10.1038/nrm1570. [DOI] [PubMed] [Google Scholar]
- 3.Pawson T. Dynamic control of signaling by modular adaptor proteins. Curr. Opin. Cell Biol. 2007;19:112–116. doi: 10.1016/j.ceb.2007.02.013. [DOI] [PubMed] [Google Scholar]
- 4.Komarova NL, Zou X, Nie Q, Bardwell L. A theoretical framework for specificity in cell signaling. Mol. Syst. Biol. 2005;1 doi: 10.1038/msb4100031. 2005.0023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jin J, Pawson T. Modular evolution of phosphorylation-based signalling systems. Philos. Trans. R. Soc. London Ser. B. 2012;367:2540–2555. doi: 10.1098/rstb.2012.0106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schrum AG, Gil D. Robustness and specificity in signal transduction via physiologic protein interaction networks. Clin. Exp. Pharmacol. 2012;2 doi: 10.4172/2161-1459.S3-001. S3.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kuroda K, Kato M, Mima J, Ueda M. Systems for the detection and analysis of protein–protein interactions. Appl. Microbiol. Biotechnol. 2006;71:127–136. doi: 10.1007/s00253-006-0395-5. [DOI] [PubMed] [Google Scholar]
- 8.Mueller M, Martens L, Apweiler R. Annotating the human proteome: Beyond establishing a parts list. Biochim. Biophys. Acta. 2007;1774:175–191. doi: 10.1016/j.bbapap.2006.11.011. [DOI] [PubMed] [Google Scholar]
- 9.Rual J-F, Venkatesan K, Hao T, Hirozane-Kishikawa T, Dricot A, Li N, Berriz GF, Gibbons FD, Dreze M, Ayivi-Guedehoussou N, Klitgord N, Simon C, Boxem M, Milstein S, Rosenberg J, Goldberg DS, Zhang LV, Wong SL, Franklin G, Li S, Albala JS, Lim J, Fraughton C, Llamosas E, Cevik S, Bex C, Lamesch P, Sikorski RS, Vandenhaute J, Zoghbi HY, Smolyar A, Bosak S, Sequerra R, Doucette-Stamm L, Cusick ME, Hill DE, Roth FP, Vidal M. Towards a proteome-scale map of the human protein–protein interaction network. Nature. 2005;437:1173–1178. doi: 10.1038/nature04209. [DOI] [PubMed] [Google Scholar]
- 10.Morell M, Avilés FX, Ventura S. Detecting and interfering protein interactions: Towards the control of biochemical pathways. Curr. Med. Chem. 2009;16:362–379. doi: 10.2174/092986709787002709. [DOI] [PubMed] [Google Scholar]
- 11.Menche J, Sharma A, Kitsak M, Ghiassian SD, Vidal M, Loscalzo J, Barabási AL. Disease networks. Uncovering disease-disease relationships through the incomplete interactome. Science. 2015;347:1257601. doi: 10.1126/science.1257601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Morris GP, Allen PM. How the TCR balances sensitivity and specificity for the recognition of self and pathogens. Nat. Immunol. 2012;13:121–128. doi: 10.1038/ni.2190. [DOI] [PubMed] [Google Scholar]
- 13.Roncagalli R, Hauri S, Fiore F, Liang Y, Chen Z, Sansoni A, Kanduri K, Joly R, Malzac A, Lähdesmäki H, Lahesmaa R, Yamasaki S, Saito T, Malissen M, Aebersold R, Gstaiger M, Malissen B. Quantitative proteomics analysis of signalosome dynamics in primary T cells identifies the surface receptor CD6 as a Lat adaptor–independent TCR signaling hub. Nat. Immunol. 2014;15:384–392. doi: 10.1038/ni.2843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Won JH, Goldberger O, Shen-Orr SS, Davis MM, Olshen RA. Significance analysis of xMap cytokine bead arrays. Proc. Natl. Acad. Sci. U.S.A. 2012;109:2848–2853. doi: 10.1073/pnas.1112599109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Langfelder P, Horvath S. WGCNA: An R package for weighted correlation network analysis. BMC Bioinformatics. 2008;9:559. doi: 10.1186/1471-2105-9-559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Linsley PS, Brady W, Grosmaire L, Aruffo A, Damle NK, Ledbetter JA. Binding of the B cell activation antigen B7 to CD28 costimulates T cell proliferation and interleukin 2 mRNA accumulation. J. Exp. Med. 1991;173:721–730. doi: 10.1084/jem.173.3.721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Boomer JS, Green JM. An enigmatic tail of CD28 signaling. Cold Spring Harb. Perspect. Biol. 2010;2:a002436. doi: 10.1101/cshperspect.a002436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Petukhova L, Duvic M, Hordinsky M, Norris D, Price V, Shimomura Y, Kim H, Singh P, Lee A, Chen WV, Meyer KC, Paus R, Jahoda CAB, Amos CI, Gregersen PK, Christiano AM. Genome-wide association study in alopecia areata implicates both innate and adaptive immunity. Nature. 2010;466:113–117. doi: 10.1038/nature09114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gilhar A, Etzioni A, Paus R. Alopecia areata. N. Engl. J. Med. 2012;366:1515–1525. doi: 10.1056/NEJMra1103442. [DOI] [PubMed] [Google Scholar]
- 20.Altan-Bonnet G, Germain RN. Modeling T cell antigen discrimination based on feedback control of digital ERK responses. PLOS Biol. 2005;3:e356. doi: 10.1371/journal.pbio.0030356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Guy CS, Vignali DAA. Organization of proximal signal initiation at the TCR:CD3 complex. Immunol. Rev. 2009;232:7–21. doi: 10.1111/j.1600-065X.2009.00843.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Longabaugh WJR. Combing the hairball with BioFabric: A new approach for visualization of large networks. BMC Bioinformatics. 2012;13:275. doi: 10.1186/1471-2105-13-275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schrum AG, Gil D, Dopfer EP, Wiest DL, Turka LA, Schamel WWA, Palmer E. High-sensitivity detection and quantitative analysis of native protein-protein interactions and multiprotein complexes by flow cytometry. Sci. STKE. 2007;2007:l2. doi: 10.1126/stke.3892007pl2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bida AT, Gil D, Schrum AG. Multiplex IP-FCM (immunoprecipitation-flow cytometry): Principles and guidelines for assessing physiologic protein–protein interactions in multiprotein complexes. Methods. 2012;56:154–160. doi: 10.1016/j.ymeth.2011.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Southwell AL, Smith SEP, Davis TR, Caron NS, Villanueva EB, Xie Y, Collins JA, Ye ML, Sturrock A, Leavitt BR, Schrum AG, Hayden MR. Ultrasensitive measurement of huntingtin protein in cerebrospinal fluid demonstrates increase with Huntington disease stage and decrease following brain huntingtin suppression. Sci. Rep. 2015;5:12166. doi: 10.1038/srep12166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kane LP, Lin J, Weiss A. Signal transduction by the TCR for antigen. Curr. Opin. Immunol. 2000;12:242–249. doi: 10.1016/s0952-7915(00)00083-2. [DOI] [PubMed] [Google Scholar]
- 27.Seet BT, Berry DM, Maltzman JS, Shabason J, Raina M, Koretzky GA, McGlade CJ, Pawson T. Efficient T-cell receptor signaling requires a high-affinity interaction between the Gads C-SH3 domain and the SLP-76 RxxK motif. EMBO J. 2007;26:678–689. doi: 10.1038/sj.emboj.7601535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Smith-Garvin JE, Koretzky GA, Jordan MS. T cell activation. Annu. Rev. Immunol. 2009;27:591–619. doi: 10.1146/annurev.immunol.021908.132706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Davis FP, Braberg H, Shen MY, Pieper U, Sali A, Madhusudhan MS. Protein complex compositions predicted by structural similarity. Nucleic Acids Res. 2006;34:2943–2952. doi: 10.1093/nar/gkl353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Armstrong JD, Pocklington AJ, Cumiskey MA, Grant SG. Reconstructing protein complexes: From proteomics to systems biology. Proteomics. 2006;6:4724–4731. doi: 10.1002/pmic.200500895. [DOI] [PubMed] [Google Scholar]
- 31.Shoemaker BA, Panchenko AR. Deciphering protein-protein interactions. Part II. Computational methods to predict protein and domain interaction partners. PLOS Comput. Biol. 2007;3:e43. doi: 10.1371/journal.pcbi.0030043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kholodenko B, Yaffe MB, Kolch W. Computational approaches for analyzing information flow in biological networks. Sci. Signal. 2012;5:re1. doi: 10.1126/scisignal.2002961. [DOI] [PubMed] [Google Scholar]
- 33.Srihari S, Leong HW. A survey of computational methods for protein complex prediction from protein interaction networks. J. Bioinform. Comput. Biol. 2013;11:1230002. doi: 10.1142/S021972001230002X. [DOI] [PubMed] [Google Scholar]
- 34.Tyers M, Mann M. From genomics to proteomics. Nature. 2003;422:193–197. doi: 10.1038/nature01510. [DOI] [PubMed] [Google Scholar]
- 35.Uhlén M, Fagerberg L, Hallström BM, Lindskog C, Oksvold P, Mardinoglu A, Sivertsson Å, Kampf C, Sjöstedt E, Asplund A, Olsson I, Edlund K, Lundberg E, Navani S, Szigyarto CA-K, Odeberg J, Djureinovic D, Takanen JO, Hober S, Alm T, Edqvist P-H, Berling H, Tegel H, Mulder J, Rockberg J, Nilsson P, Schwenk JM, Hamsten M, von Feilitzen K, Forsberg M, Persson L, Johansson F, Zwahlen M, von Heijne G, Nielsen J, Pontén F. Proteomics. Tissue-based map of the human proteome. Science. 2015;347:1260419. doi: 10.1126/science.1260419. [DOI] [PubMed] [Google Scholar]
- 36.Ermann J, Rao DA, Teslovich NC, Brenner MB, Raychaudhuri S. Immune cell profiling to guide therapeutic decisions in rheumatic diseases. Nat. Rev. Rheumatol. 2015;11:541–551. doi: 10.1038/nrrheum.2015.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Abraham RT, Weiss A. Jurkat T cells and development of the T-cell receptor signalling paradigm. Nat. Rev. Immunol. 2004;4:301–308. doi: 10.1038/nri1330. [DOI] [PubMed] [Google Scholar]
- 38.Werlen G, Hausmann B, Palmer E. A motif in the αβT-cell receptor controls positive selection by modulating ERK activity. Nature. 2000;406:422–426. doi: 10.1038/35019094. [DOI] [PubMed] [Google Scholar]
- 39.Clark RA, Chong BF, Mirchandani N, Yamanaka K-I, Murphy GF, Dowgiert RK, Kupper TS. A novel method for the isolation of skin resident T cells from normal and diseased human skin. J. Invest. Dermatol. 2006;126:1059–1070. doi: 10.1038/sj.jid.5700199. [DOI] [PubMed] [Google Scholar]
- 40.Smith SEP, Neier SC, Davis TR, Pittelkow MR, Gil D, Schrum AG. Signalling protein complexes isolated from primary human skin-resident T cells can be analysed by Multiplex IP-FCM. Exp. Dermatol. 2014;23:272–273. doi: 10.1111/exd.12362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schrum AG. Visualization of multiprotein complexes by flow cytometry. Curr. Protoc. Immunol. 2009;Chapter 5(Unit 5.9) doi: 10.1002/0471142735.im0509s87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Langfelder P, Horvath S. Fast R Functions for Robust Correlations and Hierarchical Clustering. J. Stat. Softw. 2012;46:i11. [PMC free article] [PubMed] [Google Scholar]
- 43.Cowley MJ, Pinese M, Kassahn KS, Waddell N, Pearson JV, Grimmond SM, Biankin AV, Hautaniemi S, Wu J. PINA v2.0: Mining interactome modules. Nucleic Acids Res. 2012;40:D862–D865. doi: 10.1093/nar/gkr967. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.