Abstract
Alkylation chemotherapy is one of the most widely used systemic therapies for cancer. While somewhat effective, clinical responses and toxicities of these agents are highly variable. A major contributing factor for this variability is the numerous distinct lesions that are created upon alkylation damage. These adducts activate multiple repair pathways. There is mounting evidence that the individual pathways function cooperatively, suggesting that coordinated regulation of alkylation repair is critical to prevent toxicity. Furthermore, some alkylating agents produce adducts that overlap with newly discovered methylation marks, making it difficult to distinguish between bona fide damaged bases and so called ‘epigenetic’ adducts. We discuss new efforts aimed at deciphering the mechanisms that regulate these repair pathways, emphasizing their implications for cancer chemotherapy.
Keywords: Alkylation chemotherapy, MGMT, AlkB, base excision repair
The Complexity of DNA Alkylation Damage
The vital importance of genome maintenance is underscored by the evolution of multiple highly conserved repair mechanisms, each of which function on a specific type or class of damaged DNA. Even within a specific category of DNA damage, the machinery utilized for repair varies due to the chemistry of the damage, the damage context, differential expression or post-translational modification of repair factors, the chromatin state and the cell cycle. This complexity is particularly evident for DNA alkylation damage. Compounds that transfer alkyl groups to nucleic acids are typically broadly reactive, each capable of producing several distinct DNA lesions [1]. While the sources of endogenous alkylation damage are not clearly defined, S-adenosylmethionine may directly methylate DNA at a level that is biologically significant [2]. To counter both endogenous and exogenous alkylation, multiple distinct alkylation repair pathways have evolved, giving the cell a wide-ranging arsenal to protect its genome from such damage. In turn, chemotherapeutics that induce alkylation damage are one of the most commonly used for cancer, highlighting the clinical relevance of alkylation repair [1, 3].
The simplest type of DNA alkylation – the transfer of a single methyl group to a DNA base – demonstrates the diverse nature of such damage. Methyl donors may react with ring nitrogen (N) or oxygen (O) atoms to generate twelve distinct base lesions in DNA [1]. The proportion of each lesion depends partly on the chemical nature of the methyl donor (e.g., SN1 versus SN2 nucleophilic substitution) and the DNA substrate (single-stranded versus double-stranded). Due to its high nucleophilicity, the predominant methylation adduct produced is N7-methylguanine (7meG), which accounts for ∼75% of the total methylation lesions in DNA [4]. While relatively innocuous on its own, 7meG is prone to depurination, leading to the formation of an abasic site (see Glossary), which is potentially toxic and mutagenic [5]. By contrast, many of the remaining lesions, such as N3-methyladenine (3meA) or N1-methyladenine (1meA), are intrinsically cytotoxic due to their ability to block replicative DNA polymerases [6]. However, translesion polymerases, such as DNA polymerase zeta (ζ), can bypass such lesions but are error prone [7]. Further, lesions such as 3meA can initiate methylation-induced recombination in order to tolerate these normally toxic, polymerase blocking lesions [8]. Others adducts, such as O6-methylguanine (O6meG), are mutagenic because they readily mispair during replication [9]. It is therefore not surprising that combating simple methylation damage requires at least three distinct repair pathways. These include direct demethylation by O6-methylguanine DNA methyltransferase (MGMT) and the AlkB family of enzymes, as well as base excision repair (BER; see Box 1). Larger alkylation adducts that induce DNA helix distortion activates nucleotide excision repair (NER) and bifunctional agents such as cyclophosphamide, melphalan, and busulphan may produce interstrand crosslinks, activating the Fanconi anemia (FA) pathway (these two pathways are reviewed elsewhere in [10, 11]). The complex nature of the lesions created by alkylation and the numerous pathways involved in their reversal necessitates coordination for proper repair. In the past decade, many studies have revealed critical insights into the regulatory mechanisms that govern alkylation damage repair. These mechanisms include transcriptional and epigenetic regulation of the repair enzymes, as well as post-translational and metabolite-mediated control mechanisms. Here we discuss these regulatory mechanisms, many of which have important implications for improving personalized cancer therapy as well as for minimizing chemotherapeutic side effects.
Box 1. Primary methylation/alkylation damage repair pathways.
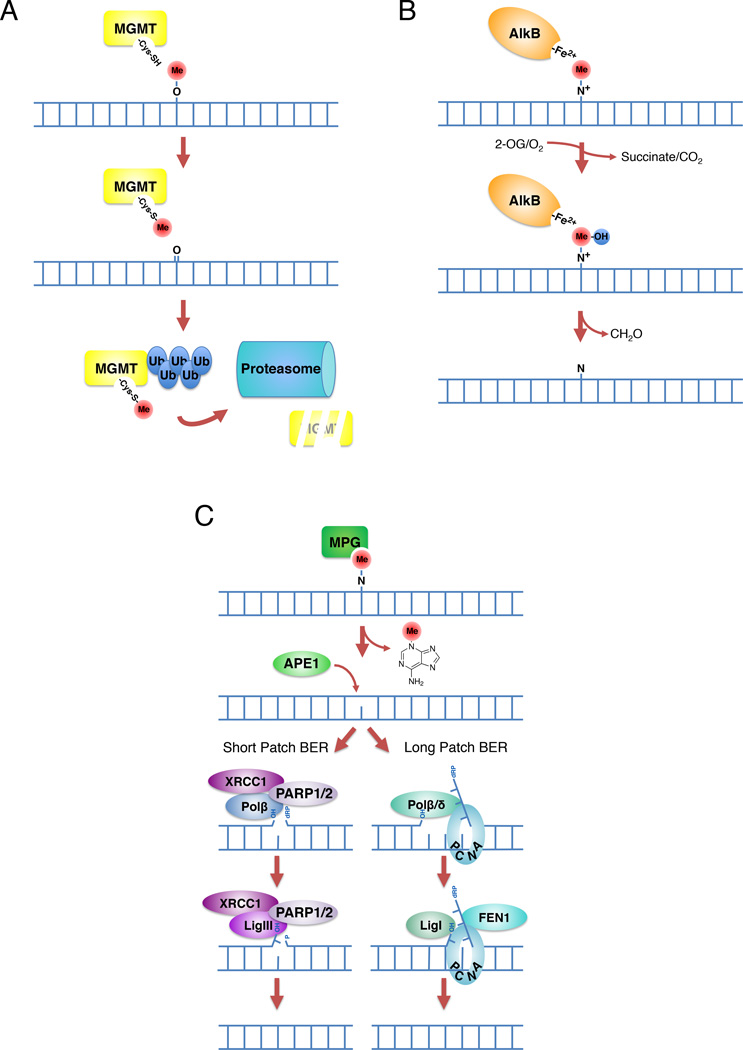
Depending on the lesion, DNA methylation damage may be reversed by MGMT, the AlkB demethylases, or base excision repair (BER). MGMT repairs O-linked lesions such as O6meG induced by most clinically tractable alkylating agents, including TMZ, dacarbazine, streptozotocin, procarbazine (Figure IA). The similarly cytotoxic MGMT substrate O6-ClEtG (modified with a chloroethyl adduct) is produced by BCNU and other chlorethylating agents [13, 102]. MGMT is not a true enzyme but instead functions by direct transfer of a methyl/alkyl group from the base to a cysteine in its catalytic site through an irreversible reaction. Once MGMT has removed an alkyl group, it is ubiquitinated and targeted for degradation by the proteasome [103]. Because this reaction is not enzymatic, the capacity for repair by MGMT is directly related to the abundance of available MGMT protein. In contrast, the AlkB family of demethylases repair N-linked adducts occurring on the Watson-Crick base-pairing interface, such as 1meA and 3meC lesions, which are minor lesions induced by SN2 alkylators (Figure IB) [1]. Two mammalian AlkB members, ALKBH2 and ALKBH3, directly repair these DNA lesions by an iron and 2-oxoglutarate (2-OG) dependent oxidative demethylation reaction that reverts the base to its unmodified state [104]. This reaction produces succinate and carbon dioxide, with the methyl group being released as formaldehyde [105, 106]. The predominant N-linked methylation lesion, 3meA, is not repaired by the AlkB pathway but instead relies on base excision repair (BER; Figure IC). BER operates through a multistep mechanism that repairs the damaged base by the recognition and removal of a single nucleotide (short patch BER) or a small stretch of nucleotides (long patch BER) [107]. Both pathways are initiated by removal of the aberrant base by a DNA glycosylase. While there are 11 distinct mammalian glycosylases [108], the predominant glycosylase relevant for alkylation repair is AAG. The removal of the base results in the formation of an abasic site. Next, the DNA backbone is nicked by the AP endonuclease APE1. Short patch BER repair is completed by gap filling with DNA polymerase β (Polβ) in association with its scaffolding partner XRCC1, and ligation with DNA ligase I or III (LIG 1/3). While not essential for BER, Poly(ADP-ribose)-polymerase (PARP) may serve to facilitate recruitment of certain BER factors [109]. Long patch BER repair is completed by gap filling with DNA polymerase δ/ε (in proliferating cells) or Polβ, removal of the small stretch of bases by flap endonuclease 1 (FEN1) / proliferating cell nuclear antigen (PCNA), and ligation with LIG1 [109].
Figure I. Pathways of methylation/alkylation damage reversal. (A) Direct reversal of O-linked lesions by methylguanine methyltransferase (MGMT). The methyl group is transferred to a catalytic cysteine, after which MGMT is ubiquitinated and targeted to the proteasome. (B) AlkB-mediated demethylation of N-linked lesions. Initial oxidation of the methyl moiety leads to its hydrolysis to formaldehyde. (C) Repair of N-linked lesions by base excision repair (BER).
MGMT expression status
A major cytotoxic DNA lesion induced by alkylating agents such as temozolomide (TMZ) and similar agents is O6meG, which is repaired by MGMT (also known as AGT; Box 1 and Figure IA). Increased MGMT expression provides resistance to TMZ and similar alkylating agents due to increased removal of the O6-adducts on guanine [12, 13]. Thus, treatment with TMZ and similar agents is more effective for tumors that are deficient in MGMT expression (Figure 1A). Loss of MGMT expression in tumors is generally the result of methylation and inactivation of the MGMT promoter [14]. As such, methylation of the MGMT promoter in glioma is a useful predictor of the responsiveness of the tumor to alkylating agents [15]. The development of methylation-specific PCR (MSP) provides for a highly sensitive and accurate measurement of the methylation status of the MGMT promoter within the tumor tissue. Methylation at discrete regions within CpG islands of a given gene promoter results in gene silencing [16], an epigenetic marker commonly seen in human cancers [17]. MSP is now the method of choice for analysis of promoter methylation and gene silencing, with predictive power for determining TMZ responsiveness and survival in glioma [18].
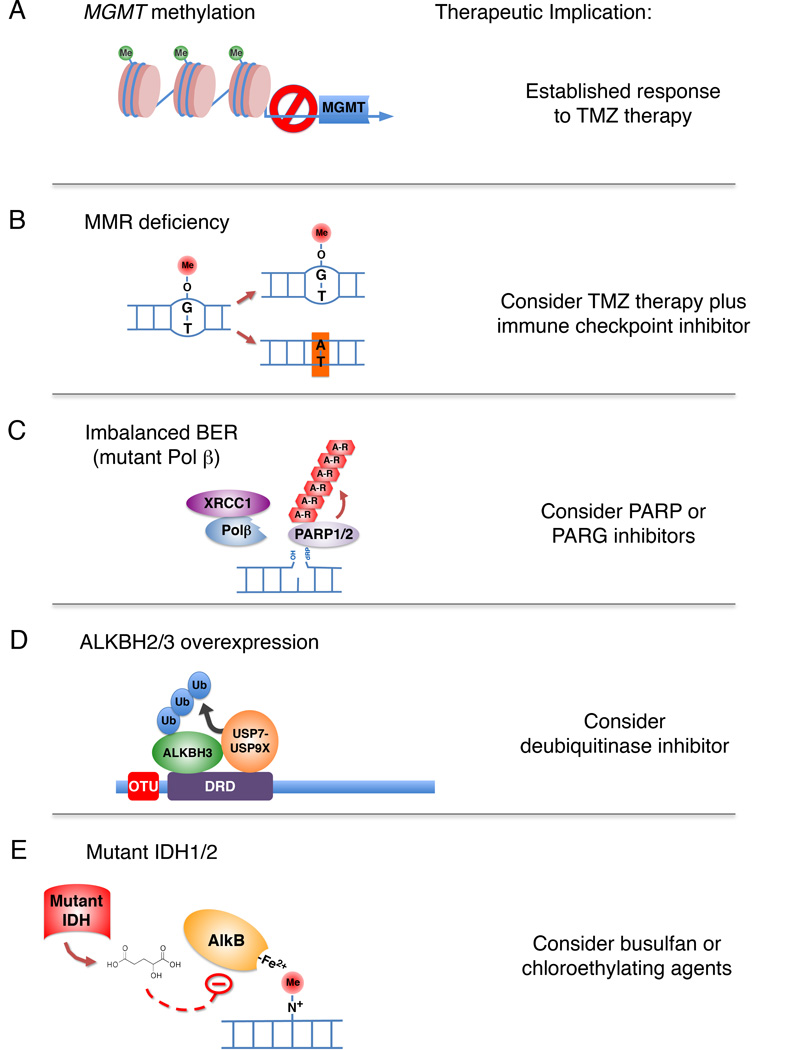
Figure 1. Biochemical mechanisms of alkylation damage repair regulation relevant to tumor therapy.
(A) MGMT gene silencing by DNA methylation leads to improved tumor responses to monomethylating agents such as TMZ. (B) Loss of mismatch repair leads to increased transition mutations, which may create higher rates of neoantigen expression upon alkylation damage. A combination of alkylation chemotherapy and immune checkpoint blockade may be effective. (C) Expression of mutant DNA Polβ may result in an imbalance in BER. In certain cases of imbalanced BER, inhibition of PARP or PARG may be a viable therapeutic. (D) Increased expression of ALKBH2/3 due to the OTUD4/USP7/USP9X deubiquitinase pathway may be countered using small molecule inhibitors against USP7 or USP9X. (E) IDH mutant tumors express the oncometabolite 2-hydroxyglutarate, which inhibits AlkB type demethylases. In these tumors the alkylating agent busulfan or chlorethylating alkylating agents may be more effective.
As a chemotherapeutic approach to cancer, numerous strategies have been developed to limit the repair of the O6meG lesion, either by depleting or inhibiting MGMT. MGMT activity can be successfully blocked by addition of free guanine base derivatives, with alkyl groups at the O6 position, which act as a pseudosubstrate and lead to MGMT depletion [19]. Two of the most promising MGMT-specific drugs are O6-benzylguanine (BG) and O6-(4-bromothenyl)guanine (Patrin, PaTrin-2, Lomeguatrib) [20–22]. It remains to be seen whether these agents have clinical efficacy against tumors, or whether they merely increase the toxicity of alkylating agents.
Mismatch repair and MGMT modulation of DNA lesion toxicity
As previously mentioned, the O6meG DNA adduct is not inherently cytotoxic. Instead, its cytotoxicity requires the presence of an intact mismatch repair (MMR) pathway (reviewed in [1]). Normally, the O6meG adduct is stable if not repaired by MGMT. However, either cytosine or thymine may be inserted opposite the O6meG DNA, resulting in potential G:C to A:T transition mutations [9]. This would be consistent with increased frequency of point mutations globally, including those of oncogenes and tumor suppressor genes, in tumors with a loss of MGMT expression [23]. Interestingly, the formation of these mutations or ‘neoantigens’ may be the cause for immune checkpoint selectivity for MMR defective tumors [24–26]. The cytotoxicity of the O6meG adduct stems from the replication-dependent formation of the O6meG:T mispair and the recognition of this mispair by the MMR machinery. This cytotoxicity may be explained by two different models. In the futile cycle model, the MutSα complex recognizes and binds to the O6meG:T mispair, recruiting the MutLα complex to the mispair to initiate repair [27–29]. Since this process involves removal and resynthesis of the T-containing DNA strand, the O6meG:T mispair is regenerated, activating MMR once again. It is proposed that continued rounds of repair may lead to the formation of double-strand breaks and eventually cell death [27–30]. Recently, it has also been suggested that DNA Polymerase ß (Polß) may play a role in the processing of these MMR substrates, providing a level of crosstalk between MMR and BER [31, 32]. In the direct DNA damage signaling model, MutSα binds to the O6meG:T mispair and without processing, recruits MutLα and the DNA damage response proteins ATR and ATRIP [NN1]to initiate DNA damage checkpoints [33]. This may lead to cell cycle arrest and apoptosis, but this in vitro study has not been confirmed. Regardless of the precise mechanism utilized by MMR proteins, their presence is required for TMZ-mediated cytotoxicity.
Loss of MMR function is an important event in the development of certain tumors, such as colorectal and gastric carcinomas [34–37]. In addition, impairment of mismatch repair by epigenetic inactivation of one or more MMR genes may play an important role in determining the responsiveness of malignant gliomas to adjuvant therapy [38] (Figure 1B). Whereas improved prognosis has been reported in tumors with loss of MGMT expression due to promoter methylation, poor prognosis is observed when MMR capacity is compromised by methylation of the promoter for essential MMR genes in glioma [39], as well as mutations in key MMR genes [40]. For example, loss of expression or inactivating mutations in MSH6 have been observed in TMZ-resistant gliomas and in recurrent tumors following TMZ therapy [41]. Interestingly, a clinical study suggested that MMR deficiency as measured by microsatelite instability (MSI), was not responsible for TMZ resistance in malignant glioma [42]. However, a more recent study demonstrated that modest reductions in MMR factors were sufficient to induce TMZ resistance without causing MSI [43]. It is likely that several mechanisms, including MGMT levels, MMR status, as well as homologous recombination [44] contribute to alkylation damage responses.
Coordination of the steps within Base Excision Repair
Unlike the direct reversal mechanisms, the multi-step nature of BER creates several repair intermediates for each alkylated base that is repaired (Box 1 and Figure IC). These include an abasic site, 5’-deoxyribose phosphate, and a single-stranded break (SSB), all of which are themselves potentially cytotoxic and mutagenic lesions [45]. Therefore, the steps in BER need to be tightly coordinated with one another to avoid the production and accumulation of these undesirable intermediates. Genetic studies in numerous systems, including yeast, human cells, and mouse transgenic models, have demonstrated that increased expression of individual BER factors can be highly detrimental due to loss of this coordination [31, 46–51]. For example, overexpression of alkyladenine glycosylase (AAG; also known as methylpurine glycosylase or MPG) paradoxically increases cellular sensitivity to the alkylating agent methyl-methane sulphonate (MMS) [46]. This results in higher rates of frameshift mutagenesis and microsatellite instability. Mechanistically, this is caused by an increase in abasic site production and/or DNA breaks triggering cell death mediated by PARP1 hyper-activation [46]. Hypersensitivity to alkylating agents can also be observed in cells where AP endonuclease (APE1) is inhibited, or upon reduced expression of DNA Polβ [52]. When AP site repair is blocked with the DNA modifying agent methoxyamine, the AP site is chemically modified and blocks further BER mediated repair, sensitizing cells to alkylating agents [53]. Interestingly, overexpression of Polβ prevent the sensitization effect of methoxyamine, suggesting that either BER is a tightly linked process or that the in vivo substrate for methoxyamine is the cleaved abasic site that is normally a substrate for Polβ [52]. Loss of AAG in PolB-deficient mouse cells rescues alkylation hypersensitivity, strongly suggesting that this phenotype is due to the absence of downstream processing of AAG-induced abasic sites [48, 54]. Deficiency of XRCC1, a scaffold factor involved in BER, also causes alkylation damage sensitivity downstream of the initial steps in BER, resulting in SSB accumulation [55].
The rapid and coordinated assembly of these factors during alkylation damage is therefore crucial for proper BER, and poly-ADP-ribosylation has been proposed to be a post-translational modification that promotes assembly of BER factors downstream of APE1-induced incision [45]. PARP binds to SSBs, where it is activated, and adds PAR chains to itself and other BER factors, promoting recruitment of XRCC1, Polβ, and DNA ligase III [56]. While poly-ADP-ribosylation is not absolutely required for BER, Parp1−/− mice are sensitive to alkylating agents [57], and it was suggested as early as 1977 that PARP inhibitors may be potentially useful for increasing sensitization of tumors to chemotherapy [58]. However, such an approach has to be carefully integrated into chemotherapy as certain tumors already have an imbalance in BER, exemplified by mutations in Polβ commonly seen in colorectal and other cancers [59, 60]. Reduced Polβ activity sensitizes tumors to alkylating agents, and inhibition of PARP in this context largely provides short-term rescue of the induced necrosis, suggesting that PARylation also functions to signal cell death when a BER intermediate accumulates [46]. However, this imbalance in BER that produces an elevation of PARP1 signaling (discussed later) also promotes long-term sensitization to PARP inhibitors and likely PARG inhibitors [61–63]. Therefore, identifying a preexisting BER imbalance within a tumor may be critical for determining whether PARP inhibition will be useful or counterproductive in combination with alkylating chemotherapy (Figure 1C). Further, targeting PARG, which removes PAR chains from substrates, would appear to be an alternative option in tumors with an intrinsic BER imbalance [52, 64]. Regardless, knowing the BER status of a given tumor will provide important insight for rational chemosensitizing strategies.
Importantly, ubiquitylation is also a key regulator of the steps of BER. As shown recently, Polβ is ubiquitylated on K206/K244 when not bound to XRCC1 in a process that is regulated by the cell cycle and DNA damage [65]. In turn, XRCC1 is then a substrate for either HSP90 or the E3 ligase CHIP. It is therefore interesting to speculate that BER may also be targeted (e.g., in homologous recombination defective tumors) via selective regulation of ubiquitylation to induce a loss of Polβ, XRCC1 or both of these critical BER proteins.
Relationship between base excision repair and metabolism
There is an intimate relationship between (i) the mechanism of BER; (ii) the synthesis and degradation of PAR followed by signaling via ADP-ribosylation and (iii) the biosynthesis and availability of the coenzyme nicotinamide adenine dinucleotide (NAD+). PARP1 promotes BER by facilitating strand-break recognition and signaling to recruit XRCC1 and Polβ to the site of the lesion (Figure IC) [65]. In turn, the activation of PARP1 requires NAD+, a small metabolite coenzyme that is an essential substrate for ADP(ribosyl)ation reactions mediated by the ADP-ribosyltransferase enzymes (ARTDs/PARPs) and for deacetylation reactions catalyzed by the sirtuins (SIRTs). As summarized recently [66], the major ARTD/PARP family member, PARP1 (ARTD1), is involved both in DNA repair and functions as a trigger of cell death resulting from its hyperactivation and the resulting energy depletion. Cell death due to PARP1 activation was originally suggested to involve energy metabolite (NAD+ and ATP) depletion [67]. This PARP1 hyperactivation-induced cell death is especially evident upon DNA damage of cells deficient in base excision repair. The failure of cells to complete repair once exposed to alkylating agents results in uncontrolled PARP1 activation and enhanced cell death [46]. This has also been demonstrated in mice following ischemia reperfusion injury [49], nephrotoxicity [68] or alkylation-induced tissue damage [50]. This appears to be the result of PARP1 activation, which triggers both a loss of NAD+, which mediates a defect in oxidative phosphorylation (mitochondrial dysfunction), as well as a poly-ADP-ribose mediated block to glycolysis [69]. This latter report proposes a working model in which PARP1 hyper-activation leads to inhibition of hexokinase (HK1), a reduction in cellular glycolysis and depletion in cellular ATP pools. This effect of PARP1 activity coupled with NAD+ depletion might explain why unrepaired DNA strand breaks and BER intermediates increase cell sensitivity to DNA alkylation damage and PARP1 activation.
In turn, the elevated PARP1 activation observed following DNA damaging agent treatment (when BER is defective) may also offer a selective approach for tumor cell killing in BER defective tumors. For example, the dependence on NAD+ for the hyper-activation of PARP1 would imply that NAD+ biosynthesis is essential. In support of this, it has been demonstrated that clinically available chemical inhibitors of NAD+ biosynthesis enhances the cell killing effect of DNA damaging agents when BER is defective or inhibited [70]. More recently, inhibitors of NAD+ biosynthesis have been suggested to be considered for cancer treatment modalities in general [71].
Cooperation between BER and AlkB dealkylases
The existence of multiple alkylation damage repair pathways provides potential redundancy to ensure repair even if one pathway is defective. The AlkB homologues ALKBH2 and ALKBH3 can repair a similar range of lesions (see Box 1), although ALKBH2 repairs lesions in double-stranded DNA and ALKBH3 prefers single-stranded substrates [72]. However, the association of ALKBH3 with the ASCC3 DNA helicase may expand the substrate range of ALKBH3 to include dsDNA [73]. Alkbh2−/− mice accumulate spontaneous genomic 1meA lesions while Alkbh3−/− mice do not have this phenotype, suggesting that ALKBH2 is the major demethylase for endogenous 1meA [74]. Interestingly, subsequent studies showed that Alkbh2−/− Alkbh3−/− double-knockout (KO) mice are more susceptible to alkylation-induced tumor development relative to Alkbh2−/− mice, suggesting that both enzymes play an essential role in alkylation resistance, albeit to different degrees [75]. This study also demonstrated that Aag−/−Alkbh2−/− Alkbh3−/− triple KO mice are highly sensitive to the inflammatory agent dextran sodium sulfate (DSS), which induces exocyclic ε-base lesion production. Consistently, these triple KO mice accumulated significantly greater εA (1,N6-ethanoadenine) and εG (1,N2-ethanoguanine) lesions relative to Aag−/− mice. This in vivo work supports previous studies that both BER and the AlkB demethylases are capable of repairing these more complex alkylation lesions in vitro [76–78].
Although AAG and the AlkB proteins function redundantly for certain DNA lesions, accumulating evidence suggests that they may compete for other adducts. In comparison to other etheno lesions, AAG binds relatively tightly to εC (3,N4-ethenocytosine) but is incapable of excising this base [79, 80]. ALKBH2 demethylates this lesion in vitro, but the presence of AAG inhibits this activity in a competitive fashion [77]. Whether this competition is relevant in vivo is unclear. The AAG-εC complex may serve as a signal to recruit ALKBH2 or other relevant factors. However, the AAG-sC complex may itself be highly toxic as it could block the replication or transcription machinery. This complex is reminiscent of the alkyltransferase-like 1 (Atl1) protein in yeast, which is homologous to MGMT but is catalytically inactive [81]. By having higher affinity to bulkier O6-alkylguanine lesions, Atl1 arrests RNA polymerase and diverts the repair pathway to transcription-coupled nucleotide excision repair [81]. It is tempting to speculate that AAG recognition of εC functions similarly as Atl1, to signal recruitment of certain alkylation repair factors, such as ALKBH2, and exclude others in order to properly match the repair activity with the lesion.
Regulation of human AlkB homologues by ubiquitination
Understanding the regulation of ALKBH2 and ALKBH3 is critical as both are overexpressed in certain tumors, such as prostate adenocarcinoma and non-small cell lung carcinoma [82, 83]. While alkylating agents are not typically used for these two types of cancer, the differential expression of these proteins may be factors that determine the potential success of alkylation chemotherapy in other tumors. Overexpression of these demethylases may promote resistance to treatment by promoting more extensive alkylation repair [84]. Recent work has demonstrated that both of these enzymes are regulated by the ubiquitin-proteasome system. Both enzymes are modified by K48-linked ubiquitination [85], which nominally targets proteins for proteasomal degradation [86]. A complex of deubiquitinases positively regulates these dealkylases by countering this ubiquitination. Central to this pathway is the deubiquitinase OTUD4, whose catalytic activity is dispensable for stabilizing ALKBH2/ALKBH3. Instead, OTUD4 functions as a scaffold to link the AlkB homologues to two additional deubiquitinases, USP7 and USP9X [85]. In turn, loss of USP7 or USP9X destabilizes the dealkylases resulting in alkylation hypersensitivity. Unlike OTUD4, however, the deubiquitinase activities of USP7 and USP9X were observed to be important for their stabilization function. Interestingly, MGMT stabilization also appears to depend upon the OTUD4/USP7/USP9X pathway (N.M., unpublished observations). This suggests that human cells may have a master regulatory complex for enzymes involved in the direct demethylation repair of DNA. A number of small molecule deubiquitinase inhibitors that target USP7 and USP9X are already available [87, 88]; these may provide a novel approach for chemotherapy sensitization of tumors that targets multiple repair pathways with a single agent (Figure 1D).
Metabolite regulation of the AlkB dealkylases
The AlkB dealkylases belong to a large superfamily of Fe+2 and 2-oxoglutarate (2-OG) dependent dioxygenases, which includes the JmjC histone demethylases and the TET family of DNA 5-methylcytosine hydroxylases [89, 90]. These chromatin-modifying enzymes are susceptible to inhibition by D-2-hydroxyglutarate (D-2HG), an oncometabolite that accumulates in tumor cells harboring mutations in IDH [91]. By blocking the demethylation and hydroxylation activities of the JmjC and TET enzymes, respectively, lineage-specific cell differentiation is blocked, contributing to the malignancy of IDH-mutant tumors. Through a similar mechanism, D-2HG inhibits the alkylation damage repair activities of ALKBH2 and ALKBH3 in vitro [91]. Consistently, IDH-mutant expressing cells display significantly slower kinetics for the repair of AlkB substrates such as 1meA, which appear to evolve into DNA double-stranded breaks and cause apoptosis. In addition, IDH-mutant cells have significantly increased sensitivity to several alkylating agents, including busulfan and CCNU, which are commonly used to treat chronic myelogenous leukemia and glioma, respectively [91]. Therefore, the production of D-2HG by IDH-mutant tumors may be a double-edged sword; while contributing to the cancerous phenotype, it also results in increased susceptibility to certain alkylating agents by inhibiting the AlkB enzymes. Thus, patients harboring IDH-mutant tumors may benefit from these alkylating agents but may also require lower doses for tumor responses, reducing their toxic side effects (Figure 1E). It will be interesting to determine whether other tumor-associated metabolic changes, such as decreased oxygen tension in the tumor vicinity, is of a sufficient degree to also inhibit the AlkB dioxygenases and therefore increase alkylation sensitivity.
Physiological versus pathological methylation
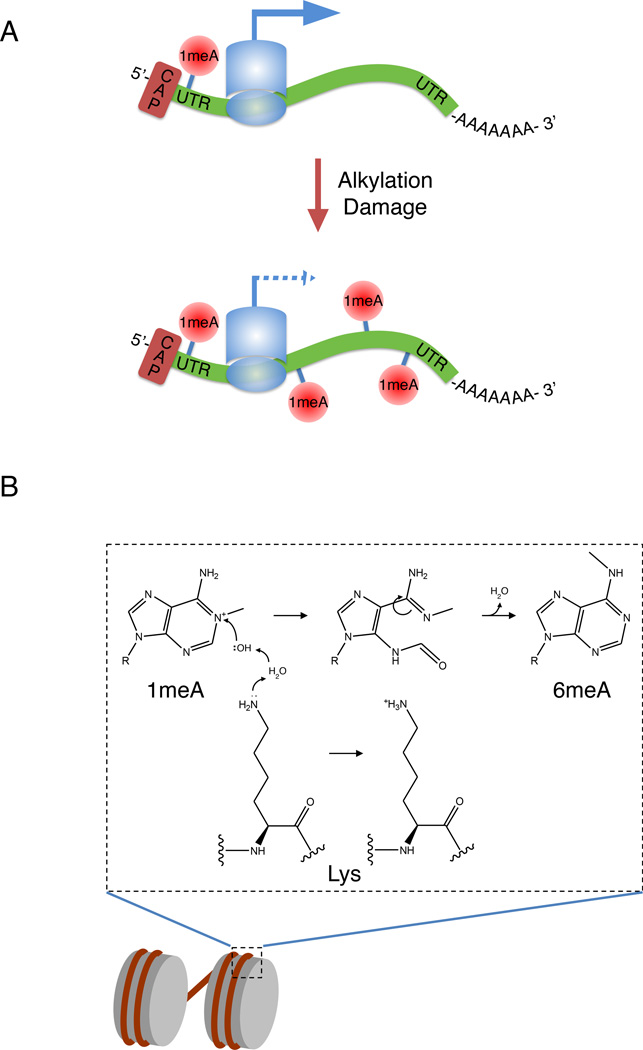
While the canonical AlkB homologues ALKBH2 and ALKBH3 are responsible for the reversal of bona fide alkylation adducts, some of their substrates, such as 1meA, may serve physiological functions, at least in RNA, where it associates with the 5’-UTRs of actively translated mRNAs [92]. The fact that ALKBH3 can demethylate 1meA in RNA may therefore serve to regulate the physiological function of this RNA modification, as well as to protect RNA from alkylation damage [72, 93]. Whether cells have the capacity to make this distinction between physiological versus alkylating agent-induced 1meA is not clear. The induction of 1meA by an alkylating agent likely induces this modification in an inappropriate region of an mRNA, potentially leading to inhibition of translation or ribosomal miscoding, a phenomenon previously demonstrated for O6meG in mRNA [94]. We term this blurring between what is considered damage-induced methylation versus physiological or epigenetic methylation as ‘epigenetic confusion’ (Figure 2A). This may also come into play with 6-methyladenine (6meA), which was recently shown to exist in the genomes of higher eukaryotes, including human cells [95]. While methyltransferases that catalyze the formation of 6meA in DNA have been found in certain eukaryotes such as Caenorhabditis elegans [96], they have yet to be discovered in mammals, for which its epigenetic function is currently unclear. Previous in vitro studies demonstrated that 6meA is produced from 1meA through the base-catalyzed Dimroth rearrangement [97]. Although this has not yet been shown to occur in vivo, the packaging of DNA into chromatin provides a basic environment, where the presence of lysine and arginine residues in the histones may promote this isomerization (Figure 2B). In cells where 1meA is not efficiently repaired, a fraction of the 6meA may be derived from 1meA in vivo. Therefore the rapid recruitment of ALKBH2/ALKBH3 would be essential to prevent this potential inter-conversion, which would leave an epigenetic mark where it may not be desirable. At least one mechanism of ALKBH2 recruitment has been reported; a direct interaction between ALKBH2 and PCNA targets this repair protein to replication foci [98, 99]. This interaction increases during DNA replication, suggesting that ALKBH2 recruitment is a cell cycle regulated process. Whether other alkylation repair factors are mobilized specifically in response to alkylation damage is unclear. Certain BER factors, such as XRCC1 and DNA Polβ form nuclear foci upon damage, suggesting that such a mechanism may exist [65]. However, whether this response is lesion specific or simply downstream of single-stranded break formation is unknown. Further investigation will be needed to determine the upstream signaling responsible for alkylation repair factor recruitment.
Figure 2. Potential mechanisms of alkylation damage-induced ‘epigenetic confusion’.
(A) 1-meA is associated with 5’ UTRs of actively translated mRNAs. Induction of alkylation damage may hinder translation if 1-meA is present in the mRNA coding region. (B) Formation of 6meA from 1meA through Dimroth rearrangement. In the chromatin environment, residues such as lysine may promote nucleophilic attack of 1meA, leading to a ring-opened intermediate, which rearranges to form 6meA. As 6meA may have an epigenetic role in DNA, this conversion may lead to its inappropriate placement and function.
Concluding remarks
The multifactorial nature of alkylation damage, its repair and regulation poses significant challenges for determining the optimal chemotherapeutic treatment for a given tumor, and may explain why the responses of tumors to alkylating agents are often highly variable. Clearly, a quantitative measurement of a single repair protein has predictive power for alkylation therapy outcomes, as has been shown for MGMT in human glioblastoma [100]. However, considering expression analysis of multiple alkylation repair factors and regulators may provide more information regarding how an individual tumor may respond to alkylating chemotherapy. This type of analysis may also be beneficial for determining which type of alkylating agent will be more useful, and whether related therapies such as PARP inhibitors should be included. An alternative may be high-throughput functional assays, which simultaneously measure DNA repair capacity for multiple pathways in the same cell [101]. Future work is needed to determine which approach provides greater predictive ability for determining chemotherapeutic agent responses for personalized cancer treatment.
Outstanding Questions.
Are the individual alkylation damage repair pathways coordinated with one another?
Cells encode 1meA and 3meC RNA methyltransferases. How do cells regulate these enzymes to prevent off-target effects?
Bacteria have an adaptive response to alkylation damage. Is there an alkylation specific DNA damage response in metazoans?
Can the inhibition of alkylation repair pathways improve responses to alkylation therapy in combination with other anti-cancer therapies such as immune checkpoint inhibitors?
Is RNA alkylation damage reversal critical for tumor responses to alkylation therapy?
Trends Box.
Alkylation damage repair involves multiple partially redundant pathways, which include direct reversal by MGMT, the ALKB family of demethylases, and base excision repair.
Myriad regulatory mechanisms of these pathways exist. These include epigenetic control, post-translational modifications of repair proteins, and cellular metabolic status.
Recent work has highlighted the importance of coordination within the individual alkylation repair pathways, as well as cooperation between them.
Alkylation adducts repaired by these pathways overlap with newly discovered methylation marks with possible epigenetic functions, highlighting the importance of regulating these repair pathways.
Understanding the molecular mechanisms of alkylation repair regulation will likely provide opportunities for improved cancer chemotherapy.
Acknowledgments
We wish to thank Joshua Brickner, Hani Zaher, and Arne Klungland for critical reading and feedback on this manuscript. J.M.S. is supported by the Monsanto Graduate Program Fellowship. This work was supported by the NIH (R01 CA148629 to R.W.S.; K08 CA158133 and R01 CA193318 to N.M.), the American Cancer Society (IRG-58-010-56 to N.M.), the Siteman Cancer Center (to N.M.), and the Children’s Discovery Institute of St. Louis Children’s Hospital (MC-II-2015-453 to N.M.). R.W.S. is an Abraham A. Mitchell Distinguished Investigator at the Mitchell Cancer Institute and is a scientific consultant for Trevigen, Inc.
Glossary
- Abasic site
A lesion in DNA or RNA where neither a purine nor a pyrimidine base is present
- AP
Apurinic/Apyrimidinic site
- ATR
Ataxia Telangiectasia and Rad3-related protein kinase
- ATRIP
ATR Interacting Protein
- ε-base
Ethenobase, a carcinogenic alkylation that consists of a two-carbon bridge
- IDH
Isocitrate dehydrogenase enzyme that converts isocitrate to α-ketoglutarate and is encoded by IDH1 or IDH2
- MutLα complex
A heterodimeric complex consisting of the two MMR proteins, MLH1 and PMS2
- MutSα complex
A heterodimeric complex consisting of the two MMR proteins, MSH2 and MSH6
- PAR
Poly(ADP-ribose), a nucleotide polymer consisting of adenosine diphosphate ribose monomers
- PARP
Poly(ADP-ribose) polymerase, a single-stranded break binding protein which catalyzes the addition of PAR chains to itself and other BER factors
- PARG
poly(ADP-ribose) glycohydrolase, an enzyme which removes PAR chains from substrates
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Fu D, et al. Balancing repair and tolerance of DNA damage caused by alkylating agents. Nat Rev Cancer. 2012;12:104–120. doi: 10.1038/nrc3185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rydberg B, Lindahl T. Nonenzymatic methylation of DNA by the intracellular methyl group donor S-adenosyl-L-methionine is a potentially mutagenic reaction. The EMBO journal. 1982;1:211–216. doi: 10.1002/j.1460-2075.1982.tb01149.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Drablos F, et al. Alkylation damage in DNA and RNA--repair mechanisms and medical significance. DNA Repair (Amst) 2004;3:1389–1407. doi: 10.1016/j.dnarep.2004.05.004. [DOI] [PubMed] [Google Scholar]
- 4.Beranek DT. Distribution of methyl and ethyl adducts following alkylation with monofunctional alkylating agents. Mutat Res. 1990;231:11–30. doi: 10.1016/0027-5107(90)90173-2. [DOI] [PubMed] [Google Scholar]
- 5.Gentil A, et al. Mutagenicity of a unique apurinic/apyrimidinic site in mammalian cells. J Mol Biol. 1992;227:981–984. doi: 10.1016/0022-2836(92)90513-j. [DOI] [PubMed] [Google Scholar]
- 6.Larson K, et al. Methylation-induced blocks to in vitro DNA replication. Mutat Res. 1985;150:77–84. doi: 10.1016/0027-5107(85)90103-4. [DOI] [PubMed] [Google Scholar]
- 7.Johnson RE, et al. A role for yeast and human translesion synthesis DNA polymerases in promoting replication through 3-methyl adenine. Mol Cell Biol. 2007;27:7198–7205. doi: 10.1128/MCB.01079-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hendricks CA, et al. The S. cerevisiae Mag1 3-methyladenine DNA glycosylase modulates susceptibility to homologous recombination. DNA Repair (Amst) 2002;1:645–659. doi: 10.1016/s1568-7864(02)00072-1. [DOI] [PubMed] [Google Scholar]
- 9.Warren JJ, et al. The structural basis for the mutagenicity of O(6)-methyl-guanine lesions. Proc Natl Acad Sci U S A. 2006;103:19701–19706. doi: 10.1073/pnas.0609580103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Spivak G. Nucleotide excision repair in humans. DNA Repair (Amst) 2015;36:13–18. doi: 10.1016/j.dnarep.2015.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kim H, D’Andrea AD. Regulation of DNA cross-link repair by the Fanconi anemia/BRCA pathway. Genes Dev. 2012;26:1393–1408. doi: 10.1101/gad.195248.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Christmann M, et al. O(6)-Methylguanine-DNA methyltransferase (MGMT) in normal tissues and tumors: enzyme activity, promoter methylation and immunohistochemistry. Biochim Biophys Acta. 2011;1816:179–190. doi: 10.1016/j.bbcan.2011.06.002. [DOI] [PubMed] [Google Scholar]
- 13.Gerson SL. MGMT: its role in cancer aetiology and cancer therapeutics. Nat Rev Cancer. 2004;4:296–307. doi: 10.1038/nrc1319. [DOI] [PubMed] [Google Scholar]
- 14.Weller M, et al. MGMT promoter methylation in malignant gliomas: ready for personalized medicine? Nat Rev Neurol. 2010;6:39–51. doi: 10.1038/nrneurol.2009.197. [DOI] [PubMed] [Google Scholar]
- 15.Esteller M, et al. Inactivation of the DNA-repair gene MGMT and the clinical response of gliomas to alkylating agents. N Engl J Med. 2000;343:1350–1354. doi: 10.1056/NEJM200011093431901. [DOI] [PubMed] [Google Scholar]
- 16.Esteller M, et al. Inactivation of the DNA repair gene O6-methylguanine-DNA methyltransferase by promoter hypermethylation is a common event in primary human neoplasia. Cancer Res. 1999;59:793–797. [PubMed] [Google Scholar]
- 17.Feinberg AP, et al. The epigenetic progenitor origin of human cancer. Nat Rev Genet. 2006;7:21–33. doi: 10.1038/nrg1748. [DOI] [PubMed] [Google Scholar]
- 18.Hegi ME, et al. MGMT gene silencing and benefit from temozolomide in glioblastoma. N Engl J Med. 2005;352:997–1003. doi: 10.1056/NEJMoa043331. [DOI] [PubMed] [Google Scholar]
- 19.Kreklau EL, et al. Prolonged inhibition of O(6)-methylguanine DNA methyltransferase in human tumor cells by O(6)-benzylguanine in vitro and in vivo. J Pharmacol Exp Ther. 1999;291:1269–1275. [PubMed] [Google Scholar]
- 20.Dolan ME, et al. Depletion of mammalian O6-alkylguanine-DNA alkyltransferase activity by O6-benzylguanine provides a means to evaluate the role of this protein in protection against carcinogenic and therapeutic alkylating agents. Proc Natl Acad Sci U S A. 1990;87:5368–5372. doi: 10.1073/pnas.87.14.5368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Turriziani M, et al. O6-(4-bromothenyl)guanine (PaTrin-2), a novel inhibitor of O6-alkylguanine DNA alkyl-transferase, increases the inhibitory activity of temozolomide against human acute leukaemia cells in vitro. Pharmacol Res. 2006;53:317–323. doi: 10.1016/j.phrs.2005.12.001. [DOI] [PubMed] [Google Scholar]
- 22.McMurry TB. MGMT inhibitors--The Trinity College-Paterson Institute experience, a chemist’s perception. DNA Repair (Amst) 2007;6:1161–1169. doi: 10.1016/j.dnarep.2007.03.015. [DOI] [PubMed] [Google Scholar]
- 23.Esteller M, Herman JG. Generating mutations but providing chemosensitivity: the role of O6-methylguanine DNA methyltransferase in human cancer. Oncogene. 2004;23:1–8. doi: 10.1038/sj.onc.1207316. [DOI] [PubMed] [Google Scholar]
- 24.Chabanon RM, et al. Mutational Landscape and Sensitivity to Immune Checkpoint Blockers. Clin Cancer Res. 2016 doi: 10.1158/1078-0432.CCR-16-0903. [DOI] [PubMed] [Google Scholar]
- 25.Westdorp H, et al. Opportunities for immunotherapy in microsatellite instable colorectal cancer. Cancer Immunol Immunother. 2016 doi: 10.1007/s00262-016-1832-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Duval A, et al. The mutator pathway is a feature of immunodeficiency-related lymphomas. Proc Natl Acad Sci U S A. 2004;101:5002–5007. doi: 10.1073/pnas.0400945101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Quiros S, et al. Processing of O6-methylguanine into DNA double-strand breaks requires two rounds of replication whereas apoptosis is also induced in subsequent cell cycles. Cell Cycle. 2010;9:168–178. doi: 10.4161/cc.9.1.10363. [DOI] [PubMed] [Google Scholar]
- 28.Mojas N, et al. Mismatch repair-dependent processing of methylation damage gives rise to persistent single-stranded gaps in newly replicated DNA. Genes Dev. 2007;21:3342–3355. doi: 10.1101/gad.455407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.York SJ, Modrich P. Mismatch repair-dependent iterative excision at irreparable O6-methylguanine lesions in human nuclear extracts. J Biol Chem. 2006;281:22674–22683. doi: 10.1074/jbc.M603667200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ochs K, Kaina B. Apoptosis induced by DNA damage O6-methylguanine is Bcl-2 and caspase-9/3 regulated and Fas/caspase-8 independent. Cancer Res. 2000;60:5815–5824. [PubMed] [Google Scholar]
- 31.Simonelli V, et al. Crosstalk between mismatch repair and base excision repair in human gastric cancer. Oncotarget. 2016 doi: 10.18632/oncotarget.10185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Guo J, et al. MutSbeta promotes trinucleotide repeat expansion by recruiting DNA polymerase beta to nascent (CAG)n or (CTG)n hairpins for error-prone DNA synthesis. Cell Res. 2016;26:775–786. doi: 10.1038/cr.2016.66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yoshioka K, et al. ATR kinase activation mediated by MutSalpha and MutLalpha in response to cytotoxic O6-methylguanine adducts. Mol Cell. 2006;22:501–510. doi: 10.1016/j.molcel.2006.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Aaltonen LA, et al. Clues to the pathogenesis of familial colorectal cancer. Science. 1993;260:812–816. doi: 10.1126/science.8484121. [DOI] [PubMed] [Google Scholar]
- 35.Liu B, et al. Mismatch repair gene defects in sporadic colorectal cancers with microsatellite instability. Nat Genet. 1995;9:48–55. doi: 10.1038/ng0195-48. [DOI] [PubMed] [Google Scholar]
- 36.Liu B, et al. Analysis of mismatch repair genes in hereditary non-polyposis colorectal cancer patients. Nat Med. 1996;2:169–174. doi: 10.1038/nm0296-169. [DOI] [PubMed] [Google Scholar]
- 37.Goel A, et al. Characterization of sporadic colon cancer by patterns of genomic instability. Cancer Res. 2003;63:1608–1614. [PubMed] [Google Scholar]
- 38.Alonso M, et al. Microsatellite instability occurs in distinct subtypes of pediatric but not adult central nervous system tumors. Cancer Res. 2001;61:2124–2128. [PubMed] [Google Scholar]
- 39.Fukushima T, et al. Promoter hypermethylation of mismatch repair gene hMLH1 predicts the clinical response of malignant astrocytomas to nitrosourea. Clin Cancer Res. 2005;11:1539–1544. doi: 10.1158/1078-0432.CCR-04-1625. [DOI] [PubMed] [Google Scholar]
- 40.Hunter C, et al. A hypermutation phenotype and somatic MSH6 mutations in recurrent human malignant gliomas after alkylator chemotherapy. Cancer Res. 2006;66:3987–3991. doi: 10.1158/0008-5472.CAN-06-0127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Xie C, et al. Association of MSH6 mutation with glioma susceptibility, drug resistance and progression. Molecular and clinical oncology. 2016;5:236–240. doi: 10.3892/mco.2016.907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Maxwell JA, et al. Mismatch repair deficiency does not mediate clinical resistance to temozolomide in malignant glioma. Clin Cancer Res. 2008;14:4859–4868. doi: 10.1158/1078-0432.CCR-07-4807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.McFaline-Figueroa JL, et al. Minor Changes in Expression of the Mismatch Repair Protein MSH2 Exert a Major Impact on Glioblastoma Response to Temozolomide. Cancer Res. 2015;75:3127–3138. doi: 10.1158/0008-5472.CAN-14-3616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Short SC, et al. Rad51 inhibition is an effective means of targeting DNA repair in glioma models and CD133+ tumor-derived cells. Neuro-oncology. 2011;13:487–499. doi: 10.1093/neuonc/nor010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Almeida KH, Sobol RW. A unified view of base excision repair: lesion-dependent protein complexes regulated by post-translational modification. DNA Repair (Amst) 2007;6:695–711. doi: 10.1016/j.dnarep.2007.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tang JB, et al. Bioenergetic metabolites regulate base excision repair-dependent cell death in response to DNA damage. Molecular cancer research : MCR. 2010;8:67–79. doi: 10.1158/1541-7786.MCR-09-0411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Klapacz J, et al. Frameshift mutagenesis and microsatellite instability induced by human alkyladenine DNA glycosylase. Mol Cell. 2010;37:843–853. doi: 10.1016/j.molcel.2010.01.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sobol RW, et al. Regulated over-expression of DNA polymerase beta mediates early onset cataract in mice. DNA Repair (Amst) 2003;2:609–622. doi: 10.1016/s1568-7864(03)00026-0. [DOI] [PubMed] [Google Scholar]
- 49.Ebrahimkhani MR, et al. Aag-initiated base excision repair promotes ischemia reperfusion injury in liver, brain, and kidney. Proc Natl Acad Sci U S A. 2014;111:E4878–E4886. doi: 10.1073/pnas.1413582111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Calvo JA, et al. Aag DNA glycosylase promotes alkylation-induced tissue damage mediated by Parp1. PLoS genetics. 2013;9:e1003413. doi: 10.1371/journal.pgen.1003413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Meira LB, et al. Aag-initiated base excision repair drives alkylation-induced retinal degeneration in mice. Proc Natl Acad Sci U S A. 2009;106:888–893. doi: 10.1073/pnas.0807030106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tang JB, et al. N-methylpurine DNA glycosylase and DNA polymerase beta modulate BER inhibitor potentiation of glioma cells to temozolomide. Neuro-oncology. 2011;13:471–486. doi: 10.1093/neuonc/nor011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Liu L, et al. Pharmacologic disruption of base excision repair sensitizes mismatch repair-deficient and -proficient colon cancer cells to methylating agents. Clin Cancer Res. 1999;5:2908–2917. [PubMed] [Google Scholar]
- 54.Sobol RW, et al. Base excision repair intermediates induce p53-independent cytotoxic and genotoxic responses. J Biol Chem. 2003;278:39951–39959. doi: 10.1074/jbc.M306592200. [DOI] [PubMed] [Google Scholar]
- 55.Thompson LH, et al. Molecular cloning of the human XRCC1 gene, which corrects defective DNA strand break repair and sister chromatid exchange. Mol Cell Biol. 1990;10:6160–6171. doi: 10.1128/mcb.10.12.6160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Masson M, et al. XRCC1 is specifically associated with poly(ADP-ribose) polymerase and negatively regulates its activity following DNA damage. Mol Cell Biol. 1998;18:3563–3571. doi: 10.1128/mcb.18.6.3563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.de Murcia JM, et al. Requirement of poly(ADP-ribose) polymerase in recovery from DNA damage in mice and in cells. Proc Natl Acad Sci U S A. 1997;94:7303–7307. doi: 10.1073/pnas.94.14.7303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Smulson ME, et al. A putative role for nicotinamide adenine dinucleotide-promoted nuclear protein modification in the antitumor activity of N-methyl-N-nitrosourea. Cancer Res. 1977;37:3006–3012. [PubMed] [Google Scholar]
- 59.Wallace SS, et al. Base excision repair and cancer. Cancer letters. 2012;327:73–89. doi: 10.1016/j.canlet.2011.12.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Nemec AA, et al. Colon cancer-associated DNA polymerase beta variant induces genomic instability and cellular transformation. J Biol Chem. 2012;287:23840–23849. doi: 10.1074/jbc.M112.362111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Prasad R, et al. Suicidal cross-linking of PARP-1 to AP site intermediates in cells undergoing base excision repair. Nucleic Acids Res. 2014;42:6337–6351. doi: 10.1093/nar/gku288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Horton JK, et al. Base excision repair defects invoke hypersensitivity to PARP inhibition. Molecular cancer research : MCR. 2014;12:1128–1139. doi: 10.1158/1541-7786.MCR-13-0502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Murai J, et al. Trapping of PARP1 and PARP2 by Clinical PARP Inhibitors. Cancer Res. 2012;72:5588–5599. doi: 10.1158/0008-5472.CAN-12-2753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Nakadate Y, et al. Silencing of poly(ADP-ribose) glycohydrolase sensitizes lung cancer cells to radiation through the abrogation of DNA damage checkpoint. Biochem Biophys Res Commun. 2013;441:793–798. doi: 10.1016/j.bbrc.2013.10.134. [DOI] [PubMed] [Google Scholar]
- 65.Fang Q, et al. HSP90 regulates DNA repair via the interaction between XRCC1 and DNA polymerase beta. Nat Commun. 2014;5:5513. doi: 10.1038/ncomms6513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Fouquerel E, Sobol RW. ARTD1 (PARP1) activation and NAD(+) in DNA repair and cell death. DNA Repair (Amst) 2014;23:27–32. doi: 10.1016/j.dnarep.2014.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Jacobson MK, et al. Effect of carcinogenic N-alkyl-N-nitroso compounds on nicotinamide adenine dinucleotide metabolism. Cancer Res. 1980;40:1797–1802. [PubMed] [Google Scholar]
- 68.Calvo JA, et al. Parp1 protects against Aag-dependent alkylation-induced nephrotoxicity in a sex-dependent manner. Oncotarget. 2016 doi: 10.18632/oncotarget.10440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Fouquerel E, et al. ARTD1/PARP1 negatively regulates glycolysis by inhibiting hexokinase 1 independent of NAD+ depletion. Cell Rep. 2014;8:1819–1831. doi: 10.1016/j.celrep.2014.08.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Goellner EM, et al. Overcoming temozolomide resistance in glioblastoma via dual inhibition of NAD+ biosynthesis and base excision repair. Cancer Res. 2011;71:2308–2317. doi: 10.1158/0008-5472.CAN-10-3213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Roulston A, Shore GC. New strategies to maximize therapeutic opportunities for NAMPT inhibitors in oncology. Molecular & cellular oncology. 2016;3:e1052180. doi: 10.1080/23723556.2015.1052180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Aas PA, et al. Human and bacterial oxidative demethylases repair alkylation damage in both RNA and DNA. Nature. 2003;421:859–863. doi: 10.1038/nature01363. [DOI] [PubMed] [Google Scholar]
- 73.Dango S, et al. DNA unwinding by ASCC3 helicase is coupled to ALKBH3-dependent DNA alkylation repair and cancer cell proliferation. Mol Cell. 2011;44:373–384. doi: 10.1016/j.molcel.2011.08.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Ringvoll J, et al. Repair deficient mice reveal mABH2 as the primary oxidative demethylase for repairing 1meA and 3meC lesions in DNA. EMBO J. 2006;25:2189–2198. doi: 10.1038/sj.emboj.7601109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Calvo JA, et al. DNA repair is indispensable for survival after acute inflammation. J Clin Invest. 2012;122:2680–2689. doi: 10.1172/JCI63338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Delaney JC, et al. AlkB reverses etheno DNA lesions caused by lipid oxidation in vitro and in vivo. Nat Struct Mol Biol. 2005;12:855–860. doi: 10.1038/nsmb996. [DOI] [PubMed] [Google Scholar]
- 77.Fu D, Samson LD. Direct repair of 3,N(4)-ethenocytosine by the human ALKBH2 dioxygenase is blocked by the AAG/MPG glycosylase. DNA Repair (Amst) 2012;11:46–52. doi: 10.1016/j.dnarep.2011.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Singer B, et al. Both purified human 1,N6-ethenoadenine-binding protein and purified human 3-methyladenine-DNA glycosylase act on 1,N6-ethenoadenine and 3-methyladenine. Proc Natl Acad Sci U S A. 1992;89:9386–9390. doi: 10.1073/pnas.89.20.9386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Lingaraju GM, et al. Structural basis for the inhibition of human alkyladenine DNA glycosylase (AAG) by 3,N4-ethenocytosine-containing DNA. J Biol Chem. 2011;286:13205–13213. doi: 10.1074/jbc.M110.192435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Gros L, et al. Hijacking of the human alkyl-N-purine-DNA glycosylase by 3,N4-ethenocytosine, a lipid peroxidation-induced DNA adduct. J Biol Chem. 2004;279:17723–17730. doi: 10.1074/jbc.M314010200. [DOI] [PubMed] [Google Scholar]
- 81.Latypov VF, et al. Atl1 regulates choice between global genome and transcription-coupled repair of O(6)-alkylguanines. Mol Cell. 2012;47:50–60. doi: 10.1016/j.molcel.2012.04.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Tasaki M, et al. ALKBH3, a human AlkB homologue, contributes to cell survival in human non-small-cell lung cancer. British journal of cancer. 2011;104:700–706. doi: 10.1038/sj.bjc.6606012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Konishi N, et al. High expression of a new marker PCA-1 in human prostate carcinoma. Clinical cancer research : an official journal of the American Association for Cancer Research. 2005;11:5090–5097. doi: 10.1158/1078-0432.CCR-05-0195. [DOI] [PubMed] [Google Scholar]
- 84.Johannessen TC, et al. The DNA repair protein ALKBH2 mediates temozolomide resistance in human glioblastoma cells. Neuro-oncology. 2013;15:269–278. doi: 10.1093/neuonc/nos301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Zhao Y, et al. Noncanonical regulation of alkylation damage resistance by the OTUD4 deubiquitinase. EMBO J. 2015;34:1687–1703. doi: 10.15252/embj.201490497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Komander D, Rape M. The ubiquitin code. Annu Rev Biochem. 2012;81:203–229. doi: 10.1146/annurev-biochem-060310-170328. [DOI] [PubMed] [Google Scholar]
- 87.Chauhan D, et al. A small molecule inhibitor of ubiquitin-specific protease-7 induces apoptosis in multiple myeloma cells and overcomes bortezomib resistance. Cancer Cell. 2012;22:345–358. doi: 10.1016/j.ccr.2012.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Kapuria V, et al. Deubiquitinase inhibition by small-molecule WP1130 triggers aggresome formation and tumor cell apoptosis. Cancer Res. 2010;70:9265–9276. doi: 10.1158/0008-5472.CAN-10-1530. [DOI] [PubMed] [Google Scholar]
- 89.Mosammaparast N, Shi Y. Reversal of histone methylation: biochemical and molecular mechanisms of histone demethylases. Annu Rev Biochem. 2010;79:155–179. doi: 10.1146/annurev.biochem.78.070907.103946. [DOI] [PubMed] [Google Scholar]
- 90.Sundheim O, et al. AlkB demethylases flip out in different ways. DNA repair. 2008;7:1916–1923. doi: 10.1016/j.dnarep.2008.07.015. [DOI] [PubMed] [Google Scholar]
- 91.Wang P, et al. Oncometabolite D-2-Hydroxyglutarate Inhibits ALKBH DNA Repair Enzymes and Sensitizes IDH Mutant Cells to Alkylating Agents. Cell Rep. 2015;13:2353–2361. doi: 10.1016/j.celrep.2015.11.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Dominissini D, et al. The dynamic N(1)-methyladenosine methylome in eukaryotic messenger RNA. Nature. 2016;530:441–446. doi: 10.1038/nature16998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Li X, et al. Transcriptome-wide mapping reveals reversible and dynamic N-methyladenosine methylome. Nat Chem Biol. 2016 doi: 10.1038/nchembio.2040. [DOI] [PubMed] [Google Scholar]
- 94.Hudson BH, Zaher HS. O6-Methylguanosine leads to position-dependent effects on ribosome speed and fidelity. Rna. 2015;21:1648–1659. doi: 10.1261/rna.052464.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Wu TP, et al. DNA methylation on N(6)-adenine in mammalian embryonic stem cells. Nature. 2016;532:329–333. doi: 10.1038/nature17640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Greer EL, et al. DNA Methylation on N6-Adenine in C. elegans. Cell. 2015;161:868–878. doi: 10.1016/j.cell.2015.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Macon JB, Wolfenden R. 1-Methyladenosine. Dimroth rearrangement and reversible reduction. Biochemistry. 1968;7:3453–3458. doi: 10.1021/bi00850a021. [DOI] [PubMed] [Google Scholar]
- 98.Gilljam KM, et al. Identification of a novel, widespread, and functionally important PCNA-binding motif. The Journal of cell biology. 2009;186:645–654. doi: 10.1083/jcb.200903138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Fu D, et al. The interaction between ALKBH2 DNA repair enzyme and PCNA is direct, mediated by the hydrophobic pocket of PCNA and perturbed in naturally-occurring ALKBH2 variants. DNA Repair (Amst) 2015;35:13–18. doi: 10.1016/j.dnarep.2015.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Hegi ME, et al. Clinical trial substantiates the predictive value of O-6-methylguanine-DNA methyltransferase promoter methylation in glioblastoma patients treated with temozolomide. Clin Cancer Res. 2004;10:1871–1874. doi: 10.1158/1078-0432.ccr-03-0384. [DOI] [PubMed] [Google Scholar]
- 101.Nagel ZD, et al. Multiplexed DNA repair assays for multiple lesions and multiple doses via transcription inhibition and transcriptional mutagenesis. Proc Natl Acad Sci U S A. 2014;111:E1823–E1832. doi: 10.1073/pnas.1401182111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Gerson SL. Clinical relevance of MGMT in the treatment of cancer. J Clin Oncol. 2002;20:2388–2399. doi: 10.1200/JCO.2002.06.110. [DOI] [PubMed] [Google Scholar]
- 103.Xu-Welliver M, Pegg AE. Degradation of the alkylated form of the DNA repair protein, O(6)-alkylguanine-DNA alkyltransferase. Carcinogenesis. 2002;23:823–830. doi: 10.1093/carcin/23.5.823. [DOI] [PubMed] [Google Scholar]
- 104.Duncan T, et al. Reversal of DNA alkylation damage by two human dioxygenases. Proc Natl Acad Sci U S A. 2002;99:16660–16665. doi: 10.1073/pnas.262589799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Falnes PO, et al. AlkB-mediated oxidative demethylation reverses DNA damage in Escherichia coli. Nature. 2002;419:178–182. doi: 10.1038/nature01048. [DOI] [PubMed] [Google Scholar]
- 106.Trewick SC, et al. Oxidative demethylation by Escherichia coli AlkB directly reverts DNA base damage. Nature. 2002;419:174–178. doi: 10.1038/nature00908. [DOI] [PubMed] [Google Scholar]
- 107.Robertson AB, et al. DNA repair in mammalian cells: Base excision repair: the long and short of it. Cellular and molecular life sciences : CMLS. 2009;66:981–993. doi: 10.1007/s00018-009-8736-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Svilar D, et al. Base excision repair and lesion-dependent subpathways for repair of oxidative DNA damage. Antioxidants & redox signaling. 2011;14:2491–2507. doi: 10.1089/ars.2010.3466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Fortini P, Dogliotti E. Base damage and single-strand break repair: mechanisms and functional significance of short- and long-patch repair subpathways. DNA Repair (Amst) 2007;6:398–409. doi: 10.1016/j.dnarep.2006.10.008. [DOI] [PubMed] [Google Scholar]