Abstract
The Escherichia coli cell envelope is a protective barrier at the frontline of interaction with the environment. Fidelity of envelope biogenesis must be monitored to establish and maintain a contiguous barrier. Indeed, the envelope must also be repaired and modified in response to environmental assaults. Envelope stress responses (ESRs) sense envelope damage or defects and alter the transcriptome to mitigate stress. We will review recent insights into stress sensing mechanisms of the σE and Cpx systems and the interaction of these ESRs. Small RNAs (sRNAs) are increasingly prominent regulators of the transcriptional response to stress. These fast-acting regulators also provide avenues for inter-ESR regulation that could be important when cells face multiple contemporaneous stresses, as is the case during infection.
Keywords: Outer membrane, Cpx, σ E, sRNA
Monitoring the cell envelope
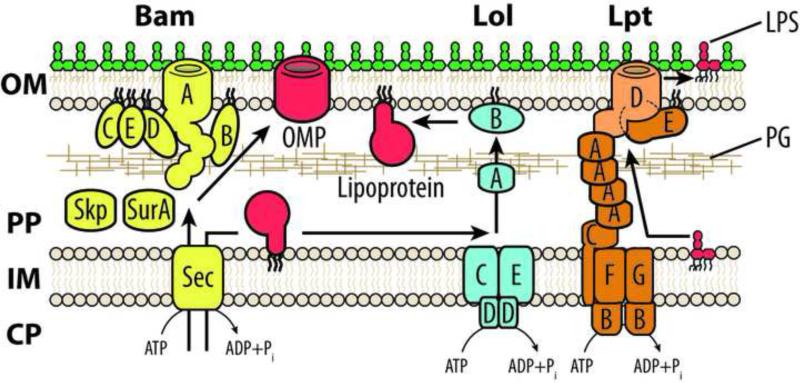
Gram-negative bacteria such as Escherichia coli are diderm (see Glossary) cells with an outer membrane (OM) that is separated from the inner membrane (IM) by an aqueous periplasmic space which houses the peptidoglycan cell wall [1] (Figure 1). The OM is an asymmetric lipid bilayer where lipopolysaccharide (LPS) forms the surface-exposed leaflet and phospholipids form the periplasmic leaflet [2]. While it remains unclear how phospholipids reach the OM, LPS is delivered by the transenvelope Lpt complex [3] (Figure 1). There are two classes of OM proteins: transmembrane β-barrel proteins (termed OMPs) [4]; and lipoproteins whose acylation anchors them in the bilayer [5]. These hydrophobic proteins require chaperones for transit across the periplasm. For OMPs, the major chaperone in E. coli is SurA, with Skp and the chaperone-protease DegP having back-up roles [6]; for lipoproteins, the chaperone is LolA [5]. Chaperones deliver OMPs to the β-barrel assembly machine (Bam) complex for assembly into the OM [4] (Figure 1). Lipoproteins are transferred from LolA to LolB for anchoring into the OM [5] (Figure 1). The envelope protects cells against environmental assaults, the immune system, and antibiotics [2]. Given the complexity of envelope biogenesis, several envelope stress responses (ESRs) are tasked with monitoring for defects or damage and restoring homeostasis. For example, the σE response is potently activated by OMP assembly defects, while the Cpx two-component system (TCS) responds to periplasmic or IM protein misfolding [7,8].
Figure 1. Cell envelope structure and OM biogenesis machines.
OMPs synthesized in the cytoplasm (CP) are secreted (via the Sec translocase) into the periplasm (PP) where chaperones such as SurA and Skp prevent their misfolding. The Bam complex (BamABCDE) receives OMPs from chaperones before folding them and facilitating their insertion into the OM. Lipoproteins are also secreted but enter the Lol pathway that extracts the acylated lipoproteins from the IM (via LolCDE) and transfers them to the LolA chaperone. LolB receives lipoproteins at the OM from LolA and anchors them into the bilayer. LPS is transported across the PP by a transenvelope bridge formed by the Lpt complex. LPS is synthesized in the IM before being extracted from the bilayer by the LptBBFG ABC transporter and transferred to LptC. Sequential rounds of ATP hydrolysis move LPS from LptC onto and across the bridge formed by a polymer of LptA. At the OM, the OMP LptD and the lipoprotein LptE act in concert to insert LPS into the outer leaflet of the OM. The PG cell wall resides in the PP. A Substrate OMP for the Bam complex, a substrate lipoprotein for the Lol pathway, and a substrate LPS molecule are colored in red.
Understanding how signals from multiple stresses are integrated remains an ongoing challenge. Efforts in cataloging ESR small RNAs (sRNAs) suggest these molecules may interface between ESRs. Intriguingly, the σE and Cpx signaling pathways are highly interconnected. Here, we review recent progress in understanding the stress-sensing mechanisms of these two ESRs, as well as their sRNA effectors. While this review focuses on σE and Cpx responses, it is notable that other signal transduction systems also respond to envelope damage and significantly remodel the envelope. Both the RcsBCDF phosphorelay and the PhoPQ TCS sense LPS layer damage inflicted by cationic antimicrobial peptides (CAMPs) and respond by inducing extracellular polysaccharide production and LPS modifying enzymes, respectively, to fortify the OM [9,10]. The EnvZ/OmpR TCS responds to osmotic challenge by regulating major OMP porins [11] .
The σE response: a two-signal model for activation
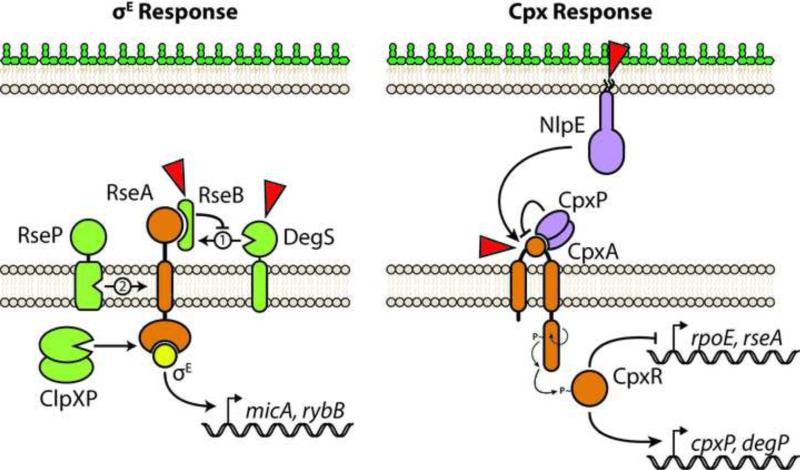
E. coli produces an alternate σ factor (σE, RpoE) that enables RNA polymerase to increase transcription of genes involved in cell envelope biogenesis [12]. Many of the steps leading to σE activation have been described in molecular detail [7] (Figure 2). Under non-inducing conditions, the IM anti-σ factor RseA sequesters σE to the plasma membrane [13]. The presence of unfolded OMPs in the periplasm is the most thoroughly characterized signal for σE activation [14,15]. Signal sensing triggers a cascade of regulated proteolysis steps that degrade RseA and release σE into the cytosol where it can promote transcription [7]. σE-dependent promoters increase production of periplasmic OMP chaperones, the Bam complex, and other factors required for OM homeostasis [12]. The negative regulatory arm of the response relies on sRNAs that prevent OMPs synthesis until stress is relieved (discussed later).
Figure 2. The σE and Cpx envelope stress responses of E. coli.
Left, the σE response is activated by a series of proteolysis steps that degrade the anti-σ factor RseA and liberate σE into the cytosol where it can increase target gene transcription. The RseA periplasmic domain is first cleaved by DegS. RseB binds RseA and inhibits DegS activity. DegS senses unfolded OMP cues. RseB is proposed to be displaced from RseA in response to LPS binding. RseB displacement allows DegS to cleave RseA and initiate the response. RseA is subsequently cleaved by the intramembrane protease RseP. A cytoplasmic portion of RseA is then degraded by the ClpXP protease to release σE. Right, the Cpx TCS responds to numerous signals detected by the periplasmic sensory domain of CpxA. CpxP inhibits CpxA activation likely by interacting with the sensory domain directly. NlpE can transduce cell adhesion signals across the envelope to activate the Cpx response. Stimulated CpxA histidine kinase autophosphorylates and transfers the phosphate to the CpxR response regulator, which alters gene expression. Sensory inputs are illustrated by red triangles.
Unfolded OMPs are sensed by DegS, an IM serine protease [16]. The periplasmic PDZ domain of DegS maintains the protease in an inactive state [14]; this inhibition is relieved when the PDZ domain binds peptides corresponding to C-terminal residues of OMPs [14]. Such sequences are inaccessible in properly folded OMPs, but they accumulate in the periplasm if OMP folding is defective [13]. Peptide binding by DegS reorients the active site to facilitate catalysis [17]. Two consecutive proteolytic cleavages of RseA are required to release σE. DegS removes a periplasmic domain of RseA and this cleavage is the rate-limiting step of σE activation [18].
Following degradation by DegS, the remaining RseA protein becomes sensitive to proteolysis by RseP, a zinc metalloprotease belonging to the S2P group of intra-membrane proteases (RIPs) [19]. The periplasmic domain of full length RseA acts as a size exclusion filter that prevents cleavage of the protein by RseP. [20]. The RseP active site is within the IM bilayer; recent insights demonstrate how an unusual RseP intramembrane β-strand unfolds the RseA target site to make it amenable for proteolysis [20]. Degradation by RseP releases a soluble fragment of RseA—still clutching σE—into the cytosol [21]. Cytoplasmic proteases, most notably ClpXP, subsequently liberate σE [21].
A periplasmic protein, RseB, negatively regulates σE activation by binding RseA and preventing cleavage by DegS [7]. In vivo, discreet OMP peptides (with sequences N-terminal to the DegS-activating peptide) can displace RseB from RseA; but, these peptides were insufficient in an in vitro reconstituted system and an additional RseB signal was hypothesized [15]. Recent in vitro work proposes the acyl chains of LPS bind RseB and are the signal that displaces it from RseA [22]. In vivo, mutations that either reduce LPS acylation or truncate the LPS core polysaccharide induce σE [22]. Moreover, a folding-defective variant of LptD (the OMP that inserts LPS into the OM [3]) induces σE even in the absence of the OMP-sensing DegS PDZ domain, suggesting LPS transport defects are sensed [22]. A two-signal hypothesis for σE activation is currently proposed: full activation requires sensing of both unfolded OMPs (to activate DegS) and periplasmic LPS (to displace RseB from RseA) [22].
A lipophilic signal for RseB had been anticipated given the protein's structural similarity to LolA and LolB, the proteins involved in OM lipoprotein transport and insertion, respectively [15,23,24]. Yet, the most potent σE-activating mutations truncate the LPS core polysaccharide, which is distal to the lipophilic LPS components [22]. What conditions might cause production of truncated, σE-inducing LPS molecules in vivo remains an unresolved question. In wildtype cells, the lipid and core components are produced stoichiometrically and yield a consistent LPS structure [25]. One suggested possibility is that truncated LPS might be poorly transported by the Lpt transenvelope complex, causing it to accumulate in the periplasm [22]. σE induction by an LptD mutant seems to support this model [22]. However, evidence that LPS can accumulate in the periplasm in a manner that exposes its acyl chains to RseB is lacking. LPS is extremely lipophilic and poorly soluble; indeed, the Lpt bridge (Figure 1) is needed to provide a conduit for the lipophilic moieties as they pass through the aqueous periplasm [26]. The bridge is only formed when the OM Lpt components are capable of receiving LPS [27]. Depletion of Lpt proteins causes LPS accumulation within the IM and results in abnormal membrane structures [28,29]. It seems unlikely that LPS is excreted directly into the periplasm. How, then, might RseB encounter LPS acyl chains? Perhaps some accessory protein can receive LPS from defective Lpt machinery and transfer the molecule to RseB, akin to the lipoprotein transfer that occurs between LolA and LolB [30]. Clearly, a complete understanding of the RseB signal remains an important challenge.
Stress sensing by the Cpx response
The core of the Cpx ESR relies on a canonical TCS: CpxA is the IM sensory histidine kinase; CpxR is the DNA-binding response-regulator (Figure 2). Cpx responds to a broad set of conditions that include elevated pH, high salt concentrations, and alterations in IM lipid composition [8]. Recent work also suggest Cpx plays a role in sensing and responding to peptidoglycan cell wall defects [31-33]. One common theme among inducing conditions is that they cause protein misfolding. Sensing is nonetheless specific and misfolded OMPs fail to activate Cpx [34]. Misfolded IM and periplasmic proteins, and defects in protein translocation across the IM are all sensed by Cpx [8]. Increasingly, it appears that Cpx is primarily tasked with defending IM integrity [8].
Existing evidence suggests that CpxA directly senses stress [35,36]. CpxA consists of two transmembrane domains with a periplasmic loop that acts as a sensory domain [36]. CpxA activation leads to autophosphorylation of its cytoplasmic histidine kinase domain, which allows for phosphotransfer to the receiver domain of CpxR [36]. Phosphorylated CpxR binds cognate DNA sequences to regulate gene expression [36]. CpxA has both kinase and phosphatase activity, enabling it to rapidly control the extent of CpxR phosphorylation (and hence the strength of the response) [36].
A recent structure of the Vibrio parahaemolyticus CpxA sensory domain shows it forms a PAS domain of five β-strands and three α-helices [37]. Mutations in the sensory domain can activate CpxA. For example, the well-studied cpxA24 mutation deletes 32 residues and entirely removes a C-terminal sensory domain α-helix [36,37]. CpxA24 is constitutively activated and signal blind, suggesting that disrupting proper sensor domain folding may directly trigger CpxA kinase activity [36,38].
Two auxiliary signaling proteins modulate CpxA activation: the positive regulator NlpE and the negative regulator CpxP [39,40]. NlpE is an OM lipoprotein that activates Cpx upon E. coli adhesion to abiotic surfaces; such signaling is important since both NlpE and the Cpx response are required for efficient adhesion to hydrophobic surfaces [41]. Unfolded NlpE is proposed to directly contact CpxA from the OM to induce signaling [42]. How cell surface adhesion might unfold NlpE in the periplasm is not yet clear.
Among the most upregulated genes of the Cpx response is cpxP [43]. The periplasmic CpxP protein completes a negative-feedback loop by inhibiting CpxA kinase activation [44]. Based on its periplasmic localization, CpxP is suggested to directly block the CpxA sensory domain to inhibit signaling [44]. However, detecting direct CpxP-CpxA interaction has proven challenging [8,37,45]. CpxP forms a dimer with a large charged surface which is proposed to mediate electrostatic interaction with CpxA [45,46]. Indeed, high salt concentrations seem to displace CpxP [45]. Mutations that alter the CpxP surface impair CpxA inhibition and—for at least the D61 residue—even conservative substitutions are not tolerated, hinting at specific biochemical interactions [45-47]. Recently, CpxP-CpxA cross-linking and affinity purifications in vivo have provided evidence supporting direct interaction [48]. But, as CpxP levels in wildtype cells are extremely low, these interaction studies rely on overexpression of both proteins above their physiological levels [48]. Detecting a dynamic interaction between CpxA and CpxP at native levels remains an ongoing challenge.
CpxP has weak chaperone activity that is important for its role in clearing misfolded P pilus subunits from the periplasm [49]. CpxP is titrated by misfolded pilins and it delivers them to the periplasmic protease DegP so that both CpxP and its cargo are degraded [49]. Displacement of CpxP from CpxA is not itself a mechanism for sensing. In fact, CpxA is activated by alkaline pH and misfolded pilins even in the absence of CpxP [38]. The regulatory and effector functions of CpxP must be important during to the Cpx response since production of the cpxP mRNA transcript is so highly induced. Astonishingly, that abundant transcript also encodes for another Cpx effector, an sRNA that has only recently been discovered.
Small RNAs packed with big responsibilities
sRNAs are deployed by each of the major ESRs. sRNAs act as important regulatory molecules that can rapidly alter gene expression profiles. Typically, sRNAs bind multiple target mRNA transcripts and act negatively to either prevent translation or promote mRNA degradation, or both [50]. Some sRNAs do act positively on target mRNAs, for instance by relieving RNA structures that inhibit translation [51]. The general RNA chaperone Hfq binds to sRNAs to stabilize them, aid in target binding, and promote recruitment of RNA degradation machinery [50]. A number of new sRNAs have been identified as effectors for each of the ESRs.
Cpx and σE sRNAs prevent OMP synthesis
By virtue of being a component of the RNA polymerase, σE can activate transcription but is unable to directly repress gene expression (though a recent example does illustrate that a σ factor can increase transcription of an overlapping non-coding RNA as a means of preventing gene expression [52]). Nonetheless, σE activation causes a marked decrease in OMP levels [53]. Strong σE-dependent promoters produce the sRNAs MicA and RybB that reduce OMP synthesis. MicA regulates both ompA and lamB production [54-56]. RybB regulates ompC and ompX expression [57]. The importance of reducing OMP levels under stress conditions is underscored by convergent targeting of ompA, lamB, ompW, and tsx transcripts by both MicA and RybB at non-overlapping sites of the mRNA [58]. With regards to envelope stress, the old adage from Will Rogers applies: “If you find yourself in a hole, stop digging.” Remarkably, MicA also provides an interface between stress responses by directly regulating the phoPQ transcript that encodes the PhoPQ TCS responsible for modifying LPS under certain stress conditions [56,58]. Additional, non-OMP MicA and RybB targets have been identified, but their contribution to σE responses remains to be characterized [58].
sRNA components of the Cpx response have only recently been recognized [59,60]. CpxR increases levels of the sRNAs OmrA, OmrB and MicF, which all belong to the EnvZ/OmpR TCS regulon [61,62]. Cpx induces these sRNAs by producing an IM protein, MzrA, that then directly stimulates the EnvZ histidine kinase and so connects the Cpx response to EnvZ/OmpR [63,64]. Indeed, OmrA/B is known to be induced by Cpx via MzrA, and it is presumed (though untested) that MicF is similarly regulated [63].
OmrA/B redundantly target several OMP-encoding transcripts for degradation (including cirA, fecA, fepA, and ompT) [65]. MicF negatively regulates the major porin OmpF [62]. Hence, the Cpx response engages EnvZ-OmpR and its sRNAs to further downregulate OMP production. The sRNAs also regulate signaling: OmrA/B target the ompR-envZ transcript in a negative-feedback loop [65]; while MicF targets the cpxRA transcript to lower CpxR and CpxA levels [60,66]. Curiously, while MicF lowers CpxR levels it does not reduce transcription from strong CpxR-dependent promoters [60]. An intriguing hypothesis is that MicF disproportionately affects weaker CpxR promoters to sculpt the extent of the regulon invoked during stress [60]. In any case, MicF is remarkable for establishing a negative-feedback loop that is wired through two signal transduction systems.
RprA: an sRNA of the Rcs and Cpx responses
The RprA sRNA is a highly induced component of the Rcs response (Box 1) [67]. RprA promotes production of σS, the master regulator of general stress, by relieving an inhibitory structure in rpoS mRNA that hinders translation [51,68]. A key role for σS is to promote the transition from planktonic growth to a program of biofilm development [69]. The transcription factor CsgD is central to this transition since it promotes production of cellulose and curli fimbrae. σS directly increases csgD expression but also acts indirectly by upregulating the diguanylate cyclase YdaM (DgcM) whose c-di-GMP production stimulates csgD transcription [70]. The RprA sRNA plays an important regulatory role in this biofilm circuit. On the one hand, RprA promotes σS production but it impedes production of both YdaM and CsgD [71]. This curious regulatory arrangement is suggested to allow cells to switch off the massive production and secretion of cellulose and curli if envelope defects are detected [69]. The csgD mRNA transcript is an amazingly complex nexus for regulatory inputs from as many as six sRNAs, including ESR regulated OmrA/B and RprA [72]. Moreover, csgD expression is not only regulated by both σS and Rcs (through RprA), but is also directly repressed directly by CpxR and activated by the osmolarity-sensing EnvZ/OmpR system [73,74].
CpxR was recently shown to bind the rprA promoter and increase production of the sRNA [60]. RprA overexpression establishes a negative-feedback loop that reduces transcription of strong CpxR-dependent promoters [60]. Hence, it seems RprA induction could allow the Rcs response to potently regulate any output of the Cpx response. It remains unclear how RprA achieves Cpx feedback, though it clearly requires CpxR [60]. A tantalizing possibility is that RprA regulates an undiscovered auxiliary protein capable of modulating CpxR activity. Such proteins are known in other signaling circuits. For example, CheZ dephosphorylates the chemotaxis response regulator CheY [75]; and TorI binds the TorR response regulator to inhibit recruitment of RNA polymerase to promoters [76].
New ESR sRNAs derived from transcript 3’ untranslated regions (UTRs)
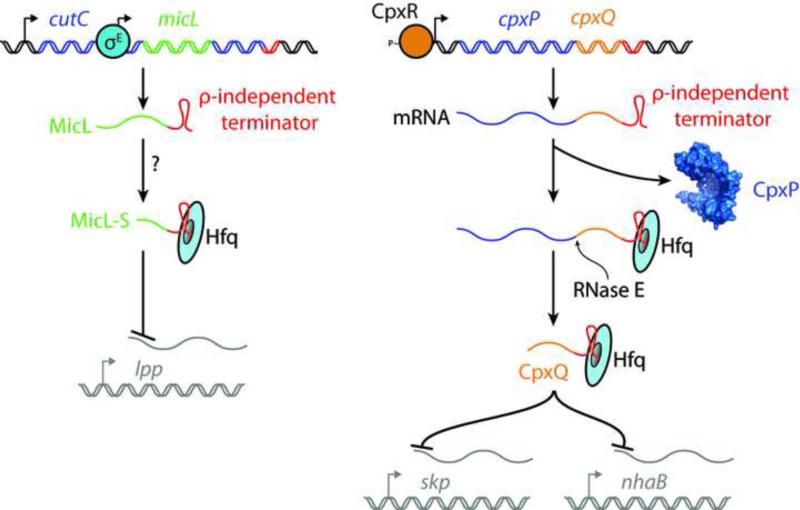
A recent breakthrough investigation of RNAs bound to Hfq discovered several new sRNAs that originated from the 3’ UTR of transcripts, revealing an overlooked source of sRNAs [77]. Indeed, a new Cpx response sRNA, CpxQ, was found to originate from the 3’ UTR of a highly induced mRNA transcript [78,79] (Figure 3). In a separate study, a new σE sRNA, MicL-S, was found encoded in the 3’ UTR of an annotated gene, though in this case the sRNA is transcribed from a dedicated promoter within that gene's coding sequence and then further processed [80] (Figure 3).
Figure 3. Biogenesis of 3’ UTR sRNAs MicL and CpxQ.
Left, the micL gene is entirely contained within 3’ UTR of the gene annotated as cutC. micL transcription ends at the native cutC ρ-independent terminator. MicL is transcribed from a σE-dependent promoter within the cutC locus and then processed into a shorter product (MicL-S). Hfq binds the sRNA. Both MicL and MicL-S inhibit production of Lpp. Right, the CpxQ sRNA is a product of normal cpxP transcript degradation mediated by RNase E. CpxQ is stabilized by Hfq and is able to prevent expression from several mRNAs in trans, including skp and nhaB.
A recent examination of the transcriptomic response to σE overproduction identified MicL as the third sRNA of this ESR [80]. MicL and its dedicated σE-dependent promoter are encoded entirely in the 3’ region of the gene annotated as cutC. MicL is processed into a smaller product (MicL-S), though the mechanism involved is unknown [80]. In any case, both MicL and MicL-S are unique in having only a single mRNA target: the lpp transcript, preventing its translation and promoting its turnover [80]. MicL now accounts for the longstanding observation of reduced lpp mRNA levels during σE overproduction [12]. Lpp is an OM lipoprotein that forms covalent linkages between the OM and the PG cell wall and is the most abundant protein produced by E. coli [81]. Lpp plays no role in OMP or LPS biogenesis, so it is perhaps puzzling why σE deploys MicL as an effector. One proposal is that reducing Lpp production may ease demand on the Lol pathway that delivers lipoproteins to the OM; in doing so, MicL could increase Lol pathway capacity for delivering lipoproteins such as BamD and LptE that do have essential roles in OMP and LPS biogenesis, respectively (Figure 1) [80]. Lpp belongs to a growing cohort of “surface-exposed” lipoproteins, since a population of Lpp is detectable outside the cell [82]. The Bam complex has been implicated in surface-exposure of several lipoproteins and is currently the only mechanism described in E. coli for lipoprotein translocation to the surface [83-86]. Although Lpp surface-exposure is yet to be directly demonstrated as being Bam-dependent, the prospect raises a curious alternate hypothesis for MicL regulation: perhaps MicL reduces Lpp synthesis to decrease demand on the Bam complex for lipoprotein translocation and allow it to become more dedicated to OMP folding, a process that σE monitors intently. Given the homology of RseB to Lol pathway proteins, MicL provides another fascinating link between the σE response and lipoproteins that awaits further exploration.
CpxQ is a product of the 3’ UTR of cpxP mRNA and must therefore be highly abundant during Cpx stress [43,78]. Unlike MicL, CpxQ does not have its own promoter [78]. Rather, normal cpxP mRNA decay liberates CpxQ which is stabilized against degradation by Hfq [78]. In Salmonella, CpxQ production does not affect CpxP levels [78]. But, in E. coli, the presence of CpxQ in the 3’ UTR causes a reduction in cellular CpxP levels [79]. CpxQ negatively regulates several targets in trans, most notably NhaB (an IM sodium-proton antiporter) and the periplasmic chaperone Skp [78,79]. Despite their distinct localizations, NhaB and Skp appear to be regulated by CpxQ for the same purpose: to protect the proton-motive force that is maintained across the IM, which is a source of cellular energy in Gram-negative bacteria [78,79].. NhaB overexpression permeates the IM to protons and its downregulation by CpxQ protects the cell from chemical agents that disrupt the PMF [78]. Skp can challenge the PMF by mislocalizing OMPs into the IM, as discussed later. Multiple Cpx effectors seem to protect the PMF. Another Cpx sRNA, CyaR, acts in a positive-feedback loop to overproduce YqaE, a CpxR-induced IM protein with homology to eukaryotic proteins that modulate membrane potential [60]. The cpxPQ RNA transcript specifies two stress effectors directed to different cellular compartments. Despite a common origin, CpxP and CpxQ appear to have distinct roles in combatting stress since there is no condition identified to-date that requires both effectors [78,79]. sRNAs typically fine-tune transcriptional responses; how sRNAs influence the fitness of cells experiencing stress has been better assessed for some sRNAs than others. A more complete appreciation of how newly identified sRNAs contribute to alleviating stress is an ongoing goal.
A hierarchical Cpx-σE regulatory axis
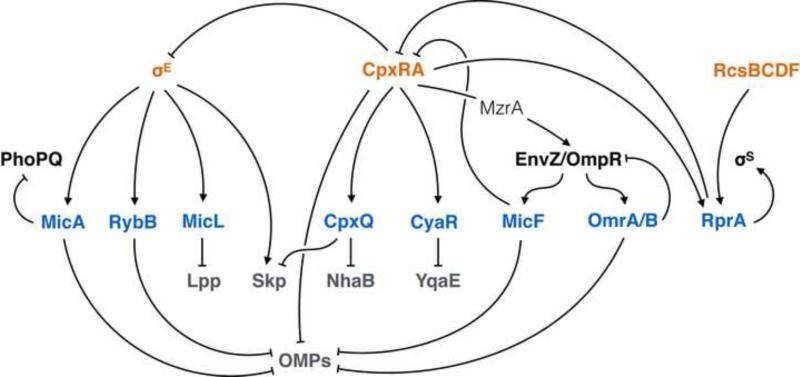
A key question of ESR signal transduction pathways is how they interact with one another. With further characterization of the Cpx sRNAs, it is notable that Cpx and σE share multiple linkages for inter-ESR communication (Figure 4). Whereas the σE response aims to restore OMP folding by inducing chaperone production to maintain nascent OMPs in folding-competent states and by increasing expression of Bam machinery, the Cpx response appears to act antagonistically to σE at several points. For example: CpxR represses the rpoE-rseA-rseB operon to prevent σE production [43,87]; Cpx directly represses major OMP production; deploys sRNAs to further inhibit OMP synthesis [63,88]; and CpxQ inhibits production of the σE-induced chaperone Skp [78,79].
Figure 4. ESR sRNAs, their targets and the interconnected σE-Cpx network.
The Cpx response can directly inhibit σE by CpxR binding and repression upstream of the rpoE gene. CpxR directly and potently represses expression of abundant OMPs, including OmpC and OmpF. CpxR also represses OMP synthesis indirectly using OmpR-regulated sRNAs; CpxR does this by inducing MzrA to stimulate the EnvZ/OmpR signal transduction pathway. CpxQ production inhibits synthesis of the σE effector Skp. MicA, RprA, and MicF each interconnect multiple signal transductions systems. Major ESRs are denoted in orange; other signal transduction systems are in black; sRNA products are in blue; gene targets are in grey.
Several findings hint that Cpx may function as a failsafe mechanism to protect against prolonged σE responses. While the Bam complex catalyzes OMP folding in vivo, direct folding of OMPs into membranes in vitro is well established [89]. Skp, unlike the major chaperone SurA, can promote direct OMP insertion into membranes [90,91]. Defects in the primary SurA-Bam OMP pathway activate σE in order to maintain OMPs in folding-competent states within the periplasm, including by up-regulating Skp [53]. From the periplasm, Skp could assist direct OMP insertion into the OM or could mislocalize OMPs into the IM. OMP mislocalization introduces an ion permeable pore through the IM and is toxic since it collapses the PMF [92]. Indeed, tethering an OMP to the IM results in Skp-dependent toxicity that bears the hallmarks of PMF depletion [79]. The Cpx response combats OMP mislocalization by inhibiting Skp synthesis via CpxQ [78,79].
An OMP “β-sequence” has been proposed to mark nascent proteins in the periplasm as Bam complex substrates that require β-barrel folding [93]. Additionally, OMPs with mutated C termini are poor substrates for Bam, but retain their inherent ability to directly fold into membranes [89]. These OMP mutants might be more likely to attempt direct membrane insertion because they fail to effectively engage Bam. Expressing such OMP mutants in vivo potently activates Cpx responses, despite Cpx not being sensitive to misfolded OMP cues [94].
In all, the Cpx-σE regulatory axis appears designed to allow initial attempts at OMP assembly recovery via the σE response. However, in an ultimate effort to protect the IM and the energy generating capability of the cell, Cpx is engaged to halt σE responses, prevent OMP folding, and—by overproducing the periplasmic chaperone-protease DegP—degrade remaining unfolded or misfolded OMPs.
Concluding Remarks
Simple models of stress have been invaluable in identifying ESRs and their effectors. Nonetheless, even in the most thoroughly characterized ESRs, questions remain about how stress is sensed (see Outstanding Questions). During transitions between ecological niches or during infection, bacteria encounter continuous, complex environmental changes that could trigger multiple stress inputs and it remains to be seen how cells interpret these complex signals to mount a coherent response against this onslaught. The ESR sRNAs have clear functions in regulating effectors as well as feedback functions in regulating responses. Notably, the sRNAs appear to provide numerous avenues for inter-ESR signaling, including by controlling transcripts specifying proteins at the apex of stress signaling circuits (Figure 4). We suggest that sRNAs will prove to play important roles in coordinating the highly interconnected stress responsive network.
BOX 1 The Rcs ESR is a complex TCS phosphorelay system.
At the core of the Rcs system is the IM histidine kinase RcsC and response regulator RcsB. Stress activates the RcsC kinase activity to ultimately phosphorylate the IM phosphotransferase RcsD which then passes the phosphate to RcsB [67]. Part of the Rcs regulon is controlled by RcsB directly, but regulation of some genes additionally requires the auxiliary transcription factor RcsA [95]. The IM protein IgaA is a negative regulator that inhibits RcsC activation [96]. Well characterized Rcs inducing cues include damage to the LPS cell surface layer caused by cationic antimicrobial peptides (cAMPs) and peptidoglycan cell wall biogenesis defects [97,98]. The OM lipoprotein RcsF is the sensor of these stresses at the cell surface and in the periplasm [9,84,86]. It is proposed that RcsF stress sensing allows the protein to interact with IgaA and thereby relieve inhibition of RcsC, initiating the signaling cascade. A major outcome of the Rcs response is overproduction of the exopolysaccharide colanic acid (which aids in resisting cAMPs) and the system also acts to represses flagella production. Differential regulation of colanic acid and flagella is important for Rcs to promote biofilm maturation [67].
Outstanding questions.
What is the nature of the LPS cue that binds RseB and activates σE response? Are σE-inducing mutant LPS forms poorly transported to the OM? Do transport defects lead to LPS excretion directly into the periplasm? Does RseB receive LPS directly, or is the glycolipid transferred from an auxiliary protein?
Can the interaction between CpxA and CpxP be further defined? How does CpxP inhibit CpxA activation? How can CpxA detect such a diverse range of stress conditions?
Does MicL regulate Lpp to prioritize Lol pathway transport of essential lipoproteins? Is Lpp regulation important during σE-inducing stress conditions?
How do MicF and RprA sRNAs fulfil negative feedback on the Cpx response?
Does Cpx protect the IM from OMP mislocalization? How are such events sensed?
How are activating signals from multiple activated ESRs integrated into a protective output response?
The ESR sRNA network provides extensive interfacing between stress systems, are these connections important during complex stress or during infection?
Trends box.
Bacterial cell envelope biogenesis and integrity is continuously monitored by dedicated stress responses.
In Escherichia coli, the σE and Cpx responses maintain homeostasis of the outer membrane and the inner membrane, respectively.
A proposed lipid signal for σE activation has expanded the sensing repertoire of this response.
sRNAs have become prominent regulators of stress responses, providing effector functions as well as interfacing with other signal transduction systems.
The σE and Cpx signalling exhibits extensive signalling linkages and appear to be antagonistic, perhaps because the cell prioritizes protecting the energy generating functions of the inner membrane.
Acknowledgements
This work was supported by NIGMS grant GM34821 to T.J.S.
Glossary
- Diderm
Cells that enveloped by two membranes
- β-barrel proteins (termed OMPs)
A transmembrane OM protein that is formed by wrapping a series of β-sheets into a cylindrical structure. The lumen of the cylinder of OMPs that function as porins forms an aqueous pore through the OM bilayer that allows diffusion of hydrophilic molecules, including nutrients, into the cell
- Lipopolysaccharide (LPS).
A glycolipid produced by Gram-negative bacteria consisting of a hexa-acylated diglucosamine to which a series of sugars are sequentially added. These additional sugars are collectively termed the LPS core polysaccharide
- Lipoprotein
A protein that is triacylated in the IM at an invariant cysteine residue. Lipoproteins destined for the OM are marked by the absence of an aspartate residue adjacent to the cysteine, which permits their entry into the Lol pathway
- Two component system (TCS)
A common bacterial signal transduction system consisting of an IM histidine kinase that detects sensory input and phosphorylates a cognate DNA-binding response regulator. Many TCSs control gene expression
- Small RNAs (sRNAs)
Short RNA products that base-pair with multiple target mRNA transcripts in order to regulate their stability or translation
- PAS domain
A signal sensory domain that is common in signaling proteins throughout bacteria, archea, and eukaryotes
- Curli fimbrae
Amyloid fibrils of curli protein produced and secreted by E. coli
- Proton motive force (PMF)
A gradient of H+ concentration across the IM, with a higher H+ concentration in the periplasm in comparison to the cytosol. The gradient is maintained to establish an electrical potential across the IM. Regulated movement of H+ into the cytosol provides energy to a number of cellular processes, including generation of ATP
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Silhavy TJ, et al. The bacterial cell envelope. Cold Spring Harb. Perspect. Biol. 2010;2:a000414. doi: 10.1101/cshperspect.a000414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nikaido H. Molecular basis of bacterial outer membrane permeability revisited. Microbiol. Mol. Biol. Rev. 2003;67:593–656. doi: 10.1128/MMBR.67.4.593-656.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Okuda S, et al. Lipopolysaccharide transport and assembly at the outer membrane: the PEZ model. Nat. Rev. Microbiol. 2016;14:337–345. doi: 10.1038/nrmicro.2016.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ricci DP, Silhavy TJ. The Bam machine: a molecular cooper. Biochim. Biophys. Acta. 2012;1818:1067–1084. doi: 10.1016/j.bbamem.2011.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Okuda S, Tokuda H. Lipoprotein sorting in bacteria. Annu Rev Microbiol. 2011;65:239–259. doi: 10.1146/annurev-micro-090110-102859. [DOI] [PubMed] [Google Scholar]
- 6.Plummer AM, Fleming KG. From Chaperones to the Membrane with a BAM! Trends Biochem Sci. 2016;41:872–882. doi: 10.1016/j.tibs.2016.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Barchinger SE, Ades SE. Regulated proteolysis: control of the Escherichia coli σE-dependent cell envelope stress response. Subcell. Biochem. 2013;66:129–160. doi: 10.1007/978-94-007-5940-4_6. [DOI] [PubMed] [Google Scholar]
- 8.Raivio TL. Everything old is new again: an update on current research on the Cpx envelope stress response. Biochim. Biophys. Acta. 2014;1843:1529–1541. doi: 10.1016/j.bbamcr.2013.10.018. [DOI] [PubMed] [Google Scholar]
- 9.Konovalova A, et al. A lipoprotein/β-barrel complex monitors lipopolysaccharide integrity transducing information across the outer membrane. Elife. 2016;5:e15276. doi: 10.7554/eLife.15276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dalebroux ZD, Miller SI. Salmonellae PhoPQ regulation of the outer membrane to resist innate immunity. Curr. Opin. Microbiol. 2014;17:106–113. doi: 10.1016/j.mib.2013.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Egger LA, et al. Signal transduction via the histidyl-aspartyl phosphorelay. Genes Cells. 1997;2:167–184. doi: 10.1046/j.1365-2443.1997.d01-311.x. [DOI] [PubMed] [Google Scholar]
- 12.Rhodius VA, et al. Conserved and variable functions of the sigmaE stress response in related genomes. PLoS Biol. 2006;4:e2. doi: 10.1371/journal.pbio.0040002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ades SE, et al. The Escherichia coli σE-dependent extracytoplasmic stress response is controlled by the regulated proteolysis of an anti-σ factor. Genes Dev. 1999;13:2449–2461. doi: 10.1101/gad.13.18.2449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Walsh NP, et al. OMP peptide signals initiate the envelope-stress response by activating DegS protease via relief of inhibition mediated by its PDZ domain. Cell. 2003;113:61–71. doi: 10.1016/s0092-8674(03)00203-4. [DOI] [PubMed] [Google Scholar]
- 15.Chaba R, et al. Signal integration by DegS and RseB governs the σE-mediated envelope stress response in Escherichia coli. Proc. Natl. Acad. Sci. U.S.A. 2011;108:2106–2111. doi: 10.1073/pnas.1019277108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Alba BM, et al. degS (hhoB) is an essential Escherichia coli gene whose indispensable function is to provide σE activity. Mol. Microbiol. 2001;40:1323–1333. doi: 10.1046/j.1365-2958.2001.02475.x. [DOI] [PubMed] [Google Scholar]
- 17.Wilken C, et al. Crystal structure of the DegS stress sensor: How a PDZ domain recognizes misfolded protein and activates a protease. Cell. 2004;117:483–494. doi: 10.1016/s0092-8674(04)00454-4. [DOI] [PubMed] [Google Scholar]
- 18.Grigorova IL, et al. Fine-tuning of the Escherichia coli σE envelope stress response relies on multiple mechanisms to inhibit signal-independent proteolysis of the transmembrane anti-σ factor, RseA. Genes Dev. 2004;18:2686–2697. doi: 10.1101/gad.1238604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Akiyama Y, et al. RseP (YaeL), an Escherichia coli RIP protease, cleaves transmembrane sequences. EMBO J. 2004;23:4434–4442. doi: 10.1038/sj.emboj.7600449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Akiyama K, et al. Roles of the membrane-reentrant β-hairpin-like loop of RseP protease in selective substrate cleavage. Elife. 2015;4:e08928. doi: 10.7554/eLife.08928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Flynn JM, et al. Modulating substrate choice: the SspB adaptor delivers a regulator of the extracytoplasmic-stress response to the AAA+ protease ClpXP for degradation. Genes Dev. 2004;18:2292–2301. doi: 10.1101/gad.1240104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lima S, et al. Dual molecular signals mediate the bacterial response to outer- membrane stress. Science. 2013;340:837–841. doi: 10.1126/science.1235358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cezairliyan BO, Sauer RT. Inhibition of regulated proteolysis by RseB. Proc. Natl. Acad. Sci. U.S.A. 2007;104:3771–3776. doi: 10.1073/pnas.0611567104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wollmann P, Zeth K. The structure of RseB: a sensor in periplasmic stress response of E. coli. J. Mol. Biol. 2007;372:927–941. doi: 10.1016/j.jmb.2007.06.039. [DOI] [PubMed] [Google Scholar]
- 25.Raetz CRH, Whitfield C. Lipopolysaccharide endotoxins. Annu. Rev. Biochem. 2002;71:635–700. doi: 10.1146/annurev.biochem.71.110601.135414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chng S-S, et al. Proteins required for lipopolysaccharide assembly in Escherichia coli form a transenvelope complex. Biochemistry. 2010;49:4565–4567. doi: 10.1021/bi100493e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Freinkman E, et al. Regulated assembly of the transenvelope protein complex required for lipopolysaccharide export. Biochemistry. 2012;51:4800–4806. doi: 10.1021/bi300592c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sperandeo P, et al. Functional analysis of the protein machinery required for transport of lipopolysaccharide to the outer membrane of Escherichia coli. J. Bacteriol. 2008;190:4460–4469. doi: 10.1128/JB.00270-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ruiz N, et al. Identification of two inner-membrane proteins required for the transport of lipopolysaccharide to the outer membrane of Escherichia coli. Proc. Natl. Acad. Sci. U.S.A. 2008;105:5537–5542. doi: 10.1073/pnas.0801196105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Okuda S, Tokuda H. Model of mouth-to-mouth transfer of bacterial lipoproteins through inner membrane LolC, periplasmic LolA, and outer membrane LolB. 2009;106:5877–5882. doi: 10.1073/pnas.0900896106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Evans KL, et al. Eliminating a set of four penicillin binding proteins triggers the Rcs phosphorelay and Cpx stress responses in Escherichia coli. J. Bacteriol. 2013;195:4415–4424. doi: 10.1128/JB.00596-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Delhaye A, et al. Fine-tuning of the Cpx envelope stress response is required for cell wall homeostasis in Escherichia coli. mBio. 2016;7:e00047–16. doi: 10.1128/mBio.00047-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bernal-Cabas M, et al. The Cpx envelope stress response modifies peptidoglycan cross-linking via the L,D-transpeptidase LdtD and the novel protein YgaU. J. Bacteriol. 2015;197:603–614. doi: 10.1128/JB.02449-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Raivio TL, Silhavy TJ. The σE and Cpx regulatory pathways: overlapping but distinct envelope stress responses. Curr. Opin. Microbiol. 1999;2:159–165. doi: 10.1016/S1369-5274(99)80028-9. [DOI] [PubMed] [Google Scholar]
- 35.Fleischer R, et al. Purification, reconstitution, and characterization of the CpxRAP envelope stress system of Escherichia coli. J. Biol. Chem. 2007;282:8583–8593. doi: 10.1074/jbc.M605785200. [DOI] [PubMed] [Google Scholar]
- 36.Raivio TL, Silhavy TJ. Transduction of envelope stress in Escherichia coli by the Cpx two-component system. J. Bacteriol. 1997;179:7724–7733. doi: 10.1128/jb.179.24.7724-7733.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kwon E, et al. The crystal structure of the periplasmic domain of Vibrio parahaemolyticus CpxA. Protein Sci. 2012;21:1334–1343. doi: 10.1002/pro.2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.DiGiuseppe PA, Silhavy TJ. Signal detection and target gene induction by the CpxRA two-component system. J. Bacteriol. 2003;185:2432–2440. doi: 10.1128/JB.185.8.2432-2440.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Snyder WB, et al. Overproduction of NlpE, a new outer membrane lipoprotein, suppresses the toxicity of periplasmic LacZ by activation of the Cpx signal transduction pathway. J. Bacteriol. 1995;177:4216–4223. doi: 10.1128/jb.177.15.4216-4223.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Danese PN, Silhavy TJ. CpxP, a stress-combative member of the Cpx regulon. J. Bacteriol. 1998;180:831–839. doi: 10.1128/jb.180.4.831-839.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Otto K, Silhavy TJ. Surface sensing and adhesion of Escherichia coli controlled by the Cpx-signaling pathway. Proc. Natl. Acad. Sci. U.S.A. 2002;99:2287–2292. doi: 10.1073/pnas.042521699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hirano Y, et al. Structural studies of the Cpx pathway activator NlpE on the outer membrane of Escherichia coli. Structure. 2007;15:963–976. doi: 10.1016/j.str.2007.06.014. [DOI] [PubMed] [Google Scholar]
- 43.Price NL, Raivio TL. Characterization of the Cpx regulon in Escherichia coli strain MC4100. J. Bacteriol. 2009;191:1798–1815. doi: 10.1128/JB.00798-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Raivio TL, et al. The Cpx envelope stress response is controlled by amplification and feedback inhibition. J. Bacteriol. 1999;181:5263–5272. doi: 10.1128/jb.181.17.5263-5272.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhou X, et al. Structural basis for two-component system inhibition and pilus sensing by the auxiliary CpxP protein. J. Biol. Chem. 2011;286:9805–9814. doi: 10.1074/jbc.M110.194092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Thede GL, et al. Structure of the periplasmic stress response protein CpxP. J. Bacteriol. 2011;193:2149–2157. doi: 10.1128/JB.01296-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Buelow DR, Raivio TL. Cpx signal transduction is influenced by a conserved N- terminal domain in the novel inhibitor CpxP and the periplasmic protease DegP. J. Bacteriol. 2005;187:6622–6630. doi: 10.1128/JB.187.19.6622-6630.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tschauner K, et al. Dynamic interaction between the CpxA sensor kinase and the periplasmic accessory protein CpxP mediates signal recognition in E. coli. PLoS ONE. 2014;9:e107383. doi: 10.1371/journal.pone.0107383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Isaac DD, et al. The extracytoplasmic adaptor protein CpxP is degraded with substrate by DegP. Proc. Natl. Acad. Sci. U.S.A. 2005;102:17775–17779. doi: 10.1073/pnas.0508936102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Vogel J, Luisi BF. Hfq and its constellation of RNA. Nat. Rev. Microbiol. 2011;9:578–589. doi: 10.1038/nrmicro2615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Majdalani N, et al. Regulation of RpoS by a novel small RNA: the characterization of RprA. Mol. Microbiol. 2001;39:1382–1394. doi: 10.1111/j.1365-2958.2001.02329.x. [DOI] [PubMed] [Google Scholar]
- 52.Zafar MA, et al. Transcriptional occlusion caused by overlapping promoters. Proc. Natl. Acad. Sci. U.S.A. 2014;111:1557–1561. doi: 10.1073/pnas.1323413111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Dartigalongue C, et al. Characterization of the Escherichia coli σE regulon. J. Biol. Chem. 2001;276:20866–20875. doi: 10.1074/jbc.M100464200. [DOI] [PubMed] [Google Scholar]
- 54.Bossi L, Figueroa-Bossi N. A small RNA downregulates LamB maltoporin in Salmonella. Mol. Microbiol. 2007;65:799–810. doi: 10.1111/j.1365-2958.2007.05829.x. [DOI] [PubMed] [Google Scholar]
- 55.Rasmussen AA, et al. Regulation of ompA mRNA stability: the role of a small regulatory RNA in growth phase-dependent control. Mol. Microbiol. 2005;58:1421–1429. doi: 10.1111/j.1365-2958.2005.04911.x. [DOI] [PubMed] [Google Scholar]
- 56.Coornaert A, et al. MicA sRNA links the PhoP regulon to cell envelope stress. Mol. Microbiol. 2010;76:467–479. doi: 10.1111/j.1365-2958.2010.07115.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Johansen J, et al. Conserved small non-coding RNAs that belong to the σE regulon: role in down-regulation of outer membrane proteins. J. Mol. Biol. 2006;364:1–8. doi: 10.1016/j.jmb.2006.09.004. [DOI] [PubMed] [Google Scholar]
- 58.Gogol EB, et al. Small RNAs endow a transcriptional activator with essential repressor functions for single-tier control of a global stress regulon. Proc. Natl. Acad. Sci. U.S.A. 2011;108:12875–12880. doi: 10.1073/pnas.1109379108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Raivio TL, et al. The Escherichia coli Cpx envelope stress response regulates genes of diverse function that impact antibiotic resistance and membrane integrity. J. Bacteriol. 2013;195:2755–2767. doi: 10.1128/JB.00105-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Vogt SL, et al. The Cpx envelope stress response regulates and is regulated by small noncoding RNAs. J. Bacteriol. 2014;196:4229–4238. doi: 10.1128/JB.02138-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Guillier M, Gottesman S. Remodelling of the Escherichia coli outer membrane by two small regulatory RNAs. Mol. Microbiol. 2006;59:231–247. doi: 10.1111/j.1365-2958.2005.04929.x. [DOI] [PubMed] [Google Scholar]
- 62.Mizuno T, et al. A unique mechanism regulating gene expression: translational inhibition by a complementary RNA transcript (micRNA). Proc. Natl. Acad. Sci. U.S.A. 1984;81:1966–1970. doi: 10.1073/pnas.81.7.1966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Gerken H, et al. MzrA: a novel modulator of the EnvZ/OmpR two-component regulon. Mol. Microbiol. 2009;72:1408–1422. doi: 10.1111/j.1365-2958.2009.06728.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Gerken H, Misra R. MzrA-EnvZ Interactions in the Periplasm Influence the EnvZ/OmpR Two-Component Regulon. J. Bacteriol. 2010;192:6271–6278. doi: 10.1128/JB.00855-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Guillier M, Gottesman S. The 5′ end of two redundant sRNAs is involved in the regulation of multiple targets, including their own regulator. Nucleic Acids Res. 2008;36:6781–6794. doi: 10.1093/nar/gkn742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Holmqvist E, et al. A mixed double negative feedback loop between the sRNA MicF and the global regulator Lrp. Mol. Microbiol. 2012;84:414–427. doi: 10.1111/j.1365-2958.2012.07994.x. [DOI] [PubMed] [Google Scholar]
- 67.Majdalani N, Gottesman S. The Rcs phosphorelay: a complex signal transduction system. Annu Rev Microbiol. 2005;59:379–405. doi: 10.1146/annurev.micro.59.050405.101230. [DOI] [PubMed] [Google Scholar]
- 68.Majdalani N, et al. Regulation and mode of action of the second small RNA activator of RpoS translation, RprA. Mol. Microbiol. 2002;46:813–826. doi: 10.1046/j.1365-2958.2002.03203.x. [DOI] [PubMed] [Google Scholar]
- 69.Mika F, Hengge R. Small RNAs in the control of RpoS, CsgD, and biofilm architecture of Escherichia coli. RNA biology. 2014;11:494–507. doi: 10.4161/rna.28867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Weber H, et al. Cyclic-di-GMP-mediated signalling within the sigma network of Escherichia coli. Mol. Microbiol. 2006;62:1014–1034. doi: 10.1111/j.1365-2958.2006.05440.x. [DOI] [PubMed] [Google Scholar]
- 71.Mika F, et al. Targeting of csgD by the small regulatory RNA RprA links stationary phase, biofilm formation and cell envelope stress in Escherichia coli. DOI. 2012 doi: 10.1111/j.1365-2958.2012.08002.x. 10.1111/j.1365-2958.2012.08002.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Boehm A, Vogel J. The csgD mRNA as a hub for signal integration via multiple small RNAs. Mol. Microbiol. 2012;84:1–5. doi: 10.1111/j.1365-2958.2012.08033.x. [DOI] [PubMed] [Google Scholar]
- 73.Prigent-Combaret C, et al. Complex regulatory network controls initial adhesion and biofilm formation in Escherichia coli via regulation of the csgD gene. J. Bacteriol. 2001;183:7213–7223. doi: 10.1128/JB.183.24.7213-7223.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Gerstel U, et al. Complex regulation of csgD promoter activity by global regulatory proteins. Mol. Microbiol. 2003;49:639–654. doi: 10.1046/j.1365-2958.2003.03594.x. [DOI] [PubMed] [Google Scholar]
- 75.Hess JF, et al. Protein phosphorylation is involved in bacterial chemotaxis. Proc. Natl. Acad. Sci. U.S.A. 1987;84:7609–7613. doi: 10.1073/pnas.84.21.7609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Ansaldi M, et al. TorI, a response regulator inhibitor of phage origin in Escherichia coli. Proc. Natl. Acad. Sci. U.S.A. 2004;101:9423–9428. doi: 10.1073/pnas.0401927101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Chao Y, et al. An atlas of Hfq-bound transcripts reveals 3' UTRs as a genomic reservoir of regulatory small RNAs. EMBO J. 2012;31:4005–4019. doi: 10.1038/emboj.2012.229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Chao Y, Vogel J. A 3' UTR-derived small RNA provides the regulatory noncoding arm of the inner membrane stress response. Mol Cell. 2016;61:352–363. doi: 10.1016/j.molcel.2015.12.023. [DOI] [PubMed] [Google Scholar]
- 79.Grabowicz M, et al. The CpxQ sRNA negatively regulates Skp to prevent mistargeting of β-Barrel outer membrane proteins into the cytoplasmic membrane. mBio. 2016;7:e00312–16. doi: 10.1128/mBio.00312-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Guo MS, et al. MicL, a new σE-dependent sRNA, combats envelope stress by repressing synthesis of Lpp, the major outer membrane lipoprotein. Genes Dev. 2014;28:1620–1634. doi: 10.1101/gad.243485.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Braun V, Sieglin U. The covalent murein-lipoprotein structure of the Escherichia coli cell wall. The attachment site of the lipoprotein on the murein. Eur J Biochem. 1970;13:336–346. doi: 10.1111/j.1432-1033.1970.tb00936.x. [DOI] [PubMed] [Google Scholar]
- 82.Cowles CE, et al. The free and bound forms of Lpp occupy distinct subcellular locations in Escherichia coli. Mol. Microbiol. 2011;79:1168–1181. doi: 10.1111/j.1365-2958.2011.07539.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Webb CT, et al. Dynamic association of BAM complex modules includes surface exposure of the lipoprotein BamC. J. Mol. Biol. 2012;422:545–555. doi: 10.1016/j.jmb.2012.05.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Konovalova A, et al. Transmembrane domain of surface-exposed outer membrane lipoprotein RcsF is threaded through the lumen of β-barrel proteins. Proc. Natl. Acad. Sci. U.S.A. 2014;111:E4350–8. doi: 10.1073/pnas.1417138111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Lenhart TR, Akins DR. Borrelia burgdorferi locus BB0795 encodes a BamA orthologue required for growth and efficient localization of outer membrane proteins. Mol. Microbiol. 2010;75:692–709. doi: 10.1111/j.1365-2958.2009.07015.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Cho S-H, et al. Detecting envelope stress by monitoring β-barrel assembly. Cell. 2014;159:1652–1664. doi: 10.1016/j.cell.2014.11.045. [DOI] [PubMed] [Google Scholar]
- 87.De Wulf P, et al. Genome-wide profiling of promoter recognition by the two-component response regulator CpxR-P in Escherichia coli. J. Biol. Chem. 2002;277:26652–26661. doi: 10.1074/jbc.M203487200. [DOI] [PubMed] [Google Scholar]
- 88.Batchelor E, et al. The Escherichia coli CpxA-CpxR envelope stress response system regulates expression of the porins ompF and ompC. J. Bacteriol. 2005;187:5723–5731. doi: 10.1128/JB.187.16.5723-5731.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Gessmann D, et al. Outer membrane β-barrel protein folding is physically controlled by periplasmic lipid head groups and BamA. Proc. Natl. Acad. Sci. U.S.A. 2014;111:5878–5883. doi: 10.1073/pnas.1322473111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.McMorran LM, et al. Dissecting the effects of periplasmic chaperones on the in vitro folding of the outer membrane protein PagP. J. Mol. Biol. 2013;425:3178–3191. doi: 10.1016/j.jmb.2013.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Burmann BM, et al. Conformation and dynamics of the periplasmic membrane- protein-chaperone complexes OmpX-Skp and tOmpA-Skp. Nat Struct Mol Biol. 2013;20:1265–1272. doi: 10.1038/nsmb.2677. [DOI] [PubMed] [Google Scholar]
- 92.Carlson JH, Silhavy TJ. Signal sequence processing is required for the assembly of LamB trimers in the outer membrane of Escherichia coli. J. Bacteriol. 1993;175:3327–3334. doi: 10.1128/jb.175.11.3327-3334.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Hagan CL, et al. Inhibition of the β-barrel assembly machine by a peptide that binds BamD. Proc. Natl. Acad. Sci. U.S.A. 2015;112:2011–2016. doi: 10.1073/pnas.1415955112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Gerken H, et al. Involvement and necessity of the Cpx regulon in the event of aberrant β-barrel outer membrane protein assembly. Mol. Microbiol. 2010;75:1033–1046. doi: 10.1111/j.1365-2958.2009.07042.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Wehland M, Bernhard F. The RcsAB box. Characterization of a new operator essential for the regulation of exopolysaccharide biosynthesis in enteric bacteria. J. Biol. Chem. 2000;275:7013–7020. doi: 10.1074/jbc.275.10.7013. [DOI] [PubMed] [Google Scholar]
- 96.Cano DA, et al. Regulation of capsule synthesis and cell motility in Salmonella enterica by the essential gene igaA. Genetics. 2002;162:1513–1523. doi: 10.1093/genetics/162.4.1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Laubacher ME, Ades SE. The Rcs phosphorelay is a cell envelope stress response activated by peptidoglycan stress and contributes to intrinsic antibiotic resistance. J. Bacteriol. 2008;190:2065–2074. doi: 10.1128/JB.01740-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Farris C, et al. Antimicrobial peptides activate the Rcs regulon through the outer membrane lipoprotein RcsF. J. Bacteriol. 2010;192:4894–4903. doi: 10.1128/JB.00505-10. [DOI] [PMC free article] [PubMed] [Google Scholar]