Abstract
Factors that stimulate the migration of fallopian tube epithelial (FTE)-derived high-grade serous ovarian cancer (HGSOC) to the ovary are poorly elucidated. This study characterized the effect of the ovarian hormone, activin A, on normal FTE and HGSOC. Activin A and TGFβ1 induced an epithelial-to-mesenchymal transition in murine oviductal epithelial (MOE) cells, but only activin A increased migration. The migratory effect of activin A was independent of Smad2/3 and required phospho-AKT, phospho-ERK, and Rac1. Exogenous activin A stimulated migration of the HGSOC cell line OVCAR3 through a similar mechanism. Activin A signaling inhibitors, SB431542 and follistatin, reduced migration in OVCAR4 cells, which expressed activin A subunits (encoded by INHBA). Murine superovulation increased phospho-Smad2/3 immunostaining in the FTE. In Oncomine, transcripts for the activin A receptors (ACVR1B and ACVR2A) were higher in serous tumors relative to the normal ovary, while inhibitors of activin A signaling (INHA and TGFB3) were lower. High expression of both INHBA and ACVR2A, but not TGFβ receptors or co-receptors, was associated with shorter disease-free survival in serous cancer patients. These results suggest activin A stimulates migration of FTE-derived tumors to the ovary.
1. Introduction
Ovarian cancer is the fifth leading cause of cancer death in women and the most lethal gynecologic malignancy [1]. Epithelial ovarian cancers were historically thought to arise from the ovarian surface epithelium (OSE). However, it is now clear that high-grade serous ovarian cancer (HGSOC), the most common and deadly histotype, can also arise from the fallopian tube epithelium (FTE, analogous to oviductal epithelium in non-primate species). As evidence of an FTE origin, several studies identified putative precursor lesions in the fallopian tube of women undergoing prophylactic oophorectomy called p53 signatures, which were patches of secretory epithelium that express mutant p53 protein [2–4]. The transcriptomic profile of HGSOC tumors significantly correlated with that of the fallopian tube [5], and the genomic methylation patterns of HGSOC were more similar to the FTE than the OSE [6]. A recent retrospective analysis found salpingectomy was associated with a 45% decrease in ovarian cancer risk [7]. Our laboratory has shown that murine oviductal epithelial (MOE) cells stably expressing PTENshRNA, PTENshRNA+KRASG12V, or PTENshRNA+TP53R273H colonized the ovary following intraperitoneal xenograft [8,9]. Several transgenic mouse models have also been developed that form fallopian tube-derived tumors that spread to the ovary [10–12].
Factors other than physical proximity likely contribute to colonization of the ovary by fallopian tube-derived tumors. For example, Coffman et al. found that when injected intravenously, HGSOC cells, but not breast cancer or lung adenocarcinoma cells, colonized the ovary [13]. The lifetime number of ovulations is positively associated with the risk of developing ovarian cancer [14]. Yang-Hartwich et al. found that superovulation, following xenografting of tumorigenic cells into the uterine lumen, increased the proportion of mice with ovarian tumors [15]. In a transgenic mouse model of fallopian tube-derived cancer, ovariectomy reduced peritoneal metastasis [10]. These data suggest that HGSOC cells preferentially colonize the ovary, a phenomena linked to ovulation, and indicate that blocking this migratory step may represent a therapeutic target in women. However, the factor(s) that draw HGSOC cells to the ovary are just beginning to be elucidated [15].
Activin A is a member of the TGFβ superfamily and a classic ovarian hormone. Activin A is composed of two inhibin βA subunits (gene symbol INHBA), while a heterodimer of inhibin α (gene symbol INHA) and inhibin βA produce inhibin A. Activin A signals by binding the type I and type II activin receptors (gene symbols ACVR1B and ACVR2A, respectively). Classically, activin A and TGFβ receptors phosphorylate Smad2/3; however, both hormones can activate other signaling pathways, such as PI3K/AKT and ERK/MEK, in a ligand and cell dependent manner. Inhibin A blocks activin signaling at the receptor level, and follistatin binds directly to activin A by sequestering it away from the receptor [16].
Activin A and TGFβ have been shown to stimulate migration and invasion of various cell types through activation of PI3K/AKT and MEK/MAPK signaling [17]. These pathways can be activated either directly or indirectly through the actions of phospho-Smad2/3. Ligand binding causes the TGFBRII receptor to interact directly with growth factor receptor-bound protein 2 (Grb2) and Shc-tranforming protein (Shc), leading to MEK/MAPK signaling [18]. Both TGFβ and activin A have been shown to activate transforming growth factor beta-activated kinase 1 (TAK1) and increase MEK/MAPK signaling [19,20]. There is also a direct interaction between TGFBRI and the p85 subunit of PI3K, leading to ligand-induced phosphorylation of AKT [21,22]. In addition, the type II activin receptor coimmunoprecipitates with p85 [23]. Activin A and TGFβ activate Rho GTPases (RhoA, Rac1, and Cdc42), which are important regulators of migration. For example, RhoA mediated activin A-induced migration in keratinocytes [24], while TGFβ stimulated migration of pancreatic ductal adenocarcinoma through Rac1 [25]. However, the specific Rho GTPase required for migration of the FTE is unknown.
Increased levels of activin A have been linked to several cancer types. For example, activin A serum concentrations are associated with tumor progression in lung cancer [26]. Additionally, activin A is overexpressed in colorectal cancer [27]. In breast cancer, activin A induces an epithelial-to-mesenchymal transition (EMT) and stimulates migration [28]. In the normal ovary, activin A plays a role in the growth of follicles and their steroidogenesis [29], with concentrations of 5–50 ng/ml of activin A being measured in follicular fluid [30]. In ovarian cancer, activin A is well known to stimulate development of stromal cell tumors, as evidenced by development of these tumors in INHA knockout mice [31]. The role of activin A in epithelial ovarian cancer is less clear. Basu et al. observed higher phospho-Smad2 immunostaining in ovarian tumors compared to normal ovaries [32], and Do et al. found that ovarian cystadenocarinoma patients with high immunostaining for INHBA tended to have shorter survival times than patients with low immunostaining [33]. Activin A stimulates migration of OCC1 and SKOV3 cells [34]. However, OCC1 cells were isolated form ovarian clear cell carcinoma, and recent genomic analysis indicated that SKOV3 cells are not representative of HGSOC [35,36].
The objectives of the current study were to 1) characterize the effects of ovarian-produced activin A on migration and proliferation of the FTE, 2) elucidate the pathways by which activin A stimulates migration of the FTE and determine if it is conserved in HGSOC cell models, and 3) explore expression of activin A (i.e. INHBA) and activin receptors (ACVR1B and ACVR2A) in HGSOC patient tumors.
2. Materials and Methods
2.1 Cell Culture
Spontaneously immortalized murine oviductal epithelial (MOE) cells were a gift from Barbara Vanderhyden at the University of Ottawa and had previously been used by our laboratory to model the FTE [8,9]. MOE cells were maintained in αMEM (10–022-CV, Cellgro, Manassas, VA) supplemented with 10% FBS (FB5001, Denville Scientific, Holliston, MA), 2 mM L-glutamine (30005068, CellGro, Manassas, VA), 10 µg/ml ITS (11074547001, Roche, Indianapolis, IN), 1.8 ng/ml EGF (100–15, Peprotech Inc, Rocky Hill, NJ), 100 U/ml penicillin-streptomycin (15140–122, Gibco, Grand Island, NY), 1 µg/ml gentamycin (30-005-CR, GellGro, Manassas, VA), and 18.2 ng/ml estradiol-17β (E1024-1G, Sigma Aldrich, St. Louis, MO). OVCAR4 cells were maintained in RPMI 1640 media (11875-093, Life Technologies, Waltham, MA) supplemented with 10% FBS, 100 U/ml penicillin-streptomycin, and 2 mM L-glutamine. OVCAR3 cells were maintained in MEM supplemented with 20% FBS, 1% non-essential amino acids (11140050, Gibco, Grand Island, NY), 1% L-glutamine, 0.1 mg/ml insulin (700112P, Gemini Bioproduct, West Sacramento, CA), and 100 U/ml penicillin-streptomycin. All cells were maintained in T-25 or T-75 flasks at 5% CO2 and 37°C in a humidified incubator. Each cell line had been authenticated by STR analysis within the past 2 years. Cells were collected with 1x trypsin/EDTA (15400-054, Life Technologies, Waltham, MA) and passed at confluence.
2.2 Hormones, Inhibitors, and RNAi
Activin A and TGFβ1 were purchased from R&D Systems (338-AC-010 and 7666-MB-005, respectively; Minneapolis, MN). TGFβ1 was dissolved in 4 mM HCl at 10 mg/ml, and activin A was dissolved in 4 mM HCl with 0.1% BSA at 10 mg/ml. Follistatin 228 (a gift from Teresa Woodruff at Northwestern University) was dissolved in PBS and used at 200 ng/ml. Rhosin was purchased from Fisher Scientific (5003, Waltham, MA). All other inhibitors (MK2206, 11593; U0126, 70970; MLS573151, 15369; SB431542, 13031; and NSC23766, 13196) were purchased from Cayman Chemical (Ann Arbor, MI). All inhibitors were dissolved in DMSO at 1,000x and added to media prior to adding treatments to each well.
The Smad2-dominant negative (Smad2DN) construct was a gift from Teresa Woodruff at Northwestern University and has been previously validated [37,38]. Cells were transfected with 500 ng/ml Smad2DN plasmid or empty vector (pcDNA3) with ViaFect (E4981, Promega, Madison, WI) following the manufacturer’s instructions. LuciferasesiRNA (LucsiRNA) and Rac1siRNA were purchased from Sigma (EHURLUC-50UG and EMU028841-20UG, respectively; St. Louis, MO). Each well was transfected with 25 nM siRNA using TransIT-X2 (MIR 6004, Mirus, Madison, WI) following the manufacturer’s instructions. The cells were incubated 24 hours post-transfection before performing experiments.
2.3 Western Blotting
Cells (150,000–450,000 per well, depending on cell line) were plated in 6-well plates in complete media. The next day the media was replaced with serum free MEM. Twenty-four hours later, media with appropriate treatments were added to each well and cell lysates were collected at indicated times in RIPA buffer containing protease and phosphatase inhibitors [39]. Protein concentrations were determined with a BCA assay (23227, Thermo Scientific, Waltham, MA), and 25 µg of protein was separated in a 10% SDS-PAGE gel by applying constant voltage (100 V) for 120 minutes. Proteins were transferred to nitrocellulose membranes, blocked for 1 hour, and incubated overnight in primary antibody. The next day the membrane was washed thee times in TBS-T, incubated with appropriate secondary antibody (anti-rabbit 1:1000 or anti-donkey 1:10,000) conjugated to horseradish peroxidase for 30 minutes at room temperature. Membranes were then washed three times with TBS-T, and developed using SuperSignal West Femto Maximum Sensitivity Substrate (34095, Thermo Scientific, Waltham, MA). Images were captured with a FluorChem E (ProteinSimple, San Jose, CA). The primary antibodies, dilutions, and blocking conditions are listed in Table S1.
2.4 Migration Assays
For scratch assays, a monolayer of cells was seeded onto 24-well plates. After ~6 hours later (OVCAR3) or the next day (MOE and OVCAR4 cells) a uniform scratch was made down the center of each monolayer with a 1000 µl pipette tip. OVCAR3 cells were cultured overnight to achieve confluence (minus the scratched area) before treatment. After scratching, the monolayer was washed two times with PBS and serum free MEM with appropriate treatment was added to each well. Images were captured at 4x at 0 and 24 hours (MOE and OVCAR4) or 0 and 48 hours (OVCAR3). The area of each scratch within the field of view was determined with ImageJ software (http://imagej.nih.gov/ij/).
For Boyden chamber assays, 0.5 ml per well of MEM with appropriate treatments was added to 24 well plates. MOE cells were collected from a T-75 flask with trypsin/EDTA and MOE media. Cells were washed two times with FBS-free MEM and 100,000 MOE cells were added to each insert (8.0 µM pores; PI8P01250, Millipore, Billerica, MA) free of any extracellular matrix proteins and placed in each well. Plates were incubated for 6 hours, and cells that had not migrated through the insert were removed with a cotton swab. Cells on the bottom of the insert were fixed with 4% PFA, permeabilized with 70% methanol, and stained with 0.2% crystal violet. Each insert was washed two times with PBS and dried. Images were captured at 4x to count the number of migrated cells. Cell number was normalized to control.
2.5 SRB Proliferation Assay
Cell proliferation was determined using the sulforhodamine B (SRB) assay [40]. Five hundred MOE cells per well were plated in 100 µl of MEM in 96-well plates. Two hours later the first plate (day 0) was collected and indicated treatment was applied to a second plate, which was collected at day 5. At collection, media was decanted, cells were fixed with 100 µl 20% trichloracetic acid (TCA), and stored at 4°C until development. Plates were rinsed four times with tap water and then 100 µl of SRB was added to each well and incubated for 30 minutes at room temperature. Plates were then washed four times with 1% acetic acid, dried, and SRB was resuspended with 200 µl of Tris buffer. Absorbance was measured at 505 nm using a Synergy 2 microplate reader (BioTek, Winooski, VT). Values are expressed as fold increase over day 0.
2.6 Murine Superovulation
All animals were treated in accordance with the National Institute of Health Guide for the Care and use of Laboratory Animals. Animals were housed in a temperature and light (12L:12D) controlled environment. Water and food were provided ad libitum. CD-1 mice were obtained from in-house breeding, and superovulation was induced as previously described [41]. Briefly, mice were treated on days 1 and 2 with PBS on both days (control) or 5 IU PMSG (day 1) followed by 5 IU hCG on day 2 (superovulation). Twelve hours after the second injection mice were sacrificed, the reproductive tract was excised and fixed in 4% PFA.
Immunohistochemical staining for phospho-Smad2/3 was carried out as previously described [38]. Reproductive tracts were fixed in 4% PFA, embedded in paraffin, and sectioned at 5 µm. Sections were probed with primary antibody against phospho-Smad2/3 (Table S1) overnight, washed, and probed with secondary antibody (1:200 biotinylated anti-rabbit; BA-1000, Vector Laboratory, Burlingame, CA). After washing, slides were mounted and images were captured at 20x or 100x with a Nikon Eclipse E600 microscope.
2.7 Oncomine™ Analysis
Oncomine™ (v4.50, IonTorrent, http://https://www.oncomine.org/resource/login.html) is a privately curated database of microarray and sequencing experiments specific to cancer biology. Oncomine™ was explored and 4 studies of interest were identified [42–45] that compared gene expression in serous ovarian tumors to normal tissue. Two of these studies used ovarian tissue as normal, one study used OSE as normal, and the fourth used peritoneal tissue as normal. In addition, four studies were also identified that compared metastasis to primary tumors. These were Adib et al. [42], Tothill et al. [46], Anglesio et al. [47], and Bittner (unpublished). In both cases, expression of genes related to TGFβ superfamily signaling (ligands and receptors) was analyzed.
2.8 Kaplan-Meier Analysis
OvMark (http://glados.ucd.ie/OvMark/index.html) [48] was used to generate Kaplan-Meier curves for genes of interest. Specifically, OvMark was used to analyze disease-free survival (DFS) in 650–1950 patients (dependent on the gene(s) analyzed) across 14 GEO datasets. While the program does not allow the exclusion of low-grade serous tumors, >2/3 of serous tumors in the datasets are high-grade [48]. Additionally, all available serous samples were used, regardless of therapy, residual tumor status, grade, or FIGO stage, in order to maximize the number of patients included in the analysis.
2.9 Copy Number and mRNA Analysis in cBioPortal
cBioPortal (http://www.cbioportal.org/index.do) was used to analyze copy number and mRNA expression for components of activin A signaling in the TCGA provisional dataset. All complete tumors (n=182) were analyzed for ACVR1B, ACVR2A, and INHBA expression. Samples were analyzed for amplification of INHBA, ACVR1B, and ACVR2A in the genome or altered mRNA expression was defined as a Z score less than −2 or greater than 2.
2.10 RT-PCR
RNA was isolated from OVCAR4 cells with TRIzol (15596026, ThermoFisher, Waltham, MA) per the manufacturer’s instructions. RNA was treated with DNase I for 20 minutes at 37°C and reverse transcribed with the iScript Reverse Transcription Supermix kit (170–8841, Bio-Rad, Hercules, CA). Each PCR reaction contained 25 ng OVCAR4 cDNA, 1x Taq Reaction Buffer, 1.25 U Taq, 200 µM dNTPs, 0.8 µM forward and reverse primers in a final volume of 25 µl. PCR conditions consisted of an initial denaturation step at 95°C for 30 seconds followed by 25 cycles of 95°C for 30 seconds, 52°C for 60 seconds, and 68°C for 30 seconds. The final extension was 68°C for 60 seconds. Products were separated on a 2% agarose cell containing ethidium bromide and images captured with a FluorChem E (ProteinSimple, San Jose, CA).
2.11 Statistical Analysis
Each experiment was replicated at least three times and data is presented as mean ± SEM. Data were analyzed by a T-test, paired T-test, or ANOVA followed by Dunnett’s post hoc analysis. P≤0.05 was considered significant. Analysis was carried out with PRISM version 6.0b.
3. Results
3.1 Activin A Stimulates Migration of the FTE
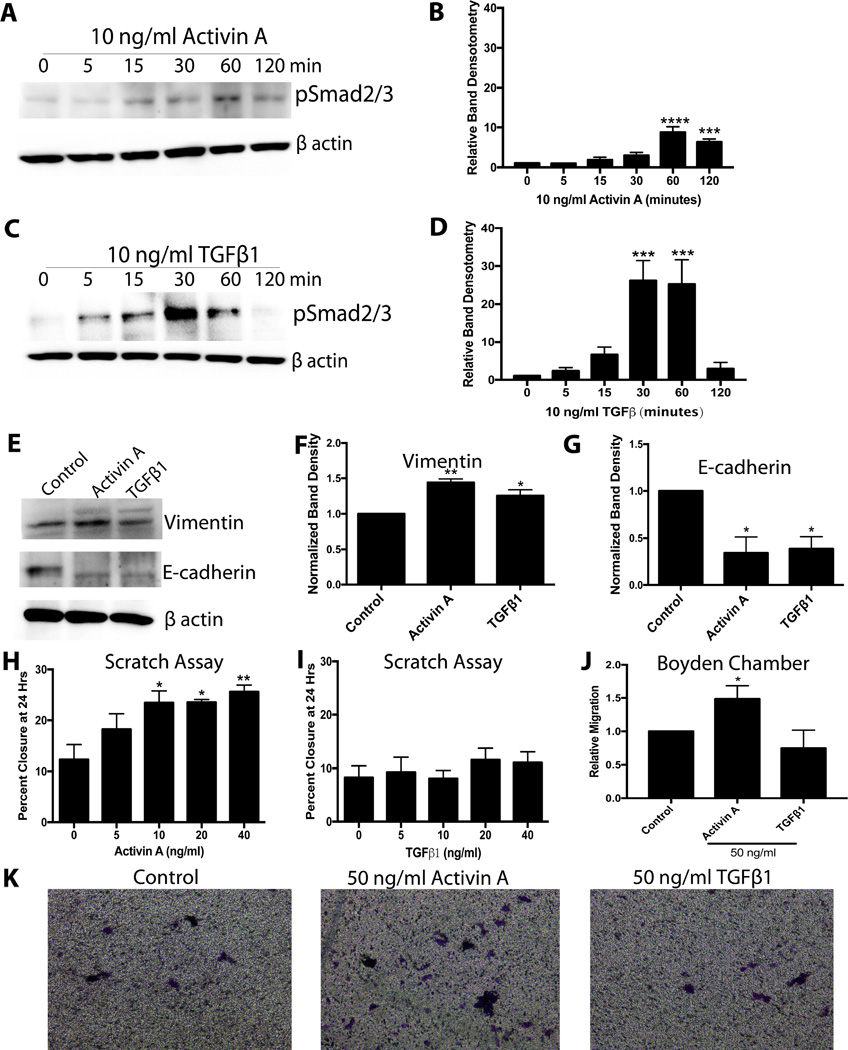
The predilection for HGSOC originating from the FTE to colonize the ovary [5,10] suggests that the ovary may secrete chemotaxic molecules. Activin A, a member of the TGFβ superfamily, is a major ovarian protein hormone produced by large follicles and the corpus luteum (CL). Activin A is well known to stimulate migration in other cancers [28]. Therefore, the ability for MOE (murine oviductal epithelial) cells to respond to activin A and TGFβ1 (as a member of the same superfamily) was tested. Activin A (10 ng/ml) induced an 8-fold increase in phospho-Smad2/3 levels in MOE cells at 60 minutes (P<0.0001), and these levels remained significantly increased (~5-fold) at 120 minutes (Figure 1A and 1B). TGFβ1 (10 ng/ml) increased phospho-Smad2/3 levels to a greater extent than activin A, reaching a ~25-fold increase at 30 and 60 minutes (P<0.05), before returning to baseline by 120 minutes (Figure 1C and 1D). To determine if activin A and TGFβ1 induce protein expression changes consistent with migration in MOE cells, E-cadherin and vimentin levels were measured in response to 24 hours of treatment. Both activin A and TGFβ1 (10 ng/ml) resulted in a 65% reduction in E-cadherin expression (P<0.05). They also increased vimentin expression (P<0.05), though activin A increased vimentin levels to a greater extent than TGFβ (44% vs 25%, Figure 1E–1G).
Figure 1.
Activin A and TGFβ1 alter expression of EMT markers in MOE cells, but only activin A stimulates migration. A) Representative western blots for phospho-Smad2/3 in MOE cells treated with activin A. B) Densitometry for phospho-Smad2/3 in response to activin A. C) Representative western blots for phospho-Smad2/3 in response to TGFβ1. D) Densitometry data for phospho-Smad2/3 in MOE cells treated with TGFβ1. E) Representative westerns for MOE cells treated with activin A or TGFβ1 for 24 hours and probed for vimentin and E-cadherin. F and G) Band densitometry for vimentin and E-cadherin. H) and I) Scratch assay for MOE cells treated with 0–40 ng/ml activin A or TGFβ1. J) Relative migration of MOE cells in response to 50 ng/ml activin A or TGFβ1 in a Boyden chamber. K) Representative images of migrated cells in a Boyden chamber. *P<0.05, **P<0.01, ***P<0.001, ****P<0.0001, n≤3.
As measured using a scratch assay, 10 ng/ml of activin A increased migration of MOE cells by 90% (P<0.05), with higher concentrations (20 and 40 ng/ml) having approximately the same effect (90% and 107%, respectively; Figure 1H; P<0.05). Surprisingly, TGFβ1 had no effect on migration at any concentration tested (5–40 ng/ml; Figure 1I). In agreement, 50 ng/ml of activin A increased migration in a Boyden chamber assay 50% over control (P<0.05), and TGFβ1 had no effect (Figure 1J and 1K). Next, the effect of activin A on proliferation was explored.
Activin A was unable to stimulate proliferation in MOE cells cultured in serum-free media. Activin A was also unable to inhibit proliferation of MOE cells in the presence of 10% FBS (Figure S1), which agrees with our previous study showing that TGFβ1 or SB431542 (activin and TGFβ inhibitor) had no effect on proliferation of murine FTE in a 3D culture system [49]. These results suggest that the TGFβ superfamily does not similarly regulate proliferation of the FTE and OSE [50]. These results also highlight a unique role for activin A to drive migration of the FTE, and possibly metastasis to the ovary.
3.2 Activin A Stimulates Migration through an AKT/MEK/RAC1 Dependent Pathway
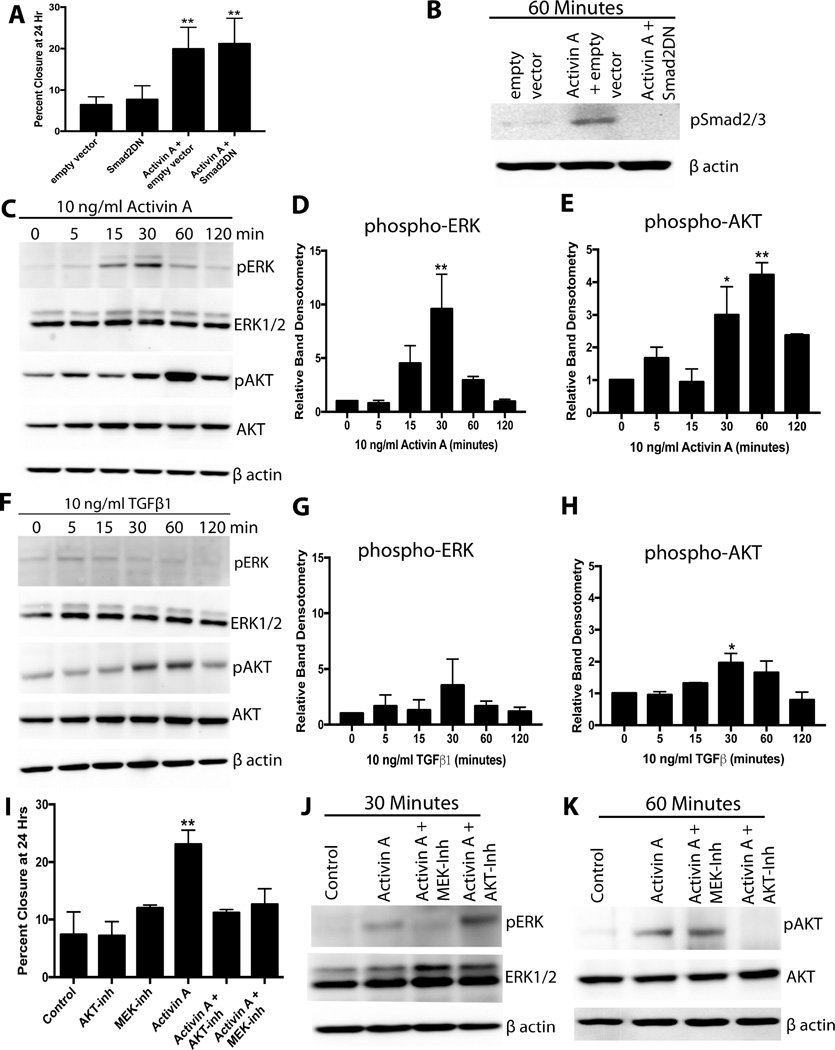
Next, the pathway by which activin A stimulates migration was explored. The finding that TGFβ1 increased phospho-Smad2/3 levels, but did not increase migration, suggested that the increased migration was Smad independent. Confirming this, a previously validated dominant negative Smad2 (Smad2DN) construct [37,38] did not abrogate the migratory effect of activin A (Figure 2A). The Smad2DN construct blocked phosphorylation of Smad2/3 (Figure 2B). Therefore, activin A stimulated migration of MOE cells through Smad-independent signaling pathways.
Figure 2.
PI3K/AKT and MEK/ERK but not Smad2/3 pathways are required for activin A-stimulated migration of MOE cells. A) Migration of MOE cells transfected with a dominant negative Smad2 (Smad2DN) construct, empty vector, and treated with 10 ng/ml activin A as indicated. B) Western blot for phospho-Smad2/3 in MOE cells treated activin A and Smad2DN construct as indicated. C–E) Example western blots (C) and densitometry for phospho-ERK (D) and phospho-AKT (E) of MOE cells treated with 10 ng/ml activin A. F–H) Example western blot (F) and densitometry of phospho-ERK (G) and phospho-AKT (H) of MOE cells treated with 10 ng/ml TGFβ1. I) Migration of MOE cells treated with activin A (10 ng/ml), an AKT inhibitor (MK2206), or a MEK inhibitor (U0126) as indicated. J and K) Example western blots for phospho-AKT and phospho-ERK in MOE cells treated with activin A for 30 (J) or 60 (K) minutes in the presence of an AKT inhibitor (MK2206) or an ERK inhibitor (U0126). *P<0.05, **P<0.01, n≤3.
TGFβ1 superfamily members can activate signaling pathways that do not rely on Smad2/3 phosphorylation in a ligand- and cell-dependent manner [17]. Therefore, the ability of activin A and TGFβ1 to stimulate PI3K/AKT and MEK/ERK signaling was evaluated via western blot. Activin A increased phospho-ERK levels by 8.5-fold at 30 minutes (P<0.05, Figure 2C and 2D). Activin A resulted in a ~3-fold increase in phospho-AKT levels after 30 minutes (P<0.05), and phospho-AKT was 4.2-fold higher at 60 minutes relative to 0 minutes (P<0.01; Figure 2C and 2E). In contrast, TGFβ1 had no effect on phospho-ERK levels at any time point examined (Figure 2F and 2G). TGFβ1 increased phospho-AKT levels, but to a lesser extent than activin A. Phospho-AKT levels were only ~2-fold higher at 30 minutes with TGFβ1 treatment (P<0.05; Figure 2F and 2H). To confirm that these pathways were required for activin A-induced migration, scratch assays were performed in the presence of MK2206 (AKT inhibitor) and U0126 (MEK inhibitor). Activin A alone significantly increased migration. Both inhibitors completely abolished activin A-stimulated migration, bringing migration values back to control levels (Figure 2I). Confirming that these inhibitors are specific in MOE cells, MK2206 completely blocked activin A-stimulated AKT phosphorylation, but not ERK phosphorylation. U0126 inhibited activin stimulated ERK phosphorylation, but not AKT (Figure 2J and 2K). These results suggested that activin A stimulated phospho-AKT and phospho-ERK in parallel of each other, and indicate both pathways are required to stimulate migration of the FTE.
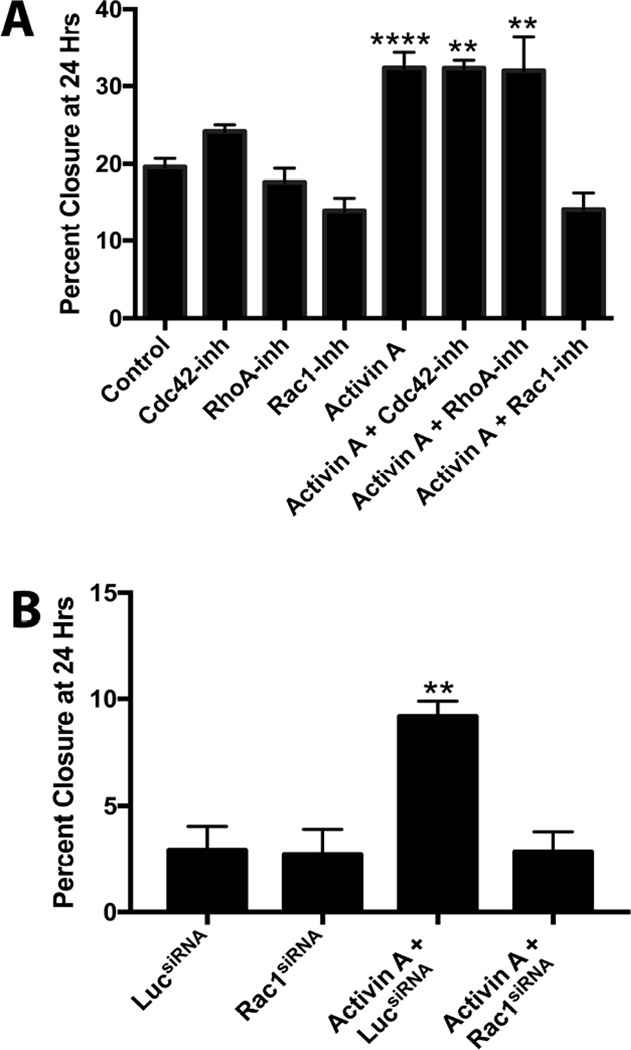
Activin A and TGFβ1 have been shown to activate members of the Rho GTPase family (mainly RhoA, Rac1, and Cdc42), which are major regulators of the Apr2/3 complex and cell migration [51]. Therefore, the ability of each of these proteins to mediate activin A-induced migration was tested using Rhosin (RhoA inhibitor), NSC23766 (Rac1 inhibitor), and MLS573151 (Cdc42 inhibitor). By themselves, none of the inhibitors changed migration. Activin A increased migration of MOE cells approximately 60% in the presence of Rhosin and MLS573151, indicating that RhoA and Cdc42 are not the primary mediators of activin A-induced migration. In contrast, NSC23766 completely abrogated migration (Figure 3A), suggesting a role for Rac1. To confirm a role for Rac1 in migration, the effect of a Rac1siRNA was evaluated. In the presence of luciferase siRNA, activin A increased migration 175% (P<0.05). In contrast, Rac1siRNA completely blocked the migratory effect of activin A (Figure 3B). These results demonstrate that Rac1 is necessary to mediate activin A-induced migration.
Figure 3.
Rac1 mediates activin A-induced migration. A) Migration of MOE cells in response to activin A, Cdc42 inhibitor (LS573151), RhoA inhibitor (Rhosin), or Rac1 inhibitor (NSC23766) as indicated. B) Migration in MOE cells treated with activin A, and luciferase siRNA, or Rac1 siRNA. **P<0.01, ****P<0.0001, n≤3.
3.3 The FTE Responds to Ovarian Activin A In Vivo
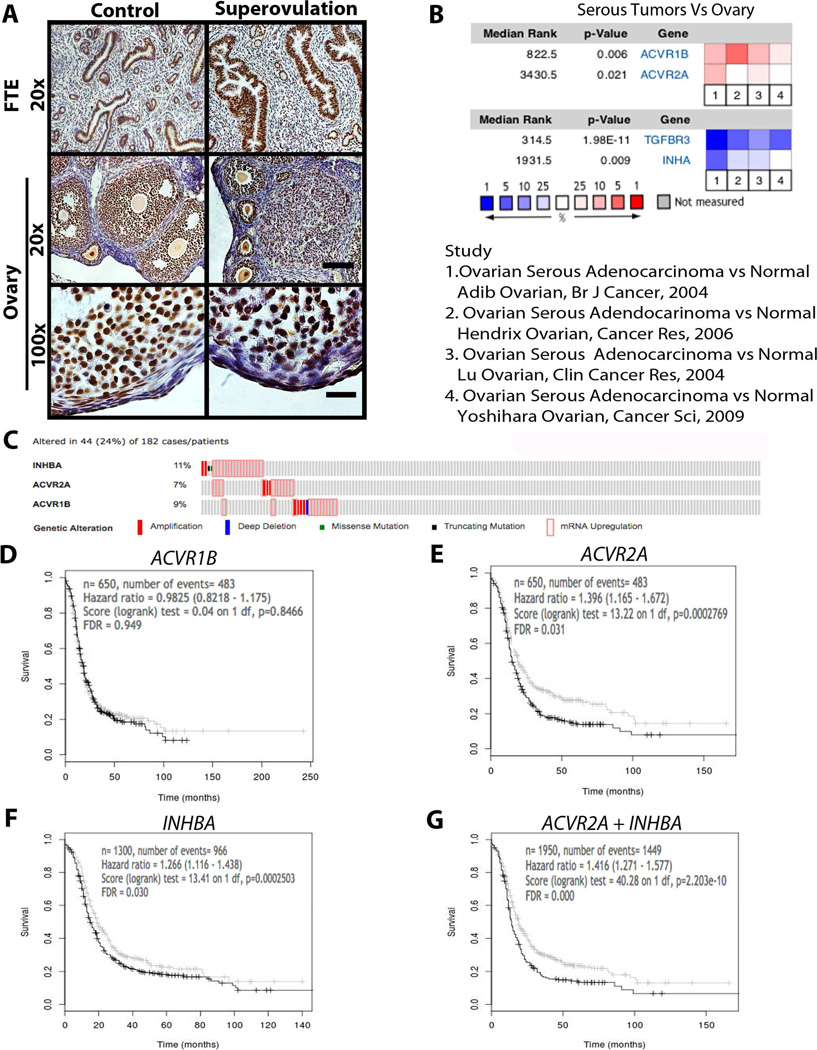
Superovulation was used to determine if ovarian factors (activin A or TGFβ) could activate receptors in the FTE (called oviducts in mice) of CD1 mice. Mice were treated with vehicle (control) or PMSG and hCG to induce superovulation, and then the ovary and fallopian tubes were evaluated via immunohistochemistry. In control mice, there was noticeable phospho-Smad2/3 immunostaining in the FTE, and superovulation increased the intensity of this immunostaining. Similarly, high-magnification of the OSE revealed Smad2/3 staining in control mice, which was increased in response to superovulation (Figure 4A).
Figure 4.
The fallopian tube epithelium responds to ovarian activin in vivo and is correlated with disease-free survival (DFS). A) Immunostaining or phospho-Smad2/3 in the fallopian tube and ovary of control and superovulated mice. Scale bar = 50 µm at 20x and 10 µm at 100x. B) Oncomine analysis for ACVR1B, ACVR2A, TGFBR3, and INHA in serous tumors relative to normal ovary across 4 studies as indicated. Top: overexpressed genes (red); Bottom: under-expressed genes (blue). C) Percentage of tumors with altered INHBA, ACVR2A, and ACVR1B in TCGA using cBioPortal. D–G) Disease Free Survival (DFS) Kaplan-Meier plots from OvMark for ACVR1B (D) and ACVR2B (E), INHBA (F), and ACVR2 + INHBA (G) in serous ovarian cancer patients.
In order to address whether human ovarian tumor samples express mRNA that supports a role for activin A in ovarian spread, the TGFβ superfamily was evaluated in HGSOCs compared to normal ovaries with Oncomine™. Four studies in Oncomine™ were identified that compared serous tumors to normal ovarian tissue [42–45]. In these 4 datasets combined, both the type I and type II activin receptors (ACVR1B and ACVR2A) were significantly overexpressed in serous tumors relative to the normal ovary (Figure 4B; P<0.05). Interestingly, ACVR2B, which can function as an activin or nodal receptor, was overexpressed in 2 of 4 studies, but this did not reach statistical significance (P=0.33). No other receptors (TGFBR1, TGFBR2, ACVR1, and ACVR1C) or ligands (INHBA, INHBB, TGFB1, TGFB2, TGFB3, NODAL) were overexpressed in serous tumors relative to normal ovaries (Figure S2). Under-expression analysis revealed that INHA and TGFB3 (also known as betaglycan), both of which reduce activin signaling, were significantly lower in serous tumors (P<0.05, Figure 4B). In addition, TGFBR2 and TGFB2 were significantly lower (P<0.05, Figure S2) in serous tumors. TGFBR1 and NODAL tended to be lower in serous tumors relative to normal ovaries but this did not reach statistical significance (P<0.01, Figure S2). Four studies were also identified that compared distant metastatic tumors to tumors located in the ovary. Confirming a role for activin A in migration of HGSOC to the ovary, but not necessarily all metastatic sites, tumors that had metastasized away from the ovary significantly over-expressed INHBA and under-expressed ACVR1B (Figure S2). These changes would make tumors less sensitive to ovarian activin. Probing the TCGA dataset using cBioPortal revealed that INBHA was altered in 11% of cases, ACVR2A in 7%, and ACVR1B in 9%. Overall, INHB, ACVR2A, or ACVR1B was altered in 24% of cases, with almost all alterations being amplification of the gene or up-regulation at the mRNA level (Figure 4C). These results support the hypothesis that activin A may encourage ovarian colonization of tumors originating in the FTE.
OvMark [48] was used to generate Kaplan-Meier plots for patients with high (above the median) or low (below the median) expression of genes involved with activin A and TGFβ signaling. While expression of ACVR1B had no effect on disease-free survival (DFS) in patients (Figure 4D), patients with high expression of ACVR2A (Figure 4E) and INHBA (Figure 4F) experienced significantly shorter DFS than patients with low expression (HR=1.39, FDR adjusted P value=0.031 and HR=1.266, FDR adjusted P value=0.030, respectively). Interestingly, combining ACVR2A and INHBA resulted in the biggest difference between high-expression and low-expression groups (Figure 4G; HR=1.416 and FDR adjusted P value <0.001), suggesting that an autocrine feedback loop with activin A may facilitate spread of serous tumors. High expression of TGFβ (TGFB1, TGFB2, or TGFB3) or TGFβ receptors (TGFBR1 and TGFBR2) was not associated with any difference in DFS (Figure S3). These results suggest a unique role for activin A in the progression of HGSOC.
3.4 Activin A Stimulates Migration in HGSOC Cell Lines
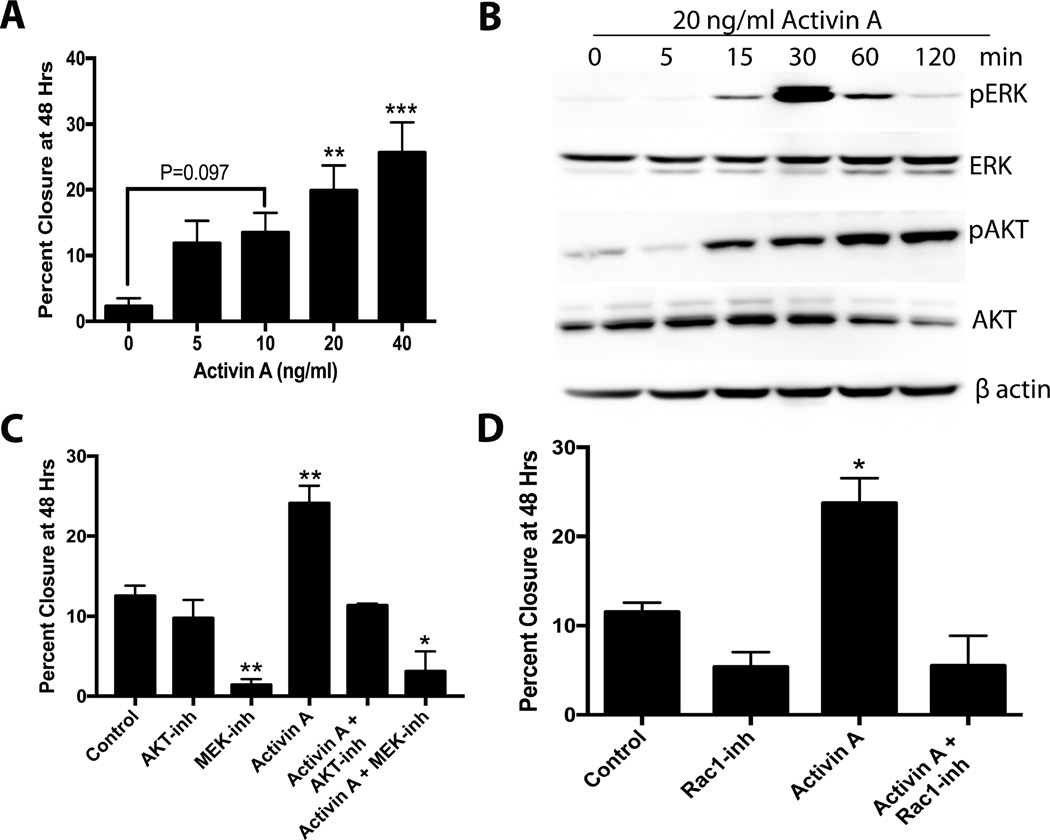
Next, the ability of activin A to drive migration of tumorigenic cells was evaluated using OVCAR3 and OVCAR4 cells, which are likely to represent HGSOC [36,52]. Activin A induced a dose-dependent increase in migration in OVCAR3 cells. Activin A at 20 and 40 ng/ml significantly increased migration (P<0.01) over control (Figure 5A). Activin A resulted in increased levels of phospho-AKT and phospho-ERK in OVCAR3 cells (Figure 5B). U0126 (ERK inhibitor), but not MK2206 (AKT inhibitor), significantly reduced basal migration in OVCAR3 cells. Activin A (20 ng/ml) significantly increased migration, which was completely abrogated by MK2206. U0126 blocked activin A-induced migration, but it also reduced migration to less than control (P<0.05, Figure 5C). NSC23766 (Rac1 inhibitor) completely blocked activin A-induced migration (Figure 5D). These data confirm that activin A stimulates migration of HGSOC in an AKT/ERK/Rac1-dependent pathway in OVCAR3.
Figure 5.
Activin A stimulates migration of OVCAR3 via AKT/MEK/RAC1. A) Migration of OVCAR3 cells in response to activin A (0–40 ng/ml). B) Example western blot for pAKT, AKT, pERK, ERK, and activin A in response to 20 ng/ml activin A over 2 hours. C) Migration in OVCAR3 in response to activin A, an AKT inhibitor (MK2206) and an ERK inhibitor (U0126) as indicated. D) Migration of OVCAR3 cell treated with activin A or a Rac1 inhibitor (NSC23766) as indicated. *P<0.05, **P<0.01, ***P<0.001. n≤3.
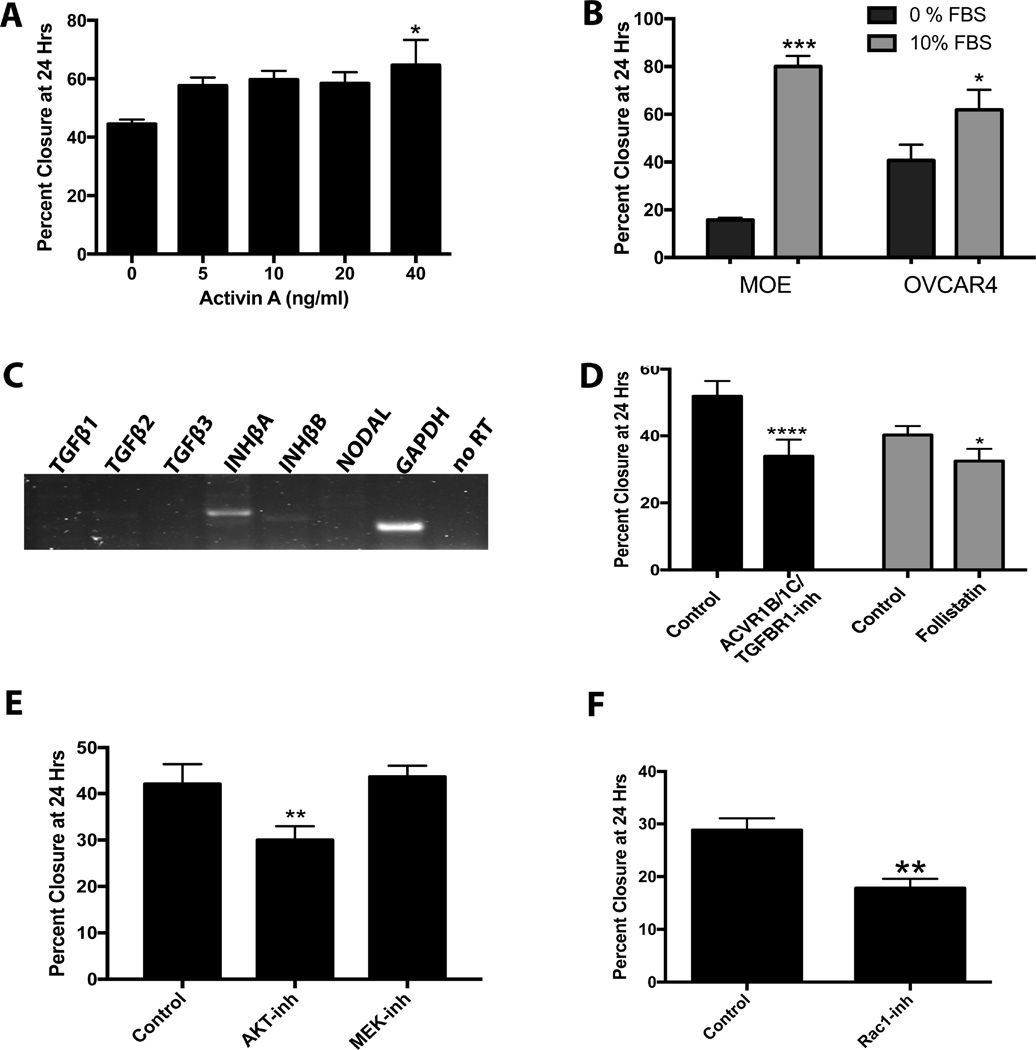
In OVCAR4 cells, activin A had less of an effect of migration with only the highest concentration of activin A (40 ng/ml) having a slight (45%), but significant, enhancement of migration (P=0.026; Figure 6A). The effect of FBS on migration of MOE cells and OVCAR4 cells (which migrate at approximately the same rate) was determined. FBS (10%) increased migration of MOE cells by 400% (P<0.0001). In contrast, 10% FBS increased migration of OVCAR4 cells by only 52% (P<0.05; Figure 6B), indicating that OVCAR4 cells are less reliant on exogenous stimulation to induce migration. RT-PCR revealed that OVCAR4 cells produce INHBA and are capable of making activin A, while TGFB1, TGFB2, TGFB3, INHBB, and Nodal were undetectable (Figure 6C). In serum-free media, SB431542 (inhibitor of TGFβ, activin A, and nodal) reduced migration of OVCAR4 cells by 35% (P<0.01), and follistatin (a specific activin inhibitor) reduced migration 20% (P<0.05; Figure 6D). Inhibition of AKT with MK2206 significantly reduced migration of OVCAR4 cells by 35%. Surprisingly, the MEK inhibitor (U0126) had no effect on migration of this cell line (Figure 6E). NSC23766 also inhibited migration (38%, P<0.05), confirming a role for Rac1. These results show that while some cell lines (MOE and OVCAR3) are dependent on exogenous activin A to stimulate migration, other cancer cell lines (OVCAR4) produce activin A to stimulate their own migration.
Figure 6.
Activin A stimulates migration of OVCAR4 cells in an autocrine manner. A) Migration of OVCAR4 cells in response to activin A (0–40 ng/ml). B) Migration of MOE and OVCAR4 cells in response to 10% FBS. C) RT-PCR bands showing OVCAR4 cells express INHBA mRNA but not TGFB1, TGFB1, TGFB3, or NODAL. D) Migration of OVCAR4 cells treated with an ACVR1B, ACVR1C and TGFBR1 inhibitor (SB431542) or a specific activin inhibitor (follistatin). E) Migration in OVCAR4 cells treated with an AKT inhibitor (MK2206) or a MEK inhibitor (U0126). F) Migration in OVCAR4 cells treated with a Rac1 inhibitor (NSC23766). *P<0.05, **P<0.01, ***P<0.001, ****P<0.0001. n≤3.
4. Discussion
The current model of ovarian cancer metastasis to the omentum is that tumorigenic cells are shed by the ovarian tumor, reach the omentum through passive flow of peritoneal fluid, and invade at milky spots, using energy stored in adjacent adipocytes [53,54]. However, much less is known about why tumors that start in the fallopian tube epithelium migrate to the ovary. It is clear that ovulation increases a woman’s risk of developing ovarian cancer [55]. However, the mechanism(s) by which ovulation increases the risk of ovarian cancer is less clear. This is important because recent evidence indicates that colonization of the ovary by FTE-derived tumors may be an important step in the progression of HGSOC [10,13] and suggests that this step may represent a therapeutic target in women at high risk of developing HGSOC [10]. The ovary produces a large number of hormones and growth factors that may stimulate migration of tumorigenic FTE cells to the ovaries. Estradiol is a major ovarian hormone, but it had no effect on migration of MOE cells [39]. In addition, steroids circulate systemically, making it unlikely that they stimulate the local migratory step from the fallopian tube to the ovary. More likely, a set of locally acting proteins from the ovary stimulates migration of tumorigenic cells. Yang-Hartwich et al. showed that stromal differentiation factor 1 (SDF-1) stimulated migration of OTIC cells [15]; however, OTICs cells have an unknown cell of origin, and therefore may not represent the FTE.
Superovulation increased phospho-Smad2/3 levels in the OSE and FTE. It is unclear if this increase was due to activin A or TGFβ1. Unfortunately, small molecule inhibitors (e.g. SB431542 or LY2109761) of the activin receptor also inhibit TGFβ receptors, making such a distinction difficult in vivo. However, reported concentrations of activin A in follicular fluid are higher than those reported for TGFβ [30,56], suggesting that activin A may play a role. More importantly, these results show that ovarian hormones reach the FTE in sufficient concentrations to activate downstream signaling pathways. Mice have an ovarian bursa which may result in a closer proximity between the ovary and FTE than would occur in humans; however, phospho-Smad2/3 has also been detected in the human FTE [57].
Here we show that activin A increases migration of MOE and OVCAR3 cells, which agrees with effects of activin A in other cancer types [28]. Activin A also resulted in increased EMT, evidenced by significantly higher expression of vimentin and lower expression of E-cadherin in MOE cells. In agreement, Basu et al. found that activin A increased mRNA expression of ZEB1, ZEB2, SNAI1 and SNAI2 in OAW-42 cells [32]. SB-431542 (an activin and TGFβ inhibitor) reduced SNAI1, TWIST1, and ZEB2 expression in spheroids grown from primary ovarian cancer ascites cells [58].
Interestingly, a woman’s risk of ovarian cancer increases after menopause. At the same time, there is a clear link between the number of ovulations and ovarian cancer risk [59]. One potential mechanism to explain this apparent paradox is that tumorigenic cells colonize the ovary prior to menopause, but tumors are not diagnosed until after menopause. In support of this hypothesis, a recent mathematical analysis calculated the latency period for ovarian cancer to be 44 years [60]. The mechanisms resulting in tumor metastasis beyond the ovary are poorly understood and are likely complex. One possibility is that tumors may become resistant to ovarian activin A prior to distant metastasis. In support of this, Oncomine data identified that distant tumors had higher levels of INHBA and lower ACVR2A, both of which would result in decreased sensitivity to activin A from the ovary. In agreement, OVCAR4 apparently stimulated their own migration via activin A secretion. Also, activin A concentrations are higher in postmenopausal women with epithelial ovarian cancer [61], and activin A levels have been shown to correlate with disease reoccurrence [62].
The omentum, a frequent site of colonization in HGSOC, also produces activin A [63]. It is unknown if ovarian cancer associated fibroblasts (CAFs) produce activin A. In colon cancer cells, CAFs enhanced transcription of TGFβ [64]. Tumor-associated myofibroblasts from the tongue produced increased amounts of activin A, which drove proliferation of oral squamous cell carcinoma cells [65]. Thus, activin A may play a role in spread of HGSOC beyond the ovary considering that it is produced by multiple sources including HGSOCs and CAFs. If further validated, activin A signaling may be an attractive target to prevent the spread of HGSOC, especially given the recent development of a small molecule activin A antagonist [66] and the ongoing phase I clinical trial using an ACVR2B soluble receptor ligand trap in patients with ovarian, fallopian tube, or endometrial cancers (ClinicalTrails.gov Identifier: NCT02262455). Importantly, these strategies may be more successful if used in patients when disease is still confined to the fallopian tube.
In the current study, activin A had no effect on proliferation of MOE cells. In agreement, neither TGFβ1 nor SB431542 had any effect on proliferation in a 3D model of the mouse FTE [49]. In contrast, activin A and TGFβ have repeatedly been shown to inhibit proliferation of OSE cells [49,50], and in vivo deletion of Smad3 resulted in increased proliferation of OSE [67]. The role of activin A and TGFβ in ovarian cancer cell lines is mixed. Steller et al. found that activin A had no effect on proliferation of OVCA429, HEY, and A2780-cp cells and increased proliferation of OCC1, SKOV3, OVCAR3, and A2780-S cells [34]. In contrast, Ramachandran et al. found that activin A inhibited proliferation of OVCAR4, NZOV2, NZOV10, NZOV5, and OVCAR3 cells, but had no effect on proliferation of COV644, OVCAR5, SKOV3, NZOV4, NZOV9, NZOV11, and NZOV13 [68]. Based on the results presented here, it is possible that the mixed response of ovarian cancer cell lines reflects their tissue of origin. It is also possible that the response of cells is dependent on changes that occur during transformation. For example, proliferation in response to TGFβ has been linked to p53 signaling [49], and p53 is mutated in almost all cases of HGSOC [69].
In conclusion, activin A (but not TGFβ1) stimulated migration of FTE and HGSOC cells through phospho-AKT and phospho-ERK signaling. In patients, serous ovarian tumors had higher levels of AVCR1B and ACVR2A compared to normal ovaries, while INHA and TGFB3 transcripts were lower. Distant ovarian tumors had higher levels of INHBA and ACVR2A relative to tumors localized to the ovary. High expression of both INHBA and ACVR2A was associated with shorter DFS in serous cancer patients, and INHBA, ACVR1B, and ACVR2B were frequently altered in HGSOC patients. These data indicate that activin A plays an important role in the early pathogenesis of HGSOC, and may contribute to the spread of FTE-derived tumors to the ovary. An increased understanding of the role of ovarian factors in the colonization of the ovary may uncover therapeutic targets for FTE-derived HGSOC.
Supplementary Material
Figure S1. Activin A has no effect on proliferation of MOE cells. SRB proliferation assays for activin A (0–40 ng/ml) in the presence (A) or absence (B) of 10% FBS.
Figure S2. Activin A signaling is associated with metastasis of ovarian tumors. A) Heat maps for genes involved with activin A and TGFβ1 signaling serous ovarian tumors compared to normal ovary. B) Genes in activin A signaling, distant tumors compared to tumors localized in the ovary. red = increased expression. blue = decreased expression.
Figure S3. Components of TGFβ signaling are not associated with disease-free survival (DFS) in serous cancer patients. DFS Kaplan-Meier plots from OvMark for TGFB1 (A), TGFB2 (B), TGFB3 (C), TGFBR1 (D), and TGFBR2 (E).
Table S1. Primary antibodies, dilution, and blocking reagent used for western blotting (WB) and immunochemistry (IHC).
Table S2. Gene symbols, primer sequences, and amplicon sizes for genes amplified via RT-PCR.
Highlights.
Activin A stimulates migration of normal fallopian tube epithelial cells.
The migratory effect of activin A is mediated by non-canonical pathways.
Activin A stimulates migration of high grade serous cells.
INHBA and ACVR2 are associated with shorter disease-free survival.
Acknowledgments
The authors thank Daniel Lantvit for his assistance with animal experiments and Ally Young for careful reading of this manuscript.
Funding
This was supported by Department of Defense grant OC130046 to J.E. Burdette and a T32 fellowship from the NCCIH T32 AT007533 to M. Dean.
Abbreviations
- FTE
fallopian tube epithelium
- HGSOC
high grade serous ovarian cancer
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Literature Cited
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2016. CA. Cancer J. Clin. 2015;66:7–30. doi: 10.3322/caac.21332. [DOI] [PubMed] [Google Scholar]
- 2.Piek JMJ, van Diest PJ, Zweemer RP, Jansen JW, Poort-Keesom RJJ, Menko FH, Gille JJP, Jongsma APM, Pals G, Kenemans P, Verheijen RHM. Dysplastic changes in prophylactically removed fallopian tubes of women predisposed to developing ovarian cancer. J. Pathol. 2001;195:451–456. doi: 10.1002/path.1000. [DOI] [PubMed] [Google Scholar]
- 3.Piek JMJ, Verheijen RHM, Kenemans P, Massuger LF, Bulten H, van Diest PJ. BRCA1/2-related ovarian cancers are of tubal origin: a hypothesis. Gynecol. Oncol. 2003;90:491. doi: 10.1016/s0090-8258(03)00365-2. [DOI] [PubMed] [Google Scholar]
- 4.Leeper K, Garcia R, Swisher E, Goff B, Greer B, Paley P. Pathologic findings in prophylactic oophorectomy specimens in high-risk women. Gynecol. Oncol. 2002;87:52–56. doi: 10.1006/gyno.2002.6779. [DOI] [PubMed] [Google Scholar]
- 5.Marquez RT, Baggerly KA, Patterson AP, Liu J, Broaddus R, Frumovitz M, Atkinson EN, Smith DI, Hartmann L, Fishman D, Berchuck A, Whitaker R, Gershenson DM, Mills GB, Bast RC, Lu KH. Patterns of gene expression in different histotypes of epithelial ovarian cancer correlate with those in normal fallopian tube, endometrium, and colon. Clin. Cancer Res. 2005;11:6116–6126. doi: 10.1158/1078-0432.CCR-04-2509. [DOI] [PubMed] [Google Scholar]
- 6.Klinkebiel D, Zhang W, Akers SN, Odunsi K, Karpf AR. DNA methylome analyses implicate fallopian tube epithelia as the origin for high-grade serous ovarian cancer. Mol. Cancer Res. 2016;14:787–794. doi: 10.1158/1541-7786.MCR-16-0097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Falconer H, Yin L, Grönberg H, Altman D. Ovarian cancer risk after salpingectomy: a nationwide population-based study. J. Natl. Cancer Inst. 2015;107 doi: 10.1093/jnci/dju410. dju410. [DOI] [PubMed] [Google Scholar]
- 8.Eddie SL, Quartuccio SM, hAinmhire EÓ, Moyle-Heyrman G, Lantvit DD, Wei J-J, Vanderhyden BC, Burdette JE. Tumorigenesis and peritoneal colonization from fallopian tube epithelium. Oncotarget. 2015;21:20500–20512. doi: 10.18632/oncotarget.3985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Quartuccio SM, Karthikeyan S, Eddie SL, Lantvit DD, Ó hAinmhire E, Modi DA, Wei J-J, Burdette JE. Mutant p53 expression in fallopian tube epithelium drives cell migration. Int. J. Cancer. 2015;137:1528–1538. doi: 10.1002/ijc.29528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Perets R, Wyant GA, Muto KW, Bijron JG, Poole BB, Chin KT, Chen JYH, Ohman AW, Stepule CD, Kwak S, Karst AM, Hirsch MS, Setlur SR, Crum CP, Dinulescu DM, Drapkin R. Transformation of the fallopian tube secretory epithelium leads to high-grade serous ovarian cancer in Brca;Tp53;Pten models. Cancer Cell. 2013;24:751–765. doi: 10.1016/j.ccr.2013.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sherman-Baust CA, Kuhn E, Valle BL, Shih I-M, Kurman RJ, Wang T-L, Amano T, Ko MS, Miyoshi I, Araki Y, Lehrmann E, Zhang Y, Becker KG, Morin PJ. A genetically engineered ovarian cancer mouse model based on fallopian tube transformation mimics human high-grade serous carcinoma development. J. Pathol. 2014;233:228–237. doi: 10.1002/path.4353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kim J, Coffey DM, Creighton CJ, Yu Z, Hawkins SM, Matzuk MM. High-grade serous ovarian cancer arises from fallopian tube in a mouse model. Proc. Natl. Acad. Sci. 2012;109:3921–3926. doi: 10.1073/pnas.1117135109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Coffman LG, Burgos-Ojeda D, Wu R, Cho K, Bai S, Buckanovich RJ. New models of hematogenous ovarian cancer metastasis demonstrate preferential spread to the ovary and a requirement for the ovary for abdominal dissemination. Transl. Res. 2016;175:92–102. doi: 10.1016/j.trsl.2016.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Merritt MA, Pari MD, Vitonis AF, Titus LJ, Cramer DW, Terry KL. Reproductive characteristics in relation to ovarian cancer risk by histologic pathways. Hum. Reprod. 2013;28:1406–1417. doi: 10.1093/humrep/des466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yang-Hartwich Y, Gurrea-Soteras M, Sumi N, Joo WD, Holmberg JC, Craveiro V, Alvero AB, Mor G. Ovulation and extra-ovarian origin of ovarian cancer. Sci. Rep. 2014;4:6116. doi: 10.1038/srep06116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Harrison CA, Gray PC, Vale WW, Robertson DM. Antagonists of activin signaling: mechanisms and potential biological applications. Trends Endocrinol. Metab. 2005;16:73–78. doi: 10.1016/j.tem.2005.01.003. [DOI] [PubMed] [Google Scholar]
- 17.Loomans HA, Andl CD. Intertwining of activin A and TGFβ signaling: dual roles in cancer progression and cancer cell invasion. Cancers. 2014;7:70–91. doi: 10.3390/cancers7010070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Galliher AJ, Schiemann WP. Src phosphorylates Tyr284 in TGF-beta type II receptor and regulates TGF-beta stimulation of p38 MAPK during breast cancer cell proliferation and invasion. Cancer Res. 2007;67:3752–3758. doi: 10.1158/0008-5472.CAN-06-3851. [DOI] [PubMed] [Google Scholar]
- 19.Watkins SJ, Jonker L, Arthur HM. A direct interaction between TGFbeta activated kinase 1 and the TGFbeta type II receptor: implications for TGFbeta signalling and cardiac hypertrophy. Cardiovasc. Res. 2006;69:432–439. doi: 10.1016/j.cardiores.2005.11.007. [DOI] [PubMed] [Google Scholar]
- 20.Safwat N, Ninomiya-Tsuji J, Gore AJ, Miller WL. Transforming Growth Factor β-Activated Kinase 1 Is a Key Mediator of Ovine Follicle-Stimulating Hormone β-Subunit Expression. Endocrinology. 2005;146:4814–4824. doi: 10.1210/en.2005-0457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yi JY, Shin I, Arteaga CL. Type I transforming growth factor beta receptor binds to and activates phosphatidylinositol 3-kinase. J. Biol. Chem. 2005;280:10870–10876. doi: 10.1074/jbc.M413223200. [DOI] [PubMed] [Google Scholar]
- 22.Lamouille S, Derynck R. Cell size and invasion in TGF-β-induced epithelial to mesenchymal transition is regulated by activation of the mTOR pathway. J. Cell Biol. 2007;178:437–451. doi: 10.1083/jcb.200611146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dupont J, McNeilly J, Vaiman A, Canepa S, Combarnous Y, Taragnat C. Activin Signaling Pathways in Ovine Pituitary and LβT2 Gonadotrope Cells. Biol. Reprod. 2003;68:1877–1887. doi: 10.1095/biolreprod.102.012005. [DOI] [PubMed] [Google Scholar]
- 24.Zhang L, Deng M, Parthasarathy R, Wang L, Mongan M, Molkentin JD, Zheng Y, Xia Y. MEKK1 transduces activin signals in keratinocytes to induce actin stress fiber formation and migration. Mol. Cell. Biol. 2005;25:60–65. doi: 10.1128/MCB.25.1.60-65.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ungefroren H, Groth S, Sebens S, Lehnert H, Gieseler F, Fändrich F. Differential roles of Smad2 and Smad3 in the regulation of TGF-β1-mediated growth inhibition and cell migration in pancreatic ductal adenocarcinoma cells: control by Rac1. Mol. Cancer. 2011;10:67. doi: 10.1186/1476-4598-10-67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hoda MA, Rozsas A, Lang E, Klikovits T, Lohinai Z, Torok S, Berta J, Bendek M, Berger W, Hegedus B, Klepetko W, Renyi-Vamos F, Grusch M, Dome B, Laszlo V. High circulating activin A level is associated with tumor progression and predicts poor prognosis in lung adenocarcinoma. Oncotarget. 2016;22:13388–13399. doi: 10.18632/oncotarget.7796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wildi S, Kleeff J, Maruyama H, Maurer CA, Büchler MW, Korc M. Overexpression of activin A in stage IV colorectal cancer. Gut. 2001;49:409–417. doi: 10.1136/gut.49.3.409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bashir M, Damineni S, Mukherjee G, Kondaiah P. Activin-A signaling promotes epithelial-mesenchymal transition, invasion, and metastatic growth of breast cancer. Npj Breast Cancer. 2015;1:15007. doi: 10.1038/npjbcancer.2015.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hillier SG, Miró F. Inhibin activin, and follistatin, Potential roles in ovarian physiology. Ann. N. Y. Acad. Sci. 1993;687:29–38. doi: 10.1111/j.1749-6632.1993.tb43850.x. [DOI] [PubMed] [Google Scholar]
- 30.Wen X, Tozer AJ, Butler SA, Bell CM, Docherty SM, Iles RK. Follicular fluid levels of inhibin A, inhibin B, and activin A levels reflect changes in follicle size but are not independent markers of the oocyte’s ability to fertilize. Fertil. Steril. 2006;85:1723–1729. doi: 10.1016/j.fertnstert.2005.11.058. [DOI] [PubMed] [Google Scholar]
- 31.Matzuk MM, Finegold MJ, Su JG, Hsueh AJ, Bradley A. Alpha-inhibin is a tumour-suppressor gene with gonadal specificity in mice. Nature. 1992;360:313–319. doi: 10.1038/360313a0. [DOI] [PubMed] [Google Scholar]
- 32.Basu M, Bhattacharya R, Ray U, Mukhopadhyay S, Chatterjee U, Roy SS. Invasion of ovarian cancer cells is induced byPITX2-mediated activation of TGF-β and Activin-A. Mol. Cancer. 2015;14:162. doi: 10.1186/s12943-015-0433-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Do T-V, Kubba LA, Antenos M, Rademaker AW, Sturgis CD, Woodruff TK. The role of activin A and Akt/GSK signaling in ovarian tumor biology. Endocrinology. 2008;149:3809–3816. doi: 10.1210/en.2007-1584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Steller MD, Shaw TJ, Vanderhyden BC, Ethier J-F. Inhibin resistance is associated with aggressive tumorigenicity of ovarian cancer cells. Mol. Cancer Res. 2005;3:50–61. [PubMed] [Google Scholar]
- 35.Wong WS, Wong YF, Ng YT, Huang PD, Chew EC, Ho TH, Chang MZ. Establishment and characterization of a new human cell line derived from ovarian clear cell carcinoma. Gynecol. Oncol. 1990;38:37–45. doi: 10.1016/0090-8258(90)90008-9. [DOI] [PubMed] [Google Scholar]
- 36.Domcke S, Sinha R, Levine DA, Sander C, Schultz N. Evaluating cell lines as tumour models by comparison of genomic profiles. Nat. Commun. 2013;4:2126. doi: 10.1038/ncomms3126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chen W, Woodruff TK, Mayo KE. Activin A-Induced HepG2 liver cell apoptosis: involvement of activin receptors and smad proteins. Endocrinology. 2000;141:1263–1272. doi: 10.1210/endo.141.3.7361. [DOI] [PubMed] [Google Scholar]
- 38.Quartuccio SM, Lantvit DD, Bosland MC, Burdette JE. Conditional inactivation of p53 in mouse ovarian surface epithelium does not alter MIS driven Smad2-dominant negative epithelium-lined inclusion cysts or teratomas. PLoS ONE. 2013;8:e65067. doi: 10.1371/journal.pone.0065067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Moyle-Heyrman G, Schipma MJ, Dean M, Davis DA, Burdette JE. Genome-wide transcriptional regulation of estrogen receptor targets in fallopian tube cells and the role of selective estrogen receptor modulators. J. Ovarian Res. 2016;9:5. doi: 10.1186/s13048-016-0213-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Skehan P, Storeng R, Scudiero D, Monks A, McMahon J, Vistica D, Warren JT, Bokesch H, Kenney S, Boyd MR. New colorimetric cytotoxicity assay for anticancer-drug screening. J. Natl. Cancer Inst. 1990;82:1107–1112. doi: 10.1093/jnci/82.13.1107. [DOI] [PubMed] [Google Scholar]
- 41.King SM, Hilliard TS, Wu LY, Jaffe RC, Fazleabas AT, Burdette JE. The impact of ovulation on fallopian tube epithelial cells: evaluating three hypotheses connecting ovulation and serous ovarian cancer. Endocr. Relat. Cancer. 2011;18:627–642. doi: 10.1530/ERC-11-0107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Adib TR, Henderson S, Perrett C, Hewitt D, Bourmpoulia D, Ledermann J, Boshoff C. Predicting biomarkers for ovarian cancer using gene-expression microarrays. Br. J. Cancer. 2004;90:686–692. doi: 10.1038/sj.bjc.6601603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yoshihara K, Tajima A, Komata D, Yamamoto T, Kodama S, Fujiwara H, Suzuki M, Onishi Y, Hatae M, Sueyoshi K, Fujiwara H, Kudo Y, Inoue I, Tanaka K. Gene expression profiling of advanced-stage serous ovarian cancers distinguishes novel subclasses and implicates ZEB2 in tumor progression and prognosis. Cancer Sci. 2009;100:1421–1428. doi: 10.1111/j.1349-7006.2009.01204.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hendrix ND, Wu R, Kuick R, Schwartz DR, Fearon ER, Cho KR. Fibroblast growth factor 9 has oncogenic activity and is a downstream target of Wnt signaling in ovarian endometrioid adenocarcinomas. Cancer Res. 2006;66:1354–1362. doi: 10.1158/0008-5472.CAN-05-3694. [DOI] [PubMed] [Google Scholar]
- 45.Lu KH, Patterson AP, Wang L, Marquez RT, Atkinson EN, Baggerly KA, Ramoth LR, Rosen DG, Liu J, Hellstrom I, Smith D, Hartmann L, Fishman D, Berchuck A, Schmandt R, Whitaker R, Gershenson DM, Mills GB, Bast RC. Selection of potential markers for epithelial ovarian cancer with gene expression arrays and recursive deescent partition analysis. Clin. Cancer Res. 2004;10:3291–3300. doi: 10.1158/1078-0432.CCR-03-0409. [DOI] [PubMed] [Google Scholar]
- 46.Tothill RW, Tinker AV, George J, Brown R, Fox SB, Lade S, Johnson DS, Trivett MK, Etemadmoghadam D, Locandro B, Traficante N, Fereday S, Hung JA, Chiew Y-E, Haviv I, Australian Ovarian Cancer Study Group Gertig D, DeFazio A, Bowtell DDL. Novel molecular subtypes of serous and endometrioid ovarian cancer linked to clinical outcome. Clin. Cancer Res. 2008;14:5198–5208. doi: 10.1158/1078-0432.CCR-08-0196. [DOI] [PubMed] [Google Scholar]
- 47.Anglesio MS, Wiegand KC, Melnyk N, Chow C, Salamanca C, Prentice LM, Senz J, Yang W, Spillman MA, Cochrane DR, Shumansky K, Shah SP, Kalloger SE, Huntsman DG. Type-specific cell line models for type-specific ovarian cancer research. PLoS ONE. 2013;8:e72162. doi: 10.1371/journal.pone.0072162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Madden SF, Clarke C, Stordal B, Carey MS, Broaddus R, Gallagher WM, Crown J, Mills GB, Hennessy BT. OvMark: a user-friendly system for the identification of prognostic biomarkers in publically available ovarian cancer gene expression datasets. Mol. Cancer. 2014;13:241. doi: 10.1186/1476-4598-13-241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ó hAinmhire E, Quartuccio SM, Cheng W, Ahmed RA, King SM, Burdette JE. Mutation or loss of p53 differentially modifies TGFβ action in ovarian cancer. PLoS ONE. 2014;9:e89553. doi: 10.1371/journal.pone.0089553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Choi KC, Kang SK, Tai CJ, Auersperg N, Leung PC. The regulation of apoptosis by activin and transforming growth factor-beta in early neoplastic and tumorigenic ovarian surface epithelium. J. Clin. Endocrinol. Metab. 2001;86:2125–2135. doi: 10.1210/jcem.86.5.7478. [DOI] [PubMed] [Google Scholar]
- 51.Welf ES, Haugh JM. Signaling pathways that control cell migration: models and analysis. Wiley Interdiscip. Rev. Syst. Biol. Med. 2011;3:231–240. doi: 10.1002/wsbm.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mitra AK, Davis DA, Tomar S, Roy L, Gurler H, Xie J, Lantvit DD, Cardenas H, Fang F, Liu Y, Loughran E, Yang J, Sharon Stack M, Emerson RE, Cowden Dahl KD, Barbolina MV, Nephew KP, Matei D, Burdette JE. In vivo tumor growth of high-grade serous ovarian cancer cell lines. Gynecol. Oncol. 2015;138:372–377. doi: 10.1016/j.ygyno.2015.05.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Nieman KM, Kenny HA, Penicka CV, Ladanyi A, Buell-Gutbrod R, Zillhardt MR, Romero IL, Carey MS, Mills GB, Hotamisligil GS, Yamada SD, Peter ME, Gwin K, Lengyel E. Adipocytes promote ovarian cancer metastasis and provide energy for rapid tumor growth. Nat. Med. 2011;17:1498–1503. doi: 10.1038/nm.2492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Clark R, Krishnan V, Schoof M, Rodriguez I, Theriault B, Chekmareva M, Rinker-Schaeffer C. Milky spots promote ovarian cancer metastatic colonization of peritoneal adipose in experimental models. Am. J. Pathol. 2013;183:576–591. doi: 10.1016/j.ajpath.2013.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Cramer DW, Hutchison GB, Welch WR, Scully RE, Ryan KJ. Determinants of ovarian cancer risk. I. reproductive experiences and family history. J. Natl. Cancer Inst. 1983;71:703–709. [PubMed] [Google Scholar]
- 56.Fried G, Wramsby H. Increase in transforming growth factor beta1 in ovarian follicular fluid following ovarian stimulation and in-vitro fertilization correlates to pregnancy. Hum. Reprod. Oxf. Engl. 1998;13:656–659. doi: 10.1093/humrep/13.3.656. [DOI] [PubMed] [Google Scholar]
- 57.Alsina-Sanchis E, Figueras A, Lahiguera Á, Vidal A, Casanovas O, Graupera M, Villanueva A, Viñals F. The TGFβ pathway stimulates ovarian cancer cell proliferation by increasing IGF1R levels. Int. J. Cancer. 2016 doi: 10.1002/ijc.30233. n/a-n/a. [DOI] [PubMed] [Google Scholar]
- 58.Rafehi S, Valdes YR, Bertrand M, McGee J, Préfontaine M, Sugimoto A, DiMattia GE, Shepherd TG. TGFβ signaling regulates epithelial-mesenchymal plasticity in ovarian cancer ascites-derived spheroids. Endocr. Relat. Cancer. 2016;23:147–159. doi: 10.1530/ERC-15-0383. [DOI] [PubMed] [Google Scholar]
- 59.Whittemore AS, Harris R, Itnyre J. Characteristics relating to ovarian cancer risk: collaborative analysis of 12 US case-control Invasive epithelial ovarian cancers in white women studies. II. Collaborative Ovarian Cancer Group. Am. J. Epidemiol. 1992;136:1184–1203. doi: 10.1093/oxfordjournals.aje.a116427. [DOI] [PubMed] [Google Scholar]
- 60.Nadler DL, Zurbenko IG, Nadler DL, Zurbenko IG. Estimating cancer latency times using a weibull model, estimating cancer latency times using a Weibull model. Adv. Epidemiol. Adv. Epidemiol. 2014;2014:2014. [Google Scholar]
- 61.Menon U, Riley SC, Thomas J, Bose C, Dawnay A, Evans LW, Groome NP, Jacobs IJ. Serum inhibin, activin and follistatin in postmenopausal women with epithelial ovarian carcinoma. BJOG Int. J. Obstet. Gynaecol. 2000;107:1069–1074. doi: 10.1111/j.1471-0528.2000.tb11102.x. [DOI] [PubMed] [Google Scholar]
- 62.Lambert-Messerlian GM, DePasquale SE, Maybruck WM, Steinhoff MM, Gajewski WH. Secretion of activin A in recurrent epithelial ovarian carcinoma. Gynecol. Oncol. 1999;74:93–97. doi: 10.1006/gyno.1999.5417. [DOI] [PubMed] [Google Scholar]
- 63.Pickering RT, Lee M-J, Jager M, Layne M, Fried SK. Secretory factors produced by stromal cultures of human omental adipose tissue inhibit adipose differentiation. FASEB J. 2016;30:126.7–126.7. [Google Scholar]
- 64.Hawinkels LJaC, Paauwe M, Verspaget HW, Wiercinska E, van der Zon JM, van der Ploeg K, Koelink PJ, Lindeman JHN, Mesker W, ten Dijke P, Sier CFM. Interaction with colon cancer cells hyperactivates TGF-β signaling in cancer-associated fibroblasts. Oncogene. 2014;33:97–107. doi: 10.1038/onc.2012.536. [DOI] [PubMed] [Google Scholar]
- 65.Sobral LM, Bufalino A, Lopes MA, Graner E, Salo T, Coletta RD. Myofibroblasts in the stroma of oral cancer promote tumorigenesis via secretion of activin A. Oral Oncol. 2011;47:840–846. doi: 10.1016/j.oraloncology.2011.06.011. [DOI] [PubMed] [Google Scholar]
- 66.Zhu J, Mishra RK, Schiltz GE, Makanji Y, Scheidt KA, Mazar AP, Woodruff TK. Virtual high-throughput screening to identify novel activin antagonists. J. Med. Chem. 2015;58:5637–5648. doi: 10.1021/acs.jmedchem.5b00753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Symonds D, Tomic D, Borgeest C, McGee E, Flaws JA. Smad 3 regulates proliferation of the mouse ovarian surface epithelium. Anat. Rec. Part A. 2003;273A:681–686. doi: 10.1002/ar.a.10090. [DOI] [PubMed] [Google Scholar]
- 68.Ramachandran A, Marshall ES, Love DR, Baguley BC, Shelling AN. Activin is a potent growth suppressor of epithelial ovarian cancer cells. Cancer Lett. 2009;285:157–165. doi: 10.1016/j.canlet.2009.05.010. [DOI] [PubMed] [Google Scholar]
- 69.Network TCGAR. Integrated genomic analyses of ovarian carcinoma. Nature. 2011;474:609–615. doi: 10.1038/nature10166. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Activin A has no effect on proliferation of MOE cells. SRB proliferation assays for activin A (0–40 ng/ml) in the presence (A) or absence (B) of 10% FBS.
Figure S2. Activin A signaling is associated with metastasis of ovarian tumors. A) Heat maps for genes involved with activin A and TGFβ1 signaling serous ovarian tumors compared to normal ovary. B) Genes in activin A signaling, distant tumors compared to tumors localized in the ovary. red = increased expression. blue = decreased expression.
Figure S3. Components of TGFβ signaling are not associated with disease-free survival (DFS) in serous cancer patients. DFS Kaplan-Meier plots from OvMark for TGFB1 (A), TGFB2 (B), TGFB3 (C), TGFBR1 (D), and TGFBR2 (E).
Table S1. Primary antibodies, dilution, and blocking reagent used for western blotting (WB) and immunochemistry (IHC).
Table S2. Gene symbols, primer sequences, and amplicon sizes for genes amplified via RT-PCR.