Abstract
Colorectal cancer (CRC), presenting the third most common malignancy worldwide. In recent years, the aberrantly upregulation or downregulation of miRNAs in CRC have been evidenced in a number of studies. In this study, our results showed that the expression of miR-429 was significantly higher in CRC tissue compared with adjacent non-tumor tissue. In addition, our findings showed that miR-429 level was significantly associated with clinicoplathological features and prognosis of patients with CRC. Moreover, our findings showed that miR-429 exerted oncogenic effect by directly targeting HOXA5, a transcription factor of HOX families that is involved in the development and progression of CRC.
Keywords: CRC, HOXA5, MiR-429, tumor growth, metastasis
Introduction
Colorectal cancer (CRC), presenting the third most common malignancy worldwide, accounts for 1.4 million new cases [1] and 50,000 deaths in 2013 (fourth cancer-related death) [2]. The incidence of CRC has been dramatically increasing in developing countries, which may be attributed to the adaption of Western lifestyle featured by increased fat intake and reduced physical activity [3,4]. Currently, standard treatment regimen for localized stage CRC is surgery combining with chemotherapy. For patients with metastatic CRC, chemotherapy is the main option. Besides conventional chemotherapy, targeted therapy has also been employed in the treatment of CRC. However, about half of the patients with CRC still die from distant metastasis (liver is the prominent organ for metastasis) within 6 years following diagnosis [5]. Therefore, it is imperative to identify novel therapeutic targets in order to develop more effective therapy for CRC patients.
Since the last decades, a lot of attention has been paid to the role of miRNAs in tumorigenicity and tumor progression [6]. MiRNAs, a family of endogenous non-coding mRNA molecules with a length of 18-25 nucleotides, can function either as oncogene or tumor suppressor through regulating the expression of target protein at the post-transcriptional level by binding the 3’-UTR portion of mRNAs through translational repression or degradation [7]. By modulating different target genes, miRNA have been found to play a role in a variety of cellular activities such as cell apoptosis, cell proliferation, invasion, migration and stem cell differentiation [8-10]. In fact, a number of miRNAs have been found to be involved in the development and progression of CRC. For instance, miR-378 is found to aberrantly downregulated in CRC tissues compared with match normal tissue and lower expression of miR-378 predicts poor clinical outcome for CRC patients [11]. In contrast, upregulation of miR-183 is closely related to advanced clinical stage, lymph node and distant metastases, and poor prognosis of CRC [12]. A previous study showed that miR-1297 was downregulated in CRC cell lines [13]. Taken together, these results suggested that miRNAs are promising therapeutic targets for CRC.
The HOX genes, a family of transcription factors firstly found to be involved in embryonic development and play a role in various biological activities of cells such as homeostasis and cell differentiation, have also been implicated in a number of malignancies [14]. Recently, HOXA family genes were found to be downregulated in primary NSCLC tissues compared to normal lung tissues [15]. As a member of HOXA family, HOXA5 is reported to promote tumor initiation and progression in breast cancer [16]. In non-small-cell lung carcinoma, HOXA5 is also able to suppress metastasis [17]. Besides solid tumor, the role of HOXA5 in hematological malignancies has also been evidenced [18]. As regards to CRC, Huelsken et al found that HOXA5 is downregulated, and its re-expression induces loss of the cancer stem cell phenotype, preventing tumor progression and metastasis [19], suggesting the tumor suppressor role of HOXA5. However, the mechanism by which HOXA5 is regulated in CRC cells remains unclear.
Materials and methods
Clinical specimen collection and analysis
The clinical study protocol was reviewed and approved by Medical Ethics Committee of Qingdao University Affiliated Hospital (Qingdao, China) and consent forms were signed by all patients or guardians. A total of 71 patients diagnosed as CRC and undergone radical resection in the Qingdao University Affiliated Hospital between June 2009 and May 2011 were enrolled in this study. None of patients had received preoperative adjuvant therapy. CRC tissue and adjacent non-cancerous tissue samples (at least 5 cm from the margin of the tumor) were collected and immediately frozen in liquid nitrogen until use. All samples were blindly examined by two senior pathologists for diagnosis and histological classification according to the 7th edition of the UICC/AJCC TNM staging system for CRC. The expression level of miR-429 in tissue was determined by quantitative RT-PCR. The expression of HOXA5 was analysed by automated capillary western blot (WES) as previously described [20].
Cell lines and cultures
Human CRC cells LOVO, HT29, SW620, HCT116 and normal human intestinal epithelial cell line (HIEC) were obtained from Cell Bank of Chinese Academy of Sciences (Shanghai, China). All cells were cultured in DMEM medium (Invitrogen, Carlsbad, CA) containing 1% penicillin/streptomycin (Thermo Fisher Scientific, Waltham, MA) and 10% fetal bovine serum (FBS; Invitrogen, Carlsbad, CA). Cells were cultured in a humidified incubator at 37°C with 5% CO2.
Transfection of miR-429 mimic or inhibitor
The lentiviral constructs of miR-429 mimics, miR-429 inhibitor and negative control (NC) were synthesized by Genepharma (Shanghai, China). CRC cells were infected with the constructed lentivirus to induce ectopic expression of miR-429 or knockdown miR-429 expression.
Quantitative real-time PCR (qRT-PCR)
Total RNA was extracted using Trizol reagent (Tiangen Biotech, Beijing, China), and miRNAs were obtained using the miRcute miRNA isolation kit (DP501) (Tiangen Biotech, Beijing, China) from cultured cells or human tissues. MiR-370 expression was quantified by real time PCR with a TaqMan Probe (Applied Biosystems, Carlsbad, CA) according to the manufacture’s instructions. Briefly, cDNA was obtained by High Capacity cDNA Archive Kit (Applied Biosystems, Foster City, CA, USA) and qRT-PCR was performed using a TaqMan PCR kit and the ABI 7500 System (Thermo Fisher Scientific, Waltham, MA). The relative expression of miR-429 in cells and tissues were normalized to that of U6. For HOXA5 mRNA detection, the primer was synthesized based on published sequence [21]. First-strand cDNA was reverse transcribed from 1 μg total RNA using the Super M-MLV Reverse transcriptase (BioTeke Co., Beijing, China). PCR reaction solution included a master mix that including SYBR GREEN mastermix (Solarbio Co., Beijing, China), forward primer, reverse primer, and 10 ng template cDNA. GADPH was used as internal control to normalize PCR results. PCR results were analyzed using the comparative ΔCt method (ABPrism software, Applied Biosystems, Foster City, CA).
Western blotting analysis
Proteins were isolated from cells and tissues by lysing frozen tissues in RIPA buffer (Sigma, St. Louis, MO). Proteins were extracted from cells after cells were lysed by lysis buffer (Beyotime, Shanghai, China) containing protease inhibitors (Sigma, St Louis, MO). The protein concentration was measured by the BCA protein assay kit (Beyotime, Shanghai, China). Extracted proteins were separated on SDS-PAGE and then transferred electrophoretically onto a polyvinylidene difluoride membrane (Millipore, Billerica, MA). Proteins were probed with specific antibodies following standard protocol. The second antibodies used in this study were goat anti-rabbit IgG-HRP, goat anti-mouse IgG-HRP and donkey anti-goat IgG-HRP (Beyotime Institute of Biotechnology, Shanghai, China). Signals were detected using chemiluminescent substrate (KPL., Guildford, UK) and the blot intensity were quantified using BandScan software (Glyko, Novato, CA).
Luciferase reporter assay
The luciferase reporter assay was performed using constructed pGL3-HOXA5-3’UTR plasmid containing 3’UTR of HOXA5 or pGL3-HOXA5-3’UTR Mut plasmid containing mutated 3’UTR of HOXA5 as previously described [22].
For luciferase assay, CRC cells were transiently co-transfected with 0.2 μg of pGL3-HOXA5-3’UTR or pGL3-HOXA5-3’UTR Mut, 0.02 μg of pRL-TK-Renilla luciferase reporter plasmids (Promega, Madison, WI) containing the Renilla-luciferase for normalization, and with 5 pmol of miR-429 mimic, inhibitor or control. 24 hours after transfection, cells were lysed and the luciferase activity was measured with a luminometer using the dual-luciferase reporter assay system, according to the manufacturer’s instructions.
Re-expression of HOXA5 in CRC cells
HOXA5 expression was restored through transfection with expressing construct for using Lipofectamine 3000 reagent (Invitrogen, Grand Island, NY) according the manufacturer’s instructions. HOXA5 overexpressing vector was constructed as previously described using a plasmid vector pGCsi-H1 [21]. Cells transfected with empty vector were used as controls. 48 hours after transfection, the cells were rinsed before resuspended in fresh culture media and the overexpression was verified by western blot analysis.
Cell proliferation assay
Cell Counting Kit-8 (Beyotime, Shanghai, China) was used to assess the cell proliferation. Briefly, a total of 1×105 cells were plated in culture plates. Following an incubation of indicated time, the viable cells were examined by measuring absorbance at 450 nm (Tecan Group Ltd, Männedorf, Switzerland).
Colony formation
Cells suspended in RPMI-1640 agarose medium were seeded in each well of a 6-well plate over a bottom layer of solidified RPMI-1640 agarose medium. Cultures were maintained for 14 days without fresh medium feeding at 37°C in a humidified atmosphere of 95% air and 5.0% CO2. Then cell colonies with over 50 cells were enumerated and stained with violet crystal before being photographed using a digital camera (Nikon DXM1200, Tokyo, Japan).
Flow cytometry
Following treatment, CRC cells were harvested and stained with Annexin V-PE and propidium iodide using an Apoptosis kit (BD Pharmingen, Franklin Lakes, NJ) according to the manufacturer’s instructions. Then the apoptotic percentage of treated cells was determined by a flow cytometer (Beckman Coulter Inc., Miami, FL).
Transwell migration assay
Transwell chambers (polycarbonate filters of 8-m porosity, Corning, NY, USA) were used in this test. The bottom chamber was filled with culture medium containing 20% FBS. Cells were suspended at a density of 5×104 cells/ml in serum-free medium and 1×104 cells were plated in the upper chamber. The cells were removed from the upper chamber by a cotton swab following 24-hour incubation. Cells penetrated and attached to the bottom of the filter were fixed with 4% formaldehyde solution for 20 minutes before stained with 0.1% crystal violet for 5 minutes. The cells were then subjected to imaging under a 20× objective using phase-contrast microscopy (Motic China Group Co., Xiamen, China). The number of cells that finally attached to the bottom dishes was counted under a 200× objective. Statistical results of cell numbers per each image field were obtained from three independent experiments averaged from five image fields.
Transwell invasion assay
24-well Transwells coated with Matrigel (8-μm pore size; BD Biosciences, San Jose, CA) were used for cell invasion assays. Equal numbers (1×105) of non-transfected cells as well as cells stably transfected were plated on separate wells. Cells were cultured overnight in serum-free medium before trypsinization and re-suspended at a density of 2×105 cells/ml in DMEM containing 1% FBS. The cells were loaded to the upper chamber, with MEM containing 10% FBS as chemoattractant in the lower chamber. The medium containing 1% FBS in the lower chamber was used as a control. The Matrigel and the cells remaining in the upper chamber were removed by cotton swabs following 24-hour incubation. The cells in the lower surface of the membrane were stained with hematoxylin after the cells were fixed with formaldehyde solution. The cells in at least five random microscopic fields (×200) were counted and photographed.
Immunoflorescence staining and confocal image
Cells were grown on glass coverslip until 80% confluent, and then fixed with 4% formaldehyde solution. Transfected and untransfected cells were then incubated with rabbit monoclonal anti-β-catenin antibody (Cell Signaling Technology, Inc., Beverly, MA, USA, 1:100) for detection of specific protein, respectively. Next, the cells were incubated with FITC-labeled goat anti-rabbit antibody (Beyotime Institute of Biotechnology, Shanghai, China, 1:100) at room temperature for 1 hour in the dark, followed by incubation with DAPI (Biosharp Biotech., Hefei, China, 1:1000) for 5 minutes before washed three times with PBS to remove excessive staining solution. The cells were then subjected to laser scanning confocal microscope (Nikon, Tokyo, Japan).
In vivo xenograft model
The protocol of animal experiments were reviewed and approved by Medical Ethics Committee of Qingdao University. For tumor growth assay, BALB/c nude mice of four-week-old were used for the CRC xenograft models (n=6 per group). 1×107 HT29 cells transfected with control vector or miR-429 mimic construct were suspended in 100 μl medium was injected subcutaneously into the lower left flank regions of mice model. The tumor volume was measured every 3 days and the mice were sacrificed after 30 days.
In the liver metastasis model, miR-429-overexpressing and empty vector control HT29 cells (1×106) in 100 mL HBSS were slowly injected into the spleen of BALB/c athymic mice under anesthesia (n=6 per group). The number and size of metastatic foci in the liver were documented.
Statistical analysis
Values were presented as the mean ± SD. The comparison of miR-429 levels in tumor and normal tissue were performed using student’s t test. Statistical comparisons between cell lines were performed by one-way ANOVA followed by Dunnett’s t-test. The overall survival of patients was assessed by Kaplan-Meier survival analysis. GraphPad Prism software (GraphPad Software Inc., La Jolla, CA) was used to analyze experimental data and a P value less than 0.05 was considered to be statistically significant.
Results
MiR-429 is aberrantly overexpressed in CRC tissues and correlates with clinicopathological parameters
The level of miR-429 in CRC tissue was determined by RT-PCR analysis. Our results revealed that miR-429 was aberrantly upregulated in CRC tissues compared with adjacent non-tumor tissues (Figure 1A). Then the association between clinicopathological parameters of CRC patients and miR-429 level was examined. CRC tissues expressing miR-429 at levels less than the median expression level was allocated to the low group while and those samples with expression above the median value were allocated to the high expression group. As shown in Table 1, the expression level of miR-429 was significantly associated with lymph node metastasis and tumor size. However, our study did not find any correlation between miR-429 level in CRC tissue and other clinicopathological parameters including TNM stage, gender, age, tumor site and histological grade.
Figure 1.
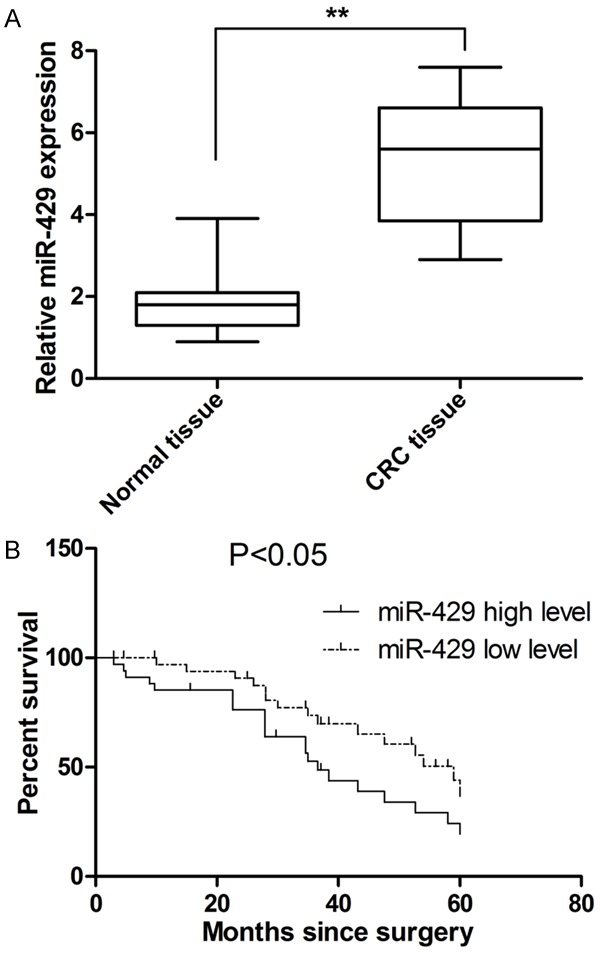
MiR-429 expression is upregulated in CRC tissue and correlates with survival of patients. A. The level of miR-429 in CRC is significantly higher than adjacent non-tumor tissue. B. MiR-429 is significantly associated with shorter patient survival. **P<0.01.
Table 1.
Correlation between miR-429 with clinicopathological features of CRC patients
| Variables | miR-429 level | P value | |
|---|---|---|---|
|
|
|||
| High (35) | Low (36) | ||
| Gender | 0.3412 | ||
| Male | 13 | 18 | |
| Female | 22 | 18 | |
| Age | 0.8128 | ||
| ≤50 | 15 | 17 | |
| >50 | 20 | 19 | |
| Site | 0.3222 | ||
| Colon | 25 | 21 | |
| Rectum | 10 | 15 | |
| Tumor size | 0.0329* | ||
| ≤5 cm | 14 | 24 | |
| >5 cm | 21 | 12 | |
| Differentiation | 0.1567 | ||
| Well, moderate | 14 | 21 | |
| Poor | 21 | 15 | |
| TNM stage | <0.001** | ||
| I and II | 11 | 26 | |
| III and IV | 24 | 10 | |
| Lymph node metastasis | 0.0321* | ||
| Absence | 11 | 21 | |
| Presence | 24 | 15 | |
P<0.05;
P<0.01.
MiR-429 level predicts prognosis of patients with CRC
To evaluate the prognostic significance of miR-429, a Kaplan-Meier survival curve was plotted. As show in Figure 1B, our results showed that patients with a high expression of miR-429 significantly correlated with shorter median survival time (34.2 months for high miR-429 group vs. 58.6 months for low miR-429 group). Furthermore, Cox multivariable analysis was performed to assess whether miR-429 level can be considered as an independent prognostic indicator for overall survival of patients with CRC. As shown in Table 2, the overall survival of patients significantly correlated with miR-429 expression, suggesting the potential of miR-429 as an independent predictor for overall survival of patients with CRC.
Table 2.
Multivariable analysis of the prognostic value of miR-429 in CRC
| Variable | Multivariable analysis | |
|---|---|---|
|
| ||
| HR (95% CI) | P value | |
| Gender (Male or Female) | 0.998 (0.492-1.965) | 0.985 |
| Age (≤50/>50) | 1.598 (0.795-3.268) | 0.425 |
| Differentiation (Well, moderate/Poor) | 1.068 (0.657-1.425) | 0.857 |
| Tumor size (≤5 cm/>5 cm) | 0.819 (0.465-1.857) | 0.857 |
| Lymph node metastasis (Absence/Presence) | 4.568 (2.987-9.245) | 0.021* |
| TNM stage (I, II/III IV) | 3.201 (2.129-7.652) | 0.008** |
| miR-429 level (high/low) | 1.852 (1.019-3.326) | 0.012* |
P<0.05;
P<0.01.
MiR-429 promotes cell proliferation and protects against apoptosis in CRC cell lines
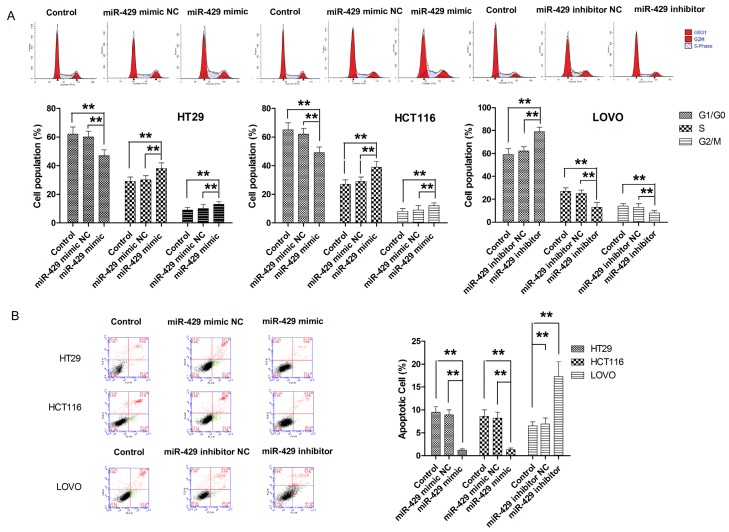
In four tested CRC cell lines, LOVO, HT29, SW620, HCT116, HT29 and HCT116 cells presented with lowest levels of miR-429 while LOVO presented with highest level of miR-429 (Supplementary Figure 1). To determine the effect of miR-429 on cell proliferation, CRC cells were transfected with miR-429 mimic, miR-429 inhibitor, or negative control (Supplementary Figure 2). The effect of miR-429 on cell proliferation was determined by CCK-8 assay. As shown in Figure 2A, ectopic overexpression miR-429 in both HT29 and HCT116 cells significantly enhanced the proliferation of both cell lines. Then colony formation assay was conducted to examine the impact of miR-429 on long-term cell proliferation. As shown in Figure 2B, transfection with miR-429 resulted in significantly more colonies in HT29 and HCT116 cells compared with parental cell lines. In contrast, LOVO cells transfected with miR-429 inhibitor showed significantly suppression in cell proliferation, as demonstrated by CCK-8 and colony formation assay (Figure 2A and 2B). Next, we examined the role of miR-429 in cell cycle progression. As shown in Figure 3A, CRC cells transfected with miR-429 mimic exhibited lower percentages of cells in the G1 phase and increased percentages of cells in the S and G2/M phases compared with parental cell lines. On the other hand, miR-429 knockdown with miR-429 inhibitor exhibited higher percentages of cells in the G1 phase and reduced percentages of cells in S and G2/M phase compared with parental cell line. Then the effect of miR-429 on apoptosis was also examined. As shown in Figure 3B, overexpression of miR-429 resulted in a significantly decrease in apoptotic cell population while knockdown of miR-429 associated with significantly higher apoptosis. To further demonstrate the role of miR-429 in tumour growth, a xenograft model of mice was used. As shown in Figure 5A, consistent with these findings in vitro, miR-429 remarkably enhanced the tumorigenic ability of CRC cells in vivo. Taken together, our results showed that miR-429 promoted the tumour growth.
Figure 2.
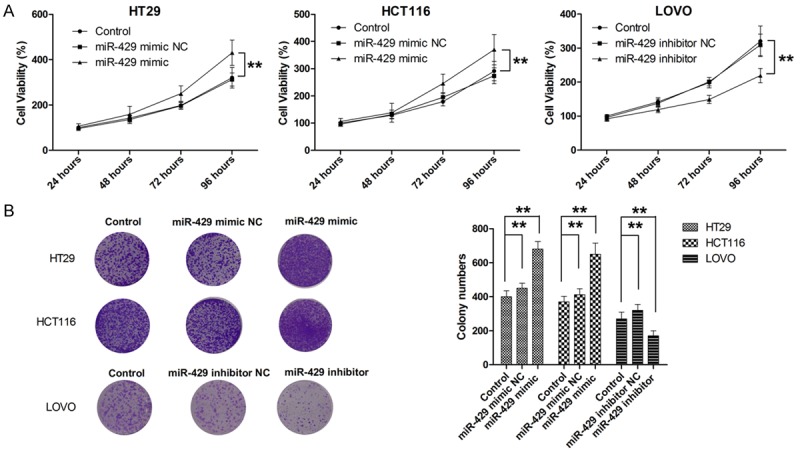
MiR-429 promotes cell proliferation. A. MiR-429 promotes cell proliferation, as determined by CCK-8 assay. B. MiR-429 promotes long term cell proliferation, as determined by colony formation. **P<0.01.
Figure 3.
MiR-429 promotes cell cycle progression and inhibits cell apoptosis. A. MiR-429 promotes cell cycle progression, as determined by flow cytometry. B. MiR-429 suppresses cell apoptosis, as determined by flow cytometry. **P<0.01.
Figure 5.
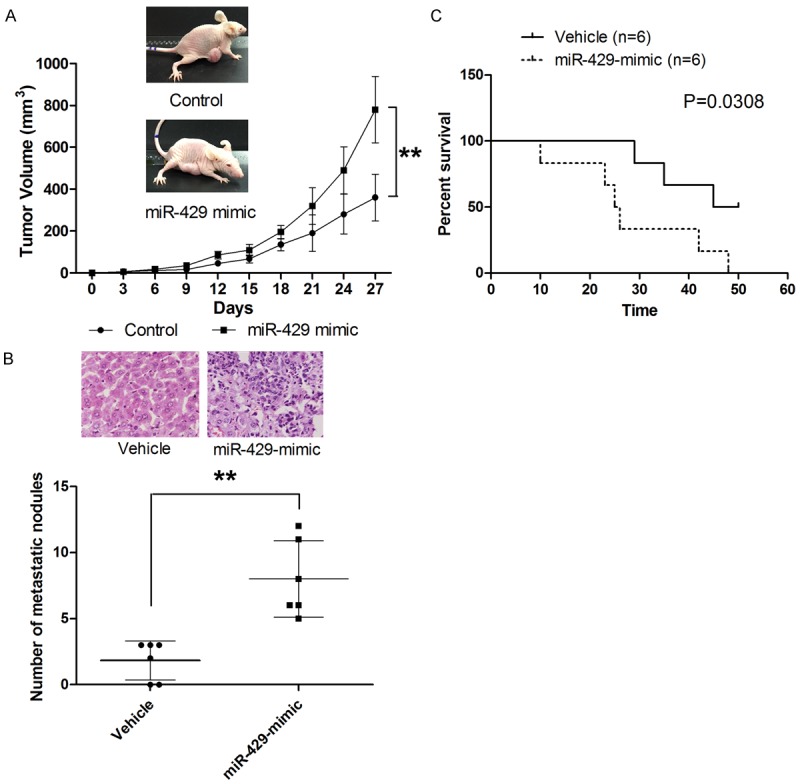
MiR-429 promotes tumor growth and metastasis in vivo. A. MiR-429 exters oncogenic effect in vivo. B. MiR-429 promotes the liver metastasis of CRC. C. MiR-429 overexpression is associated with shorter survival of metastatic model mice. **P<0.01.
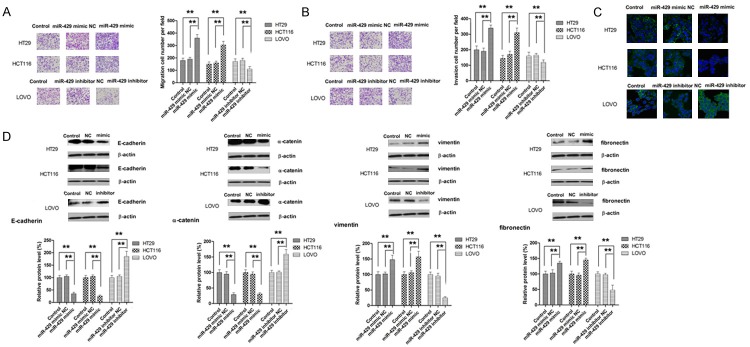
MiR-429 enhances metastasis in CRC
The effect of miR-429 on cell migration was assessed by Transwell assay. As shown in Figure 4A, miR-429 overexpression significantly enhanced the migration of HT29 and HCT116 cells while miR-429 knockdown was associated with suppressed migratory ability of LOVO cells. Correspondingly, transfection with miR-429 mimic significantly increased the invasiveness of CRC cells (Figure 4B). Given the crucial role of EMT process in the metastasis of tumour, we sought to determine whether miR-429 plays a role in EMT. Since EMT is marked by nuclear translocation of β-catenin, we examined the effect of miR-429 on translocation of β-catenin in CRC cells. As shown in Figure 4C, our results revealed that significantly more β-catenin was found in nucleus in HCT116 and HT29 cells compared to parental cells. Meanwhile, a redistribution of β-catenin from nucleus to cytoplasm was observed in LOVO cells transfected with miR-429 inhibitor (Figure 4C). To fully appreciate the role of miR-429 in EMT, the level of EMT makers including E-cadherin, α-catenin, vimentin and fibronectin was examined in cells transfected with miR-429 mimic or inhibitor. As shown in Figure 4D, epithelial markers (E-cadherin and α-catenin) was significantly repressed whereas mesenchymal markers (vimentin and fibronectin) was significantly elevated by overexpressing miR-429 in HCT116 and HT29 cells. On the other hand, miR-429 knockdown significantly suppressed the expression of both mesenchymal markers while markedly increased the expression of both epithelial markers. The role of miR-429 in metastasis was validated in liver metastasis model. As shown in Figure 5B and 5C, mice model established with HT29 cells transfected with miR-429 mimic presented with significantly more metastatic nodules and shorter overall survival. Collectively, our results showed that miR-429 promotes the metastasis of CRC.
Figure 4.
MiR-429 promotes metastatic behavior of CRC cells. A. MiR-429 promotes cell migration. B. MiR-429 promotes cell invasion. C. MiR-429 promotes the translocation of β-catenin. D. MiR-429 modulates the expression of EMT markers. **P<0.01.
HOXA5 is a direct target of miR-429
To clarify the mechanic basis for the regulatory effect of miR-429 on CRC cells, bioinformatics database, MicroCosm and Target scan, were utilized to search for candidate target genes of miR-429. Analysis of both bioinformatic algorithm indicated that HOXA5 might be a direct target of miR-429, accounting for miR-429-induced biological effects. To verify this postulation, luciferase assay was performed. As shown in Figure 6A, luciferase activity of the reporter containing wild-type 3’-UTR of HOXA5 was significantly decreased in cells transfected with miR-429 mimic while no significant change in luciferase activity of the reporter containing mutated 3’-UTR was observed, providing direct evidence that HOXA5 was directly targeted by miR-429. To further investigate the correlation between the expression levels of miR-429 and HOXA5, 20 randomly selected tissue samples were subjected to RT-PCR and simple western analysis. As shown in Figure 6B and 6C, both the protein and mRNA levels of HOXA5 as significantly inversely associated with miR-429 expression. Moreover, miR-429 overexpression led to a significant decrease in HOXA5 mRNA and protein expression compared with parental cell lines (Figure 6D and 6E). On the other hand, downregulation of miR-429 was associated with significantly higher expression of both HOXA5 mRNA and protein. Taken together, these data suggested that miR-429 might be involved in both degradation of mRNA and post-transcriptional regulation of protein expression of HOXA5.
Figure 6.
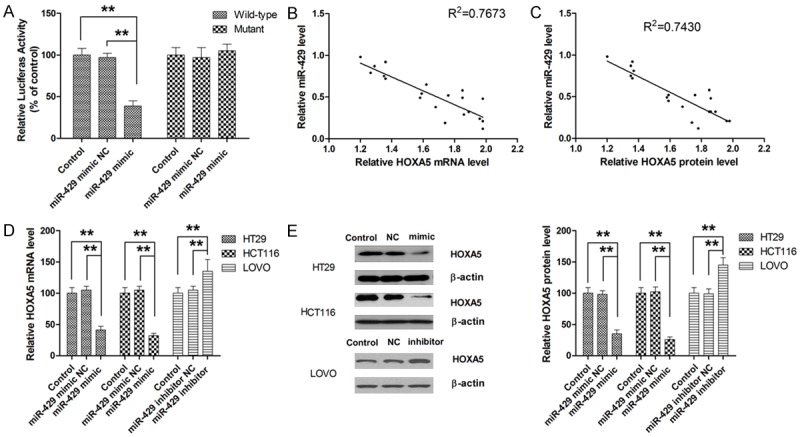
HOXA5 is a direct target of miR-429. A. MiR-429 directly binds to the 3’-UTR of HOXA5. B. HOXA5 mRNA expression is inversely associated with miR-429 expression in CRC tissue. C. HOXA5 protein expression is inversely associated with miR-429 expression in CRC tissue. D. MiR-429 mimic significantly represses the expression of HOXA5 mRNA in CRC cells. E. MiR-429 mimic significantly represses the expression of HOXA5 protein in CRC cells. **P<0.01.
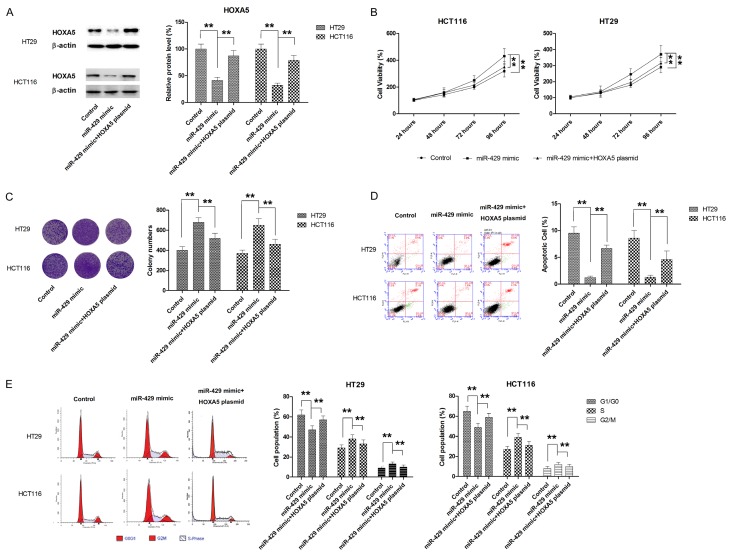
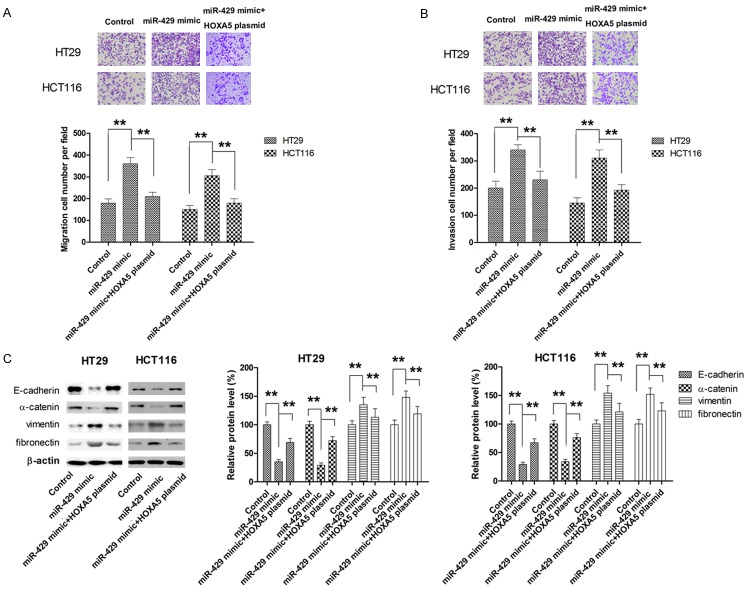
Ectopic overexpression of HOXA5 partially reverses the oncogenic effects of miR-429
To fully appreciate the role of HOXA5 as mediator for miR-429 induced effects, CRC cells were cotransfected with miR-429 mimic and HOXA5 overexpressing plasmid. The expression of HOXA5 was determined by western blot (Figure 7A). As shown in Figure 5B, HOXA5 overexpression significantly attenuated the promoting effect of miR-429 on cell proliferation (Figure 7B). Similar results were also obtained from colony formation assay (Figure 7C). Meanwhile, the apoptosis inhibition and cell cycle promotion by miR-429 mimic was also markedly abrogated by HOXA5 overexpression (Figure 7D and 7E). Transwell assay also revealed that ectopic overexpression of HOXA5 was able to partially reverse miR-429 mimic-induced migration and invasion in both HT29 and HCT116 cells (Figure 8A and 8B). Moreover, the EMT processes induced by miR-429 mimic was also significantly suppressed by transfection with HOXA5 plasmid, as demonstrated by the changes in the expression of EMT markers (Figure 8C). Taken together, our findings demonstrated that miR-429 exerted oncogenic effect, at least in part, by directly repressing HOXA5 expression.
Figure 7.
HOXA5 mediates the pro-proliferative effect of miR-429 in CRC. A. HOXA5 plasmid significantly enhanced the HOXA5 expression in CRC cells. B. HOXA5 overexpression significantly attenuated the promoting effect of miR-429 on cell proliferation. C. HOXA5 overexpression significantly decreases miR-429 induced colony formation. D. HOXA5 overexpression dampened miR-429-induced cell cycle progression. E. HOXA5 overexpression compromises the protective effect of miR-429 on apoptosis. **P<0.01.
Figure 8.
HOXA5 mediates the miR-429-induced enhancement in metastatic behavior in CRC. A. HOXA5 overexpression dampened miR-429-induced cell migration. B. HOXA5 overexpression dampened miR-429-induced cell invasion. C. HOXA5 overexpression dampened miR-429-induced EMT. **P<0.01.
Discussion
As a class of non-coding single-strand RNAs, miRNAs modulate cellular activities through posttranscriptional gene regulatory pathways [7]. Through binding to the 3’-untranslated region (3’UTR) of target mRNA, miRNAs could represses the expression of target gen by inducing transcript degradation, translational repression, and gene silencing. Dysregulation of miRNAs have been observed in a variety of human malignancies, and accumulating evidence have showed that miRNAs plays crucial roles in tumor survival, growth, apoptosis, invasion, and the capacity to metastasizem acting as either oncogenes or tumor suppressor genes. miR-429, belonging to the miR-200 family, has also been found to be involved in tumor development and progression and the dysregulation of miR-429 occurs in various types of cancer. However, conflicting evidence have been obtained about the functional role of miR-429 in cancer. In endometrioid endometrial carcinoma, aberrantly upregulation of miR-429 along with miR-200a/b has been documented compared to adjacent normal endometrial tissuesand tumor suppressor PTEN has been identified as a direct target of miR-429 in endometrioid endometrial carcinoma responsible for its oncogenic role [23]. In human prostate cancer cells, knockdown of miR-429 correlates with inhibit proliferation suppression, which is attributed to the modulation of p27Kip1 as target gene of miR-429 [24]. A recent study by Xiao et al also reported that miR-429 promotes the proliferation of non-non-small cell lung cancer cells via targeting DLC-1, further supporting the role of miR-429 as an oncogene [25]. On the contratry, the role of miR-429 as a tumor suppressor in certain cancer types has also been evidenced. Song et al (22) reported that miR-429 was frequently downregulated in pancreatic ductal adenocarcinoma (PDAC) tissues and cell lines, and lower miR-429 expression in PDAC tissues significantly correlated with shorter survival of PDAC patients. In addition, overexpression of miR-429 inhibited PDAC cell growth via targeting TANK-binding kinase 1 [26]. Ye et al (23) also documented the suppression on breast cancer cell migration and invasion by miR-429 and identified zinc finger E-box binding homeobox 1 and Crk-like as direct target of miR-429 [27]. The downregulation of miR-429 has also been detected in renal cell carcinoma and low expression of miR-429 is found to correlate with poor prognosis [28]. In vitro studies also provided evidence for the suppressing effect of miR-429 on cell growth, migration, invasion and EMT in gastric carcinoma cells [29], glioblastoma cells [30], cervical cancer cells [31] and bladder cancer cells [32]. All these studies suggest that whether miR-429 can function as an oncogene or tumor suppressor may depend on the key target genes regulated by miR-429, which may be cell specific. More interestingly, even in one single human malignancy, colorectal cancer, the role of miR-429 remains controversial. In 2013, Li et al reported that miR-429 expression was up-regulated in human colorectal cancer (CRC) tissues, and the high miR-429 expression was significantly associated with tumor size, lymph node metastasis and poor prognosis [33]. Their findings were backed up by a few of following studies which demonstrated that miR-429 functions as an oncogene in CRC [34,35]. However, a few research groups have reported that miR-429 was aberrantly dwonregulated in CRC tissue and function as tumor suppressor in CRC. For instance, Sun et al reported that miR-429 was significantly downregulated in CRC tissues and cell lines, and suggested that miR-429 inhibits the initiation of EMT and regulated expression of EMT-related markers by targeting Onecut2 [36]. Tian et al also pointed that miR-429 inhibits the migration and invasion of colon cancer cells by targeting PAK6/cofilin signaling [37]. Moreover, increase the expression of miR-429 by drugs could significantly suppress the tumor growth of CRC [38]. In present study, the expression of miR-429 was also examined in primary CRC tissues and compared with their matched normal tissues. Aberrantly upregulation of miR-429 has been revealed. Moreover, our findings showed that miR-429 positively associated with advanced stage and metastasis of CRC, supporting that miR-429 functions as oncogene in CRC. We postulated that the conflicting results regarding the role of miR-429 in CRC may result from usage of small size of clinical sample and different cell lines. To fully appreciate the role of miR-429 in CRC, a larger size of clinical samples must be examined and more CRC cell lines must be utilized to avoid bias.
A number of molecular targets have been proposed to be responsible for the oncogenic effect of miR-429 in human malignancies, including IKKβ in cervical cancer cells [31]. DLC-1 in non-small cell lung cancer cells [25], and SOX2 in colorectal cancer [33]. In our study, HOXA5 has been discovered as a novel direct target of miR-429 in CRC and forced ectopic expression of HOXA5 reversed the promoting effect of miR-429 on tumor growth and metastasis. The role of HOXA5 in cancer biology was firstly noticed in breast because that the expression of HOXA5 is lost in more than 60% of breast cancer cell lines and primary carcinomas due to promoter hypermethylation [39]. Following mechanistic studies showed that HOXA5 promoted breast cancer cell death through p53-dependent or caspase 2- and 8-activated apoptosis [13,14], and the loss of HOXA5 expression could lead to the functional activation of Twist, resulting in aberrant cell cycle regulation and the promotion of breast tumorigenesis [39-41]. Since then, the involvement of HOXA5 in other human malignancies has been reported in a variety of human cancers, including leukemia, renal cell carcinoma, oral squamous cell carcinoma, colorectal cancer and non-small cell lung cancer [17-19,42,43], although HOXA5 seems to play distinct roles in different human malignancies. Although most evidence pointed out that HOXA5 function as tumor suppressor, it does act as oncogene in leukemia, non-small cell lung cancer and hepatocellular carcinoma [18,44,45]. In the context of CRC, Ordóñez-Morán et al recently reported that HOXA5 is downregulated, and its re-expression induces loss of the cancer stem cell phenotype, preventing tumor progression and metastasis [19]. In line with their findings, our results also showed that HOXA5 expression was repressed in CRC tissue, confirming the tumor suppressor role of HOXA5 in CRC.
In summary, the results presented here provide evidence that miR-429 function as an onco-miRNA and the level of miR-429 correlate with clinical characteristics and prognosis. More important, our study shows that miR-429 promotes CRC progression and metastasis by directly targeting HOXA5.
Acknowledgements
This work was supported by National Natural Science Foundation (No. 81473384).
Disclosure of conflict of interest
None.
Supporting Information
References
- 1.Ferlay J, Shin HR, Bray F, Forman D, Mathers C, Parkin DM. Estimates of worldwide burden of cancer in 2008: GLOBOCAN 2008. Int J Cancer. 2010;127:2893–2917. doi: 10.1002/ijc.25516. [DOI] [PubMed] [Google Scholar]
- 2.Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA Cancer J Clin. 2011;61:69–90. doi: 10.3322/caac.20107. [DOI] [PubMed] [Google Scholar]
- 3.Byeon JS, Yang SK, Kim TI, Kim WH, Lau JY, Leung WK, Fujita R, Makharia GK, Abdullah M, Hilmi I, Sollano J, Yeoh KG, Wu DC, Chen MH, Kongkam P, Sung JJ Asia Pacific Working Group for Colorectal Cancer. Colorectal neoplasm in asymptomatic Asians: a prospective multinational multicenter colonoscopy survey. Gastrointest Endosc. 2007;65:1015–1022. doi: 10.1016/j.gie.2006.12.065. [DOI] [PubMed] [Google Scholar]
- 4.Xie J, Gao H, Peng J, Han Y, Chen X, Jiang Q, Wang C. Hispidulin prevents hypoxia-induced epithelial-mesenchymal transition in human colon carcinoma cells. Am J Cancer Res. 2015;5:1047–1061. [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 5.Dattatreya S. Metastatic colorectal cancer-prolonging overall survival with targeted therapies. South Asian J Cancer. 2013;2:179–185. doi: 10.4103/2278-330X.114152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lai EC. Micro RNAs are complementary to 3’ UTR sequence motifs that mediate negative post-transcriptional regulation. Nat Genet. 2002;30:363–364. doi: 10.1038/ng865. [DOI] [PubMed] [Google Scholar]
- 8.Xu N, Papagiannakopoulos T, Pan G, Thomson JA, Kosik KS. MicroRNA-145 regulates OCT4, SOX2, and KLF4 and represses pluripotency in human embryonic stem cells. Cell. 2009;137:647–658. doi: 10.1016/j.cell.2009.02.038. [DOI] [PubMed] [Google Scholar]
- 9.le Sage C, Nagel R, Egan DA, Schrier M, Mesman E, Mangiola A, Anile C, Maira G, Mercatelli N, Ciafre SA, Farace MG, Agami R. Regulation of the p27(Kip1) tumor suppressor by miR-221 and miR-222 promotes cancer cell proliferation. EMBO J. 2007;26:3699–3708. doi: 10.1038/sj.emboj.7601790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yoo AS, Staahl BT, Chen L, Crabtree GR. MicroRNA-mediated switching of chromatin-remodelling complexes in neural development. Nature. 2009;460:642–646. doi: 10.1038/nature08139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang GJ, Zhou H, Xiao HX, Li Y, Zhou T. MiR-378 is an independent prognostic factor and inhibits cell growth and invasion in colorectal cancer. BMC Cancer. 2014;14:109. doi: 10.1186/1471-2407-14-109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhou T, Zhang GJ, Zhou H, Xiao HX, Li Y. Overexpression of microRNA-183 in human colorectal cancer and its clinical significance. Eur J Gastroenterol Hepatol. 2014;26:229–233. doi: 10.1097/MEG.0000000000000002. [DOI] [PubMed] [Google Scholar]
- 13.Chen P, Wang BL, Pan BS, Guo W. MiR-1297 regulates the growth, migration and invasion of colorectal cancer cells by targeting cyclo-oxygenase-2. Asian Pac J Cancer Prev. 2014;15:9185–9190. doi: 10.7314/apjcp.2014.15.21.9185. [DOI] [PubMed] [Google Scholar]
- 14.Kelly ZL, Michael A, Butler-Manuel S, Pandha HS, Morgan RG. HOX genes in ovarian cancer. J Ovarian Res. 2011;4:16. doi: 10.1186/1757-2215-4-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Plowright L, Harrington KJ, Pandha HS, Morgan R. HOX transcription factors are potential therapeutic targets in non-small-cell lung cancer (targeting HOX genes in lung cancer) Br J Cancer. 2009;100:470–475. doi: 10.1038/sj.bjc.6604857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Teo WW, Merino VF, Cho S, Korangath P, Liang X, Wu RC, Neumann NM, Ewald AJ, Sukumar S. HOXA5 determines cell fate transition and impedes tumor initiation and progression in breast cancer through regulation of E-cadherin and CD24. Oncogene. 2016;35:5539–5551. doi: 10.1038/onc.2016.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang CC, Su KY, Chen HY, Chang SY, Shen CF, Hsieh CH, Hong QS, Chiang CC, Chang GC, Yu SL, Chen JJ. HOXA5 inhibits metastasis via regulating cytoskeletal remodelling and associates with prolonged survival in non-small-cell lung carcinoma. PLoS One. 2015;10:e0124191. doi: 10.1371/journal.pone.0124191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li N, Jia X, Wang J, Li Y, Xie S. Knockdown of homeobox A5 by small hairpin RNA inhibits proliferation and enhances cytarabine chemosensitivity of acute myeloid leukemia cells. Mol Med Rep. 2015;12:6861–6866. doi: 10.3892/mmr.2015.4331. [DOI] [PubMed] [Google Scholar]
- 19.Ordonez-Moran P, Dafflon C, Imajo M, Nishida E, Huelsken J. HOXA5 counteracts stem cell traits by inhibiting wnt signaling in colorectal cancer. Cancer Cell. 2015;28:815–829. doi: 10.1016/j.ccell.2015.11.001. [DOI] [PubMed] [Google Scholar]
- 20.Ridnour LA, Cheng RY, Weiss JM, Kaur S, Soto-Pantoja DR, Basudhar D, Heinecke JL, Stewart CA, DeGraff W, Sowers AL, Thetford A, Kesarwala AH, Roberts DD, Young HA, Mitchell JB, Trinchieri G, Wiltrout RH, Wink DA. NOS inhibition modulates immune polarization and improves radiation-induced tumor growth delay. Cancer Res. 2015;75:2788–2799. doi: 10.1158/0008-5472.CAN-14-3011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Berube-Simard FA, Prudhomme C, Jeannotte L. YY1 acts as a transcriptional activator of Hoxa5 gene expression in mouse organogenesis. PLoS One. 2014;9:e93989. doi: 10.1371/journal.pone.0093989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang Y, Xu L, Jiang L. miR-1271 promotes non-small-cell lung cancer cell proliferation and invasion via targeting HOXA5. Biochem Biophys Res Commun. 2015;458:714–719. doi: 10.1016/j.bbrc.2015.02.033. [DOI] [PubMed] [Google Scholar]
- 23.Yoneyama K, Ishibashi O, Kawase R, Kurose K, Takeshita T. miR-200a, miR-200b and miR-429 are onco-miRs that target the PTEN gene in endometrioid endometrial carcinoma. Anticancer Res. 2015;35:1401–1410. [PubMed] [Google Scholar]
- 24.Ouyang Y, Gao P, Zhu B, Chen X, Lin F, Wang X, Wei J, Zhang H. Downregulation of microRNA-429 inhibits cell proliferation by targeting p27Kip1 in human prostate cancer cells. Mol Med Rep. 2015;11:1435–1441. doi: 10.3892/mmr.2014.2782. [DOI] [PubMed] [Google Scholar]
- 25.Xiao P, Liu W, Zhou H. miR-429 promotes the proliferation of non-small cell lung cancer cells via targeting DLC-1. Oncol Lett. 2016;12:2163–2168. doi: 10.3892/ol.2016.4904. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 26.Song B, Zheng K, Ma H, Liu A, Jing W, Shao C, Li G, Jin G. miR-429 determines poor outcome and inhibits pancreatic ductal adenocarcinoma growth by targeting TBK1. Cell Physiol Biochem. 2015;35:1846–1856. doi: 10.1159/000373995. [DOI] [PubMed] [Google Scholar]
- 27.Ye ZB, Ma G, Zhao YH, Xiao Y, Zhan Y, Jing C, Gao K, Liu ZH, Yu SJ. miR-429 inhibits migration and invasion of breast cancer cells in vitro. Int J Oncol. 2015;46:531–538. doi: 10.3892/ijo.2014.2759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Machackova T, Mlcochova H, Stanik M, Dolezel J, Fedorko M, Pacik D, Poprach A, Svoboda M, Slaby O. MiR-429 is linked to metastasis and poor prognosis in renal cell carcinoma by affecting epithelial-mesenchymal transition. Tumour Biol. 2016;37:14653–14658. doi: 10.1007/s13277-016-5310-9. [DOI] [PubMed] [Google Scholar]
- 29.Zhang M, Dong BB, Lu M, Zheng MJ, Chen H, Ding JZ, Xu AM, Xu YH. miR-429 functions as a tumor suppressor by targeting FSCN1 in gastric cancer cells. Onco Targets Ther. 2016;9:1123–1133. doi: 10.2147/OTT.S91879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang Z, Zhou Q, Miao Y, Tian H, Li Y, Feng X, Song X. MiR-429 induces apoptosis of glioblastoma cell through Bcl-2. Tumour Biol. 2015 doi: 10.1007/s13277-015-4291-4. [DOI] [PubMed] [Google Scholar]
- 31.Fan JY, Fan YJ, Wang XL, Xie H, Gao HJ, Zhang Y, Liu M, Tang H. miR-429 is involved in regulation of NF-kappaB activity by targeting IKKbeta and suppresses oncogenic activity in cervical cancer cells. FEBS Lett. 2017;591:118–128. doi: 10.1002/1873-3468.12502. [DOI] [PubMed] [Google Scholar]
- 32.Wu CL, Ho JY, Chou SC, Yu DS. MiR-429 reverses epithelial-mesenchymal transition by restoring E-cadherin expression in bladder cancer. Oncotarget. 2016;7:26593–26603. doi: 10.18632/oncotarget.8557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li J, Du L, Yang Y, Wang C, Liu H, Wang L, Zhang X, Li W, Zheng G, Dong Z. MiR-429 is an independent prognostic factor in colorectal cancer and exerts its anti-apoptotic function by targeting SOX2. Cancer Lett. 2013;329:84–90. doi: 10.1016/j.canlet.2012.10.019. [DOI] [PubMed] [Google Scholar]
- 34.Liu H, Huang C, Wu L, Wen B. Effect of evodiamine and berberine on miR-429 as an oncogene in human colorectal cancer. Onco Targets Ther. 2016;9:4121–4127. doi: 10.2147/OTT.S104729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dong SJ, Cai XJ, Li SJ. The clinical significance of MiR-429 as a predictive biomarker in colorectal cancer patients receiving 5-fluorouracil treatment. Med Sci Monit. 2016;22:3352–3361. doi: 10.12659/MSM.900674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sun Y, Shen S, Liu X, Tang H, Wang Z, Yu Z, Li X, Wu M. MiR-429 inhibits cells growth and invasion and regulates EMT-related marker genes by targeting Onecut2 in colorectal carcinoma. Mol Cell Biochem. 2014;390:19–30. doi: 10.1007/s11010-013-1950-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tian X, Wei Z, Wang J, Liu P, Qin Y, Zhong M. MicroRNA-429 inhibits the migration and invasion of colon cancer cells by targeting PAK6/cofilin signaling. Oncol Rep. 2015;34:707–714. doi: 10.3892/or.2015.4039. [DOI] [PubMed] [Google Scholar]
- 38.Suliman MA, Zhang Z, Na H, Ribeiro AL, Zhang Y, Niang B, Hamid AS, Zhang H, Xu L, Zuo Y. Niclosamide inhibits colon cancer progression through downregulation of the Notch pathway and upregulation of the tumor suppressor miR-200 family. Int J Mol Med. 2016;38:776–784. doi: 10.3892/ijmm.2016.2689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Raman V, Martensen SA, Reisman D, Evron E, Odenwald WF, Jaffee E, Marks J, Sukumar S. Compromised HOXA5 function can limit p53 expression in human breast tumours. Nature. 2000;405:974–978. doi: 10.1038/35016125. [DOI] [PubMed] [Google Scholar]
- 40.Chen H, Chung S, Sukumar S. HOXA5-induced apoptosis in breast cancer cells is mediated by caspases 2 and 8. Mol Cell Biol. 2004;24:924–935. doi: 10.1128/MCB.24.2.924-935.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Stasinopoulos IA, Mironchik Y, Raman A, Wildes F, Winnard P Jr, Raman V. HOXA5-twist interaction alters p53 homeostasis in breast cancer cells. J Biol Chem. 2005;280:2294–2299. doi: 10.1074/jbc.M411018200. [DOI] [PubMed] [Google Scholar]
- 42.Yoo KH, Park YK, Kim HS, Jung WW, Chang SG. Epigenetic inactivation of HOXA5 and MSH2 gene in clear cell renal cell carcinoma. Pathol Int. 2010;60:661–666. doi: 10.1111/j.1440-1827.2010.02578.x. [DOI] [PubMed] [Google Scholar]
- 43.Rodini CO, Xavier FC, Paiva KB, De Souza Setúbal Destro MF, Moyses RA, Michaluarte P, Carvalho MB, Fukuyama EE Head And Neck Genome Project Gencapo. Tajara EH, Okamoto OK, Nunes FD. Homeobox gene expression profile indicates HOXA5 as a candidate prognostic marker in oral squamous cell carcinoma. Int J Oncol. 2012;40:1180–1188. doi: 10.3892/ijo.2011.1321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kim DS, Kim MJ, Lee JY, Lee SM, Choi JY, Yoon GS, Na YK, Hong HS, Kim SG, Choi JE, Lee SY, Park JY. Epigenetic inactivation of Homeobox A5 gene in nonsmall cell lung cancer and its relationship with clinicopathological features. Mol Carcinog. 2009;48:1109–1115. doi: 10.1002/mc.20561. [DOI] [PubMed] [Google Scholar]
- 45.Kanai M, Hamada J, Takada M, Asano T, Murakawa K, Takahashi Y, Murai T, Tada M, Miyamoto M, Kondo S, Moriuchi T. Aberrant expressions of HOX genes in colorectal and hepatocellular carcinomas. Oncol Rep. 2010;23:843–851. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.