Abstract
Rationale
ER stress causes accumulation of misfolded proteins in the ER, activating the transcription factor, ATF6, which induces ER stress response genes. Myocardial ischemia induces the ER stress response; however, neither the function of this response nor whether it is mediated by ATF6 is known.
Objective
Here, we examined the effects of blocking the ATF6-mediated ER stress response on ischemia/reperfusion (I/R) in cardiac myocytes and mouse hearts.
Methods and Results
Knockdown of ATF6 in cardiac myocytes subjected to I/R increased ROS and necrotic cell death, which were mitigated by ATF6 overexpression. Under non-stressed conditions, WT and ATF6 knockout (KO) mouse hearts were similar. However, compared to WT, ATF6 KO hearts showed increased damage and decreased function upon I/R. Mechanistically, gene array analysis showed that ATF6, which is known to induce genes encoding ER proteins that augment ER protein-folding, induced numerous oxidative stress response genes not previously known to be ATF6-inducible. Many of the proteins encoded by the ATF6-induced oxidative stress genes identified here reside outside the ER, including catalase, which is known to decrease damaging ROS in the heart. Catalase was induced by the canonical ER stressor, tunicamycin, and by I/R in cardiac myocytes from WT but not in cardiac myocytes from ATF6 KO mice. ER stress response elements were identified in the catalase gene and were shown to bind ATF6 in cardiac myocytes, which increased catalase promoter activity. Overexpression of catalase, in vivo, restored ATF6 KO mouse heart function to WT levels in a mouse model of I/R, as did AAV9-mediated ATF6 overexpression.
Conclusions
ATF6 serves as a previously unappreciated link between the ER stress and oxidative stress gene programs, supporting a novel mechanism by which ATF6 decreases myocardial I/R damage.
Keywords: Endoplasmic reticulum stress (ER stress), protein folding, oxidative stress, ATF6, catalase, cardiac myocyte, ischemia, ischemia/reperfusion injury, stress
INTRODUCTION
Cellular function depends on protein homeostasis, also known as proteostasis 1. Proteostasis requires the efficient folding of newly synthesized proteins, as well as protein quality control and degradation, which decrease the accumulation of misfolded, potentially toxic proteins 1. At least 1/3 of all proteins, including calcium handling proteins, transmembrane receptors, growth factors, and hormones are synthesized, modified, and folded in the endoplasmic reticulum (ER), then trafficked to various membrane compartments, or secreted 2. Thus, the environment in the ER must be optimal for efficient synthesis and folding of these important proteins 3–5.
A variety of diseases, including many that affect the heart challenge ER protein folding capacity 6–9. Such challenges can be due to mutations in ER proteins, which can affect their folding or targeting, or to disease-related perturbations of the ER environment 10, which lead to imbalanced proteostasis, in extreme cases causing ER stress. ER stress contributes to pathology by impeding the production of critical ER proteins, and by increasing the accumulation of potentially toxic misfolded proteins.
ER protein misfolding activates the ER stress response, a conserved signaling system that initiates multiple processes, including gene programs that initially restore proteostasis, thus serving adaptive roles. However, if the ER stress is not resolved by these initial processes, continued ER stress leads to cell death 8, 11, 12. Since the ER stress response can be adaptive or maladaptive, the roles for the ER stress response in either mitigating or causing damage during diseases, including ischemic heart disease are not well understood13. Therefore, defining the molecular processes in the diseased heart that are regulated by ER stress, as well as understanding how and when ER stress is activated in the heart, are required to determine whether to use ER stress activating or inhibiting strategies to improve cardiac function during pathology.
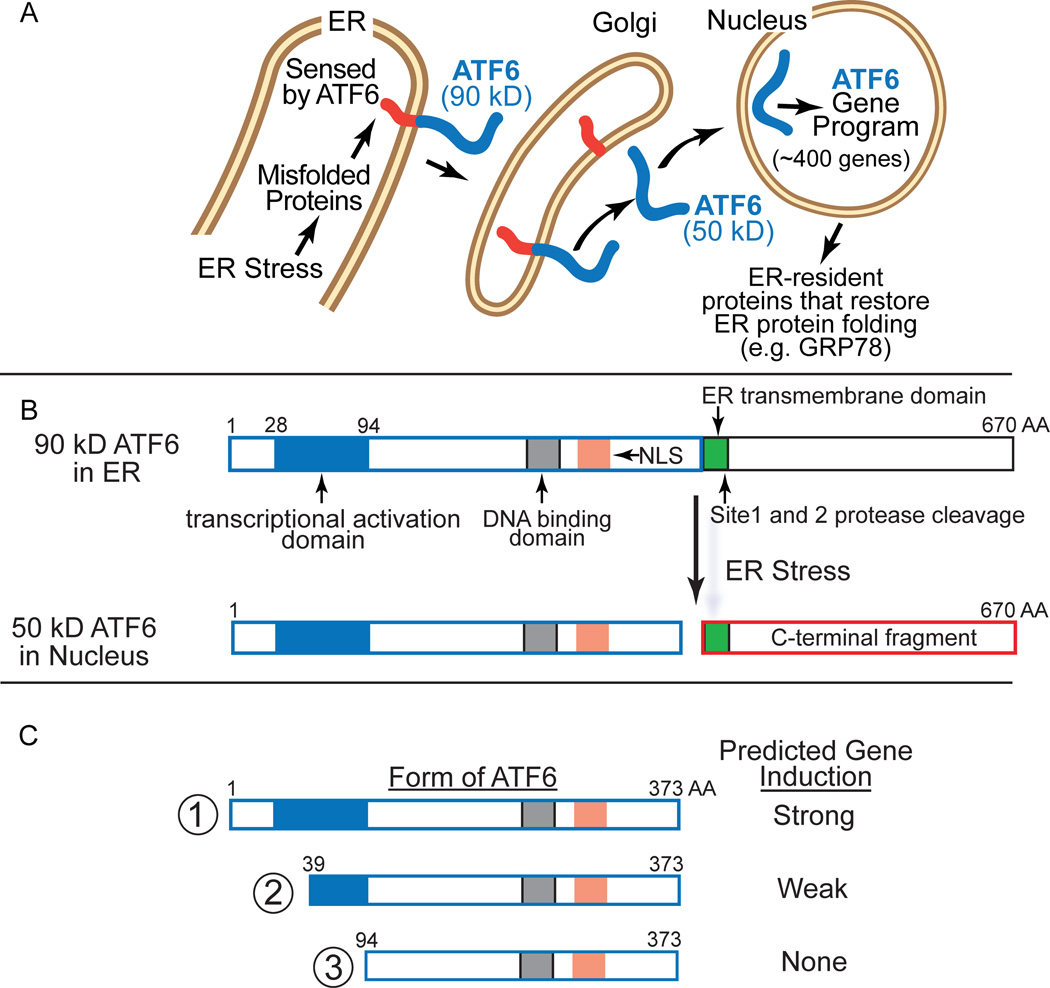
ER stress response genes are regulated by several transcription factors, including ATF6α, abbreviated here as ATF6, a 670 amino acid, ER-transmembrane protein 14, 15. Although not well studied in cardiac myocytes, in model cell lines ER protein misfolding triggers the translocation of ATF6 to the nucleus, where it induces ER stress response genes (Fig. 1A)15. Most of these genes encode proteins, such as the well-studied ER chaperone, glucose regulated protein 78 kD (GRP78), that are localized to the ER, where they enhance the ER protein-folding environment and contribute to resolving the ER stress. To examine roles for ectopically expressed ATF6 in the heart, we previously generated a transgenic mouse that expresses a conditionally activated form of ATF6 in cardiac myocytes; in these mice we showed that activation of ATF6 increased numerous genes, including Grp78, and decreased myocardial ischemia/reperfusion (I/R) damage16, 17. While this showed that ectopically expressed ATF6 can be adaptive in the heart, the function of endogenous ATF6 in the heart has not been studied. Moreover, since most known ATF6-induced proteins localize to the ER, where they enhance protein-folding18, it is unclear how ATF6-regulated genes could decrease I/R damage in the heart, most of which is caused by reactive oxygen species (ROS) generated outside the ER19, 20. Here, we examined the effects of gene-targeted disruption of endogenous ATF6 and showed that it increased myocardial I/R damage. Mechanistically, we found that during I/R, ATF6 is required for the induction of numerous oxidative stress response genes that encode antioxidant proteins, many of which reside outside the ER. This is the first study of ATF6 gene deletion in the heart, and the first to show, in any cell or tissue type that ATF6 is a key transcriptional inducer of oxidative stress response genes encoding proteins outside the ER that decrease damaging ROS during I/R.
Figure 1. ATF6 Activation by ER Stress.
A, Inactive ATF6 is a 90 kD ER transmembrane protein that is released from the ER upon protein misfolding, translocates to the Golgi, where it is cleaved by site 1 and site 2 proteases, to liberate a 50 kD N-terminal fragment, which translocates to the nucleus and acts as a transcription factor to regulate the ATF6 gene program. B, Shown are the relative locations of the ATF6 transcriptional activation domain (blue), DNA binding domain, nuclear localization sequence (NLS), site 1 and site 2 protease cleavage sites, and ER transmembrane domains in full-length, p90 ATF6 (670 amino acids), as well as the location where ATF6 is cleaved upon ER stress. C, Shown are the N-terminal truncated forms of active ATF6 used in this study. Form 1 represents the native ATF6 with the transcriptional activation, DNA binding and nuclear localization domains intact, while forms 2 and 3 have progressive truncations through the transcriptional activation domain. Also shown is the predicted relative strength of gene induction and of each form.
METHODS
Further details on the Methods can be found in the Online Supplement.
Laboratory animals
The research reported in this paper has been reviewed and approved by the SDSU Institutional Animal Care and Use Committee and it conforms to the Guide for the Care and Use of Laboratory Animals published by the National Research Council.
ATF6 knockout mice
There are two isoforms of ATF6, ATF6α and ATF6β. The mice used in this study were generated so that ATF6α, which is expressed in all cell types, has been globally deleted in all tissues21. In that study, it was found that ATF6α deletion had no effect on mouse development through at least 4 months of age. Since ATF6β was not examined in this study, in most cases in this paper we use the term ATF6 to mean ATF6α. ATF6α floxed mice are unavailable. The ATF6 knockout mice used in this study were 10 week-old males.
Statistics
Unless otherwise stated, values shown are mean ± SEM and statistical treatments are either t-test or one-way ANOVA followed by Newman-Keuls post hoc analysis.
RESULTS
Though not well studied in cardiac myocytes, in model cell lines it has been shown that the 90 kD ER form of ATF6 senses misfolded proteins in the ER, then translocates to the Golgi where it is cleaved to liberate an N-terminal fragment in the cytosol that has a nuclear localization sequence (NLS) and a DNA binding domain (Fig. 1B). The N-terminal 50 kD fragment of ATF6, which is about 400 amino acids, translocates to the nucleus where it functions as a transcription factor22. Using HeLa cells as a model, we previously mapped the transcriptional activation domain of ATF6 to a region between amino acids 29–9423. However, studies examining the ATF6 transcriptional activation domain have not been done in cardiac myocytes, or in the heart. Here, we generated expression constructs that encode three forms of ATF6 that we predicted from our studies with HeLa cells to exhibit a range of transcriptional activities (Fig. 1C).
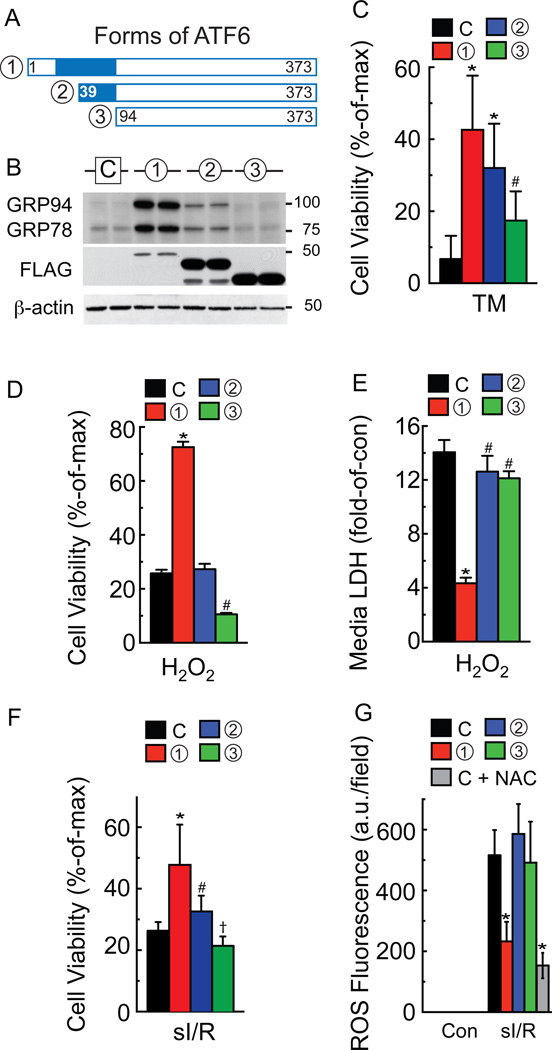
Initial studies in neonatal rat ventricular myocytes (NRVM) showed that the three forms of ATF6 (Fig. 2A), exhibited different abilities to increase expression of the well-known ATF6-induced proteins, GRP94 and GRP78. The native N-terminal fragment of ATF6, ATF6(1–373), called form 1 here, which was expressed at approximately similar levels as endogenous ATF6 (Online Fig. IA), mimics the cleaved form of ATF6 and was the most potent inducer, while ATF6(39–373), form 2, which has about half of the transcriptional activation domain missing, was partially active, and ATF6(94–373), form 3, which lacks all of the transcriptional activation domain was inactive (Fig. 2B GRP94 and GRP78). None of the three forms of ATF6 affected the other two branches of the ER stress response, as they did not increase PERK phosphorylation, nor did they induce XBP1 splicing, an indicator of IRE1 activation (Online Fig. IB). We also previously showed in HeLa cells that ATF6 exhibits a novel degraded-when-active property, such that the more transcriptionally active forms of ATF6 exhibit the shortest half-lifes23, 24. Consistent with those studies were our findings in NRVM showing that the transcriptional activity of ATF6 was inversely related to the expression level of ATF6 (Fig. 2B FLAG).
Figure 2. Effects of ATF6 overexpression on ER- and oxidative stress in cultured cardiac myocytes.
A, Diagram of the forms of ATF6 used in this study. B, Neonatal rat ventricular myocytes (NRVM) were infected with adenovirus (AdV) encoding either no protein (C), or one of the three forms of ATF6 shown, then immunoblotted using a FLAG antibody to detect the overexpressed ATF6, a KDEL antibody, which detects GRP94 and 78, or β-actin antibody. C-G, NRVM were infected with either control (C) or each of the ATF6-expressing AdV shown in A, then treated for 48h with TM (40 µg/ml) (C), 37.5 µM H2O2 for 8h (D, E), sI/R (F, G), and 5 mM N-acetyl cysteine (NAC) (G), followed by viability determination using an MTT assay, or by media enzyme assay to determine LDH activity, or ROS measurement with CellROX, as shown. * # † p<0.05 different than other values by ANOVA.
Given the differential effects of these forms of ATF6 on ER stress gene induction, they are potentially valuable reagents with which to discover cardiac myocyte functions that are regulated by ATF6-induced genes. For example, if these forms of ATF6 affect a particular myocyte function, such as survival, with the same rank-order that they affect ATF6 target gene expression, then it is likely that at least some ATF6 target genes contribute to those functions. As an example, we showed that in NRVM treated with the prototypical ER stressor, tunicamycin (TM), which induces ER protein misfolding by inhibiting protein glycosylation, the three forms of ATF6 decreased ER stress-induced cell death with the same rank-order that they induce ER proteins that reduce ER protein misfolding, e.g. GRP94 and GRP78 (Fig. 2C).
Next, we examined the effects of other stressors that are not typically considered to be inducers of ER protein misfolding and the canonical ER stress response, such as H2O2, which induces oxidative stress by increasing ROS levels. While ATF6 was expected to protect against ER stress-induced cell death, which is primarily apoptotic, we anticipated that it would be ineffective against oxidative stress-induced cell death, which is primarily necrotic. Surprisingly, form 1 of ATF6 protected myocytes from H202-induced cell death (Fig. 2D red), form 2 was without effect (Fig. 2D blue), while form 3, which exerts dominant-negative effects on endogenous ATF6 signaling, actually increased H2O2-induced myocyte death (Fig. 2D green). It is unclear why the relative effects of the three forms of ATF6 are somewhat different in response to TM vs H2O2, but one possibility is that the effects of ATF6 on H2O2-induced myocyte death might be due to induction of proteins that do not reside in the ER and do not directly affect ER protein folding, such as antioxidant genes. Since ATF6 is not known for its ability to affect the expression of antioxidant genes, we focused on the antioxidant effects of ATF6 in more depth.
It has been shown that H2O2 kills myocytes by necrosis25; accordingly, we examined the effects of the three forms of ATF6 on the media levels of LDH and HMGB1, which are measures of necrosis25, 26. Again, form 1 of ATF6 strongly decreased media levels of LDH and HMGB1, while the other forms had less of an effect (Fig. 2E; Online Fig. IC), indicating that activated ATF6 can inhibit H2O2-induced necrosis of cardiac myocytes. Consistent with this was our finding that H2O2 had no effect on PARP cleavage, a measure of apoptosis27 (Online Fig. ID and IE). Since the deleterious effects of H2O2 are due to ROS, we tested the three forms of ATF6 in NRVM subjected to simulated ischemia/reperfusion (sI/R), a pathophysiological maneuver that increases mitochondrial-derived ROS (Online Fig. IF). Similar to the findings with H2O2, form 1 of ATF6 exhibited the most significant ability to protect cardiac myocytes from sI/R-mediated death (Fig. 2F), and to decrease ROS generation during sI/R (Fig. 2G and Online Fig. IG). Moreover, N-acetyl cysteine (NAC), a well-characterized ROS scavenger, mimicked the ability of form 1 of ATF6 to decrease ROS when examined using two different assays, as recently recommended28 (Fig. 2G and Online Fig. IG, sI/R NAC), indicating that ATF6 acted functionally like a ROS scavenger.
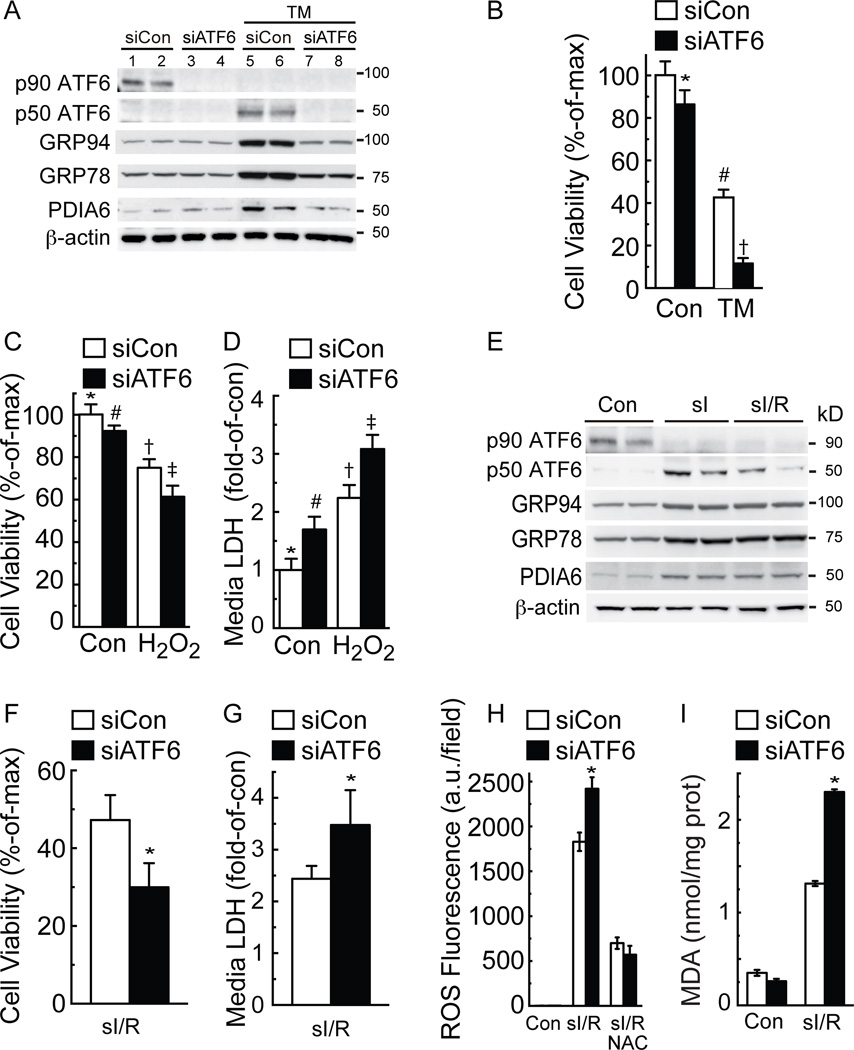
These findings suggested that there might be a previously unappreciated link between ER stress signaling by ATF6 and ROS reduction; moreover, since the damaging ROS produced during reperfusion are mostly generated in mitochondria29, the link could involve ATF6-mediated decreases in total cellular ROS. To examine whether this link also existed between endogenous ATF6 and ROS levels, NRVM were treated with siRNA to knockdown endogenous ATF6. Immunoblots showed that two different ATF6 siRNAs decreased the levels of full-length, p90 ATF6 and resulted in the absence of cleaved, p50 ATF6 upon TM treatment (Fig. 3A, p90 and p50 ATF6 lanes 5–8; Online Fig. IIA). Importantly, siRNA-mediated ATF6 knockdown also blunted the induction of ATF6-inducible target genes, Grp94, Grp78 and PDIA6 (Fig. 3A GRP94, GRP78, PDIA6, lanes 5–8; Online Fig. IIA), but had no effect on activation of the other two branches of the ER stress response that are mediated by PERK and IRE-1 (Online Fig. IIB). Endogenous ATF6 is difficult to detect by immunoblotting. Accordingly, to ensure that the bands observed in Figure 3A; Online Figure IIA were bona fide endogenous p90 and p50 ATF6, we validated a variety of commercially available antibodies using known controls in which we knew p90 and p50 ATF6 were expressed (Online Figure III).
Figure 3. Effects of ATF6 knockdown on ER stress and oxidative stress in cultured cardiac myocytes.
A, NRVM were transfected with a non-targeted siRNA (siCon), or an siRNA targeted to rat ATF6 (siATF6), and then treated without or with TM (10 µg/ml) for 24h, then immunoblotted for endogenous ATF6 (p90 and p50 ATF6), GRP94, GRP78, PDIA6 and β-actin. Note that this figure is replicated in Online Figure IIA with the addition of a second siRNA to ATF6. NRVM were treated similarly with siCon or siATF6 for all subsequent experiments in this figure, except (E). B, NRVM were treated for 48h without or with TM (40 µg/ml) followed by MTT for cell viability. * # † p<0.05 different from other values by ANOVA. C and D, NRVM were treated for 8h with H2O2, then examined by MTT for cell viability (C), or media assayed for LDH activity (D). *#†‡ p<0.05 different than other values by ANOVA. E, NRVM were subjected to Con, sI or sI/R, then extracts were immunoblotted for the proteins shown. F–I, NRVM were treated with sI/R then examined by calcein blue AM for cell viability, media LDH activity, ROS using CellRox, and malondialdehyde (MDA). * p<0.05 different from siCon by t-test.
ATF6 knockdown decreased cell viability in NRVM treated with either TM (Fig. 3B) or H2O2 (Fig. 3C). Moreover, ATF6 knockdown increased necrotic cell death in response to H2O2 treatment, as determined by increased media levels of LDH (Fig. 3D) and HMGB1 (Online Fig. IIC). Simulated ischemia was shown to activate ATF6 and downstream genes in NRVM, as evidenced by the conversion of p90 ATF6 to p50 ATF6 and the increased levels of canonical ATF6 target proteins, GRP94, GRP78 and PDIA6 (Fig. 3E sI). ATF6 activation appeared to persist during sI/R (Fig. 3E sI/R). Moreover, immunocytoflourescence (ICF) of NRVM showed that under control conditions, ATF6 was found in a diffuse staining pattern, consistent with an SR/ER, non-nuclear localization, while after sI (not shown) or sI/R (Online Figure IV), ATF6 was found almost exclusively in nuclei. ATF6 knockdown decreased viability in NRVM subjected to I/R, increased media levels of LDH and HMGB1, increased ROS levels and increased malondialdehyde (MDA), the latter of which is a measure of ROS-associated lipid peroxidation30 (Fig. 3F–I; Online Fig. IID-G). Treatment with NAC verified that ROS were generated upon sI/R (Fig. 3H; Online Fig. IIF). Thus, endogenous ATF6 protected NRVM from the maladaptive effects of prolonged ER protein misfolding and ER stress by TM, as well as from the damaging effects of oxidative stress induced by H2O2 and sI/R.
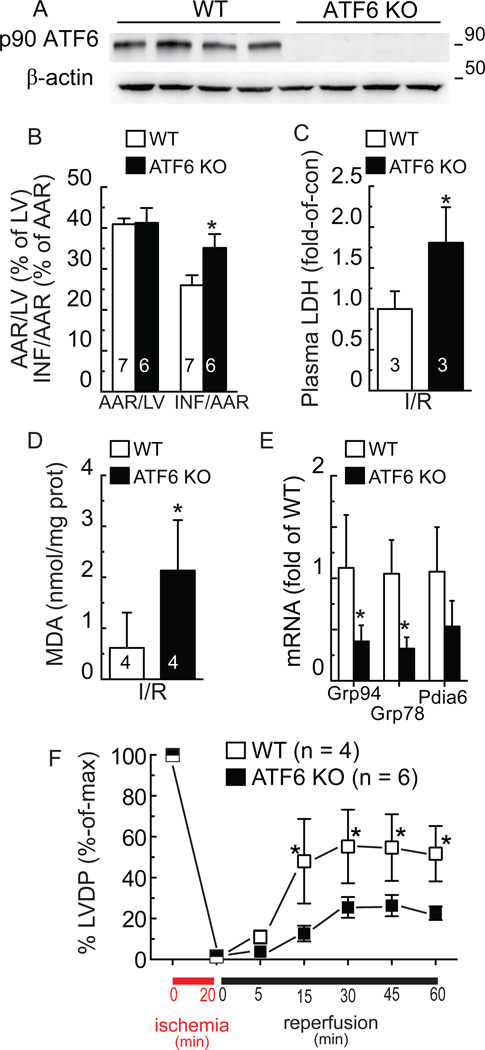
The effects of ATF6 deletion in the mouse heart have not been previously examined; therefore, to assess the effects of deleting ATF6, in vivo, ATF6 knockout mice (KO)21 were used;. Immunoblots confirmed the absence of p90 ATF6 in ATF6 KO mouse hearts compared to wild type (WT) mouse hearts (Fig. 4A). The ATF6 KO mice do not exhibit any overt phenotype under non-stressed conditions, developing normally through adulthood21. Here, echocardiography confirmed that this was the case in the heart, demonstrating that all cardiac dimensions and contractile properties of the ATF6 KO mice were the same as WT mice (Online Table I). When mice were subjected to a model of surgical transient coronary artery occlusion, for 30 min followed by reperfusion for 24h, the area-at-risk was the same for both lines, however, infarct sizes were significantly larger in ATF6 KO mice following I/R (Fig. 4B). Moreover, after I/R, plasma LDH levels and tissue MDA levels were greater in ATF6 KO mice than in WT mice (Fig. 4C and 4D). Immunocytofluorescence and immunoblots of mouse heart tissue sections showed that I/R increased ATF6 and GRP78 in WT mice, but not in ATF6KO mice (Online Figs. V-VII). Moreover, I/R did not activate PERK or IRE-1, as shown by the absence of phosphorylated PERK and spliced XBP1 (Online Fig. VII). Additionally, examination of heart extracts for transcript levels of canonical ATF6 regulated genes showed that, compared to WT mouse hearts, there was 50% or less Grp94, Grp78 and PDIA6 mRNA in ATF6 KO mouse hearts (Fig. 4E).
Figure 4. Effect of ATF6 gene deletion in hearts subjected to ischemia/reperfusion.
A, WT (n = 4) or ATF6 KO (n = 4) mouse heart extracts were examined for ATF6 and β-actin by immunoblotting. To detect p90 ATF6 in mouse heart extracts, the antibody raised against the C-terminal of ATF6 was used. B, WT (n = 7) or ATF6 KO (n = 6) mice were subjected to in vivo I/R, then hearts were assessed for damage; AAR = area at risk; LV = left ventricle; INF = infarcted area, * p<0.05 different than WT INF/AAR by t-test, C, plasma from WT (n = 3) and ATF6 KO (n = 3) mice assessed for LDH, * p<0.05 different than WT by t-test, or D, heart extracts from WT (n = 4) and ATF6 KO (n = 4) mice assessed for MDA ,* p<0.05 different than WT by t-test. E, WT (n = 3) and ATF6 KO (n = 4) mouse hearts were subjected to in vivo I/R after which heart extracts were analyzed for Grp94, Grp78, and Pdia6 mRNA levels, normalized to β-actin mRNA by qRT-PCR, * p<0.05 different than WT for each gene target by t-test. F, WT (n = 4) and ATF6 KO (n = 6) mouse hearts were subjected to ex vivo global ischemia/reperfusion and left ventricular developed pressure (LVDP) was determined before and after ischemia and presented as % of maximal function observed during equilibration for each mouse line, * p<0.05 different than ATF6 KO by t-test.
We also examined whether the effects of ATF6 deletion seen in vivo could be recapitulated in isolated hearts subjected to ex vivo I/R. Compared to WT mouse hearts, ATF6 KO mouse hearts were more susceptible to a loss of cardiac function following global ischemia, exhibiting a significantly reduced recovery of left ventricular developed pressure (LVDP) upon reperfusion (Fig. 4F). Thus, the effects of global ATF6 deletion on I/R damage were autonomous to the heart.
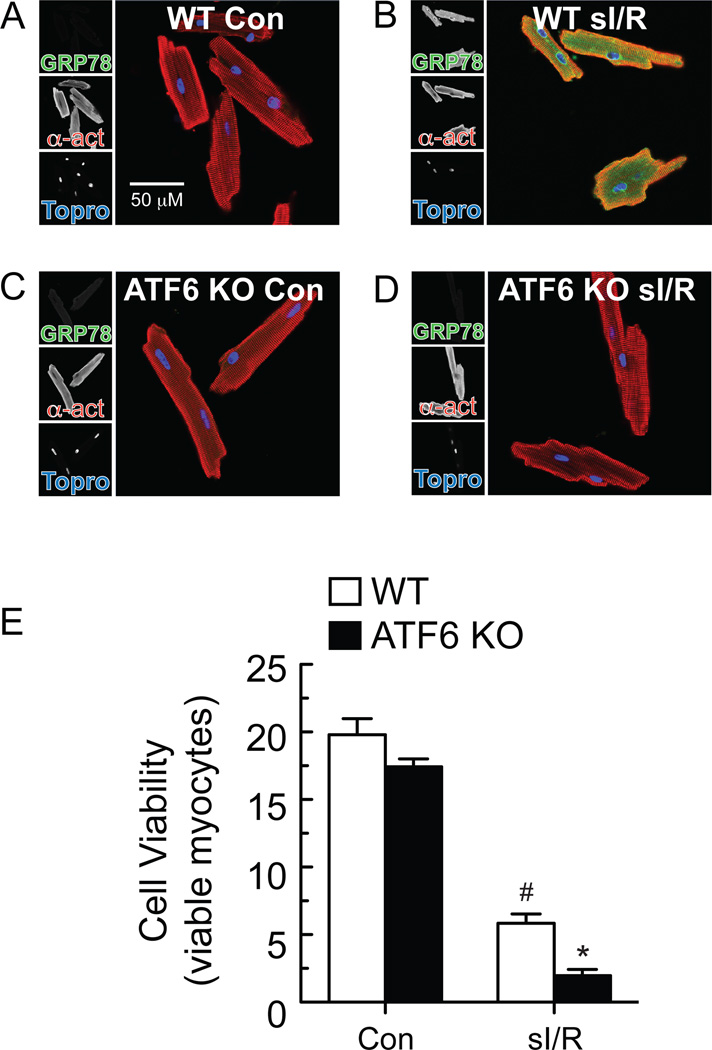
Next we examined the effects of ATF6 deletion in cardiac myocytes isolated from ATF6 KO mouse hearts. ICF showed that myocytes from WT mouse hearts exhibited low levels of ATF6 under basal conditions; however, upon TM treatment, ATF6 increased in WT mouse myocytes and localized throughout the cell as well as in the nucleus (Online Figure VIII, A and B). In contrast, ATF6 KO mouse myocytes did not exhibit any detectible ATF6 under basal conditions, or when treated with TM (Online Figure VIII, C and D). Examination of the ATF6 target gene product, GRP78, was consistent with these results; in WT mouse myocytes, GRP78 levels increased upon treatment with TM (Online Figure IX, A and B), or sI/R (Fig. 5A and 5B). In contrast, myocytes isolated from ATF6 KO mouse hearts showed no induction of GRP78 in response to TM (Online Figure IX, C and D), or sI/R (Fig. 5C and D). Moreover, compared to myocytes from WT mouse hearts, myocytes from ATF6 KO mouse hearts exhibited greater death in response to sI/R (Fig. 5E). Thus, the deleterious effects of ATF6 deletion on ATF6 target gene induction by TM or sI/R, as well as sI/R-mediated cell death, was observed in ATF6 KO mouse hearts in vivo and ex vivo, as well as in myocytes isolated from ATF6 KO mouse hearts.
Figure 5. Effect of ATF6 deletion on GRP78 expression and cell viability of isolated adult mouse ventricular myocytes.
A-D, Myocytes were isolated from adult WT or ATF6 KO mice, subjected to sI/R, then fixed and examined by ICF for GRP78 (green), α-actinin (red) or TOPRO (blue). E, Myocytes were isolated from adult WT or ATF6 KO mice, then subjected to sI/R followed by determination of cell viability using calcein blue AM staining. n = 3 cultures for each treatment; shown is a representative experiment of three independent experiments, *# p<0.05 different than all other groups by ANOVA.
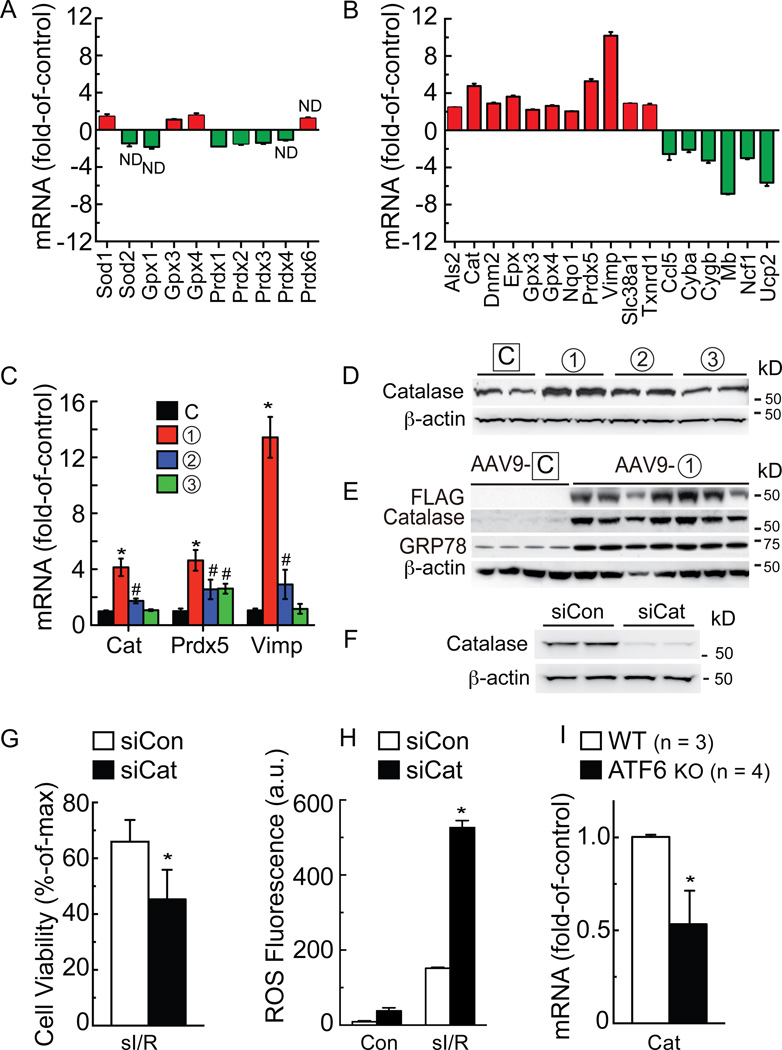
Since ATF6 reduced ROS levels and protected cardiac myocytes and hearts against oxidative stress, we examined whether ATF6 overexpression in NRVM affected the levels of typical oxidative stress response genes. An initial survey of expression levels of several typical antioxidant genes, i.e. superoxide dismutase 1 and 2 (SOD1/2), glutathione peroxidase 1, 3 and 4 (Gpx1/3/4), and peroxiredoxin 1–4 and 6 (Prdx1-4/6) showed that compared to control, ATF6 had a very small effect on the expression of any of these genes (Fig. 6A). Accordingly, a wider range of oxidative stress response genes was assessed using a PCR gene array. Amongst the 84 genes represented in the array, ATF6 significantly changed the levels of 17 genes; eleven genes were increased by ATF6, including catalase (Cat), peroxiredoxin 5 (Prdx5) and Vimp (Fig. 6B red), which encode antioxidant proteins, while 6 genes were decreased by ATF6 (Fig. 6B green), most of which respond to oxidative stress, but do not encode antioxidants. Details of the proteins encoded by the genes shown in Figure 6B can be found in Online Table II. PCR was used to examine the expression of several key genes in the array upon which ATF6 had the most robust induction effects, Cat, Prdx5 and Vimp. Form 1 of ATF6 was the strongest inducer of all three genes, with much less induction being observed for forms 2 and 3 (Fig. 6C). These results suggested that ATF6 might directly transcriptionally induce these genes.
Figure 6. Analysis of oxidative stress gene expression.
A and B, NRVM were infected with AdV-Con or AdV-ATF6 (form 1), then RNA was subjected to qRT-PCR for the genes shown (A), or analyzed with an oxidative stress gene array (B). Shown are only genes that were increased (red) or decreased (green) by ATF6. In A and B all values were p<0.05 different than control by t-test, unless marked ND (no difference). C and D, NRVM were infected with the AdV-ATF6 shown, then RNA was isolated and analyzed for catalase (Cat), peroxiredoxin 5 (Prdx5), and Vimp mRNA by qRT-PCR (C), * # p<0.05 different than all other values for each target gene, and protein was analyzed for catalase levels by immunoblotting (D). E, WT mice were injected with AAV9-control or AAV9-CMV-FLAG-ATF6 form 1. Two weeks later, mice were sacrificed and hearts were analyzed for FLAG-ATF6, catalase, GRP78 and β-actin by immunoblotting. F, NRVM were transfected with siCon or siCat RNAs; 48h later, culture extracts were examined for catalase and β-actin. G, H, NRVM were transfected with siCon or siCat RNA, subjected to sI/R, then analyzed for cell viability by calcein blue AM staining (G), or for ROS by Amplex Red (H). I, following I/R, WT (n = 3) and ATF6 KO (n = 4) mouse heart extracts were analyzed for Cat mRNA levels, normalized to β-actin mRNA by qRT-PCR. * p<0.05 different from siCon (G, H) or WT (I) by t-test.
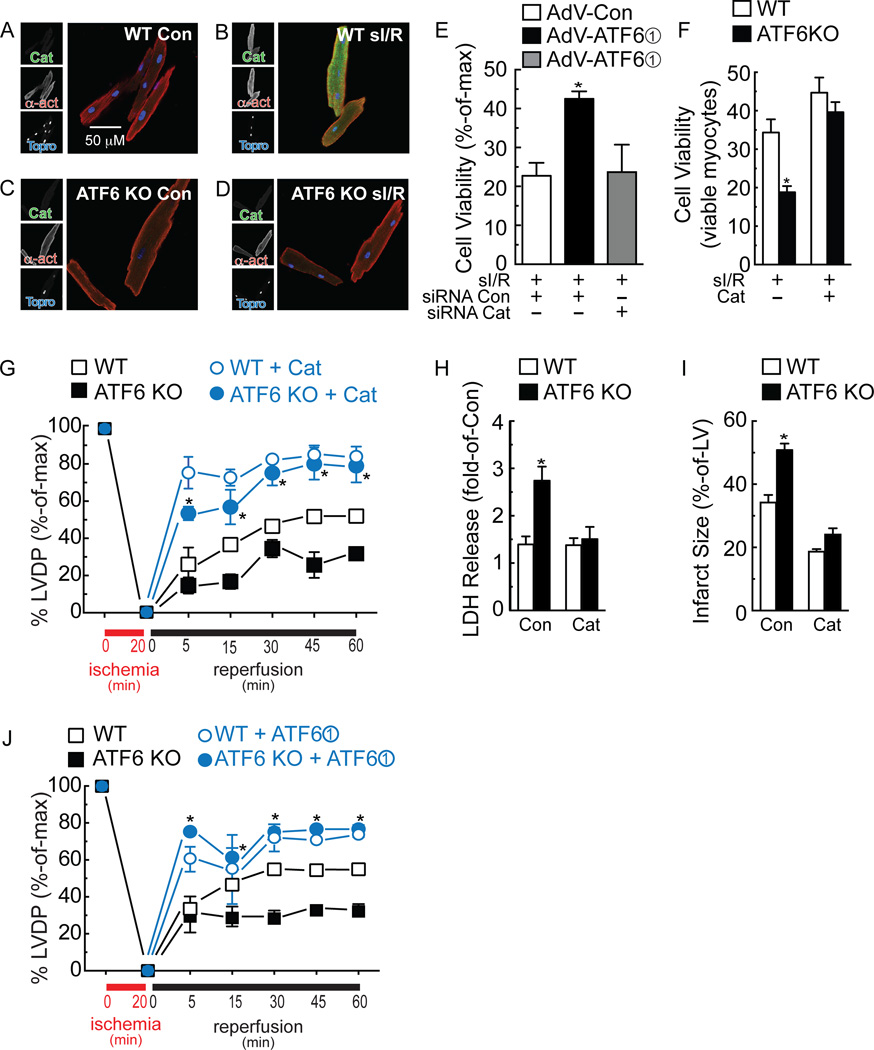
Since catalase is known to decrease I/R damage in the heart31, we probed deeper into the mechanism by which ATF6 mediated the induction of catalase, which has not been studied before. Immunoblots of NRVM treated with the three forms of ATF6 showed that catalase protein levels were increased most by form 1, followed by form 2 and then not at all by form 3 (Fig. 6D), consistent with the effects of these forms of ATF6 on catalase mRNA (Fig. 6C). Moreover, when mice were injected with a recombinant AAV9 that expresses form 1 of ATF6 in cardiac myocytes (Fig. 6E FLAG), immunoblots of heart extracts showed that catalase was robustly induced by ATF6 overexpression in the mouse heart, in vivo (Fig. 6E Catalase). As a control, we showed that AAV9-ATF6 also induced the known ATF6 target gene, GRP78 (Fig. 6E GRP78). Knocking down catalase in NRVM with siRNA (Fig. 6F) decreased cell viability and increased ROS generation in response to sI/R (Fig. 6G and 6H). Quantitative RT-PCR of mouse hearts subjected to I/R showed that, compared to WT mouse hearts, catalase was about half in ATF6 KO mouse hearts (Fig. 6I). While myocytes isolated from WT mouse hearts exhibited a strong induction of catalase by TM (Online Fig. X, A and B) or sI/R (Fig. 7A and 7B), catalase did not increase in myocytes isolated from ATF6 KO mouse hearts treated with either TM (Online Fig. X, C and D) or sI/R (Fig. 7C and 7D). ICF of mouse heart tissue sections showed that catalase increased in the hearts of WT mouse hearts subjected to I/R, in vivo, but not in ATF6 KO mouse hearts (Online Fig. XI A-D). Immunoblots verified the impaired induction of catalase in the hearts of ATF6 KO mice subjected to I/R, in vivo (Online Fig. XI, E and F). In NRVM, catalase siRNA decreased the beneficial effects of ATF6 form 1, indicating that catalase is a major contributor of ATF6-mediated protection from I/R-induced cell death in cardiac myocytes (Fig. 7E). Moreover, the increased susceptibility of ATF6 KO myocytes to death upon I/R was rescued by treating them with a cell-permeable form of catalase, PEG-catalase, as previously described32, after which there was no difference in viability between myocytes from WT and ATF6 KO mouse hearts (Fig. 7F). Treatment of mice with PEG-catalase in vivo, as previously described33, resulted in a similar rescue of LVDP in ATF6 KO mouse hearts subjected to ex vivo I/R, such that the performance of WT and ATF6 KO mouse hearts was indistinguishable (Fig. 7G). This catalase-mediated rescue was also seen when LDH release and infarct size were measured in WT vs ATF6 KO mouse hearts subjected to ex vivo I/R (Fig. 7H and 7I). Moreover, the PEG-catalase rescue effect was recapitulated by AAV9-mediated ATF6 overexpression in ATF6 KO mouse hearts (Fig. 7J).
Figure 7. Effects of ATF6 deletion on catalase.
A-D, Myocytes were isolated from adult WT (A, B) or ATF6 KO (C, D) mice, subjected to sI/R, then fixed and examined by ICF for catalase (green), α-actinin (red) or TOPRO (blue). E, NRVM were treated with siCon or siCat RNAs, as well as infection with the AdVs shown, then subjected to sI/R followed by determination of cell viability by calein blue AM staining. * p<0.05 different from all other values by ANOVA. F, Myocytes isolated from WT or ATF6KO mouse hearts were treated with vehicle or PEG-catalase, followed by sI/R, the viability was determined by calein blue AM staining. * p<0.05 different from WT by t-test. G-I, Mice were injected with vehicle or PEG-Cat for 16h, then hearts were subjected to ex vivo I/R and LVDP, LDH release, and infarct size were measured. In G, * p<0.001 different from WT vehicle or ATF6 KO vehicle at a given reperfusion time by two-way ANOVA. In H and I, * p<0.001 different from WT Con, by t-test. J, Mice were injected with AAV9-Con or AAV9-CMV-FLAG-ATF6 form 1 for 2d, then hearts were subjected to ex vivo I/R and LVDP was measured. * p<0.001 different from WT AAV9-Con or ATF6 KO AAV9-Con at a given reperfusion time by two-way ANOVA.
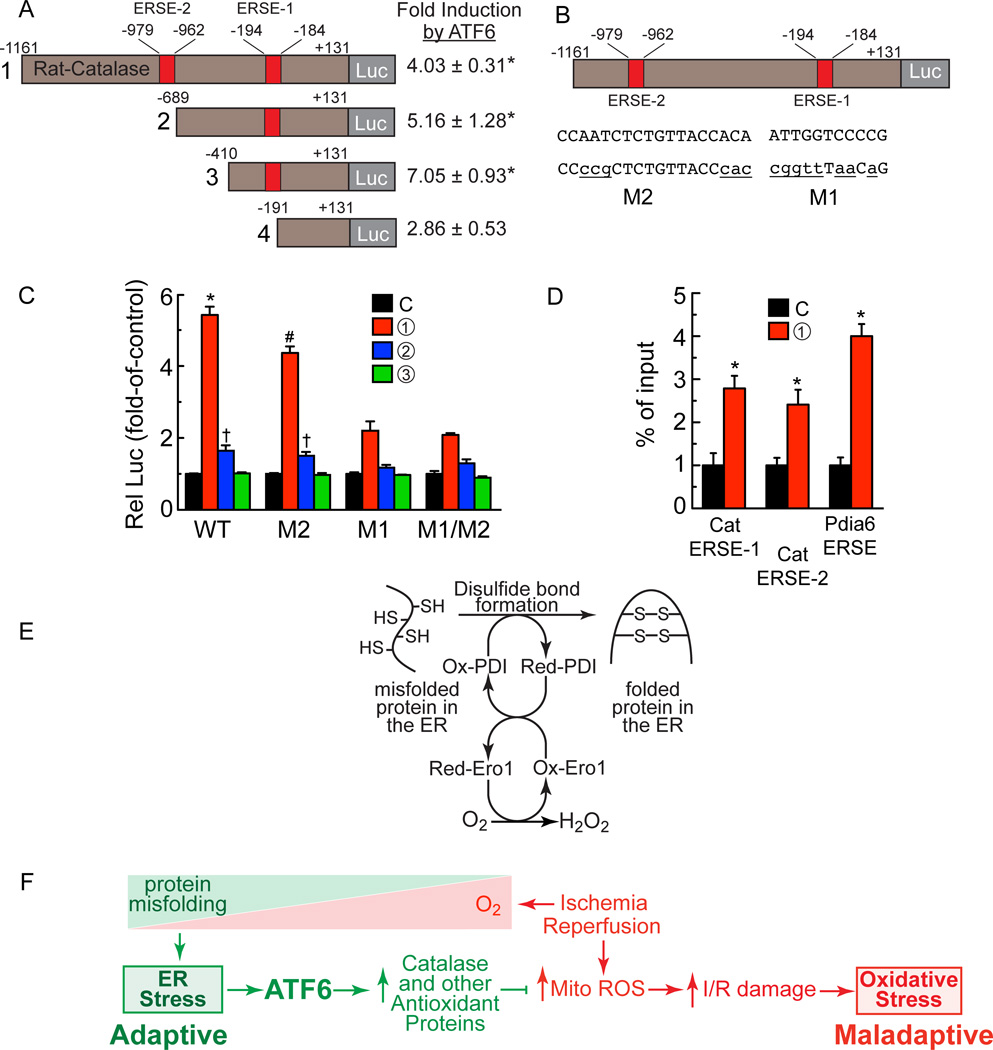
To further examine the mechanism by which ATF6 induces catalase, the 5’-flanking sequence (5’-FS) of the rat Cat gene was scanned for a DNA sequence that might bind ATF6, so called ER stress response element (ERSE). Two such elements were found within 1,000 nucleotides 5’ of the Cat mRNA start site; we named these ERSE-1 (-194 to -184) and ERSE-2 (-979 to -962). To examine whether the rat Cat 5’-flanking sequences conferred transcriptional induction in response to ATF6, truncated versions of the rat Cat 5’-FS were cloned in front of firefly luciferase and the abilities of co-transfected ATF6 form 1 to induce luciferase were examined in NRVM. Truncating from -1161 to -689 and to -410, which removed ERSE-2, had little effect on ATF6 induction (Fig. 8A, constructs 1 – 3). However, a truncated form of rat Cat that removed ERSE-1 resulted in a significant decrease in ATF6-mediated luciferase induction (Fig. 8A, construct 4). In a second series of experiments, ERSE-1 and ERSE-2 were mutated in ways predicted to inhibit ATF6 binding (Fig. 8B, M1 and M2). When these reporters were co-transfected with the various forms of ATF6 into NRVM, the mutation in ERSE-2, i.e. M2, moderately decreased Cat promoter activity, while the mutation in ERSE-1, i.e. M1, or in both ERSE-1 and ERSE-2, i.e. M1/M2, showed greater reductions of ATF6-meidated transcriptional induction (Fig. 8C). Finally, a chromatin immunoprecipitation experiment showed that form 1 of ATF6 was able to bind to either ERSE-1 or ERSE-2 in myocytes (Fig. 8D). Thus, activated ATF6 can bind to putative ERSEs in the rat Cat gene regulatory region and confer transcriptional induction, demonstrating one mechanism by which ATF6 could protect cardiac myocytes from oxidative stress during I/R.
Figure 8. Effects of ATF6 on the catalase promoter.
A, Truncations of the catalase 5’-flanking sequence driving luciferase, as shown (left), i.e. rat-cat(-1161/+131)-Luc, rat-cat(-689/+131)-Luc, rat-cat(-410/+131)-Luc and rat-cat(-191/+131)-Luc were tramsfected into NRVM which were then infected with AdV encoding form 1 of ATF6 or with a control AdV. Luciferase enzyme activity in AdV-ATF6-infected cells was normalized to luciferase enzyme activity in AdV-Con-infected cells to determine the fold-induction by ATF6 (right), * p<0.05 different from control by t-test. B, Diagram of the locations of ERSE-2 and 1 in the catalase 5’-flanking region, their sequences (upper case), and the mutations to those sequences (lower case). C, NRVM were transfected with plasmids encoding rat-cat(-1161/+131)-Luc WT, M2, M1 or M1/M2 and then infected with the ATF6-expressing AdV, as shown; then, 48h later, luciferase levels were measured in extracts, *#† p<0.05 different than other values in WT or M2 by ANOVA. D, NRVM were infected with AdV encoding control or FLAG-ATF6 (form 1), and then ATF6 binding to endogenous ERSE-1 1 ERSE-2, as well as to the endogenous Pdia6 ERSE, previously studied and used here as a control, was examined by ChIP, * p<0.05 different than control by t-test. E, O2 is required for protein disulfide bond formation and protein folding in the ER. F, ATF6 links the ER stress response with the oxidative stress response. As O2 decreases during ischemia (red triangle), protein misfolding increases (green triangle), leading to adaptive ER stress, ATF6 activation and induction of catalase and other antioxidant proteins that decrease ROS generated in the mitochondria during reperfusion.
DISCUSSION
ATF6 links the ER stress response and the oxidative stress response
This study provides evidence supporting a newly described role for ATF6 as a molecular link between the ER stress and oxidative stress gene programs. Initially, we thought this role for ATF6 fit well with a potential connection between ER stress and oxidative stress involving protein disulfide bond formation. Protein disulfide bond formation, which takes place only in the ER, requires oxygen to fuel the redox couple between ER oxidoreductase 1 (Ero1) and the final enzyme involved in protein disulfide bond formation, protein disulfide isomerase (PDI)19, 34, 35 (Fig. 8E). This ER redox system results in the conversion of molecular oxygen to H2O2, which contributes to total cellular ROS, although a very small amount compared to the ROS generated by mitochondria36. There are three ER proteins thought to be responsible for neutralizing the H2O2 generated during protein disulfide bond formation in the ER; peroxiredoxin 4 (Prx4), and glutathione perioxidases 7 and 8 (GPx7/8)34. Accordingly, when we first discovered that ATF6 had antioxidant activity, since ATF6 induces mostly ER proteins, we thought that ATF6 most likely exerted its antioxidant effects by inducing antioxidant proteins that reside in the ER, such as Prx4 and/or GPx7/8. However, our results showed that these genes were minimally affected by ATF6, and that ATF6 had more robust effects on expression of 11 other genes, all but one of which encode antioxidant proteins that reside outside of the ER (Fig. 6 and Online Table II). Thus, ATF6 can have widespread effects on antioxidant protein expression, even outside the ER, which increases the scope of the functional impact of ATF6 well beyond improving ER protein folding. It might be of clinical relevance to acutely activate ATF6 during reperfusion, in vivo, but this would likely require the development of new methods that do not depend on ATF6 overexpression. Although many of the antioxidant proteins induced by ATF6 may contribute in some way to reducing myocardial damage during I/R, we focused the mechanistic aspects of this study on catalase because of its ability to neutralize large quantities of ROS generated in various cellular locations, and because it has not previously been previously shown to be induced in an ATF6- and ER stress-dependent manner in any cell or tissue.
Catalase is an example of how ATF6 regulates antioxidant protein expression
Catalase is a 527 amino acid protein (rat) that resides mainly in peroxisomes. Peroxisomes generate and utilize H2O2 for oxidative purposes, including peroxidative detoxification and β-oxidation of fatty acids. Catalase is an important component of peroxisomes, because it neutralizes H2O2 that remains after the required peroxidative reactions have taken place. Catalase can also oxidize peroxynitrate, nitric oxide and organic peroxides. In addition to peroxisomes, catalase has also been found in the cytosol and in cardiac mitochondria, however it has not been found in the ER. Moreover, it has been shown that in the heart, catalase overexpression in cardiac myocytes decreases I/R injury by reducing ROS levels31.
Here, we showed that, in cardiac myocytes, ischemia, which impairs ER protein disulfide bond formation and increases misfolded proteins, activates ATF6, which binds to specific elements in the regulatory regions of the catalase gene and increases catalase transcription. We also showed that ATF6 can increase expression of other antioxidant proteins, supporting the hypothesis that, together with catalase these antioxidants decrease ROS and moderate myocardial damage during reperfusion (Figure 8F). In further support of this hypothesis was our finding that, compared to WT mouse hearts, the induction of catalase during I/R was impaired in ATF6KO mouse hearts. To the best of our knowledge, this is the first demonstration that catalase is induced in the heart by I/R in an ATF6-dependent manner. Since we showed that ATF6 also induced two other antioxidant genes encoding proteins residing outside the ER, it appears that, during ER stress induced by ischemia, ATF6 is likely to be a direct transcriptional inducer of numerous antioxidant genes.
Catalase is an ATF6-inducible ER stress response gene
Previous studies, mostly in non-cardiac myocytes, have shown that catalase expression is regulated by the transcription factors Sp1, NF-Y and Foxo3a37. However, there have been no studies examining whether ATF6 can regulate catalase transcription. Here, we showed that there are two sites in the catalase regulatory region to which ATF6 can bind, ERSE-1 and ERSE-2. Mutating ERSE-2 had less of an effect on catalase promoter activity than mutating ERSE-1, suggesting that ERSE-1, which lies proximal to the catalase promoter, is the major site through which ATF6 confers transcriptional induction in cardiac myocytes (Fig. 8A–D). Moreover, since catalase is induced upon treatment with the prototypical ER stressor, TM, and by the pathological ER stress, I/R, and since the ER stress transcription factor, ATF6, binds to and induces catalase transcription, we posit that catalase should be categorized as an ER stress response gene. Consistent with the identification of catalase as an ER stress response gene is the apparent reduction of its expression as a function of development in the heart. We previously showed that the expression of several canonical ER stress response genes was much higher in neonatal rat cardiac myocytes than in adult rat cardiac myocytes38. The same appears to be true for catalase; here, immunoblotting showed relatively high levels of basal catalase in NRVM compared to adult mouse hearts (Figs. 6D and 6E). Therefore, like some other ER stress response genes, catalase expression appears to be relatively low in the adult heart, compared to the neonatal heart. This is underscored further by our finding that the ATF6 dependence of catalase expression was quite evident when adult mouse cardiac myocytes isolated from WT and ATF6 KO mice were treated with TM or sI/R and then immunostained for catalase (Fig. 7 and Online Figure X).
Global effects of ATF6 on antioxidant protein expression
The antioxidant genes that were induced the most by ATF6 in this study were catalase, peroxiredoxin 5 (Prdx5) and Vimp. ATF6-mediated induction of these genes was validated using the three forms of ATF6; each gene was induced with a profile consistent with the direct effects of ATF6 on transcription (Fig. 6C). Like catalase, Prdx5 is not located in the ER lumen; however, it is unusual amongst other peroxiredoxins, in that it has been found in several cellular areas, including mitochondria, peroxisomes, cytosol and nucleus39 (Online Table II). Prdx5 is a 213 amino acid protein (rat) that catalyzes the glutathione-mediated reduction of potentially damaging peroxides, providing a new mechanism by which the ATF6 branch of the ER stress response could contribute to decreasing ROS during myocardial reperfusion.
Vimp is a 189 amino acid ER-transmembrane protein (rat) configured with most of its structure, including its catalytic domain on the cytosolic face of the ER40. Vimp, also known as SelS, is believed to interact with Hrd1 and other ER stress, ATF6-regulated ER-transmembrane proteins that are involved in ER associated degradation, a process by which terminally misfolded proteins in the ER are degraded to mitigate their toxic effects. Vimp is one of only 24 selenoprotein genes in the mouse genome; in general, selenoproteins are known for their antioxidant roles as ROS scavengers41. However, the functions of selenoproteins and, in particular Vimp have not been studied in the heart. It is interesting to note that five of the 11 ATF6-inducible oxidative stress response genes identified in this study are selenoproteins. This is the first demonstration in any cell or tissue type that ATF6 has a global effect on selenoprotein expression, which provides even further linkage between the ATF6 branch of the ER stress response and the oxidative stress response.
In summary, previous studies showed that ectopic expression of activated ATF6 can decrease I/R damage in the heart16. However, neither the mechanism of this effect, nor whether endogenous ATF6 plays a role in myocardial I/R damage had been examined. Here, we determined functions for endogenous ATF6 in the heart, and found evidence of a previously unappreciated role for ATF6 as an inducer of antioxidant genes, which establishes a mechanistic link by which ATF6 can decrease myocardial I/R damage (Fig. 8F).
Supplementary Material
Novelty and Significance.
What Is Known?
Myocardial ischemia causes endoplasmic reticulum (ER) stress and the misfolding of proteins in the ER of cardiac myocytes, which activates the ER stress response transcription factor, ATF6 (activating transcription factor 6 alpha).
ATF6 increases expression of ER proteins, such as chaperones that restore ER protein folding, thus avoiding the toxic effects of misfolded proteins.
Ischemia/reperfusion (I/R) damage in the heart, which is caused mainly by increased reactive oxygen species (ROS), is decreased by ATF6; however, the mechanism is unknown.
What New Information Does This Article Contribute?
ATF6 induces numerous antioxidative stress genes, such as catalase, which is known to decrease ROS and reduce I/R damage in the heart.
ATF6-mediated catalase induction contributes to the ability of ATF6 to protect against I/R damage.
ATF6 links ER stress to oxidative stress in the heart
The ER is the site of secreted and membrane protein synthesis and folding, thus serving a critical role in cardiac myocyte function. ER protein folding is impaired in the ischemic heart, which contributes to pathology. Impaired ER protein folding activates the ER stress transcription factor, ATF6, which decreases I/R damage in the heart. However, since most known ATF6-induced proteins function to enhance protein-folding in the ER, it is unknown how ATF6 can decrease myocardial I/R damage, most of which is caused by ROS. Here we used ATF6 gain- and loss-of-function approaches to show that during I/R, ATF6 is not only required for induction of ER-resident proteins that enhance protein folding in the ER, but surprisingly is also responsible for inducing numerous oxidative stress response genes that encode antioxidant proteins, such as catalase, which do not reside the ER. ATF6-mediated catalase induction was found to contribute to the protective effects of ATF6. These results show that antioxidant proteins are induced by ATF6 during ER stress, and reveal a new function for ATF6 as a link between the ER stress and oxidative stress in the heart.
Acknowledgments
SOURCES OF FUNDING
JKJ was supported by an American Heart Association, Post-doctoral Fellowship 16POST27510010.
SD was supported an Excellence Grant from the German Centre for Cardiovascular Research (DZHK).
CCG was supported by National Institutes of Health (NIH) grants R01 HL75573, R01 HL104535, and P01 HL085577.
RJK was supported by NIH/NCI grants R37DK042394, R01DK103183, R24DK110973, R01CA128814, and the Sanford Burnham Prebys NCI Cancer Center Grant P30 CA030199.
Nonstandard Abbreviations and Acronyms
- AAV
adeno-associated virus
- AdV
adenovirus
- ANOVA
analysis of variance
- ATF6
activating transcription factor 6 alpha
- ATF6 KO
ATF6 alpha knockout
- ER
endoplasmic reticulum
- ERAD
endoplasmic reticulum associated degradation
- Grp94
94 kilodalton glucose-regulated protein
- Grp78
78 kilodalton glucose-regulated protein
- HR
heart rate
- HW
heart weight
- ICF
immunocytofluorescence
- I/R
ischemia/reperfusion
- LV
left ventricle
- LVEDV
left ventricular end diastolic volume
- LVESV
left ventricular end systolic volume
- LVIDD
left ventricular inner diameter in diastole
- LVIDS
left ventricular inner diameter in systole
- Ox-Ero1
oxidized ER oxidoreductin 1
- Ox-PDI
oxidized protein disulfide isomerase
- PARP
NAD(+) ADP-ribosyltransferase
- Pdia6
protein disulfide isomerase family A, member 6
- PWTD
left ventricular posterior wall thickness in diastole
- PWTS
left ventricular posterior wall thickness in systole
- Red-Ero1
reduced ER oxidoreductin 1
- Red-PDI
reduced protein disulfide isomerase
- ROS
reactive oxygen species
- sI/R
simulated ischemia/reperfusion
- SR
sarcoplasmic reticulum
- TL
tibia length
- TM
tunicamycin
Footnotes
DISCLOSURES
None.
REFERENCES
- 1.Balch WE, Morimoto RI, Dillin A, Kelly JW. Adapting proteostasis for disease intervention. Science. 2008;319(5865):916–919. doi: 10.1126/science.1141448. [DOI] [PubMed] [Google Scholar]
- 2.Glembotski CC. Roles for the sarco-/endoplasmic reticulum in cardiac myocyte contraction, protein synthesis, and protein quality control. Physiology (Bethesda) 2012;27(6):343–350. doi: 10.1152/physiol.00034.2012. [DOI] [PubMed] [Google Scholar]
- 3.Gidalevitz T, Stevens F, Argon Y. Orchestration of secretory protein folding by ER chaperones. Biochim Biophys Acta. 2013;1833(11):2410–2424. doi: 10.1016/j.bbamcr.2013.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Amm I, Sommer T, Wolf DH. Protein quality control and elimination of protein waste: the role of the ubiquitin-proteasome system. Biochim Biophys Acta. 2014;1843(1):182–196. doi: 10.1016/j.bbamcr.2013.06.031. [DOI] [PubMed] [Google Scholar]
- 5.Guerriero CJ, Brodsky JL. The delicate balance between secreted protein folding and endoplasmic reticulum-associated degradation in human physiology. Physiol Rev. 2012;92(2):537–576. doi: 10.1152/physrev.00027.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dickhout JG, Carlisle RE, Austin RC. Interrelationship between cardiac hypertrophy, heart failure, and chronic kidney disease: endoplasmic reticulum stress as a mediator of pathogenesis. Circ Res. 2011;108(5):629–642. doi: 10.1161/CIRCRESAHA.110.226803. [DOI] [PubMed] [Google Scholar]
- 7.Minamino T, Komuro I, Kitakaze M. Endoplasmic reticulum stress as a therapeutic target in cardiovascular disease. Circ Res. 2010;107(9):1071–1082. doi: 10.1161/CIRCRESAHA.110.227819. [DOI] [PubMed] [Google Scholar]
- 8.Doroudgar S, Glembotski CC. New concepts of endoplasmic reticulum function in the heart: Programmed to conserve. J Mol Cell Cardiol. 2013 doi: 10.1016/j.yjmcc.2012.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Millott R, Dudek E, Michalak M. The endoplasmic reticulum in cardiovascular health and disease. Can J Physiol Pharmacol. 2012;90(9):1209–1217. doi: 10.1139/y2012-058. [DOI] [PubMed] [Google Scholar]
- 10.Olzmann JA, Kopito RR, Christianson JC. The Mammalian Endoplasmic Reticulum-Associated Degradation System. Cold Spring Harb Perspect Biol. 2012 doi: 10.1101/cshperspect.a013185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Malhotra JD, Kaufman RJ. The endoplasmic reticulum and the unfolded protein response. Semin Cell Dev Biol. 2007;18(6):716–731. doi: 10.1016/j.semcdb.2007.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Glembotski CC. Endoplasmic reticulum stress in the heart. Circ Res. 2007;101(10):975–984. doi: 10.1161/CIRCRESAHA.107.161273. [DOI] [PubMed] [Google Scholar]
- 13.Walter P, Ron D. The unfolded protein response: from stress pathway to homeostatic regulation. Science. 2011;334(6059):1081–1086. doi: 10.1126/science.1209038. [DOI] [PubMed] [Google Scholar]
- 14.Zhu C, Johansen FE, Prywes R. Interaction of ATF6 and serum response factor. Mol Cell Biol. 1997;17(9):4957–4966. doi: 10.1128/mcb.17.9.4957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Haze K, Yoshida H, Yanagi H, Yura T, Mori K. Mammalian transcription factor ATF6 is synthesized as a transmembrane protein and activated by proteolysis in response to endoplasmic reticulum stress. Mol Biol Cell. 1999;10(11):3787–3799. doi: 10.1091/mbc.10.11.3787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Martindale JJ, Fernandez R, Thuerauf D, Whittaker R, Gude N, Sussman MA, Glembotski CC. Endoplasmic reticulum stress gene induction and protection from ischemia/reperfusion injury in the hearts of transgenic mice with a tamoxifen-regulated form of ATF6. Circ Res. 2006;98(9):1186–1193. doi: 10.1161/01.RES.0000220643.65941.8d. [DOI] [PubMed] [Google Scholar]
- 17.Belmont PJ, Tadimalla A, Chen WJ, Martindale JJ, Thuerauf DJ, Marcinko M, Gude N, Sussman MA, Glembotski CC. Coordination of growth and endoplasmic reticulum stress signaling by regulator of calcineurin 1 (RCAN1), a novel ATF6-inducible gene. J Biol Chem. 2008;283(20):14012–14021. doi: 10.1074/jbc.M709776200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yamamoto K, Sato T, Matsui T, Sato M, Okada T, Yoshida H, Harada A, Mori K. Transcriptional induction of mammalian ER quality control proteins is mediated by single or combined action of ATF6alpha and XBP1. Dev Cell. 2007;13(3):365–376. doi: 10.1016/j.devcel.2007.07.018. [DOI] [PubMed] [Google Scholar]
- 19.Cao SS, Kaufman RJ. Endoplasmic reticulum stress and oxidative stress in cell fate decision and human disease. Antioxid Redox Signal. 2014;21(3):396–413. doi: 10.1089/ars.2014.5851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Granger DN, Kvietys PR. Reperfusion injury and reactive oxygen species: The evolution of a concept. Redox Biol. 2015;6:524–551. doi: 10.1016/j.redox.2015.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wu J, Rutkowski DT, Dubois M, Swathirajan J, Saunders T, Wang J, Song B, Yau GD, Kaufman RJ. ATF6alpha optimizes long-term endoplasmic reticulum function to protect cells from chronic stress. Dev Cell. 2007;13(3):351–364. doi: 10.1016/j.devcel.2007.07.005. [DOI] [PubMed] [Google Scholar]
- 22.Yoshida H, Okada T, Haze K, Yanagi H, Yura T, Negishi M, Mori K. ATF6 activated by proteolysis binds in the presence of NF-Y (CBF) directly to the cis-acting element responsible for the mammalian unfolded protein response. Mol Cell Biol. 2000;20(18):6755–6767. doi: 10.1128/mcb.20.18.6755-6767.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Thuerauf DJ, Morrison LE, Hoover H, Glembotski CC. Coordination of ATF6-mediated transcription and ATF6 degradation by a domain that is shared with the viral transcription factor, VP16. J Biol Chem. 2002;277(23):20734–20739. doi: 10.1074/jbc.M201749200. [DOI] [PubMed] [Google Scholar]
- 24.Thuerauf DJ, Morrison L, Glembotski CC. Opposing roles for ATF6alpha and ATF6beta in endoplasmic reticulum stress response gene induction. J Biol Chem. 2004;279(20):21078–21084. doi: 10.1074/jbc.M400713200. [DOI] [PubMed] [Google Scholar]
- 25.Marshall KD, Edwards MA, Krenz M, Davis JW, Baines CP. Proteomic mapping of proteins released during necrosis and apoptosis from cultured neonatal cardiac myocytes. Am J Physiol Cell Physiol. 2014;306(7):C639–C647. doi: 10.1152/ajpcell.00167.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Siman R, McIntosh TK, Soltesz KM, Chen Z, Neumar RW, Roberts VL. Proteins released from degenerating neurons are surrogate markers for acute brain damage. Neurobiol Dis. 2004;16(2):311–320. doi: 10.1016/j.nbd.2004.03.016. [DOI] [PubMed] [Google Scholar]
- 27.van Wijk SJ, Hageman GJ. Poly(ADP-ribose) polymerase-1 mediated caspase-independent cell death after ischemia/reperfusion. Free Radic Biol Med. 2005;39(1):81–90. doi: 10.1016/j.freeradbiomed.2005.03.021. [DOI] [PubMed] [Google Scholar]
- 28.Griendling KK, Touyz RM, Zweier JL, Dikalov S, Chilian W, Chen YR, Harrison DG, Bhatnagar A. Measurement of Reactive Oxygen Species, Reactive Nitrogen Species, and Redox-Dependent Signaling in the Cardiovascular System: A Scientific Statement From the American Heart Association. Circ Res. 2016;119(5):e39–e75. doi: 10.1161/RES.0000000000000110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Santos CX, Raza S, Shah AM. Redox signaling in the cardiomyocyte: From physiology to failure. Int J Biochem Cell Biol. 2016;74:145–151. doi: 10.1016/j.biocel.2016.03.002. [DOI] [PubMed] [Google Scholar]
- 30.Pompella A, Maellaro E, Casini AF, Ferrali M, Ciccoli L, Comporti M. Measurement of lipid peroxidation in vivo: a comparison of different procedures. Lipids. 1987;22(3):206–211. doi: 10.1007/BF02537304. [DOI] [PubMed] [Google Scholar]
- 31.Li G, Chen Y, Saari JT, Kang YJ. Catalase-overexpressing transgenic mouse heart is resistant to ischemia-reperfusion injury. Am J Physiol. 1997;273(3 Pt 2):H1090–H1095. doi: 10.1152/ajpheart.1997.273.3.H1090. [DOI] [PubMed] [Google Scholar]
- 32.Nakagami H, Takemoto M, Liao JK. NADPH oxidase-derived superoxide anion mediates angiotensin II-induced cardiac hypertrophy. J Mol Cell Cardiol. 2003;35(7):851–859. doi: 10.1016/s0022-2828(03)00145-7. [DOI] [PubMed] [Google Scholar]
- 33.Schroder K, Zhang M, Benkhoff S, Mieth A, Pliquett R, Kosowski J, Kruse C, Luedike P, Michaelis UR, Weissmann N, Dimmeler S, Shah AM, Brandes RP. Nox4 is a protective reactive oxygen species generating vascular NADPH oxidase. Circ Res. 2012;110(9):1217–1225. doi: 10.1161/CIRCRESAHA.112.267054. [DOI] [PubMed] [Google Scholar]
- 34.Delaunay-Moisan A, Appenzeller-Herzog C. The antioxidant machinery of the endoplasmic reticulum: Protection and signaling. Free Radic Biol Med. 2015;83:341–351. doi: 10.1016/j.freeradbiomed.2015.02.019. [DOI] [PubMed] [Google Scholar]
- 35.Higa A, Chevet E. Redox signaling loops in the unfolded protein response. Cell Signal. 2012;24(8):1548–1555. doi: 10.1016/j.cellsig.2012.03.011. [DOI] [PubMed] [Google Scholar]
- 36.Tu BP, Weissman JS. Oxidative protein folding in eukaryotes: mechanisms and consequences. J Cell Biol. 2004;164(3):341–346. doi: 10.1083/jcb.200311055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Glorieux C, Zamocky M, Sandoval JM, Verrax J, Calderon PB. Regulation of catalase expression in healthy and cancerous cells. Free Radic Biol Med. 2015;87:84–97. doi: 10.1016/j.freeradbiomed.2015.06.017. [DOI] [PubMed] [Google Scholar]
- 38.Doroudgar S, Volkers M, Thuerauf DJ, Khan M, Mohsin S, Respress JL, Wang W, Gude N, Muller OJ, Wehrens XH, Sussman MA, Glembotski CC. Hrd1 and ER-Associated Protein Degradation, ERAD, are Critical Elements of the Adaptive ER Stress Response in Cardiac Myocytes. Circ Res. 2015;117(6):536–546. doi: 10.1161/CIRCRESAHA.115.306993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Knoops B, Goemaere J, Van der Eecken V, Declercq JP. Peroxiredoxin 5: structure, mechanism, and function of the mammalian atypical 2-Cys peroxiredoxin. Antioxid Redox Signal. 2011;15(3):817–829. doi: 10.1089/ars.2010.3584. [DOI] [PubMed] [Google Scholar]
- 40.Liu J, Rozovsky S. Membrane-bound selenoproteins. Antioxid Redox Signal. 2015;23(10):795–813. doi: 10.1089/ars.2015.6388. [DOI] [PubMed] [Google Scholar]
- 41.Reeves MA, Hoffmann PR. The human selenoproteome: recent insights into functions and regulation. Cell Mol Life Sci. 2009;66(15):2457–2478. doi: 10.1007/s00018-009-0032-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.