Abstract
Hepatitis B virus (HBV) has an important role in the development of human hepatocellular carcinoma (HCC). Accumulated evidence has shown that HBV-encoded X protein (HBx) can induce both genetic alterations in tumor suppressor genes and oncogenes, as well as epigenetic aberrations in HCC pathogens. Non-coding RNAs (ncRNAs) mainly include microRNAs and long non-coding RNAs (lncRNAs). Although ncRNAs cannot code proteins, growing evidence has shown that they have various important biological functions in cell proliferation, cell cycle control, anti-apoptosis, epithelial–mesenchymal transition, tumor invasion and metastasis. This review summarizes the current knowledge regarding the mechanisms and emerging roles of ncRNAs in the pathogenesis of HBV-related HCC. Accumulated data have shown that ncRNAs regulated by HBx have a crucial role in HBV-associated hepatocarcinogenesis. The findings of these studies will contribute to more clinical applications of HBV-related ncRNAs as potential diagnostic markers or as molecular therapeutic targets to prevent and treat HBV-related HCC.
Introduction
Hepatocellular carcinoma (HCC) is the fifth most prevalent life-threatening cancer and the third leading cause of cancer deaths worldwide.1 Hepatitis B virus (HBV) infection has the most important role in the development of HCC, accounting for more than half of the cases worldwide and notably 80% or more of the cases in Southeast Asia and sub-Saharan Africa.2 Although many studies have shown a strong relationship between chronic infection of HBV and the development of HCC, the molecular mechanisms underlying HBV-induced hepatocarcinogenesis are still unclear. However, it has been suggested that HBV may promote HCC through both cis-acting oncogenes or tumor suppressor genes as well as trans-acting cellular genes.3 The HBV genome has four known genes (C, S, X and P) that encode four proteins: the core protein, the envelope protein, the X protein and polymerase. Hepatitis B virus X protein (HBx) is a 154 amino acid long protein that has an important role in trans-activating virus gene expression at the transcriptional level. A previous study has revealed that the HBx protein acts as a multifunctional oncoprotein in the development of HCC, and that it can localize to both the cell cytoplasm and the nucleus.4 Accumulated data have suggested that HBx, which acts as an oncogene or a cofactor in the development of HCC, can facilitate hepatocarcinogenesis via changing the physiological mechanisms of the normal cell. HBx has been shown to trigger viral and cellular promoters or enhancers indirectly by interacting with various cytokines and growth factors, such as nuclear factor kappa B, or by modulating the activation of many cytoplasmic signaling pathways, such as mitogen-activated protein kinase (MAPK) pathways, that are associated with cell growth and survival.5, 6, 7, 8 All the known functions of HBx indicate that HBx has an important role in the tumorigenesis of host liver cells. However, despite extensive studies, the underlying mechanism of how HBx facilitates the development of HCC remains unclear.9
Over the past decade, accumulated data have suggested that HBx can induce both genetic alterations in tumor suppressor genes and oncogenes and epigenetic aberrations in HCC, and these findings have largely changed our understanding of HCC.10, 11, 12, 13, 14 The precise mechanisms and roles of the epigenetic modifications have been comprehensively described in previous years.15, 16, 17, 18 Abnormal DNA methylation, regulated by DNA methyltransferases, and histone modifications, altered by histone-modifying enzymes, are the most widely understood mechanisms of epigenetic processes. Both of these mechanisms could lead to chromatin remodeling and in turn changes in the expression of regional encoded genes and non-coding RNAs (ncRNAs).19 Recent evidence has shown that microRNA (miRNA) and long non-coding RNA (lncRNA), both of which are types of ncRNAs, have a great influence on the epigenome.20, 21, 22, 23 Aberrant epigenetic modifications can be regulated by many factors, such as infection, physical agents and chemical factors. However, HBx has been shown to have the most important role in the epigenetic modification of hepatocytes in HBV-induced HCC; HBx can activate oncogenes or depress tumor suppression genes to facilitate or promote hepatocarcinogenesis. Indeed, studies of epigenetic mechanisms have found that HBx-related modifications are crucial for gene replication, transcription and differentiation of HCC cells.
ncRNAs account for ~90% of the genome and can be divided into the following two categories: lncRNAs (200 nt to 100 kb) and small ncRNAs (18–200 nt). ncRNAs include miRNAs, small interfering and Piwi-interacting RNAs.24, 25 Although these ncRNAs cannot code proteins, increasing evidence has demonstrated that they have a variety of essential biological functions, such as modulating cell proliferation, the cell cycle, apoptosis, invasion and metastasis.26 The dysfunction of miRNAs and lncRNAs is extensively associated with many human disease processes, including tumourigenesis.27, 28, 29, 30 HBx can also facilitate HBV-related hepatocarcinogenesis by regulating the expression of multiple ncRNAs because some of these ncRNAs are known to act as tumor suppressors, whereas others are oncogenes.31 As a multifunctional protein, although HBx lacks the DNA-binding domain, it modulates ncRNAs indirectly by interacting with numerous transcription factors, such as p53, c-myc, survivin and nuclear factor kappa B, and activates cellular signal transduction pathways,32, 33, 34, 35, 36, 37, 38, 39 which changes host gene expression and leads to the development of HCC. More importantly, HBx can transactivate cellular and viral genes by regulating ncRNAs through other epigenetic modifications, such as genomic methylation or acetylation status.40, 41, 42 Recently, it was found that HBx could downregulate miRNA through lncRNA.43 In this review, we will briefly discuss the known regulation and functions of ncRNAs in HBx-related HCC and suggest reasonable strategies for future studies.
miRNAs and hbx-related HCC
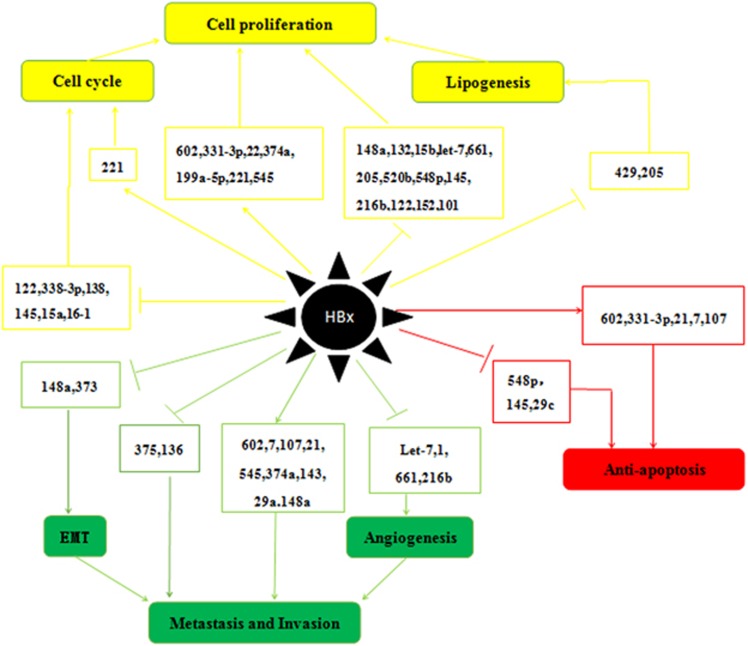
miRNAs (19–25 nt long) regulate ~30% of human gene expression at the transcriptional and post-transcriptional levels,44 usually leading to gene silencing via translational repression or catalyzing the cleavage of target gene messenger RNA (mRNA) by binding to the complementary sequences (mostly in the 3′-untranslated region).45, 46 Since the early 1990s, >1000 miRNAs have been found in human cells. Some miRNAs can function as oncogenes or tumor suppressor genes to enhance or inhibit cancer development, respectively.47, 48 Deviations from normal miRNA expression patterns have a part in many cancers. Over the past decade, many studies have demonstrated that HBx can enhance HCC progression and metastasis by up- or downregulating different miRNAs, as shown in Table 1. What is important is that the discovery of these miRNAs has not only changed our concept of gene expression regulation but has also provided a new opportunity for developing more effective early diagnosis strategies and anticancer therapies for HCC.49, 50 Here we will briefly summarize five known mechanisms of miRNAs that are modulated by HBx in HCC, including cell proliferation, cell cycle control, anti-apoptosis, epithelial-to-mesenchymal transition (EMT) and invasion and metastasis (Figure 1).
Table 1. Targets and functions of miRNAs regulated by HBx in HCC.
| MiRNA | Expression | Target genes | Functions of miRNA | References |
|---|---|---|---|---|
| miR-143 | Upregulation | FNDC3B | Promoting cell migration, invasion and metastasis | 39 |
| miR-148a | Upregulation | PTEN/PI3K/Akt/β-catenin | Promoting cell proliferation, cell cycle progression, migration and anchorage-independent growth | 92 |
| miR-199a-5p | Upregulation | CHC | Promoting cell proliferation | 63 |
| miR-21 | Upregulation | PDCD4 | Promoting cell anti-apoptosis | 78 |
| miR-221 | Upregulation | ERα | Promoting cell cycle progression and cell proliferation | 58 |
| miR-29a | Upregulation | PTEN/PI3K/Akt/MMP-2 | Enhancing cell migration | 90 |
| miR-331-3p | Upregulation | ING5 | Promoting cell proliferation and decreasing cell apoptosis | 85 |
| miR-545,374a | Upregulation | ESRRG | Enhancing cell proliferation and cell migration | 60 |
| miR-602 | Upregulation | RASSF1a | Decreasing cell apoptosis and promoting proliferation | 57 |
| miR-7 | Upregulation | EGFR/Raf/EKERK/PI3K-Akt | Inhibiting cell growth | 128 |
| miR-7,21,107 | Upregulation | Maspin | Enhancing cell migration and anti-apoptosis | 77 |
| let-7 | Downregulation | STAT3 | Enhancing angiogenesis, cell proliferation and migration | 33, 51 |
| miR-1 | Downregulation | EDN1/PI3K/Akt | Enhancing angiogenesis, cell migration and invasion | 94 |
| miR-101 | Downregulation | DNMT3A/RASSF1/PRDM2/GSTP1 | Promoting cell proliferation, cell migration and invasion | 124 |
| miR-122 | Downregulation | β-catenin/E-cadherin | Promoting EMT, cell migration, invasion and metastasis | 43 |
| miR-122 | Downregulation | CCNG1 | Promoting cell cycle progression | 42 |
| miR-132 | Downregulation | Akt | Enhancing cell proliferation | 40 |
| miR-138 | Downregulation | CCND3 | Promoting cell cycle progression | 63 |
| miR-145 | Downregulation | MAP3K | Enhancing cell proliferation | 65 |
| miR-145 | Downregulation | CUL5 | Promoting cell cycle progression and anti-apoptosis | 76 |
| miR-148a | Downregulation | HPIP/AKT/ERK/FOXO4/ATF5/mTOR | Enhancing cell proliferation, EMT and cell migration | 32 |
| miR-152 | Downregulation | DNMT1/GSTP/CDH1 | Enhancing cell proliferation, cell migration and invasion | 123 |
| miR-15a/16-1 | Downregulation | CCND1 | Promoting cell cycle progression | 37, 75 |
| miR-15b | Downregulation | FUT2/GloboH | Enhancing cell proliferation | 61 |
| miR-205 | Downregulation | ACSL4 | Accelerating lipogenesis and promoting cell proliferation | 41, 53 |
| miR-21(D) | Downregulation | PTEN/PIP3/Akt | Inhibiting cell proliferation | 65 |
| miR-216b | Downregulation | IGF2BP2/IGF2/AKT/mTOR/MAPK/ERK | Enhancing cell proliferation and cell migration | 36 |
| miR-222(D) | Downregulation | p27 | Inhibiting cell cycle progression | 65 |
| miR-29c | Downregulation | Bcl-2/Mcl-1 | Decreasing apoptosis | 79 |
| miR-338-3p | Downregulation | CCND1 | Promoting cell cycle progression | 68 |
| miR-373 | Downregulation | E-cadherin | Promoting EMT, cell migration, invasion and metastasis | 88 |
| miR-375,136 | Downregulation | AEG-1 | Promoting cell migration, invasion and metastasis | 91 |
| miR-429 | Downregulation | Rab18 | Promoting dysregulation of lipogenesis and cell proliferation | 52 |
| miR-520b | Downregulation | HBXIP | Enhancing cell proliferation | 34 |
| miR-548p | Downregulation | HBXIP | Enhancing cell proliferation and decreasing apoptosis | 54 |
| miR-661 | Downregulation | MTA1/NK-κB/iNOS/NO | Enhancing angiogenesis, cell proliferation and migration | 62 |
Abbreviations: ACSL4, acyl-CoA synthetase long-chain family member 4; AEG-1: astrocyte elevated gene-1; Akt: protein kinase B; CCND1, cyclin D1; CCNG1, cyclin G1; CUL5, Cullin-RING E3 ubiquitin family; Cullin-5 D, HBV genotype D; EDN1, Endothelin 1; EGFR, epidermal growth factor receptor; ERa, estrogen receptor a; ESRRG, Estrogen-related receptor gamma; EZH2, a histone lysine methyltransferase; FNDC3B, fibronectin type III domain containing 3B; FUT2, fucosyltransferase 2; Globo H, a cancer-associated carbohydrate antigen; HBXIP, Mammalian hepatitis B X-interacting protein; HPIP, hematopoietic pre-B-cell leukemia transcription factor-interacting protein; IL-1a, interleukin-1; IL-6, interleukin-6; ING5, inhibitor of growth 5; MAP3K, mitogen-activated protein kinase kinase kinase; Maspin, mammary serine protease inhibitor; MTA1, metastatic tumor antigen 1; MyD88, Toll-like receptor (TLR) adaptor protein; PDCD4, tumor suppressor programmed cell death 4; PTEN, phosphatase and tensin homolog; PTPRT, a member of the protein tyrosine phosphatases family; Rab18, a member of Ras family; STAT, signal transducer and activator of transcription; URG11, a gene upregulated by HBx.
Figure 1.
Roles of miRNAs regulated by HBx in HCC. The mechanisms include the up- or downregulation of miRNAs and resultant alterations of in cell proliferation, cell cycle control, apoptosis, EMT, invasion and metastasis. (let-7,33, 51 miR-1,94 miR-7,77, 128 miR-15b,61 miR-15a,37, 75 miR-16-1,37, 75 miR-21,65, 77, 78 miR-22,121 miR-29c,79 miR-29a,90 miR-101,124 miR-107,77 miR-122,42, 43 miR-132,40 miR-136,91 miR-138,63 miR-143,39 miR-145,65, 76 miR-148a,32, 92 miR-152,123 miR-199a-5p,63 miR-205,41 miR-216b,36 miR-221,58 miR-331-3p,85 miR-338-3p,68 miR-373,88 miR-374a,60 miR-375,91 miR-429,52 miR-520b,34 miR-545,60 miR-548p,54 miR-60257 and miR-661.62).
miRNAs and proliferation
To date, many miRNAs have been found to have a role in promoting cell proliferation in HBx-associated HCC. Let-7a was the first established miRNA downregulated by HBx, and several members of the Let-7 miRNA family that are downregulated by HBx are known to suppress tumorigenesis. The suppression of Let-7 supports cell proliferation by targeting signal transducer and activator of transcription 3 (STAT3), ras and c-myc.51 A recent study by Wu et al. suggested that a HBx-c-Myc-lin-28 homolog B (LIN28B) axis mediated the repression of let-7 in HepG2 cells. They also found another double-negative feedback loop between c-Myc/Lin28B and let-7, all of which caused the development of HCC through cell proliferation.33
A mouse model of liver cancer showed that HBx could control the expression of miR-148a by inhibiting tumor suppressor p53. In addition, repressed miR-148a increases the expression of hematopoietic pre-B-cell leukemia transcription factor-interacting protein (HPIP), which causes the activation of Akt and extracellular signal-regulated kinase (ERK)-induced mammalian target of rapamycin (mTOR) activation, including the AKT/ERK/Fork head box protein O4 (FOXO4)/ATF5 pathways. Downregulation of miR-148a induces HCC cell proliferation, EMT, invasion and metastasis by the HPIP/AKT/ERK/FOXO4/ATF5/mTOR pathways.32 HBx also suppresses p53-mediated activation of miR-216b, which targets IGF2BP2 and subsequently facilitates the activation of downstream IGF2, AKT/mTOR and MAPK/ERK signaling pathways. Reduced miR-216b induces cell proliferation, migration and invasion of HCC by upregulating IGF2BP2.36
HBx can induce epigenetic repression of miR-132 by methylating DNA. The hypermethylation status of miR-132 facilitates the Akt signaling pathway, which induces the growth of hepatoma cells. This result suggests that the DNA-hypermethylation-dependent repression of the tumor-suppressive function of miRNAs may provide another method for cancer diagnostics and therapeutics.40 In addition, a previous study has suggested that HBx can inhibit miR-205 to enhance HCC tumor cell proliferation by inducing hypermethylation of the miR-205 promoter during their interaction.41
HBx can also enhance hepatocarcinogenesis by dysregulating lipogenesis and promoting the proliferation of hepatoma cells via the HBx/miR-429/Rab18 pathway. miR-429, one of the miRNAs downregulated by HBx, can directly target oncogene Rab18 to induce lipogenesis dysregulation, which in turn leads to HCC tumor cell proliferation.52 Another miRNA that can cause lipogenesis dysregulation was found by Cui et al. in 2014. HBx accelerated lipogenesis and proliferation by decreasing the expression of miR-205. This effect resulted in the overexpression of acyl-CoA synthetase long-chain family member 4, which is a direct target of miR-205, and led to abnormal lipid metabolism through the accumulation of cholesterol in hepatoma cells.53 These results show that miR-205 is a tumor suppressor miRNA, and its downregulation could enhance hepatocarcinogenesis.
miR-520b and miR-548p are also decreased by HBx, both of which induce the expression of oncoprotein mammalian hepatitis B X-interacting protein (HBXIP) and promote tumorigenesis and progression.34, 54 HBXIP was initially identified as a binding protein of HBx and was subsequently shown to serve as a regulatory component for the activation of mTOR complex 1, which regulates cell growth.55, 56 Zhang et al. showed that HBx promotes the development of HCC with its partners survivin and transcription factor Sp1 by downregulating tumor suppressor miR-520b and decreasing the combination of miR-520b and 3′-untranslated region of HBXIP mRNA, which increases the expression of HBXIP. However, HBx can induce demethylation of the HBXIP promoter with its partner survivin, which can also result in the upregulation of HBXIP.34 Recently, Hu et al. proposed a model for the HBx/HNF4A/miR-548p/HBXIP signaling pathway that regulates HCC cell growth. Hepatocyte nuclear factor-4a (HNF4A) is considered an important transcriptional regulator of hepatocyte differentiation and hepatocellular carcinogenesis,35 and has been shown to be repressed by HBx. This HBx repressed HNF4A downregulates the expression of miR-548p, which in turn induces the transcription of HBXIP.54 Therefore, all these factors, such as survivin, HNF4A, HBXIP and HBx, could act as molecular therapeutic targets in HCC.
In addition, HBx-regulated miR-602 is overexpressed during hepatocarcinogenesis. As its target, RASSF1A, a putative tumor suppressor, is decreased and then facilitates tumor proliferation and migration.57 Chen et al.58 found that HBx could downregulate estrogen receptor a (Era) expression via upregulating the expression of miR-221. ERa is considered a direct target of miR-221. Previous evidence has indicated that ERa has protective effects in HCC,59 and that HBx promotes cell growth and accelerates G1–S transition via the upregulation of miR-221 and downregulation of ERa in HepG2 cells.58 Estrogen-related receptor gamma (ESRRG) is closely related to the ER family, which has been identified as a potential target gene of miR-545. The miR-545/374a cluster, located in the Ftx lncRNA, is overexpressed in HBx-related HCC and promotes HCC cell proliferation, cell migration and invasion via decreased ESRRG expression.60 Wu et al.61 found that HBx downregulates the expression of miR-15b, which directly targets fucosyltransferase 2 and then induces Globo H (a cancer-associated carbohydrate antigen) expression and ultimately enhances HCC cell proliferation. HBx-downregulated miR-661, which targets metastatic tumor antigen 1 (MTA1), induces angiogenesis and tumor cell growth by the MTA1/NK-κB/iNOS pathway.62 Wang et al.63 showed that miR-199a-5p could be downregulated by HBx in HBV-associated HCC, and that clathrin heavy chain is suggested as the direct target of miR-199a-5p. Clathrin heavy chain functions as a coactivator for p53 and its overexpression promotes HCC proliferation.64 miR-145 is also suppressed by HBx, which results in cell growth and proliferation by targeting MAP3K (Raf1), which has a great role in HCC tumor cell proliferation via regulating the downstream signaling cascade.65
miRNAs and the cell cycle
miRNAs can also participate in the regulation of cell cycle progression. Control of the cell cycle is often modulated through the cyclin (CCN) and cyclin-dependent kinase (CDK) complexes and cyclin-dependent kinase inhibitors (CDKIs).66, 67 In HCC tumor cells, miRNAs mediated by HBx have been shown to accelerate the process of the cell cycle, especially the transition from the G1 phase to S phase due to the accumulation of CCN and CDK complexes or the reduction of CDKIs, promoting tumor cell proliferation and anti-apoptosis.37, 58, 68 For example, HBx could downregulate ERa expression via upregulating the expression of miR-221,58 which leads to cell growth and G1–S transition via inhibiting the expression of the cell cycle controllers CDKN1B/p27 and CDKN1C/p57 (members of CDKIs) in HepG2 cells.69, 70 Fu et al.68 also reported another miRNA, miR-338-3p, was decreased by HBx that could modulate cyclin D1 (CCND1). Compared with other cyclins, CCND1 is more sensitive to cell cycle modulation.71 Overexpressed CCND1 could activate CDKs, namely, CDK4 and CDK6, to promote the progression of the G1/S cell cycle phase, which has an important role in tumor growth.72 Wang et al.63 reported that downregulation of miR-138 by HBx could promote cell cycle progression by targeting CCND3, which is expressed in nearly all proliferating cells and can induce cell cycle progression.73, 74
Even more importantly, Wu et al. found that HBx could downregulate the expression of the tumor suppressor miR-15a/16 in HepG2 cells and its suppression was mediated by Myc. miR-15a/16 belongs to the miR-16 family and this family functions as a key regulator of the cell cycle checkpoint from the G1 phase to S phase by targeting CCND1, which is depressed by HBx in HepG2 cells, which in turn enhances HCC cell proliferation.37 In addition, another study has shown that viral HBx RNA directly mediates the downregulation of miR-15a/miR-16-1, which indicates that both HBx protein and RNA may act in a synergistic way to regulate the cellular miRNA program in HBV-infected cells.75 In addition, Song et al. found that peroxisome proliferator-activated receptor-gamma (a ligand-activated transcription factor) and retinoid X receptor alpha complex-mediated miR-122 expression is suppressed by HBx through recruitment of the co-repressors, such as the nuclear receptor corepressor protein and the silencing mediator of retinoid and thyroid hormone receptors, and H3K9 histone methyltransferase (SUV39H1) in HCC cells. miR-122 is known to downregulate cyclin G1 and the suppressed miR-122 by HBx could lead to the promotion of the cell cycle and cause HCC cell proliferation. Therefore, downregulation of miR-122 by HBx in HCC is a complicated process that involves the peroxisome proliferator-activated receptor-gamma/retinoid X receptor alpha complex, nuclear receptor corepressor protein/silencing mediator of retinoid and thyroid hormone receptor co-repressor complex, histone acetylation and histone H3K9 methylation.42
Recently, Gao et al.76 reported that downregulated miRNA by HBx miR-145 could release the inhibition of Cullin-5 (CUL5, Cullin-RING E3 ubiquitin family) and then influence numerous cellar activities involving the cell cycle. However, the details of this mechanism are not fully understood and need further exploration.
miRNAs and apoptosis
Apoptosis is a process of type I programmed cell death that occurs in multicellular organisms. Once cell apoptosis is suppressed, it often leads to malignant proliferation. The dysregulation of miRNAs could also repress apoptosis and induce the development of HCC. Li et al. reported that increased miR-21 could be mediated by the HBx-induced interleukin-6 (IL-6)–STAT3 pathway. HBx could trigger the IL-6 pathway followed by phosphorylation and activation of transcriptional factor STAT3, which induces the expression of miR-21.38 A recent report regarding HBx-induced miRNAs involving miR-7, -21 and -107 also showed that HBx may be anti-apoptotic in HCC. Chen et al.77 found that upregulated miR-7, -21 and -107 could directly target mammary serine protease inhibitor (Maspin, also named serpin B5) mRNA, leading to its protein downregulation, which could promote HCC tumor cell anti-apoptosis and metastasis.
In addition, Qiu et al.78 showed that HBx could promote anti-apoptosis of HepG2 cells by upregulating miR-21, which decreases the levels of tumor suppressor programmed cell death 4 expression. This finding suggests that HBx may have an important role in anti-apoptosis of HCC development. miR-29c has been suggested to be downregulated by carboxy-terminal truncated HBx, which could contribute to anti-apoptosis in the development and progression of HBV-related HCC by targeting myeloid cell leukemia sequence 1 and Bcl-2, which are both known anti-apoptotic molecules.79, 80 Recently, Cao et al. showed that miR-331-3p is upregulated by HBx and promotes the proliferation of HCC cells through the repression of inhibitor of growth 5 expression. It has been reported that inhibitor of growth 5 has tumor suppressor functions, and that it induces apoptosis and inhibits cell growth by decreasing the number of cells in S phase.81, 82, 83, 84 Therefore, the overexpression of miR-331-3p decreases tumor cell apoptosis in HBx-related HCC85.
As previous studies have shown, HBx-upregulated miR-602, which targets tumor suppressor RASSF1A, facilitates cell proliferation and tumor migration. miR-602 is upregulated through the development of HBV-related HCC, namely, from chronic HBV infection and liver cirrhosis to HCC. Therefore, miR-602 may be involved at an early stage of hepatocarcinogenesis, which indicates that miR-602 may be a potential very early diagnostic marker for HCC and may serve as a molecular biomarker for the treatment of HBV-related HCC.57 miR-548p, downregulated by HBx, suppresses cell proliferation and promotes cell apoptosis. Repressed miR-548p decreased the ability of cell apoptosis via the HBx/HNF4A/miR-548p/HBXIP signaling pathway.54 HBx-suppressed miR-145 also decreased HCC tumor cell apoptosis by targeting CUL5. Other than its role in controlling the cell cycle, CUL5 has also been shown to be involved in modulating apoptosis by regulating MAPK.76, 86
miRNAs and EMT
EMT is when epithelial cells turn into mesenchymal stem cells. Epithelial cells have cell polarity and express high levels of E-cadherin, which have an important role in cell–cell adhesion. However, mesenchymal cells lose these abilities and express N-cadherin, fibronectin and vimentin, which indicates that EMT occurs in the initiation of invasion and metastasis for tumor progression. Therefore, the reduction of E-cadherin or the induction of vimentin could both lead to the process of EMT.87 It has been reported that HBx downregulates E-cadherin by several mechanisms, one of which is by HBx-mediated downregulation of miR-373.88 Normally, miR-373 activates CDH1, the E-cadherin gene, by binding to a target site in the CDH1 promoter. HBx-suppressed miR-373 decreases the expression of E-cadherin, which is a hallmark of EMT and induces EMT that is characterized by acquisition of a motile mesenchymal phenotype.87 Another miRNA leading to EMT is miR-148a, which has been shown above. HBx represses p53-mediated activation of miR-148a, which in turn activates the HPIP/AKT/ERK/FOXO4/ATF5/mTOR pathways. Inhibition of miR-148a could not only induce HCC tumor cell proliferation but also cause EMT, invasion and metastasis by these pathways.32
miRNAs and invasion or metastasis
Poor prognosis and patient survival of HBV-related HCC are predominantly due to tumor cell invasion and metastasis, which could cause a much higher mortality rate.89 Modification of some adhesion molecules, such as matrix metalloproteinases,90, 91, in HCC has been characterized and these modifications can contribute to the migration of tumor cells in HBx-related HCC.88 Meanwhile, several recent studies have also shown other mechanisms by which HBx regulates miRNAs to enhance HCC cell metastasis. These findings have provided us with an in-depth understanding of hepatocarcinogenesis.
As shown above, both miR-373 and miR-148a can be suppressed by HBx to induce the progression of EMT, which enhances tumor cell invasion and metastasis.32, 88 By contrast, miR-148a has been reported to be upregulated by HBx, and its targets, that is, tumor suppressor phosphatase and tensin homolog (PTEN), are inactivated in the process of HCC development. Yuan et al. have suggested that upregulation of URG11 (upregulated gene 11) and miR-148a by HBx may provide two mechanisms that block PTEN activity, leading to the activation of PI3K/Akt/GSK3 (glycogen synthase kinase-3)/β-catenin signaling pathway. β-Catenin is known to be a part of Wnt signaling and has a great role in cell–cell adhesion by contacting E-cadherin, whose activation results in tumor cell proliferation, invasion and metastasis.92 In addition, Kong et al.90 reported another miRNA that could target PTEN. Overexpression of miR-29a by HBx enhances HCC tumor cell migration via targeting PTEN, and activates both phosphorylated Akt and matrix metalloproteinase-2 (MMP-2), both of which have a crucial role in cell adhesion and movement. A study by Zhao et al.91 demonstrated that HBx is able to decrease the expression of miR-375 and miR-136 and in turn elevate the oncoprotein astrocyte elevated gene-1/MMP-9 level to promote cell migration. MMP-9 can also account for the invasion and metastasis of tumors by facilitating the degradation of environment barriers.93 However, the precise mechanisms for cell migration between HBx and astrocyte elevated gene-1/MMP-9 remain to be determined, for there is considerable crosstalk and there are overlaps in modulating various signaling pathways among them.91
Angiogenesis is a necessary early event for tumor invasion and metastasis. A study by Lu et al. found that miR-1 is suppressed by HBx and enhances migration via targeting Endothelin 1 in HCC tissues. END1 has a crucial role in HCC development by activating the PI3K/Akt signaling pathways, which promotes angiogenesis, tumor growth and migration.94 Downregulated Let-7 by HBx can also result in angiogenesis and migration by activating STAT3 or other oncogenes such as Ras, c-myc and Lin28B.51, 95 miR-661, which is suppressed by HBx and targets metastatic tumor antigen 1 (MTA1), can induce angiogenesis, tumor cell growth and metastasis through the MTA1/NK-κB/iNOS pathway.62
In addition, miR-143 has been reported to be transcriptionally activated by nuclear factor kappa B in HBx-related HCC. Overexpression of miR-143 enhances cancer cell invasion and migration by inhibiting the expression of fibronectin type III domain containing 3B.39 Recently, mammary serine protease inhibitor (Maspin), which is a member of the serine protease inhibitor (serpin) superfamily, has been reported to be downregulated by HBx-dependent induction of miR-7, miR-107 and miR-21. The repressed Maspin could facilitate HCC cell metastasis involving angiogenesis, cell adhesion and cell motility.77 In addition, miR-216b, downregulated by HBx, could also induce HCC tumor cell proliferation and metastasis via IGF2BP2/IGF2/AKT/mTOR and MAPK/ERK signaling pathways.36 HBx-upregulated miR-602, which targets RASSF1a, could also facilitate tumor proliferation and migration. RASSF1a, known as a tumor suppressor gene, affects several cellular functions, such as cell migration, proliferation and apoptosis.57 Overexpression of miR-545/374a in HBx-related HCC could also enhance tumor cell proliferation, cell invasion and metastasis by targeting ESRRG. However, the precise mechanisms need to be further investigated.60
lncRNAs and hbx-related HCC
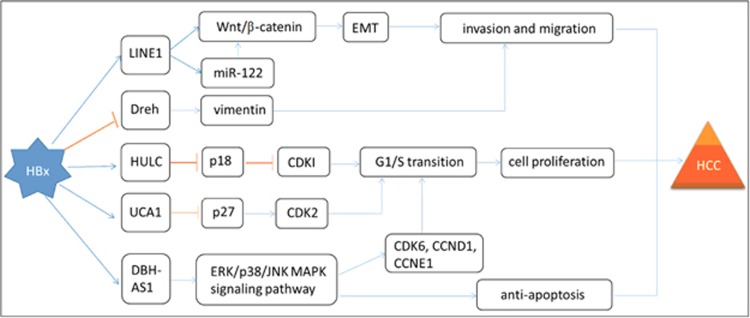
Although regulatory lncRNAs used to be disregarded as transcriptional ‘noise', a growing number of studies have shown that they are involved in different biological processes, including carcinogenesis.96 Compared with miRNAs, lncRNAs have much more complicated functions, including chromatin modification, transcription and post-transcriptional processing. lncRNAs also have many important cellular signal transduction regulations through epigenetic silencing, mRNA splicing, lncRNA–miRNA interactions, lncRNA–protein interactions and lncRNA–mRNA interactions.30, 97, 98, 99, 100, 101 Recently, several studies have demonstrated that various lncRNAs, such as HULC, UCA1, DBH-AS1, Dreh and LINE1, are involved in the development of hepatocarcinogenesis, including cell proliferation, apoptosis and metastasis.43, 102, 103, 104, 105, 106 The dysregulation of lncRNAs gradually became a primary feature of most human cancers, including HCC.98 Therefore, lncRNAs can potentially be used as diagnostic biomarkers or as therapeutic targets for HCC patients in clinical applications. However, only a few functional lncRNAs have been identified in HBx-related HCC so far. More information about how HBx participates in tumorigenesis through lncRNAs needs to be elucidated. Here we will briefly summarize five known lncRNAs regulated by HBx and their functions in the development of HCC (Figure 2).
Figure 2.
Regulatory networks of selected lncRNAs involved in the development of HBx-HCC. The mechanisms involved in lncRNAs are related to intermediate filament protein, the Wnt/β-catenin signaling pathway, the ERK/p38/JNK MAPK signaling pathway and CCNs/CDKs/CDKIs complexes, which cause cell proliferation, aberrant cell cycle control, anti-apoptosis, epithelial-to-mesenchymal transition, invasion and metastasis.
HULC
HULC was first suggested as a novel mRNA-like lncRNA that was overexpressed markedly in HCC.107 It was reported by Wang et al. that HULC is activated by the transcription factor cyclic AMP-responsive element-binding protein (CREB).99 Du et al. found that the abnormal expression of lncRNA HULC was markedly elevated in HBx-induced HCC. They showed that HBx increased the expression of lncRNA HULC through the CREB-activated promoter of HULC via a luciferase reporter gene assay and chromatin immunoprecipitation assay.102 As shown in a previous report, CREB activates the HULC promoter, which contains the CREB-binding site in HCC cells.99 It has been suggested that lncRNAs located near protein-coding genes may have a role in regulating the expression of their neighboring protein-coding genes. Indeed, Du et al. showed that overexpressed HULC downregulates a downstream gene, p18, which promotes HCC cell proliferation.102 P18 is known as a member of CDKIs), which can regulate cell cycle progression and functions in different signaling pathways, including ATM/ATR and p53.108 miRNAs have been widely accepted to act as potential biomarkers for HCC diagnosis and therapy.109, 110 Similarly, it has also been suggested that lncRNAs can be used as biomarkers for early diagnosis and new therapeutic targets for HBx-related HCC.
UCA1
A recent report by Hu et al. demonstrated that the signaling pathway of the HBx-lnc-UCA1/EZH2-p27Kip1 axis is a new mechanism of hepatocarcinogenesis. They found that HBx-upregulated lnc-UCA1 was physically associated with the chromatin-modifying complexes PRC2 component EZH2 (enhancer of zeste homolog 2, a histone methyltransferase), which could suppress p27Kip1 (a member of CDKIs) through histone methylation of the p27Kip1 promoter. Silenced p27 enhances the expression of CDK2, which belongs to cyclin-dependent kinases (CDKs) and has a key role in the G1-to-S phase transition. Therefore, UCA1 could accelerate cell cycle progression, enhance cell proliferation and decrease cell apoptosis in HBx-related HCC.105
DBH-AS1
Another novel lncRNA DBH-AS1 has also been reported to be involved in HBx-mediated hepatocarcinogenesis. HBx can upregulate the expression of DBH-AS1, which induces cell cycle progression by accelerating the G1/S and G2/M transitions via activating the ERK/p38/JNK MAPK signaling pathway.106 Once activated, ERK/p38/JNK MAPK can upregulate CDK6 (a member of cyclin-dependent kinases), CCND1 and CCNE1 (members of cyclins), and downregulate p16, p21 and p27 (members of CDKIs), thus inducing the G1/S and G2/M transitions and cell proliferation.111, 112, 113 However, DBH-AS1 was also shown to protect HCC cells from serum starvation-induced apoptosis, which in turn enhanced cell survival.106 Some studies have also reported that activation of the ERK/p38/JNK MAPK pathways resist apoptosis and enhance cell proliferation in HCC.114
LINE1
Another lncRNA HBx-LINE1, which was reported by Lau et al., can activate the Wnt/β-catenin signaling pathway to promote hepatocarcinogenesis. Aberrant activation of Wnt signaling is a major pathway in the development of HBV-related HCC.115 β-Catenin forms a membranous complex with E-cadherin, which functions as a cell adhesion protein and contributes to the EMT. Once activated, β-catenin dissociates from the E-cadherin complex and sufficient accumulation of β-catenin in the cell causes it to be translocated to the nucleus, where it activates the Wnt signaling pathway.116 They also showed that the expression of c-Myc, cyclin D1 and ZEB1 can be spontaneously modulated by HBx-LINE1 through the Wnt/β-catenin pathway, which results in the promotion of cell metastasis via the EMT process.104 ZEB1, a transcriptional regulator, is implicated in EMT not only by targeting E-cadherin repression but also by activating other mesenchymal genes.117 Recently, HBx-LINE1 was also found by Liang et al.43 to effectively deplete cellular miR-122, which could promote hepatic cell EMT-like changes and enhance cell migration. By downregulating miR-122, HBx-LINE1 induces hepatic cell Wnt/β-catenin activation, E-cadherin suppression and in turn enhances HCC tumor cell proliferation, invasion and migration.
Dreh
Huang et al. also reported an lncRNA that downregulated expression by HBx (termed lncRNA-Dreh) and could be decreased by HBx protein. Dreh, acting as a tumor suppressor in the process of HBx-related hepatocarcinogenesis, could combine with the intermediate filament protein vimentin and downregulate its expression, further changing the normal cytoskeleton structure and thus inhibiting cell invasion and migration. Therefore, HBx inhibits the function of Dreh by repressing its expression and facilitating HCC. This finding supports a role of lncRNA-Dreh in tumor suppression and survival prediction in HCC patients, which provided new ideas and targets for the treatment of HBx-related HCC. The results of this study provide a possible basis for developing epigenetic therapies that use synthetic lncRNAs for the treatment of HBx-related HCC.103 The observed relationships between HBx and lncRNAs suggest that HBx-related modulation of gene expression might be due to lncRNA-related mechanisms of epigenetic change. Therefore, further studies are needed to explain how HBx changes the expression of host genes via the alteration of the lncRNAs.
Perspective
It is evident that aberrant ncRNA changes induced by HBx have an important role in the process of HBV-related HCC, including cell proliferation, cell cycle, apoptosis, EMT and metastasis. A detailed and complete understanding of their mechanisms may provide a theoretical basis for the diagnoses and treatment of HBx-related HCC. Of note, epigenetic gene silencing is different from gene mutations. Epigenetic changes are reversible, whereas gene mutations are irreversible, and the former occurs even earlier than the latter at a premalignant stage. Therefore, the potentially reversible epigenetic changes present possible molecular therapeutic targets for preventing and treating this tumor. In addition, the molecular changes involving ncRNAs and their downstream targets may serve as markers for early diagnosis, risk assessment and prognosis estimation.88, 118, 119 For example, miR-602 may be a new diagnostic marker for HBx-HCC at a very early preneoplastic stage, including chronic HBV hepatitis and cirrhosis, and may function as a molecular therapeutic target for HBV-related HCC.57
A notable feature of HBV-induced HCC is male predominance. Therefore, many studies have paid attention to the relationship between androgen receptor (AR) or ER and the development of HCC. Zhu et al.120 found that HBx correlated with high levels of AR in HCC cases, which occurs via the hypermethylation of the p16INK4A promoter, and correlated negatively with the hypermethylation of the AR promoter. Previous evidence has shown that ERα has protective roles in HCC.59 Indeed, Jiang et al. and Chen et al. found that HBx downregulates ERα expression by upregulating the expression of miR-22 and miR-221.58, 121 miR-22 overexpression in males occurs more often than in females; miR-22 could suppress ERα and result in the upregulation of IL-1α and IL-6, causing males to be susceptible to HBV-HCC. Moreover, these indices may be valuable markers in the clinical diagnosis of HBx-associated HCC. In particular, IL-1α could also predict the direction of chronic liver inflammation; thus, IL-1α can be an early diagnostic marker of HBV-HCC.121 Another study of miR-545 showed that ESRRG was inversely associated with miR-545 expression and was associated with a poor prognosis in HCC patients. However, the comparatively small change in miR-545 expression in female patients with HCC could function as a protective mechanism. This result also suggested that miR-545 and ESRRG may serve as prognostic biomarkers of HBx-related HCC.60 These data indicate that HBx may have a negative role in ERa signaling, and that ERa agonists could be developed as a new therapeutic target for HCC therapy.
Although different epigenetic studies seem scattered, we can find there is a direct and interdependent link between miRNA and other epigenetic alterations,122 which illustrates the complexity of epigenetic changes in HCC. Growing evidence suggests that miRNAs act as both targets and effectors in dysregulated mechanisms of DNA methylation. For example, miR-152/miR-101 is frequently downregulated and regulates DNMT1/DNMT3A in HBV-related HCC, which causes the silencing of some tumor suppressor genes.123, 124 By contrast, Wei et al.124 and Zhang et al.41 showed that HBx could induce the repression of miR-132/miR-205 expression through DNA hypermethylation, which also contributes to HCC tumor cell proliferation. Recently, it has also been suggested that there is a close relationship between lncRNAs and miRNAs. Some lncRNAs, such as Ftx lncRNA, which encodes four miRNAs in its introns, including miR-545/374a (which causes HCC by targeting ESRRG), act as miRNA precursors.60 In addition, another HBx-LINE1 was also found to effectively deplete cellular miR-122, which leads to EMT in HCC.43 In this regard, further investigations are needed to explore more aspects of epigenetic control and the connections among them to find more effective ways to diagnosis or treat this disease. For example, the use of hypermethylated CpG islands in DNA sequences as tumor markers may enable the early detection of HCC. The DNA-hypermethylation-dependent repression of tumor-suppressive functions of miR-132/miR-205 in HBx-HCC could provide another new method for early diagnostics and therapeutics.40, 41 It has been suggested that a number of stable miRNAs may be present in human serum/plasma, which could be used as a crucial noninvasive diagnostic or prognostic marker for HBx-HCC patients in the clinic.125 Currently, some approaches are directed at DNA methylation or histone modification and several important drugs (including demethylating agents 5-azacytidine (Vidaza), decitabine and histone deacetylase inhibitors (SAHA)) have been studied preclinically in different cancers, including HCC. Studies of antagomirs or recombinant miRNA compounds are also underway to provide new therapeutics for HBx-HCC patients in the future.
However, HBx could control multiple miRNAs in different manners to enhance the development of HCC, and the up- or downregulation of the miRNAs is not absolute.65 For a given miRNA, it can be upregulated or be downregulated by HBx, which could function as an oncogene or a tumor suppressor in different cells by targeting different genes. For example, miR-148a can be induced by HBx and target PTEN, which promotes HCC cell migration.92 By contrast, PTEN can be downregulated, by which miR-148a could also facilitate HCC progression via the HPIP/AKT/ERK/FOXO4/ATF5/mTOR pathways.32 In addition, miR-21 and miR-222 have been shown to be upregulated by HBx and to induce the development of HCC via targeting tumor suppressor programmed cell death 4 and p27.78, 126 However, Bandopadhyay et al.65 showed that miR-21 and miR-222 were inhibited in HepG2 cells transfected with HBx-plasmid (genotype D) and with full-length HBV genome (genotype D), which caused the inhibition of the development of HCC. This result may be attributed to the different genotypes of HBV. Notably, until now, HBV could be categorized into 10 genotypes, named A–J.127 Therefore, further studies are needed to validate the results in other different genotypes. However, the exact role of HBx in hepatocarcinogenesis is not exactly clear, and sometimes it may cause different outcomes, such as cell apoptosis or cell growth. Chen et al.128 found that HBx could suppress the expression of EGFR by upregulating miR-7, which resulted in a slow-growth behavior of HCC cells. In addition, several other studies also supported the anti-growth activity of HBx in hepatoma cells.129, 130, 131, 132 These results suggest that it is crucial to determine the relationship between these obviously contradictory data and the precise mechanisms of HBx-related epigenetic changes in the development of HCC. Only in this way can we find more effective treatments for patients with HBV-HCC.
In the future, more extensive studies will be needed to develop a precise profile of HBx regulation of ncRNAs and their roles in different phases of HCC development and progression. Finally, both epigenetic modifications and genetic events are important in the development of different cancers. Therefore, we also need to combine genetics with epigenetic events in the clinical management of this disease. Further comprehensive studies, including various HCC subtypes at different levels, such as at the genomic and epigenomic levels and different stages of the development of cancer, are necessary in the future.
Acknowledgments
This work was supported by grants from the National Natural Science Foundation of China (no. 81641100 and 81472266) and the Excellent Youth Foundation of Jiangsu Province, China (BK20140032).
Footnotes
The authors declare no conflict of interest.
References
- Siegel R, Ma J, Zou Z, Jemal A. Cancer statistics. 2014CA Cancer J Clin 2014; 64: 9–29. [DOI] [PubMed] [Google Scholar]
- Kew MC. Epidemiology of chronic hepatitis B virus infection, hepatocellular carcinoma, and hepatitis B virus-induced hepatocellular carcinoma. Pathol Biol 2010; 58: 273–277. [DOI] [PubMed] [Google Scholar]
- Arbuthnot P, Kew M. Hepatitis B virus and hepatocellular carcinoma. Int J Exp Pathol 2001; 82: 77–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang XD, Wang Y, Ye LH. Hepatitis B virus X protein accelerates the development of hepatoma. Cancer Biol Med 2014; 11: 182–190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edamoto Y, Hara A, Biernat W, Terracciano L, Cathomas G, Riehle HM et al. Alterations of RB1, p53 and Wnt pathways in hepatocellular carcinomas associated with hepatitis C, hepatitis B and alcoholic liver cirrhosis. Int J Cancer 2003; 106: 334–341. [DOI] [PubMed] [Google Scholar]
- Tommasi S, Pinto R, Pilato B, Paradiso A. Molecular pathways and related target therapies in liver carcinoma. Curr Pharm Des 2007; 13: 3279–3287. [DOI] [PubMed] [Google Scholar]
- Kew MC. Hepatitis B virus x protein in the pathogenesis of hepatitis B virus-induced hepatocellular carcinoma. J Gastroenterol Hepatol 2011; 26: 144–152. [DOI] [PubMed] [Google Scholar]
- McCubrey JA, Steelman LS, Abrams SL, Lee JT, Chang F, Bertrand FE et al. Roles of the RAF/MEK/ERK and PI3K/PTEN/AKT pathways in malignant transformation and drug resistance. Adv Enzyme Regul 2006; 46: 249–279. [DOI] [PubMed] [Google Scholar]
- Diao J, Garces R, Richardson CD. X protein of hepatitis B virus modulates cytokine and growth factor related signal transduction pathways during the course of viral infections and hepatocarcinogenesis. Cytokine Growth Factor Rev 2001; 12: 189–205. [DOI] [PubMed] [Google Scholar]
- Hussain SP, Schwank J, Staib F, Wang XW, Harris CC. TP53 mutations and hepatocellular carcinoma: insights into the etiology and pathogenesis of liver cancer. Oncogene 2007; 26: 2166–2176. [DOI] [PubMed] [Google Scholar]
- Nault JC, Zucman-Rossi J. Genetics of hepatobiliary carcinogenesis. Semin Liver Dis 2011; 31: 173–187. [DOI] [PubMed] [Google Scholar]
- Nishida N, Goel A. Genetic and epigenetic signatures in human hepatocellular carcinoma: a systematic review. Curr Genomics 2011; 12: 130–137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tischoff I, Tannapfe A. DNA methylation in hepatocellular carcinoma. World J Gastroenterol 2008; 14: 1741–1748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herceg Z, Paliwal A. Epigenetic mechanisms in hepatocellular carcinoma: how environmental factors influence the epigenome. Mutat Res 2011; 727: 55–61. [DOI] [PubMed] [Google Scholar]
- Baylin SB. DNA methylation and gene silencing in cancer. Nat Clin Pract Oncol 2005; 2(Suppl 1): S4–S11. [DOI] [PubMed] [Google Scholar]
- Herceg Z. Epigenetics and cancer: towards an evaluation of the impact of environmental and dietary factors. Mutagenesis 2007; 22: 91–103. [DOI] [PubMed] [Google Scholar]
- Sawan C, Vaissière T, Murr R, Herceg Z. Epigenetic drivers and genetic passengers on the road to cancer. Mutat Res 2008; 642: 1–13. [DOI] [PubMed] [Google Scholar]
- Vaissière T, Sawan C, Herceg Z. Epigenetic interplay between histone modifications and DNA methylation in gene silencing. Mutat Res 2008; 659: 40–48. [DOI] [PubMed] [Google Scholar]
- Li B, Carey M, Workman JL. The role of chromatin during transcription. Cell 2007; 128: 707–719. [DOI] [PubMed] [Google Scholar]
- Malone CD, Hannon GJ. Small RNAs as guardians of the genome. Cell 2009; 136: 656–668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carthew RW, Sontheimer EJ. Origins and Mechanisms of miRNAs and siRNAs. Cell 2009; 136: 642–655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moazed D. Small RNAs in transcriptional gene silencing and genome defence. Nature 2009; 457: 413–420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kogo R, Shimamura T, Mimori K, Kawahara K, Imoto S, Sudo T et al. Long noncoding RNA HOTAIR regulates polycomb-dependent chromatin modification and is associated with poor prognosis in colorectal cancers. Cancer Res 2011; 71: 6320–6326. [DOI] [PubMed] [Google Scholar]
- Nana-Sinkam SP, Croce CM. Non-coding RNAs in cancer initiation and progression and as novel biomarkers. Mol Oncol 2011; 5: 483–491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 2004; 116: 281–297. [DOI] [PubMed] [Google Scholar]
- Lee YS, Dutta A. MicroRNAs in cancer. Annu Rev Pathol 2009; 4: 199–227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matouk IJ, DeGroot N, Mezan S, Ayesh S, Abu-lail R, Hochberg A et al. The H19 non-coding RNA is essential for human tumor growth. PLoS ONE 2007; 2: e845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mercer TR, Dinger ME, Mattick JS. Long non-coding RNAs: insights into functions. Nat Rev Genet 2009; 10: 155–159. [DOI] [PubMed] [Google Scholar]
- Wilusz JE, Sunwoo H, Spector DL. Long noncoding RNAs: functional surprises from the RNA world. Genes Dev 2009; 23: 1494–1504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta RA, Shah N, Wang KC, Kim J, Horlings HM, Wong DJ et al. Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature 2010; 464: 1071–1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian Y, Yang W, Song J, Wu Y, Ni B. Hepatitis B virus X protein-induced aberrant epigenetic modifications contributing to human hepatocellular carcinoma pathogenesis. Mol Cell Biol 2013; 33: 2810–2816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu X, Fan Z, Kang L, Han J, Jiang C, Zheng X et al. Hepatitis B virus X protein represses miRNA-148a to enhance tumorigenesis. J Clin Invest 2013; 123: 630–645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu G, Huang P, Ju X, Li Z, Wang Y. Lin28B over-expression mediates the repression of let-7 by hepatitis B virus X protein in hepatoma cells. Int J Clin Exp Med 2015; 8: 15108–15116. [PMC free article] [PubMed] [Google Scholar]
- Zhang W, Lu Z, Kong G, Gao Y, Wang T, Wang Q et al. Hepatitis B virus X protein accelerates hepatocarcinogenesis with partner survivin through modulating miR-520b and HBXIP. Mol Cancer 2014; 13: 128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin C, Wang PQ, Xu WP, Yang Y, Zhang Q, Ning BF et al. Hepatocyte nuclear factor-4α reverses malignancy of hepatocellular carcinoma through regulating miR-134 in the DLK1-DIO3 region. Hepatology 2013; 58: 1964–1976. [DOI] [PubMed] [Google Scholar]
- Liu FY, Zhou SJ, Deng YL, Zhang ZY, Zhang EL, Wu ZB et al. MiR-216b is involved in pathogenesis and progression of hepatocellular carcinoma through HBx-miR-216b-IGF2BP2 signaling pathway. Cell Death Dis 2015; 6: e1670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu G, Yu F, Xiao Z, Xu K, Xu J, Tang W et al. Hepatitis B virus X protein downregulates expression of the miR-16 family in malignant hepatocytes in vitro. Br J Cancer 2011; 105: 146–153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li CH, Xu F, Chow S, Feng L, Yin D, Ng TB et al. Hepatitis B virus X protein promotes hepatocellular carcinoma transformation through interleukin-6 activation of microRNA-21 expression. Eur J Cancer 2014; 50: 2560–2569. [DOI] [PubMed] [Google Scholar]
- Zhang X, Liu S, Hu T, Liu S, He Y, Sun S. Up-regulated microRNA-143 transcribed by nuclear factor kappa B enhances hepatocarcinoma metastasis by repressing fibronectin expression. Hepatology 2009; 50: 490–499. [DOI] [PubMed] [Google Scholar]
- Wei X, Tan C, Tang C, Ren G, Xiang T, Qiu Z et al. Epigenetic repression of miR-132 expression by the hepatitis B virus x protein in hepatitis B virus-related hepatocellular carcinoma. Cell Signal 2013; 25: 1037–1043. [DOI] [PubMed] [Google Scholar]
- Zhang T, Zhang J, Cui M, Liu F, You X, Du Y et al. Hepatitis B virus X protein inhibits tumor suppressor miR-205 through inducing hypermethylation of miR-205 promoter to enhance carcinogenesis. Neoplasia 2013; 15: 1282–1291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song K, Han C, Zhang J, Lu D, Dash S, Feitelson M et al. Epigenetic regulation of MicroRNA-122 by peroxisome proliferator activated receptor-gamma and hepatitis b virus X protein in hepatocellular carcinoma cells. Hepatology 2013; 58: 1681–1692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang HW, Wang N, Wang Y, Wang F, Fu Z, Yan X et al. Hepatitis B virus-human chimeric transcript HBx-LINE1 promotes hepatic injury via sequestering cellular microRNA-122. J Hepatol 2016; 64: 278–291. [DOI] [PubMed] [Google Scholar]
- Fu LL, Wen X, Bao JK, Liu B. MicroRNA-modulated autophagic signaling networks in cancer. Int J Biochem Cell Biol 2012; 44: 733–736. [DOI] [PubMed] [Google Scholar]
- Ambros V. The functions of animal microRNAs. Nature 2004; 431: 350–355. [DOI] [PubMed] [Google Scholar]
- Meister G, Tuschl T. Mechanisms of gene silencing by double-stranded RNA. Nature 2004; 431: 343–349. [DOI] [PubMed] [Google Scholar]
- Budhu A, Jia HL, Forgues M, Liu CG, Goldstein D, Lam A et al. Identification of metastasis-related microRNAs in hepatocellular carcinoma. Hepatology 2008; 47: 897–907. [DOI] [PubMed] [Google Scholar]
- Dong Q, Meng P, Wang T, Qin W, Qin W, Wang F et al. MicroRNA let-7a inhibits proliferation of human prostate cancer cells in vitro and in vivo by targeting E2F2 and CCND2. PLoS ONE 2010; 5: e10147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou J, Lin L, Zhou W, Wang Z, Ding G, Dong Q et al. Identification of miRNomes in human liver and hepatocellular carcinoma reveals miR-199a/b-3p as therapeutic target for hepatocellular carcinoma. Cancer Cell 2011; 19: 232–243. [DOI] [PubMed] [Google Scholar]
- Giordano S, Columbano A. MicroRNAs: new tools for diagnosis, prognosis, and therapy in hepatocellular carcinoma? Hepatology 2013; 57: 840–847. [DOI] [PubMed] [Google Scholar]
- Wang Y, Lu Y, Toh ST, Sung WK, Tan P, Chow P et al. Lethal-7 is down-regulated by the hepatitis B virus x protein and targets signal transducer and activator of transcription 3. J Hepatol 2010; 53: 57–66. [DOI] [PubMed] [Google Scholar]
- You X, Liu F, Zhang T, Li Y, Ye L, Zhang X. Hepatitis B virus X protein upregulates oncogene Rab18 to result in the dysregulation of lipogenesis and proliferation of hepatoma cells. Carcinogenesis 2013; 34: 1644–1652. [DOI] [PubMed] [Google Scholar]
- Cui M, Wang Y, Sun B, Xiao Z, Ye L, Zhang X. MiR-205 modulates abnormal lipid metabolism of hepatoma cells via targeting acyl-CoA synthetase long-chain family member 1 (ACSL1) mRNA. Biochem Biophys Res Commun 2014; 444: 270–275. [DOI] [PubMed] [Google Scholar]
- Hu XM, Yan XH, Hu YW, Huang JL, Cao SW, Ren TY et al. MicroRNA-548p suppresses hepatitis B virus X protein associated hepatocellular carcinoma by downregulating oncoprotein HBXIP. Hepatol Res 2016; 46: 804–815. [DOI] [PubMed] [Google Scholar]
- Melegari M, Scaglioni PP, Wands JR. Cloning and characterization of a novel hepatitis B virus x binding protein that inhibits viral replication. J Virol 1998; 72: 1737–1743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bar-Peled L, Schweitzer LD, Zoncu R, Sabatini DM. Ragulator is a GEF for the rag GTPases that signal amino acid levels to mTORC1. Cell 2012; 150: 1196–1208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L, Ma Z, Wang D, Zhao W, Chen L, Wang G. MicroRNA-602 regulating tumor suppressive gene RASSF1A is overexpressed in hepatitis B virus-infected liver and hepatocellular carcinoma. Cancer Biol Ther 2010; 9: 803–808. [DOI] [PubMed] [Google Scholar]
- Chen JJ, Tang YS, Huang SF, Ai JG, Wang HX, Zhang LP. HBx protein-induced upregulation of microRNA-221 promotes aberrant proliferation in HBV-related hepatocellular carcinoma by targeting estrogen receptor-α. Oncol Rep 2015; 33: 792–798. [DOI] [PubMed] [Google Scholar]
- Naugler WE, Sakurai T, Kim S, Maeda S, Kim K, Elsharkawy AM et al. Gender disparity in liver cancer due to sex differences in MyD88-dependent IL-6 production. Science 2007; 317: 121–124. [DOI] [PubMed] [Google Scholar]
- Zhao Q, Li T, Qi J, Liu J, Qin C. The miR-545/374a cluster encoded in the Ftx lncRNA is overexpressed in HBV-related hepatocellular carcinoma and promotes tumorigenesis and tumor progression. PLoS ONE 2014; 9: e109782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu CS, Yen CJ, Chou RH, Chen JN, Huang WC, Wu CY et al. Downregulation of microRNA-15b by hepatitis B virus X enhances hepatocellular carcinoma proliferation via fucosyltransferase 2-induced Globo H expression. Int J Cancer 2014; 134: 1638–1647. [DOI] [PubMed] [Google Scholar]
- Bui-Nguyen TM, Pakala SB, Sirigiri DR, Martin E, Murad F, Kumar R. Stimulation of inducible nitric oxide by hepatitis B virus transactivator protein HBx requires MTA1 coregulator. J Biol Chem 2010; 285: 6980–6986. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Wang W, Zhao LJ, Tan YX, Ren H, Qi ZT. Identification of deregulated miRNAs and their targets in hepatitis B virus-associated hepatocellular carcinoma. World J Gastroenterol 2012; 18: 5442–5453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohata H, Ota N, Shirouzu M, Yokoyama S, Yokota J, Taya Y et al. Identification of a function-specific mutation of clathrin heavy chain (CHC) required for p53 transactivation. J Mol Biol 2009; 394: 460–471. [DOI] [PubMed] [Google Scholar]
- Bandopadhyay M, Banerjee A, Sarkar N, Panigrahi R, Datta S, Pal A et al. Tumor suppressor micro RNA miR-145 and onco micro RNAs miR-21 and miR-222 expressions are differentially modulated by hepatitis B virus X protein in malignant hepatocytes. BMC Cancer 2014; 14: 721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonelli P, Tuccillo FM, Borrelli A, Schiattarella A, Buonaguro FM. CDK/CCN and CDKI alterations for cancer prognosis and therapeutic predictivity. Biomed Res Int 2014; 2014: 361020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malumbres M, Barbacid M. Cell cycle, CDKs and cancer: a changing paradigm. Nat Rev Cancer 2009; 9: 153–166. [DOI] [PubMed] [Google Scholar]
- Fu X, Tan D, Hou Z, Hu Z, Liu G. miR-338-3p is down-regulated by hepatitis B virus X and inhibits cell proliferation by targeting the 3′-UTR region of CyclinD1. Int J Mol Sci 2012; 13: 8514–8539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gramantieri L, Ferracin M, Fornari F, Veronese A, Sabbioni S, Liu CG et al. Cyclin G1 is a target of miR-122a, a microRNA frequently down-regulated in human hepatocellular carcinoma. Cancer Res 2007; 67: 6092–6099. [DOI] [PubMed] [Google Scholar]
- le Sage C, Nagel R, Egan DA, Schrier M, Mesman E, Mangiola A et al. Regulation of the p27(Kip1) tumor suppressor by miR-221 and miR-222 promotes cancer cell proliferation. EMBO J 2007; 26: 3699–3708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imoto M, Doki Y, Jiang W, Han EK, Weinstein IB. Effects of cyclin D1 overexpression on G1 progression-related events. Exp Cell Res 1997; 236: 173–180. [DOI] [PubMed] [Google Scholar]
- Fu M, Wang C, Li Z, Sakamaki T, Pestell RG. Minireview: cyclin D1: normal and abnormal functions. Endocrinology 2004; 145: 5439–5447. [DOI] [PubMed] [Google Scholar]
- Lin J, Jinno S, Okayama H. Cdk6-cyclin D3 complex evades inhibition by inhibitor proteins and uniquely controls cell's proliferation competence. Oncogene 2001; 20: 2000–2009. [DOI] [PubMed] [Google Scholar]
- Grillo M, Bott MJ, Khandke N, McGinnis JP, Miranda M, Meyyappan M et al. Validation of cyclin D1/CDK4 as an anticancer drug target in MCF-7 breast cancer cells: effect of regulated overexpression of cyclin D1 and siRNA-mediated inhibition of endogenous cyclin D1 and CDK4 expression. Breast Cancer Res Treat 2006; 95: 185–194. [DOI] [PubMed] [Google Scholar]
- Wang Y, Jiang L, Ji X, Yang B, Zhang Y, Fu XD. Hepatitis B viral RNA directly mediates down-regulation of the tumor suppressor microRNA miR-15a/miR-16-1 in hepatocytes. J Biol Chem 2013; 288: 18484–18493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao F, Sun X, Wang L, Tang S, Yan C. Downregulation of microrna-145 caused by hepatitis B virus X protein promotes expression of CUL5 and contributes to pathogenesis of hepatitis B virus-associated hepatocellular carcinoma. Cell Physiol Biochem 2015; 37: 1547–1559. [DOI] [PubMed] [Google Scholar]
- Chen WS, Yen CJ, Chen YJ, Chen JY, Wang LY, Chiu SJ et al. miRNA-7/21/107 contribute to HBx-induced hepatocellular carcinoma progression through suppression of maspin. Oncotarget 2015; 6: 25962–25974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu X, Dong S, Qiao F, Lu S, Song Y, Lao Y et al. HBx-mediated miR-21 upregulation represses tumor-suppressor function of PDCD4 in hepatocellular carcinoma. Oncogene 2013; 32: 3296–3305. [DOI] [PubMed] [Google Scholar]
- Xiong Y, Fang JH, Yun JP, Yang J, Zhang Y, Jia WH et al. Effects of microRNA-29 on apoptosis, tumorigenicity, and prognosis of hepatocellular carcinoma. Hepatology 2010; 51: 836–845. [DOI] [PubMed] [Google Scholar]
- Yip WK, Cheng AS, Zhu R, Lung RW, Tsang DP, Lau SS et al. Carboxyl-terminal truncated HBx regulates a distinct microRNA transcription program in hepatocellular carcinoma development. PLoS ONE 2011; 6: e22888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiseki M, Nagashima M, Pedeux RM, Kitahama-Shiseki M, Miura K, Okamura S et al. p29ING4 and p28ING5 bind to p53 and p300, and enhance p53 activity. Cancer Res 2003; 63: 2373–2378. [PubMed] [Google Scholar]
- Xing YN, Yang X, Xu XY, Zheng Y, Xu HM, Takano Y et al. The altered expression of ING5 protein is involved in gastric carcinogenesis and subsequent progression. Hum Pathol 2011; 42: 25–35. [DOI] [PubMed] [Google Scholar]
- Li X, Nishida T, Noguchi A, Zheng Y, Takahashi H, Yang X et al. Decreased nuclear expression and increased cytoplasmic expression of ING5 may be linked to tumorigenesis and progression in human head and neck squamous cell carcinoma. J Cancer Res Clin Oncol 2010; 136: 1573–1583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cengiz B, Gunduz E, Gunduz M, Beder LB, Tamamura R, Bagci C et al. Tumor-specific mutation and downregulation of ING5 detected in oral squamous cell carcinoma. Int J Cancer 2010; 127: 2088–2094. [DOI] [PubMed] [Google Scholar]
- Cao Y, Chen J, Wang D, Peng H, Tan X, Xiong D et al. Upregulated in Hepatitis B virus-associated hepatocellular carcinoma cells, miR-331-3p promotes proliferation of hepatocellular carcinoma cells by targeting ING5. Oncotarget 2015; 6: 38093–38106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Dort C, Zhao P, Parmelee K, Capps B, Poel A, Listenberger L et al. VACM-1, a cul-5 gene, inhibits cellular growth by a mechanism that involves MAPK and p53 signaling pathways. Am J Physiol Cell Physiol 2003; 285: C1386–C1396. [DOI] [PubMed] [Google Scholar]
- Iwatsuki M, Mimori K, Yokobori T, Ishi H, Beppu T, Nakamori S et al. Epithelial-mesenchymal transition in cancer development and its clinical significance. Cancer Sci 2010; 101: 293–299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arzumanyan A, Friedman T, Kotei E, Ng IO, Lian Z, Feitelson MA. Epigenetic repression of E-cadherin expression by hepatitis B virus x antigen in liver cancer. Oncogene 2012; 31: 563–572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lara-Pezzi E, Gómez-Gaviro MV, Gálvez BG, Mira E, Iñiguez MA, Fresno M et al. The hepatitis B virus X protein promotes tumor cell invasion by inducing membrane-type matrix metalloproteinase-1 and cyclooxygenase-2 expression. J Clin Invest 2002; 110: 1831–1838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong G, Zhang J, Zhang S, Shan C, Ye L, Zhang X. Upregulated microRNA-29a by hepatitis B virus X protein enhances hepatoma cell migration by targeting PTEN in cell culture model. PLoS ONE 2011; 6: e19518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J, Wang W, Huang Y, Wu J, Chen M, Cui P et al. HBx elevates oncoprotein AEG-1 expression to promote cell migration by downregulating miR-375 and miR-136 in malignant hepatocytes. DNA Cell Biol 2014; 33: 715–722. [DOI] [PubMed] [Google Scholar]
- Yuan K, Lian Z, Sun B, Clayton MM, Ng IO, Feitelson MA. Role of miR-148a in hepatitis B associated hepatocellular carcinoma. PLoS ONE 2012; 7: e35331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woessner JF Jr. Matrix metalloproteinases and their inhibitors in connective tissue remodeling. FASEB J 1991; 5: 2145–2154. [PubMed] [Google Scholar]
- Lu JW, Liao CY, Yang WY, Lin YM, Jin SL, Wang HD et al. Overexpression of endothelin 1 triggers hepatocarcinogenesis in zebrafish and promotes cell proliferation and migration through the AKT pathway. PLoS ONE 2014; 9: e85318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barh D, Malhotra R, Ravi B, Sindhurani P. MicroRNA let-7: an emerging next-generation cancer therapeutic. Curr Oncol 2010; 17: 70–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Struhl K. Transcriptional noise and the fidelity of initiation by RNA polymerase II. Nat Struct Mol Biol 2007; 14: 103–105. [DOI] [PubMed] [Google Scholar]
- Johnsson P, Ackley A, Vidarsdottir L, Lui WO, Corcoran M, Grandér D et al. A pseudogene long-noncoding-RNA network regulates PTEN transcription and translation in human cells. Nat Struct Mol Biol 2013; 20: 440–446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin R, Maeda S, Liu C, Karin M, Edgington TS. A large noncoding RNA is a marker for murine hepatocellular carcinomas and a spectrum of human carcinomas. Oncogene 2007; 26: 851–858. [DOI] [PubMed] [Google Scholar]
- Wang J, Liu X, Wu H, Ni P, Gu Z, Qiao Y et al. CREB up-regulates long non-coding RNA, HULC expression through interaction with microRNA-372 in liver cancer. Nucleic Acids Res 2010; 38: 5366–5383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hämmerle M, Gutschner T, Uckelmann H, Ozgur S, Fiskin E, Gross M et al. Posttranscriptional destabilization of the liver-specific long noncoding RNA HULC by the IGF2 mRNA-binding protein 1 (IGF2BP1). Hepatology 2013; 58: 1703–1712. [DOI] [PubMed] [Google Scholar]
- Yoon JH, Abdelmohsen K, Srikantan S, Yang X, Martindale JL, De S et al. LincRNA-p21 suppresses target mRNA translation. Mol Cell 2012; 47: 648–655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du Y, Kong G, You X, Zhang S, Zhang T, Gao Y et al. Elevation of highly up-regulated in liver cancer (HULC) by hepatitis B virus X protein promotes hepatoma cell proliferation via down-regulating p18. J Biol Chem 2012; 287: 26302–26311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang JF, Guo YJ, Zhao CX, Yuan SX, Wang Y, Tang GN et al. Hepatitis B virus X protein (HBx)-related long noncoding RNA (lncRNA) down-regulated expression by HBx (Dreh) inhibits hepatocellular carcinoma metastasis by targeting the intermediate filament protein vimentin. Hepatology 2013; 57: 1882–1892. [DOI] [PubMed] [Google Scholar]
- Lau CC, Sun T, Ching AK, He M, Li JW, Wong AM et al. Viral-human chimeric transcript predisposes risk to liver cancer development and progression. Cancer Cell 2014; 25: 335–349. [DOI] [PubMed] [Google Scholar]
- Hu JJ, Song W, Zhang SD, Shen XH, Qiu XM, Wu HZ et al. HBx-upregulated lncRNA UCA1 promotes cell growth and tumorigenesis by recruiting EZH2 and repressing p27Kip1/CDK2 signaling. Sci Rep 2016; 6: 23521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang JL, Ren TY, Cao SW, Zheng SH, Hu XM, Hu YW et al. HBx-related long non-coding RNA DBH-AS1 promotes cell proliferation and survival by activating MAPK signaling in hepatocellular carcinoma. Oncotarget 2015; 6: 33791–33804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panzitt K, Tschernatsch MM, Guelly C, Moustafa T, Stradner M, Strohmaier HM et al. Characterization of HULC, a novel gene with striking up-regulation in hepatocellular carcinoma, as noncoding RNA. Gastroenterology 2007; 132: 330–342. [DOI] [PubMed] [Google Scholar]
- Park BJ, Kang JW, Lee SW, Choi SJ, Shin YK, Ahn YH et al. The haploinsufficient tumor suppressor p18 upregulates p53 via interactions with ATM/ATR. Cell 2005; 120: 209–221. [DOI] [PubMed] [Google Scholar]
- Toffanin S, Villanueva A, Llovet JM. miRNA delivery: emerging therapy for hepatocellular carcinoma. Gastroenterology 2010; 138: 1202–1204. [DOI] [PubMed] [Google Scholar]
- Borel F, Konstantinova P, Jansen PL. Diagnostic and therapeutic potential of miRNA signatures in patients with hepatocellular carcinoma. J Hepatol 2012; 56: 1371–1383. [DOI] [PubMed] [Google Scholar]
- Meloche S, Pouysségur J. The ERK1/2 mitogen-activated protein kinase pathway as a master regulator of the G1- to S-phase transition. Oncogene 2007; 26: 3227–3239. [DOI] [PubMed] [Google Scholar]
- Liu D, Liu J, Lin B, Liu S, Hou R, Hao Y et al. Lewis y regulate cell cycle related factors in ovarian carcinoma cell RMG-I in vitro via ERK and Akt signaling pathways. Int J Mol Sci 2012; 13: 828–839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherr CJ, Roberts JM. CDK inhibitors: positive and negative regulators of G1-phase progression. Genes Dev 1999; 13: 1501–1512. [DOI] [PubMed] [Google Scholar]
- Min L, He B, Hui L. Mitogen-activated protein kinases in hepatocellular carcinoma development. Semin Cancer Biol 2011; 21: 10–20. [DOI] [PubMed] [Google Scholar]
- Whittaker S, Marais R, Zhu AX. The role of signaling pathways in the development and treatment of hepatocellular carcinoma. Oncogene 2010; 29: 4989–5005. [DOI] [PubMed] [Google Scholar]
- Polette M, Mestdagt M, Bindels S, Nawrocki-Raby B, Hunziker W, Foidart JM et al. Beta-catenin and ZO-1: shuttle molecules involved in tumor invasion-associated epithelial-mesenchymal transition processes. Cells Tissues Organs 2007; 185: 61–65. [DOI] [PubMed] [Google Scholar]
- Sánchez-Tilló E, de Barrios O, Siles L, Cuatrecasas M, Castells A, Postigo A. β-catenin/TCF4 complex induces the epithelial-to-mesenchymal transition (EMT)-activator ZEB1 to regulate tumor invasiveness. Proc Natl Acad Sci USA 2011; 108: 19204–19209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park IY, Sohn BH, Yu E, Suh DJ, Chung YH, Lee JH et al. Aberrant epigenetic modifications in hepatocarcinogenesis induced by hepatitis B virus X protein. Gastroenterology 2007; 132: 1476–1494. [DOI] [PubMed] [Google Scholar]
- Zheng DL, Zhang L, Cheng N, Xu X, Deng Q, Teng XM et al. Epigenetic modification induced by hepatitis B virus X protein via interaction with de novo DNA methyltransferase DNMT3A. J Hepatol 2009; 50: 377–387. [DOI] [PubMed] [Google Scholar]
- Zhu R, Zhang JS, Zhu YZ, Fan J, Mao Y, Chen Q et al. HBx-induced androgen receptor expression in HBV-associated hepatocarcinoma is independent of the methylation status of its promoter. Histol Histopathol 2011; 26: 23–35. [DOI] [PubMed] [Google Scholar]
- Jiang R, Deng L, Zhao L, Li X, Zhang F, Xia Y et al. miR-22 promotes HBV-related hepatocellular carcinoma development in males. Clin Cancer Res 2011; 17: 5593–5603. [DOI] [PubMed] [Google Scholar]
- Lopez-Serra P, Esteller M. DNA methylation-associated silencing of tumor-suppressor microRNAs in cancer. Oncogene 2012; 31: 1609–1622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J, Wang Y, Guo Y, Sun S. Down-regulated microRNA-152 induces aberrant DNA methylation in hepatitis B virus-related hepatocellular carcinoma by targeting DNA methyltransferase 1. Hepatology 2010; 52: 60–70. [DOI] [PubMed] [Google Scholar]
- Wei X, Xiang T, Ren G, Tan C, Liu R, Xu X et al. miR-101 is down-regulated by the hepatitis B virus x protein and induces aberrant DNA methylation by targeting DNA methyltransferase 3A. Cell Signal 2013; 25: 439–446. [DOI] [PubMed] [Google Scholar]
- Chen X, Ba Y, Ma L, Cai X, Yin Y, Wang K et al. Characterization of microRNAs in serum: a novel class of biomarkers for diagnosis of cancer and other diseases. Cell Res 2008; 18: 997–1006. [DOI] [PubMed] [Google Scholar]
- Galardi S, Mercatelli N, Giorda E, Massalini S, Frajese GV, Ciafrè SA et al. miR-221 and miR-222 expression affects the proliferation potential of human prostate carcinoma cell lines by targeting p27Kip1. J Biol Chem 2007; 282: 23716–23724. [DOI] [PubMed] [Google Scholar]
- Chu CJ, Lok AS. Clinical significance of hepatitis B virus genotypes. Hepatology 2002; 35: 1274–1276. [DOI] [PubMed] [Google Scholar]
- Chen YJ, Chien PH, Chen WS, Chien YF, Hsu YY, Wang LY et al. Hepatitis B virus-encoded X protein downregulates EGFR expression via inducing microRNA-7 in hepatocellular carcinoma cells. Evid Based Complement Alternat Med 2013; 2013: 682380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuo CY, Wu CC, Hsu SL, Hwang GY. HBx inhibits the growth of CCL13-HBX-stable cells via the GSK-3beta/beta-catenin cascade. Intervirology 2008; 51: 130–136. [DOI] [PubMed] [Google Scholar]
- Slagle BL, Lee TH, Medina D, Finegold MJ, Butel JS. Increased sensitivity to the hepatocarcinogen diethylnitrosamine in transgenic mice carrying the hepatitis B virus X gene. Mol Carcinog 1996; 15: 261–269. [DOI] [PubMed] [Google Scholar]
- Terradillos O, Billet O, Renard CA, Levy R, Molina T, Briand P et al. The hepatitis B virus X gene potentiates c-myc-induced liver oncogenesis in transgenic mice. Oncogene 1997; 14: 395–404. [DOI] [PubMed] [Google Scholar]
- Yu DH, Lin J, Qu JH, Zhu Z, Li FM, Ni CR et al. Hepatitis B virus X protein inhibits hepatoma cell growth in vitro through p14(ARF)-dependent and p14(ARF)-independent pathways. Nan Fang Yi Ke Da Xue Xue Bao 2009; 29: 1089–1093. [PubMed] [Google Scholar]