Abstract
Reliable methods for sampling the nasal mucosa provide clinical researchers with key information regarding respiratory biomarkers of exposure and disease. For quick and noninvasive sampling of the nasal mucosa, nasal lavage (NL) collection has been widely used as a clinical tool; however, limitations including volume variability, sample dilution, and storage prevent NL collection from being used in nonlaboratory settings and analysis of low abundance biomarkers. In this study, we optimize and validate a novel methodology using absorbent Leukosorb paper cut to fit the nasal passage to extract epithelial lining fluid (ELF) from the nasal mucosa. The ELF sampling method limits the dilution of soluble mediators, allowing quantification of both high- and low-abundance soluble biomarkers such as IL-1β, IL-8, IL-6, interferon gamma-induced protein 10 (IP-10), and neutrophil elastase. Additionally, we demonstrate that this method can successfully detect the presence of respiratory pathogens such as influenza virus and markers of antibiotic-resistant bacteria in the nasal mucosa. Efficacy of ELF collection by this method is not diminished in consecutive-day sampling, and percent recovery of both recombinant IL-8 and soluble mediators are not changed despite freezing or room temperature storage for 24 h. Our results indicate that ELF collection using Leukosorb paper sampling of ELF provides a sensitive, easy-to-use, and reproducible methodology to collect concentrated amounts of soluble biomarkers from the nasal mucosa. Moreover, the methodology described herein improves upon the standard NL collection method and provides researchers with a novel tool to assess changes in nasal mucosal host defense status.
Keywords: nasal mucosa, biomarkers, innate immune status, epithelial lining fluid, storage conditions
scientific technologies have advanced throughout recent history to permit a variety of noninvasive, but extremely informative biological sample collection and analysis techniques to be used in field research. Rapid sample collection with minimal risk to human subjects permits more frequent sampling and more detailed analyses throughout a time course. The development of sample collection techniques with flexible storage requirements also facilitates investigations in clinical and population studies. Dried blood spots, for example, provide a method to detect systemic markers of inflammation, infection, and disease (10, 16). However, an adequate method for detecting respiratory inflammation or infection has not yet been developed. Here, we describe a novel nasal mucosal sampling method to analyze mucosal biomarkers that are suitable for use in clinical and epidemiological studies. This method uses a wettable, fibrous, synthetic matrix that can be stably stored at room temperature for later analysis.
Sampling the nasal mucosa by nasal lavage (NL), or irrigation of the nasal passage with isotonic saline, has emerged as the current noninvasive standard for collection of soluble markers in epithelial lining fluid (ELF) of the upper airways and was optimized and validated in 2014 (17). NL has been used extensively in peer-reviewed publications, and was present in 2,559 instances in PubMed when “nasal lavage” was used as a search term and 1,843 instances when “nasal lavage and humans” was used as a search term as of August 2016. The first mention of NL in PubMed dates to 1947 in a study by Atlas (3). NL is less expensive to obtain and process, and much less invasive than bronchoalveolar lavage (BAL), which is currently used to sample the lower airways, but requires subjects to undergo a more invasive bronchoscopy. NL collection is also less time consuming and labor intensive for the subject and investigators than induced sputum, another technique currently used to sample the lower airways. Induced sputum is less invasive than BAL collection (2) but involves significant dilution of samples with saline and processing with reducing reagents such as dithiothreitol for analysis, which can interfere with the functionality and detection accuracy of inflammatory mediators in commercially available ELISA kits (35). NL is attractive to researchers because samples can be quickly collected and stored for long periods of time (months to years) at −80°C for bulk analysis. However, there are limitations to this method, including excessive sample dilution with saline, variability in recovered NL volume between subjects, potential contamination with blood during repeated expulsion of saline from the nose, and the need for freezers for long-term storage.
Our method is an alternative to conventional NL collection and uses Leukosorb medium (Pall Scientific, Port Washington, NY), described by the manufacturer as an absorbent, fibrous matrix designed for the isolation of leukocytes from whole blood (http://www.pall.com/main/oem-materials-and-devices/product.page?id=47512). As in previous studies (9, 15, 20, 32), we use the absorbent matrix to isolate biomarkers and soluble mediators of respiratory inflammation (i.e., cytokines, proteases, and others) that are present at the nasal mucosal surface. To refine the previously published technique, we fabricated ergonomic Leukosorb strips that were designed to easily fit within nasal passages. In addition to reliably and reproducibly assessing levels of soluble mediators in the nasal mucosa, we also show that this method can successfully detect the presence of pathogens, such as influenza virus and markers of antibiotic-resistant bacteria, within the nasal mucosa, and allows the strips to be stored at various conditions without affecting mediator recovery. In addition, our method is also easier to administer than similar studies that used a sponge in place of Leukosorb paper (26), because a medical professional is not needed to insert and remove the Leukosorb strips. Taken together, the experiments described herein suggest that this method will be effective for assessing biomarkers in the nasal mucosa in field studies and other settings where traditional NL is more difficult to reliably obtain or where access to freezers for storage might be limited.
MATERIALS AND METHODS
Subject recruitment and sample collection.
Healthy adult subjects aged 22–43 yr were recruited. The mean age of subjects was 27.2 ± 2.3 yr (see Table 1 for demographic information), and the median age was 24 yr. Exclusion criteria for this study included current symptoms of allergic rhinitis, asthma, FEV1 less than 75% predicted at screen, chronic obstructive pulmonary disorder, cardiac disease, any chronic cardiorespiratory condition, bleeding disorders, recent nasal surgery, immunodeficiency, tobacco use, or current pregnancy. Subjects were recruited for a total of two visits, the second visit being at least 24 h but not more than 2 wk from the first visit. During visit 1, after subjects provided consent, demographic information, pregnancy tests (for women), vital signs, nasal lavage fluid (NLF), and ELF were collected. During visit 2, NLF and ELF were collected. The protocol was submitted to and approved by the University of North Carolina at Chapel Hill Biomedical Institutional Review Board.
Table 1.
Subject demographics
| Descriptor | Value |
|---|---|
| Body mass index | 24.5 ± 0.7 |
| Age, yr | 27.2 ± 2.3 |
| Sex, female/male | 4/6 |
| Race/ethnicity, White/African American/Asian | 8/1/1 |
Body mass index and age are presented as means ± SE, n = 10.
NLF collection.
NLF was obtained as described in previous studies (19, 27–31). Briefly, nostrils were repeatedly sprayed with 0.9% sterile, normal saline irrigation solution (a total of 4 ml per nostril) and expelled into collection cups. The cellular component was then isolated from the NLF using filtration (40 μm cell strainer) and centrifugation as described previously (19, 27–31). Cell-free NLF and NLF cells were then stored at −80°C until analysis.
ELF collection on Leukosorb strips.
Strips were cut from sheets of Leukosorb medium using a laser cutter to the dimensions shown in Fig. 1, B and C. Strips and their respective 1.5-ml microcentrifuge containers were preweighed. Strips were removed from their tubes and inserted into the anterior part of the inferior nasal turbinate of each nostril. Before the strips were inserted, each nostril was briefly moistened with ~100 μl of 0.9% sterile, normal saline solution. Leukosorb strips were then inserted into each nostril until the indicator mark was at or close to the base of each nare. After insertion, nostrils were clamped shut using a padded nose clip for 2 min. Strips were then removed from the nostril and collected in their respective 1.5-ml collection tubes, and again weighed and stored at −20°C, unless evaluating storage conditions, until ELF elution.
Fig. 1.
Percent recovery of recombinant IL-8 from Leukosorb strips using various storage conditions. A: flowchart describing methods for detecting the percent recovery of IL-8 recombinant protein from Leukosorb strips. B: image of a Leukosorb strip and its relative size. C: diagram and measurements of cut Leukosorb strips. D: percent recovery of recombinant IL-8 from Leukosorb strips at varying storage conditions (n = 5). A known amount of recombinant IL-8 with no carrier was spiked onto Leukosorb strips. The strips were then stored in triplicate in four storage conditions (immediate elution, −20°C, room temperature, or 37°C). Protein was then eluted from strips and the eluant was stored at −20°C until analysis (at least 24 h). The strips were then batch-analyzed by ELISA and compared with protein of a known concentration. Compared with the strips that were immediately eluted, there was a significant difference in percent recovery only in the 37°C group. Values are presented as means ± SE. *P ≤ 0.05.
Evaluating percent recovery of protein from Leukosorb strips.
Varying known amounts of IL-8 recombinant protein (PeproTech, Rocky Hill, NJ) suspended in double-deionized H2O (100 μl total volume) was applied to strips of Leukosorb paper and allowed to absorb. The strips were then stored in one of four storage conditions [immediate elution, −20°C, room temperature (21 ± 2°C), or 37°C] for 24 h. Strips were then eluted and analyzed via ELISA (see flowchart, Fig. 1A).
Evaluating variability.
Variability within (nare to nare and day to day) and between subjects was evaluated by collecting strips from both nares of subjects on 3 consecutive days and storing them at −20°C until elution and analysis via ELISA (Fig. 2).
Fig. 2.
Intranare, day-to-day, and interindividual variability of IL-8 production in epithelial lining fluid (ELF). A: flowchart describing methods for determining variability in ELF samples within nares, from day to day, and between individuals using Leukosorb paper and IL-8 ELISA. B: intranare variability in IL-8 production using the average of the right nare over 3 days of sampling and left nare over 3 days of sampling (biological replicates = 3). There was no significant difference in IL-8 concentration intranare. Means are graphed for each individual. C: day-to-day variability in IL-8 production. The left and right nare were averaged for each day for a biological replicate of two. There was no significant variability in IL-8 concentration over the 3 days. Means are graphed for each individual at each day. D: interindividual variability in IL-8 production. Right and left nare measurements for each of the 3 days is graphed for each individual. There is significant interindividual variability. There is also a significant sex difference, IL-8 production is greater in men than it is in women. Values are presented as means ± SE. In B–D, n = 5; open squares indicate men, closed circles indicate women. ***P ≤ 0.001.
Evaluating storage conditions.
The Leukosorb strips were stored in four different storage conditions (−20°C, room temperature (21 ± 2°C), immediate elution, and 37°C) according to the flowchart in Fig. 3A after collection. The two strips collected at each visit were randomized to either protocol A or protocol B for visit 1 and stored in the alternate protocol for visit 2. For the storage conditions at room temperature or 37°C, strips were removed from their 1.5-ml tube and placed in covered Petri dishes (Nalge Nunc International, Rochester, NY) in either a vented storage container at room temperature or in a heated incubator (Innova 4000; New Brunswick Scientific, Edison, NJ) for 24 h. After the 24-h period, the strips at room temperature and 37°C (both conditions yielded completely dried strips) were returned to their 1.5-ml tube for subsequent elution.
Fig. 3.
Effectiveness of a variety of storage conditions for collection and analysis of ELF for biomarkers of immune status from Leukosorb strips. A: flowchart describing methods for determining the effectiveness of varying storage conditions on analysis of ELF from Leukosorb strips for biomarkers of immune status. B: IL-1β collected per strip. Levels were lower in the 37°C group than in the immediate-elution group. C: IL-6 collected per strip. D: IL-8 collected per strip. Levels were lower in the 37°C group than the immediate-elution group. E: IP-10 collected per strip. F: neutrophil elastase collected per strip. Groups of strips stored at room temperature (RT) and 37°C had lower values of neutrophil elastase than the group that underwent immediate elution. G: IL-8 concentration resultant from elution of Leukosorb strips. The groups stored at RT and 37°C were dried out during storage. They were thus eluted in a lower volume of liquid, making the resultant protein more concentrated. The concentration of the eluant per milliliter is shown. The eluent at room temperature was more concentrated than the group that underwent immediate elution (Immed. Elution) and thus would be better for detecting low-abundant protein than the more dilute eluant of the immediate-elution group. Values are presented as means ± SE. In B–G, n = 10. Statistical analysis included one-way ANOVA with a Dunnett’s post hoc test, comparing all groups with the immediate-elution group. *P ≤ 0.05, ***P ≤ 0.001.
ELF elution from Leukosorb strips.
For ELF elution, strips were placed in 650-μl microcentrifuge tubes, which were modified with an 18-gauge needle to have a hole at the tip of the tube. Tubes containing the strips were placed in a 1.5-ml microcentrifuge tube and 100 μl of a 1% BSA + 0.05% Triton X-100 in Dulbecco’s PBS (GIBCO/Thermo Fischer, Waltham, MD) was added to the strip. Strips were centrifuged twice at 13,000 revolutions per minute for 2 min to elute the ELF from the strip into the 1.5-ml tube. The ELF was then stored at −20°C until analysis.
ELISA of biomarkers of immunological status.
Cell-free NLF and ELF were used to analyze IL-8, IL-6, IL-1β, interferon γ-induced protein 10 (IP-10), and neutrophil elastase via commercially available ELISA kits (BD OptEIA, Franklin Lakes, NJ for IL-8, IL-6, IP-10, and IL-1β; eBioscience Human PMN-Elastase Platinum ELISA, San Diego, CA, for neutrophil elastase) (see Figs. 1–4). Absorbance was measured using a CLARIOstar microplate reader (BMG Labtech, Ortenberg, Germany).
Fig. 4.
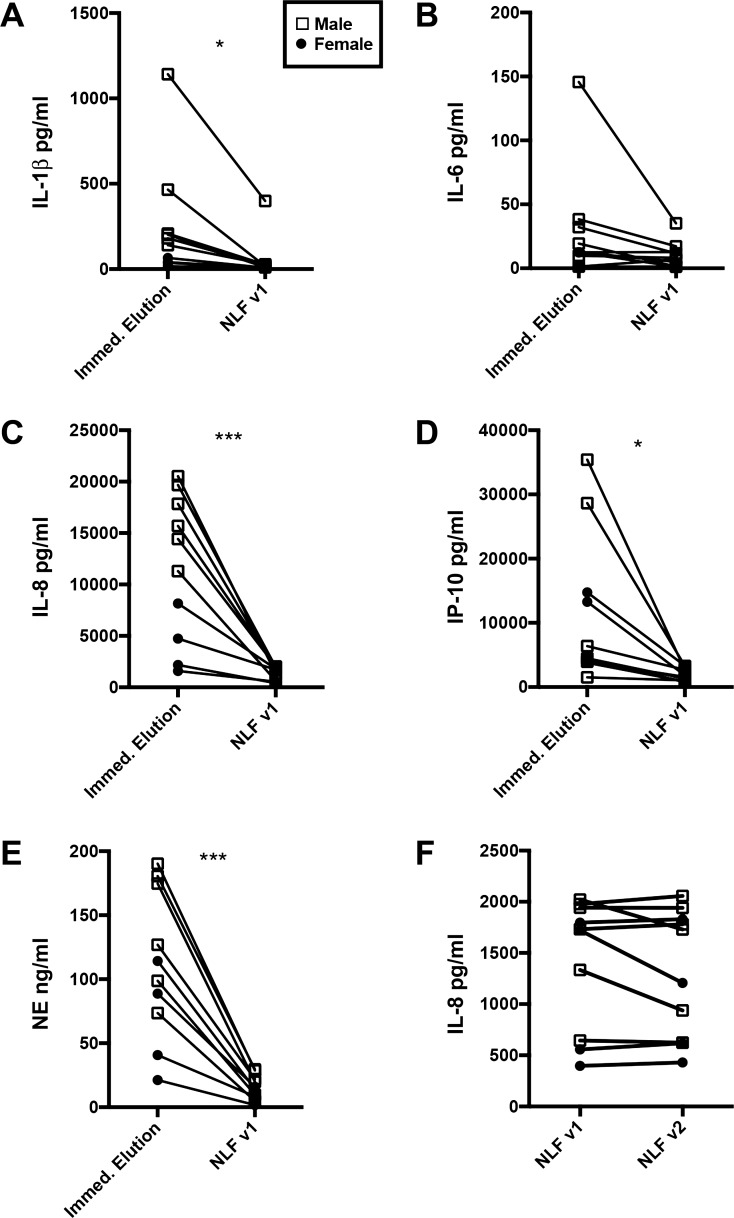
Comparison of ELF from Leukosorb strips and traditional nasal lavage fluid (NLF) for detection of biomarkers of immune status. The immediate-elution group from the ELF storage condition study was compared with visit 1 (v1) NLF from the same subjects. A: IL-1β levels in ELF vs. NLF. Significantly more IL-1β was detected in the ELF than the NLF. B: IL-6 levels in ELF vs. NLF. C: IL-8 levels in ELF vs. NLF. Significantly more IL-8 was detected in ELF than in NLF. In addition, there was a significant sex difference in the immediate-elution group, in which IL-8 levels were higher in men than in women. D: IP-10 levels in ELF vs. NLF. Significantly more IP-10 was detected in ELF than in NLF. E: neutrophil elastase (NE) levels in ELF vs. NLF. Significantly more NE was detected in ELF than NLF. In addition, there was a significant sex difference in the immediate elution group, in which NE levels were higher in men than in women. F: day-to-day variability in IL-8 NLF. NLF from the 2 days of collection were compared, and there was no variability detected between the 2 days; n = 10; v2 indicates visit 2. Statistical analysis included a paired t-test comparing ELF with NLF. Sex difference was detected using a two-way ANOVA with sex and storage condition as factors with a Dunnett’s post hoc test comparing all groups with the immediate elution group. *P ≤ 0.05, ***P ≤ 0.001.
Detection of pathogens.
Subjects were given a standard dose of live attenuated influenza virus (LAIV) vaccine (MedImmune Astra Zeneca, Gaithersburg, MD) in both nostrils to simulate an influenza virus infection, which allowed us to assess a controlled disease population mimic. ELF and NLF samples were collected before exposure (day 0) and 1 day (day 1) after LAIV exposure. The schematic of experimental design is summarized in Fig. 5A. ELF and cells isolated from NLF were subjected to further analysis. Informed consent was obtained from all subjects, and the protocol was submitted to and approved by the University of North Carolina at Chapel Hill Biomedical Institutional Review Board.
Fig. 5.
Ability of ELF to measure viral markers of disease in subjects given the live attenuated influenza virus (LAIV) vaccine and the presence of bacteria. A: flowchart describing methods for administration of LAIV to healthy volunteers. B: measurement of influenza FluB gene expression before LAIV administration (day 0) and 1 day after exposure (day 1) from samples collected through both ELF and NLF sampling methods. C: measurement of gene expression of the antibiotic-resistant bacteria marker mecA before and after LAIV in ELF and NLF samples; n = 6.
Total RNA was isolated from the Leukosorb strips using the Pure Link RNA Mini Kit (Life Technologies, Carlsbad, CA) by submerging the strips in 300 μl of Pure Link RNA Mini Kit Lysis Buffer for 15 min, vortexed every 5 min, and collecting the eluate through centrifugation as described above. First-strand cDNA preparation and real-time quantitative PCR (qPCR) were performed as previously described (1, 2) using the following primer/probe pairs specific for the M1 gene of the LAIV Influenza B Ann Arbor/1/66 master donor strain: 5′-FAM-CCCTCTTGTTGTTGCCGC-TAMRA-3′ (probe), 5′-GGGTGCAGATGCAACGATT-3′ (sense), and 5′-AATATCAAGTGCAAGATCCCAATG-3′ (antisense); and for the methicillin-resistance (mecA) gene commonly found in antibiotic-resistant bacteria, such as Staphylococcus aureus: 5′-FAM-AGATCTTATGCAAACTTAATTGGCAAATCC-TAMRA-3′ (probe), 5′-GGCAATATTACCGCACCTCA-3′ (sense), and 5′-GTCTGCCACTTTCTCCTTGT-3′ (antisense). Differences in expression were determined using the ΔΔCt method and β-actin mRNA expression for normalization (21, 22).
Statistical analysis.
Prism 6 software (GraphPad, La Jolla, CA) was used to visualize and analyze all data sets. A paired t-test was used to test for an effect of nare, day, and the difference between ELF and NLF. A repeated-measures one-way ANOVA was used to test for an effect of day, storage condition, or interindividual variability. A Dunnett’s test was used for post hoc testing to compare all groups with the immediate elution group. A value of P < 0.05 was considered significant.
RESULTS
Percent recovery of recombinant protein from Leukosorb paper.
Recovery of recombinant IL-8 for all four storage conditions (see flowchart, Fig. 1A) was found to be 94.8 ± 11.9% at immediate elution, 69.3 ± 10.6% at −20°C, 83.9 ± 5.2% at room temperature, and 57.1 ± 3.2% at 37°C. There was a statistical difference only between the immediate elution and 37°C groups (Fig. 1D).
Intranare, day-to-day, and interindividual variability.
Intranare, day-to-day, and interindividual variability was assessed using methods summarized in the flowchart in Fig. 2A. There was no statistical difference between nares (Fig. 2B) or day-to-day (Fig. 2C), however, there was a significant interindividual variability (Fig. 2D).
Evaluating storage conditions.
Leukosorb strips were collected and stored as summarized in Fig. 3A. Figure 3, B–F shows the amount of each cytokine, chemokine, or protease recovered per strip. There were no significant differences between groups for cytokines and chemokines compared with the immediate-elution group, except IL-1β and IL-8 at 37°C (Fig. 3, B–E). There was a significant difference in neutrophil elastase between the room-temperature and 37°C groups compared with the immediate-elution group (Fig. 3F). The volume of eluant recovered from the groups varied; ~200 μl was recovered from the immediate-elution and −20°C groups, and 100 μl was recovered from the room-temperature and 37°C groups. This difference resulted from the strips at room temperature and 37°C drying out during the 24 h of storage, during which all moisture was removed from the strip, whereas the immediate-elution and −20°C groups retained moisture of the mucous from the nasal passage in addition to the volume of buffer used to elute immune mediators from the strips. The eluant from strips maintained at room temperature and 37°C was therefore twice as concentrated as that in the immediate-elution and −20°C groups, which is reflected in the eluant concentration depicted in Fig. 3G. This resulted in IL-8 concentrations within the room-temperature group being significantly greater than the immediate-elution group when considering the concentration of the eluant (Fig. 3G) rather than the amount of mediator recovered from each strip (Fig. 3, A–F).
ELF vs. NLF comparison.
ELF from Leukosorb strips provided significantly more concentrated mediators compared with traditional NLF collection for all mediators analyzed except IL-6 (Fig. 4, A–E). There was no day-to-day variability in NLF (Fig. 4F), therefore, all comparisons were made between the immediate-elution group from Leukosorb paper and visit 1 NLF.
In addition to a significant difference between ELF and NLF, sex differences in IL-8 and neutrophil elastase were detected in the immediate-elution group. IL-8 levels were significantly higher in men than in women (significant main effect of storage condition, sex, and their interaction) (Fig. 4C). Neutrophil elastase levels were also higher in men than in women (significant main effect of storage condition and sex) (Fig. 4E).
Detection of pathogens.
To determine whether the presence of viral and bacterial RNA from potential diseased populations is measurable, we used the ELF sampling method to retrieve samples from healthy volunteers inoculated with LAIV, thereby mimicking a diseased population in a controlled fashion. We then analyzed the samples for expression of influenza B and mecA, a gene associated with antibiotic-resistant bacteria. Viral and bacterial RNA content was assessed in matched samples acquired using the ELF sampling method and the standard NLF collection method. As expected, minimal to no viral RNA was detectable before LAIV administration on day 0. At day 1 after administration, virus was detectable in all ELF samples and NLF cell samples (Fig. 5B). Bacterial RNA was detected in ELF samples from two subjects, regardless of sampling day, but could not be detected in NLF cells (Fig. 5C).
DISCUSSION
The goal of this study was to develop a rapid, noninvasive method for the collection and storage of ELF from the nasal passages of human subjects that would be comparable to ease of collection and storage of dried blood spots. The use of Leukosorb paper to evaluate soluble mediators in the nasal mucosa was previously described, when two types of absorptive matrices, Accuwik Ultra and Leukosorb, were used to recover cytokines and evaluate percent protein recovery (14, 20). Accuwik Ultra has been previously compared with other matrices and shown to be adequate for isolation of cytokines, but it is no longer available, and Leukosorb was suggested as an adequate alternative (14, 20, 32). Other matrices of similar quality may be available but were not tested in this study. We show that cytokines, chemokines, and proteases can be reliably recovered from Leukosorb paper that was frozen, dried at room temperature, or dried at 37°C for 24 h. This method (ELF) also provides more concentrated samples and subsequent increased sensitivity of biomarker analysis, especially when dried at room temperature, than NLF (several orders of magnitude times more concentrated, depending on the mediator), suggesting that difficult-to-measure respiratory biomarkers could be assessed using this strategy. Moreover, our data indicate that Leukosorb strips can be used to assess the presence of respiratory pathogens such as influenza and bacteria, thus providing a reliable and noninvasive method to assess markers of immune status in the nasal mucosa of human volunteers.
The results of our study illustrate the day-to-day reproducibility, intranare similarities, and expected interindividual variability in nasal mucosal mediator levels. There is no statistically significant day-to-day or intranare variability, suggesting that repeated sampling over multiple days can be used with little to no irritation of the nasal epithelium (Fig. 2). The consistency of day-to-day and intranare measures does not mitigate the ability to detect interindividual variability (Fig. 2C). In fact, the concentration of ELF samples permits increased detection sensitivity for differences between individuals, treatments, exposures, etc. Increased detection sensitivity is further demonstrated by our ability to reproducibly detect inflammatory mediators of low abundance (IL-1β), medium abundance (IL-6 and IP-10), and high abundance (IL-8), as well as protease (neutrophil elastase) in the nasal mucosa of healthy individuals. In contrast, IL-1β and IL-6 were at the lower limits of detection in NL samples (Fig. 4). Considering that variability of mediator measurements increases at the lower limit of detection, interindividual differences will likely be more detectable using the ELF methodology and potentially reveal differences between different study groups that were previously undetected. For example, the observation that there is a sex difference in baseline nasal mucosal IL-8 levels has not been previously reported. Here we report that nasal IL-8 levels were significantly higher in men than in women and there was a significant effect of sex in baseline neutrophil elastase in the immediate elution group, in which men had a higher amount of neutrophil elastase than women (Fig. 2D). Increased levels of both IL-8 and neutrophil elastase suggest a possible increased prevalence of neutrophils or improved ability to prime neutrophils to respond to infection (36, 37) in the nasal mucosa of men compared with women. The enhanced ability to detect this baseline sex difference may be due to the reduced variability in recovery volume of the ELF methodology (100–200 μl) compared with the highly variable recovery volume of NLF (2–6 ml), as well as an increased concentration of mediators within the eluate. However, because the sample size was small and the results may have been influenced by other outside/clinical factors such as occupational toxicant exposure, rural vs. urban habitation, recreational drug use, etc., such conclusions are limited and should be further investigated in future studies.
Additionally, the increased concentration and recovery of cytokines and chemokines from Leukosorb paper does not dissipate when strips are stored at room temperature for 24 h (Fig. 3, A–E). In fact, drying strips further concentrates the soluble mediators in the ELF, making it possible to detect low-abundance mediators (Fig. 3G). Some degradation of neutrophil elastase occurred in the room-temperature and 37°C groups (Fig. 3F), and some degradation of IL-1β and IL-8 occurred in the 37°C group; however, all groups that exhibited some degradation were still within the limits of detection and of greater abundance in the ELF than the NLF tested. An additional benefit to drying strips can be inferred from the dried blood spot literature, in which it has been demonstrated that drying samples can damage viral caspids (6, 33), thus reducing the potential for contamination of laboratory staff who process the samples without compromising the ability to detect virus in analysis (6, 33). It also permits samples to be shipped via mail with little to no risk to the general public (24). Hence, the use of Leukosorb strips is not limited to storage in refrigerated or frozen conditions, making this method advantageous for nasal biomarker collection studies conducted outside a laboratory setting.
Although our study was limited to 24 h of storage at room temperature, other studies have indicated that many mucosal mediators are stable for more extended periods at temperatures higher than −80°C and over at least one freeze-thaw cycle (4, 11). Studies using dried blood spots have also shown that the stability of proteins varies depending on the protein; however, generally, most proteins are stable at room temperature up to 1 wk (7). The literature (25) also suggests that drying cytokines reduces molecular mobility, thereby delaying degradation for a more extended period than if they were in solution. Specific degradation timelines for cytokines or other proteins of interest should be validated before using this method in future studies, because a limited number of cytokines and only one time point were investigated here. Our study was also limited by the relatively small sample size, and the results of this study should be interpreted as proof of principle and should be further validated with a larger cohort for use in clinical studies. Therefore, for nonlaboratory setting studies, the use of Leukosorb strips to collect nasal mucosal mediators and the ability to store samples at room temperature would present a novel, noninvasive, and reproducible methodology to determine respiratory biomarkers of infection or inflammation.
In addition to quantitatively assessing soluble mediators, ELF collected on Leukosorb strips can also be used to determine the presence of respiratory pathogens such as influenza virus and antibiotic-resistant bacteria, such S. aureus, thus making it a potentially valuable method for assessing diseases within a given population and studying the airway microbiome. Leukosorb strips obtained from subjects before and after administration of LAIV (a well-controlled diseased population mimic because it replicates only in the upper airway) tested positive for viral RNA on the day following LAIV vaccine (Fig. 5B) (28). Notably, the samples obtained using the ELF collection method yielded similar RNA levels compared with NLF cell collection, highlighting the fact that ELF allows measurement of shed virus particles present in the nasal mucosa. These samples were also used to detect the presence of the mecA gene, which is commonly associated with antibiotic-resistant S. aureus, a pathogen known to colonize the nasal passages in ~80% of the population (20–30% being permanent carriers and 60% intermittent) (8, 23). Two out of the six ELF samples tested positive for mecA expression, which is within the range of antibiotic-resistant S. aureus found in epidemiological studies (1, 12, 13, 18), whereas none were detectable in NLF cells (Fig. 5C). Assessing the nasal mucosa for respiratory pathogens has clinical relevance because the nose is the predominant entry point for many respiratory pathogens, including viruses and bacteria (5), demonstrating that the ELF method can be used to assess immune status or the presence of pathogens in diseased populations. The ability to detect bacterial genes from ELF also suggests that ELF could be used to assess the microbiome and potentially the mycobiome. As such, local changes of inflammatory mediators and microbiota and mycobiota within the nasal mucosa may be evaluated similarly to our method as early indicators of disease (34).
In conclusion, Leukosorb sampling of nasal ELF may be a reliable and reproducible method to assess biomarkers of innate immune status in the airway that results in more concentrated and less variable sample of the nasal mucosa than with traditional NL. Furthermore, this method may aid in the detection of pathogens such as influenza virus and antibiotic-resistant bacteria within the nasal mucosa. Validation of the novel ELF collection method also suggests that samples can be stored in a variety of conditions without compromising sample quality and biomarker detectability. Taken together, these data indicate that ELF collected via Leukosorb strip is effective in assessing nasal mucosal biomarkers of innate immune status and will be useful in both field studies and in populations in which traditional NL is more difficult to reliably obtain and store.
GRANTS
This work was supported by National Institutes of Health (NIH) Grants P50 HL-12010004, T32 ES-00712634, and R01 ES-013611). Research reported in this publication was in part supported by NIH and the Food and Drug Administration (FDA) Center for Tobacco Products. The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH or the FDA.
DISCLOSURES
No conflicts of interest, financial or otherwise, are declared by the authors.
AUTHOR CONTRIBUTIONS
M.E.R., A.M.S., P.W.C., and I.J. conceived and designed research; M.E.R. and A.M.S. performed experiments; M.E.R., A.M.S., and I.J. analyzed data; M.E.R., A.M.S., P.W.C., and I.J. interpreted results of experiments; M.E.R., A.M.S., and I.J. prepared figures; M.E.R., A.M.S., P.W.C., and I.J. drafted manuscript; M.E.R., A.M.S., P.W.C., and I.J. edited and revised manuscript; M.E.R., A.M.S., P.W.C., and I.J. approved final version of manuscript.
ACKNOWLEDGMENTS
We thank the Periodontal Microbiology Laboratory at the Ohio State University for suggesting the tube-with-a-hole method for facilitating elution of ELF from Leukosorb strips. We also thank Dr. Brian Button for use of the laser cutter.
REFERENCES
- 1.Abou Shady HM, Bakr AE, Hashad ME, Alzohairy MA. Staphylococcus aureus nasal carriage among outpatients attending primary health care centers: a comparative study of two cities in Saudi Arabia and Egypt. Braz J Infect Dis 19: 68–76, 2015. doi: 10.1016/j.bjid.2014.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alexis NE, Hu SC, Zeman K, Alter T, Bennett WD. Induced sputum derives from the central airways: confirmation using a radiolabeled aerosol bolus delivery technique. Am J Respir Crit Care Med 164: 1964–1970, 2001. doi: 10.1164/ajrccm.164.10.2104051. [DOI] [PubMed] [Google Scholar]
- 3.Atlas LT. Description of apparatus and method for obtaining nasal washings. J Lab Clin Med 32: 1016–1023, 1947. [PubMed] [Google Scholar]
- 4.Aziz N, Nishanian P, Mitsuyasu R, Detels R, Fahey JL. Variables that affect assays for plasma cytokines and soluble activation markers. Clin Diagn Lab Immunol 6: 89–95, 1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Beule AG. Physiology and pathophysiology of respiratory mucosa of the nose and the paranasal sinuses. GMS Curr Top Otorhinolaryngol Head Neck Surg 9: Doc07, 2010. doi: 10.3205/cto000071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cassol S, Salas T, Gill MJ, Montpetit M, Rudnik J, Sy CT, O’Shaughnessy MV. Stability of dried blood spot specimens for detection of human immunodeficiency virus DNA by polymerase chain reaction. J Clin Microbiol 30: 3039–3042, 1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chambers AG, Percy AJ, Yang J, Camenzind AG, Borchers CH. Multiplexed quantitation of endogenous proteins in dried blood spots by multiple reaction monitoring-mass spectrometry. Mol Cell Proteomics 12: 781–791, 2013. doi: 10.1074/mcp.M112.022442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chaves-Moreno D, Wos-Oxley ML, Jáuregui R, Medina E, Oxley AP, Pieper DH. Exploring the transcriptome of Staphylococcus aureus in its natural niche. Sci Rep 6: 33174, 2016. doi: 10.1038/srep33174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chawes BL, Edwards MJ, Shamji B, Walker C, Nicholson GC, Tan AJ, Folsgaard NV, Bonnelykke K, Bisgaard H, Hansel TT. A novel method for assessing unchallenged levels of mediators in nasal epithelial lining fluid. J Allergy Clin Immuno 125: 1387–1389.e3, 2010. doi: 10.1016/j.jaci.2010.01.039. [DOI] [PubMed] [Google Scholar]
- 10.Crimmins E, Kim JK, McCreath H, Faul J, Weir D, Seeman T. Validation of blood-based assays using dried blood spots for use in large population studies. Biodemogr Soc Biol 60: 38–48, 2014. doi: 10.1080/19485565.2014.901885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.de Jager W, Bourcier K, Rijkers GT, Prakken BJ, Seyfert-Margolis V. Prerequisites for cytokine measurements in clinical trials with multiplex immunoassays. BMC Immunol 10: 52, 2009. doi: 10.1186/1471-2172-10-52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.DeLeo FR, Otto M, Kreiswirth BN, Chambers HF. Community-associated methicillin-resistant Staphylococcus aureus. Lancet 375: 1557–1568, 2010. doi: 10.1016/S0140-6736(09)61999-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Duran N, Yildirim Y, Duran GG, Pasa O, Kilinc C, Yildirim I, Eryilmaz N, Bayraktar S. Virulence factors in staphylococci isolated from nasal cavities of footballers. Am J Med Sci 351: 279–285, 2016. doi: 10.1016/j.amjms.2015.12.016. [DOI] [PubMed] [Google Scholar]
- 14.Feneley A, Rainer M, Haider F, Narayanswamy B, Stenning J, Mordin C, Greenaway S, Clarke G. The nose as a research tool: intra-subject variability in nasal sampling. Eur Respir J 38, Suppl 55: 4071, 2011. http://erj.ersjournals.com/content/38/Suppl_55/p4071. [Google Scholar]
- 15.Følsgaard NV, Chawes BL, Rasmussen MA, Bischoff AL, Carson CG, Stokholm J, Pedersen L, Hansel TT, Bønnelykke K, Brix S, Bisgaard H. Neonatal cytokine profile in the airway mucosal lining fluid is skewed by maternal atopy. Am J Respir Crit Care Med 185: 275–280, 2012. doi: 10.1164/rccm.201108-1471OC. [DOI] [PubMed] [Google Scholar]
- 16.Grüner N, Stambouli O, Ross RS. Dried blood spots—preparing and processing for use in immunoassays and in molecular techniques. J Vis Exp 13: 2015. doi: 10.3791/52619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hentschel J, Müller U, Doht F, Fischer N, Böer K, Sonnemann J, Hipler C, Hünniger K, Kurzai O, Markert UR, Mainz JG. Influences of nasal lavage collection-, processing- and storage methods on inflammatory markers—evaluation of a method for non-invasive sampling of epithelial lining fluid in cystic fibrosis and other respiratory diseases. J Immunol Methods 404: 41–51, 2014. doi: 10.1016/j.jim.2013.12.003. [DOI] [PubMed] [Google Scholar]
- 18.Hernandez DR, Newton DW, Ledeboer NA, Buchan B, Young C, Clark AE, Connoly J, Wolk DM. Multicenter evaluation of MRSASelect II chromogenic agar for identification of methicillin-resistant Staphylococcus aureus from wound and nasal specimens. J Clin Microbiol 54: 305–311, 2016. doi: 10.1128/JCM.02410-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Horvath KM, Herbst M, Zhou H, Zhang H, Noah TL, Jaspers I. Nasal lavage natural killer cell function is suppressed in smokers after live attenuated influenza virus. Respir Res 12: 102, 2011. doi: 10.1186/1465-9921-12-102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jackson D, Clements Y, Johnston S, Hansel T. P178 validation of a novel synthetic absorptive matrix (SAM) for sampling nasal mucosal lining fluid. Thorax 65, Suppl 4: A153–A153, 2010. doi: 10.1136/thx.2010.151043.29. [DOI] [Google Scholar]
- 21.Jaspers I, Ciencewicki JM, Zhang W, Brighton LE, Carson JL, Beck MA, Madden MC. Diesel exhaust enhances influenza virus infections in respiratory epithelial cells. Toxicol Sci 85: 990–1002, 2005. doi: 10.1093/toxsci/kfi141. [DOI] [PubMed] [Google Scholar]
- 22.Jaspers I, Zhang W, Fraser A, Samet JM, Reed W. Hydrogen peroxide has opposing effects on IKK activity and IkappaBalpha breakdown in airway epithelial cells. Am J Respir Cell Mol Biol 24: 769–777, 2001. doi: 10.1165/ajrcmb.24.6.4344. [DOI] [PubMed] [Google Scholar]
- 23.Kluytmans J, van Belkum A, Verbrugh H. Nasal carriage of Staphylococcus aureus: epidemiology, underlying mechanisms, and associated risks. Clin Microbiol Rev 10: 505–520, 1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lehmann S, Delaby C, Vialaret J, Ducos J, Hirtz C. Current and future use of “dried blood spot” analyses in clinical chemistry. Clin Chem Lab Med 51: 1897–1909, 2013. doi: 10.1515/cclm-2013-0228. [DOI] [PubMed] [Google Scholar]
- 25.Lipiäinen T, Peltoniemi M, Sarkhel S, Yrjönen T, Vuorela H, Urtti A, Juppo A. Formulation and stability of cytokine therapeutics. J Pharm Sci 104: 307–326, 2015. doi: 10.1002/jps.24243. [DOI] [PubMed] [Google Scholar]
- 26.Lü FX, Esch RE. Novel nasal secretion collection method for the analysis of allergen specific antibodies and inflammatory biomarkers. J Immunol Methods 356: 6–17, 2010. doi: 10.1016/j.jim.2010.03.004. [DOI] [PubMed] [Google Scholar]
- 27.Martin EM, Clapp PW, Rebuli ME, Pawlak EA, Glista-Baker E, Benowitz NL, Fry RC, Jaspers I. E-cigarette use results in suppression of immune and inflammatory-response genes in nasal epithelial cells similar to cigarette smoke. Am J Physiol Lung Cell Mol Physiol 311: L135–L144, 2016. doi: 10.1152/ajplung.00170.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Noah TL, Zhang H, Zhou H, Glista-Baker E, Müller L, Bauer RN, Meyer M, Murphy PC, Jones S, Letang B, Robinette C, Jaspers I. Effect of broccoli sprouts on nasal response to live attenuated influenza virus in smokers: a randomized, double-blind study. PLoS One 9: e98671, 2014. doi: 10.1371/journal.pone.0098671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Noah TL, Zhou H, Jaspers I. Alteration of the nasal responses to influenza virus by tobacco smoke. Curr Opin Allergy Clin Immunol 12: 24–31, 2012. doi: 10.1097/ACI.0b013e32834ecc80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Noah TL, Zhou H, Monaco J, Horvath K, Herbst M, Jaspers I. Tobacco smoke exposure and altered nasal responses to live attenuated influenza virus. Environ Health Perspect 119: 78–83, 2011. doi: 10.1289/ehp.1002258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Noah TL, Zhou H, Zhang H, Horvath K, Robinette C, Kesic M, Meyer M, Diaz-Sanchez D, Jaspers I. Diesel exhaust exposure and nasal response to attenuated influenza in normal and allergic volunteers. Am J Respir Crit Care Med 185: 179–185, 2012. doi: 10.1164/rccm.201103-0465OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Scadding GW, Calderon MA, Bellido V, Koed GK, Nielsen N-C, Lund K, Togias A, Phippard D, Turka LA, Hansel TT, Durham SR, Wurtzen PA. Optimisation of grass pollen nasal allergen challenge for assessment of clinical and immunological outcomes. J Immunol Methods 384: 25–32, 2012. doi: 10.1016/j.jim.2012.06.013. [DOI] [PubMed] [Google Scholar]
- 33.Villar LM, de Oliveira JC, Cruz HM, Yoshida CF, Lampe E, Lewis-Ximenez LL. Assessment of dried blood spot samples as a simple method for detection of hepatitis B virus markers. J Med Virol 83: 1522–1529, 2011. doi: 10.1002/jmv.22138. [DOI] [PubMed] [Google Scholar]
- 34.Vissers M, de Groot R, Ferwerda G. Severe viral respiratory infections: are bugs bugging? Mucosal Immunol 7: 227–238, 2014. doi: 10.1038/mi.2013.93. [DOI] [PubMed] [Google Scholar]
- 35.Woolhouse IS, Bayley DL, Stockley RA. Effect of sputum processing with dithiothreitol on the detection of inflammatory mediators in chronic bronchitis and bronchiectasis. Thorax 57: 667–671, 2002. doi: 10.1136/thorax.57.8.667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wright HL, Moots RJ, Bucknall RC, Edwards SW. Neutrophil function in inflammation and inflammatory diseases. Rheumatology (Oxford) 49: 1618–1631, 2010. doi: 10.1093/rheumatology/keq045. [DOI] [PubMed] [Google Scholar]
- 37.Zeilhofer HU, Schorr W. Role of interleukin-8 in neutrophil signaling. Curr Opin Hematol 7: 178–182, 2000. doi: 10.1097/00062752-200005000-00009. [DOI] [PubMed] [Google Scholar]