Abstract
Tissue extracellular matrix (ECM) provides structural support and creates unique environments for resident cells (Bateman JF, Boot-Handford RP, Lamandé SR. Nat Rev Genet 10: 173–183, 2009; Kjaer M. Physiol Rev 84: 649–98, 2004). However, the identities of cells responsible for creating specific ECM components have not been determined. In striated muscle, the identity of these cells becomes important in disease when ECM changes result in fibrosis and subsequent increased tissue stiffness and dysfunction. Here we describe a novel approach to isolate and identify cells that maintain the ECM in both healthy and fibrotic muscle. Using a collagen I reporter mouse, we show that there are three distinct cell populations that express collagen I in both healthy and fibrotic skeletal muscle. Interestingly, the number of collagen I-expressing cells in all three cell populations increases proportionally in fibrotic muscle, indicating that all cell types participate in the fibrosis process. Furthermore, while some profibrotic ECM and ECM-associated genes are significantly upregulated in fibrotic muscle, the fibrillar collagen gene expression profile is not qualitatively altered. This suggests that muscle fibrosis in this model results from an increased number of collagen I-expressing cells and not the initiation of a specific fibrotic collagen gene expression program. Finally, in fibrotic muscle, we show that these collagen I-expressing cell populations differentially express distinct ECM proteins—fibroblasts express the fibrillar components of ECM, fibro/adipogenic progenitors cells differentially express basal laminar proteins, and skeletal muscle progenitor cells differentially express genes important for the satellite cell.
Keywords: collagen, extracellular matrix, fibrosis, skeletal muscle
chronic skeletal muscle pathologies such as muscular dystrophy and cerebral palsy as well as acute conditions such as trauma are often characterized by skeletal muscle fibrosis (33, 38, 61). Muscle fibrosis is defined as the abnormal accumulation of extracellular matrix (ECM) protein, primarily in the form of type I collagen (33). Skeletal and cardiac muscle fibrosis are significant clinical problems that result when regeneration pathways are disrupted and healthy muscle is replaced by fibrous connective tissue. The resulting fibrous scar reduces muscle strength, decreases cardiac function and limits joint range of motion and mobility of patients (6, 32, 68).
Muscle fibrosis is typically characterized by biochemical and histological detection of collagen in the extracellular space (13, 33, 43). More recently, the characterization of fibrosis by mechanical testing has provided functional definitions based on specific values of tissue stiffness (13, 43, 49, 56). While various methods used to describe ECM and fibrosis provide valuable insights, less is known about the cells responsible for the production of fibrotic scars, especially in skeletal muscle. A thorough understanding of the cells involved is vital to developing effective therapies against fibrosis. Traditionally, attention has focused on fibroblasts and myofibroblasts, as these cells are a primary source of collagen in fibrosis (7, 18, 21, 33, 63). However, other mononuclear cells exist in skeletal muscle that may be involved in fibrosis. Mainly, it has been shown that the fibro/adipogenic progenitor (FAP) cell becomes activated upon muscle injury (26, 58). Additionally, myoblasts have been implicated, albeit to a lesser extent, in producing collagen I and contributing to muscle fibrosis (1). The contribution of different cell types to collagen production in both healthy and diseased muscle is thus unclear.
We used a strategy that labeled all collagen I-expressing cells and determined their identity using fluorescence-activated cell sorting (FACS) and RNA-sequencing techniques. In addition to the identity of these cells not being well known, it is also unclear how collagen-expressing cells differ in a healthy versus a fibrotic environment in terms of gene expression. Therefore, the purpose of this study was to create a model to label cells responsible for collagen deposition directly in both healthy and diseased muscle to gain further understanding of ECM regulation in skeletal muscle.
MATERIALS AND METHODS
Mice.
A model of chronic skeletal muscle fibrosis, the nesprin-desmin double knockout (DKO) mouse (strain: mixed Black Swiss and 129/SvJ background), was used (13). Expression of collagen I, the main contributor to fibrotic tissue in skeletal muscle, was identified using a mouse line expressing GFP under the control of the collagen-α1(I) promoter (strain: C3H/C57B1) (64). This permitted direct identification of any cell that actively expresses type I collagen based on fluorescence. To study collagen-expressing cells in a model of fibrosis, we created nesprin-desmin double knockout mice that express GFP under the control of the collagen-α1(I) promoter by crossing these two mouse lines. Mice heterozygous for both nesprin 1 and desmin were bred with collagen-GFP mice. The resulting male and female nesprin 1+/−, desmin+/−, Col-GFP+ mice were bred to create nesprin 1+/+, desmin+/+, Col-GFP+ [wild type (WT)] and nesprin 1−/−, desmin−/−, Col-GFP+ (DKO) littermates. Eight-week-old male and female mice were used for experiments. All experiments were approved by the Animal Care Program at the University of California, San Diego. A total of 16 WT and 14 DKO mice were used as biological replicates in all experiments.
The presence of GFP in each mouse ear punch was determined using a fluorescent microscope (Leica MZFL III, Leica Microsystems, Wetzlar, Germany). Genotyping of nesprin 1 and desmin were performed by PCR on DNA from mouse ear punches using the following primer sets: desmin wild-type (forward, 5′-CTCGTCCAGCCAGCGCGTGT-3′; reverse, 5′-GCCCTTGAGCCGGTTGACCTC-3′), desmin knockout (forward, 5′-TCCGCCCAGCCAGCCTCGTC-3′; reverse, 5′-CCATGGCGATGCCTGCTTGC), nesprin wild-type (forward, 5′-TGGTAGTCATCAAGATGCTGGCTTGGG-3′; reverse, 5′-CTTTCTAAGTCTACAGTGGTGGGCTC-3′), and nesprin knockout (forward, 5′-GTAATATTTGTGGGACCGAGTTCTCTGAG-3′; reverse, 5′-CTTTCTAAGTCTACAGTGGTGGGCTC-3′).
Histology.
Extensor digitorum longus (EDL) muscles were dissected from WT and DKO collagen-GFP mice. Muscles were washed in PBS, pinned and fixed in 0.5% paraformaldehyde at room temperature for 2 h. EDL muscles were then placed in a 20% sucrose solution overnight at 4°C. Muscles were frozen in liquid nitrogen-cooled isopentane and stored at −80°C until further processing. Cross-sectional sections (10 μm thick) were cut from OCT-embedded samples at −20°C using a Microm HM500 cryostat (Waldorf, Germany). Since GFP loses its fluorescence during fixation, an antibody against GFP was used to visualize GFP. Nonspecific binding was blocked with 1% BSA, and tissue sections were stained with a rabbit polyclonal anti-GFP primary antibody (dilution = 1:500; A-11122; Life Technologies, Eugene, OR), followed by staining with an Alexa-Fluor 488 goat anti-rabbit secondary antibody (dilution = 1:200; A-11008; Invitrogen, Eugene, OR). To determine the position of GFP+ cells relative to the basal lamina, a rabbit polyclonal anti-laminin primary antibody (dilution = 1:500; L9393; Sigma-Aldrich, St. Louis, MO) was used. Before laminin labeling, the anti-laminin antibody was conjugated to an Alexa-Fluor (AF) 568 fluorophore using an APEX Antibody Labeling Kit (Life Technologies) according to manufacturer’s instructions. Briefly, APEX antibody labeling tip resin was hydrated using wash buffer (0.1 M PBS, pH 7.5, 2 mM azide) and 20 μg of anti-laminin antibody were loaded onto the resin. The AF-568 fluorophore was dissolved in DMSO and 50 mM borate buffer, and 10 μl of this reactive dye solution were added to the resin and incubated for 2 h at room temperature. To elute unreacted dye, wash buffer was passed through the resin. Labeled anti-laminin was then eluted from the resin using 0.2 M acetic acid, pH 3.3, and neutralized with 1 M Tris, pH 9.0. The AF-568-anti-laminin conjugate was diluted 1:500 in 0.1% BSA and incubated on the GFP-labeled tissue sections for 2 h at room temperature. Nuclei in each section were stained using DRAQ5 (dilution = 1:1,000; no. 4084; Cell Signaling Technology, Danvers, MA). Samples were then imaged under a ×63.3 objective with glycerol immersion using an inverted confocal microscope (Leica SP5, Wetzlar, Germany).
Fluorescence-activated cell sorting.
Mice (n = 16 WT; n = 14 DKO) were anesthetized with 2% isoflurane at 2 l/min and then euthanized by cervical dislocation. Muscle samples were prepared for FACS as previously described (43). The quadriceps, tibialis anterior, and gastrocnemius muscles were dissected and trimmed of all visible tendon. Muscles were weighed, cut into small pieces, and incubated at 37°C in digestive solution (in DMEM: 0.27% type I collagenase, 0.06 U/ml dispase II, 50 U/ml streptomycin, 50 U/ml penicillin) for 50 min. Muscles were then mechanically broken with forceps and then incubated at 37°C for 30 min. Samples were further broken down by pipetting, and then incubated for another 10 min. Muscle cell suspensions were then passed through a 70-μm filter followed by a 40-μm filter. Cells were then centrifuged and resuspended in FACS buffer consisting of 2.5% normal goat serum and 1 mM EDTA in PBS. Isolated cells were then labeled for 20 min on ice with primary antibodies, centrifuged, and resuspended in FACS buffer. Fluorescence-minus-one controls were created by combining cell samples with appropriate antibodies. Additionally, a portion of the cell population was left unstained to serve as the negative control.
Antibodies used were as follows: CD31-Pacific blue (to identify endothelial cells) (102422; Biolegend, San Diego, CA), CD45-Pacific blue (to identify hematopoietic cells) (103126; Biolegend), α-7 integrin-PE [to identify skeletal muscle progenitor cells (SMPs)] (53–0010–05; AbLab, Vancouver, BC, Canada), and Sca-1-APC/Cy [to identify FAPs) (108126; Biolegend). Unlabeled cells (CD31−, CD45−, Sca-1−, α-7 integrin−) were further investigated to determine their identity by isolating them from the hindlimbs of WT mice (n = 5). These cells were fixed in 70% ethanol for 20 min followed by permeabilization in a solution of 2% BSA, 5% FBS, 0.2% Triton X-100, and 0.1% sodium azide. Cells were stained with fibroblast and myofibroblast markers ER-TR7 (9) (sc-73355 PE; Santa Cruz Biotechnology, Dallas, TX) and α-smooth muscle actin (α-SMA) (20) (LS-C124868; LSBio, Seattle, WA), respectively, both conjugated to the phycoerythrin (PE) fluorophore), and analyzed by flow cytometry. Dilution for all FACS antibodies was 1:200.
Cells were sorted using the BD FACS Aria II Special Order Research Project (BD Biosciences, San Jose, CA) with four lasers (405 nm, 100 mW; 488 nm, 50 mW; 561 nm, 50 mW; 640 nm, 40 mW). Cells were first gated on size based on side versus forward scatter plots to eliminate cellular debris. Clumped cells were then eliminated by gating on side scatter area versus side scatter width. To eliminate all endothelial (CD31+) and hematopoietic (CD45+) cells, antibodies for both CD31 and CD45 conjugated to Pacific blue were used, and then we excluded all Pacific blue-positive cells. Remaining cells, henceforth referred to as lineage negative (lin−) cells, were gated on GFP expression. GFP+ cells were then sorted into three collection vials based on staining for Sca-1 and α-7 integrin: Sca-1+, α-7 integrin−; Sca-1−, α-7 integrin+; Sca-1−, α-7 integrin−. The fluorescent antibodies were detected at the following wavelengths: 450 ± 25 nm (Pacific blue), 525 ± 25 nm (GFP), 582 ± 7.5 nm (PE), and 780 ± 30 nm (APC/Cy7). GFP+ cells were sorted into three different collection vials based on antibody staining filled with FACS buffer. Cells were centrifuged and lysed in Buffer RLT (Qiagen, Valencia, CA) and TRIzol (Invitrogen, Carlsbad, CA). All data were collected using BD FACSDiva software (BC Biosciences, San Jose, CA) and analyzed with FlowJo software (version 10.0.7, FlowJo, Ashland, OR).
RNA isolation.
To obtain sufficient RNA for RNA sequencing, samples were pooled. WT cell populations were pooled from three different mice, and DKO populations were pooled from two different mice. RNA was then extracted from these pooled samples using an RNeasy Mini Kit (Qiagen). After cell lysis with 1.0 ml of Buffer RLT/TRIzol, 0.3 ml of chloroform were added and the sample was vortexed for 15 s then centrifuged for 15 min at 12,000 g. The supernatant was removed and mixed with equal parts 70% ethanol. Samples were then added to an RNeasy Mini spin column and centrifuged. The column was washed with RW1 followed by RPE buffer. After washes, DEPC-treated water was added to the column and RNA was eluted from the columns with centrifugation. RNA concentrations were determined by sample absorbance at 260 nm using an ND-1000 Spectrophotometer (Thermo Scientific, Waltham, MA).
RNAseq.
RNA sequencing was performed on Illumina Hi-seq 2500 (rapid run mode). Four biological replicates for each cell type [FAP, SMP and fibroblast (FB)] under each condition (DKO and WT) were multiplexed across two lanes (total samples = 24). One hundred base-paired reads per sample with an average of 22 million reads per sample were obtained. RNA read mapping and the initial quality control was performed using the OSA algorithm (24) to the mouse reference genome version mm10, with Refseq annotation. Four samples (2 SMP DKO and 2 SMP WT) with <75% uniquely mapped reads were eliminated from further analysis. All RNA sequencing data generated for this study have been deposited in NCBI’s Gene Expression Omnibus (GEO) (19) repository and can be accessed using the GEO Series accession no. GSE89633 (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE89633).
The BAM files obtained after RNA read mapping for each of the 20 samples were used to construct the counts matrix using the “SummarizeOverlaps” function of the “GenomicAlignments” package available through R/Bioconductor (29). The “makeTranscriptDbFromUCSC” from the “GenomicFeatures” package was used to build the gene model (corresponding to refgene annotation under version mm10) for counting reads (29). The resulting count matrix consisted of 24,393 genes across 20 samples.
Count matrix normalization and differential analysis was performed using the “DEseq2” package available through R/Bioconductor as outlined in their vignette (36). DESeq models count data as negative binomial distribution, with variance and mean linked by local regression (2). DESeq2 builds on the previous package by utilizing shrinkage estimate for dispersions and fold changes, improving stability and interpretability. The design matrix for our analysis was constructed to accommodate all possible comparisons across cell types and conditions, resulting in nine possible comparisons. An absolute log2-based-fold change cut-off >0.5 with a Benjamini-Hochberg false adjusted P < 0.05 was used as a threshold of significance for identifying differential expression, for each comparison. Heatmaps were generated using “pheatmap” library in R. Functional enrichment analysis of differentially expressed genes was performed using an open source tool–Database for Annotation, Visualization and Integrated Discovery (DAVID) (25). The colors on the heatmap represent the expression counts scaled (to one), row-wise using the built-in “scale” function available through “pheatmap,” ECM and ECM-related genes were identified by cross-referencing all of the differentially expressed genes in this study with the “Matrisome” ECM annotations proposed by Hynes and collaborators (44). The matrisome is split into the “core matrisome” (collagens, ECM glycoproteins, and roteoglycans) and “matrisome-associated” genes (ECM regulators, secreted factors, and ECM-affiliated proteins). Comparisons were made between cell types within a genotype (WT:FB vs. FAP vs. SMP; DKO: FB vs. FAP vs. SMP) and among the same cell types in each genotype (WT FBs vs. DKO FBs; WT FAPs vs. DKO FAPs; WT SMPs vs. DKO SMPs).
RESULTS
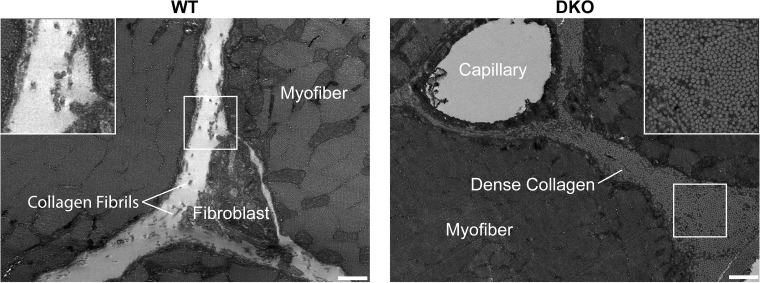
Our work demonstrated previously that significant skeletal muscle fibrosis resulted when both cytoskeletal proteins desmin and nesprin 1 were knocked out (DKO mice) (13). In the present study, we used transmission electron microscopy to further characterize ECM changes in DKO skeletal muscle (Fig. 1). These images confirm our previous findings and clearly demonstrate a significant alteration to the ECM in DKO skeletal muscle. The amount and quality of the collagen fibrils is clearly increased in the DKO model as previously suggested by the biochemical data (12, 13).
Fig. 1.
Extensive fibrosis is present in double knockout (DKO) skeletal muscle. Transverse transmission electron micrographs of the extracellular space of wild-type (WT) and nesprin-1/desmin DKO (13) tibialis anterior muscle show a significant increase in collagen content in DKO muscle. Inset shows that DKO collagen is more densely packed compared with WT collagen, with collagen fibrils roughly aligned with the muscle fiber long axis. Scale, 500 nm.
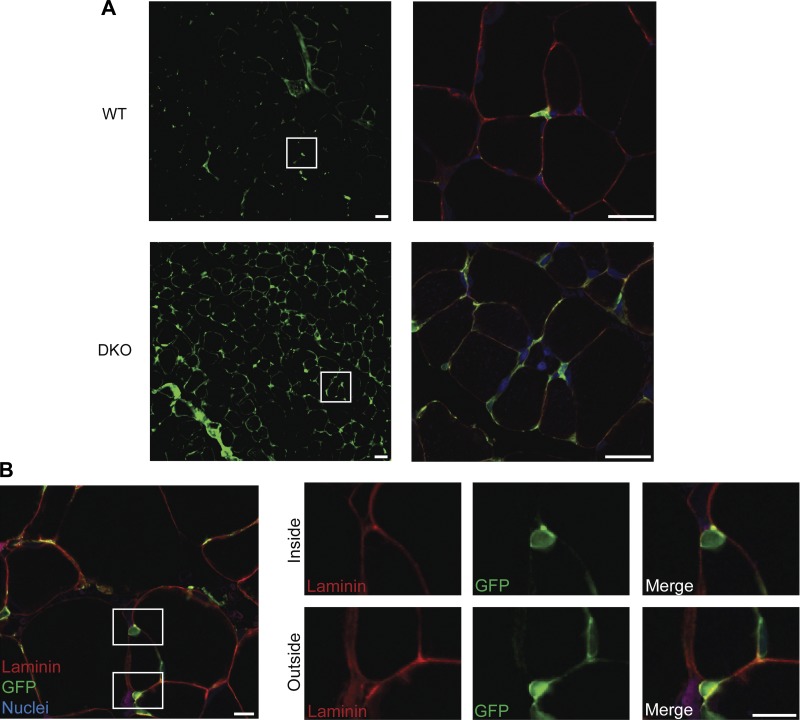
To provide insights into the cellular mechanism behind this ECM alteration, we bred DKO mice with collagen I reporter mice (64). These reporter mice allow direct identification of collagen expressing cells based on green fluorescence. Compared with healthy WT muscle, DKO muscles had an increase in GFP+ collagen-expressing cells (Fig. 2A). In addition to the increased number of collagen-expressing cells in DKO muscle, the GFP+ cells in fibrotic DKO muscle also had extensive cellular projections, a possible indication of being in an activated state (5, 38). Importantly, we observed collagen-expressing cells that were located both beneath and outside of the basal lamina in WT and DKO muscle (Fig. 2B). This shows that the collagen-expressing cell population is heterogeneous and consists of mononuclear cells associated with both the skeletal muscle fibers and the extracellular matrix. We thus performed FACS to determine the identities of collagen-expressing cells and show how the collagen expressing cell populations are altered in fibrosis.
Fig. 2.
Morphological evidence of increased type I collagen-expressing cells. A: collagen-expressing cells are more abundant in DKO skeletal muscle sections compared with WT. White boxes in the left panel correspond to the area in the right panel. In addition to an increased number of collagen expressing cells in DKO muscle, high-magnification images reveal an increase in cellular projections of collagen-expressing cells in DKO muscle. Scale, 25 μm. B: collagen-expressing cells (GFP+) were found both inside and outside of the muscle cell. White boxes in left figure depict enlarged areas on the right. In right figure, the top panel clearly shows a GFP+ collagen-expressing cell lying completely beneath the basal lamina. The bottom panel illustrates a collagen-expressing cell outside of the basal lamina nestled between two muscle fibers. This shows that collagen-expressing cells are a heterogeneous cell population. Scale, 10 μm. Red = laminin, green = GFP+ collagen cells and blue = DRAQ5 nuclear staining.
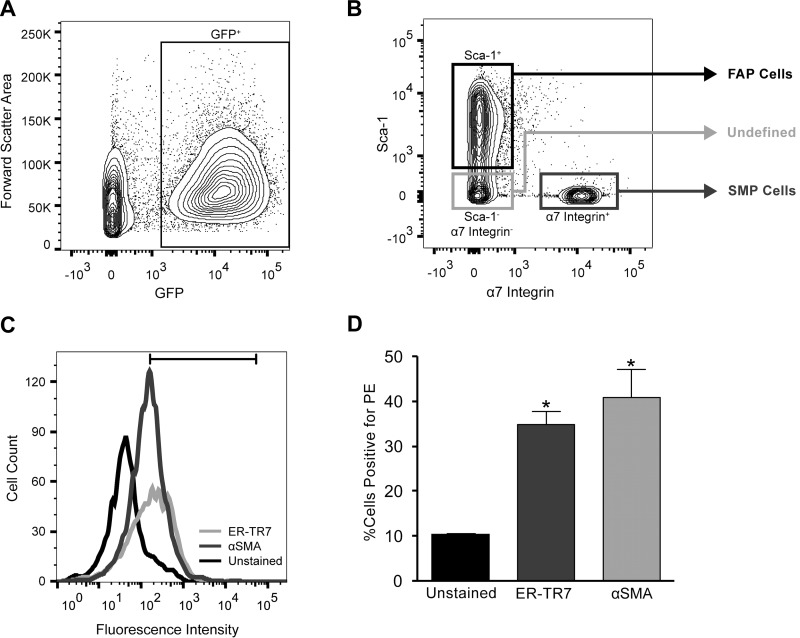
Collagen-expressing mononuclear cells isolated from WT and DKO muscles were identified by the presence of GFP (Fig. 3A). On the basis of labeling with Sca-1 and α-7 integrin, GFP+ cells were identified as either SMPs (CD31−, CD45−, Sca-1− α-7 integrin+) or FAPs (CD31−, CD45−, Sca-1+, α-7 integrin−) (Fig. 3B). These same cell population definitions have been previously validated (26). A portion of the collagen-expressing cells remained unlabeled. We hypothesized that the unstained cells were likely fibroblasts, but to characterize this population, these cells were stained with additional markers. A large proportion of this population stained positively for either ER-TR7 or α-SMA, suggesting that this population is primarily composed of fibroblasts and activated fibroblasts, which will be referred to as FB cells (Fig. 3, C and D). Since PE was used to separate cell populations in the first sort, there was small amount of PE detected in the unlabeled negative control population (Fig. 3, C and D).
Fig. 3.
Cell sorting strategy of collagen-expressing cells from skeletal muscle reveals three distinct cell populations. Quadriceps, tibialis anterior, and gastrocnemius muscles were dissected from hindlimbs of collagen-GFP reporter mice and prepared for fluorescence-activated cell sorting (FACS). Cells were labeled with antibodies against CD31 (Pacific blue), CD45 (Pacific blue), α-7 integrin (PE), and Sca-1 (APC/Cy7). Cells were gated first on the basis of size using forward scatter–area versus side scatter–area plots to eliminate tissue and cellular debris. Single cells were then subjected to a lineage negative (lin−) gate to eliminate CD31 (endothelial cells) and CD45 (hematopoietic cells) positive cells. A: these lineage negative cells were then gated on the basis of GFP expression level. B: finally, the GFP+ cell population was divided into three separate collection vials for later processing: GFP+, α-7 integrin+, Sca-1− [skeletal muscle progenitor (SMP) cells]; GFP+, α-7 integrin−, Sca-1+ [fibro/adipogenic progenitor (FAP) cells]; GFP+, α-7 integrin−, Sca-1−. To further characterize the cell population that was negative for both Sca-1 and α-7 integrin, cells were fixed and stained with fibroblast and myofibroblasts markers, ER-TR7 and α-smooth muscle actin (SMA), respectively. C and D: a large proportion of the GFP+, α-7 integrin−, Sca-1− cell population stained positively for either ER-TR7 and α-SMA, suggesting that this population is primarily composed of fibroblasts and activated fibroblasts (FB).
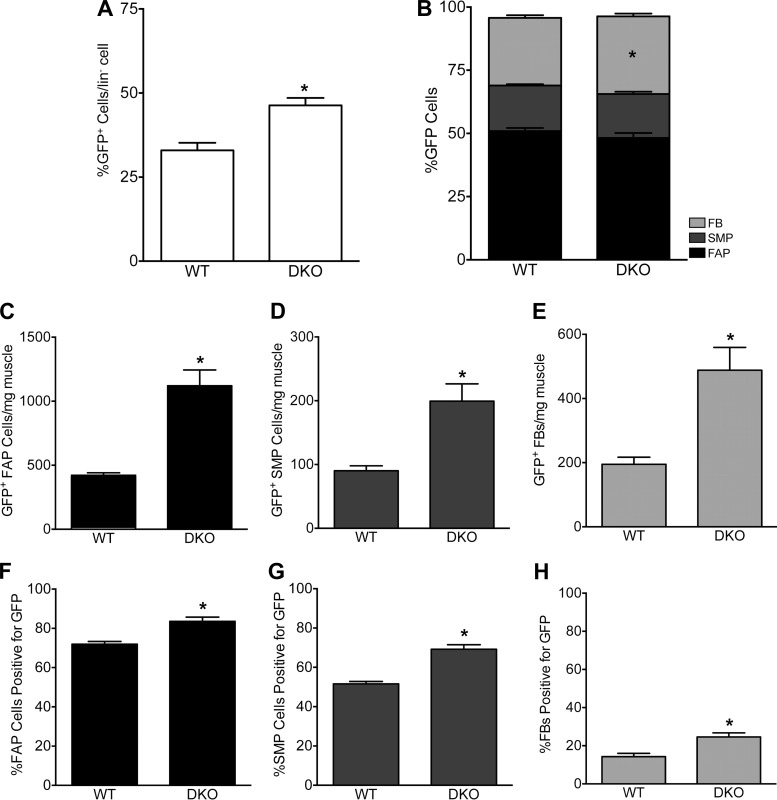
When collagen-expressing cells are examined as a whole, FACS data demonstrated that the proportion of GFP+ cells that make up the mononuclear cell population was significantly increased in DKO mice (46.3 ± 2.25%) compared with WT mice (33.0 ± 2.27%; P = 0.0003; Fig. 4A). Within this GFP+ cell population, we identified FAPs, SMPs, and FBs. GFP+ FAPs made up the largest population of GFP+ cells in both WT and DKO animals, with ~50% of GFP+ cells being positive for Sca-1 (P = 0.15; Fig. 4B). There were also no differences in the proportion of GFP+ cells that were SMPs which made up ~18% of the WT and DKO GFP+ cell population (P = 0.56; Fig. 4B). The remaining GFP+ cells (FBs) comprised ~30% of the GFP+ cell population in WT and DKO mice (P = 0.017; Fig. 4B). FBs made up a slightly higher percentage of collagen-expressing cells in DKO mice, suggesting a small shift toward FBs in fibrotic muscle. However, with the significant increase in the numbers of GFP+ cells in DKO mice relative to WT mice (Fig. 4A), the proportion of each cell type within the GFP+ population remained relatively constant.
Fig. 4.
Multiple collagen I-expressing cell types increase in number to produce fibrosis. A: the percentage of collagen-expressing cells among lin− cells increases with fibrosis. In fibrotic skeletal muscle, the percentage of collagen-expressing cells among lin− cells was significantly elevated compared with WT mice (P = 0.0003, unpaired t-test). B: distribution of collagen-expressing cells among FAPs, SMPs, and FBs remains relatively constant between WT and DKO muscle, where the majority of GFP+ cells were FAPs. The proportion of skeletal muscle progenitor cells (α-7 integrin+) and fibro/adipogenic progenitor cells (Sca-1+) positive for GFP was constant between WT and DKO mice (P = 0.56 and P = 0.15, respectively). There was a significant increase in the proportion of FBs (GFP+, Sca-1−, α-7 integrin−) in DKO over WT muscle (P = 0.017). C–E: the concentration of GFP+ cells (cells/mg muscle) increased in all cell types (P < 0.0001). F–H: increased percentage of SMPs, FAPs and FBs are GFP+, and thus expressing collagen I, in fibrotic skeletal muscle. F: percentage of SMPs expressing collagen was significantly elevated in DKO skeletal muscle compared with WT (P < 0.0001). Over 50% of SMPs in both WT and DKO muscle are GFP+. G: the percentage of FAPs cells positive for GFP is elevated in DKO muscles over WT (P < 0.0001). Note that the vast majority of FAPs in both healthy and fibrotic skeletal muscle are actively expressing collagen (GFP+). H: percentage of FBs that express collagen increases in DKO compared with WT muscle (P < 0.001). All figures: n = 16 (WT); n = 14 (DKO).
The abundance of GFP- cells in WT and DKO mice was not significantly affected (1,914 ± 135 cells/mg vs. 2,329 ± 287 cells/mg, respectively; P = 0.18). Thus, although the GFP− cell count in WT and DKO mice was constant, the cell counts per muscle mass (i.e., cellular concentration) of GFP+ FAPs, SMPs, and FBs all significantly increased in DKO mice compared with WT (Fig. 4, C−E). The concentration of GFP+ FAPs more than doubled in DKO muscle relative to WT muscle (Fig. 4C, P < 0.0001). The concentration of SMPs and FBs positive for GFP more than doubled as well (Fig. 4, D and E, P < 0.0001). Although DKO skeletal muscles were significantly smaller than WT muscles (304.7 ± 17.6 mg vs. 398.1 ± 27.55 mg, respectively; P < 0.05), the 20% drop in muscle mass did not account for the >120% increase in GFP+ cell concentrations in DKO muscle. These data demonstrate that the dramatic rise in collagen content in DKO muscle is related to an increased number of collagen-expressing FBs, FAPs, and SMPs. Additionally, these data demonstrate the heterogeneity of collagen I-expressing cells in both healthy and fibrotic muscle. Furthermore, while the abundance of GFP+ cells increased in DKO mice, the proportion of FAPs, SMPs, and FBs that comprise the GFP+ cell population did not overwhelmingly shift, demonstrating that all three populations participated proportionally in the fibrotic response.
We next examined the FAP, SMP, and FB cell populations individually to determine how collagen expression in these populations was affected in fibrosis. When examining the FAP population further, we found a significant increase in the percentage of FAP cells positive for GFP in DKO (83.6 ± 2.13%) versus WT (71.9 ± 1.4%; P < 0.0001) mice (Fig. 4F). This was also found in SMP cells positive for GFP in DKO (69.2 ± 2.33%) versus WT (51.6 ± 1.18%; P < 0.0001) skeletal muscle (Fig. 4G). The proportion of FB cells positive for GFP was also increased in DKO (24.6 ± 2.16%) versus WT (14.3 ± 1.68%; P < 0.001) mice (Fig. 4H). These data suggest that more FAP, SMP and FB cells are being recruited to express collagen in DKO muscle.
To understand differential function among these cell types, gene expression in each cell type was examined from both WT and DKO muscles using RNA-seq. When comparing the number of differentially expressed genes, the primary difference observed was between cell types (FB vs. FAP vs. SMP) and not between the same cell types in WT versus DKO (Table 1 and Supplemental Table S1; Supplemental Material for this article is available at the journal website). Ontology analysis of differentially expressed genes revealed a strong enrichment for cellular components associated with the ECM and the extracellular region for all comparisons made (P < 0.05; Supplemental Table S1).
Table 1.
Summary of all differentially regulated genes and matrisome genes between cell types and genotypes
| Genotype | SMP vs. FAP | SMP vs. FB | FAP vs. FB |
|---|---|---|---|
| All Genes | |||
| WT | 4462 | 4047 | 1912 |
| DKO | 3677 | 3760 | 1499 |
| Matrisome Genes | |||
| WT | 398 | 386 | 269 |
| DKO | 369 | 354 | 237 |
| Cell Type | WT vs. DKO |
|---|---|
| All Genes | |
| SMP | 303 |
| FAP | 133 |
| FB | 356 |
| Matrisome Genes | |
| SMP | 44 |
| FAP | 35 |
| FB | 74 |
SMP, skeletal muscle progenitor; FAP, fibro/adipogenic progenitor; FB, fibroblast; WT, wild type; DKO, double knockout.
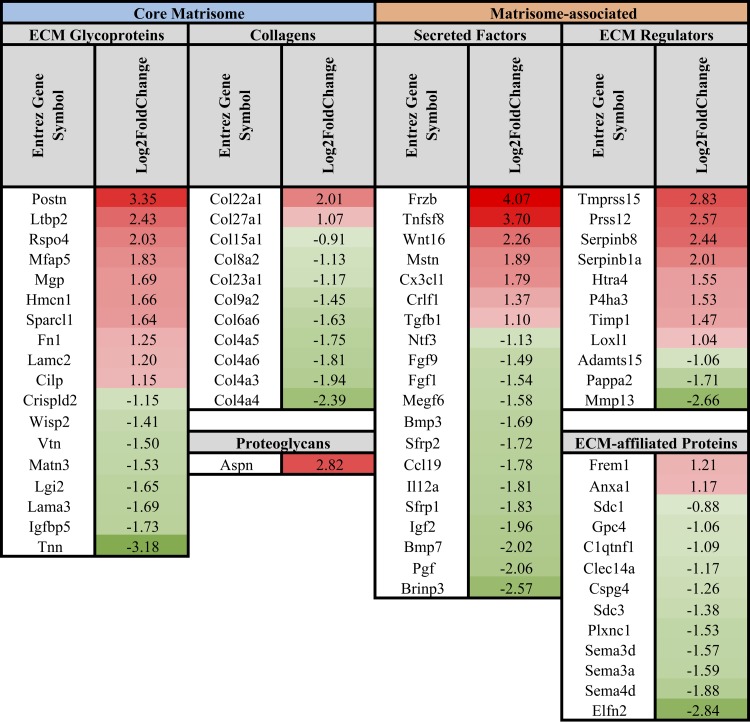
Differentially expressed genes between WT and DKO FBs, FAPs, and SMPs were cross-referenced with the matrisome gene list to investigate how ECM gene expression is affected by fibrosis. Numerous ECM and ECM-related genes were affected by fibrosis (Figs. 5–7). Differential gene expression analysis comparing WT with DKO mice demonstrated that fibrosis did not dramatically affect the expression of fibrillar collagen genes (Figs. 5–7). Notably, collagen I gene expression as well as many other fibrillar collagen genes (collagens II, III, V, XXIV) were not differentially expressed in each cell type when comparing cells from WT and DKO muscles (log2 fold change < 0.5 and Benjamini-Hochberg false adjusted P > 0.05). A scaled expression heatmap of these genes, as well as other fibrillar and nonfibrillar genes, can be found in Supplemental Table S1 (tab 29), demonstrating how a majority of fibrillar collagen genes are not significantly affected by fibrosis. While differential expression of fibrillar collagen genes did not dominate fibrosis development in this model, differential gene expression in FBs favored fibrillar ECM formation in DKO muscle (Fig. 5). This is accomplished by decreased expression of nonfibrillar ECM genes (Col4a3, Col4a4, Col4a5, Col4a6, Col8a2, Col15a1, Lama3) (46), increased expression of fibrillar ECM glycoproteins [Postn (35), Ltbp2 (23), Mfap5 (45), Sparcl1 (57), Hmcn1 (62), Rspo4 (65), Fn1 (30)], increased expression of genes important in fibrillar ECM formation [Aspn (42), Frzb, Wnt16, Mstn, Tgfb1) (38)], decreased expression of Tgf-β antagonists [Bmp3 (3), Bmp7 (66), Fgf1 (51)], increased inhibition of collagen I breakdown [Timp1 (10)], and increased expression of genes associated with ECM remodeling [Prss12 (31), Serpinb8 (47), Serpinb1a (47), Loxl1 (16)] (Fig. 5).
Fig. 5.
List of differentially regulated core matrisome and matrisome-associated genes in DKO fibroblasts compared with WT. Differential gene expression in fibroblasts shows shift toward fibrillar ECM gene expression in DKO vs. WT muscle. This is accomplished by decreased expression of nonfibrillar ECM genes, increased expression of fibrillar ECM glycoproteins, increased expression of genes important in fibrillar ECM formation, decreased expression of Tgf-β antagonists, increased inhibition of collagen I breakdown, and increased expression of genes associated with ECM remodeling. Data are presented as log2 fold change in DKO relative to WT fibroblasts.
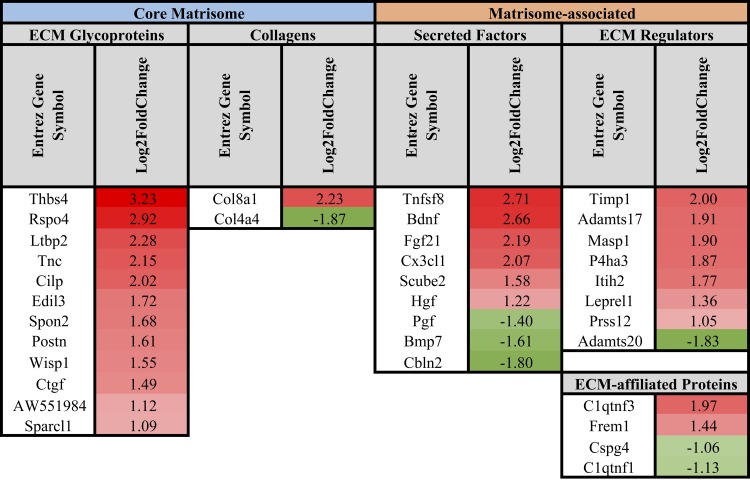
Fig. 7.
List of differentially regulated core matrisome and matrisome-associated genes in DKO skeletal muscle progenitor cells compared with WT. Only a few core matrisome genes were significantly changed in DKO SMPs compared with WT. Similar to FBs and FAPs, SMPs in fibrotic muscle demonstrated an upregulation of ECM remodeling genes. Data are presented as log2 fold change in DKO relative to WT SMPs.
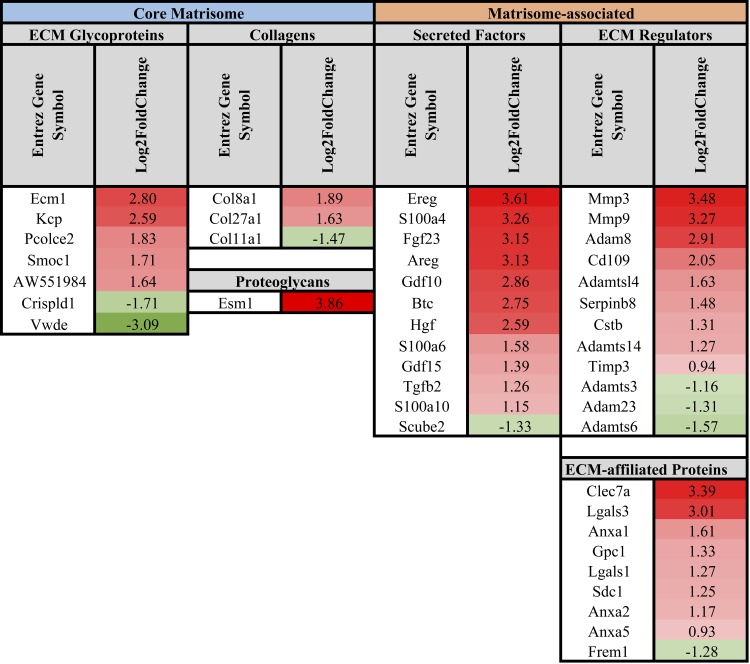
In terms of collagen ECM genes, FAPs in DKO muscle only had two differentially regulated collagen genes when compared with WT FAPs, where Col4a4 was downregulated and Col8a1 was upregulated in DKO compared with WT (Fig. 6). Fewer ECM genes were differentially regulated in FAPs compared with FBs, but similar to FBs, gene expression in FAPs demonstrated a shift in gene expression to favor fibrillar ECM formation in cells from DKO muscle (Fig. 6). This is demonstrated through increased expression of fibrillar ECM glycoproteins [Postn, Ltbp2, Sparcl1, Rspo4, Tnc (21)], increased expression of genes important in fibrillar ECM formation [Ctgf (50), Wisp1 (28)], decreased expression of Tgf-β antagonist (Bmp7), increased inhibition of collagen I breakdown (Timp1), and increased expression of genes associated with ECM remodeling [Masp1 (37), Prss12] (Fig. 6).
Fig. 6.
List of differentially regulated core matrisome and matrisome-associated genes in DKO fibro/adipogenic progenitor cells compared with WT. Gene expression in FAPs demonstrated a shift to favor fibrillar ECM formation in cells from DKO muscle. This is demonstrated through increased expression of fibrillar ECM glycoproteins, increased expression of genes important in fibrillar ECM formation, decreased expression of a Tgf-β antagonist, increased inhibition of collagen I breakdown, and increased expression of genes associated with ECM remodeling. Data are presented as log2 fold change in DKO relative to WT FAPs.
SMPs in DKO muscle showed an upregulation of just two collagen genes (Col8a1 and Col27a1) and downregulation of one (Col11a1) compared with WT SMPs (Fig. 7). In addition to these collagen genes, only a few core matrisome genes were significantly altered (Fig. 7). When examining matrisome-associated genes, similar to both FBs and FAPs, SMPs in fibrotic muscle demonstrated an upregulation of ECM remodeling (Mmp3, Mmp9, Adamtsl4, Serpinb8, Timp3, Adamts14).
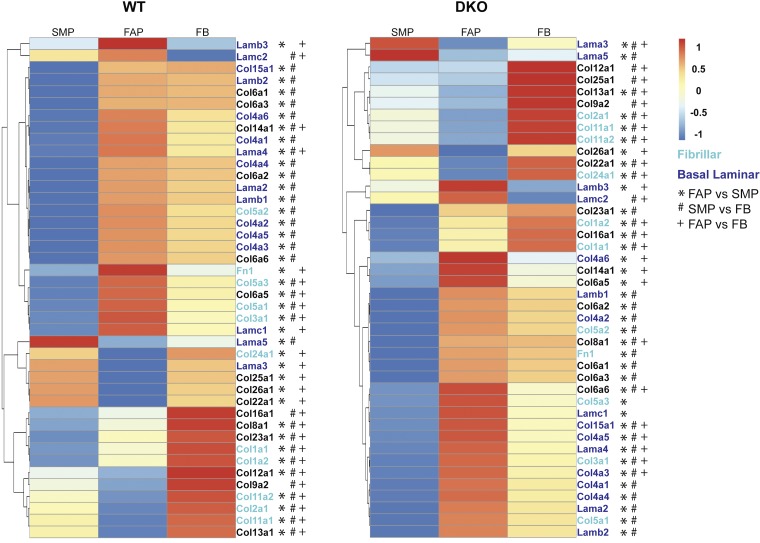
When SMPs versus FAPs versus FBs within each genotype were compared, gene ontology revealed a strong enrichment for genes associated with the ECM (P ≪ 10−10; Supplemental Table S1). With the use of the matrisome database, a further analysis of the differentially expressed genes associated within these comparisons revealed numerous ECM and ECM-related genes to be differentially regulated between cell types (Supplemental Table S1, tabs 11–28). Despite all cells being GFP+, and thus expressing collagen I, RNAseq data showed differential expression of collagen I in both genotypes (Fig. 8). In WT and DKO skeletal muscle, the highest collagen I expression was in the FB cell population, followed by FAPs and then SMP cells. Further analysis of differentially regulated core matrisome ECM genes in SMPs, FAPs, and FBs in DKO animals suggested that each cell type expresses ECM genes for different components of skeletal muscle ECM (Fig. 8 and Supplemental Table S1). FBs showed elevated expression of fibrillar ECM proteins [inter alia: fibrillar collagen genes: Col1a1, Col1a2, Col2a1, Col11a1, Col11a2; fibrillar ECM glycoproteins: Tnc, Postn, Ltbp2, Ctgf, Thbs2, Thbs4; regulators of fibrillar ECM: Lox, Loxl3, Loxl4, Tgfb1, Tgfb3, Pdgfa, Mstn, Frzb; see Supplemental Table S1 (tabs 22 and 28) for a complete list of all differentially regulated core-matrisome and matrisome-associated genes], while FAP cells exhibited elevated expression of basal laminar ECM proteins (inter alia: collagen basement membrane genes: Col4a3, Col4a5, Col4a6, Col15a1; basement membrane proteins: Lamb3, Lama4, Lamc2, Fbn1 (48), Nid2 (46); regulator of basement membrane ECM: Mmp3; see Supplemental Table S1 (tabs 22 and 25). Finally, SMP cells in both WT and DKO muscle expressed lower levels of a majority of ECM proteins included in the core matrisome (Fig. 8 and Supplemental Table S1). Lama5, a gene associated with stem cell renewal, however, was found to be highly expressed in WT and DKO SMPs compared with FBs and FABs (Supplemental Table S1, tabs 25 and 28) (17). Although similar patterns were present in healthy WT animals, FBs and FAP cells did not show differential expression of some core basement membrane genes (Col4a3, Col4a5, Col4a6), suggesting this specific gene expression arises in the pathological state (Fig. 8 and Supplemental Table S1, tabs 11–19). These findings demonstrate that ECM production in fibrotic skeletal muscle represents an effort among different cell types to achieve a fully functional composite of ECM proteins.
Fig. 8.
Specialization of ECM expression in each of the three cell types during fibrosis. In both WT and DKO mice, collagen I expression is highest in FB followed by FAPs and then SMPs, suggesting that each population does not contribute equally to collagen I expression in both healthy and fibrotic skeletal muscle. Compared with SMP and FAP cells in WT muscle, FBs showed elevated expression of fibrillar ECM proteins, denoted by green lettering. FBs and FAP cells showed elevated expression of basal laminal ECM proteins over SMPs, denoted by blue lettering. Finally, SMP cells appeared to have the lowest amount of ECM expression compared with the other two cell types. Scaled gene expression counts of SMP, FAP and FB cells in DKO animals reveals a distinct role for each cell type in fibrosis. Compared with SMP and FAP cells, FBs showed elevated expression of fibrillar ECM proteins. FAP cells showed elevated expression of basal laminal ECM proteins. Finally, SMP cells had the lowest amount of ECM expression compared with the other two cell types. However, there were a few genes, such as Lama5, that were expressed at a higher level compared with the other cell types. This suggests that these proteins may be important for maintaining the ECM around SMP cells. Symbols depict differentially expressed genes: *differential expression between SMP and FAP, #differential expression between SMP and FB, +differential expression between FAP and FB. Data are presented as row-normalized scaled gene expression counts ranging between −1 and 1.
DISCUSSION
Overall, this study demonstrates that muscle fibrosis results from an overabundance of collagen-expressing cells, and not an increase in fibrillar ECM expression from one cell type. Furthermore, this study shows that collagen I-expressing cells in muscle are a diverse population composed of fibroblasts, fibro/adipogenic progenitor cells, and skeletal muscle progenitor cells (Fig. 3). Finally, these experiments identified a distinct ECM expression profile for each of these cell types in fibrotic skeletal muscle (Fig. 8 and Supplemental Table S1). These findings were observed using a series of histological, fluorescence activated cell sorting and RNA sequencing experiments on a collagen I reporter mouse.
The concentration of all collagen I-expressing cells dramatically increased in DKO fibrotic muscle, while the percentage of each population expressing collagen only increased modestly. This apparent discrepancy can be explained by the fact that Fig. 4, C–E, shows that the number of collagen-expressing cells in each group increases dramatically, while Fig. 4, F–H depicts the percentage of cells that express collagen in each cell type. The graphs shown in Fig. 4, C–E, and Fig. 4, F–H, represent different expressions of the data. Theoretically there could be a doubling of cell number with no change in the percentage of cells that are GFP+ if the entire cell population (GFP+ and GFP− portions) grew equally with fibrosis, but here we show that there is actually a shift in the population such that the GFP+ population has become a larger portion of the entire cell population. The discrepancy in effect size between Fig. 4, C–E, and Fig. 4, F–H, can thus be explained by the fact that both the GFP+ and GFP− portions of each population are increasing in DKO mice. Therefore, we can conclude that the collagen-expressing cell populations are increasing in number and that a larger portion of each population has begun expressing collagen.
The fact that the concentration of each cell type expressing collagen in muscle increased to about the same extent (Fig. 4, C–E) suggests that there is a common factor and/or pathway that is activated in DKO muscle that stimulates all collagen-expressing cells equally such as an inflammatory signaling cascade. This is consistent with previous reports that suggest that FAPs are activated upon muscle injury, and in turn, these activated FAPs increase skeletal muscle progenitor differentiation (26). Additionally, it has been shown that activated satellite cells have significantly elevated levels of collagen I gene expression over quiescent cells (59). Furthermore, FBs are known to proliferate during the progression of fibrosis in skeletal muscle (31a). Therefore, the fibrosis that occurs in DKO muscle most likely results from FB, FAP, and SMP proliferation. However, to fully understand this mechanism, cellular proliferation assays would have to be completed. Nevertheless, the activation and proliferation of these cells would correspond to an increased number of GFP+ cells as well as a population shift in percentage of GFP+ cells within the FB, FAP, and SMP populations, which is consistent with the data in our study. Furthermore, the lack of differences between WT and DKO in terms of fibrotic gene expression supports the notion that the fibrotic phenotype seen in DKO skeletal muscle is more likely related to the proliferation of collagen-expressing cells as opposed to an increase in “fibrotic” gene expression program by a specific cell type.
Surprisingly, the expression of fibrotic collagen genes (collagens I, II, III, V, and XXIV) was not dramatically affected in collagen-expressing cells from DKO muscle; instead, there was actually a downregulation of most collagen genes. Interestingly, this downregulation was in genes that are primarily associated with the basement membrane (collagen IV and XV). Downregulation of these basement membrane genes is also seen in several types of cancers, characterized by stiff microenvironments where the basement membrane is degraded allowing for increased cellular infiltration and formation of a stiffer ECM (14, 15, 37). A similar mechanism could be in play in this model of skeletal muscle fibrosis where the downregulation of basement membrane genes allows infiltration of inflammatory cells and ECM-producing cells (such as fibroblasts, myofibroblasts, fibro/adipogenic progenitors, etc.) to create a robust and stiff fibrotic scar.
Gene expression data show differential expression of collagen I between FBs, FAPs, and SMPs in both WT and DKO muscle (Fig. 8). These expression data coupled with data on collagen-expressing cell population composition (Fig. 4B) show that all cells do not contribute equally to collagen I expression in healthy and fibrotic muscle. Taken together, these data show that FBs and FAPs likely contribute more to the production of collagen I as compared with SMPs. SMPs have the lowest collagen I expression and make up the smallest portion of collagen-expressing cells, suggesting that collagen I production by SMPs is not a significant source of collagen in fibrosis.
It should be noted that the labeling strategy for fibroblasts in this study is not ideal. Fibroblasts and myofibroblasts in this study were labeled with ER-TR7 and α-smooth muscle actin, respectively. Although the labeling of fibroblasts has been a source of debate and scientific confusion, these two markers have been used in previous studies to label fibroblasts and myofibroblasts (11, 38, 43). While the staining of these fibroblast populations is difficult, these two stains in combination serve to label a large portion of fibroblasts in skeletal muscle. In addition to ER-TR7 and α-SMA, the use of transcription factor 4 (Tcf4) has also been used to stain for fibroblasts (39). However, Tcf4 has been found to be expressed in myoblasts, making its application for this study not ideal. Tcf-4 is expressed at a lower level in myoblasts, and genetic labeling techniques do exist for the identification of only fibroblasts (i.e., cells expressing high levels of Tcf-4), but those techniques genetically label Tcf4-high expressing cells with GFP (39). This GFP labeling would interfere with the use of the collagen I-GFP reporter mouse in this study.
While the GFP labeling strategy employed in this study is ideal for identifying mononuclear cells that express collagen I in a majority of tissues, in skeletal muscle this approach presents a slight limitation. Since skeletal muscle cells are multinucleated, this means that the collagen-expressing cell labeling strategy is limited and FACS does not identify post-mitotic multinucleated myofibers that could be producing collagen. Given that SMPs made up a small portion of collagen-expressing cells and that these cells had the lowest expression of collagen I, the amount of collagen expression in myofibers is most likely insignificant. In fact, collagen I expression has been shown to dramatically fall upon satellite cell differentiation and fusion with muscle fibers, further demonstrating that the amount of collagen I expression in mature muscle fibers is trivial (60). Finally, no GFP signal was detected in muscle fiber cytoplasm during histological analysis, which further confirms an undetectable amount of collagen expression in muscle fibers (Fig. 2).
Because this study focuses on 8-wk-old animals, we are unable to exactly determine whether differences between healthy and fibrotic muscle are responsible for or the consequence of fibrosis. In the current study, we presented data from a single time point after the process of fibrosis has already begun. However, a previous study examining the progression of muscle fibrosis in desmin single knockout animals showed that the animals are born the same, but fibrosis develops as the desmin knockout animals age (43). This suggests that the cellular differences seen in this study of desmin/nesprin 1 double knockout animals are responsible for the development of this fibrosis. In desmin knockout mice, ECM properties developed over the life of the animal, suggesting that changes in cell populations may proceed changes in the ECM. However, a detailed time course examining changes in cell populations and ECM properties is needed to definitively address this issue.
Muscle passive mechanical properties are significantly affected by fibrosis (13, 33, 43, 55). Although collagen content and passive stiffness both increase in a variety of fibrotic models, a causal relationship has yet to be determined (12, 55). The current study shows that, in fibrotic muscle, fibroblasts change their ECM expression profiles to favor fibrillar ECM (Fig. 5). With the downregulation of basal laminal ECM proteins and increased degradation of the basal lamina there is the potential for significant structural changes in the ECM. While these data do not directly explain the dramatic increase in fibrillar collagen production, particularly collagen I production, found previously (13), we now have evidence that FB gene expression is altered in a way that remodels the ECM, favoring a stiffer fibrillar structure. These structural changes could explain why collagen concentration does not correlate perfectly with tissue mechanical properties. Of course, additional experiments must be performed to confirm this hypothesis, but it is clear that the passive mechanical changes that occur in muscle fibrosis are more complicated than simply increased collagen content.
Although defining a specific mechanism for the increased concentration of collagen-expressing cells observed here is beyond the scope of this study, the RNA-sequencing data suggest that an increase could occur by a combined increase in cellular proliferation and dysregulation of apoptosis. Gene expression shows that FBs increase expression of genes associated with cellular proliferation [GO:0042127: Pthlh, Notch1, Edn1, Anxa1, Kctd11, Tgfb1, F2r, Mki67 (GO:0008283); Supplemental Table S1], while FAPs increased expression of genes associated with the negative regulation of apoptosis [GO:0043066: Bdnf, Hgf, Cx3cl1, Timp1, F2r (Supplemental Table S1)]. Furthermore, SMPs increase expression of genes positively regulating cell division (GO:0051781: S100a6, Ereg, Btc, Tgfb2; Supplemental Table S1). Given these changes in gene expression, additional experiments examining the exact mechanisms behind how the collagen-expressing cell populations grow are warranted. This knowledge could help prevent the onset of fibrosis in several myopathies.
The results presented here provide insights that can help in the creation of antifibrotic therapies in muscle. The development of therapies to combat skeletal muscle fibrosis is important for the success of treating debilitating diseases such as muscular dystrophy. In cases such as Duchenne muscular dystrophy, cell-based therapies aimed at replenishing the muscle’s missing dystrophin protein have not been tremendously successful in treating the disease (22, 40, 52–54). This can be partly attributed to the fact that cells do not engraft efficiently into a stiff (i.e., fibrotic) microenvironment (8, 41). Because fibrosis in this model develops from an increase in the number of collagen-expressing cells as opposed to an increased fibrotic gene program, a potential target for antifibrotic therapies should aim at targeting excessive cell proliferation and alterations in apoptosis. Future studies are needed in this area to identify therapeutic targets that prevent excessive cellular expansion. Alternatively, methods aimed at targeting cell populations with apoptosis could be used as previously reported in mdx mice (7).
Conclusions
The findings in this study describe and present a cellular mechanism behind the formation of skeletal muscle fibrosis in a murine cytoskeletal knockout model. FACS and RNA sequencing methods revealed that the large increase in collagen content seen in DKO muscle was primarily the result of an increased number of collagen I-expressing cells. Surprisingly, gene expression of fibrillar ECM genes, specifically fibrillar collagens, revealed no differential expression between healthy and fibrotic muscle. Although many profibrotic genes were upregulated in DKO muscle, these were genes associated with promoting fibrosis and not the major structural ECM genes associated with fibrotic scars in muscle.
GRANTS
Research was supported by the National Institute of Arthritis and Musculoskeletal and Skin Diseases of the National Institutes of Health (Award Numbers AR061303 and T32AR0607) and by the National Institute of Child Health and Human Development (Award Number HD050837). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. We acknowledge the National Science Foundation for an NSF-Graduate Research Fellowship (to M. A. Chapman).
DISCLOSURES
No conflicts of interest, financial or otherwise, are declared by the author(s).
AUTHOR CONTRIBUTIONS
M.A.C. performed experiments; M.A.C., K.M., and R.L.L. analyzed data; M.A.C., K.M., S.S., and R.L.L. interpreted results of experiments; M.A.C., K.M., and R.L.L. prepared figures; M.A.C. and K.M. drafted manuscript; M.A.C., K.M., S.S., D.B., and R.L.L. edited and revised manuscript; M.A.C., K.M., S.S., D.B., and R.L.L. approved final version of manuscript.
Supplementary Material
ACKNOWLEDGMENTS
The authors thank Rachel Meza for providing TEM, Dr. Adam Engler for providing laboratory space/supplies, and Dr. Gretchen Meyer and Shannon Bremner for technical assistance.
Present address for M. A. Chapman: Karolinska Institute, von Eulers väg 8, 171 77 Stockholm, Sweden.
REFERENCES
- 1.Alexakis C, Partridge T, Bou-Gharios G. Implication of the satellite cell in dystrophic muscle fibrosis: a self-perpetuating mechanism of collagen overproduction. Am J Physiol Cell Physiol 293: C661–C669, 2007. doi: 10.1152/ajpcell.00061.2007. [DOI] [PubMed] [Google Scholar]
- 2.Anders S, Huber W. Differential expression analysis for sequence count data. Genome Biol 11: R106, 2010. doi: 10.1186/gb-2010-11-10-r106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bahamonde ME, Lyons KM. BMP3: to be or not to be a BMP. J Bone Joint Surg Am 83-A, Suppl 1: S56–S62, 2001. [PubMed] [Google Scholar]
- 4.Bateman JF, Boot-Handford RP, Lamandé SR. Genetic diseases of connective tissues: cellular and extracellular effects of ECM mutations. Nat Rev Genet 10: 173–183, 2009. doi: 10.1038/nrg2520. [DOI] [PubMed] [Google Scholar]
- 5.Baum J, Duffy HS. Fibroblasts and myofibroblasts: what are we talking about? J Cardiovasc Pharmacol 57: 376–379, 2011. doi: 10.1097/FJC.0b013e3182116e39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Biernacka A, Frangogiannis NG. Aging and cardiac fibrosis. Aging Dis 2: 158–173, 2011. [PMC free article] [PubMed] [Google Scholar]
- 7.Bo Li Z, Zhang J, Wagner KR. Inhibition of myostatin reverses muscle fibrosis through apoptosis. J Cell Sci 125: 3957–3965, 2012. doi: 10.1242/jcs.090365. [DOI] [PubMed] [Google Scholar]
- 8.Boldrin L, Zammit PS, Morgan JE. Satellite cells from dystrophic muscle retain regenerative capacity. Stem Cell Res 14: 20–29, 2015. doi: 10.1016/j.scr.2014.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brack AS, Conboy MJ, Roy S, Lee M, Kuo CJ, Keller C, Rando TA. Increased Wnt signaling during aging alters muscle stem cell fate and increases fibrosis. Science 317: 807–810, 2007. [DOI] [PubMed] [Google Scholar]
- 10.Carmeli E, Moas M, Reznick AZ, Coleman R. Matrix metalloproteinases and skeletal muscle: a brief review. Muscle Nerve 29: 191–197, 2004. doi: 10.1002/mus.10529. [DOI] [PubMed] [Google Scholar]
- 11.Chapman MA, Meza R, Lieber RL. Skeletal muscle fibroblasts in health and disease. Differentiation 92: 108–115, 2016. doi: 10.1016/j.diff.2016.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chapman MA, Pichika R, Lieber RL. Collagen crosslinking does not dictate stiffness in a transgenic mouse model of skeletal muscle fibrosis. J Biomech 48: 375–378, 2015. doi: 10.1016/j.jbiomech.2014.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chapman MA, Zhang J, Banerjee I, Guo LT, Zhang Z, Shelton GD, Ouyang K, Lieber RL, Chen J. Disruption of both nesprin 1 and desmin results in nuclear anchorage defects and fibrosis in skeletal muscle. Hum Mol Genet 23: 5879–5892, 2014. doi: 10.1093/hmg/ddu310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chopra A, Maitra B, Korman NJ. Decreased mRNA expression of several basement membrane components in basal cell carcinoma. J Invest Dermatol 110: 52–56, 1998. doi: 10.1046/j.1523-1747.1998.00089.x. [DOI] [PubMed] [Google Scholar]
- 15.Cox TR, Erler JT. Remodeling and homeostasis of the extracellular matrix: implications for fibrotic diseases and cancer. Dis Model Mech 4: 165–178, 2011. doi: 10.1242/dmm.004077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Csiszar K. Lysyl oxidases: a novel multifunctional amine oxidase family. Prog Nucleic Acid Res Mol Biol 70: 1–32, 2001. doi: 10.1016/S0079-6603(01)70012-8. [DOI] [PubMed] [Google Scholar]
- 17.Domogatskaya A, Rodin S, Boutaud A, Tryggvason K. Laminin-511 but not -332, -111, or -411 enables mouse embryonic stem cell self-renewal in vitro. Stem Cells 26: 2800–2809, 2008. doi: 10.1634/stemcells.2007-0389. [DOI] [PubMed] [Google Scholar]
- 18.Dulauroy S, Di Carlo SE, Langa F, Eberl G, Peduto L. Lineage tracing and genetic ablation of ADAM12(+) perivascular cells identify a major source of profibrotic cells during acute tissue injury. Nat Med 18: 1262–1270, 2012. doi: 10.1038/nm.2848. [DOI] [PubMed] [Google Scholar]
- 19.Edgar R, Domrachev M, Lash AE. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res 30: 207–210, 2002. doi: 10.1093/nar/30.1.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Eyden B. The myofibroblast: phenotypic characterization as a prerequisite to understanding its functions in translational medicine. J Cell Mol Med 12: 22–37, 2008. doi: 10.1111/j.1582-4934.2007.00213.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gillies AR, Lieber RL. Structure and function of the skeletal muscle extracellular matrix. Muscle Nerve 44: 318–331, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gussoni E, Blau HM, Kunkel LM. The fate of individual myoblasts after transplantation into muscles of DMD patients. Nat Med 3: 970–977, 1997. doi: 10.1038/nm0997-970. [DOI] [PubMed] [Google Scholar]
- 23.Hayes AJ, Gibson MA, Shu C, Melrose J. Confocal microscopy demonstrates association of LTBP-2 in fibrillin-1 microfibrils and colocalisation with perlecan in the disc cell pericellular matrix. Tissue Cell 46: 185–197, 2014. doi: 10.1016/j.tice.2014.04.002. [DOI] [PubMed] [Google Scholar]
- 24.Hu J, Ge H, Newman M, Liu K. OSA: a fast and accurate alignment tool for RNA-Seq. Bioinformatics 28: 1933–1934, 2012. doi: 10.1093/bioinformatics/bts294. [DOI] [PubMed] [Google Scholar]
- 25.Huang W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc 4: 44–57, 2008. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 26.Joe AWB, Yi L, Natarajan A, Le Grand F, So L, Wang J, Rudnicki MA, Rossi FM. Muscle injury activates resident fibro/adipogenic progenitors that facilitate myogenesis. Nat Cell Biol 12: 153–163, 2010. doi: 10.1038/ncb2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kjaer M. Role of extracellular matrix in adaptation of tendon and skeletal muscle to mechanical loading. Physiol Rev 84: 649–698, 2004. doi: 10.1152/physrev.00031.2003. [DOI] [PubMed] [Google Scholar]
- 28.Königshoff M, Kramer M, Balsara N, Wilhelm J, Amarie OV, Jahn A, Rose F, Fink L, Seeger W, Schaefer L, Günther A, Eickelberg O. WNT1-inducible signaling protein-1 mediates pulmonary fibrosis in mice and is upregulated in humans with idiopathic pulmonary fibrosis. J Clin Invest 119: 772–787, 2009. doi: 10.1172/JCI33950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lawrence M, Huber W, Pagès H, Aboyoun P, Carlson M, Gentleman R, Morgan MT, Carey VJ. Software for computing and annotating genomic ranges. PLoS Comput Biol 9: e1003118, 2013. doi: 10.1371/journal.pcbi.1003118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Leask A, Abraham DJ. TGF-beta signaling and the fibrotic response. FASEB J 18: 816–827, 2004. doi: 10.1096/fj.03-1273rev. [DOI] [PubMed] [Google Scholar]
- 31.Levy AD, Omar MH, Koleske AJ. Extracellular matrix control of dendritic spine and synapse structure and plasticity in adulthood. Front Neuroanat 8: 116, 2014. doi: 10.3389/fnana.2014.00116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31a.Li ZB, Kollias HD, Wagner KR. Myostatin directly regulates skeletal muscle fibrosis. J Biol Chem 283: 19371–19378, 2008. doi: 10.1074/jbc.M802585200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lieber RL, Fridén J. Spasticity causes a fundamental rearrangement of muscle-joint interaction. Muscle Nerve 25: 265–270, 2002. doi: 10.1002/mus.10036. [DOI] [PubMed] [Google Scholar]
- 33.Lieber RL, Ward SR. Cellular mechanisms of tissue fibrosis. 4. Structural and functional consequences of skeletal muscle fibrosis. Am J Physiol Cell Physiol 305: C241–C252, 2013. doi: 10.1152/ajpcell.00173.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lin S-L, Kisseleva T, Brenner DA, Duffield JS. Pericytes and perivascular fibroblasts are the primary source of collagen-producing cells in obstructive fibrosis of the kidney. Am J Pathol 173: 1617–1627, 2008. doi: 10.2353/ajpath.2008.080433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lorts A, Schwanekamp JA, Baudino TA, McNally EM, Molkentin JD. Deletion of periostin reduces muscular dystrophy and fibrosis in mice by modulating the transforming growth factor-β pathway. Proc Natl Acad Sci USA 109: 10978–10983, 2012. doi: 10.1073/pnas.1204708109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15: 550, 2014. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lu P, Takai K, Weaver VM, Werb Z. Extracellular matrix degradation and remodeling in development and disease. Cold Spring Harb Perspect Biol 3: a005058, 2011. doi: 10.1101/cshperspect.a005058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mann CJ, Perdiguero E, Kharraz Y, Aguilar S, Pessina P, Serrano AL, Muñoz-Cánoves P. Aberrant repair and fibrosis development in skeletal muscle [Online]. Skelet Muscle 1: 21, 2011. doi: 10.1186/2044-5040-1-21. http://www.skeletalmusclejournal.com/content/1/1/21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mathew SJ, Hansen JM, Merrell AJ, Murphy MM, Lawson JA, Hutcheson DA, Hansen MS, Angus-Hill M, Kardon G. Connective tissue fibroblasts and Tcf4 regulate myogenesis. Development 138: 371–384, 2011. doi: 10.1242/dev.057463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mendell JR, Kissel JT, Amato AA, King W, Signore L, Prior TW, Sahenk Z, Benson S, McAndrew PE, Rice R, Nagaraja H, Stephens R, Lantry L, Morris G, Burghes A. Myoblast transfer in the treatment of Duchenne’s muscular dystrophy. N Engl J Med 333: 832–838, 1995. doi: 10.1056/NEJM199509283331303. [DOI] [PubMed] [Google Scholar]
- 41.Meng J, Bencze M, Asfahani R, Muntoni F, Morgan JE. The effect of the muscle environment on the regenerative capacity of human skeletal muscle stem cells. Skelet Muscle 5: 11, 2015. doi: 10.1186/s13395-015-0036-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Merline R, Schaefer RM, Schaefer L. The matricellular functions of small leucine-rich proteoglycans (SLRPs). J Cell Commun Signal 3: 323–335, 2009. doi: 10.1007/s12079-009-0066-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Meyer GA, Lieber RL. Skeletal muscle fibrosis develops in response to desmin deletion. Am J Physiol Cell Physiol 302: C1609–C1620, 2012. doi: 10.1152/ajpcell.00441.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Naba A, Clauser KR, Ding H, Whittaker CA, Carr SA, Hynes RO. The extracellular matrix: tools and insights for the “omics” era. Matrix Biol 49: 10–24, 2016. doi: 10.1016/j.matbio.2015.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Penner AS, Rock MJ, Kielty CM, Shipley JM. Microfibril-associated glycoprotein-2 interacts with fibrillin-1 and fibrillin-2 suggesting a role for MAGP-2 in elastic fiber assembly. J Biol Chem 277: 35044–35049, 2002. doi: 10.1074/jbc.M206363200. [DOI] [PubMed] [Google Scholar]
- 46.Randles MJ, Humphries MJ, Lennon R. Proteomic definitions of basement membrane composition in health and disease. Matrix Biol S0945-053X(16)30117-2, 2016. doi: 10.1016/j.matbio.2016.08.006. [DOI] [PubMed] [Google Scholar]
- 47.Rawlings ND, Tolle DP, Barrett AJ. Evolutionary families of peptidase inhibitors. Biochem J 378: 705–716, 2004. doi: 10.1042/bj20031825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sakai LY, Keene DR, Engvall E. Fibrillin, a new 350-kD glycoprotein, is a component of extracellular microfibrils. J Cell Biol 103: 2499–2509, 1986. doi: 10.1083/jcb.103.6.2499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sato EJ, Killian ML, Choi AJ, Lin E, Esparza MC, Galatz LM, Thomopoulos S, Ward SR. Skeletal muscle fibrosis and stiffness increase after rotator cuff tendon injury and neuromuscular compromise in a rat model. J Orthop Res 32: 1111–1116, 2014. doi: 10.1002/jor.22646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Serrano AL, Muñoz-Cánoves P. Regulation and dysregulation of fibrosis in skeletal muscle. Exp Cell Res 316: 3050–3058, 2010. doi: 10.1016/j.yexcr.2010.05.035. [DOI] [PubMed] [Google Scholar]
- 51.Shimbori C, Bellaye P-S, Xia J, Gauldie J, Ask K, Ramos C, Becerril C, Pardo A, Selman M, Kolb M. Fibroblast growth factor-1 attenuates TGF-β1-induced lung fibrosis. J Pathol 240: 197–210, 2016. doi: 10.1002/path.4768. [DOI] [PubMed] [Google Scholar]
- 52.Skuk D, Goulet M, Roy B, Chapdelaine P, Bouchard JP, Roy R, Dugré FJ, Sylvain M, Lachance JG, Deschênes L, Senay H, Tremblay JP. Dystrophin expression in muscles of duchenne muscular dystrophy patients after high-density injections of normal myogenic cells. J Neuropathol Exp Neurol 65: 371–386, 2006. doi: 10.1097/01.jnen.0000218443.45782.81. [DOI] [PubMed] [Google Scholar]
- 53.Skuk D, Goulet M, Roy B, Piette V, Côté CH, Chapdelaine P, Hogrel J-Y, Paradis M, Bouchard J-P, Sylvain M, Lachance J-G, Tremblay JP. First test of a “high-density injection” protocol for myogenic cell transplantation throughout large volumes of muscles in a Duchenne muscular dystrophy patient: eighteen months follow-up. Neuromuscul Disord 17: 38–46, 2007. doi: 10.1016/j.nmd.2006.10.003. [DOI] [PubMed] [Google Scholar]
- 54.Skuk D, Roy B, Goulet M, Chapdelaine P, Bouchard JP, Roy R, Dugré FJ, Lachance JG, Deschênes L, Senay H, Sylvain M, Tremblay JP. Dystrophin expression in myofibers of Duchenne muscular dystrophy patients following intramuscular injections of normal myogenic cells. Mol Ther 9: 475–482, 2004. doi: 10.1016/j.ymthe.2003.11.023. [DOI] [PubMed] [Google Scholar]
- 55.Smith LR, Barton ER. Collagen content does not alter the passive mechanical properties of fibrotic skeletal muscle in mdx mice. Am J Physiol Cell Physiol 306: C889–C898, 2014. doi: 10.1152/ajpcell.00383.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Smith LR, Lee KS, Ward SR, Chambers HG, Lieber RL. Hamstring contractures in children with spastic cerebral palsy result from a stiffer extracellular matrix and increased in vivo sarcomere length. J Physiol 589: 2625–2639, 2011. doi: 10.1113/jphysiol.2010.203364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sullivan MM, Barker TH, Funk SE, Karchin A, Seo NS, Höök M, Sanders J, Starcher B, Wight TN, Puolakkainen P, Sage EH. Matricellular hevin regulates decorin production and collagen assembly. J Biol Chem 281: 27621–27632, 2006. doi: 10.1074/jbc.M510507200. [DOI] [PubMed] [Google Scholar]
- 58.Uezumi A, Ito T, Morikawa D, Shimizu N, Yoneda T, Segawa M, Yamaguchi M, Ogawa R, Matev MM, Miyagoe-Suzuki Y, Takeda S, Tsujikawa K, Tsuchida K, Yamamoto H, Fukada S. Fibrosis and adipogenesis originate from a common mesenchymal progenitor in skeletal muscle. J Cell Sci 124: 3654–3664, 2011. doi: 10.1242/jcs.086629. [DOI] [PubMed] [Google Scholar]
- 59.Urciuolo A, Quarta M, Morbidoni V, Gattazzo F, Molon S, Grumati P, Montemurro F, Tedesco FS, Blaauw B, Cossu G, Vozzi G, Rando TA, Bonaldo P. Collagen VI regulates satellite cell self-renewal and muscle regeneration. Nat Commun 4: 1964, 2013. doi: 10.1038/ncomms2964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Velleman SG, McFarland DC. Myotube morphology, and expression and distribution of collagen type I during normal and low score normal avian satellite cell myogenesis. Dev Growth Differ 41: 153–161, 1999. doi: 10.1046/j.1440-169x.1999.00422.x. [DOI] [PubMed] [Google Scholar]
- 61.Voermans NC, Bönnemann CG, Huijing PA, Hamel BC, van Kuppevelt TH, de Haan A, Schalkwijk J, van Engelen BG, Jenniskens GJ. Clinical and molecular overlap between myopathies and inherited connective tissue diseases. Neuromuscul Disord 18: 843–856, 2008. doi: 10.1016/j.nmd.2008.05.017. [DOI] [PubMed] [Google Scholar]
- 62.Whittaker CA, Hynes RO. Distribution and evolution of von Willebrand/integrin A domains: widely dispersed domains with roles in cell adhesion and elsewhere. Mol Biol Cell 13: 3369–3387, 2002. doi: 10.1091/mbc.E02-05-0259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wynn TA. Cellular and molecular mechanisms of fibrosis. J Pathol 214: 199–210, 2008. doi: 10.1002/path.2277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Yata Y, Scanga A, Gillan A, Yang L, Reif S, Breindl M, Brenner DA, Rippe RA. DNase I-hypersensitive sites enhance alpha1(I) collagen gene expression in hepatic stellate cells. Hepatology 37: 267–276, 2003. doi: 10.1053/jhep.2003.50067. [DOI] [PubMed] [Google Scholar]
- 65.Yoon JK, Lee J-S. Cellular signaling and biological functions of R-spondins. Cell Signal 24: 369–377, 2012. doi: 10.1016/j.cellsig.2011.09.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zeisberg M, Hanai J, Sugimoto H, Mammoto T, Charytan D, Strutz F, Kalluri R. BMP-7 counteracts TGF-beta1-induced epithelial-to-mesenchymal transition and reverses chronic renal injury. Nat Med 9: 964–968, 2003. doi: 10.1038/nm888. [DOI] [PubMed] [Google Scholar]
- 68.Zumstein MA, Jost B, Hempel J, Hodler J, Gerber C. The clinical and structural long-term results of open repair of massive tears of the rotator cuff. J Bone Joint Surg Am 90: 2423–2431, 2008. doi: 10.2106/JBJS.G.00677. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.