Abstract
Objective:
Coronary artery disease (CAD) is a common, complex, and progressive disorder characterized by the accumulation of lipids and fibrous elements in the arteries. It is one of the leading causes of death in industrialized nations. Oxidative modification of low-density lipoprotein (LDL) in the arterial wall plays an important role in the initiation and progression of atherosclerosis. Paraoxonase1 (PON1) is involved in lipid metabolism and is believed to protect LDL oxidation. In our study, we aimed to clarify the relationship between PON1 gene L55M polymorphism and the extent and severity of CAD.
Methods:
In total, 114 patients (54 males, mean age: 56.7±12.0 years; 60 females, mean age: 55.7±13.2 years) with stable angina or angina equivalent symptoms were enrolled in this prospective study. Cardiological evaluation was performed with electrocardiogram and transthoracic echocardiogram. The presence of hypertension, dyslipidemia, diabetes, and smoking status were ascertained. The patients were grouped according to their Gensini scores and gender. Genetic analysis of the PON1 gene L55M polymorphism was performed by polymerase chain reaction-restriction fragment length polymorphism.
Results:
We determined that the LL genotype was more prevalent in patients with Gensini score higher than or equal to 20 (p=0.026) and that this correlated with severe atherosclerotic coronary artery lesions in both gender groups, reaching a statistical significance in the female subjects (p=0.038).
Conclusion:
It was thought that the PON1 gene L55M polymorphism plays a significant role in CAD progression, especially in females.
Keywords: paraoxonase1, coronary artery disease, atherosclerosis, L55M polymorphism
Introduction
Coronary artery disease (CAD) is a common, complex, and progressive disorder that is characterized by the accumulation of lipids and fibrous elements in the arteries. It is one of the major causes of death in industrialized nations (1). The etiology of the atherosclerotic process is multifactorial, and various risk factors have been implicated in the development of CAD, such as sex hormones, serum lipid profiles, obesity, hypertension, lifestyle, and environmental factors. Epidemiological and experimental studies point toward a gender bias; females encounter clinically symptomatic atherosclerosis approximately one decade later than males (2). Higher levels of estrogen are related to higher levels of high-density lipoprotein (HDL) and lower levels of low-density lipoprotein (LDL) cholesterol (2). The oxidative modification of LDL in the arterial wall plays an important role in the initiation and progression of atherosclerosis, and the inflammatory response generated by the macrophages to oxidized LDL is a critical trigger in the pathophysiology of atherosclerosis (3). On the other hand, HDL plays an antiatherogenic role. The protective capacity of HDL is primarily attributed to its ability to remove excess cholesterol from peripheral tissues in the reverse cholesterol transport pathway. Paraoxonase1 (PON1) is a 43-kDa, Ca+2-dependent enzyme, which is synthesized in the liver and secreted into circulation. It remains associated with apolipoprotein (apo) A-I and apolipoprotein J (ApoJ) on HDL (4), which is believed to protect LDL and HDL from oxidation and thereby from the risk of CAD. PON1 is also involved in lipid metabolism and in the elimination of carcinogenic lipid-soluble radicals (5). In vivo, its enzymatic activity is substrate dependent, and PON1 shows wide interindividual differences due to both dietary and lifestyle factors and functional genetic polymorphisms (6). Lower PON1 enzymatic activity is associated with lower serum HDL cholesterol levels. Moreover, there is a close relationship between lower serum PON1 activity and CAD, myocardial infarction, familial hypercholesterolemia, and diabetes mellitus (DM) (7).
PON1 gene, located on chromosome 7 (7q21.3), has two main coding polymorphisms affecting PON1 enzymatic activity; one is glutamine to arginine substitution at codon 192 (Q192R, rs662), and the other is leucine to methionine substitution at codon 55 (L55M, rs854560) (8). It was shown that the L55M variant modulates circulating enzyme levels. LL individuals have the highest PON1 level among genotypes. On the other hand, the Q192R variant modulates enzymatic activity; hence, RR individuals have higher activity (9). These two polymorphisms do not act in isolation; other risk factors such as diet, smoking, alcohol intake, age, as well as other coexisting polymorphisms in the same gene, such as rs705379 and PON1_304A/G or other gene polymorphisms can alter the outcome (10). Consequently, there are contradictory results in the field on the association between PON1 polymorphisms and CAD, where some studies show an association (11-13) and others do not (14-16).
In the present study, we investigated the association of PON1 Leu55Met polymorphism with the extent and severity of atherosclerotic CAD in the Turkish population and whether there is a gender difference.
Methods
In this prospective study, 150 patients with stable angina or angina equivalent symptoms were screened between 2007 and 2011. In total, 114 patients (54 males, mean age: 56.7±12.0 years; 60 females, mean age: 55.7±13.2 years) with stable angina or angina-equivalent symptoms were enrolled in the study. Cardiological evaluation was performed with electrocardiogram and transthoracic echocardiogram. The presence of hypertension, hyperlipidemia, diabetes, and smoking status were ascertained. The patients were grouped according to their Gensini scores and gender. The genetic analysis of the PON1 gene L55M polymorphism was performed by polymerase chain reaction-restriction fragment length polymorphism (PCR-RFLP). The study design was approved by the Kırıkkale University Ethics Committee (2009/148).
Exclusion criteria were acute coronary syndromes, systolic heart failure (EF<50%), congenital heart disease, moderate and severe valvular heart disease, acute or chronic renal dysfunction (serum creatinine level>1.5 mg/dL), malignancies, morbid obesity (body mass index≥40 kg/m2), asthma or chronic obstructive lung disease, connective tissue disorders, neurological problems, psychiatric diseases (psychosis, major depression, and anxiety disorders), endocrine diseases other than DM, alcohol and drug abuse, use of medications for hormonal treatment, and pregnancy or lactation. A total of 114 subjects (age 56.2±12.6 years old) were enrolled to the study after 36 were excluded for having one or more of the exclusion criteria.
Cardiological evaluation
Following a detailed medical history, physical examination was performed for all subjects. Twelve-channel electrocardiography (ECG) (Cardioline Delta1 plus, Remco; Italy) recordings and transthoracic echocardiography (GE-Vivid 7 Pro, General Electric; FL, USA) were performed. Coronary angiography was performed for patients who had pathological ST-segment and T-wave changes in their ECG recordings or systolic wall motion abnormalities in the analysis of transthoracic echocardiographic images (Integris-V, 2007, Philips; Amsterdam, Netherland). Hypertension (HT) criteria were systolic blood pressure equal to or higher than 140 mm Hg and diastolic blood pressure equal to or higher than 90 mm Hg.
Coronary angiography
The 114 patients underwent diagnostic quantitative coronary angiography (CAG) to assess CAD severity. Coronary angiography was conducted using standardized projections (Integris V-Philips, 2007). Same angle and skew of the gantry, sequence of standard angiographic views, contrast agent (Iomeron® 400, Bracco-Patheon, Italy), and cardiac catheterization suite were used for all the subjects. All angiograms were filmed at 25 frames/s. The cinefilm reviewers were blinded to the subject groups, and the interpretations of the angiograms were made at the clinical center. Two blinded experienced cardiologists examined the angiographs. When there was a disagreement between them, a third investigator was admitted. Inter- and intra-observer variability for the repeated evaluations of angiograms of 16 randomly selected patients was low (for Gensini score: inter-observer variability 3.9±1.8%; intra-observer variability 4.5±1.8%). Three consecutive frames from the same phase of the cardiac cycle (preferably end-diastole) in the optimal single-plane projection that identified the stenosis in its greatest severity were chosen for quantitative angiographic analysis with a previously described and validated automated edge-detection algorithm. The severity of coronary stenotic lesions was assessed with the Gensini score based on the degree of luminal narrowing and its geographic importance (17).
For statistical evaluation, the patients were categorized according to the results of Gensini scoring: Gensini score group 1: patients with a normal coronary angiogram [CAD (-)]. Gensini score group 2: patients with Gensini score lower than 20; mild, non-severe CAD [CAD (+)]. Gensini score group 3: patients with Gensini score equal to or higher than 20; severe CAD [CAD (+)].
We also statistically analyzed gender differences for the association of the paraoxonase 1 gene L55M polymorphism with the extent and severity of CAD.
Laboratory Biochemical and hormonal analyses
A fasting blood sample was drawn between 09:00 and 10:00. Laboratory work-up involved a detailed biochemical analysis. Total cholesterol, HDL cholesterol and triglyceride, fasting blood glucose, urea, creatinine, ALT and AST levels were measured; complete blood count was performed (Olympus AU 610 autoanalyzer, Olympus Diagnostics GmbH, Hamburg, Germany). The LDL cholesterol level was calculated by Friedewald formula (18). The patients with total cholesterol level higher than 200 mg/dL, LDL cholesterol higher than 160 mg/dL, or triglyceride level higher than 200 mg/dL were categorized as dyslipidemic. DM is defined as having fasting blood glucose level equal to or higher than 126 mg/dL at two consecutive days or measurements.
PON1 polymorphism screening
DNA samples were isolated from 5 mL venous blood samples according to the manufacturer’s instructions (Invisorb® Spin DNA Extraction Kit, STRATEC Biomedical AG Birkenfeld, Germany). PCR was conducted in a total volume of 25 µL. Briefly, 50-100 ng of DNA was subjected to PCR at 1.5 mM MgCl2 concentration using 1 U Taq DNA polymerase (Biotools DNA polymerase, Biotools B&M Labs, S.A.) with 0.1 nmoL of the following primers: sense primer 5’- GAA GAG TGA TGT ATA GCC CCA G-3’ and antisense primer 5’- TTT AAT CCA GAG CTA ATG AAA GCC-3’. After denaturing the DNA for 5 min at 95°C, the reaction mixture was subjected to 30 cycles of 1 min of denaturation at 94°C, 30 s of annealing at 61°C, and 1 min of extension at 72°C. The PCR product (170 bp) was digested with the restriction enzyme NlaIII (New England Biolabs, Ipswich, UK) (37°C, overnight), and the digested products were separated by 2% agarose gel electrophoresis. The L genotype (leucine) does not contain an NlaIII site, whereas the M genotype (methionine) does, giving rise to 126 and 44 bp-sized products (19). The image of the agarose gel after PCR and restriction enzyme digestion is shown in Figure 1. For statistical analyses, the patients were grouped according to the PON1 L55M genotype.
Figure 1.
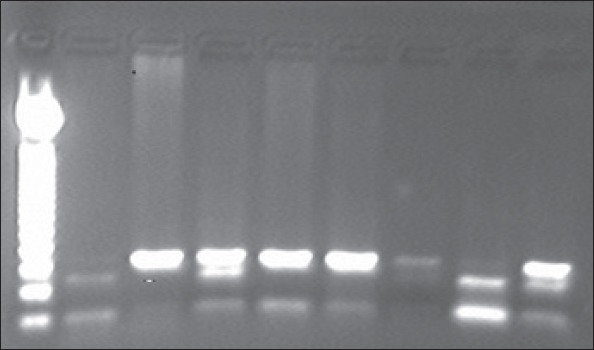
The image of the agarose gel after polymerase chain reaction and restriction enzyme digestion (PCR-RFLP). Lane 1: 50 bp DNA marker, lanes 2 and 8: MM; lanes 3, 5, 6, and 7: LL; lanes 4 and 9: LM
Statistical analysis
All statistical analyses were performed using SPSS version 15.0 (SPSS; Chicago, IL, USA). All variables were tested for normal distribution using the Kolmogorov-Smirnov test. Data with normal distribution were presented as mean±standard deviation (SD). Categorical variables were expressed as percentages and were compared with the chi-square test or Fisher’s exact test.
In the comparison of continuous variables, Student’s-t and one-way analysis of variance (ANOVA) with Bonferroni adjustment were used for comparing normally distributed data. Mann–Whitney U and Kruskal-Wallis tests were used for comparing data with skewed distribution. Partial correlation analysis was utilized to evaluate the correlations between the Gensini score and other continuous variables. Univariate analysis was also performed to evaluate important factors in association with the Gensini score and PON1 polymorphism. Additionally, we performed linear regression analysis with the backward method to determine the linear relationships between the PON1 L55Met polymorphism and atherosclerotic risk factors. A p value<0.05 was accepted as statistically significant.
Results
Patient characteristics
In the present study a total of 114 participants (54 males, mean age: 56.7±12.0 years; 60 females, mean age: 55.7±13.2 years) were enrolled. They were divided into three groups according to their Gensini score. The clinical characteristics of the 114 patients according to their genotype are given in Table 1. No statistically significant difference was detected in anthropometric values, biochemical tests, blood pressure values, and atherosclerotic risk factors among the PON1 polymorphism groups.
Table 1.
Clinical characteristics of the subjects according to their genotype
| Parameters | Group 1 LL n=55 | Group 2 LM n=52 | Group 3 MM n=7 | P* | |||
|---|---|---|---|---|---|---|---|
| Anthropometry | |||||||
| Age, years | 54.1±13.2 | 57.6±12.3 | 61.7±6.2 | NS | |||
| Weight, kg | 70.3±13.2 | 76.3±16.2 | 77.0±9.9 | NS | |||
| Body mass index, kg/m2 | 26.9±5.3 | 27.9±5.9 | 33.2±5.7 | NS | |||
| Blood pressure | |||||||
| Systolic blood pressure, mm Hg | 130.0±20.5 | 130.3±15.5 | 137.8±18.2 | NS | |||
| Diastolic blood pressure, mm Hg | 78.6±10.2 | 79.2±10.2 | 78.6±9.0 | NS | |||
| Biochemistry | |||||||
| Fasting blood glucose, mg/dL | 107.2±18.9 | 111.4±21.9 | 115.9±35.8 | NS | |||
| Total cholesterol, mg/dL | 197.3±42.5 | 190.0±41.1 | 188.0±18.6 | NS | |||
| LDL cholesterol, mg/dL | 119.5±33.9 | 111.2±32.4 | 108.2±23.9 | NS | |||
| HDL cholesterol, mg/dL | 46.7±11.4 | 44.4±10.1 | 42.2±6.3 | NS | |||
| Triglyceride, mg/dL | 168.9±89.7 | 171.1±100.3 | 195.7±75.6 | NS | |||
| n (%) Risk factors and number of patients | |||||||
| Hypertension | HT (+) | 44 (38.6) | 19 | 25 | 3 | NS | |
| HT (-) | 67 (58.8) | 36 | 27 | 4 | |||
| Dyslipidemia | DL (+) | 79 (69.3) | 31 | 38 | 6 | NS | |
| DL (-) | 35 (30.7) | 24 | 14 | 1 | |||
| Diabetes mellitus | DM (+) | 31 (27.2) | 14 | 16 | 1 | NS | |
| DM (-) | 83 (72.8) | 41 | 36 | 6 | |||
| Smoking | SM (+) | 59 (51.8) | 27 | 27 | 5 | NS | |
| SM (-) | 55 (48.2) | 28 | 25 | 2 | |||
ANOVA test, HDL - high-density lipoprotein; HT - hypertension; LDL - low-density lipoprotein
Laboratory results
In the study group, there were 55 (48.2%) subjects with the LL genotype, 52 (45.6%) with the LM genotype, and 7 (6.1%) with the MM genotype. Genotypic distribution was in Hardy-Weinberg equilibrium. The genotypic distribution of the PON1 L55M allele was analyzed according to the dominant genetic model suggesting that the M allele is a risk factor for coronary heart disease; therefore, the LM and MM genotypes were pooled. The distribution of subjects according to their Gensini score groups, gender, and genotypes are given in Table 2. Statistically significant differences were detected between the Gensini score groups and PON1 polymorphism groups (p=0.026, chi-square test). When we examined gender differences on the genotypic distribution in the Gensini score groups, we found a statistically significant difference in females but not in males (p=0.038 and p>0.05; respectively). In our cohort, the LL (Leu55Leu) genotype and L allele were more prevalent in subjects with more widespread and severe CAD (Gensini score≥20) in both genders and in total, although this did not reach statistical significance in males (Table 2).
Table 2.
Gensini score groups and gender according to the genotypes
| Gensini group | ||||||
|---|---|---|---|---|---|---|
| Gender | PON 1 | Normal | Gensini<20 | Gensini ≥20 | Total | P |
| Female | LL | 18 (54.5%) | 3 (23%) | 10 (71.4%) | 31 (51.7%) | 0.038¶ |
| LM+MM | 15 (45.5%) | 10 (77%) | 4 (28.6%) | 29 (48.3%) | ||
| Total, n | 33 | 13 | 14 | 60 | ||
| Allele frequencies | L | 0.74 | 0.5 | 0.86 | 0.72 | |
| M | 0.26 | 0.5 | 0.14 | 0.28 | ||
| Male, M | LL | 5 (41.7%) | 5 (31.3%) | 14 (53.8%) | 24 (44.4%) | 0.351¶ |
| LM+MM | 7 (58.3%) | 11 (68.7%) | 12 (46.2%) | 30 (55.6%) | ||
| Total, n | 12 | 16 | 26 | 54 | ||
| Allele frequencies | L | 0.63 | 0.66 | 0.77 | 0.7 | |
| M | 0.37 | 0.34 | 0.23 | 0.3 | ||
| F+M | LL | 23 (51.1%) | 8 (27.6%) | 24 (60%) | 55 (48.2%) | 0.026¶ |
| LM+MM | 22 (48.9%) | 21 (72.4%) | 16 (40%) | 59 (51.8%) | ||
| Total, n | 45 | 29 | 40 | 114 | ||
| Allele frequencies | L | 0.71 | 0.59 | 0.8 | 0.71 | |
| M | 0.29 | 0.41 | 0.2 | 0.29 |
Chi-square test
The comparison of baseline measures among the Gensini score groups gave significant differences for systolic blood pressure, HT, serum levels of HDL cholesterol, and smoking habit (ANOVA) (Table 3). After Bonferroni correction, we detected that in the CAD (+) and Gensini score>20 group, systolic blood pressure, HT, and smoking habits were higher than those in other groups; on the other hand, in the CAD (-) group, the HDL cholesterol levels were higher.
Table 3.
The comparison of baseline measures among Gensini score groups
| Parameters | CAD (-) Normal coronary n=45 | CAD (+) Gensini<20 n=29 | CAD (+) Gensini>20 n=40 | P* | |
|---|---|---|---|---|---|
| Age, years | 53.0±11.8 | 56.5±13.9 | 59.5±11.7 | NS | |
| Weight, kg | 74.5±15.6 | 71.2±15.4 | 74.7±14.3 | NS | |
| Body mass index, kg/m2 | 28.2±5.5 | 27.1±5.4 | 27.5±4.7 | NS | |
| Blood pressure | |||||
| Systolic blood pressure, mm Hg | 124.2±13.2 | 133.4±16.9 | 135.8±21.6 | 0.009 | |
| Diastolic blood pressure, mm Hg | 77.1±8.6 | 79.8±9.9 | 80.1±11.4 | NS | |
| Biochemistry | |||||
| Fasting blood glucose, mg/dL | 104.3±17.7 | 110.0±25.5 | 114.6±22.7 | NS | |
| Total cholesterol, mg/dL | 189.1±40.4 | 190.4±35.9 | 200.5±44.2 | NS | |
| LDL cholesterol, mg/dL | 108.9±31.8 | 114.2±29.7 | 122.4±35.0 | NS | |
| HDL cholesterol, mg/dL | 49.2±11.2 | 43.5±9.8 | 42.7±9.4 | 0.009 | |
| Triglyceride, mg/dL | 160.5±83.5 | 165.9±104.1 | 171.6±93.5 | NS | |
| Risk factors and number of patients | |||||
| Hypertension | HT (+) | 8 | 17 | 21 | <0.001 |
| HT (-) | 37 | 12 | 19 | ||
| Dyslipidemia | DL (+) | 29 | 16 | 28 | NS |
| DL (-) | 16 | 13 | 12 | ||
| Diabetes mellitus | DM (+) | 9 | 9 | 15 | NS |
| DM (-) | 36 | 20 | 25 | ||
| Smoking | SM (+) | 11 | 12 | 36 | <0.001 |
| SM (-) | 34 | 17 | 4 | ||
ANOVA test, p<0.05. CAD - coronary artery disease; HDL - high-density lipoprotein; LDL - low-density lipoprotein
For the determination of factors associated with the Gensini score groups, univariate ANOVA was used. PON1 L55M polymorphism, gender, age, weight, height, body mass index, smoking, systolic and diastolic blood pressures, presence or absence of HT, fasting blood glucose, presence or absence of DM, and serum total cholesterol, LDL cholesterol, HDL cholesterol, and triglyceride levels, and statin usage were analyzed. In this univariate analysis model, we were unable to detect any statistically significant association between the PON1 L55M polymorphism and Gensini score groups (F: 0.959, p>0.05). However, we were able to detect a statistically significant association between the PON1 L55M polymorphism and Gensini score in females (F: 472.288, p=0.002) when we ran the same analysis for gender groups separately.
For the identification of the factors that are in linearly correlated with the Gensini score, linear regression analysis with the backward method was used. The PON1 L55M polymorphism, age, weight, height, body mass index, systolic and diastolic blood pressures, smoking status, presence or absence of HT, presence or absence of DM, and fasting blood glucose, serum total cholesterol, LDL cholesterol, HDL cholesterol, and triglyceride levels, and statin usage were included in the analysis. The linear regression analysis of the PON1 L55M polymorphism and important atherosclerotic risk factors that affected the Gensini score are shown in Table 4. Our results indicate that the PON1 L55M polymorphism had a statistically significant linear association with the Gensini score in our cohort and with females when analyzed in gender groups separately.
Table 4.
The linear regression analysis of PON1 L55M polymorphism and important atherosclerotic risk factors that affect Gensini scores
| Atherosclerotic risk factors | All participants | ||
|---|---|---|---|
| Beta-value | T-value | P | |
| Diabetes mellitus | 0.364 | 3.661 | 0.001 |
| Systolic blood pressure | 0.283 | 2.866 | 0.006 |
| Smoking | 0.294 | 2.737 | 0.008 |
| PON1 polymorphism | 0.228 | 2.447 | 0.017 |
| Females | |||
| Beta-value | T-value | P | |
| Diabetes mellitus | 0.529 | 5.224 | <0.001 |
| PON1 polymorphism | 0.412 | 4.081 | 0.001 |
| Smoking | 0.356 | 3.286 | 0.004 |
| Total cholesterol | 2.169 | 2.799 | 0.012 |
| LDL cholesterol | 1.364 | 2.672 | 0.016 |
| Triglyceride | 1.388 | 2.755 | 0.014 |
| HDL cholesterol | 0.767 | 2.594 | 0.019 |
| Males | |||
| Beta-value | T-value | P | |
| Systolic blood pressure | 0.427 | 2.798 | 0.009 |
| Fasting blood glucose | 0.401 | 2.626 | 0.014 |
P<0.05. HDL - high-density lipoprotein; LDL - low-density lipoprotein
Discussion
In this study, we demonstrated that there is a statistically significant association between the PON1 L55M polymorphism and extent and severity of CAD in all participants, especially in females.
Atherosclerotic CAD is a multifactorial disorder, and genetic and environmental factors such as composition of the diet, smoking, gender, age, and presence of HT and/or DM play key roles in its development. Therefore, factors defining susceptibility among individuals, such as proteins that orchestrate inflammation, endothelial cell function (aggregant and antiaggregant equilibrium), serum lipid profiles (levels and functionality of HDL and LDL cholesterol), and oxidative stress (20), vary tremendously.
Epidemiological studies showed that elevated LDL cholesterol and triglyceride levels and low HDL levels are risk factors for atherosclerotic heart disease. For HDL levels, it was suggested that the number of particles is not the only factor that determines the protective role (21-24). HDL can become proatherogenic as a result of oxidative changes, dysfunction, and/or the lack of protective molecules in its composition.
The PON1 enzyme, which is encoded by the PON1 gene located on 7q21.3, is an arylesterase that mainly hydrolyzes paraoxon to p-nitrophenol. It possesses antioxidant/anti-inflammatory properties and is an HDL-associated enzyme shown to be responsible for the antioxidative property of HDL (24). Low or absent PON1 activity reduces the capacity of HDL to prevent LDL oxidation and may lead to CAD (24). Therefore, it is important to evaluate the quality as well as they quantity of HDL. Polymorphisms in this gene are thought to be a risk factor for CAD. In this study, we investigated the role of the L55M polymorphism on CAD severity. The M isoform is known to decrease enzyme concentration, and MM individuals have the lowest serum concentration (25). In the studies investigating the effect of PON1 gene polymorphisms such as L55M and Q192R on PON1 enzyme activity, it was reported that individuals with LL and RR genotypes have the highest PON1 activity and that these polymorphisms are protective against atherosclerotic diseases (26). However, in other studies, the opposite was reported (27). In summary, there are a lot of contradicting results reporting the effects of PON1 genotypes on PON1 enzyme activity and atherosclerosis. Some of them, including our study, report that patients with LL genotypes are more prone to the development of atherosclerosis (13, 28-30). Some studies have reported that there is no association between PON1 gene polymorphisms and atherosclerotic development (31-35). In light of these reports, it can be seen that the relationship between atherosclerosis and PON1 genotypes is quite complex because of the multiplicity of genotype-genotype, genotype-phenotype, and genotype-environment interactions. In the Austrian Stroke Prevention Study, the 55LL genotype was found to be associated with the progression of cerebral white matter lesions, which is a marker of small-vessel disease and the frequency and severity of carotid stenosis (13, 36). Multiple studies have shown that the development of CAD and association of CAD with intermediate biochemical variations and physiological risk factors are dependent on gender. However, most of the studies so far have not taken gender differences into account or have been conducted in only in men. To date, only few studies assessed the relationship between PON1 polymorphisms and CAD in women. Van Himbergen et al. (6) investigated 17,357 middle-aged women for the effect of PON1 genotype and activity on the incidence of CAD and acute myocardial infarction (AMI) and found no relationship between PON1 genetic variants (-107C>T, L55M, R192Q), PON1 activity, and CAD or AMI. Mukamal et al. (33) investigated males and females for the association between PON1 genotypes and CAD and found no association in either sex.
In our study, we found that the LL genotype was more prevalent in patients with the Gensini score higher than or equal to 20, which shows more severe atherosclerotic coronary artery lesions in both gender groups, although this was not statistically significant in men. Moreover, the PON1 L55M polymorphism in females correlated with a higher predisposition to severe atherosclerotic lesions. After adjusting for age, body mass index, lipid profile, blood pressure, presence of HT and/or DM, and statin use, a statistically significant association between the PON1 L55M polymorphism and Gensini score remained in the females. We concluded that gender and PON1 L55M polymorphism interactions vary. Our results are in agreement with the study by Rios et al. (37), which showed that PON1 55LL genotype increases CAD risk among female Caucasian-Brazilians, irrespective of other CAD risk factors. We cannot precisely explain the gender effect of PON1 on CAD development, but hormonal differences, diet, lifestyle factors, and other metabolism differences between genders may elucidate our results. Rios et al. (37) speculated that hormonal differences between genders influence lipoprotein oxidation and therefore, the PON1 effect on atherosclerosis. In a recent study, Yunoki et al. (38) found that plasma myeloperoxidase levels show a significant inverse correlation with serum PON1 concentrations. We can assume that like myeloperoxidase, there are other mediators that affect PON1 activity and atherosclerotic process in a gender-specific way.
We believe that contradicting results in literature for PON1 alleles and atherosclerotic process are explained by the positive selection of males for CAD studies, thereby ignoring gender differences. Future studies focusing on females have the potential to reveal different mechanisms for the role of PON1 in atherosclerosis.
Study limitations
The small sample size and unavailability of plasma activity of PON1 are the major limitations of our study, which make it difficult to interpret a significant correlation. Although we indicated a gender difference, we were unable to get the hormonal status of the female patients, which could be the reason for the difference. The genotyping of other functional polymorphisms of the PON1 gene could be also useful for investigating the effect of PON1 on CAD. Our study should be considered as a pilot study, and additional studies that consider gender differences should be performed.
Conclusion
Besides the limitations, we believe our results will help clarify the role of PON1 polymorphism on CAD development. We showed a statistically significant association of PON1 L55M polymorphism with the Gensini score. The patients with PON1 55LL genotype had more severe CAD than others, especially in women.
Acknowledgement:
The study was supported through a grant (2009/27) by Kırıkkale University Scientific Research Projects Coordination Unit.
Footnotes
Conflict of interest: None declared.
Peer-review: Externally peer-reviewed.
Authorship contributions: Concept - F.Ç.; Design - F.Ç., D.B.S.K., M.T.D.; Supervision - D.B.S.K., M.T.D.; Funding - F.Ç., D.B.S.K., N.Y., S.E.; Materials - M.T.D., V.Ş., Y.Ç., S.A.B., D.B.S.K.; Data collection &/or processing - D.B.S.K., M.T.D., V.Ş., Y.Ç., S.A.B.; Literature search - M.T.D., D.B.S.K., S.E.; Writing - D.B.S.K., M.T.D.; Critical review - D.B.S.K., M.T.D., N.Y., S.E.
References
- 1.Finegold JA, Asaria P, Francis DP. Mortality from ischaemic heart disease by country, region, and age:Statistics from World Health Organisation and United Nations. Int J Cardiol. 2013;168:934–45. doi: 10.1016/j.ijcard.2012.10.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Thomas JL, Braus PA. Coronary artery disease in women. A historical perspective. Arch Intern Med. 1998;158:333–7. doi: 10.1001/archinte.158.4.333. [DOI] [PubMed] [Google Scholar]
- 3.Maiolino G, Rossitto G, Caielli P, Bisogni V, Rossi GP, Calò LA. The role of oxidized low-density lipoproteins in atherosclerosis:the myths and the facts. Mediators Inflamm 2013. 2013:714653. doi: 10.1155/2013/714653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gugliucci A, Menini T. Paraoxonase 1 and HDL maturation. Clin Chim Acta. 2015;439C:5–13. doi: 10.1016/j.cca.2014.09.016. [DOI] [PubMed] [Google Scholar]
- 5.Mackness B, Mackness MI, Arrol S, Turkie W, Durrington PN. Effect of the human serum paraoxonase 55 and 192 genetic polymorphisms on the protection by high density lipoprotein against low density lipoprotein oxidative modification. FEBS Lett. 1998;423:57–60. doi: 10.1016/s0014-5793(98)00064-7. [DOI] [PubMed] [Google Scholar]
- 6.van Himbergen TM, van der Schouw YT, Voorbij HA, van Tits LJ, Stalenhoef AF, Peeters PH, et al. Paraoxonase (PON1) and the risk for coronary heart disease and myocardial infarction in a general population of Dutch women. Atherosclerosis. 2008;199:408–14. doi: 10.1016/j.atherosclerosis.2007.11.018. [DOI] [PubMed] [Google Scholar]
- 7.Jayakumari N, Thejaseebai G. High prevalence of low serum paraoxonase-1 in subjects with coronary artery disease. J Clin Biochem Nutr. 2009;45:278–84. doi: 10.3164/jcbn.08-255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Primo-Parmo SL, Sorenson RC, Teiber J, La Du BN. The human serum paraoxonase/arylesterase gene (PON1) is one member of a multigene family. Genomics. 1996;33:498–507. doi: 10.1006/geno.1996.0225. [DOI] [PubMed] [Google Scholar]
- 9.Bhattacharyya T, Nicholls SJ, Topol EJ, Zhang R, Yang X, Schmitt D, et al. Relationship of paraoxonase 1 (PON1) gene polymorphisms and functional activity with systemic oxidative stress and cardiovascular risk. JAMA. 2008;299:1265–76. doi: 10.1001/jama.299.11.1265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yuzhalin AE, Kutikhin AG. Common genetic variants in the myeloperoxidase and paraoxonase genes and the related cancer risk:a review. J Environ Sci Health C Environ Carcinog Ecotoxicol Rev. 2012;30:287–322. doi: 10.1080/10590501.2012.731957. [DOI] [PubMed] [Google Scholar]
- 11.Chen Q, Reis SE, Kammerer CM, McNamara DM, Holubkov R, Sharaf BL, et al. WISE Study Group. Association between the severity of angiographic coronary artery disease and paraoxonase gene polymorphisms in the National Heart, Lung, and Blood Institute-sponsored Women’s Ischemia Syndrome Evaluation (WISE) study. Am J Hum Genet. 2003;72:13–22. doi: 10.1086/345312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Serrato M, Marian AJ. A variant of human paraoxonase/arylesterase (HUMPONA) gene is a risk factor for coronary artery disease. J Clin Invest. 1995;96:3005–8. doi: 10.1172/JCI118373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schmidt H, Schmidt R, Niederkorn K, Gradert A, Schumacher M, Watzinger N, et al. Paraoxonase PON1 polymorphism leu-Met54 is associated with carotid atherosclerosis:results of the Austrian Stroke Prevention Study. Stroke. 1998;29:2043–8. doi: 10.1161/01.str.29.10.2043. [DOI] [PubMed] [Google Scholar]
- 14.Ko YL, Ko YS, Wang SM, Hsu LA, Chang CJ, Chu PH, et al. The Gln-Arg 191 polymorphism of the human paraoxonase gene is not associated with the risk of coronary artery disease among Chinese in Taiwan. Atherosclerosis. 1998;141:259–64. doi: 10.1016/s0021-9150(98)00179-8. [DOI] [PubMed] [Google Scholar]
- 15.Antikainen M, Murtomäki S, Syvänne M, Pahlman R, Tahvanainen E, Jauhiainen M, et al. The Gln-Arg191 polymorphism of the human paraoxonase gene (HUMPONA) is not associated with the risk of coronary artery disease in Finns. J Clin Invest. 1996;98:883–5. doi: 10.1172/JCI118869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Aynacıoğlu AS, Kepekçi Y. The human paraoxonase Gln-Argl92 (Q/R) polymorphism in Turkish patients with coronary artery disease. Int J Cardiol. 2000;74:33–7. doi: 10.1016/s0167-5273(00)00242-4. [DOI] [PubMed] [Google Scholar]
- 17.Gensini GG. A more meaningful scoring system for determining the severity of coronary heart disease. Am J Cardiol. 1983;51:606. doi: 10.1016/s0002-9149(83)80105-2. [DOI] [PubMed] [Google Scholar]
- 18.Friedewald WT, Levy RI, Fredrickson DS. Estimation of the concentration of low-density lipoprotein cholesterol in plasma, without use of the preparative ultracentrifuge. Clin Chem. 1972;18:499–502. [PubMed] [Google Scholar]
- 19.Mackness B, Durrington PN, Abuashia B, Boulton AJ, Mackness MI. Low paraoxonase activity in type II diabetes mellitus complicated by retinopathy. Clin Sci (Lond) 2000;98:355–63. [PubMed] [Google Scholar]
- 20.Li HL, Liu DP, Liang CC. Paraoxonase gene polymorphisms, oxidative stress, and diseases. J Mol Med (Berl) 2003;81:766–79. doi: 10.1007/s00109-003-0481-4. [DOI] [PubMed] [Google Scholar]
- 21.Zheng L, Nukuna B, Brennan ML, Sun M, Goormastic M, Settle M, et al. Apolipoprotein A-I is a selective target for myeloperoxidase-catalyzed oxidation and functional impairment in subjects with cardiovascular disease. J Clin Invest. 2004;114:529–41. doi: 10.1172/JCI21109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Aviram M. Does paraoxonase play a role in susceptibility to cardiovascular disease? Mol Med Today. 1999;5:381–6. doi: 10.1016/s1357-4310(99)01546-4. [DOI] [PubMed] [Google Scholar]
- 23.Navab M, Hama SY, Cooke CJ, Anantharamaiah GM, Chaddha M, Jin L, et al. Normal high density lipoprotein inhibits three steps in the formation of mildly oxidized low density lipoprotein:step 1. J Lipid Res. 2000;41:1481–94. [PubMed] [Google Scholar]
- 24.Navab M, Hama SY, Anantharamaiah GM, Hassan K, Hough GP, Watson AD, et al. Normal high density lipoprotein inhibits three steps in the formation of mildly oxidized low density lipoprotein:steps 2 and 3. J Lipid Res. 2000;41:1495–508. [PubMed] [Google Scholar]
- 25.Rainwater DL, Rutherford S, Dyer TD, Rainwater ED, Cole SA, Vandeberg JL, et al. Determinants of variation in human serum paraoxonase activity. Heredity (Edinb) 2009;102:147–54. doi: 10.1038/hdy.2008.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Haj Mouhamed D, Ezzaher A, Mechri A, Neffati F, Omezzine A, Bouslama A, et al. Effect of cigarette smoking on paraoxonase 1 activity according to PON1 L55M and PON1 Q192R gene polymorphisms. Environ Health Prev Med. 2012;17:316–21. doi: 10.1007/s12199-011-0256-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schiavon R, Turazzini M, De Fanti E, Battaglia P, Targa L, Del Colle R, et al. PON1 activity and genotype in patients with arterial ischemic stroke and in healthy individuals. Acta Neurol Scand. 2007;116:26–30. doi: 10.1111/j.1600-0404.2006.00765.x. [DOI] [PubMed] [Google Scholar]
- 28.Özkök E, Aydın M, Babalık E, Özbek Z, İnce N, Kara İ. Combined impact of matrix metalloproteinase-3 and paraoxanase 1 55/192 gene variants on coronary artery disease in Turkish Patients. Med Sci Monit. 2008;14:CR536–42. [PubMed] [Google Scholar]
- 29.Kaman D, İlhan N, Metin K, Akbulut M, Üstündağ B. A preliminary study of human paraoxanase and PON 1 L/M 55 –PON1 Q/R 192 polymorphisms in Turkish patients with coronary artery disease. Cell Biochem Funct. 2009;27:88–92. doi: 10.1002/cbf.1539. [DOI] [PubMed] [Google Scholar]
- 30.Taşkıran P, Çam SF, Şekuri C, Tüzün N, Alioğlu E, Altıntaş N, et al. The relationship between paraoxonase gene Leu-Met (55) and Gln-Arg (192) polymorphisms and coronary artery disease. Arch Turk Soc Cardiol. 2009;37:473–8. [PubMed] [Google Scholar]
- 31.Wheeler JG, Keavney BD, Watkins H, Collins R, Danesh J. Four paraoxonase gene polymorphisms in 11212 cases of coronary heart disease and 12786 controls:meta-analysis of 43 studies. Lancet. 2004;363:689–95. doi: 10.1016/S0140-6736(04)15642-0. [DOI] [PubMed] [Google Scholar]
- 32.Arca M, Ombres D, Montali A, Campagna F, Mangieri E, Tanzilli G, et al. PON1 L55M polymorphism is not a predictor of coronary atherosclerosis either alone or in combination with Q192R polymorphism in an Italian population. Eur J Clin Invest. 2002;32:9–15. doi: 10.1046/j.1365-2362.2002.00935.x. [DOI] [PubMed] [Google Scholar]
- 33.Mukamal KJ, Pai JK, Jensen MK, Rimm EB. Paraoxonase 1 polymorphisms and risk of myocardial infarction in women and men. Circ J. 2009;73:1302–7. doi: 10.1253/circj.cj-08-0912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang M, Lang X, Zou L, Huang S, Xu Z. Four genetic polymorphisms of paraoxonase gene and risk of coronary heart disease:a meta-analysis based on 88 case-control studies. Atherosclerosis. 2011;214:377–85. doi: 10.1016/j.atherosclerosis.2010.11.028. [DOI] [PubMed] [Google Scholar]
- 35.Hazar A, Dilmeç F, Göz M, Koçarslan A, Aydın MS, Demirkol AH. The paraoxonase 1 (PON1) gene polymorphisms in coronary artery disease in the Southeastern Turkish population. Turk J Med Sci. 2011;41:895–902. [Google Scholar]
- 36.Schmidt R, Schmidt H, Fazekas F, Kapeller P, Roob G, Lechner A, et al. MRI cerebral white matter lesions and paraoxonase PON1 polymorphisms:three-year follow-up of the austrian stroke prevention study. Arterioscler Thromb Vasc Biol. 2000;20:1811–6. doi: 10.1161/01.atv.20.7.1811. [DOI] [PubMed] [Google Scholar]
- 37.Rios DL, D’Onofrio LO, Cerqueira CC, Bonfim-Silva R, Carvalho HG, Santos-Filho A, et al. Paraoxonase 1 gene polymorphisms in angiographically assessed coronary artery disease:evidence for gender interaction among Brazilians. Clin Chem Lab Med. 2007;45:874–8. doi: 10.1515/CCLM.2007.136. [DOI] [PubMed] [Google Scholar]
- 38.Yunoki K, Naruko T, Inaba M, Inoue T, Nakagawa M, Sugioka K, et al. Gender-specific correlation between plasma myeloperoxidase levels and serum high-density lipoprotein-associated paraoxonase-1 levels in patients with stable and unstable coronary artery disease. Atherosclerosis. 2013;231:308–14. doi: 10.1016/j.atherosclerosis.2013.08.037. [DOI] [PubMed] [Google Scholar]


