Abstract
The composition of the multisubunit eukaryotic RNA exosome was described more than a decade ago, and structural studies conducted since that time have contributed to our mechanistic understanding of factors that are required for 3′-to-5′ RNA processing and decay. This chapter describes the organization of the eukaryotic RNA exosome with a focus on presenting results related to the noncatalytic nine-subunit exosome core as well as the hydrolytic exo- and endoribonuclease Rrp44 (Dis3) and the exoribonuclease Rrp6. This is achieved in large part by describing crystal structures of Rrp44, Rrp6, and the nine-subunit exosome core with an emphasis on how these molecules interact to endow the RNA exosome with its catalytic activities.
1. Introduction
3′ to 5′ RNA decay is an evolutionarily conserved process in all known kingdoms of life and the family of enzymes that catalyze RNA decay share mechanistic and structural relationships. In eukaryotes, nuclear and cytoplasmic 3′ to 5′ decay is catalyzed by an essential multi-subunit complex termed the RNA exosome [1, 2] (Fig. 3.1). The RNA exosome includes a non-catalytic core formed by six subunits (Rrp41, Rrp45, Rrp42, Rrp43, Mtr3 and Rrp46) that share similarity to RNase PH and three subunits (Csl4, Rrp4 and Rrp40) that share similarity to proteins containing S1/KH RNA binding domains. The exosome core associates with two hydrolytic endo- and exoribonucleases (Rrp44 and Rrp6) that catalyze processive and distributive 3′ to 5′ exoribonuclease activities as well as endoribonuclease activities.
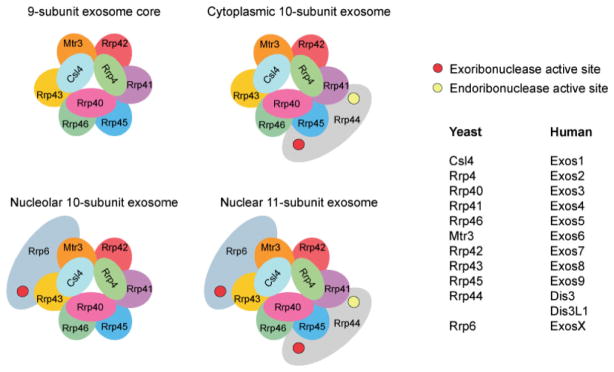
Figure 3.1.
Schematics of eukaryotic exosomes. Cartoon schematics depicting subunit compositions and general architecture of the 9-subunit exosome core (upper left), the cytoplasmic 10-subunit exosome (upper right), the nucleolar 10-subunit exosome (lower left), and nuclear 11-subunit exosome (lower right). Subunits are labeled and color coded and include the PH-like ring subunits Mtr3 (orange), Rrp42 (red), Rrp41 (purple), Rrp45 (blue), Rrp46 (green) and Rrp43 (yellow); the S1/KH domain proteins Csl4 (light blue), Rrp4 (green) and Rrp40 (pink); the catalytic subunits Rrp44 (gray) and Rrp6 (gray-blue). The S1/KH protein ring is shown on the top of the PH-like ring with Rrp44 shown below the PH-like ring to reflect structural models of the complex. Rrp6 is shown on the other side of the complex below the PH-like ring, although there is no definitive structural data for this complex. The exoribonuclease active sites are depicted by red circles in Rrp44 and Rrp6, and the endoribonuclease active site of Rrp44 is depicted with a yellow circle. The names of subunits are indicated on the right under headings for yeast and human although human proteins are often referred to by the corresponding yeast nomenclature.
The RNA exosome is essential in budding yeast and its subunit composition is largely conserved from yeast to human [3–5]. In yeast, nuclear RNA exosomes include the nine-subunit exosome core, Rrp44 and Rrp6, while cytoplasmic exosomes appear to include only the exosome core and Rrp44 [3, 6]. Interestingly, human encodes two Rrp44 homologs as well as Rrp6, and while each can associate with the RNA exosome core, they exhibit distinct subcellular localizations [7]. This observation suggests that RNA exosome subunit composition may be dynamically regulated or that distinct exosomes exist for distinct functions. For instance, subcellular localization patterns suggest the existence of a nucleolar human exosome that includes the RNA exosome core and Rrp6.
Components of the eukaryotic RNA exosome share evolutionary relationships to bacterial and archaeal factors that catalyze 3′–5′ RNA decay. In bacteria 3′-to-5′ RNA decay is catalyzed by RNase II and RNase R, two processive hydrolytic enzymes that share similarity to Rrp44; RNase D, a distributive hydrolytic enzyme that shares similarity to Rrp6; and PNPase, a processive phosphorolytic exoribonuclease that shares similarities to the non-catalytic human RNA exosome core [8, 9]. PNPase is a multi-domain protein that homooligomerizes as a trimer to form a two-ring structure that features a prominent central channel. The top ring is formed by S1/KH domains, while the bottom ring is formed by PH-domains that harbor the 3′-to-5′ processive phosphorolytic active sites. RNA must pass through the central channel to enter the phosphorolytic chamber [10].
Archaeal exosomes are also processive phosphorolytic enzymes [11] but are composed of up to four individually encoded proteins that oligomerize to form a two-ring structure analogous to PNPase. In this case, six PH-domain subunits form the bottom ring while three S1/KH domain proteins form the top ring [12, 13]. No RNase II or RNase D family members have yet been identified in archaea [14, 15]. Analogous to PNPase, archaeal exosome rings possess a central channel that guides RNA substrates into the phosphorolytic chamber and active sites [16].
The eukaryotic RNA exosome core is structurally related to bacterial PNPase and archaeal exosomes, although it is composed of nine distinct subunits. Furthermore, unlike PNPase and archaeal exosomes, the nine-subunit eukaryotic exosome core is devoid of catalytic or phosphorolytic activities [17, 18]. Therefore, it seems that the RNA exosome core has diverged mechanistically from its bacterial and archaeal cousins, dropping phorphorolytic catalytic capacity in its core in favor of interactions with the hydrolytic endo- and exoribonuclease Rrp44 and the hydrolytic exoribonuclease Rrp6 as well as protein cofactors such as the TRAMP and SKI complexes [2], which presumably add additional layers of regulation to 3′-to-5′ decay pathways in eukaryotes. Structural comparisons between the eukaryotic RNA exosome core and enzymes from bacteria and archaea have been extensively discussed elsewhere [19], so this chapter focuses on the structure and functions of the eukaryotic exosome core; its associated ribonucleases, Rrp44 and Rrp6; and their role in forming catalytically competent cytoplasmic, nuclear, and nucleolar exosomes.
2. Global architecture of the eukaryotic exosome core
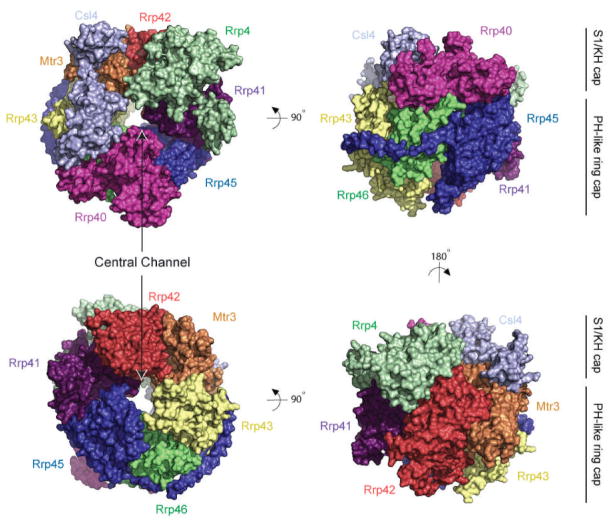
The structure of the human nine-subunit exosome core (Exo9) revealed a pseudo-hexameric six-component ring composed of the RNase PH-like proteins Rrp41, Rrp45, Rrp42, Rrp43, Mtr3 and Rrp46 that is capped by a three-component ring formed by the S1/KH-domain proteins Csl4, Rrp4 and Rrp40 (Fig. 3.2; [17]). This structure revealed overall architectural similarities to bacterial PNPase [21–23] and archaeal exosomes [12, 16] including a prominent central channel. In the human exosome structure, Rrp4 bridges Rrp41 and Rrp42, Rrp40 bridges Rrp45 and Rrp46, and Csl4 contacts Mtr3 and, to lesser extent, Rrp43. While archaeal exosomes form stable and catalytically active six-subunit RNase PH subunit rings, eukaryotic exosomes require at least one cap protein to form stable complexes in vitro. The general architecture and subunit composition of the human Exo9 core is likely conserved across eukaryotic phylogeny based on sequence analysis and conservation of individual subunits in organisms ranging from budding yeast to man [17]. Furthermore, some human exosome core subunits can complement deletion of the corresponding yeast genes [24, 25].
Figure 3.2.
Structure of the human nine-subunit exosome core. Surface representations of the human exosome core (PDB 2NN6) from the top (upper left) and bottom (lower left) in addition to two orthogonal views depicting the structure from side views to enable visualization of each exosome subunit in the complex. The subunits are labeled and color coded as in Fig. 3.1 showing Mtr3 (orange), Rrp42 (red), Rrp41 (purple), Rrp45 (blue), Rrp46 (green) and Rrp43 (yellow), and the S1/KH-domain proteins Csl4 (light blue), Rrp4 (green), and Rrp40 (pink). The central channel is apparent in the left panels and indicated by a label and arrows. The positions of the S1/KH cap and PH-like ring are indicated in the right panels with lines and labels. All structure depictions generated with the program PyMol [20].
3. RNase PH-like domains comprise a PH-like ring in eukaryotic exosomes
RNase PH domains are comprised of a βαβα-fold and are conserved in RNase PH, and PNPase in prokaryotes, archaeal exosomes, and eukaryotic exosomes (Fig. 3.3). Exosome subunits with structural homology to RNase PH are thus referred to as “PH-like” proteins and include Rrp41, Rrp42, Rrp43, Rrp45, Rrp46, and Mtr3. The six-component PH-like ring in eukaryotes consists of three distinct heterodimer pairs that are arranged in a head-to-tail configuration. Although eukaryotic organisms have diverged to encode these subunits in six distinct genes, Rrp41, Mtr3 and Rrp46 share higher sequence and structural similarity to archaeal Rrp41 or PNPase RNase PH 2-like proteins while Rrp42, Rrp43 and Rrp45 share greater similarity to archaeal Rrp42 or PNPase RNase PH 1-like proteins. In eukaryotes, Rrp41 pairs with Rrp45, Rrp43 pairs with Rrp46, and Mtr3 pairs with Rrp42 (Fig. 3.3).
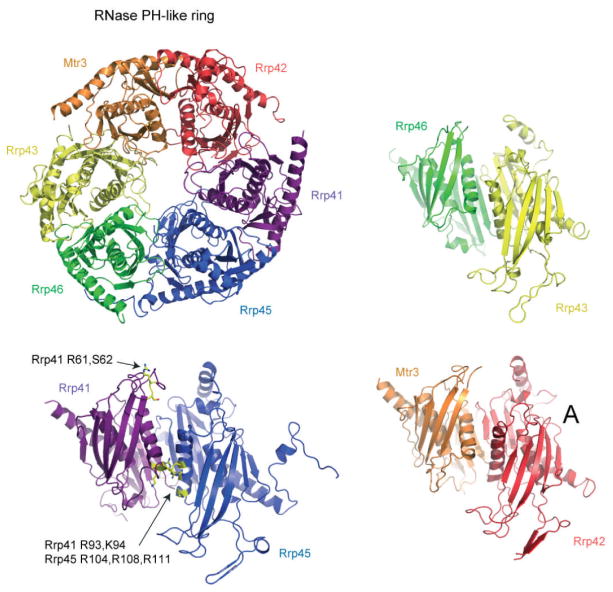
Figure 3.3.
The human PH-like subunit ring. Ribbon diagram of the human PH-like ring (PDB 2NN6) depicting β strands as arrows, α helices as coiled ribbons, and connecting elements as thin tubes (upper left). The view of the intact PH-like ring is from the “top” as presented in Figs. 3.1 and 3.2 with subunits labeled and color coded with Mtr3 (orange), Rrp42 (red), Rrp41 (purple), Rrp45 (blue), Rrp46 (green) and Rrp43 (yellow). The Rrp41/Rrp45 heterodimer is shown lower left in a side view as if the viewer were inside the central channel looking outward. The subunits are labeled and color coded as before with amino acid side chains implicated in RNA-binding interactions labeled and side chains colored yellow for Rrp41 Arg61, Ser62, Arg93, and Lys94 and Rrp45 Arg104, Arg108, and Arg111. The Rrp46/Rrp43 and Mtr3/Rrp42 heterodimers are shown in a similar orientation with subunits labeled and color coded as before.
The majority of amino acid side chains that form the phosphorolytic active site and RNA-binding surfaces in bacterial PNPase and the archaeal exosome are not conserved in eukaryotic exosomes; however, amino acid side chains that comprise one potential RNA-binding surface are conserved in the Rrp41–Rrp45 heterodimer interface (Fig. 3.2). The location of this putative RNA-binding site was evident in sequence alignments and by location of a tungstate ion in crystals of the human exosome core [17]. Furthermore, point mutations that disrupt this anion-binding site (Rrp41 R95E/R96E and Rrp41 K62E/S63D) attenuate RNA decay activity of cytoplasmic exosomes (Exo9 core plus Rrp44) in vitro [26]. These residues are not required in vivo as these and other point mutations in the central channel and RNA-binding surface do not lead to a growth defect in Saccharomyces cerevisiae [18, 27]. Given the structural similarities between bacterial and archaeal exosomes, it remains possible that additional residues within the PH-like ring contribute to RNA binding as several basic residues not involved in intersubunit contacts face the central channel and are conserved across eukaryotes [17].
4. S1 and KH domains cap the PH-like ring
Rrp4, Rrp40, and Csl4 form a ring and cap the PH-like ring of the eukaryotic exosome core. Each of these subunits contain an N-terminal domain (NTD) that makes extensive contacts to their respective RNase PH-like 2 binding partner, tethering the cap proteins onto the PH-like ring (Fig. 3.4; [17]). The Rrp4 NTD interacts with Rrp41, the Rrp40 NTD binds to Rrp46, and the Csl4 NTD contacts Mtr3. Rrp4 and Rrp40 include two putative RNA binding domains, KH type I and S1, while Csl4 contains an S1 domain and C-terminal zinc ribbon fold. KH type I domains [29] feature a β1-α1-α2-β2-β3-α3 secondary structure topology and tertiary structure that consists of three β-strands that form a sheet and pack against three α helices. Single-stranded RNA (ssRNA) typically binds KH type I domains via surfaces formed by residues within helix α1, a conserved GXXG motif between helices α1 and α2; helix α2, the variable loop between strands β2 and β3; and residues within strand β2 [30]. However, this motif is not conserved in eukaryotic Rrp4 and Rrp40; instead, a unique and conserved GXNG motif is found between strands β7 and β8. The GXNG motif is buried at the interface between the S1 and KH domains, and unless RNA binding induces a dramatic conformational change to expose the GXNG motif, this surface likely contributes to the structural stability of the protein rather than to RNA binding.
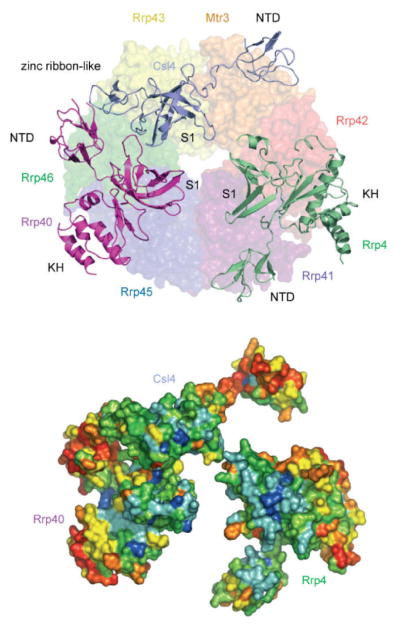
Figure 3.4.
The human S1/KH protein ring. Ribbon diagram of the human S1/KH proteins on top of a transparent surface representation of the PH-like ring shown from the “top” with subunits labeled and color coded as in previous figures with Csl4 (light blue), Rrp4 (green), and Rrp40 (pink) (top). The N-terminal domain (NTD), S1, KH, and zinc ribbon-like domains are labeled. Note that the S1 domains from each S1/KH protein face the central channel. The bottom panel depicts just the S1/KH proteins in surface representation in the same orientation as in the top panel indicating sequence conservation colored from red (variable) to blue (conserved) as calculated by ConSurf [28] from manually assembled sequence alignments [17]. Note that the most conserved surfaces are located on the S1 domains that face the central channel.
S1 domains [31] contain an OB (oligonucleotide/oligosaccharide binding) fold with a five-stranded β-sheet coil to form a closed β-barrel [32]. OB domains generally bind nucleic acid through surfaces composed of positively charged and hydrophobic residues on the solvent exposed β-sheet [33]. Rrp40 exhibits the typical β-barrel, with long loops between β3 and β4 that project into the central channel [17,34]. A comparably long loop between β3 and β4 is also present in Rrp4; however, both of these regions are disordered in the structure of the human exosome. These loops project toward the central channel and contain several basic residues that are highly conserved throughout eukaryotes and thus may be responsible for binding or guiding RNA into the central channel.
The zinc ribbon domain of Csl4 contains a three-stranded β-sheet and is structurally related to archaeal Csl4; however, the four cysteine residues that coordinate Zn2+ in archaea are not conserved in eukaryotes, nor is Zn2+ coordinated in human Csl4. While this domain in not essential in vivo, its deletion results in conditional defects in nonsense-mediated decay in budding yeast [35]. Furthermore, while Csl4 is essential in vivo, budding yeast strains expressing Csl4 truncations that lack either the NTD or S1 domain are still viable. These data are perhaps consistent with the observation that Csl4 is not stably associated with exosomes reconstituted in vitro [36]. In contrast, each of the Rrp4 and Rrp40 NTD, S1, and KH domains is essential in budding yeast [35].
The arrangement of the cap subunits on the PH-like six-subunit ring in the human exosome positions conserved and positively charged putative RNA-binding S1-domain surfaces towards the central channel while the NTDs and KH domains are located on the periphery of the complex (Fig. 3.4). While RNA binding cannot be detected in vitro for Rrp40 [34], reconstituted exosomes lacking one cap subunit exhibit weaker RNase protection patterns compared to a complete Exo9 core [36]. Alignment of multiple crystal structures of the archaeal exosome from Sulfolobus solfataricus indicates that the cap proteins, specifically Rrp4, sometimes deviate from the three fold symmetry observed in other crystal structures, suggesting limited conformational flexibility [13,37]. Furthermore, Csl4 and Rrp40, and not the PH-like subunits, display increased flexibility when the crystal structure of the human exosome core is compared to a 14 Å cryo-electron microscopy (EM) reconstruction of reconstituted Exo10 from budding yeast [17,36]. It remains unclear if these differences are biologically relevant or if they simply reflect subtle differences between human and yeast RNA exosomes.
5. Rrp44, a hydrolytic endoribonuclease and processive exoribonuclease
Rrp44 includes five domains, an N-terminal Pilus-forming N-terminus (PIN) domain that contains an active site capable of endoribonuclease activity [38], two cold-shock domains (CSD1 and CSD2), a central ribonuclease domain (RNB) that catalyzes processive 3′-to-5′ exoribonuclease activity and a C-terminal S1 domain (Fig. 3.5). Humans and other higher eukaryotes encode three homologs of budding yeast Rrp44, including Dis3 and Dis3L, which are localized to the nucleus and cytoplasm, respectively [7, 39]. The third paralog, Dis3L2, lacks a PIN domain and has not been extensively characterized to date. Human Dis3 is most similar to yeast Rrp44, and budding yeast strains depleted of Rrp44 can be partially rescued by human Dis3, but not Dis3L [7]. Dis3 and Dis3L associate with exosomes in vivo as evidenced by their ability to pull down other exosome core subunits as well as Mtr4 in lysate from human cells [7, 39].
Figure 3.5.
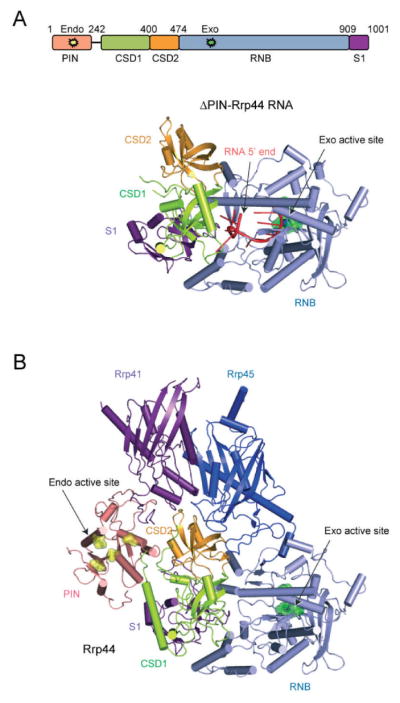
The endo- and exoribonuclease Rrp44. (A) Schematic representation of the Rrp44 polypeptide indicating the PIN, CSD1, CSD2, RNB, and S1 domains with labels and color coded pink, green, orange, blue, and purple, respectively. Amino acid numbering is for Saccharomyces cerevisiae Rrp44. Below the schematic is the structure of yeast Rrp44 bound to RNA (PDB 2VNU) in cartoon ribbon representation with β strands as arrows, loops as ribbons and helices as solid tubes. The domains are labeled and color coded as in the schematic. The exoribonuclease active site is indicated by a label and arrow with key residues colored green in stick and surface representation. RNA is shown as a red ribbon with bases indicated as sticks with the 5′ end labeled and indicated by an arrow; the 3′ end is buried in the exoribonuclease active site. (B) Structure of full-length yeast Rrp44 in complex with yeast Rrp41 and Rrp45 (PDB 2WP8) in cartoon ribbon representation color coded as in the top panel with the PIN domain in pink. The endoribonuclease active site is labeled and indicated by an arrow with key residues colored yellow in stick and surface representation.
Rrp44 is structurally and mechanistically related to bacterial RNase II and RNase R [40], and a crystal structure of budding yeast Rrp44 lacking the PIN domain determined in complex with RNA [41] revealed many overall similarities to RNase II in apo- and RNA-bound states (Fig. 3.5; [42]) including conservation of CSD1, CSD2, RNB, and S1 domains. Comparison of the structures reveals that Rrp44 and RNase II RNB active sites make similar contacts to the four ribonucleotides upstream of the 3′ OH. Based on structural similarities within their active sites, it is likely that Rrp44 employs the same two-metal-ion catalytic mechanism as RNase II to hydrolyze RNA 3′-to-5′, resulting in the release of 5′ nucleotide monophosphates [19]. One of two catalytic magnesium ions is visible in the Rrp44 structure and is coordinated by Asp543 and Asp552. Although the second magnesium is not detected in the Rrp44-RNA structure, it is likely to be coordinated by Asp551 and Asp549. In fact, mutation of Asp551 to Asn retains RNA binding, but leads to loss of exoribonuclease activity in vitro, and a slow growth phenotype in vivo [18].
Several basic residues within the RNB domain line the active site and contact RNA via contacts to the RNA backbone. An exception is Tyr595, which participates in base stacking interactions with the 3′ most nucleotide, while bases 1–5 stack with each other [41]. The prominence of protein contacts to the RNA backbone and absence of base-specific interactions are consistent with the enzyme’s lack of sequence specificity; however, several direct or water-mediated hydrogen bonds between protein residues and the 2′ hydroxyl groups of nucleotides 2, 3, 4, 6, and 8 impart specificity in recognition of ssRNA rather than ssDNA as Rrp44 binds ssRNA 50-fold better than ssDNA [41]. Rrp44 can degrade duplex RNAs with 3′ single-stranded overhangs as short as four nucleotides but is most efficient with overhangs of at least 10 nucleotides [41], similar to bacterial RNase R [43]. The requirement of a 3′ single-stranded overhang is consistent with the structures insofar as the channel to the RNB active site is only large enough to accommodate ssRNA.
Many similarities are shared between Rrp44 and RNase II; nevertheless, differences are readily apparent with respect to how they engage RNA via the CSDs and S1 domain. In RNase II, RNA threads through a channel formed by the CSDs and S1 domain into the RNB active site. In addition to contacts to the 3′ end via the RNB domain, CSD2 and the S1 domain make additional contacts to single stranded RNA that contribute to the enzyme’s processive activity [42]. In contrast, the Rrp44-RNA complex shows the 5′ end of RNA in a path perpendicular to that observed in RNase II in a channel formed at the interface between the RNB domain and CSD1 which is 15 Å closer to the RNB domain compared to RNase II. The mode of RNA binding by RNase II explains its processive activities, while the binding mode observed for Rrp44 may facilitate alternative routes for RNA ingress including substrates exiting the central channel of the eukaryotic exosome core. Although it is unclear if the S1/KH domain ring of the eukaryotic Exo9 core functions in a manner analogous to the CSDs and S1 domain in RNase II with respect to ssRNA binding, it is interesting to note that the S1/KH ring and CSDs/S1 ring share overall architectural similarities, particularly with respect to their positions over an extended channel in the Rrp44 RNB domain and the exosome PH-like ring.
The structure of the yeast Rrp44 PIN domain was revealed in a crystal structure of full-length Rrp44 bound to Rrp41 and Rrp45 (Fig. 3.5; [26]). The PIN domain consists of a central twisted five-stranded β-sheet flanked by α helices and is similar to the overall fold observed in other RNase H family members [44, 45]. PIN domains that catalyze nuclease activity include four conserved acidic residues that coordinate two divalent cations to cleave nucleic acid via a similar mechanism involving two-metal-ion catalysis [46–49]. The endoribonucleolytic active site of the Rrp44 PIN domain from budding yeast consists of Asp91, Glu120, Asp171, and Asp198 (Fig. 3.5).
Rrp44 endoribonuclease activity is dependent on the integrity of the PIN active site and can be observed in vitro in the presence of millimolar manganese concentrations; however, an active site point mutation (D171N) that abolishes endonuclease activity in vitro does not display a growth defect in vivo [38]. These observations make the biological function of the endoribonuclease activity unclear, but it is important to note that a combination of mutations (D551N/D171N) that simultaneously disrupt exoribonuclease and endoribonuclease activities results in synthetic lethality [35, 38]. The four acidic residues are conserved in human Dis3 but not in Dis3L, as it only retains two of the four acidic residues. Consistent with this observation, only Dis3 appears catalytically competent for endoribonucleolytic function [7]. In addition to its catalytic activities, the PIN domain interacts directly with Rrp41/Rrp45 heterodimer as exemplified in the structure of Rrp44 in complex with Rrp41 and Rrp45 (Fig. 3.5). While the endoribonuclease activity is not essential, deletion of the PIN domain is lethal in budding yeast suggesting that contacts to Rrp41/Rrp45 and presumably the exosome core are essential for growth [35].
6. Rrp44 and the 10-component exosome
Models of the eukaryotic 10-component cytoplasmic exosome Exo1044 have been proposed based on the X-ray structure of the human nine-component exosome core [17], the crystal structure of the budding yeast Rrp41–Rrp45-Rrp44 trimer (Fig. 3.5; [26]), and negative-stain and cryo-EM structures of apo budding yeast Rrp44 bound to the core exosome (Exo1044) [36,50]. These models reveal that Rrp44 is anchored to the bottom of exosome core through extensive interactions between the PIN domain of Rrp44 and Rrp41/Rrp45 in addition to contacts between the Rrp44 CSD1 and Rrp43 (Fig. 3.6).
Figure 3.6.
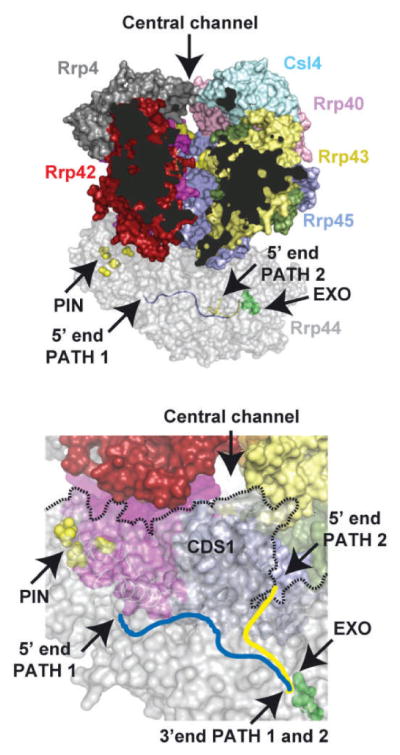
Model of the 10-subunit cytoplasmic exosome. The top panel depicts a side view of the 10-subunit exosome in surface representation that was constructed by aligning Rrp41/Rrp45 from the yeast Rrp41/Rrp45/Rrp44 and human nine-subunit exosome core structures. To enable visualization of the exosome core central channel, the Mtr3 subunit was removed from the complex and parts of Rrp42 and Rrp43 were removed from view (dark areas). The subunits are colored as before except Rrp4 is now depicted in gray. The central channel is labeled and indicated by an arrow at the top of the channel. Rrp44 is depicted in transparent gray with endoribonuclease (yellow) and exoribonuclease (green) sites labeled with side chains in surface representation. Two RNA paths are depicted in Rrp44, one derived from the structure of RNase II in complex with RNA (blue; PDB 2I×1) which passes through the CSDs and S1 domain and the other derived from the dPIN-Rrp44/RNA complex (yellow; PDB 2NVU) which passes by CSD1 and the RNB domain. Bottom panel shows an orthogonal view looking up into the exosome central channel indicating the position of CDS1 which appears to block a direct path from the exosome central channel to the RNB active site (green). RNA paths, PIN and EXO active sites are labeled as in the top panel.
How does RNA engage the activities of Rrp44 when Rrp44 is associated with the exosome core? Biochemical studies revealed that charge-swap mutations in Rrp41 and the central channel diminished Rrp44 activity in the presence of the exosome core [26]. Furthermore, a 12-Å resolution cryo-EM structure of gold-labeled RNA bound to a catalytically inactive Exo1044 from budding yeast showed the gold label to be located in the center of the exosome coincident with the location of the conserved channel and additional density not present in apo reconstructions was observed within the channel of RNA-bound structures [36]. These data are consistent with the hypothesis that RNA transits through the exosome core and central channel to engage the Rrp44 exoribonuclease active site.
The path of RNA to the Rrp44 exoribonuclease active site can be further explored through alignment of crystal structures of human Exo9 [17], yeast Rrp44 in complex with RNA [41], and yeast Rrp44-Rrp41-Rrp45 [26]. In this model, RNA could transit through and exit the central channel of the exosome to reach Rrp44, yet would require a ~45° turn around CSD1 before entering the RNB channel and active site (Fig. 3.6). While this path is consistent with RNase protection assays that indicate RNA substrates require at least 31–34 single-stranded nucleotides at the 3′ end to be engaged by a 10-component exosome [26], it is also conceivable that RNA binding and/or additional protein-protein contacts between Rrp44 and Exo9 might induce conformational changes to facilitate a more direct RNA path through the exosome central channel to the Rrp44 RNB active site.
Although this model is attractive, the importance of the central channel for exosome function in vivo remains unclear as channel lining mutations exhibit no apparent phenotype [18, 27] and because models of Rrp44 in complex with Exo9 indicate that both endoribonuclease and exoribonuclease active sites remain exposed to solvent (Fig. 3.6). These observations suggest that the channel is not essential in vivo or that alternative routes exist for RNA to access the Rrp44 active sites. In fact, recent biochemical studies revealed that the exosome core modulates both Rrp44 endo- and exoribonuclease activities because both activities are attenuated in Rrp44-associated ten-subunit exosomes [27]. Furthermore, Rrp44 endo- and exoribonuclease activities are dependent on the integrity of the central channel as channel-occluding mutations severely diminish both RNase activities and binding to RNA. Existing models for Exo1044 cannot fully explain these observations, and further structural work will be required to understand how the exosome core regulates access to both Rrp44 exo- and endoribonuclease active sites.
7. Rrp6, a eukaryotic exosome subunit with distributive hydrolytic activities
Rrp6 is associated with the nuclear exosome (Exo1144/6) in budding yeast and humans, although recent evidence suggests the existence of a nucleolar exosome in human cells consisting of Rrp6 and the Exo9 core which we denote Exo106 [7]. Rrp6 is the only nonessential subunit of the exosome; however, budding and fission yeast strains lacking Rrp6 exhibit a temperature-sensitive growth phenotype and accumulate many nuclear RNA precursors [51, 52]. Rrp6 is involved in 3′ end processing of snRNAs, snoRNAs [52, 53], pre-rRNAs [54], destruction of aberrant nuclear RNAs, and the degradation of cryptic unstable transcripts that result from bidirectional transcription [55, 56]. In fission yeast, Rrp6 and the nuclear exosome cooperate with the RITS complex to induce constitutive heterochromatin spreading at centromeres [57, 58] and to silence meiotic genes in vegetative cells [59, 60]. Evidence for exosome-independent functions of Rrp6 have been reported in budding yeast, but these findings are restricted to nuclear RNAs [61, 62]. In other species, including trypanosomes [63], Drosophila [5], and humans [7, 64], Rrp6 has been detected in the cytoplasm, although the biological implications of these findings have not been fully characterized.
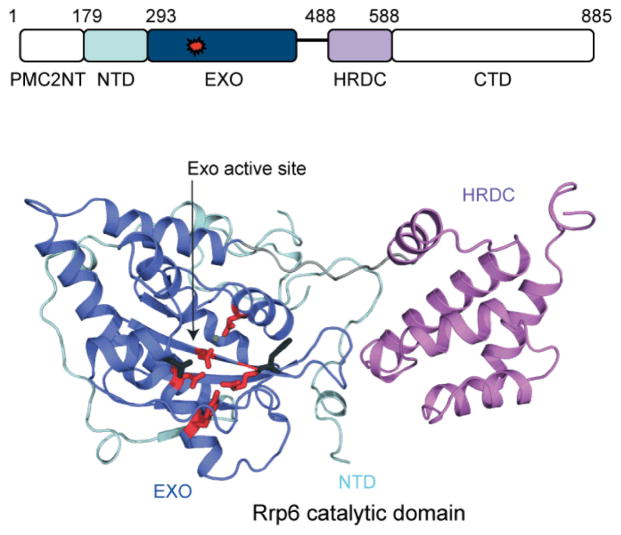
Rrp6 is composed of a PMC2NT domain, a NTD, a DEDD-Y exoribonuclease domain (EXO), a helicase and RNase D carboxy terminal (HRDC) domain, and a C-terminal domain (CTD). The PMC2NT domain is required for interaction with its nuclear cofactor, Rrp47 [65]. The EXO domain of Rrp6 is related to RNase D and members of the DEDD-Y nuclease family that are so named for four conserved acidic residues, DEDD, and a conserved tyrosine that coordinate two metals to catalyze distributive, hydrolytic 3′-to-5′ exoribonuclease activity via a two-metal-ion mechanism [66]. HRDC domains are posited to bind nucleic acid. While it remains unclear if the Rrp6 HRDC binds RNA, deletion of this domain in human Rrp6 results in diminished catalytic activity [67]. The CTD contains a nuclear localization sequence and is not necessary for interaction with the exosome [61]. At present, no structural information exists for the PMC2NT and CTD domains and both domains are predicted to be unstructured.
Crystal structures of Rrp6 fragments that include the NTD, EXO, and HRDC domains were determined from budding yeast and human (Fig. 3.7; [66, 67]). The NTD wraps around the EXO domain and forms a platform with a linker that connects the EXO and HRDC domains. The EXO core shares the α/β fold observed in the Klenow fragment of DNA Polymerase I from Escherichia coli [68] as well as conserved DEDD residues in the EXO domain that are required for coordination of the divalent metal ions for two-metal-ion catalysis. In Rrp6, the conserved tyrosine side chain activates a nucleophilic water molecule for cleavage of the phosphodiester. Mutating any of the conserved active site residues either abolishes or severely attenuates exoribonuclease activity in vitro [69]. The human and yeast Rrp6 structures show the EXO domain and active site to be solvent-exposed, thus potentially explaining Rrp6 distributive activity as these structures lack channels that could bind or guide single-stranded RNA into the active site. The HRDC domain resembles the first HRDC domain of E. coli RNase D [70] and consists of five α helices. The HRDC confers substrate specificity, as disruption of the EXO/HRDC interface in budding yeast Rrp6 (D457A) results in deficiencies in 3′ end processing of nuclear RNAs, such as snRNAs, but not in the clearance of the 5′ ETS fragment of prerRNA and maturation of 5.8S rRNA [71]. Although Rrp6 fragments containing the NTD/EXO/HRDC domains exhibit similar catalytic activities when compared to full-length Rrp6, it is worth noting that Rrp6 fragments that include the NTD, EXO, and HRDC do not complement growth defects observed in budding yeast strains lacking RRP6 [66].
Figure 3.7.
The exoribonuclease Rrp6. Schematic of the Rrp6 polypeptide indicating the PMC2NT (white), NTD (light blue), EXO (dark blue), HRDC (purple) and C-terminal (CTD; white) domains with the exoribonuclease active site colored red. Amino acid numbering is for human Rrp6. Lower panel depicts a cartoon ribbon representation of the human Rrp6 catalytic domain structure with α helices in cartoon ribbon, loops as thin ribbons, and β strands as arrows (PDB 3SAF). Domains are colored and labeled as in the schematic with the linker between the EXO and HRDC domain in gray. The EXO active site is labeled and indicated with an arrow with key side chains colored red and shown in stick representation; the magnesium ion is shown as a small green sphere.
Although the structures of the catalytic domains from budding yeast and human share many similarities throughout the EXO and HRDC domains, the human Rrp6 active site appears more solvent exposed when compared to the active site in yeast Rrp6 [67]. This structural difference is attributed to a difference in linker length between the EXO and HRDC domains – in yeast and lower eukaryotes, the linker is 26 residues, but in humans, the linker is only 10 residues long [66, 67]. The net effect of a shorter linker is a more solvent exposed human Rrp6 active site that it is able to degrade structured RNAs more efficiently than yeast Rrp6 in vitro, presumably because these larger substrates can access the human Rrp6 active site [67].
8. Rrp44, Rrp6, and the 11-component nuclear exosome
The 11-subunit nuclear exosome of budding yeast (Exo1144/6) is the best characterized Rrp6-associated exosome complex and includes the Exo9 core, Rrp44, and Rrp6 [3]. Although Rrp6 is not essential in budding yeast, mutation of the Rrp44 endoribonuclease site (D171N) in combination with rrp6Δ leads to a synthetic growth defect [38], and inactivation of the Rrp44 exoribonuclease site (D551N) with rrp6Δ results in synthetic lethality [18]. These data suggest overlapping functions for Rrp6 and Rrp44 activities in vivo.
Insights to how Rrp6 contributes to the activities of the exosome are beginning to emerge. As described above, association of Rrp44 with the Exo9 core attenuates the RNA binding and exoribonuclease activities of Rrp44 in Exo1044 in vitro in a manner dependent on the exosome core and central channel [27]. Interestingly, addition of Rrp6 in Exo1144/6 stimulates the endoribonuclease and endoribonuclease activities of Rrp44, independent of Rrp6 catalytic activity, and addition of Rrp44 that contains an inactivating mutation in its exoribonuclease domain severely inhibits Rrp6 in Exo1144exo-/6 [27]. These observations suggest that the exosome core and central channel mediate a dynamic interplay between Rrp44 and Rrp6 activities.
How does Rrp6 interact with the exosome? Structural details regarding this issue remain unknown; however, yeast two-hybrid studies suggest that Rrp6 interacts with the PH-like ring proteins Rrp41, Rrp43, Rrp46, and Mtr3 [72], and a 35-Å resolution negative-stain EM structure of the Leishmania tarentolae exosome purified from native sources positions Rrp6 towards the top of exosome core interacting with the cap proteins [73]. It is unknown if the Rrp6-binding surface is conserved from trypanosomes to humans. Also in question is whether Exo1144/6 is the sole nuclear exosome in humans, as the endogenous stoichiometry of human Dis3 to Rrp6 was roughly estimated to be 1:10 in human cells [7]. Thus, it remains unclear if Rrp6-associated exosome cores represent a predominant species, or if free Rrp6 is abundant in human cells. Reconstitution of Rrp6 associated 10 or 11 subunit nuclear exosomes from human or other higher eukaryote has not been reported to date. Further studies are required to address how the exosome core modulates Rrp6 activity and how Rrp6 stimulates Rrp44 ribonuclease activities. Structural studies addressing the aforementioned points could provide key insights to the biological functions of nuclear and nucleolar exosomes.
9. Conclusions
Structures and models for the eukaryotic exosome core illustrate striking architectural similarities to RNA-degrading enzymes in bacteria and archaea with respect to PH-like and S1/KH-domain rings that stack to form a prominent central channel wide enough to accommodate ssRNA substrates. In bacterial PNPase and archaeal exosomes, this channel harbors both RNA-binding surfaces and phosphorolytic active sites that confer processivity to these complexes by providing at least two RNA-binding surfaces that prevent RNA substrates from diffusing away from the complex between successive rounds of cleavage.
Eukaryotic exosomes appear to use the same strategy to engage RNA substrates by utilizing a noncatalytic core to bind and guide ssRNA substrates through the central channel to engage the RNase activities of Rrp44 and Rrp6, at least for the yeast exosome [27]. It remains unknown if this feature is conserved among all eukaryotic exosomes or if all RNA substrates engage the exosome core in a similar manner. Although it is clear that the exosome core can modulate the activities of Rrp44 and Rrp6, very little is known regarding how factors such as the TRAMP and SKI complexes change or affect substrate specificity or the activities of the RNA exosome. While much has been learned since discovery of the eukaryotic exosome, it will remain a significant challenge to determine the structural basis for eukaryotic exosome RNase activities as well as its association with cofactors that modulate its nuclear and cytoplasmic functions in RNA processing and decay.
Acknowledgments
Research reported in this publication was supported by the National Institute of General Medical Sciences of the National Institutes of Health under award numbers F31GM097910 (E.V.W) and R01 GM079196 (C.D.L). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
References
- 1.Mitchell P, Petfalski E, Tollervey D. The 3′ end of yeast 5.8S rRNA is generated by an exonuclease processing mechanism. Genes Dev. 1996;10:502–513. doi: 10.1101/gad.10.4.502. [DOI] [PubMed] [Google Scholar]
- 2.Houseley J, LaCava J, Tollervey D. RNA-quality control by the exosome. Nat Rev Mol Cell Biol. 2006;7:529–539. doi: 10.1038/nrm1964. [DOI] [PubMed] [Google Scholar]
- 3.Allmang C, Petfalski E, Podtelejnikov A, Mann M, Tollervey D, Mitchell P. The yeast exosome and human PM-Scl are related complexes of 3′ --> 5′ exonucleases. Genes Dev. 1999;13:2148–2158. doi: 10.1101/gad.13.16.2148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schneider C, Anderson JT, Tollervey D. The exosome subunit Rrp44 plays a direct role in RNA substrate recognition. Mol Cell. 2007;27:324–331. doi: 10.1016/j.molcel.2007.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Graham AC, Kiss DL, Andrulis ED. Differential distribution of exosome subunits at the nuclear lamina and in cytoplasmic foci. Mol Biol Cell. 2006;17:1399–1409. doi: 10.1091/mbc.E05-08-0805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mitchell P, Petfalski E, Shevchenko A, Mann M, Tollervey D. The exosome: a conserved eukaryotic RNA processing complex containing multiple 3′-->5′ exoribonucleases. Cell. 1997;91:457–466. doi: 10.1016/s0092-8674(00)80432-8. [DOI] [PubMed] [Google Scholar]
- 7.Tomecki R, Kristiansen MS, Lykke-Andersen S, Chlebowski A, Larsen KM, Szczesny RJ, Drazkowska K, Pastula A, Andersen JS, Stepien PP, Dziembowski A, Jensen TH. The human core exosome interacts with differentially localized processive RNases: hDIS3 and hDIS3L. EMBO J. 2010;29:2342–2357. doi: 10.1038/emboj.2010.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Carpousis AJ. The Escherichia coli RNA degradosome: structure, function and relationship in other ribonucleolytic multienzyme complexes. Biochem Soc Trans. 2002;30:150–155. [PubMed] [Google Scholar]
- 9.Symmons MF, Jones GH, Luisi BF. A duplicated fold is the structural basis for polynucleotide phosphorylase catalytic activity, processivity, and regulation. Structure. 2000;8:1215–1226. doi: 10.1016/s0969-2126(00)00521-9. [DOI] [PubMed] [Google Scholar]
- 10.Nurmohamed S, Vaidialingam B, Callaghan AJ, Luisi BF. Crystal structure of Escherichia coli polynucleotide phosphorylase core bound to RNase E, RNA and manganese: implications for catalytic mechanism and RNA degradosome assembly. J Mol Biol. 2009;389:17–33. doi: 10.1016/j.jmb.2009.03.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Evguenieva-Hackenberg E, Walter P, Hochleitner E, Lottspeich F, Klug G. An exosome-like complex in Sulfolobus solfataricus. EMBO Rep. 2003;4:889–893. doi: 10.1038/sj.embor.embor929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lorentzen E, Walter P, Fribourg S, Evguenieva-Hackenberg E, Klug G, Conti E. The archaeal exosome core is a hexameric ring structure with three catalytic subunits. Nat Struct Mol Biol. 2005;12:575–581. doi: 10.1038/nsmb952. [DOI] [PubMed] [Google Scholar]
- 13.Büttner K, Wenig K, Hopfner KP. Structural framework for the mechanism of archaeal exosomes in RNA processing. Mol Cell. 2005;20:461–471. doi: 10.1016/j.molcel.2005.10.018. [DOI] [PubMed] [Google Scholar]
- 14.Mian IS. Comparative sequence analysis of ribonucleases HII, III, II PH and D. Nucleic Acids Res. 1997;25:3187–95. doi: 10.1093/nar/25.16.3187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zuo Y, Deutscher MP. Exoribonuclease superfamilies: structural analysis and phylogenetic distribution. Nucleic Acids Res. 2001;29:1017–1026. doi: 10.1093/nar/29.5.1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lorentzen E, Dziembowski A, Lindner D, Seraphin B, Conti E. RNA channelling by the archaeal exosome. EMBO Rep. 2007;8:470–476. doi: 10.1038/sj.embor.7400945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liu Q, Greimann JC, Lima CD. Reconstitution, activities, and structure of the eukaryotic RNA exosome. Cell. 2006;127:223–1237. doi: 10.1016/j.cell.2006.10.037. [DOI] [PubMed] [Google Scholar]
- 18.Dziembowski A, Lorentzen E, Conti E, Séraphin B. A single subunit, Dis3, is essentially responsible for yeast exosome core activity. Nat Struct Mol Biol. 2007;14:15–22. doi: 10.1038/nsmb1184. [DOI] [PubMed] [Google Scholar]
- 19.Januszyk K, Lima CD, Jensen TH, editors. Structural Components and architectures of RNA exosomes. Vol. 702. Landes Bioscience and Springer Science; New York: 2010. pp. 9–28. [Google Scholar]
- 20.Delano WL. The PyMOL Moleculare Graphics System. DeLano Scientific; San Carlos, CA: 2002. http://www.pymol.org. [Google Scholar]
- 21.Ishii R, Nureki O, Yokoyama S. Crystal structure of the tRNA processing enzyme RNase PH from Aquifex aeolicus. J Biol Chem. 2003;278:32397–32404. doi: 10.1074/jbc.M300639200. [DOI] [PubMed] [Google Scholar]
- 22.Harlow LS, Kadziola A, Jensen KF, Larsen S. Crystal structure of the phosphorolytic exoribonuclease RNase PH from Bacillus subtilis and implications for its quaternary structure and tRNA binding. Protein Sci. 2004;13:668–677. doi: 10.1110/ps.03477004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shi Z, Yang WZ, Lin-Chao S, Chak KF, Yuan HS. Crystal structure of Escherichia coli PNPase: central channel residues are involved in processive RNA degradation. RNA. 2008;14:2361–2371. doi: 10.1261/rna.1244308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Brouwer R, Allmang C, Raijmakers R, van Aarssen Y, Egberts WV, Petfalski E, van Venrooij WJ, Tollervey D, Pruijn GJ. Three novel components of the human exosome. J Biol Chem. 2001;276:6177–6184. doi: 10.1074/jbc.M007603200. [DOI] [PubMed] [Google Scholar]
- 25.Mitchell P, Tollervey D. Musing on the structural organization of the exosome complex. Nat Struct Mol Biol. 2000;7:843–846. doi: 10.1038/82817. [DOI] [PubMed] [Google Scholar]
- 26.Bonneau F, Basquin J, Ebert J, Lorentzen E, Conti E. The yeast exosome functions as a macromolecular cage to channel RNA substrates for degradation. Cell. 2009;139:547–559. doi: 10.1016/j.cell.2009.08.042. [DOI] [PubMed] [Google Scholar]
- 27.Wasmuth EV, Lima CD. The exo- and endoribonucleolytic activities of yeast cytoplasmic and nuclear RNA exosomes are dependent on the non-catalytic core and central channel. Mol Cell. 2012 doi: 10.1016/j.molcel.2012.07.012. http://dx.doi.org/10.1016/j.molcel.2012.07.012. [DOI] [PMC free article] [PubMed]
- 28.Landau M, Mayrose I, Rosenberg Y, Glaser F, Martz E, Pupko T, Ben-Tal N. ConSurf 2005: the projection of evolutionary conservation scores of residues on protein structures. Nucleic Acids Res. 2005;33:W299–W302. doi: 10.1093/nar/gki370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Siomi H, Matunis MJ, Michael WM, Dreyfuss G. The pre-mRNA binding K protein contains a novel evolutionarily conserved motif. Nucleic Acids Res. 1993;21:1193–1198. doi: 10.1093/nar/21.5.1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Valverde R, Edwards L, Regan L. Structure and function of KH domains. FEBS J. 2008;275:2712–2726. doi: 10.1111/j.1742-4658.2008.06411.x. [DOI] [PubMed] [Google Scholar]
- 31.Subramanian AR. Structure and functions of ribosomal protein S1. Prog Nucleic Acid Res Mol Biol. 1983;28:101–142. doi: 10.1016/s0079-6603(08)60085-9. [DOI] [PubMed] [Google Scholar]
- 32.Worbs M, Bourenkov GP, Bartunik HD, Huber R, Wahl MC. An extended RNA binding surface through arrayed S1 and KH domains in transcription factor NusA. Mol Cell. 2001;7:1177–1189. doi: 10.1016/s1097-2765(01)00262-3. [DOI] [PubMed] [Google Scholar]
- 33.Schubert M, Edge RE, Lario P, Cook MA, Strynadka NC, Mackie GA, McIntosh LP. Structural characterization of the RNase E S1 domain and identification of its oligonucleotide-binding and dimerization interfaces. J Mol Biol. 2004;341:37–54. doi: 10.1016/j.jmb.2004.05.061. [DOI] [PubMed] [Google Scholar]
- 34.Oddone A, Lorentzen E, Basquin J, Gasch A, Rybin V, Conti E, Sattler M. Structural and biochemical characterization of the yeast exosome component Rrp40. EMBO Rep. 2007;8:63–69. doi: 10.1038/sj.embor.7400856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schaeffer D, Tsanova B, Barbas A, Reis FP, Dastidar EG, Sanchez-Rotunno M, Arraiano CM, van Hoof A. The exosome contains domains with specific endoribonuclease, exoribonuclease and cytoplasmic mRNA decay activities. Nat Struct Mol Biol. 2009;16:56–62. doi: 10.1038/nsmb.1528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Malet H, Topf M, Clare DK, Ebert J, Bonneau F, Basquin J, Drazkowska K, Tomecki R, Dziembowski A, Conti E, Saibil HR, Lorentzen E. RNA channelling by the eukaryotic exosome. EMBO Rep. 2010;11:936–942. doi: 10.1038/embor.2010.164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lu C, Ding F, Ke A. Crystal structure of the S. solfataricus archaeal exosome reveals conformational flexibility in the RNA-binding ring. PLoS One. 2010;5:e8739. doi: 10.1371/journal.pone.0008739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lebreton A, Tomecki R, Dziembowski A, Séraphin B. Endonucleolytic RNA cleavage by a eukaryotic exosome. Nature. 2008;456:993–996. doi: 10.1038/nature07480. [DOI] [PubMed] [Google Scholar]
- 39.Staals RH, Bronkhorst AW, Schilders G, Slomovic S, Schuster G, Heck AJ, Raijmakers R, Pruijn GJ. Dis3-like 1: a novel exoribonuclease associated with the human exosome. EMBO J. 2010;29:2358–2367. doi: 10.1038/emboj.2010.122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cheng ZF, Deutscher MP. Purification and characterization of the Escherichia coli exoribonuclease RNase R. Comparison with RNase II. J Biol Chem. 2002;277:21624–21629. doi: 10.1074/jbc.M202942200. [DOI] [PubMed] [Google Scholar]
- 41.Lorentzen E, Basquin J, Tomecki R, Dziembowski A, Conti E. Structure of the active subunit of the yeast exosome core, Rrp44: diverse modes of substrate recruitment in the RNase II nuclease family. Mol Cell. 2008;29:717–728. doi: 10.1016/j.molcel.2008.02.018. [DOI] [PubMed] [Google Scholar]
- 42.Frazão C, McVey CE, Amblar M, Barbas A, Vonrhein C, Arraiano CM, Carrondo MA. Unravelling the dynamics of RNA degradation by ribonuclease II and its RNA-bound complex. Nature. 2006;443:110–114. doi: 10.1038/nature05080. [DOI] [PubMed] [Google Scholar]
- 43.Vincent HA, Deutscher MP. Insights into how RNase R degrades structured RNA: analysis of the nuclease domain. J Mol Bio. 2009;387:570–583. doi: 10.1016/j.jmb.2009.01.068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Arcus VL, Bäckbro K, Roos A, Daniel EL, Baker EN. Distant structural homology leads to the functional characterization of an archaeal PIN domain as an exonuclease. J Biol Chem. 2004;279:16471–16478. doi: 10.1074/jbc.M313833200. [DOI] [PubMed] [Google Scholar]
- 45.Nowotny M, Gaidamakov SA, Crouch RJ, Yang W. Crystal structures of RNase H bound to an RNA/DNA hybrid: substrate specificity and metal-dependent catalysis. Cell. 2005;121:1005–1016. doi: 10.1016/j.cell.2005.04.024. [DOI] [PubMed] [Google Scholar]
- 46.Steitz TA, Steitz JA. A general two-metal-ion mechanism for catalytic RNA. Proc Natl Acad Sci USA. 1993;90:6498–6502. doi: 10.1073/pnas.90.14.6498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.De Vivo M, Dal Peraro M, Klein ML. Phosphodiester cleavage in ribonuclease H occurs via an associative two-metal-aided catalytic mechanism. J Am Chem Soc. 2008;130:10955–10962. doi: 10.1021/ja8005786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Huntzinger E, Kashima I, Fauser M, Saulière J, Izaurralde E. SMG6 is the catalytic endonuclease that cleaves mRNAs containing nonsense codons in metazoan. RNA. 2008;14:2609–2617. doi: 10.1261/rna.1386208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Eberle AB, Lykke-Andersen S, Mühlemann O, Jensen TH. SMG6 promotes endonucleolytic cleavage of nonsense mRNA in human cells. Nat Struct Mol Biol. 2009;16:49–55. doi: 10.1038/nsmb.1530. [DOI] [PubMed] [Google Scholar]
- 50.Wang HW, Wang J, Ding F, Callahan K, Bratkowski MA, Butler JS, Nogales E, Ke A. Architecture of the yeast Rrp44 exosome complex suggests routes of RNA recruitment for 3′ end processing. Proc Natl Acad Sci USA. 2007;104:16844–16849. doi: 10.1073/pnas.0705526104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kim DU, Hayles J, Kim D, Wood V, Park HO, Won M, Yoo HS, Duhig T, Nam M, Palmer G, Han S, Jeffery L, Baek ST, Lee H, Shim YS, Lee M, Kim L, Heo KS, Noh EJ, Lee AR, Jang YJ, Chung KS, Choi SJ, Park JY, Park Y, Kim HM, Park SK, Park HJ, Kang EJ, Kim HB, Kang HS, Park HM, Kim K, Song K, Song KB, Nurse P, Hoe KL. Analysis of a genome-wide set of gene deletions in the fission yeast Schizosaccharomyces pombe. Nat Biotechnol. 2010;28:617–623. doi: 10.1038/nbt.1628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Allmang C, Kufel J, Chanfreau G, Mitchell P, Petfalski E, Tollervey D. Functions of the exosome in rRNA, snoRNA and snRNA synthesis. EMBO J. 1999;18:5399–410. doi: 10.1093/emboj/18.19.5399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.van Hoof A, Lennertz P, Parker R. Yeast exosome mutants accumulate 3′-extended polyadenylated forms of U4 small nuclear RNA and small nucleolar RNAs. Mol Cell Biol. 2000;20:441–452. doi: 10.1128/mcb.20.2.441-452.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Allmang C, Mitchell P, Petfalski E, Tollervey D. Degradation of ribosomal RNA precursors by the exosome. Nucleic Acids Res. 2000;28:1684–1691. doi: 10.1093/nar/28.8.1684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Neil H, Malabat C, d’Aubenton-Carafa Y, Xu Z, Steinmetz LM, Jacquier A. Widespread bidirectional promoters are the major source of cryptic transcripts in yeast. Nature. 2009;457:1038–1042. doi: 10.1038/nature07747. [DOI] [PubMed] [Google Scholar]
- 56.Wyers F, Rougemaille M, Badis G, Rousselle JC, Dufour ME, Boulay J, Régnault B, Devaux F, Namane A, Séraphin B, Libri D, Jacquier A. Cryptic pol II transcripts are degraded by a nuclear quality control pathway involving a new poly(A)polymerase. Cell. 2005;121:725–737. doi: 10.1016/j.cell.2005.04.030. [DOI] [PubMed] [Google Scholar]
- 57.Bühler M, Haas W, Gygi SP, Moazed D. RNAi-dependent and -independent RNA turnover mechanisms contribute to heterochromatic gene silencing. Cell. 2007;129:707–721. doi: 10.1016/j.cell.2007.03.038. [DOI] [PubMed] [Google Scholar]
- 58.Reyes-Turcu FE, Zhang K, Zofall M, Chen E, Grewal SI. Defects in RNA quality control factors reveal RNAi-independent nucleation of heterochromatin. Nat Struct Mol Biol. 2011;18:1132–1138. doi: 10.1038/nsmb.2122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Harigaya Y, Tanaka H, Yamanaka S, Tanaka K, Watanabe Y, Tsutsumi C, Chikashige Y, Hiraoka Y, Yamashita A, Yamamoto M. Selective elimination of messenger RNA prevents an incidence of untimely meiosis. Nature. 2006;442:45–50. doi: 10.1038/nature04881. [DOI] [PubMed] [Google Scholar]
- 60.Zofall M, Yamanaka S, Reyes-Turcu FE, Zhang K, Rubin C, Grewal SI. RNA elimination machinery targeting meiotic mRNAs promotes facultative heterochromatin formation. Science. 2012;335:96–100. doi: 10.1126/science.1211651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Callahan KP, Butler JS. Evidence for core exosome independent function of the nuclear exoribonuclease Rrp6p. Nucleic Acids Res. 2008;36:6645–55. doi: 10.1093/nar/gkn743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Callahan KP, Butler JS. TRAMP complex enhances RNA degradation by the nuclear exosome component Rrp6. J Biol Chem. 2010;285:3540–3547. doi: 10.1074/jbc.M109.058396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Haile S, Cristodero M, Clayton C, Estévez AM. The subcellular localisation of trypanosome RRP6 and its association with the exosome. Mol Biochem Parasitol. 2007;151:52–58. doi: 10.1016/j.molbiopara.2006.10.005. [DOI] [PubMed] [Google Scholar]
- 64.Lejeune F, Li X, Maquat LE. Nonsense-mediated mRNA decay in mammalian cells involves decapping, deadenylating, and exonucleolytic activities. Mol Cell. 2003;12:675–687. doi: 10.1016/s1097-2765(03)00349-6. [DOI] [PubMed] [Google Scholar]
- 65.Stead JA, Costello JL, Livingstone MJ, Mitchell P. The PMC2NT domain of the catalytic exosome subunit Rrp6p provides the interface for binding with its cofactor Rrp47p, a nucleic acid-binding protein. Nucleic Acids Res. 2007;35:5556–5567. doi: 10.1093/nar/gkm614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Midtgaard SF, Assenholt J, Jonstrup AT, Van LB, Jensen TH, Brodersen DE. Structure of the nuclear exosome component Rrp6p reveals an interplay between the active site and the HRDC domain. Proc Natl Acad Sci USA. 2006;103:11898–11903. doi: 10.1073/pnas.0604731103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Januszyk K, Liu Q, Lima CD. Activities of human RRP6 and structure of the human RRP6 catalytic domain. RNA. 2011;17:1566–1577. doi: 10.1261/rna.2763111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Ollis DL, Brick P, Hamlin R, Xuong NG, Steitz TA. Structure of large fragment of Escherichia coli DNA polymerase I complexed with dTMP. Nature. 1985;313:762–766. doi: 10.1038/313762a0. [DOI] [PubMed] [Google Scholar]
- 69.Assenholt J, Mouaikel J, Andersen KR, Brodersen DE, Libri D, Jensen TH. Exonucleolysis is required for nuclear mRNA quality control in yeast THO mutants. RNA. 2008;14:2305–2313. doi: 10.1261/rna.1108008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zuo Y, Wang Y, Malhotra A. Crystal structure of Escherichia coli RNase D, an exoribonuclease involved in structured RNA processing. Structure. 2005;13:973–984. doi: 10.1016/j.str.2005.04.015. [DOI] [PubMed] [Google Scholar]
- 71.Phillips S, Butler JS. Contribution of domain structure to the RNA 3′ end processing and degradation functions of the nuclear exosome subunit Rrp6p. RNA. 2003;9:1098–1107. doi: 10.1261/rna.5560903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Lehner B, Sanderson CM. A protein interaction framework for human mRNA degradation. Genome Res. 2004;14:1315–1323. doi: 10.1101/gr.2122004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Cristodero M, Böttcher B, Diepholz M, Scheffzek K, Clayton C. The Leishmania tarentolae exosome: purification and structural analysis by electron microscopy. Mol Biochem Parasitol. 2008;159:24–29. doi: 10.1016/j.molbiopara.2007.12.012. [DOI] [PubMed] [Google Scholar]