Abstract
Objective:
Coronary artery disease (CAD), which develops from complex interactions between genetic and enviromental factors, is a leading cause of death worldwide. Based on genome-wide association studies (GWAS), the chromosomal region 9p21 has been identified as the most relevant locus presenting a strong association with CAD in different populations. The aim of the present study was to investigate the association of two SNPs on chromosome 9p21 on susceptibility to CAD and the effect of these SNPs along with cardiovascular risk factors on the severity of CAD in the Turkish population.
Methods:
This study had an observational case-control design. We genotyped 460 subjects, aged 30-65 years, to investigate the association of 2 SNPs (rs1333049, rs2383207) on chromosome 9p21 and CAD risk in Turkish population. Real-time polymerase chain reaction (RT-PCR) was used to analyze the 2 SNPs in CAD patients and healthy controls. The genotype and allelic variations of these SNPs with the severity of CAD was also assessed using semi-quantitative methods such as the Gensini score. Student’s t test and multiple regression analysis were used for statistical analysis.
Results:
The SNPs rs1333049 and rs2383207 were found to be associated with CAD with an adjusted OR of 1.81 (95% Cl 1.05-3.12) and 2.12 (95% CI 1.19-4.10) respectively. After adjustment of CAD risk factors such as smoking, family history of CAD and diabetes, the homozygous AA genotype for rs2383207 increased the CAD risk with an OR 3.69. Also a very strong association was found between rs1333049 and rs2383207 and Gensini scores representing the severity of CAD (p<0.001).
Conclusion:
The rs2383207 and rs1333049 SNPs on 9p21 chromosome were significantly associated with the risk and severity of CAD in the Turkish population.
Keywords: coronary artery disease, single nucleotide polymorphism, 9p21 chromosome, Turkish population
Introduction
Coronary artery disease (CAD) is the leading cause of mortality and morbidity worldwide (1, 2). The clinical and pathological spectrum of CAD varies from chronic stable angina to myocardial infarction and sudden cardiac death. Recently, many epidemiological studies demonstrated that in addition to conventional risk factors of CAD including age, male sex, hypertension, diabetes mellitus, obesity, hypercholesterolemia, smoking and family history, both lifestyle and environmental factors increase the susceptibility of CAD (3). Apart from these factors, genetic predisposition is also thought to play an important role in the pathogenesis of CAD as illustrated by twin and family studies (4). Although large population studies have reported that multiple genetic variations contribute to the inherited risk of CAD, the exact identity of the candidate genes and the quantity of their effect on disease pathogenesis is not well known. High throughput large scale genome-wide association studies (GWAS) using dense genotyping chips, containing sets of thousands of single nucleotide polymorphisms (SNPs) for whole genome assessment of genetic variants associated with common complex disease traits, and multiple epigenetic studies have identified several chromosomal loci associated with the risk of CAD.
The chromosome 9p21 has been identified as the most relevant locus presenting a strong association with CAD and acute myocardial infarction (AMI) (5-7). The combined analysis of the Wellcome Trust Case-Control Consortium (WTCCC) study (8) and the German Myocardial Infarction (MI) Family Study (9) has revealed additional loci at 9p21, 6q25.1 and 2q36.3 as loci for increased susceptibility and risk of CAD with the leading SNPs; being rs1333049, rs6922269, and rs2943634 respectively. Similar associations between SNPs in the 9p21 region and CAD have been obtained from other GWAS conducted in Canadians (rs10757274, rs2383206) (6) and in an Icelandic population (rs1333040, rs2383207, rs10116277 and rs10757278) (5). Associations between novel SNPs in the 9p21 genomic region with CAD have also been replicated in large scale studies conducted in Caucasian (5, 9, 10) Korean (11-13), Japanese (14, 15), Chinese (16-20), Italian (21), Indian (22), German (9), Swedish (23), British (24, 25) and other European populations (26-30). Hence, although multiple candidate SNPs on chromosome 9p21, which are risk alleles for CAD have been identified by GWAS in different large populations, the underlying mechanisms leading to CAD and AMI remain to be elucidated.
The 9p21 genomic region spans 58.000 base pairs, contains a noncoding ribonucleic acid antisense sequence (ANRIL) with unknown function, and maps near two identified tumor suppressor genes cyclin-dependent kinase A and B (CDKN2A and CD-KN2B), encoding proteins p16INK4a, p15INK4b, and methylthio-adenosine (MTAP) involved in the regulation of cell proliferation, TGF-β induced growth inhibition, cell aging and apoptosis (24). This region has also been associated with other important diseases such as abdominal aortic and intracranial aneurysms (25), type 2 diabetes, metabolic syndrome (31) and stroke (32).
In our best knowledge, this is the first study evaluating the association and investigating the relationship between 9p21 and CAD in Turkish population. The aim of the present study was to investigate the relationship between two SNPs (rs1333049, rs2383207) on chromosome 9p21 for susceptibility to CAD, the effect of these SNPs on cardiovascular risk factors and severity of CAD in Turkish population.
Methods
Study design
This study had an observational case-control design. Power analyses of the study were performed with PS: Power and sample size calculation software (Version 3.0 2009 Vanderbilt University, TN, USA) (33). The statistical power ranged from 0.45 to 0.892.
Study population
The study was performed in university hospital between June 2010 and July 2011. Participants enrolled in the study were selected among patients admitted to the cardiology outpatient clinic for symptoms of angina, dyspnea and chest discomfort at the time of diagnosis. Study population included 220 patients (mean age 53.78±6.91 years, 67.7% male) with CAD and 240 control subjects (mean age 52.78±7.70 years, 39.6% male) without CAD.
The exclusion criteria of the present study were defined as follows: >55 years of in male subjects and > 65 years of age in female subjects, auto-immune disease, severe kidney and hepatic diseases, cancer, dementia and Alzheimer’s disease, gout disease, glioma, malign melanoma, ischemic stroke, pregnancy and subjects who had contraindications for heparin and contrast agents.
The study complies with the Declaration of Helsinki and the trial protocol was approved by the local Ethics Committee. The written informed consent was obtained from all participants, who were included in this study.
Study protocol
Patients with CAD and control subjects without CAD included to the study. Five milliliters of blood per patient has been collected for biochemical analyses. The blood was clotted at room temperature and subsequently centrifuged at 3000 rpm. Serum was separated, aliquoted, and stored at -20°C. For analysis delayed for a longer period of time, serum was stored at -80°C. Also, peripheral venous blood sample was collected from each subject into tubes containing the anticoagulant EDTA for DNA isolation and PCR procedure. Samples were stored in aliquots at -20°C until they were analyzed. Also, Real-time Polymerase Chain Reaction (RT-PCR) method was used to genotype samples for analyses of two SNPs (rs1333049, rs2383207).
Study variables
The variables of the study were as follows: age, gender, weight, height, smoking status, body mass index (BMI), waist circumference, pulse rate, systolic and diastolic blood pressure, fasting serum glucose, plasma lipids (i.e., triglyceride, HDL cholesterol, total cholesterol, and LDL cholesterol concentrations). In this study, presence of CAD was a primary predictor variable, the outcome variables were two SNPs (rs1333049, rs2383207).
Diagnosis and definitions
CAD was defined as 50% stenosis in the left main coronary artery, or multiple significant stenosis (≥70%) in more than one coronary artery as documented by coronary angiography, history of prior cardiac bypass surgery, history of prior percutaneous coronary intervention or AMI. Early-onset CAD was defined as clinical symptoms of CAD occurring at ≤55 years of age in males or ≤65 years of age in female patients. The diagnosis of AMI was confirmed with ’’2007 European Society of Cardiology Universal Definition of Myocardial Infarction Guideline’’ criteria (21). Grading of coronary artery stenosis was carried out by two independent cardiologists who did not participate in this study. The extent and severity of CAD were determined with quantitative angiographic scores using the Gensini score system (34). CAD risk factors such as age, smoking habits, body mass index (BMI), history of diabetes and hypertension were identified from the information obtained through the patient’s health questionnaire. Subjects with systolic blood pressure (SBP) ≥140 mm Hg, diastolic blood pressure (DBP) ≥90 mm Hg and/or those using antihypertensive medication were classified as hypertensive. Hyperlipidemia was diagnosed in subjects when the 12 hours fasting blood total cholesterol (TC) was ≥220 mg/dL, low density lipoprotein (LDL) was ≥100 mg/dL and triglyceride (TG) was ≥150 mg/dL and/or in subjects taking antihyperlipidemic drugs. Diabetes mellitus in subjects was defined as fasting glucose levels ≥126 mg/dL by two consecutive fasting plasma glucose measurements, hemoglobin (Hb) A1c ≥6.5% or those who were being treated with oral anti-diabetic drugs or insulin. The height and weight were recorded and BMI was calculated as the ratio of weight in kilograms divided by the square of height in meters.
The control group consisted of 240 healthy subjects from the same geographical area, who were admitted to the outpatient clinic with mild-moderate chest pain and had positive exertional test without any stenosis in their coronary arteries as documented by coronary angiography. Also, the patients in control group were underwent further noninvasive tests to exclude syndrome X.
DNA extraction and genotyping
Since no information of the genomic variation was available in Turkish populations, we selected two SNPs (rs1333049, rs2383207) which had the strongest association with CAD or MI in previous GWAS conducted in European and Caucasian populations in the screening phase (5, 6, 9, 10, 18, 26-30). The linkage disequilibrium (LD) data of some 9p21 SNPs for both Caucasian and European populations were reviewed from the HapMap database (International HapMap Consortium 2005).
Real-time Polymerase Chain Reaction (RT-PCR) method was used to genotype samples for analyses of two SNPs (rs1333049, rs2383207). PCR procedure includes three steps: Human genomic DNA was extracted from blood leukocytes by using High Pure PCR Template Preparation Kit (Roche Diagnostics, Mannheim, Germany). DNA samples were amplified with hybridization probes in RT-PCR reactions for initial denaturation at 95°C for 10 min, 45 cycles of denaturation at 95°C for 10 s, annealing at 60°C for 10 s and extension at 72°C for 15 s, followed by a melting reaction at 95°C for 30 s and at 40°C for 2 min. In addition, cooling was performed at 40°C for 30 s. Finally, amplification products were determined by the melting temperatures (Tm) of each allele. For rs1333049 the Tm of G allele is 53.23°C, and the Tm of C allele is 63.12°C. For rs2383207 the Tm of A allele is 44.96°C and the Tm of G allele is 55.43°C. LightCycler 1.5® system was used to perform SNP genotyping using hybridization probes consisting of 3’-fluorescein and a 5’-LightCycler® red labeled pair of oligonucleotide probes (TIB MOLBIOL GmbH, Berlin, Germany). Primers for rs1333049 polymorphism were AgATgTTTAAATgTCgAATTATTg and AATgTgACTgCTTCTgCAT, and the probes were ggCTgCTTTTCAACTgTTgA FL and 640-CATATggTTAgTATgAggAATgACAgTAgggTp For the rs2383207 polymorphism, the primers were ggAggACATATTTTATATgTAACAAT and gAgAATgTTgAgCATACACTCTA, and the probes were AATCATgCTTAgCCgAAgggA-FL and 640-CCgAACAggAgTAAAAAATggTCCCAA p. Genotyping was performed in a 20 μL volume containing 2.0 μL of LightCycler® FastStart DNA Master HybProbe (Roche Diagnostics, Mannheim, Germany), 1.0 μL Reagent Mix, 3.0 mM MgCl2 and 50 ng of genomic DNA. The quality of SNP genotyping was ensured by independently replicating the genotyping of randomly selected samples. The results from quality control were 100% in agreement with the initial genotyping results.
Statistical analysis
All statistical analyses were performed using the SPSS for Windows software (SPSS for Windows, Version 16.0. Chicago, SPSS Inc.). The variables were investigated using visual (histograms, probability plots) and analytical methods (Kolmogorov-Smirnov test) to determine the normal distribution. Descriptive analyses are presented using means and standard deviations. The categorical variables are expressed as numbers and percentages. Numerical variables were compared using the Student’s t-test. Categorical data were compared with the chi-square test. The relationships of the two polymorphisms with the presence of CAD were determined with an Odds ratio (OR) and 95% confidence interval (CI) for the risk alleles by multiple logistic regression analysis after adjustment for CAD risk factors as an age, gender, family history of CAD, diabetes mellitus and smoking habits. P value <;0.05 was considered statistically significant. The linkage disequilibrium (LD), whereby genetic variants are associated between the tested SNPs was calculated by the Haploview software package (version 4.2-Broad Institute of MIT and Harvard, Massachusetts, USA) (23).
Results
Baseline demographic and clinical characteristics of patients and controls were presented in Table 1. There were 220 CAD patients and 240 healthy controls enrolled in this study. No significant differences were observed between patients and control groups in terms of age (p=0.147) and BMI (p=0.058). However, CAD patients had significantly greater weight (p=0.001), height (p=0.001), SBP (p=0.001), DBP (p=0.001), pulse rate (p=0.001), fasting blood glucose (p=0.001), serum total cholesterol, triglycerides, LDL and HDL cholesterol (all p=0.001) than controls. Also, the prevalence of hypertension, diabetes mellitus, hyperlipidemia, metabolic syndrome, family history of CAD, smoking habit and male gender were higher in patients compared with the control group (p<0.001).
Table 1.
Baseline demographic and clinical characteristics of CAD patients and controls
| CAD (n=220) | Control (n=240) | *P | |
|---|---|---|---|
| Age, years | 53.78±6.91 | 52.78±7.70 | 0.147 |
| Gender (M/F), n (%) | 149/71 (67.7/32.3) | 95/145 (39.6/60.4) | <0.001 |
| Weight, kg | 78.17±11.25 | 73.87±10.26 | <0.001 |
| Height, cm | 168.49±7.74 | 166.02±7.35 | <0.001 |
| BMI, kg/m2 | 27.55±3.72 | 26.87±3.95 | 0.058 |
| SBP, mm Hg | 139.15±20.18 | 131.69±18.05 | <0.001 |
| DBP, mm Hg | 82.61±13.04 | 77.31±10.83 | <0.001 |
| Hypertension, n (%) | 172 (78.2) | 62 (25.8) | <0.001 |
| Diabetes, n (%) | 68 (30.9) | 40 (16.7) | <0.001 |
| Hyperlipidemia, n (%) | 99 (45) | 41 (17.1) | <0.001 |
| MetS, n (%) | 90 (40.9) | 27 (11.25) | <0.001 |
| Family history, n (%) | 116 (52.7) | 31 (12.9) | <0.001 |
| Smoking, n (%) | 141(64.1) | 66 (27.5) | <0.001 |
| Pulse rate, beat/min | 80.70±11.24 | 77.17±10.99 | 0.001 |
| Fasting glucose, mg/dL | 124.83±38.99 | 106.27±19.25 | <0.001 |
| TC, mg/dL | 185.84±43.40 | 157.32±35.14 | <0.001 |
| LDL-C, mg/dL | 118.55±39.58 | 95.37±31.79 | <0.001 |
| HDL-C, mg/dL | 39.02±12.97 | 55.38±29.76 | <0.001 |
| TG, mg/dL | 171.94±69.08 | 134.34±51.36 | <0.001 |
Values were presented as mean±SD or number or percentage of patients.
Student’s t-test, chi-square tests.
BMI - body mass index; CAD - coronary artery disease; DBP - diastolic blood pressure; HDL-C - high-density lipoprotein, cholesterol; LDL-C - low-density lipoprotein cholesterol; MetS - metabolic syndrome; SD - standart deviation; SBP - systolic blood pressure; TC - total cholesterol; TG - triglycerides
The genotype and allele frequencies of two SNPs (rs2383207 and rs1333049) in CAD patients and healthy controls were presented in Table 2A and 2B. The genotypes frequencies of rs2383207 and rs1333049 were in accordance with the Hardy-Weinberg equilibrium among the patients and controls (p>0.05). Significant differences were observed in genotype frequencies of rs2383207 and rs1333049 variants between CAD patients and healthy controls (p=0.030 and p=0.037, respectively). For rs2383207 and rs1333049 variants at the 9p21 chromosome, both the risk genotypes AA and GG were associated with an increased risk of CAD (OR of 2.12, 95% CI 1.19-4.10 and 1.81, 95% Cl 1.05-3.12, respectively). A significant difference was also observed between gender and genotype distributions of rs2383207 and rs1333049 variants. AA genotype for the rs2383207 variant and the GG genotype for the rs1333049 variant were found to be more frequent in the female CAD patients than in controls (p=0.023; OR=3.99, 95% Cl 1.41-11.30 for rs2383207 and p=0.084; OR=2.35, 95% Cl 1.06-5.18 for rs1333049). However, there was no significant relationship between the genotype distribution of rs2383207 and rs1333049 in male CAD patients (p=0.532 and p=0.512, respectively). Furthermore, risk allele A of rs2383207 and risk allele G of rs1333049 polymorphisms were found to be present in 39.3% (p=0.043) and 48.2% (p=0.021) in CAD patients, respectively. In female patients, for rs2383207, allele A in rs2383207 and allele G in rs1333049 increased the risk of CAD 1.59 times (95% Cl 1.08-2.35) (p=0.019); and 1.49 times (95% Cl 1.03-2.15), respectively in comparison to controls (p=0.034). In male patients, both allele A in rs2383207 and allele G in rs1333049 insignificantly increased the risk of CAD 1.13 times (95% Cl 0.75-1.71, p=0.562 and 95% Cl 0.75-1.69, p=0.556 respectively) in comparison to controls.
Table 2A.
The genotype frequencies of rs2383207 and rs1333049 variants on chromosome 9p21 in CAD patients and control groups
| RS2383207 | RS1333049 | |||||
|---|---|---|---|---|---|---|
| Control | CAD | OR (95%CI) | Control | CAD | OR (95%CI) | |
| AA | 20/8.3 | 36/16.4 | 2.12 (1.19-4.10) | CC 85/35.4 | 54/24.5 | Reference |
| AG | 118/49.2 | 101/45.9 | 1.05 (0.71-1.55) | CG 115/47.9 | 120/54.5 | 1.64 (1.07-2.51) |
| GG | 102/42.5 | 83/37.7 | Reference | GG 40/16.7 | 46/20.9 | 1.81 (1.05-3.12) |
| P* | 0.030 | 0.037 | ||||
| Female | ||||||
| AA | 5/5.3 | 25/16.8 | 3.99 (1.41-11.30) | CC 32/33.7 | 33/22.1 | Reference |
| AG | 46/48.4 | 69/46.3 | 1.20 (0.69-2.06) | CG 49/51.6 | 82/55.0 | 1.62 (0.89-2.96) |
| GG | 44/46.3 | 55/36.9 | Reference | GG 14/14.7 | 34/22.8 | 2.35 (1.06-5.18) |
| P* | 0.023 | 0.084 | ||||
| Male | ||||||
| AA | 15/10.3 | 11/15.5 | 1.52 (0.62-3.73) | CC 53/36.6 | 21/29.6 | Reference |
| AG | 72/49.7 | 32/45.1 | 0.92 (0.49-1.70) | CG 66/45.5 | 38/53.5 | 1.45 (0.76-2.76) |
| GG | 58/40.0 | 28/39.4 | Reference | GG 26/17.9 | 12/16.9 | 1.16 (0.49-2.72) |
| P* | 0.532 | 0.512 | ||||
chi-square test
Table 2B.
The allele frequencies of rs2383207 and rs1333049 variants on chromosome 9p21 in CAD patients and control groups
| RS2383207 | RS1333049 | |||||
|---|---|---|---|---|---|---|
| Control | CAD | OR (95% CI) | Control | CAD | OR (95% CI) | |
| A | 158/32.9 | 173/39.3 | 1.32 (1.01-1.73) | C 285/59.4 | 228/51.8 | Reference |
| G | 322/67.1 | 267/60.7 | Reference | G 195/40.6 | 212/48.2 | 1.36 (1.05-1.76) |
| P* | 0.043 | 0.021 | ||||
| Female | ||||||
| A | 56/29.5 | 119/39.9 | 1.59 (1.08-2.35) | C 113/59.5 | 148/49.7 | Reference |
| G | 134/70.5 | 179/60.1 | Reference | G 77/40.5 | 150/50.3 | 1.49 (1.03-2.15) |
| P* | 0.019 | 0.034 | ||||
| Male | ||||||
| A | 102/35.2 | 54/38.0 | 1.13 (0.75-1.71) | C 172/59.3 | 80/56.3 | Reference |
| G | 188/64.8 | 88/62.0 | Reference | G 118/40.7 | 62/43.7 | 1.13 (0.75-1.69) |
| P* | 0.562 | 0.556 | ||||
chi-square test
The possible selective effects of the two SNPs on CAD were investigated using multiple logistic regression analysis after adjusting for some risk factors such as smoking habit, family history and diabetes mellitus. Smoking habit (p<0.001), having a family history (p<0.001), diabetes (p=0.035) and AA variant of rs2383207 (p=0.022) were found to be important risk factors for CAD (Table 3). After age and gender adjustments, logistic regression analyses revealed that having a homozygous AA genotype increased the risk of CAD with an OR 2.34 for rs2383207 (p<0.01, OR= 2.34, 95% Cl 1.23-4.47). However, homozygous GG and heterozygous CG genotypes for rs1333049 did not increase CAD significantly (p=0.064). In males, the GG genotype at rs1333049 increased the risk of CAD with an OR 2.32 (p=0.037, 95% Cl 1.05-5.12).
Table 3.
Multiple logistic regression analysis of CAD predictors
| B | P | OR | 95% CI | ||
|---|---|---|---|---|---|
| RS2383207 | GG | 0.067 | 1 | Reference | |
| AA | 1.305 | 0.022 | 3.69 | 1.21-11.23 | |
| AG | 0.194 | 0.510 | 1.21 | 0.68-2.16 | |
| RS1333049 | CC | 0.014 | 1 | Reference | |
| CG | 1.199 | 0.115 | 1.64 | 0.89-3.03 | |
| GG | 0.703 | 0.166 | 0.49 | 0.18-1.34 | |
| Smoking | 1.489 | <0.001 | 4.43 | 2.77-7.08 | |
| Family history | 1.786 | <0.001 | 5.96 | 3.58-9.94 | |
| Diabetes | 0.590 | 0.035 | 1.80 | 1.04-3.12 |
The association of two SNPs with the extension and severity of CAD in terms of median Gensini scores were also investigated in the present study. The severity of CAD was found to be significantly higher in GG and AA carriers of rs1333049 and rs2383207 respectively (all p values were <0.001) (Fig.1).
Figure 1.
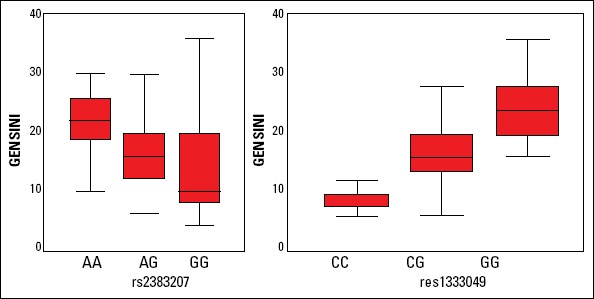
The associations between the rs2383207 and rs1333049 variants and severity of coronary artery disease (CAD) with respect to median Gensini score
Haplotype analysis was conducted to probe the relationship between the SNP patterns and CAD. Case-control ratio for the GC haplotype was 0.499-0.537 (χ2=1.346, p=0.246), the AG haplotype was 0.374-0.273 (χ2=10.81, p=0.001), the GG haplotype was 0.108-0.133 (χ2=1.43, p=0.2317), and the AC haplotype was 0.019-0.056 (χ2=8.622, p=0.0033). The frequencies for GC, AG, GG, AC haplotypes were 51.9%, 32.1%, 12.1% and 3.9% respectively (LOD=60.78, r2= 0.462, D’=0.808). While the AG haplotype conferred a significant risk effect on CAD, the AC haplotype provided a protection from this disease for these two variants.
Discussion
The main findings of the present study were as follows: 1) This is the first study to demonstrate an association between common SNPs (rs2383207 and rs1333049) on 9p21 chromosome and CAD in a Turkish population consistent with previous studies with differences in risk genotypes and alleles, 2) In contrast to some population studies in European and Asian descents, which CC was found to be a risk genotype for rs1333049, GG was found to be a risk genotype for this SNP in CAD patients in Turkish population, 3) For the SNP rs2383207, the risk allele A increased CAD risk with an OR 1.3 in contrast to other study, which the G risk allele increased CAD risk with an OR 1.25, 4) A very strong relation was found between two variants (rs1333049 and rs2383207) and Gensini scores representing the severity of CAD in Turkish subjects consistent with previous studies.5) In multiple logistic regression analysis, smoking habit, family history and AA variant of rs2383207 were found to be an independent determinant of CAD.
CAD is a complex and multi-factorial cardiovascular disorder caused by the interaction of genetic and environmental risk factors. GWAS provides an excellent resolution for whole-genome assessment of genomic variants associated with common complex disorders including CAD (5-8). Recently, many independent multiple large scale studies have identified the strongest genomic association between the chromosomal locus 9p21 and CAD and/or AMI (5, 8-10). This relationship has since then been replicated in various different population studies (5, 9-18). In the present study, a similar association has been reported in Turkish subjects. In four different Caucasian populations, McPherson et al. (6) presented that 20-25% of homozygotes for the risk allele face an increased risk of approximately 30-40% to CAD. In European descents, Helgadottir et al. (5) and Schunkert et al. (9) detected that nearly 50% of the subjects were carrying 1 high risk allele and other 20-25% were carrying 2 alleles had a 20-40% and 40-70% greater CAD risk respectively (6-8). In this study, risk allele A of rs2383207 and risk allele G of rs1333049 variants were found to occur in 39.3% and 48.2% of CAD patients, respectively. These risk alleles increased the susceptibility to CAD by 35% and 42% respectively in our population. These differences are possibly due to the genetic diversity of Turkish population.
The associations of ten candidate coronary heart disease risk genetic variants [rs2144300 (GALNT2), rs599839 (CELSR2/PSRC1/SORT1), rs780094 (GCKR), rs499818 (6p24.1), rs10757278 (9p21), rs1333049 (9p21), rs2383206 (9p21), rs2383207 (9p21), rs17228212 (SMAD3), and rs2549513 (16q23.1)] and subclinical atherosclerosis were investigated in European American, African American, American Indian (AI), and Mexican American ancestry in the Population Architecture using Genomics and Epidemiology (PAGE) study (35). While none of the ten SNPs was significantly related with subclinical atherosclerosis measured by ankle brachial index and common or internal carotid intimamedia thickness, a significant association was found between rs780094 and carotid plaque only in AI populations (OR=1.32, 95% confidence interval: 1.17, 1.49, p=1.08 ×10-5).
In this study, the SNPs, 1333049 and rs2383207 were found to be associated with CAD in the Turkish population (5, 9-20) with an adjusted OR of 1.81 and 2.12 respectively. While GG was found to be a risk genotype for the rs1333049 in CAD patients, CC was found to be a risk genotype for this SNP in different populations (5, 9-20). The risk allele (G) frequency for rs1333049 (0.48) and the risk allele (A) frequency for rs2383207 (0.39) found in our study were different from previous studies in European and Asian descents (5, 9-20). In particular, the C risk allele increased susceptibility to CAD for rs1333049 in European (9) and Asian (12) population studies with an OR of 1.29 and 1.47 respectively; and the G risk allele increased this risk with an OR of 1.36 in Turkish subjects. This lower risk allele frequency in the two variants is possibly due to the ethnic differences and genetic heterogeneity of our population.
For the SNP rs2383207, the risk allele a increased CAD risk with an OR 1.3 in contrast to the observation that the G risk allele increased CAD risk with an OR 1.25 as per the other European study by Helgadottir et al. (9).
After age and gender adjustments, logistic regression analyses revealed that having a homozygous AA genotype increased the risk of CAD with an OR of 2.34 for the rs2383207 variant. After adjusting for other CAD risk factors such as smoking, family history of CAD and diabetes mellitus with multiple logistic regression analysis, the data indicated the homozygous AA genotype further increased CAD risk with an OR 3.69 for the same variant. However, rs1333049 had lost its significance on risk of CAD with an OR of 0.49. This observation was different from other population studies (5, 9-20), in which the homozygous genotype increased the risk of CAD after adjustment for similar risk factors as listed above.
The association between the 9p21 variants and the severity of CAD was also investigated by using the Gensini score system in the present study. Although, Patel et al. (27) reported a positive correlation between rs10757278 and severity of CAD, Chen et al. (30) did not find any relation between the gene dosage for rs10757278, rs2383207, rs2383206, rs1333049 and disease severity. Two other studies have also demonstrated an important association between 9p21 and CAD burden with number of diseased vessels and/or semi-quantitative methods such as the Gensini score (28, 33, 36). We found a very strong association between these two variants (rs1333049 and rs2383207) and Gensini scores representing the severity of CAD in our subjects consistent with previous studies. These data suggest that, Turkish patients homozygous for the 9p21 risk allele may suffer from more severe CAD.
Study limitations
The present study focused on both CAD patients and control subjects. The limitations of this study included a small sample size and data from a single center.
Conclusion
Nevertheless, to our knowledge, this is the first study to demonstrate an association between common SNPs (rs2383207 and rs1333049) on 9p21 chromosome and CAD in a Turkish population consistent with previous studies with differences in risk genotypes and alleles. Complex cardiovascular diseases such as CAD have variable gene to gene and gene to environment interactions in different populations. While multiple and different SNPs in this chromosomal region should be studied to elucidate the exact identity of the candidate genes and the quantity of their effect on the CAD pathogenesis, different genetic polymorphisms should be studied prospectively in a wider population to fully understand the molecular mechanism underlying the association and to assign a pathophysiological role to the 9p21 chromosome variants in the etiopathology of CAD in populations of Turkish descent.
Acknowledgements:
We would like to thank all those who participated in the study. The present study was supported financially by Scientific Research Projects Coordination Unit of Istanbul University, Turkey (Project No: 4060). We are also grateful to the health nurses and laboratory technicians for their contribution.
Footnotes
Conflict of interest: None declared.
Peer-review: Externally peer-reviewed.
Authorship contributions: Concept - H.A.Ç.; Design - H.A.Ç., B.B., G.C.; Supervision - A.V. A., H.Y., M.C.; Resource - B.B., E.D., B.K.; Material - B.K., E.D.; Data collection &/or processing - E.D., B.B., B.K.; Analysis &/or interpretation - G.C.; Literature search - H.A.Ç., B.B.; Writing - H.A.Ç.; Critical review - H.Y., V.A.V., G.C., M.C.

References
- 1.Chen Z, Qian Q, Ma G, Wang J, Zhang X, Feng Y, et al. A common variant on chromosome 9p21 affects the risk of early-onset coronary artery disease. Mol Biol Rep. 2009;36:889–93. doi: 10.1007/s11033-008-9259-7. [CrossRef] [DOI] [PubMed] [Google Scholar]
- 2.Thom TJ, Epstein FH. Heart disease, cancer, and stroke mortality trends and their interrelations. An international perspective. Circulation. 1994;90:574–82. doi: 10.1161/01.cir.90.1.574. [CrossRef] [DOI] [PubMed] [Google Scholar]
- 3.Wilson PW. Established risk factors and coronary artery disease: The Framingham Study. Am J Hypertens. 1994;7:7–12. doi: 10.1093/ajh/7.7.7s. [DOI] [PubMed] [Google Scholar]
- 4.Shea S, Ottman R, Gabrieli C, Stein Z, Nichols A. Family history as an independent risk factor for coronary artery disease. J Am Coll Cardiol. 1984;4:793–801. doi: 10.1016/s0735-1097(84)80408-8. [CrossRef] [DOI] [PubMed] [Google Scholar]
- 5.Helgadottir A, Thorleifsson G, Manolescu A, Gretarsdottir S, Blondal T, Jonasdottir A, et al. A common variant on chromosome 9p21 affects the risk of myocardial infarction. Science. 2007;316:1491–3. doi: 10.1126/science.1142842. [CrossRef] [DOI] [PubMed] [Google Scholar]
- 6.McPherson R, Pertsemlidis A, Kavaslar N, Stewart A, Roberts R, Cox DR, et al. A common allele on chromosome 9 associated with coronary heart disease. Science. 2007;316:1488–91. doi: 10.1126/science.1142447. [CrossRef] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Christensen K, Murray JC. What genome-wide association studies can do for medicine. N Engl J Med. 2007;356:1094–7. doi: 10.1056/NEJMp068126. [CrossRef] [DOI] [PubMed] [Google Scholar]
- 8.Wellcome Trust Case Control Consortium. Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature. 2007;447:661–78. doi: 10.1038/nature05911. [CrossRef] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schunkert H, Gotz A, Braund P, McGinnis R, Tregouet DA, Mangino M, et al. Cardiogenics Consortium. Repeated replication and a prospective meta-analysis of the association between chromosome 9p21.3 and coronary artery disease. Circulation. 2008;117:1675–84. doi: 10.1161/CIRCULATIONAHA.107.730614. [CrossRef] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Samani NJ, Erdmann J, Hall AS, Hengstenberg C, Mangino M, Mayer B, et al. WTCCC and the Cardiogenics Consortium. Genomewide association analysis of coronary artery disease. N Engl J Med. 2007;357:443–53. doi: 10.1056/NEJMoa072366. [CrossRef] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shen GQ, Li L, Rao S, Abdullah KG, Ban JM, Lee BS, et al. Four SNPs on chromosome 9p21 in a South Korean population implicate a genetic locus that confers high cross-race risk for development of coronary artery disease. Arterioscler Thromb Vasc Biol. 2008;28:360–5. doi: 10.1161/ATVBAHA.107.157248. [CrossRef] [DOI] [PubMed] [Google Scholar]
- 12.Hinohara K, Nakajima T, Takahashi M, Hohda S, Sasaoka T, Nakahara K, et al. Replication of the association between a chromosome 9p21 polymorphism and coronary artery disease in Japanese and Korean populations. J Hum Genet. 2008;53:357–9. doi: 10.1007/s10038-008-0248-4. [CrossRef] [DOI] [PubMed] [Google Scholar]
- 13.Zhou LT, Qin L, Zheng DC, Song ZK, Ye L. Meta-analysis of genetic association of chromosome 9p21 with early-onset coronary artery disease. Gene. 2012;510:185–8. doi: 10.1016/j.gene.2012.09.003. [CrossRef] [DOI] [PubMed] [Google Scholar]
- 14.Hiura Y, Fukushima Y, Yuno M, Sawamura H, Kokubo Y, Okamura T, et al. Validation of the association of genetic variants on chromosome 9p21 and 1q41 with myocardial infarction in a Japanese population. Circ J. 2008;72:1213–7. doi: 10.1253/circj.72.1213. [CrossRef] [DOI] [PubMed] [Google Scholar]
- 15.Zhou L, Zhang X, He M, Cheng L, Chen Y, Hu FB, et al. Associations between single nucleotide polymorphisms on chromosome 9p21 and risk of coronary heart disease in Chinese Han population. Arterioscler Thromb Vasc Biol. 2008;28:2085–9. doi: 10.1161/ATVBAHA.108.176065. [CrossRef] [DOI] [PubMed] [Google Scholar]
- 16.Xie F, Chu X, Wu H, Sun W, Shen M, Yang L, et al. Replication of putative susceptibility loci from genome-wide association studies associated with coronary atherosclerosis in Chinese Han population. PLoS One. 2011;6:e20833. doi: 10.1371/journal.pone.0020833. [CrossRef] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang F, Xu CQ, He Q, Cai JP, Li XC, Wang D, et al. Genome-wide association identifies a susceptibility locus for coronary artery disease in the Chinese Han population. Nat Genet. 2011;43:345–9. doi: 10.1038/ng.783. [CrossRef] [DOI] [PubMed] [Google Scholar]
- 18.Guo J, Li W, Liu X, Wang XY, Wang Y, Liu LS. Association of single nucleotide polymorphisms on chromosome!p13 and 9p21 with acute myocardial infarction in a Chinese population: the AMI study in China. Acad J Sec Mil Med Univ. 2011;32:822–9. [CrossRef] [Google Scholar]
- 19.Zhang Q, Wang XF, Cheng SS, Wan XH, Cao FF, Li L, et al. Three SNPs on chromosome 9p21 confer increased risk of myocardial infarction in Chinese subjects. Atherosclerosis. 2009;207:26–8. doi: 10.1016/j.atherosclerosis.2009.04.017. [CrossRef] [DOI] [PubMed] [Google Scholar]
- 20.Lu X, Wang L, Chen S, He L, Yang X, Shi Y, et al. Coronary Artery DIsease Genome-Wide Replication And Meta-Analysis (CARDIoGRAM) Consortium. Genome-wide association study in Han Chinese identifies four new susceptibility loci for coronary artery disease. Nat Genet. 2012;44:890–4. doi: 10.1038/ng.2337. [CrossRef] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gori F, Specchia C, Pietri S, Crociati L, Barlera S, Franciosi M, et al. GISSI Prevenzione Investigators;SIBioC-GISSI Prevenzione Group. Common genetic variants on chromosome 9p21 are associated with myocardial infarction and type 2 diabetes in an Italian population. BMC Med Genet. 2010;11:60. doi: 10.1186/1471-2350-11-60. [CrossRef] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Maitra A, Dash D, John S, Sannappa PR, Das AP, Shanker J, et al. A common variant in chromosome 9p21 associated with coronary artery disease in Asian Indians. J Genet. 2009;88:113–8. doi: 10.1007/s12041-009-0017-y. [CrossRef] [DOI] [PubMed] [Google Scholar]
- 23.Diabetes Genetics Initiative of Broad Institute of Harvard and MIT, Lund University, and Novartis Institutes of BioMedical Research. Saxena R, Voight BF, Lyssenko V, Burtt NP, de Bakker NP, Chen H, et al. Genome-wide association analysis identifies loci for type 2 diabetes and triglyceride levels. Science. 2007;316:1331–6. doi: 10.1126/science.1142358. [CrossRef] [DOI] [PubMed] [Google Scholar]
- 24.Broadbent HM, Peden JF, Lorkowski S, Goel A, Ongen H, Green F, et al. PROCARDIS consortium. Susceptibility to coronary artery disease and diabetes is encoded by distinct, tightly linked SNPs in the ANRIL locus on chromosome 9p. Hum Mol Genet. 2008;17:806–14. doi: 10.1093/hmg/ddm352. [CrossRef] [DOI] [PubMed] [Google Scholar]
- 25.Helgadottir A, Thorleifsson G, Magnusson KP, Grétarsdottir S, Steinthorsdottir V, Manolescu A, et al. The same sequence variant on 9p21 associates with myocardial infarction, abdominal aortic aneurysm and intracranial aneurysm. Nat Genet. 2008;40:217–24. doi: 10.1038/ng.72. [CrossRef] [DOI] [PubMed] [Google Scholar]
- 26.Muendlein A, Saely CH, Rhomberg S, Sonderegger G, Loacker S, Rein P, et al. Evaluation of the association of genetic variants on the chromosomal loci 9p21.3, 6q25.1, and 2q36.3 with angiographically characterized coronary artery disease. Atherosclerosis. 2009;205:174–80. doi: 10.1016/j.atherosclerosis.2008.10.035. [CrossRef] [DOI] [PubMed] [Google Scholar]
- 27.Patel RS, Su S, Neeland IJ, Ahuja A, Veledar E, Zhao J, et al. The chromosome 9p21 risk locus is associated with angiographic severity and progression of coronary artery disease. Eur Heart J. 2010;31:3017–23. doi: 10.1093/eurheartj/ehq272. [CrossRef] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dandona S, Stewart AF, Chen L, Williams K, So D, O’Brien E, et al. Gene dosage of the common variant 9p21 predicts severity of coronary artery disease. J Am Coll Cardiol. 2010;56:479–86. doi: 10.1016/j.jacc.2009.10.092. [CrossRef] [DOI] [PubMed] [Google Scholar]
- 29.Gong Y, Beitelshees AL, Cooper-DeHoff RM, Lobmeyer MT, Langaee TY, Wu J. Chromosome 9p21 haplotypes and prognosis in white and black patients with coronary artery disease. Circ Cardiovasc Genet. 2011;4:169–78. doi: 10.1161/CIRCGENETICS.110.959296. [CrossRef] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chen SN, Ballantyne CM, Gotto AM, jr, Marian AJ. The 9p21 susceptibility locus for coronary artery disease and the severity of coronary atherosclerosis. BMC Cardiovasc Disord. 2009;9:3. doi: 10.1186/1471-2261-9-3. [CrossRef] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bayoglu B, Cakmak HA, Yuksel H, Can G, Karadag B, Ulutin T, et al. Chromosome 9p21 rs10757278 polymorphism is associated with the risk of metabolic syndrome. Mol Cell Biochem. 2013;379:77–85. doi: 10.1007/s11010-013-1629-3. [CrossRef] [DOI] [PubMed] [Google Scholar]
- 32.Ding H, Xu Y, Wang X, Wang Q, Zhang L, Tu Y, et al. 9p21 is a shared susceptibility locus strongly for coronary artery disease and weakly for ischemic stroke in Chinese Han population. Circ Cardiovasc Genet. 2009;2:338–46. doi: 10.1161/CIRCGENETICS.108.810226. [CrossRef] [DOI] [PubMed] [Google Scholar]
- 33.Dupont WD, Plummer WD., Jr Power and sample size calculations. A review and computer program. Control Clin Trials. 1990;11:116–28. doi: 10.1016/0197-2456(90)90005-m. [CrossRef] [DOI] [PubMed] [Google Scholar]
- 34.Gensini GG. A more meaningful scoring system for determining the severity of coronary heart disease. Am J Cardiol. 1983;51:606. doi: 10.1016/s0002-9149(83)80105-2. [CrossRef] [DOI] [PubMed] [Google Scholar]
- 35.Zhang L, Buzkova PWassel CL, Roman MJ, North KE, Crawford DC, et al. Lack of associations of ten candidate coronary heart disease risk genetic variants and subclinical atherosclerosis in four US populations: the Population Architecture using Genomics and Epidemiology (PAGE) study. Atherosclerosis. 2013;228:390–9. doi: 10.1016/j.atherosclerosis.2013.02.038. [CrossRef] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chan K, Motterle A, Laxton RC, Ye S. Common variant on chromosome 9p21 predicts severity of coronary artery disease. J Am Coll Cardiol. 2011;57:1497–8. doi: 10.1016/j.jacc.2010.09.078. [CrossRef] [DOI] [PubMed] [Google Scholar]


