Summary
LKB1 is activated by forming a heterotrimeric complex with STRAD and MO25. Recent studies suggest that LKB1 has pro-oncogenic functions, besides acting as a tumor-suppressor. How the LKB1 activity is maintained and how LKB1 regulates cancer development are largely unclear. Here we show that K63-linked LKB1 polyubiquitination by Skp2-SCF ubiquitin ligase is critical for LKB1 activation by maintaining LKB1-STRAD-MO25 complex integrity. We further demonstrate that oncogenic Ras acts upstream of Skp2 to promote LKB1 polyubiquitination by activating Skp2-SCF ubiquitin ligase. Moreover, Skp2-mediated LKB1 polyubiquitination is required for energy stress-induced cell survival. We also detected overexpression of Skp2 and LKB1 in late-stage hepatocellular carcinoma (HCC), and their overexpression predicts poor survival outcomes. Finally, we show that Skp2-mediated LKB1 polyubiquitination is important for HCC tumor growth in vivo. Our study provides new insights into the upstream regulation of LKB1 activation and suggests a potential target, the Ras/Skp2/LKB1 axis, for cancer therapy.
Introduction
Cancer cells display dramatic alterations in cellular metabolism to meet their needs of increased growth and proliferation. In the last decade, cancer research has brought these pathways into focus, and one emerging issue that has come to attention is that many oncogenes and tumor-suppressors are intimately linked to metabolic regulation. One of the key tumor-suppressors involved in metabolism is LKB1 (also called STK11). LKB1 is a master kinase that plays diverse roles in multiple cellular processes, including energy metabolism, proliferation, apoptosis, and cell polarity, by regulating various substrates (Alessi et al., 2006). It can phosphorylate and activate at least 12 AMP-activated protein kinase (AMPK)-related kinases (Lizcano et al., 2004), and among those, AMPK is the best-characterized substrate of LKB1. AMPK is an evolutionarily conserved sensor of intracellular energy levels. Under low-energy conditions, activation of the LKB1/AMPK pathway restores intracellular ATP levels by stimulating catabolic pathways while inhibiting anabolic pathways (Hardie, 2007). Therefore, activation of the LKB1/AMPK pathway provides a survival advantage under energy stress, and inactivation of the LKB1/AMPK pathway renders cells more susceptible to energy stress-induced cell death (Narbonne and Roy, 2009; Shaw et al., 2004; van der Velden et al., 2011).
LKB1 was first identified as a tumor-suppressor gene, because its mutations have been linked to Peutz-Jeghers syndrome (PJS), a cancer-prone inherited disorder (Hemminki et al., 1998). Later on, somatic mutations of LKB1 were identified in sporadic cancers (Sanchez-Cespedes, 2007), and LKB1 heterozygous mice were found to develop gastrointestinal polyps resembling the PJS phenotype. Although genetic evidence supports the tumor-suppressive role of LKB1 (Ollila and Makela, 2011), other evidence has revealed that LKB1 may also exhibit pro-oncogenic functions. Bardeesy et al. demonstrated that LKB1-deficient cells are resistant to oncogenic transformation, which may account for the lack of malignant tumors in PJS patients and in many mouse models with LKB1 deficiency (Bardeesy et al., 2002; Jeon et al., 2012). Moreover, knockdown of LKB1 or AMPK in breast cancer cells attenuates breast tumor development due to failure to inhibit acetyl Co-A carboxylase (ACC) activity and to maintain intracellular NADPH levels (Jeon et al., 2012). Because tumors mostly reside in a metabolic stress environment, it is possible that cancer cells could use LKB1/AMPK signaling for their survival. Accordingly, LKB1 may be a “double-edged sword” in terms of cancer suppression and promotion, and which role LKB1 plays may be determined by distinct cell types and nutrient availability within tumor microenvironments. Whether LKB1 has a stage-specific function in cancer progression remains to be determined.
Unlike AMPK and other kinases that are activated by phosphorylation at their activation loops, LKB1 is predominantly activated through complex assembly (Boudeau et al., 2004). LKB1 forms a heterotrimeric complex with STRAD and MO25 (Baas et al., 2003; Boudeau et al., 2003). Binding of these two proteins to LKB1 promotes and stabilizes the activated conformation of LKB1 (Zeqiraj et al., 2009a); thus, both are critical for LKB1 kinase activity. However, how the LKB1-STRAD-MO25 complex is maintained remains unclear, and the regulatory mechanisms of LKB1 activity and activation are poorly understood. Although several studies demonstrated that phosphorylation of LKB1 regulates its function in cell-cycle arrest, tumor suppression and cell polarity, such posttranslational modification does not directly impact LKB1 activity (Sebbagh et al., 2011). Therefore, it is of great interest to address whether other types of posttranslational modification on LKB1 can modulate LKB1 activity directly.
In this study, we aimed to decipher the mechanism by which LKB1 kinase activity is maintained and to explore the functional role of LKB1 in tumor progression/maintenance, thereby shedding new insights into understanding the complexity of LKB1’s role in tumorigenesis and offering potentially therapeutic strategies for cancer therapy.
Results
Skp2 regulates the LKB1/AMPK pathway
Ubiquitination is a posttranslational modification that functions in a wide variety of cellular processes, and the Skp1/Cullin/F-box (SCF) ubiquitin ligase complex is evolutionally conserved from yeast to human. Previously, a series of findings regarding yeast AMPK—Snf1p led to identification of LKB1 as the mammalian upstream kinase of AMPK on the basis of homology (Sutherland et al., 2003; Woods et al., 2003). It was found that Grr1p, yeast Skp2-related F-box protein, regulates glucose metabolism (Flick and Johnston, 1991) and plays a role in morphogenesis as Elm1p (Blacketer et al., 1995), yeast AMPK kinase closely related to LKB1. In mammals, both Skp2-knockout (Skp2-KO) and LKB1-knockout (LKB1-KO) mouse embryonic fibroblasts (MEFs) were shown to exhibit resistance to oncogenic transformation (Bardeesy et al., 2002; Lin et al., 2010), and both proteins were demonstrated to be involved in the maintenance of hematopoietic stem cells (Gurumurthy et al., 2010; Wang et al., 2011). Moreover, Skp2 deficiency caused severe hyperglycemia in mice (Zhong et al., 2007), which was also observed in mice with LKB1 deletion in the liver (Shaw et al., 2005). All these findings imply that Skp2 may be linked to the LKB1/AMPK pathway.
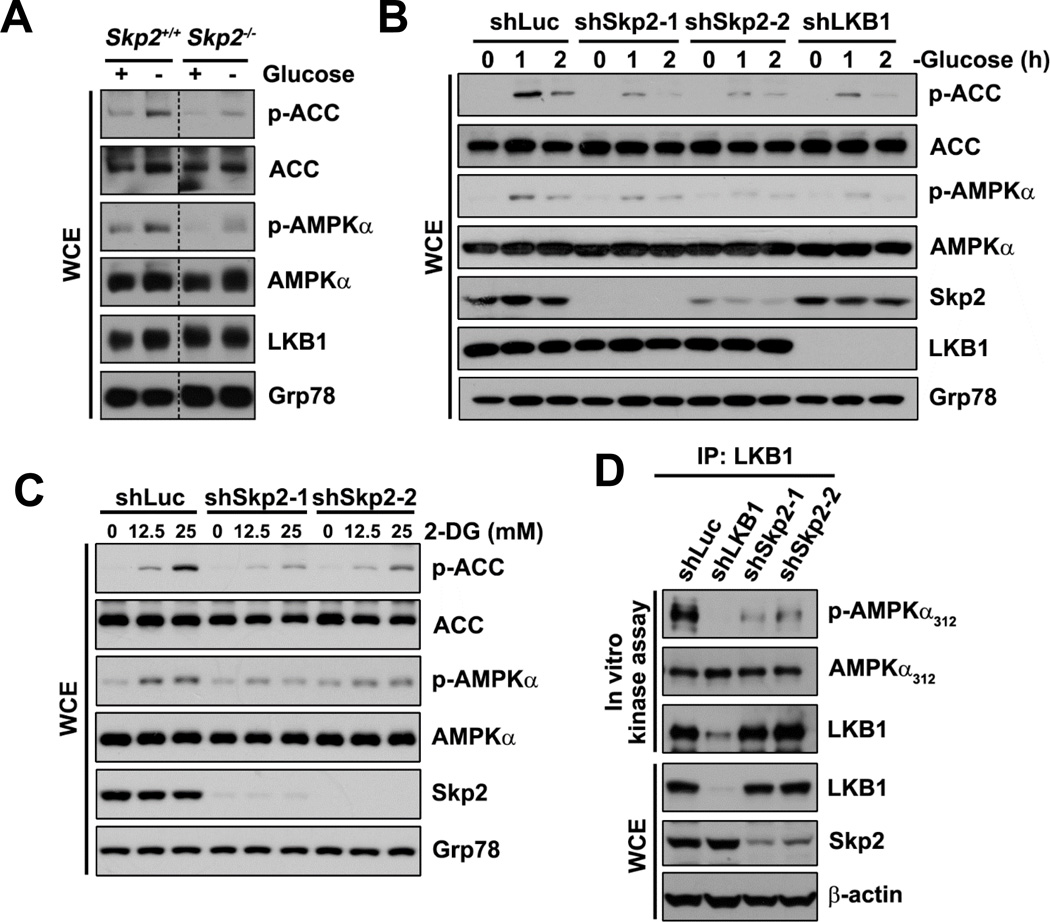
To determine whether Skp2 is involved in the LKB1/AMPK pathway, we first examined energy stress-induced activation of the LKB1/AMPK signaling upon Skp2 deficiency. Phosphorylation of AMPKα at Thr172 and ACC at Ser79, a well-established readout of AMPKα activity, were analyzed. As expected, phosphorylation of AMPKα and ACC were induced by glucose deprivation in wild-type (WT) primary MEFs; however, in Skp2-KO MEFs, glucose deprivation induced much lower levels of AMPKα and ACC phosphorylation (Figure 1A). Moreover, Skp2 knockdown in multiple cancer cell lines (Hep3B, BT-474, and MCF-7) impaired glucose deprivation-induced phosphorylation of AMPKα and/or ACC (Figures 1B, S1A and S1B). Similarly, AMPKα and ACC phosphorylation induced by the glycolysis inhibitor 2-deoxyglucose (2-DG) or the AMPK activator AICAR were compromised by Skp2 knockdown (Figures 1C and S1C). These results indicate that Skp2 is critical for activating the LKB1/AMPK signaling.
Figure 1. The LKB1/AMPK pathway and LKB1 activity are downregulated upon Skp2 deficiency.
(A) WT (Skp2+/+) and Skp2-KO (Skp2−/−) MEFs cultured in the presence (+) or absence (−) of glucose were subjected to immunoblotting (IB). WCE, whole cell extract.
(B) Hep3B cells with control (shLuc), Skp2 or LKB1 knockdown cultured in the absence of glucose for the indicated time points were subjected to IB.
(C) HEK293 cells with control or Skp2 knockdown after treatment with 2-DG at the indicated concentrations for 15 minutes were subjected to IB.
(D) Immunoprecipitates (IPs) by anti-LKB1 antibody from HEK293 cells with control, Skp2 or LKB1 knockdown were subjected to in vitro LKB1 kinase assay followed by IB.
See also Figure S1.
We next examined whether Skp2 is critical for maintaining LKB1 activity by measuring LKB1 kinase activity toward AMPKα in vitro. Strikingly, LKB1 protein isolated from cells with Skp2 knockdown displayed a decreased ability to phosphorylate AMPKα, compared with LKB1 derived from control cells (Figure 1D). This indicates that Skp2 deficiency impairs the ability of LKB1 to activate AMPK, thereby downregulating the LKB1/AMPK signaling. Our results suggest that Skp2 is a novel LKB1 regulator that is required for maintaining LKB1 activity.
Skp2 induces K63-linked polyubiquitination of LKB1
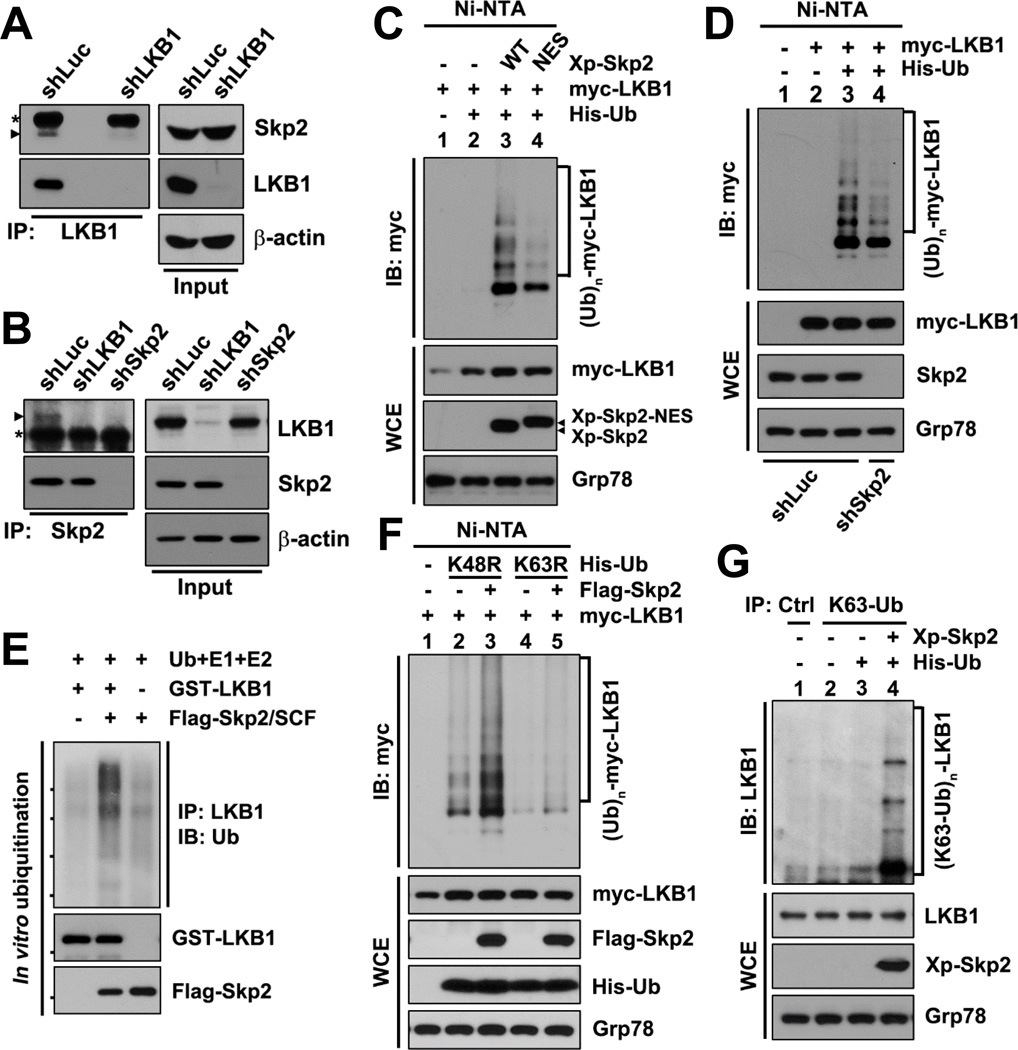
To gain a further insight into how Skp2 regulates LKB1 activity, we examined the interaction between endogenous Skp2 and LKB1 using reciprocal co-immunoprecipitation (co-IP) assays. We found that Skp2 and LKB1 were co-immunoprecipitated by each other (Figures 2A and 2B). The specificity of their interaction was verified in cells with LKB1 or Skp2 knockdown, and both LKB1 knockdown and Skp2 knockdown compromised the Skp2-LKB1 co-IP efficiency (Figures 2A and 2B). Because Skp2 is a substrate recognition component of the SCF complex, our finding that Skp2 interacts with LKB1 raises the question of whether Skp2 can promote LKB1 ubiquitination. Using in vivo ubiquitination assays, we found that overexpression of WT Skp2 promoted LKB1 polyubiquitination in the absence of the proteasome inhibitor MG132 (Figure 2C). In contrast, the ubiquitin ligase-defective mutant of Skp2 (Skp2-NES), which does not form the Skp2-SCF complex (Lin et al., 2009), failed to promote LKB1 polyubiquitination, although Skp2-NES still bound to LKB1 as efficiently as WT Skp2 (Figures 2C and S2A). Consistently, administration of the specific Skp2 inhibitor, which impairs the ubiquitin ligase activity of the Skp2-SCF complex by preventing the Skp2-Skp1 binding (Chan et al., 2013), diminished Skp2-mediated LKB1 polyubiquitination (Figure S2B). Moreover, Skp2 knockdown reduced LKB1 polyubiquitination (Figure 2D). To verify whether Skp2-SCF is a bona fide ubiquitin ligase toward LKB1, we performed the in vitro ubiquitination assay. The purified Skp2-SCF was capable of ubiquitinating recombinant LKB1 in vitro (Figure 2E), confirming that the Skp2-SCF is a direct ubiquitin ligase for LKB1. Similar to genetic inhibition of Skp2, pharmacological inactivation of Skp2 by the Skp2 inhibitor downregulated the AICAR-induced LKB1/AMPK signaling as determined by decreased phosphorylation of ACC in a dose-dependent manner (Figure S2C). Thus, our data indicate that Skp2 orchestrates LKB1 activity through maintaining polyubiquitination of LKB1.
Figure 2. Skp2-SCF ubiquitinates LKB1 via K63-linkage.
(A, B) IPs by anti-LKB1 (A) or anti-Skp2 (B) antibody from HEK293 cells with control, Skp2 or LKB1 knockdown were subjected to IB. The asterisks indicate heavy chains of the antibodies and the arrowheads indicate the bands corresponding to Skp2 (A) or LKB1 (B).
(C, D, F) In vivo ubiquitination assays in HEK293T cells (C and F) or HEK293 cells with control or Skp2 knockdown (D) transfected with the indicated plasmids were followed by IB. Ni-NTA, nickel bead precipitates.
(E) In vitro ubiquitination assay, in which recombinant GST-LKB1 was incubated with purified Flag-Skp2/SCF along with recombinant ubiquitin (Ub), E1 and E2 enzymes for reaction, was followed by IP and IB.
(G) IPs by anti-IgG control or anti-K63-Ub antibody from HEK293T cells transfected with the indicated plasmids were subjected to IB.
See also Figure S2.
A recent study demonstrated that the ubiquitin ligase CHIP is involved in LKB1 degradation (Gaude et al., 2012). However, we did not observe significant changes in LKB1 protein levels upon Skp2 knockdown or overexpression in our experiments, so we speculated that Skp2 mediates non-degradative polyubiquitination of LKB1. K48-linked ubiquitin chains are known as a major targeting signal for proteasomal degradation, whereas K63-linked ubiquitin chains have non-proteolytic functions in many cellular processes, such as kinase activation, DNA repair, and protein trafficking (Yang et al., 2010). Accordingly, to confirm that Skp2-mediated polyubiquitination of LKB1 is non-proteolytic, we applied two mutant forms of ubiquitin to our in vivo ubiquitination assay: ubiquitin-K48R and ubiquitin-K63R, which exclusively eliminate K48-linked and K63-linked polyubiquitination, respectively. Strikingly, we found that Ub-K63R, but not Ub-K48R, blocked Skp2-mediated LKB1 polyubiquitination (Figure 2F). Furthermore, Skp2-mediated K63-linked polyubiquitination of endogenous LKB1 was verified by using anti-Lys63-specific ubiquitin (K63-Ub) antibody (Figure 2G). Because the linkage specificity of polyubiquitin chains is determined by E2 ubiquitin-conjugating enzymes, we also investigated the effect of K63-Ub-specific E2 enzymes Ubc13 and Ubc5C on Skp2-mediated LKB1 polyubiquitination. Knockdown of either Ubc13 or Ubc5C decreased Skp2-mediated LKB1 polyubiquitination (Figure S2D). Hence, Skp2-mediated LKB1 polyubiquitination primarily occurs through non-degradative K63-linked ubiquitination for non-proteolytic regulation, such as kinase activation, which is consistent with our above finding that Skp2 affects LKB1 activity rather than stability.
LKB1 polyubiquitination is critical for LKB1 activation
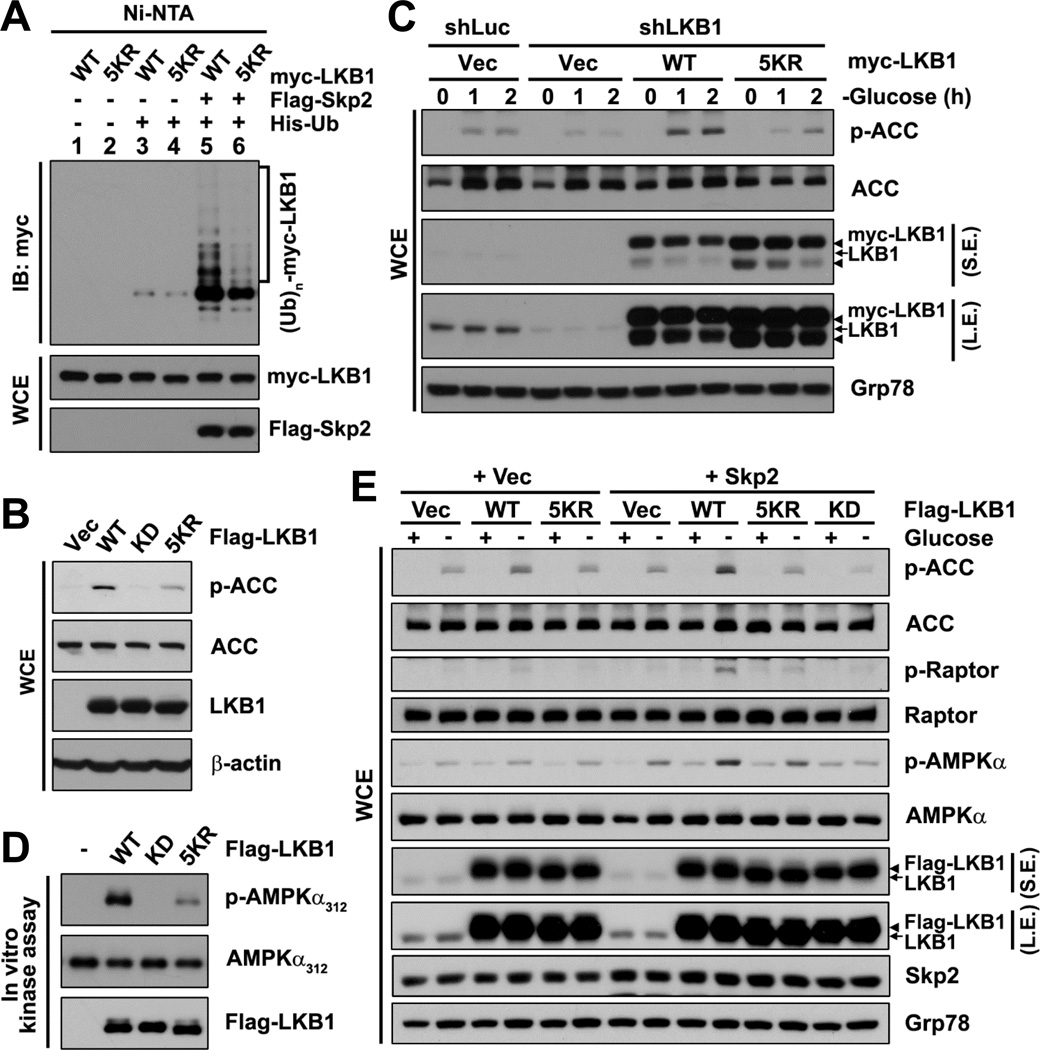
To further characterize the role of Skp2-mediated LKB1 polyubiquitination in LKB1 activation, we attempted to identify the site(s) on LKB1 where Skp2-mediated polyubiquitination takes place. Human LKB1 contains 32 Lys residues, and 26 of them are highly conserved among species. We first screened various fragments of LKB1 using in vivo ubiquitination assays to locate Skp2-mediated LKB1 polyubiquitination. By comparing polyubiquitination among the LKB1 fragments, we concluded that Skp2-mediated polyubiquitination occurred most intensively within the N-terminus of LKB1 (amino acids 1–243 and 1–87; Figures S3A–S3C). Following further screening within the region by mutagenesis, we found that none of a single Lys-to-Arg (K-to-R) substitution significantly affected LKB1 polyubiquitination (Figure S3D). By screening using multiple K-to-R substitutions, we identified that the 5KR mutant of LKB1 (LKB1–5KR), which carries 5 K-to-R substitutions at the position 41, 44, 48, 62 and 64, displayed much less Skp2-mediated polyubiquitination and basal polyubiquitination than WT LKB1 (Figures 3A and S3E), although LKB1–5KR bound to Skp2 comparably to WT LKB1 (Figure S3F).
Figure 3. Skp2-mediated polyubiquitination of LKB1 is critical for LKB1 kinase activity.
(A) In vivo ubiquitination assay in HEK293T cells transfected with the indicated plasmids was followed by IB.
(B) LKB1-deficient HeLa cells with stable restoration of the indicated Flag-LKB1 were subjected to IB.
(C) Control- or LKB1-knockdown Hep3B cells with stable transduction of the indicated shRNA non-targeting myc-LKB1 cultured in the absence of glucose for the indicated time points were subjected to IB.
(D) IPs of exogenous Flag-LKB1 by anti-Flag antibody from HEK293T cells transfected with the indicated Flag-LKB1 were subjected to in vitro LKB1 kinase assay followed by IB.
(E) Hep3B cells with stable transduction of vector (Vec) or Skp2 along with the indicated Flag-LKB1 cultured in the presence (+) or absence (−) of glucose were subjected to IB.
S.E., short exposure; L.E., long exposure.
See also Figure S3.
To understand whether Skp2-mediated K63-linked ubiquitination of LKB1 is important for LKB1 activity, we examined the ability of the ubiquitination-deficient mutant LKB1–5KR to activate the downstream AMPK signaling in cells. When stably expressed in LKB1-deficient HeLa cells, WT LKB1, but not kinase-dead LKB1 (LKB1-KD), strongly activated the AMPK signaling, showing robust induction of ACC phosphorylation compared with the vector control (Figure 3B). However, the 5KR mutant significantly reduced LKB1’s ability to induce the AMPK signaling, as determined by less phosphorylation of ACC compared with WT LKB1 (Figure 3B). Also, under the low-energy conditions—glucose deprivation or 2-DG treatment, LKB1–5KR exhibited a compromised ability to induce ACC phosphorylation in LKB1-deficient A549 cells and LKB1-KO MEFs in comparison with WT LKB1 (Figures S3G and S3H). Similarly, LKB1–5KR failed to rescue the defect of LKB1 knockdown in glucose deprivation-induced phosphorylation of ACC in Hep3B cells (Figure 3C). To confirm the impact of Skp2-mediated K63-linked polyubiquitination on LKB1 activity, we performed in vitro kinase assays. Consistently, LKB1–5KR protein isolated from cells and LKB1 protein derived from cells with Ubc13 knockdown, both of which displayed attenuated LKB1 polyubiquitination, showed decreased kinase activity toward AMPKα compared with their control counterparts (Figures 3D and S3I). Additionally, the 5KR mutant displayed a decreased ability to phosphorylate another LKB1’s downstream substrate SAD-B besides AMPKα (Figure S3J). To further demonstrate that the 5 Lys residues corresponding to the 5KR mutation are not critical for LKB1 kinase activity per se, we compared the in vitro kinase activity between recombinant LKB1–5KR and WT LKB1 proteins. Notably, there was no significant difference in the in vitro kinase activity between them (Figure S3K), suggesting that mutations of the Lys residues do not directly interfere with LKB1 kinase activity. Moreover, co-overexpression of Skp2 at a low level along with WT LKB1, but not LKB1–5KR, in LKB1-proficient cells could synergistically enhance the LKB1/AMPK signaling, as determined by increased phosphorylation of AMPKα, ACC and Raptor (at Ser792) (another AMPK substrate) in comparison with the vector counterparts (Figures 3E and S3L). Altogether, our data suggest that Skp2-mediated K63-linked ubiquitination of LKB1 serves as a novel regulatory mechanism for LKB1 activation.
LKB1 polyubiquitination is crucial for maintaining LKB1 complex integrity
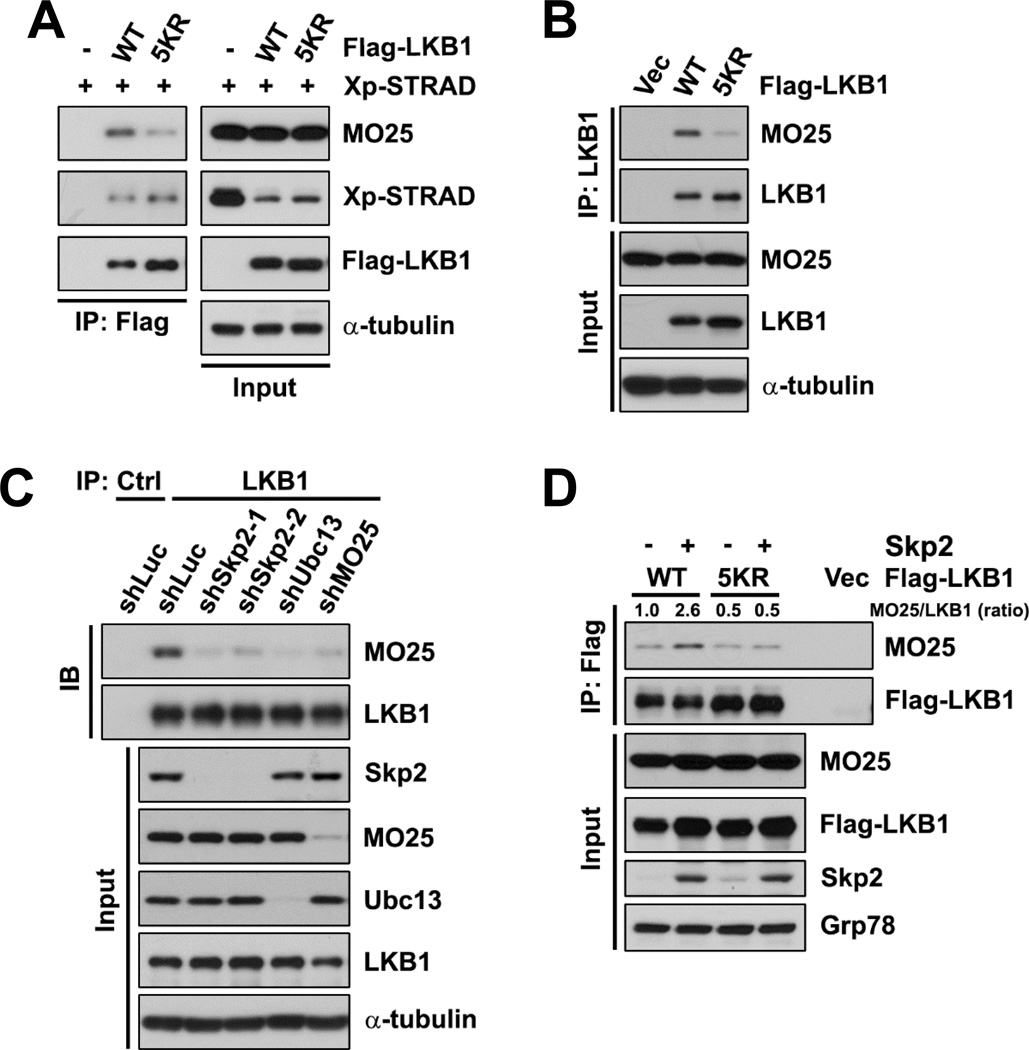
Next, we sought to determine the molecular mechanism of how Skp2-mediated K63-linked ubiquitination of LKB1 regulates LKB1 activity. First, we examined the ability of LKB1 to bind to its substrates. Although LKB1 polyubiquitination affected LKB1-dependent phosphorylation of its substrates, binding of LKB1 to any of its substrates was not affected by either LKB1–5KR mutant or Skp2 knockdown in co-IP assays (Figures S4A–S4C). Considering that LKB1 is mainly activated through the formation of the LKB1-STRAD-MO25 complex, we then investigated the integrity of the LKB1 heterotrimeric complex using co-IP assays. No significant differences were noticed in the LKB1-STRAD interaction between WT LKB1 and LKB1–5KR; however, the ubiquitination-deficient mutant LKB1–5KR displayed an impaired ability to bind to MO25 under either normal condition or glucose starvation (Figures 4A and S4D). Attenuated interaction between LKB1 and MO25 was also observed in multiple LKB1-deficient cell lines reconstituted with LKB1–5KR (Figures 4B, S4E, and S4F). Furthermore, Skp2 knockdown and Ubc13 knockdown, both of which attenuated LKB1 polyubiquitination, impaired the LKB1-MO25 interaction (Figures 4C and S4G). In contrast, overexpression of Skp2, which induced polyubiquitination of WT LKB1 but not LKB1–5KR, enhanced the association of WT LKB1, but not LKB1–5KR, with MO25 (Figures 4D and S4H). Since STRAD, but not MO25, has been demonstrated to facilitate nucleocytoplasmic shuttling of LKB1 (Dorfman and Macara, 2008), our result that LKB1 ubiquitination does not influence the LKB1-STRAD binding could explain our own observation that Skp2-mediated polyubiquitination of LKB1 does not affect the subcellular localization of LKB1 (Figures S4I–S4L). In addition, there was no significant difference in the protein half-life between WT LKB1 and LKB1–5KR (Figure S4M). Taken together, Skp2-mediated K63-linked polyubiquitination of LKB1 modulates LKB1 activity by maintaining the integrity of the LKB1 complex.
Figure 4. Skp2-mediated polyubiquitination of LKB1 is crucial for maintaining LKB1 complex integrity.
(A) IPs by anti-Flag antibody from cytosolic fractions of HEK293T cells transfected with Xp-STRAD and the indicated Flag-LKB1 were subjected to IB.
(B) IPs by anti-LKB1 antibody from cytosolic fractions of LKB1-deficient HeLa cells with stable reconstitution of the indicated Flag-LKB1 were subjected to IB.
(C) IPs by anti-IgG control or anti-LKB1 antibody from cytosolic fractions of HEK293T cells with control, Skp2, Ubc13 or MO25 knockdown were subjected IB.
(D) IPs by anti-Flag antibody from Hep3B cells with stable transduction of vector or Skp2 along with the indicated Flag-LKB1 were subjected to IB. The ratio of MO25-LKB1 binding is shown.
See also Figure S4.
Oncogenic Ras activates the Skp2-SCF to induce LKB1 polyubiquitination and activation
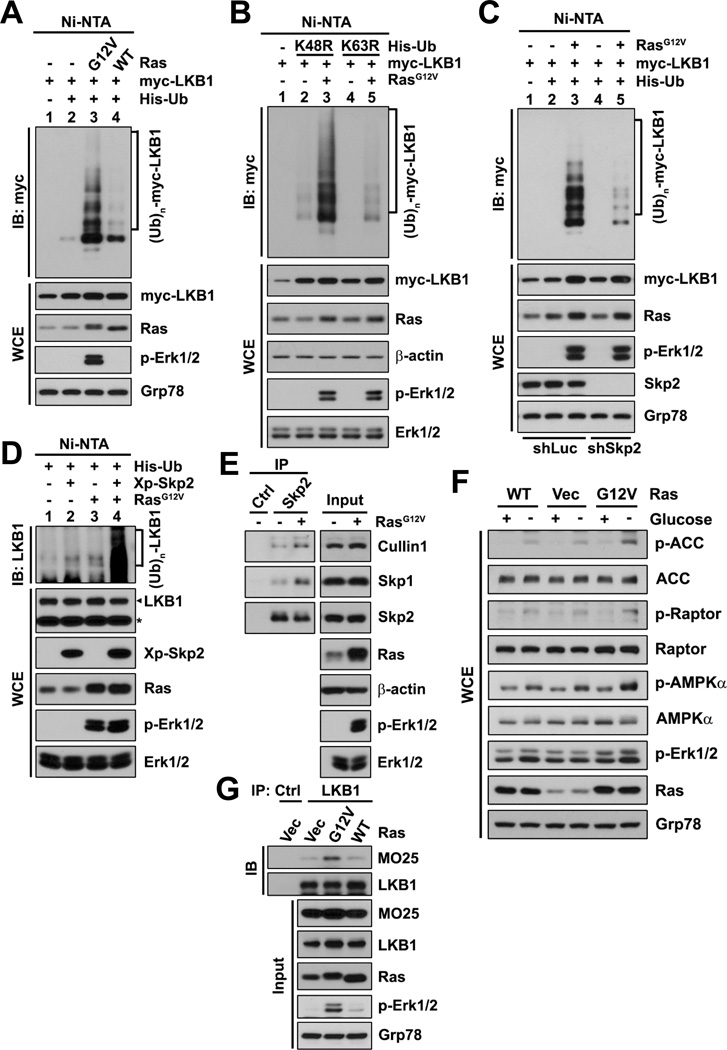
We further attempted to identify the upstream regulator(s) that can induce Skp2-mediated polyubiquitination and activation of LKB1. Surprisingly, we found that hyperactivation of Ras by overexpression of constitutively active H-Ras (H-RasG12V) promoted polyubiquitination of exogenous and endogenous LKB1 to a much greater extent than did overexpression of WT H-Ras (Figures 5A and S5A), and Ras-induced LKB1 polyubiquitination occurred mainly via K63 linkage, as assayed by using ubiquitin mutants and Ubc13 knockdown (Figures 5B and S5B). To further assess whether Ras-induced LKB1 polyubiquitination is dependent on Skp2, Skp2 knockdown, the Skp2 inhibitor and the ubiquitination-deficient mutant of LKB1 were used in the in vivo ubiquitination assays. Either upon Skp2 knockdown or in the presence of the Skp2 inhibitor, where Skp2-SCF E3 ligase activity is inhibited, active Ras no longer promoted LKB1 polyubiquitination efficiently (Figures 5C and S5C). A similar defect in Ras-mediated LKB1 polyubiquitination was also observed on the LKB1–5KR mutant (Figure S5D). In contrast, when co-overexpressed with Skp2, active Ras displayed a robust synergistic effect on polyubiquitination of endogenous LKB1 (Figure 5D). Notably, overexpression of active Ras promoted the assembly of the Skp2-SCF complex (Figure 5E; the Skp2-Skp1-Cullin1 interaction), indicating that Ras can induce Skp2-SCF activity.
Figure 5. Oncogenic Ras induces K63-linked polyubiquitination of LKB1 and activation of the LKB1/AMPK signaling via Skp2.
(A–D) In vivo ubiquitination assays in HEK293T cells (A, B and D) or HEK293T cells with control or Skp2 knockdown (C) transfected with the indicated plasmids were followed by IB. The asterisk in (D) indicates non-specific bands (as loading controls), and the arrowhead indicates the bands corresponding to LKB1.
(E) IPs by anti-IgG control or anti-Skp2 antibody from HEK293T cells transfected with vector or H-Ras were subjected to IB.
(F) WT MEFs with stable transduction of vector or the indicated H-Ras cultured in the presence (+) or absence (−) of glucose were subjected to IB.
(G) IPs by anti-IgG control or anti-LKB1 antibody from Hep3B cells with stable transduction of vector or the indicated H-Ras were subjected to IB.
Phosphorylation of Erk1/2 indicates Ras activation.
See also Figure S5.
We next investigated whether hyperactivation of Ras has effect on LKB1 activation. We found that, compared to WT Ras, overexpression of active Ras further upregulated the LKB1/AMPK signaling as indicated by elevated phosphorylation of AMPKα, ACC and Raptor (Figure 5F), and also promoted the LKB1-MO25 binding (Figure 5G). We then determined whether Ras-mediated activation of the LKB1/AMPK signaling involves Skp2-dependent LKB1 ubiquitination. Skp2 knockdown and LKB1 knockdown diminished Ras-mediated activation of the LKB1/AMPK signaling, as determined by reduced induction of ACC and Raptor phosphorylation in comparison with control knockdown (Figure S5E), and Ras co-overexpressed with WT LKB1, but not LKB1–5KR or LKB1-KD, further induced the LKB1/AMPK signaling (Figures S5F and S5G). Thus, our results suggest that oncogenic Ras induces K63-linked polyubiquitination of LKB1 and activation of the LKB1/AMPK signaling via activation of the Skp2-SCF complex.
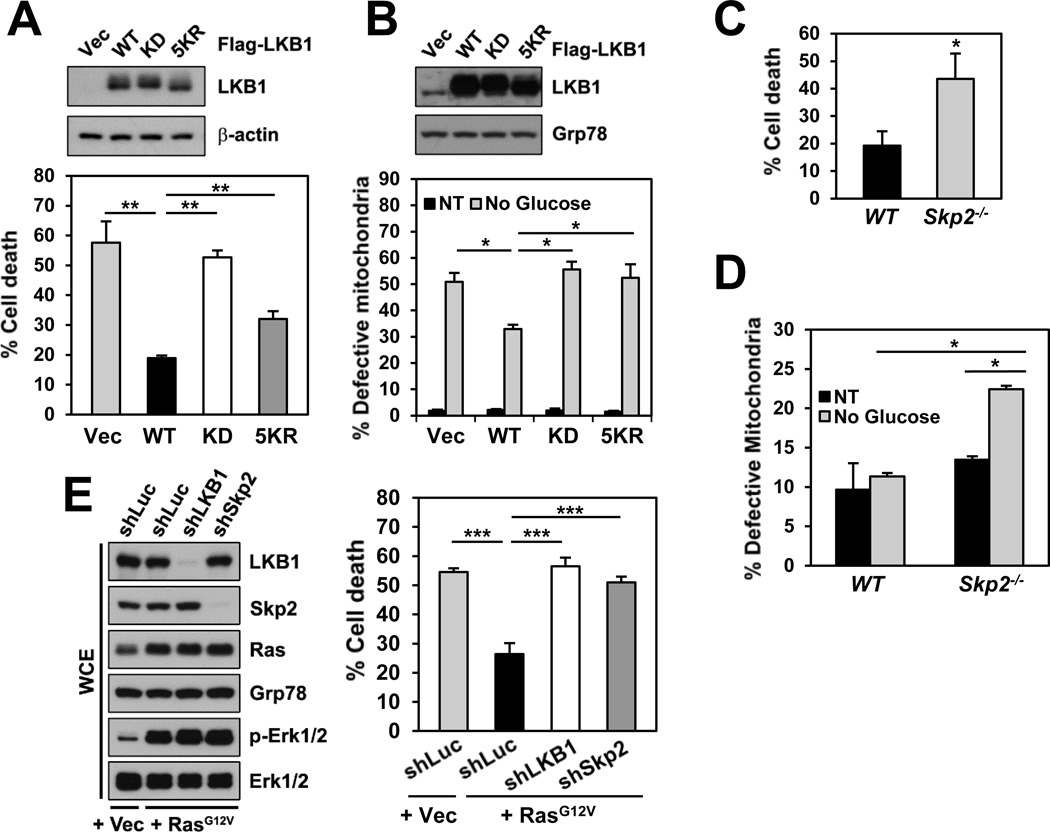
LKB1 polyubiquitination regulates LKB1’s function in cell survival under energy stress
As kinase activity has been shown to be critical for LKB1 to execute its biological function via the LKB1/AMPK axis during energy stress, we therefore investigated whether Skp2-mediated K63-linked polyubiquitination of LKB1 regulates LKB1 function. Activation of the LKB1/AMPK pathway plays an important role in protecting cells from apoptosis under metabolic stress. Accordingly, the effect of LKB1–5KR on energy stress-induced cell survival was examined. Consistent with previous studies (Shaw et al., 2004), compared with the vector control, WT LKB1 exhibited a protective effect on cell viability in LKB1-deficient cells under metabolic stress induced by glucose deprivation (Figure 6A) or treatment of AMP analogue AICAR (Figure S6A). In contrast, LKB1–5KR, which had compromised kinase activity, failed to protect LKB1-deficient cells from metabolic stress-induced cell death compared to WT LKB1 (Figures 6A and S6A). Recent studies demonstrated that the AMPK/ULK1 signaling and LKB1 have function in mitophagy, a selective form of autophagy that degrades damaged mitochondria following stresses (Egan et al., 2011; Shackelford et al., 2013). We then assessed the role of LKB1 polyubiquitination in mitochondrial homeostasis. Glucose deprivation caused increases in dysfunctional mitochondria, as assayed by flow cytometry following co-staining of cells with MitoTracker Green and Red (Figure S6B). Notably, under glucose deprivation, ectopic expression of WT LKB1, but not LKB1–5KR, led to a reduced number of defective mitochondria (Figures 6B and S6B) and increased phosphorylation of ULK1 at Ser555 (Figure S6C), which is required for mitophagy and cell survival under starvation (Egan et al., 2011). The data imply that overexpression of WT LKB1 may induce functional mitophagy. Similar results were obtained in Skp2-KO MEFs, showing that Skp2 deficiency, which lacks a functional LKB1/AMPK signaling, resulted in elevated cell death (Figure 6C) and accumulation of defective mitochondria (Figure 6D) under metabolic stress. Furthermore, we found that either LKB1 knockdown or Skp2 knockdown sensitized cancer cells to the mitochondrial inhibitor phenformin (Figures S6D and S6E), and pharmacological inactivation of Skp2 by the Skp2 inhibitor, which diminishes Skp2-mediated LKB1 polyubiquitination, enhanced the sensitivity of cancer cells to phenformin treatment (Figure S6F). These results implicate that Skp2-mediated K63-linked polyubiquitination of LKB1 is critical for LKB1 function in cell survival during energy stress.
Figure 6. Skp2-mediated polyubiquitination of LKB1 is critical for LKB1’s function in cell survival under energy stress.
(A) LKB1-deficient A549 cells stably restored with the indicated Flag-LKB1 were subjected to glucose starvation for 48 hours, and then cell death was determined by DAPI staining (n=3). Whole cell extracts of the untreated cells were subjected to IB (upper panel).
(B) Mitochondria of Hep3B cells with stable transduction of the indicated Flag-LKB1 cultured in the presence (NT, non-treated) or absence of glucose for 10 hours were analyzed by flow cytometry with MitoTracker staining (n=2). Whole cell extracts of the untreated cells were subjected to IB (upper panel).
(C) WT and Skp2-KO MEFs were subjected to treatment with 2mM AICAR for 24 hours. Cell viability was determined by trypan blue exclusion assay, and cell death is expressed as a percentage of the untreated controls (n=3).
(D) Mitochondria of MEFs as in (C) cultured in the presence (NT) or absence of glucose for 15 hours were analyzed as in (B) (n=2).
(E) Hep3B cells with stable transduction of vector or H-Ras along with the indicated stable knockdown were subjected to glucose starvation for 8 hours, and then cell death was determined by DAPI staining (n=4). Whole cell extracts of the untreated cells were subjected to IB (left panel).
All data are shown as means ± s.d. *P<0.05; **P<0.01; ***P<0.005.
See also Figure S6.
Given our above finding that Ras hyperactivation induces Skp2-depedent polyubiquitination and activation of LKB1, we reasoned that Ras may activate the LKB1/AMPK pathway to protect cancer cells against metabolic stress-induced cell death. In line with this notion, we observed that Ras hyperactivation reduced glucose deprivation-induced cell death (Figure 6E). However, either LKB1 knockdown or Skp2 knockdown abolished Ras-mediated protective effect on cell survival under glucose starvation (Figure 6E). Our results underscore the critical role of the Ras/Skp2/LKB1 axis in regulating cell survival during energy stress.
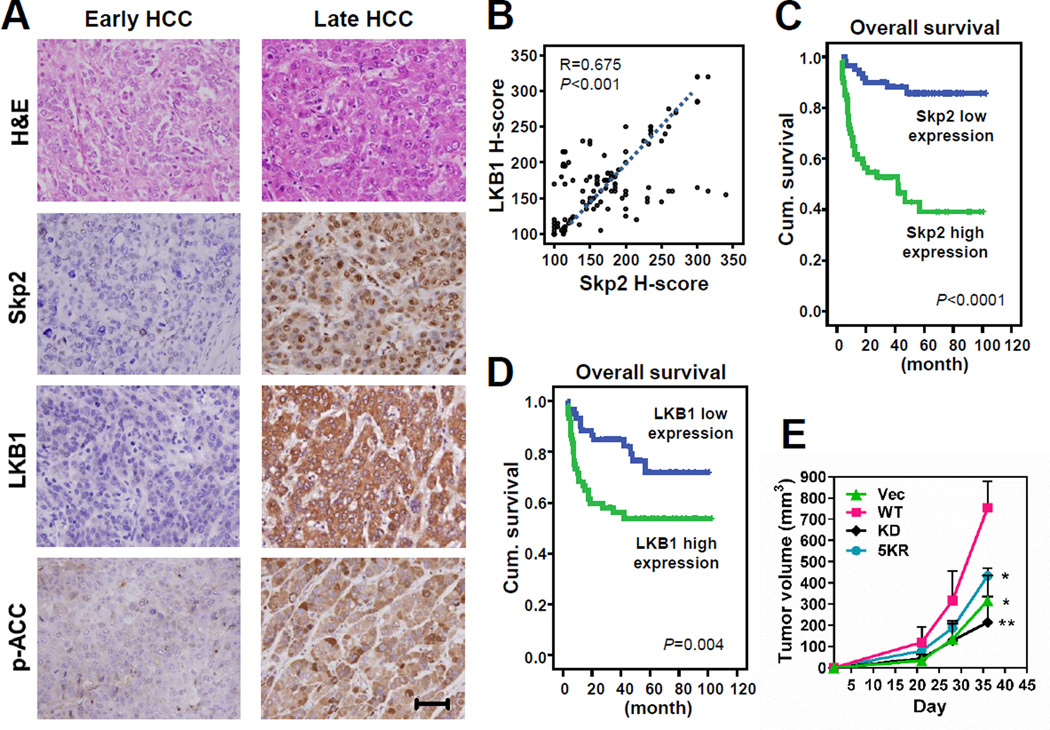
LKB1 is overexpressed and oncogenic in HCC
Skp2 displays oncogenic activity in vitro and in vivo, and overexpression of Skp2 has been reported in a variety of human cancers (Chan et al., 2010), including HCC (Calvisi et al., 2009; Lu et al., 2009). Our findings that Skp2 triggers K63-linked polyubiquitination and activation of LKB1 raise the possibility that LKB1 may also exhibit oncogenic activity in certain tissues like liver. To test this idea, we therefore assessed the expression of Skp2 and LKB1 and their correlation in human HCC patients. First, we analyzed the expression-profiling dataset of HCC tissues versus normal liver tissues from Gene Expression Omnibus, and found that both LKB1 (STK11) and Skp2 transcripts were upregulated and correlated in HCC (Figure S7A). Consistently, in our HCC patient cohorts, we detected that both Skp2 and LKB1 were overexpressed in late-stage HCC (Figure 7A). Our result is in agreement with the previous report showing that LKB1 is overexpressed in human HCC (Martinez-Lopez et al., 2012). In our patient cohorts, the increased expression of both Skp2 and LKB1 was significantly associated with numerous adverse clinical features (Table S1). Notably, Skp2 expression was also significantly related to liver cancer stages (Figure 7A and Table S1) and positively correlated with LKB1 expression (Figure 7B). Moreover, univariate survival analyses reveal that high expression of Skp2 or LKB1 was remarkably correlated with both overall and local recurrence-free survival (Figures 7C, 7D, S7B, S7C and Table S2). In multivariate survival analyses, Skp2 or LKB1 overexpression was independently predictive for worse overall survival, and only LKB1 expression level was prognostically significant for local recurrence-free survival (Table S3). We also found that phosphorylation of ACC, which is downstream of LKB1/AMPK and the readout of AMPK activity, was upregulated in late-stage HCC (Figure 7A) and associated with Skp2 and LKB1 expression (Figures S7D, S7E and Table S1). Importantly, the high level of ACC phosphorylation predicted poor patient survival (Figures S7F, S7G, Tables S2 and S3). Altogether, our data not only show that both Skp2 and LKB1 are overexpressed in HCC patients and their overexpression serves as prognostic markers for poor survival outcome, but also indicate the existence of the upregulated Skp2/LKB1/AMPK axis in HCC.
Figure 7. Both Skp2 and LKB1 are overexpressed in HCC and their overexpression predicts poor survival outcomes.
(A) Representative images of histological analysis of Skp2, LKB1 and p-ACC staining in early (left panel) and late-stage (right panel) HCC. Scale bar, 100 µm.
(B) Scatter plot of Skp2 expression versus LKB1 expression in HCC samples.
(C, D) Kaplan-Meier plots show that high expression of Skp2 (C) or LKB1 (D) is significantly predictive for inferior overall survival.
(E) Hep3B cells with stable transduction of the indicated Flag-LKB1 were subcutaneously injected into nude mice. Tumor size was measured by the caliper (n=3). The result is shown as means ± s.d. *P<0.05; **P<0.01.
See also Figure S7, and Tables S1, S2 and S3.
To further determine whether overexpression of LKB1 is oncogenic in HCC, we stably overexpressed LKB1 and its mutants in the human HCC cell line Hep3B and then investigated tumorigenicity of the cells in vivo in mouse subcutaneous tumor models. Notably, overexpression of WT LKB1 promoted HCC tumor growth in vivo, whereas overexpression of LKB1-KD or LKB1–5KR failed to do so (Figure 7E). Hence, our data suggest that LKB1 displays oncogenic activity in HCC in a manner dependent on its polyubiquitination and kinase activity.
Discussion
LKB1 has been suggested to be constitutively active (Sebbagh et al., 2011), and its activation is mediated by an allosteric mechanism dependent on complex assembly with STRAD and MO25 but independent of phosphorylation of the activation loop. All studies undertaken to date have underlined the intrinsic interaction between each component within the complex for LKB1 activation (Boudeau et al., 2004; Milburn et al., 2004; Zeqiraj et al., 2009a; Zeqiraj et al., 2009b). However, how the LKB1 activity is maintained and whether the complex is regulated by other extrinsic proteins (e.g., other unidentified LKB1 regulators) remain unclear. In this study, we uncover that Skp2 is a novel regulator of LKB1 activation by promoting K63-linked polyubiquitination of LKB1. This posttranslational modification is important for LKB1 to bind to one of its subunits, MO25, but not the other, STRAD. Co-overexpression of LKB1 and STRAD enhances LKB1 activity by about 3–5 times, whereas the coexistence of MO25 in the LKB1-STRAD complex further boosts LKB1 activity by about additional 5–10 times (Boudeau et al., 2003; Zeqiraj et al., 2009b). Because the ubiquitination-deficient mutant of LKB1 we identified only compromises the LKB1-MO25 interaction but not LKB1-STRAD interaction, the LKB1–5KR mutant still retains some residual kinase activity compared with the kinase-dead mutant of LKB1. The 5 K-to-R substitutions are clustered at the N-terminus of LKB1; on the basis of the reported crystal structure of the LKB1-STRAD-MO25 complex (Zeqiraj et al., 2009a), the 5 Lys residues of LKB1 are not located within the key catalytic motifs. Therefore, the 5KR mutation supposedly would not affect the LKB1 kinase activity owing to amino acid substitution. In addition, the possibility that the 5KR mutation causes unanticipated structural alterations in LKB1 could be largely ruled out by the fact that the LKB1–5KR mutant binds to LKB1’s substrates, STRAD and Skp2 comparably to WT LKB1 and has a basal kinase activity toward AMPK comparable to WT LKB1 when purified alone for the in vitro kinase assay.
To our knowledge, there have been no studies to date demonstrating LKB1 complex activation under any stimulating conditions (Sebbagh et al., 2011). Consistent with this notion, we did not observe that, any physiologically relevant stimuli tested (e.g., glucose deprivation, hypoxia or growth factor treatment) could markedly induce more polyubiquitination or higher kinase activity of LKB1 compared to the untreated controls. The impacts of Skp2 loss and the LKB1–5KR mutant on LKB1 ubiquitination, kinase activity and complex integrity could all be observed under normal conditions, implicating that Skp2-mediated K63-linked polyubiquitination of LKB1 modulates the integrity and the activity of the LKB1 complex in a ‘steady-state’ manner regardless of distinct physiological conditions. Unexpectedly, we found that the oncogenic insult by Ras hyperactivation (by expression of oncogenic H-Ras) could promote Skp2-mediated LKB1 polyubiquitination and complex formation, thereby activating the LKB1/AMPK signaling for cancer cell survival under energy stress. It has been reported that AMPK activation by oncogenic events is required for maintaining tumor cell growth and survival in subcutaneous tumor xenografts (Laderoute et al., 2006; Rios et al., 2013), and elevated AMPK activity has been found in mouse and human brain tumors (Jang et al., 2011; Rios et al., 2013). In line with this notion, we observed the positive correlation of Skp2 overexpression with LKB1 expression and AMPK activity (assessed by ACC phosphorylation) in our in-house HCC patient samples, which further implicates the existence of the upregulated Skp2/LKB1/AMPK axis in HCC. Hence, we speculate that, during cancer pathogenesis, cancer cells may gain genetic alterations such as Ras mutations or Skp2 overexpression to survive against diverse stresses by activating the LKB1/AMPK pathway.
Our finding that LKB1 is oncogenic in HCC appears paradoxical because LKB1 has been shown to display the tumor-suppressive activity in various genetic mouse models (Ollila and Makela, 2011). In this study, we propose that during tumor development, activation of the Ras/Skp2/LKB1 axis is required for cancer cell survival during energy stress. Therefore, our results are in line with the current accepted understanding of LKB1 function in stress-induced cell survival (Shaw et al., 2004). Most LKB1 genetic mouse models reported to date used tissue-specific knockout strategies; however, knockouts with chronic inactivation of LKB1 could not address the temporal requirement of LKB1 throughout the course of cancer development, especially at the later stage where severe metabolic stress takes place within the tumor microenvironment. Thus, the role of LKB1 in cancer maintenance and progression is largely undefined. Our result showing the enhanced expression of LKB1 and Skp2 in late-stage HCC implicates the oncogenic function of LKB1 in liver cancer progression rather than initiation.
Given that Ras-driven tumors are highly aggressive and likely experiencing severe metabolic stress inside the tumors, it is therefore conceivable that in order for Ras-driven tumors to develop into a full-blown disease, the cancer cells must activate a survival program to counteract metabolic stress-induced cell death. Our findings may therefore shed light on potential therapeutic implications for targeting a new branch of the Ras signaling networks (i.e., the Ras/Skp2/LKB1 axis). A recent study has underscored the significance of a functional LKB1/AMPK pathway in cancer cell survival in response to metabolic stress despite the presence of other tumor mutations including K-Ras mutations (Shackelford et al., 2013). Shackelford et al. showed that in a mouse model, non-small cell lung cancers which lack a functional LKB1/AMPK pathway are more sensitive to the treatment with the metabolic drug phenformin, which inhibits mitochondrial complex I and results in the elevation of intracellular AMP levels. In support of their notion, we observed that LKB1 knockdown and inactivation of Skp2 either genetically by Skp2 knockdown or pharmacologically by the Skp2 inhibitor, which all downregulate the LKB1/AMPK signaling, enhanced the sensitivity of Hep3B cancer cells to phenformin treatment. We also found that administration of the Skp2 inhibitor, which interrupts the Ras/Skp2/LKB1 cascade, further sensitized the Ras-overexpressing Hep3B cells to phenformin. Thus, we reasoned that a combination therapy with the Skp2 inhibitor and phenformin (or other metabolic drugs) may be an effective strategy for treating liver cancer and Ras-driven cancers.
In summary, our study shows that Skp2-mediated K63-linked polyubiquitination of LKB1 is critical for maintaining the integrity of the LKB1-STRAD-MO25 complex, thereby modulating LKB1 activity and cellular function in stress-induced cell survival responses, and that Ras acts upstream of this Skp2/LKB1 pathway. Identification of the Ras/Skp2/LKB1 axis in this study not only provides great insight into how LKB1 kinase activity is maintained, but also offers new therapeutic strategies for targeting Ras-driven cancers.
Experimental Procedures
Cell culture and reagents
Under normal conditions, cells were cultured in regular medium supplemented with 10% fetal bovine serum (FBS). For glucose deprivation, cells were washed with PBS once and then incubated in glucose-free DMEM supplemented with 10% dialyzed FBS for the indicated times. 2-deoxyglucose (2-DG), 5-aminoimidazole-4-carboxamide ribonucleotide (AICAR) and phenformin were purchased from Sigma. See also Supplemental Experimental Procedures.
In vitro LKB1 kinase assay
Endogenous LKB1 or exogenous Flag-LKB1 immunoprecipitated from cells was incubated with recombinant His-AMPKα1–312 for kinase reaction. LKB1 kinase activity was determined by measuring Thr172 phosphorylation of recombinant AMPKα1–312. See also Supplemental Experimental Procedures.
In vivo and in vitro ubiquitination assay
In vivo and in vitro ubiquitination assays were performed as described previously (Chan et al., 2012). See also Supplemental Experimental Procedures.
Cell viability assay
Cell viability was determined by using the trypan blue exclusion assay or DAPI staining. See also Supplemental Experimental Procedures.
Detection of damaged mitochondria
Mitochondria were analyzed by MitoTracker staining followed by flow cytometry analysis. See also Supplemental Experimental Procedures.
In vivo tumorigenesis assay
3×106 of Hep3B cells with stable LKB1 overexpression were subcutaneously injected into the flanks of nude mice. The tumor size was measured by the caliper, and the tumor volume (in mm3) was calculated by the equation: volume = (width)2 x length/2. All animal experiments were performed under Institutional Animal Care and Use Committee approval protocol.
Patients, tissue specimens and immunohistochemistry
This retrospective study had been approved by the Institutional Review Board of the Chi-Mei Medical Center (10311006). Immunoexpression was assessed on 120 consecutively treated primary hepatocellular carcinoma underwent surgical resection with curative intent between 1997 and 2002. The clinicopathologic variables evaluated from the 120 patients are listed in Table S1. The procedures of immunohistochemistry were identical to previously described (Chan et al., 2012). The slides were incubated with primary antibodies against Skp2 (1:100; Zymed), LKB1/STK11 (1:50; Abcam) or phospho-ACC (Ser79) (1:500; Cell Signaling). Primary antibodies were detected using the ChemMate DAKO EnVision kit (DAKO, K5001). The slides were incubated with the secondary antibody for 30 minutes and developed with 3,3-diaminobenzidine for 5 minutes. Immunostaining was scored by two pathologists (C.-F.L. and H.-Y.H.) by using a multiheaded microscope to reach a consensus for each case on the H-score as previously described (Chan et al., 2012).
Statistical analysis
All data are shown as means ± s.d. for at least three independent experiments, unless otherwise indicated. All statistical significance was determined by unpaired two-tailed Student’s t-tests, and P-values less than 0.05 were considered statistically significant.
Supplementary Material
Acknowledgments
We thank Drs. Gary D. Lopaschuk, Christelle Forcet, Nabeel Bardeesy, Franck Polleux and Kun-Liang Guan for reagents. We thank all the members of the Lin’s lab for their valuable comments and suggestions. We especially thank Yuan Gao in the Lin’s lab for reading and editing the manuscript. We also thank Zhenbo Han and Su Zhang in the Department of Molecular and Cellular Oncology for their technical support. This work was supported by NIH grants, CPRIT grant, MD Anderson Prostate SPORE Development Award, MD Anderson Prostate Cancer Moon Shot grant, R. Lee Clark Award, and China Medical University Fund to H.-K.L.
Footnotes
Author Contributions
S.-W.L. and H.-K.L. conceived the project, designed the experiments, analyzed and evaluated the data, and wrote the paper. S.-W.L. performed the experiments with assistance from G.J., Z.C., F.H., C.-H.C., W.-L.Y., B.-K.L. and A.H.R. C.-F.L. and H.-Y.H. designed and performed the immunohistochemistry experiments with human subjects, and analyzed and evaluated the data.
References
- Alessi DR, Sakamoto K, Bayascas JR. LKB1-dependent signaling pathways. Annu Rev Biochem. 2006;75:137–163. doi: 10.1146/annurev.biochem.75.103004.142702. [DOI] [PubMed] [Google Scholar]
- Baas AF, Boudeau J, Sapkota GP, Smit L, Medema R, Morrice NA, Alessi DR, Clevers HC. Activation of the tumour suppressor kinase LKB1 by the STE20-like pseudokinase STRAD. EMBO J. 2003;22:3062–3072. doi: 10.1093/emboj/cdg292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bardeesy N, Sinha M, Hezel AF, Signoretti S, Hathaway NA, Sharpless NE, Loda M, Carrasco DR, DePinho RA. Loss of the Lkb1 tumour suppressor provokes intestinal polyposis but resistance to transformation. Nature. 2002;419:162–167. doi: 10.1038/nature01045. [DOI] [PubMed] [Google Scholar]
- Blacketer MJ, Madaule P, Myers AM. Mutational analysis of morphologic differentiation in Saccharomyces cerevisiae. Genetics. 1995;140:1259–1275. doi: 10.1093/genetics/140.4.1259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boudeau J, Baas AF, Deak M, Morrice NA, Kieloch A, Schutkowski M, Prescott AR, Clevers HC, Alessi DR. MO25alpha/beta interact with STRADalpha/beta enhancing their ability to bind, activate and localize LKB1 in the cytoplasm. EMBO J. 2003;22:5102–5114. doi: 10.1093/emboj/cdg490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boudeau J, Scott JW, Resta N, Deak M, Kieloch A, Komander D, Hardie DG, Prescott AR, van Aalten DM, Alessi DR. Analysis of the LKB1-STRAD-MO25 complex. J Cell Sci. 2004;117:6365–6375. doi: 10.1242/jcs.01571. [DOI] [PubMed] [Google Scholar]
- Calvisi DF, Ladu S, Pinna F, Frau M, Tomasi ML, Sini M, Simile MM, Bonelli P, Muroni MR, Seddaiu MA, et al. SKP2 and CKS1 promote degradation of cell cycle regulators and are associated with hepatocellular carcinoma prognosis. Gastroenterology. 2009;137:1816–1826. e1811–e1810. doi: 10.1053/j.gastro.2009.08.005. [DOI] [PubMed] [Google Scholar]
- Chan CH, Lee SW, Wang J, Lin HK. Regulation of Skp2 expression and activity and its role in cancer progression. ScientificWorldJournal. 2010;10:1001–1015. doi: 10.1100/tsw.2010.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan CH, Li CF, Yang WL, Gao Y, Lee SW, Feng Z, Huang HY, Tsai KK, Flores LG, Shao Y, et al. The Skp2-SCF E3 ligase regulates Akt ubiquitination, glycolysis, herceptin sensitivity, and tumorigenesis. Cell. 2012;149:1098–1111. doi: 10.1016/j.cell.2012.02.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan CH, Morrow JK, Li CF, Gao Y, Jin G, Moten A, Stagg LJ, Ladbury JE, Cai Z, Xu D, et al. Pharmacological inactivation of Skp2 SCF ubiquitin ligase restricts cancer stem cell traits and cancer progression. Cell. 2013;154:556–568. doi: 10.1016/j.cell.2013.06.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorfman J, Macara IG. STRADalpha regulates LKB1 localization by blocking access to importin-alpha, and by association with Crm1 and exportin-7. Mol Biol Cell. 2008;19:1614–1626. doi: 10.1091/mbc.E07-05-0454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egan DF, Shackelford DB, Mihaylova MM, Gelino S, Kohnz RA, Mair W, Vasquez DS, Joshi A, Gwinn DM, Taylor R, et al. Phosphorylation of ULK1 (hATG1) by AMP-activated protein kinase connects energy sensing to mitophagy. Science. 2011;331:456–461. doi: 10.1126/science.1196371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flick JS, Johnston M. GRR1 of Saccharomyces cerevisiae is required for glucose repression and encodes a protein with leucine-rich repeats. Mol Cell Biol. 1991;11:5101–5112. doi: 10.1128/mcb.11.10.5101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaude H, Aznar N, Delay A, Bres A, Buchet-Poyau K, Caillat C, Vigouroux A, Rogon C, Woods A, Vanacker JM, et al. Molecular chaperone complexes with antagonizing activities regulate stability and activity of the tumor suppressor LKB1. Oncogene. 2012;31:1582–1591. doi: 10.1038/onc.2011.342. [DOI] [PubMed] [Google Scholar]
- Gurumurthy S, Xie SZ, Alagesan B, Kim J, Yusuf RZ, Saez B, Tzatsos A, Ozsolak F, Milos P, Ferrari F, et al. The Lkb1 metabolic sensor maintains haematopoietic stem cell survival. Nature. 2010;468:659–663. doi: 10.1038/nature09572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardie DG. AMP-activated/SNF1 protein kinases: conserved guardians of cellular energy. Nat Rev Mol Cell Biol. 2007;8:774–785. doi: 10.1038/nrm2249. [DOI] [PubMed] [Google Scholar]
- Hemminki A, Markie D, Tomlinson I, Avizienyte E, Roth S, Loukola A, Bignell G, Warren W, Aminoff M, Hoglund P, et al. A serine/threonine kinase gene defective in Peutz-Jeghers syndrome. Nature. 1998;391:184–187. doi: 10.1038/34432. [DOI] [PubMed] [Google Scholar]
- Jang T, Calaoagan JM, Kwon E, Samuelsson S, Recht L, Laderoute KR. 5’-AMP-activated protein kinase activity is elevated early during primary brain tumor development in the rat. International journal of cancer. Journal international du cancer. 2011;128:2230–2239. doi: 10.1002/ijc.25558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeon SM, Chandel NS, Hay N. AMPK regulates NADPH homeostasis to promote tumour cell survival during energy stress. Nature. 2012;485:661–665. doi: 10.1038/nature11066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laderoute KR, Amin K, Calaoagan JM, Knapp M, Le T, Orduna J, Foretz M, Viollet B. 5’-AMP-activated protein kinase (AMPK) is induced by low-oxygen and glucose deprivation conditions found in solid-tumor microenvironments. Mol Cell Biol. 2006;26:5336–5347. doi: 10.1128/MCB.00166-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin HK, Chen Z, Wang G, Nardella C, Lee SW, Chan CH, Yang WL, Wang J, Egia A, Nakayama KI, et al. Skp2 targeting suppresses tumorigenesis by Arf-p53-independent cellular senescence. Nature. 2010;464:374–379. doi: 10.1038/nature08815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin HK, Wang G, Chen Z, Teruya-Feldstein J, Liu Y, Chan CH, Yang WL, Erdjument-Bromage H, Nakayama KI, Nimer S, et al. Phosphorylation-dependent regulation of cytosolic localization and oncogenic function of Skp2 by Akt/PKB. Nat Cell Biol. 2009;11:420–432. doi: 10.1038/ncb1849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lizcano JM, Goransson O, Toth R, Deak M, Morrice NA, Boudeau J, Hawley SA, Udd L, Makela TP, Hardie DG, et al. LKB1 is a master kinase that activates 13 kinases of the AMPK subfamily, including MARK/PAR-1. EMBO J. 2004;23:833–843. doi: 10.1038/sj.emboj.7600110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu M, Ma J, Xue W, Cheng C, Wang Y, Zhao Y, Ke Q, Liu H, Liu Y, Li P, et al. The expression and prognosis of FOXO3a and Skp2 in human hepatocellular carcinoma. Pathol Oncol Res. 2009;15:679–687. doi: 10.1007/s12253-009-9171-z. [DOI] [PubMed] [Google Scholar]
- Martinez-Lopez N, Garcia-Rodriguez JL, Varela-Rey M, Gutierrez V, Fernandez-Ramos D, Beraza N, Aransay AM, Schlangen K, Lozano JJ, Aspichueta P, et al. Hepatoma cells from mice deficient in glycine N-methyltransferase have increased RAS signaling and activation of liver kinase B1. Gastroenterology. 2012;143:787–798. e781–e713. doi: 10.1053/j.gastro.2012.05.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milburn CC, Boudeau J, Deak M, Alessi DR, van Aalten DM. Crystal structure of MO25 alpha in complex with the C terminus of the pseudo kinase STE20-related adaptor. Nat Struct Mol Biol. 2004;11:193–200. doi: 10.1038/nsmb716. [DOI] [PubMed] [Google Scholar]
- Narbonne P, Roy R. Caenorhabditis elegans dauers need LKB1/AMPK to ration lipid reserves and ensure long-term survival. Nature. 2009;457:210–214. doi: 10.1038/nature07536. [DOI] [PubMed] [Google Scholar]
- Ollila S, Makela TP. The tumor suppressor kinase LKB1: lessons from mouse models. J Mol Cell Biol. 2011;3:330–340. doi: 10.1093/jmcb/mjr016. [DOI] [PubMed] [Google Scholar]
- Rios M, Foretz M, Viollet B, Prieto A, Fraga M, Costoya JA, Senaris R. AMPK activation by oncogenesis is required to maintain cancer cell proliferation in astrocytic tumors. Cancer Res. 2013;73:2628–2638. doi: 10.1158/0008-5472.CAN-12-0861. [DOI] [PubMed] [Google Scholar]
- Sanchez-Cespedes M. A role for LKB1 gene in human cancer beyond the Peutz-Jeghers syndrome. Oncogene. 2007;26:7825–7832. doi: 10.1038/sj.onc.1210594. [DOI] [PubMed] [Google Scholar]
- Sebbagh M, Olschwang S, Santoni MJ, Borg JP. The LKB1 complex-AMPK pathway: the tree that hides the forest. Fam Cancer. 2011;10:415–424. doi: 10.1007/s10689-011-9457-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shackelford DB, Abt E, Gerken L, Vasquez DS, Seki A, Leblanc M, Wei L, Fishbein MC, Czernin J, Mischel PS, et al. LKB1 inactivation dictates therapeutic response of non-small cell lung cancer to the metabolism drug phenformin. Cancer Cell. 2013;23:143–158. doi: 10.1016/j.ccr.2012.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw RJ, Kosmatka M, Bardeesy N, Hurley RL, Witters LA, DePinho RA, Cantley LC. The tumor suppressor LKB1 kinase directly activates AMP-activated kinase and regulates apoptosis in response to energy stress. Proc Natl Acad Sci U S A. 2004;101:3329–3335. doi: 10.1073/pnas.0308061100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw RJ, Lamia KA, Vasquez D, Koo SH, Bardeesy N, Depinho RA, Montminy M, Cantley LC. The kinase LKB1 mediates glucose homeostasis in liver and therapeutic effects of metformin. Science. 2005;310:1642–1646. doi: 10.1126/science.1120781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutherland CM, Hawley SA, McCartney RR, Leech A, Stark MJ, Schmidt MC, Hardie DG. Elm1p is one of three upstream kinases for the Saccharomyces cerevisiae SNF1 complex. Curr Biol. 2003;13:1299–1305. doi: 10.1016/s0960-9822(03)00459-7. [DOI] [PubMed] [Google Scholar]
- van der Velden YU, Wang L, Zevenhoven J, van Rooijen E, van Lohuizen M, Giles RH, Clevers H, Haramis AP. The serine-threonine kinase LKB1 is essential for survival under energetic stress in zebrafish. Proc Natl Acad Sci U S A. 2011;108:4358–4363. doi: 10.1073/pnas.1010210108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Han F, Wu J, Lee SW, Chan CH, Wu CY, Yang WL, Gao Y, Zhang X, Jeong YS, et al. The role of Skp2 in hematopoietic stem cell quiescence, pool size, and self-renewal. Blood. 2011;118:5429–5438. doi: 10.1182/blood-2010-10-312785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woods A, Johnstone SR, Dickerson K, Leiper FC, Fryer LG, Neumann D, Schlattner U, Wallimann T, Carlson M, Carling D. LKB1 is the upstream kinase in the AMP-activated protein kinase cascade. Curr Biol. 2003;13:2004–2008. doi: 10.1016/j.cub.2003.10.031. [DOI] [PubMed] [Google Scholar]
- Yang WL, Zhang X, Lin HK. Emerging role of Lys-63 ubiquitination in protein kinase and phosphatase activation and cancer development. Oncogene. 2010;29:4493–4503. doi: 10.1038/onc.2010.190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeqiraj E, Filippi BM, Deak M, Alessi DR, van Aalten DM. Structure of the LKB1-STRAD-MO25 complex reveals an allosteric mechanism of kinase activation. Science. 2009a;326:1707–1711. doi: 10.1126/science.1178377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeqiraj E, Filippi BM, Goldie S, Navratilova I, Boudeau J, Deak M, Alessi DR, van Aalten DM. ATP and MO25alpha regulate the conformational state of the STRADalpha pseudokinase and activation of the LKB1 tumour suppressor. PLoS Biol. 2009b;7:e1000126. doi: 10.1371/journal.pbio.1000126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong L, Georgia S, Tschen SI, Nakayama K, Bhushan A. Essential role of Skp2-mediated p27 degradation in growth and adaptive expansion of pancreatic beta cells. J Clin Invest. 2007;117:2869–2876. doi: 10.1172/JCI32198. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.