Abstract
P bodies and stress granules are RNA-containing structures governing mRNA degradation and translational arrest, respectively. Saccharomyces cerevisiae Pbp1 protein localizes to stress granules and promotes their formation and is involved in proper polyadenylation, suppression of RNA-DNA hybrids, and preventing aberrant rDNA recombination. A genetic screen for Aspergillus nidulans mutants aberrant in secondary metabolism identified the Pbp1 homolog, PbpA. Using Dcp1 (mRNA decapping) as a marker for P-body formation and FabM (Pab1, poly-A binding protein) to track stress granule accumulation, we examine the dynamics of RNA granule formation in A. nidulans cells lacking pub1, edc3, and pbpA. Although PbpA acts as a functional homolog of yeast PBP1, PbpA had little impact on either P-body or stress granule formation in A. nidulans in contrast to Pub1 and Edc3. However, we find that PbpA is critical for sexual development and its loss increases the production of some secondary metabolites including the carcinogen sterigmatocystin.
Keywords: stress response, RNA granule, secondary metabolism, sexual development
1. Introduction
Regulation of mRNA levels are critical processes by which cells may rapidly respond to flux in inter- or extracellular conditions. This regulation is achieved by such means as mRNA localization, mRNA stability, and mRNA translation. Recent evidence indicates that these processes can be co-regulated by segregation of nontranslating mRNA molecules and their associated proteins into distinct structures within the cytoplasm, termed mRNP granules (Buchan, 2014). These mRNP granules include both P-bodies and stress granules, which perform separate but overlapping functions, and share a number of constituents (Kedersha and Anderson, 2009). Formation of mRNP granules is interdependent, as that P-body formation is required for normal stress granule assembly (Buchan et al., 2008). mRNA trafficking and sequestration in fungi have been associated with such diverse processes as stress response (Keller, 2015; Ren et al., 2016; Wang et al., 2015), polar growth (Becht et al., 2006; Inglis et al., 2013; Tey et al., 2005), nutrient acquisition (Morozov et al., 2010b), and morphological switching (Göhre et al., 2013).
While both P-bodies and stress granules contribute to the regulation of translation, P-bodies are more commonly associated with mRNA decay, and are defined by an enrichment of proteins involved in this process. In yeast, these include decapping enzymes (Dcp1/2), activators of decapping (Edc3 and Lsm1–7), and an exonuclease (Xrn1). P-bodies are normally present in low numbers, but can increase after translation inhibition or exposure to certain environmental stresses (reviewed in (Parker and Sheth, 2007). In Aspergillus nidulans, P-bodies components have been linked to response to nitrogen sufficiency by facilitating turnover of the transcription factor AreA (Morozov et al., 2010a; Morozov et al., 2010b). Deletion of the P-body component Edc3 in this system led to severe depletion of P-bodies (as monitored by Dcp1-GFP) and a defect in the rate of both global and targeted RNA turnover, although no significant growth phenotype was noted.
Stress granules are much less common under standard cellular conditions, and normally form under strong inhibition of translation, as induced by translational inhibitors or other environmental stimuli (Kedersha et al., 2005). mRNA arrest in these stress granules can lead to reintroduction of the mRNAs back into the translating pools at a later time. Although the exact composition of stress granules can vary depending on their cause of formation and species, in S. cerevisiae protein components can include Poly-A binding proteins (Pab1 and Pub1), elongation factors (eIF4G, eIF4E), proteins involved in RNA destabilization (Ngr1), and Pbp1, a Poly-A binding protein (PAB1) binding protein (Buchan et al., 2008; Hoyle et al., 2007). Removal of both pub1 and pbp1 have been shown to limit stress granule formation in S. cerevisiae (Buchan et al., 2008), while the essential gene PAB1 is often used as a marker (Buchan et al., 2008; Kozubowski et al., 2011). In A. oryzae, various stresses led to accumulation of the PAB1 homolog (Huang et al., 2013). Presumed limitation of stress granule formation by deletion of the pub1 homolog led to greatly increased sensitivity to stress, suggesting that the integrity of these granules may be critical to respond to adverse conditions (Huang et al., 2013).
Numerous previous studies have shown a link between stress and fungal secondary metabolism (Keller, 2015; Ren et al., 2016; Wang et al., 2015). We have reported on the applicability of using a genetic suppressor screen to look for genes involved in secondary metabolism including RsmA, a bZIP transcription factor linking both secondary metabolism and sexual development with the stress response in A. nidulans (Shaaban et al., 2010; Yin et al., 2012). From this same screen, we now present our identification of an A. nidulans homolog of the S. cerevisiae Pab1 binding protein Pbp1 as also playing an unexpected role in secondary metabolism and sexual development. In both yeast and mammalians cells, Ppb1 and its mammalian homolog Ataxin-2 have been shown to be involved in assembly of stress granules (Buchan et al., 2008), with mutations in ATXN2, the gene encoding Ataxin-2, associated with the neurodegenerative disease spinocerebellar ataxia type 2 (Stevanin et al., 2000). Deletion of PBP1 in S. cerevisiae leads to significant decreases in stress granule formation under glucose deprivation, while P-bodies are unaffected. In line with its role in stress granule assembly, PBP1 has also been implicated in resistance to various stresses, including caffeine, cycloheximide, hydroxyurea (Kapitzky et al., 2010), and recovery from ethanol stress (Kato et al., 2011), as well as regulation of poly-A tail length (Mangus et al., 1998; Mangus et al., 2004), and cell growth (Kimura and Irie, 2013). Here we show that AN1325, here called pbpA, is a homolog of yeast PBP1 and is required for normal sexual sporulation and has a repressive effect on secondary metabolism but has no major role in either P-body or stress granule biology in A. nidulans.
2. Materials and Methods
2.1 Sequence analysis
Genomic DNA sequence and translation of AN1325.4 (pbpA) gene was obtained from the Aspergillus Genome Database (www.aspgd.org). fabM (AN4000 (Marhoul and Adams, 1996)), edc3 and dcp1 (AN6893 and AN7746 (Morozov et al., 2010b), and pub1 (AN10164 (Huang et al., 2013)) have been previously described.
Alignment was performed using the NCBI’s Conserved Domain Detection tools (Marchler-Bauer et al., 2015) and COBALT (Papadopoulos and Agarwala, 2007) with the following Genbank accession sequences: Homo sapiens ATXN2: AAI14547; Aspergillus nidulans: XP_658929; Saccharomyces cerevisiae: CAA97204; Schizosaccharomyces pombe: CAB57927; Candida albicans: XP_717736; and Cryptococcus neoformans: XP_571007. Percent identity was calculated using Clustal Omega (Sievers et al., 2011). Amino acid alignments are included in Fig. S1.
2.2 Culture conditions, Southern, and northern analysis
All strains (Table S1) were propagated at 37°C on glucose minimum medium (GMM) with appropriate supplements. Fungal DNA was isolated as previously described (Shimizu and Keller, 2001). DNA manipulations, Southern, and northern analysis were conducted according to standard procedures (Sambrook and Russell, 2001).
2.3 Yeast complementation assay
PCR was used to amplify either ScPBP1 + 1 kb flanks or pbpA and PBP1 flanks. Yeast recombineering was used to insert these fragments into the backbone of pGAD424 (Chien et al., 1991). These plasmids were transformed into BY4741 and BY4741 ΔPBP1. For assessment of the petite negative phenotype, overnight cultures were grown in YPD. Five-fold dilutions of these cultures were performed and plated on YPD and YPD + 40 µg/ml ethidium bromide (Hwang et al., 2007).
2.4 Construction of mutant pbpA strains
One kb flanks upstream and downstream of pbpA were amplified and fused to an A. parasiticus pyrG – A. nidulans gpdA fusion cassette from pJMP9 (Soukup et al., 2012b) using double joint PCR (Yu et al., 2004). The resulting knockout construct was transformed into RJMP1.1 as previously described (Szewczyk et al., 2006). Transformants were examined for targeted replacement of the native locus by PCR and Southern blotting (Fig. S2A,B), and confirmed by northern blot of the appropriate transcript (Fig. S2C). Prototrophic overexpression strains were obtained by crossing the transformants with RTMH207.13 or DVARI (Kim et al., 2002). Desired recombinants were confirmed by PCR screening.
For pub1 deletion strains, 1 kb of flanking regions were amplified and fused to A. parasiticus pyrG from pJW24 (Calvo et al., 2004) using double joint PCR. The resulting knockout construct was transformed into TJMP1.1. Transformants were examined for targeted replacement of the native locus by PCR and Southern blotting. Prototrophic deletion strains were obtained by crossing transformants with RTMH207.13 or DVARI.
For complementation of pbpA, the coding region and 1 kb upstream and downstream flanks were amplified using PCR and cloned into the NotI sites of pJW53. The resulting knockout constructs was transformed into RJMP1.59 as previously described (Szewczyk et al., 2006). Transformants were examined for integration by PCR and Southern blotting (Fig. S2D). These were crossed to TAAS110.7 to produce prototrophic pbpA complements.
2.5 Northern analysis
Fifty milliliter cultures of liquid GMM were inoculated with 1 × 106 spores per ml and incubated at 250 rpm and 37°C for 36 hours under light. Mycelia were harvested, lyophilized overnight, and total RNA was extracted using Isol-RNA Lysis Reagent (5 Prime) according to manufacturer’s recommendations. Subsequent northern analysis was done using radiolabeled probes for the corresponding transcript (primers are listed in Table S2).
2.6 Fluorescent strain construction
For initial C-terminal tagging of pbpA, dcp1, and fabM, 1 kb of the 3’ end of the gene and 1 kb downstream of the gene of interest were amplified and fused to either pXDRFP4 (RFP) or pFNO3 (GFP)(Yang et al., 2004) via double joint PCR (Yu et al., 2004). The resulting constructs was transformed into RJMP1.1 as previously described (Szewczyk et al., 2006). Transformants were examined for targeted integration at the native locus by PCR and Southern blotting (Fig. S4). To obtain the final florescent strains sequential crosses were performed between TAAS228.16 and PW1 to yield RAAS235.6. This was crossed to TAAS227.6 to yield RAAS236.1. RAAS236.1 (fabM::gfp::pyrG; dcp1::rfp::pyrG; metG1; biA1) was crossed to the appropriate transformants. Desired strains were confirmed via PCR. The fluorescent prototroph (RAAS237.2: fabM::gfp::pyrG; dcp1::rfp::pyrG) was compared to WT (RJMP103.5) to confirm functionality of tagged proteins (Fig. S5).
For construction of stcS-GFP, 1 kb of the C-terminal coding region and 1 kb downstream of stcS was amplified and fused to gfp:pyroA from plasmid pHL84 (Liu et al., 2009) via double joint PCR (Yu et al., 2004). The resulting constructs was transformed into RJMP1.59 as previously described (Szewczyk et al., 2006). Transformants were examined for targeted integration at the native locus by PCR and Southern blotting (Fig. S4). To obtain the final florescent strains subsequent crosses were performed between TAAS245.27 and PW1 or TAAS110.1 to yield RAAS246.9 and RAAS247.1, respectively. Desired strains were confirmed via PCR.
2.7 Phenotypic characterization and SM analysis on solid media
Secondary metabolite production was assessed by thin-layer chromatography (TLC). For TLC, 10 µl of 1 × 103 spore/µl was point-inoculated on the center of glucose minimal medium (GMM) and cultured for 72 hr at 37°C. An agar plug of the center of colonies was removed and SMs extracted with ethyl acetate according to the Smedsgaard’s method (Smedsgaard, 1997). Extracts (10 µl/sample) were loaded onto silica TLC plates (Whatman, PE SIL, Maidstone, Kent, England) and metabolites were separated in the developing solvent toluene:ethyl acetate:glacial acetic acid (TEA, 8:1:1). Images were taken following exposure to UV radiation at 366 nm.
2.8 Analysis of spore production
Quantification was performed on overlay inoculated cultures set up by pipetting 1 × 106 conidia into GMM or CHAMPS medium with 0.75% molten agar that was subsequently poured over 1.5% solid agar petri dishes of the corresponding medium. Cultures were incubated at 37°C in the dark for 3 or 5 days and agar cores were taken from the plates with a 1 cm cork borer. After homogenization, ascospores and conidia were quantified using a hemacytometer and represented as spores per square millimeter. 3 replicates were performed for each strain and condition.
2.9 Antibacterial bioassay
For the penicillin bioassay, 50 milliliter cultures of liquid GMM were inoculated with 1 × 106 spores per ml and incubated at 250 rpm and 37°C for 72 hours under light. 10 ml culture was used to perform the bioassay against Micrococcus luteus as described in Bok and Keller (2004).
2.10 Microscopy
Ten milliliters of ammonium minimal media (AMM) was inoculated with A. nidulans spore suspensions at a concentration of 104 spores/mL in a 60 mm diameter petri dish containing a sterile 1.5 mm coverslip. Cultures were allowed to grow for 17 hours at room temperature. To induce glucose stress, 17 hours post inoculation media was exchanged for 10 mL AMM lacking glucose and left at room temperature for one hour. For cycloheximide treatment during glucose starvation, cycloheximide (Sigma) was added to a final concentration of 200 µg/mL as previously described (Huang et al., 2013) 30 minutes before media exchange. At 17 hours post inoculation, 10 mL of AMM lacking glucose containing 200 µg/mL cycloheximide was used to exchange media. Cultures were placed at room temperature for one hour before imaging.
Coverslips were removed from the petri dishes, mounted on a glass slide, and sealed using clear fingernail polish. Images were collected using a Nikon Eclipse Ti inverted microscope equipped with a 60x Plan Apo VC Oil DIC N2 objective. Images were analyzed using the Nikon NIS Elements Advanced Research software package (v. 4.30.01) and scale bars in all figures represents 20 um. Stress granule and P body counts (n=10) were standardized to germling length and analyzed for significance using the Student’s T Test within the Graphpad Prism statistical package. For P body quantification, a fluorescent threshold value was used to develop a binary image from which P bodies were counted. Specifically, images were loaded into ImageJ and a color threshold specified using the IsoData thresholding method within ImageJ, with the brightness threshold values set to range from 115–255. Binary images were then used to count the number of P bodies (Fig. S6). Error bars represent standard error. *p=<0.05 **=p<0.01 ***=p<0.001 ns= not significant
3. Results
3.1 AN1325 encodes a homolog of S. cerevisiae PBP1
We previously described a genetic screen where we identified A. nidulans mutants that impacted secondary metabolism as determined by pigmentation of the mutant strain on common growth media (details in (Shaaban et al., 2010). Two proteins characterized from this screen included the previously mentioned RsmA, as well as EsaA, a histone 4 acetyltransferase (Soukup et al., 2012a). A third gene identified by this screen is AN1325. BLAST analysis (Altschul et al., 1997) of the predicted product of AN1325 revealed a 29.3% identity to S. cerevisiae Pbp1 (Mangus et al., 1998), 28.3% identity to Schizosaccharomyces pombe Ath1 (Wang et al., 2012), 31.3% to Cryptococcus neoformans Pbp1 (Park et al., 2016) and 25.5% identity to the mammalian ortholog Ataxin-2 (ATXN2 (Ralser et al., 2005). AN1325 (pbpA) encodes a 1047 amino acid long protein containing the expected ataxin-2 domain. All orthologs contain an N-terminal Lsm associated domain, and the majority (with the exception of C. neoformans) contain the above mentioned Ataxin 2 similar domain (Fig. 1A and S1).
Fig. 1.
A. Diagram of PbpA orthologs and their protein domain structure. Blue: Ataxin 2 like domain. Green: Lsm Associated Domain. Red: PAB1 binding domain. An: A. nidulans. Sc: S. cerevisiae. Schp: S. pombe. Cn: C. neoformans. Hs: H. sapiens. B. PbpA functionally complements a S. cerevisiae PBP1 deletion in a petite negative assay. Colonies represent 10 fold dilutions. BY4741, the ΔPBP1 parental strain, grows normally on both YPD and YPD + 40 µg/ml ethidium bromide, which induces loss of the mitochondrial genome. Lower densities of the ΔPBP1 strain are unable to survive this loss. Addition of either PBP1 or pbpA does not affect growth or survival in a wild type background, but restores normal growth to the ΔPBP1 parent.
3.2 Complementation of S. cerevisiae Δpbp1
Previous studies have shown the S. cerevisiae PBP1 deletant to have a petite negative phenotype (Dunn and Jensen, 2003), which is observed as an inability to survive loss of the mitochondrial genome on rich medium. In order to determine whether PbpA could serve to functionally complement a S. cerevisiae PBP1 deletion and its petite negative phenotype, we constructed complementation vectors containing either yeast PBP1 or pbpA genomic sequence under the yeast PBP1 promoter and selectable URA3 marker. We chose to examine growth of wild type and a Δpbp1 strain transformed with these vectors on YPD medium with and without 40 µg/ml ethidium bromide (EtBr), which induces mitochondrial loss (Hwang et al., 2007). As expected, Δpbp1 shows significantly decreased growth in the presence of EtBr, which can be restored by the addition of PBP1 (Fig. 1B). Introduction of pbpA also led to normal growth on this medium, suggesting that A. nidulans pbpA can functionally complement Δpbp1 and prevent mitochondrial loss under these conditions.
3.3 Deletion of pbpA does not suppress ΔfabM lethality
In both S. cerevisiae and A. nidulans, PAB1 and its homolog fabM are essential genes (Marhoul and Adams, 1996; Sachs et al., 1987). In yeast, the requirement for PAB1 is bypassed by deletion of both PAB1 and PBP1. We therefore sought to determine if this was also the case in A. nidulans. A ΔpbpA strain was constructed through transformation and homologous recombination of the pbpA gene with the pyrG gene of A. parasiticus and confirmed via Southern blot (Fig S1A). The construct to delete fabM by replacement with the A. fumigatus riboB gene was then transformed into both wild type and ΔpbpA backgrounds. Multiple transformants were obtained in both cases, with many displaying slow growth and sparse hyphal density (Fig. S3B). Southern blots were performed to confirm the successful integration of the deletion construct (Fig. S3 A). In all transformants, regardless of genetic background, bands indicating either (i) the presence of wild type fabM or (ii) wild type fabM and the deletion construct were seen, suggesting that all of the transformants which correctly integrated the deletion construct were heterokaryons with respect to fabM. Use of the heterokaryon rescue technique (Osmani et al., 2006) confirmed that fabM was essential in both backgrounds (data not shown).
3.4 PbpA does not localize to stress granules in A. nidulans under glucose deprivation
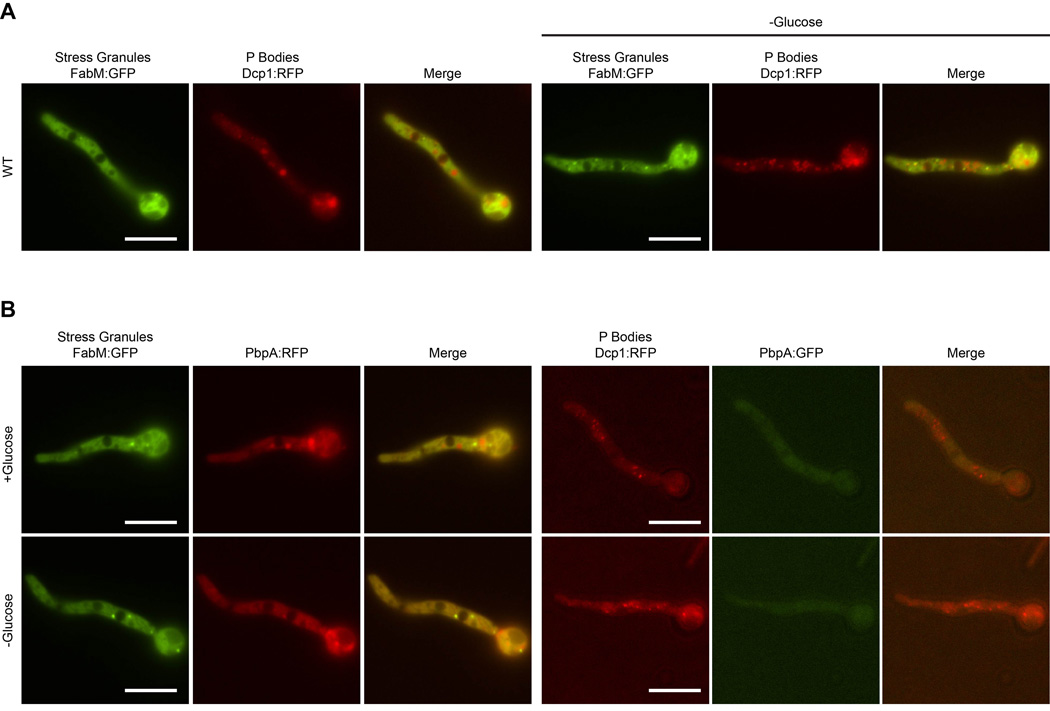
Pbp1, Ath1, and their mammalian ortholog, Ataxin-2, accumulate in stress granules upon glucose deprivation or arsenite stress, respectively (Buchan et al., 2008; Nonhoff et al., 2007; Wang et al., 2012). In order to determine if this was also the case in A. nidulans, we constructed fluorescent tagged versions of stress granule and P-body components based on previous studies (Buchan et al., 2008; Huang et al., 2013; Kozubowski et al., 2011; Morozov et al., 2010b), and confirmed normal vegetative growth of these strains (Fig. S5). Dcp1-RFP was used as a marker for P-bodies, and FabM-GFP as a marker for stress granules. RFP and GFP tagged PbpA were generated, and crossed into the appropriate backgrounds for comparison (see methods).
Under normal growth conditions, Dcp1-RFP localizes to multiple cytoplasmic foci (Fig 2A), which we will refer to as P-bodies. Addition of stress through glucose increases the number of these foci approximately twofold. In contrast, FabM-GFP is distributed throughout the cytoplasm, with very few foci of high intensity under normal growth conditions. Upon shifting to media lacking glucose, both the number and intensity of these stress granule foci increase, as seen in yeast (Buchan et al., 2008). PbpA-GFP is present throughout the cytoplasm under low stress conditions (Fig. 2B), with no clear foci present. Glucose deprivation does not change the intensity or distribution of the GFP signal. No clear colocalization is seen with PbpA and either Dcp1 or FabM. Therefore, under the conditions tested, PbpA does not accumulate in either P-bodies or stress granules upon stress.
Fig. 2.
A. Visualization of P-body and stress granule assembly. Dcp1-RFP serves as a marker for P-bodies, while FabM-GFP depicts stress granule assembly. B. Strains were imaged under normal growth conditions and after 1 hour glucose deprivation. Fluorescently labelled PbpA is present throughout the cytoplasm and does not coalesce into P-bodies or stress granules under the conditions tested. Scale bar represents 20 µm.
3.5 Deletion of pub1, but not pbpA, affects stress granule formation in A. nidulans
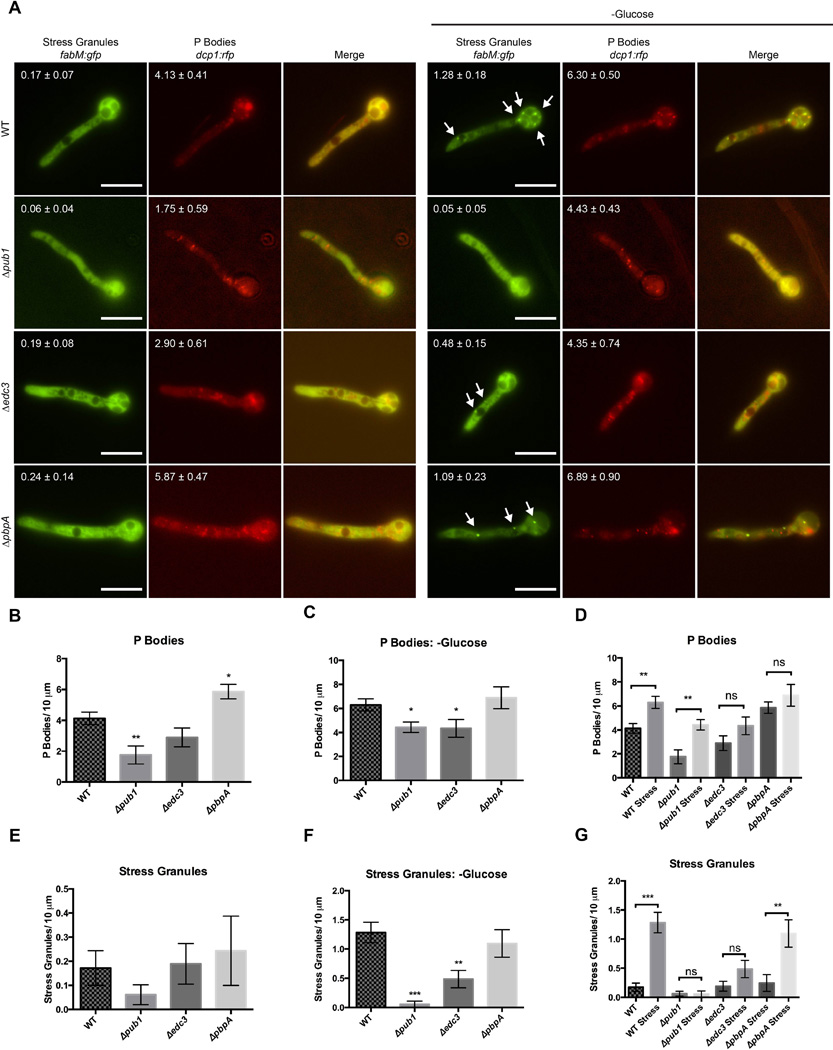
Previous studies in S. cerevisiae have shown that P-bodies help promote formation of stress granules, although mutations that affect stress granule assembly generally do not alter P-body formation (Buchan et al., 2008), although the requirements for assembly components of both types of granules vary amonst different species (Wang et al., 2012). In order to determine the requirements for mRNP particle formation in A. nidulans, we examined the effects of deleting edc3, pub1, and pbpA on stress granule and P-body formation. Edc3 (enhancer of decapping) has been shown to deplete P-body assembly under certain conditions in S. cerevisiae (Decker et al., 2007) and A. nidulans (Morozov et al., 2010b), but not S. pombe (Wang et al., 2013). Pub1 (Poly-uridylate binding protein) binds polyadenylated RNA and colocalizes with FabM in numerous fungi, although its requirement for stress granule and P-body formation varies (Anderson et al., 1993; Buchan et al., 2008; Wang et al., 2012; Zhang et al., 2014). Loss of Pbp1 homologs has been shown to either deplete P-body formation (Buchan et al., 2008), or have no effect (Wang et al., 2013).
Shown in Fig. 2, under normal growth conditions the presence of P-bodies is apparent as indicated by Dcp1-RFP. FabM-GFP localized throughout the cell, with very few foci of high intensity. Upon stress by glucose deprivation, the number of P-bodies approximately doubled, and several FabM containing stress granules formed per germling (Fig. 3). Deletion of pub1 almost completely ablated formation of stress granules, and caused a decrease in P-body formation under both normal and stressed conditions. A decrease in P-body formation was also seen in Δedc3 strains under stress conditions. Deletion of pbpA did not cause a significant decrease in either P-bodies or stress granules under the conditions tested, although a minor increase in P-bodies was seen under normal conditions. Addition of cycloheximide, which traps mRNAs in polysomes due to a translational block, abolishes formation of both P bodies and stress granules (Fig. S7; (Kedersha et al., 2005). These data suggest A. nidulans mRNP granule formation presents both unique and shared characteristics with S. pombe and S. cerevisiae.
Fig. 3.
Effect of various mutants on P-body and stress granule assembly. A. Strains were imaged under normal growth conditions and after 1 hour of glucose deprivation. Values represent average number of foci over a 10 micrometer distance of hyphae +/- standard error (n=10). B. Quantification of Dcp1-RFP foci. Images were subjected to color thresholding (see Material and Methods) to consistently quantify P body foci (Supplemental Figure S4). Edc3 cells contain approximately half the number of P-bodies. C. Quantification of Dcp1-RFP foci after 1 hour without glucose. D. Comparison of number of P-bodies in strains with and without stress. E. Quantification of FabM-GFP foci. F. Quantification of FabM-GFP foci after 1 hour without glucose. G. Comparison of number of stress granules in strains with and without stress. Error bars represent standard error and scale bar represents 20 µm. *p=<0.05 **=p<0.01 ***=p<0.001 ns= not significant. Arrows have been used to identify stress granules which were only identified under glucose starvation.
3.6 Deletion of pbpA greatly reduces meiotic spore production and alters secondary metabolism
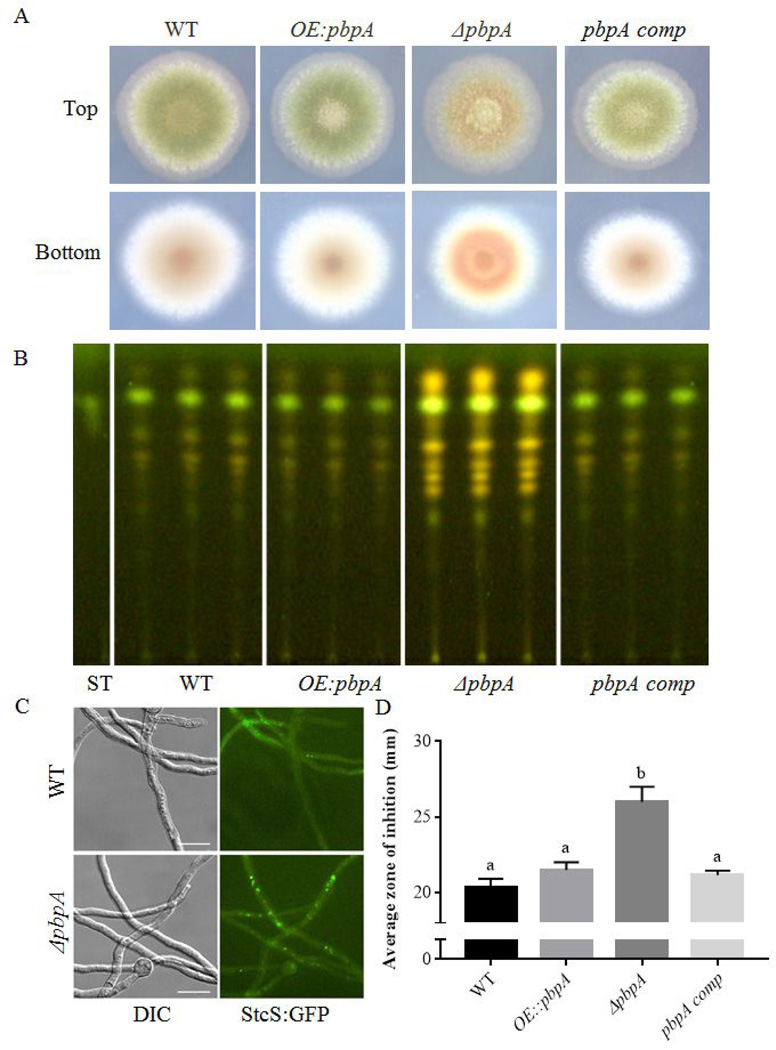
Yeast PBP1 has been shown to contribute to resistance to a number of stresses in S. cerevisiae, including caffeine, cycloheximide, hydroxyurea (Kapitzky et al., 2010), and recovery from ethanol stress (Kato et al., 2011). We examined the effects of both pbpA deletion and overexpression on resistance to a number of different stresses, including sodium chloride, menadione, cycloheximide, sorbitol, and hydroxyurea. Under all conditions tested, no significant differences were seen in relative growth rates among strains (Fig. S8). However, we noted that under normal growth conditions, ΔpbpA showed increased pigmentation (the phenotype from the original suppressor screen) as well as altered sporulation patterns (Fig. 4A), while Δpub1 and Δedc3 did not have any noticeable macroscopic phenotypes (data not shown).
Fig. 4.
Loss of pbpA results in alterations in sexual development and secondary metabolism. A. Phenotypes of strains grown for 3 days at 37°C on GMM. ΔpbpA displays increased pigmentation and altered sporulation patterns. B. pbpA deletion mutants fail to produce ascospores. Phenotypically, no significant differences are seen among among strain under conditions promoting sexual (CHAMPS medium, 5 days growth in dark) or asexual (GMM, 3 days growth in light). C. When spore number is quantified under each condition, ΔpbpA strains display a significant decrease in ascospore number, producing ∼3% WT numbers. D. TLC analysis of organic extracts. Increased intensity of numerous bands, including ST, is seen in the pbpA mutant. E. Localization of StcS-GFP in WT and pbpA mutant backgrounds. F. Penicillin assay quantifying the zone of inhibition produced from culture supernatants against Micrococcus luteus.
We thus compared asexual spore production, sexual spore production and secondary metabolite output of the ΔpbpA mutant to wild type, its complement, and an overexpression strain. Whereas the null strain showed no difference in the production of the asexual spores (conidia), it was greatly impacted in sexual spore production, only producing ∼3% as many ascospores as wild type (Fig. 4B,C).
Sexual development is frequently linked with changes in secondary metabolism (Calvo and Cary, 2015), and thus we next investigated any potential changes in secondary metabolism of the pbpA mutant. TLC analysis of mutant strain extracts revealed that pbpA loss led to increased intensity of numerous bands, including that corresponding to sterigmatocystin (ST) and ST precursors (Fig. 4D). Examination of StcS-GFP (a p450-monooxygenase required for converting the precursor versicolorin A to ST, (Keller et al., 1995) showed an increased intensity of StcS foci in the ΔpbpA background, supporting increased production of sterigmatocystin (Fig. 4D). In order to determine if antibacterial secondary metabolites were impacted by pbpA loss, an assay was performed using culture supernatants against Micrococcus luteus (Fig. 4F). An increased zone of inhibition seen in the samples suggests that there are increased levels of penicillin, or another antibacterial, present in the ΔpbpA supernatant.
4. Discussion
Genetic screens remain powerful tools for identifying genes involved in the feature of interest. The results can be unexpected and provide valuable insight to cellular processes involved in particular genetic traits. In genetic screens for genes affecting secondary metabolism, we have characterized a bZIP transcription factor RsmA, as well as two proteins, EsaA and MvlA, involved in chromatin remodeling in the genetic model Aspergillus nidulans (Bok et al., 2013; Shaaban et al., 2010; Soukup et al., 2012a). Along with altering secondary metabolite synthesis, both RsmA and EsaA also affected sexual spore production, highlighting the close linkage of these two features. Isolated from the same screen as RsmA and EsaA, we now demonstrate that PbpA also is involved in the regulation of both sexual and chemical development in A. nidulans.
Aspergillus nidulans PbpA shares a number of similarities with its S. cerevisiae homolog PBP1, as suggested by its similar domain structure and ability to functionally complement the petite negative phenotype of the pbp1 mutant (Hwang et al., 2007) and restore growth on medium containing ethidium bromide (Fig. 1). Although ΔpbpA was unable to rescue the lethality of a fabM deletion in A. nidulans, unlike rescue of Δpab1 by a Δpbp1 deletion in S. cerevisiae (Mangus et al., 1998), heterokaryon exhibited phenotypic alterations including a decrease in pigmentation (Fig. S3). As pigment production and stress response have previously been linked in a number of fungi (Atanasova et al., 2013; Avalos and Carmen Limón, 2015; Rangel et al., 2006; Yang et al., 2013), this may suggest that fabM+/− ΔpbpA+/− heterokaryons, while unable to completely suppress lethality, undergo a decreased stress response relative to ΔfabM+/− heterokaryons.
Consistent with what has been seen in other species, P-bodies containing Dcp1-RFP are present at low levels under normal growth conditions, and are induced upon stress (Jung and Kim, 2011; Kozubowski et al., 2011), as has previously been shown in A. nidulans (Morozov et al., 2010b). FabM, the Pab1 homolog, is primarily distributed throughout the cytoplasm under normal growth conditions, but also localizes to distinct stress granules upon stress. Unlike the primary stress granule localized near the hyphal tip seen in A. oryzae (Huang et al., 2013), multiple stress granules form and are distributed throughout the cell. Also, in contrast to other fungi, clear co-localization of PbpA and FabM in stress granules is not apparent (Buchan et al., 2008; Jung and Kim, 2011; Park et al., 2016; Wang et al., 2012). PbpA is also present in areas lacking FabM signal, suggesting that PbpA may have functions independent of FabM such as its significant impact on sexual development.
In both S. cerevesiae and C. albicans, deletion of edc3 decreases P-body formation (Buchan et al., 2008; Jung and Kim, 2011). Our studies show a similar phenotype where the edc3 deletion strains show a decreased number of P-bodies present after glucose deprivation (Fig. 3), and a subsequent decrease in stress granules. However, unlike these two other studies, P-body numbers did not change under normal growth conditions in the edc3 mutant. This is similar to what has been seen in S. pombe, where Edc3 was not essential for P-body assembly (Wang et al., 2013). In A. nidulans, an increased number of P-bodies is seen in ammonium rather that nitrate containing medium (Morozov et al., 2010b). These results reflect different requirements for P-body assembly that are both species and condition dependent.
In A. oryzae, deletion of pub1 led to growth defects under stress conditions (Huang et al., 2013), potentially due to a disruption in stress granule formation. Here we show not only a drastic decrease of stress granules in the Δpub1 mutant under stress conditions, but also a decrease in P-body formation under both normal and stress conditions. This decrease in stress granules assembly in the pub1 mutant has also been documented in S. cerevisiae, although not S. pombe. Thus, A. nidulans displays a novel combination of requirements for both P-body and stress granule assembly.
Although studies in yeast suggest that P-body formation is independent of stress granule formation, but not vice versa (Buchan et al., 2008), work on a human cell line suggest that transcripts are first sent to stress granules for sorting, followed by trafficking to P-bodies (Kedersha et al., 2005). The body of work investigating the interrelatedness of these structures is ongoing (Stoecklin and Kedersha, 2013). In our study, mutants affecting either P-body or stress granule formation resulted in defects in the other category. This may reflect response to the particular stress of glucose deprivation, or may be generalized to other forms of stress as well. Under these conditions, deletion of pbpA did not affect formation of either category of mRNP granule, although a minor increase in P-body number was seen under normal growth conditions. This may reflect the particular nature of the response to glucose deprivation, where PbpA is not required. Previous studies in mammalian cells show altered responses of both P-bodies and stress granules to different types of stress (reviewed in (Kedersha and Anderson, 2009), and we propose that PbpA aids in coordinating development in response to unique stresses. As granules can have different activities based on their protein components (Shah et al., 2016), and germlings likely differ in their constitution from more differentiated cell, cellular responses to stress could well vary during the life of the cell.
Deletion of pbpA led to numerous developmental responses in A. nidulans, including a severe reduction in meiotic spores and increased production of several characterized and uncharacterized secondary metabolites. Analysis of the ST pathway by both TLC analysis and imaging of an enzyme required for synthesis showed increased production of this common metabolite in the deletion strain. Increases in additional metabolites or sterigmatocystin intermediates are also seen in the mutant, suggesting an increase in multiple secondary metabolite pathways. An antimicrobial assay also showed increased production of antibacterial activity as seen by inhibition of the bacterium Micrococcus luteus. This enhanced production was partially ablated by the addition of penicillinase suggesting that penicillin synthesis was increased in the mutant (data not shown).
Pbp1 and its homologs have previously been associated with sexual reproduction through regulation of mating type switching in S. cerevisiae (Tadauchi et al., 2004), through post-transcriptional regulation of HO endonuclease mRNA. More recently, Cryptococcus neoformans pbp1 mutants have been shown to be impaired in pheromone production, as well as impaired in sexual development (Park et al., 2016). We confirm this requirement for effective sexual reproduction in A. nidulans. Additionally, this study confirms the integration of sexual development and secondary metabolite production as seen in numerous other mutants isolated from screens identifying regulators of secondary metabolism (Bok et al., 2013; Ramamoorthy et al., 2012; Shaaban et al., 2010; Soukup et al., 2012a).
Secondary metabolites have already been shown to be associated with sexual development as pigments of fruiting body and/or sexual spores (Brown et al., 2012; Schindler and Nowrousian, 2014; Studt et al., 2012; Szewczyk et al., 2008). This coordination of metabolite production and development may be mediated through overlapping transcription factors, as has been seen to be the case in asexual spore development (Lim et al., 2014). Alternatively, trafficking of signaling molecules, precursors, or intermediates in other pathways may all be impacted coordinately. A previous study has highlighted the impact of misregulation of primary metabolism and its impact on sexual development (Palmer et al., 2010), demonstrating pleiotropic effects of metabolism on development.
Although Pbp1 and its homologs have been implicated in a plethora of processes, including regulation of poly-A tail length (Mangus et al., 1998; Mangus et al., 2004), resistance to stress (Kapitzky et al., 2010; Kato et al., 2011; Park et al., 2016), survival without mitochondrial DNA (Dunn and Jensen, 2003), sexual development (Park et al., 2016), and elongation or termination of translation (Tadauchi et al., 2004), little is known about the precise mechanism through which Pbp1 functions. Further studies have shown deletions of pbp1 to suppress defects and or/lethality associated with mutations of other key genes (Kimura and Irie, 2013; Mangus et al., 1998; Woolstencroft et al., 2006). This is particularly interesting in light of the lack of severe phenotypes seen in pbp1 mutants (Kimura and Irie, 2013). Given Pbp1’s role in regulating HO translation (Tadauchi et al., 2004), and pheromone production (Park et al., 2016), it is a formal possibility that PbpA is required for proper expression and/or translation of mating type (MAT) genes. A previous study demonstrated that deletion of the MAT loci ablated ascospore production, while allowing formation of cleistothecia, and not affecting vegetative growth or asexual sporulation, phenotypes also seen in the ΔpbpA strain. Future experiments investigating mRNA levels and localization of key sexual regulators will shed further light upon the mechanism of action coordinating these critical developmental events.
Supplementary Material
Highlights.
AnpbpA encodes a functional homolog of Saccharomyces cerevisiae PBP1
Requirements for mRNA granule synthesis vary among species
AnpbpA is required for normal sexual reproduction and secondary metabolism
Acknowledgments
This work was supported by NIH grant PO1GM084077 from the National Institute of General Medical Sciences to N.P.K., by NIH T32 GM07133 to A.A.S., and by and NIH NRSA AI55397 to A.A.S. and G.J.F.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
7. References
- Altschul SF, et al. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson JT, et al. PUB1 is a major nuclear and cytoplasmic polyadenylated RNA-binding protein in Saccharomyces cerevisiae . Mol Cell Biol. 1993;13:6102–6113. doi: 10.1128/mcb.13.10.6102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atanasova L, et al. The polyketide synthase gene pks4 of Trichoderma reesei provides pigmentation and stress resistance. Eukaryot Cell. 2013;12:1499–1508. doi: 10.1128/EC.00103-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avalos J, Carmen Limón M. Biological roles of fungal carotenoids. Curr Genet. 2015;61:309–324. doi: 10.1007/s00294-014-0454-x. [DOI] [PubMed] [Google Scholar]
- Becht P, et al. The RNA-binding protein Rrm4 is essential for polarity in Ustilago maydis and shuttles along microtubules. J Cell Sci. 2006;119:4964–4973. doi: 10.1242/jcs.03287. [DOI] [PubMed] [Google Scholar]
- Bok JW, Keller NP. LaeA, a regulator of secondary metabolism in Aspergillus spp . Eukaryot Cell. 2004;3:527–535. doi: 10.1128/EC.3.2.527-535.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bok JW, et al. VeA and MvlA repression of the cryptic orsellinic acid gene cluster in Aspergillus nidulans involves histone 3 acetylation. Mol Microbiol. 2013;89:963–974. doi: 10.1111/mmi.12326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown DW, et al. Identification of gene clusters associated with fusaric acid, fusarin, and perithecial pigment production in Fusarium verticillioides . Fungal Genet Biol. 2012;49:521–532. doi: 10.1016/j.fgb.2012.05.010. [DOI] [PubMed] [Google Scholar]
- Buchan JR. mRNP granules. RNA Biol. 2014;11:1019–1030. doi: 10.4161/15476286.2014.972208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchan JR, et al. P bodies promote stress granule assembly in Saccharomyces cerevisiae . J Cell Biol. 2008;183:441–455. doi: 10.1083/jcb.200807043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calvo AM, Cary JW. Association of fungal secondary metabolism and sclerotial biology. Front Microbiol. 2015;6:62. doi: 10.3389/fmicb.2015.00062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chien CT, et al. The two-hybrid system: a method to identify and clone genes for proteins that interact with a protein of interest. Proc Natl Acad Sci U S A. 1991;88:9578–9582. doi: 10.1073/pnas.88.21.9578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Decker CJ, et al. Edc3p and a glutamine/asparagine-rich domain of Lsm4p function in processing body assembly in Saccharomyces cerevisiae . J Cell Biol. 2007;179:437–449. doi: 10.1083/jcb.200704147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunn CD, Jensen RE. Suppression of a defect in mitochondrial protein import identifies cytosolic proteins required for viability of yeast cells lacking mitochondrial DNA. Genetics. 2003;165:35–45. doi: 10.1093/genetics/165.1.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Göhre V, et al. RNA biology in fungal phytopathogens. PLoS Pathog. 2013;9:e1003617. doi: 10.1371/journal.ppat.1003617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoyle NP, et al. Stress-dependent relocalization of translationally primed mRNPs to cytoplasmic granules that are kinetically and spatially distinct from P-bodies. J Cell Biol. 2007;179:65–74. doi: 10.1083/jcb.200707010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang HT, et al. Aspergillus oryzae AoSO is a novel component of stress granules upon heat stress in filamentous fungi. PLoS One. 2013;8:e72209. doi: 10.1371/journal.pone.0072209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang DK, et al. Tim54p connects inner membrane assembly and proteolytic pathways in the mitochondrion. J Cell Biol. 2007;178:1161–1175. doi: 10.1083/jcb.200706195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inglis DO, et al. Comprehensive annotation of secondary metabolite biosynthetic genes and gene clusters of Aspergillus nidulans, A. fumigatus, A. niger and A. oryzae . BMC Microbiol. 2013;13:91. doi: 10.1186/1471-2180-13-91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung JH, Kim J. Accumulation of P-bodies in Candida albicans under different stress and filamentous growth conditions. Fungal Genet Biol. 2011;48:1116–1123. doi: 10.1016/j.fgb.2011.10.003. [DOI] [PubMed] [Google Scholar]
- Kapitzky L, et al. Cross-species chemogenomic profiling reveals evolutionarily conserved drug mode of action. Mol Syst Biol. 2010;6:451. doi: 10.1038/msb.2010.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato K, et al. Severe ethanol stress induces assembly of stress granules in Saccharomyces cerevisiae . Yeast. 2011;28:339–347. doi: 10.1002/yea.1842. [DOI] [PubMed] [Google Scholar]
- Kedersha N, Anderson P. Regulation of translation by stress granules and processing bodies. Prog Mol Biol Transl Sci. 2009;90:155–185. doi: 10.1016/S1877-1173(09)90004-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kedersha N, et al. Stress granules and processing bodies are dynamically linked sites of mRNP remodeling. J Cell Biol. 2005;169:871–884. doi: 10.1083/jcb.200502088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller NP. Translating biosynthetic gene clusters into fungal armor and weaponry. Nat Chem Biol. 2015;11:671–677. doi: 10.1038/nchembio.1897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller NP, et al. stcS, a putative P-450 monooxygenase, is required for the conversion of versicolorin A to sterigmatocystin in Aspergillus nidulans . Appl Environ Microbiol. 1995;61:3628–3632. doi: 10.1128/aem.61.10.3628-3632.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H, et al. The veA gene activates sexual development in Aspergillus nidulans . Fungal Genet Biol. 2002;37:72–80. doi: 10.1016/s1087-1845(02)00029-4. [DOI] [PubMed] [Google Scholar]
- Kimura Y, Irie K. Pbp1 is involved in Ccr4- and Khd1-mediated regulation of cell growth through association with ribosomal proteins Rpl12a and Rpl12b. Eukaryot Cell. 2013;12:864–874. doi: 10.1128/EC.00370-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozubowski L, et al. Calcineurin colocalizes with P-bodies and stress granules during thermal stress in Cryptococcus neoformans . Eukaryot Cell. 2011;10:1396–1402. doi: 10.1128/EC.05087-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim FY, et al. Co-ordination between BrlA regulation and secretion of the oxidoreductase FmqD directs selective accumulation of fumiquinazoline C to conidial tissues in Aspergillus fumigatus . Cell Microbiol. 2014 doi: 10.1111/cmi.12284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu HL, et al. The three fungal transmembrane nuclear pore complex proteins of Aspergillus nidulans are dispensable in the presence of an intact An-Nup84-120 complex. Mol Biol Cell. 2009;20:616–630. doi: 10.1091/mbc.E08-06-0628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mangus DA, et al. Pbp1p, a Factor Interacting with Saccharomyces cerevisiae Poly(A)-Binding Protein, Regulates Polyadenylation. Mol Cell Biol. 1998:7383–7396. doi: 10.1128/mcb.18.12.7383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mangus DA, et al. Identification of factors regulating poly(A) tail synthesis and maturation. Mol Cell Biol. 2004;24:4196–4206. doi: 10.1128/MCB.24.10.4196-4206.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchler-Bauer A, et al. CDD: NCBI’s conserved domain database. Nucleic Acids Res. 2015;43:D222–D226. doi: 10.1093/nar/gku1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marhoul JF, Adams TH. Aspergillus fabM encodes an essential product that is related to poly(A)-binding proteins and activates development when overexpressed. Genetics. 1996;144:1463–1470. doi: 10.1093/genetics/144.4.1463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morozov IY, et al. CUCU modification of mRNA promotes decapping and transcript degradation in Aspergillus nidulans . Mol Cell Biol. 2010a;30:460–469. doi: 10.1128/MCB.00997-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morozov IY, et al. Distinct roles for Caf1, Ccr4, Edc3 and CutA in the co-ordination of transcript deadenylation, decapping and P-body formation in Aspergillus nidulans . Mol Microbiol. 2010b;76:503–516. doi: 10.1111/j.1365-2958.2010.07118.x. [DOI] [PubMed] [Google Scholar]
- Nonhoff U, et al. Ataxin-2 interacts with the DEAD/H-box RNA helicase DDX6 and interferes with P-bodies and stress granules. Mol Biol Cell. 2007;18:1385–1396. doi: 10.1091/mbc.E06-12-1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osmani AH, et al. Identification and analysis of essential Aspergillus nidulans genes using the heterokaryon rescue technique. Nat Protoc. 2006;1:2517–2526. doi: 10.1038/nprot.2006.406. [DOI] [PubMed] [Google Scholar]
- Palmer JM, et al. Telomere position effect is regulated by heterochromatin-associated proteins and NkuA in Aspergillus nidulans . Microbiology. 2010;156:3522–3531. doi: 10.1099/mic.0.039255-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papadopoulos JS, Agarwala R. COBALT: constraint-based alignment tool for multiple protein sequences. Bioinformatics. 2007;23:1073–1079. doi: 10.1093/bioinformatics/btm076. [DOI] [PubMed] [Google Scholar]
- Park HS, et al. Calcineurin Targets Involved in Stress Survival and Fungal Virulence. PLoS Pathog. 2016;12:e1005873. doi: 10.1371/journal.ppat.1005873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker R, Sheth U. P bodies and the control of mRNA translation and degradation. Mol Cell. 2007;25:635–646. doi: 10.1016/j.molcel.2007.02.011. [DOI] [PubMed] [Google Scholar]
- Ralser M, et al. An integrative approach to gain insights into the cellular function of human ataxin-2. J Mol Biol. 2005;346:203–214. doi: 10.1016/j.jmb.2004.11.024. [DOI] [PubMed] [Google Scholar]
- Ramamoorthy V, et al. veA-dependent RNA-pol II transcription elongation factorlike protein, RtfA, is associated with secondary metabolism and morphological development in Aspergillus nidulans . Mol Microbiol. 2012;85:795–814. doi: 10.1111/j.1365-2958.2012.08142.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rangel DE, et al. Mutants and isolates of Metarhizium anisopliae are diverse in their relationships between conidial pigmentation and stress tolerance. J Invertebr Pathol. 2006;93:170–182. doi: 10.1016/j.jip.2006.06.008. [DOI] [PubMed] [Google Scholar]
- Ren A, et al. Hydrogen-rich water regulates effects of ROS balance on morphology, growth and secondary metabolism via glutathione peroxidase in Ganoderma lucidum . Environ Microbiol. 2016 doi: 10.1111/1462-2920.13498. [DOI] [PubMed] [Google Scholar]
- Sachs AB, et al. A single domain of yeast poly(A)-binding protein is necessary and sufficient for RNA binding and cell viability. Mol Cell Biol. 1987;7:3268–3276. doi: 10.1128/mcb.7.9.3268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J, Russell DW. Molecular cloning : a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 2001. [Google Scholar]
- Schindler D, Nowrousian M. The polyketide synthase gene pks4 is essential for sexual development and regulates fruiting body morphology in Sordaria macrospora . Fungal Genet Biol. 2014;68:48–59. doi: 10.1016/j.fgb.2014.04.008. [DOI] [PubMed] [Google Scholar]
- Shaaban MI, et al. Suppressor mutagenesis identifies a velvet complex remediator of Aspergillus nidulans secondary metabolism. Eukaryot Cell. 2010;9:1816–1824. doi: 10.1128/EC.00189-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah KH, et al. A Hybrid-Body Containing Constituents of Both P-Bodies and Stress Granules Forms in Response to Hypoosmotic Stress in Saccharomyces cerevisiae . PLoS One. 2016;11:e0158776. doi: 10.1371/journal.pone.0158776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimizu K, Keller NP. Genetic involvement of a cAMP-dependent protein kinase in a G protein signaling pathway regulating morphological and chemical transitions in Aspergillus nidulans . Genetics. 2001;157:591–600. doi: 10.1093/genetics/157.2.591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sievers F, et al. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol Syst Biol. 2011;7:539. doi: 10.1038/msb.2011.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smedsgaard J. Micro-scale extraction procedure for standardized screening of fungal metabolite production in cultures. J Chromatogr A. 1997;760:264–270. doi: 10.1016/s0021-9673(96)00803-5. [DOI] [PubMed] [Google Scholar]
- Soukup AA, et al. Overexpression of the Aspergillus nidulans histone 4 acetyltransferase EsaA increases activation of secondary metabolite production. Mol Microbiol. 2012a;86:314–330. doi: 10.1111/j.1365-2958.2012.08195.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soukup AA, et al. NosA, a transcription factor important in Aspergillus fumigatus stress and developmental response, rescues the germination defect of a laeA deletion. Fungal Genet Biol. 2012b doi: 10.1016/j.fgb.2012.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevanin G, et al. Clinical and molecular advances in autosomal dominant cerebellar ataxias: from genotype to phenotype and physiopathology. Eur J Hum Genet. 2000;8:4–18. doi: 10.1038/sj.ejhg.5200403. [DOI] [PubMed] [Google Scholar]
- Stoecklin G, Kedersha N. Relationship of GW/P-bodies with stress granules. Adv Exp Med Biol. 2013;768:197–211. doi: 10.1007/978-1-4614-5107-5_12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Studt L, et al. Biosynthesis of fusarubins accounts for pigmentation of Fusarium fujikuroi perithecia. Appl Environ Microbiol. 2012;78:4468–4480. doi: 10.1128/AEM.00823-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szewczyk E, et al. Identification and characterization of the asperthecin gene cluster of Aspergillus nidulans . Appl Environ Microbiol, United States. 2008:7607–7612. doi: 10.1128/AEM.01743-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szewczyk E, et al. Fusion PCR and gene targeting in Aspergillus nidulans. Nat Protoc. 2006;1:3111–3120. doi: 10.1038/nprot.2006.405. [DOI] [PubMed] [Google Scholar]
- Tadauchi T, et al. Posttranscriptional regulation of HO expression by the Mkt1-Pbp1 complex. Mol Cell Biol. 2004;24:3670–3681. doi: 10.1128/MCB.24.9.3670-3681.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tey WK, et al. Polarized gene expression determines woronin body formation at the leading edge of the fungal colony. Mol Biol Cell. 2005;16:2651–2659. doi: 10.1091/mbc.E04-10-0937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang CY, et al. Pdc1 functions in the assembly of P bodies in Schizosaccharomyces pombe . Mol Cell Biol. 2013;33:1244–1253. doi: 10.1128/MCB.01583-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang CY, et al. Analysis of stress granule assembly in Schizosaccharomyces pombe . RNA. 2012;18:694–703. doi: 10.1261/rna.030270.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, et al. The bZIP transcription factor PfZipA regulates secondary metabolism and oxidative stress response in the plant endophytic fungus Pestalotiopsis fici . Fungal Genet Biol. 2015;81:221–228. doi: 10.1016/j.fgb.2015.03.010. [DOI] [PubMed] [Google Scholar]
- Woolstencroft RN, et al. Ccr4 contributes to tolerance of replication stress through control of CRT1 mRNA poly(A) tail length. J Cell Sci. 2006;119:5178–5192. doi: 10.1242/jcs.03221. [DOI] [PubMed] [Google Scholar]
- Yang L, et al. Rapid production of gene replacement constructs and generation of a green fluorescent protein-tagged centromeric marker in Aspergillus nidulans . Eukaryot Cell. 2004;3:1359–1362. doi: 10.1128/EC.3.5.1359-1362.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Q, et al. Involvement of BcVeA and BcVelB in regulating conidiation, pigmentation and virulence in Botrytis cinerea . Fungal Genet Biol. 2013;50:63–71. doi: 10.1016/j.fgb.2012.10.003. [DOI] [PubMed] [Google Scholar]
- Yin WB, et al. An Aspergillus nidulans bZIP response pathway hardwired for defensive secondary metabolism operates through aflR. Mol Microbiol. 2012;83:1024–1034. doi: 10.1111/j.1365-2958.2012.07986.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu JH, et al. Double-joint PCR: a PCR-based molecular tool for gene manipulations in filamentous fungi. Fungal Genet Biol. 2004;41:973–981. doi: 10.1016/j.fgb.2004.08.001. [DOI] [PubMed] [Google Scholar]
- Zhang C, et al. Only a subset of the PAB1-mRNP proteome is present in mRNA translation complexes. Protein Sci. 2014;23:1036–1049. doi: 10.1002/pro.2490. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.