Abstract
Whether respiratory syncytial virus (RSV) induces severe infantile pulmonary disease may depend on viral strain and expression of types I and III interferons (IFNs). These IFNs impact disease severity by inducing expression of many anti-viral IFN-stimulated genes (ISGs). To investigate the impact of RSV strain on IFN and ISG expression, we stimulated human monocyte-derived DCs (MDDCs) with either RSV A2 or Line 19 and measured expression of types I and III IFNs and ISGs. At 24h, A2 elicited higher ISG expression than Line 19. Both strains induced MDDCs to express genes for IFN-β, IFN-α1, IFN-α8, and IFN-λ1–3, but only A2 induced IFN-α2, -α14 and -α21. We then show that IFN-α8 and IFN-α14 most potently induced MDDCs and bronchial epithelial cells (BECs) to express ISGs. Our findings demonstrate that RSV strain may impact patterns of types I and III IFN expression and the magnitude of the ISG response by DCs and BECs.
Keywords: respiratory syncytial virus, interferon subtype, interferon, interferon stimulated gene, dendritic cell, bronchial epithelial cell, innate immunity
Introduction
Respiratory Syncytial Virus (RSV) is the most frequent cause of viral bronchiolitis and pneumonia in infants. In addition to the immediate morbidity and mortality associated with severe RSV lower respiratory tract infection, it may also predispose infants to childhood asthma, and even mild RSV infection exacerbates existing asthma (Lotz et al., 2013). RSV infection occurs via inhalation of virus-containing droplets, which infect bronchial epithelial cells in small foci (Johnson et al., 2007). In the upper airway, dendritic cells reside beneath the epithelium and extend their dendrites towards the surface to sample inhaled antigens. Once DCs detect viral infection, they serve two functions: migration to local lymph nodes to initiate adaptive immunity, and expression of inflammatory mediators, including types I and III interferons (IFNs).
Types I and III IFNs are key inducers of an anti-viral state in response to viral infection. Human type I IFNs include IFN-β and functional genes for 12 subtypes of IFN-α that share 70–80% amino acid identity (Genin et al., 2009). All type I IFNs signal through the IFNAR1/2 receptor complex, which is expressed ubiquitously. Type III IFNs include IFN-λ1, -λ2, -λ3, and the recently described IFN-λ4 (Lazear et al., 2015; O’Brien et al., 2014). IFN-λ2 and IFN-λ3 share 96% amino acid identity, but are divergent from IFN-λ1, and IFN-λ4 shares only approximately 29% identity with other IFN-λs (Lazear et al., 2015; O’Brien et al., 2014). Type III IFNs signal through a receptor dimer comprised of the IFN-λR1 and IL-10R2 chains; expression of IFNLR1 is largely limited to cells of epithelial lineage (Sommereyns et al., 2008). Although they signal through distinct receptors, types I and III interferons share a common signaling pathway that induces autocrine and paracrine expression of more than 600 genes, collectively referred to as interferon-stimulated genes (ISG) (Schoggins et al., 2011). Many ISGs code for proteins that directly inhibit viral replication and protect against spread of viral infection to neighboring cells. The importance of types I and III IFNs during RSV infection is emphasized by the strong inhibition of IFN induction and signaling mediated by the two earliest transcribed of the eleven RSV gene products, NS1 and NS2 (Barik, 2013; Spann et al., 2004).
Human monocyte-derived DCs (MDDCs) are often used as a model for myeloid DCs (mDCs). Similar to mDCs, MDDCs can be infected with RSV in vitro and produce IFNs and pro-inflammatory cytokines (Chi et al., 2006; Guerrero-Plata et al., 2006). Previously we demonstrated that the A2 strain of RSV induces human MDDCs to express types I and III IFNs (Chi et al., 2006), and that these together may affect the adaptive response to RSV. In two studies in mice, immune responses to two commonly used RSV strains were compared (Lukacs et al., 2006; Moore et al., 2009). The authors showed that strains A2 and Line 19 induced high and low expression of IFN-α respectively (Moore et al., 2009). Here, using the same two RSV strains, we asked which types I and III IFNs, ISGs and pro-inflammatory cytokines were induced in human MDDCs in response to RSV and whether the specific types I and III IFNs produced by MDDCs affect the anti-viral ISGs expressed by these cells as well as airway epithelial cells.
Materials and Methods
Viruses
RSV strains A2 (Lewis et al., 1961) and Line 19 (Herlocher et al., 1999) were grown in Vero cells for 6–8 days at an input multiplicity of infection (MOI) of 0.01 PFU/cell using OptiPro serum free medium (Life Technologies, Grand Island, NY) supplemented with 4 mM L-glutamine (Life Technologies). Procedures for isolating RSV from the cells were as described in Le Nouën et al. (Le Nouen et al., 2009) unless otherwise stated. Cells were harvested by scraping, vortexed to liberate surface-associated virus and clarified by centrifugation. The virus was purified by centrifugation through discontinuous sucrose gradients in a Beckman SW32Ti rotor at 121000 x g for 1.5 hours to remove cytokines and other cell derived macromolecules. Virus was harvested from the interface between the sucrose layers, diluted in RPMI 1640 (Life Technologies) supplemented with 2 mM L-glutamine and pelleted at 8,000 x g for 2 hours in a Sorvall SS-34 rotor to remove the sucrose. Pellets were snap frozen in RPMI 1640 and stored at −80°C until use. Viral titer was determined by plaque assay on Vero cells. RSV was UV-inactivated using a dose of 2400 mJ/cm2 with a UVC 500 crosslinker (Hoefer Instruments, Holliston, MA).
Cells
Elutriated monocytes from healthy adult donors were obtained from the NIH Clinical Center Blood Bank and purified using CD14+ MACS beads (Miltenyi Biotech, Auburn, CA). CD14+ cells were differentiated into MDDCs by culturing them at 3x105 cells/ml with IL-4 (1000 U/ml, Peprotech, Rocky Hill, NJ) and GM-CSF (800 U/ml, Immunex, Seattle, WA) for seven days in medium consisting of RPMI 1640 (Life Technologies) with 10% fetal bovine serum (FBS, Hyclone, Logan, UT) and 20 μg/ml gentamicin (Life Technologies). After seven days, MDDCs were stained with monoclonal antibodies against HLA-DR Alexa 680 (clone IV.3, conjugated in house), CD11c PE (clone B-ly6), and CD209 PerCP-Cy 5.5 (clone DCN43, both BD Biosciences, San Jose, CA) to check for purity. Data were acquired using an LSRII flow cytometer (BD Biosciences) and analyzed with Flowjo 9.0 (Treestar Software, Ashland, OR). After seven day of differentiation, cells were >95% CD11c+HLA-DR+.
MDDCs were harvested after seven days, washed three times in medium to remove cytokines and re-cultured at 1x106 cells/ml alone or with live RSV A2 or Line 19 at an MOI of 3, an equal volume of UV-inactivated virus or the indicated concentrations of type I (PBL, Piscataway, NJ) or type III IFNs (R&D Systems, Minneapolis, MN). In some experiments recombinant B18R (the vaccinia virus soluble type I IFN receptor, eBioscience, San Diego, CA) was added at 0.1 μg/ml to block type I IFNs from binding to their receptor. MDDCs were incubated at 37°C for the time points specified in the text and figure legends and then harvested and stained for flow cytometry or lysed using RLT buffer for RNA isolation (Qiagen). Cell culture supernatants were also harvested and stored at −80°C until use. This study was approved by the Institutional Review Boards of the NIH and the U.S. Food and Drug Administration.
The BEAS-2B bronchial epithelial cell line (CRL 9609, ATCC, Manassas, VA) was grown using the BEGM Bullet Kit (Lonza, Walkersville, MD) according to the manufacturer’s instructions and maintained without antibiotics. Cells were seeded in 12-well plates with 20 μg/ml gentamicin and grown until 90% confluence. They were incubated at 37°C with type I or type III IFNs at the concentrations indicated for 24 hours and harvested as described for MDDCs.
Quantitative Real-time PCR
RNA was isolated using RNeasy spin columns including on column DNase digestion (Qiagen, Germantown, MD). RNA was either reverse transcribed using the RT2 first strand kit (SA Biosciences, Frederick, MD) and gene expression detected using toll like receptor (TLR) signaling pathway RT2 profiler plates with SYBR green master mix (SA Biosciences), or using Superscript III Supermix (Life Technologies) followed by gene expression assays (Life Technologies). All qRT-PCR was performed using the 7900HT or Viia 7 system (both Applied Biosystems) and fold change of gene expression was calculated using the ΔΔCq method. Gene expression of the twelve functional type I IFN subtypes and the three type III IFNs was measured individually using the novel qRT-PCR assay developed in the lab, as previously described (Hillyer et al., 2012). Succinate dehydrogenase complex, subunit A (SDHA) and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) were the two most consistent housekeeping genes (HKG) in MDDCs, data not shown and (Hillyer et al., 2012; Mane et al., 2008) GAPDH and ubiquitin C (UBC) were the most consistent in epithelial cells. GAPDH was used to determine comparative levels of expression for the RT2 profiler studies in MDDCs (SDHA was not available in the arrays) and for all PCR experiments on epithelial cells, while SDHA was used for gene expression and IFN assays in MDDCs.
RSV RNA synthesis in MDDCs was measured using a qRT-PCR assay specific for conserved sequences within the M genes of both strains (forward primer: 5′-GCAAATATGGAAACATACGTGAACAA-3′, nt 3,255-80; reverse primer: 5′-GGCACCCATATTGTAAGTGATGCA-3′, nt 3,370-47; probe: 5′FAM-CTTCACGAAGGCTCCACATATAMRA-3′, nt 3,282-301; nucleotide numbering refers to the RSV A2 sequence (Genbank accession number M74568). This method measures both genomic and antigenomic RNA, as well as mRNA expression of the RSV M gene.
Measurement of cytokine proteins
Cytokines were measured using a Milliplex Map human cytokine/chemokine kit (Millipore, Billerica, MA). Data were acquired using Bioplex (Biorad, Hercules, CA) and concentrations determined from a standard curve (Prism 5.0, Graphpad, La Jolla, CA). IFN-α (all subtypes except IFN-α21), IFN-β, and IFN-λ (all three types) were measured by ELISA (PBL Interferon Source, Piscataway, NJ). In all assays, the lower limit of detection was calculated as: blank+ (2 x SD of blank).
Statistics
Differences in gene expression of each IFN type/subtype at single time points (i.e., at 4 hours, 8 hours, and 24 hours) were evaluated using a mixed effects model with the donor as a random effect and the virus (control, live or UV-killed RSV strains A2 or Line 19) as the fixed effect. Pairwise comparisons were made among seven pairs of interest: between control and live A2, control and Line 19, control and UV A2, control and UV Line 19, UV A2 and live A2, UV Line 19 and Line 19, and live A2 and Line 19. No comparisons to the control were performed for RSV-M gene since these cells were uninfected and the gene was unquantifiable. The false discovery rate was controlled at 0.05 to adjust for multiplicity at each time point using the method of (Benjamini and Hochberg, 1995).
Statistical differences in cytokine, chemokine and interferon secretion were determined with Prism 6.0 (Graphpad, La Jolla, CA) using the Friedman test with Dunn’s correction for multiple comparisons.
Results
The A2 RSV strain induces higher expression of innate immune response genes in primary human MDDCs than the Line 19 RSV strain
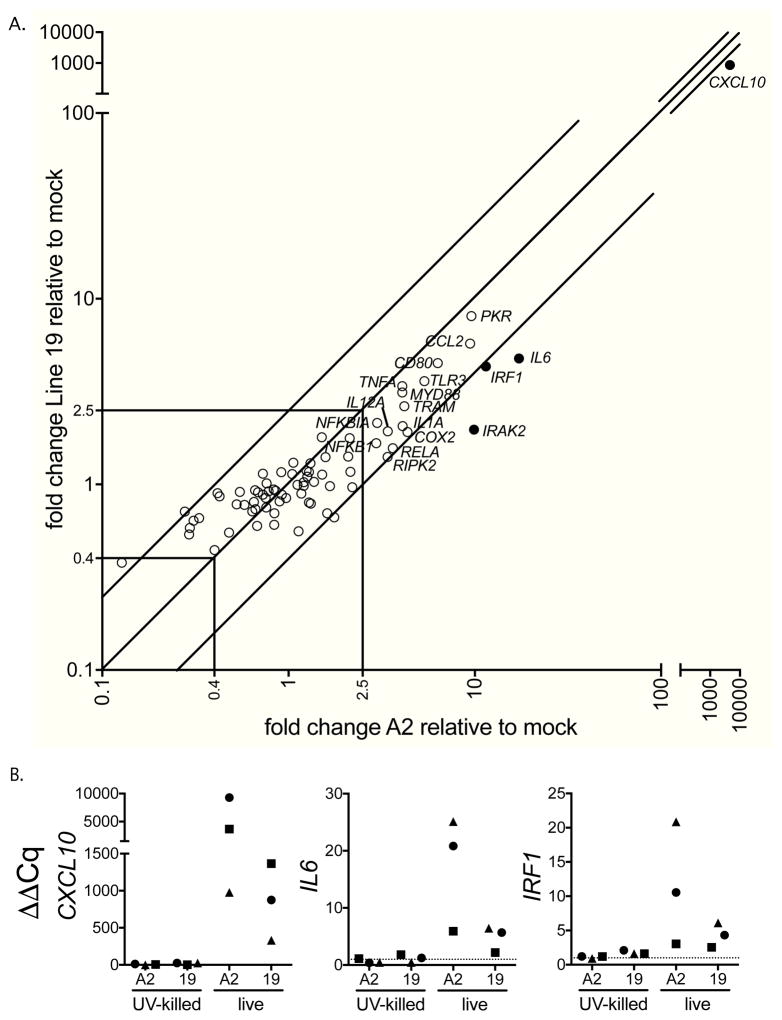
To first determine MDDC expression of innate immune genes in response to RSV infection, we stimulated MDDCs from three donors with live or UV-killed RSV strains A2 or Line 19, or medium (mock) for 24 hours. We then measured expression of a panel of 84 innate immune response genes with a qRT-PCR based array. SDHA was the most consistent housekeeping gene (HKG) in MDDCs(data not shown and (Hillyer et al., 2012; Mane et al., 2008)). However, SDHA was not available in the array kit used for Figure 1, so GAPDH; the next most consistent HKG was used for normalization in this one experiment, SDHA is used for MDDCs in all other figures. Figure 1A shows that live virus of each strain induced expression of a similar set of genes that included DC activation markers, TLR pathway signaling intermediaries, inflammatory mediators, and chemokines. UV killed virus of either strain induced little to no gene expression by MDDCs (Figure 1B). Among genes expressed 2.5-fold above mock in response to RSV, IRF1, IL6, and CXCL10 were consistently expressed at higher levels in response to A2 than Line 19 (Figure 1B) suggesting that A2 induced a greater IFN and pro-inflammatory response in MDDCs than Line 19.
Figure 1. At 24 hours, innate immune gene expression is higher in response to A2 than Line 19.
MDDCs from three donors were infected with either live or UV-inactivated RSV strain A2 or Line 19 (MOI = 3) for 24h. Gene expression was measured by qRT-PCR using a TLR pathway gene array. Data are expressed as mean fold change from mock-infected cells normalized to GAPDH. A. Live A2 and live Line 19 induce expression of similar genes, but live A2 induces higher expression of some TLR pathway genes in MDDCs compared to live Line 19. Each point represents one gene of 84 examined, and shows the mean of the data for the three donors. Genes upregulated > 2.5 fold by RSV infection (either strain) are labeled, diagonal lines represent 2.5 fold change in gene expression between the two strains of RSV examined, specific genes expressed > 2.5 fold higher in response to A2 compared to Line 19 are indicated as filled circles. B. Live virus is required to induce gene expression. Data shown are from the same three donors in response to live A2 or Line 19, or UV-inactivated A2 or Line 19. Each individual donor is represented by a different symbol. The dotted line represents gene expression in mock-infected cells.
To further investigate the responses to the two RSV strains, we performed a more detailed analysis of MDDCs from 18 donors, including the original three. We previously showed that only a small proportion of MDDCs (2 – 15%) are robustly infected with RSV (Chi et al., 2006; Le Nouen et al., 2009) and most of the RSV particles in MDDC cultures are cell-associated rather than in the supernatant (Chi et al., 2006). In addition, RSV binding to the cell surface and/or internalization is required to stimulate pattern recognition receptors on MDDCs. For these reasons, rather than comparing the kinetics of A2 and Line 19 virus particle production, we compared the levels of the virus gene expression by qRT-PCR of the two strains using the RSV matrix (M) gene as a target.
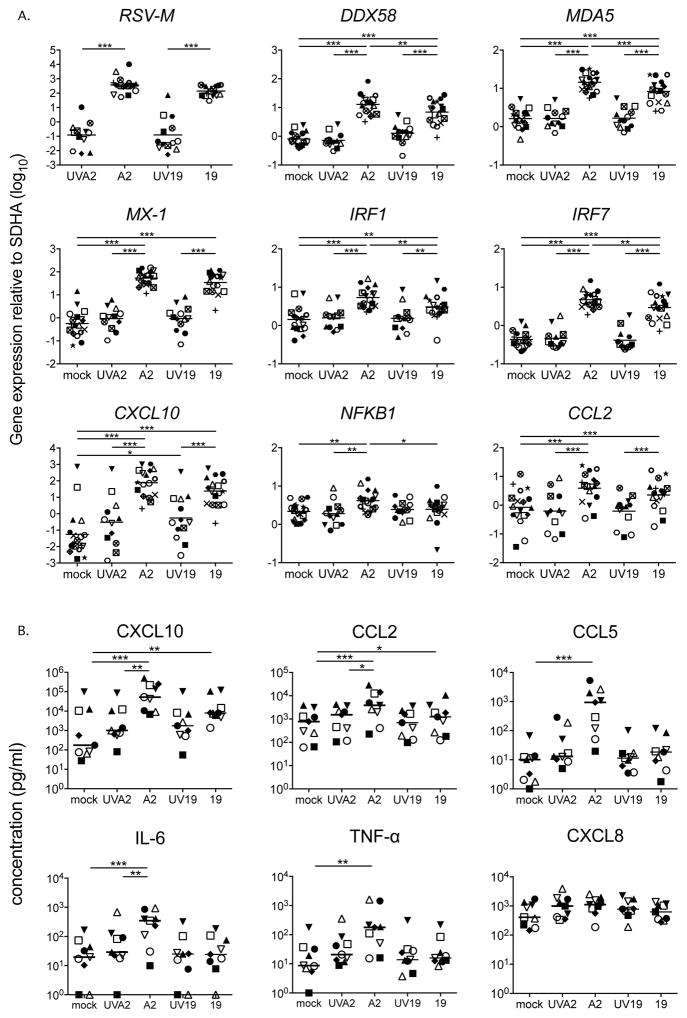
As shown in Figure 2A, at 24 hpi RSV A2-infected MDDCs expressed similar levels of RSV-M to those infected with Line 19. However, despite considerable donor variability, A2 induced statistically higher expression of a set of ISGs (DDX58, MDA-5, IRF1 and IRF7) and NFKB1 (presented in Figure 2A, all on a log10 scale). Analysis of the MDDC supernatants revealed that while CXCL10 and CCL2 protein expression increased significantly above mock in response to RSV A2 or Line 19, expression was generally higher in each donor in response to A2, and that CCL5, IL-6 and TNF-α proteins were significantly induced only in response to infection by the A2 strain (Figure 2B). Although CXCL8 expression was slightly increased in several donors in response to A2, this was not statistically significant. Additionally, RSV A2 induced higher production of CXCL10, CCL2 and IL-6 than did UV-killed A2 virus, indicating that live virus is necessary to induce expression of these pro-inflammatory mediators. Neither of the RSV strains induced expression of IL-10, IFN-γ, or IL-12 p70 (data not shown) or enhanced surface expression of CD80, CD83, or HLA-DR (data not shown).
Figure 2. At 24 hours, expression of ISGs and inflammatory cytokines are higher in A2 compared to Line 19 infected MDDCs.
MDDCs from 18 donors were infected with either live or UV-inactivated RSV A2 or Line 19 (MOI = 3) for 24h. A. Gene expression by MDDCs from all 18 donors (UV-A2 12 donors, UV-Line 19 13 donors) was quantified by real time PCR. Gene expression was calculated relative to the housekeeping gene SDHA and is plotted on a log10 scale. Each shape represents one donor. All p values were calculated using a mixed effects model (see methods for further details). Adjusted p values * p < 0.05, ** p < 0.01, *** p < 0.001. B. Cytokines and chemokines were quantified by multiplex bead assay in 9 of 18 donors. Each shape represents one donor. Horizontal line indicates the median. Statistical differences were determined by Friedman test with Dunn’s multiple comparison test. * p < 0.05, ** p < 0.01, *** p < 0.001.
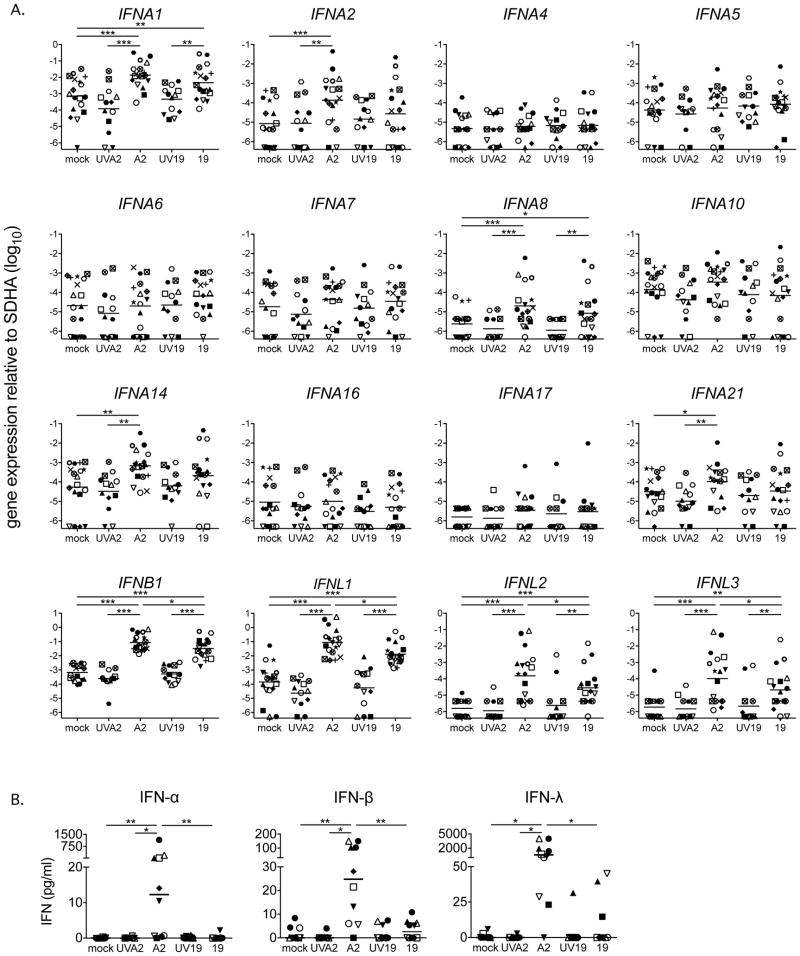
We then measured expression of types I and III IFN mRNA transcripts in all 18 donors and found that both RSV strains induced levels of expression of IFNA1, IFNA8, IFNB1 and IFNL1-3 significantly above the mock-infected controls (Figure 3A). While these IFN genes were more highly expressed in response to A2 than Line 19 in many individual donors, only IFNB1, IFNL1, IFNL2 and IFNL3 were statistically different between the strains (Figure 3A). Only A2 significantly induced expression of genes for three IFN-α subtypes: IFNA2, IFNA14 and IFNA21 (Figure 3A). Line 19 induced expression of these three subtypes above mock levels in some but not all donors, this reflects greater donor to donor variability in the IFN-α response to Line 19. The seven remaining IFN-α subtypes were not induced above mock by either strain of RSV (Figure 3A). We therefore conclude that the IFN-α gene expression patterns we observe may be induced by both RSV strains with greater variability than IFN-β and -λs. We detected IFN-α, IFN-β and IFN-λ protein in MDDC supernatants only in response to A2 (Figure 3B), again with considerable donor variability.
Figure 3. At 24 hours, RSV induces expression of a subset of type I and III IFNs.
A. IFN subtype expression by MDDCs from 18 donors after exposure to A2 or Line 19 was quantified using a qRT-PCR assay that discriminates among each of the twelve unique subtypes of IFN-α and detects IFN-β and the three IFN-λ subtype genes. Expression of IFNA1, IFNA8, IFNB1, IFNL1, IFNL2 and IFNL3 was increased in response to both RSV strains. IFNA2, IFNA14 and IFNA21 were significantly increased in response to A2, and in many donors, in response to Line 19. Gene expression was calculated relative to SDHA and is shown on a log10 scale. Each shape represents one donor. Mock, A2, line 19, n=18, UVA2 and UV19 n=13. Statistical differences were calculated using a mixed effects model (see methods for further details). Adjusted p values * p < 0.05, ** p < 0.01, *** p < 0.001. B. The levels of type I and III IFNs were measured by ELISA. RSV strain A2, but not Line 19 induced IFN-α, -β and -λ expression. Each shape represents one donor. Horizontal line indicates the median. All statistical differences were determined by Friedman test with Dunn’s multiple comparison test. * p < 0.05, ** p < 0.01.
The kinetics of ISG expression differ according to RSV strain
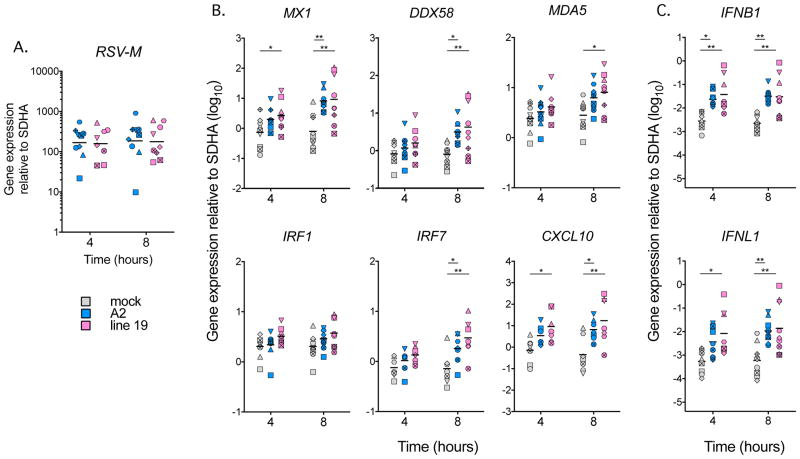
Since the innate response is initiated earlier than 24 hpi, we explored the timing of ISG and IFN induction in response to live A2 or live Line 19. In a subset of nine of the 18 donors used for Figures 2 and 3, we measured responses to RSV at 4 and 8 hpi. Figure 4A and B show that while there were no differences in RSV-M gene expression between the two RSV strains, Line 19 induced earlier expression of a subset of ISGs than A2, and earlier expression of IFNB1 and IFNL1 as well (Figure 4C). Taken together with the 24 hpi data shown in Figures 2 and 3 these analyses show earlier induction of a subset of ISGs and IFNs in response to Line 19, with a later but stronger induction in response to A2.
Figure 4. The kinetics of RSV-M, ISG and IFN expression differ dependent on RSV strain.
MDDCs from nine donors (eight donors at 4 hpi and 24 hpi Line 19) were exposed to live virus of each strain (MOI = 3) for 4, 8 or 24 hours and the RNA harvested. qRT-PCR was used to measure gene expression over time relative to SDHA. Each shape represents a different donor. Data are shown on a log10 scale. A. RSV-M, B. ISG, C. IFNB1 and IFNL1 expression. Statistical differences were calculated using a mixed effects model at 4h and 8h as shown * p < 0.05, ** p < 0.01, *** p < 0.001 (see methods for further details).
IFN-α subtypes differentially induce ISG expression
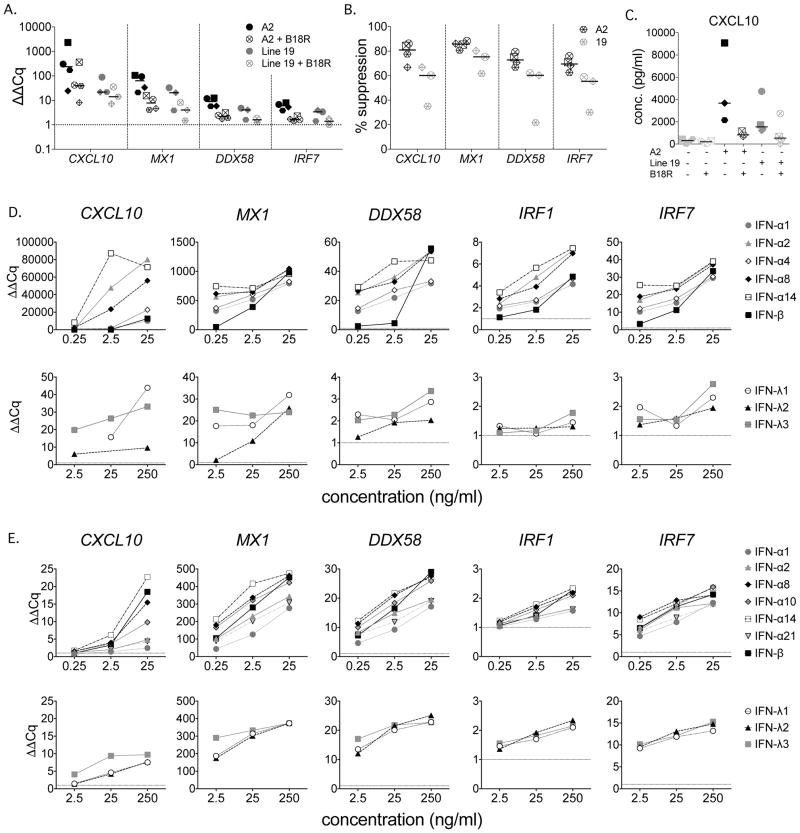
RSV induced expression of both types I and III IFNs, but functional IFNλR is generally restricted to epithelial cell rich tissues (Sommereyns et al., 2008; Witte et al., 2009). We first confirmed the primary importance of type I IFNs by measuring ISG expression at 24 hpi in MDDCs cultured with A2 or Line 19 (MOI = 3) in the presence or absence of recombinant B18R, a soluble type I IFN receptor which is encoded by vaccinia virus. In each of the four donors, B18R suppressed ISG expression (Figure 5A), however this suppression was not complete (Figure 5B) and was generally greater for A2 than Line 19 (e.g. CXCL10, A2 median suppression 81%, range 66.9 – 86.1%, Line 19 median suppression 60.1%, range 35.1 – 66.9%). MX1, DDX58 and IRF7 also had similarly non-overlapping ranges of suppression. The lesser and more variable suppression observed in response to Line 19 was likely due to overall lower expression of ISGs compared to A2 (Figure 5A and B). Similar results were shown for CXCL10 protein expression (Figure 5C) in response to A2 (median suppression 81.8%, range 61.8 – 87.2%) or Line 19 (median suppression 71.0%, range 42.1 – 98.5%). In the four donors tested, B18R did not affect levels of A2 RSV- M (median 104.9%, range 82.8–117.9%), but enhanced levels of Line 19 RSV-M (median 119.5%, range 107.8–142.8%).
Figure 5. Type I IFNs are the major inducers of ISGs by MDDCs.
A – C. MDDCs from four donors were stimulated with live RSV of each strain (MOI = 3) for 24 hours in the presence or absence of B18R, a vaccinia virus encoded soluble type I IFN receptor. Gene expression was measured by qRT-PCR. Data were normalized to SDHA and expressed as ΔΔCq (fold change) compared to unstimulated MDDCs (A), or as percent suppression in the presence of B18R compared to RSV alone (B). A2 n=4, Line 19, n=3. CXCL10 protein expression was quantified by multiplex bead assay (C). RSV A2 n=3, Line 19 n=4. D-E. MDDCs and BEAS-2B cells were stimulated with the indicated levels of each IFN for 24 hours and gene expression measured by qRT-PCR. The dotted horizontal line indicates expression in unstimulated cultures. D. MDDCs, Data were normalized to SDHA and expressed as ΔΔCq (fold change) compared to unstimulated MDDCs. Data shown is one representative donor of four analyzed. E. BEAS-2B cells, data were normalized to GAPDH and expressed as ΔΔCq (fold change) compared to unstimulated BEAS-2B cells. Data shown is one representative experiment of two analyzed.
We then asked whether the IFNs induced by RSV (namely IFN-α1, IFN-α2, IFN-α8, IFN-α14, and IFN-β) differentially induce expression of ISGs in MDDCs by stimulating them for 24 hours with increasing doses of each of these rhIFNs, or IFN-α4 for comparison to a non-RSV responsive IFN. Repetition of these experiments showed that IFN-α2, -α8 and -α14 were consistently the most potent IFNs tested (Figure 5D). Surprisingly, IFN-β, which has the highest affinity for IFNAR1 and IFNAR2 (Schreiber and Piehler, 2015), was generally less potent than these IFNs at low doses. Consistent with the low expression of functional IFN-λR on immune cells (Witte et al., 2009), the type III IFNs were less potent inducers of ISG expression than the type I IFNs (Figure 5D).
We then investigated whether the hierarchy of IFN potency was applicable to bronchial epithelial cells, the primary target for RSV infection. We stimulated BEAS-2B bronchial epithelial cells with a panel of rhIFNs that included IFNα10 and IFNα21 as high and low affinity comparators, respectively. Figure 5E shows that the overall hierarchy of expression of ISGs by BEAS-2B bronchial epithelial cells among the IFN-α subtypes was similar to that in MDDCs, but that comparative potency of IFN-β was higher and more consistent with its known receptor affinity. Consistent with the high expression of membrane bound IFN-λR on lung epithelial cells (Mordstein et al., 2010; Sommereyns et al., 2008), IFNλs, at 10-fold higher concentrations, stimulated the BEAS-2B cells to express most of the ISGs at similar levels as the less potent type I IFNs (Figure 5E).
In summary, using human MDDCs, we demonstrated that 1) RSV induces expression of a subset of IFN subtypes, 2) The type I IFNs expressed in response to RSV are potent inducers of ISGs in both DCs and epithelial cells, 3) ISG expression was higher in response to A2 than line 19 at 24 hpi, 4) Type I and III IFN and ISG expression levels and kinetics differed dependent on donor and RSV strain, and 5) Overall a greater diversity of IFNs was induced in response to RSV A2 compared to Line 19 (Figures 3 and 5).
Discussion
We explored early innate immune responses of human MDDCs to RSV and characterized expression of types I and III IFNs, ISGs and pro-inflammatory cytokines in response to two commonly used RSV strains. We used RSV strains A2 and Line 19 to explore the recent observation that among these two strains, Line 19 induced severe disease in inbred mice which was characterized by lower expression of IFN-α, and higher expression of mucin and IL-13 dependent airway hyperreactivity than the RSV A2-infected mice (Lukacs et al., 2006; Moore et al., 2009). The RSV strains, A2 and Line 19, were isolated from infected infants over 50 years ago. RSV A2 was isolated in 1961 in Australia (Lewis et al., 1961), is well characterized, and has been used extensively in vitro and in animal models. RSV Line 19 was first described in 1999 (Herlocher et al., 1999) as a several decades old clinical isolate at the University of Michigan. However, due to its sequence similarity, Line 19 may have been originally derived from the Long strain, (Melero and Moore, 2013) a 1957 isolate from Baltimore (Chanock et al., 1957). While these two laboratory strains have been serial passaged over many years, phenotypic characterization of recent clinical isolates in mice confirm that RSV A2 is representative of clinical isolates 3–12 and 3–2, which induced little mucin and did not induce IL-13 and that Line 19 is representative of clinical isolates 2–20, 12–35 and 3–4, which induce IL-13, high levels of mucin expression and airway hyper-reactivity in mice (Stokes et al., 2011). This study demonstrates that A2 and Line 19 are at opposite ends of a wide spectrum of host responses to different RSV strains.
Line 19 may elicit early expression of IFN and ISG genes because it is detected by PRRs better or more quickly than A2, infects cells more quickly, replicates more quickly, is less efficient at suppressing IFNs or a combination of these. Moore and colleagues showed that mice infected with a strain A2 virus chimera that expresses the Line 19 F (fusion) protein had higher viral load at day 4 and lower expression of IFN-α at 24 hpi than those infected with A2 with A2F (Moore et al., 2009). This suggests that that the Line 19 F protein permits more rapid infection and viral replication, perhaps associated with inefficient activation or better suppression of IFN-α, which may at later time points lead to a disconnect between viral load and the type I IFN response. The effects of this disconnect appear to be sustained, since despite equal viral titers by Day 6, the A2-Line 19F chimera-infected mice expressed higher levels of IL-13, and had more mucus secretion and airway hyperreactivity than those with A2 F protein. In a second study, these authors showed that higher fusion activity of RSV A2-Line19 mutants correlated with increased early viral load. In addition, two amino acid mutations unique to Line 19F were strongly associated with higher fusion activity (Hotard et al., 2015).
The inverse relationship between IFN-α and IL-13 observed in these reports is consistent with evidence of mutual antagonism between IFN-α and the type 2 response (Essayan et al., 1999; Huber et al., 2010; Schandene et al., 1996). For example, asthmatic children with fewer exacerbations of wheezing expressed the highest levels of IFN-α in response to rhinovirus ex vivo (Teach et al., 2015). In addition, STAT1 deficient mice, which cannot respond to type I, II or III IFNs, have augmented Th2 responses after RSV infection (Durbin et al., 2002). Mechanisms by which IFN-α suppress type 2 responses include suppressed expression of GATA3 (Huber et al., 2010) and the high-affinity IgE receptor (Gill et al., 2010), and suppression of STAT6 signaling by SOCS1, a classical ISG (Fukuyama et al., 2009) (Dickensheets et al., 1999). Conversely, IL-4 or IL-13 may induce expression of SOCS1, which suppresses signaling of STAT1 and STAT2 (Hebenstreit et al., 2005; Hebenstreit et al., 2003). Once T cells are polarized, however, type I IFNs enhance expression of IL-4 (Hillyer et al., 2013), and may therefore adversely amplify type 2 inflammation associated with a viral respiratory infection.
We and others have previously observed that RSV infects MDDCs and myeloid DCs (mDCs) in vitro (Bartz et al., 2003; Chi et al., 2006; de Graaff et al., 2005; Le Nouen et al., 2009). Similar to epithelial cells, only live virus stimulates maturation (Bartz et al., 2003; de Graaff et al., 2005; Le Nouen et al., 2009), expression of inflammatory cytokines and chemokines (Figure 2 and (Chi et al., 2006; Guerrero-Plata et al., 2006; Munir et al., 2008), and types I and III IFNs (Figure 3 and (Bartz et al., 2003; Chi et al., 2006; Munir et al., 2008). Here we show that RSV induces IFNB1, IFNL1-3, and a select subset of IFN-α subtypes that is similar to those expressed by the human U937 cell line in response to Sendai virus (Zaritsky et al., 2015).
In vivo, mDCs reside adjacent to the respiratory epithelium (Holt, 2005) where they respond to infection by expressing pro-inflammatory cytokines and types I and III IFNs before migrating to local lymph nodes to initiate an adaptive response. Types I and III IFNs canonically signal through STAT1/STAT2/IRF9 and induce transcription of a similar pattern of ISGs (Bolen et al., 2014; Jilg et al., 2014). Recent evidence, however, supports non-redundant functional roles for each, particularly at sub-saturating levels, which are likely more relevant to initial exposure of the respiratory mucosa to small doses of a viral pathogen contained in micro-droplets. For example, sub-saturating combinations of IFN-β and IFN-λ1 synergize towards suppression of vesicular stomatitis virus in vitro (Voigt and Yin, 2015), and additively enhance and sustain expression of many ISGs (Novatt et al., 2016). In RSV infection of MDDCs, using B18R to block type I IFNs (Figure 5), we have shown that the majority of ISG induction relies on the presence of type I IFNs, however this does not exclude the possibility that optimal ISG expression requires -α, -β and -λ IFNs in combination. In nasopharyngeal wash samples from infants hospitalized for RSV associated bronchiolitis, IFN-λ expression correlates positively both with anti-viral ISG expression and with the clinical score index of disease severity, but not with viral load. It should be noted however, that type I IFNs were not measured in this study, so it is unclear whether IFN-λ or another IFN was responsible for ISG expression (Selvaggi et al., 2014).
One impediment towards dissecting individual roles for IFN-α subtypes is that they are often expressed in groups rather than individually. Recently, Zaritsky and colleagues reported that after infection with Sendai virus, the U937 human monocytic cell line expresses multiple subsets of the 12 IFN-α, depending on the infectious dose used and autocrine/paracrine IFN stimulation through IFNAR1/2 (Zaritsky et al., 2015). One subset was IFNAR-independent and was comprised of IFN-α1, -2, and -α8 (three of the five IFN-α subtypes we observed), and another subset was IFNAR-dependent and included IFN-α14 and -α21 (the fourth and fifth IFN-α subtypes we observed). In this context, our comparison of RSV A2 and Line 19 infection of MDDCs suggests that at biologically relevant infectious doses of virus, the later waves of IFN-α subtypes may be a response to the early IFN-β autocrine/paracrine stimulation and ongoing viral replication.
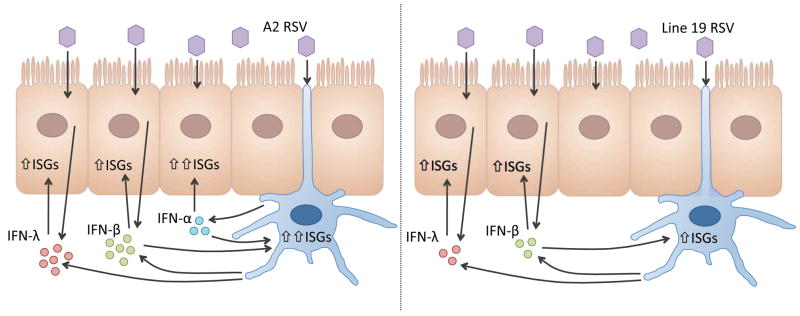
Finally, we observed that expression of a set of anti-viral ISGs was most potently induced by IFN-α2, -α8, and -α14 in MDDCs, and by IFN-α8, -α14 and IFN-β in respiratory epithelial cells. As reported by Lavoie et al, this hierarchy of potency is not necessarily consistent with the known affinities of the IFN-α subtypes for IFNAR1 or IFNAR2 (Lavoie et al., 2011). Instead, relative anti-viral potency varies according to cell type and pathogen. In addition, since respiratory epithelial cells do not express IFN-α in response to RSV (Okabayashi et al., 2011; Spann et al., 2004), it is likely that their expression by mDC enhance the local anti-viral state. As modeled in Figure 6, the greater consistency by which these IFN-α subtypes are induced in response to A2 strain may explain why similar clinical strains are associated with a milder disease phenotype than those represented by Line 19.
Figure 6. Strain dependent IFN expression by DCs.
RSV A2 (left panel) induces DCs to express high levels of IFN-β and IFN-λ1, and to express three IFN-α subtypes: IFN-α2, -α8, and -α14. These IFNs induce high expression of ISGs by DCs with a generalized hierarchy of IFN-α14 > -α2 > -α8 > -β and by bronchial epithelial cells with a hierarchy of IFN-α14 > -α8 > -β > -λ3 > -λ1, -λ2 and -α2. In contrast, RSV line 19 induces lower expression of IFN-β and −λ1, and IFN-α1, which may not be optimal for inducing an antiviral response in the epithelial cells, or for the DCs to mediate an appropriate adaptive response.
Highlights.
RSV induces expression of a subset of IFN subtypes in human DCs
The type I IFNs expressed are potent inducers of ISGs in DCs and BEAS-2B cells
ISG and IFN expression levels and kinetics differed dependent on RSV strain
Acknowledgments
Funding Information
This work was supported by NIH intramural funds, FDA/CBER intramural funds and the FDA Medical Countermeasures Initiative. PH, VPM, MBS, LMS, RES were supported by appointment to the Research Participation Program of the Center for Biologics Evaluation and Research administered by the Oak Ridge Institute for Science and Education through an interagency agreement between the U.S. Department of Energy and the U.S. FDA.
The authors would like to thank Manuel Osorio for assistance with the multiplex bead assays, Steven Carbone and Matthew Carroll at PBL for performing the IFN ELISAs on our culture supernatants and Ray Donnelly for careful review of the manuscript.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Barik S. Respiratory syncytial virus mechanisms to interfere with type 1 interferons. Current topics in microbiology and immunology. 2013;372:173–191. doi: 10.1007/978-3-642-38919-1_9. [DOI] [PubMed] [Google Scholar]
- Bartz H, Turkel O, Hoffjan S, Rothoeft T, Gonschorek A, Schauer U. Respiratory syncytial virus decreases the capacity of myeloid dendritic cells to induce interferon-gamma in naive T cells. Immunology. 2003;109:49–57. doi: 10.1046/j.1365-2567.2003.01629.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. Journal of the royal statistical society. Series B (Methodological) 1995:289–300. [Google Scholar]
- Bolen CR, Ding S, Robek MD, Kleinstein SH. Dynamic expression profiling of type I and type III interferon-stimulated hepatocytes reveals a stable hierarchy of gene expression. Hepatology. 2014;59:1262–1272. doi: 10.1002/hep.26657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chanock R, Roizman B, Myers R. Recovery from infants with respiratory illness of a virus related to chimpanzee coryza agent (CCA). I. Isolation, properties and characterization. Am J Hyg. 1957;66:281–290. doi: 10.1093/oxfordjournals.aje.a119901. [DOI] [PubMed] [Google Scholar]
- Chi B, Dickensheets HL, Spann KM, Alston MA, Luongo C, Dumoutier L, Huang J, Renauld JC, Kotenko SV, Roederer M, Beeler JA, Donnelly RP, Collins PL, Rabin RL. Alpha and lambda interferon together mediate suppression of CD4 T cells induced by respiratory syncytial virus. J Virol. 2006;80:5032–5040. doi: 10.1128/JVI.80.10.5032-5040.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Graaff PM, de Jong EC, van Capel TM, van Dijk ME, Roholl PJ, Boes J, Luytjes W, Kimpen JL, van Bleek GM. Respiratory syncytial virus infection of monocyte-derived dendritic cells decreases their capacity to activate CD4 T cells. J Immunol. 2005;175:5904–5911. doi: 10.4049/jimmunol.175.9.5904. [DOI] [PubMed] [Google Scholar]
- Dickensheets HL, Venkataraman C, Schindler U, Donnelly RP. Interferons inhibit activation of STAT6 by interleukin 4 in human monocytes by inducing SOCS-1 gene expression. Proc Natl Acad Sci U S A. 1999;96:10800–10805. doi: 10.1073/pnas.96.19.10800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durbin JE, Johnson TR, Durbin RK, Mertz SE, Morotti RA, Peebles RS, Graham BS. The role of IFN in respiratory syncytial virus pathogenesis. J Immunol. 2002;168:2944–2952. doi: 10.4049/jimmunol.168.6.2944. [DOI] [PubMed] [Google Scholar]
- Essayan DM, Krishnaswamy G, Oriente A, Lichtenstein LM, Huang SK. Differential regulation of antigen-induced IL-4 and IL-13 generation from T lymphocytes by IFN-alpha. J Allergy Clin Immunol. 1999;103:451–457. doi: 10.1016/s0091-6749(99)70470-7. [DOI] [PubMed] [Google Scholar]
- Fukuyama S, Nakano T, Matsumoto T, Oliver BG, Burgess JK, Moriwaki A, Tanaka K, Kubo M, Hoshino T, Tanaka H, McKenzie AN, Matsumoto K, Aizawa H, Nakanishi Y, Yoshimura A, Black JL, Inoue H. Pulmonary suppressor of cytokine signaling-1 induced by IL-13 regulates allergic asthma phenotype. Am J Respir Crit Care Med. 2009;179:992–998. doi: 10.1164/rccm.200806-992OC. [DOI] [PubMed] [Google Scholar]
- Genin P, Vaccaro A, Civas A. The role of differential expression of human interferon--a genes in antiviral immunity. Cytokine Growth Factor Rev. 2009;20:283–295. doi: 10.1016/j.cytogfr.2009.07.005. [DOI] [PubMed] [Google Scholar]
- Gill MA, Bajwa G, George TA, Dong CC, Dougherty II, Jiang N, Gan VN, Gruchalla RS. Counterregulation between the FcepsilonRI pathway and antiviral responses in human plasmacytoid dendritic cells. J Immunol. 2010;184:5999–6006. doi: 10.4049/jimmunol.0901194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guerrero-Plata A, Casola A, Suarez G, Yu X, Spetch L, Peeples ME, Garofalo RP. Differential response of dendritic cells to human metapneumovirus and respiratory syncytial virus. Am J Respir Cell Mol Biol. 2006;34:320–329. doi: 10.1165/rcmb.2005-0287OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hebenstreit D, Luft P, Schmiedlechner A, Duschl A, Horejs-Hoeck J. SOCS-1 and SOCS-3 inhibit IL-4 and IL-13 induced activation of Eotaxin-3/CCL26 gene expression in HEK293 cells. Mol Immunol. 2005;42:295–303. doi: 10.1016/j.molimm.2004.09.004. [DOI] [PubMed] [Google Scholar]
- Hebenstreit D, Luft P, Schmiedlechner A, Regl G, Frischauf AM, Aberger F, Duschl A, Horejs-Hoeck J. IL-4 and IL-13 induce SOCS-1 gene expression in A549 cells by three functional STAT6-binding motifs located upstream of the transcription initiation site. J Immunol. 2003;171:5901–5907. doi: 10.4049/jimmunol.171.11.5901. [DOI] [PubMed] [Google Scholar]
- Herlocher ML, Ewasyshyn M, Sambhara S, Gharaee-Kermani M, Cho D, Lai J, Klein M, Maassab HF. Immunological properties of plaque purified strains of live attenuated respiratory syncytial virus (RSV) for human vaccine. Vaccine. 1999;17:172–181. doi: 10.1016/s0264-410x(98)00155-8. [DOI] [PubMed] [Google Scholar]
- Hillyer P, Mane VP, Schramm LM, Puig M, Verthelyi D, Chen A, Zhao Z, Navarro MB, Kirschman KD, Bykadi S, Jubin RG, Rabin RL. Expression profiles of human interferon-alpha and interferon-lambda subtypes are ligand- and cell-dependent. Immunol Cell Biol. 2012;90:774–783. doi: 10.1038/icb.2011.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hillyer P, Raviv N, Gold DM, Dougherty D, Liu J, Johnson TR, Graham BS, Rabin RL. Subtypes of type I IFN differentially enhance cytokine expression by suboptimally stimulated CD4 T cells. European journal of immunology. 2013 doi: 10.1002/eji.201243288. [DOI] [PubMed] [Google Scholar]
- Holt PG. Pulmonary dendritic cells in local immunity to inert and pathogenic antigens in the respiratory tract. Proc Am Thorac Soc. 2005;2:116–120. doi: 10.1513/pats.200502-017AW. [DOI] [PubMed] [Google Scholar]
- Hotard AL, Lee S, Currier MG, Crowe JE, Jr, Sakamoto K, Newcomb DC, Peebles RS, Jr, Plemper RK, Moore ML. Identification of residues in the human respiratory syncytial virus fusion protein that modulate fusion activity and pathogenesis. J Virol. 2015;89:512–522. doi: 10.1128/JVI.02472-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huber JP, Ramos HJ, Gill MA, Farrar JD. Cutting edge: Type I IFN reverses human Th2 commitment and stability by suppressing GATA3. J Immunol. 2010;185:813–817. doi: 10.4049/jimmunol.1000469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jilg N, Lin W, Hong J, Schaefer EA, Wolski D, Meixong J, Goto K, Brisac C, Chusri P, Fusco DN, Chevaliez S, Luther J, Kumthip K, Urban TJ, Peng LF, Lauer GM, Chung RT. Kinetic differences in the induction of interferon stimulated genes by interferon-alpha and interleukin 28B are altered by infection with hepatitis C virus. Hepatology. 2014;59:1250–1261. doi: 10.1002/hep.26653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson JE, Gonzales RA, Olson SJ, Wright PF, Graham BS. The histopathology of fatal untreated human respiratory syncytial virus infection. Mod Pathol. 2007;20:108–119. doi: 10.1038/modpathol.3800725. [DOI] [PubMed] [Google Scholar]
- Lavoie TB, Kalie E, Crisafulli-Cabatu S, Abramovich R, DiGioia G, Moolchan K, Pestka S, Schreiber G. Binding and activity of all human alpha interferon subtypes. Cytokine. 2011;56:282–289. doi: 10.1016/j.cyto.2011.07.019. [DOI] [PubMed] [Google Scholar]
- Lazear HM, Nice TJ, Diamond MS. Interferon-lambda: Immune Functions at Barrier Surfaces and Beyond. Immunity. 2015;43:15–28. doi: 10.1016/j.immuni.2015.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Nouen C, Munir S, Losq S, Winter CC, McCarty T, Stephany DA, Holmes KL, Bukreyev A, Rabin RL, Collins PL, Buchholz UJ. Infection and maturation of monocyte-derived human dendritic cells by human respiratory syncytial virus, human metapneumovirus, and human parainfluenza virus type 3. Virology. 2009;385:169–182. doi: 10.1016/j.virol.2008.11.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis FA, Rae ML, Lehmann NI, Ferris AA. A syncytial virus associated with epidemic disease of the lower respiratory tract in infants and young children. Med J Aust. 1961;2:932. [Google Scholar]
- Lotz MT, Moore ML, Peebles RS., Jr Respiratory syncytial virus and reactive airway disease. Current topics in microbiology and immunology. 2013;372:105–118. doi: 10.1007/978-3-642-38919-1_5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukacs NW, Moore ML, Rudd BD, Berlin AA, Collins RD, Olson SJ, Ho SB, Peebles RS., Jr Differential immune responses and pulmonary pathophysiology are induced by two different strains of respiratory syncytial virus. Am J Pathol. 2006;169:977–986. doi: 10.2353/ajpath.2006.051055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mane VP, Heuer MA, Hillyer P, Navarro MB, Rabin RL. Systematic method for determining an ideal housekeeping gene for real-time PCR analysis. J Biomol Tech. 2008;19:342–347. [PMC free article] [PubMed] [Google Scholar]
- Melero JA, Moore ML. Influence of respiratory syncytial virus strain differences on pathogenesis and immunity. Current topics in microbiology and immunology. 2013;372:59–82. doi: 10.1007/978-3-642-38919-1_3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore ML, Chi MH, Luongo C, Lukacs NW, Polosukhin VV, Huckabee MM, Newcomb DC, Buchholz UJ, Crowe JE, Jr, Goleniewska K, Williams JV, Collins PL, Peebles RS., Jr A chimeric A2 strain of respiratory syncytial virus (RSV) with the fusion protein of RSV strain line 19 exhibits enhanced viral load, mucus, and airway dysfunction. J Virol. 2009;83:4185–4194. doi: 10.1128/JVI.01853-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mordstein M, Neugebauer E, Ditt V, Jessen B, Rieger T, Falcone V, Sorgeloos F, Ehl S, Mayer D, Kochs G, Schwemmle M, Gunther S, Drosten C, Michiels T, Staeheli P. Lambda interferon renders epithelial cells of the respiratory and gastrointestinal tracts resistant to viral infections. J Virol. 2010;84:5670–5677. doi: 10.1128/JVI.00272-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munir S, Le Nouen C, Luongo C, Buchholz UJ, Collins PL, Bukreyev A. Nonstructural proteins 1 and 2 of respiratory syncytial virus suppress maturation of human dendritic cells. J Virol. 2008;82:8780–8796. doi: 10.1128/JVI.00630-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novatt H, Theisen TC, Massie T, Massie T, Simonyan V, Voskanian-Kordi A, Renn LA, Rabin RL, Teach SJ, Gill MA, Togias A, Sorkness CA, Arbes SJ, Jr, Calatroni A, Wildfire JJ, Gergen PJ, Cohen RT, Pongracic JA, Kercsmar CM, Khurana Hershey GK, Gruchalla RS, Liu AH, Zoratti EM, Kattan M, Grindle KA, Gern JE, Busse WW, Szefler SJ, Lavoie TB, Kalie E, Crisafulli-Cabatu S, Abramovich R, DiGioia G, Moolchan K, Pestka S, Schreiber G, Moll HP, Maier T, Zommer A, Lavoie T, Brostjan C. Distinct Patterns of Expression of Transcription Factors in Response to Interferonbeta and Interferonlambda1 Preseasonal treatment with either omalizumab or an inhaled corticosteroid boost to prevent fall asthma exacerbations Binding and activity of all human alpha interferon subtypes The differential activity of interferon-alpha subtypes is consistent among distinct target genes and cell types. J Interferon Cytokine Res. 2016;36:589–598. doi: 10.1089/jir.2016.0031. [DOI] [PubMed] [Google Scholar]
- O’Brien TR, Prokunina-Olsson L, Donnelly RP. IFN-lambda4: the paradoxical new member of the interferon lambda family. J Interferon Cytokine Res. 2014;34:829–838. doi: 10.1089/jir.2013.0136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schandene L, Del Prete GF, Cogan E, Stordeur P, Crusiaux A, Kennes B, Romagnani S, Goldman M. Recombinant interferon-alpha selectively inhibits the production of interleukin-5 by human CD4+ T cells. J Clin Invest. 1996;97:309–315. doi: 10.1172/JCI118417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoggins JW, Wilson SJ, Panis M, Murphy MY, Jones CT, Bieniasz P, Rice CM. A diverse range of gene products are effectors of the type I interferon antiviral response. Nature. 2011;472:481–485. doi: 10.1038/nature09907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schreiber G, Piehler J. The molecular basis for functional plasticity in type I interferon signaling. Trends Immunol. 2015;36:139–149. doi: 10.1016/j.it.2015.01.002. [DOI] [PubMed] [Google Scholar]
- Selvaggi C, Pierangeli A, Fabiani M, Spano L, Nicolai A, Papoff P, Moretti C, Midulla F, Antonelli G, Scagnolari C. Interferon lambda 1–3 expression in infants hospitalized for RSV or HRV associated bronchiolitis. J Infect. 2014;68:467–477. doi: 10.1016/j.jinf.2013.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sommereyns C, Paul S, Staeheli P, Michiels T. IFN-lambda (IFN-lambda) is expressed in a tissue-dependent fashion and primarily acts on epithelial cells in vivo. PLoS Pathog. 2008;4:e1000017. doi: 10.1371/journal.ppat.1000017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spann KM, Tran KC, Chi B, Rabin RL, Collins PL. Suppression of the induction of alpha, beta, and lambda interferons by the NS1 and NS2 proteins of human respiratory syncytial virus in human epithelial cells and macrophages [corrected] J Virol. 2004;78:4363–4369. doi: 10.1128/JVI.78.8.4363-4369.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stokes KL, Chi MH, Sakamoto K, Newcomb DC, Currier MG, Huckabee MM, Lee S, Goleniewska K, Pretto C, Williams JV, Hotard A, Sherrill TP, Peebles RS, Jr, Moore ML. Differential pathogenesis of respiratory syncytial virus clinical isolates in BALB/c mice. J Virol. 2011;85:5782–5793. doi: 10.1128/JVI.01693-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teach SJ, Gill MA, Togias A, Sorkness CA, Arbes SJ, Jr, Calatroni A, Wildfire JJ, Gergen PJ, Cohen RT, Pongracic JA, Kercsmar CM, Khurana Hershey GK, Gruchalla RS, Liu AH, Zoratti EM, Kattan M, Grindle KA, Gern JE, Busse WW, Szefler SJ. Preseasonal treatment with either omalizumab or an inhaled corticosteroid boost to prevent fall asthma exacerbations. J Allergy Clin Immunol. 2015;136:1476–1485. doi: 10.1016/j.jaci.2015.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voigt EA, Yin J. Kinetic Differences and Synergistic Antiviral Effects Between Type I and Type III Interferon Signaling Indicate Pathway Independence. J Interferon Cytokine Res. 2015 doi: 10.1089/jir.2015.0008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witte K, Gruetz G, Volk HD, Looman AC, Asadullah K, Sterry W, Sabat R, Wolk K. Despite IFN-lambda receptor expression, blood immune cells, but not keratinocytes or melanocytes, have an impaired response to type III interferons: implications for therapeutic applications of these cytokines. Genes Immun. 2009;10:702–714. doi: 10.1038/gene.2009.72. [DOI] [PubMed] [Google Scholar]
- Zaritsky LA, Bedsaul JR, Zoon KC. Virus Multiplicity of Infection Affects Type I Interferon Subtype Induction Profiles and Interferon-Stimulated Genes. J Virol. 2015;89:11534–11548. doi: 10.1128/JVI.01727-15. [DOI] [PMC free article] [PubMed] [Google Scholar]