Abstract
The metabolome describes the full complement of the tens to hundreds of thousands of low molecular weight metabolites present within a biological system. Identification of the metabolome is critical for discovering the maximum amount of biochemical knowledge from metabolomics datasets. Yet no exhaustive experimental characterisation of any organismal metabolome has been reported to date, dramatically contrasting with the genome sequencing of thousands of plants, animals and microbes. Here we review the status of metabolome annotation and describe advances in the analytical methodologies being applied. In part through new international coordination, we conclude that we are now entering a new era of metabolome annotation.
Introduction
Metabolomics is the multidisciplinary field of research concerned with the study of metabolomes, the complement of naturally-occurring and exogenous (e.g. environmental pollutants), low-molecular-weight (typically <1500 Da) metabolites present within biological systems [1]. Comprising of precursors, intermediates and products of biochemical pathways, metabolites constitute some of the terminal products of higher cellular processes and collectively provide a ‘fingerprint’ of the complex interplay between genome and environment. From an analytical perspective, both the measurement and identification of whole metabolomes presents a considerable challenge, not least due to the vast structural heterogeneity of metabolites, their large number (e.g. an estimated 200,000 structurally-distinct secondary metabolites across the plant kingdom [2]) and their wide concentration ranges (estimated to span 12 orders of magnitude [3]). As a point of clarity, the formal definitions of metabolite annotation and identification, as developed by the Chemical Analysis Working Group of the Metabolomics Standards Initiative (MSI) [4], are shown in Table 1. The categorical scoring system defines ‘identification’ as the most rigorous (level 1) while ‘annotation’ does not require such exhaustive analytical validation (levels 2 and 3). Currently there are no completed lists of experimentally-derived metabolites that describe the metabolome of any model organism, not even as putative annotations.
Table 1.
Summary of levels of confidence in metabolite ‘identification’, as defined by the Chemical Analysis Working Group of the Metabolomics Standards Initiative [4].
| Level of confidence |
Description | Requisite analytical data |
|---|---|---|
| Level 1 | ‘Identified metabolites’ |
Two orthogonal analytical techniques applied to the analysis of both the metabolite of interest and to a chemical reference standard of suspected structural equivalence, with all analyses performed under identical analytical conditions within the same laboratory. Examples of appropriate orthogonal data: accurate mass via MS with retention time; accurate mass MS and fragmentation data or isotopic pattern; 2D NMR spectra; full 1H and/or 13C NMR spectra etc. |
| Level 2 | ‘Putatively annotated compounds’ |
As for levels 3 and 4, including spectral (NMR and/or MS) similarity with public or commercial libraries. |
| Level 3 | ‘Putatively characterised compound classes’ |
As for level 4, plus spectral and/or physicochemical properties consistent with a particular class of organic compounds. |
| Level 4 | ‘Unknown’ | A discernible spectral signal (NMR, MS or other) that can be reproducibly detected and quantified. |
Meaningful biological inferences may only be drawn from metabolomics datasets where peaks can be structurally identified as named metabolites, i.e. it is only when we are empowered to move beyond discussing unidentified peaks to rigorously identified metabolites that we can fully engage in describing metabolic pathways and integrate metabolism with other levels of biological hierarchy. For over a decade, molecular identification has remained the principal technical bottleneck in metabolomics [5, 6]. Hence, for metabolomics to deliver its full potential in fields from medicine to ecology, innovations in analytical workflows are urgently required. Yet based on the literature, it is readily apparent that the core metabolomics workflow has changed little over the past 15 years, typically comprising of sampling, measurement of metabolites by mass spectrometry (MS) and/or nuclear magnetic resonance (NMR) spectroscopy, data processing and statistical analyses, with a view to discovering peaks of biological importance [7, 8]. Those peaks are typically searched against databases, providing limited putative annotation. Rarely do investigators undertake the challenging and time consuming step of identifying peaks that are not present in databases [9], using methods that are common to natural products chemistry such as fractionation, high resolution accurate mass MS, and 1D and 2D NMR for structure determination. Typically, a significant proportion of detected peaks are not annotated or identified, dependent on the analytical platform used and sample type. Hence it is appropriate to conclude that all experimental metabolomics studies to date would have generated additional biological insights were metabolite identification a more tractable process, that in turn may have allowed for more complete metabolome network derivation. Metabolite identification remains a colossal challenge and a step change is needed. Here we review the status of metabolome annotation, introduce the important role of model organisms, and describe the analytical methodologies being applied.
Can model organisms help metabolome annotation?
A critical question is how such a transformative change will occur in metabolomics, to address this more than decade long problem. We believe a combination of approaches is required, including new analytical strategies, computational algorithms and database resources, and also a concerted effort by the metabolomics community to solve this bottleneck. This latter point has recently been recognised with the formation, in 2015, of a scientific task group of the international Metabolomics Society to progress the characterisation of metabolomes by initially focusing on a few model organisms [10]. The value of model organisms across biology and medicine is huge [11]. While seemingly disparate, research into bacteria, yeast, insects, worms, fish, rodents and plants has shown that the core biochemical operating principles have been conserved across all living organisms. Hence findings derived from non-mammalian model animals, for example, can shed light on biological processes in humans (Table 2).
Table 2.
Species-specific metabolome databases available for the model organisms prioritised for deeper investigation by the Model Organism Metabolomes task group [10].
| Latin name | Common name | Database | Content |
|---|---|---|---|
| Escherichia coli | - |
E. coli Metabolome Database (ECMDB) http://ecmdb.ca/ |
3,755 small molecules from textbooks, scientific journals, metabolic reconstructions and electronic databases |
|
Saccharomyces cerevisiae |
yeast | Yeast Metabolome Database (YMDB) http://www.ymdb.ca/ |
16,042 small molecules from textbooks, scientific journals, metabolic reconstructions and electronic databases |
|
Caenorhabditis elegans |
nematode | Small Molecule Identifiers (SMIDs) http://smid-db.org/ |
ca. 180 experimentally identified metabolites |
| Daphnia magna | water flea | None currently | - |
|
Drosophila melanogaster |
fruit fly | None currently | - |
| Danio rerio | zebrafish | None currently | - |
| Mus musculus | mouse | Mouse Multiple tissue Metabolome DataBase (MMMDB) http://mmmdb.iab.keio.ac.jp/ |
Contains ca. 200 known metabolites per tissue and many unknown peaks |
|
Arabidopsis thaliana |
thale cress | AraCyc http://www.plantcyc.org/databases/aracyc/14.0 |
Metabolic pathway reconstruction and experimental data, contains 2802 compounds |
|
Medicago truncatula |
barrel medic | MedicCyc 2.0 http://mediccyc.noble.org/ |
Metabolic pathway reconstruction, contains >400 pathways with related genes, enzymes and metabolites |
| Oryza sativa | rice | OryzaCyc 4.0 http://www.plantcyc.org/databases/oryzacyc/4.0 |
Metabolic pathway reconstruction, contains 2487 compounds |
|
Solanum lycopersicum |
tomato | TomatoCyc 2.0 http://www.plantcyc.org/databases/tomatocyc/2.0 |
Metabolic pathway reconstruction, contains 2550 compounds |
The Model Organism Metabolomes (MOM) task group’s philosophy is to leverage upon the critical mass of research activity and knowledge that exists for model organisms, i.e. to encourage the community to focus their metabolite identification efforts on systems we know the most about already (i.e. have species-specific metabolite databases [12–14]), that have sequenced genomes (hence can create genome-wide metabolic reconstructions to predict metabolism; [15]), and that when the metabolomes are successfully identified this knowledge will be of greatest value to the community [10]. The two primary aims of the MOM task group are to integrate disparate model organism-focused research groups into an interactive community, and to share, discuss and develop the analytical and bioinformatics strategies to progress the identification of model organism metabolomes, resulting in best practice documents. Ultimately, this task group has set a grand challenge: to identify and map all ‘system’ metabolites onto metabolic pathways, to develop quantitative metabolic models for model organisms, and to relate organism metabolic pathways within the context of evolutionary metabolomics, i.e. phylometabolomics [10]. Efforts have begun to optimise analytical methods for metabolome identification, e.g. in Escherichia coli [16], Saccharomyces cerevisiae [17] and Caenorhabditis elegans [18], as well as to mine the literature for existing knowledge, e.g. in S. cerevisiae [19]. An atlas of tissue-specific metabolomes has also been initiated for Drosophila melanogaster, including both polar and lipophilic metabolites [20].
Extending our analytical strategies to progress metabolome identification
With the ambition to more deeply characterise the complete metabolomes of model organisms, what recent developments in analytical chemistry have been applied? Unlike for genomics, where disruptive technologies are relatively common [21], the analytical methods used in metabolomics have changed relatively little over the last decade. What has occurred recently is a considerable growth of targeted methods for studying swathes of metabolism, likely driven by the very frustration of limited peak annotation in non-targeted metabolomics, as discussed above. For example, a number of targeted LC-MS/MS assays have been developed to profile from a few tens to a couple of hundred metabolites in rice [22–24] and mammalian samples [25, 26]. While benefitting from yielding metabolic data that is identified and often quantitative, all of these studies only scrape the surface of the thousands of metabolites estimated to comprise a metabolome. Hence, to an extent, this shift to targeted assays is a distraction (except for cases where the targets are already known) from solving the real challenge of metabolite identification and understanding biology in greater detail utilising non-targeted metabolomics.
So what is the current status of non-targeted metabolomics for fully characterising metabolomes? Both gas chromatography (GC) and liquid chromatography (LC) methods continue to be developed, including multi-dimensional chromatography and the application of multiple columns for the separation of different classes of metabolites. For example, a “broad spectrum” GC-MS method has been developed to measure non-volatile metabolites in tropical fruits [27]; a total of 92 peaks were detected of which the authors identified only 45. Utilising a comprehensive GCxGC approach, coupled with headspace solid phase microextraction (HS SPME) and a time of flight (ToF) MS, Alves et al. reported the identification of 257 volatile metabolites from S. cerevisiae distributed over more than a dozen chemical families [28]. For a recent review of advanced multi-dimensional separations in mass spectrometry, see [29]. The inherent requirements of GC-MS for the thermal stability and volatility of the analytes, or derivatives thereof, means that, alone, this technique is unable to facilitate comprehensive metabolome annotation. Instead, the majority of non-targeted metabolomics studies continue to employ LC, and there is an increasing trend towards the application of several column types in a given study; e.g. Tufi et al. not only used two LC columns (C18 and HILIC) but also GC-MS to study a freshwater snail Lymnaea stagnalis [30]. This more comprehensive analytical approach was applied specifically to obtain a broader picture of the hydrophilic and lipophilic metabolome.
The application of multiple analytical platforms, as applied to L. stagnalis [30] is an emerging trend in metabolomics. Bundy and co-workers [31] applied three different platforms to analyse C. elegans, including 1D 1H NMR spectroscopy, GC/MS and UPLC-MS. The deeper integration of NMR and MS data in automated metabolite identification pipelines is an emerging topic [32]. An even broader range of metabolites were measured in 31 varieties of rice using HS SPME GC-MS, primary polar metabolites by GC-ToF-MS, both polar and semi-polar compounds by 1H NMR and direct infusion MS, and multi-elemental analysis using ICP-MS [33]. While more time intensive and costly, deeper characterisation of organismal metabolomes currently requires such a multi-platform strategy. Fortunately, as long as the metabolic knowledge is captured in relevant open access databases, such as MetaboLights [34], then this strategy only needs to be conducted once. A related project to deeply annotate the metabolome of the NIH model species Daphnia magna (waterflea) is underway in the primary authors’ laboratory, applying multiple extraction methods, LC-MS/MS and MSn methods, GC-Orbitrap MS, and 1D and 2D NMR spectroscopy. Progress has also been reported in the integration of several platforms to enable metabolite identification by UHPLC-SPE-NMR-MS [35]. In addition, approaches such as ion mobility mass spectrometry [36] and ultrahigh resolution mass spectrometry [37] hold considerable promise for contributing to metabolome annotation projects.
Another methodology that has considerable potential for aiding metabolite identification is stable isotope labelling, for example to probe the sulfur metabolome of Arabidopsis [38, 39]. Isotopic ratio outlier analysis (IROA) is another isotope labelling technology that is designed to generate specific 13C isotopic patterns in metabolites for both high resolution LC-MS and GC-MS [17, 40–43]. Unlike other stable isotope labelling methods, rather than utilising natural abundance and 98–99% enrichment for the control and experimental populations, respectively [44–48], IROA uses an enrichment level of 95% and 5% 13C. This leads to more observable isotopic peaks in the mass spectra in predictable and diagnostic patterns. Recent studies have demonstrated the promise of IROA for metabolic phenotyping in model organisms, including for prototrophic S. cerevisiae [17, 49] and C. elegans [43]; the latter was grown in liquid culture with 13C-labeled E. coli that was first grown in M9 minimal media on either 95% or 5% 13C glucose, creating labelled C. elegans. These 95% 13C and 5% 13C glucose labelling experiments, when extracted and combined, show distinctive IROA patterns: 12C -derived molecules, 13C-derived molecules, artifacts (lack IROA patterns) and derivatives of exogenously applied compounds. Only metabolites of biological origin will have mirrored 12C and 13C metabolite peaks at the same retention time. Furthermore, the abundance of the heavy isotopologues in the 5% 13C samples (M+1, M+2, etc., the 12C envelope) or light isotopologues in the 95% 13C samples (M−1, M−2, etc., the 13C envelope), follows the binomial distribution for 13C in metabolite products based on the initial substrate enrichment. The mass difference between the 12C monoisotopic peak and the 13C monoisotopic peak indicates the number of carbons in the metabolite. Uniquely, the accurate mass IROA-GC/MS protocol developed, using both chemical ionization (CI) and electron ionization (EI), extends the information acquired from the isotopic peak patterns for molecular formulae generation. The process has been formulated as an algorithm, in which the numbers of carbons, methoximations and silylations are used as search constraints, and an accurate mass CI IROA library with retention times based on the Fiehn protocol has been published [17]. The combination of CI and EI IROA protocols affords a metabolite identification procedure that can identify co-eluting metabolites [17]. In summary, non-targeted stable isotope metabolite profiling using IROA reduces the complexity for global stable isotope metabolite identification [50], and can extend metabolome analysis by identifying “known unknowns” with an IROA mirror pattern, and generating the number of carbons in the unknown metabolite.
Conclusions
The hugely beneficial impact of the Human Genome Project on 21st century science is undeniable [51, 52]. No such large-scale experimental characterisations of organism metabolomes have been reported and many of the studies published to date describe only a fraction of the estimated size of a metabolome. That said, efforts are now underway from text mining to novel experimental approaches that offer to accelerate this process, and coordination of some of these activities is being achieved through the Metabolomics Society’s task group. Activity in metabolome identification is therefore expected to increase over the next couple of years with significant returns on this investment within 5–10 years. Looking further ahead, challenges will include developing analytical strategies to quantify several thousand known metabolites (simultaneously) as well as the spatial localisation of these compounds, e.g. using MS imaging [53, 54]. Ultimately, a better understanding of the parts list is going to facilitate growth of several fields, including phylometabolomics, the study of organism metabolic pathways in the context of evolution.
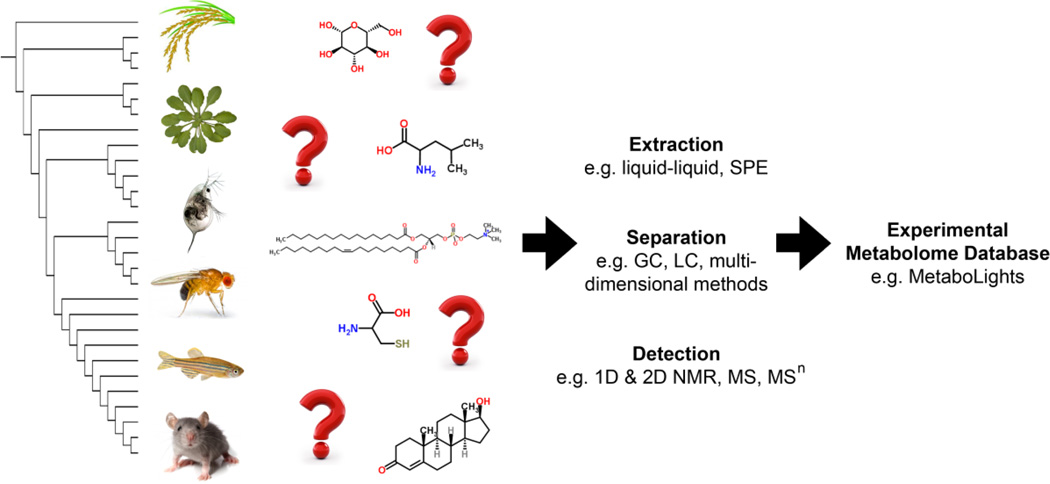
Figure 1.
Schematic workflow for the deep experimental characterisation of the metabolomes of model organisms. The creation of such knowledge in open-access metabolome databases would greatly accelerate the study of metabolism in an evolutionary context, i.e. phylometabolomics.
Highlights.
No exhaustive experimental characterisation of an organism’s metabolome reported
Deep metabolome annotation strategies based on multi-platform assays
International task group formed to help coordinate metabolome annotation
Entering new era of annotation focused on model organisms
Acknowledgments
This work was in part supported by the UK Natural Environment Research Council (NE/J017442/1), Thermo Fisher Scientific (CASE award) and by an Einstein-Mount Sinai Diabetes Research Center Grant (P60DK020541).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Oliver SG, Winson MK, Kell DB, Baganz F. Systematic functional analysis of the yeast genome. Trends in Biotechnology. 1998;16:373–378. doi: 10.1016/s0167-7799(98)01214-1. [DOI] [PubMed] [Google Scholar]
- 2.Hartmann T. From waste products to ecochemicals: Fifty years research of plant secondary metabolism. Phytochemistry. 2007;68:2831–2846. doi: 10.1016/j.phytochem.2007.09.017. [DOI] [PubMed] [Google Scholar]
- 3.Lei ZT, Huhman DV, Sumner LW. Mass Spectrometry Strategies in Metabolomics. Journal of Biological Chemistry. 2011;286:25435–25442. doi: 10.1074/jbc.R111.238691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sumner LW, Amberg A, Barrett D, Beale MH, Beger R, Daykin CA, Fan TWM, Fiehn O, Goodacre R, Griffin JL, et al. Proposed minimum reporting standards for chemical analysis. Metabolomics. 2007;3:211–221. doi: 10.1007/s11306-007-0082-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wishart DS. Advances in metabolite identification. Bioanalysis. 2011;3:1769–1782. doi: 10.4155/bio.11.155. [DOI] [PubMed] [Google Scholar]
- 6.Dunn WB, Erban A, Weber RJM, Creek DJ, Brown M, Breitling R, Hankemeier T, Goodacre R, Neumann S, Kopka J, et al. Mass appeal: metabolite identification in mass spectrometry-focused untargeted metabolomics. Metabolomics. 2013;9:S44–S66. [Google Scholar]
- 7.Dunn WB, Broadhurst D, Begley P, Zelena E, Francis-McIntyre S, Anderson N, Brown M, Knowles JD, Halsall A, Haselden JN, et al. Procedures for large-scale metabolic profiling of serum and plasma using gas chromatography and liquid chromatography coupled to mass spectrometry. Nature Protocols. 2011;6:1060–1083. doi: 10.1038/nprot.2011.335. [DOI] [PubMed] [Google Scholar]
- 8.Southam AD, Weber RJM, Engel J, Jones MR, Viant MR. A complete workflow for high-resolution spectral-stitching nanoelectrospray direct infusion mass spectrometry-based metabolomics and lipidomics. Nature Protocols. doi: 10.1038/nprot.2016.156. (accepted) [DOI] [PubMed] [Google Scholar]
- 9.Gillard J, Frenkel J, Devos V, Sabbe K, Paul C, Rempt M, Inze D, Pohnert G, Vuylsteke M, Vyverman W. Metabolomics Enables the Structure Elucidation of a Diatom Sex Pheromone. Angewandte Chemie-International Edition. 2013;52:854–857. doi: 10.1002/anie.201208175. [DOI] [PubMed] [Google Scholar]
- 10. Edison AS, Hall RD, Junot C, Karp PD, Kurland IJ, Mistrik R, Reed LK, Saito K, Salek RM, Steinbeck C, et al. The Time Is Right to Focus on Model Organism Metabolomes. Metabolites. 2016;6 doi: 10.3390/metabo6010008. * Describes the launch of the first internationally coordinated action to accelerate research into the deep characterisation of model organism metabolomes. A team of scientists have laid down the challenge "to identify and map all ‘system’ metabolites onto metabolic pathways, to develop quantitative metabolic models for model organisms, and to relate organism metabolic pathways within the context of evolutionary metabolomics."
- 11.Fontana L, Partridge L. Promoting Health and Longevity through Diet: From Model Organisms to Humans. Cell. 2015;161:106–118. doi: 10.1016/j.cell.2015.02.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wishart DS, Tzur D, Knox C, Eisner R, Guo AC, Young N, Cheng D, Jewell K, Arndt D, Sawhney S, et al. HMDB: the human metabolome database. Nucleic Acids Research. 2007;35:D521–D526. doi: 10.1093/nar/gkl923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jewison T, Knox C, Neveu V, Djoumbou Y, Guo AC, Lee J, Liu P, Mandal R, Krishnamurthy R, Sinelnikov I, et al. YMDB: the Yeast Metabolome Database. Nucleic Acids Research. 2012;40:D815–D820. doi: 10.1093/nar/gkr916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Guo AC, Jewison T, Wilson M, Liu YF, Knox C, Djoumbou Y, Lo P, Mandal R, Krishnamurthy R, Wishart DS. ECMDB: The E-coli Metabolome Database. Nucleic Acids Research. 2013;41:D625–D630. doi: 10.1093/nar/gks992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kim T, Dreher K, Nilo-Poyanco R, Lee I, Fiehn O, Lange BM, Nikolau BJ, Sumner L, Welti R, Wurtele ES, et al. Patterns of Metabolite Changes Identified from Large-Scale Gene Perturbations in Arabidopsis Using a Genome-Scale Metabolic Network. Plant Physiology. 2015;167 doi: 10.1104/pp.114.252361. 1685-U1890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shen Y, Fatemeh T, Tang LH, Cai ZW. Quantitative metabolic network profiling of Escherichia coli: An overview of analytical methods for measurement of intracellular metabolites. Trac-Trends in Analytical Chemistry. 2016;75:141–150. [Google Scholar]
- 17. Qiu Y, Moir R, Willis I, Beecher C, Tsai YH, Garrett TJ, Yost RA, Kurland IJ. Isotopic Ratio Outlier Analysis of the S. cerevisiae Metabolome Using Accurate Mass Gas Chromatography/Time-of-Flight Mass Spectrometry: A New Method for Discovery. Anal Chem. 2016;88:2747–2754. doi: 10.1021/acs.analchem.5b04263. ** First report using IROA technology in combination with accurate mass GC/TOF-MS, used to examine the S. cerevisiae metabolome. An accurate mass CI IROA library containing 126 metabolites with retention times based on the Fiehn protocol was established. The combination of CI and EI IROA protocols identifies co-eluting metabolites and differentiates metabolite spectra from artifacts, and "known unknown" metabolites were easily separated, with the number of carbons identified from the IROA patterns.
- 18.Stupp GS, Clendinen CS, Ajredini R, Szewc MA, Garret T, Menger RF, Yost RA, Beecher C, Edison AS. Isotopic Ratio Outlier Analysis Global Metabolomics of Caenorhabditis elegans. Analytical Chemistry. 2013;85:11858–11865. doi: 10.1021/ac4025413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nobata C, Dobson PD, Iqbal SA, Mendes P, Tsujii J, Kell DB, Ananiadou S. Mining metabolites: extracting the yeast metabolome from the literature. Metabolomics. 2011;7:94–101. doi: 10.1007/s11306-010-0251-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chintapalli VR, Al Bratty M, Korzekwa D, Watson DG, Dow JAT. Mapping an Atlas of Tissue-Specific Drosophila melanogaster Metabolomes by High Resolution Mass Spectrometry. Plos One. 2013;8 doi: 10.1371/journal.pone.0078066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mikheyev AS, Tin MMY. A first look at the Oxford Nanopore MinION sequencer. Molecular Ecology Resources. 2014;14:1097–1102. doi: 10.1111/1755-0998.12324. [DOI] [PubMed] [Google Scholar]
- 22.Cao ZY, Sun LH, Mou RX, Zhang LP, Lin XY, Zhu ZW, Chen MX. Profiling of phytohormones and their major metabolites in rice using binary solid-phase extraction and liquid chromatography-triple quadrupole mass spectrometry. Journal of Chromatography A. 2016;1451:67–74. doi: 10.1016/j.chroma.2016.05.011. [DOI] [PubMed] [Google Scholar]
- 23.Hu CY, Tohge T, Chan SA, Song Y, Rao J, Cui B, Lin H, Wang L, Fernie AR, Zhang DB, et al. Identification of Conserved and Diverse Metabolic Shifts during Rice Grain Development. Scientific Reports. 2016;6 doi: 10.1038/srep20942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Schafer M, Brutting C, Baldwin IT, Kallenbach M. High-throughput quantification of more than 100 primary- and secondary-metabolites, and phytohormones by a single solid-phase extraction based sample preparation with analysis by UHPLC-HESI-MS/MS. Plant Methods. 2016;12 doi: 10.1186/s13007-016-0130-x. * Excellent demonstration of the quantitatitive capabilities of targeted metabolite analysis, here assaying more than 100 primary and secondary metabolites simultaneously following a single extraction of plant material. Such an approach is highly recommended when the metabolite 'targets' are already known, and hence non-targeted metabolomics is not required.
- 25.Chen JH, Hou W, Han B, Liu GH, Gong J, Li YM, Zhong DM, Liao QF, Xie ZY. Target-based metabolomics for the quantitative measurement of 37 pathway metabolites in rat brain and serum using hydrophilic interaction ultra-high-performance liquid chromatography-tandem mass spectrometry. Anal Bioanal Chem. 2016;408:2527–2542. doi: 10.1007/s00216-016-9352-z. [DOI] [PubMed] [Google Scholar]
- 26.Jain M, Nilsson R, Sharma S, Madhusudhan N, Kitami T, Souza AL, Kafri R, Kirschner MW, Clish CB, Mootha VK. Metabolite Profiling Identifies a Key Role for Glycine in Rapid Cancer Cell Proliferation. Science. 2012;336:1040–1044. doi: 10.1126/science.1218595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Khakimov B, Mongi RJ, Sorensen KM, Ndabikunze BK, Chove BE, Engelsen SB. A comprehensive and comparative GC-MS metabolomics study of non-volatiles in Tanzanian grown mango, pineapple, jackfruit, baobab and tamarind fruits. Food Chemistry. 2016;213:691–699. doi: 10.1016/j.foodchem.2016.07.005. [DOI] [PubMed] [Google Scholar]
- 28.Alves Z, Melo A, Figueiredo AR, Coimbra MA, Gomes AC, Rocha SM. Exploring the Saccharomyces cerevisiae Volatile Metabolome: Indigenous versus Commercial Strains. Plos One. 2015;10 doi: 10.1371/journal.pone.0143641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.May JC, McLean JA. Advanced Multidimensional Separations in Mass Spectrometry: Navigating the Big Data Deluge. In: Bohn PW, Pemberton JE, editors. Annual Review of Analytical Chemistry, Vol 9. 2016. pp. 387–409. Annual Review of Analytical Chemistry, vol 9.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Tufi S, Lamoree MH, De Boer J, Leonards PEG. Cross-platform metabolic profiling: application to the aquatic model organism Lymnaea stagnalis. Anal Bioanal Chem. 2015;407:1901–1912. doi: 10.1007/s00216-014-8431-2. * Good example of the application of several separation technologies - both GC and LC - with the latter using two different column chemistries (C18 and HILIC). This approach was used to increased the breadth of experimental measurement of the snail metabolome.
- 31. Geier FM, Want EJ, Leroi AM, Bundy JG. Cross-Platform Comparison of Caenorhabditis elegans Tissue Extraction Strategies for Comprehensive Metabolome Coverage. Analytical Chemistry. 2011;83:3730–3736. doi: 10.1021/ac2001109. * Good example of the application of multiple analytical platforms to enable a deeper characterisation of this important model organism's metabolome. Multiple tissue extraction strategies were examined and an 80% methanol solution was determined to be the best.
- 32.Bingol K, Bruschweiler R. Knowns and unknowns in metabolomics identified by multidimensional NMR and hybrid MS/NMR methods. Current Opinion in Biotechnology. 2016;43:17–24. doi: 10.1016/j.copbio.2016.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Mumm R, Hageman JA, Calingacion MN, de Vos RCH, Jonker HH, Erban A, Kopka J, Hansen TH, Laursen KH, Schjoerring JK, et al. Multi-platform metabolomics analyses of a broad collection of fragrant and non-fragrant rice varieties reveals the high complexity of grain quality characteristics. Metabolomics. 2016;12 doi: 10.1007/s11306-015-0925-1. ** An exemplary paper, demonstrating through hierarchical cluster analysis, both the complementarity and distinct merits of multiple analytical platforms in the study of rice variant metabolome constituents. Importantly, data is interrogated in a way that aligns well with the vision of the MOM task group, i.e. phylometabolomic comparisons, for a crop species of fundamental societal importance.
- 34.Haug K, Salek RM, Conesa P, Hastings J, de Matos P, Rijnbeek M, Mahendraker T, Williams M, Neumann S, Rocca-Serra P, et al. MetaboLights-an open-access general-purpose repository for metabolomics studies and associated metadata. Nucleic Acids Research. 2013;41:D781–D786. doi: 10.1093/nar/gks1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gu WY, Li N, Leung ELH, Zhou H, Yao XJ, Liu L, Wu JL. Rapid identification of new minor chemical constituents from Smilacis Glabrae Rhizoma by combined use of UHPLC-Q-TOF-MS, preparative HPLC and UHPLC-SPE-NMR-MS techniques. Phytochemical Analysis. 2015;26:428–435. doi: 10.1002/pca.2577. [DOI] [PubMed] [Google Scholar]
- 36.May JC, Branum RLG, McLean JA. Targeting the untargeted in molecular phenomics with structurally-selective ion mobility-mass spectrometry. Current Opinion in Biotechnology. 2016;39:192–197. doi: 10.1016/j.copbio.2016.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Romano S, Dittmar T, Bondarev V, Weber RJM, Viant MR, Schulz-Vogt HN. Exo-Metabolome of Pseudovibrio sp FO-BEG1 Analyzed by Ultra-High Resolution Mass Spectrometry and the Effect of Phosphate Limitation. Plos One. 2014;9 doi: 10.1371/journal.pone.0096038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Glaser K, Kanawati B, Kubo T, Schmitt-Kopplin P, Grill E. Exploring the Arabidopsis sulfur metabolome. Plant Journal. 2014;77:31–45. doi: 10.1111/tpj.12359. [DOI] [PubMed] [Google Scholar]
- 39.Nakabayashi R, Tsugawa H, Mori T, Saito K. Automation of chemical assignment for identifying molecular formula of S-containing metabolites by combining metabolomics and chemoinformatics with 34S labeling. Metabolomics. 2016 [Google Scholar]
- 40.Clendinen CS, Stupp GS, Ajredini R, Lee-McMullen B, Beecher C, Edison AS. An overview of methods using (13)C for improved compound identification in metabolomics and natural products. Front Plant Sci. 2015;6:611. doi: 10.3389/fpls.2015.00611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.de Jong FA, Beecher C. Addressing the current bottlenecks of metabolomics: Isotopic Ratio Outlier Analysis, an isotopic-labeling technique for accurate biochemical profiling. Bioanalysis. 2012;4:2303–2314. doi: 10.4155/bio.12.202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Edison AS, Clendinen CS, Ajredini R, Beecher C, Ponce FV, Stupp GS. Metabolomics and Natural-Products Strategies to Study Chemical Ecology in Nematodes. Integr Comp Biol. 2015;55:478–485. doi: 10.1093/icb/icv077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Stupp GS, Clendinen CS, Ajredini R, Szewc MA, Garrett T, Menger RF, Yost RA, Beecher C, Edison AS. Isotopic ratio outlier analysis global metabolomics of Caenorhabditis elegans. Anal Chem. 2013;85:11858–11865. doi: 10.1021/ac4025413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Giavalisco P, Kohl K, Hummel J, Seiwert B, Willmitzer L. 13C isotope-labeled metabolomes allowing for improved compound annotation and relative quantification in liquid chromatography-mass spectrometry-based metabolomic research. Anal Chem. 2009;81:6546–6551. doi: 10.1021/ac900979e. [DOI] [PubMed] [Google Scholar]
- 45.Wu L, Mashego MR, van Dam JC, Proell AM, Vinke JL, Ras C, van Winden WA, van Gulik WM, Heijnen JJ. Quantitative analysis of the microbial metabolome by isotope dilution mass spectrometry using uniformly 13C-labeled cell extracts as internal standards. Anal Biochem. 2005;336:164–171. doi: 10.1016/j.ab.2004.09.001. [DOI] [PubMed] [Google Scholar]
- 46.Bennett BD, Yuan J, Kimball EH, Rabinowitz JD. Absolute quantitation of intracellular metabolite concentrations by an isotope ratio-based approach. Nat Protoc. 2008;3:1299–1311. doi: 10.1038/nprot.2008.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Weindl D, Wegner A, Jager C, Hiller K. Isotopologue ratio normalization for non-targeted metabolomics. J Chromatogr A. 2015;1389:112–119. doi: 10.1016/j.chroma.2015.02.025. [DOI] [PubMed] [Google Scholar]
- 48.Blank LM, Desphande RR, Schmid A, Hayen H. Analysis of carbon and nitrogen co-metabolism in yeast by ultrahigh-resolution mass spectrometry applying 13C- and 15N-labeled substrates simultaneously. Anal Bioanal Chem. 2012;403:2291–2305. doi: 10.1007/s00216-012-6009-4. [DOI] [PubMed] [Google Scholar]
- 49.VanderSluis B, Hess DC, Pesyna C, Krumholz EW, Syed T, Szappanos B, Nislow C, Papp B, Troyanskaya OG, Myers CL, et al. Broad metabolic sensitivity profiling of a prototrophic yeast deletion collection. Genome Biol. 2014;15:R64. doi: 10.1186/gb-2014-15-4-r64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Weindl D, Wegner A, Hiller K. Metabolome-Wide Analysis of Stable Isotope Labeling-Is It Worth the Effort? Front Physiol. 2015;6:344. doi: 10.3389/fphys.2015.00344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lander ES, Int Human Genome Sequencing C, Linton LM, Birren B, Nusbaum C, Zody MC, Baldwin J, Devon K, Dewar K, Doyle M, et al. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 52.Venter JC, Adams MD, Myers EW, Li PW, Mural RJ, Sutton GG, Smith HO, Yandell M, Evans CA, Holt RA, et al. The sequence of the human genome. Science. 2001;291 1304-+ [Google Scholar]
- 53.Boughton BA, Thinagaran D, Sarabia D, Bacic A, Roessner U. Mass spectrometry imaging for plant biology: a review. Phytochemistry Reviews. 2016;15:445–488. doi: 10.1007/s11101-015-9440-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Palmer A, Trede D, Alexandrov T. Where imaging mass spectrometry stands: here are the numbers. Metabolomics. 2016;12 [Google Scholar]