Abstract
Cells alter the proteome to respond to environmental and developmental cues. Global analysis of proteomic responses is of limited value in heterogeneous environments, where there is no “average” cell. Advances in sequencing, protein labeling, mass spectrometry, and data analysis have fueled recent progress in the investigation of specific subpopulations of cells in complex systems. Here we highlight recently developed chemical tools that enable cell-selective proteomic analysis of complex biological systems, from bacterial pathogens to whole animals.
Keywords: cell-selective, proteome, TRAP, CTAP, BONCAT, SORT, OP-Puro, APEX, newly-synthesized, subcellular, click chemistry
Graphical Abstract
Introduction
Cellular protein synthesis changes rapidly in response to internal and external cues in ways that vary from cell to cell. Global proteomic analyses of microbial communities, tissues and organisms have provided important insights into the behavior of such systems, but can obscure the diversity of responses characteristic of different cellular subpopulations (Figure 1). Cell-selective methods for the analysis of protein synthesis are being developed to resolve proteomic changes in space and time.
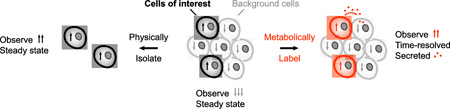
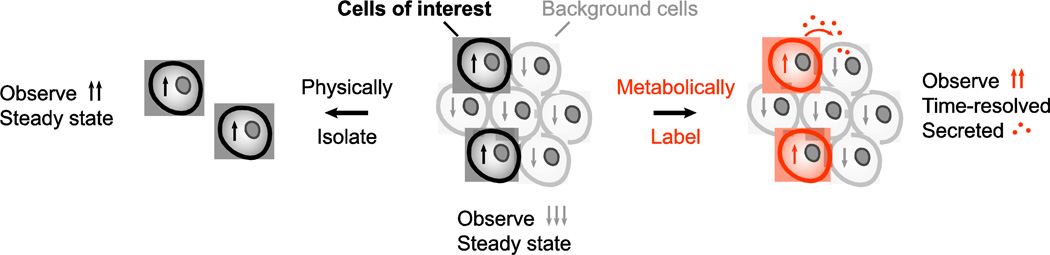
Figure 1.
The importance of cell-type-specific proteomics. Bulk measurements of complex tissues can obscure proteomic changes that occur in specific sub-populations of cells. A protein that is highly expressed (up arrows) in the cells of interest might be detected at low abundance overall due to low expression (down arrows) in background cells. Cells of interest must be physically isolated or tagged to measure the cell-specific proteome. Physical isolation measures steady-state levels of intracellular proteins, whereas labeling methods can be time-resolved and used to identify secreted proteins.
Cell-type-specific transcriptomics experiments have revealed mRNA expression patterns in a wide array of biological systems, but mRNA and protein levels are often dissonant [1]. Moreover, some important elements of proteome dynamics, including posttranslational modification, degradation, and localization, cannot be addressed by mRNA measurements alone [2,3]. Until recently, changes in protein abundance in specific cells could be measured only in targeted, low-throughput experiments, but innovations in mass spectrometry and computational algorithms have facilitated the identification and quantification of thousands of proteins simultaneously from complex biological samples [4–6].
In this Opinion, we highlight recent developments in determining cell-type-specific proteomes and recommend experimental design strategies that are guided by the question at hand.
Cell-selective translatomics and ribosome profiling
Translatomic studies, which select for ribosome-associated transcripts, have yielded stronger correlations between transcript and protein abundances than experiments that measure steady-state mRNA levels [7]. Cell-type-specific studies have been enabled by translating ribosome affinity purification (TRAP), a method in which epitope-tagged ribosomes and their associated transcripts are captured, enriched and subjected to amplification and deep sequencing [8]. TRAP can be rendered cell-specific by placing expression of the tagged ribosome under control of a selective promoter.
More recently, Ingolia and Weissman have developed ribosome profiling, which identifies ribosome-protected mRNA footprints and allows investigators to determine ribosome occupancy with positional specificity. This information can be used to measure translation levels and locate non-canonical start sites [7]. Gonzalez et al. used TRAP to cell-selectively purify ribosome-bound transcripts, and employed ribosome profiling to identify the translatome of gliomas and to reveal decreased translation in glial progenitors compared to the tumor microenvironment [9]. Ribosome profiling is a powerful technique that we expect to find increasing use upon further development of cell-specific methods.
While translatomic studies provide greater depth of coverage than current proteomic measurements, ribosome binding does not ensure that a transcript is undergoing active translation [10].
Separating cells for steady-state proteomic analysis
The earliest strategies to determine cell-specific proteomes relied on separating and purifying the cells of interest prior to analysis. Cells can be sorted on the basis of expression of a transgene under control of a cell-specific promoter or by antibody staining of marker epitopes. These tools are well established and have been thoughtfully reviewed [10,11]. Physical methods have been used for years to isolate cell types from mammalian tissues for subsequent downstream analyses [12,13]. More recently these methods have been used to measure growth rates and elucidate proteomic signatures of Salmonella during murine infection [14].
Physical separations remain the best method for analyzing clinical specimens and genetically intractable organisms. However, imperfect separations and long sample processing times can diminish selectivity and increase the likelihood of artifacts. Furthermore, such methods intrinsically yield steady-state proteomic information. In contrast, metabolic labeling strategies enable cell-specific proteomic analysis to be accomplished in time-resolved fashion.
Metabolic labeling: trade-offs between sensitivity and perturbation
Metabolic labeling methods are temporally resolved and use an arsenal of amino acid isotopologs, non-canonical amino acids, and analogs of protein synthesis inhibitors (Figure 2). Each of these strategies can be placed under control of cell-specific genetic elements to afford cellular resolution. The choice of promoter(s) is key for these systems, and the degree of protein labeling needs to be weighed against the possibility of perturbing the system. Results should be validated via independent assays because labels may affect protein expression, stability, and/or function.
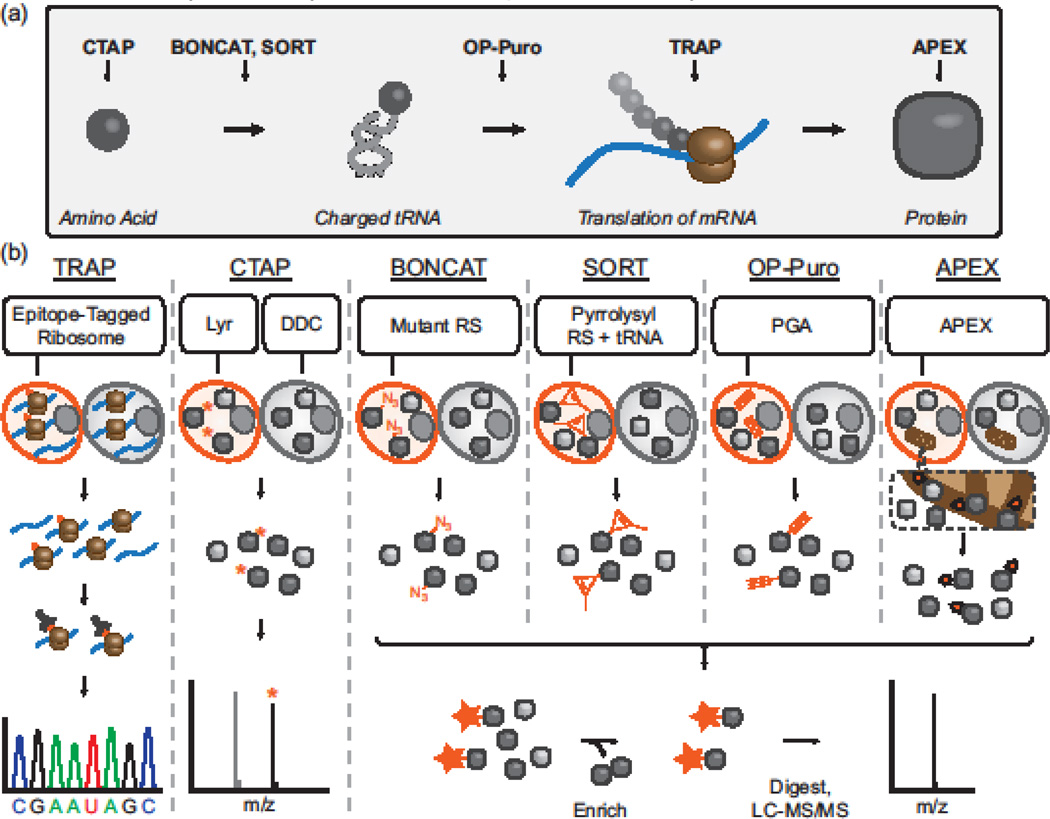
Figure 2.
Labeling strategies for cell-selective proteomics. a) The process by which amino acids are incorporated into proteins, and the step exploited by each of the labeling methods discussed in this Opinion. b) Schematic of each technique.
Translating ribosome affinity purification: TRAP; Cell type-specific labeling using amino acid precursors: CTAP; Bio-orthogonal non-canonical amino acid tagging: BONCAT; Stochastic orthogonal recoding of translation: SORT; O-propargyl puromycin: OP-Puro; ascorbate peroxidase: APEX; Lysine racemase:Lyr; diaminopimelate decarboxylase: DDC; aminoacyl-tRNA synthetase: RS; penicillin-G-acylase: PGA.
Cell-type-specific labeling using amino acid precursors (CTAP)
Stable isotope labeling by amino acids in cell culture (SILAC) relies on the incorporation of isotopically labeled amino acids into proteins. To make SILAC cell-selective, Gauthier et al. introduced cell-type-specific labeling using amino acid precursors (CTAP), a method that exploits the fact that lysine is an essential amino acid in mammalian cells [15]. Cell-selective expression of biosynthetic enzymes allows L-lysine isotopologs to be synthesized in situ starting from isotope-labeled precursors. Only minor differences in gene expression resulted from feeding the heavy precursor to cells expressing the biosynthetic machinery versus supplementing cells directly with L-lysine.
In principle, both exchange of L-lysine between cells and extracellular processing of the precursor can compromise the cell-specificity of the CTAP method. When Lavis and coworkers employed an analogous strategy to unmask fluorophores in targeted cells, they noted that the unmasked small molecule diffused through gap junctions. This effect can be exploited to study cell-cell connectivity, but would confound cell-specific protein labeling if the small molecule were to diffuse to cells lacking the decaging enzyme [16]. To address these concerns, Tape et al. optimized CTAP for eukaryotic cell types and achieved ~90% cell-specific labeling in ten-day co-cultures [17]. Using their optimized method, Tape et al. combined CTAP with phosphoproteomics to study heterocellular KRASG12D signaling in pancreatic ductal adenocarcinoma cells [18]. By restricting their proteomic analysis to cells that expressed KRASG12D, the authors showed that the oncogene regulates AKT through reciprocal signaling – not through the accepted cell-autonomous pathway.
Bio-orthogonal Non Canonical Amino acid Tagging (BONCAT)
CTAP is most suitable for cell-specific experiments conducted in culture on timescales of 3–7 days [19]. For studies that require better time resolution, the bio-orthogonal non-canonical amino acid tagging (BONCAT) method, introduced by Dieterich and coworkers, offers a good alternative [20,21]. In its original form, BONCAT exploits the capacity of the endogenous aminoacyl-tRNA synthetases to charge non-canonical amino acids (ncAAs) to their cognate tRNAs for incorporation into proteins. ncAAs bearing bio-orthogonal chemical handles, often azides or alkynes, enable conjugation to affinity tags and separation of tagged proteins from the rest of the protein pool. The methionine surrogates azidohomoalanine (Aha) and homopropargylglycine (Hpg) have been used to probe proteome dynamics in bacterial [22–26] and mammalian [27] systems, and notably, to enrich and quantify secreted proteins [28]. Depletion of cellular methionine is not necessary for Aha labeling; Bagert et al. showed that a 30:1 ratio of Aha to Met yielded excellent protein labeling while minimizing perturbations that might be expected to arise from methionine starvation [29]. Other studies have shown that ncAA labeling for periods of up to two days do not perturb embryonic growth in live mice [30]. In designing a BONCAT experiment, the investigator should choose concentrations of the ncAA label and its natural counterpart that reflect the relative rates of activation of the amino acids by the cognate synthetase.
In 2009, Ngo and coworkers developed a cell-selective version of BONCAT by engineering an E. coli methionyl-tRNA synthetase (EcMetRS) variant that activates azidonorleucine (Anl). Because Anl is a poor substrate for wild-type EcMetRS, labeling is essentially restricted to cells that express the mutant synthetase. In the first example of the cell-specific BONCAT method, Ngo et al. reported specific labeling of E. coli cells co-cultured with murine alveolar macrophages [31]. Grammel et al. expanded on this method by enriching for proteins synthesized during Salmonella typhimurium infection [32], and Mahdavi and coworkers used BONCAT to determine the order in which Yersinia enterocolitica effector proteins are injected into HeLa cells in the course of infection [33].
Cell-selective BONCAT has now been extended to proteomic analysis in live animals, highlighting its potential utility in creating cell-specific proteomic “atlases”. In 2015 we reported a mutant phenylalanyl-tRNA synthetase (PheRS) that enables the use of p-azidophenylalanine (Azf) as a BONCAT probe in Caenorhabditis elegans [34]. Combining cell-selective BONCAT with stable isotope labeling, we used the myo-2 promoter to direct expression of the mutant synthetase to the 20 pharyngeal muscle cells of the worm. We were able to quantify 2270 proteins by this method, and to verify the pharyngeal expression patterns of several previously uncharacterized proteins.
Dieterich and coworkers have adapted cell-selective BONCAT labeling to Drosophila melanogaster through controlled expression of the DmMetRS L262G mutant [35]. Chronic administration of Anl in developing flies expressing the mutant synthetase caused slight impairments in larval growth and behavior, but shorter (48 h) labeling times led to no noticeable defects. Importantly, administration of the amino acid in flies that did not express the mutant MetRS caused no discernible effect. Using this strategy, Niehues et al. measured reduced neuronal protein synthesis rates in a Drosophila model of Charcot-Marie-Tooth (CMT) neuropathy. Mahdavi et al. and Muller et al. have employed the analogous (L274G) mouse synthetase in mammalian cell culture and in a neuronglia co-culture system, respectively [36,37]. The latter experiments enabled the investigators to monitor changes in the astrocytic proteome in response to treatment with brain-derived neurotrophic factor (BDNF).
Split synthetases have been developed to enable cell-selective analysis of systems in which no single promoter restricts expression of the mutant enzyme to the cells of interest [38]. Notably, all amino acids and enrichment media needed for BONCAT experiments are commercially available.
Stochastic Orthogonal Recoding of Translation (SORT)
Chin and coworkers have developed a residue-specific ncAA-labeling technology termed stochastic orthogonal recoding of translation (SORT), which – like BONCAT – allows chemoselective modification and enrichment of newly synthesized cellular proteins. SORT relies on expression of a pyrrolysyl-tRNA synthetase and its cognate tRNA [39,40]. Using this method, Elliott et al. cell-selectively labeled and identified proteins made during different stages of larval growth in Drosophila. Importantly, SORT allows the anticodon of the cognate tRNA to be changed to direct the ncAA to different sets of codons in the labeled proteins. Elliott et al. have characterized the enrichment process and found that tagging at different codons leads to the enrichment of overlapping, but distinct sets of proteins [41]. The authors noted that simultaneous expression of multiple tRNAs (i.e., tRNA-Ala, -Ser and -Met) increases labeling efficiency. Furthermore, Elliott et al. found that enrichment after tagging improves detection of low-abundance proteins.
Cell-selective O-propargyl-puromycin (OP-Puro) labeling
The O-propargyl-puromycin (OP-Puro) method also incorporates “clickable” handles into nascent proteins [42]. Cohen and coworkers recently achieved cell-targeted OP-puromycin labeling by using a phenylacetyl-caged analog that is uncaged by cell-selective expression of penicillin G acylase (PGA) [43]. The OP-puro method is the fastest of the metabolic labeling methods and the best suited for studies requiring ultra-short labeling times [44]. Prolonged labeling with OP-puro would be expected to perturb cellular behavior through inhibition of global translation. Furthermore, premature truncation renders this method ineffective for the identification of secreted proteins.
Spatially restricted & subcellular proteomics
Ting and coworkers first used a mutant ascorbate peroxidase (APEX) to selectively tag proteins localized to the mitochondrial matrix [45,46]. Unlike the cell-selective metabolic labeling methods just described, this method labels all proteins, including pre-existing proteins, within a subcellular volume. Chen et al. used this elegant strategy to characterize multiple cell types in Drosophila, including the mitochondrial matrix of muscle tissue [47]. The Weissman laboratory has combined the APEX labeling method with ribosome profiling to characterize localized protein synthesis in yeast [48,49]; extension of their method to cell-selective analysis is readily imagined.
Choosing a cell-selective proteomic method
The choice of a cell-selective method of proteomic analysis should reflect careful consideration of the advantages and disadvantages of each of the available approaches (Table 1).
Table.
Advantages and disadvantages of cell-specific proteomic methods.
| Cell- Specific Method |
Biomolecule identified |
Organisms demonstrated in? |
Temporal Resolution | Secreted? | PTM? | Advantages | Disadvantages | References | |
|---|---|---|---|---|---|---|---|---|---|
| Translatomics | TRAP | mRNA | Prokaryotes, eukaryotes |
Snapshot of translation | No | No | High sequence coverage, able to combine with ribosome profiling |
Requires expression of tagged ribosome, miss translational control |
[7–11] |
|
Cell Separation |
Manual | mRNA, Protein | Prokaryotes, eukaryotes, clinical samples |
Steady-state proteome | No | Yes | Straightforward, inexpensive | Possible artifacts from sample preparation, time and labor intensive |
[3], [12–13] |
| FACS | mRNA, Protein | Prokaryotes, eukaryotes, clinical samples |
Steady-state proteome | No | Yes | High-throughput | Requires dissociation of cells, need expression of transgene or recognizable epitope, need specialized equipment |
[14] | |
| Metabolic | CTAP | Protein | Cell culture | Up to 10 days continuous cell culture |
Yes | Yes | Quantitative, compatible with long-term cell culture |
Requires expression of Lyr/DDC, cells must be auxotrophic for lysine, restricted to cell culture |
[15], [17–19] |
| BONCAT | Protein | Prokaryotes, eukaryotes, cell culture |
Newly synthesized proteins in minutes (prokaryotic, cell culture) to days (whole animal) |
Yes | Yes | Commercially available reagents, high degree of temporal resolution |
Requires expression of synthetase; only Met/Phe residue replacement available currently, requires delivery of the ncAA |
[20–38], [44] | |
| SORT | Protein | Eukaryotes, cell culture |
Newly synthesized proteins in minutes (cell culture) to days (whole animal) |
Yes | Yes | Easy to change the residue of non-canonical amino acid incorporation, high degree of temporal resolution |
Requires expression of synthetase/tRNA pair, reagents not currently commercially available, requires delivery of the ncAA |
[39–41] | |
| OP-Puro | Protein | Cell culture | Newly synthesized proteins in minutes (cell culture) |
No | No | Not residue-dependent, highest degree of temporal resolution |
Requires expression of PGA, reagents not currently commercially available |
[42–44] | |
| Spatial | APEX | mRNA, Protein | Eukaryotes, cell culture |
Subcellular, steady- stateproteome |
No | Yes | High degree of spatial resolution (subcellular) |
Requires expression of APEX/HRP, no temporal resolution, requires delivery of peroxide + biotin-phenol reagent, possible toxicity of peroxide over longer timescales, needs oxidative environment |
[45–49] |
Physical sorting methods allow straightforward characterization of the steady-state proteome of the cell type of interest. However, removing cells from their natural environments prior to analysis raises concerns about artifacts, leads to limited temporal information, and sacrifices information about secreted proteins.
Ribosome profiling, when combined with cell-selective TRAP, provides significantly higher coverage of the gene expression profile than any direct proteomic measurement. But ribosome profiling is not a perfect proxy for protein synthesis and yields no information regarding protein secretion [50]. Moreover, only direct proteomic methods allow detection of post-translational modifications.
CTAP simplifies quantitative proteomic measurements for samples of relatively low complexity, but enrichment-based strategies (i.e., BONCAT, SORT or OP-Puro) are likely to be superior for short labeling times or for analysis of rare cells in complex tissues. Only APEX yields snapshots of the steady-state proteome with sub-cellular resolution. All cell-selective, enrichment-based experiments require the use of genetically tractable organisms.
Optimization of enrichment-based strategies requires careful consideration of alternative purification chemistries. Attachment to the resin used for purification can be accomplished either by direct covalent ligation or by a two-step process of affinity-tagging (e.g., with biotin reagents) and non-covalent binding (e.g., to streptavidin resins). Following appropriate washing steps, samples can be released from the resin by competitive binding, by proteolysis, or by selective cleavage of the affinity reagent. APEX appends biotin to surrounding molecules, so streptavidin-based resins are used to enrich for labeled proteins [46]. OP-Puro requires an azide-based affinity handle or resin for enrichment [43]. SORT uses cyclopropene labels and tetrazine linkers in a ligation reaction reported to be 100 to 1000 times faster than the strain-promoted azide-alkyne cycloaddition [41]. BONCAT labels with either alkynes or azides, and enriches with complementary azide or alkyne reagents. A special consideration arises in the analysis of lysates labeled with azides: Free thiols, which are known to react with cyclooctynes, must be blocked with capping reagents such as iodoacetamide or N-ethylmaleimide to avoid high background [34]. Many azide and alkyne resins and linkers are commercially available, and tetrazine-based reagents are beginning to appear on the market.
If the investigator wishes to identify the sites at which protein labeling has occurred, linkers with cleavable moieties can be used [51]. For many experiments, though, identification of labeling sites is not necessary, and on-bead digestion of enriched proteins is often simpler and more straightforward. In our hands, directly conjugating azide-labeled lysates to cyclooctyne resins has allowed us to identify larger numbers of relevant proteins [34]. Because enrichments are never perfect, running mock enrichments of unlabeled sample along with labeled samples provides a useful indication of background reactivity and non-specific protein contamination. Samples with abundant contaminating biopolymers such as pectin, serum proteins, or mucin may need an additional step to remove or degrade these contaminants and facilitate successful enrichment.
Conclusions and future outlook
Recent years have witnessed the introduction of powerful techniques that allow investigators to monitor protein synthesis with unprecedented resolution in space and time. Cell-specific proteomic analyses will play a key role in the identification of the mechanisms that govern cell specialization and that allow complex organisms to respond to changing environments.
Highlights.
Cell-selective proteomics is important in complex, heterocellular environments
Innovative chemical tools enable unbiased cell-type-specific interrogation of translation
Labeling methods including TRAP, CTAP, BONCAT, SORT, OP-Puro and APEX have been developed for cell-selective analysis
Sequencing and mass spectrometry-based strategies complement one other in the study of protein synthesis
The strengths and limitations of each analytical method must be considered carefully in the context of the biological question to be addressed
Acknowledgments
Funding: Caltech research on cell-specific proteomic analysis has been supported by NIH grants R01-GM062523 and R21-AI121890, and by the Institute for Collaborative Biotechnologies through grant W911NF-09-0001 from the U.S. Army Research Office.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Vogel C, Marcotte EM. Insights into the regulation of protein abundance from proteomic and transcriptomic analyses. Nat Rev Gen. 2012;13:227–232. doi: 10.1038/nrg3185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Maier T, Guell M, Serrano L. Correlation of mRNA and protein in complex biological samples. FEBS Lett. 2009;583:3966–3973. doi: 10.1016/j.febslet.2009.10.036. [DOI] [PubMed] [Google Scholar]
- 3.Kislinger T, Cox B, Kannan A, Chung C, Hu PZ, Ignatchenko A, Scott MS, Gramolini AO, Morris Q, Hallett MT, et al. Global survey of organ and organelle protein expression in mouse: Combined proteomic and transcriptomic profiling. Cell. 2006;125:173–186. doi: 10.1016/j.cell.2006.01.044. [DOI] [PubMed] [Google Scholar]
- 4.Zhang YY, Fonslow BR, Shan B, Baek MC, Yates JR. Protein Analysis by Shotgun/Bottom-up Proteomics. Chem Rev. 2013;113:2343–2394. doi: 10.1021/cr3003533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Aebersold R, Mann M. Mass-spectrometric exploration of proteome structure and function. Nature. 2016;537:347–355. doi: 10.1038/nature19949. [DOI] [PubMed] [Google Scholar]
- 6.Yuet KP, Tirrell DA. Chemical Tools for Temporally and Spatially Resolved Mass Spectrometry-Based Proteomics. Ann Biomed Eng. 2014;42:299–311. doi: 10.1007/s10439-013-0878-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ingolia NT. Ribosome Footprint Profiling of Translation throughout the Genome. Cell. 2016;165:22–33. doi: 10.1016/j.cell.2016.02.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sanz E, Yang L, Su T, Morris DR, McKnight GS, Amieux PS. Cell-type-specific isolation of ribosome-associated mRNA from complex tissues. Proc Natl Acad Sci. 2009;106:13939–13944. doi: 10.1073/pnas.0907143106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gonzalez C, Sims JS, Hornstein N, Mela A, Garcia F, Lei L, Gass DA, Amendolara B, Bruce JN, Canoll P, et al. Ribosome Profiling Reveals a Cell-Type-Specific Translational Landscape in Brain Tumors. J Neurosci. 2014;34:10924–10936. doi: 10.1523/JNEUROSCI.0084-14.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Handley A, Schauer T, Ladurner AG, Margulies CE. Designing Cell-Type-Specific Genome-wide Experiments. Mol Cell. 2015;58:621–631. doi: 10.1016/j.molcel.2015.04.024. •This review delineates recent advances in cell-type-specific genomic methods, and thoughtfully recommends experimental approaches and controls.
- 11.Okaty BW, Sugino K, Nelson SB. Cell Type-Specific Transcriptomics in the Brain. J Neurosci. 2011;31:6939–6943. doi: 10.1523/JNEUROSCI.0626-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sharma K, Schmitt S, Bergner CG, Tyanova S, Kannaiyan N, Manrique-Hoyos N, Kongi K, Cantuti L, Hanisch UK, Philips MA, et al. Cell type- and brain region-resolved mouse brain proteome. Nat Neuro. 2015;18:1819–1831. doi: 10.1038/nn.4160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Azimifar SB, Nagaraj N, Cox J, Mann M. Cell-Type-Resolved Quantitative Proteomics of Murine Liver. Cell Metab. 2014;20:1076–1087. doi: 10.1016/j.cmet.2014.11.002. [DOI] [PubMed] [Google Scholar]
- 14.Claudi B, Sprote P, Chirkova A, Personnic N, Zankl J, Schurmann N, Schmidt A, Bumann D. Phenotypic Variation of Salmonella in Host Tissues Delays Eradication by Antimicrobial Chemotherapy. Cell. 2014;158:722–733. doi: 10.1016/j.cell.2014.06.045. [DOI] [PubMed] [Google Scholar]
- 15.Gauthier NP, Soufi B, Walkowicz WE, Pedicord VA, Mavrakis KJ, Macek B, Gin DY, Sander C, Millerephrin ML. Cell-selective labeling using amino acid precursors for proteomic studies of multicellular environments. Nat Meth. 2013;10:768-+. doi: 10.1038/nmeth.2529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tian L, Yang YL, Wysocki LM, Arnold AC, Hu A, Ravichandran B, Sternson SM, Looger LL, Lavis LD. Selective esterase-ester pair for targeting small molecules with cellular specificity. Proc Natl Acad Sci. 2012;109:4756–4761. doi: 10.1073/pnas.1111943109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tape CJ, Norrie IC, Worboys JD, Lim L, Lauffenburger DA, Jorgensen C. Cell-specific Labeling Enzymes for Analysis of Cell-Cell Communication in Continuous Co-culture. Mol Cell Prot. 2014;13:1866–1876. doi: 10.1074/mcp.O113.037119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Tape CJ, Ling S, Dimitriadi M, McMahon KM, Worboys JD, Leong HS, Norrie IC, Miller CJ, Poulogiannis G, Lauffenburger DA, et al. Oncogenic KRAS Regulates Tumor Cell Signaling via Stromal Reciprocation. Cell. 2016;165:910–920. doi: 10.1016/j.cell.2016.03.029. •The authors combine cell-specific proteomic labeling (CTAP) and multivariate phosphoproteomics to study pancreatic ductal adenocarcinomas in heterocellular contexts. This study reveals reciprocal KRASG12D signaling, which would have gone undetected if the carcinomas were studied in isolation.
- 19.Tape CJ. Systems Biology Analysis of Heterocellular Signaling. Trends in Biotech. 2016;34:627–637. doi: 10.1016/j.tibtech.2016.02.016. [DOI] [PubMed] [Google Scholar]
- 20.Dieterich DC, Link AJ, Graumann J, Tirrell DA, Schuman EM. Selective identification of newly synthesized proteins in mammalian cells using bioorthogonal noncanonical amino acid tagging (BONCAT) Proc Natl Acad Sci. 2006;103:9482–9487. doi: 10.1073/pnas.0601637103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dieterich DC, Lee JJ, Link AJ, Graumann J, Tirrell DA, Schuman EM. Labeling, detection and identification of newly synthesized proteomes with bioorthogonal non-canonical amino-acid tagging. Nat Prot. 2007;2:532–540. doi: 10.1038/nprot.2007.52. [DOI] [PubMed] [Google Scholar]
- 22.Bagert JD, van Kessel JC, Sweredoski MJ, Feng L, Hess S, Bassler BL, Tirrell DA. Time-resolved proteomic analysis of quorum sensing in Vibrio harveyi. Chem Sci. 2015;7:1797–1806. doi: 10.1039/c5sc03340c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Feng L, Rutherford ST, Papenfort K, Bagert JD, van Kessel JC, Tirrell DA, Wingreen NS, Bassler BL. A qrr noncoding RNA deploys four different regulatory mechanisms to optimize quorum-sensing dynamics. Cell. 2015;160:228–240. doi: 10.1016/j.cell.2014.11.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kramer G, Sprenger RR, Back J, Dekker HL, Nessen MA, van Maarseveen JH, de Koning LJ, Hellingwerf KJ, de Jong L, de Koster CG. Identification and quantitation of newly synthesized proteins in Escherichia coli by enrichment of azidohomoalanine-labeled peptides with diagonal chromatography. Mol Cell Prot. 2009;8:1599–1611. doi: 10.1074/mcp.M800392-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sinai L, Rosenberg A, Smith Y, Segev E, Ben-Yehuda S. The molecular timeline of a reviving bacterial spore. Mol Cell. 2015;57:695–707. doi: 10.1016/j.molcel.2014.12.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hatzenpichler R, Connon SA, Goudeau D, Malmstrom RR, Woyke T, Orphan VJ. Visualizing in situ translational activity for identifying and sorting slow-growing archaeal-bacterial consortia. Proc Natl Acad Sci. 2016;113:E4069–E4078. doi: 10.1073/pnas.1603757113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Howden AJ, Geoghegan V, Katsch K, Efstathiou G, Bhushan B, Boutureira O, Thomas B, Trudgian DC, Kessler BM, Dieterich DC, et al. QuaNCAT: quantitating proteome dynamics in primary cells. Nat Meth. 2013;10:343–346. doi: 10.1038/nmeth.2401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Eichelbaum K, Winter M, Diaz MB, Herzig S, Krijgsveld J. Selective enrichment of newly synthesized proteins for quantitative secretome analysis. Nat Biotech. 2012;30:984-+. doi: 10.1038/nbt.2356. [DOI] [PubMed] [Google Scholar]
- 29.Bagert JD, Xie YSJ, Sweredoski MJ, Qi YT, Hess S, Schuman EM, Tirrell DA. Quantitative, Time-Resolved Proteomic Analysis by Combining Bioorthogonal Noncanonical Amino Acid Tagging and Pulsed Stable Isotope Labeling by Amino Acids in Cell Culture. Mol Cell Prot. 2014;13:1352–1358. doi: 10.1074/mcp.M113.031914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Calve S. Incorporation of non-canonical amino acids into the developing murine proteome. Sci Rep. 2016 doi: 10.1038/srep32377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ngo JT, Champion JA, Mahdavi A, Tanrikulu IC, Beatty KE, Connor RE, Yoo TH, Dieterich DC, Schuman EM, Tirrell DA. Cell-selective metabolic labeling of proteins. Nat Chem Biol. 2009;5:715–717. doi: 10.1038/nchembio.200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Grammel M, Zhang MZM, Hang HC. Orthogonal Alkynyl Amino Acid Reporter for Selective Labeling of Bacterial Proteomes during Infection. Angew Chem. 2010;49:5970–5974. doi: 10.1002/anie.201002050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mahdavi A, Szychowski J, Ngo JT, Sweredoski MJ, Graham RLJ, Hess S, Schneewind O, Mazmanian SK, Tirrell DA. Identification of secreted bacterial proteins by noncanonical amino acid tagging. Proc Natl Acad Sci. 2014;111:433–438. doi: 10.1073/pnas.1301740111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Yuet KP, Doma MK, Ngo JT, Sweredoski MJ, Graham RLJ, Moradian A, Hess S, Schuman EM, Sternberg PW, Tirrell DA. Cell-specific proteomic analysis in Caenorhabditis elegans. Proc Natl Acad Sci. 2015;112:2705–2710. doi: 10.1073/pnas.1421567112. •The authors present a mutant PheRS to label and visualize proteins expressed in various cell types in the nematode Caenorhabitis elegans. The authors combine BONCAT and SILAC to enrich, identify and quantify proteins produced in the 20 pharyngeal muscle cells of C. elegans.
- 35. Erdmann I, Marter K, Kobler O, Niehues S, Abele J, Muller A, Bussmann J, Storkebaum E, Ziv T, Thomas U, et al. Cell-selective labelling of proteomes in Drosophila melanogaster. Nat Comm. 2015;6 doi: 10.1038/ncomms8521. •This study utilizes a mutant MetRS to perform cell-type-specific BONCAT analysis in Drosophila.
- 36.Mahdavi A, Hamblin GD, Jindal GA, Bagert JD, Dong C, Sweredoski MJ, Hess S, Schuman EM, Tirrell DA. Engineered Aminoacyl-tRNA Synthetase for Cell-Selective Analysis of Mammalian Protein Synthesis. J Am Chem Soc. 2016;138:4278–4281. doi: 10.1021/jacs.5b08980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Muller A, Stellmacher A, Freitag CE, Landgraf P, Dieterich DC. Monitoring Astrocytic Proteome Dynamics by Cell Type-Specific Protein Labeling. PLoS One. 2015;10 doi: 10.1371/journal.pone.0145451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mahdavi A, Segall-Shapiro TH, Kou SZ, Jindal GA, Hoff KG, Liu S, Chitsaz M, Ismagilov RF, Silberg JJ, Tirrell DA. A Genetically Encoded AND Gate for Cell-Targeted Metabolic Labeling of Proteins. J Am Chem Soc. 2013;135:2979–2982. doi: 10.1021/ja400448f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Elliott TS, Townsley FM, Bianco A, Ernst RJ, Sachdeva A, Elsasser SJ, Davis L, Lang K, Pisa R, Greiss S, et al. Proteome labeling and protein identification in specific tissues and at specific developmental stages in an animal. Nat Biotech. 2014;32:465-U186. doi: 10.1038/nbt.2860. This study introduces the use of SORT to label, image and identify proteins from germ cells in Drosophila melanogaster ovaries.
- 40.Elliott TS, Bianco A, Chin JW. Genetic code expansion and bioorthogonal labelling enables cell specific proteomics in an animal. Curr Opin Chem Biol. 2014;21:154–160. doi: 10.1016/j.cbpa.2014.07.001. [DOI] [PubMed] [Google Scholar]
- 41.Elliott TS, Bianco A, Townsley FM, Fried SD, Chin JW. Tagging and Enriching Proteins Enables Cell-Specific Proteomics. Cell Chem Biol. 2016;23:805–815. doi: 10.1016/j.chembiol.2016.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Liu J, Xu YQ, Stoleru D, Salic A. Imaging protein synthesis in cells and tissues with an alkyne analog of puromycin. Proc Natl Acad Sci. 2012;109:413–418. doi: 10.1073/pnas.1111561108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Barrett RM, Liu HW, Jin HH, Goodman RH, Cohen MS. Cell-specific Profiling of Nascent Proteomes Using Orthogonal Enzyme-mediated Puromycin Incorporation. ACS Chem Biol. 2016;11:1532–1536. doi: 10.1021/acschembio.5b01076. •This study describes cell-type-specific metabolic labeling by O-propargyl-puromycin.
- 44.Dieck ST, Kochen L, Hanus C, Heumueller M, Bartnik I, Nassim-Assir B, Merk K, Mosler T, Garg S, Bunse S, et al. Direct visualization of newly synthesized target proteins in situ. Nat Meth. 2015;12:411-+. doi: 10.1038/nmeth.3319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rhee HW, Zou P, Udeshi ND, Martell JD, Mootha VK, Carr SA, Ting AY. Proteomic Mapping of Mitochondria in Living Cells via Spatially Restricted Enzymatic Tagging. Science. 2013;339:1328–1331. doi: 10.1126/science.1230593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hung V, Udeshi ND, Lam SS, Loh KH, Cox KJ, Pedram K, Carr SA, Ting AY. Spatially resolved proteomic mapping in living cells with the engineered peroxidase APEX2. Nat Prot. 2016;11:456–475. doi: 10.1038/nprot.2016.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Chen CL, Hu YH, Udeshi ND, Lau TY, Wirtz-Peitz F, He L, Ting AY, Carr SA, Perrimon N. Proteomic mapping in live Drosophila tissues using an engineered ascorbate peroxidase. Proc Natl Acad Sci. 2015;112:12093–12098. doi: 10.1073/pnas.1515623112. •This study reports the development of APEX, an engineered ascorbate peroxidase that biotinylates proximal proteins. The authors further report a database that inventories mitochondrial proteins annotated at the sub-compartment level.
- 48.Jan CH, Williams CC, Weissman JS. Principles of ER cotranslational translocation revealed by proximity-specific ribosome profiling. Science. 2014;346:716-+. doi: 10.1126/science.1257521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Williams CC, Jan CH, Weissman JS. Targeting and plasticity of mitochondrial proteins revealed by proximity-specific ribosome profiling. Science. 2014;346:748–751. doi: 10.1126/science.1257522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tian RJ. Exploring intercellular signaling by proteomic approaches. Proteomics. 2014;14:498–512. doi: 10.1002/pmic.201300259. [DOI] [PubMed] [Google Scholar]
- 51.Szychowski J, Mahdavi A, Hodas JJL, Bagert JD, Ngo JT, Landgraf P, Dieterich DC, Schuman EM, Tirrell DA. Cleavable Biotin Probes for Labeling of Biomolecules via Azide-Alkyne Cycloaddition. J Am Chem Soc. 2010;132:18351–18360. doi: 10.1021/ja1083909. [DOI] [PMC free article] [PubMed] [Google Scholar]