Abstract
APOBEC3G (A3G) is a cytidine deaminase with potent antiviral activity that is antagonized by Vif. A3G is expressed in a cell type-specific manner and some semi-permissive cells, including A3.01, express A3G but fail to block replication of Vif-null HIV-1. Here we explored the semi-permissive nature of A3.01 cells and found it to be defined exclusively by the levels of A3G. Indeed, minor changes in A3G levels rendered A3.01 cells either fully permissive or non-permissive for Vif-null HIV-1. Our data indicate that A3.01 cells express sub-lethal levels of catalytically active A3G that affects Vif-null HIV-1 at the proviral level but does not completely block virus replication due to purifying selection. Attempts to use the selective pressure exerted by such sub-lethal levels of A3G to select for APOBEC-resistant Vif-null virus capable of replicating in H9 cells failed despite passaging virus for five months, demonstrating that Vif is a critical viral accessory protein.
Keywords: APOBEC3G, cytidine deaminase, hypermutation, HIV-1, Vif, antiviral resistance, deamination, A3.01 cells, viral evolution, purifying selection
1. Introduction
The viral infectivity factor (Vif) is an HIV accessory protein that is critical for viral replication in vivo. It primarily antagonizes the antiviral activity of APOBEC3G (A3G) (Sheehy et al., 2002). A3G is a cytidine deaminase that is packaged into retroviral particles where it can mutagenize the viral genome during reverse transcription (reviewed in (Goila-Gaur and Strebel, 2008)). Vif inhibits the packaging of A3G into progeny virions at least in part by inducing proteasomal degradation of the deaminase (Conticello et al., 2003; Kao et al., 2003; Kao et al., 2004; Marin et al., 2003; Mehle et al., 2004; Sheehy et al., 2003; Stopak et al., 2003; Yu et al., 2003). A3G is not ubiquitously expressed in all cell lines and Vif-dependence of HIV-1 replication is therefore, at least in vitro, cell line-dependent. Based on the level of restriction of Vif-null HIV-1, cell types are categorized as non-permissive (e.g. PBMC, macrophages, H9, MT2), semi-permissive (e.g. A3.01, CEMx174), or permissive (e.g. Jurkat, CEM-SS, SupT1) (Borman et al., 1995; Gabuzda et al., 1992; Hoglund et al., 1994; Ma et al., 1994; Sakai et al., 1993).
Previous reports indicate that expression of A3G in vivo can vary in a donor-specific manner (Cho et al., 2006; Jin et al., 2005). Also, A3F, A3DE (also referred to as A3D), and A3H have been shown to affect HIV-1 replication in a Vif-sensitive manner (Chaipan et al., 2013; Dang et al., 2006; Li et al., 2010; OhAinle et al., 2006; Wiegand et al., 2004; Zhen et al., 2010; Zheng et al., 2004). It is therefore conceivable that variation in their expression contributes to the non-permissive or semi-permissive phenotype of the host cells. The identification of natural Vif variants with reduced A3G antagonizing potency could be an indication of donor- or tissue-specific variations in the expression of A3G and other cytidine deaminases (Binka et al., 2012; Fourati et al., 2010). Nevertheless, as far as cell line-specific differences in Vif dependence observed in tissue culture are concerned, it is currently not clear whether they are due to differences in the relative expression of A3G, differential expression of additional cytidine deaminases, or a combination of both. While natural Vif variants can differ in their ability to target A3G, A3F, or A3H (Kataropoulou et al., 2009; Peng et al., 2013; Porcellini et al., 2009; Simon et al., 2005; Vallanti et al., 2005), there are no known primary replication competent viruses that completely lack expression of a Vif protein. This suggests that Vif-null viruses are replication incompetent in vivo making Vif an interesting target for antiviral therapy. Yet, there are currently no drugs in clinical use that specifically target Vif.
Here, we studied replication of Vif-null HIV-1 NL4-3 in A3.01 cells to understand in more detail the reasons for the semi-permissive phenotype of these cells. Among possible contributing factors we explored (i) heterogeneous expression of A3G (i.e. mixed population), (ii) polymorphisms in the A3G gene potentially affecting its catalytic activity, and (iii) differences in cellular expression and packaging of A3G into progeny virions. We found that A3.01 cells represent a homogeneous population with virtually all of the cells expressing A3G. Furthermore, sequence comparison of A3G expressed in A3.01 cells and H9 cells failed to reveal sequence polymorphisms. In contrast, we found that the cellular expression of A3G protein in A3.01 cells was somewhat lower than in H9 cells, and progeny virions produced in A3.01 cells contained approximately 1/3 of the A3G packaged into virus produced from H9 cells. To understand the impact of these differences on HIV-1 replication we either reduced A3G expression in A3.01 cells by shRNA-mediated gene silencing or increased A3G production by transduction of cells with an A3G-expression vector. Interestingly, silencing of A3G rendered A3.01 cells fully permissive for Vif-null HIV-1 suggesting that the semi-permissive nature of A3.01 cells is primarily, if not exclusively, associated with A3G expression. Importantly, increasing the levels of A3G in A3.01 cells to levels similar to those in H9 cells rendered the cells fully non-permissive. Our results indicate that A3.01 cells express sub-lethal levels of A3G that cause mutation of proviral sequences but allow the virus to survive by a mechanism of purifying selection. Of note, small changes to the levels of A3G in A3.01 cells were sufficient to change their phenotype from semi-permissive to either fully permissive or fully restrictive. Moreover, even though A3.01 cells have been reported to express A3F and A3DE (Dang et al., 2006; Rose et al., 2005), our data suggest that these deaminases contribute little if anything to the Vif-restrictive phenotype of these cells. Finally, taking advantage of the selective pressure exerted by A3.01 cells, we tried to select APOBEC-resistant variants of Vif-null HIV-1. However, despite passaging virus for a total of 5 months, initially in A3.01 cells and then in mixed populations of A3.01 and H9 cells, we were unable to passage NL4-3 Vif-null into H9 cells. We conclude that Vif-null HIV-1 can survive sub-lethal levels of A3G. However, in cells that exceed a threshold level of A3G or potentially encode additional Vif-sensitive deaminases, virus replication remains strictly Vif-dependent.
2. Results
2.1 Vif is required for virus replication in CD4+ T cell lines in a cell line-dependent manner
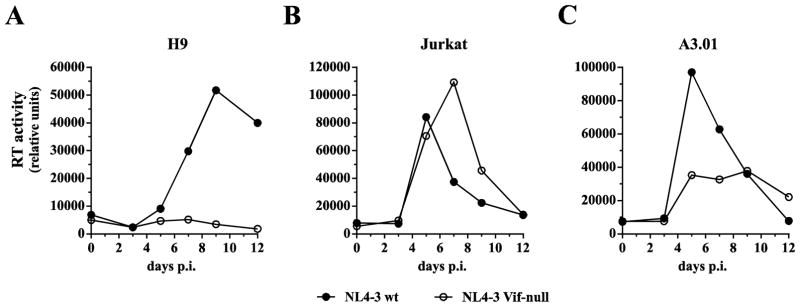
As mentioned in the introduction, the requirement of Vif for HIV-1 replication, at least in vitro, is cell line-dependent. This is illustrated in Fig. 1. In A3G-positive H9 cells, Vif-null virus is unable to establish a spreading infection (Fig. 1A) whereas in A3G-negative Jurkat cells, replication of Vif-null HIV-1 is not significantly impaired (Fig. 1B). Finally, A3.01 cells efficiently support replication of wild type virus; however, replication of Vif-null virus is less efficient and is characterized by lower levels of peak virus production and a more drawn-out replication profile (Fig. 1C). This indicates that A3.01 cells express levels of restrictive deaminases that diminish but do not completely block replication of Vif-null HIV-1.
Fig. 1.
Comparison of HIV-1 replication in H9, Jurkat, and A3.01 cells. Wild type NL4-3 or NL4-3 Vif-null virus stocks were produced from transiently transfected 293T cells and used to infect non-permissive H9 cells (A), permissive Jurkat cells (B), or semi-permissive A3.01 cells (C). Virus replication was monitored for 12 days by measuring the virus-associated reverse transcriptase activity (Willey et al., 1988). Results were plotted as a function of time. Shown is a representative experiment. Similar analyses were performed multiple times (n > 20) with similar results in the context of other unrelated studies.
2.2 H9 and A3.01 cells have similar A3G characteristics
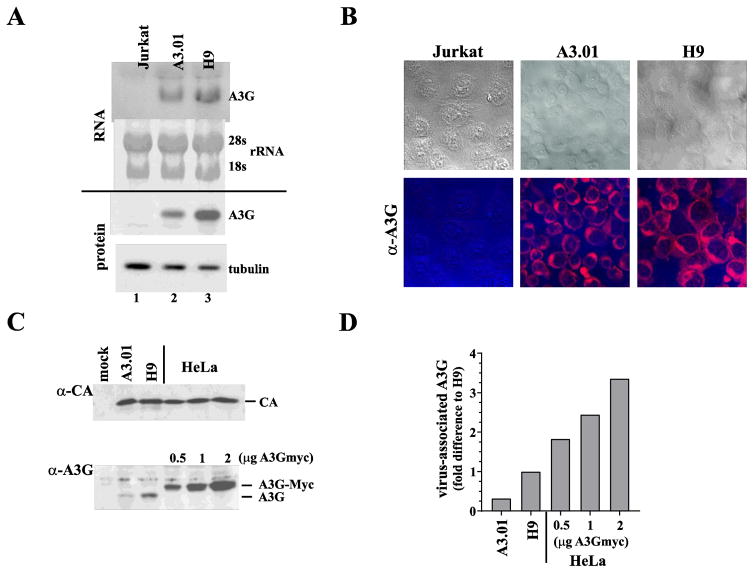
The partial restriction of Vif-null HIV-1 in A3.01 cells could be due to the expression of lower levels of A3G in these cells. Alternatively, A3.01 cells could represent a mixed population of A3G positive and negative cells or could express a polymorphic A3G allele with reduced antiviral potency. To better understand why A3.01 cells exhibit a semi-permissive phenotype for HIV-1 replication, we first compared the relative expression of A3G in Jurkat, A3.01 and H9 cells at the mRNA as well as at the protein level (Fig. 2A). RNA and protein extracts were prepared as described in the Materials and Methods section and subjected to northern blotting (Fig. 2A, top panel) and immunoblotting (Fig. 2A, lower panel), respectively. As expected, Jurkat cells did not express detectable levels of A3G mRNA or A3G protein (Fig. 2A, lane 1) whereas A3.01 and H9 cells both expressed A3G mRNA as well as A3G protein (Fig. 2A, lanes 2 & 3). Of note, A3G mRNA and protein levels were about 2-fold lower in A3.01 cells when compared to those of H9 cells. Levels of A3G protein in activated primary CD4+ T cells were comparable to those in H9 cells (data not shown).
Fig. 2.
H9 and A3.01 cells have similar A3G characteristics. (A) Expression of endogenous A3G mRNA in Jurkat, A3.01, and H9 cells was determined by northern blot analysis (RNA). Twenty μg total RNA were loaded in each lane. To control for total RNA in the samples, ribosomal RNAs (28s and 18s) were visualized on the membrane by methylene blue staining before probing with a [32P]-labeled A3G-specific DNA probe. Results were quantified by phosphor-image analysis. A3G protein expression was determined by immunoblotting using an A3G-specific antibody (protein). To control for total protein in the samples, expression of α-tubulin was assessed by reprobing the A3G blot with a tubulin-specific monoclonal antibody. Protein gels were quantified by densitometry. (B) Expression of A3G in Jurkat, A3.01, and H9 cells as determined by immunocytochemistry. Cells were immobilized on glass slides by cytocentrifugation and fixed in methanol. Cells were stained with an antibody to A3G and visualized using a Texas Red-conjugated secondary antibody. Images were acquired by confocal microscopy (lower panels). Corresponding bright field images are shown at the top. (C) Comparison of virus-associated A3G. VSV G-pseudotyped stocks of NL4-3 Vif-null virus were used to infect A3.01 or H9 cells. Medium was replaced at 5 h and 12 h after infection and virus produced between 12 and 24 h post-infection was harvested, filtered, and concentrated by pelleting through a 4 ml cushion of 20% sucrose. Concentrated virus was quantified by reverse transcriptase assay and equal reverse transcriptase units were loaded on two 12.5% gels. Viruses produced from transfected HeLa cells in the presence of increasing amounts of A3Gmyc vector (0.5, 1, 2 μg) were analyzed in parallel. Proteins were transferred to PVDF membranes and probed with a capsid-specific monoclonal antibody (α-CA) or a polyclonal antibody to A3G (α-A3G). Proteins were visualized by ECL using HRP-conjugated second antibodies. (D) Signals from the immunoblots in panel C were quantified and relative amounts of A3G in the virus samples were determined by normalizing for equal CA signals. The A3G signal obtained from H9 derived virus was defined as 1. A3G signals for the other viruses were calculated as fold-difference relative to the H9 virus population.
While the lower expression of A3G in A3.01 cells could explain at least in part the semi-permissive nature of these cells, we wanted to rule out possible A3G polymorphisms as a contributing factor. To do so, we cloned A3G mRNA from H9 and A3.01 cells. Sequence comparison of the resulting cDNA clones (2 H9 clones; 2 A3.01 clones) revealed that the A3G sequences in H9- and A3.01 cells were identical to the published CEM-derived A3G sequence (Genbank accession NM_021822).
Next, we performed immunocytochemistry to control for the possibility that A3.01 cells represent a mixed population of A3G-positive and A3G-negative cells. Such a situation could explain the lower levels of A3G relative to H9 cells detected in panel A and account for the delayed replication phenotype. To determine the fraction of A3G-positive cells in each cell type, uninfected A3.01 cells - and for comparison H9 cells and Jurkat cells- were stained for A3G as described in Materials and Methods and analyzed by confocal microscopy. We found that 100% of the A3.01 and H9 cell populations expressed A3G. As expected, Jurkat cells were negative for A3G staining. A3G in both H9 and A3.01 cells was identified primarily in the cytoplasm (Fig. 2B). From these results, we conclude that the semi-permissive phenotype of A3.01 cells is not due to mixed cell populations or sequence polymorphisms.
A previous study assessing the stoichiometry of A3G in Vif-deficient HIV-1 produced in PBMC concluded that only 7 +/− 4 molecules of A3G are packaged into primary T lymphocytes in the absence of Vif indicating that very few molecules of A3G are sufficient to effectively inactivate the virus (Xu et al., 2007). Even packaging of as little as 1–2 molecules of A3G was reported to significantly reduce viral infectivity (Browne et al., 2009). To investigate if the semi-permissive nature of A3.01 cells is correlated with reduced packaging of A3G, we compared the amounts of endogenous A3G packaged from H9 and A3.01 cells into Vif-null viruses. This was done by infecting 1x107 A3.01 and H9 cells at an MOI of ~1 with VSV G-pseudotyped NL4-3 Vif-null produced in A3G-negative HeLa cells. Residual input virus was removed by replacing the culture medium twice: first 5 h after infection and again 12 h post infection. Newly synthesized virus was then harvested 24 h post-infection, filtered, and concentrated by pelleting through a 4 ml cushion of 20% sucrose. To test for possible limitations as to how much A3G can be packaged into HIV-1 virions, packaging of exogenously expressed A3G into Vif-null virions was analyzed in parallel. For that purpose, HeLa cells were transfected with 4 μg of NL4-3 Vif-null DNA together with varying amounts (0.5, 1, 2 μg) of pcDNA-A3Gmyc. Virus-containing supernatants were harvested 24 h later and concentrated through a 20% sucrose cushion. Viral extracts were separated by SDS-PAGE. Gag and A3G were visualized by immunoblotting using antibodies to HIV-1 CA or A3G (Fig. 2C). Protein bands were quantified by densitometry and the relative amounts of A3G in the virus preparations was calculated based on the immunoblot signals taking into consideration variations in the relative capsid levels (Fig. 2D). Our results indicate that Vif-null virions produced from H9 cells contained about 3-times more A3G molecules than viruses produced from A3.01 cells. Cotransfection of A3G and NL4-3 Vif-null into HeLa cells resulted in the packaging of significantly higher levels of A3G per virion indicating that there is no defined upper limit for packaging of A3G. Rather, packaging of A3G into Vif-null virions appeared to be directly proportional to cellular expression levels.
2.3 Increasing A3G levels in A3.01 cells induces a non-permissive phenotype
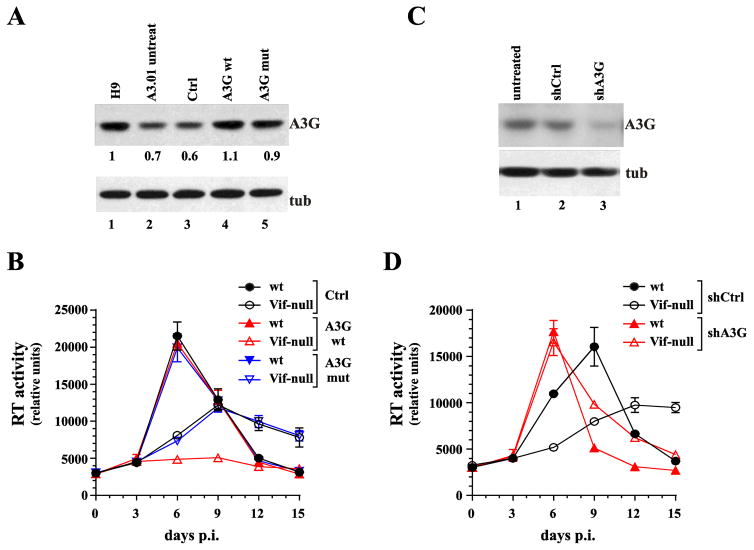
We have ruled out allelic variation or mixed population as cause for the semi-permissive nature of A3.01 cells, leaving the relatively modest difference in cellular A3G expression and virion association observed for A3.01 and H9 cells as the only measurable difference. It is conceivable that there is an upper limit to levels of A3G that can be tolerated by Vif-null HIV-1 without suffering lethal mutagenesis. To test this hypothesis, we transduced A3.01 cells with a lentiviral construct expressing untagged wild type A3G (A3G wt) to increase the total cellular levels of catalytically active A3G. As a control we transduced A3.01 cells with an empty vector (Ctrl) or a vector expressing a deaminase-defective A3G construct (A3G mut). After puromycin selection of the transduced cells, A3G protein levels were determined by immunoblotting (Fig. 3A). As expected, transduction of A3.01 cells with an empty vector had no effect on cellular A3G expression (Fig. 3A, compare lanes 2 & 3). In contrast, transduction of A3.01 cells with vectors expressing A3G wt or A3G mut resulted in a slight increase in the total levels of A3G (Fig. 3A, lanes 4 & 5). Levels of A3G in transduced A3.01 cells were comparable to those in untreated H9 cells (Fig. 3A, compare lanes 1, 4, 5). We next infected these cells with NL4-3 wt or NL4-3 Vif-null and determined the replication profiles over a 15-day period (Fig. 3B). As expected, NL4-3 wt replicated with identical kinetics in all three cell lines, irrespective of whether they had been transduced with empty vector or vector encoding deaminase-defective (A3G mut) or wild type A3G (A3G wt) (Fig. 3B, solid symbols). Furthermore, cells transduced with either empty vector or the deaminase-defective A3G supported replication of NL4-3 Vif-null with delayed kinetics typical for the semi-permissive phenotype of A3.01 cells (Fig. 3B, black and blue open symbols). In contrast, NL4-3 Vif-null was unable to replicate in cells transduced with A3G wt (Fig. 3B, red open triangles). Thus, the subtle increase in A3G expression in cells transduced with A3G wt was sufficient to render these cells non-permissive to Vif-null HIV-1.
Fig. 3.
Increasing A3G levels in A3.01 cells induces a non-permissive phenotype whereas silencing of A3G produces a fully permissive phenotype. (A) A3.01 cells were transduced with lentiviral vectors encoding untagged wild type A3G (lane 4) or a deaminase-defective mutant (A3G C288S, C291A) (lane 5). Cells transduced with empty vector were analyzed in parallel (lane 3). Untreated H9 cells (lane 1) and A3.01 cells (lane 2) were analyzed in parallel. After transduction, cells were cultured for 48 h prior to puromycin selection. Puromycin-resistant A3.01 cells were amplified and A3G expression was determined by immunoblotting using untreated parental A3.01 cells and endogenous tubulin (tub) as reference. Protein gels were quantified by densitometry. (B) Transduced A3.01 cells were infected with wild type and Vif-null virus and replication profiles were determined by measuring supernatant reverse transcriptase levels over a 15-day period. Data are presented as mean +/− SD calculated from triplicate infections. (C) A3G was knocked down by transducing A3.01 cells with A3G shRNA encoding lentiviral vectors (lane 3). As a control, cells were treated with a vector encoding a non-silencing shRNA (lane 2). Puromycin-resistant cells were selected as described for panel A. Successful knockdown of A3G was verified by immunoblotting using an A3G specific polyclonal antibody. Parental A3.01 cells (untreated) served as specificity control (lane 1). The blot was reprobed with antibodies to α-tubulin to control for sample loading. (D) Puromycin-resistant knock-down cells and control cells were infected with wild type or NL4-3 Vif-null and replication profiles were determined as in panel B. Data are presented as mean +/− SD calculated from triplicate infections.
2.4 Silencing A3G in A3.01 cells induces a fully permissive phenotype
Aside from A3G, other members of the APOBEC3 family of cytidine deaminases, in particular A3F and A3DE, have been reported to also inhibit HIV-1 in a Vif-dependent manner (reviewed in (Goila-Gaur and Strebel, 2008)). However, the in vivo importance of A3F and A3DE to HIV-1 restriction remains unclear (Bishop et al., 2006; Dang et al., 2006; Holmes et al., 2007; Luo et al., 2007; Miyagi et al., 2010; Zennou and Bieniasz, 2006). A3F and A3DE mRNA are both expressed at comparable levels in H9 and A3.01 cells (Dang et al., 2006; Rose et al., 2005). To assess the possible contribution of these deaminases to the semi-permissive phenotype of A3.01 cells, we silenced endogenous A3G in A3.01 cells via transduction of A3G shRNA (shA3G). Cells transduced with a non-silencing shRNA (shCtrl) were used as a control. Following puromycin selection, successful silencing of A3G was monitored by immunoblotting using non-transduced A3.01 cells (untreated) as reference (Fig. 3C). Cells were infected with NL4-3 wt or NL4-3 Vif-null and the viral replication profiles were monitored over a 15-day period (Fig. 3D). As expected, treatment of A3.01 cells with a non-silencing shRNA had no impact on the semi-permissive phenotype of the transduced cells (Fig. 3D, black symbols). In contrast, shRNA silencing of A3G rendered the A3.01 cells fully permissive resulting in virtually identical replication profiles for wild type and NL4-3 Vif-null (Fig. 3D, red symbols). Indeed, virus production for both wild type and Vif-null viruses peaked several days earlier than wild type virus in the control cells suggesting that A3G imposes a selective pressure even on wild type virus. We conclude that A3G is largely, if not entirely, responsible for the semi-permissive phenotype of A3.01 cells suggesting that A3F and A3DE do not noticeably contribute to the restriction of HIV-1 in A3.01 cells.
2.5 Purifying selection limits the accumulation of A3G-induced mutations in viral RNA of Vif-null HIV-1 produced in A3.01 cells
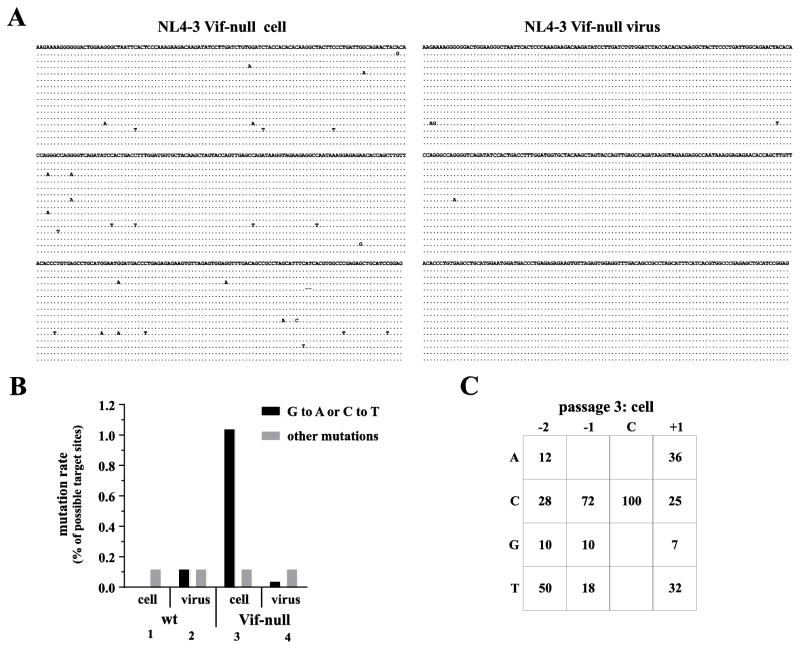
The delayed replication kinetics of NL4-3 Vif-null in A3.01 cells is likely caused by an A3G-imposed selective pressure that reduces viral fitness through sub-lethal mutagenesis. Indeed, the phenomenon of purifying selection of naturally occurring Vif variants was previously demonstrated in semi-permissive CEM cells (Russell et al., 2009). Our next aim was to assess the extent of A3G-induced hypermutation of Vif-null HIV-1 genomes during replication in A3.01 cells. Wild type NL4-3 and NL4-3 Vif-null virus stocks were produced in 293T cells and used to infect A3.01 cells. Infected cells and virus-containing supernatants were harvested at peak virus production. Wild type and Vif-null virus from this first passage was normalized for equal RT activity and re-passaged twice onto fresh A3.01 cultures. In each case, infected cells and virus-containing supernatants were harvested at peak infection. Extracts from infected cells from passage 3 were used for PCR amplification of proviral DNA using a 3′-LTR specific primer set. Resulting products were analyzed not only for G to A changes, reflecting cytidine deamination on minus strand cDNA, but also for C to T changes which can result from the fact that the U3 region of the LTR becomes transiently single stranded during plus-strand synthesis (Navarro and Landau, 2004). Furthermore, viral genomic RNA extracted from culture supernatants at peak of infection was subjected to one-step RT-PCR using the same 3′-LTR-specific primers. Fifteen clones each were sequenced to assess APOBEC-induced hypermutation of the proviral or viral genomes (Fig. 4A; only sequences from NL4-3 Vif-null-infected cultures are shown). As expected, passaging wild-type NL4-3 did not result in the accumulation of APOBEC-induced mutations (Fig. 4B, columns 1 & 2). In contrast, Vif-null proviral DNA revealed significant levels of A3G-induced mutations (Fig. 4B, column 3). Indeed, > 1% of all G or C residues of the 15 clones analyzed had been mutated. Interestingly, analysis of viral genomic RNA revealed noticeably lower levels of A3G-induced mutations (< 0.1%) (Fig. 4B, column 4). These results indicate that replication of Vif-null HIV-1 in A3.01 cells results in the accumulation of mutated integrated proviral genomes while replicating virus accumulates only few A3G-induced mutations. We conclude that A3G expressed in A3.01 cells is catalytically active and can cause deamination-induced mutation of viral genomes. However, Vif-null HIV-1 can replicate in these cells presumably due to sub-lethal mutagenesis allowing for purifying selection of replication-competent virus. Analysis of the sequence context of mutations in the proviral genomes revealed that 72% of the mutations had footprints typical of A3G (CC) activity (Fig. 4C).
Fig. 4.
Purifying selection limits the accumulation of A3G-induced mutations in viral RNA of Vif-null HIV-1 produced in A3.01 cells. Wild type and Vif-null virus stocks were produced in 293T cells and used to infect A3.01 cells. Virus-containing supernatants were collected at peak virus production and passaged two more times on fresh cultures of A3.01 cells. Cells and virus-containing supernatants from the third passage collected at peak infection were used for cloning of proviral DNA and viral genomic RNA. (A) A 377 b fragment from the 3-LTR region from viral genomic RNA and from integrated proviral DNA was cloned and 15 individual clones were sequenced as described in Materials and Methods. Sequences were aligned against the sequence of the molecular clone NL4-3. Sequences obtained from integrated proviruses are shown on the left; sequences obtained from RT-PCR of viral genomic RNA are shown on the right. Only sequences obtained from Vif-null virus are shown. (B) Results from panel A were quantified for wild type and Vif-null viruses. Solid bars represent the total number of G to A (mutations introduced on minus-strand cDNA) and C to T mutations (mutations introduced on plus-strand cDNA) expressed as percentage of the sum of all available G and C residues. Gray bars represent all other mutations. (C) Sequences from panel A were analyzed for A3G or A3F footprints by determining the sequence context (positions −2, −1, and +1 relative to the target site) around each G to A or C to T mutation. A3G preferentially targets cytidines in the context of a CC dinucleotide (i.e. C at position −1) whereas A3F prefers cytidines in a TC dinucleotide context (i.e. T at position −1) (Liddament et al., 2004; Wiegand et al., 2004).
2.6 Attempts to select A3G resistant Vif-null HIV-1 from A3.01 cells
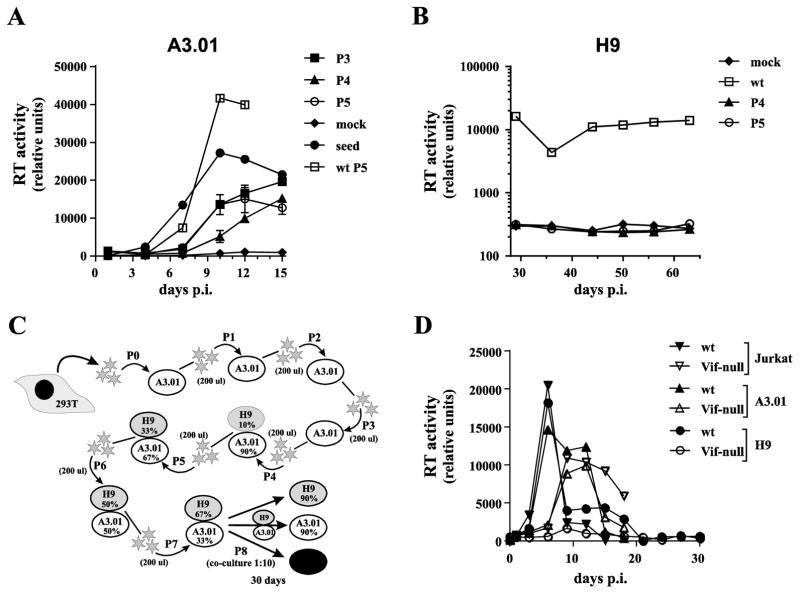
A previous study reported the identification of A3G-resistant Vif-null HIV-1 variants that were obtained by passaging the virus on CEM-SS cells stably transduced to express A3G (Hache et al., 2008). We hypothesized that the selection pressure imposed by A3G on virus replication in A3.01 cells would offer a similar opportunity to isolate A3G-resistant HIV-1 variants. We tested this hypothesis, by subjecting NL4-3 Vif-null to multiple passages in A3.01 cells. For this purpose, A3.01 cells were infected at low MOI with NL4-3 Vif-null using NL4-3 wt as control. Culture media were replaced every 2–3 days and virus replication was monitored by reverse transcriptase assay for up to 15 days. Virus containing supernatants were harvested at or near the peak of virus production (typically around day 10–13). Supernatants were then normalized for RT units and passaged onto fresh cultures of uninfected A3.01 cells. In total, virus was passaged 5 times over a period of more than 60 days. To test if the replication fitness of the Vif-null virus population had changed, virus from peak infection of passages 3, 4, and 5 was normalized for RT activity and used for parallel triplicate infections of A3.01 cells. NL4-3 wt from passage 5 and Vif-null seed virus (passage 0) were included as references. Viral replication profiles were monitored for 15 days (Fig. 5A). We found that passaging virus in A3.01 cells for a combined total of more than 60 days did not improve the replication fitness of Vif-null HIV-1 in A3.01 cells (Fig. 5A). If anything, the replication fitness of the passaged virus was reduced when compared to the non-passaged seed virus (Fig. 5A, compare open circles to closed circles). Not surprisingly, inoculation of H9 cells with Vif-null virus from passages 4 & 5 did not result in productive infection even though cells were monitored for more than 60 days to allow for the outgrowth of slow growing virus (Fig. 5B). On the other hand, infection of H9 cells with wt NL4-3 from passage 5 induced chronic infection of H9 cells (Fig. 5B, open squares).
Fig. 5.
Failure to select A3G resistant Vif-null HIV-1 from A3.01 cells.(A) Wild type or NL4-3 Vif-null virus stocks were used to infect A3.01 cells. Virus production was monitored by reverse transcriptase assay. Viruses were passaged 5 times for a combined total of more than 60 days with virus collected at peak virus production serving as input virus for each subsequent passage. Finally, virus from each passage, normalized for reverse transcriptase activity, was used to initiate a parallel infection of A3.01 cells to allow for the identification of changes in viral replication fitness. Vif-null seed virus (HeLa supernatant) and wild type NL4-3 from passage 5 was analyzed in parallel and mock infected cells were cultured alongside (mock). Virus replication was monitored for 15 days and virus production was plotted as a function of time. Analysis of passages P3, P4, and P5 were performed in triplicate and error bars represent the SEM. (B) Vif-null virus from passages 4 & 5 as well as wild type virus from passage 5 were used to infect H9 cells. Virus replication was monitored for 63 days as described for panel A. (C & D) Attempt to isolate A3G-resistant HIV-1 using A3.01/H9 mixed cultures. (C) Schematic outline of the experimental strategy. Virus was initially passaged three times on A3.01 cells (P1-P3) followed by four additional passages on a mixture of A3.01 and H9 cells with increasing ratios of H9 to A3.01 cells (10% to 67% H9 cells). The final passage (P8) shown in panel D was not initiated by cell-free virus but by co-cultivation of mixed cultures from passage 7 (10%) with H9, A3.01 or Jurkat cells (90% each). Overall, the virus was passaged in this experiment for 153 days. (D) Cells from passage 7 were collected 30 days post-infection and co-cultivated with H9, A3.01, or Jurkat cells at a 1:10 ratio. Virus replication was monitored for another 30 days by RT assay and results were plotted as a function of time.
To improve our chances to isolate an A3G-resistant NL4-3 Vif-null variant, we modified our experimental approach as outlined in Fig. 5C. In this experiment, wt and Vif-null virus stocks were prepared by transfection of 293T cells. Cell-free virus was passaged three times on A3.01 cells (Fig. 5C, P1–P3) using 200 μl of cell-free virus from the peak of infection of each passage to infect 1x106 fresh cells. Cell-free virus was then passaged onto a mixture of A3.01 and H9 cells for an additional 4 passages (Fig. 5C, P4–P7). The selection pressure was gradually increased by changing the ratio of H9 to A3.01 cells with each passage from 10% H9 (P4) to 67% H9 (P7). The rationale for infecting mixtures of A3.01 and H9 cells was to allow the virus to enter H9 cells either as cell-free virus, via cell-to-cell transmission, or even through syncytia formation, thereby increasing the chance of transmission/adaptation through continuous exposure of H9 cells to virus-producing A3.01 cells. Altogether, virus was passaged for 123 days using this experimental setup. For the final passage (P8), the H9/A3.01 culture was mixed at a 1:10 ratio with uninfected H9, A3.01, or Jurkat cells and virus replication was monitored for another 30 days (Fig. 5D). As expected, wild type virus was able to replicate in all three co-cultures with similar efficiency (Fig. 5D, solid symbols). Similarly, Vif-null virus was recovered from Jurkat and A3.01 co-cultures (Fig. 5D, open triangles) indicating that after 123 days of passaging there was replication-competent Vif-null virus remaining in the culture. However, we were unable to recover virus from the H9 co-culture Fig. 5D, open circles) indicating that despite increased selection pressure, Vif-null HIV-1 was unable to develop Vif-independence against H9 cells.
3. Discussion
A3G and other members of the APOBEC3 family of cytidine deaminases are part of the innate immune system that acts as a first-line defense against viral or microbial infections. Although A3G is constitutively expressed in PBMC and macrophages as well as various HIV-susceptible T cell lines, its expression varies in a cell type-specific manner and can be upregulated by interferon-α treatment in some cell types, including resting CD4+ T cells, macrophages, and plasmacytoid dendritic cells (Chen et al., 2006; Peng et al., 2006; Sarkis et al., 2006; Wang et al., 2008a). In primary cells and some T cell lines such as H9, A3G is powerful enough to effectively block replication of Vif-null HIV-1. However, in other cell types such as A3.01, the reduction in A3G levels while not dramatic in nature can be sufficient to allow replication of Vif-null HIV-1. Since A3G restricts virus infectivity by deaminating the viral genome during reverse transcription subsequent to infection of a new target cell, the level of restriction is determined by the amounts of A3G packaged into virions. While the absolute number of A3G molecules packaged into virions from A3.01 cells is not clear, one previous study reported a 10-fold difference relative to viruses produced from H9 cells (Rose et al., 2005). Our data are more consistent with an about 3-fold difference (Fig. 2D). Irrespective of whether the difference is ten-fold or three-fold, it is clear from other reports that packaging of very few molecules of A3G can have strong antiviral effects (Browne et al., 2009; Xu et al., 2007). Consistent with that, our experiments indicate that replication in A3.01 cells does put mutagenic pressure on Vif-null virus and leads to the accumulation of deaminase-induced mutations in integrated proviral genomes during spreading virus replication.
While we cannot formally rule out that additional Vif-sensitive deaminases such as A3F and A3DE contribute to the restriction of Vif-null HIV-1 in H9 cells, we consider such a scenario unlikely for the following reasons: (i) silencing of A3G in A3.01 cells rendered the cells not only fully permissive for Vif-null HIV-1 but even improved the replication of wild type HIV-1 (Fig. 3D), suggesting that A3F and A3DE do not significantly contribute to the restriction of Vif-null HIV-1 in A3.01 cells. Since H9 cells express similar levels of A3F and A3DE than A3.01 cells (Dang et al., 2006; Rose et al., 2005), we infer that they also do not significantly contribute to the non-permissive phenotype in H9 cells. This conclusion is consistent with our previous observation that stably expressed A3F has overall negligible antiviral activity when compared to A3G (Miyagi et al., 2010) although there is no unified opinion on that in the literature (Albin et al., 2014; Bishop et al., 2004; Chaipan et al., 2013; Holmes et al., 2007; Liddament et al., 2004; Mulder et al., 2010). (ii) Increasing A3G expression in A3.01 cells to levels comparable to H9 or CD4+ T cells using wild type A3G effectively blocked replication of Vif-null HIV-1 suggesting that a relatively modest increase in A3G expression is sufficient to completely restrict Vif-null virus (Fig. 3). Importantly, increasing A3G expression using a deaminase-defective variant of A3G did not alter the semi-permissive phenotype of the A3.01 cells. These results suggest that viral restriction is associated with increased A3G deaminase activity consistent with previous reports showing that efficient inhibition of HIV-1 requires enzymatically active A3G (Browne et al., 2009; Kobayashi et al., 2014; Miyagi et al., 2007; Schumacher et al., 2008). On the other hand, a previous study proposed that A3G-mediated viral restriction requires a cellular cofactor that may be lacking in certain cell lines such as CEM-T4 (Han et al., 2008). However, others suggested that CEM-T4 cells represent a mixed population with respect to A3G expression allowing Vif-null virus to replicate in a sub-population of A3Glow cells (Hache and Harris, 2009) although the issue remains under debate (Zhou T, 2009). As far as our A3.01 culture is concerned, we used immunocytochemistry to rule out a mixed population phenomenon as a possible reason for the semi-permissive nature of these cells. In addition, we have evidence that replication of Vif-null virus in our A3.01 culture results in the accumulation of A3G-induced mutations in the proviral genomes (Fig. 4) indicating that virus is indeed exposed to A3G activity in these cultures.
Our attempts to isolate A3G-resistant virus capable of replicating in non-permissive cells was motivated by our observation that replication of Vif-null HIV-1 in A3.01 cells causes sub-lethal mutagenesis that should impose enough of a mutagenic pressure to allow the virus to evolve and potentially develop resistance (Fig. 5A). We were also encouraged by a previous study reporting the identification of an A3G-resistant HIV-1 isolate capable of replicating in an artificial A3G-expressing CEM-SS cell line (Hache et al., 2008). The virus that emerged in that study carried a promoter mutation and a Vpr null mutation. A3G resistance was subsequently attributed to increased virus production, which presumably reduced amounts of virion-associated A3G due to a dilution effect (Hache et al., 2009). Importantly, however, despite optimal translation initiation of the revertants, the “A3G resistant” virus emerging from the CEM-SS-A3G cell line remained unable to replicate in naturally A3G-positive cells (e.g. CEM, H9) and the authors concluded that “something other than APOBEC3G was still inhibiting virus replication” (Hache et al., 2008). Our own strategy to isolate APOBEC-resistant (or rather, Vif-independent) virus was a brute force approach involving long-term passaging of Vif-null virus in A3.01/H9 co-cultures with the ratio of H9 cells changing in favor of H9 cells over time to gradually increase the selection pressure on the virus. We chose this strategy to allow the virus to enter H9 cells via cell-free infection, cell-to-cell transmission, or even cell fusion (syncytia formation). One of the scenarios we envisioned was the outgrowth of a virus with the ability to prevent the encapsidation of A3G into its cores. The fact that passaging Vif-null virus under these conditions for a total of five months did not result in outgrowth of an APOBEC-resistant Vif-null virus capable of replicating in H9 cells attests to the power of A3G as a restriction factor. It suggests that A3G packaging into virions is either non-specific and thus cannot be blocked or that signals required for packaging have other critical functions in the viral life cycle and therefore cannot mutate without being lethal to the virus (Bach et al., 2008; Khan et al., 2007; Khan et al., 2005; Wang et al., 2007; Wang et al., 2008b). The overall conclusion from our study is that it may not be possible to select Vif-null viruses capable of replicating in relevant HIV target cells in vivo thus highlighting the critical role of Vif for HIV-1 replication. On a positive note, the absolute requirement for Vif for replication of HIV-1 in non-permissive cells makes Vif a very strong candidate as a target for the development of novel antivirals and efforts to develop inhibitors of Vif should be encouraged.
4. Materials and methods
4.1 Cell culture
A3.01 cells are a derivative of CEM, a human T-cell line derived from the peripheral blood buffy coat of a four-year-old Caucasian female with acute lymphoblastic leukemia. The cells were derived by subcloning CEM cultures and selection of individual cell clones for sensitivity to infection with HIV-1 LAV and susceptibility to cytopathic effects when infected (Folks et al., 1985). Infection of A3.01 cells by NL4-3 Vif-null revealed a semi-permissive phenotype (Strebel et al., 1987). A3.01, H9, and Jurkat cells were cultured in RPMI 1640 medium containing 10% fetal bovine serum (FBS). HeLa, 293T, and 293TN cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM) containing 10% FBS. TransIT-LT1 (Mirus Corp., Madison WI) or Lipofectamine Plus reagents (Life Technologies, Grand Island NY) were used for transfection of plasmid DNAs.
4.2 Plasmids
The full-length molecular clone pNL4-3 (Adachi et al., 1986) and its Vif-null derivative pNL4-3Vif (-), carrying a 182 by NdeI/PflMI out-of-frame deletion (Karczewski and Strebel, 1996) were used for the preparation of virus stocks. The construction of vectors for the expression of untagged and C-terminally Myc-tagged wild type A3G and catalytically inactive A3G C288S/C291A in the backbone of pcDNA3.1(-) (Invitrogen Corp, Carlsbad CA) have been described previously (Kao et al., 2003; Opi et al., 2006). For transduction of T cell lines, A3G was cloned into pCDH-MSCV-MCS-EF1-puro (Systems Biosciences, Mountain View CA), which allows A3G expression under the control of the Murine Stem Cell Virus (MSCV) promoter. For that purpose, pcDNA-A3G (wt) was first linearized with HindIII and the ends blunted by treatment with Klenow DNA polymerase in the presence of dNTPs (fill-in reaction). The plasmid was then cut with EcoRI and the purified insert was cloned into the EcoRI and SwaI sites of pCDH-MSCV-MCS-EF1-puro resulting in pCDH-A3G(wt). For the construction of the corresponding deaminase-defective variant, pCDH-A3G(mut), an EcoRI/PmlI fragment from pcDNA-A3G (C288S, C291A) was transferred into the pCDH-A3G vector. Plasmid pHCMV-G contains the vesicular stomatitis virus (VSV) glycoprotein G (VSV-G) gene expressed from the immediate-early gene promoter of human cytomegalovirus (Yee et al., 1994) and was used for the production of VSV-G pseudotypes. An A3G shRNA encoding vector (Sigma-Aldrich, Inc., St. Louis MO; Cat# TRCN0000052188) was employed to make A3G shRNA lentiviral particles. A vector encoding non-specific shRNA (Sigma-Aldrich, Inc., St. Louis MO; Cat# SHC016) was employed to make control shRNA lentiviral particles.
4.3 Antisera and proteins
A3G was identified using a polyclonal rabbit serum against a synthetic peptide comprising the 17 C-terminal residues of A3G (anti-ApoC17; available through the NIH AIDS Research and Reference Reagent Program; Cat # 10082). A monoclonal antibody to α-tubulin (Sigma-Aldrich, Inc., St. Louis MO; Cat # T-9026) was used to detect α-tubulin in cell extracts and served as loading control. HIV CA protein was identified using a Gag-specific monoclonal antibody (mAb #24-3), obtained from Michael Malim through the NIH AIDS Research and Reference Reagent Program (cat # 6458 (Simon et al., 1997)).
4.4 Virus preparation and infection
HIV-1 virus stocks were prepared by transfecting 25 cm2 flasks of 293T cells with 5 μg of pNL4-3 or pNL4-3 Vif-null. VSV G-pseudotyped virus stocks were prepared by cotransfecting 0.5 μg of pHCMV-G. Virus-containing supernatants were harvested 48 h later, clarified by centrifugation (5 min, 1500 rpm), and filtered through 0.45 μm membrane filters to remove residual cellular debris. For preparation of lentiviral particles 0.6 μg of pCDH-A3G(wt) or pCDH-A3G(mut) were transfected with 6.7 μl of pPACKH-1 packaging mix (Systems Biosciences, Mountain View CA) into 293TN cells (Systems Biosciences). Four hours later the culture medium was changed to RPMI 1640 medium containing 10% FBS. For production of A3G shRNA-containing lentiviral particles, 1 μg of A3G shRNA plasmid was mixed with 10 μl of packaging mix (Sigma-Aldrich, Inc., St. Louis MO; Cat # SHP001) and added to 293TN cells. Four hours later, the culture medium was changed to RPMI 1640 medium containing 10% FBS. Lentiviral particle-containing supernatants were collected 48 hours after transfection. The supernatants were harvested and cellular debris was removed by centrifugation (5 min, 800 rpm). Clarified supernatants were filtered (0.45 μM) to remove residual cellular debris and stored at −80°C.
For viral replication studies, Jurkat, A3.01, or H9 cells were infected with the same amount of HIV-1 wt or HIV-1 Vif-null viruses as determined by reverse transcriptase (RT) activity (Willey et al., 1988). For mixed cultures of A3.01 and H9 cells, a total of 1x106 cells in 5 ml were infected with 200 μl of culture supernatants from the peak of virus production from the preceding passage. Viral supernatants were harvested every 2 to 3 days and virus replication profiles were determined by quantifying the virus-associated reverse transcriptase activity in the supernatants. To detect virion associated A3G, filtered virus stocks were further purified by pelleting through a 20% sucrose cushion (75 min, 4°C at 35,000 rpm in an SW41 rotor). Virus pellets were then subjected to immunoblotting.
4.5 Immunoblotting
Cell lysates were prepared as follows: Cells were washed once with PBS, suspended in PBS and mixed with an equal volume of sample buffer (4% sodium dodecyl sulfate, 125 mM Tris-HCl, pH 6.8, 10% 2-mercaptoethanol, 10% glycerol, and 0.002% bromophenol blue). Similarly, virus pellets were suspended in PBS and mixed with an equal volume of sample buffer. Proteins were solubilized by heating 10 to 15 min at 95°C with occasional vortexing. Cell and virus lysates were subjected to SDS-PAGE; proteins were transferred to PVDF membranes and reacted with appropriate antibodies as described in the text. Membranes were then incubated with horseradish peroxidase-conjugated secondary antibodies (GE Healthcare Bio-Science, Pittsburgh PA) and proteins were visualized using ECL (GE Healthcare Bio-Science, Pittsburgh PA) or Immobilon Western HRP substrate (Millipore, Bedford MA).
4.6 Northern blotting
Cells were washed twice with PBS and RNA was extracted using the RNeasy mini kit according to the manufacturer’s instructions (QIAGEN Inc., Germantown MD). RNA samples were electrophoresed on denaturing 1% agarose gels and capillary blotted onto a nylon membrane using a Turbo blotter (Schleicher & Schuell, Inc., Keene NH). After UV cross-linking with a UV Stratalinker 2400 (Agilent Technologies, Santa Clara CA) using the auto-crosslink feature, the membrane was stained for ribosomal RNA (rRNA) by soaking the membrane in 5% acetic acid for 15 min at RT followed by 5 min incubation at RT in a solution of 0.5 M sodium acetate (pH 5.2) and 0.04% methylene blue. The membrane was rinsed in water for 10 min and the stained membrane was photographed. Following the staining, the membrane was pre-hybridized with 10 ml of QuickHyb Hybridization Solution (Agilent Technologies, Santa Clara CA) for 1 h at 68°C. The membrane was then hybridized with probes for 5 h at 68°C. The probe for A3G mRNA was a 562 bp BamHI/HindIII restriction fragment derived from pcDNA-A3G (nucleotides 592-1155 of the A3G gene (Kao et al., 2003)). Probes were labeled with [32P]-dTTP using a BcaBESTTM labeling kit (Takara Bio Inc., Mountain View CA) and 1 x 107 cpm of the probe was added after premixing with 100 μl of sonicated salmon sperm DNA (10 mg/ml; Agilent Technologies, Santa Clara CA), heating at 95°C for 5 min, and chilling on ice. Following hybridization, membranes were washed twice with wash buffer (2x SSPE, 0.1% SDS) for 15 min at room temperature, followed by a 15 min wash in 0.2x SSPE, 0.1% SDS at 65°C. RNA bands were visualized by autoradiography.
4.7 Immunocytochemistry
Uninfected Jurkat, A3.01, or H9 cells were centrifuged onto microscope slides using a cytocentrifuge (4 min, 3000 rpm). Samples were air-dried for 5 min before fixing in ice-cold methanol (10 min, −20°C). For antibody staining, slides were incubated in a humid chamber at 37°C for 1 h with A3G-specific antibody (1:100) in 1% BSA in PBS. Slides were washed once in PBS (5 min, room temperature) and incubated with Texas red-conjugated secondary antibody (1:100; Jackson Immuno Research Laboratories, West Grove PA) for 30 min at 37°C in a humid chamber. Slides were then washed twice with PBS and covered with cover slips using glycerol gelatin containing 0.1 M N-propyl gallate (Sigma-Aldrich, Inc., St. Louis MO) to minimize photo bleaching. Images were acquired on a Zeiss LSM410 inverted laser scanning microscope equipped with a krypton-argon mixed-gas laser using a Plan-Apochromat 63x/1.4 oil immersion objective (Zeiss). Additional magnification was achieved using the LSM zoom feature. Bright-field images (Nomarski optics) were collected in parallel.
4.8 Sequence analysis of 3′LTR region
To identify A3G-induced mutations, proviral DNA was extracted from HIV-infected cells near the peak of infection using a DNeasy tissue kit (QIAGEN Inc., Germantown MD). A 377 nucleotide region of the 3′LTR was then amplified by DNA PCR using the Expand Long Template PCR System (Roche Diagnostics Corp., Indianapolis IN) and primers LTR-5′ (CTG TAG ATC TTA GCC ACT TTT TAA) and LTR-3′ (TCG ATG TCA GCA GTT CTT GAA GTA). To identify mutations in replicating virus, viral RNA was extracted at the same time from cell-free virus-containing supernatants using a viral RNA extraction kit (QIAGEN Inc., Germantown MD). Using the same primers, RNA was reverse transcribed and amplified by SuperScript One-step RT-PCR kit with platinum Taq polymerase (Thermo Fisher Scientific, Waltham MA). PCR fragments were cloned into the pGEM-T vector (Promega, Corp., Madison WI) and up to 15 individual clones for each condition were sequenced to measure the G to A or C to T mutations.
4.9 Knock-down of A3G in A3.01 cells by shRNA or increase of A3G expression by transduction of A3G vectors
A3G- or control-knockdown cell lines were established by infection of cells with A3G shRNA or control shRNA lentiviral particles, respectively, described above. Cells were cultured for 2 days in the absence of drug before puromycin selection (3 μg/ml). Puromycin-resistant cells were amplified and successful A3G knock-down was assessed by immunoblotting. Cells were subsequently maintained in RPMI without puromycin. To increase A3G expression in A3.01 cells, cells were transduced with lentiviral vectors encoding wild type (A3G wt) or deaminase-defective A3G (A3G mut). Again, cells were cultured for 2 days in the absence of drug and then selected for puromycin resistance. Increased A3G expression was confirmed by immunoblotting.
Research Highlights.
Vif-null HIV-1 can replicate in semi-permissive cell types such as A3.01
The semi-permissive phenotype of A3.01 cells is determined by levels of APOBEC3G
Subtle changes in APOBEC3G levels dramatically affect restriction of Vif-null HIV-1
Replication in A3.01 cells depends on purifying selection of Vif-null HIV-1
It is not possible for Vif-null HIV-1 to develop complete resistance to APOBEC3G
Vif is critical for HIV-1 in vivo and thus is a promising target for antiviral therapy
Acknowledgments
We thank Sayaka Sukegawa, Sarah Welbourn, and Angela Yoo for helpful discussions and critical comments on the manuscript. The following reagents were obtained through the NIH AIDS Reagent Program, Division of AIDS, NIAID, NIH: TZM-bl cells from Dr. John C. Kappes, Dr. Xiaoyun Wu and Tranzyme Inc. (Cat #8129) and HIV-1 immunoglobulin (Cat #3957), which was used for immunoblot analyses. This work was supported by the Intramural Research Program of the NIH, NIAID (1 Z01 AI000669).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Adachi A, Gendelman HE, Koenig S, Folks T, Willey R, Rabson A, Martin MA. Production of acquired immunodeficiency syndrome-associated retrovirus in human and nonhuman cells transfected with an infectious molecular clone. J Virol. 1986;59:284–291. doi: 10.1128/jvi.59.2.284-291.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albin JS, Brown WL, Harris RS. Catalytic activity of APOBEC3F is required for efficient restriction of Vif-deficient human immunodeficiency virus. Virology. 2014;450–451:49–54. doi: 10.1016/j.virol.2013.11.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bach D, Peddi S, Mangeat B, Lakkaraju A, Strub K, Trono D. Characterization of APOBEC3G binding to 7SL RNA. Retrovirology. 2008;5:54. doi: 10.1186/1742-4690-5-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Binka M, Ooms M, Steward M, Simon V. The activity spectrum of Vif from multiple HIV-1 subtypes against APOBEC3G, APOBEC3F, and APOBEC3H. J Virol. 2012;86:49–59. doi: 10.1128/JVI.06082-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bishop KN, Holmes RK, Malim MH. Antiviral potency of APOBEC proteins does not correlate with cytidine deamination. J Virol. 2006;80:8450–8458. doi: 10.1128/JVI.00839-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bishop KN, Holmes RK, Sheehy AM, Davidson NO, Cho SJ, Malim MH. Cytidine deamination of retroviral DNA by diverse APOBEC proteins. Curr Biol. 2004;14:1392–1396. doi: 10.1016/j.cub.2004.06.057. [DOI] [PubMed] [Google Scholar]
- Borman AM, Quillent C, Charneau P, Dauguet C, Clavel F. Human immunodeficiency virus type 1 Vif- mutant particles from restrictive cells: role of Vif in correct particle assembly and infectivity. J Virol. 1995;69:2058–2067. doi: 10.1128/jvi.69.4.2058-2067.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Browne EP, Allers C, Landau NR. Restriction of HIV-1 by APOBEC3G is cytidine deaminase-dependent. Virology. 2009;387:313–321. doi: 10.1016/j.virol.2009.02.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaipan C, Smith JL, Hu WS, Pathak VK. APOBEC3G Restricts HIV-1 to a Greater Extent than APOBEC3F and APOBEC3DE in Human Primary CD4+ T Cells and Macrophages. J Virol. 2013;87:444–453. doi: 10.1128/JVI.00676-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen K, Huang J, Zhang C, Huang S, Nunnari G, Wang FX, Tong X, Gao L, Nikisher K, Zhang H. Alpha interferon potently enhances the anti-human immunodeficiency virus type 1 activity of APOBEC3G in resting primary CD4 T cells. J Virol. 2006;80:7645–7657. doi: 10.1128/JVI.00206-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho SJ, Drechsler H, Burke RC, Arens MQ, Powderly W, Davidson NO. APOBEC3F and APOBEC3G mRNA levels do not correlate with human immunodeficiency virus type 1 plasma viremia or CD4+ T-cell count. J Virol. 2006;80:2069–2072. doi: 10.1128/JVI.80.4.2069-2072.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conticello SG, Harris RS, Neuberger MS. The Vif protein of HIV triggers degradation of the human antiretroviral DNA deaminase APOBEC3G. Curr Biol. 2003;13:2009–2013. doi: 10.1016/j.cub.2003.10.034. [DOI] [PubMed] [Google Scholar]
- Dang Y, Wang X, Esselman WJ, Zheng YH. Identification of APOBEC3DE as Another Antiretroviral Factor from the Human APOBEC Family. J Virol. 2006;80:10522–10533. doi: 10.1128/JVI.01123-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Folks T, Benn S, Rabson A, Theodore T, Hoggan MD, Martin M, Lightfoote M, Sell K. Characterization of a continuous T-cell line susceptible to the cytopathic effects of the acquired immunodeficiency syndrome (AIDS)-associated retrovirus. Proc Natl Acad Sci U S A. 1985;82:4539–4543. doi: 10.1073/pnas.82.13.4539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fourati S, Malet I, Binka M, Boukobza S, Wirden M, Sayon S, Simon A, Katlama C, Simon V, Calvez V, Marcelin AG. Partially active HIV-1 Vif alleles facilitate viral escape from specific antiretrovirals. AIDS. 2010;24:2313–2321. doi: 10.1097/QAD.0b013e32833e515a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabuzda DH, Lawrence K, Langhoff E, Terwilliger E, Dorfman T, Haseltine WA, Sodroski J. Role of vif in replication of human immunodeficiency virus type 1 in CD4+ T lymphocytes. J Virol. 1992;66:6489–6495. doi: 10.1128/jvi.66.11.6489-6495.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goila-Gaur R, Strebel K. HIV-1 Vif, APOBEC, and intrinsic immunity. Retrovirology. 2008;5:51. doi: 10.1186/1742-4690-5-51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hache G, Abbink TE, Berkhout B, Harris RS. Optimal translation initiation enables Vif-deficient human immunodeficiency virus type 1 to escape restriction by APOBEC3G. J Virol. 2009;83:5956–5960. doi: 10.1128/JVI.00045-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hache G, Harris RS. CEM-T4 cells do not lack an APOBEC3G cofactor. PLoS Pathog. 2009;5:e1000528. doi: 10.1371/journal.ppat.1000528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hache G, Shindo K, Albin JS, Harris RS. Evolution of HIV-1 isolates that use a novel Vif-independent mechanism to resist restriction by human APOBEC3G. Curr Biol. 2008;18:819–824. doi: 10.1016/j.cub.2008.04.073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han Y, Wang X, Dang Y, Zheng YH. APOBEC3G and APOBEC3F require an endogenous cofactor to block HIV-1 replication. PLoS Pathog. 2008;4:e1000095. doi: 10.1371/journal.ppat.1000095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoglund S, Ohagen A, Lawrence K, Gabuzda D. Role of vif during packing of the core of HIV-1. Virology. 1994;201:349–355. doi: 10.1006/viro.1994.1300. [DOI] [PubMed] [Google Scholar]
- Holmes RK, Koning FA, Bishop KN, Malim MH. APOBEC3F can inhibit the accumulation of HIV-1 reverse transcription products in the absence of hypermutation. Comparisons with APOBEC3G. J Biol Chem. 2007;282:2587–2595. doi: 10.1074/jbc.M607298200. [DOI] [PubMed] [Google Scholar]
- Jin X, Brooks A, Chen H, Bennett R, Reichman R, Smith H. APOBEC3G/CEM15 (hA3G) mRNA levels associate inversely with human immunodeficiency virus viremia. J Virol. 2005;79:11513–11516. doi: 10.1128/JVI.79.17.11513-11516.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kao S, Khan MA, Miyagi E, Plishka R, Buckler-White A, Strebel K. The human immunodeficiency virus type 1 Vif protein reduces intracellular expression and inhibits packaging of APOBEC3G (CEM15), a cellular inhibitor of virus infectivity. J Virol. 2003;77:11398–11407. doi: 10.1128/JVI.77.21.11398-11407.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kao S, Miyagi E, Khan MA, Takeuchi H, Opi S, Goila-Gaur R, Strebel K. Production of infectious human immunodeficiency virus type 1 does not require depletion of APOBEC3G from virus-producing cells. Retrovirology. 2004;1:27. doi: 10.1186/1742-4690-1-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karczewski MK, Strebel K. Cytoskeleton association and virion incorporation of the human immunodeficiency virus type 1 Vif protein. J Virol. 1996;70:494–507. doi: 10.1128/jvi.70.1.494-507.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kataropoulou A, Bovolenta C, Belfiore A, Trabatti S, Garbelli A, Porcellini S, Lupo R, Maga G. Mutational analysis of the HIV-1 auxiliary protein Vif identifies independent domains important for the physical and functional interaction with HIV-1 reverse transcriptase. Nucleic Acids Res. 2009;37:3660–3669. doi: 10.1093/nar/gkp226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan MA, Goila-Gaur R, Opi S, Miyagi E, Takeuchi H, Kao S, Strebel K. Analysis of the contribution of cellular and viral RNA to the packaging of APOBEC3G into HIV-1 virions. Retrovirology. 2007;4:48. doi: 10.1186/1742-4690-4-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan MA, Kao S, Miyagi E, Takeuchi H, Goila-Gaur R, Opi S, Gipson CL, Parslow TG, Ly H, Strebel K. Viral RNA is required for the association of APOBEC3G with human immunodeficiency virus type 1 nucleoprotein complexes. J Virol. 2005;79:5870–5874. doi: 10.1128/JVI.79.9.5870-5874.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi T, Koizumi Y, Takeuchi JS, Misawa N, Kimura Y, Morita S, Aihara K, Koyanagi Y, Iwami S, Sato K. Quantification of deaminase activity-dependent and -independent restriction of HIV-1 replication mediated by APOBEC3F and APOBEC3G through experimental-mathematical investigation. J Virol. 2014;88:5881–5887. doi: 10.1128/JVI.00062-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li MM, Wu LI, Emerman M. The range of human APOBEC3H sensitivity to lentiviral Vif proteins. J Virol. 2010;84:88–95. doi: 10.1128/JVI.01344-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liddament MT, Brown WL, Schumacher AJ, Harris RS. APOBEC3F properties and hypermutation preferences indicate activity against HIV-1 in vivo. Curr Biol. 2004;14:1385–1391. doi: 10.1016/j.cub.2004.06.050. [DOI] [PubMed] [Google Scholar]
- Luo K, Wang T, Liu B, Tian C, Xiao Z, Kappes J, Yu XF. Cytidine Deaminases APOBEC3G and APOBEC3F Interact with Human Immunodeficiency Virus Type 1 Integrase and Inhibit Proviral DNA Formation. J Virol. 2007;81:7238–7248. doi: 10.1128/JVI.02584-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma XY, Sova P, Chao W, Volsky DJ. Cysteine residues in the Vif protein of human immunodeficiency virus type 1 are essential for viral infectivity. J Virol. 1994;68:1714–1720. doi: 10.1128/jvi.68.3.1714-1720.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marin M, Rose KM, Kozak SL, Kabat D. HIV-1 Vif protein binds the editing enzyme APOBEC3G and induces its degradation. Nat Med. 2003;9:1398–1403. doi: 10.1038/nm946. [DOI] [PubMed] [Google Scholar]
- Mehle A, Strack B, Ancuta P, Zhang C, McPike M, Gabuzda D. Vif overcomes the innate antiviral activity of APOBEC3G by promoting its degradation in the ubiquitin-proteasome pathway. J Biol Chem. 2004;279:7792–7798. doi: 10.1074/jbc.M313093200. [DOI] [PubMed] [Google Scholar]
- Miyagi E, Brown CR, Opi S, Khan M, Goila-Gaur R, Kao S, Walker RC, Jr, Hirsch V, Strebel K. Stably expressed APOBEC3F has negligible antiviral activity. J Virol. 2010;84:11067–11075. doi: 10.1128/JVI.01249-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyagi E, Opi S, Takeuchi H, Khan M, Goila-Gaur R, Kao S, Strebel K. Enzymatically active APOBEC3G is required for efficient inhibition of human immunodeficiency virus type 1. J Virol. 2007;81:13346–13353. doi: 10.1128/JVI.01361-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulder LC, Ooms M, Majdak S, Smedresman J, Linscheid C, Harari A, Kunz A, Simon V. Moderate influence of human APOBEC3F on HIV-1 replication in primary lymphocytes. J Virol. 2010;84:9613–9617. doi: 10.1128/JVI.02630-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navarro F, Landau NR. Recent insights into HIV-1 Vif. Curr Opin Immunol. 2004;16:477–482. doi: 10.1016/j.coi.2004.05.006. [DOI] [PubMed] [Google Scholar]
- OhAinle M, Kerns JA, Malik HS, Emerman M. Adaptive evolution and antiviral activity of the conserved mammalian cytidine deaminase APOBEC3H. J Virol. 2006;80:3853–3862. doi: 10.1128/JVI.80.8.3853-3862.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Opi S, Takeuchi H, Kao S, Khan MA, Miyagi E, Goila-Gaur R, Iwatani Y, Levin JG, Strebel K. Monomeric APOBEC3G is catalytically active and has antiviral activity. J Virol. 2006;80:4673–4682. doi: 10.1128/JVI.80.10.4673-4682.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng G, Lei KJ, Jin W, Greenwell-Wild T, Wahl SM. Induction of APOBEC3 family proteins, a defensive maneuver underlying interferon-induced anti-HIV-1 activity. J Exp Med. 2006;203:41–46. doi: 10.1084/jem.20051512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng J, Ao Z, Matthews C, Wang X, Ramdahin S, Chen X, Li J, Chen L, He J, Ball B, Fowke K, Plummer F, Embree J, Yao X. A Naturally Occurring Vif Mutant (I107T) Attenuates Anti-APOBEC3G Activity and HIV-1 Replication. J Mol Biol. 2013;425:2840–2852. doi: 10.1016/j.jmb.2013.05.015. [DOI] [PubMed] [Google Scholar]
- Porcellini S, Alberici L, Gubinelli F, Lupo R, Olgiati C, Rizzardi GP, Bovolenta C. The F12-Vif derivative Chim3 inhibits HIV-1 replication in CD4+ T lymphocytes and CD34+-derived macrophages by blocking HIV-1 DNA integration. Blood. 2009;113:3443–3452. doi: 10.1182/blood-2008-06-158790. [DOI] [PubMed] [Google Scholar]
- Rose KM, Marin M, Kozak SL, Kabat D. Regulated production and anti-HIV type 1 activities of cytidine deaminases APOBEC3B, 3F, and 3G. AIDS Res Hum Retroviruses. 2005;21:611–619. doi: 10.1089/aid.2005.21.611. [DOI] [PubMed] [Google Scholar]
- Russell RA, Moore MD, Hu WS, Pathak VK. APOBEC3G induces a hypermutation gradient: purifying selection at multiple steps during HIV-1 replication results in levels of G-to-A mutations that are high in DNA, intermediate in cellular viral RNA, and low in virion RNA. Retrovirology. 2009;6:16. doi: 10.1186/1742-4690-6-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakai H, Shibata R, Sakuragi J, Sakuragi S, Kawamura M, Adachi A. Cell-dependent requirement of human immunodeficiency virus type 1 Vif protein for maturation of virus particles. J Virol. 1993;67:1663–1666. doi: 10.1128/jvi.67.3.1663-1666.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarkis PT, Ying S, Xu R, Yu XF. STAT1-independent cell type-specific regulation of antiviral APOBEC3G by IFN-alpha. J Immunol. 2006;177:4530–4540. doi: 10.4049/jimmunol.177.7.4530. [DOI] [PubMed] [Google Scholar]
- Schumacher AJ, Hache G, Macduff DA, Brown WL, Harris RS. The DNA deaminase activity of human APOBEC3G is required for Ty1, MusD, and human immunodeficiency virus type 1 restriction. J Virol. 2008;82:2652–2660. doi: 10.1128/JVI.02391-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheehy AM, Gaddis NC, Choi JD, Malim MH. Isolation of a human gene that inhibits HIV-1 infection and is suppressed by the viral Vif protein. Nature. 2002;418:646–650. doi: 10.1038/nature00939. [DOI] [PubMed] [Google Scholar]
- Sheehy AM, Gaddis NC, Malim MH. The antiretroviral enzyme APOBEC3G is degraded by the proteasome in response to HIV-1 Vif. Nat Med. 2003;9:1404–1407. doi: 10.1038/nm945. [DOI] [PubMed] [Google Scholar]
- Simon JH, Fouchier RA, Southerling TE, Guerra CB, Grant CK, Malim MH. The Vif and Gag proteins of human immunodeficiency virus type 1 colocalize in infected human T cells. J Virol. 1997;71:5259–5267. doi: 10.1128/jvi.71.7.5259-5267.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon V, Zennou V, Murray D, Huang Y, Ho DD, Bieniasz PD. Natural variation in Vif: differential impact on APOBEC3G/3F and a potential role in HIV-1 diversification. PLoS Pathog. 2005;1:e6. doi: 10.1371/journal.ppat.0010006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stopak K, de Noronha C, Yonemoto W, Greene WC. HIV-1 Vif blocks the antiviral activity of APOBEC3G by impairing both its translation and intracellular stability. Mol Cell. 2003;12:591–601. doi: 10.1016/s1097-2765(03)00353-8. [DOI] [PubMed] [Google Scholar]
- Strebel K, Daugherty D, Clouse K, Cohen D, Folks T, Martin MA. The HIV ‘A’ (sor) gene product is essential for virus infectivity. Nature. 1987;328:728–730. doi: 10.1038/328728a0. [DOI] [PubMed] [Google Scholar]
- Vallanti G, Lupo R, Federico M, Mavilio F, Bovolenta C. T Lymphocytes transduced with a lentiviral vector expressing F12-Vif are protected from HIV-1 infection in an APOBEC3G-independent manner. Mol Ther. 2005;12:697–706. doi: 10.1016/j.ymthe.2005.05.014. [DOI] [PubMed] [Google Scholar]
- Wang FX, Huang J, Zhang H, Ma X. APOBEC3G upregulation by alpha interferon restricts human immunodeficiency virus type 1 infection in human peripheral plasmacytoid dendritic cells. J Gen Virol. 2008a;89:722–730. doi: 10.1099/vir.0.83530-0. [DOI] [PubMed] [Google Scholar]
- Wang T, Tian C, Zhang W, Luo K, Sarkis PT, Yu L, Liu B, Yu Y, Yu XF. 7SL RNA mediates virion packaging of the antiviral cytidine deaminase APOBEC3G. J Virol. 2007;81:13112–13124. doi: 10.1128/JVI.00892-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang T, Zhang W, Tian C, Liu B, Yu Y, Ding L, Spearman P, Yu XF. Distinct viral determinants for the packaging of human cytidine deaminases APOBEC3G and APOBEC3C. Virology. 2008b;377:71–79. doi: 10.1016/j.virol.2008.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiegand HL, Doehle BP, Bogerd HP, Cullen BR. A second human antiretroviral factor, APOBEC3F, is suppressed by the HIV-1 and HIV-2 Vif proteins. EMBO J. 2004;23:2451–2458. doi: 10.1038/sj.emboj.7600246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willey RL, Smith DH, Lasky LA, Theodore TS, Earl PL, Moss B, Capon DJ, Martin MA. In vitro mutagenesis identifies a region within the envelope gene of the human immunodeficiency virus that is critical for infectivity. J Virol. 1988;62:139–147. doi: 10.1128/jvi.62.1.139-147.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu H, Chertova E, Chen J, Ott DE, Roser JD, Hu WS, Pathak VK. Stoichiometry of the antiviral protein APOBEC3G in HIV-1 virions. Virology. 2007;360:247–256. doi: 10.1016/j.virol.2006.10.036. [DOI] [PubMed] [Google Scholar]
- Yee JK, Miyanohara A, LaPorte P, Bouic K, Burns JC, Friedmann T. A general method for the generation of high-titer, pantropic retroviral vectors: highly efficient infection of primary hepatocytes. Proc Natl Acad Sci U S A. 1994;91:9564–9568. doi: 10.1073/pnas.91.20.9564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu X, Yu Y, Liu B, Luo K, Kong W, Mao P, Yu XF. Induction of APOBEC3G ubiquitination and degradation by an HIV-1 Vif-Cul5-SCF complex. Science. 2003;302:1056–1060. doi: 10.1126/science.1089591. [DOI] [PubMed] [Google Scholar]
- Zennou V, Bieniasz PD. Comparative analysis of the antiretroviral activity of APOBEC3G and APOBEC3F from primates. Virology. 2006;349:31–40. doi: 10.1016/j.virol.2005.12.035. [DOI] [PubMed] [Google Scholar]
- Zhen A, Wang T, Zhao K, Xiong Y, Yu XF. A single amino acid difference in human APOBEC3H variants determines HIV-1 Vif sensitivity. J Virol. 2010;84:1902–1911. doi: 10.1128/JVI.01509-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng YH, Irwin D, Kurosu T, Tokunaga K, Sata T, Peterlin BM. Human APOBEC3F Is Another Host Factor That Blocks Human Immunodeficiency Virus Type 1 Replication. J Virol. 2004;78:6073–6076. doi: 10.1128/JVI.78.11.6073-6076.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou TYS, Han Y, Wang X, Dang Y, Zheng YH. An APOBEC3G cofactor. Reader comment to “CEM-T4 cells do not lack an APOBEC3G cofactor”. PLoS Pathog. 2009;5:e1000528. doi: 10.1371/journal.ppat.1000528. [DOI] [PMC free article] [PubMed] [Google Scholar]