Abstract
This study was designed to determine how aspirin influences the growth kinetics and characteristics of cultured colorectal cancer (CRC) cells that harbor a variety of different mutational backgrounds, including PIK3CA and KRAS activating mutations and the presence or absence of microsatellite instability. CRC cell lines (HCT116, HCT116+Chr3/5, RKO, SW480, HCT15, CACO2, HT29, and SW48) were treated with pharmacologically relevant doses of aspirin (0.5–10 mM) and evaluated for proliferation and cell cycle distribution. These parameters were fitted to a mathematical model to quantify the effects and understand the mechanism(s) by which aspirin modifies growth in CRC cells. We also evaluated the effects of aspirin on key G0/G1 cell cycle genes that are regulated by PI3K-Akt pathway. Aspirin decelerated growth rates and disrupted cell cycle dynamics more profoundly in faster growing CRC cell lines, which tended to be PIK3CA-mutants. Additionally, microarray analysis of 151 CRC cell lines identified important cell cycle regulatory genes downstream targets of PIK3, which were dysregulated by aspirin treatment cycle genes (PCNA and RB1, p<0.01). Our study demonstrated what clinical trials have only speculated, that PIK3CA-mutant CRCs are more sensitive to aspirin. Aspirin inhibited cell growth in all CRC cell lines regardless of mutational background, but the effects were exacerbated in cells with PIK3CA mutations. Mathematical modeling combined with bench science revealed that cells with PIK3CA mutations experience significant G0/G1 arrest and explains why patients with PIK3CA-mutant CRCs may benefit from aspirin use after diagnosis.
Keywords: Aspirin, chemoprevention, acetylsalicylic acid, colorectal cancer, cell cycle, PIK3CA
INTRODUCTION
Colorectal cancer (CRC) affects nearly 5% of the population, and was responsible for ~140,000 new diagnoses and ~50,000 American deaths in 2013. Epidemiological and clinical studies suggest that cyclo-oxygenase (COX)-2 inhibitors (1) and non-steroidal anti-inflammatory drugs (NSAIDs) (2), including aspirin, are effective at prolonging CRC patient survival after initial diagnosis (3–10). However, aspirin is associated with dose-related side effects, particularly gastrointestinal bleeding. Therefore, aspirin has the potential for wider use for preventing relapse in CRC patients if benefits can be predicted. The multiplicity of cellular effects of aspirin and the heterogeneity of the genetic backgrounds among all CRC patients (11) have challenged its routine implementation to prevent disease reoccurrence.
The CAPP2 trial by Burn et al. demonstrated that 600 mg/day of aspirin for 2 years resulted in 63% reduction in CRC incidence, and 59% mitigation in CRC-associated mortality during a 55.7 month follow-up (12). Interestingly, no change in CRC or adenoma risk was detected in the trial’s first four years (13) suggesting that aspirin prevents CRC after extended exposure, but not adenoma development following a short-term treatment duration. These findings suggest that aspirin may more profoundly affect the mutational events occurring at later stages of the multistep genetic model for CRC carcinogenesis. This model postulates that tumorigenesis proceeds initially through the adenoma-to-carcinoma transition (14) by disabling WNT signaling (most often through APC inactivation), followed by progression to the intermediate adenoma stage by triggering activating mutations in the KRAS or BRAF genes (15). This process is followed by further loss of the SMAD genes or gain of PIK3CA function through activating mutations (15,16), and finally undergoing the adenoma-to-carcinoma transition, often through biallelic loss of p53 (17). The inactivation of DNA mismatch repair (MMR) genes in CRCs provokes a distinct downstream series of mutational events that also contribute to tumorigenesis (18,19).
A molecular-pathological epidemiological study concluded that aspirin improves survival and inhibits recurrence in CRC patients who harbor activating mutations in the PIK3CA gene and suggests that patients with wild-type tumors may not benefit from aspirin use (20). Aspirin’s effectiveness against PIK3CA-mutant tumors raises questions about the involvement of upstream activators of the PI3K-Akt pathway in colorectal carcinogenesis. To the best of our knowledge no study has compared aspirin’s relative effectiveness between cells with activating PIK3CA mutations vs. wild-type CRC cells, and no study has speculated on the mechanisms involved in aspirin-mediated chemoprevention in such a scenario.
The present study was designed to elucidate aspirin’s cellular growth inhibitory effects on cell cycle dynamics in a panel of CRC cell lines with dysfunctional DNA MMR, KRAS mutations, or constitutively active PIK3-Akt pathway. Our goals were to obtain systematic and comprehensive data on cellular kinetics of aspirin-treated CRC cells, and fit these cellular responses in a mathematical model that quantifies these effects of aspirin within the context of different mutational backgrounds, and propose a mechanism that might help explain why aspirin is effective in a specific CRC patient population vs. others. We hypothesized that aspirin inhibits CRC cell growth by disrupting the expression of cell cycle regulatory genes to varying degrees based on specific mutational backgrounds. Improved understanding of the molecular mechanisms by which aspirin prolongs survival (post diagnosis) and exerts its chemopreventive effects is critical to identifying whether a specific subset of CRC patients may benefit more from its prophylactic use – an observation that has significant clinical implications in managing this fatal malignancy.
MATERIALS & METHODS
Cell Lines and viability measurements
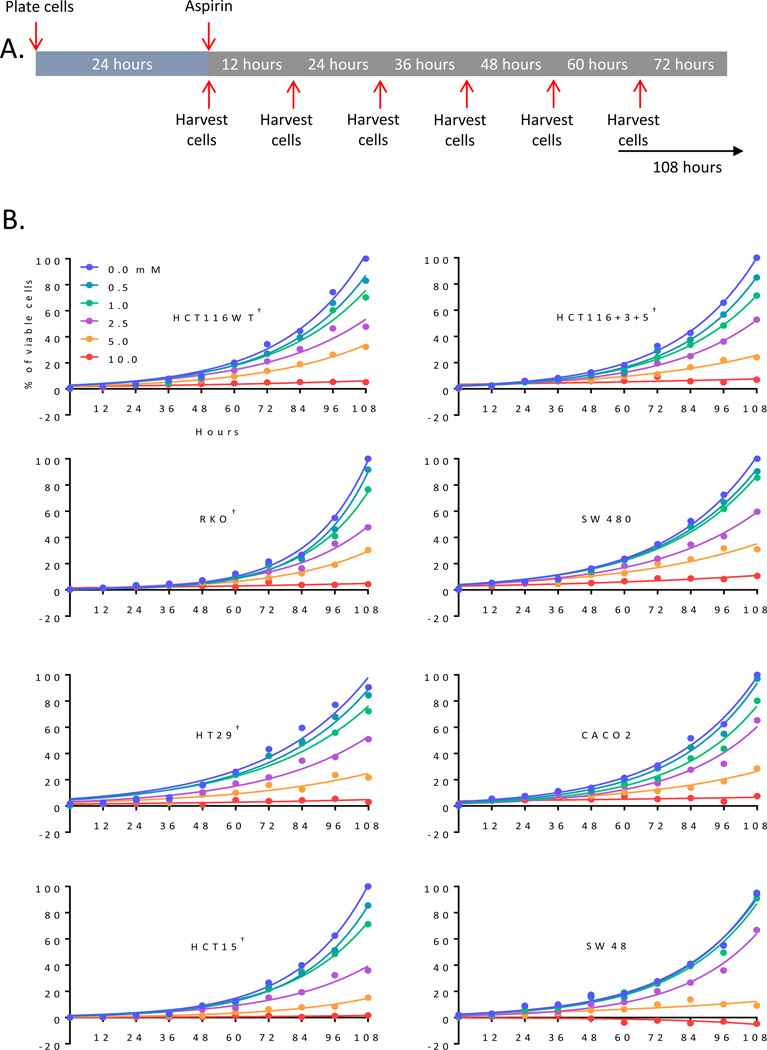
A panel of eight CRC cell lines (HCT116, HCT116+Chr3/5, RKO, SW480, HCT15, Caco2, HT29, and SW48) with known mutational backgrounds (21–23) were obtained from American Type Culture Collection (Table 1). HCT116+Chr3/5 cells were corrected for MMR deficiency by stable transfer of chromosome 3 and 5 and in parental HCT116 cells (24). All cells were authenticated by genetic profiling. HCT116 cells with PIK3CA kinase domain mutant allele (H1047R) knockout were purchased from Horizon discovery (Cambridge, UK). Cells were grown as monolayers in Iscove’s Modified Dulbecco’s Medium (IMDM) (Life Technologies, Carlsbad, CA) supplemented with 10% fetal calf serum (Life Technologies), and 1X penicillin, streptomycin (Life Technologies) at 37°C in 5% CO2. Cells were trypsinized (Life Technologies), harvested and washed with ice cold PBS (Life Technologies) every 12 hours up to 108 hours (Figure 1). For cell viability measurements, cells were plated at a density of 12,000 cells/well 24 hours before aspirin treatment and dead and live cell numbers were determined via trypan blue exclusion assay using an automated cell counter, Countess II (Life Technologies). All experiments were performed in triplicates and each experiment was repeated at least three times.
Table 1.
Mutational background of CRC cell panel
| Cell Line | MMR genes | PIK3CA mut?* | KRAS mut?* |
|---|---|---|---|
| HCT116 | MLH1−/MSH3− | Yes | Yes |
| HCT116+Chr3/5 | Corrected MLH1/MLH3 | Yes | Yes |
| RKO | Hypermethylated MLH1 | Yes | No |
| SW480 | No | Yes | |
| HT29 | Yes | No | |
| Caco2 | No | No | |
| SW48 | Hypermethylated MLH1 | No | No |
| HCT15 | MSH6− | Yes | Yes |
Mutation according to the American Type Culture Collection
Figure 1.
Aspirin-mediated growth inhibition is dose dependent for all cell lines studied. A) Experiment timeline of aspirin treatment and cell line harvesting. B) Growth curves for 8 CRC cell lines, 6 concentrations of aspirin, and 10 time points. Each point represents average of three experiments. †, PIK3CA-mutant cell lines.
Antibodies and reagents
Aspirin (Sigma-Aldrich, St. Louis, MO) was dissolved in cell culture media at a dose range of biologically relevant and previously reported doses (0–10 mM). Similarly 5-FU (Sigma-Aldrich) was dissolved in DMSO (Sigma-Aldrich) and diluted in cell culture media at a dose range of 0–10 µM). All experiments were performed in triplicate. Total cell lysates were obtained by lysing monolayers of cells in RIPA buffer (50mM Tris base, pH 7.5, 150 mM NaCl, 0.1% sodium dodecyl sulfate, 0.5% sodium deoxycholate, NP-40 and 1 mM EDTA (Sigma-Aldrich)), with protease inhibitors (1 mM phenylmethanesulfonyl fluoride, aprotinin and Leupeptin; Sigma-Aldrich), and phosphatase inhibitor cocktail 3 (Sigma-Aldrich). Protein lysates were clarified by centrifugation at 15,000 rpm for 15 minutes at 4°C and then sonicated for 10 seconds on ice using Sonifier 150 (Branson Ultrasonics, Danbury, CT). Western blots were performed by denaturing equal amounts of proteins (30 µg) in SDS-sample buffer and resolved on 10% Acrylamide/Bis gels (Bio-Rad Laboratories, Hercules, CA). Proteins were transferred onto a Hybond-P (Amersham Pharmacia Biotech, Uppsala, Sweden) PVDF membranes in Tris Glycine buffer and 20% methanol for 16 hours. Membranes were blocked in 5% nonfat milk and phosphate-buffered saline (PBS) for 1 hour, and stained with primary antibodies and specific horseradish peroxidase (HRP)-conjugated secondary antibody following manufacturers’ protocols. Chemoluminescence was detected using Pierce ECL Plus (Thermo Fisher Scientific, Waltham, MA). Antibodies were diluted in PBS containing 5% nonfat milk and 0.05% NaN3. The following antibodies were used: anti-β-actin (AC-15) mouse monoclonal antibody (mAb), anti-cyclin D1 (A-12) mouse mAb, anti-ATM (2C1) mouse mAb, and anti-PCNA (PC10) mouse mAb. All antibodies were purchased from Santa Cruz Biotech (Santa Cruz, CA) except for anti-retinoblastoma protein mouse mAb (BD Pharmingen, Sigma-Aldrich).
Flow Cytometry Analysis
Cells were harvested every 12 hours, washed in PBS, and fixed in 70% ethanol at −20°C for more than 3 hours; subsequently, fixed cells were removed from ethanol by centrifuging for 5 min at 5,000 rpm and resuspended in PBS, and then re-spun and once again suspended in propidium iodide following manufacturer’s instructions (Merck Millipore, Billerica, MA). Cells were stained at room temperature in the dark for 30 min. DNA content and cycle distribution were evaluated via flow cytometry using the Muse® Cell Analyzer (Merck Millipore).
Mathematical Modeling
The model, explained in the Results section, was fit to the experimental data and the best fitting parameters were determined by employing the standard least squares procedures using Berkeley-Madonna™ software (Berkeley, CA). Determining the best fitting parameters requires an independent estimate of the parameter α, the rate at which stained dead cells become undetectable in culture. Cells begin to disappear in solution once they develop apoptotic bodies and begin to fragment. Therefore, our assay will no longer ‘count’ these cells because the cell fragments would become too small. Quantity 1/α is the mean time for a cell to go from being detected as apoptotic to disappearing from the culture; a reasonable range for this quantity is between 2 and 24 hours (25). We implemented the following approach to estimate the values of α. First, the parameter γ (the net growth rate of cells) was found for each cell line and condition by the simultaneous linear regression of the data on both live and dead cells. The constant ŷ was determined such that the function ŷeγt provided the best exponential approximation of the dynamics of the dead cells. The dynamics of y comprises two processes: the initial influx of dead cells caused by plating and subsequent continuous production of dead cells from the population of live cells (with constant removal at rate α). The values of parameter α measure how long the population of dead cells takes to lose information about their initial condition and settle to their exponential behavior; this follows from examining the analytical solution for y(t) given by Based on this idea, we can estimate parameter α by measuring how long it takes for the population of dead cells to approach an exponential.
For each cell line, we constructed the relative difference between the true dynamics and the exponential dynamics, Q=(y- ŷeγt)/(ŷeγt), where y stands for the measured data. Determining the time this quantity takes to decay to relatively small numbers depends on our choice of the smallness threshold, s. Then for each data point for a given cell line we can count how many points (starting from t=0) satisfy Q>s. Since the measurements were performed every 12 hours we can calculate the characteristic decay time, 1/α. This procedure was repeated for a range of s values from 0 to 1. Therefore, an estimate for the parameter α is between 0.06 and 0.08.
qRT-PCR
RNA was extracted using RNeasy® Plus Mini Kit (Qiagen, Valencia, CA). qRT-PCR was performed using the QuantStudio 6 Flex Real-Time PCR system (Life Technologies) with the Fast SYBR Green Master Mix and High Capacity Reverse Transcription Kit (Applied Biosystems, Waltham, MA). All reactions were carried out in 10 µL volumes in duplicate. The relevant gene expression was determined by normalizing the values obtained to β-actin expression. Primer sequences were generated using PerlPrimer software (26) (Supplemental Table) and were evaluated in silico using the UCSC Genome Browser (http://genome.ucsc.edu/) (27).
Microarray analysis
Microarray expression data (GSE59857) were uploaded via the GEOquery package in R programming language (28) to comprehensively analyze gene expression in 151 CRC cell lines and then to identify key cell cycle genes frequently dysregulated by the PIK3-Akt pathway. Uploaded data were loess normalized using the R Lumi package (29). Additional filtering was conducted via present in at least one (PALO) filter with a 0.01 threshold as well as low varying probes (SD <0.25). One way ANOVA model was conducted to model the “8 or 9 conditions” for differential gene expression. Linear contrasts were used to conduct the various hypothesis tests between “conditions”.
Xenograft experiment
The 7 week-old male athymic nude mice (Envigo, Houston, TX) were housed under controlled conditions of light and fed ad libitum. CRC derived xenograft tumors were generated by subcutaneously injecting 2×106 HCT116 or HCT116 PIK3CA kinase domain mutant allele (H1047R) knockout cells suspended in Matrigel matrix (BD Biosciences, Franklin Lake, NJ) into flanks of mice using 27-gauge needle (n = 12 per group). Tumor size was measured every day by calipers for 12 days. Mice were then gavaged daily with aspirin and vehicle (100 mg/kg, 300 mg/kg body weight aspirin suspended in water with 1% methyl cellulose) daily. Tumor volume was calculated using the following formula: 1/2(length x width x height), then normalized to vehicle treated animals as percentage. The animal protocol was approved by the Institutional Animal Care and Use Committee, Baylor Research Institute, Dallas, Texas.
Statistical Analysis
Graphpad Prism 6.0 software (Graphpad Software, San Diego, CA) and Microsoft Excel were used to generate p-values and sigmoidal dose-response curves. All data was analyzed using Student’s T-test and a p-value of <0.05 was considered statistically significant.
RESULTS
Effect of aspirin on the kinetics of cell growth
Aspirin treatment was assessed in vitro using increasing doses (0.0, 0.5, 1.0, 2.5, 5.0, and 10 mM) and several time points over a duration of 108 hours (Figure 1A). These doses were chosen empirically from the half-maximal effective concentrations (EC50) of aspirin (Supplemental Figure 1). The proportion of viable cells as a function of time is plotted in Figure 1B for all cell lines treated with various doses of aspirin. We calculated net growth rates for all cell lines and doses by measuring the number of viable cells as a function of time (Supplemental Figure 2); which led us to determine the maximum aspirin dose for subsequent experiments.
To quantify the kinetic parameters that characterized the effect of aspirin on cellular growth, a mathematical model was developed to fit these data. The model was given by a set of ordinary differential equations that described the time evolution of live and dead cells:
| (1) |
where x represented the number of live cells and y for the number of dead cells stained with trypan blue. Parameters were: r, the division rate of cells; d, the death rate of cells; and α, the rate at which dead cells fragment and become too small to count using an automated cell counter. When fitting the model to the data, the residual sum of squares error between observation and prediction was minimized, yielding the best fitting parameters (see Materials & Methods for details).
The net growth rate of the cells was given by γ=r-d, and was plotted against aspirin dose for all experiments in Supplemental Figure 3. The cellular growth rates and aspirin dose were highly significant, but demonstrated an inverse correlation (Supplemental Figure 3A). Most of this negative correlation was explained by the variations in the rate of cell division, r, which also displayed a similarly significant negative correlation with aspirin dose (Supplemental Figure 3B). Overall, cell death rates and aspirin doses yielded a significant positive correlation (Supplemental Figure 3C).
Correlates of aspirin sensitivity
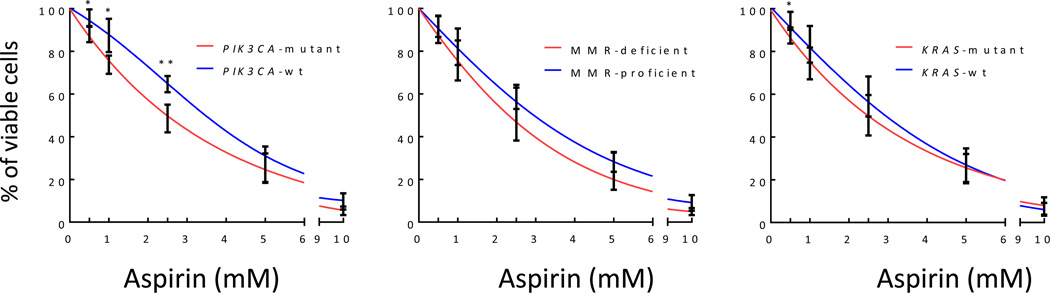
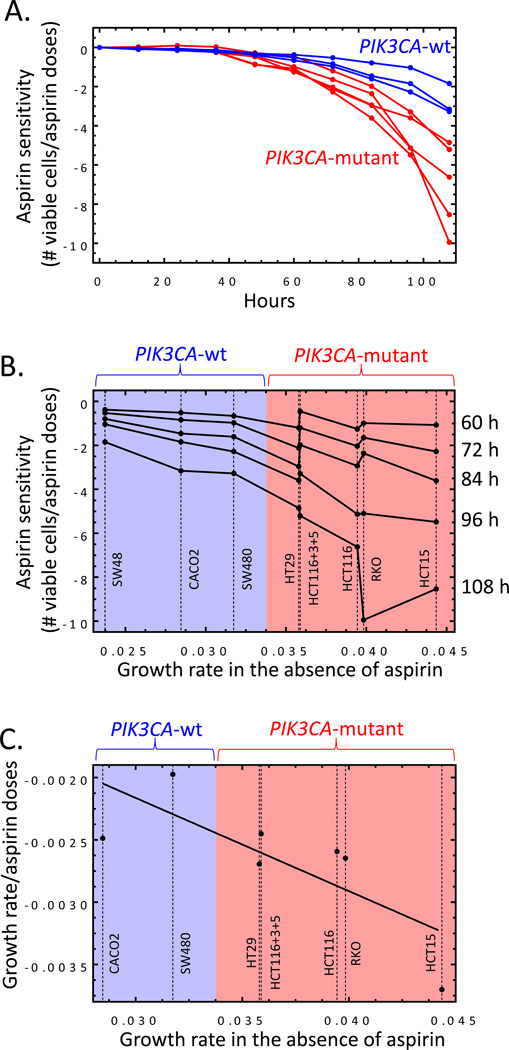
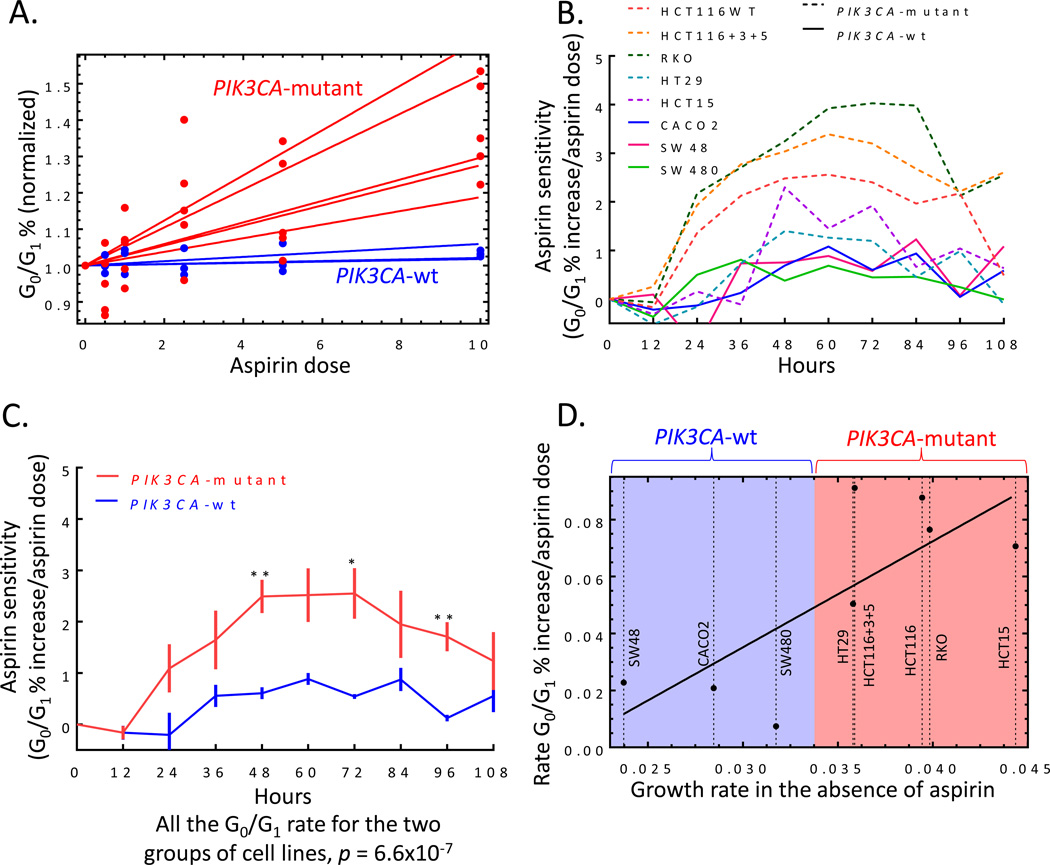
We investigated the extent to which cell lines were sensitive to aspirin and identified the determinants involved. PIK3CA gene mutated cell lines (HCT116, HCT116+Chr3/5, RKO, HT29, and HCT15) were significantly more responsive than wild-type cells (SW480, Caco2, and SW48), regardless of MMR status or KRAS gene mutations (Figure 2A). These data illustrated that aspirin appeared to affect cancer cells differently depending on PIK3CA gene mutational status, however it only represented the final time point. To compare both groups of cell lines at all ten time points and determine at which time point the PIK3CA-mutant cells lines begin to demonstrate higher sensitivity to aspirin, the effects of aspirin at all doses needed to be condensed into a single metric. This calculation was import to perform because we needed to illustrate our unique mathematical model and combine many different factors to comprehensively describe the effects of aspirin on CRC. A single metric would then allow the overall effect of aspirin to be plotted against time. This was done by plotting the number of live cells against all aspirin doses and generating a separate trend line for each time point and cell line. The value of the corresponding slope was, therefore, a measure of cell line sensitivity (or ‘aspirin sensitivity’): the higher the value, the more pronounced the negative effect of aspirin on cell growth. The aspirin sensitivity was then plotted for each cell line at every time point (Figure 3A). Aspirin sensitivity became significantly different between PIK3CA-mutant and wild-type cells starting at hour 72 (p=0.008; Figure 3A). Likewise, Figure 3B depicts aspirin sensitivity (as defined in Figure 3A) as a function of the cell growth rate in the absence of aspirin, for the eight cell lines. The five curves correspond to the last five time-points in the experiment, beginning at hour 60. The natural growth rate of each cell line (in the absence of aspirin) appeared to dictate the magnitude of response to aspirin. Therefore, we calculated the aspirin-related reduction of net growth rates in various cell lines (parameter γ, see Materials & Methods) and discovered that it correlated with the cells’ growth rate in the absence of aspirin (Figure 3C). In other words, the faster the net growth rate (in the absence of treatment) the more responsive the cell line was to aspirin.
Figure 2.
Increased sensitivity to aspirin in PIK3CA-mutated cell lines. Differences in MMR and KRAS status did not result in differential aspirin response; however, PIK3CA-mutant cells were more sensitive than wild-type cells at low doses (0.5, 1.0, and 2.5 mM) of aspirin after 108 hours of treatment. Student’s T-test was performed to determine significance. Error bars represent ± standard error of the mean (SEM) of grouped cell lines. *, p value <0.05 and **, p<0.01.
Figure 3.
The effect of aspirin on the number of viable cells. A) The slope values of the number of viable cells with respect to all aspirin doses (aspirin sensitivity) normalized with the number of viable cells in the absence of aspirin and plotted at each time point. The different curves correspond to different cell lines. The red curves are mutant PIK3CA cell lines and the blue lines are wild-type. B) Aspirin sensitivity compared to the natural growth rate in the absence of aspirin. The different curves correspond to different time points. For all of these time-points, the negative correlation is statistically significant with p-value <0.05 (starting with t=72 h, p<0.01). The cell lines are labeled next to their growth rates. C) The effect of aspirin on the calculated net growth of cells (parameter γ, described in the Results) as a function of the growth rate in the absence of aspirin. Each point corresponds to a single cell line. The linear regression line is also plotted (p<0.05). All cell lines are included except line SW48, which fails to grow at high aspirin doses. Student’s T-test was performed to determine significance. Error bars represent ± SEM of grouped cell lines. *, p value <0.05 and **, p<0.01.
Next, in order to confirm that aspirin was indeed more effective against CRC cell lines with PIK3CA mutations, and did not only target faster growing cells, we investigated the effects of various doses of 5-fluorouracil (5-FU), a common DNA damaging chemotherapeutic agent, on cellular proliferation (Supplementary Fig. 4A). When cells lines were grouped by PIK3CA mutational status, no differences were observed between mutated vs. wild type groups of cell lines (Supplementary Fig. 4B). This suggested that proliferation rates are not the sole determinant of exogenous growth inhibition. Furthermore, we tested aspirin’s sensitivity in isogenic pairs of HCT116 cells with or without PIK3CA-mutant (Supplementary Fig. 5A). As expected, HCT116 cells with mutant PIK3CA had higher sensitivity to aspirin when compared to those with wild type PIK3CA (Supplementary Fig. 5B). Collectively, these data support our finding that cells with mutant PIK3CA are more sensitive to aspirin.
Aspirin inhibits PIK3CA mutant tumors more effectively than the wild type tumors in an animal model
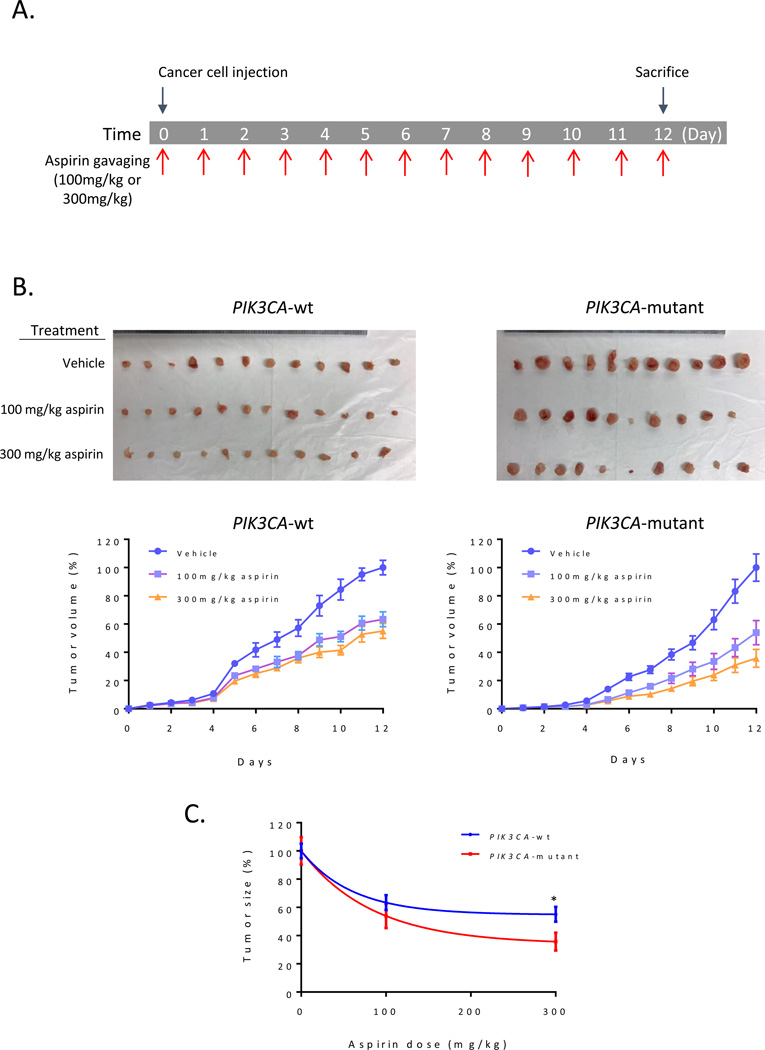
In order to confirm our in vitro findings, we generated xenograft tumors using HCT116 cells with or with PIK3CA-mutation. We then gavaged the animals with two different doses of aspirin (100 and 300 mg/kg body weight) every day (Figure 4A). Aspirin significantly inhibited tumor growth in both PIK3CA-mutant and wild type tumors (Figure 4B). In particular, aspirin more significantly inhibited tumor growth of PIK3CA mutant tumors compared to tumors with wild type PIK3CA (Figure 4C). These data collectively support our in vitro finding that aspirin is more effective against PIK3CA-mutant tumors in vivo.
Figure 4.
Aspirin is more sensitive to PIK3CA-mutated tumors. A) Graphical representation of aspirin treatment strategy B) Representation of tumor size (top) and progressive tumor volume increase during treatment period (bottom) for wild type and mutant tumors. C) PIK3CA-mutant cells were more sensitive than wild-type cells at (100 mg/kg aspirin) after 12 days of treatment. *, p value <0.05
Amount of cell cycle arrest explains the effect of aspirin on cell growth
Our analysis found that aspirin significantly reduced the growth of all CRC cell lines and this effect of aspirin was greatest on the faster growing cell lines with PIK3CA gene mutations. To further explore these findings, we analyzed cell cycle dynamics at every time point and dose of aspirin for each cell line. Next, we plotted the percent of cells in G0/G1 cell cycle phase as a function of the aspirin dose for each cell line and for each time point. The Figure 5A and Supplemental Figure 6 show the percentage of cells in G0/G1 phase at a particular time (96 h) as a function of the aspirin dose for each cell line. Calculating the slope of the linear regression over time demonstrates how strongly aspirin affects the percentage of G0/G1 cells. Figure 5B illustrates these slope values for all the cell lines over all time points. As shown, we observed distinct clustering of cell lines depending upon their PIK3CA mutational status; whereby, PIK3CA-mutant cell lines were significantly more responsive to aspirin compared to the wild-type cell lines. We next determined the statistical significance of grouping the cells by their PIK3CA status; again, we observed a more significant response to aspirin in cell lines with a PIK3CA mutation vs. wild-type status (Figure 5C). On similar lines, Figure 5D highlights how the magnitude of the effect of aspirin correlates with the natural growth rate of cells in the absence of aspirin. Corroborating our earlier findings, cells with a PIK3CA gene mutation had a faster growth rate in the absence of aspirin compared with wild-type cell lines. All PIK3CA-mutant cell lines demonstrated a dose dependent increase in G0/G1 cell cycle arrest, while the PIK3CA-wt cell lines did not, and a more pronounced cell cycle arrest was observed for cell lines that grew faster in the absence of treatment, which tended to be the PIK3CA-mutants.
Figure 5.
The effect of aspirin on the proportion of cells in G0/G1 arrest. A) The slope of the % of cells in G0/G1 cell cycle phase with respect to all aspirin doses at t=96 hours normalized with the % of cells in G0/G1 phase in the absence of aspirin for all cell lines. B) Slopes (aspirin sensitivity) of G0/G1 arrest per aspirin dose plotted at each time point withPIK3CA-mutant cell lines (dotted lines) and PIK3CA-wt (solid lines). C) Grouping the cell lines in (B) into two classes according to their PIK3CA status. D) The effect of aspirin on the % of cells in G0/G1. Each point corresponds to a single cell line. The linear regression is also plotted, with p=0.029. Student’s T-test was performed to determine significance. Error bars represent ± SEM of grouped cell lines. *, p value <0.05 and **, p<0.01.
PI3K-Akt pathway dysregulation results in differential regulation of cell cycle regulatory genes
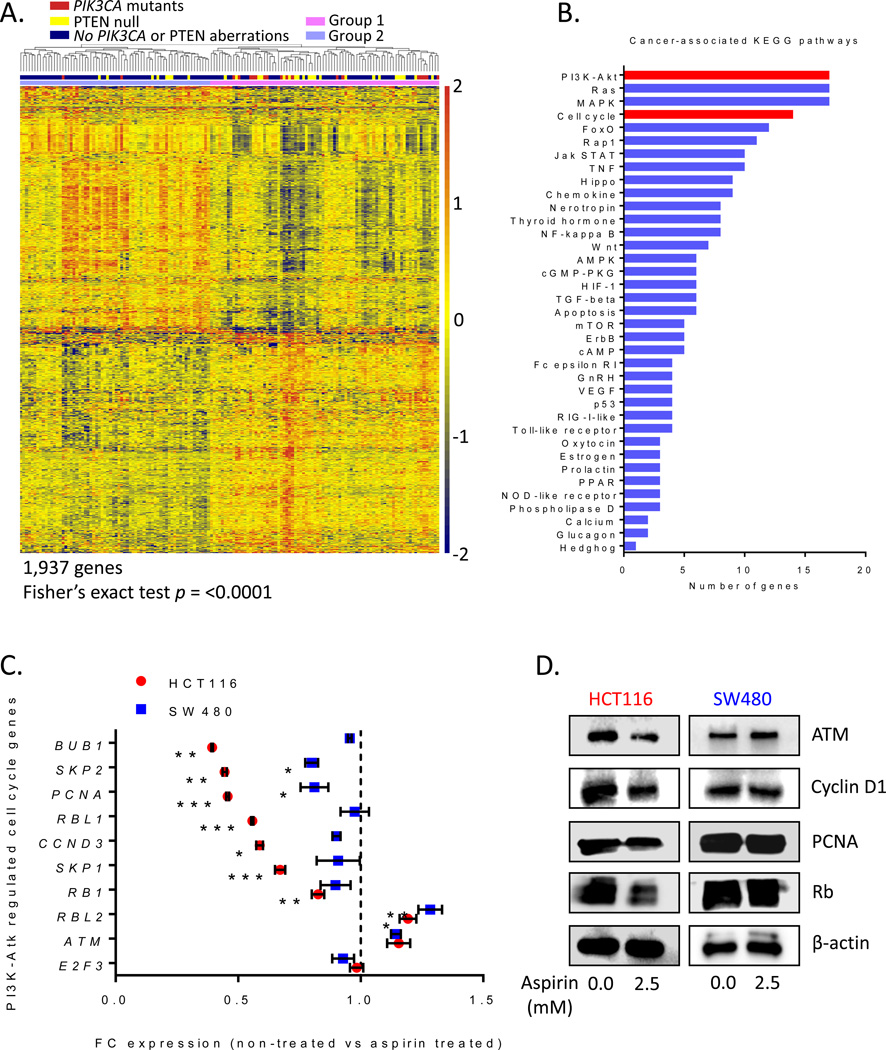
The phosphatidylinositol 3-kinase (PI3K)-Akt pathway regulates cell survival through cellular proliferation and metabolic processes and it is constitutively activated in over a third of CRCs due to mutations in coding exons of PIK3CA (30) or mutational silencing of the PTEN gene (31). A comprehensive explanation of mechanisms that contribute to aspirin sensitivity in cells with PIK3CA mutations is yet to be established. To help answer this question, we used a bioinformatics approach to explore cancer-associated pathways differentially expressed in PIK3CA mutated vs. wild-type CRC cells. PTEN normally antagonizes the PI3K-Akt-mTOR pathway (32); therefore, to fully explore cancer-associated gene dysregulation by the PI3K-Akt axis, we extended our analysis to include PTEN null CRC cells for determining the role of the PTEN gene in response to aspirin. The GSE59857 dataset was obtained from NCBI’s Gene Expression Omnibus (GEO) (33) to identify cell cycle genes differentially regulated in the context of PI3K-Akt dysregulation. This dataset comprised of expression data for 151 CRC cell lines, 30 of which harbored PIK3CA mutations, 21 were PTEN null, and two had concurrent mutations in both genes. The microarray expression data (GSE59857) were uploaded via the GEOquery package in R programming language, normalized, and filtered to detect differential gene expression (See Materials and Materials & METHODS; Figure 6A). Differentially expressed genes were further filtered with a cut-off of ±0.3 fold-change and p<0.05. Hierarchically clustering segregated the cell lines into two major groups. Fisher’s exact test confirmed that ‘Group 2’ was indeed enriched for cell lines with PIK3CA/PTEN mutations while ‘Group 1’ was not. Differentially expressed genes were interpreted biologically for higher-level system functions by mapping into the KEGG (Kyoto Encyclopedia of Genes and Genomes) Pathway database (34). Our analysis identified PI3K-Akt among the top ranked cancer-associated pathways, which confirmed the appropriateness of our gene selection criteria (Figure 6B). Our analysis also identified several bona fide cell cycle genes regulated by the PI3K-Akt signaling cascade, such as ATM (35), BUB1 (36), CCND3 (37), E2F3 (38), PCNA (39), RB1 (40) /RBL1/RBL2, SKP1, and SKP2 (41). Analysis of this dataset identified several cell cycle-related genes that are down-stream targets of the PI3K-Akt pathway.
Figure 6.
Differentially expressed cell cycle genes in CRC cells with PIK3CA/PTEN gene mutations. A) Heat map of hierarchically-clustered differentially-expressed genes filtered with ± 0.3 fold-change and p<0.05 cut-off. Fisher’s exact test determined enrichment of PIK3CA/PTEN mutant CRC cell lines in a single group. B) Higher-level system biological interpretation of filtered genes using the KEGG Pathway database (34). Data displayed as the number of genes mapped to each cancer-associated pathway. (C) PI3K-Akt regulated cell cycle genes are downregulated by aspirin. Transcriptional changes of key cell cycle genes were measured after 48 hours of 2.5 mM aspirin treatment. Data displayed as fold-change (FC) from non-treated conditions. D) Western blots of PCNA, cyclin D1, ATM and retinoblastoma proteins show downregulation at the protein level in PIK3CA-mutant (HCT116) and WT (SW480) cell lines. β-actin served as a loading control. Error bars represent ± SEM of grouped cell lines. *, p value <0.05; **, p<0.01; and ***, p<0.001.
Aspirin dysregulates PI3K-Akt-regulated cell cycle genes
Aspirin induces changes to ribosomal gene expression in PIK3CA-mutant HT-29 cells (42). To determine whether aspirin potentially targets cell cycle-related genes regulated by the PI3K-Akt pathway, transcriptional changes of identified targets were measured using 2.5 mM aspirin treatment for 48 hours in HCT116 and SW480 cells. This concentration was deemed optimal for determining the difference in responses between PIK3CA mutant and wild-type cells (see Figure 2). Several cell cycle associated genes (BUB1, SKP2, PCNA, RBL1, CCND3, SKP1, and RB1 were downregulated significantly in HCT116 cells upon aspirin treatment, much more than that seen in cells with wild-type PIK3CA (Figure 6C). It should be noted that SKP2 and PCNA were also downregulated in SW480 cells, however this effect was much less pronounced than in HCT116 cells. RBL2 was upregulated in both cell lines, while ATM and E2F3 did not change significantly. The results of this experiment demonstrated that cell cycle genes regulated by the PI3K-Akt pathway were significantly more sensitive to aspirin in PIK3CA mutated HCT116 cells, compared to PIK3CA wild-type SW480 cells. Western blots were performed to determine if protein expression in response to aspirin was affected in these two cell lines. We noted that PCNA and Rb oncogenes were both downregulated in response to aspirin treatment in the HCT116 cells (Figure 6D). Likewise, the expression of cyclin D1 was markedly downregulated in both cell lines following aspirin exposure, further supporting the observations for observed cell cycle arrest in these cell lines. Taken together, we observed a significant concordance between gene and protein expression for various cell cycle-related genes in both cell lines treated with aspirin.
DISCUSSION
This study was a unique effort to comprehensively understand aspirin-induced chemoprevention in CRC, the underlying kinetics of cell growth inhibition, and the impact of aspirin responsiveness on differing molecular backgrounds. Herein, we firstly demonstrated that aspirin negatively affected the growth of a comprehensive panel of tumor cell lines and clearly revealed that the extent of growth inhibition depends on the molecular characteristics of each cell line. These in vitro findings were subsequently validated in a xenograft animal model. Furthermore, we determined that pharmacologically relevant doses of aspirin reduced cell growth primarily by decreasing the rate of cell division, increasing G0/G1 arrest, and causing cell death. Most profoundly, this effect of aspirin inhibition of cell growth was more pronounced in cells that inherently grow faster, and these very same cells had PIK3CA gene mutations. Greater sensitivity to aspirin upon rapidly dividing cells is biologically relevant to drug interference, because faster growing tumors commonly respond better to chemotherapy than slow growing tumors. It is possible that heightened sensitivity to aspirin in cells with PIK3CA gene mutations was a consequence more rapid cell division. Alternatively, mutations in the PIK3CA gene may directly affect cell lines by rendering them more sensitive to aspirin inhibition. However, this question could not be resolved in the present study. An intriguing aspect of our results was that relatively low doses of aspirin inhibited cell growth, which was most pronounced in the PIK3CA-mutant cell lines. These observations corroborate the findings of Liao et al. who noted that patients with PIK3CA-mutated cancers survived longer than those with wild-type PIK3CA tumors when taking aspirin. This study highlighted that heterogeneity in cellular responses to aspirin may be very important to better tailor aspirin use in cancer prevention and treatment.
Cell lines with PIK3CA mutations underwent more acute G0/G1 arrest upon aspirin treatment when compared to those without similar mutations. Our findings strongly suggest that aspirin profoundly inhibits cells with mutations that occur in late adenoma development (15). Furthermore, our mathematical modeling approach supports the hypothesis that specific genetic variants or mutational backgrounds are major determinants of aspirin’s effectiveness and indicates that patients with unregulated PIK3-Akt activity may have a more desirable risk-benefit response from aspirin.
The relationship between genetic variants and the chemopreventive use of aspirin is not limited to patients with CRC with PIK3CA mutations. A genome-wide analysis of the interactions between single-nucleotide polymorphisms (SNPs) and aspirin use in relation to CRC risk identified two SNPs on chromosome 12 and 15 that differentially correlate with aspirin and NSAID effects on CRC prevention. One is located upstream from the phosphatidylinositol-4-phosphate 3-kinase catalytic subunit type 2 gamma (PIK3C2G), which belongs to the PI3K family (43) and therefore is particularly relevant to our study. Aspirin and/or NSAID use associated with reduced risk of CRC among individuals with this genotype.
We must account for the limitations in our experimental methods. KRAS mutations may activate the PI3K-Akt pathway, thus, it diminishes our ability to use microarray to discern genes that are dysregulated by constitutively active PI3K-Akt alone. Amplifications of PIK3CA have been reported (44); however, our cell line panel did not include any with PIK3CA amplifications; therefore we cannot comment on the effect of aspirin on CRCs with similar alterations. We cannot discount the possibility that contact inhibition and spatial restrictions of the cells confound growth dynamics at later time points. We minimized these confounders by seeding each cell line at the lowest possible concentration that could still be assessed for viability, cell numbers, and cycle analysis. While the present study focused primarily on the effects of aspirin on PIK3CA mutations, aspirin has been known to modulate several key molecular pathways including COX-mediated inhibition of PGE2, and serves as an immune-modulator (45,46). Also, in vitro experiments may not recapitulate in vivo studies, although certainly the results from the human studies (3–5,7–9,47) mentioned above are strong proof that our findings are relevant in humans.
The potential short-term or long-term toxicity of aspirin has discouraged routine clinical use despite many studies demonstrating its benefits for patients after a diagnosis of CRC. The doses used in our study are clinically relevant because they correspond to salicylate concentrations found in the plasma of human patients who use aspirin to control arthritis (48): salicylate and other salicylate metabolites derived from the rapid hydrolyzation of aspirin under in vivo (49) and in vitro (50) conditions. However, this comparison is relatively ersatz because in vitro systems are inadequate substitutes for replicating the metabolic conditions found in epithelial and tumor tissues. Furthermore, we acknowledge that the aspirin concentrations used in the study might not reflect physiological doses used for clinical purposes as a chemopreventive agent. However, the primary purpose of this study was to investigate the effects of aspirin on CRC cell proliferation, where we were not surprised that a high dose of aspirin was needed to alter the cancer cell growth. Nevertheless, the concentrations used in the study were consistent to that of previous studies (51–54). In addition, the time of aspirin exposure to cancer cells in our model is shorter than that practiced during long-term clinical use.
Our study elegantly represented the effect of aspirin on an already growing tumor cell population. It is possible that the reduction of an already growing tumor cell population constitutes an important mechanism by which aspirin delays the growth of tumors to undetectable levels. Aspirin might, however, also act by modulating cellular processes in healthy tissues, reducing the probability that a successfully growing tumor is generated in the first place. This is one aspect of the action of aspirin we currently are investigating. It will be important to merge the effect of aspirin on these two phases of carcinogenesis in order to obtain a greater understanding of the role of aspirin in chemoprevention. It is, however, currently unclear whether the PIK3CA mutation status per se that determines cellular sensitivity to aspirin or whether it primarily is driven by the natural division rate of tumor cells. This needs to be investigated further, perhaps investigating the effect of aspirin on cell growth in tumor cells with other mutations that are carcinogenic in future studies.
Supplementary Material
Acknowledgments
Authors would like to thank Divya Pasham for her technical assistance.
Grant Funding: The present work was supported by the grants R01 CA72851, CA181572, CA184792 and U01 CA187956 from the National Cancer Institute, National Institute of Health, pilot grants from the Baylor Sammons Cancer Center and Foundation, as well as funds from the Baylor Research Institute to A.G.
Footnotes
Disclosure of Potential Conflicts of interst: The authors disclose no potential conflicts of interest.
REFERENCES
- 1.Ng K, Meyerhardt JA, Chan AT, Sato K, Chan JA, Niedzwiecki D, et al. Aspirin and COX-2 inhibitor use in patients with stage III colon cancer. Journal of the National Cancer Institute. 2015;107(1):345. doi: 10.1093/jnci/dju345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Din FV, Theodoratou E, Farrington SM, Tenesa A, Barnetson RA, Cetnarskyj R, et al. Effect of aspirin and NSAIDs on risk and survival from colorectal cancer. Gut. 2010;59(12):1670–1679. doi: 10.1136/gut.2009.203000. [DOI] [PubMed] [Google Scholar]
- 3.Baron JA, Cole BF, Sandler RS, Haile RW, Ahnen D, Bresalier R, et al. A randomized trial of aspirin to prevent colorectal adenomas. The New England journal of medicine. 2003;348(10):891–899. doi: 10.1056/NEJMoa021735. [DOI] [PubMed] [Google Scholar]
- 4.Rothwell PM, Fowkes FG, Belch JF, Ogawa H, Warlow CP, Meade TW. Effect of daily aspirin on long-term risk of death due to cancer: analysis of individual patient data from randomised trials. Lancet. 2011;377(9759):31–41. doi: 10.1016/S0140-6736(10)62110-1. [DOI] [PubMed] [Google Scholar]
- 5.Rothwell PM, Wilson M, Elwin CE, Norrving B, Algra A, Warlow CP, et al. Long-term effect of aspirin on colorectal cancer incidence and mortality: 20-year follow-up of five randomised trials. Lancet. 2010;376(9754):1741–1750. doi: 10.1016/S0140-6736(10)61543-7. [DOI] [PubMed] [Google Scholar]
- 6.Sandler RS, Halabi S, Baron JA, Budinger S, Paskett E, Keresztes R, et al. A randomized trial of aspirin to prevent colorectal adenomas in patients with previous colorectal cancer. The New England journal of medicine. 2003;348(10):883–890. doi: 10.1056/NEJMoa021633. [DOI] [PubMed] [Google Scholar]
- 7.Rothwell PM, Wilson M, Price JF, Belch JF, Meade TW, Mehta Z. Effect of daily aspirin on risk of cancer metastasis: a study of incident cancers during randomised controlled trials. Lancet. 2012;379(9826):1591–1601. doi: 10.1016/S0140-6736(12)60209-8. [DOI] [PubMed] [Google Scholar]
- 8.Bastiaannet E, Sampieri K, Dekkers OM, de Craen AJ, van Herk-Sukel MP, Lemmens V, et al. Use of aspirin postdiagnosis improves survival for colon cancer patients. British journal of cancer. 2012;106(9):1564–1570. doi: 10.1038/bjc.2012.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chan AT, Ogino S, Fuchs CS. Aspirin use and survival after diagnosis of colorectal cancer. JAMA. 2009;302(6):649–658. doi: 10.1001/jama.2009.1112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Reimers MS, Bastiaannet E, van Herk-Sukel MP, Lemmens VE, van den Broek CB, van de Velde CJ, et al. Aspirin use after diagnosis improves survival in older adults with colon cancer: a retrospective cohort study. Journal of the American Geriatrics Society. 2012;60(12):2232–2236. doi: 10.1111/jgs.12033. [DOI] [PubMed] [Google Scholar]
- 11.Comprehensive molecular characterization of human colon and rectal cancer. Nature. 2012;487(7407):330–337. doi: 10.1038/nature11252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Burn J, Gerdes AM, Macrae F, Mecklin JP, Moeslein G, Olschwang S, et al. Long-term effect of aspirin on cancer risk in carriers of hereditary colorectal cancer: an analysis from the CAPP2 randomised controlled trial. Lancet. 2011;378(9809):2081–2087. doi: 10.1016/S0140-6736(11)61049-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Burn J, Bishop DT, Mecklin JP, Macrae F, Moslein G, Olschwang S, et al. Effect of aspirin or resistant starch on colorectal neoplasia in the Lynch syndrome. The New England journal of medicine. 2008;359(24):2567–2578. doi: 10.1056/NEJMoa0801297. [DOI] [PubMed] [Google Scholar]
- 14.Hadac JN, Leystra AA, Paul Olson TJ, Maher ME, Payne SN, Yueh AE, et al. Colon Tumors with the Simultaneous Induction of Driver Mutations in APC, KRAS, and PIK3CA Still Progress through the Adenoma-to-carcinoma Sequence. Cancer prevention research. 2015;8(10):952–961. doi: 10.1158/1940-6207.CAPR-15-0003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fearon ER, Vogelstein B. A genetic model for colorectal tumorigenesis. Cell. 1990;61(5):759–767. doi: 10.1016/0092-8674(90)90186-i. [DOI] [PubMed] [Google Scholar]
- 16.Pino MS, Chung DC. The chromosomal instability pathway in colon cancer. Gastroenterology. 2010;138(6):2059–2072. doi: 10.1053/j.gastro.2009.12.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Boland CR, Sato J, Appelman HD, Bresalier RS, Feinberg AP. Microallelotyping defines the sequence and tempo of allelic losses at tumour suppressor gene loci during colorectal cancer progression. Nature medicine. 1995;1(9):902–909. doi: 10.1038/nm0995-902. [DOI] [PubMed] [Google Scholar]
- 18.Boland CR, Goel A. Microsatellite instability in colorectal cancer. Gastroenterology. 2010;138(6):2073–2087. doi: 10.1053/j.gastro.2009.12.064. e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Goel A, Boland CR. Epigenetics of colorectal cancer. Gastroenterology. 2012;143(6):1442–1460. doi: 10.1053/j.gastro.2012.09.032. e1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liao X, Lochhead P, Nishihara R, Morikawa T, Kuchiba A, Yamauchi M, et al. Aspirin use, tumor PIK3CA mutation, and colorectal-cancer survival. The New England journal of medicine. 2012;367(17):1596–1606. doi: 10.1056/NEJMoa1207756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gayet J, Zhou XP, Duval A, Rolland S, Hoang JM, Cottu P, et al. Extensive characterization of genetic alterations in a series of human colorectal cancer cell lines. Oncogene. 2001;20(36):5025–5032. doi: 10.1038/sj.onc.1204611. [DOI] [PubMed] [Google Scholar]
- 22.Barretina J, Caponigro G, Stransky N, Venkatesan K, Margolin AA, Kim S, et al. The Cancer Cell Line Encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature. 2012;483(7391):603–607. doi: 10.1038/nature11003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ahmed D, Eide PW, Eilertsen IA, Danielsen SA, Eknaes M, Hektoen M, et al. Epigenetic and genetic features of 24 colon cancer cell lines. Oncogenesis. 2013;2:e71. doi: 10.1038/oncsis.2013.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Haugen AC, Goel A, Yamada K, Marra G, Nguyen TP, Nagasaka T, et al. Genetic instability caused by loss of MutS homologue 3 in human colorectal cancer. Cancer research. 2008;68(20):8465–8472. doi: 10.1158/0008-5472.CAN-08-0002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Poon IK, Chiu YH, Armstrong AJ, Kinchen JM, Juncadella IJ, Bayliss DA, et al. Unexpected link between an antibiotic, pannexin channels and apoptosis. Nature. 2014;507(7492):329–334. doi: 10.1038/nature13147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Marshall OJ. PerlPrimer: cross-platform, graphical primer design for standard, bisulphite and real-time PCR. Bioinformatics. 2004;20(15):2471–2472. doi: 10.1093/bioinformatics/bth254. [DOI] [PubMed] [Google Scholar]
- 27.Kent WJ, Sugnet CW, Furey TS, Roskin KM, Pringle TH, Zahler AM, et al. The human genome browser at UCSC. Genome research. 2002;12(6):996–1006. doi: 10.1101/gr.229102. Article published online before print in May 2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Davis S, Meltzer PS. GEOquery: a bridge between the Gene Expression Omnibus (GEO) and BioConductor. Bioinformatics. 2007;23(14):1846–1847. doi: 10.1093/bioinformatics/btm254. [DOI] [PubMed] [Google Scholar]
- 29.Du P, Kibbe WA, Lin SM. lumi: a pipeline for processing Illumina microarray. Bioinformatics. 2008;24(13):1547–1548. doi: 10.1093/bioinformatics/btn224. [DOI] [PubMed] [Google Scholar]
- 30.Samuels Y, Wang Z, Bardelli A, Silliman N, Ptak J, Szabo S, et al. High frequency of mutations of the PIK3CA gene in human cancers. Science. 2004;304(5670):554. doi: 10.1126/science.1096502. [DOI] [PubMed] [Google Scholar]
- 31.Guanti G, Resta N, Simone C, Cariola F, Demma I, Fiorente P, et al. Involvement of PTEN mutations in the genetic pathways of colorectal cancerogenesis. Human molecular genetics. 2000;9(2):283–287. doi: 10.1093/hmg/9.2.283. [DOI] [PubMed] [Google Scholar]
- 32.Maehama T, Dixon JE. The tumor suppressor, PTEN/MMAC1, dephosphorylates the lipid second messenger, phosphatidylinositol 3,4,5-trisphosphate. The Journal of biological chemistry. 1998;273(22):13375–13378. doi: 10.1074/jbc.273.22.13375. [DOI] [PubMed] [Google Scholar]
- 33.Medico E, Russo M, Picco G, Cancelliere C, Valtorta E, Corti G, et al. The molecular landscape of colorectal cancer cell lines unveils clinically actionable kinase targets. Nature communications. 2015;6:7002. doi: 10.1038/ncomms8002. [DOI] [PubMed] [Google Scholar]
- 34.Ogata H, Goto S, Sato K, Fujibuchi W, Bono H, Kanehisa M. KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res. 1999;27(1):29–34. doi: 10.1093/nar/27.1.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Savitsky K, Bar-Shira A, Gilad S, Rotman G, Ziv Y, Vanagaite L, et al. A single ataxia telangiectasia gene with a product similar to PI-3 kinase. Science. 1995;268(5218):1749–1753. doi: 10.1126/science.7792600. [DOI] [PubMed] [Google Scholar]
- 36.Kiyomitsu T, Obuse C, Yanagida M. Human Blinkin/AF15q14 is required for chromosome alignment and the mitotic checkpoint through direct interaction with Bub1 and BubR1. Developmental cell. 2007;13(5):663–676. doi: 10.1016/j.devcel.2007.09.005. [DOI] [PubMed] [Google Scholar]
- 37.Cato MH, Chintalapati SK, Yau IW, Omori SA, Rickert RC. Cyclin D3 is selectively required for proliferative expansion of germinal center B cells. Molecular and cellular biology. 2011;31(1):127–137. doi: 10.1128/MCB.00650-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wu L, Timmers C, Maiti B, Saavedra HI, Sang L, Chong GT, et al. The E2F1-3 transcription factors are essential for cellular proliferation. Nature. 2001;414(6862):457–462. doi: 10.1038/35106593. [DOI] [PubMed] [Google Scholar]
- 39.Mathews MB, Bernstein RM, Franza BR, Jr, Garrels JI. Identity of the proliferating cell nuclear antigen and cyclin. Nature. 1984;309(5966):374–376. doi: 10.1038/309374a0. [DOI] [PubMed] [Google Scholar]
- 40.Mihara K, Cao XR, Yen A, Chandler S, Driscoll B, Murphree AL, et al. Cell cycle-dependent regulation of phosphorylation of the human retinoblastoma gene product. Science. 1989;246(4935):1300–1303. doi: 10.1126/science.2588006. [DOI] [PubMed] [Google Scholar]
- 41.Schulman BA, Carrano AC, Jeffrey PD, Bowen Z, Kinnucan ER, Finnin MS, et al. Insights into SCF ubiquitin ligases from the structure of the Skp1-Skp2 complex. Nature. 2000;408(6810):381–386. doi: 10.1038/35042620. [DOI] [PubMed] [Google Scholar]
- 42.Yin H, Xu H, Zhao Y, Yang W, Cheng J, Zhou Y. Cyclooxygenase-independent effects of aspirin on HT-29 human colon cancer cells, revealed by oligonucleotide microarrays. Biotechnology letters. 2006;28(16):1263–1270. doi: 10.1007/s10529-006-9084-9. [DOI] [PubMed] [Google Scholar]
- 43.Nan H, Hutter CM, Lin Y, Jacobs EJ, Ulrich CM, White E, et al. Association of aspirin and NSAID use with risk of colorectal cancer according to genetic variants. JAMA. 2015;313(11):1133–1142. doi: 10.1001/jama.2015.1815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jehan Z, Bavi P, Sultana M, Abubaker J, Bu R, Hussain A, et al. Frequent PIK3CA gene amplification and its clinical significance in colorectal cancer. The Journal of pathology. 2009;219(3):337–346. doi: 10.1002/path.2601. [DOI] [PubMed] [Google Scholar]
- 45.Tsujii M, Kawano S, Tsuji S, Sawaoka H, Hori M, DuBois RN. Cyclooxygenase regulates angiogenesis induced by colon cancer cells. Cell. 1998;93(5):705–716. doi: 10.1016/s0092-8674(00)81433-6. [DOI] [PubMed] [Google Scholar]
- 46.Tougeron D, Sha D, Manthravadi S, Sinicrope FA. Aspirin and colorectal cancer: back to the future. Clin Cancer Res. 2014;20(5):1087–1094. doi: 10.1158/1078-0432.CCR-13-2563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chan AT, Ogino S, Fuchs CS. Aspirin and the risk of colorectal cancer in relation to the expression of COX-2. The New England journal of medicine. 2007;356(21):2131–2142. doi: 10.1056/NEJMoa067208. [DOI] [PubMed] [Google Scholar]
- 48.Pachman LM, Olufs R, Procknal JA, Levy G. Pharmacokinetic monitoring of salicylate therapy in children with juvenile rheumatoid arthritis. Arthritis and rheumatism. 1979;22(8):826–831. doi: 10.1002/art.1780220804. [DOI] [PubMed] [Google Scholar]
- 49.Gao J, Kashfi K, Liu X, Rigas B. NO-donating aspirin induces phase II enzymes in vitro and in vivo. Carcinogenesis. 2006;27(4):803–810. doi: 10.1093/carcin/bgi262. [DOI] [PubMed] [Google Scholar]
- 50.Levy G, Tsuchiya T. Salicylate accumulation kinetics in man. The New England journal of medicine. 1972;287(9):430–432. doi: 10.1056/NEJM197208312870903. [DOI] [PubMed] [Google Scholar]
- 51.Gao M, Kong Q, Hua H, Yin Y, Wang J, Luo T, et al. AMPK-mediated up-regulation of mTORC2 and MCL-1 compromises the anti-cancer effects of aspirin. Oncotarget. 2016;7(13):16349–16361. doi: 10.18632/oncotarget.7648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Jung YR, Kim EJ, Choi HJ, Park JJ, Kim HS, Lee YJ, et al. Aspirin Targets SIRT1 and AMPK to Induce Senescence of Colorectal Carcinoma Cells. Mol Pharmacol. 2015;88(4):708–719. doi: 10.1124/mol.115.098616. [DOI] [PubMed] [Google Scholar]
- 53.Moon CM, Kwon JH, Kim JS, Oh SH, Jin Lee K, Park JJ, et al. Nonsteroidal anti-inflammatory drugs suppress cancer stem cells via inhibiting PTGS2 (cyclooxygenase 2) and NOTCH/HES1 and activating PPARG in colorectal cancer. Int J Cancer. 2014;134(3):519–529. doi: 10.1002/ijc.28381. [DOI] [PubMed] [Google Scholar]
- 54.Goel A, Chang DK, Ricciardiello L, Gasche C, Boland CR. A novel mechanism for aspirin-mediated growth inhibition of human colon cancer cells. Clin Cancer Res. 2003;9(1):383–390. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.