SUMMARY
Lack of sensitive single-cell analysis tools has limited the characterization of metabolic activity in cancer stem cells. By hyperspectral stimulated Raman scattering imaging of single living cells and mass spectrometry analysis of extracted lipids, we report here significantly increased levels of unsaturated lipids in ovarian cancer stem cells (CSCs) as compared to non-CSCs. Higher lipid unsaturation levels were also detected in CSC-enriched spheroids compared to monolayer cultures of ovarian cancer cell lines or primary cells. Inhibition of lipid desaturases effectively eliminated CSCs, suppressed sphere formation in vitro, and blocked tumor initiation capacity in vivo. Mechanistically, we demonstrate that NF-κB directly regulates the expression levels of lipid desaturases and that inhibition of desaturases blocks NF-κB signaling. Collectively, our findings reveal that increased lipid unsaturation is a metabolic marker for ovarian CSCs and a target for CSC-specific therapy.
Keywords: lipid desaturation, ovarian cancer, cancer stem cells, Raman spectroscopic imaging, NF-κB
Graphical abstract
INTRODUCTION
Cancer stem cells (CSCs), also recognized as tumor initiating cells, represent a small population of cancer cells that have the ability to self-renew and initiate tumors in vivo (Bjerkvig et al., 2005). CSCs are resistant to conventional therapies and are responsible for tumor relapse after chemotherapy (Jordan et al., 2006; Pattabiraman and Weinberg, 2014) and development of distant metastases (Jordan et al., 2006). Understanding their unique characteristics and vulnerabilities will enable the development of CSC-targeting therapies with the ultimate goal of overcoming tumor relapse and metastasis.
Recent studies have focused on blocking signaling pathways or genetic programs that fuel cellular “stemness”. For example, epithelial to mesenchymal transition (EMT) is emerging as a key program in CSCs required for initiation of metastasis (Visvader and Lindeman, 2008a). Related signaling pathways, like Wnt and transforming growth factor β (TGF-β), are recognized as new targets for CSC-specific therapy (Pattabiraman and Weinberg, 2014). The Hedgehog and Notch pathways implicated in the self-renewal of CSCs (Zhou et al., 2009) are being targeted and specific inhibitors have recently entered clinical development. In ovarian cancer, the Mullerian Inhibiting Substance was proposed as a potential targeting strategy for chemotherapy resistant CSCs (Meirelles et al., 2012; Szotek et al., 2006). However, as the development of technologies enabling the study of rare populations has lagged, one underexplored niche remains understanding the CSC metabolism.
So far, only limited studies have begun to address this niche. A recent report suggested that glucose played an important role maintaining the side population in non-small lung and colon cancer models and that inhibition of glycolysis blocked this population (Liu et al., 2014). A few studies have linked lipogenesis to CSCs. Specifically, inhibition of fatty acid synthase was shown to suppress the growth of breast cancer stem-like cells in vivo (Pandey et al., 2011); the peroxisome proliferator-activated receptor γ (PPARγ) pathway was found important in maintaining the CSC properties of ERBB2-positive breast cancer cells partly by upregulating the de novo lipogenic pathway (Wang et al., 2013), and increased numbers of lipid droplets were identified in colorectal CSCs compared to differentiated cancer cells (Tirinato et al., 2015). Collectively, these studies point to lipogenesis as a potentially altered metabolic process in CSCs, but the precise mechanism by which lipids regulate stemness remains unknown.
In this study, we identify and characterize lipid unsaturation in ovarian CSCs by chemical imaging of single living cells through recently developed hyperspectral stimulated Raman scattering (SRS) microscopy (Cheng and Xie, 2015; Zhang et al., 2015). This single-cell imaging study and mass spectrometry analysis show a significantly increased level of lipid unsaturation in flow-sorted ovarian CSCs (ALDH+/CD133+) compared to non-CSCs (ALDH−/CD133−) and in ovarian cancer (OC) cells growing as spheres compared to monolayers. Inhibition of lipid desaturases, either Δ9 (SCD1) or Δ6, impaired cancer cell stemness, suppressed sphere formation, and prevented tumor formation in vivo. We further identified the NF-κB pathway as a critical mechanism through which lipid desaturase inhibitors perturb the functions of CSCs. Collectively, our findings put forward lipid desaturation as a metabolic marker of ovarian CSCs and as a new target for CSC-specific therapy.
RESULTS
Increased lipid unsaturation in isolated ovarian CSCs compared to non-CSCs
We employed hyperspectral SRS microscopy to quantitatively analyze the composition of intracellular lipids inside single cells. ALDH+/CD133+ cells, which have been previously described as cells possessing CSC characteristics (Flesken-Nikitin et al., 2014; Foster et al., 2013), were isolated from COV362, an ovarian cancer (OC) cell line. By tuning the Raman shift frame by frame, 50 images of individual ALDH−/CD133− and ALDH+/CD133+ COV362 cells were recorded at the C-H vibration region from 2800 to 3050 cm−1 with a step size of ~5 cm−1 (Supplemental movie 1, 2). Inside each cell, the C-H bond rich lipid droplets were highlighted in the context of relatively weaker signals from protein and nucleotides. Initial analysis of the images at 2850 cm−1, where the CH2 symmetric stretch vibration resides, revealed an increase of total amount of lipid droplets in ALDH+/CD133+ cells. An in-depth comparison revealed that lipid droplets in ALDH+/CD133+ cells had a stronger signal at 3002 cm−1 than those in ALDH−/CD133− cells (Fig. 1A). As the peak at 3002 cm−1 is known to be from the vibration of =C-H bonds mostly in unsaturated lipids (Movasaghi et al., 2007), our data suggests that ALDH+/CD133+ CSC cells have a higher level of unsaturated lipids. To eliminate the influence of the total number of C-H bonds, we used the peak around 2900 cm−1, contributed by C-H stretch vibrations in all the lipids (Movasaghi et al., 2007), to normalize the SRS intensity at 3002 cm−1. After normalization, the ratio of SRS intensity at 3002 cm−1 to that at 2900 cm−1 (I3002/I2900) was found to be significantly higher in ALDH+/CD133+ cells than that in ALDH−/CD133− cells (Fig. 1A). The difference of the peak intensity at 3002 cm−1 became more obvious when we plotted together the SRS spectra from different lipid droplets (Fig. 1B). Quantitatively, we found a statistically significant difference between the two groups in terms of lipid unsaturation level (Fig. S1A).
Figure 1. Increased lipid unsaturation level in sorted ALDH+/CD133+ ovarian cancer cells.
(A) Representative hyperspectral SRS images of flow-sorted ALDH−/CD133− and ALDH+/CD133+ COV362 cells. Images at 2900 cm−1, 3002 cm−1 and the intensity ratio image between 3002 and 2900 cm−1 are shown. Scale bars: 10 μm. (B) Average SRS spectra from the lipid droplets in ALDH−/CD133− (n=3) and ALDH+/CD133+ cells (n=8). Shaded area indicates the standard deviation (SD) of SRS spectral measurement from different cells. (C) Spontaneous Raman spectra taken from LDs in ALDH−/CD133− and ALDH+/CD133+ sorted COV362 cells. The spectra were normalized by the height of the Raman peak at 1450 cm−1. The differences at 1264 cm−1, 1660 cm−1, and 3002 cm−1 were highlighted in gray. (D) Scatter plot of Raman spectra height ratio between the peaks at 3002 cm−1 and 1450 cm−1 in ALDH−/CD133− and ALDH+/CD133+ COV362 cells. Each dot represents a single cell, and the bars indicate means ± SEM; P = 0.0005. (E) Raman spectra taken from LDs in ALDH−/CD133− and ALDH+/CD133+ OVCAR5 cells. The spectra were normalized by the height of peak at 1450 cm−1. The differences at 1264 cm−1, 1660 cm−1, and 3002 cm−1 were highlighted in gray. (F) Scatter plot of Raman spectra height ratio between the peaks at 3002 cm−1 and 1450 cm−1 in ALDH−/CD133− and ALDH+/CD133+ OVCAR5 cells. Each dot represents a single cell, and the bars indicate means ± SEM; P = 0.0012. See also Figure S1, Movie S1, and Movie S2.
To confirm the above observation, we acquired full Raman spectra from individual LDs in both C-H stretch and fingerprint vibration regions. After normalization by the CH2 bending vibration peak at 1450 cm−1, the Raman spectra clearly showed that the peak at 1264 cm−1 (in plane bending motion of =C-H), the peak at 1660 cm−1 (stretching vibration of C=C), and the peak at 3002 cm−1 (vibration of =C-H) in ALDH+/CD133+ cells were higher than those in ALDH−/CD133− cells (Fig. 1C). In consistence with the hyperspectral SRS imaging data, the height ratios of the peak at 3002 cm−1 to the peak at 1450 cm−1 (Fig. 1D), 1660 cm−1 to 1450 cm−1 (Fig. S1B), and 1264 cm−1 to 1450 cm−1 (Fig. S1C) all showed a significant increase of lipid unsaturation level in ALDH+/CD133+ cells. To ensure that the findings are not cell line specific, we compared ALDH+/CD133+ and ALDH−/CD133− populations isolated from OVCAR5 cells. After normalization by the 1450 cm−1 peak, higher peak intensities at 1264 cm−1, 1660 cm−1 and 3002 cm−1 were noted in ALDH+/CD133+ OVCAR5 cells compared to ALDH−/CD133− OVCAR5 cells (Fig. 1E). Quantitatively, the height ratio of the peak at 3002 cm−1 to the peak at 1450 cm−1 was significantly higher in CSCs compared to non-CSCs (Fig. 1F). These results collectively demonstrate increased lipid unsaturation in ovarian CSCs compared to non-CSCs.
Increased lipid unsaturation in spheroids compared to monolayer cultures
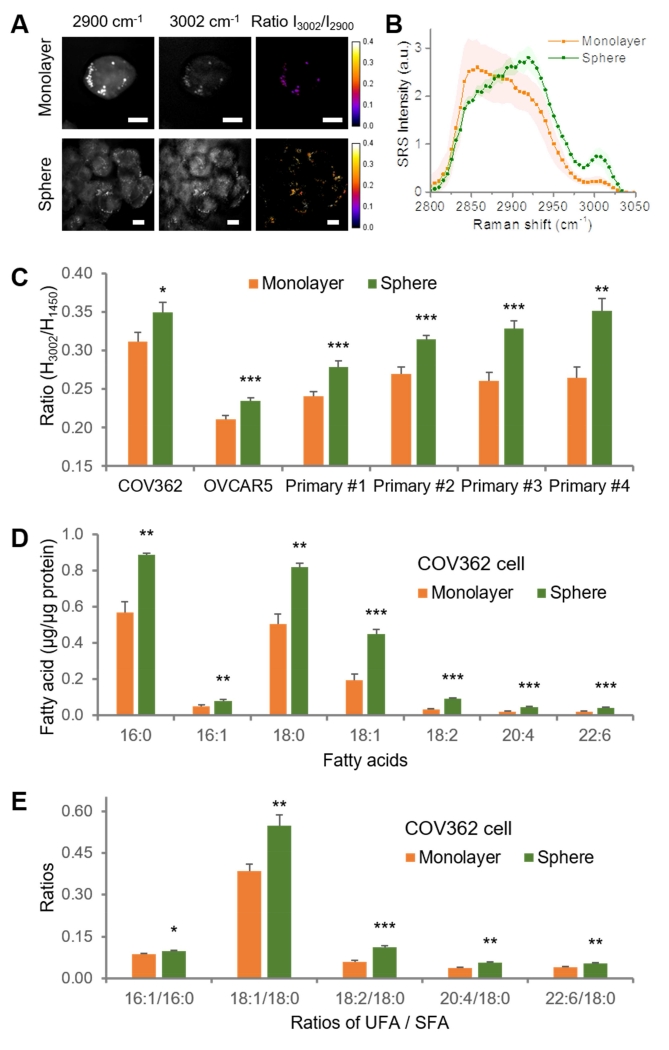
It is known that CSCs have the ability to proliferate as spheres when cultured under non-adherent conditions (Ponti et al., 2005; Zhou and Zhang, 2008). Thus, we utilized CCS-rich spheroids as a second model to validate our observations. Hyperspectral SRS imaging was conducted to analyze lipid composition of COV362 cells grown either as monolayers or as spheres under similar serum-free conditions (Supplemental movie 3, 4). Using the intensity ratio between 3002 cm−1 and 2900 cm−1, we observed an increased unsaturation level in spheres compared to monolayer cultures (Fig. 2A). As most cells in spheroids showed higher levels of unsaturation, it is possible that both CSCs and other progenitor cells present in spheres are rich in unsaturated lipids (Ponti et al., 2005). Comparison of SRS spectral profile (Fig. 2B) and quantitative analysis of the intensity ratio (I3002/I2900) (Fig. S2A) further confirmed the difference in lipid unsaturation levels between monolayers and spheres. Based on the height ratio of the peak at 3002 cm−1 to the peak at 1450 cm−1, Raman spectral analysis of COV362, OVCAR5, and primary cells derived from malignant OC ascites (total 4 patients, Table S1) quantitatively demonstrates elevated lipid unsaturation levels in spheres compared to monolayers cells (Fig. 2C).
Figure 2. Increased lipid unsaturation level in spheres compared to monolayer cultures.
(A) Representative hyperspectral SRS images of COV362 cells grown as monolayers and spheres. Images at 2900 cm−1, 3002 cm−1 and the intensity ratio between 3002 cm−1 and 2900 cm−1 are shown. Scale bars: 10 μm. (B) Average SRS spectra from the lipid droplets in COV362 monolayer (n=6) and spheres (n=6). Shaded area indicates the standard deviation of SRS spectral measurement from different cells. (C) Quantitation of the height ratio between the =C-H peak at 3002 cm−1 and the C-H bending peak at 1450 cm−1 based on Raman spectral measurements in monolayers and spheres derived from COV362, OVCAR5 and primary cells isolated from malignant OC ascites of four patients. The data are shown as means + SEM; n ≥ 10. (D) LC-MS measurement of fatty acids saponfied from all lipids extracted from OVCAR5 monolayer cultures and spheres. Fatty acids levels were normalized by total protein amount extracted from an equal number of cells. (E) Quantitation of the ratios of unsaturated fatty acid (UFA) to saturated fatty acid (SFA) in OVCAR5 monolayer and spheres. The data are shown as means + SD; n = 3. * P < 0.05, ** P < 0.01, *** P < 0.001. See also Figure S2, Table S1, Movie S3, and Movie S4.
Although the SRS imaging and Raman spectral analysis focused on lipid droplets, which are storage sites of neutral lipids mainly in the form of triglycerides and sterol esters, we speculated that the increased unsaturation in lipid droplets was part of an altered lipid metabolic network in CSCs. To test the hypothesis, we performed whole lipid analysis by mass spectrometry. First, we extracted free fatty acids from cells grown as monolayers or spheres (Ma and Xia, 2014) and analyzed by Electrospray Ionization Mass Spectrometry (ESI-MS). As shown in the mass spectra (Fig. S2B), the major components of free fatty acids in both monolayer and sphere cultures derived from OVCAR5 cells were 18:1 (e.g., oleic acid) and 18:0 (e.g., stearic acid). When normalized by the peak intensity of 18:0, the peak intensity of 18:1 in spheres was found to be significantly higher than that in cells grown as monolayers (Fig. S2C). Consistent results were obtained in COV362 (Fig. S2D) and primary OC cells isolated from malignant OC ascites (Fig. S2E, F). Furthermore, using Liquid Chromatography-Mass Spectrometry (LC-MS), we performed a more quantitative and comprehensive analysis of fatty acids saponified from all lipids (Kamphorst et al., 2013). The levels of all measured saponfied fatty acids, including palmitic (16:0), palmitoleic (16:1), stearic (18:0), oleic (18:1), linoleic (18:2), arachidonic (20:4), and docosahexaenoic acid (22:6), were significantly increased in cells grown as spheres compared to monolayer cultures (Fig. 2D, S2G). These data are consistent with the proposed phenomenon of increased de novo lipogenesis in CSCs. Moreover, the ratio of unsaturated fatty acids (UFA) to saturated fatty acids (SFA) was significantly increased in COV362 (Fig. 2E) and OVCAR5 (Fig. S2H) cells grown as spheres, confirming the increased unsaturation level in lipid droplets measured by Raman spectral analysis. These results suggest that unsaturation level in lipid droplets can be used as a readout of intracellular activities of desaturases. Taken together, the increased lipid unsaturation in ovarian CSCs was validated through hyperspectral SRS imaging, Raman spectroscopy, and mass spectrometry, in different CSC models derived from cell lines and primary OC cells.
The increased lipid unsaturation is mediated by lipid desaturases
Intracellular lipids originate either from the uptake of extracellular lipids or through de novo synthesis. We have shown higher levels of lipid unsaturation in both flow-sorted ALDH+CD133+ cells grown in the presence of serum (Fig. 1 and Fig. S1) and in spheroids grown in serum-free medium (Fig. 2 and Fig. S2). To determine the source of increased unsaturated lipids in CSCs, we examined the contribution of de novo synthesis, which is mediated by three fatty acid desaturases Δ9, Δ6, and Δ5 (Santos and Schulze, 2012). Among the three desaturases, Δ9 (stearoyl-CoA desaturase-1, SCD1) catalyzes the synthesis of monounsaturated fatty acids by adding one double bond to saturated fatty acids (mostly stearic acid), while the Δ6 and Δ5 desaturases are involved predominantly in the synthesis of polyunsaturated fatty acids (Nakamura and Nara, 2004). While the lipid desaturases are expressed in the majority of ovarian tumors (data not shown) and other cancers (Roongta et al., 2011), they are enriched in CSCs. SCD1 mRNA expression level was significantly higher in ALDH+/CD133+ vs. ALDH− /CD133− cells in both OVCAR5 (Fig. 3A) and COV362 (Fig. 3B) cell lines.
Figure 3. Increased desaturation in CSCs is due to lipid desaturases.
(A, B) Quantitative RT-PCR measurement of SCD1 expression levels in ALDH−/CD133− and ALDH+/CD133+ OVCAR5 and COV362 cells. The data are shown as means + SEM; n = 3. (C, D) Raman spectral measurement of lipid unsaturation level in primary spheres treated with the SCD1 inhibitor CAY10566 or Δ6 inhibitor SC-26196 at 1.0 μM for 6 days. The data are shown as means + SEM; n ≥ 10. (E-H) LC-MS analysis of relative abundance of 13C incorporation into fatty acids 16:0, 16:1, 18:0 and 18:1 saponified from all lipids in OVCAR5 spheroids treated with DMSO or 1 μM CAY10566 for 6 days. The data are shown as means + SD; n = 3. * P < 0.05, ** P < 0.01, *** P < 0.001. See also Figure S3.
Small molecule inhibitors, CAY10566 which blocks SCD1 (Liu et al., 2007) and SC-26196 which blocks Δ6 (Obukowicz et al., 1998) and shRNA based downregulation were used to study further the function of desaturases in ovarian CSCs. Both inhibitors significantly reduced the lipid unsaturation levels in OC spheroids (Fig. 3C, D). The SCD1 inhibitor was more potent than the Δ6 inhibitor, perhaps because monounsaturated rather than polyunsaturated lipids contribute more significantly to the increased unsaturation levels in CSCs. To measure the effects of lipid desaturase inhibitors on metabolic flux from SFAs to UFAs, we deployed metabolic tracing with 13C labeled glucose and glutamine (Kamphorst et al., 2013). The relative abundances of 13C labeled fatty acids saponfied from all lipids, including palmitic acid (16:0), palmitoleic acid (16:1), stearic acid (18:0), and oleic acid (18:1), were quantitatively analyzed by mass spectrometry. As shown in Fig. 3E-H, CAY10566 significantly suppressed the incorporation of 13C into UFAs (FA 16:1 and FA 18:1), but not in SFAs (FA 16:0 and FA 18:0) in OVCAR5 spheroids. Similar effects were observed in cells in which SCD1 was knocked-down by shRNA compared to shRNA control transfected cells (Fig. S3A-D). Collectively, these data support an essential role of lipid desaturases for the synthesis of UFAs in CSCs.
Lipid desaturases contribute to the maintenance of cancer cell stemness
To test the functional significance of lipid desaturases relative to the known traits of CSCs, we measured the expression levels of CSC markers by Q-RT-PCR after desaturase inhibitor treatment or shRNA based downregulation. Both CAY10566 and SC-26196 reduced ALDH1A1 mRNA expression levels in OVCAR5, COV362 and primary ovarian cancer spheroids (Fig. 4A). Additionally, the expression levels of CSC-associated transcription factors Sox2, Nanog, and Oct-4 were decreased by treatment with the inhibitors in OVCAR5, COV362, and primary spheroids (Fig. 4B, S4A, B). Similarly, knock-down of SCD1 by shRNA reduced the mRNA expression levels of ALDH1A1, Nanog, Sox2, and Oct-4, in both OVCAR5 (Fig. 4C and Fig. S4C) and COV362 spheroids (Fig. 4D and Fig. S4D). The reduction of stemness was further confirmed by a reduction in the ALDH+ cell population after treatment with the inhibitors as measured by flow cytometry (Fig. 4E). Overall, these data support an important role of lipid desaturases in CSC maintenance.
Figure 4. Inhibition of lipid desaturation impairs ovarian cancer cell stemness.
(A) RT-PCR measurement of ALDH1A1 mRNA expression level in OVCAR5, COV362, and primary spheroids. (B) RT-PCR measurement of Sox2 mRNA expression level in OVCAR5, COV362, and primary spheroids. Treatment with CAY10566 or SC-26196 in spheroids was done at a concentration of 1.0 μM for 6 days. (C, D) RT-PCR measurement of SCD1, ALDH1A1 mRNA expression level in OVCAR5, COV362 cells stably transfected with scrambled control shRNA (shCtr) or SCD1 shRNA (shSCD). The data were shown as means + SEM; n = 3. * P < 0.05, ** P < 0.01, *** P < 0.001. (E) Flow cytometry analysis of ALDH+ cells sorted from OVCAR5, COV362 and primary cells after CAY10566 or SC-26196 treatment at a concentration of 1.0 μM for 6 days. DEAB was used as negative control. See also Figure S4.
Inhibition of lipid desaturases prevents sphere formation in vitro and tumorigenesis in vivo
To determine the effect of desaturase inhibitors on the functions of CSCs, we examined the capacity of CSCs to form spheroids in vitro (Visvader and Lindeman, 2008a) and initiate tumors in vivo. Both CAY10566 (Fig. 5A) and SC-26196 (Fig. 5B) effectively suppressed sphere formation in OVCAR5, COV362, and primary OC cells. The suppression of sphere proliferation was further confirmed by using the CCK-8 assay in OVCAR5, COV362, and primary OC cells (Fig. S5A, B). Suppression of sphere formation was also observed in OVCAR5 (Fig. S5C) and COV362 cells (Fig. S5D), in which SCD1 was stably knocked down compared to shRNA control transfected cells. In contrast, OVCAR5 cells grown as monolayers (containing fewer CSCs) (Fig. S5E) and normal human dermal fibroblasts (NHF544) (Fig. S5F) were less sensitive to desaturase inhibitors compared to spheroids, supporting that lipid desaturation represents a CSC-specific target.
Figure 5. Inhibition of lipid desaturation prevents sphere growth in vitro and tumor formation in vivo.
(A, B) Representative images of OVCAR5, COV362 and primary spheres treated with CAY10566 or SC-26196 at indicated concentrations for 6 days. (C, D) Sphere proliferation of COV362 cells supplemented with fatty acid (C) 18:0 and (D) 18:1 at indicated concentrations and treated with CAY10566 or SC-26196 at 1 μM for 6 days. Comparisons were performed between inhibitors treated group and DMSO treated group under the same concentrations of fatty acid supplementation (P value indicated by *), or between DMSO groups under different concentrations of fatty acid supplementation (P value indicated by #). The data are normalized to the blank control (no fatty acid supplementation and treated with DMSO) and shown as means + SEM; n = 4. (E, F) Growth curves of xenografts derived from (E) DMSO (n = 7) or CAY10566 (n = 6), and (F) DMSO (n = 8) or SC-26196 (n = 8) pretreated OVCAR5 cells grown in spheroids. The data are presented as means + SEM. Inset: tumor size comparison at the end of study (42 days after tumor cells inoculation). Upper: DMSO; lower: treated. * P < 0.05, ** P < 0.01, *** P < 0.001. (G) Tumor initiation assay using serial dilution of OVCAR5 cells pretreated with DMSO or CAY10566 at 1 μM. Cells were counted and then cultured for 6 days with the presence of DMSO or CAY10566 to allow sphere formation before injected into the mice. 5 mice were used for each group. See also Figure S5, Figure S6, and Table S2.
Next, to investigate the contribution of exogenous lipid uptake to ovarian CSCs proliferation, we measured sphere formation in the presence of several exogenous fatty acids (e.g., FA 16:0, 16:1, 18:0, 18:1, 18:2, 20:4, and 22:6). Fatty acids supplementation did not alter (positively or negatively) sphere proliferation and addition of exogenous lipids only partially rescued the inhibition exerted by desaturase inhibitors on sphere formation (Fig. 5C-D, Fig. S6A-E). These data suggest that CSCs rely on the de novo fatty acid synthesis pathway, rather than on exogenous lipid uptake.
To measure the effects of desaturase inhibitors on tumor formation in vivo, we inoculated DMSO or inhibitor pretreated CSCs in the flanks of nude mice. Time to tumor formation and tumor volumes were measured. Pretreatment of ALDH+CD133+ flow sorted OVCAR5 cells with the desaturase inhibitors CAY10566 and SC-26196 significantly delayed the median time to tumor initiation from 14 days (DMSO) to 39 days and 23 days (Table S2), respectively. Pretreatment with CAY10566 (Fig. 5E) or SC-20196 (Fig. 5F) significantly suppressed tumor growth. Tumor weights (mean ± SEM) were significantly reduced from 827 ± 154 mg (DMSO) to 22 ± 12 mg by CAY10566 (Fig. S6F), and from 562 ± 160 mg (DMSO) to 219 ± 31 mg by SC-26196 (Fig. S6G). To verify the effects of the inhibitors on stemness, a tumor initiation assay was subsequently performed by using serial dilutions of CSCs flow sorted from OVCAR5 cell line. CAY10566 pretreatment reduced the frequency of tumor initiation to 20% (1 out of 5 mice) when ~500 ALDH+/CD133+ cells were used, 40% (2 out of 5 mice) for ~1,000 CSCs, 80% (4 out of 5 mice) for ~5,000 and ~10,000 CSCs, whereas tumors formed in all control groups (Fig. 5G). Collectively, these data support that blockade of lipid desaturation selectively suppresses the sphere formation and tumor initiation capacity of ovarian CSCs.
Inhibition of lipid desaturases downregulates the NF-κB pathway
Having shown the effects of desaturase inhibitors on sphere formation and tumorigenesis, we next explored the mechanism by which lipid desaturation alters the functions of CSCs. We first screened the effects of the inhibitors on various CSC-related signaling mechanisms by using pathway specific Q-RT-PCR arrays. Both CAY10566 and SC-26196 downregulated key stem cell-related signaling networks in OC spheroids, particularly the NF-κB, PI3K/Akt, Hedgehog, Notch, self-renewal, asymmetric division, migration and metastasis pathways (Figs 6A, B). One of the most responsive pathways to desaturase inhibition was NF-κB signaling, a key survival pathway in cancer, which has been also linked to cancer stemness (Shostak and Chariot, 2011). To confirm the suppression of NF-κB by desaturase inhibitors, we used an NF-κB reporter assay. Both CAY 10566 and SC-26196 significantly suppressed NF-κB transcriptional activity in OVCAR5, COV362 and primary OC spheres (Fig. 6C). Confirming suppression of NF-κB activity, the mRNA expression levels of IL-6 (Fig. S7A) and IL-8 (Fig. S7B), known NF-κB target genes, were significantly downregulated by desaturase inhibitors in OC cells.
Figure 6. Inhibition of lipid desaturation down-regulates the NF-κB pathway.
CSC genes preferentially down-regulated by (A) CAY10566 treatment compared to control (DMSO) (> 2.0 fold) or (B) SC-26196 treatment compared to control (DMSO) (> 2.0 fold) were quantified by RT2 Profiler PCR array in OVCAR5 spheroids treated with CAY10566 at 1 μM for 6 days. Pie chart analysis showing the downregulated genes (% of total) for each represented pathway in control vs. treated spheres. (C) NF-κB promoter activity measured by gene reporter assay in OVCAR5, COV362 and primary OC cells spheroids treated with CAY10566 or SC-26196 at 1 μM for 6 days. (D) Proliferation of OVCAR5 and COV362 cells stably transduced with empty vector (pQCXIP) or with pQCXIP/p65 and grown as spheroids. The data are shown as means + SEM; n = 4. (E) RT-PCR measurement of ALDH1A1 in COV362 spheroids stably transduced with empty vector (pQCXIP) or with pQCXIP/p65. (F) RT-PCR measurement of ALDH1A1 mRNA levels in primary OC spheroids treated with NF-κB inhibitor, DMAPT, at indicated concentrations. The data are shown as means + SEM; n = 3. * P < 0.05, ** P < 0.01, *** P < 0.001. See also Figure S7A-B.
To investigate the link between NF-κB and the stem cell phenotype, p65 (relA), the active subunit of NF-κB, was overexpressed in ovarian cancer cells and the stem cell characteristics were evaluated. Indeed, overexpression of p65 increased ovarian cancer sphere proliferation in OVCAR5 and COV362 cells (Fig. 6D). Quantitative RT-PCR demonstrated that the overexpression of p65 increased ALDH1A1 expression in COV362 cells (Fig. 6E). Consistently, a small molecule inhibitor of NF-κB, dimethylaminoparthenolide (DMAPT) (Shanmugam et al., 2011), reduced ALDH1A1 expression in primary OC spheroids (Fig. 6F). Together, these data support that NF-κB, as an important pathway downregulated by desaturases inhibition, is invovled in stem cell regulation.
NF-κB and ALDH regulate lipid unsaturation
To further understand the mechanism by which NF-κB is linked to lipid desaturation and ovarian cancer cell stemness, ovarian cancer spheres were treated with the NF-κB inhibitor. DMAPT decreased both lipid unsaturation levels (Fig. 7A) as well as SCD1 mRNA expression levels in primary ovarian cancer spheres (Fig. 7B). Next, to investigate whether NF-κB directly regulates the expression of desaturases, a promoter search motif identified two putative p65 (relA) binding sites at position −215 to −206 and at position +179 to 188 in the SCD1 promoter (Fig. S7C). The direct interaction between p65 and the SCD1 promoter region was then confirmed by chromatin immunoprecipitation (ChIP) using a p65 antibody in OVCAR5 cells stably transduced with pQCXIP vector or pQCXIP/p65 (Fig. 7C). Consistent with the proposed direct transcriptional regulatory role of NF-κB, the overexpression of p65 significantly upregulated the expression levels of SCD1 in OVCAR5 and COV362 spheroids (Fig. 7D), IL8, a known p65 target gene was used as a positive control in these experiments (Fig. S7D). Together these results demonstrate that NF-κB directly regulates SCD1 expression and promotes CSC characteristics.
Figure 7. NF-κB and ALDH1A1 promote lipid unsaturation.
(A) Raman spectral measurement of lipid unsaturation level in OC spheroids treated with the NF-κB inhibitor, DMAPT, at indicated concentrations. n ≥ 10. (B) RT-PCR measurement of SCD1 mRNA levels in primary OC spheroids treated with DMAPT at indicated concentrations. n = 3. (C) ChIP demonstrates that p65 binds the SCD1 promoter region. Immunoprecipitated chromatin with an antibody against p65 was used for quantitative PCR amplification. Primers flanking two predicted p65-binding regions of the SCD1 promoter (−215 to −206 and +179 to +188 bp) were used. Positive control was p65 antibody-immunoprecipitated chromatin amplified with primers for the IL8 promoter, a known p65 target. Negative controls were chromatin immunoprecipitated with IgG and amplified with SCD1 promoter primers and chromatin immunoprecipitated with p65 antibody and amplified with primers to a region in the SCD1 promoter located upstream of the predicted p65 binding sites. (D) Quantitative RT-PCR measured SCD1 mRNA expression levels in COV362 spheroids transfected with pQCXIP vector or pQCXIP/p65. The data are shown as means + SEM; n = 3. (E) Raman spectral measurement of lipid unsaturation level in COV362 spheroids treated with ALDH1A1 inhibitor, CM037, or CM037 and retinoic acid for 6 days. The data are shown as means + SEM; n ≥ 10. * P < 0.05, ** P < 0.01, *** P < 0.001, n.s: not significant. (F) The proposed mechanism by which lipid desaturation is linked to cancer cell stemness. See also Figure S7C-H.
We also detected increased lipid unsaturation in ALDH+ (single marker) OC cells (Fig. S7E). ALDH1A1 functions as a detoxification enzyme and is implicated in the conversion of retinol to retinoic acid (Ma and Allan, 2011; Marcato et al., 2011). ALDH1A1 is recognized as a stem cell marker in several tumor types, including ovarian cancer (Silva et al., 2011), but its involvement remains unclear. It has been proposed that its role in the maintenance of CSCs is related to regulation of cell differentiation mediated by retinoic acid signaling (Chute et al., 2006; Ginestier et al., 2009). To determine the potential mechanism by which ALDH1A1 may be involved in regulating lipid desaturation, we used a recently identified and highly specific ALDH1A1 inhibitor, CM037 (Condello et al., 2015). Treatment with CM037 potently decreased the mRNA expression levels of SCD1 in primary OC spheres (Fig. S7F), supporting a link between the two pathways. Additionally, CM037 decreased lipid unsaturation levels in COV362 (Fig. 7E), OVCAR5 (Fig. S7G) and primary OC spheres (Fig. S7H). Because ALDH1A1 regulates retinoic acid signaling (Ma and Allan, 2011), we next tested whether its effects on lipid unsaturation can be reversed by retinoic acid. The lipid unsaturation levels reduced by CM037 were successfully rescued by the addition of retinoic acid in COV362 (Fig. 7E), OVCAR5 (Fig. S7G) and primary OC spheres (Fig. S7H). Taken together, these data suggest a role of ALDH1A1 in the regulation of lipid desaturation in ovarian CSCs mediated through retinoic acid signaling.
As summarized in Fig. 7F, our studies revealed a functional role of altered lipid metabolism in ovarian CSCs. Through de novo lipogenesis and subsequent lipid desaturation, the unsaturated fatty acids promote activation of NF-κB, a key pathway involved in maintaining the CSC phenotype. In turn, the NF-κB pathway modulates the expression levels of lipid desaturases at the transcriptional level. Together, our data support a positive feedback loop between lipid metabolism and NF-κB signaling in regulating OC cell stemness.
DISCUSSION
Since the first use of CD34 and CD38 as surface markers of leukemia stem cells, efforts to search for bona fide markers of CSCs have intensified. The lack of reliable methods to identify and isolate CSCs has remained one of the biggest challenges facing CSC research (Clevers, 2011). Current methods to enrich or isolate CSCs include surface marker labeling for flow cytometry sorting, side population sorting, and sphere culture (Zhou and Zhang, 2008). Surface markers have been widely used as the standard method to isolate CSCs. Some intracellular markers, such as aldehyde dehydrogenase (ALDH) have also been used alone or in combination with other markers. However, many of the CSC markers selected based on empirical assays are not necessarily linked to clear functional roles (Clevers, 2011; Visvader and Lindeman, 2008b). Recently, Heeschen and colleagues reported that a strong intracellular autofluorescence signal, originating from riboflavin accumulation, can be used as a biomarker for epithelial CSCs (Miranda-Lorenzo et al., 2014). However, the precise function of the accumulated fluorophore remains unknown and the level of the autofluorescence signal can be easily altered by environmental changes, limiting its use as a reliable marker for CSCs.
Our study proposes a stable, universal and functional metabolic marker for ovarian CSCs. By using Raman spectroscopic imaging, we identified an increase of lipid unsaturation levels in the lipid droplets of single ovarian CSCs derived from cell lines and human specimens. As the site for neutral lipid storage, LDs buffer the synthesis and consumption of lipids, which renders their composition relatively stable over time. Since unsaturated lipids are primarily derived from de novo synthesis, they are not easily subject to environmental changes. Indeed, we observed consistently increased lipid unsaturation in CSCs regardless of culture conditions (e.g. serum enriched or serum free media) in OC cell lines and human primary malignant cells. Raman spectral measurement is based on the ratio of unsaturated lipids to total lipids, instead of the absolute amount of unsaturated lipids, eliminating the change induced by the total lipids amount. It is possible that increased unsaturated lipids are detectable in CSCs from other cancer types (our unpublished data in pancreatic and leukemia cells) and future studies will address this concept more broadly. Combining this marker with Raman spectroscopic imaging could lead to a reliable and universal method for in situ detection of CSCs. Further refinement of Raman spectroscopic imaging based techniques, such as coherent Raman scattering flow cytometry (Camp et al., 2009; Camp et al., 2011; Wang et al., 2008), would allow fast, noninvasive detection and isolation of CSCs.
As shown by single cell spectroscopic imaging, CSCs are characterized by a higher conversion rate from saturated to unsaturated lipids, compared to differentiated cancer cells, a process mediated by fatty acid desaturases. We further observed that lipid desaturases are enriched in CSCs and can be targeted. By measuring key characteristics of stem cells in vitro and in vivo, including sphere formation and tumor initiation, we showed that lipid desaturases are critical to maintaining CSCs, suggesting that unlike most surface markers, unsaturated lipids possess definitive functions. Our data support further investigation of lipid desaturases as CSC specific targets and development of combination strategies using these inhibitors with standard chemotherapy targeting the non-CSC population to improve disease control.
Finally, we identified a positive feedback loop involving the NF-κB pathway, ALDH1A1 and lipid desaturases in ovarian CSCs. A functional link between NF-κB as the master regulator of inflammation converging pathways and cancer has been established for other tumor models (Karin et al., 2002; Shostak and Chariot, 2011) and activation of NF-κB in stem cells has been recently reported (Jia et al., 2015). Here, we link NF-κB, lipid desaturases, and CSCs. Our data support that NF-κB regulates SCD1 at the transcriptional level, that NF-κB activation promotes ovarian sphere proliferation, and that in turn desaturase inhibition blocks NF-κB activity and reduces cancer stemness. Interestingly, we show that IL-6, one of the known targets genes of NF-κB, is potently downregulated by the inhibitors. IL-6 has been recently linked to cancer stemness in multiple cancer models (Sansone et al., 2016; van der Zee et al., 2015). The exact mechanism by which NF-κB is inactivated by desaturase inhibitors in CSCs remains undefined. A possible explanation could be that the reduction of unsaturated lipids induced by the inhibitors directly blocks NF-κB given that unsaturated fatty acids, such as arachidonic acid (Camandola et al., 1996), oleic and linoleic acid (Poletto et al., 2015), are known activators of the complex. Besides the NF-κB pathway, our PCR array assay showed that disruption of lipid desaturation also impaired other stem cell associated pathways, which may contribute additionally to inhibiting CSCs. Unsaturated lipids are also known to modulate membrane fluidity (Nakamura and Nara, 2004), which could contribute to altered oncogenic signaling and impairment of stemness.
In all, our results discovered a new functional marker of ovarian CSCs, intimately linked to the NF-κB survival pathway and lipid metabolism. We propose that this marker represents a targetable metabolic vulnerability of recalcitrant stem cells, which should be exploited to improve the outcome of existing therapies.
STAR METHODS
CONTACT FOR REAGENT AND RESOURCE SHARING
Further information and requests may be directed to, and will be fulfilled by the Lead Contact, Ji-Xin Cheng (jcheng@purdue.edu).
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Cell lines
Human ovarian cancer cell lines COV362 and OVCAR5 were a gift from Dr. Kenneth P. Nephew (Indiana University, Bloomington, IN). Both cell lines were tested to be mycoplasma negative and OVCAR5 cell line was authenticated by Short Tandem Repeat (STR) analysis. COV362 cells were cultured in DMEM high glucose medium supplemented with 10% FBS and 100 units/mL penicillin and 100 μg/mL streptomycin. OVCAR5 cells were grown in D-MEM (high glucose), 10% fetal bovine serum (FBS), 0.1 mM Non-Essential Amino Acids (NEAA), 2 mM L-glutamine, and antibiotics. All cells were cultured at 37°C in a humidified incubator with 5% CO2 supply.
Primary human cells
De-identified malignant ascites fluid specimens from ovarian cancer patients (n = 4) were obtained at the Indiana University Simon Cancer Center (IUSCC) under an IRB approved protocol (IUCRO#505). Additional information regarding subjects’ clinical history is provided in Table S1. The normal human dermal fibroblasts were obtained and provided by Dr. Dan Spandau (IUSCC, Indianapolis, IN) after isolation from foreskin tissue. The procedures related to human tissue samples procurement were approved by the Indiana University School of Medicine IRB.
Tumor cells were purified as previously described (Condello et al., 2015). After centrifugation at 1,200 rpm for 5 min, 25,000 ascites derived tumor cells were cultured as monolayers in DMEM medium supplemented with 10% FBS and antibiotics or suspended in Mammocult Complete medium (StemCell Technologies) and plated in ultra-low adherent surfaces to allow spheroid formation.
Subcutaneous xenograft mouse model
All animal experiments were conducted following protocols approved by Purdue Animal Care and Use Committee (PACUC). 5~6 week-old female athymic nude mice (strain Hsd:Athymic Nude-Foxn1nu) (Envigo) were used to establish the tumor subcutaneous xenograft model. Housing and husbandry of the nude mice were conducted by the staffs at the animal facility of Purdue University following PACUC regulations.
METHOD DETAILS
Sphere formation assay
FACS-sorted ALDH+/CD133+ OC cell lines or primary cells were seeded as single cell suspension at a concentration of 10,000 cells/well in 96-well ultra-low attachment plates (Corning, Corning, NY, USA) in Mammocult complete medium (StemCell Technologies). After 6 days, spheroids were counted after centrifugation at 300×g for 5 minutes. Cell numbers were also quantified by the CCK-8 assay (Dojindo Molecular Technologies, Rockville, MD). All assays were performed in four replicates.
Aldefluor assay and Fluorescence-Activated Cell Sorting
ALDH and CD133 double positive or negative cells were isolated by fluorescence-activated cell sorting (FACS) using the Aldefluor assay kit (Stemcell Technologies) and the allophycocyanin (APC) conjugated CD133/1 (AC133) antibody (Miltenyi Biotec, Auburn, CA, USA). Briefly, dissociated monolayer single cells were resuspended in Aldefluor assay buffer containing the ALDH1 substrate, bodipyaminoacetaldehyde (BAAA), at 1.5mM. After incubation for 45 min at 37 °C, the cells were centrifuged at 1,200 rpm for 5 min at 4 °C, washed twice with Aldefluor assay buffer and incubated in the same buffer supplemented with 0.5% bovine serum albumin BSA and anti-CD133/1 (AC133) antibody (1:10 dilution) for 30 min on ice. The test ALDH+/CD133+ population was gated using control cells incubated under an identical condition in the presence of a 10-fold molar excess of the ALDH inhibitor (DEAB), and anti-mouse IgG1 isotype control APC-conjugated antibody (1:10 dilution) (Miltenyi Biotec). FACS was performed using a FACS Aria II flow cytometer (BD Biosciences, San Jose, CA) under sterile conditions.
Hyperspectral stimulated Raman scattering (SRS) imaging
Hyperspectral SRS imaging was done with a spectral focusing method following previously published protocol (Fu et al., 2013; Liu et al., 2015). Briefly, the Raman shift is tuned by controlled the temporal delay between two chirped femtosecond pulses. In our scheme, a femtosecond laser (Coherent) operating at 80 MHz provided the pump and Stokes laser source. With pump beam tuned to 810 nm, Stokes beam was tuned to 1055 nm to cover the C-H vibration region. The Stokes beam was modulated at 2.3 MHz by an acousto-optic modulator (1205-C, Isomet). After combination, both beams were chirped by two 12.7 cm long SF57 glass rods and then sent to a laser-scanning microscope. A 60× water immersion objective (NA = 1.2, UPlanApo/IR, Olympus) was used to focus the light on the sample, and an oil condenser (NA = 1.4, U-AAC, Olympus) was used to collect the signal.
To obtain a hyperspectral SRS image, a stack of 50 images at different pump-Stokes temporal delay was recorded. The temporal delay was controlled by an automatic stage, which moved forward with a step size of 10 μm, corresponding to ~5 cm−1. To calibrate the Raman shift to the temporal delay, standard chemicals with known Raman peaks in C-H region from 2800 to 3050 cm−1, including DMSO, methanol, oleic acid, and linolenic acid, were used. To eliminate the power difference at different Raman shift, SRS signal was normalized by two-photon absorption signal from Rhodamine 6G, the cross-section of which is considered constant in this small region. The spectral resolution of the system was estimated to be ~25 cm−1. Hyperspectral SRS images were analyzed using software ImageJ.
Spontaneous Raman spectroscopy
Confocal Raman spectral analysis from individual LDs was performed as described previously (Slipchenko et al., 2009). A 5-picosecond laser at 707 nm was used as excitation beam for Raman spectral acquisition. Acquisition time for a typical spectrum from individual LDs was 20 seconds, with the beam power maintained around 15 mW at the sample. For each specimen, at least 10 spectra from individual LDs in different locations or cells were obtained. The spectra were analyzed using software Origin 8.5. The background was removed manually, and peak height was measured.
Mass spectrometry analysis of fatty acids
OVCAR5, COV362 and primary human OC cells were cultured under serum-free conditions either as monolayers or spheres. For the 13C tracing experiments, cells were seeded in ultra-low adherence plates and grown in sphere culture medium (containing glucose and glutamine) supplemented with additional 25 mM Glucose (U-13C6, 99%) and 4 mM L-Glutamine (U-13C5, 99%). The cells were grown as spheres for 6 days with or without inhibitor treatment or SCD1 shRNA knock-down. Spheres were collected by centrifuging at 200 rpm for 1 min to separate from floating single cells. All the experiments were conducted in three replicated sets of samples.
Free fatty acid extraction was performed following previous protocol (Ma and Xia, 2014). Cell pellets were resuspended in 300 μL PBS, mixed with 1 ml methanol and acidified with HCl to 25 mM final concentration. After addition of 1 ml isooctane was added, and samples were vortexed and centrifuged at 3,000 g for 1 min. The top layer was transferred to a glass tube and dried under vacuum. Extracted lipids were re-dissolved in 100 μl acetone: water (1:1). Electrospray Ionization-Mass Spectrometry (ESI-MS) analysis of fatty acids was conducted.
Extraction and preparation of saponified fatty acids were performed following previous protocol (Kamphorst et al., 2013; Yang et al., 2007). Briefly, cells were lysed in 1 ml of cold 50:50 MeOH/H2O solution with 0.1 M HCl, mixed with 0.5 ml chloroform, and centrifuged at 16,000 g for 5 min. The lower phase was transferred to glass vials and dried under N2 flow. Dried lipid extract was reconstituted into 90:10 MeOH/H2O containing 0.3 M KOH, saponified at 80 °C for 1 h, and then acidified with 0.1 mL of formic acid. Fatty acids were extracted twice with 1 mL of hexane, dried under N2 flow. For quantitative analysis, each fatty acid (Sigma) at known concentration were used as internal standards. Liquid Chromatography-Mass Spectrometry (LC-MS) analysis of fatty acids was performed following previously described protocol (Yang et al., 2007). For 13C tracing experiments, fatty acids (e.g., FA 16:0, 16:1, 18:0, 18:1) with various numbers of 13C labeling were analyzed. The natural isotopic abundances were correlated following described protocol (Tumanov et al., 2015).
Tumor initiation assays in subcutaneous xenograft mouse model
For tumor formation and growth experiment, FACS-isolated ALDH+/CD133+ OVCAR5 cells were seeded at a density of 20,000 cells/well in non-adherent ultra-low plates in Mammocult complete medium (StemCell Technologies), and treated with DMSO, CAY10566 or SC26196 respectively at a concentration of 1 μM for 6 days. Spheres were collected and mixed with equal volume of Matrigel Matrix (Corning, Cat# 356234) prior to subcutaneous injection. An equal number of DMSO and inhibitors treated spheres were injected subcutaneously into the left and right flank of 5~6 week-old female athymic nude mice (Envigo), respectively, with 8 mice randomly assigned to each group. Tumors were measured twice a week using calipers. Tumor volume was calculated as 1/2 × L × W2, where L stands for the length, and W for the width measured by a caliper in mm. Time to tumor formation was defined as the time from cell inoculation to the time when tumors were first detected. At the end of the study (e.g. when at least one tumor reached 2,000 mm3), mice were euthanized, tumors were harvested, measured and weighed.
For tumor initiation assay, FACS-isolated ALDH+/CD133+ OVCAR5 cells were seeded in non-adherent ultra-low 96-well plates at densities of 20000, 10000, 5000, 1000, and 500 cells per well. The cells were treated with DMSO or CAY10566 at a concentration of 1 μM and grown as spheroids for 6 days. Tumor cell inoculation was performed as described above with 5 mice randomly assigned to each group. Tumor initiation was determined ~30 days after tumor cell inoculation.
Generation of SCD1 stably knock-down cells
OVCAR5 and COV362 cells were transfected with SCD1 targeting shRNA lentiviral particles (Santa Cruz Biotechnology, Inc., Dallas, TX, sc-36464-V) following the manufacturer’s protocol. Scrambled shRNA lentiviral particles (Santa Cruz, sc-108080) were used as a control. Stably transfected cells were selected with 0.5 μg/ml puromycin.
Stable retroviral transduction
The active p65 (Rel A) subunit of NF-κB subcloned into the pQCXIP retroviral vector (Chua et al., 2007) was transduced in OVCAR5 and COV362 cells and pooled colonies were selected with puromycin. Transduction of the empty vector was performed in parallel.
Gene reporter assay
Dual-Luciferase Assay (Promega, Madison, WI) quantified NF-κB activity in COV362, OVCAR5 and primary cells grown as spheroids, according to the manufacturer’s instructions. In brief, cells were plated in ultra-low adherence plates in Mammocult complete medium and transiently co-transfected with the NF-κB promoter luciferase and renilla plasmids at the ratio of 10:1 by using DreamFect Gold transfection reagent (OZ Biosciences, Marseille, France). After 24 h, cells were treated with the SCD1 inhibitor CAY10566 or the Δ6 inhibitor SC26196 at a concentration of 1.0 μM for 48 h. Luminescence was measured by using the TD-20/20 Luminometer (Turner Biosystems, Madison, WI). Experiments were performed in triplicate and repeated two times in independent conditions. To control for transfection efficiency, NF-κB luminescence was normalized to renilla activity.
Reverse transcription-PCR (RT-PCR)
Total RNA was extracted from OC cell lines or primary cells by using RNA STAT-60 (Tel-Test Inc., Friendswood, TX) and reverse transcribed using iScript cDNA Synthesis Kit (Bio-Rad, Hercules, CA). The reverse transcriptase product (1 μL) and primers (Table S3) were heated at 94 °C for 10 min, followed by 40 cycles of amplification at 94 °C for 15 secs and 60 °C for 1 min. At the end of the PCR reaction a melting curve was generated and the cycle threshold (Ct) was recorded for the reference and control genes. The relative expression of different transcripts was calculated as ΔCt and normalized by subtracting the Ct of target genes from that of the housekeeping control (18S). Real-time PCR was carried out on an ABI Prism 7900 platform (Applied Biosystems, Grand Island, NY) using the iTaq universal SYBR Green super mix (Bio-Rad, Hercules, CA, USA). Human IL6 gene expression was detected using TaqMan® Gene Expression Assay (Hs00985639_m1) and TaqMan® Gene Expression Master Mix (Applied Biosystems). Results are presented as means + SEM of replicates. Measurements were performed in duplicate and experiments were run three times in independent conditions.
The human cancer stem cells RT2 Profiler PCR Array was purchased from SA Bioscience and real-time PCR was performed on ABI Prism 7900 HT (Applied Biosystems), according to the manufacturer’s instructions. Data analysis was performed based on the ΔΔCt method with normalization of the raw data to the housekeeping genes using a Microsoft excel algorithm provided by the manufacturer. An ontology classification assignment for each gene was performed, and fold-changes were calculated and expressed as percent of composition for each represented pathway in control versus treated spheres.
Chromatin immunoprecipitation (ChIP)
Chromatin immunoprecipitation was performed by using a kit from EMD Millipore (Billerica, MA USA) according to the manufacturer’s instructions, with some modifications. In brief, 107 OVCAR5 cells stably transduced with pQCXIP and pQCXIP/p65 were fixed in 1% formaldehyde for 10 minutes at 37°C to cross-link histones to DNA. Cells were washed twice with ice-cold phosphate-buffered saline (10 mmol/L Na2HPO4, 2 mmol/L NaH2PO4, 137 mmol/L NaCl, 2.7 mmol/L KCl) containing protease inhibitors (1 μg/mL aprotinin, 1 μg/mL leupeptin, and 1 mmol/L phenylmethylsulfonyl fluoride) and lysed for 10 minutes in SDS buffer (1% SDS, 50 mM Tris [pH 8.1], 10 mM EDTA). Cell lysates were sonicated on ice with 3 sets of 10-second pulses using a Sonic Dismembrator Model 100 (Fisher Scientific, Pittsburgh, PA) set at 30% of maximum power. The soluble chromatin was then centrifuged, resuspended in ChIP dilution buffer (final concentration of 15 mmol/L Tris-HCl [pH 8.1], 2 mmol/L EDTA, 150 mmol/L NaCl, 0.1% SDS, 1% Triton X-100) containing protease inhibitors, and one tenth of the supernatant was used as DNA input control. The remaining volume of ChIP solution was pre-cleared by incubation with sheared salmon sperm DNA/protein A–agarose 50% slurry and incubated with rabbit immunoglobulin G (IgG) or rabbit polyclonal anti-NF-κB p65 antibody (Cell Signaling Technology Inc., 1:100 dilution) overnight at 4°C with agitation. The immunoprecipitated complexes were incubated with protein A slurry and then washed successively with low-salt buffer (0.1% SDS, 1% Triton X-100, 2 mM EDTA, 20 mM Tris-HCl [pH 8.1], 150 mM NaCl), high-salt buffer (500 mM NaCl), LiCl buffer (0.25 M LiCl, 1% IGEPAL-CA630, 1% deoxycholate, 1 mM EDTA, 10 mM Tris-HCl [pH 8.1]), and Tris-EDTA (pH 8.0), and then were eluted with 1% SDS, 100 mM NaHCO3 buffer. The cross-linking of protein-DNA complexes was reversed by incubation with 5 M NaCl at 65°C for 4 h, and DNA was digested with 10 mg of proteinase K (Sigma)/ml for 1 h at 45°C before DNA extraction.
To detect whether NF-κB p65 binds to the SCD1 promoter, input and immunoprecipitated chromatin was extracted with QIAquick PCR purification kit (QIAGEN, Valencia, CA) by following the manufacturer’s protocol and subjected to PCR amplification using primers designed for the putative p65 binding region 1 (SCD F1/R1) and putative p65 binding region 2 (SCD F2/R2) in the SCD1 promoter (Table S3). The PCR products were analyzed by real-time PCR and normalized by subtracting the Ct of target genes from that of the input/starting material. The expression levels of target genes were calculated as fold changes compared to the signal of DNA immunoprecipitated with IgG (control). As a positive control, DNA immunoprecipitated with p65 antibody was amplified using primers for the known sequence of the IL8 promoter (Table S3) (Li et al., 2012), known NF-κB target gene. As a negative control, DNA immunoprecipitated with p65 antibody was amplified with primers designed for the SCD1 promoter, upstream of the putative p65 binding sites (SCD upstream). All the measurements were performed in triplicates.
QUANTIFICATION AND STATISTICAL ANALYSIS
Hyperspectral SRS images were quantitatively analyzed using ImageJ software. Raman spectroscopy data were analyzed using Origin 8.5 software. Mass spectrometry data were analyzed using Agilent MassHunter Workstation Software Quantitative Analysis. One-way ANOVA or Student’s t test were used for comparisons between groups. Results are represented as means +/± SEM or as indicated. Each experiment was performed with a minimum of three biological replicates; exact numbers are indicated in associated figure legends. Significant differences were considered at * P < 0.05, ** P < 0.01, and *** P < 0.001. n.s. means not significant.
DATA AND SOFTWARE AVAILABILITY
Not applicable.
ADDITIONAL RESOURCES
Not applicable.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank Leelyn Chong, Drs. Bin Liu, Chi Zhang, and Bakhtiyor Yakubov for technical support. The authors acknowledge the use of the metabolite profiling facility of the Bindley Bioscience Center at Purdue University. This work was supported by VA Merit Award (BX000792-05A1) and NCI grant CA198409 to D.M., NCI grant CA182608 and Walther Cancer Foundation to J-X.C., and by NIH under grant P30CA023168. J.L. and J-X.C. are co-founders of Resarci Therapeutics LLC.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
STAR METHODS
- CONTACT FOR REAGENT AND RESOURCE SHARING
-
EXPERIMENTAL MODEL AND SUBJECT DETAILS
-
○Cell lines
-
○Primary ovarian carcinoma cells
-
○Subcutaneous xenograft mouse model
-
○
-
METHOD DETAILS
-
○Sphere formation assay
-
○Aldefluor assay and Fluorescence-Activated Cell Sorting
-
○Hyperspectral stimulated Raman scattering (SRS) imaging
-
○Spontaneous Raman spectroscopy
-
○Mass spectrometry analysis of fatty acids
-
○Tumor initiation assays in subcutaneous xenograft mouse model
-
○Generation of SCD1 stably knock-down cells
-
○Stable retroviral transduction
-
○Gene reporter assay
-
○Reverse transcription-PCR (RT-PCR)
-
○Chromatin immunoprecipitation (ChIP)
-
○
- QUANTIFICATION AND STATISTICAL ANALYSIS
- DATA AND SOFTWARE AVAILABILITY
- ADDITIONAL RESOURCES
Supplemental information includes seven figures, three tables, and four movies and can be found with this article online.
AUTHOR CONTRIBUTIONS
Conceptualization, J-X.C. and D.M.; Methodology, J.L., S.C., D.M., and J-X.C.; Investigation, J.L., S.C., and X.M.; Formal Analysis, J.L. and S.C.; Resources, J.T-P., T.D., and Y.X.; Writing – Original Draft, J.L.; Writing – Review & Editing, all; Visualization, J.L., S.C., D.M., and J-X.C.; Funding Acquisition, J-X.C. and D.M..
References
- Bjerkvig R, Tysnes BB, Aboody KS, Najbauer J, Terzis AJA. The origin of the cancer stem cell: current controversies and new insights. Nat Rev Cancer. 2005;5:995–995. doi: 10.1038/nrc1740. [DOI] [PubMed] [Google Scholar]
- Camandola S, Leonarduzzi G, Musso T, Varesio L, Carini R, Scavazza A, Chiarpotto E, Baeuerle PA, Poli G. Nuclear factor kB is activated by arachidonic acid but not by eicosapentaenoic acid. Biochem Biophys Res Commun. 1996;229:643–647. doi: 10.1006/bbrc.1996.1857. [DOI] [PubMed] [Google Scholar]
- Camp CH, Yegnanarayanan S, Eftekhar AA, Sridhar H, Adibi A. Multiplex coherent anti-Stokes Raman scattering (MCARS) for chemicallysensitive, label-free flow cytometry. Opt Express. 2009;17:22879–22889. doi: 10.1364/OE.17.022879. [DOI] [PubMed] [Google Scholar]
- Camp JCH, Yegnanarayanan S, Eftekhar AA, Adibi A. Label-free flow cytometry using multiplex coherent anti-Stokes Raman scattering (MCARS) for the analysis of biological specimens. Opt Lett. 2011;36:2309–2311. doi: 10.1364/OL.36.002309. [DOI] [PubMed] [Google Scholar]
- Cheng J-X, Xie XS. Vibrational spectroscopic imaging of living systems: An emerging platform for biology and medicine. Science. 2015;350:1054. doi: 10.1126/science.aaa8870. [DOI] [PubMed] [Google Scholar]
- Chute JP, Muramoto GG, Whitesides J, Colvin M, Safi R, Chao NJ, McDonnell DP. Inhibition of aldehyde dehydrogenase and retinoid signaling induces the expansion of human hematopoietic stem cells. Proc Natl Acad Sci U S A. 2006;103:11707–11712. doi: 10.1073/pnas.0603806103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clevers H. The cancer stem cell: premises, promises and challenges. Nat Med. 2011:313–319. doi: 10.1038/nm.2304. [DOI] [PubMed] [Google Scholar]
- Condello S, Morgan CA, Nagdas S, Cao L, Turek J, Hurley TD, Matei D. [beta]-Catenin-regulated ALDH1A1 is a target in ovarian cancer spheroids. Oncogene. 2015;34:2297–2308. doi: 10.1038/onc.2014.178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flesken-Nikitin A, Odai-Afotey AA, Nikitin AY. Role of the stem cell niche in the pathogenesis of epithelial ovarian cancers. Mol Cell Oncol. 2014;1:e963435. doi: 10.4161/23723548.2014.963435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foster R, Buckanovich RJ, Rueda BR. Ovarian cancer stem cells: Working towards the root of stemness. Cancer Lett. 2013;338:147–157. doi: 10.1016/j.canlet.2012.10.023. [DOI] [PubMed] [Google Scholar]
- Ginestier C, Wicinski J, Cervera N, Monville F, Finetti P, Bertucci F, Wicha MS, Birnbaum D, Charafe-Jauffret E. Retinoid signaling regulates breast cancer stem cell differentiation. Cell Cycle. 2009;8:3297–3302. doi: 10.4161/cc.8.20.9761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia D, Yang W, Li L, Liu H, Tan Y, Ooi S, Chi L, Filion LG, Figeys D, Wang L. [beta]-Catenin and NF-[kappa]B co-activation triggered by TLR3 stimulation facilitates stem cell-like phenotypes in breast cancer. Cell Death Differ. 2015;22:298–310. doi: 10.1038/cdd.2014.145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jordan CT, Guzman ML, Noble M. Cancer Stem Cells. N Engl J Med. 2006;355:1253–1261. doi: 10.1056/NEJMra061808. [DOI] [PubMed] [Google Scholar]
- Kamphorst JJ, Cross JR, Fan J, de Stanchina E, Mathew R, White EP, Thompson CB, Rabinowitz JD. Hypoxic and Ras-transformed cells support growth by scavenging unsaturated fatty acids from lysophospholipids. Proc Natl Acad Sci U S A. 2013;110:8882–8887. doi: 10.1073/pnas.1307237110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karin M, Cao Y, Greten FR, Li ZW. NF-kappaB in cancer: from innocent bystander to major culprit. Nat Rev Cancer. 2002;2:301–310. doi: 10.1038/nrc780. [DOI] [PubMed] [Google Scholar]
- Liu G, Lynch JK, Freeman J, Liu B, Xin Z, Zhao H, Serby MD, Kym PR, Suhar TS, Smith HT, et al. Discovery of Potent, Selective, Orally Bioavailable Stearoyl-CoA Desaturase 1 Inhibitors. J Med Chem. 2007;50:3086–3100. doi: 10.1021/jm070219p. [DOI] [PubMed] [Google Scholar]
- Liu PP, Liao J, Tang ZJ, Wu WJ, Yang J, Zeng ZL, Hu Y, Wang P, Ju HQ, Xu RH, et al. Metabolic regulation of cancer cell side population by glucose through activation of the Akt pathway. Cell Death Differ. 2014;21:124–135. doi: 10.1038/cdd.2013.131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma I, Allan A. The Role of Human Aldehyde Dehydrogenase in Normal and Cancer Stem Cells. Stem Cell Rev. 2011;7:292–306. doi: 10.1007/s12015-010-9208-4. [DOI] [PubMed] [Google Scholar]
- Ma X, Xia Y. Pinpointing Double Bonds in Lipids by Paternò-Büchi Reactions and Mass Spectrometry. Angew Chem Int Ed Engl. 2014;126:2630–2634. doi: 10.1002/anie.201310699. [DOI] [PubMed] [Google Scholar]
- Marcato P, Dean CA, Giacomantonio CA, Lee PWK. Aldehyde dehydrogenase: Its role as a cancer stem cell marker comes down to the specific isoform. Cell Cycle. 2011;10:1378–1384. doi: 10.4161/cc.10.9.15486. [DOI] [PubMed] [Google Scholar]
- Meirelles K, Benedict LA, Dombkowski D, Pepin D, Preffer FI, Teixeira J, Tanwar PS, Young RH, MacLaughlin DT, Donahoe PK, et al. Human ovarian cancer stem/progenitor cells are stimulated by doxorubicin but inhibited by Mullerian inhibiting substance. Proc Natl Acad Sci U S A. 2012;109:2358–2363. doi: 10.1073/pnas.1120733109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miranda-Lorenzo I, Dorado J, Lonardo E, Alcala S, Serrano AG, Clausell-Tormos J, Cioffi M, Megias D, Zagorac S, Balic A, et al. Intracellular autofluorescence: a biomarker for epithelial cancer stem cells. Nat Methods. 2014;11:1161–1169. doi: 10.1038/nmeth.3112. [DOI] [PubMed] [Google Scholar]
- Movasaghi Z, Rehman S, Rehman IU. Raman spectroscopy of biological tissues. Appl Spectrosc Rev. 2007;42:493–541. [Google Scholar]
- Nakamura MT, Nara TY. Structure, function, and dietary regulation of delta6, delta5, and delta9 desaturases. Annu Rev Nutr. 2004;24:345–376. doi: 10.1146/annurev.nutr.24.121803.063211. [DOI] [PubMed] [Google Scholar]
- Obukowicz MG, Welsch DJ, Salsgiver WJ, Martin-Berger CL, Chinn KS, Duffin KL, Raz A, Needleman P. Novel, selective delta6 or delta5 fatty acid desaturase inhibitors as antiinflammatory agents in mice. J Pharmacol Exp Ther. 1998;287:157–166. [PubMed] [Google Scholar]
- Pandey PR, Okuda H, Watabe M, Pai SK, Liu W, Kobayashi A, Xing F, Fukuda K, Hirota S, Sugai T, et al. Resveratrol suppresses growth of cancer stem-like cells by inhibiting fatty acid synthase. Breast Cancer Res Treat. 2011;130:387–398. doi: 10.1007/s10549-010-1300-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pattabiraman DR, Weinberg RA. Tackling the cancer stem cells: what challenges do they pose? Nat Rev Drug Discov. 2014;13:497–512. doi: 10.1038/nrd4253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poletto AC, Furuya DT, David-Silva A, Ebersbach-Silva P, Lellis Santos C, Corrêa-Giannella ML, Passarelli M, Machado UF. Oleic and linoleic fatty acids downregulate Slc2a4/GLUT4 expression via NFKB and SREBP1 in skeletal muscle cells. Mol Cell Endocrinol. 2015;401:65–72. doi: 10.1016/j.mce.2014.12.001. [DOI] [PubMed] [Google Scholar]
- Ponti D, Costa A, Zaffaroni N, Pratesi G, Petrangolini G, Coradini D, Pilotti S, Pierotti MA, Daidone MG. Isolation and In vitro Propagation of Tumorigenic Breast Cancer Cells with Stem/Progenitor Cell Properties. Cancer Res. 2005;65:5506–5511. doi: 10.1158/0008-5472.CAN-05-0626. [DOI] [PubMed] [Google Scholar]
- Roongta UV, Pabalan JG, Wang X, Ryseck RP, Fargnoli J, Henley BJ, Yang WP, Zhu J, Madireddi MT, Lawrence RM, et al. Cancer cell dependence on unsaturated fatty acids implicates stearoyl-CoA desaturase as a target for cancer therapy. Mol Cancer Res. 2011;9:1551–1561. doi: 10.1158/1541-7786.MCR-11-0126. [DOI] [PubMed] [Google Scholar]
- Sansone P, Ceccarelli C, Berishaj M, Chang Q, Rajasekhar VK, Perna F, Bowman RL, Vidone M, Daly L, Nnoli J, et al. Self-renewal of CD133hi cells by IL6/Notch3 signalling regulates endocrine resistance in metastatic breast cancer. Nat Commun. 2016;7 doi: 10.1038/ncomms10442. doi:10.1038/ncomms10442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos CR, Schulze A. Lipid metabolism in cancer. FEBS J. 2012;279:2610–2623. doi: 10.1111/j.1742-4658.2012.08644.x. [DOI] [PubMed] [Google Scholar]
- Shanmugam R, Kusumanchi P, Appaiah H, Cheng L, Crooks P, Neelakantan S, Peat T, Klaunig J, Matthews W, Nakshatri H, et al. A water soluble parthenolide analog suppresses in vivo tumor growth of two tobacco-associated cancers, lung and bladder cancer, by targeting NF-κB and generating reactive oxygen species. Int J Cancer. 2011;128:2481–2494. doi: 10.1002/ijc.25587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shostak K, Chariot A. NF-kappaB, stem cells and breast cancer: the links get stronger. Breast Cancer Res. 2011;13:214–220. doi: 10.1186/bcr2886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva IA, Bai S, McLean K, Yang K, Griffith K, Thomas D, Ginestier C, Johnston C, Kueck A, Reynolds RK, et al. Aldehyde Dehydrogenase in Combination with CD133 Defines Angiogenic Ovarian Cancer Stem Cells That Portend Poor Patient Survival. Cancer Res. 2011;71:3991–4001. doi: 10.1158/0008-5472.CAN-10-3175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szotek PP, Pieretti-Vanmarcke R, Masiakos PT, Dinulescu DM, Connolly D, Foster R, Dombkowski D, Preffer F, MacLaughlin DT, Donahoe PK. Ovarian cancer side population defines cells with stem cell-like characteristics and Mullerian Inhibiting Substance responsiveness. Proc Natl Acad Sci U S A. 2006;103:11154–11159. doi: 10.1073/pnas.0603672103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tirinato L, Liberale C, Di Franco S, Candeloro P, Benfante A, La Rocca R, Potze L, Marotta R, Ruffilli R, Rajamanickam VP, et al. Lipid droplets: a new player in colorectal cancer stem cells unveiled by spectroscopic imaging. Stem Cells. 2015;33:35–44. doi: 10.1002/stem.1837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Zee M, Sacchetti A, Cansoy M, Joosten R, Teeuwssen M, Heijmans-Antonissen C, Ewing-Graham PC, Burger CW, Blok LJ, Fodde R. IL6/JAK1/STAT3 Signaling Blockade in Endometrial Cancer Affects the ALDHhi/CD126+ Stem-like Component and Reduces Tumor Burden. Cancer Res. 2015;75:3608–3622. doi: 10.1158/0008-5472.CAN-14-2498. [DOI] [PubMed] [Google Scholar]
- Visvader JE, Lindeman GJ. Cancer stem cells in solid tumours: accumulating evidence and unresolved questions. Nat Rev Cancer. 2008a;8:755–768. doi: 10.1038/nrc2499. [DOI] [PubMed] [Google Scholar]
- Visvader JE, Lindeman GJ. Cancer stem cells in solid tumours: accumulating evidence and unresolved questions. Nat Rev Cancer. 2008b;8:755–768. doi: 10.1038/nrc2499. [DOI] [PubMed] [Google Scholar]
- Wang H-W, Bao N, Le TL, Lu C, Cheng J-X. Microfluidic CARS cytometry. Opt Express. 2008;16:5782–5789. doi: 10.1364/oe.16.005782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Sun Y, Wong J, Conklin DS. PPAR[gamma] maintains ERBB2-positive breast cancer stem cells. Oncogene. 2013;32:5512–5521. doi: 10.1038/onc.2013.217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C, Zhang D, Cheng J-X. Coherent Raman Scattering Microscopy in Biology and Medicine. Annu Rev Biomed Eng. 2015;17:415–445. doi: 10.1146/annurev-bioeng-071114-040554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou BB, Zhang H, Damelin M, Geles KG, Grindley JC, Dirks PB. Tumour-initiating cells: challenges and opportunities for anticancer drug discovery. Nat Rev Drug Discov. 2009;8:806–823. doi: 10.1038/nrd2137. [DOI] [PubMed] [Google Scholar]
- Zhou J, Zhang Y. Cancer stem cells: Models, mechanisms and implications for improved treatment. Cell Cycle. 2008;7:1360–1370. doi: 10.4161/cc.7.10.5953. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.