Abstract
Metastatic uveal melanoma (UM) patients usually die within one year of diagnosis, emphasizing an urgent need to develop new treatment strategies. The liver is the most common site of metastasis. Mitogen-activated protein kinase kinase (MEK) inhibitors improve survival in V600 BRAF-mutated cutaneous melanoma patients but have limited efficacy in UM patients. Our previous work showed that HGF signaling elicits resistance to MEK inhibitors in metastatic UM. In this study, we demonstrate that expression of two BH3-only family proteins, Bim-EL and BMF, contributes to HGF-mediated resistance to MEK inhibitors. Targeting HGF-cMET signaling with LY2875358, a neutralizing and internalizing anti-cMET bivalent antibody, and LY2801653, a dual cMET/RON inhibitor, overcomes resistance to trametinib provided by exogenous HGF and by conditioned medium from primary hepatic stellate cells. We further determined that activation of PI3Kα/γ/δ isoforms mediates the resistance to MEK inhibitors by HGF. Combination of LY2801653 with trametinib decreases AKT phosphorylation and promotes pro-apoptotic PARP cleavage in metastatic UM explants. Together, our data support the notion that selectively blocking cMET signaling or PI3K isoforms in metastatic UM may break the intrinsic resistance to MEK inhibitors provided by factors from stromal cells in liver.
Keywords: melanoma/skin cancers, signal transduction, uveal melanoma, MEK, liver
Introduction
Uveal melanoma (UM) is the most common intraocular malignant tumor in adults and comprises approximately 5% of all melanomas (1). Even after treatment of the primary tumor, 20–50% of patients succumb to metastatic disease. The liver is the predominant organ of metastasis. Standard chemotherapies and immune checkpoint blockers rarely induce clinical responses in patients with macro-metastasis and their 1-year survival is <30% (2). This knowledge emphasizes an urgent unmet need for effective therapeutic strategies for advanced UM.
Activating mutations in GNAQ and GNA11 (typically Q209 and less commonly R183), which encode alpha subunits of the heterotrimeric G proteins, Gαq and Gα11, are found in 80–90% of UM (3–6). Silencing GNAQ induces apoptosis in mutant but not wild-type UM cells (4, 7). Mutant Gαq and Gα11 activate phospholipase C-β (PLC-β), which regulates several pathways including MEK-ERK1/2 and protein kinase C (PKC) signaling (8). In addition, PLC-β-independent activation of Trio, a guanine nucleotide exchange factor, transduces signaling downstream from Gαq to activate Yes-Associated Protein 1 (YAP) and promote UM tumorigenesis (9–11). In UM, GNAQ and GNA11 mutations occur early in disease progression; however, additional alterations are required (5, 12). Mutations in the tumor-suppressor BRCA1-associated protein 1 (BAP1) on chromosome 3 are found in 32–50% of primary melanomas (13). BAP1 mutations associate with aggressive disease and higher likelihood of metastasis. Silencing BAP1 in primary UM cell lines results in a gain of stem-like properties with little/no effect on proliferation and invasion (14). Additional genes mutated in UM are SF3B1 and EIF1AX, which associate with a favorable prognosis (15–17), PLCB4 (18) and CYSLTR2 (19).
A major effector pathway downstream of mutant Gαq and Gα11 is RAF-MEK1/2-ERK1/2 signaling. Inhibition of MEK1/2 with trametinib or selumetinib induces either cell cycle arrest or apoptosis in UM cell lines (7, 20); however, clinical studies in advanced stage UM patients indicate that MEK inhibitors have limited clinical benefit. Trametinib was ineffective in a phase I trial cohort of 16 metastatic UM patients (21). A phase II trial with selumetinib in 120 patients showed a 9-week improvement in progression-free survival compared with standard chemotherapy, but no improvement in overall survival (22). In the most recent phase III trial with 129 patients (23), a combination of selumetinib and standard chemotherapy dacarbazine failed to improve progression-free survival compared with chemotherapy alone. Thus, while MEK inhibitor may form part of a therapeutic approach for advanced-stage UM, further investigation is required to identify inhibitors to act in combination.
The majority of UM patients with overt metastasis show primary resistance to MEK inhibitors, which may be mediated by factors from the tumor microenvironment. In UM cell monocultures, hepatocyte growth factor (HGF) provides resistance to MEK inhibitors (20). HGF is secreted by quiescent hepatic stellate cells. Consistent with the presence of HGF in tumor microenvironment, the majority of UM liver metastases express phosphorylated/activated cMET (20). Together, these results suggest that MEK inhibitors in combination with cMET targeting agents may have utility in advanced UM. In this study, we explored the molecular mechanism of HGF-mediated resistance to MEK inhibitors in UM cells and pre-clinically evaluated the efficacy of co-targeting cMET with MEK inhibitor in metastatic cell lines and ex vivo explants. Our data show that down-regulation in the BH3-only proteins, Bim-EL and Bmf, contribute to HGF-mediated protective effect in metastatic UM cells. Clinical grade cMET targeting agents effectively overcome the resistance provided by exogenous HGF as well as factors derived from hepatic stellate cells. Combined inhibition of cMET and MEK1/2 enhances apoptotic signal in cell lines and an ex vivo explant model of metastatic UM. Together, these data provide a pre-clinical basis for combinational therapies targeting mutant Gαq/11 signaling and signaling initiated by factors from tumor microenvironment in advanced-stage UM patients.
Materials and Methods
Cell culture
UM001 and UM004 cells were derived from liver and orbital metastases of human UM, respectively; both harbor GNAQ Q209L mutations (20, 24). UM001 cells were cultured in RPMI 1640 medium supplemented with 10% heat-inactivated FBS, 10% non-essential amino acids, 2 mM L-glutamine and 10 mM Hepes buffer. UM004 cells were maintained in MEM medium containing 10% heat-inactivated FBS and 2 mM L-glutamine. LX-2 human hepatic stellate cell line was purchased from EMD Millipore (Billerica, MA) and cultured in DMEM medium containing 2% FBS and 2 mM L-glutamine according to manufacturer’s protocol. Human hepatic stellate cells (HHSteC) were cultured in basal medium, 2% FBS and 1% stellate cell growth supplement according to manufacturer’s protocol (ScienCell Research Laboratories. Carlsbad, CA). HHSteC conditioned medium is collected from HHSteC cultures immediately before sub-culture. Medium conditioned by early passage (<6 passages) HHSteC cultures was used for functional co-culture experiments with UM001 and UM004 cells.
Cell line validation
UM001 and UM004 cells were confirmed as harboring GNAQ mutations as determined by Sanger DNA sequencing. UM001 and UM004 cells were analyzed by STR analysis on January 15th, 2015; The UM001 and UM004 profiles were unique, although the latter had a 94% match with 3 changed alleles to MDA-MB-330 cells on the DSMZ resource.
Inhibitors, growth factors and function-blocking antibodies
Trametinib (GSK1120212), MK2206 (PubChem compound database (CID, 24964624)), GDC0032 (25), TGX221 (26), BYL710 (25) and IPI145 (25) were purchased from Selleck Chemicals (Houston, TX). Recombinant human HGF was purchased from PeproTech (Rocky Hill, NJ) and used at 10 ng/ml based on our previous studies (20). The neutralizing and internalizing anti-cMET antibody, LY2875358, and the cMET/RON inhibitor, LY2801653 (27), were provided by Eli Lilly and Company (Indianapolis, IN).
Short-interfering RNA (siRNA) and transfection
UM004 cells (3 × 105) were seeded in 6-well plates overnight before transfection with chemically synthesized siRNAs at a final concentration of 25 nM using Lipofectamine™ RNAiMAX (Invitrogen, Carlsbad, CA) as previously described (28). Bim-EL specific siRNAs (GACCGAGAAGGUAGACAAUUGTT and CAAUUGUCUACCUUCUCGGUCTT) were purchased from Cell Signaling Technology (Danvers, MA). Bmf specific siRNA (GAGUAACAGAUAACGAUUA) was purchased from Dharmacon Inc. (Lafayette, CO). A non-targeting siRNA (UAGCGACUAAACACAUCAAUU) was used as a control.
Western blotting
Cells were washed in cold PBS and lysed directly in Laemmli sample buffer. Lysates were resolved by SDS polyacrylamide gel electrophoresis and transferred to polyvinylidene difluoride membranes. Membranes were blocked with 1% BSA and incubated with indicated primary antibodies overnight at 4°C. Proteins were detected using the horseradish peroxidase-conjugated secondary antibodies followed by development using chemiluminescence substrate (Pierce, Rockford, IL). The following primary antibodies were used: ERK2 (D-2), Cyclin A and Noxa from Santa Cruz Biotech. Inc. (Santa Cruz, CA); Bcl-2, Bcl-xl, Mcl-1, Bax, Bad, Bid, Puma, cleaved caspase 3, cleaved PARP, RB, phospho-RB (S780), cMET, phospho-cMET Y1234/1235 (D26), phospho-cMET Y1349, AKT, phospho-AKT T308 (C31E5E), phospho-AKT S473 (D9E), phospho-ERK1/2 (D13.14.4E), Stat3, phospho-Stat3 Y705, p110α-PI3K, p110β-PI3K and p110δ-PI3K from Cell Signaling Tech; cyclin D1 and Bcl-w from BD Pharmingen, p110δ-PI3K, α-SMA and FAP from Abcam (Cambridge, MA); Bim-EL and BMF from Enzo Life Sciences (Farmingdale, NY); Actin from Sigma-Aldrich (St Louis, MO) and Hsp90 (clone 4F3.E8) from StressMarq Biosciences (Victoria BC). Chemiluminescence was visualized on a ChemiDoc Imaging System and quantitated using Image Lab 4.1 software (Bio-Rad, Hercules, CA).
EdU incorporation assay
UM001 and UM004 cells were treated with DMSO, 50 nM trametinib, 10 ng/ml HGF or trametinib plus HGF for 32 hours before the addition of 10 µM EdU for another 16 hours. Cells were then processed using the Click-iT Plus EdU Alexa Fluor 647 Flow Cytometry Assay kit (Invitrogen) according to manufacturer’s protocol. Experiments were performed in triplicate and statistical analysis was performed using a two-tailed t test assuming unequal variance with error bars representing SD.
Annexin V/PI staining
Cells were trypsinized, washed with cold PBS and resuspended in 0.1 ml binding buffer. Cells were then stained with 5 µl of Annexin V-APC and 2 µl of propidium iodide (PI) for 30 minutes at room temperature. Staining was then analyzed by flow cytometry on a BD FACSCalibur flow cytometer (BD Biosciences, Franklin Lakes, NJ). Data were analyzed by FlowJo software (Tree Star, Inc., Ashland, OR). Experiments were performed in triplicate with statistical analysis as in EdU incorporation assay.
Crystal violet staining
Exponentially growing UM001 and UM004 cells were plated in 6-well or 12-well dishes and treated as described in figure legends. Cells were then stained with crystal violet solution (1% crystal violet, 10% buffered formalin) for 30 minutes, washed and air dried. Plates were imaged by scanning while pictures were taken at ×100 magnification on the Nikon™ Eclipse Ti inverted microscope with NIS-Elements AR 3.00 software (Nikon, Tokyo, Japan). Crystal violet staining images were quantitated using Image J. Experiments were performed in triplicate and statistical analysis performed using a two-tailed t test assuming unequal variance
Enzyme-linked immunosorbent assay (ELISA)
HGF levels in conditioned medium collected from stellate cell cultures were measured with ELISA kits (Invitrogen), according to the manufacturer’s instructions.
Migration and invasion assays
Sub-confluent UM001 and UM004 cells were cultured overnight in serum-free medium. For migration assays, 1 × 104 cells in serum-free medium were placed inside 8.0 µm pore-size cell culture inserts (BD Biosciences, San Jose, CA). For invasion assays, the inserts were first coated with 0.75 mg/ml Matrigel (BD Biosciences) for one hour before plating cells inside each chamber. Cells were allowed to migrate or invade for 16 hours towards an attractant of stellate cell culture medium or conditioned medium from passage 3 stellate cells. Chamber filters were fixed in buffered formalin and stained with crystal violet. Cells in the inner chamber were removed. Images were taken with a Nikon™ Eclipse Ti inverted microscope at 100 × magnification.
Reverse phase protein array (RPPA) analysis
UM001 and UM004 cells were plated in 6-well dishes at 4 × 105 cells per well. Cells were treated with unconditioned medium or passage 2 HHSteC conditioned medium for either 1 hour or 48 hours. Cells were lysed and prepared as previously described (29) and analyzed at the MD Anderson Functional Proteomic core facility (Houston, TX). Serial dilutions of samples were arrayed on nitrocellulose-coated slides and run against 295 validated antibodies. Spot density was determined using MiroVigene and analyses of triplicate normalized data were performed using SuperCurve. Hierarchical clustering of the 295 antibodies was performed via the clustergram function in Matlab® (version 2015b) on RPPA median-centered log2 expression values. The samples were pre-sorted based on cell line, treatment type, time point and replicate number.
Ex vivo UM explants
Human metastatic UM tissue was collected following patient consent at Thomas Jefferson University Hospital under an IRB-approved protocol (#02.9014R). Less than 16 hours post-surgery, excess adipose and stromal tissue was removed and the tumor (Explant of Patient No.4/Ex-Pt#4) was cut into 1 mm3 pieces. Vetspon absorbable hemostatic gelatin 1 cm3 sponges (Novartis, Basel, Switzerland) were pre-soaked in 12-well plates for 15 minutes at 37°C in 500 µL of DMEM medium containing 10% FBS, penicillin-streptomycin and drugs. DMSO was used as a vehicle control. To avoid concerns of intra-tumoral heterogeneity, up to four ~1 mm3 pieces from different locations of the original tumor were placed per sponge per treatment condition. Samples were treated for 48 hours with medium being replaced after 24 hours. Tumor pieces for western blotting were homogenized in lysis buffer with phosphatase and protease inhibitors (PhosSTOP and cOmplete tablets, Roche, Basel, Switzerland). Laemmli sample buffer was added and samples were heated at 99°C for 5 minutes.
Statistical Analysis
Statistical analyses were performed using SPSS v22.0 (IBM, Armonk, NY, USA). One-way ANOVA analyses were performed on normalized data from groups of equal sizes. No outliers were identified during inspection of boxplots. All of the groups were determined to be normally distributed using Shapiro–Wilk’s test (P > 0.05). There was homogeneity of variances among all groups, as determined by Levene’s test of equality of variances (P > 0.05). Dunnett’s one-tailed multiple comparison post-hoc tests were performed to determine statistical significance.
Results
HGF promotes G1/S cell cycle progression and decreases cytotoxicity of trametinib-treated cells
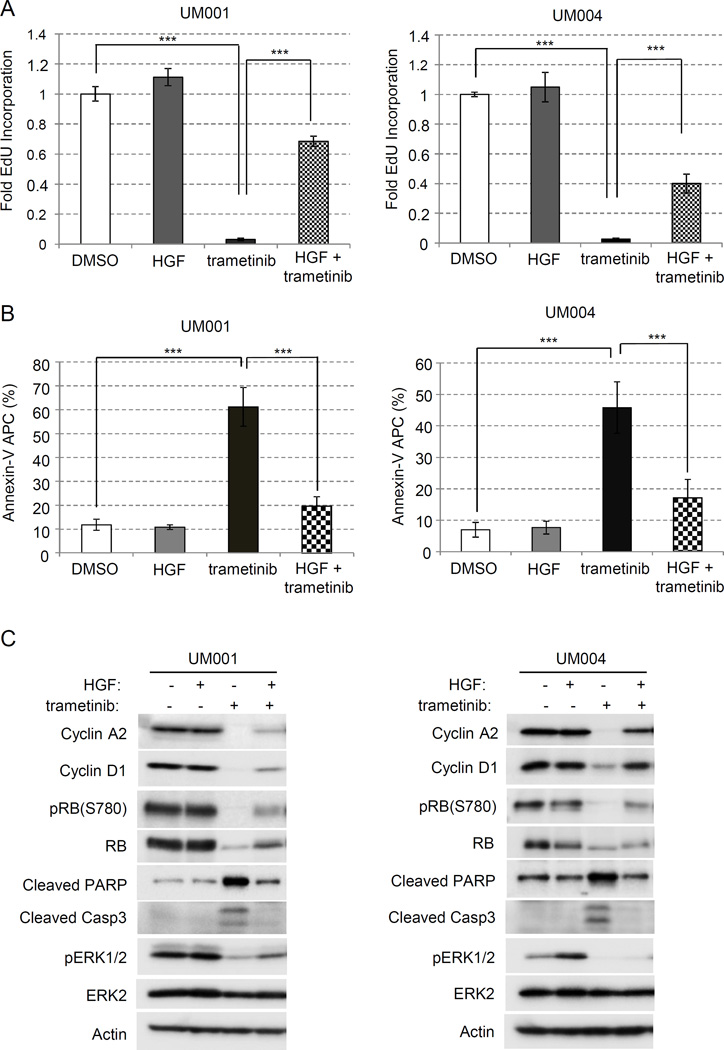
We determined whether HGF-mediated resistance to MEK inhibitors in UM cells was associated with effects on S-phase entry (EdU incorporation) and/or apoptosis (annexin V/PI staining). HGF was used at 10 ng/ml based on our previous studies (20). In UM001 and UM004 cells, trametinib treatment resulted in > 90% decrease in EdU incorporation (Fig 1A) and 45–60% increase in annexin V staining (Fig 1B). Treatment with HGF in the absence of MEK inhibition elicited minimal effect on S-phase entry and apoptosis. By contrast in trametinib-treated UM001 and UM004 cells, HGF restored S-phase entry by 70% and 40% respectively compared with DMSO controls (Fig 1A). Additionally, HGF significantly reduced the annexin V population in trametinib-treated UM001 and UM004 cells (Fig 1B; Suppl Fig 1).
Figure 1. HGF promotes cell cycle progression of trametinib-treated UM cells.
(A) HGF promotes S-phase entry in trametinib-treated cells. 3.0 × 105 UM001 cells and UM004 cells were seeded in triplicates and treated with DMSO, 50 nM trametinib, 10 ng/ml HGF, and a combination of 50 nM trametinib and HGF, respectively, for a total of 32 hours. A final concentration of 10 µM EdU was allowed to incorporate for 16 hours before processing. S-phase entry is normalized to DMSO condition, and data points are indicative of 3 experimental repeats. *P<0.05, **P<0.01, ***P<0.001. (B) HGF inhibits trametinib-induced apoptosis in UM cells. UM001 and UM004 cells were treated with DMSO, 10 ng/ml HGF, 50 nM trametinib, a combination of HGF and trametinib, respectively, for 48 hours. Cells were then subjected to annexin V/PI staining. The percentage of annexin V-positive cells is graphed. *P<0.05, **P<0.01, ***P<0.001. (C) HGF modulates expression of cell cycle regulators in trametinib-treated cells. UM001 cells (left) and UM004 cells (right) were treated with 50 nM trametinib, 10 ng/ml HGF, 2.5 µM MK2206, or combinations as indicated for 48 hours. Cell lysates were probed for levels of cell cycle regulators cyclin A2, cyclin D1, phospho-RB, RB, cleaved PARP and cleaved caspase 3, respectively. Levels of pAKT and pERK were also determined by Western blotting.
We next analyzed the cell cycle profiles of UM001 and UM004 cells treated with HGF, trametinib or the combination of HGF plus trametinib. Trametinib treatment was associated with changes in G1/S regulators including lower expression of cyclin A2 and cyclin D1 and reduced retinoblastoma (RB) phosphorylation (Fig 1C). Down-regulation of total RB expression following trametinib treatment was also detected, an effect previously observed in breast cancer cell lines following inhibition of cell cycle progression with CDK4/6 inhibitors (30, 31). MEK inhibition increased expression of the apoptotic markers, cleaved PARP and cleaved caspase 3. Notably, trametinib-treated cells treated with HGF showed a partial recovery of cyclin A2, cyclin D1 and phospho-RB levels. HGF also modestly increased levels of phosho-ERK1/2 in trametinib-treated cells. Additionally, the induction of cleaved PARP and cleaved caspase 3 was mitigated by HGF in trametinib-treated cells (Fig 1C). Together, these data indicate that HGF promotes the growth of trametinib-treated cells through restoration of cell cycle progression and inhibition of apoptosis.
Down-regulation of Bim-EL and Bmf contributes to HGF-mediated resistance to trametinib in UM cells
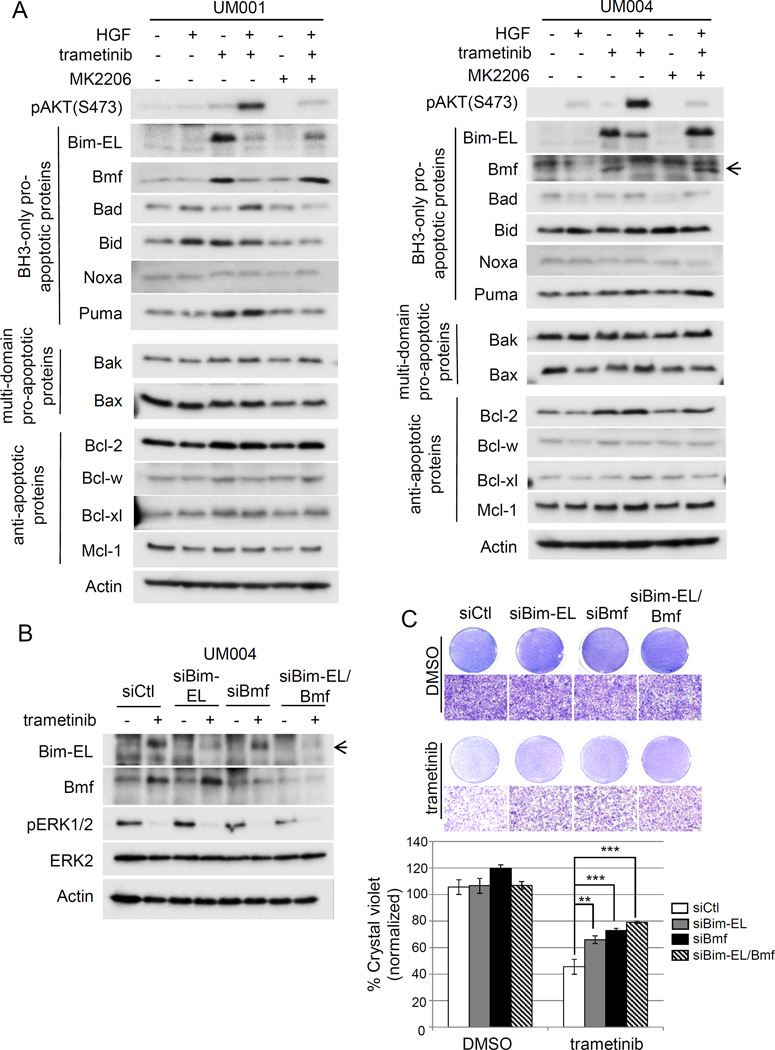
To molecularly understand how HGF counteracts trametinib-mediated apoptosis, we compared the levels of Bcl-2 family proteins in UM cells treated with HGF, trametinib, or the combination of both. We also treated cells with MK2206 to evaluate the role of AKT activity. HGF promoted the phosphorylation of AKT in the presence of trametinib, an effect that was diminished by MK2206 (Fig 2A). Trametinib treatment did not alter expression of anti-apoptotic proteins, Bcl-w, Bcl-xl, multi-domain pro-apoptotic proteins, Bak and Bax, or BH3-only proteins Bad, Bid and Noxa in UM001 and UM004 cells (Fig 2A). By contrast, the BH3-only pro-apoptotic proteins, Bcl-2-interacting mediator of cell death-extra large (Bim-EL) and Bcl-2 modifying factors (Bmf) were up-regulated in trametinib-treated cells. The induction of Bim-EL and Bmf was diminished or markedly reduced when HGF was supplemented to trametinib-treated UM cells (Fig 2A). Notably, Bim-EL and Bmf levels were re-induced in cells treated with a combination of trametinib, HGF and MK2206, suggesting that HGF activation of AKT mediates the resistance to trametinib. A modest up-regulation of the pro-survival protein Bcl-2 and the BH3-only pro-apoptotic protein, Puma, was detected with trametinib treatment in one (Puma) or both (Bcl-2) cell lines (Fig 2A).
Figure 2. Downregulation of Bim-EL and BMF contributes to HGF-mediated resistance to trametinib in UM cells.
(A) HGF inhibits trametinib-induced Bim-EL and Bmf expression in UM cells. UM001 cells (left) and UM004 cells (right) were treated with 50 nM trametinib, 10 ng/ml HGF, 2.5 µM MK2206, or combinations as indicated for 48 hours. Cell lysates were analyzed for expression of indicated BH3-only pro-apoptotic proteins (Bim-EL, Bmf, Bad, Bid, Noxa and Puma), multi-domain pro-apoptotic proteins (Bax and Bak) and anti-apoptotic proteins (Bcl-2, Bcl-w, Bcl-xl and Mcl-1). Actin was used as loading control. (B) Silencing of Bim-EL and Bmf in UM cells. UM004 cells were transfected with 20 nM control siRNA, Bim-EL siRNA, Bmf siRNA or Bim-EL/Bmf siRNA. Expression of Bim-EL and Bmf was examined by Western blotting with indicated antibodies. (C) Silencing of Bim-EL and Bmf renders UM cells resistant to trametinib-induced apoptosis. UM cells were transfected with siRNAs, as above. 48 hours post-transfection, cells were treated with DMSO or 50 nM trametinib for another 48 hours. Cell growth was determined by crystal violet staining. Representative images of the cells at 100× magnification are shown. The scale bar is equal to 100 µm. Quantitation of crystal violet staining is presented as mean of percentage crystal violet from triplicate experiments following normalization to siCtl. *P<0.05, **P<0.01, ***P<0.001.
To determine whether up-regulation of Bim-EL and Bmf is required for trametinib-induced inhibition of cell viability, Bim-EL and/or Bmf-silenced UM004 cells were treated with trametinib (Fig 2B) and evaluated by crystal violet staining (Fig 2C). In comparison to controls, trametinib decreased cell viability by ~60% (Fig 2C). Individual knockdown of Bim-EL and Bmf each partially rescued cells from trametinib with cell viability inhibited by 32–39%; however, simultaneous silencing of Bim-EL and Bmf further restored the viability of trametinib-treated cells with cell growth decreased by ~26% of the control (Fig 2C). To examine whether Bim-EL and Bmf are sufficient to promote UM cell apoptosis, UM001 and UM004 cells were infected with adenoviruses to express Bim-EL, Bmf and enhanced green fluorescence protein (eGFP), as a control. Ectopic expression of Bim-EL and/or Bmf significantly increased apoptosis in UM cells, while expression of eGFP showed little effect (Suppl Fig 2). These results suggest that Bim-EL and Bmf are sufficient to induce apoptosis and are down-regulated in HGF-mediated resistance to MEK inhibitors.
LY2801653 and LY2875358 abrogates HGF-mediated resistance to trametinib in UM cells
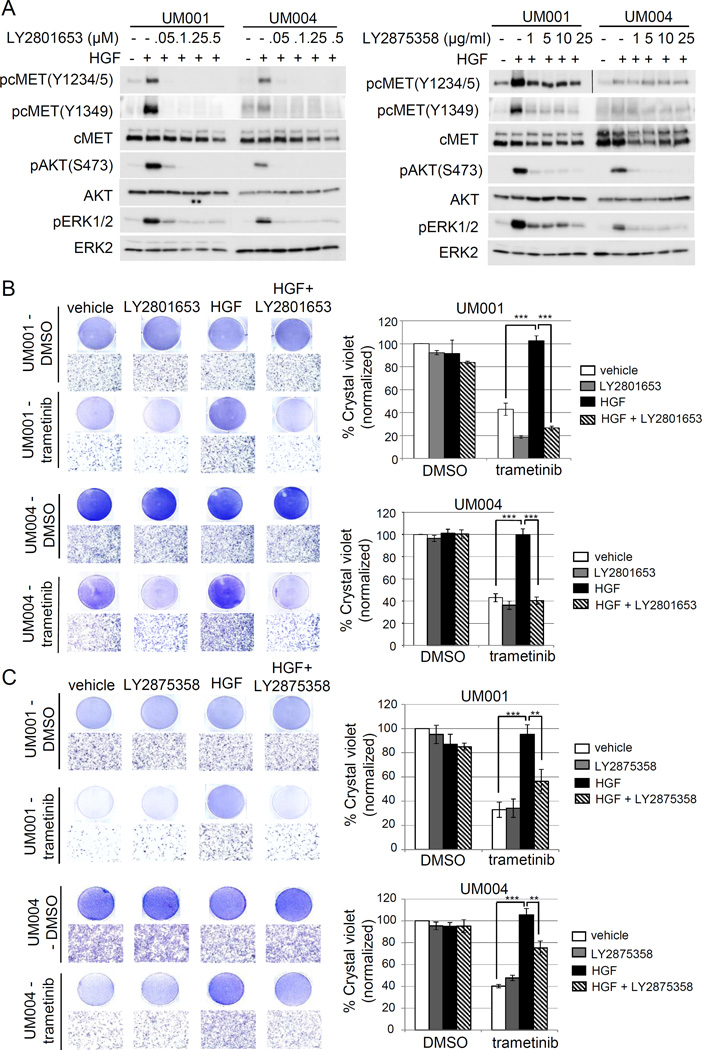
To inhibit HGF-mediated signaling, we utilized two cMET targeting agents that are being tested in clinical trials for UM patients with liver metastasis as well as other advanced cancers. Of these two agents, LY2801653 is a type II kinase inhibitor with cMET as one of its target and displays anti-tumor activity in non-small cell lung carcinoma and cholangiocarcinoma preclinical models (32–34). LY2875358 is a neutralizing and internalizing anti-cMET bivalent antibody that showed potent anti-tumor activity in both HGF-dependent and cMET amplified preclinical tumor models (35). Initially, UM001 and UM004 cells were treated with increasing doses of LY2801653 and LY2875358 followed by HGF stimulation. Both cMET inhibitors effectively blocked HGF-induced phosphorylation of ERK1/2, AKT and cMET at tyrosine 1349 (Fig 3A and 3B). Phosphorylation at tyrosine 1349 in the cMET cytoplasmic domain provides a direct binding site for Gab1 (36), which promotes AKT pathway activation. Of note, LY2875358 had minimal effect on HGF-induced phosphorylation of cMET at tyrosine 1234/5 (Fig 3A and 3B), critical sites for kinase activation. We evaluated the ability of these two cMET targeting agents in overcoming HGF-mediated resistance to trametinib in UM cells. LY2801653 alone did not significantly alter UM001 and UM004 cell growth at 100 nM; however, growth of trametinib-treated UM cells which decreased by ~57% compared to the vehicle control was further inhibited when treated with LY2801653 (Fig 3B). The viability of trametinib/LY2801653 co-treated UM001 and UM004 cells decreased by 81% and 64%, respectively, of the vehicle control. Importantly, HGF-mediated growth protection from trametinib treatment was abrogated by LY2801653 (Fig 3B). Similarly, LY2875358 alone had little effects on UM001 and UM004 cell growth. Although LY2875358 did not further inhibit growth of trametinib-treated cells, LY2875358 blocked HGF-mediated protection from trametinib (Fig 3C). The viability of trametinib-treated UM001 and UM004 cells was increased to levels similar to the vehicle control when cells were treated with HGF; an effect that was decreased with LY2875358 by 44% in UM001 and 25% in UM004 compared to vehicle control (Fig 3C). Together, these data demonstrate that targeting HGF signaling with clinical grade cMET neutralizing antibody and inhibitor overcomes HGF-mediated resistance to trametinib in metastatic UM cells.
Figure 3. Clinical grade cMET targeting agents, LY2801653 and LY2875358, overcome HGF-mediated resistance to trametinib in UM cells.
(A) A dual cMET/RON small-molecule inhibitor LY2801653 and a bivalent cMET monoclonal antibody LY2875358 block HGF/cMET signaling. UM001 cells and UM004 cells were pretreated with increasing doses of LY2801653 (left) and LY2875358 (right) overnight. The next day cells were stimulated with 10 ng/ml HGF for 30 minutes. Phosphorylation of cMET, AKT and ERK1/2 was assessed by Western blotting. (B) HGF-induced resistance to trametinib is reversed by LY2801653. UM001 cells (top) and UM004 cells (bottom) were treated with DMSO or 50 nM trametinib, in combination with 10 ng/ml HGF and/or 100 nM of LY2801643 as indicated for 48 hours (UM004) or 72 hours (UM001). Cells were washed and stained with crystal violet. Images were taken (100 × magnification). Scale bar is equal to 100 µm. The mean of percentage crystal violet from triplicate experiments following normalization to vehicle control is shown. *P<0.05, **P<0.01, ***P<0.001.
(C) HGF-induced resistance to trametinib is reversed by LY2875358. UM001 cells (top) and UM004 cells (bottom) were first treated with 10 µg/ml LY2875358 for 45 min, followed by 10 ng/ml HGF and 50 nM trametinib for 48 hours (UM004) or 72 hours (UM001). Cells were stained with crystal violet. Representative microscopic images are shown (100 × magnification). Scale bar is equal to 100 µm. The percentage of crystal violet is normalized to vehicle control and the mean of percentage crystal violet from triplicate experiments is shown. *P<0.05, **P<0.01, ***P<0.001.
Primary hepatic stellate cell medium protects UM cells against trametinib through HGF-cMET signaling
Basal phosphorylation of cMET and downstream signaling is low in UM lines and understanding the communication between cancer cells and the stroma in the metastatic site is necessary for the development of optimal therapeutic regimens. UM frequently metastasizes to the liver. Hepatic stellate cells are intralobular connective tissue cells that are quiescent in a healthy liver, but transition into myofibroblast-like cells and become activated during liver fibrosis and hepatocellular carcinomas (37). Current available stellate cell lines are either immortalized by hTERT or become activated due to long time culture and therefore at least partially lose characteristics of their primary origins (38). Therefore we utilized primary stellate cells that were isolated from human liver. These cells were cultured for up to six passages to minimize their activation, passages at which they did not express the fibroblast markers, α-SMA and FAP (Suppl Fig 3A).
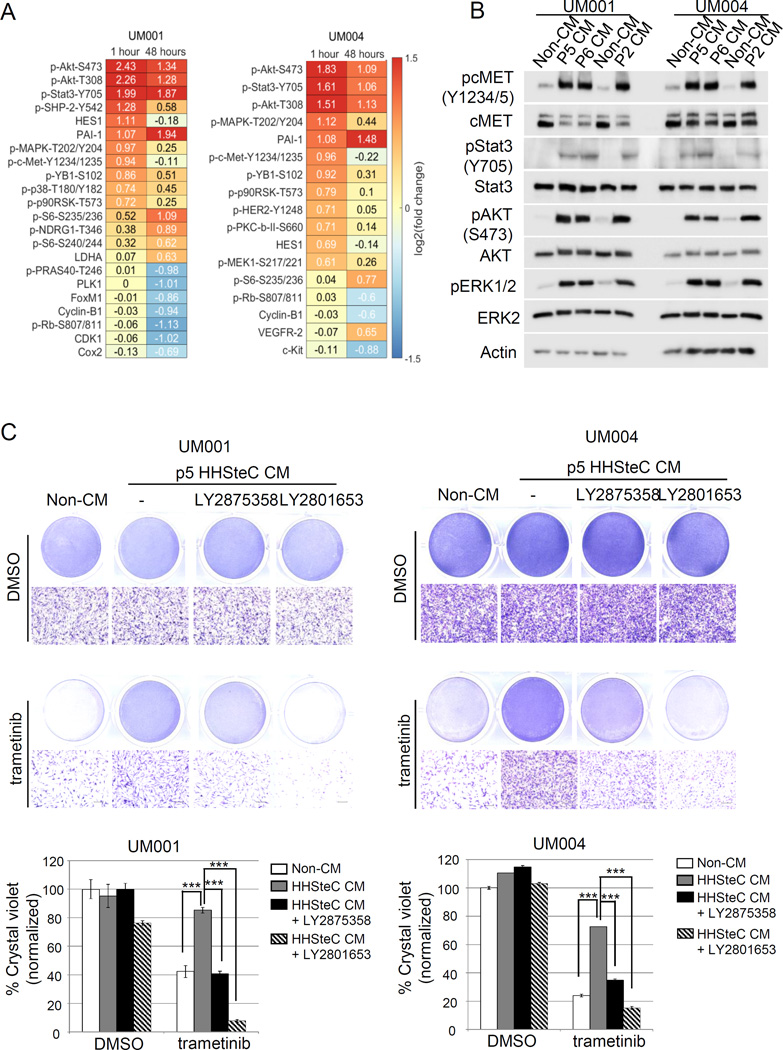
To better understand the effects of hepatic stellate cells on UM cells, we first performed high-throughput antibody-based RPPA analysis on UM001 and UM004 cells incubated for 1 hour or 48 hours with either unconditioned medium or stellate cell conditioned medium. Supervised clustering of proteins that were regulated by stellate cell conditioned medium and further significance analysis of microarrays (SAM) identified several proteins that were differentially regulated by addition of stellate cell conditioned medium (Fig 4A; Suppl Fig 3B). In both UM001 and UM004 cells, PI3K/AKT and ERK/MAPK signaling were the most activated pathways by stellate cell medium (Fig 4A). We performed Western blot analysis to validate the RPPA findings. In UM001 and UM004 cells, conditioned medium from stellate cells rapidly induced phosphorylation of ERK1/2, AKT, cMET, and Stat3 (Fig 4B). We also demonstrated that HGF was present at ng/ml levels in the conditioned medium from early passages of primary hepatic stellate cells by ELISA (Suppl Fig 3C). In contrast, the conditioned medium from an immortalized human hepatic stellate cell line did not induce cMET activation (Suppl Fig 3D). Consistent with the known role of HGF (39), we showed that stellate cell conditioned medium promoted the migration and invasion of UM001 and UM004 cells (Suppl Fig 4).
Figure 4. Human hepatic stellate cells promote resistance to trametinib through HGF-cMET signaling in UM cells.
(A) UM001 and UM004 cells were incubated with unconditioned medium or passage 2 human hepatic stellate cells (HHSteC) conditioned medium for 1 hour or a total of 48 hours. Cell lysates were subjected to RPPA analysis against 295 validated antibodies. Significance analysis of microarray for RPPA of UM001 cells (left) and UM004 cells (right) growing in unconditioned medium or stellate cells conditioned medium was shown. (B) Stellate cells conditioned medium stimulates cMET signaling cascades. UM001 and UM004 cells were incubated with unconditioned medium and stellate cells conditioned (passage 2, 5 and 6) for one hour. Levels of pcMET, pStat3, pAKT and pERK1/2 were assessed by Western blotting. (C) HHSteC conditioned medium renders resistance to trametinib through cMET in UM cells. UM001 cells (left) and UM004 cells (right) were cultured in unconditioned medium or passage 5 (p5) HHSteC conditioned medium and treated with 50 nM trametinib, with or without 25 ng/ml LY2875358 and 100 nM LY2801653 for 48 hours (UM004) or 72 hours (UM001). Cell growth was determined by crystal violet staining. Representative microscopic images are shown (100 × magnification). Scale bar is equal to 100 µm. The percentage of crystal violet is normalized to non-CM control and the mean of percentage crystal violet from triplicate experiments is shown. *P<0.05, **P<0.01, ***P<0.001.
To determine whether conditioned medium from stellate cells drives resistance to trametinib though HGF-cMET pathway activation, we cultured UM001 and UM004 cells in either unconditioned medium or stellate cell conditioned medium. Factors from stellate cells protected UM cells from trametinib-induced growth inhibition as the viability of UM cells cultured in conditioned medium increased by 2–3 fold compared to trametinib treatment/non conditioned medium conditions (Fig 4C). Importantly, stellate cell conditioned medium protection to trametinib was restored by LY2875358 and LY2801653, with LY2801653 being more potent in sensitizing UM cells (85–90% reduction in cell viability compared to the vehicle control). This suggests that LY2801653 is a more effective cMET inhibitor and/or signaling molecules other than cMET may play a role in response to trametinib in UM cells (Fig 4C). However, together these data indicate that factors from hepatic stellate cells elicit innate resistance to trametinib at least partially through HGF-cMET signaling.
HGF-mediated growth protection from MEK inhibitors is reversed by PI3Kβ-sparing inhibitors
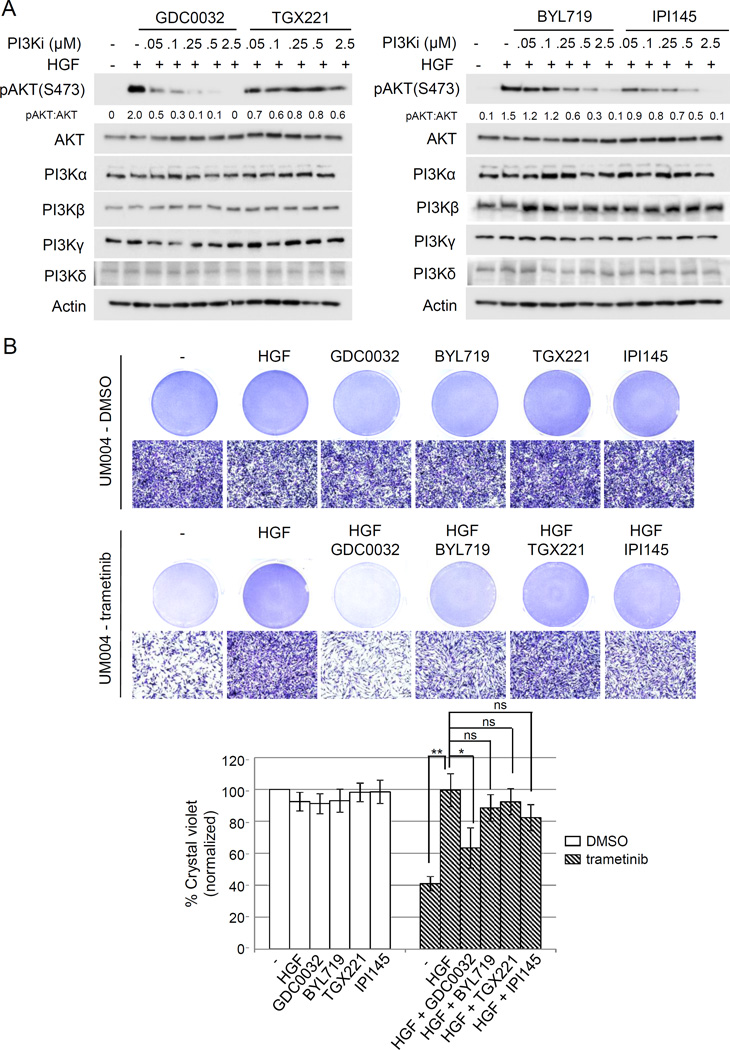
Since PI3K-AKT is a major pathway activated by HGF, we examined the dependency of PI3K isoforms on HGF-mediated AKT phosphorylation and HGF-mediated resistance to trametinib in UM cells. We utilized PI3K isoform specific inhibitors: GDC0032 is a PI3Kβ-sparing isoform inhibitor targeting PI3Kα/δ/γ; TGX221 is a p110β-specific inhibitor; BYL719 is a selective PI3Kα inhibitor; and IPI145 is a selective PI3K δ/γ inhibitor. We first pre-treated UM001 (Suppl Fig 5) and UM004 cells (Fig 5) with increasing doses of individual inhibitors followed by HGF stimulation. All these four p110 isoforms were expressed in both UM001 and UM004 cells and expression was unchanged by HGF/inhibitor treatments (Fig 5A, Suppl Fig 5A). The PI3Kβ-sparing isoform inhibitor GDC0032 effectively blocked HGF-mediated AKT phosphorylation at 250 nM in UM001 cells and 50 nM in UM004 cells. PI3Kα inhibitor BYL719 and PI3K δ/γ inhibitor IPI145 significantly inhibited HGF-mediated AKT phosphorylation at 500 nM, whereas the PI3Kβ specific inhibitor TGX221 did not block AKT phosphorylation even at 2.5 µM (Fig 5A, Suppl. Fig 5A). We evaluated the ability of PI3K isoform specific inhibitors to overcome HGF-mediated resistance to trametinib in these cells. Individual inhibitors at 500 nM elicited no effect or minimal effect on cell growth (Fig 5B, Suppl Fig 5B). HGF-mediated growth protection in trametinib-treated UM001 cells (Suppl Fig 5B) and UM004 cells (Fig 5B) was reverted by GDC0032 (37% reduction in UM004 cell growth compared to vehicle control) and partially reverted by BYL719 and IPI145 (12–17% inhibition of UM004 cell growth compared to vehicle control). Additionally, PI3Kβ-specific inhibitor TGX221 failed to markedly induce growth inhibition in HGF and trametinib treated cells. These data suggest PI3Kα/δ/γ, but not PI3Kβ, account for HGF-mediated AKT phosphorylation and resistance to MEK inhibition in UM cells.
Figure 5. HGF-mediated resistance to trametinib is dependent on PI3Kβ-sparing isoforms in UM cells.
(A) Isoform specific PI3K inhibitors differentially block HGF-mediated activation of AKT in UM cells. UM004 cells were pretreated with increasing doses of GDC0032, TGX221, BYL719 and IPI145 for 6 hours. Cells were then stimulated with 10 ng/ml HGF for 30 minutes. Cell lysates were probed with pAKT, AKT, PI3Kα, PI3Kβ, PI3Kγ, PI3Kδ and actin antibodies. (B) PI3Kβ-sparing inhibitor GDC0032, but not PI3Kβ inhibitor TGX221, abrogates HGF-mediated resistance to trametinib in UM cells. UM004 cells were treated 50 nM trametinib, 10 ng/ml HGF, 0.5 µM of GDC0032, TGX221, BYL719 or IPI145 respectively or in combination for 48 hours. Cells were stained with crystal violet. Representative microscopic images are shown (100× magnification). Scale bar is equal to 100 µm. The percentage of crystal violet is normalized to vehicle control and the mean of percentage crystal violet from triplicate experiments is shown. *P<0.05, **P<0.01, ***P<0.001, ns: not significant.
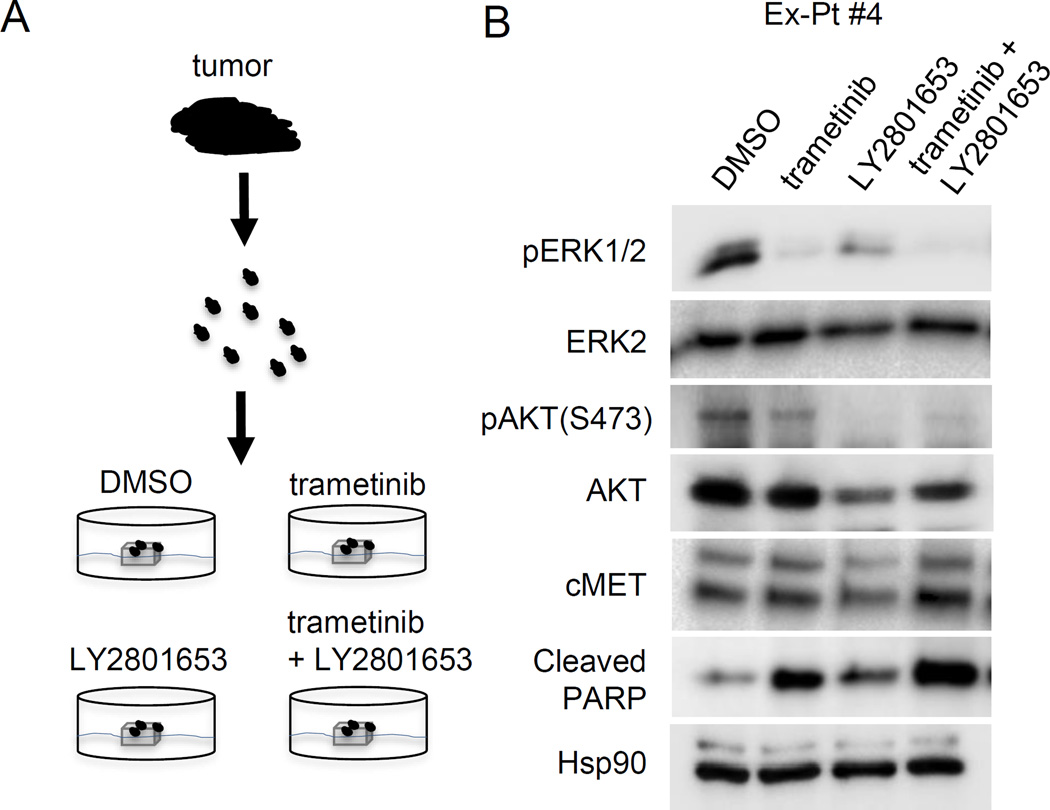
LY2875358 combines with MEK1/2 targeting to promote apoptotic index in metastatic UM explants
To test whether combined therapies targeting MEK1/2 and HGF-cMET signaling improves the response in metastatic UM, We next extended our study to analyze a mutant GNAQ harboring UM patient sample using an ex vivo treatment approach (Fig 6A Supplementary Table S1). Tumor tissue pieces were treated with DMSO, trametinib, LY2875358, or a combination of trametinib and LY2875358. As expected, treatment with trametinib inhibited the phosphorylation of ERK1/2 (Fig 6B), suggesting that ex vivo treatment of patient-derived explants is a feasible strategy for testing drug response in UM. Ex vivo treatment with trametinib also promoted apoptosis, as evidenced by an increased expression of cleaved-PARP. Interestingly, combination of trametinib with LY2875358 further upregulated the expression of cleaved-PARP. These data are supportive that a combined therapy with MEK and cMET inhibition may represent a novel and effective strategy in treating metastatic UM patients.
Figure 6. Trametinib and LY2875358 combine to inhibit AKT activation and promote apoptosis in UM explants.
(A) Schematic diagram of ex vivo treatment of UM explants. Fresh UM tumors obtained from surgery were cut into approximately 1 mm3 pieces and plated on Vetspon absorbable hemostatic gelatin sponges for 48 hours. The sponges were pre-soaked in medium containing DMSO, 50 nM trametinib, 100 nM LY2801653 or a combination of trametinib and LY2801653, respectively. Medium was replenished every 24 hours. (B) Metastatic UM specimen Ex-Pt#4 was treated, as above. Tumor samples were lysed in RPPA lysis buffer after 48 hours of treatment. Lysates were probed with pERK1/2, ERK2, pAKT, AKT, cleaved-PARP and Hsp90 antibodies.
Discussion
The majority of UM metastases show a tropism for the liver and are highly resistance to targeted therapies such as MEK inhibitors. How the tumor microenvironment regulates the response in UM to targeted inhibitors is poorly understood. Here we utilized cell lines derived from metastatic UM and conditioned medium derived from stromal cells in the liver microenvironment. We provide evidence that the use of cMET targeting agents as a part of combinational approach may counteract tumor microenvironment-mediated primary resistance to MEK inhibitors in mutant GNAQ/11 metastatic UM.
Recent results from the phase III, randomized trial (NCT01974752) of the MEK inhibitor, selumetinib, in combination with dacarbazine in patients with metastatic UM were disappointing with only 3 out 97 patients treated with the combination eliciting a partial response based on a central review. These results are in contrast to findings in cutaneous melanoma, which led to the FDA approval of trametinib for the treatment of BRAF V600E/K unresectable or metastatic cutaneous melanoma (40). HGF is abundant in the liver microenvironment and, when supplied exogenously, rescues the growth of MEK-inhibited mutant GNAQ human metastatic UM cell lines (20). Our data herein indicate that HGF-mediated resistance to MEK inhibitors in UM cells involves silencing of the pro-apoptotic, Bim-EL and Bmf. These data are similar to the role of Bim-EL and Bmf in resistance to the BRAF inhibitor, PLX4720, in cutaneous melanoma cells (41).
To investigate the effect of liver microenvironment on response to MEK inhibitors in UM cells, we examined factors derived from human hepatic stellate cells. Early passage human stellate cells do not display activation markers and do secrete HGF indicating that they may be an appropriate model for studying stromal contributions from the metastatic UM tumor microenvironment. Pro-HGF is subsequently cleaved to form HGF, which acts as a growth factor for hepatocytes (42). cMET is expressed in both primary and metastatic UMs, but metastatic lesions tend to have higher cMET expression levels (43), which is activated in the majority of UM liver metastases (20). Indeed, cMET signaling is constitutively activated in UM cells when cultured in conditioned medium from stellate cells. These data support a role for tumor microenvironment in regulating HGF-cMET signaling in metastatic UM, which is mediated by stellate cell - cancer cell communication in the liver.
While UM cells are sensitive to trametinib in regular growth medium, they are resistant when grown in conditioned medium derived from stellate cells. Importantly, resistance is overcome by cMET targeting agents. These data suggest that innate/intrinsic resistance of UM to MEK inhibitors is driven, at least in part, by HGF from stellate cells in the liver microenvironment. We demonstrate that cMET targeting agents such as LY2801653 and LY2875358 may improve the response to MEK inhibitors in metastatic UM patients. We extended our studies to analyze a UM surgical specimen in an ex vivo treatment approach, which maintains the tumor microenvironment. Interestingly, we observed that LY2801653 treatment promoted the expression of cleaved PARP, an indicator of apoptosis. We acknowledge that further studies using preclinical models are important to address the effect of combinational MEK1/2 and cMET based target therapy in metastatic UM.
The main activated downstream pathway of HGF-cMET is PI3K-AKT signaling. In the presence of trametinib, HGF promotes the activation of PI3K/AKT, which compensates for loss of MEK-ERK1/2 activity in UM cells (20). Despite the evidence highlighting the importance of the PI3K pathway activation in the development of resistance to target therapy in melanoma, initial testing of class I PI3K inhibitors in patients has not produced dramatic results mainly due to the overlapping toxicities with MEK inhibitors that limits their effective dosing (44). One possible way to overcome this limitation is to utilize PI3K isoform specific inhibitors. We identified that PI3Kα/γ/δ isoforms, but not PI3Kβ, are responsible for HGF-mediated AKT activation and HGF-mediated resistance to MEK inhibitors. These data suggest that the use of PI3Kβ-sparing inhibitors may represent a useful strategy to overcome HGF-mediated resistance and subsequently improve responses to MEK inhibitors in metastatic UM. Of note, in cutaneous BRAF-mutated GEM melanoma models, the combination of MEK inhibitor plus the PI3Kβ-sparing inhibitor enhanced initial tumor regression and forestalled the onset of tumor resistance (45).
In summary, the data presented here show for the first time that stellate cells from the liver provide innate resistance to MEK inhibitors in metastatic UM though HGF-cMET signaling. We have provided evidence that down-regulation of the BH3-only proteins, Bim-EL and Bmf, contributes to HGF-mediated resistance. Blocking HGF signaling with either clinical-grade cMET targeting agents or PI3Kα/γ/δ inhibitors in UM cells overcome resistance to MEK inhibitors mediated by stellate cells or exogenous HGF. Ongoing efforts include testing anti-cMET monoclonal antibodies in combination with MEK inhibitors in preclinical UM studies. In addition, profiling other factors within hepatic cellular architecture that regulate response to targeted therapy may identify novel targets for more effective therapeutic strategies.
Supplementary Material
Acknowledgments
Dr. Mike Davies has received grants from GlaxoSmithKline, AstraZeneca, Roche/Genentech, and Sanofi-Aventis and serves as a consultant/ advisory board member for Novartis, GlaxoSmithKline, Roche/Genentech, and Sanofi-Aventis and Vaccinex. Dr. Tamaki Sato has received research support from Eli Lilly and Company and Guerbet LLC, and is a consultant/advisory board member for Immunocore. Dr. Andrew Aplin discloses a grant from Pfizer Inc.
LY2801653 and LY2875358 were generously provided by Eli Lilly and Company.
Financial Support: This project was funded by a Dr. Ralph and Marian Falk Medical Research Trust Catalyst Award, a Programmatic Initiative Award from the Provost’s office at Thomas Jefferson University, and CURE award funding from Pennsylvania Department of Health. The Sidney Kimmel Cancer Center Flow Cytometry and Genomics core facilities are supported by National Institutes of Health/National Cancer Institute Support Grant (P30 CA056036). The RPPA studies were supported by the Dr. Miriam and Sheldon G. Adelson Medical Research Foundation and were performed at the Functional Proteomics Core Facility at M.D. Anderson Cancer Center, which is supported by a National Cancer Institute (NCI) Cancer Center Support Grant (CA-16672).
Footnotes
Conflict of interest: No other conflicts of interest were disclosed.
References
- 1.Singh AD, Bergman L, Seregard S. Uveal melanoma: epidemiologic aspects. Ophthalmol Clin North Am. 2005;18:75–84. viii. doi: 10.1016/j.ohc.2004.07.002. [DOI] [PubMed] [Google Scholar]
- 2.Luke JJ, Triozzi PL, McKenna KC, Van Meir EG, Gershenwald JE, Bastian BC, et al. Biology of advanced uveal melanoma and next steps for clinical therapeutics. Pigment Cell Melanoma Res. 2014;28:135–147. doi: 10.1111/pcmr.12304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Van Raamsdonk CD, Griewank KG, Crosby MB, Garrido MC, Vemula S, Wiesner T, et al. Mutations in GNA11 in uveal melanoma. N Engl J Med. 2009;363:2191–2199. doi: 10.1056/NEJMoa1000584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Van Raamsdonk CD, Bezrookove V, Green G, Bauer J, Gaugler L, O'Brien JM, et al. Frequent somatic mutations of GNAQ in uveal melanoma and blue naevi. Nature. 2009;457:599–602. doi: 10.1038/nature07586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Harbour JW. The genetics of uveal melanoma: an emerging framework for targeted therapy. Pigment Cell Melanoma Res. 2012;25:171–181. doi: 10.1111/j.1755-148X.2012.00979.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shoushtari AN, Carvajal RD. GNAQ and GNA11 mutations in uveal melanoma. Melanoma Res. 2014;24:525–534. doi: 10.1097/CMR.0000000000000121. [DOI] [PubMed] [Google Scholar]
- 7.Khalili JS, Yu X, Wang J, Hayes BC, Davies MA, Lizee G, et al. Combination small molecule MEK and PI3K inhibition enhances uveal melanoma cell death in a mutant GNAQ- and GNA11-dependent manner. Clin Cancer Res. 2012;18:4345–4355. doi: 10.1158/1078-0432.CCR-11-3227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Patel M, Smyth E, Chapman PB, Wolchok JD, Schwartz GK, Abramson DH, et al. Therapeutic implications of the emerging molecular biology of uveal melanoma. Clin Cancer Res. 2011;17:2087–2100. doi: 10.1158/1078-0432.CCR-10-3169. [DOI] [PubMed] [Google Scholar]
- 9.Feng X, Degese MS, Iglesias-Bartolome R, Vaque JP, Molinolo AA, Rodrigues M, et al. Hippo-independent activation of YAP by the GNAQ uveal melanoma oncogene through a trio-regulated rho GTPase signaling circuitry. Cancer Cell. 2014;25:831–845. doi: 10.1016/j.ccr.2014.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Vaque JP, Dorsam RT, Feng X, Iglesias-Bartolome R, Forsthoefel DJ, Chen Q, et al. A genome-wide RNAi screen reveals a Trio-regulated Rho GTPase circuitry transducing mitogenic signals initiated by G protein-coupled receptors. Mol Cell. 2013;49:94–108. doi: 10.1016/j.molcel.2012.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yu FX, Luo J, Mo JS, Liu G, Kim YC, Meng Z, et al. Mutant Gq/11 promote uveal melanoma tumorigenesis by activating YAP. Cancer Cell. 2014;25:822–830. doi: 10.1016/j.ccr.2014.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Harbour JW, Chao DL. A molecular revolution in uveal melanoma: implications for patient care and targeted therapy. Ophthalmology. 2014;121:1281–1288. doi: 10.1016/j.ophtha.2013.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Harbour JW, Onken MD, Roberson ED, Duan S, Cao L, Worley LA, et al. Frequent mutation of BAP1 in metastasizing uveal melanomas. Science. 2010;330:1410–1413. doi: 10.1126/science.1194472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Matatall KA, Agapova OA, Onken MD, Worley LA, Bowcock AM, Harbour JW. BAP1 deficiency causes loss of melanocytic cell identity in uveal melanoma. BMC Cancer. 2013;13:371. doi: 10.1186/1471-2407-13-371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Harbour JW, Roberson ED, Anbunathan H, Onken MD, Worley LA, Bowcock AM. Recurrent mutations at codon 625 of the splicing factor SF3B1 in uveal melanoma. Nat Genet. 2013;45:133–135. doi: 10.1038/ng.2523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Furney SJ, Pedersen M, Gentien D, Dumont AG, Rapinat A, Desjardins L, et al. SF3B1 mutations are associated with alternative splicing in uveal melanoma. Cancer Discov. 2013;3:1122–1129. doi: 10.1158/2159-8290.CD-13-0330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Martin M, Masshofer L, Temming P, Rahmann S, Metz C, Bornfeld N, et al. Exome sequencing identifies recurrent somatic mutations in EIF1AX and SF3B1 in uveal melanoma with disomy 3. Nat Genet. 2013;45:933–936. doi: 10.1038/ng.2674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Johansson P, Aoude LG, Wadt K, Glasson WJ, Warrier SK, Hewitt AW, et al. Deep sequencing of uveal melanoma identifies a recurrent mutation in PLCB4. Oncotarget. 2015;7:4624–4631. doi: 10.18632/oncotarget.6614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Moore AR, Ceraudo E, Sher JJ, Guan Y, Shoushtari AN, Chang MT, et al. Recurrent activating mutations of G-protein-coupled receptor CYSLTR2 in uveal melanoma. Nat Genet. 2016;48:675–680. doi: 10.1038/ng.3549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cheng H, Terai M, Kageyama K, Ozaki S, McCue PA, Sato T, et al. Paracrine effect of NRG1 and HGF drives resistance to MEK inhibitors in metastatic uveal melanoma. Cancer Res. 2015;75:2737–2748. doi: 10.1158/0008-5472.CAN-15-0370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Falchook GS, Lewis KD, Infante JR, Gordon MS, Vogelzang NJ, DeMarini DJ, et al. Activity of the oral MEK inhibitor trametinib in patients with advanced melanoma: a phase 1 dose-escalation trial. Lancet Oncol. 2012;13:782–789. doi: 10.1016/S1470-2045(12)70269-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Carvajal RD, Sosman JA, Quevedo JF, Milhem MM, Joshua AM, Kudchadkar RR, et al. Effect of selumetinib vs chemotherapy on progression-free survival in uveal melanoma: a randomized clinical trial. JAMA. 2014;311:2397–2405. doi: 10.1001/jama.2014.6096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Carvajal RD, Schwartz GK, Mann H, Smith I, Nathan PD. Study design and rationale for a randomised, placebo-controlled, double-blind study to assess the efficacy of selumetinib (AZD6244; ARRY-142886) in combination with dacarbazine in patients with metastatic uveal melanoma (SUMIT) BMC Cancer. 2015;15:467. doi: 10.1186/s12885-015-1470-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yoshida M, Selvan S, McCue PA, DeAngelis T, Baserga R, Fujii A, et al. Expression of insulin-like growth factor-1 receptor in metastatic uveal melanoma and implications for potential autocrine and paracrine tumor cell growth. Pigment Cell Melanoma Res. 2014;27:297–308. doi: 10.1111/pcmr.12206. [DOI] [PubMed] [Google Scholar]
- 25.Jabbour E, Ottmann OG, Deininger M, Hochhaus A. Targeting the phosphoinositide 3-kinase pathway in hematologic malignancies. Haematologica. 99:7–18. doi: 10.3324/haematol.2013.087171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chen R, Zhao Y, Huang Y, Yang Q, Zeng X, Jiang W, et al. Nanomicellar TGX221 blocks xenograft tumor growth of prostate cancer in nude mice. Prostate. 2015;75:593–602. doi: 10.1002/pros.22941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yan SB, Peek VL, Ajamie R, Buchanan SG, Graff JR, Heidler SA, et al. LY2801653 is an orally bioavailable multi-kinase inhibitor with potent activity against MET, MST1R, and other oncoproteins, and displays anti-tumor activities in mouse xenograft models. Invest New Drugs. 31:833–844. doi: 10.1007/s10637-012-9912-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Boisvert-Adamo K, Aplin AE. B-RAF and PI-3 kinase signaling protect melanoma cells from anoikis. Oncogene. 2006;25:4848–4856. doi: 10.1038/sj.onc.1209493. [DOI] [PubMed] [Google Scholar]
- 29.Tibes R, Qiu Y, Lu Y, Hennessy B, Andreeff M, Mills GB, et al. Reverse phase protein array: validation of a novel proteomic technology and utility for analysis of primary leukemia specimens and hematopoietic stem cells. Mol Cancer Ther. 2006;5:2512–2521. doi: 10.1158/1535-7163.MCT-06-0334. [DOI] [PubMed] [Google Scholar]
- 30.Dean JL, Thangavel C, McClendon AK, Reed CA, Knudsen ES. Therapeutic CDK4/6 inhibition in breast cancer: key mechanisms of response and failure. Oncogene. 2010;29:4018–4032. doi: 10.1038/onc.2010.154. [DOI] [PubMed] [Google Scholar]
- 31.Geng Y, Yu Q, Sicinska E, Das M, Bronson RT, Sicinski P. Deletion of the p27Kip1 gene restores normal development in cyclin D1-deficient mice. Proc Natl Acad Sci USA. 2001;98:194–199. doi: 10.1073/pnas.011522998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wu W, Bi C, Credille KM, Manro JR, Peek VL, Donoho GP, et al. Inhibition of tumor growth and metastasis in non-small cell lung cancer by LY2801653, an inhibitor of several oncokinases, including MET. Clin Cancer Res. 2013;19:5699–5710. doi: 10.1158/1078-0432.CCR-13-1758. [DOI] [PubMed] [Google Scholar]
- 33.Kawada I, Hasina R, Arif Q, Mueller J, Smithberger E, Husain AN, et al. Dramatic antitumor effects of the dual MET/RON small-molecule inhibitor LY2801653 in non-small cell lung cancer. Cancer Res. 2014;74:884–895. doi: 10.1158/0008-5472.CAN-12-3583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Barat S, Bozko P, Chen X, Scholta T, Hanert F, Gotze J, et al. Targeting c-MET by LY2801653 for treatment of cholangiocarcinoma. Mol Carcinog. 2016 doi: 10.1002/mc.22449. [DOI] [PubMed] [Google Scholar]
- 35.Liu L, Zeng W, Wortinger MA, Yan SB, Cornwell P, Peek VL, et al. LY2875358, a neutralizing and internalizing anti-MET bivalent antibody, inhibits HGF-dependent and HGF-independent MET activation and tumor growth. Clin Cancer Res. 2014;20:6059–6070. doi: 10.1158/1078-0432.CCR-14-0543. [DOI] [PubMed] [Google Scholar]
- 36.Schaeper U, Gehring NH, Fuchs KP, Sachs M, Kempkes B, Birchmeier W. Coupling of Gab1 to c-Met, Grb2, and Shp2 mediates biological responses. J Cell Biol. 2000;149:1419–1432. doi: 10.1083/jcb.149.7.1419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yin C, Evason KJ, Asahina K, Stainier DY. Hepatic stellate cells in liver development, regeneration, and cancer. J Clin Invest. 2013;123:1902–1910. doi: 10.1172/JCI66369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Herrmann J, Gressner AM, Weiskirchen R. Immortal hepatic stellate cell lines: useful tools to study hepatic stellate cell biology and function? J Cell Mol Med. 2007;11:704–722. doi: 10.1111/j.1582-4934.2007.00060.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Birchmeier C, Birchmeier W, Gherardi E, Vande Woude GF. Met, metastasis, motility and more. Nat Rev Mol Cell Biol. 2003;4:915–925. doi: 10.1038/nrm1261. [DOI] [PubMed] [Google Scholar]
- 40.Flaherty KT, Robert C, Hersey P, Nathan P, Garbe C, Milhem M, et al. Improved survival with MEK inhibition in BRAF-mutated melanoma. N Engl J Med. 2012;367:107–114. doi: 10.1056/NEJMoa1203421. [DOI] [PubMed] [Google Scholar]
- 41.Shao Y, Aplin AE. BH3-only protein silencing contributes to acquired resistance to PLX4720 in human melanoma. Cell Death Differ. 2012;19:2029–2039. doi: 10.1038/cdd.2012.94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Taub R. Liver regeneration: from myth to mechanism. Nat Rev Mol Cell Biol. 2004;5:836–847. doi: 10.1038/nrm1489. [DOI] [PubMed] [Google Scholar]
- 43.Gardner FP, Serie DJ, Salomao DR, Wu KJ, Markovic SN, Pulido JS, et al. c-MET expression in primary and liver metastases in uveal melanoma. Melanoma Res. 2014;24:617–620. doi: 10.1097/CMR.0000000000000118. [DOI] [PubMed] [Google Scholar]
- 44.Cheng H, Aplin AE. Game of isoforms: PI3K beta-sparing inhibitor is coming. Pigment Cell Melanoma Res. 2015;28:133–134. doi: 10.1111/pcmr.12350. [DOI] [PubMed] [Google Scholar]
- 45.Deuker MM, Marsh Durban V, Phillips WA, McMahon M. PI3'-kinase inhibition forestalls the onset of MEK1/2 inhibitor resistance in BRAF-mutated melanoma. Cancer Discov. 2015;5:143–153. doi: 10.1158/2159-8290.CD-14-0856. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.