Abstract
Single-molecule nanodot arrays, in which a biomolecule of choice (protein, nucleic acid, etc.) is bound to a metallic nanoparticle on a solid substrate, are becoming an increasingly important tool in the study of biomolecular and cellular interactions. We have developed an on-chip measurement protocol to monitor and control the molecular occupancy of nanodots. Arrays of widely spaced nanodots and nanodot clusters were fabricated on glass surfaces by nanolithography and functionalized with fluorescently labeled proteins. The molecular occupancy was determined by monitoring individual fluorophore bleaching events, while accounting for fluorescence quenching effects. We found that the occupancy can be interpreted as a packing problem, and depends on nanodot size and binding ligand concentration, where the latter is easily adjusted to compensate the flexibility of dimension control in nanofabrication. The results are scalable with nanodot cluster size, extending to large area close packed arrays. As an example, the nanoarray platform was used to probe the geometric requirement of T-cell activation at the single-molecule level.
Keywords: molecular occupancy, stoichiometry, single-molecule assays, photobleaching, fluorescence quenching, electron beam lithography
Graphical abstract
Single-molecule nanoarrays, in which individual molecules are presented on a surface, have become an important tool for a broad range of studies and applications, including genomics,1,2 proteomics,3,4 protein–DNA interactomics,5,6 and biodiagnostics7 based on DNA–DNA,8,9 protein–protein,10 antigen–antibody,11,12 or aptamer-protein13,14 binding. Miniaturization from conventional microassays to single-molecule nanoarrays leads to significant improvements in sensitivity, specificity, speed and throughput.15 Various approaches have been taken to create single-molecule nanoarrays. The most straightforward is the simple deposition from a dilute solution onto bare glass16,17 or polyethylene glycol (PEG) coated glass,18,19 relying on the sparse distribution of molecules on the surface to achieve single-molecule resolution. A more sophisticated technique uses zero mode waveguides (ZMWs),2 nanoapertures20 or nanogrids,21 which restricts the imaging volume, so that, on average, only a single molecular interaction is probed at each site. Nanoarrays with more orderly arrangements have been made by direct patterning of biomolecules by dip-pen nanolithography (DPN)22–24 or surface chemistry patterning by electron beam lithography (EBL) or nanoimprint lithography (NIL).25,26
A particular type of single-molecule nanoarray that is seeing increasing utility has a biomolecule of choice immobilized on a surface-bound metallic nanoparticle or nanodot, typically ∼2– 10 nm in diameter. By tuning the nanodot surface chemistry relative to the background, very high selectivity can be achieved. The most common approach involves the formation of a stable self-assembled monolayer (SAM) of alkylthiol on a variety of metals,27 among which Au, AuPd, and Ag10 have been the most widely used. These nanodot arrays are finding increasing application in a variety of cellular studies, providing insight into the crucial role of geometric organization of ligands for transmembrane receptors involved in critical cellular functions, such as adhesion and spreading,28–31 differentiation,32 apoptosis,33 immune recognition,34–37 and the maintenance of healthy tissue.38,39 (They have also been used for the directed assembly of a variety of functional nanomaterials, including quantum dots (QDs),40 carbon nanotubes,41 and DNA origami.42,43)
The basic assumption in the use of these nanoarrays is that each nanodot is occupied by a single molecule (ligand or receptor, whichever is larger), as the nature of the individual interactions being probed is generally sensitive to stoichiometry. For large receptors, for example, integrins (the head is ∼8–12 nm), single-molecular occupancy is assured as long as the nanodot is smaller than the receptor, even if each nanodot presents multiple ligands.28–31 For small receptors, for example, the T-cell receptor (TCR, a heterodimer with two ∼40–50 kDa chains44), single ligand occupancy is more critical, whereas a rough control probably contributes to a great variation in probing the T-cell activation among different platforms.34–36 This is a challenging task, requiring ultrahigh resolution techniques to image at the single-molecule level. Transmission electron microscopy (TEM) has been used to directly image protein-bound QDs on gold nanoparticles (AuNPs) of different size created by dip pen nanolithography,45 showing that the number of bound QDs scales with the nanoparticle size.46 This sort of measurement requires the use of electron microscopy and is not compatible with surface based nanoarrays, especially in the context of biological experiments. Other strategies based on super-resolution fluorescence microscopy such as photoactivated localization microscopy (PALM) have been applied to gold nanoarrays, but the measured functionalization efficiency in those studies was very low38 (possibly as a result of fluorescence quenching).
In this work, we demonstrate an on-chip technique for determining the molecular occupancy of single-molecule nanoarrays based on the observation of individual fluorescence photobleaching events using common epifluorescence microscopy, and we describe how the occupancy can be controlled. The key to the technique is the fabrication of ordered arrays of nanodots spaced apart sufficiently so that the intensity signal and discrete bleaching events from each one can be clearly distinguished. Equally important is highly selective functionalization of the nanodots, in order to minimize the background signal. This approach is based upon our previous work in which we monitored DNA cleavage by a restriction enzyme5 and single QD blinking events40 on similar nanoarrays. Here, smaller nanodots in the range of ∼5–15 nm were used so that this platform could be optimized to ensure true single-molecule control.
A variety of technologies have been developed to create metallic nanoarrays, from bottom-up approach such as block copolymer micelle nanolithography (BCML),29–36,43,47–50 and self-assembled nanosphere lithography (NSL),9–11,51–53 to topdown approaches, including photolithography,54 DPN,45,46 EBL, and NIL.5,28,37,40–42,55 The so-called top-down approaches enable heterogeneous arrays on the same surface, with arbitrary geometry and various dot sizes. This is important for both the molecular occupancy analysis and cell assays, because the integration of various patterns on the same sample enables direct comparison between them, avoiding the variations from sample to sample. As a demonstration of the efficacy of this technique, our nanoarray platform was used to determine the geometric requirement of T-cell activation. It not only ensures on-chip single-molecule control but also provides a uniform surface background among various patterns, which is essential as the T-cell activation is affected by adhesion.56
RESULTS AND DISCUSSION
Molecular Occupancy Measurement by Photobleaching
The process for creating the nanoarray platform used in this work is outlined in Supporting Information Figure S1. The final step in the process is a thermal anneal of ultrathin metallic films at the patterned sites, transforming them into spherical nanoparticles with a smaller diameter and a high degree of uniformity (Figure 1a). Next, a SAM mixture (alkylthiol and biotin-alkylthiol) is formed on the nanodots, where the mole fraction could be adjusted. This is followed by passivation of the background unpatterned glass surface with PEG-silane to prevent nonspecific binding and improve the signal-to-background ratio (SBR). For this study, AuPd was used instead of pure Au (as discussed in the Supporting Information). A fluorescently labeled streptavidin was bound to the biotin in the SAM. We chose streptavidin because it is commonly used to attach a wide variety of molecules to metallic nanoparticles owing to its reliable binding to any moiety that can be biotinylated.5,28,37,40 The results, however, can be generalized to other molecular species bound to the nanoparticles, either directly or through a streptavidin (or other molecular) bridge. We developed a molecular model based on protein data bank (PDB) files according to the functionalization process (Figure 1b). When the nanoarray is under constant exposure to excitation light, the fluorophores bleach over time, thus providing a signal for measurement (Figure 1c).
Figure 1.
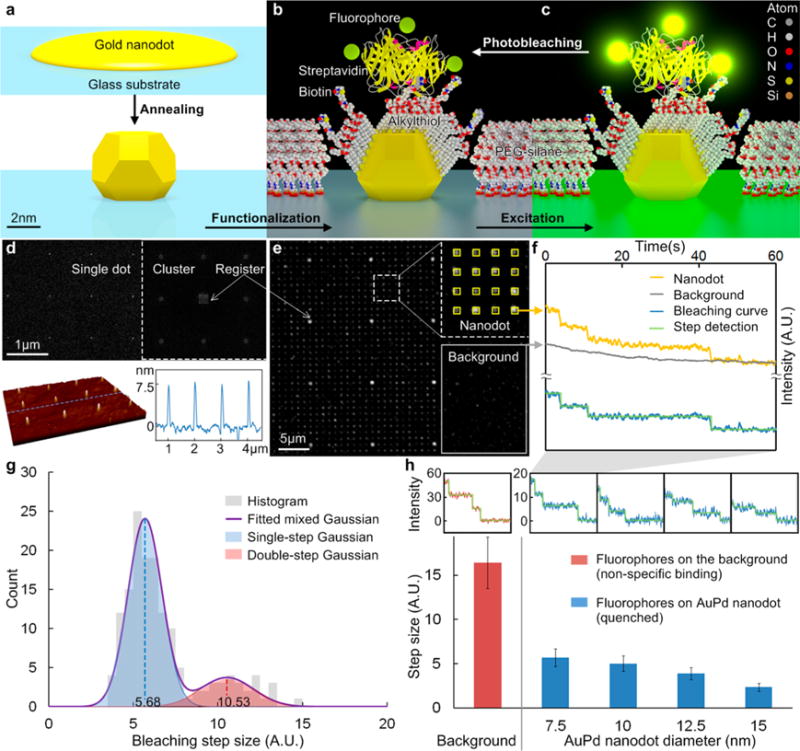
Schematic diagram of the metallic nanodot array: (a) annealing, (b) functionalization, (c) excitation and photobleaching of the fluorophores labeled on a streptavidin (only the SAM in the cross-section is shown for clarity). (d) SEM image of the NIL mold and AFM image of the AuPd nanodot (D = 7.5 nm) array. (e) Fluorescence (Alexa Fluor 555) image with ROIs of both nanodot (heptamer) and background. (f) Fluorescence intensity vs time. The bleaching curve was obtained by subtracting the background from the signal on nanodots. The size of the steps is close, indicating the each step represents the bleaching of a single fluorophore. (g) Bleaching step size histogram of 7.5 nm nanodot. (h) Average bleaching step size vs AuPd nanodot size. The insets show typical bleaching curves for corresponding nanodot size.
A typical nanodot array, with an interdot spacing of 1 μm, is shown in Figure 1d. Scanning electron microscopy (SEM) of the mold used to fabricate the nanodot arrays shows both single nanodots and nanodot clusters (Figure 1d, upper). The atomic force microscopy (AFM) scan in Figure 1d (lower) shows an example of an AuPd nanoarray with a uniform height profile of ∼7.5 nm. Because the nanodots are spherical, this is also their lateral size.55 In order to achieve a sufficiently high SBR per site, multiple fluorophore/protein (F/P) ratio and nanodot clusters, such as the 1 μm-spaced heptamers interspersed with 10 μm-spaced registration arrays (inset), were used for continuous fluorescence imaging. We used a standard epi-fluorescence microscope with high magnification coupled to an EM-CCD camera that was able to detect single bleaching events. Figure 1e shows the fluorescence signal from a functionalized nanodot array, where two types of regions of interest (ROIs) are selected: nanodots (inset) and background. By plotting the fluorescence intensity vs time, and then subtracting the background from the nanodot signal, a typical bleaching curve is obtained, where discrete steps represent bleaching events of individual fluorophores (Figure 1f). Given the F/P ratio, the molecular occupancy could be determined from the number of steps for each site (heptamer, in this case). Since the initial fluorescence intensity is linear with the number of fluorophores,57 it is unnecessary to count step numbers for the entire set of bleaching curves in order to efficiently obtain statistical results. Alternatively, the average molecular occupancy can be estimated by
| (1) |
where N is the molecular occupancy, that is, the number of molecules per nanodot; I is the initial intensity of the nanodot cluster; h is the bleaching step size, that is, the intensity of a single fluorophore signal; r is the F/P ratio of the labeled molecule, that is, the number of fluorophores per molecule; and c is the cluster size, that is, the number of nanodots per cluster.
Figure 1g shows a histogram of step sizes for 7.5 nm nanodots, derived from the bleaching curves (e.g., Figure 1f); this histogram was fitted to a set of Gaussian distributions. The center of the second peak is approximately twice the value of the first, which indicates simultaneous bleaching events (observed especially in the early stages of the bleaching curves), and must be taken into consideration in determining the average value. The overlapping area of the two Gaussians was determined by numerical integration, assuming equal probability of one or two fluorophores, which is not necessarily the same as the probability of bleaching steps. It indicates that the probability of ambiguous fluorophore number (one or two) at the same intensity is only 5.72%. However, this probability increases with increased fluorophore number because the variances of individual fluorophore intensities are added, which is a common problem in quantitative fluorescence microscopy.57 Therefore, a large sample size is required for reliable measurements using eq 1. Our method provides sufficient sample size by monitoring thousands of sites in parallel. In addition, the intensity distribution of a single fluorophore or molecule was used to fit the histogram of the initial cluster intensity and to estimate the probability distribution of molecular occupancy, which is more accurate than merely calculating average values.
Fluorescence Quenching Effects
It is well known that the fluorescence from a molecule in close proximity to a metal nanoparticle is affected by radiative and nonradiative energy transfer, leading to excited-state enhancement, or quenching, which depends on the material, shape, structure, and dimensions of the metal nanoparticle, as well as the distance from the fluorophore to the particle surface.58–60 Figure 1h shows typical bleaching curves and the average step sizes for nanodots with diameters up to 15 nm, which are different in the absence of quenching effect. Dielectric materials (e.g., metal oxides) could eliminate the quenching effect and simplify the bleaching measurements, but the binding chemistry (e.g., alkyl phosphate SAM61) for these materials is not sufficiently selective for single-molecule resolution.
In this study, streptavidins were used in which the fluorophores bind to the primary amines on lysine residues. The molecular model in Figure 2a shows the nitrogen atoms on primary amines, which are used to approximate the fluorophore locations. The distance histogram from these locations to the nanodot surface, considering all four binding sites of biotin, indicates that 69% of the fluorophores are within 4.3–6.6 nm of the nanodot. Other possible positions are closer, but the fluorophores are unlikely to locate there because this would block the biotin binding in the first place. Recently, both theoretical and experimental works have determined that the fluorescence quenching efficiency increases with the AuNP size in the range of our interest.62 A size-dependent nanometal surface energy transfer (NSET) model was developed by Breshike et al.63 to explain the dependence of quenching efficiency on both distance and nanodot size. Figure 2b shows schematic curves based on this model. In light of the fourth power dependence on distance, a narrow distribution of fluorophore distances from the nanodot is essential for the step size measurement. The experimental results of quenching efficiency (Figure 2b inset) were obtained by normalizing bleaching step sizes with that of nonspecifically bound fluorophores on the background (as in Figure 1h). Within the same distance range, the quenching efficiency increases and the dispersion decreases with the nanodot size, which is in agreement with the model.
Figure 2.
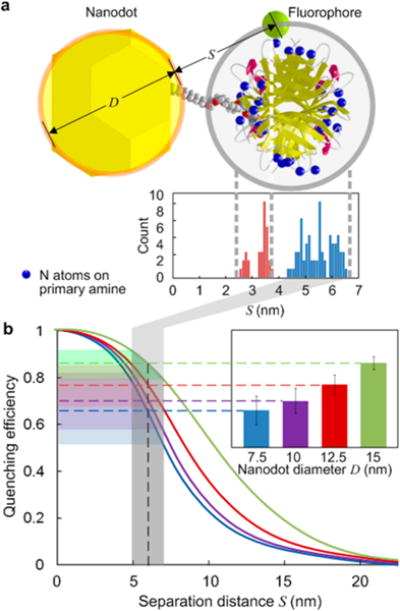
(a) Molecular model showing the distance distribution from the fluorophores (approximated by the nitrogen atoms on primary amine of lysine) on streptavidin to the nanodot surface. (b) Schematic curves of quenching efficiency vs separation distance for various nanodot size. The inset shows the experimental results of quenching efficiency based on bleaching step size from Figure 1h.
Because the intensity of nonspecifically bound fluorophores on the background is much larger than for the nanodots in the absence of quenching, control of nonspecific binding on the glass substrate (i.e., noise) is critical for these measurements. PEG passivation is widely used in single-molecule studies, but it is typically most effective at low (nanomolar) concentrations,64 which is suitable for deposition from a dilute solution (20–100 pM18). Single-molecule nanoarrays, on the other hand, require a much higher concentration (50 nM46–10 μM28) to ensure functionalization efficiency, therefore making the passivation more challenging. We used a 15 nm thick layer of cured hydrogen silsesquioxane (HSQ) to cover common impurities found on glass coverslip surfaces, thereby reducing the defects and nonspecifically bound proteins in the PEG passivation layer. As a result, the nonspecifically bound streptavidin density was counted as only 0.42 ± 0.08 molecules/μm2, ensuring not only sufficient SBR for 1 μm spaced nanoarrays, but also biological inertness for most applications.
Factors Affecting Molecular Occupancy
Figure 3a shows an example of a heterogeneous array with nanodots ranging from 7.5 to 17.5 nm. Its fluorescence intensity profile indicates the dependence of molecular occupancy on nanodot size (Figure 3b). A statistical analysis based on eq 1 indicates that the occupancy increases with both the nanodot size and the biotin-alkylthiol concentration (Figure 3c). This could serve as a guide for the functionalization of nanodot arrays using streptavidin–biotin binding. It introduces a degree of flexibility and alleviates the need to produce nanodots at the same size as the molecule in order to achieve single-molecule occupancy with a pure SAM of biotin-alkylthiol (mole fraction 100%).46 Instead, nanodots with a diameter of 7.5 or 12.5 nm at a mole fraction of 50% or 25%, respectively, could also yield single-molecule occupancy, albeit with increased dispersion (see the error bars in Figure 3c).
Figure 3.
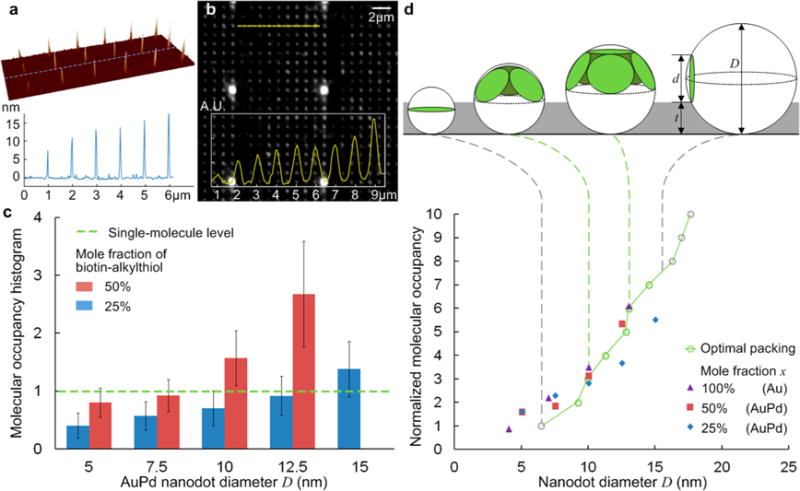
Heterogeneous nanoarray with various dot sizes: (a) AFM image and (b) fluorescence image. (c) Histogram of the average molecular occupancy vs nanodot size (n > 1500). (d) Molecular occupancy normalized by the mole fraction of biotin-alkylthiol. Data for mole fractions of 50% and 25% from (c) on AuPd nanodots, data for 100% from ref 46 on AuNPs (error bars removed for clarity). Theoretical trend line is given by the optimal packing model. Gray dash lines show the range of validity.
The dependence of molecular occupancy on nanodot size can be interpreted as “the optimal packing of circles on a sphere”.65 Molecular models and scanning tunneling microscopy (STM) measurements indicate that streptavidin has a two-dimensional size of ∼5.4 nm × 6. nm,66 so the circle diameter is approximated as 6.5 nm. Additionally, the nanodots are surrounded by the protein-repellent PEG layer (MW = 5000), whose thickness (Gaussian chain length) is estimated as 4.5 nm.67 For the dot size of interest (∼5–15 nm), the streptavidins tend to bind only on the top hemisphere of the nanodot. Therefore, the model is further simplified as “the packing of circles on a hemisphere”,68 as plotted in Figure 3d, whose trend line shows good agreement with the molecular occupancy normalized by the mole fraction. Thus, the molecular occupancy can be estimated by an empirical equation
| (2) |
where x is the mole fraction of binding ligands in the mixed SAMs, d is the diameter of the target molecule which binds to the ligands, D is the diameter of the nanodot, p(d/D) is the optimal packing number of circles of diameter d on a hemisphere of diameter D (Supporting Information Table 1), and t is the thickness of the passivation layer. This estimation may be generalized for other functionalization schemes using mixed SAMs on metallic nanodots surrounded by protein-repellent surfaces. Equation 2 can also be used to determine the mole fraction x required to achieve a single-molecule occupancy for a given nanodot size, as adjusting the reagent concentration is much easier than controlling the nanodot size in practice.
In the given range of interest (d < D < d + 2t), the molecular occupancy appears to scale linearly with the mole fraction of biotin-alkylthiol. This can be explained by the phase separation of the mixed SAMs,69 that is, they tend to be segregated into nanometer scale domains with and without biotin ends, respectively, rather than forming a uniform mixture. The streptavidin molecule only binds to the domains with biotin ends, so the occupancy is linear with the area percentage, approximately equal to the mole fraction. When the nanodot is larger and the mole fraction is lower, there may be a larger portion of biotin domains too small to merge with each other and support the streptavidin binding. This could explain the lower occupancy and larger dispersion when d = 12.5, 15 nm, x = 25%. When D > d + 2 t, the molecular occupancy would be higher, due to an increased chance of binding on the lower hemisphere, but molecules there are not as effective for further applications. Planar nanostructures, discs or apertures, would probably be preferable in this case. On the other hand, when D is close to or below d, the nanodot sphericity is highly underestimated by the packing model, which could accommodate more molecules than predicted by eq 2.
Scalability and Single Nanodot/Molecule Array
The results presented up to this point were obtained on arrays of heptameric clusters, as in the inset of Figure 1d. In order to verify its scalability for different cluster sizes, we created a heterogeneous array comprising clusters containing 1–19 nanodots with a diameter of 12.5 nm (Figure 4a). A biotin fraction of 25% was used to achieve approximately single molecule occupancy on each nanodot, as per Figure 3c. A profile of the fluorescence image shows that the intensity increases essentially monotonically with cluster size (Figure 4b). The nanodots in each cluster were in hexagonal arrangement with 60 nm spacing, which is robust in fabrication and ensures that the fluorescence is not quenched by adjacent nanodots. But the total fluorescence intensity of a cluster is also affected by the interdot spacing, for a given ROI (Supporting Information Figure S3). Therefore, a spatial correction based on the measured point-spread function (PSF) and 1D approximation was applied (Supporting Information Table 2). The corrected measurement leads to an average molecular occupancy per cluster that scales approximately linearly with the cluster size (Figure 4c) and is in good agreement with a single molecule per nanodot for all cluster sizes.
Figure 4.
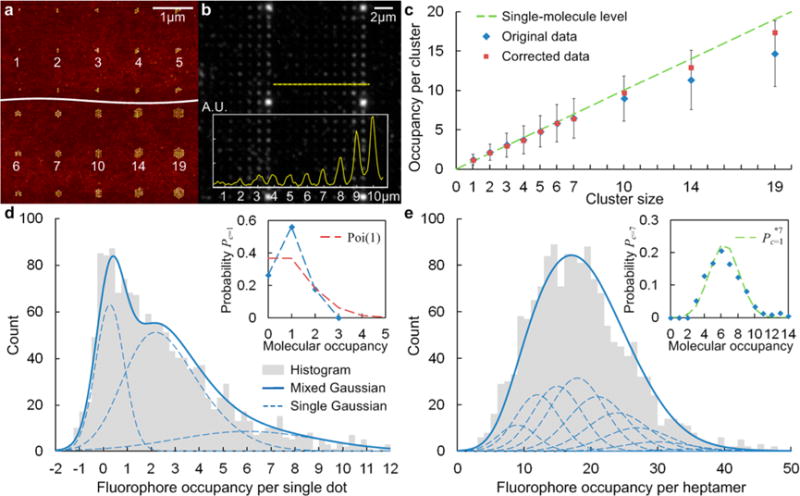
Heterogeneous nanoarray with various cluster sizes: (a) AFM image and (b) fluorescence image. (c) Histogram of the molecular occupancy per cluster vs cluster size. Fluorophore and molecular occupancy distributions for (d) single dot and (e) heptamer arrays. A mixed Gaussian fit was applied to the fluorophore occupancy histogram to estimate the probability of molecular occupancy. The set of single Gaussians are the nth convolution powers based on the Gaussian approximation of the binomial distribution (fluorophore number per streptavidin with an average of F/P ratio r = 3).
For the smaller clusters (c ≤ 4), the signal was quite low and insufficient for reliable statistics. This is why heptamers were chosen for the analysis, rather than a single nanodot, particularly in the presence of quenching effect. Despite this, the fluorophore occupancy histogram for a single dot array (Figure 4d) was analyzed in addition to a heptamer array (Figure 4e). It is commonly accepted that, at low and moderate probe to protein ratios, the number of probes within the protein follows binomial distribution.70 With an average of three fluorophores (r = 3) on 28 binding sites (the average of 24–32 lysines for each homotetramer), the fluorophore numbers per streptavidin should follow a binomial distribution B(28, 0.1071). Based on this, a mixed Gaussian fit was applied to the fluorophore occupancy histogram to estimate the probability of molecular occupancy (inset). This analysis revealed details of the molecular occupancy distribution with an average N ∼ 0.9: Nearly 60% of the single nanodots had one bound molecule, nearly 20% had two bound molecules, and about 20% had none. This distribution has a smaller dispersion than the Poisson distribution Poi(1), which is generally found in other forms of single-molecule arrays such as nanoapertures20 and nanogrids.21 The actual functionalization efficiency may be higher, considering a small portion of unlabeled molecules, as well as initial bleaching that might not have been captured in the videos. This loss is negligible for the measurements based on heptamers, given the large number of fluorophores and their slow average bleaching rate (Supporting Information Figure S2b). The molecular occupancy distribution per heptamer Pc=7 is in good agreement with the seventh convolution power of the single dot distribution Pc=1*7, which further confirms the scalability of our platform.
T-Cell Stimulation by Single-Molecule Nanoarrays
In the immune recognition process, a T-cell engages with an antigen presenting cell (APC), forming an elaborate arrangement of signaling, adhesion and costimulatory molecules organized into a stereotypic geometric structure, known as the immunological synapse (IS).71 A central cluster of TCR and antigenic peptide bound to major histocompatibility complex (pMHC) is surrounded by a ring of leukocyte function-associated antigen 1 (LFA-1) and intercellular adhesion molecule 1 (ICAM-1). This specific geometric structure has inspired major research efforts to probe the minimum geometric requirements for T-cell stimulation (at the single-molecule level). This has clear potential applications in adoptive immunotherapy.34–37 Due to the small dimension of the TCR, controlling the molecular occupancy of its ligands bound to nanodots is critical to accurately assessing the spatial and/or density threshold for activation. We used a biotinylated UCHT1 Fab′ (a single binding site of TCR) as the secondary molecule bound to the streptavidin linker. The surrounding surface was functionalized with a his-tag ICAM-1 linked to a PEG-silane via a nickel-NTA bridge to facilitate binding of LFA-1, which enhances cell adhesion.37 Figure 5a schematically shows a live T-cell on this bifunctional surface. Bleaching analysis of 1 μm spaced heptamer arrays revealed that nanodots (D = 7.5 nm, x = 50%) had a Fab′ occupancy of ∼1 (Nstreptavidin = 0.92 ± 0.28; NFab′ = 0.98 ± 0.58, Supporting Information Figure S4), albeit with a higher coefficient of variation (CV) than streptavidin. Although the streptavidin has three binding pockets available for the Fab′, the actual binding may be limited by the streptavidin orientation on the nanodots and/or possible interactions between Fab′ molecules. We have already shown that the molecular occupancy is scalable to larger cluster sizes (Figure 4c), and indeed, we find the fluorescence intensity of both streptavidin and Fab′ to be linear with the nanodot density, as shown in Figure 5b. Note again that the Fab′ has larger CVs than streptavidin (Figure 5b inset). This is especially useful for cellular assays, which frequently utilize dense patterns such as extended hexagonal arrays (Supporting Information Figure S5a).
Figure 5.
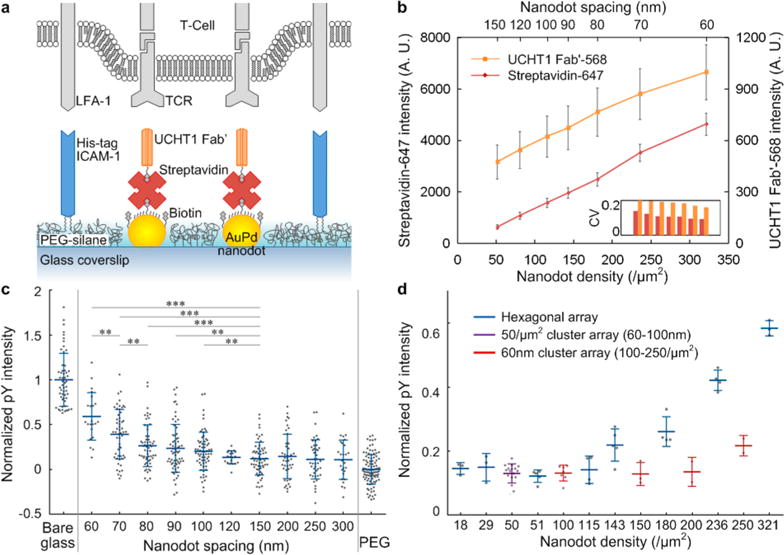
(a) Schematic diagram of the bifunctional nanoarray surface with a live T-cell. (b) Fluorescence intensity of both streptavidin-647 and UCHT1 Fab′-568 for hexagonal arrays with various spacings. Normalized pY intensity after 5 min stimulation: (c) plot against nanodot spacing, individual cells shown as dots, *** = p < 0.001, ** = p < 0.01 (Wilcoxon rank sum test); positive control, nonspecifically bound Fab′ and ICAM-1 on bare glass (maximum concentration); negative control, PEG passivation background (minimized concentration). (d) Plot against nanodot density, individual experiments shown as dots.
Primary human CD4+ T-cell response to the nanoarrays was assessed by monitoring the phosphotyrosine (pY) intensity using total internal reflection fluorescence microscopy (TIRF) 5 min after plating. For hexagonal arrays with different spacing from 60 to 300 nm, the average pY intensity decreased with increasing spacing to a threshold of ∼100 nm spacing (corresponding to a density of ∼115 TCR binding sites/μm2), at which point the signal drops to nearly that of cells on the PEG passivation background (outside the pattern area, a negative control) (Figure 5c). This trend is similar to that reported previously,34–36 but verified single binding sites with refined spacing ranges result in a more accurate threshold (Supporting Information Table 3). The variation among different platforms is not observed in integrin mediated adhesion, probably because the larger size of the extracellular head of the integrin (Supporting Information Table 4).28–31
In addition to hexagonal arrays, our platform also included cluster arrays, which allowed us to decouple the effects of local interdot spacing and global density (Figure 5d). This is not generally doable with arrays made by BCML. Heptamer arrays at a constant density of 50 /μm2, but with spacings ranging from 60 to 90 nm (Supporting Information Figure S5b) showed a negative response (i.e., pY signal equal to the negative control), as shown in Figure 5d, implying that in the absence of sufficient ICAM-1 (the density was <50 /μm2 in our experiments, due to a low grafting yield of nitrilotriacetic acid (NTA) on the PEG background), costimulatory, and other factors, it is global density, and not local spacing that determines TCR stimulation, with a threshold ∼115 TCR binding sites/μm2. This requirement is much higher than stimulation by APCs72 or artificial APC surfaces.21 We speculate that without sufficient ICAM-1 density, the TCR serves a dual role of antigen recognition and adhesion receptor, which requires additional TCR molecules. This is in agreement with the finding that adhesion mediated by TCR requires a high density of TCR agonist.73 Meanwhile, 60 nm heptamer arrays at a global density from 100 to 250 binding sites/μm2 (Supporting Information Figure S5c) had a lower response than hexagonal arrays with a similar density, suggesting a secondary effect of local spacing and cluster size. The interdot spacing within each heptamer was kept at 60 nm, but some of the spacings between nanodots from adjacent clusters are larger than the spacing threshold of 100 nm. This implies that, at a similar level of TCR-mediated adhesion, a larger cluster size (>7) is preferable in stimulation.
CONCLUSION
In conclusion, we have developed a technique to determine the molecular occupancy of metallic nanodot arrays by observing discrete steps in the intensity time profiles, which represent individual fluorophore bleaching events. The technique takes into account both the nanodot size and the binding ligand concentration. The quenching efficiency of single fluorophores on a single nanodot of various size was also determined in the measurement. The technique employs conventional epifluorescence microscopy and is compatible with on-chip, in situ measurement, so that it can be incorporated into real single-molecule experiments. A thin layer of thermally cured HSQ before conventional PEG-silane passivation reduced the background noise of nonspecific binding effectively.
For streptavidins on AuPd nanodots ranging from 5 to 15 nm, the histogram of occupancy was obtained for different mole fractions of biotin-alkylthiol. Nanodots with a diameter of 7.5 or 12.5 nm at mole fractions of 50% or 25% will yield single-molecule occupancy, respectively. The occupancy distribution with ∼80% functionalization efficiency was narrower than the Poisson distribution on similar platforms. The results are scalable for various cluster sizes and can be reasonably extended to dense close packed arrays. We found that the molecular occupancy depends on the spatial packing and scales approximately linearly with the binding ligand concentration. This can be generalized to other molecular species bound to the nanoparticles, either directly or through a streptavidin (or other molecular) bridge. Biotinylated UCHT1 Fab′ through a streptavidin bridge was shown as an example. Since the binding ligand concentration of the mixed SAMs can be easily adjusted, it introduces great flexibility in creating single-molecule arrays. Photobleaching analysis is also a general method for the on-chip measurement of molecular occupancy on nanodot arrays.
Finally, single-molecule occupancy was demonstrated to be crucial to improving the accuracy of cellular assays. Our platform also provides arbitrary geometries and better comparison between different patterns on a same chip. A density threshold of ∼115 TCR binding sites/μm2 has been determined for human T-cell stimulation (with low ICAM-1 concentration), possibly because the TCR has to play dual roles of both immune activity and adhesion. Overall, nanoscale geometric organization affects T-cell activation and behavior, which has potential applications in adoptive immunotherapy. Further investigation would require an improved presentation of ICAM-1, in order to better mimic the APC surface.
METHODS
Nanoarray Fabrication
The choice of AuPd alloy over pure Au is discussed in the Supporting Information. AuPd nanodot arrays were fabricated on standard glass coverslips (22 × 22 mm, no. 1.5, Corning) using EBL or NIL, with self-aligned pattern transfer,37,55 which is shown schematically in Supporting Information Figure S1a. In brief, PMMA (MW 495 K, Microchem) was patterned by direct EBL (NanoBeam nB4), with a thin conductive layer of Aquasave, which was rinsed by DI water before development in MIBK/IPA 3:1 at 4 °C and ultrasonication for 1 min. Alternatively, PMMA (MW 35K, Micro Resist Technology) was used for thermal NIL (Nanonex BX-200) at 500 psi and 180 °C for 5 min, in order to improve the throughput. NIL templates were made of ∼25 nm HSQ (Dow Corning XR-1541) on Si substrate, developed in MFCD-26 for 4 min, then annealed on hot plate at 540 °C for 1 h, followed by antiadhesion coating (Nanonex Ultra-100). In both cases, the PMMA thickness was ∼60 nm. After lithography, a 12 nm Ti hard mask was evaporated (Semicore SC2000) at an angle of 30°, which protects the thin PMMA layer in oxygen plasma (Oxford PlasmaLab 80), exposing only the nanodot regions. AuPd (60/40) with Ti adhesion layer was evaporated, forming nanodot arrays after lift-off in boiling acetone for 5 min. An annealing step at 540 °C for 2 h caused the nanodots to agglomerate into spheres whose size was coded by the lithography. The samples were inspected by SEM (Hitachi 4700) and AFM (Park XE-100). In order to reduce the background noise caused by nonspecific binding, a 15 nm HSQ layer was spun on the glass coverslips and thermally cured at 540 °C for 2 h before the nanoarray fabrication.
Functionalization
The functionalization process is shown in Supporting Information Figure S1b. First of all, nanoarray samples were cleaned by immersion in 1.5 h-aged piranha solution (3:1 H2SO4:H2O2) for 3 min, followed by exposure to an oxygen plasma for 5 min. This process also activates the glass surface for binding of PEG-silane by creating hydroxyl groups. Samples were immediately immersed in a freshly prepared 1 mM mixture of HS-C11-EG6-Biotin and HS-C11-EG3–OH (ProChimia Surfaces) solution in ethanol for ∼18 h (overnight). The biotin concentration on the SAM formed on AuPd nanodots was thereby adjusted by the mole fraction of biotin-alkylthiol. The thiolated samples were rinsed by ethanol, dried and then incubated in a solution of 2 mg mPEG-Silane (MW 5000, Laysan Bio) in 25 mL anhydrous toluene with 30 μL acetic acid (as a catalyst) for 48 h. The long PEGylation time was found to improve the passivation against nonspecific binding. Samples were rinsed with acetone then ethanol and blew dry. They were incubated for 2 h in 10 μg/mL streptavidin (Streptavidin, Alexa Fluor 555 Conjugate, F/P 3:1, Life Technologies) solution in 1.5 mL Phosphate-buffered saline (PBS, Gibco DPBS 1 ×) with 1 mg/mL Albumin. In this way, streptavidin molecules bind to AuPd nanodots, whose occupancy can be adjusted by both dot size and mole fraction of biotin-alkylthiol.
In the case of T-cell assays,37 1 mg silane-PEG-NHS (MW 5000, Nanocs) solution in 25 mL anhydrous methanol with 1 mM NTA-L-lysine was incubated overnight to synthesize silane-PEG-NTA, which was used in place of mPEG-silane. After PEGylation, samples were first incubated in 40 mM aqueous NiSO4 for 1 h to adsorb Ni(II) to the NTA groups, followed by 1 μg/mL His-tag ICAM-1 solution in PBS for 2 h. Then the samples were incubated in streptavidin solution in PBS for 30 min, followed by 2 μg/mL biotinylated UCHT1 Fab-568 (F/P 0.45) for another 30 min.
Microscopy and Cell Assays
Nanoarray samples were mounted in a chamber for replaceable coverslips (CSC-22 × 22, Bioscience Tools) and covered with buffer solution (PBS). For molecular occupancy of streptavidin-555, the measurements were done by conventional epi-fluorescence microscopy (Olympus IX81) using a DSRed filter for excitation and emission. The nanoarray fluorescence was collected by an oil-immersion objective (100 ×, 1.49NA, Olympus) with an additional 2× magnification, and recorded by an EM-CCD camera (Cascade II 512, Photometrics) at 50 frames per second for 210 s.
Fresh human CD4+ T-cells (106 cells per mL) were plated on the nanoarrays and incubated for 5 min at 37 °C and 0% CO2. Cells were fixed (warm 2% paraformaldehyde in PHEMO buffer (10 mM EGTA, 2 mM MgCl2, 60 mM Pipes, 25 mM HEPES, pH 7.2), 10 min) and permeabilized (0.1% Triton in PBS, 3 min). Cells were then stained with the pY binding monoclonal antibody 488-PY20 (Biolegend) and imaged using TIRF (100×, 1.49NA, Nikon Eclipse Ti) with a back illuminated EM-CCD camera (AndorDU897).
Data Analysis
The FIJI software package was used for image processing and data analysis. Both the signals of nanodot array and the background noises of nonspecific bound fluorophores were detected by the “Mosaic” plug-in,74 and then separated by a self-written macro according to the nanodot geometric arrangement (Supporting Information Figure S3a). A 3 × 3 pixel ROI was used based on the PSF (Supporting Information Figure S3b). The noises were counted in a 1600 μm2 area of five random locations in order to measure the density of nonspecific bound fluorophores. The signals on nanoarrays were used for the bleaching analysis.
A step detection algorithm for molecular motors75 was adopted to fit the bleaching curves, so that the total number of step and the size of each step were extracted using Matlab. The histogram of step size with double peaks was fitted by a mixed Gaussian distribution using expectation maximization algorithm. The fluorescence average over the first 1 s exposure was used as the initial intensity. A set of single Gaussians (nth convolution powers) based on the fitted single fluorophore intensity (bleaching step size) distribution was used to fit the continuous histogram of intensity. The resulted discrete probability distribution provided an estimation on the fluorophore occupancy (Supporting Information Figure S4b). Similarly, a Gaussian approximation to the binomial distribution of the fluorophore number per streptavidin was used to estimate the molecular occupancy (Figure 4d,e). This method provides important details on the occupancy distribution, and is more accurate than merely calculating average values with eq 1, due to the large intensity variance of a single fluorophore/molecule.
For the T-cell assays on various nanoarrays (200 × 200 μm2 for each pattern), the average pY intensity was measured for the ROI of each cell, with subtraction of a surrounding background ROI. The integration of various patterns on the same sample enables direct comparison. For the comparison between samples, the data was normalized based on the positive control with nonspecifically bound Fab′ and ICAM-1 on bare glass, and negative control on PEG passivation background. The density of his-tag ICAM-1 linked to the PEG background was estimated by comparing its fluorescence intensity with a supported lipid bilayer containing ICAM-1 of known density, assuming a linear relationship between the fluorescence intensity and molecule density.
Supplementary Material
Acknowledgments
The authors thank Dr. M. Palma for guidance on the single fluorophore bleaching measurements and Dr. Silvia Curado for coordinating T-cell experiments. This work was supported primarily by the National Science Foundation (NSF) under award no. CMMI-1300590 and by the National Institutes of Health (NIH) Common Fund Nanomedicine program grant PN2 EY016586. H.W. was supported by a Marie Curie International Outgoing Fellowship within the Seventh European Commission Framework Programme (PIOF-GA-2012-332045). The authors are grateful to the Columbia Nano Initiative for providing cleanroom and other facilities used in this work.
Footnotes
Supporting Information
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acsnano.5b07425.
Fabrication and functionalization, molecular model and quenching effect, optimal packing problem, ROI selection and spatial effect correction, T-Cell stimulation by single-molecule nanoarrays. (PDF)
Author Contributions
H.C. performed experiments and data analysis and wrote the manuscript. H.W. performed fluorescence bleaching experiments. D.D. prepared antibodies and performed T-cell assays. M.L.D. and M.P.S. designed cell assays, analyzed data, and participated in preparation of the manuscript. S.J.W. designed the experiments, analyzed data and wrote the manuscript.
Notes
The authors declare no competing financial interest.
References
- 1.Drmanac R, Sparks AB, Callow MJ, Halpern AL, Burns NL, Kermani BG, Carnevali P, Nazarenko I, Nilsen GB, Yeung G, Dahl F, Fernandez A, Staker B, Pant KP, Baccash J, Borcherding AP, Brownley A, Cedeno R, Chen LS, Chernikoff D;, et al. Human Genome Sequencing Using Unchained Base Reads on Self-Assembling DNA Nanoarrays. Science. 2010;327:78–81. doi: 10.1126/science.1181498. [DOI] [PubMed] [Google Scholar]
- 2.Eid J, Fehr A, Gray J, Luong K, Lyle J, Otto G, Peluso P, Rank D, Baybayan P, Bettman B, Bibillo A, Bjornson K, Chaudhuri B, Christians F, Cicero R, Clark S, Dalal R, Dewinter A, Dixon J, Foquet M, et al. Real-Time DNA Sequencing from Single Polymerase Molecules. Science. 2009;323:133–138. doi: 10.1126/science.1162986. [DOI] [PubMed] [Google Scholar]
- 3.Kingsmore SF. Multiplexed Protein Measurement: Technologies and Applications of Protein and Antibody Arrays. Nat Rev Drug Discovery. 2006;5:310–320. doi: 10.1038/nrd2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lynch M, Mosher C, Huff J, Nettikadan S, Johnson J, Henderson E. Functional Protein Nanoarrays for Biomarker Profiling. Proteomics. 2004;4:1695–1702. doi: 10.1002/pmic.200300701. [DOI] [PubMed] [Google Scholar]
- 5.Palma M, Abramson JJ, Gorodetsky AA, Penzo E, Gonzalez RL, Sheetz MP, Nuckolls C, Hone J, Wind SJ. Selective Biomolecular Nanoarrays for Parallel Single-Molecule Investigations. J Am Chem Soc. 2011;133:7656–7659. doi: 10.1021/ja201031g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bulyk ML, Gentalen E, Lockhart DJ, Church GM. Quantifying DNA-Protein Interactions by Double-Stranded DNA Arrays. Nat Biotechnol. 1999;17:573–577. doi: 10.1038/9878. [DOI] [PubMed] [Google Scholar]
- 7.Rosi NL, Mirkin CA. Nanostructures in Biodiagnostics. Chem Rev. 2005;105:1547–1562. doi: 10.1021/cr030067f. [DOI] [PubMed] [Google Scholar]
- 8.Taton TA, Mirkin CA, Letsinger RL. Scanometric DNA Array Detection with Nanoparticle Probes. Science. 2000;289:1757–1760. doi: 10.1126/science.289.5485.1757. [DOI] [PubMed] [Google Scholar]
- 9.Li M, Cushing SK, Liang HY, Suri S, Ma DL, Wu NQ. Plasmonic Nanorice Antenna on Triangle Nanoarray for Surface-Enhanced Raman Scattering Detection of Hepatitis B Virus DNA. Anal Chem. 2013;85:2072–2078. doi: 10.1021/ac303387a. [DOI] [PubMed] [Google Scholar]
- 10.Haes AJ, Van Duyne RP. A Nanoscale Optical Blosensor: Sensitivity and Selectivity of an Approach Based on the Localized Surface Plasmon Resonance Spectroscopy of Triangular Silver Nanoparticles. J Am Chem Soc. 2002;124:10596–10604. doi: 10.1021/ja020393x. [DOI] [PubMed] [Google Scholar]
- 11.Haes AJ, Hall WP, Chang L, Klein WL, Van Duyne RP. A Localized Surface Plasmon Resonance Biosensor: First Steps toward an Assay for Alzheimer’s Disease. Nano Lett. 2004;4:1029–1034. [Google Scholar]
- 12.Lee KB, Kim EY, Mirkin CA, Wolinsky SM. The Use of Nanoarrays for Highly Sensitive and Selective Detection of Human Immunodeficiency Virus Type 1 in Plasma. Nano Lett. 2004;4:1869–1872. [Google Scholar]
- 13.Guo LH, Ferhan AR, Lee K, Kim DH. Nanoarray-Based Biomolecular Detection Using Individual Au Nanoparticles with Minimized Localized Surface Plasmon Resonance Variations. Anal Chem. 2011;83:2605–2612. doi: 10.1021/ac200432c. [DOI] [PubMed] [Google Scholar]
- 14.McCauley TG, Hamaguchi N, Stanton M. Aptamer-Based Biosensor Arrays for Detection and Quantification of Biological Macromolecules. Anal Biochem. 2003;319:244–250. doi: 10.1016/s0003-2697(03)00297-5. [DOI] [PubMed] [Google Scholar]
- 15.Wingren C, Borrebaeck CAK. Progress in Miniaturization of Protein Arrays – a Step Closer to High-Density Nanoarrays. Drug Discovery Today. 2007;12:813–819. doi: 10.1016/j.drudis.2007.08.003. [DOI] [PubMed] [Google Scholar]
- 16.Peterson EM, Harris JM. Quantitative Detection of Single Molecules in Fluorescence Microscopy Images. Anal Chem. 2010;82:189–196. doi: 10.1021/ac901710t. [DOI] [PubMed] [Google Scholar]
- 17.Hanley DC, Harris JM. Quantitative Dosing of Surfaces with Fluorescent Molecules: Characterization of Fractional Monolayer Coverages by Counting Single Molecules. Anal Chem. 2001;73:5030–5037. doi: 10.1021/ac010572h. [DOI] [PubMed] [Google Scholar]
- 18.Roy R, Hohng S, Ha T. A Practical Guide to Single-Molecule Fret. Nat Methods. 2008;5:507–516. doi: 10.1038/nmeth.1208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Morimatsu M, Mekhdjian AH, Adhikari AS, Dunn AR. Molecular Tension Sensors Report Forces Generated by Single Integrin Molecules in Living Cells. Nano Lett. 2013;13:3985–3989. doi: 10.1021/nl4005145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kinz-Thompson CD, Palma M, Pulukkunat DK, Chenet D, Hone J, Wind SJ, Gonzalez RL. Robustly Passivated, Gold Nanoaperture Arrays for Single-Molecule Fluorescence Microscopy. ACS Nano. 2013;7:8158–8166. doi: 10.1021/nn403447s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Manz BN, Jackson BL, Petit RS, Dustin ML, Groves J. T-Cell Triggering Thresholds Are Modulated by the Number of Antigen within Individual T-Cell Receptor Clusters. Proc Natl Acad Sci U S A. 2011;108:9089–9094. doi: 10.1073/pnas.1018771108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nam JM, Han SW, Lee KB, Liu XG, Ratner MA, Mirkin CA. Bioactive Protein Nanoarrays on Nickel Oxide Surfaces Formed by Dip-Pen Nanolithography. Angew Chem, Int Ed. 2004;43:1246–1249. doi: 10.1002/anie.200353203. [DOI] [PubMed] [Google Scholar]
- 23.Lee KB, Park SJ, Mirkin CA, Smith JC, Mrksich M. Protein Nanoarrays Generated by Dip-Pen Nanolithography. Science. 2002;295:1702–1705. doi: 10.1126/science.1067172. [DOI] [PubMed] [Google Scholar]
- 24.Demers LM, Ginger DS, Park SJ, Li Z, Chung SW, Mirkin CA. Direct Patterning of Modified Oligonucleotides on Metals and Insulators by Dip-Pen Nanolithography. Science. 2002;296:1836–1838. doi: 10.1126/science.1071480. [DOI] [PubMed] [Google Scholar]
- 25.Gopinath A, Rothemund PWK. Optimized Assembly and Covalent Coupling of Single-Molecule DNA Origami Nano Arrays. ACS Nano. 2014;8:12030–12040. doi: 10.1021/nn506014s. [DOI] [PubMed] [Google Scholar]
- 26.Hoff JD, Cheng LJ, Meyhofer E, Guo LJ, Hunt AJ. Nanoscale Protein Patterning by Imprint Lithography. Nano Lett. 2004;4:853–857. [Google Scholar]
- 27.Love JC, Estroff LA, Kriebel JK, Nuzzo RG, Whitesides GM. Self-Assembled Monolayers of Thiolates on Metals as a Form of Nanotechnology. Chem Rev. 2005;105:1103–1169. doi: 10.1021/cr0300789. [DOI] [PubMed] [Google Scholar]
- 28.Schvartzman M, Palma M, Sable J, Abramson J, Hu X, Sheetz MP, Wind SJ. Nanolithographic Control of the Spatial Organization of Cellular Adhesion Receptors at the Single-Molecule Level. Nano Lett. 2011;11:1306–12. doi: 10.1021/nl104378f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Arnold M, Cavalcanti-Adam EA, Glass R, Blummel J, Eck W, Kantlehner M, Kessler H, Spatz JP. Activation of Integrin Function by Nanopatterned Adhesive Interfaces. ChemPhysChem. 2004;5:383–388. doi: 10.1002/cphc.200301014. [DOI] [PubMed] [Google Scholar]
- 30.Cavalcanti-Adam EA, Volberg T, Micoulet A, Kessler H, Geiger B, Spatz JP. Cell Spreading and Focal Adhesion Dynamics Are Regulated by Spacing of Integrin Ligands. Biophys J. 2007;92:2964–2974. doi: 10.1529/biophysj.106.089730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Huang JH, Grater SV, Corbellini F, Rinck S, Bock E, Kemkemer R, Kessler H, Ding JD, Spatz JP. Impact of Order and Disorder in Rgd Nanopatterns on Cell Adhesion. Nano Lett. 2009;9:1111–1116. doi: 10.1021/nl803548b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jaehrling S, Thelen K, Wolfram T, Pollerberg GE. Nanopatterns Biofunctionalized with Cell Adhesion Molecule Dm-Grasp Offered as Cell Substrate: Spacing Determines Attachment and Differentiation of Neurons. Nano Lett. 2009;9:4115–4121. doi: 10.1021/nl9023325. [DOI] [PubMed] [Google Scholar]
- 33.Ranzinger J, Krippner-Heidenreich A, Haraszti T, Bock E, Tepperink J, Spatz JP, Scheurich P. Nanoscale Arrangement of Apoptotic Ligands Reveals a Demand for a Minimal Lateral Distance for Efficient Death Receptor Activation. Nano Lett. 2009;9:4240–4245. doi: 10.1021/nl902429b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Deeg J, Axmann M, Matic J, Liapis A, Depoil D, Afrose J, Curado S, Dustin ML, Spatz JP. T Cell Activation Is Determined by the Number of Presented Antigens. Nano Lett. 2013;13:5619–26. doi: 10.1021/nl403266t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Matic J, Deeg J, Scheffold A, Goldstein I, Spatz JP. Fine Tuning and Efficient T Cell Activation with Stimulatory Acd3 Nanoarrays. Nano Lett. 2013;13:5090–5097. doi: 10.1021/nl4022623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Delcassian D, Depoil D, Rudnicka D, Liu ML, Davis DM, Dustin ML, Dunlop IE. Nanoscale Ligand Spacing Influences Receptor Triggering in T Cells and NK Cells. Nano Lett. 2013;13:5608–5614. doi: 10.1021/nl403252x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cai H, Depoil D, Palma M, Sheetz MP, Dustin ML, Wind SJ. Bifunctional Nanoarrays for Probing the Immune Response at the Single-Molecule Level. Journal of vacuum science and technology B, Nanotechnology & microelectronics: materials, processing, measurement, & phenomena: JVST B. 2013;31:6F902. doi: 10.1116/1.4823764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lohmuller T, Triffo S, O’Donoghue GP, Xu Q, Coyle MP, Groves JT. Supported Membranes Embedded with Fixed Arrays of Gold Nanoparticles. Nano Lett. 2011;11:4912–8. doi: 10.1021/nl202847t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lohmuller T, Xu Q, Groves JT. Nanoscale Obstacle Arrays Frustrate Transport of Epha2-Ephrin-A1 Clusters in Cancer Cell Lines. Nano Lett. 2013;13:3059–3064. doi: 10.1021/nl400874v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Abramson J, Palma M, Wind SJ, Hone J. Quantum Dot Nanoarrays: Self-Assembly with Single-Particle Control and Resolution. Adv Mater. 2012;24:2207–11. doi: 10.1002/adma.201104216. [DOI] [PubMed] [Google Scholar]
- 41.Penzo E, Palma M, Wang R, Cai H, Zheng M, Wind SJ. Directed Assembly of End-Functionalized Single Wall Carbon Nanotube Segments. Nano Lett. 2015;15:6547. doi: 10.1021/acs.nanolett.5b02220. [DOI] [PubMed] [Google Scholar]
- 42.Wang RS, Palma M, Penzo E, Wind SJ. Lithographically Directed Assembly of One-Dimensional DNA Nanostructures via Bivalent Binding Interactions. Nano Res. 2013;6:409–417. [Google Scholar]
- 43.Pearson AC, Pound E, Woolley AT, Linford MR, Harb JN, Davis RC. Chemical Alignment of DNA Origami to Block Copolymer Patterned Arrays of 5 Nm Gold Nanoparticles. Nano Lett. 2011;11:1981–1987. doi: 10.1021/nl200306w. [DOI] [PubMed] [Google Scholar]
- 44.Weiss A. Structure and Function of the T-Cell Antigen Receptor. J Clin Invest. 1990;86:1015–1022. doi: 10.1172/JCI114803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chai JA, Huo FW, Zheng ZJ, Giam LR, Shim W, Mirkin CA. Scanning Probe Block Copolymer Lithography. Proc Natl Acad Sci U S A. 2010;107:20202–20206. doi: 10.1073/pnas.1014892107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chai JA, Wong LS, Giam L, Mirkin CA. Single-Molecule Protein Arrays Enabled by Scanning Probe Block Copolymer Lithography. Proc Natl Acad Sci U S A. 2011;108:19521–19525. doi: 10.1073/pnas.1116099108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Aydin D, Schwieder M, Louban I, Knoppe S, Ulmer J, Haas TL, Walczak H, Spatz JP. Micro-Nanostructured Protein Arrays: A Tool for Geometrically Controlled Ligand Presentation. Small. 2009;5:1014–1018. doi: 10.1002/smll.200801219. [DOI] [PubMed] [Google Scholar]
- 48.Glass R, Arnold M, Blümmel J, Küller A, Möller M, Spatz JP. Micro-Nanostructured Interfaces Fabricated by the Use of Inorganic Block Copolymer Micellar Monolayers as Negative Resist for Electron-Beam Lithography. Adv Funct Mater. 2003;13:569–575. [Google Scholar]
- 49.Glass R, Möller M, Spatz JP. Block Copolymer Micelle Nanolithography. Nanotechnology. 2003;14:1153–1160. [Google Scholar]
- 50.Spatz JP, Mossmer S, Hartmann C, Möller M, Herzog T, Krieger M, Boyen HG, Ziemann P, Kabius B. Ordered Deposition of Inorganic Clusters from Micellar Block Copolymer Films. Langmuir. 2000;16:407–415. [Google Scholar]
- 51.Anker JN, Hall WP, Lyandres O, Shah NC, Zhao J, Van Duyne RP. Biosensing with Plasmonic Nanosensors. Nat Mater. 2008;7:442–453. doi: 10.1038/nmat2162. [DOI] [PubMed] [Google Scholar]
- 52.Haynes CL, Van Duyne RP. Nanosphere Lithography: A Versatile Nanofabrication Tool for Studies of Size-Dependent Nanoparticle Optics. J Phys Chem B. 2001;105:5599–5611. [Google Scholar]
- 53.Hulteen JC, Vanduyne RP. Nanosphere Lithography – a Materials General Fabrication Process for Periodic Particle Array Surfaces. J Vac Sci Technol, A. 1995;13:1553–1558. [Google Scholar]
- 54.Sun SQ, Mendes P, Critchley K, Diegoli S, Hanwell M, Evans SD, Leggett GJ, Preece JA, Richardson TH. Fabrication of Gold Micro- and Nanostructures by Photolithographic Exposure of Thiol-Stabilized Gold Nanoparticles. Nano Lett. 2006;6:345–350. doi: 10.1021/nl052130h. [DOI] [PubMed] [Google Scholar]
- 55.Schvartzman M, Wind SJ. Robust Pattern Transfer of Nanoimprinted Features for Sub-5-Nm Fabrication. Nano Lett. 2009;9:3629–34. doi: 10.1021/nl9018512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bachmann MF, McKallFaienza K, Schmits R, Bouchard D, Beach J, Speiser DE, Mak TW, Ohashi PS. Distinct Roles for Lfa-1 and Cd28 During Activation of Naive T Cells: Adhesion versus Costimulation. Immunity. 1997;7:549–557. doi: 10.1016/s1074-7613(00)80376-3. [DOI] [PubMed] [Google Scholar]
- 57.Heider EC, Peterson EM, Barhoum M, Gericke KH, Harris JM. Quantitative Fluorescence Microscopy to Determine Molecular Occupancy of Phospholipid Vesicles. Anal Chem. 2011;83:5128–5136. doi: 10.1021/ac200129n. [DOI] [PubMed] [Google Scholar]
- 58.Holzmeister P, Pibiri E, Schmied JJ, Sen T, Acuna GP, Tinnefeld P. Quantum Yield and Excitation Rate of Single Molecules Close to Metallic Nanostructures. Nat Commun. 2014;5:5356. doi: 10.1038/ncomms6356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Dulkeith E, Morteani AC, Niedereichholz T, Klar TA, Feldmann J, Levi SA, van Veggel FCJM, Reinhoudt DN, Möller M, Gittins DI. Fluorescence Quenching of Dye Molecules near Gold Nanoparticles: Radiative and Nonradiative Effects. Phys Rev Lett. 2002;89:203002. doi: 10.1103/PhysRevLett.89.203002. [DOI] [PubMed] [Google Scholar]
- 60.Hu M, Chen JY, Li ZY, Au L, Hartland GV, Li XD, Marquez M, Xia YN. Gold Nanostructures: Engineering Their Plasmonic Properties for Biomedical Applications. Chem Soc Rev. 2006;35:1084–1094. doi: 10.1039/b517615h. [DOI] [PubMed] [Google Scholar]
- 61.Hofer R, Textor M, Spencer ND. Alkyl Phosphate Monolayers, Self-Assembled from Aqueous Solution onto Metal Oxide Surfaces. Langmuir. 2001;17:4014–4020. [Google Scholar]
- 62.Chhabra R, Sharma J, Wang HN, Zou SL, Lin S, Yan H, Lindsay S, Liu Y. Distance-Dependent Interactions between Gold Nanoparticles and Fluorescent Molecules with DNA as Tunable Spacers. Nanotechnology. 2009;20:485201. doi: 10.1088/0957-4484/20/48/485201. [DOI] [PubMed] [Google Scholar]
- 63.Breshike CJ, Riskowski RA, Strouse GF. Leaving Forster Resonance Energy Transfer Behind: Nanometal Surface Energy Transfer Predicts the Size-Enhanced Energy Coupling between a Metal Nanoparticle and an Emitting Dipole. J Phys Chem C. 2013;117:23942–23949. [Google Scholar]
- 64.Hua BY, Han KY, Zhou RB, Kim HJ, Shi XH, Abeysirigunawardena SC, Jain A, Singh D, Aggarwal V, Woodson SA, Ha T. An Improved Surface Passivation Method for Single-Molecule Studies. Nat Methods. 2014;11:1233. doi: 10.1038/nmeth.3143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Clare BW, Kepert DL. The Optimal Packing of Circles on a Sphere. J Math Chem. 1991;6:325–349. [Google Scholar]
- 66.Cooper JM, Shen J, Young FM, Connolly P, Barker JR, Moores G. The Imaging of Streptavidin and Avidin Using Scanning-Tunneling-Microscopy. J Mater Sci: Mater Electron. 1940;5:106–110. [Google Scholar]
- 67.Liu YL, Shipton MK, Ryan J, Kaufman ED, Franzen S, Feldheim DL. Synthesis, Stability, and Cellular Internalization of Gold Nanoparticles Containing Mixed Peptide-Poly(Ethylene Glycol) Monolayers. Anal Chem. 2007;79:2221–2229. doi: 10.1021/ac061578f. [DOI] [PubMed] [Google Scholar]
- 68.Appelbaum J, Weiss Y. The Packing of Circles on a Hemisphere. Meas Sci Technol. 1999;10:1015–1019. [Google Scholar]
- 69.Stranick SJ, Parikh AN, Tao YT, Allara DL, Weiss PS. Phase-Separation of Mixed-Composition Self-Assembled Monolayers into Nanometer-Scale Molecular Domains. J Phys Chem. 1994;98:7636–7646. [Google Scholar]
- 70.Skinner JP, Chi LL, Ozeata PF, Ramsay CS, O’Hara RL, Calfin BB, Tetin SY. Introduction of the Mass Spread Function for Characterization of Protein Conjugates. Anal Chem. 2012;84:1172–1177. doi: 10.1021/ac202239j. [DOI] [PubMed] [Google Scholar]
- 71.Grakoui A, Bromley SK, Sumen C, Davis MM, Shaw AS, Allen PM, Dustin ML. The Immunological Synapse: A Molecular Machine Controlling T Cell Activation. Science. 1999;285:221–227. doi: 10.1126/science.285.5425.221. [DOI] [PubMed] [Google Scholar]
- 72.Irvine DJ, Purbhoo MA, Krogsgaard M, Davis MM. Direct Observation of Ligand Recognition by T Cells. Nature. 2002;419:845–849. doi: 10.1038/nature01076. [DOI] [PubMed] [Google Scholar]
- 73.Dustin ML, Miller JM, Ranganath S, Vignali DAA, Viner NJ, Nelson CA, Unanue ER. Tcr-Mediated Adhesion of T Cell Hybridomas to Planar Bilayers Containing Purified Mhc Class Ii Peptide Complexes and Receptor Shedding During Detachment. J Immunol. 1996;157:2014–2021. [PubMed] [Google Scholar]
- 74.Sbalzarini IF, Koumoutsakos P. Feature Point Tracking and Trajectory Analysis for Video Imaging in Cell Biology. J Struct Biol. 2005;151:182–195. doi: 10.1016/j.jsb.2005.06.002. [DOI] [PubMed] [Google Scholar]
- 75.Aggarwal T, Materassi D, Davison R, Hays T, Salapaka M. Detection of Steps in Single Molecule Data. Cell Mol Bioeng. 2012;5:14–31. doi: 10.1007/s12195-011-0188-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


