Abstract
Acid soils limit agricultural production worldwide. Major reason of crop losses in acid soils is the toxicity of aluminum (Al). In the present work, we investigated expression alterations of microRNAs in flax (Linum usitatissimum L.) plants under Al stress. Flax seedlings of resistant (TMP1919 and G1071/4_k) and sensitive (Lira and G1071/4_o) to Al cultivars and lines were exposed to AlCl3 solution for 4 and 24 hours. Twelve small RNA libraries were constructed and sequenced using Illumina platform. In total, 97 microRNAs from 18 conserved families were identified. miR319, miR390, and miR393 revealed expression alterations associated with Al treatment of flax plants. Moreover, for miR390 and miR393, the alterations were distinct in sensitive and resistant to Al genotypes. Expression level changes of miR319 and miR390 were confirmed using qPCR analysis. In flax, potential targets of miR319 are TCPs, miR390–TAS3 and GRF5, and miR393–AFB2-coding transcripts. TCPs, TAS3, GRF5, and AFB2 participate in regulation of plant growth and development. The involvement of miR319, miR390, and miR393 in response to Al stress in flax was shown here for the first time. We speculate that these microRNAs play an important role in Al response via regulation of growth processes in flax plants.
1. Introduction
Acid soils result in decrease of agricultural production all over the world [1]. Toxicity of aluminum (Al) is a major reason of crop losses in acid soils [2]. Different mechanisms of plant response to Al stress were identified: organic acid exudation by roots to chelate Al ions in soil, detoxification of Al in plants via chelation or transportation into the vacuole, modifications of cell wall to alter Al binding with its components, and so forth [3–5].
MicroRNA (miRNA) negatively regulates gene expression and in this way controls numerous biological processes in plants [6], including stress response [7–9]. Gene expression regulation via miRNA was revealed as one of the mechanisms of response to Al in different plant species [10–12]. However, there is no data on involvement of miRNAs in response to Al stress in important agricultural plant, flax (Linum usitatissimum L.). Flax fiber is utilized in textile industry; flax seeds are used for production of oil, linoleum, food, and pharmaceutical products [13–15]. Flax genetics and epigenetics are in the focus of research interest [16–20]. In the previous works on flax, the involvement of miRNAs in response to saline and alkaline stresses [21] and excessive or deficient nutrition [22–24] was shown.
In the present work, we performed high-throughput sequencing of flax small RNAs under normal conditions and Al exposure to identify miRNAs, whose expression was altered in response to aluminum stress, and suggested potential targets of these miRNAs in flax to speculate on affected signaling pathways.
2. Materials and Methods
2.1. Plant Material
L. usitatissimum plants of resistant (TMP1919 and G1071/4_k) and sensitive (Lira and G1071/4_o) to aluminum cultivars and lines were used in the present study. Seeds germinated on filter paper soaked with distilled water for 5 days. Then seedlings were transferred to falcon tubes with filter paper soaked with a 0.5 mM CaCl2 solution at pH 4.5 for 24 h before being exposed to a 0.5 mM CaCl2 solution (pH 4.5) containing 0 (N) or 500 μM AlCl3 for 4 (Al-4) and 24 (Al-24) hours. Roots were cut off and immediately frozen in liquid nitrogen. Plant samples were stored at −70°C.
Total RNA was extracted from roots of flax plants using RNA MicroPrep kit (Zymo Research, USA). RNA quality and concentration were determined by Qubit 2.0 fluorometer (Life Technologies, USA) and Agilent 2100 Bioanalyzer (Agilent Technologies, USA). For further analysis, only RNA samples with RNA Integrity Number (RIN) value not less than 8.0 were used.
2.2. Flax Small RNA Sequencing
Library preparation was performed using Illumina TruSeq small RNA preparation kit (Illumina, USA) in compliance with manufacturer's protocol. Twelve libraries from pooled plant samples were obtained: N, Al-4, and Al-24 for each of four cultivars/lines (TMP1919, G1071/4_k, G1071/4_o, and Lira). Library quality was evaluated using Agilent 2100 Bioanalyzer (Agilent Technologies). The sequencing was performed on Genome Analyzer IIx (Illumina).
2.3. Bioinformatics Analysis of miRNAs
Low-quality reads and adapter reads were removed from raw sequencing data using Trimmomatic [25]. For further analysis, we used cleaned reads with abundance six or more. To identify conserved miRNAs in flax, small RNA sequences were aligned with known matured miRNA sequences from miRBase 21.0 [26]. Prediction of miRNA targets was performed using psRNATarget server [27] with default parameters using identified L. usitatissimum transcripts [16, 28].
miRNA levels were normalized to obtain reads per million (RPM) values. The comparison of miRNA expression levels in Al-4 and Al-24 libraries with N library was performed using fold change parameter: FC = RPM in Al-4 or Al-24/RPM in N. P values were calculated using χ2 test with Benjamini-Hochberg multiple testing correction. Changes were considered significant if FC or 1/FC were 1.5 or higher, that is, absolute value of log2FC ≥ 0.58.
2.4. Quantitative PCR (qPCR) Analysis of miRNA Expression
We performed qPCR analysis to evaluate expression of miR319 and miR390. TaqMan MicroRNA Assays aau-miR319 and ath-miR390a (Thermo Fisher Scientific, USA) were used. Reverse transcription was performed in 15 μL reaction containing 1x RT primer (Thermo Fisher Scientific), 200 U of RevertAid Reverse Transcriptase (Thermo Fisher Scientific), 1x Reverse Transcription Buffer, 250 nM of dNTPs, and 10 ng of total RNA using the following program: 16°C for 30 min, 42°C for 30 min, and 85°C for 5 min. QPCR was performed using the 7500 Real-Time PCR System in a 20 μL reaction mix containing 1x PCR mix (GenLab, Russia), 250 nM of dNTPs, 2 U of polymerase (GenLab), Rox dye, and RT product using the following program: 95°C for 15 min, 40 cycles of 95°C for 15 s, and 60°C for 60 s. Three technical replicates were performed. For the evaluation of expression level alterations, ΔΔCteff values, which are directly proportional to the expression level changes, were calculated [29, 30]. ETIF3H and ETIF3E were chosen as the reference genes for the qPCR data analysis [24, 29, 31]. All the calculations were done using the Analysis of Transcription of Genes software [24, 32]. Correlation between high-throughput sequencing (log2FC) and qPCR (ΔΔCteff) expression data was evaluated using Spearman's correlation coefficient.
3. Results and Discussion
Seedlings of resistant (TMP1919 and G1071/4_k) and sensitive (Lira and G1071/4_o) to Al flax cultivars and lines were exposed to Al for 4 and 24 hours. Twelve small RNA libraries were constructed and sequenced on Illumina GAIIx. In total, about 40 million raw reads were obtained. All the sequences were deposited in the European Nucleotide Archive, accession number PRJEB15342.
Search for flax miRNAs using miRBase sequences led to identification of 97 potential flax miRNAs from 18 conserved families: miR156, miR157, miR159, miR160, miR162, miR164, miR165, miR166, miR167, miR168, miR171, miR319, miR390, miR393, miR394, miR396, miR398, and miR408 (Supplementary Table 1 in Supplementary Material available online at https://doi.org/10.1155/2017/4975146). Among these miRNAs, the search for Al responsive miRNAs was performed. To reveal common trends specific to flax plants, we evaluated expression alterations after 4 and 24 hours of Al treatment using pooled data for all examined cultivars and lines (log2FC values are represented in Table 1).
Table 1.
Expression alterations of miRNAs after 4 and 24 hours of Al treatment in resistant (TMP1919 and G1071/4_k) and sensitive (G1071/4_o and Lira) flax cultivars and lines.
| miRNA family | log2FC | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| TMP1919 | G1071/4_k | G1071/4_o | Lira | All samples | ||||||
| Al-4 | Al-24 | Al-4 | Al-24 | Al-4 | Al-24 | Al-4 | Al-24 | Al-4 | Al-24 | |
| miR156 | 0.77 | 0.29 | −1.79 | −2.16 | 0.20 | −1.58 | 0.74 | −0.87 | −0.42 | −1.44 |
| miR157 | 0.79 | 0.36 | −1.40 | −1.51 | 0.59 | −1.93 | −0.52 | 0.20 | −0.26 | −0.90 |
| miR159 | −2.81 | −1.57 | −0.11 | −1.97 | −0.51 | −2.17 | 0.11 | 0.66 | −0.62 | −1.13 |
| miR160 | −0.23 | −1.00 | 12.13 | 0.00 | −0.30 | −2.28 | −1.22 | −0.74 | −0.35 | −1.40 |
| miR162 | 1.09 | −0.86 | −0.92 | −0.29 | −0.88 | −1.94 | −1.43 | −1.78 | −0.47 | −1.04 |
| miR164 | 1.27 | −1.18 | 0.15 | −1.72 | 0.50 | −2.40 | −2.49 | 0.46 | 0.61 | −0.98 |
| miR165 | 1.62 | −0.84 | −1.98 | −3.16 | 0.19 | −1.10 | 0.54 | −1.86 | −0.34 | −2.13 |
| miR166 | 1.13 | −1.19 | −1.75 | −2.23 | 0.27 | −1.60 | −0.39 | −2.33 | −0.36 | −1.98 |
| miR167 | −0.65 | −1.46 | −1.84 | −15.07 | −0.32 | −2.34 | −2.07 | −0.22 | −0.78 | −1.54 |
| miR168 | 0.20 | −1.14 | −0.39 | −1.81 | −0.30 | −1.60 | −0.60 | −1.25 | −0.28 | −1.41 |
| miR171 | 0.94 | 0.31 | −2.36 | −1.78 | −1.22 | −2.60 | 13.02 | 0.00 | −0.01 | −0.95 |
| miR319 | 1.16 | −0.65 | 0.22 | −2.23 | 0.28 | −2.04 | 0.45 | −1.29 | 0.58 | −1.40 |
| miR390 | 1.27 | −0.01 | 0.85 | −0.84 | −0.78 | −3.04 | −0.35 | −1.77 | 0.17 | −1.38 |
| miR393 | 1.66 | 0.20 | 1.10 | 0.07 | −0.46 | −1.43 | −0.10 | 0.51 | 0.61 | −0.33 |
| miR394 | 1.54 | −0.18 | 13.71 | 12.33 | −0.44 | −1.99 | −0.83 | −1.24 | 0.71 | −0.68 |
| miR396 | 0.40 | −1.45 | −0.99 | −1.90 | −0.23 | −1.51 | −1.00 | −1.26 | −0.32 | −1.45 |
| miR398 | −0.06 | −1.22 | 0.00 | 0.00 | 0.38 | −0.51 | 0.56 | −2.83 | 0.47 | −1.78 |
| miR408 | −0.17 | −0.37 | −0.35 | −1.86 | 1.40 | 1.98 | −2.20 | −3.61 | −0.82 | −0.86 |
After 4 hours of Al exposure, we observed significant (absolute value of log2FC ≥ 0.58) upregulation of miR164, miR319, miR393, and miR394 and downregulation of miR159, miR167, and miR408. After 24 hours of Al exposure, significant expression decrease was revealed for all 18 miRNA families except miR393. Thus, after 4 hours of Al treatment, miRNA levels of different families were increased, decreased, or stable. However, after 24 hours of Al exposure, expression was decreased for almost all miRNA families.
We also performed analysis of expression alterations of miRNA families in individual cultivars and lines to identify trends in resistant and sensitive to Al flax plants (Supplementary Table 2). log2FC values are represented in Table 1. Some of the miRNAs showed opposite directions of expression alterations in studied flax cultivars and lines. As seen from Table 1, after 4 hours of Al exposure, miR156, miR157, miR162, miR165, miR166, and miR171 were significantly upregulated in one of the resistant genotypes and significantly downregulated in the other one. The same was observed for miR171 and miR408 in sensitive genotypes. We suggested that these miRNAs with opposite regulation under Al stress in resistant or sensitive cultivars and lines do not play the key role in flax response to Al. After 24 hours of Al treatment, the majority of miRNAs was downregulated in all 4 cultivars and lines.
Definite regularities were revealed for expression alterations of miR319, miR390, and miR393 families. The level of miR319 was changed in a similar way in resistant and sensitive to Al cultivars and lines: expression was increased after 4 hours of Al exposure (log2FC varied from 0.22 to 1.16) and decreased after 24 hours (log2FC varied from −0.65 to −2.23; Figure 1). miR390 level was decreased after 4 hours of Al exposure in sensitive to Al flax genotypes (log2FC was −0.78 for G1071/4_o and −0.35 for Lira), but increased in resistant genotypes (log2FC was 1.27 in TMP1919 and 0.85 in G1071/4_k; Figure 1). Moreover, after 24 hours of Al exposure, we revealed retention or moderate downregulation of miR390 in resistant to Al cultivar and line (log2FC was −0.01 for TMP1919 and −0.84 for G1071/4_k), but strong downregulation in sensitive cultivar and line (log2FC was −3.04 for G1071/4_o and −1.77 for Lira). miR393 was upregulated in resistant to Al genotypes (log2FC was 1.66 for TMP1919 and 1.10 for G1071/4_k), but slightly decreased in sensitive to Al genotypes (log2FC was −0.46 for G1071/4_o and −0.10 for Lira) after 4 hours of Al exposure (Figure 1). After 24 hours of Al exposure, miR393 level was stable in resistant to Al genotypes (log2FC was 0.20 for TMP1919 and 0.07 for G1071/4_k) and deregulated in sensitive to Al genotypes (log2FC was −1.43 for G1071/4_o and 0.51 for Lira).
Figure 1.

Expression alterations of miR319, miR390, and miR393 in resistant and sensitive flax cultivars and lines under Al stress.
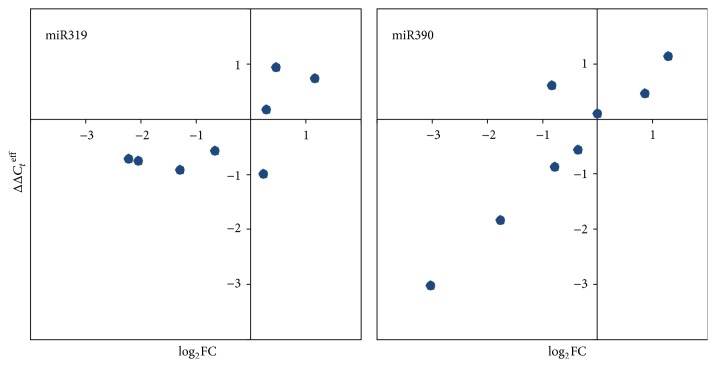
For validation of high-throughput sequencing data, expression of miR319 and miR390 was evaluated using qPCR. RNA samples of flax plants, which were used for high-throughput sequencing and were taken into qPCR analysis. The data obtained by qPCR and high-throughput sequencing methods were highly consistent: Spearman's correlation coefficient was 0.68 for miR319 and 0.76 for miR390 (P < 0.05; Figure 2).
Figure 2.
Correlation between qPCR (ΔΔCteff) and high-throughput sequencing (log2FC) expression data for miR319 and miR390 in flax cultivars and lines under Al stress.
In plants, miR319, miR390, and miR393 families have been identified as Al responsive [10, 11]. For miR319, both upregulation [33] and downregulation [34] were revealed in Medicago truncatula in response to Al stress. miR390 was upregulated in wild soybean under Al exposure [35]. However, in M. truncatula, miR390 was slightly downregulated after short-term Al treatment and was significantly upregulated after long-term one [34]. For miR393, upregulation was revealed in common bean roots in response to Al treatment for 24 hours [36], and downregulation in rice roots after 8 hours of Al treatment [37]. Opposite directions of the alterations of miRNA levels could be associated with different time of Al treatment or diverse resistance of examined genotypes to Al stress.
In previous flax studies, potential targets for some miRNAs from miR319, miR390, and miR393 families were predicted [21, 38, 39]. Here, we performed target prediction for flax highly-expressed miRNAs from miR319, miR390, and miR393 families (Supplementary Table 3).
For miR319, the following targets were predicted by us in flax: transcripts encoding cysteine-rich secretory proteins, antigen 5, pathogenesis-related 1 protein; myb domain protein 65 (MYB65); Teosinte Branched/Cycloidea/PCF transcription factor 3 (TCP3) superfamily protein; TCP family transcription factor 4; ABC transporter. Within predicted targets of miR319, Lus10002195 and Lus10000463 transcripts encoding TCP3 and TCP4 are the most interesting. It was previously shown that miR319 targets mRNA of TCP transcription factors, which control plant growth and development [40–42]. We suggest that the most probable target of miR319 in flax is also TCPs.
For miR390, predicted targets in flax were transcripts encoding leucine-rich receptor-like protein kinase family protein; protein kinase superfamily protein; growth-regulating factor 5 (GRF5); root hair specific 10 (RHS10); poor homologous synapsis 1 (PHS1); leucine-rich repeat (LRR) family protein; TAS3. Transcript Lus10009533 encoding GRF5 was one of the potential targets of miR390. GRFs play important role in plant developmental processes and growth under adverse environments and could be regulated by TCP4 [43]. TAS3-coding transcript, genolin_c19878 from L. usitatissimum unigene library, was also predicted as potential target of miR390. In other plant species, miR390 initiates tasiRNA (trans-acting small interfering RNA) biogenesis via cleavage or interaction with TAS3 transcript. TAS3 tasiRNAs negatively regulate ARFs (auxin response factors) that is necessary for proper plant development [44–48]. We suppose that TAS3 and GRF5 are the most likely targets of miR390 in flax.
Auxin signaling F-box 2 (AFB2), zinc finger (C3HC4-type RING finger) family protein, and pol-like 5 (PLL5) transcripts were predicted as targets of miR393 in flax. Lus10031991 and Lus10035160 encoding AFB2 are the most interesting targets of miR393. It was previously reported that targets of miR393 are AFB1 (auxin f-box protein1), AFB2, and AFB3, which are involved in auxin signaling [32, 49–51]. We speculate that miR393 probably regulates AFB2 expression in flax.
Thus, expression alterations of miR319, miR390, and miR393, which were revealed as Al-responsive in flax, could affect expression level of a number of key transcripts involved in plant growth and development.
4. Conclusions
High-throughput sequencing and qPCR analyses of flax small RNAs allowed us to reveal miRNAs with expression alterations under Al exposure. Our results suggest the involvement of miR319, miR390, and miR393 in Al response in L. usitatissimum plants. Moreover, we revealed diverse alterations of miR390 and miR393 levels in resistant and sensitive to Al genotypes. We concluded that, in flax, potential targets of miR319 are TCPs, miR390–TAS3 and GRF5, and miR393–AFB2. Thus, we speculate that miR319, miR390, and miR393 play an important role in Al stress response via regulation of growth and development processes in flax plants.
Supplementary Material
MicroRNAs in flax plants under aluminum stress (Table 1: Conserved microRNAs; Table 2: MicroRNA expression; Table 3: MicroRNA targets).
Acknowledgments
This work was financially supported by the Russian Science Foundation, Grant 16-16-00114. The authors thank Lomonosov Moscow State University for the help with plant growing and All-Russian Research Institute for Flax for the selection and provision of seeds. This work was performed using the equipment of “Genome” center of Engelhardt Institute of Molecular Biology (http://www.eimb.ru/rus/ckp/ccu_genome_c.php).
Competing Interests
The authors declare that there is no conflict of interests regarding the publication of this paper.
Authors' Contributions
Alexey A. Dmitriev, Anna V. Kudryavtseva, and Nadezhda L. Bolsheva contributed equally to this work.
References
- 1.von Uexküll H. R., Mutert E. Global extent, development and economic impact of acid soils. Plant and Soil. 1995;171(1):1–15. doi: 10.1007/BF00009558. [DOI] [Google Scholar]
- 2.Kochian L. V., Hoekenga O. A., Piñeros M. A. How do crop plants tolerate acid soils? Mechanisms of aluminum tolerance and phosphorous efficiency. Annual Review of Plant Biology. 2004;55:459–493. doi: 10.1146/annurev.arplant.55.031903.141655. [DOI] [PubMed] [Google Scholar]
- 3.Kochian L. V., Piñeros M. A., Liu J., Magalhaes J. V. Plant adaptation to acid soils: the molecular basis for crop aluminum resistance. Annual Review of Plant Biology. 2015;66:571–598. doi: 10.1146/annurev-arplant-043014-114822. [DOI] [PubMed] [Google Scholar]
- 4.Panda S. K., Baluska F., Matsumoto H. Aluminum stress signaling in plants. Plant Signaling and Behavior. 2009;4(7):592–597. doi: 10.4161/psb.4.7.8903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sade H., Meriga B., Surapu V., et al. Toxicity and tolerance of aluminum in plants: tailoring plants to suit to acid soils. BioMetals. 2016;29(2):187–210. doi: 10.1007/s10534-016-9910-z. [DOI] [PubMed] [Google Scholar]
- 6.Jones-Rhoades M. W., Bartel D. P., Bartel B. MicroRNAs and their regulatory roles in plants. Annual Review of Plant Biology. 2006;57:19–53. doi: 10.1146/annurev.arplant.57.032905.105218. [DOI] [PubMed] [Google Scholar]
- 7.Koroban N. V., Kudryavtseva A. V., Krasnov G. S., et al. The role of microRNA in abiotic stress response in plants. Molecular Biology. 2016;50(3):337–343. doi: 10.1134/S0026893316020102. [DOI] [PubMed] [Google Scholar]
- 8.Kumar R. Role of microRNAs in biotic and abiotic stress responses in crop plants. Applied biochemistry and biotechnology. 2014;174(1):93–115. doi: 10.1007/s12010-014-0914-2. [DOI] [PubMed] [Google Scholar]
- 9.Shriram V., Kumar V., Devarumath R. M., Khare T. S., Wani S. H. MicroRNAs as potential targets for abiotic stress tolerance in plants. Frontiers in Plant Science. 2016;7:p. 817. doi: 10.3389/fpls.2016.00817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.He H., He L., Gu M. Role of microRNAs in aluminum stress in plants. Plant Cell Reports. 2014;33(6):831–836. doi: 10.1007/s00299-014-1565-z. [DOI] [PubMed] [Google Scholar]
- 11.Mendoza-Soto A. B., Sánchez F., Hernández G. MicroRNAs as regulators in plant metal toxicity response. Frontiers in Plant Science. 2012;3, article 105 doi: 10.3389/fpls.2012.00105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Min Yang Z., Chen J. A potential role of microRNAs in plant response to metal toxicity. Metallomics. 2013;5(9):1184–1190. doi: 10.1039/c3mt00022b. [DOI] [PubMed] [Google Scholar]
- 13.Muir A. D., Westcott N. D. Flax: The Fenus Linum. London, UK: Taylor & Francis; 2003. [Google Scholar]
- 14.Goyal A., Sharma V., Upadhyay N., Gill S., Sihag M. Flax and flaxseed oil: an ancient medicine & modern functional food. Journal of Food Science and Technology. 2014;51(9):1633–1653. doi: 10.1007/s13197-013-1247-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Singh K. K., Mridula D., Rehal J., Barnwal P. Flaxseed: a potential source of food, feed and fiber. Critical Reviews in Food Science and Nutrition. 2011;51(3):210–222. doi: 10.1080/10408390903537241. [DOI] [PubMed] [Google Scholar]
- 16.Wang Z., Hobson N., Galindo L., et al. The genome of flax (Linum usitatissimum) assembled de novo from short shotgun sequence reads. Plant Journal. 2012;72(3):461–473. doi: 10.1111/j.1365-313x.2012.05093.x. [DOI] [PubMed] [Google Scholar]
- 17.Melnikova N. V., Kudryavtseva A. V., Zelenin A. V., et al. Retrotransposon-based molecular markers for analysis of genetic diversity within the Genus Linum. BioMed Research International. 2014;2014:14. doi: 10.1155/2014/231589.231589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bolsheva N. L., Zelenin A. V., Nosova I. V., et al. The diversity of karyotypes and genomes within section syllinum of the genus linum (linaceae) revealed by molecular cytogenetic markers and RAPD analysis. PLoS ONE. 2015;10(4) doi: 10.1371/journal.pone.0122015.e0122015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang N., Deyholos M. K. RNASeq analysis of the shoot apex of flax (Linum usitatissimum) to identify phloem fiber specification genes. Frontiers in Plant Science. 2016;7, article no. 950 doi: 10.3389/fpls.2016.00950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Johnson C., Moss T., Cullis C. Environmentally induced heritable changes in flax. Journal of Visualized Experiments. 2011;47 doi: 10.3791/2332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yu Y., Wu G., Yuan H., et al. Identification and characterization of miRNAs and targets in flax (Linum usitatissimum) under saline, alkaline, and saline-alkaline stresses. BMC Plant Biology. 2016;16(1, article 124) doi: 10.1186/s12870-016-0808-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Melnikova N. V., Belenikin M. S., Bolsheva N. L., et al. Flax Inorganic Phosphate Deficiency Responsive miRNAs. Journal of Agricultural Science. 2013;6(1):156–160. doi: 10.5539/jas.v6n1p156. [DOI] [Google Scholar]
- 23.Melnikova N. V., Dmitriev A. A., Belenikin M. S., et al. Excess fertilizer responsive miRNAs revealed in Linum usitatissimum L. Biochimie. 2015;109:36–41. doi: 10.1016/j.biochi.2014.11.017. [DOI] [PubMed] [Google Scholar]
- 24.Melnikova N. V., Dmitriev A. A., Belenikin M. S., et al. Identification, expression analysis, and target prediction of flax genotroph microRNAs under normal and nutrient stress conditions. Frontiers in Plant Science. 2016;7, article 399 doi: 10.3389/fpls.2016.00399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bolger A. M., Lohse M., Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30(15):2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kozomara A., Griffiths-Jones S. miRBase: integrating microRNA annotation and deep-sequencing data. Nucleic Acids Research. 2011;39, S1:D152–D157. doi: 10.1093/nar/gkq1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dai X., Zhao P. X. PsRNATarget: a plant small RNA target analysis server. Nucleic Acids Research. 2011;39(2):W155–W159. doi: 10.1093/nar/gkr319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fenart S., Ndong Y.-P. A., Duarte J., et al. Development and validation of a flax (Linum usitatissimum L.) gene expression oligo microarray. BMC Genomics. 2010;11(1, article 592) doi: 10.1186/1471-2164-11-592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dmitriev A. A., Krasnov G., Rozhmina T., et al. Glutathione S-transferases and UDP-glycosyltransferases are involved in response to aluminum stress in flax. Frontiers in Plant Science. 2016;7:p. 1920. doi: 10.3389/fpls.2016.01920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dmitriev A. A., Kudryavtseva A. V., Krasnov G. S., et al. Gene expression profiling of flax (Linum usitatissimum L.) under edaphic stress. BMC Plant Biology. 2016;16(3) doi: 10.1186/s12870-016-0927-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Huis R., Hawkins S., Neutelings G. Selection of reference genes for quantitative gene expression normalization in flax (Linum usitatissimum L.) BMC Plant Biology. 2010;10, article no. 71 doi: 10.1186/1471-2229-10-71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Krasnov G. S., Oparina N. Y., Dmitriev A. A., et al. RPN1, a new reference gene for quantitative data normalization in lung and kidney cancer. Molecular Biology. 2011;45(2):211–220. doi: 10.1134/S0026893311020129. [DOI] [PubMed] [Google Scholar]
- 33.Zhou Z. S., Huang S. Q., Yang Z. M. Bioinformatic identification and expression analysis of new microRNAs from Medicago truncatula. Biochemical and Biophysical Research Communications. 2008;374(3):538–542. doi: 10.1016/j.bbrc.2008.07.083. [DOI] [PubMed] [Google Scholar]
- 34.Chen L., Wang T., Zhao M., Tian Q., Zhang W.-H. Identification of aluminum-responsive microRNAs in Medicago truncatula by genome-wide high-throughput sequencing. Planta. 2012;235(2):375–386. doi: 10.1007/s00425-011-1514-9. [DOI] [PubMed] [Google Scholar]
- 35.Zeng Q.-Y., Yang C.-Y., Ma Q.-B., Li X.-P., Dong W.-W., Nian H. Identification of wild soybean miRNAs and their target genes responsive to aluminum stress. BMC Plant Biology. 2012;12, article no. 182 doi: 10.1186/1471-2229-12-182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mendoza-Soto A. B., Naya L., Leija A., Hernández G. Responses of symbiotic nitrogen-fixing common bean to aluminum toxicity and delineation of nodule responsive microRNAs. Frontiers in Plant Science. 2015;6, article 587 doi: 10.3389/fpls.2015.00587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lima J. C., Arenhart R. A., Margis-Pinheiro M., Margis R. Aluminum triggers broad changes in microRNA expression in rice roots. Genetics and Molecular Research. 2011;10(4):2817–2832. doi: 10.4238/2011.November.10.4. [DOI] [PubMed] [Google Scholar]
- 38.Neutelings G., Fénart S., Lucau-Danila A., Hawkins S. Identification and characterization of miRNAs and their potential targets in flax. Journal of Plant Physiology. 2012;169(17):1754–1766. doi: 10.1016/j.jplph.2012.06.011. [DOI] [PubMed] [Google Scholar]
- 39.Moss T. Y., Cullis C. A. Computational prediction of candidate micrornas and their targets from the completed Linum ussitatissimum genome and EST database. Journal of Nucleic Acids Investigation. 2012;3(1, article no. e2) doi: 10.4081/jnai.2012.e2. [DOI] [Google Scholar]
- 40.Palatnik J. F., Allen E., Wu X., et al. Control of leaf morphogenesis by microRNAs. Nature. 2003;425(6955):257–263. doi: 10.1038/nature01958. [DOI] [PubMed] [Google Scholar]
- 41.Schommer C., Debernardi J. M., Bresso E. G., Rodriguez R. E., Palatnik J. F. Repression of cell proliferation by miR319-regulated TCP4. Molecular Plant. 2014;7(10):1533–1544. doi: 10.1093/mp/ssu084. [DOI] [PubMed] [Google Scholar]
- 42.Nag A., King S., Jack T. miR319a targeting of TCP4 is critical for petal growth and development in Arabidopsis. Proceedings of the National Academy of Sciences of the United States of America. 2009;106(52):22534–22539. doi: 10.1073/pnas.0908718106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Omidbakhshfard M. A., Proost S., Fujikura U., Mueller-Roeber B. Growth-regulating factors (GRFs): a small transcription factor family with important functions in plant biology. Molecular Plant. 2015;8(7, article 76):998–1010. doi: 10.1016/j.molp.2015.01.013. [DOI] [PubMed] [Google Scholar]
- 44.Axtell M. J., Jan C., Rajagopalan R., Bartel D. P. A two-hit trigger for siRNA biogenesis in plants. Cell. 2006;127(3):565–577. doi: 10.1016/j.cell.2006.09.032. [DOI] [PubMed] [Google Scholar]
- 45.Allen E., Xie Z., Gustafson A. M., Carrington J. C. microRNA-directed phasing during trans-acting siRNA biogenesis in plants. Cell. 2005;121(2):207–221. doi: 10.1016/j.cell.2005.04.004. [DOI] [PubMed] [Google Scholar]
- 46.Adenot X., Elmayan T., Lauressergues D., et al. DRB4-Dependent TAS3 trans-Acting siRNAs Control Leaf Morphology through AGO7. Current Biology. 2006;16(9):927–932. doi: 10.1016/j.cub.2006.03.035. [DOI] [PubMed] [Google Scholar]
- 47.Fahlgren N., Montgomery T. A., Howell M. D., et al. Regulation of AUXIN RESPONSE FACTOR3 by TAS3 ta-siRNA Affects Developmental Timing and Patterning in Arabidopsis. Current Biology. 2006;16(9):939–944. doi: 10.1016/j.cub.2006.03.065. [DOI] [PubMed] [Google Scholar]
- 48.Montgomery T. A., Howell M. D., Cuperus J. T., et al. Specificity of ARGONAUTE7-miR390 interaction and dual functionality in TAS3 trans-acting siRNA formation. Cell. 2008;133(1):128–141. doi: 10.1016/j.cell.2008.02.033. [DOI] [PubMed] [Google Scholar]
- 49.Navarro L., Dunoyer P., Jay F., et al. A plant miRNA contributes to antibacterial resistance by repressing auxin signaling. Science. 2006;312(5772):436–439. doi: 10.1126/science.1126088. [DOI] [PubMed] [Google Scholar]
- 50.Wójcik A. M., Gaj M. D. miR393 contributes to the embryogenic transition induced in vitro in Arabidopsis via the modification of the tissue sensitivity to auxin treatment. Planta. 2016;244(1):231–243. doi: 10.1007/s00425-016-2505-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mockaitis K., Estelle M. Auxin receptors and plant development: a new signaling paradigm. Annual Review of Cell and Developmental Biology. 2008;24:55–80. doi: 10.1146/annurev.cellbio.23.090506.123214. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
MicroRNAs in flax plants under aluminum stress (Table 1: Conserved microRNAs; Table 2: MicroRNA expression; Table 3: MicroRNA targets).