Abstract
Gastric cancer (GC) is one of the most prominent global cancer-related health threats. Genes play a key role in the precise mechanisms of gastric cancer. SNPs in mi-RNAs could affect mRNA expression and then affect the risk and prognosis of GC. Firstly, we have decided to perform a case-control study which included 897 GC patients and 992 controls to evaluate the association of miR-219-1 rs213210, miR-938 rs2505901, miR-34b/c rs4938723, and miR-218 rs11134527 polymorphisms with gastric cancer susceptibility. Secondly, among the 897 GC patients above, 755 cases underwent a radical operation, without distant metastasis and with negative surgical margins included in the survival analysis to evaluate the association of the four SNPs above with gastric cancer prognosis. The C/T or C/C genotypes of rs213210 were related to a lower GC risk (OR = 0.76, 95% CI: 0.62–0.93, P = 0.009) compared to the T/T genotype. Rs11134527 in miR-218 was associated with GC survival, and the G/A and G/G genotypes of rs11134527 resulted in a decreased risk of death when compared with the A/A genotype (HR = 0.75, 95% CI: 0.61–0.95, P = 0.016). This study found that miR-219-1 rs213210 polymorphism was associated with GC susceptibility and rs11134527 in miR-218 was positively correlated with GC prognosis.
1. Introduction
Gastric cancer (GC) is one of the most prominent global cancer-related health threats [1, 2]. A recent report from the National Central Cancer Registry of China showed that GC was the second most common malignant cancer and the third most common cause of cancer-related death in China [3].
MicroRNAs (miRNA) area class of evolutionarily conserved small noncoding RNA that negatively regulates gene expression at a posttranscriptional level by inhibiting translation through binding to the complementary site of a targeted mRNA [4, 5]. Aberrant expression of miRNA may characterize many diseases, including cancer, and these patterns could be used as predictors of susceptibility and prognosis in cancer patients [6, 7]. Various studies have indicated that single nucleotide polymorphisms (SNPs) in the 3′-untranslated region affected the expression levels or the mature sequence of miRNAs and further impacted mRNA and protein expressions [8–10].
Various studies linked miRNA genetic variations in both miRNA encoding genes and in their target genes to GC risk and prognosis. Single nucleotide polymorphisms were the common genetic variations; SNPs in miRNAs could affect mRNA expression by altering the transcription of the primary miRNA transcript and the interaction of miRNA with mRNA [11, 12]. SNPs in different candidate miRNAs (i.e., let-7e rs8111742, miR-365b rs121224, miR-4795 rs1002765, miR-499 rs3746444, and miR-146a rs2910164) have been associated with the susceptibility or prognosis of GC [10, 13, 14]. More recently, Arisawa et al. found that rs2505901 in miRNA-938 was associated with susceptibility to gastric cancer in a Japanese population [15]. Other studies found that miR-34b/c rs4938723 polymorphism was associated with breast cancer and cervical cancer risk [16, 17]. A study by Pardini et al. showed that rs213210 in miR-219-1 was associated with clinical outcomes of colorectal cancer patients [18] and a meta-analysis indicated that miR-218 rs11134527 polymorphism may have an association with many types of cancer [19].
Some of the SNPs above have been studied in some other cancers or populations, but had not been examined in Chinese Han population with regard to gastric cancer. In the present study, we took advantage of these interesting results to investigate whether miR-219-1 rs213210, miR-938 rs2505901, miR-34b/c rs4938723, and miR-218 rs11134527 polymorphisms were related to the risk and clinical outcomes of gastric cancers in a Chinese Han population.
2. Materials and Methods
2.1. Study Population
Gastric cancer patients were enrolled from July 2008 to December 2013 in the Gastric and Colorectal Surgery Department of the First Hospital, Jilin University, Changchun, China. All cases were histologically diagnosed as GC by a pathologist, and no patients had received preoperative chemotherapy or radiotherapy. In total, 897 GC patients were included in the association study between SNPs and GC development. Additionally, tumor-free controls were recruited from the Physical Examination Center in the First Hospital of Jilin University. In all, 992 frequency age- (±5 years) and gender-matched control participants were included in the study.
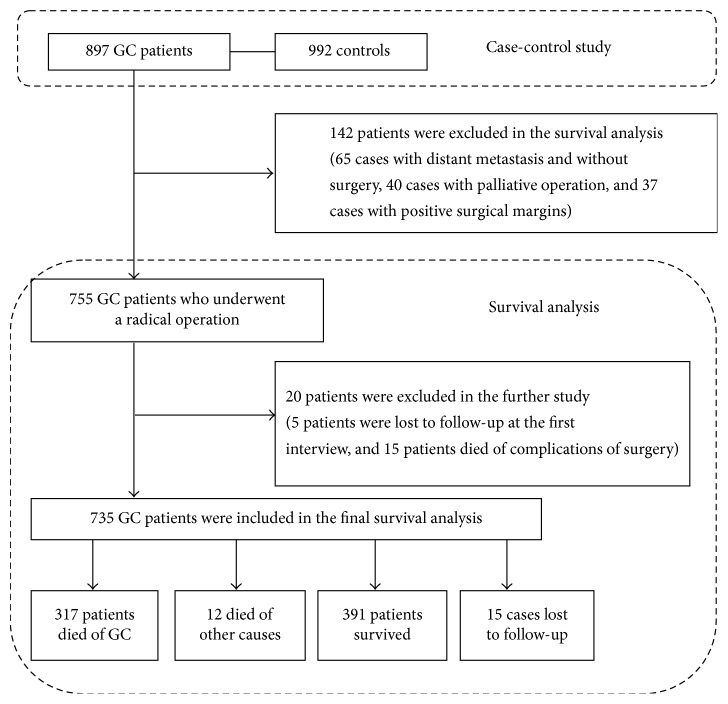
Patients with distant metastasis and positive surgical margins were more susceptible to worse outcomes. To assess the association between SNPs and longer survival status in GC patients who underwent tumor curative resection, patients with distant metastasis and without surgery, with palliative operation, and with positive surgical margins were excluded in the survival analysis. As a result, 755 patients were included in the survival analysis within the 897 GC patients mentioned above, and the flow chart of our study design was shown in Figure 1. The study was approved by the ethics committee of the First Hospital of Jilin University. All subjects in this study signed a written informed consent form.
Figure 1.
The flow chart of study design.
2.2. Data Collection
Clinicopathologic parameters (including tumor sizes, histological grade, WHO classification, vascular invasion, neural invasion, depth invasion, lymph metastasis, clinical stages, and postoperational chemotherapy) were collected from the medical records. The pathological classification of tumors was determined according to the AJCC/UICC, 2010 classifications. Postoperational chemotherapy was defined as at least three cycles of chemotherapy that were received after surgery and that were divided into FOLFOX-4 (a combination of 5-fluorouracil, leucovorin and oxaliplatin); XELOX (a combination of capecitabine and oxaliplatin); other (such as capecitabine or 5-fluorouracilalone); or none.
Gastric cancer patients were followed up on the third month, sixth month, and first year after the tumorectomy and every subsequent year until death or the end of our study. Survival time was defined as the duration from the date of surgery to the date of death (if patients died) or to the date of the last successful interview (if patients were alive or lost to follow-up). Patients with palliative operation, distant metastasis, or positive surgical margins were excluded in the survival analysis. Patients who died from complications of the surgical operation in the preoperative period or were lost to follow-up at the first time of interview were also excluded from the survival analysis.
2.3. Tests of Helicobacter pylori (H. pylori) Infection
Enzyme-linked immune absorbent assay kit (Biohit, Finland) was used to determinate the H. pylori infection. Titers higher than 30 EIU were classified as positive for H. pylori infection according to the manufacturer's instruction.
2.4. Genotyping
Four SNPs in miRNAs (miR-219-1 rs213210, miR-938 rs2505901, miR-34b/c rs4938723, and miR-218 rs11134527) were selected based on previous studies that documented associations between SNPs and GC or other cancers in other ethnicities. The minor allele frequency (MAF) of all the SNPs was greater than 0.05 in the Chinese Han population.
Blood samples of GC cases were collected before the gastric cancer resection, and fasting blood samples of controls were collected during the same period with GC groups. Both case and control blood samples were stored at −80°C in EDTA tubes. Genomic DNA was extracted following the manufacturer's instructions (AxyPrep Blood Genomic DNA Miniprepkit, Axygen, Union City, CA, USA). Genotyping of each SNP was conducted using the MassARRAY technology platform (Sequenom, CA, USA) and determined by BGI tech (Beijing, China). The detection rates for the SNPs miR-219-1 rs213210, miR-938 rs2505901, miR-34b/c rs4938723, and miR-218 rs11134527 were 99.3%, 99.7%, 99.6%, and 99.7%, respectively. 1% of samples were randomly selected and tested repeatedly, and the concordance rates were 100% for all the four SNPs.
2.5. Statistical Analyses
Continuous variables with a normal distribution were described as the mean ± standard deviation and compared with Student's t-test. Discrete variables were described as frequency (percentage) and compared using the χ2 test. For each SNP, test of the Hardy-Weinberg disequilibrium (HWD) was conducted. Associations between the SNPs and GC were computed using logistic regression model adjusted for age, gender, and H. pylori infection. The Kaplan–Meier method was used to plot the survival curves, and the log-rank test was conducted for comparing the survival curves between different genotypes within each SNP. Univariate and multivariate Cox regression models were performed to assess the hazard ratios (HRs) and 95% CIs of the possible prognostic factors. All of the analyses were conducted using the SPSS program (version 17.0, Chicago, IL, USA), and P < 0.05 was considered to be statistically significant.
3. Results
3.1. Subject Characteristics
A total of 1889 participants (897 gastric cancer cases and 992 tumor-free controls) were enrolled in our study. The characteristics of the subjects were shown in Table 1. The gender ratio between the gastric cancer group and the control group was not significantly different (P = 0.898). As a result of frequency matching by age (±5 years), the average age was two years younger in the control group compared to the GC group. The H. pylori-positive rate was significantly higher in GC patients than that in controls (P < 0.05).
Table 1.
Characteristics, genotype distributions, and allele frequencies of SNPs between GC and controls.
| Characteristics | Gastric cancer (n = 897) | Controls (n = 992) | P |
|---|---|---|---|
| Age | 61.04 ± 11.34 | 59.15 ± 10.04 | <0.001 |
| Gender | |||
| Male | 648 (72.2%) | 714 (72.0%) | 0.898 |
| Female | 249 (27.8%) | 278 (28.0%) | |
| H. pylori infection | |||
| Yes | 603 (67.6%) | 487 (49.1%) | <0.001 |
| No | 289 (32.4%) | 505 (50.9%) | |
| Rs213210 | |||
| T/T | 266 (30.0%) | 241 (24.4%) | 0.020 |
| C/T | 422 (47.6%) | 519 (52.5%) | |
| C/C | 199 (22.4%) | 229 (23.1%) | |
| T | 954 (53.8%) | 1001 (50.6%) | 0.052 |
| C | 820 (46.2%) | 977 (47.9%) | |
| Rs2505901 | |||
| T/T | 587 (65.5%) | 635 (64.3%) | 0.853 |
| C/T | 274 (30.6%) | 313 (31.7%) | |
| C/C | 35 (3.9%) | 40 (4.0%) | |
| T | 1448 (80.8%) | 1583 (80.1%) | 0.593 |
| C | 344 (19.9%) | 393 (19.9%) | |
| Rs4938723 | |||
| T/T | 405 (45.4%) | 476 (48.1%) | 0.287 |
| C/T | 396 (44.3%) | 430 (43.4%) | |
| C/C | 92 (10.3%) | 84 (8.5%) | |
| T | 1206 (67.5%) | 1382 (69.8%) | 0.133 |
| C | 580 (32.5%) | 598 (30.2%) | |
| Rs11134527 | |||
| A/A | 345 (38.6%) | 394 (39.8%) | 0.737 |
| A/G | 412 (46.1%) | 439 (44.3%) | |
| G/G | 137 (15.3%) | 158 (15.9%) | |
| A | 1102 (61.6%) | 1227 (61.9%) | 0.867 |
| G | 686 (38.4%) | 755 (38.2%) |
Data were described as mean ± SD or N (%).
3.2. Allele Frequency Comparisons
The distributions of 4 SNPs (rs213210, rs2505901, rs4938723, and rs11134527) were all in Hardy-Weinberg equilibrium (HWE) in the control group. The comparisons of genotype distributions and allele frequencies of the 4 SNPs between GC and controls were shown in Table 1. Allele frequency differences between the two groups were not observed for any of the 4 SNPs. A significant difference was only revealed in the rs213210 genotype distributions between the two groups (P = 0.020). Additionally, in the control group, the C/C genotype of rs213210 was associated with lower risk of H. pylori infection (OR = 0.66, 95% CI: 0.46–0.95, P = 0.025, Table S1 in Supplementary Material available online at https://doi.org/10.1155/2017/4731891).
3.3. Association of SNPs with Risk of Gastric Cancer
Genotype distribution of codominant and dominant models of four SNPs was shown in Table 2. Compared to the T/T genotype, the C/T or C/C genotypes of rs213210 were associated with a lower GC risk (OR = 0.76, 95% CI = 0.62–0.93, P = 0.009) after adjusting for the confounding factors of age, gender, and H. pylori infection.
Table 2.
Genotype distribution comparisons and odds ratio (OR) estimates of 4 SNPs for GC.
| Genotype | Case | Control | OR (95% CI)a | P a |
|---|---|---|---|---|
| Rs213210 | ||||
| T/T | 266 (30.0%) | 241 (24.4%) | 1.00 | 0.026 |
| C/T | 422 (47.6%) | 519 (52.5%) | 0.74 (0.60–0.92) | |
| C/C | 199 (22.4%) | 229 (23.1%) | 0.80 (0.62–1.04) | |
| C/T-C/C | 621 (70.0%) | 748 (75.6%) | 0.76 (0.62–0.93) | 0.009 |
| Rs2505901 | ||||
| T/T | 587 (65.5%) | 635 (64.3%) | 1.00 | 0.870 |
| C/T | 274 (30.6%) | 313 (31.7%) | 0.95 (0.78–1.16) | |
| C/C | 35 (3.9%) | 40 (4.0%) | 0.93 (0.58–1.49) | |
| C/T-C/C | 309 (34.5%) | 353 (35.7%) | 0.95 (0.79–1.15) | 0.600 |
| Rs4938723 | ||||
| T/T | 405 (45.4%) | 476 (48.1%) | 1.00 | 0.230 |
| C/T | 396 (44.3%) | 430 (43.4%) | 1.09 (0.90–1.32) | |
| C/C | 92 (10.3%) | 84 (8.5%) | 1.32 (0.95–1.82) | |
| C/T-C/C | 488 (54.6%) | 514 (51.9%) | 1.13 (0.94–1.35) | 0.200 |
| Rs11134527 | ||||
| A/A | 345 (38.6%) | 394 (39.8%) | 1.00 | 0.700 |
| A/G | 412 (46.1%) | 439 (44.3%) | 1.07 (0.88–1.31) | |
| G/G | 137 (15.3%) | 158 (15.9%) | 0.97 (0.74–1.28) | |
| A/G-G/G | 549 (61.4%) | 597 (60.2%) | 1.05 (0.87–1.26) | 0.063 |
ORs and P values were adjusted by age, gender, and H. pylori infection using logistic regression model. Here, a means adjusted P value.
3.4. Association between SNPs and Survival of Gastric Cancer
Among the 897 GC patients mentioned above, 65 cases were with distant metastasis and without surgery, 40 cases with palliative operation, and 37 cases with positive surgical margins. These cases were excluded from the survival analysis. Among the remaining 755 cases, 5 patients were lost to follow-up at the first interview, 15 patients died of complications of surgery, and these 20 (2.2%) cases were also excluded from survival analysis. Finally, 735 patients were included in the final survival analysis. The median follow-up time was 51.64 months (ranging from 1.61 to 94.32 months). During the follow-up, 317 (43.1%) patients died of GC, 12 (1.6%) died of other causes, 391 (53.2%) patients survived, and 15 (2.0%) cases were lost to follow-up; see Figure 1.
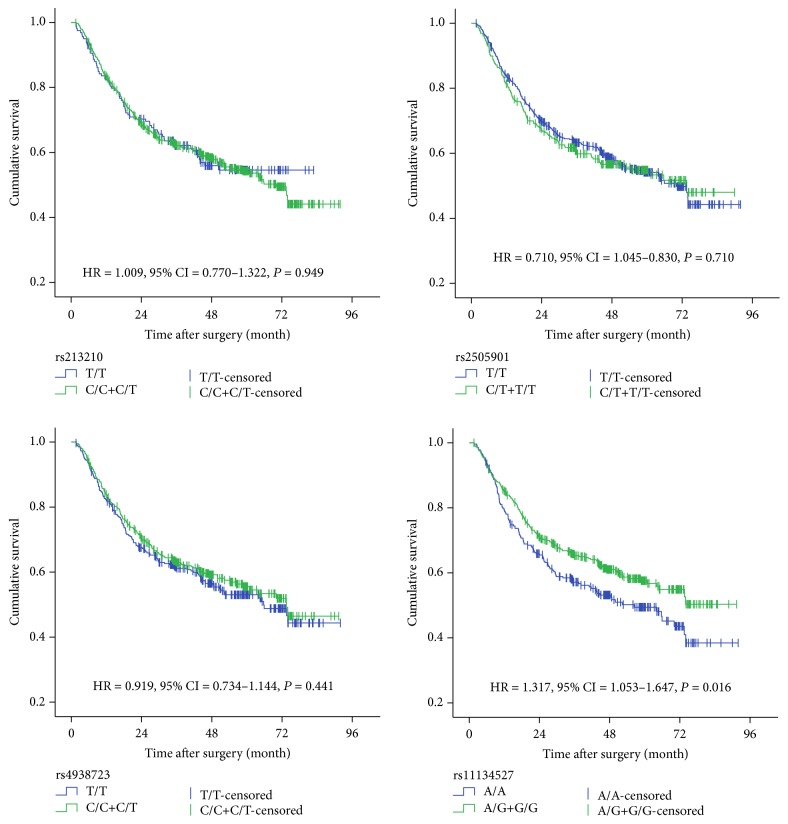
Survival analysis was performed to evaluate the associations of clinicopathologic parameters and SNP genotypes on survival of gastric cancer using the dominant model. As shown in Table 3, tumor size ≥5 cm, high histological grade, T3/T4 depth of invasion, N1/N2/N3 lymph metastasis, and higher TNM stage were associated with worse survival of GC. In this study, 48.5% of patients carrying the rs11134527 A/A genotype died of GC compared to only 39.5% of cases with A/G-G/G genotypes. Patients bearing genotypes A/G-G/G of rs11134527 were found to live longer than those bearing genotype A/A (HR = 0.75, 95% CI: 0.61–0.95, P = 0.016) (Figure 2). No significant associations were observed between SNPs and other clinic pathological parameters such as tumor size, histological grade, WHO classification, TNM stage, distant metastasis, and chemotherapy (data not shown).
Table 3.
Univariate analysis for prognostic factors of gastric cancer.
| Characteristic | Patient n | Death n (%) | Mean OS∗ (months) | P † |
|---|---|---|---|---|
| Age (year) | ||||
| ≤45 years | 65 | 29 (44.6) | 54.01 | 0.935 |
| >45 years | 670 | 287 (42.8) | 57.42 | |
| Gender | ||||
| Male | 539 | 237 (44.0) | 56.55 | 0.343 |
| Female | 196 | 79 (40.3) | 59.35 | |
| Tumor size | ||||
| <5 cm | 422 | 144 (34.1) | 64.89 | <0.001 |
| ≥5 cm | 302 | 165 (54.6) | 47.22 | |
| Histological grade | ||||
| low grade | 309 | 118 (38.2) | 61.85 | 0.006 |
| high grade | 373 | 177 (47.5) | 52.74 | |
| WHO classification | ||||
| Tubular | 569 | 238 (41.8) | 58.55 | 0.167 |
| Signet-ring cell | 36 | 18 (50.0) | 50.57 | |
| Others | 130 | 60 (46.2) | 45.23 | |
| Vascular invasion | ||||
| Negative | 217 | 39 (18.0) | 76.06 | <0.001 |
| Positive | 506 | 270 (53.4) | 49.61 | |
| Neural invasion | ||||
| Negative | 330 | 97 (29.4) | 68.19 | <0.001 |
| Positive | 393 | 212 (53.9) | 48.43 | |
| Depth of invasion | ||||
| T1/T2 | 198 | 26 (13.1) | 80.23 | <0.001 |
| T3/T4 | 515 | 278 (54.0) | 49.00 | |
| Lymph metastasis | ||||
| N0 | 214 | 33 (15.4) | 76.15 | <0.001 |
| N1/N2/N3 | 501 | 271 (54.1) | 48.79 | |
| TNM stage | ||||
| I | 136 | 12 (8.8) | 81.13 | <0.001 |
| II | 278 | 86 (30.9) | 67.90 | |
| III | 312 | 212 (67.9) | 36.55 | |
| Chemotherapy | ||||
| None | 486 | 200 (41.2) | 57.68 | 0.056 |
| FOLFOX-4 | 137 | 66 (48.2) | 52.37 | |
| XELOX | 74 | 26 (35.1) | 62.25 | |
| Others | 38 | 24 (63.2) | 42.63 | |
| Rs213210 | ||||
| T/T | 159 | 67 (42.1) | 54.24 | 0.949 |
| C/C+C/T | 566 | 245 (43.3) | 57.14 | |
| Rs2505901 | ||||
| T/T | 476 | 204 (42.9) | 57.52 | 0.710 |
| C/T-C/C | 258 | 112 (43.4) | 56.11 | |
| Rs4938723 | ||||
| T/T | 330 | 148 (44.8) | 65.71 | 0.440 |
| C/C-C/T | 401 | 166 (41.4) | 73.50 | |
| Rs11134527 | ||||
| A/A | 276 | 134 (48.5) | 52.93 | 0.016 |
| A/G-G/G | 456 | 180 (39.5) | 60.01 |
∗For most characteristics, less than half patients were dead, so mean overall survival (OS) time was presented for most of the median OS that could not be calculated. †P values were computed by the log-rank test.
Figure 2.
Survival plots for SNPs in miRNAs using the dominant model of gastric cancer patients.
As shown in Table 4, the results from the multivariate Cox regression test showed that rs11134527 was an independent prognostic factor. Patients carrying G/A or G/G genotypes lived longer than patients with the A/A genotype (HR = 0.73, 95% CI: 0.58–0.93, P = 0.010). Additionally, positive vascular invasion, tumor size ≥5 cm, and higher TNM stage (II and III) were also independently associated with worse prognosis. Receiving standard XELOX chemotherapy after tumor resection was associated with better OS (HR: 0.51, 95% CI: 0.33–0.80, P = 0.003).
Table 4.
Multivariate Cox regression test for prognostic factors of gastric cancer.
| Variable | HR | 95% CI | P |
|---|---|---|---|
| Vascular invasion | |||
| Negative | 1 | — | — |
| Positive | 1.90 | 1.27–2.76 | 0.002 |
| Tumor size | |||
| <5 cm | 1 | — | — |
| ≥5 cm | 1.41 | 1.11–1.80 | 0.005 |
| TNM stage | |||
| I | 1 | — | — |
| II | 2.86 | 1.44–5.67 | 0.003 |
| III | 9.60 | 4.82–19.02 | <0.001 |
| Chemotherapy | |||
| None | 1 | — | — |
| FOLFOX-4 | 0.90 | 0.67–1.20 | 0.444 |
| XELOX | 0.51 | 0.33–0.80 | 0.003 |
| Others | 0.99 | 0.64–1.56 | 0.988 |
| Rs11134527 | |||
| A/A | 1 | — | — |
| G/A-G/G | 0.73 | 0.58–0.93 | 0.010 |
HR: Hazard ratio. P values were calculated with multivariate Cox regression with the stepwise selection method including all the variables at initiation.
4. Discussion
In the present work, we assessed the association between four SNPs in miRNAs and susceptibility or prognosis in gastric cancer patients. Among them, rs213210 in miR-219-1 was associated with the risk of GC. Compared to the T/T genotype, patients carrying the C/T and C/C genotypes of rs213210 had a lower GC risk. On the other hand, rs11134527 in miR-218 was associated with GC survival; the G/A or G/G genotypes of rs11134527 resulted in a decreased risk of death when compared with the A/A genotype.
MicroRNAs are involved in various crucial biological processes by targeting hundreds of mRNAs that take part in cell proliferation, differentiation, apoptosis, and the progression of cancer [20]. Single nucleotide polymorphisms in miRNAs could alter the transcription of the primary miRNA transcript and the interaction of miRNA with mRNA, thus affecting mRNA expression [11, 12]. Moreover, researchers have found that half of the miRNA genes may be located in cancer-related regions [21]. As a result, SNPs in these miRNAs could be associated with cancer risk or prognosis [22]. Though there were many GWAS results have been published, but they always focused on the SNPs on coding genes, not on miRNA, and refer only to the cancer risk but not the cancer prognosis. The present study aimed to assess the association between miR-219-1 rs213210, miR-938 rs2505901, miR-34b/c rs4938723, and miR-218 rs11134527 polymorphisms not only with GC susceptibility but also with GC prognosis.
For rs213210 in miR-219-1, only a few studies have investigated the role of this polymorphism in cancer susceptibility. Song et al. reported that there was no association between the rs213210 polymorphism and risk of esophageal squamous cell carcinoma in Chinese [23]. Yoon, K. A et al. found that no significant association emerged between this polymorphism and survival of nonsmall cell lung cancer. Further, Pardini et al. thought that rs231210 could be a predictor of clinical outcomes in colorectal cancer patients [8]. However, our results suggested that rs231210 was associated with GC susceptibility but not associated with GC prognosis. To some extent, it was not suitable to compare the results of the prior studies to ours since there were different ethnicities and cancer categories involved in each. We used F-SNP online software (http://compbio.cs.queensu.ca/F-SNP/) to predict the function of rs231210 and found that this SNP is located in a region involved in transcriptional regulation. The change from T to C of rs231210 could have an effect upon the expression of miR-219-1. Research on the targeting gene of miR-219-1 found that mature miR-219-1 is involved in the pathway related to transcription regulation, regulation of RNA metabolic processes, transcription from RNA polymerase II promoter, and gene expression [18]. Another study indicated that upexpression of miR-219-1-3p induced a decrease of cell proliferation and migration in pancreatic cancer by negatively regulating expression of the mucin MUC4. The studies could partially confirm the importance of the SNPs in miR-219-1-related genes in the mechanisms of cancer risk.
The association between rs11134527 in miR-218 and cancer risk is tumor location-dependent. A meta-analysis showed that rs11134527 polymorphism was associated with risk of cervical cancer but not hepatocellular carcinoma [24] In the present study, no association was observed between rs11134527 and GC risk in all of the four genetic models (dominant model, recessive model, codominant model, and overdominant model). Jiang et al. found that the G/G genotype of rs11134527 was significantly associated with a better prognosis of esophageal squamous cell carcinoma in a codominant model [25]. In our study, we used the dominant model to assess the association between rs11134527 and GC and found that people carrying the G allele lived longer. Then, we used online software to search the target gene of miR-218. Finally, twenty genes were validated according to the miRWalk tool [26]. Some of the target genes were closely related to cancer development, such as CDH2, ZEB2, MAP3K2, and SLC24A4. Other target genes of miR-218 that have been identified by previous researchers including BMI1, LAMB3, and LASP1 and the expressions of these genes were associated with the invasion or prognosis in different cancer types [27–29]. In addition to this, we evaluated the expression levels of miR-218 among 359 GC patients in the TCGA dataset and observed that higher miR-218 expression level was associated with a better survival of GC (P = 0.002). Recent research also confirmed that lower miR-218 tissue expression level was associated with aggressive progression of gastric cancer [30]. Meanwhile, miR-218 could inhibit invasion and metastasis of gastric cancer by targeting the Robo1 receptor [31]. RNAfold prediction analysis showed that the transition from A to G of rs11134527 could alter the local second structure of miR-218 [32]. Thus, we supposed that the expression level of miR-218 may be partly relative to SNPs and more researches are needed in the future.
Another study found that rs2505901 in miR-938 was associated with susceptibility to gastric cancer in a Japanese population [15] and other studies found that miR-34b/c rs4938723 polymorphism was associated with risk of breast cancer and cervical cancer. However, the present study indicated that miR-938 rs2505901 and miR-34b/c rs4938723 were associated neither with gastric cancer nor with susceptibility or with prognosis. The different cancer types, ethnicities, and sample sizes might contribute to these discrepancies.
5. Conclusions
In conclusion, rs213210 in miR-219-1 was associated with the risk of GC. Compared to the T/T genotype, the C/T and C/C genotypes of rs213210 were associated with a lower GC risk. On the other hand, rs11134527 in miR-218 was associated with GC survival in patients without distant metastasis and without positive surgical margins. The G/A and G/G genotypes of rs11134527 resulted in decreased risk of death when compared with the A/A genotype. More laboratory study is needed to clarify the underlying mechanism of SNPs in miRNAs.
Supplementary Material
Relationship of H. pylori infection and SNPs in the control group shown that, the C/C genotype of re213210 was associated with lower risk of H. pylori infection (OR = 0.66, 95% CI: 0.46-0.95, P = 0.025).
Acknowledgments
This work was supported by the National Natural Science Foundation of China (nos. 81373084 and 81273065), the Scientific and Technological Development Program of Jilin Province (no. 20160519016JH), the Norman Bethune Program of Jilin University (no. 2013025), and the Youth Fund of the First Hospital of Jilin University (no. JDYY72016049).
Competing Interests
The authors declare that there are no competing interests regarding the publication of this paper.
Authors' Contributions
Yanhua Wu and Zhifang Jia contributed equally to this work and were listed as co-first authors.
References
- 1.Leung W. K., Wu M.-S., Kakugawa Y., et al. Screening for gastric cancer in Asia: current evidence and practice. The Lancet Oncology. 2008;9(3):279–287. doi: 10.1016/S1470-2045(08)70072-X. [DOI] [PubMed] [Google Scholar]
- 2.Van Cutsem E., Sagaert X., Topal B., Haustermans K., Prenen H. Gastric cancer. The Lancet. 2016;388(10060):2654–2664. doi: 10.1016/s0140-6736(16)30354-3. [DOI] [PubMed] [Google Scholar]
- 3.Chen W., Zheng R., Baade P. D., et al. Cancer statistics in China, 2015. CA Cancer Journal for Clinicians. 2016;66(2):115–132. doi: 10.3322/caac.21338. [DOI] [PubMed] [Google Scholar]
- 4.Kong Y. W., Ferland-McCollough D., Jackson T. J., Bushell M. MicroRNAs in cancer management. The Lancet Oncology. 2012;13(6):e249–e258. doi: 10.1016/S1470-2045(12)70073-6. [DOI] [PubMed] [Google Scholar]
- 5.Banaudha K. K., Verma M. The role of MicroRNAs in the management of liver cancer. Methods in Molecular Biology. 2012;863:241–251. doi: 10.1007/978-1-61779-612-8_14. [DOI] [PubMed] [Google Scholar]
- 6.Dalmay T., Edwards D. R. MicroRNAs and the hallmarks of cancer. Oncogene. 2006;25(46):6170–6175. doi: 10.1038/sj.onc.1209911. [DOI] [PubMed] [Google Scholar]
- 7.Schetter A. J., Leung S. Y., Sohn J. J., et al. MicroRNA expression profiles associated with prognosis and therapeutic outcome in colon adenocarcinoma. JAMA. 2008;299(4):425–436. doi: 10.1001/jama.299.4.425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pardini B., Rosa F., Barone E., et al. Variation within 3'-UTRs of base excision repair genes and response to therapy in colorectal cancer patients: a potential modulation of microRNAs binding. Clinical Cancer Research. 2013;19(21):6044–6056. doi: 10.1158/1078-0432.ccr-13-0314. [DOI] [PubMed] [Google Scholar]
- 9.Naccarati A., Pardini B., Stefano L., et al. Polymorphisms in mirna-binding sites of nucleotide excision repair genes and colorectal cancer risk. Carcinogenesis. 2012;33(7):1346–1351. doi: 10.1093/carcin/bgs172. [DOI] [PubMed] [Google Scholar]
- 10.Jiang J., Jia Z. F., Cao D. H., Wu Y. H., Sun Z. W., Cao X. Y. Association of the miR-146a rs2910164 polymorphism with gastric cancer susceptibility and prognosis. Future Oncology. 2016;12(19):2215–2226. doi: 10.2217/fon-2016-0224. [DOI] [PubMed] [Google Scholar]
- 11.Ryan B. M., Robles A. I., Harris C. C. Genetic variation in microRNA networks: the implications for cancer research. Nature Reviews Cancer. 2010;10(6):389–402. doi: 10.1038/nrc2867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Berezikov E., Guryev V., Van De Belt J., Wienholds E., Plasterk R. H. A., Cuppen E. Phylogenetic shadowing and computational identification of human microRNA genes. Cell. 2005;120(1):21–24. doi: 10.1016/j.cell.2004.12.031. [DOI] [PubMed] [Google Scholar]
- 13.Xu Q., Wu Y.-F., Li Y., et al. SNP-SNP interactions of three new pri-miRNAs with the target gene PGC and multidimensional analysis of H. pylori in the gastric cancer/atrophic gastritis risk in a Chinese population. Oncotarget. 2016;7(17):23700–23714. doi: 10.18632/oncotarget.8057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cai M., Zhang Y., Ma Y., et al. Association between microRNA-499 polymorphism and gastric cancer risk in Chinese population. Bulletin du Cancer. 2015;102(12):973–978. doi: 10.1016/j.bulcan.2015.09.012. [DOI] [PubMed] [Google Scholar]
- 15.Arisawa T., Tahara T., Shiroeda H., et al. Genetic polymorphisms of IL17A and pri-microRNA-938, targeting IL17A 3′-UTR, influence susceptibility to gastric cancer. Human Immunology. 2012;73(7):747–752. doi: 10.1016/j.humimm.2012.04.011. [DOI] [PubMed] [Google Scholar]
- 16.Sanaei S., Hashemi M., Rezaei M., Hashemi S. M., Bahari G., Ghavami S. Evaluation of the pri-miR-34b/c rs4938723 polymorphism and its association with breast cancer risk. Biomedical Reports. 2016;5(1):125–129. doi: 10.3892/br.2016.690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yuan F., Sun R., Chen P., et al. Combined analysis of pri-miR-34b/c rs4938723 and TP53 Arg72Pro with cervical cancer risk. Tumor Biology. 2016;37(5):6267–6273. doi: 10.1007/s13277-015-4467-y. [DOI] [PubMed] [Google Scholar]
- 18.Pardini B., Rosa F., Naccarati A., et al. Polymorphisms in microRNA genes as predictors of clinical outcomes in colorectal cancer patients. Carcinogenesis. 2015;36(1):82–86. doi: 10.1093/carcin/bgu224. [DOI] [PubMed] [Google Scholar]
- 19.Zhao H., Wang R. Quantitative assessment of pre-miR-218 rs11134527 polymorphism and cancer risk in Chinese population. OncoTargets and Therapy. 2015;8:1859–1862. doi: 10.2147/OTT.S88480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Abba M. L., Patil N., Leupold J. H., et al. MicroRNAs as novel targets and tools in cancer therapy. Cancer Letters. 2017;387:84–94. doi: 10.1016/j.canlet.2016.03.043. [DOI] [PubMed] [Google Scholar]
- 21.Garzon R., Marcucci G., Croce C. M. Targeting microRNAs in cancer: rationale, strategies and challenges. Nature Reviews Drug Discovery. 2010;9(10):775–789. doi: 10.1038/nrd3179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ma X. P., Zhang T., Peng B., Yu L., Jiang D. K. Association between microRNA polymorphisms and cancer risk based on the findings of 66 case-control studies. PLoS ONE. 2013;8(11) doi: 10.1371/journal.pone.0079584.e79584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Song X., You W., Zhu J., et al. A genetic variant in miRNA-219-1 is associated with risk of esophageal squamous cell carcinoma in chinese kazakhs. Disease Markers. 2015;2015:10. doi: 10.1155/2015/541531.541531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gao Y., Liu Y., Liu G.-L., et al. Association between the pre-mir-218 polymorphism and cancer risk in the Chinese population: a meta-analysis. Asian Pacific Journal of Cancer Prevention. 2014;15(6):2517–2522. doi: 10.7314/apjcp.2014.15.6.2517. [DOI] [PubMed] [Google Scholar]
- 25.Jiang L., Wang C., Sun C., et al. The impact of pri-miR-218 rs11134527 on the risk and prognosis of patients with esophageal squamous cell carcinoma. International Journal of Clinical and Experimental Pathology. 2014;7(9):6206–6212. [PMC free article] [PubMed] [Google Scholar]
- 26.Dweep H., Gretz N. MiRWalk2.0: a comprehensive atlas of microRNA-target interactions. Nature Methods. 2015;12(8):p. 697. doi: 10.1038/nmeth.3485. [DOI] [PubMed] [Google Scholar]
- 27.Yamamoto N., Kinoshita T., Nohata N., et al. Tumor suppressive microRNA-218 inhibits cancer cell migration and invasion by targeting focal adhesion pathways in cervical squamous cell carcinoma. International Journal of Oncology. 2013;42(5):1523–1532. doi: 10.3892/ijo.2013.1851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kinoshita T., Hanazawa T., Nohata N., et al. Tumor suppressive microRNA-218 inhibits cancer cell migration and invasion through targeting laminin-332 in head and neck squamous cell carcinoma. Oncotarget. 2012;3(11):1386–1400. doi: 10.18632/oncotarget.709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.He X., Dong Y., Wu C. W., et al. MicroRNA-218 inhibits cell cycle progression and promotes apoptosis in colon cancer by downregulating BMI1 polycomb ring finger oncogene. Molecular Medicine. 2012;18(12):1491–1498. doi: 10.2119/molmed.2012.00304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang X. X., Ge S. J., Wang X. L., Jiang L. X., Sheng M. F., Ma J. J. miR-218 tissue expression level is associated with aggressive progression of gastric cancer. Genetics and Molecular Research. 2016;15(2) doi: 10.4238/gmr.15027521. [DOI] [PubMed] [Google Scholar]
- 31.Tie J., Pan Y., Zhao L., et al. MiR-218 inhibits invasion and metastasis of gastric cancer by targeting the robo1 receptor. PLoS Genetics. 2010;6(3) doi: 10.1371/journal.pgen.1000879.e1000879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shi T.-Y., Chen X.-J., Zhu M.-L., et al. A pri-miR-218 variant and risk of cervical carcinoma in Chinese women. BMC Cancer. 2013;13, article 19 doi: 10.1186/1471-2407-13-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Relationship of H. pylori infection and SNPs in the control group shown that, the C/C genotype of re213210 was associated with lower risk of H. pylori infection (OR = 0.66, 95% CI: 0.46-0.95, P = 0.025).