Abstract
The neutrophil is the major phagocyte and the final effector cell of the innate immunity, with a primary role in the clearance of extracellular pathogens. Using the broad array of cytokines, extracellular traps, and effector molecules as the humoral arm, neutrophils play a crucial role in the host defense against pathogen infections. On the other hand, the pathogen has the capacity to overcome neutrophil-mediated host defense to establish infection causing human disease. Pathogens, such as S. aureus, have the potential to thwart neutrophil chemotaxis and phagocytosis and thereby succeed in evading killing by neutrophils. Furthermore, S. aureus surviving within neutrophils promotes neutrophil cytolysis, resulting in the release of host-derived molecules that promote local inflammation. Here, we provide a detailed overview of the mechanisms by which neutrophils kill the extracellular pathogens and how pathogens evade neutrophils degradation. This review will provide insights that might be useful for the development of novel therapies against infections caused by antibiotic resistant pathogens.
1. Introduction
The immune system protects the body from microbes that invade and harm the host. In humans roughly 100 billion neutrophils enter and leave circulating blood every day [1] and constitute the dominant leukocyte population in the circulation, mediate the earliest innate immune responses to infection, and play a pivotal role in the resolution of microbial infections. Neutropenia, an acquired or inherited neutropenia, and neutrophil malfunction result in recurrent, life-threatening infections with bacteria [2].
Neutrophils originate and mature in the bone marrow and are subsequently released into the peripheral vasculature. After a pathogen has breached the epithelial barriers, neutrophils are the first innate immune cells that are rapidly recruited from the bloodstream to sites of infection. Pathogens entry and replication in host tissues lead to the release of exogenous products, such as formyl peptides, lipoproteins, or peptidoglycan. Moreover, the invasive pathogen can also damage body tissues that produce inflammatory signals, for example, chemoattractants and cytokines [3]. These pathogenic products and inflammatory signals are detected by neutrophils via Toll-like receptors (TLRs), G protein-coupled receptors (GPCR), and cognate immune receptors. By sensing the receptor signal, neutrophils will respond to these stimuli, extravasate from blood vessels, and migrate towards the site of infection to phagocytose pathogens. This multistep process encompasses rolling adhesion of neutrophils on endothelial cells, firm adhesion of neutrophils, extravasation through the endothelium, chemotactic migration, and subsequent killing of invading bacterial pathogens. Following migration to the site of infection and phagocytosis, neutrophils have a repertoire of antimicrobial arsenal at their disposal to fulfil this function [4]. Neutrophils utilize a combination of NADPH oxidase-derived reactive oxygen species (ROS), cytotoxic granule components, antimicrobial peptides, and neutrophil extracellular traps (NETs) to generate a highly lethal environment that is essential for efficient microbe killing and degradation [5, 6].
On the other hand, many pathogens have evolved efficient strategies to outfox the weaponry of neutrophils. The main strategies can be divided into five categories: evading extravasation and chemotaxis, preventing opsonization and phagocytosis, surviving inside the neutrophil, inducing cell death, and avoiding killing in NETs [7, 8]. In this review, we will highlight the suite of mechanisms employed by neutrophils to clear bacterial infections and the corresponding counterattack mounted by bacterial pathogens.
2. Neutrophil-Mediated Phagocytosis of Pathogenic Microorganism
Initial elimination of invading pathogenic microorganism from human tissue is mediated by professional phagocytes. For efficient phagocytosis, neutrophils first need to leave the bloodstream and reach the site of infection, termed neutrophil recruitment. Furthermore, initiation of phagocytosis requires decoration of bacteria with opsonins that are recognized by specific surface receptors, of which process is termed opsonization of microbes. Lastly, neutrophils express numerous receptors that recognize microbe via binding its specific molecules and host proteins (such as IgG and complement), termed pathogen recognition.
2.1. Neutrophils Migrate from the Bloodstream to the Site of Infection
Upon the breach of epithelium by pathogens, as the first responder to microbial invasion, neutrophils leave the bloodstream and move to the site of infection. This recruitment process consists of three major steps: initiation of adherence to activated endothelial cells and rolling, neutrophil arrest caused by firm attachment to the endothelium, and finally migrating across the endothelial barrier to the infection site.
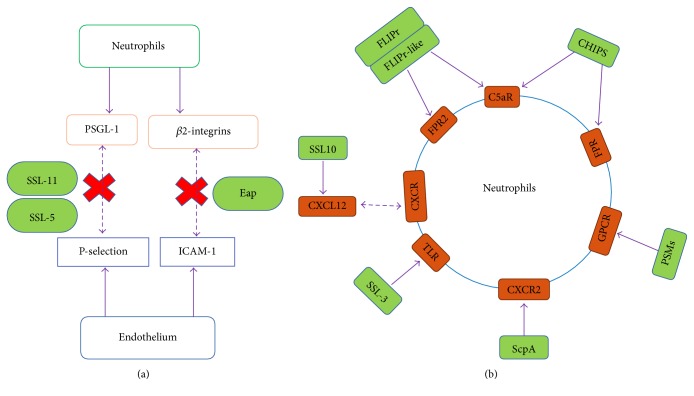
The initial step occurs through the interaction between the glycoprotein P-selectin glycoprotein ligand-1 (PSGL-1) of neutrophils and P-selectin/E-selectin of endothelial cells [9] (Figure 1(a)). Owing to this loose adhesion, neutrophils can roll along the endothelial cells. The second step is dependent on the interaction between β2 integrins (such as LFA-1 and Mac-1) present on the surface of neutrophils and intercellular adhesion molecule 1 (ICAM-1) present on endothelial cells (Figure 1(a)). The final step is triggered by chemokines released by host cells and bacterial products. Host-derived chemokine, such as IL8, GRO-α, granulocyte chemotactic protein 2, and complement component C5a/C3a, are potent proinflammatory mediators that are used to recruit additional neutrophils to areas of infection. Furthermore, neutrophils migration also can be elicited by bacteria-derived chemokine, such as lipoteichoic acid or N-formyl peptides (fMLP).
Figure 1.
Evasion of neutrophil adhesion and transmigration. (a) Mechanisms by which Staphylococcus aureus subverts neutrophil extravasation. (b) Neutrophil attack and evasion of activation.
2.2. Neutrophil Phagocytosis Is Dependent on Opsonization of Microbes
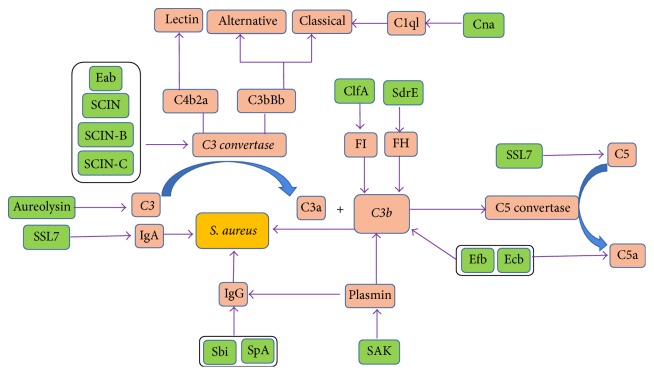
Initiation of neutrophil phagocytosis is dependent on opsonization of the target microbes that are recognized by specific surface receptors of neutrophils. Complement components and immunoglobulins (Igs) are the predominant factor in serum that enables efficient opsonization. The human complement system is composed of more than 30 proteins and is activated by any one of three routes: the classical pathway, the lectin pathway, and the alternative pathway (Figure 2). Complement system uses three independent pathways to distinguish bacteria from host cells and then can rapidly recognize and opsonize bacteria or kill gram-negative bacteria directly by formation of the membrane attack complex [10]. All three pathways converge in the assembly of a C3 convertase, which are enzyme complexes that consist of C4b2a and C3bBb (C4b2a for the classical and lectin pathways and C3bBb for the alternative pathway). The C3 convertase catalyzes the key reaction in complement activation: cleavage of complement protein C3 results in release of anaphylactic agents C3a and C3b. Most of C3b is further processed into iC3b by complement factor H and complement factor I (Figure 2). At high local concentrations of C3b, the C3 convertase is changed into a C5 convertase, which cleaves C5, resulting in release of the potent chemoattractants C5a, and C5b, which initiates the lytic pathway when deposited on gram-negative bacteria, thereby amplifying the opsonization process.
Figure 2.
Staphylococcus aureus was interfered with chemotaxis and activation of complement.
Igs, which are the second most abundant protein in serum/plasma, play an important role in opsonization of bacteria and subsequent recognition by specific Fc receptor present on the surface of neutrophils. Several Ig subtypes (IgG, IgM, and IgA) have roles in microbial infection control (Figure 2). Different subclasses of Igs display distinct differences in complement activation and Fcγ receptor (FcγR). Normal human neutrophils express two major FcγRs, FcγRII and FcγRIIIB, and do not express the FcγR1 [11]. IgG can activate the classical complement pathway and neutralize toxins or other bacterial virulence factors. IgM, owing to its polymeric nature, is particularly effective at complement activation and opsonization. In contrast to IgG and IgM, IgA does not activate the complement system.
2.3. Receptor-Mediated Pathogen Recognition and Phagocytosis
Once neutrophils migrate to the site of infection, the opsonized pathogen can be recognized and phagocytized via receptor-mediated uptake into a vacuole within the cell. Similar to other phagocytes, such as macrophages, neutrophils express a large number of receptors including pattern-recognition receptors (PRRs), G protein-coupled receptors (GPCRs), and opsonic receptors. These receptors can recognize microbe-associated molecular patterns (MAMPs) and host proteins (such as IgG and complement) which were used to opsonize the microbe (Figure 1(b)). The PRRs can recognize pathogen-associated molecular patterns (PAMPs), such as bacterial DNA, lipopolysaccharide, peptidoglycan, and lipoteichoic acids. The major types of PRRs on neutrophils include Dectin-1 (recognizing fungal β-glucan), triggering receptor expressed on myeloid cells-1 (TREM-1, recognizing bacteria and fungi) [12], and toll-like receptors (TLRs, recognizing lipids, carbohydrates, and peptide). GPCRs, which are expressed in the surface of neutrophils, can recognize bacterial products as well as endogenous molecules released during inflammation. The formyl peptide receptors 1 (FPR1) and its homologue FPR2 belong to the GPCRs family, recognize N-formylated proteins and peptides (fMLP), and consequently induce and potentiate chemotaxis, phagocytosis, and the generation of oxidative burst in neutrophils (Figure 1(b)). Invasive bacterial pathogens are opsonized with complement (e.g., C3b) and antibody (e.g., IgG), recognized by opsonic receptors, including FcγRs and the complement receptors (CRs), respectively. Activation of opsonic receptors rapidly enhances the efficiency of phagocytosis and is critical for neutrophil-mediated pathogen killing [13].
3. Pathogen Killing by Neutrophils
Neutrophils are the first line of innate immune cells arriving at the site of bacterial inoculation, where they exert diverse antimicrobial mechanisms to prevent pathogen dissemination to normally sterile sites. The process by which neutrophils kill invading pathogens depends on three primary mechanisms [14]: production of highly toxic reactive oxygen species (ROS) in the pathogen-containing vacuole; fusion of neutrophil granules, containing various antimicrobial mediators to the vacuole; NETs formation. These steps may also contribute to inflammatory diseases in which ligands are deposited on tissue components.
3.1. Phagocytic Uptake of Bacteria Triggers Production of ROS
Coincident with phagocytosis of bacteria, neutrophils produce an oxidative burst resulting in the rapid release of high levels of bactericidal reactive chemical species under the catalyzation of NADPH oxidase, myeloperoxidase (MPO), or nitric oxide (NO) synthetase [15]. NADPH oxidase is responsible for the generation of ROS, such as superoxide anion (O2−), hydrogen peroxide (H2O2), and hydroxyl radicals (HO∙). The NADPH oxidase functions by shuttling electrons across the phagosomal membrane from cytosolic NADPH to molecular oxygen to produce O2−. By superoxide dismutase (SOD), the superoxide anion is readily converted to hydrogen peroxide. H2O2 and O2− can combine to generate the highly reactive HO∙ via the Haber-Weiss reaction, which requires a metal such as iron. As a microbicidal agent, HO∙ was probably not found in intact cells because that lactoferrin inhibits the generation of HO∙ and other free radical reactions by binding free copper and iron. Against certain pathogens, such as Aspergillus, NADPH oxidase is critical for host defense independently of proteinases, and its importance is revealed in that patients who lack any one of the oxidase subunits suffer from chronic granulomatous disease (CGD) [16].
MPO converts hydrogen peroxide to primarily hypochlorous acid (HOCl). HOCl is the most bactericidal oxidant in neutrophils. Notably, hydrogen peroxide and other secondary oxygen derivatives such hydroxyl radical, chloramines, and HOCl can inactivate iron-sulphur proteins, membrane proteins, and the origin of replication site for DNA synthesis, which play a critical role in the killing of pathogenic bacteria [17]. Indeed, some patients, whose neutrophils lacked MPO, were thought to be immunodeficient [18]. And MPO knockout mice have also shown an undue susceptibility to bacterial and fungal infections [19].
Oxidative deamination of L-arginine by nitric oxide (NO) synthetase generates NO that together with superoxide anion forms reactive nitrogen intermediates with antimicrobial activity [20]. NO, a short-lived (half-life of a few seconds), highly reactive molecule, is produced by inducible nitric oxide synthase (iNOS), which is present in primary granules and is induced upon neutrophil priming (via TNF, IL-1, or IFN-γ) and during bacterial infection. NO production complements ROS production by neutrophils to exert antibacterial functions.
3.2. Phagocytic Uptake of Bacteria Triggers Production of Degranulation
Pathogens sequestered by neutrophils are trafficked to and fused with the phagosome in a process called degranulation, leading to the killing of invading pathogens in a process involving the release and action of proteinases and peptidases (Table 1). Functionally, the granules can be subdivided into three different classes based on the contents of their matrices and their integral membrane proteins: azurophilic granule, specific granule, and gelatinase granule. Neutrophils are “prepacked” with multiple types of granules that fuse with phagocytic vacuoles to facilitate pathogens destruction. Moreover, granules also help to initiate an inflammatory response and contain alkaline phosphatase, lactoferrin, lysozyme, and NADPH oxidase.
Table 1.
Mechanism of action of neutrophil antimicrobial proteins/peptide.
| Antimicrobial protein/peptide | Direct antimicrobial mechanism | Alternative antimicrobial mechanism | Subcellular localization | Ref. |
|---|---|---|---|---|
| α-Defensins | Membrane-active; inhibition of DNA, RNA, protein, bacterial cell wall synthesis | Opsonisation of bacteria/ROS formation | Primary granules, NETs | [21] |
| LL-37 | Transmembrane pore-forming | ROS formation | Secondary granules, NETs | [22] |
| BPI | Hydrolysis of bacterial phospholipids by binding to LPS | Inhibiting cytokine liberation by binding to CD14 | Primary granules | [23, 24] |
| Histones | Membrane-active | NETs formation | Nucleus, NETs | [25] |
| Lysozyme | Degrades bacterial cell wall | NETs formation | Lysosomes | [5, 26] |
| PR3 | Proteolytic activity; degrading virulence factors | NETs formation | Primary granules/NETs | [27–29] |
| NE | Proteolytic activity; degrading virulence factors | NETs formation | Primary granules/NETs | [27–29] |
| CatG | Proteolytic activity | NETs formation; ROS formation | Primary granules/NETs | [28, 29] |
| NSP4 | Trypsin-like activity | Unknown | Primary granules | [30–32] |
| Azurocidin | Membrane-active | Opsonisation of bacteria | Primary granules | [33] |
| Lactoferrin | Altering bacterial growth by binding to iron; increase in membrane permeability by binding to the lipid A | Decreasing the release IL-1, IL-2, and TNFα; Suppressing NETs release | Secondary granules/NETs | [34–37] |
| Calprotectin | Altering bacterial growth by sequestering Mn2+ and Zn2+ | Inhibition of Mn2+-dependent bacterial superoxide defenses; NETs formation | Secondary granules | [38, 39] |
| PTX3 | As a soluble pattern recognition receptor in innate immunity | NETs formation | Secondary granules/NETs | [40] |
| NADPH oxidase | Generation of superoxide anion | NETs formation | Lysosomes | [41] |
| MPO | Generation of hypochlorous acid | NETs formation | Lysosomes | [18, 42, 43] |
| Platelets | Activating neutrophils to release NETs | NETs formation | NETs | [44] |
| NGAL | Inhibit bacteria growth by capturing and depleting siderophores | Acting as a growth and differentiation factor in multiple cell type | Secondary granules | [45, 46] |
Azurophil (or primary) granules are the first to be produced and contain MPO and a spectrum of neutrophil serine proteases (NSPs): cathepsin G (CG), neutrophil elastase (NE), proteinase 3 (PR3), and the recently discovered neutrophil serine protease-4 (NSP4) [30, 90]. NSPs are critical for the effective functioning of neutrophils and greatly contribute to immune protection against bacterial infections [27]. NSPs are currently believed to have three functions. (1) NSPs can directly kill bacterial cell. NE has been shown to directly kill the gram-negative bacteria E. coil by cleavage of its outer membrane protein A, resulting in loss of membrane integrity and cell death. In vivo, the concerted action of NE, CG, and PR3 can kill S. pneumonia within phagocytic vacuole. (2) NSPs can cleave host proteins to generate antimicrobial peptides. The best-known example is that PR3 that has been shown to cleave hCAP-18 to generate the antimicrobial peptide LL-37. (3) NSPs can attenuate bacterial virulence by inactivating factors required for pathogenesis. Shigella flexneri mobility proteins IcsA and IpaA-C can be cleaved by NE, consequentially preventing its dissemination into the cytoplasm of neutrophils. Similar to NE, CG can cleave the S. aureus adhesin clumping factor A and remove its active domain. Together, these NSPs are critical for the effective functioning of neutrophils and immune protection against bacterial infections [27].
In addition, neutrophils also contain a full-length cationic antimicrobial protein, bactericidal/permeability-increasing protein (BPI) in azurophil granules [91]. BPI possesses three types of anti-infective activities: direct antimicrobial activity, neutralizing endotoxin activity through direct binding of LPS, and opsonic activity. BPI binding to LPS results in increased bacterial permeability, hydrolysis of bacterial phospholipids, and death of the bacterium. In addition to its well-documented anti-infective properties, BPI has also been shown to possess additional bioactivities, such as accelerating apoptosis, binding the vascular endothelial growth factor (VEGF), and inhibiting migration of human umbilical vein endothelial cells.
The specific granules are smaller with 0.1 μm diameter and formed after azurophilic granules. These granules do not contain MPO and are characterized by the presence of the glycoprotein lactoferrin. They primarily contain a wide range of antimicrobial compounds including calprotectin, lactoferrin, neutrophil gelatinase-associated lipocalin (NGAL), hCAP-18, and lysozymes. Calprotectin, also called S100A, is a critical factor in the innate immune response to infection and has been shown to inhibit microbial growth through chelation of nutrient Mn2+ and Zn2+, resulting in reprogramming of the bacterial transcriptome [92]. Lactoferrin, also called lactotransferrin, is an iron-binding glycoprotein present in most biological fluids of mammals and is released from neutrophil granules during inflammatory responses [93, 94]. Lactoferrin possesses a number of types of antibacterial activities: (1) blocking the entry of bacterial pathogens competitively binding onto cell receptors, such as glycosaminoglycans; (2) degrading protein virulence produced by bacteria, such as H. influenza and E. coil, through proteolysis; (3) preventing bacterial adhesion through competing bacterial adhesion sites on bacteria and host cells [95].
The tertiary granules, also named gelatinase granules, are smaller than specific granules and are both MPO- and lactoferrin-negative. These granules contain few antimicrobials but serve as a storage location for a number of metalloproteases, such as gelatinase and leukolysin. These granules may represent one end of the population of granules formed during neutrophil maturation.
3.3. Neutrophil Extracellular Traps Killing Bacteria
In addition to pathogens phagocytosis and subsequent reactive species- and enzyme-dependent pathogen destruction, neutrophils also exert antibacterial activity through neutrophil extracellular traps (NETs), which was first described by Brinkmann et al. in 2004 [96]. Sensing the entry of bacteria, neutrophils extrude a mesh-like structure consisting of DNA/histones and are peppered with granule-derived antimicrobial peptides and enzymes, a process termed NETosis. NETs are composed of DNA strands associated with histones and decorated with about 20 different proteins, including NE, CG, PR3, MPO, lactoferrin, pentraxin 3 [40], high mobility group protein B1, LL37, and buforin II [97]. Mitochondria can also serve as a source of DNA for NET formation. The NETs are capable of ensnaring microbes by localizing and trapping pathogens within a sticky meshwork of chromatin. Furthermore, NETs facilitate pathogen exposing to highly concentrate antimicrobial peptides and enzymes, such as MPO, neutrophil elastase, LL-37, S100A, and lactoferrin-chelating proteins [98]. Along with the chromatin network, these antimicrobial agents are concentrated and the potential for synergistic action is enhanced. When neutrophils extrude a meshwork of chromatin to form NETs, it is not an end point for neutrophils and anuclear neutrophils can also migrate and retain the necessary components to kill bacteria through phagocytosis and formation of mature phagosomes [99].
The molecular mechanisms details of NETs formation are tightly linked to the production of ROS. The magnitude and duration of ROS production play an important role in promoting NETs formation and may be a major role in determining the fate of the neutrophil. In addition, individuals lacking MPO and NADPH oxidase, two key enzymes in the ROS cascade, are unable to make NETs and suffer from debilitating infections [100]. However, ROS are not the only vital roles in NETs formation and decondensation of chromatin is also critical for proper NETs formation. Neutrophil elastase was shown to partially degrade histones and further leads to decondensation of chromatin, which is also a pivotal event in the process of NETs formation [101].
NETosis also has the dark side: apart from this antimicrobial function, the cytotoxicity of NETs can be harmful to the host if their release is inappropriately controlled. Excessive NETs formation is linked to various neutrophil-mediated pathologies, including vasculitis, sepsis, and systemic lupus erythematosus nephritis. NETs also induce platelet procoagulant activation, which can lead to significant thrombosis and vascular injury. Excessive NETs formation and endothelial cell activation are also associated with preeclampsia of pregnancy [102].
4. “Catch Me If You Can”: How Pathogens Evade Antibacterial Arsenal of Destruction by Neutrophils
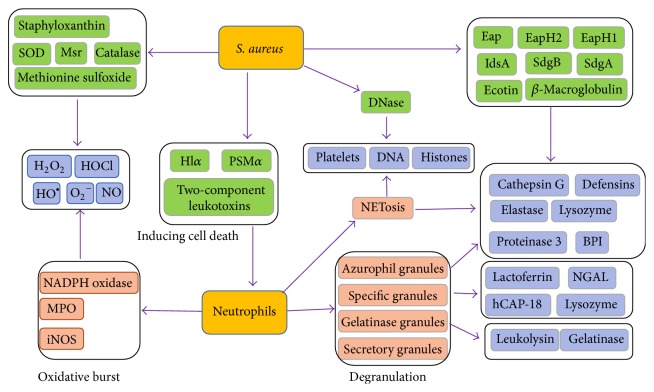
To promote its own survival within the host, bacterial pathogens have evolved an array of specific mechanisms to overcome destructions by neutrophils (Figure 3, Table 2). S. aureus, the culprit of many types of infections, exhibits many characteristics of antineutrophils pathogens [103].
Figure 3.
Direct antimicrobial mechanisms from neutrophils and the S. aureus counterattack. Neutrophils are equipped with multiple anti-infective strategies including the bacterial uptake (phagocytosis), the phagolysosomal degradation of bacteria via reactive oxygen species (oxidative burst), the release of antimicrobial molecules (degranulation), and the formation of a web-like structure composed of chromatin, histones, and antimicrobials (neutrophil extracellular traps, NETs). S. aureus is equipped with a magnitude of neutrophil resistance factors (green boxes) allowing the pathogen to uniquely counteract each antibacterial strategy of neutrophils.
Table 2.
Neutrophil antibacterial functions subverted by S. aureus. S. aureus produces a large suite of virulence factors to counteract specific neutrophil clearance mechanisms during the pathogenesis of invasive infection.
| Virulence factor | Targets | Function | Ref. |
|---|---|---|---|
| SSL-5 | PSGL1/GPCRs | Recruitment/chemotaxis inhibition | [47, 48] |
| SSl-6 | PSGL1 | Recruitment inhibition | [49] |
| SSl-11 | PSGL1 | Recruitment inhibition | [50] |
| SSl-3 | TLR2 | Chemotaxis inhibition | [51, 52] |
| SEIX | PSSG1 | Recruitment inhibition | [49] |
| ScpA | CXCR2 | Chemotaxis inhibition | [53] |
| CHIPS | FPR1, C5aR | Chemotaxis inhibition | [54, 55] |
| FLIPr | FPR2 | Chemotaxis inhibition | [56, 57] |
| FLIPrL | FPR1, FPR2 | Chemotaxis inhibition | [56, 57] |
| PSMs | FPR2 | Chemotaxis inhibition/neutrophils lysis | [56, 58] |
| Eap | ICAM1/C4b/NE/CG/PR3 | Recruitment/phagocytic inhibition | [59] |
| Aureolysin | C3 | Complement inhibition | [60] |
| SCIN | C3bBb | Complement inhibition | [61, 62] |
| SCIN-B/C | C3bBb | Complement inhibition | [61, 62] |
| Efb | C3b | Complement inhibition | [63] |
| Ecb | C3b | Complement inhibition | [64] |
| SSL7 | IgA/C5 | Phagocytosis/complement inhibition | [65] |
| SSL10 | IgG | Phagocytosis inhibition | [66] |
| SAK | C3/IgG | Phagocytosis inhibition | [67] |
| Sbi | IgG/C3/factor H | Phagocytosis inhibition | [68, 69] |
| SpA | IgG | Phagocytosis inhibition | [70] |
| ClfA | Factor I | Phagocytosis inhibition | [56] |
| SOK | Unknown | Phagocytosis inhibition | [71] |
| CP | Unknown | Phagocytosis inhibition | [72] |
| SdrE | Factor H | Complement inhibition | [73] |
| IsdH | C3b | Complement inhibition | [74] |
| Cna | C1q | Complement inhibition | [75] |
| LukAB | αM integrin | Neutrophils lysis | [76] |
| LukED | CCR5/CXCR1/CXCR2 | Neutrophils lysis | [77] |
| LukMF | Not known | Neutrophils lysis | [78] |
| PVL | C5aR | Neutrophils lysis | [79] |
| Hla | C5aR | Neutrophils lysis | [80] |
| Staphyloxanthin | Unknown | Resistance to ROS | [81] |
| KatA | Hydrogen peroxide | Resistance to ROS | [82] |
| AhpC | Hydrogen peroxide | Resistance to ROS | [82] |
| Msr | Hydrogen peroxide | Resistance to ROS | [83] |
| AdsA | Adenosine | Resistance to ROS | [84] |
| IsdA | Fibrinogen | Resistance to lactoferrin | [85] |
| OatA | Peptidoglycan | Resistance to lysozyme | [86, 87] |
| EapH1 | NSPs | Resistance to NSPs | [88] |
| EapH2 | NSPs | Resistance to NSPs | [88] |
| Nuclease | DNA | Resistance to NETs | [89] |
4.1. Inhibition of Neutrophil Recruitment
Counter measures adopted by pathogen may affect these steps to inhibit neutrophil recruitment. For instance, staphylococcal superantigen-like 5 (SSL5) can block neutrophil adhesion to endothelial cells by binding to PSGL-1 and consequently blocking its interaction with the natural ligand P-selectin [104]. SSL5 and other family members also inhibit leukocyte responses to chemokines, such as CXC, CC, CX3C, and CXCL12, and to the complement fragments C3a and C5a. Moreover, extracellular adherence protein (Eap) generated by S. aureus can bind and inhibit ICAM-1, a crucial molecule used to facilitate the neutrophils firm adhesion of endothelial cells. Furthermore, bacteria can secrete a variety of proteases, leading to degradation of chemokines. Chemotaxis inhibiting protein (CHIPS), a protein freely secreted by S. aureus, binds directly to the C5a receptor and formyl peptide receptors (FPRs) and thereby inhibits neutrophils recruitment [105, 106]. As a homologue of CHIPS in S. aureus, FPR-like 1 inhibitory proteins (FLIPr and FLIPr-like) bind and inhibit FPR1 as well as C5aR and then impair neutrophil chemotaxis. Another cysteine protease secreted by S. aureus is staphopain A, which inactivates CXCR2 chemokines by cleaving its N-terminal domain and then inhibits neutrophil activation and recruitment [53]. In addition, SSL3 specifically binds and inhibits TLR2 activation, which is critical for host defense against S. aureus [107].
4.2. Preventing Phagocytosis
S. aureus has successfully developed ways to evade the complement system by secretion of specific complement inhibitors (Figure 2, Table 2). The secreted factors described below allow bacteria to either diminish or delay the detrimental effects of an innate immune attack, thereby generating a window of opportunity to replicate and establish a microenvironment conducive to bacterial survival and disease pathogenesis [108, 109].
4.2.1. Cleavage of IgG
SSL7 binds host IgA and complement component C5, inhibiting generation of C5a, phagocytosis, and production of phagocyte reactive oxygen species. S. aureus expresses two surface-anchored proteins, staphylococcal protein A (SpA) and staphylococcal immunoglobulin-binding protein (Sbi), which impair IgG function. SpA possesses five immunoglobulin-binding repeat domains. Each domain can bind the Fc-part of IgG, thereby blocking the interaction with Fc receptors on neutrophils. Sbi consists of four small domains, of which two (Sbi-I and Sbi-II) can bind IgG [110–112].
4.2.2. Direct Inactivation of C3 Convertases
It has been shown that SCIN and its homologues (SCIN–B and SCIN–C), as strongly antiphagocytic molecules, modulate all the three complement pathways through the unique interaction with C3 convertases [61]. Extracellular fibrinogen-binding protein (Efb) and its homologue extracellular complement-binding protein (Ecb) can modulate the alternative pathway convertase by binding to the C3b molecule directly [113]. S. aureus secretes the 16 Kda Efb that binds two different plasma proteins using separate domains: the Efb N-terminus binds to fibrinogen, while the C-terminus binds complement C3b.
4.2.3. Binding or Cleavage of Human Convertase Regulators
S. aureus recruits the complement regulatory protein factor H (fH) and factor I (fI) to its surface to inhibit the alternative pathway of complement activation. The surface-associated protein SdrE, as an fH-binding protein, enhances recruitment of fH which resulted in increased iC3b generation [73]. The clumping factor A (ClfA) of S. aureus binds to complement regulator factor I and increases factor I cleavage of C3b [114–116]. Similar to ClfA, iron-regulated surface determinant protein H (IsdH) could act as a factor I-mimicking protease and directly trap factor I to the S. aureus surface, promoting cleavage of C3b [74].
4.2.4. Eliminating Opsonic Molecules from the Bacterial Surface
Staphylokinase (SAK) is a secreted protein that binds and activates surface-bound plasminogen into plasmin, which removes IgG as well as C3b from the bacterial surface, making this protein a unique antiopsonic molecule [67]. Aureolysin [60], a secreted metalloprotease, inhibits the deposition of C3b on S. aureus surfaces and the release of the chemoattractant C5a. It has been shown that aureolysin cleaves the central complement protein C3 specifically in the α-chain, close to the C3 convertase cleavage site, yielding active C3a and C3b. The antiphagocytic activity of the capsule is well established and the quantity of capsule is decisive for S. aureus and virulence. Overexpression of capsular polysaccharides type 8 renders S. aureus more resistant to phagocytosis by neutrophils in vitro [72, 117].
4.3. Surviving inside the Neutrophil
The combined action of ROS and antimicrobial proteins generated by granules creates a lethal environment for microbes. However, S. aureus harbored by neutrophils can survive in the presence of extreme environment, although not replication [118]. This is because S. aureus has evolved many means to resist oxidant damage and antimicrobial proteins degradation, as well as surviving within phagosomes.
First of all, S. aureus strains can express five types of enzymes or pigment promoting resistance to oxidative killing by stimulated neutrophils, including superoxide dismutase, catalase, staphyloxanthin, methionine sulfoxide reductases (Msr), and adenosine synthase A (AdsA). Superoxide dismutase produced by S. aureus can convert superoxide anion to H2O2, which is then consumed to yield O2 and H2O by catalase, thereby eliminating oxidants generated by stimulated neutrophils. Furthermore, S. aureus strains also produce the pigment staphyloxanthin, which consumes oxidants and renders bacteria resistant to oxidant-dependent killing, protecting bacteria from singlet oxygen via an undefined mechanism. Msr is a highly conserved enzyme that repairs oxidative damage incurred within neutrophils, contributing to survival of bacteria within neutrophils. AdsA, a cell wall-anchored enzyme, can convert adenosine monophosphate to adenosine [119]. As a critical virulence factor, AdsA promotes staphylococcal synthesis of adenosine in blood, escaping from phagocytic clearance [84]. Adenosine is also known to inhibit neutrophil degranulation, adhesion to vascular surfaces, and superoxide burst [120]. These findings indicate that phagocytosed S. aureus devote significant energy and effort to self-preservation rather than to growth and replication.
Bacteria pathogens have evolved two strategies to counteract human NSPs. The first one is modifications of bacterial NSPs substrates. For gram-positives, such as S. epidermidis and S. aureus, glycosyltransferases (SdgA and SdgB) are expressed to modify the serine-aspartate dipeptide repeats (SDR) of GLcNAc, which will protect these bacteria from proteolytic degradation by CG. The LPS of Gram-negatives, such as Neisseria meningitidis, are anchored to the outer membrane by lipid A. The lipid can be modified by phosphoethanolamine transferase to prevent proteolysis-independent killing by CG.
The second strategy is production of NSPs inhibitors. A recent study reports that S. aureus has evolved three highly specific NSPs inhibitors: extracellular adherence protein (Eap) and its smaller homologues EapH1 and EapH2 [88]. These proteins are very potent and specific inhibitors of NSPs and imply a crucial role for NSPs in the defense against S. aureus. Iron-regulated surface determinant protein H (IsdH) is present in the surface of S. aureus, which binds to lactoferrin, the most abundant antistaphylococcal polypeptide [121]. IsdA confers resistance to killing by lactoferrin. In addition, recombinant IsdA was a competitive inhibitor of lactoferrin protease activity. Thus, IsdA can protect S. aureus against lactoferrin and acts as a protease inhibitor [122].
4.4. Inducing Cell Death by Cytolytic Toxins
Following phagocytosis of bacteria pathogens, neutrophils would kill most bacteria and have initial features typical of apoptosis. However, S. aureus can survive within these neutrophils and ultimately cause cytolysis. Recent studies have provided evidence that cytolytic toxins produced by S. aureus contribute to neutrophil lysis after phagocytosis [123]. Cytolytic toxins produced by S. aureus, including the phenol soluble modulins (PSMs), alpha-hemolysin (Hlα), and two-component leukotoxins, facilitate neutrophil killing after phagocytosis [124]. PSMs were first identified in 1999 by hot phenol extraction from S. epidermidis culture filtrate, in which three peptides termed PSMα, PSMβ, and PSMγ were identified. PSMs do not have uniform charge characteristics. PSMαs of S. aureus are positively charged, while PSMβ peptides are all negatively charged, and the PSMγ is neutral [125]. In S. aureus, PSMα peptides have a pronounced ability to lyse human neutrophils, in which PSMα3 has by far the strongest activity. However, PSMγ (also named δ-toxin) has moderate cytolytic activity and the PSMβ peptides are noncytolytic. At the micromolar concentrations, PSMα has the pronounced capacity to kill human neutrophils after phagocytosis by disrupting the cytoplasmic membrane [126]. While at nanomolar concentrations, PSMα may stimulate neutrophils and initiate proinflammatory responses including neutrophil chemoattraction, activation, and the release of IL-8 [127]. Neutrophils sense PSMs via formyl peptide receptor 2 (FPR2), which may sense the amphipathic, α-helical structure of PSMs rather than a specific amino acid sequence motif.
Panton-Valentine Leukocidin (PVL) is a prophage-encoded pore-forming exotoxin, which mainly acts on neutrophils as a crucial virulence factor in necrotizing diseases. PVL is a staphylococcal bicomponent pore-forming toxin comprising the protein subunits LukS-PV and LukF-PV [128]. Initial binding of LukS-PV to the surface of target cells triggers secondary binding of LukF-PV and subsequently induces the assembly of lytic pore-forming [129]. PVL-induced pore formation is mediated by the human C5aR, which determines species specificity of PVL [79]. The C5aR can bind LukS-PV, which is a potent inhibitor of C5a-induced immune cell activation.
S. aureusα-hemolysin (α-toxin, Hla) belongs to the class of small β-barrel pore-forming cytotoxins [130]. As a water soluble monomer, α-hemolysin is capable of binding and oligomerization into a heptameric structure on neutrophils. Then, α-hemolysin exhibits the main action on pore formation and neutrophils lysis after phagocytosis [131]. In other studies, α-hemolysin has been suggested to directly disrupt the S. aureus phagosome and promote S. aureus escape to and replication in the cytoplasm [131]. S. aureusα-hemolysin facilitates the secretion of newly synthesized CXC chemokines into the airway and stimulates neutrophil homing in staphylococcus aureus pneumonia [132].
4.5. Avoiding Killing in NETs
In addition to phagocytosis and intracellular killing, neutrophils release NETs that capture and kill microbes in the extracellular space. Several bacterial pathogens have evolved sophisticated mechanisms to suppress, escape, and/or resist NETs. Expression of nucleases is one highly conserved anti-NET factor among bacteria, which can degrade NETs indicating that the chromatin functions as a scaffold and is a major component of the fibres. Interestingly, extracellular nucleases are found in several pathogenic bacteria including S. aureus, Clostridium perfringens, and S. pyogenes (group A Streptococcus, GAS) [133].
In addition to produce nucleases, GAS can also suppress NETs formation by degrading the neutrophil stimulatory chemokine IL-8 with peptidase SpyCEP or HA capsule engagement of the inhibitory neutrophil receptor Siglec-9. Other GAS resistance factors, including M1 protein, Scl-1 protein, and the GLcNAc side chain, contribute to GAS resistance to antimicrobial components.
5. Conclusion
The interaction between neutrophils and pathogens remains a fascinating subject. The host requires the action of neutrophils to fight invaders and the pathogens in turn must cope with neutrophil attacks in order to colonize the host [134]. Several pharmacological agents can be used to enhance neutrophil energy generation, antimicrobial activities, and treatment outcomes. For instance, hypoxia-inducible factor 1 (HIF-1), innate defense regulator peptides (IDRs), and vitamin B3 all enhance antimicrobial activities to provide prophylactic and therapeutic activity against bacterial pathogens in vivo. And tamoxifen [135] or anacardic acid [136] could boost NETs formation and bacterial killing of neutrophils. To overcome antibiotic resistant pathogens which harness the multifaceted antimicrobial properties of neutrophils, these host-directed strategies provide a critical new element to boost neutrophil function and minimize the risk for development of antibiotic resistance during infection.
Acknowledgments
This work was supported by the Foundation of Science and Technology Department of Henan Province under Grant 162300410233 and the Natural Science Foundation of Education Department of Henan Province under Grant 17A310015.
Competing Interests
The authors declare that they have no competing interests.
References
- 1.Borregaard N. Neutrophils, from marrow to microbes. Immunity. 2010;33(5):657–670. doi: 10.1016/j.immuni.2010.11.011. [DOI] [PubMed] [Google Scholar]
- 2.Leliefeld P. H. C., Wessels C. M., Leenen L. P. H., Koenderman L., Pillay J. The role of neutrophils in immune dysfunction during severe inflammation. Critical Care. 2016;20(1, article 73) doi: 10.1186/s13054-016-1250-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nathan C. Neutrophils and immunity: challenges and opportunities. Nature Reviews Immunology. 2006;6(3):173–182. doi: 10.1038/nri1785. [DOI] [PubMed] [Google Scholar]
- 4.Mayadas T. N., Cullere X., Lowell C. A. The multifaceted functions of neutrophils. Annual Review of Pathology: Mechanisms of Disease. 2014;9:181–218. doi: 10.1146/annurev-pathol-020712-164023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Segal A. W. How neutrophils kill microbes. Annual Review of Immunology. 2005;23:197–223. doi: 10.1146/annurev.immunol.23.021704.115653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Urban C. F., Lourido S., Zychlinsky A. How do microbes evade neutrophil killing? Cellular Microbiology. 2006;8(11):1687–1696. doi: 10.1111/j.1462-5822.2006.00792.x. [DOI] [PubMed] [Google Scholar]
- 7.Döhrmann S., Cole J. N., Nizet V. Conquering neutrophils. PLoS Pathogens. 2016;12(7) doi: 10.1371/journal.ppat.1005682.e1005682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nauseef W. M. Neutrophils, from cradle to grave and beyond. Immunological Reviews. 2016;273(1):5–10. doi: 10.1111/imr.12463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rigby K. M., DeLeo F. R. Neutrophils in innate host defense against Staphylococcus aureus infections. Seminars in Immunopathology. 2012;34(2):237–259. doi: 10.1007/s00281-011-0295-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dunkelberger J. R., Song W.-C. Complement and its role in innate and adaptive immune responses. Cell Research. 2010;20(1):34–50. doi: 10.1038/cr.2009.139. [DOI] [PubMed] [Google Scholar]
- 11.van Kesse K. P. M., Bestebroer J., van Strijp J. A. G. Neutrophil-mediated phagocytosis of Staphylococcus aureus. Frontiers in Immunology. 2014;5, article 467 doi: 10.3389/fimmu.2014.00467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ford J. W., McVicar D. W. TREM and TREM-like receptors in inflammation and disease. Current Opinion in Immunology. 2009;21(1):38–46. doi: 10.1016/j.coi.2009.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mócsai A. Diverse novel functions of neutrophils in immunity, inflammation, and beyond. The Journal of Experimental Medicine. 2013;210(7):1283–1299. doi: 10.1084/jem.20122220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Amulic B., Cazalet C., Hayes G. L., Metzler K. D., Zychlinsky A. Neutrophil function: from mechanisms to disease. Annual Review of Immunology. 2012;30:459–489. doi: 10.1146/annurev-immunol-020711-074942. [DOI] [PubMed] [Google Scholar]
- 15.Roos D., van Bruggen R., Meischl C. Oxidative killing of microbes by neutrophils. Microbes and Infection. 2003;5(14):1307–1315. doi: 10.1016/j.micinf.2003.09.009. [DOI] [PubMed] [Google Scholar]
- 16.Greenlee-Wacker M., DeLeo F. R., Nauseef W. M. How methicillin-resistant Staphylococcus aureus evade neutrophil killing. Current Opinion in Hematology. 2015;22(1):30–35. doi: 10.1097/MOH.0000000000000096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Odobasic D., Kitching A. R., Holdsworth S. R. Neutrophil-mediated regulation of innate and adaptive immunity: the role of myeloperoxidase. Journal of Immunology Research. 2016;2016:11. doi: 10.1155/2016/2349817.2349817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Klebanoff S. J. Myeloperoxidase: friend and foe. Journal of Leukocyte Biology. 2005;77(5):598–625. doi: 10.1189/jlb.1204697. [DOI] [PubMed] [Google Scholar]
- 19.Morozov V. I., Pryatkin S. A., Kalinski M. I., Rogozkin V. A. Effect of exercise to exhaustion on myeloperoxidase and lysozyme release from blood neutrophils. European Journal of Applied Physiology. 2003;89(3-4):257–262. doi: 10.1007/s00421-002-0755-5. [DOI] [PubMed] [Google Scholar]
- 20.Winterbourn C. C., Kettle A. J., Hampton M. B. Reactive oxygen species and neutrophil function. Annual Review of Biochemistry. 2016;85:765–792. doi: 10.1146/annurev-biochem-060815-014442. [DOI] [PubMed] [Google Scholar]
- 21.Cardot-Martin E., Casalegno J. S., Badiou C., et al. α-defensins partially protect human neutrophils against Panton-Valentine leukocidin produced by Staphylococcus aureus. Letters in Applied Microbiology. 2015;61(2):158–164. doi: 10.1111/lam.12438. [DOI] [PubMed] [Google Scholar]
- 22.Frasca L., Lande R. Role of defensins and cathelicidin LL37 in auto-immune and auto-inflammatory diseases. Current Pharmaceutical Biotechnology. 2012;13(10):1882–1897. doi: 10.2174/138920112802273155. [DOI] [PubMed] [Google Scholar]
- 23.Wu H., Liu L., Lin M., et al. Importance of the residue 190 on bactericidal activity of the bactericidal/permeability-increasing protein 5. Oncotarget. 2016;7(28):43088–43094. doi: 10.18632/oncotarget.9292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schuerholz T., Brandenburg K., Marx G. Antimicrobial peptides and their potential application in inflammation and sepsis. Critical Care. 2012;16(2, article 207) doi: 10.1186/cc11220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Park C. B., Yi K.-S., Matsuzaki K., Kim M. S., Kim S. C. Structure-activity analysis of buforin II, a histone H2A-derived antimicrobial peptide: the proline hinge is responsible for the cell-penetrating ability of buforin II. Proceedings of the National Academy of Sciences of the United States of America. 2000;97(15):8245–8250. doi: 10.1073/pnas.150518097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nash J. A., Ballard T. N. S., Weaver T. E., Akinbi H. T. The peptidoglycan-degrading property of lysozyme is not required for bactericidal activity in vivo. The Journal of Immunology. 2006;177(1):519–526. doi: 10.4049/jimmunol.177.1.519. [DOI] [PubMed] [Google Scholar]
- 27.Stapels D. A. C., Geisbrecht B. V., Rooijakkers S. H. M. Neutrophil serine proteases in antibacterial defense. Current Opinion in Microbiology. 2015;23:42–28. doi: 10.1016/j.mib.2014.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ginsburg I. The role of bacteriolysis in the pathophysiology of inflammation, infection and post-infectious sequelae. APMIS. 2002;110(11):753–770. doi: 10.1034/j.1600-0463.2002.1101101.x. [DOI] [PubMed] [Google Scholar]
- 29.Korkmaz B., Horwitz M. S., Jenne D. E., Gauthier F. Neutrophil elastase, proteinase 3, and cathepsin G as therapeutic targets in human diseases. Pharmacological Reviews. 2010;62(4):726–759. doi: 10.1124/pr.110.002733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kasperkiewicz P., Poreba M., Snipas S. J., et al. Design of a selective substrate and activity based probe for human neutrophil serine protease 4. PLOS ONE. 2015;10(7) doi: 10.1371/journal.pone.0132818.e0132818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Perera N. C., Schilling O., Kittel H., Back W., Kremmer E., Jenne D. E. NSP4, an elastase-related protease in human neutrophils with arginine specificity. Proceedings of the National Academy of Sciences of the United States of America. 2012;109(16):6229–6234. doi: 10.1073/pnas.1200470109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Perera N. C., Wiesmüller K.-H., Larsen M. T., et al. NSP4 is stored in azurophil granules and released by activated neutrophils as active endoprotease with restricted specificity. Journal of Immunology. 2013;191(5):2700–2707. doi: 10.4049/jimmunol.1301293. [DOI] [PubMed] [Google Scholar]
- 33.Watorek W. Azurocidin—inactive serine proteinase homolog acting as a multifunctional inflammatory mediator. Acta Biochimica Polonica. 2003;50(3):743–752. [PubMed] [Google Scholar]
- 34.Legrand D. Overview of lactoferrin as a natural immune modulator. Journal of Pediatrics. 2016;173, supplement:S10–S15. doi: 10.1016/j.jpeds.2016.02.071. [DOI] [PubMed] [Google Scholar]
- 35.Farnaud S., Evans R. W. Lactoferrin—a multifunctional protein with antimicrobial properties. Molecular Immunology. 2003;40(7):395–405. doi: 10.1016/s0161-5890(03)00152-4. [DOI] [PubMed] [Google Scholar]
- 36.Caccavo D., Pellegrino N. M., Altamura M., et al. Antimicrobial and immunoregulatory functions of lactoferrin and its potential therapeutic application. Journal of Endotoxin Research. 2002;8(6):403–417. doi: 10.1179/096805102125001000. [DOI] [PubMed] [Google Scholar]
- 37.Okubo K., Kamiya M., Urano Y., et al. Lactoferrin suppresses neutrophil extracellular traps release in inflammation. EBioMedicine. 2016;10:204–215. doi: 10.1016/j.ebiom.2016.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kehl-Fie T. E., Chitayat S., Hood M. I., et al. Nutrient metal sequestration by calprotectin inhibits bacterial superoxide defense, enhancing neutrophil killing of Staphylococcus aureus. Cell Host and Microbe. 2011;10(2):158–164. doi: 10.1016/j.chom.2011.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.D'Incà R., Pont E., Leo V., et al. Calprotectin and lactoferrin in the assessment of intestinal inflammation and organic disease. International Journal of Colorectal Disease. 2007;22(4):429–437. doi: 10.1007/s00384-006-0159-9. [DOI] [PubMed] [Google Scholar]
- 40.Daigo K., Yamaguchi N., Kawamura T., et al. The proteomic profile of circulating pentraxin 3 (PTX3) complex in sepsis demonstrates the interaction with azurocidin 1 and other components of neutrophil extracellular traps. Molecular & Cellular Proteomics. 2012;11(6) doi: 10.1074/mcp.m111.015073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Levine A. P., Segal A. W. The NADPH oxidase and microbial killing by neutrophils, with a particular emphasis on the proposed antimicrobial role of myeloperoxidase within the phagocytic vacuole. Microbiology Spectrum. 2016;4(4) doi: 10.1128/microbiolspec.MCHD-0018-2015. [DOI] [PubMed] [Google Scholar]
- 42.Theeß W., Sellau J., Steeg C., et al. Myeloperoxidase attenuates pathogen clearance during Plasmodium yoelii nonlethal infection. Infection and Immunity. 2016;85(1) doi: 10.1128/iai.00475-16.e00475-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Klebanoff S. J., Kettle A. J., Rosen H., Winterbourn C. C., Nauseef W. M. Myeloperoxidase: a front-line defender against phagocytosed microorganisms. Journal of Leukocyte Biology. 2013;93(2):185–198. doi: 10.1189/jlb.0712349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Carestia A., Kaufman T., Schattner M. Platelets: new bricks in the building of neutrophil extracellular traps. Frontiers in Immunology. 2016;7, article 271 doi: 10.3389/fimmu.2016.00271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schmidt-Ott K. M., Mori K., Jau Y. L., et al. Dual action of neutrophil gelatinase-associated lipocalin. Journal of the American Society of Nephrology. 2007;18(2):407–413. doi: 10.1681/ASN.2006080882. [DOI] [PubMed] [Google Scholar]
- 46.Chakraborty S., Kaur S., Guha S., Batra S. K. The multifaceted roles of neutrophil gelatinase associated lipocalin (NGAL) in inflammation and cancer. Biochimica et Biophysica Acta—Reviews on Cancer. 2012;1826(1):129–169. doi: 10.1016/j.bbcan.2012.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bestebroer J., Poppelier M. J. J. G., Ulfman L. H., et al. Staphylococcal superantigen-like 5 binds PSGL-1 and inhibits P-selectin-mediated neutrophil rolling. Blood. 2007;109(7):2936–2943. doi: 10.1182/blood-2006-06-015461. [DOI] [PubMed] [Google Scholar]
- 48.Bestebroer J., Van Kessel K. P. M., Azouagh H., et al. Staphylococcal SSL5 inhibits leukocyte activation by chemokines and anaphylatoxins. Blood. 2009;113(2):328–337. doi: 10.1182/blood-2008-04-153882. [DOI] [PubMed] [Google Scholar]
- 49.Fevre C., Bestebroer J., Mebius M. M., et al. Staphylococcus aureus proteins SSL6 and SElX interact with neutrophil receptors as identified using secretome phage display. Cellular Microbiology. 2014;16(11):1646–1665. doi: 10.1111/cmi.12313. [DOI] [PubMed] [Google Scholar]
- 50.Chung M. C., Wines B. D., Baker H., Langley R. J., Baker E. N., Fraser J. D. The crystal structure of staphylococcal superantigen-like protein 11 in complex with sialyl Lewis X reveals the mechanism for cell binding and immune inhibition. Molecular Microbiology. 2007;66(6):1342–1355. doi: 10.1111/j.1365-2958.2007.05989.x. [DOI] [PubMed] [Google Scholar]
- 51.Bardoel B. W., Vos R., Bouman T., et al. Evasion of Toll-like receptor 2 activation by staphylococcal superantigen-like protein 3. Journal of Molecular Medicine. 2012;90(10):1109–1120. doi: 10.1007/s00109-012-0926-8. [DOI] [PubMed] [Google Scholar]
- 52.Koymans K. J., Feitsma L. J., Brondijk T. H. C., et al. Structural basis for inhibition of TLR2 by staphylococcal superantigen-like protein 3 (SSL3) Proceedings of the National Academy of Sciences of the United States of America. 2015;112(35):11018–11023. doi: 10.1073/pnas.1502026112/-/DCSupplemental. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Laarman A. J., Mijnheer G., Mootz J. M., et al. Staphylococcus aureus Staphopain A inhibits CXCR2-dependent neutrophil activation and chemotaxis. EMBO Journal. 2012;31(17):3607–3619. doi: 10.1038/emboj.2012.212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Haas P.-J., De Haas C. J. C., Kleibeuker W., et al. N-terminal residues of the chemotaxis inhibitory protein of Staphylococcus aureus are essential for blocking formylated peptide receptor but not C5a receptor. Journal of Immunology. 2004;173(9):5704–5711. doi: 10.4049/jimmunol.173.9.5704. [DOI] [PubMed] [Google Scholar]
- 55.Haas P.-J., De Haas C. J. C., Poppelier M. J. J. C., et al. The structure of the C5a receptor-blocking domain of chemotaxis inhibitory protein of Staphylococcus aureus is related to a group of immune evasive molecules. Journal of Molecular Biology. 2005;353(4):859–872. doi: 10.1016/j.jmb.2005.09.014. [DOI] [PubMed] [Google Scholar]
- 56.Kretschmer D., Gleske A.-K., Rautenberg M., et al. Human formyl peptide receptor 2 senses highly pathogenic Staphylococcus aureus. Cell Host and Microbe. 2010;7(6):463–473. doi: 10.1016/j.chom.2010.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kobayashi S. D., Braughton K. R., Palazzolo-Ballance A. M., et al. Rapid neutrophil destruction following phagocytosis of Staphylococcus aureus. Journal of Innate Immunity. 2010;2(6):560–575. doi: 10.1159/000317134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wang R., Braughton K. R., Kretschmer D., et al. Identification of novel cytolytic peptides as key virulence determinants for community-associated MRSA. Nature Medicine. 2007;13(12):1510–1514. doi: 10.1038/nm1656. [DOI] [PubMed] [Google Scholar]
- 59.Chavakis T., Hussain M., Kanse S. M., et al. Staphylococcus aureus extracellular adherence protein serves as anti-inflammatory factor by inhibiting the recruitment of host leukocytes. Nature Medicine. 2002;8(7):687–693. doi: 10.1038/nm728. [DOI] [PubMed] [Google Scholar]
- 60.Laarman A. J., Ruyken M., Malone C. L., van Strijp J. A. G., Horswill A. R., Rooijakkers S. H. M. Staphylococcus aureus metalloprotease aureolysin cleaves complement C3 to mediate immune evasion. The Journal of Immunology. 2011;186(11):6445–6453. doi: 10.4049/jimmunol.1002948. [DOI] [PubMed] [Google Scholar]
- 61.Rooijakkers S. H. M., Ruyken M., Roos A., et al. Immune evasion by a staphylococcal complement inhibitor that acts on C3 convertases. Nature Immunology. 2005;6(9):920–927. doi: 10.1038/ni1235. [DOI] [PubMed] [Google Scholar]
- 62.Rooijakkers S. H. M., Wu J., Ruyken M., et al. Structural and functional implications of the alternative complement pathway C3 convertase stabilized by a staphylococcal inhibitor. Nature Immunology. 2009;10(7):721–727. doi: 10.1038/ni.1756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lee L. Y. L., Höök M., Haviland D., et al. Inhibition of complement activation by a secreted Staphylococcus aureus protein. Journal of Infectious Diseases. 2004;190(3):571–579. doi: 10.1086/422259. [DOI] [PubMed] [Google Scholar]
- 64.Jongerius I., Garcia B. L., Geisbrecht B. V., Van Strijp J. A. G., Rooijakkers S. H. M. Convertase inhibitory properties of staphylococcal extracellular complement-binding protein. Journal of Biological Chemistry. 2010;285(20):14973–14979. doi: 10.1074/jbc.M109.091975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Bestebroer J., Aerts P. C., Rooijakkers S. H. M., et al. Functional basis for complement evasion by staphylococcal superantigen-like 7. Cellular Microbiology. 2010;12(10):1506–1516. doi: 10.1111/j.1462-5822.2010.01486.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Walenkamp A. M. E., Boer I. G. J., Bestebroer J., et al. Staphylococcal superantigen-like 10 inhibits CXCL12-induced human tumor cell migration. Neoplasia. 2009;11(4):333–344. doi: 10.1593/neo.81508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Rooijakkers S. H. M., Van Wamel W. J. B., Ruyken M., Van Kessel K. P. M., Van Strijp J. A. G. Anti-opsonic properties of staphylokinase. Microbes and Infection. 2005;7(3):476–484. doi: 10.1016/j.micinf.2004.12.014. [DOI] [PubMed] [Google Scholar]
- 68.Haupt K., Reuter M., van den Elsen J., et al. The Staphylococcus aureus protein Sbi acts as a complement inhibitor and forms a tripartite complex with host complement Factor H and C3b. PLoS Pathogens. 2008;4(12) doi: 10.1371/journal.ppat.1000250.e1000250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Burman J. D., Leung E., Atkins K. L., et al. Interaction of human complement with Sbi, a staphylococcal immunoglobulin-binding protein: indications of a novel mechanism of complement evasion by Staphylococcus aureus. The Journal of Biological Chemistry. 2008;283(25):17579–17593. doi: 10.1074/jbc.m800265200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kim H. K., Cheng A. G., Kim H.-Y., Missiakas D. M., Schneewind O. Nontoxigenic protein A vaccine for methicillin-resistant staphylococcus aureus infections in mice. Journal of Experimental Medicine. 2010;207(9):1863–1870. doi: 10.1084/jem.20092514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Malachowa N., Kohler P. L., Schlievert P. M., et al. Characterization of a staphylococcus aureus surface virulence factor that promotes resistance to oxidative killing and infectious endocarditis. Infection and Immunity. 2011;79(1):342–352. doi: 10.1128/IAI.00736-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Nanra J. S., Buitrago S. M., Crawford S., et al. Capsular polysaccharides are an important immune evasion mechanism for Staphylococcus aureus. Human Vaccines and Immunotherapeutics. 2013;9(3):480–487. doi: 10.4161/hv.23223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Sharp J. A., Echague C. G., Hair P. S., et al. Staphylococcus aureus surface protein SdrE binds complement regulator factor H as an immune evasion tactic. PLoS ONE. 2012;7(5) doi: 10.1371/journal.pone.0038407.e38407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Visai L., Yanagisawa N., Josefsson E., et al. Immune evasion by Staphylococcus aureus conferred by iron-regulated surface determinant protein IsdH. Microbiology. 2009;155(3):667–679. doi: 10.1099/mic.0.025684-0. [DOI] [PubMed] [Google Scholar]
- 75.Zong Y., Xu Y., Liang X., et al. A ‘Collagen Hug’ model for Staphylococcus aureus CNA binding to collagen. The EMBO Journal. 2005;24(24):4224–4236. doi: 10.1038/sj.emboj.7600888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.DuMont A. L., Yoong P., Surewaard B. G. J., et al. Staphylococcus aureus elaborates leukocidin AB to mediate escape from within human neutrophils. Infection and Immunity. 2013;81(5):1830–1841. doi: 10.1128/IAI.00095-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Alonzo F., III, Kozhaya L., Rawlings S. A., et al. CCR5 is a receptor for Staphylococcus aureus leukotoxin ED. Nature. 2013;493(7430):51–55. doi: 10.1038/nature11724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Ventura C. L., Malachowa N., Hammer C. H., et al. Identification of a novel Staphylococcus aureus two-component leukotoxin using cell surface proteomics. PLoS ONE. 2010;5(7) doi: 10.1371/journal.pone.0011634.e11634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Spaan A. N., Henry T., Van Rooijen W. J. M., et al. The staphylococcal toxin panton-valentine leukocidin targets human C5a receptors. Cell Host and Microbe. 2013;13(5):584–594. doi: 10.1016/j.chom.2013.04.006. [DOI] [PubMed] [Google Scholar]
- 80.Inoshima I., Inoshima N., Wilke G. A., et al. A Staphylococcus aureus pore-forming toxin subverts the activity of ADAM10 to cause lethal infection in mice. Nature Medicine. 2011;17(10):1310–1314. doi: 10.1038/nm.2451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Clauditz A., Resch A., Wieland K.-P., Peschel A., Götz F. Staphyloxanthin plays a role in the fitness of Staphylococcus aureus and its ability to cope with oxidative stress. Infection and Immunity. 2006;74(8):4950–4953. doi: 10.1128/IAI.00204-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Cosgrove K., Coutts G., Jonsson I.-M., et al. Catalase (KatA) and alkyl hydroperoxide reductase (AhpC) have compensatory roles in peroxide stress resistance and are required for survival, persistence, and nasal colonization in Staphylococcus aureus. Journal of Bacteriology. 2007;189(3):1025–1035. doi: 10.1128/JB.01524-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Pang Y. Y., Schwartz J., Bloomberg S., Boyd J. M., Horswill A. R., Nauseef W. M. Methionine sulfoxide reductases protect against oxidative stress in staphylococcus aureus encountering exogenous oxidants and human neutrophils. Journal of Innate Immunity. 2014;6(3):353–364. doi: 10.1159/000355915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Thammavongsa V., Schneewind O., Missiakas D. M. Enzymatic properties of Staphylococcus aureus adenosine synthase (AdsA) BMC Biochemistry. 2011;12(1, article 56) doi: 10.1186/1471-2091-12-56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Cassat J. E., Skaar E. P. Metal ion acquisition in Staphylococcus aureus: overcoming nutritional immunity. Seminars in Immunopathology. 2012;34(2):215–235. doi: 10.1007/s00281-011-0294-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Bera A., Herbert S., Jakob A., Vollmer W., Götz F. Why are pathogenic staphylococci so lysozyme resistant? The peptidoglycan O-acetyltransferase OatA is the major determinant for lysozyme resistance of Staphylococcus aureus. Molecular Microbiology. 2005;55(3):778–787. doi: 10.1111/j.1365-2958.2004.04446.x. [DOI] [PubMed] [Google Scholar]
- 87.Herbert S., Bera A., Nerz C., et al. Molecular basis of resistance to muramidase and cationic antimicrobial peptide activity of lysozyme in staphylococci. PLoS Pathogens. 2007;3(7):p. e102. doi: 10.1371/journal.ppat.0030102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Stapels D. A. C., Ramyar K. X., Bischoff M., et al. Staphylococcus aureus secretes a unique class of neutrophil serine protease inhibitors. Proceedings of the National Academy of Sciences of the United States of America. 2014;111(36):13187–13192. doi: 10.1073/pnas.1407616111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Berends E. T. M., Horswill A. R., Haste N. M., Monestier M., Nizet V., Von Köckritz-Blickwede M. Nuclease expression by Staphylococcus aureus facilitates escape from neutrophil extracellular traps. Journal of Innate Immunity. 2010;2(6):576–586. doi: 10.1159/000319909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Schreiber A., Pham C. T. N., Hu Y., Schneider W., Luft F. C., Kettritz R. Neutrophil serine proteases promote IL-1β generation and injury in necrotizing crescentic glomerulonephritis. Journal of the American Society of Nephrology. 2012;23(3):470–482. doi: 10.1681/ASN.2010080892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Canny G., Levy O. Bactericidal/permeability-increasing protein (BPI) and BPI homologs at mucosal sites. Trends in Immunology. 2008;29(11):541–547. doi: 10.1016/j.it.2008.07.012. [DOI] [PubMed] [Google Scholar]
- 92.Corbin B. D., Seeley E. H., Raab A., et al. Metal chelation and inhibition of bacterial growth in tissue abscesses. Science. 2008;319(5865):962–965. doi: 10.1126/science.1152449. [DOI] [PubMed] [Google Scholar]
- 93.Embleton N. D., Berrington J. E., McGuire W., Stewart C. J., Cummings S. P. Lactoferrin: antimicrobial activity and therapeutic potential. Seminars in Fetal and Neonatal Medicine. 2013;18(3):143–149. doi: 10.1016/j.siny.2013.02.001. [DOI] [PubMed] [Google Scholar]
- 94.Mayeur S., Spahis S., Pouliot Y., Levy E. Lactoferrin, a pleiotropic protein in health and disease. Antioxidants & Redox Signaling. 2016;24(14):813–836. doi: 10.1089/ars.2015.6458. [DOI] [PubMed] [Google Scholar]
- 95.Brock J. H. Lactoferrin—50 years on. Biochemistry and Cell Biology. 2012;90(3):245–251. doi: 10.1139/o2012-018. [DOI] [PubMed] [Google Scholar]
- 96.Brinkmann V., Reichard U., Goosmann C., et al. Neutrophil extracellular traps kill bacteria. Science. 2004;303(5663):1532–1535. doi: 10.1126/science.1092385. [DOI] [PubMed] [Google Scholar]
- 97.Bestebroer J., van Kessel K. P., Azouagh H., et al. Staphylococcal SSL5 inhibits leukocyte activation by chemokines and anaphylatoxins. Blood. 2009;113(2):328–337. doi: 10.1182/blood-2008-04-153882. [DOI] [PubMed] [Google Scholar]
- 98.McDonald B., Urrutia R., Yipp B. G., Jenne C. N., Kubes P. Intravascular neutrophil extracellular traps capture bacteria from the bloodstream during sepsis. Cell Host & Microbe. 2012;12(3):324–333. doi: 10.1016/j.chom.2012.06.011. [DOI] [PubMed] [Google Scholar]
- 99.Baums C. G., von Köckritz-Blickwede M. Novel role of DNA in neutrophil extracellular traps. Trends in Microbiology. 2015;23(6):330–331. doi: 10.1016/j.tim.2015.04.003. [DOI] [PubMed] [Google Scholar]
- 100.Fuchs T. A., Abed U., Goosmann C., et al. Novel cell death program leads to neutrophil extracellular traps. Journal of Cell Biology. 2007;176(2):231–241. doi: 10.1083/jcb.200606027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Yang H., Biermann M. H., Brauner J. M., Liu Y., Zhao Y., Herrmann M. New insights into neutrophil extracellular traps: mechanisms of formation and role in inflammation. Frontiers in Immunology. 2016;7, article 302 doi: 10.3389/fimmu.2016.00302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Amulic B., Hayes G. Neutrophil extracellular traps. Current Biology. 2011;21(9):R297–R298. doi: 10.1016/j.cub.2011.03.021. [DOI] [PubMed] [Google Scholar]
- 103.Thammavongsa V., Kim H. K., Missiakas D., Schneewind O. Staphylococcal manipulation of host immune responses. Nature Reviews Microbiology. 2015;13(9):529–543. doi: 10.1038/nrmicro3521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Ley K., Laudanna C., Cybulsky M. I., Nourshargh S. Getting to the site of inflammation: the leukocyte adhesion cascade updated. Nature Reviews Immunology. 2007;7(9):678–689. doi: 10.1038/nri2156. [DOI] [PubMed] [Google Scholar]
- 105.Postma B., Poppelier M. J., Van Galen J. C., et al. Chemotaxis inhibitory protein of Staphylococcus aureus binds specifically to the C5a and formylated peptide receptor. Journal of Immunology. 2004;172(11):6994–7001. doi: 10.4049/jimmunol.172.11.6994. [DOI] [PubMed] [Google Scholar]
- 106.Van Wamel W. J. B., Rooijakkers S. H. M., Ruyken M., Van Kessel K. P. M., Van Strijp J. A. G. The innate immune modulators staphylococcal complement inhibitor and chemotaxis inhibitory protein of Staphylococcus aureus are located on β-hemolysin-converting bacteriophages. Journal of Bacteriology. 2006;188(4):1310–1315. doi: 10.1128/JB.188.4.1310-1315.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Bardoel B. W., Kenny E. F., Sollberger G., Zychlinsky A. The balancing act of neutrophils. Cell Host and Microbe. 2014;15(5):526–536. doi: 10.1016/j.chom.2014.04.011. [DOI] [PubMed] [Google Scholar]
- 108.McGuinness W., Kobayashi S., DeLeo F. Evasion of neutrophil killing by Staphylococcus aureus. Pathogens. 2016;5(1):p. 32. doi: 10.3390/pathogens5010032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Rooijakkers S. H. M., van Strijp J. A. G. Bacterial complement evasion. Molecular Immunology. 2007;44(1–3):23–32. doi: 10.1016/j.molimm.2006.06.011. [DOI] [PubMed] [Google Scholar]
- 110.Burman J. D., Leung E., Atkins K. L., et al. Interaction of human complement with Sbi, a staphylococcal immunoglobulin-binding protein: indications of a novel mechanism of complement evasion by Staphylococcus aureus. Journal of Biological Chemistry. 2008;283(25):17579–17593. doi: 10.1074/jbc.m800265200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Smith E. J., Corrigan R. M., van der Sluis T., et al. The immune evasion protein Sbi of Staphylococcus aureus occurs both extracellularly and anchored to the cell envelope by binding lipoteichoic acid. Molecular Microbiology. 2012;83(4):789–804. doi: 10.1111/j.1365-2958.2011.07966.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Smith E. J., Visai L., Kerrigan S. W., Speziale P., Foster T. J. The Sbi protein is a multifunctional immune evasion factor of Staphylococcus aureus. Infection and Immunity. 2011;79(9):3801–3809. doi: 10.1128/iai.05075-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Ko Y.-P., Kuipers A., Freitag C. M., et al. Phagocytosis escape by a Staphylococcus aureus protein that connects complement and coagulation proteins at the bacterial surface. PLOS Pathogens. 2013;9(12) doi: 10.1371/journal.ppat.1003816.e1003816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Hair P. S., Echague C. G., Sholl A. M., et al. Clumping factor A interaction with complement factor I increases C3b cleavage on the bacterial surface of Staphylococcus aureus and decreases complement-mediated phagocytosis. Infection and Immunity. 2010;78(4):1717–1727. doi: 10.1128/IAI.01065-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Hair P. S., Ward M. D., Semmes O. J., Foster T. J., Cunnion K. M. Staphylococcus aureus clumping factor A binds to complement regulator factor I and increases factor I cleavage of C3b. The Journal of Infectious Diseases. 2008;198(1):125–133. doi: 10.1086/588825. [DOI] [PubMed] [Google Scholar]
- 116.Higgins J., Loughman A., Van Kessel K. P. M., Van Strijp J. A. G., Foster T. J. Clumping factor A of Staphylococcus aureus inhibits phagocytosis by human polymorphonuclear leucocytes. FEMS Microbiology Letters. 2006;258(2):290–296. doi: 10.1111/j.1574-6968.2006.00229.x. [DOI] [PubMed] [Google Scholar]
- 117.Foster T. J., Geoghegan J. A., Ganesh V. K., Höök M. Adhesion, invasion and evasion: the many functions of the surface proteins of Staphylococcus aureus. Nature Reviews Microbiology. 2014;12(1):49–62. doi: 10.1038/nrmicro3161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Guerra F. E., Addison C. B., de Jong N. W., et al. Staphylococcus aureus SaeR/S-regulated factors reduce human neutrophil reactive oxygen species production. Journal of Leukocyte Biology. 2016;100(5):1005–1010. doi: 10.1189/jlb.4vmab0316-100rr. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Thammavongsa V., Kern J. W., Missiakas D. M., Schneewind O. Staphylococcus aureus synthesizes adenosine to escape host immune responses. The Journal of Experimental Medicine. 2009;206(11):2417–2427. doi: 10.1084/jem.20090097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Cronstein B. N., Daguma L., Nichols D., Hutchison A. J., Williams M. The adenosine/neutrophil paradox resolved: human neutrophils possess both A1 and A2 receptors that promote chemotaxis and inhibit O-2 generation, respectively. Journal of Clinical Investigation. 1990;85(4):1150–1157. doi: 10.1172/jci114547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Legrand D. Overview of lactoferrin as a natural immune modulator. The Journal of Pediatrics. 2016;173:S10–S15. doi: 10.1016/j.jpeds.2016.02.071. [DOI] [PubMed] [Google Scholar]
- 122.Clarke S. R., Foster S. J. IsdA protects Staphylococcus aureus against the bactericidal protease activity of apolactoferrin. Infection and Immunity. 2008;76(4):1518–1526. doi: 10.1128/iai.01530-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Mayer-Scholl A., Averhoff P., Zychlinsky A. How do neutrophils and pathogens interact? Current Opinion in Microbiology. 2004;7(1):62–66. doi: 10.1016/j.mib.2003.12.004. [DOI] [PubMed] [Google Scholar]
- 124.do Vale A., Cabanes D., Sousa S. Bacterial toxins as pathogen weapons against phagocytes. Frontiers in Microbiology. 2016;7, article 42 doi: 10.3389/fmicb.2016.00042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Peschel A., Otto M. Phenol-soluble modulins and staphylococcal infection. Nature Reviews Microbiology. 2013;11(10):667–673. doi: 10.1038/nrmicro3110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Surewaard B. G. J., De Haas C. J. C., Vervoort F., et al. Staphylococcal alpha-phenol soluble modulins contribute to neutrophil lysis after phagocytosis. Cellular Microbiology. 2013;15(8):1427–1437. doi: 10.1111/cmi.12130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Otto M. Phenol-soluble modulins. International Journal of Medical Microbiology. 2014;304(2):164–169. doi: 10.1016/j.ijmm.2013.11.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Genestier A.-L., Michallet M.-C., Prévost G., et al. Staphylococcus aureus Panton-Valentine leukocidin directly targets mitochondria and induces Bax-independent apoptosis of human neutrophils. Journal of Clinical Investigation. 2005;115(11):3117–3127. doi: 10.1172/JCI22684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Löffler B., Hussain M., Grundmeier M., et al. Staphylococcus aureus panton-valentine leukocidin is a very potent cytotoxic factor for human neutrophils. PLoS Pathogens. 2010;6(1) doi: 10.1371/journal.ppat.1000715.e1000715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Dhakal B. K., Mulvey M. A. The UPEC pore-forming toxin α-hemolysin triggers proteolysis of host proteins to disrupt cell adhesion, inflammatory, and survival pathways. Cell Host and Microbe. 2012;11(1):58–69. doi: 10.1016/j.chom.2011.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Berube B. J., Wardenburg J. B. Staphylococcus aureus α-toxin: nearly a century of intrigue. Toxins. 2013;5(6):1140–1166. doi: 10.3390/toxins5061140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Bartlett A. H., Foster T. J., Hayashida A., Park P. W. α-toxin facilitates the generation of CXC chemokine gradients and stimulates neutrophil homing in Staphylococcus aureus pneumonia. Journal of Infectious Diseases. 2008;198(10):1529–1535. doi: 10.1086/592758. [DOI] [PubMed] [Google Scholar]
- 133.Thammavongsa V., Missiakas D. M., Schneewind O. Staphylococcus aureus degrades neutrophil extracellular traps to promote immune cell death. Science. 2013;342(6160):863–866. doi: 10.1126/science.1242255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Spaan A. N., Surewaard B. G. J., Nijland R., van Strijp J. A. G. Neutrophils versus Staphylococcus aureus: a biological tug of war. Annual Review of Microbiology. 2013;67(1):629–650. doi: 10.1146/annurev-micro-092412-155746. [DOI] [PubMed] [Google Scholar]
- 135.Corriden R., Hollands A., Olson J., et al. Tamoxifen augments the innate immune function of neutrophils through modulation of intracellular ceramide. Nature Communications. 2015;6, article no. 8369 doi: 10.1038/ncomms9369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Hollands A., Corriden R., Gysler G., et al. Natural product anacardic acid from cashew nut shells stimulates neutrophil extracellular trap production and bactericidal activity. The Journal of Biological Chemistry. 2016;291(27):13964–13973. doi: 10.1074/jbc.m115.695866. [DOI] [PMC free article] [PubMed] [Google Scholar]